Abstract
Light and temperature are two key environmental factors that coordinately regulate plant growth and development. Although the mechanisms that integrate signaling mediated by cold and red light have been unraveled, the roles of the blue light photoreceptors cryptochromes in plant responses to cold remain unclear. In this study, we demonstrate that the CRYPTOCHROME2 (CRY2)-COP1-HY5-BBX7/8 module regulates blue light-dependent cold acclimation in Arabidopsis thaliana. We show that phosphorylated forms of CRY2 induced by blue light are stabilized by cold stress and that cold-stabilized CRY2 competes with the transcription factor HY5 to attenuate the HY5–COP1 interaction, thereby allowing HY5 to accumulate at cold temperatures. Furthermore, our data demonstrate that B-BOX DOMAIN PROTEIN7 (BBX7) and BBX8 function as direct HY5 targets that positively regulate freezing tolerance by modulating the expression of a set of cold-responsive genes, which mainly occurs independently of the C-repeat-binding factor pathway. Our study uncovers a mechanistic framework by which CRY2-mediated blue-light signaling enhances freezing tolerance, shedding light on the molecular mechanisms underlying the crosstalk between cold and light signaling pathways in plants.
Genetic and biochemical analyses combined with transcriptome analysis reveal the molecular mechanisms by which the cryptochrome CRY2 regulates freezing tolerance in Arabidopsis thaliana.
Introduction
Low temperature is a key environmental factor that limits the growth, development, and geographical distribution of plants. Over the course of their adaptation to their surroundings, temperate plants have developed the capacity to increase their freezing tolerance by pre-exposure to low but nonfreezing temperatures, so-called cold acclimation (Thomashow, 1999; Shi et al., 2018). C-repeat-binding factors (CBFs), also known as dehydration-responsive element-binding protein1s (DREB1s), play an important role in cold acclimation in Arabidopsis (Arabidopsis thaliana). Accordingly, the CBF-dependent cold signaling pathway has been extensively characterized (Jia et al., 2016; Zhao et al., 2016; Shi et al., 2018; Ding et al., 2020). Transcription of CBF genes is rapidly induced by cold stress, and the resulting cold-induced CBF proteins directly modulate a set of downstream cold-responsive (COR) genes, which in turn enhance freezing tolerance (Stockinger et al., 1997; Liu et al., 1998; Ding et al., 2020). Despite the critical role of CBFs in cold acclimation, only 10%–20% of COR genes are regulated by CBFs (Jia et al., 2016; Zhao et al., 2016), suggesting the involvement of other transcription factors independent of CBFs. However, little is known about these CBF-independent pathways and their roles in modulating cold acclimation.
Light serves as another key environmental signal that regulates plant growth, development, and responses to stress (Li et al., 2011; Legris et al., 2019; Roeber et al., 2021). Different wavelengths of light are absorbed by different families of photoreceptors. Phytochromes (Phys) are red (R) and far-red (FR) light photoreceptors that exist in vivo as two interconvertible forms: the inactive red light-absorbing Pr form and the active far-red light-absorbing Pfr form (Li et al., 2011; Legris et al., 2019). Phys are synthesized in the Pr form and localize to the cytoplasm in the dark. When exposed to light, they convert to the Pfr form and translocate to the nucleus, where they alter the expression of many light-responsive genes, eventually leading to an adaptive response (Klose et al., 2015; Legris et al., 2019). Cryptochromes (CRYs) are blue light photoreceptors that retain their ability to perform the light-dependent redox reactions of the flavin cofactor from photolyases. In the dark, the flavin of CRYs is in an inactive (FADOX) redox state and exists as a monomer in the nucleus (Hense et al., 2015). Under blue light irradiation, an electron is transferred to flavin, resulting in a neutral free radical FADH° (flavin adenine dinucleotide) redox state (Hense et al., 2015). Meanwhile, CRYs undergo homo-oligomerization and phosphorylation to become biochemically and physiologically active (Wang and Lin, 2020). Further exposure to blue light causes a second electron to be transferred to the flavin, leading to the fully reduced (FADH-) inactive redox state (Hense et al., 2015).
The biological activity of CRYs is determined by the equilibrium concentration of the active FADH° redox state under continuous illumination (Hammad et al., 2020). Phytochrome-interacting factors (PIFs) are a subfamily of basic helix–loop–helix transcription factors that physically interact with both Phys and CRYs (Ni et al., 1998; Leivar and Quail, 2011; Ma et al., 2016; Pedmale et al., 2016). Upon light exposure, photo-excited Phys and CRYs interact with PIFs and alleviate their repressive actions on light signaling pathways, either by inducing their rapid phosphorylation and degradation or by preventing their binding to the promoters of their target genes (Al-Sady et al., 2006; Leivar and Quail, 2011; Ma et al., 2016; Pedmale et al., 2016). In addition, photo-excited Phys and CRYs disrupt and inactivate the E3 ubiquitin ligase complexes to which constitutive photomorphogenic1 (COP1) and suppressor of phya-105 (SPA) proteins contribute, thus allowing for the accumulation of photomorphogenesis-promoting factors, such as the transcription factor elongated hypocoty5 (HY5) (Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011; Park et al., 2012; Lu et al., 2015; Sheerin et al., 2015).
B-BOX (BBX) domain-containing proteins are zinc-finger transcription factors that play pivotal roles in regulating photomorphogenesis (Song et al., 2020; Xu, 2020). The Arabidopsis genome encodes 32 BBX proteins (Gangappa and Botto, 2014), several of which are actively involved in HY5-mediated photomorphogenesis (Xu, 2020). BBX21 directly binds to the T/G-box cis-element in the HY5 promoter to activate its expression and promote photomorphogenesis (Xu et al., 2016). BBX21–BBX25 and BBX28 can also form heterodimers with HY5. BBX21, BBX22, and BBX23 enhance the transcriptional activity of HY5 (Datta et al., 2008; Zhang et al., 2017; Job et al., 2018), whereas BBX24, BBX25, and BBX28 repress it (Gangappa et al., 2013; Job et al., 2018; Lin et al., 2018). HY5 also binds to the BBX22 promoter and positively regulates its expression (Gangappa et al., 2013). In contrast, HY5 inhibits the expression of BBX30 and BBX31 by directly binding to their promoters (Heng et al., 2019; Yadav et al., 2019). BBX20, BBX21, and BBX22 act as rate-limiting cofactors of HY5 to regulate light signaling (Bursch et al., 2020).
Many key components of light signaling participate in plant responses to temperature fluctuations. Decreasing temperatures promote the function of the photoreceptor phytochrome B (phyB) by increasing the half-life of its Pfr form, thus acting as a thermosensor to perceive ambient temperature changes (Jung et al., 2016; Legris et al., 2016). A recent report showed that the flavin reoxidation rate of CRYs from the active state to the inactive state is lower at 15°C than at 25°C, suggesting that low temperature increases the concentrations of the active redox forms of CRYs, which may have higher biological activity (Pooam et al., 2021). Besides phyB and CRYs, whose active states change reversibly at a temperature range of 15–25°C (Legris et al., 2016; Pooam et al., 2021), the blue light receptor phototropin of the liverwort Marchantia polymorpha perceives the cold signal (4°C) by regulating the half-life of its phosphorylated active form (Fujii et al., 2017).
Moreover, photoperiod and light quality modulate plant response to cold stress via phytochromes (Franklin and Whitelam, 2007; Lee and Thomashow, 2012). Key components of light signaling, including PIF1, PIF3, PIF4, PIF5, and PIF7, negatively regulate cold responses in plants (Lee and Thomashow, 2012; Jiang et al., 2020). The expression of PIF1, PIF4, and PIF5 and PIF protein stability are repressed by cold stress (Jiang et al., 2020), whereas PIF3 is stabilized at low temperature (Jiang et al., 2017, 2020). Furthermore, PIF3 interacts with cold-induced CBF proteins, which attenuate PIF3-mediated phyB degradation; thus, phyB is stabilized by cold stress and positively regulates freezing tolerance by modulating the expression of a set of COR genes (Jiang et al., 2020). HY5 is degraded by COP1 in the dark at 22°C; however, cold induces the translocation of COP1 from the nucleus to the cytoplasm in the dark, thereby stabilizing HY5 to positively regulate freezing tolerance in plants (Catala et al., 2011).
CRYs play important roles in plant responses to high ambient temperatures (Blazquez et al., 2003; Ma et al., 2016). In particular, CRY1 interacts with PIF4 and PIF5 in a blue light-dependent manner to regulate hypocotyl elongation at high ambient temperatures (Ma et al., 2016). However, whether CRY-mediated blue-light signaling plays a role in regulating plant cold responses remains largely unknown. In this study, we demonstrate that CRY2 positively regulates plant cold acclimation independently of the CBF signaling pathway. We further show that blue light-induced phosphorylated CRY2 is stabilized by cold stress and that CRY2 competes with HY5 to interact with COP1, thereby stabilizing HY5 protein. In addition, HY5 directly targets the promoters of BBX7 and BBX8 to positively regulate their expression. Overexpression of BBX7 and BBX8 rescued the cold-sensitive phenotypes observed in Arabidopsis hy5 mutants. Finally, we show that BBX7 and BBX8 regulate plant cold tolerance without affecting the expression of CBFs or their target COR genes. Our study thus unravels the mechanism by which the CRY2–COP1–HY5–BBX7/8 module regulates blue-light-dependent cold acclimation in Arabidopsis.
Results
CRY2 positively regulates cold acclimation in Arabidopsis
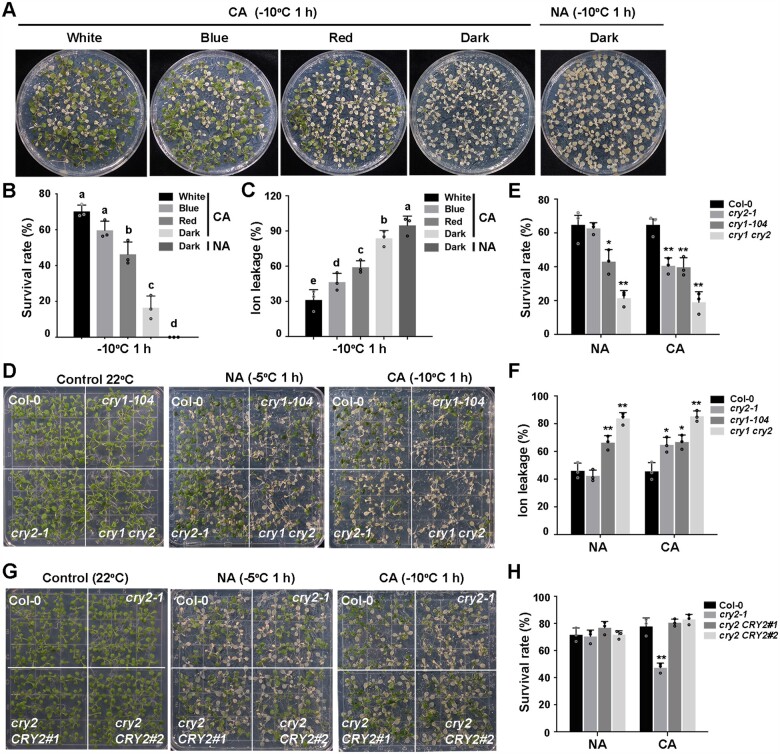
Previous studies have revealed the important role of Phy-mediated regulation of cold acclimation in plants (Franklin and Whitelam, 2007; Catala et al., 2011; Lee and Thomashow, 2012). To assess whether blue-light signaling also contributes to cold acclimation in plants, we compared the freezing tolerance of Arabidopsis wild-type (Col-0) seedlings after cold acclimation under different light conditions. Col-0 seedlings were grown in white light (long-day [LD] conditions) for 13 days and subjected to cold acclimation at 4°C for 2 days in the dark or in white-, blue-, or red light. Seedlings were then placed into a cold chamber, where temperatures were dropped by 1°C/h from 0°C until –10°C and remained for an additional 1 h. After freezing treatment, the seedlings were shifted to 4°C in the dark for 12 h and recovered at 22°C under LD conditions for additional 3 days. Cold-acclimated Col-0 seedlings under white light displayed the strongest freezing tolerance of all conditions tested, with the highest survival rate and lowest ion leakage, followed by seedlings under blue light, red light, and in the dark (Figure 1, A–C). As a control, nonacclimated Col-0 seedlings shifted to the dark for 2 days were all dead after freezing treatment (Figure 1, A–C). These results indicate that blue light plays an important role in regulating cold acclimation in Arabidopsis.
Figure 1.
CRYs positively regulates freezing tolerance in Arabidopsis. A–C, Effects of light quality on cold acclimation in 13-d-old wild-type seedlings. Seedlings were grown on half-strength Murashige-Skoog (1/2 MS) plates at 22°C under LD (16-h light/8-h dark photoperiod, 20 µmol m−2 s−1 white fluorescent light) conditions. For cold acclimation, 13-day-old seedlings were transferred to 4°C for 2 days in the dark or under white, red or blue light conditions (5 µmol m−2 s−1) and subjected to freezing treatment at –10°C for 1 h. Seedlings were treated at 22°C in the dark for 2 days as a negative control. Following recovery at 22°C for 3 days, representative photographs (A) were taken and the survival rates (B) and ion leakage (C) were measured. D–F, Freezing phenotypes (D), survival rates (E), and ion leakage (F) of Col-0, cry1-104, cry2-1, and cry1 cry2 seedlings. G and H, Freezing phenotypes (G) and survival rates (H) of Col-0, the cry2-1 mutant, and two complementation lines (cry2 CRY2#1 and cry2 CRY2#2). In (B and C), data are means of three independent experiments ± sem (Standard Error of Mean); each experiment was repeated three times (n = 50). Different letters represent significant differences at P < 0.05 (one-way ANOVA and Tukey’s multiple comparison tests). In (D–H), 13-day-old seedlings grown under LD conditions were subjected to freezing at –5°C for 1 h for NA seedlings and –10°C for 1 h for cold-acclimated seedlings (CA; 2 days at 4°C). In (E, F, and H), data are means of three independent experiments ± sem; each experiment was repeated three times (n = 30). Asterisks indicate significant differences compared with Col-0 under the same treatment (*P < 0.05, **P < 0.01, Student’s t test).
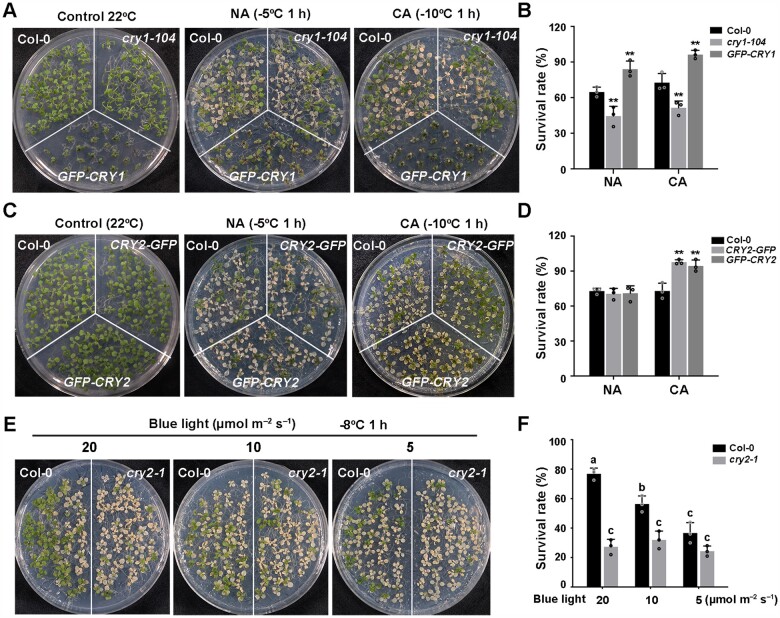
We asked whether the blue light photoreceptors CRY1 and CRY2 are involved in this process. We performed the freezing tolerance assays using null mutants of cry1, cry2, and cry1 cry2, as well as lines overexpressing each CRY. The cry1-104 mutant exhibited a reduced survival rate and increased ion leakage compared with the wild-type, whereas cry2-1 exhibited impaired freezing tolerance only after cold acclimation, with a lower survival rate and higher ion leakage than the wild-type (Figure 1, D–F). Moreover, the cry1 cry2 double mutant was more sensitive to cold stress than either cry1-104 or cry2-1 single mutants (Figure 1, D–F). The cold-sensitive phenotypes of the cry2-1 mutant were fully rescued by transformation with a CRY2 genomic fragment including the CRY2 promoter and coding region (Figure 1, G and H; Supplemental Figure S1, A–C). Conversely, transgenic seedlings overexpressing Green Fluorescent Protein (GFP)-CRY1 (Ma et al., 2016) always showed enhanced freezing tolerance with or without prior cold acclimation (Figure 2, A and B; Supplemental Figure S1D), and transgenic seedlings overexpressing GFP-CRY2 (Yu et al., 2009) and CRY2-GFP exhibited enhanced freezing tolerance only after cold acclimation (Figure 2, C and D; Supplemental Figure S1, A, E, and F). These data demonstrate that CRY1 regulates both basal and acquired freezing tolerance and that CRY2 specifically affects cold acclimation, thus regulating acquired freezing tolerance. Moreover, CRY1 and CRY2 function redundantly in acquired freezing tolerance.
Figure 2.
CRY2 positively regulates cold acclimation in Arabidopsis. A and B, Freezing phenotypes (A) and survival rates (B) of Col-0, the cry1-104 mutant, and GFP-CRY1 overexpressing transgenic plants. C and D, Freezing phenotypes (C) and survival rates (D) of Col-0 and two transgenic overexpression lines (35S:GFP-CRY2 [GFP-CRY2] or Super:CRY2-GFP [CRY2-GFP]). 13-day-old seedlings grown under LD conditions were subjected to freezing at –5°C for 1 h for NA seedlings and –10°C for 1 h for CA seedlings (CA; 2 days at 4°C). E and F, Freezing phenotypes (E) and survival rates (F) of blue light-treated Col-0 and cry2-1 seedlings. 13-day-old seedlings grown under LD conditions were subjected to different intensities of blue light (20, 10, 5 µmol m−2 s−1) for 24 h, followed by freezing at –8°C for 1 h. In (B, D, and F), data are means of three independent experiments ±sem; each experiment was repeated three times (n = 30). In (B and D), asterisks indicate significant differences compared with Col-0 under the same treatment (*P < 0.05, **P < 0.01, Student’s t test). In (F), different letters represent significant difference at P < 0.05 (one-way ANOVA and Tukey’s multiple comparison tests).
To assess whether blue light also simulates cold acclimation at 22°C, we exposed wild-type Col-0 and cry2-1 seedlings to blue light with different intensities for 24 h at 22°C and directly subjected them to freezing tolerance assays. The freezing tolerance of wild-type Col-0 was significantly enhanced with increasing in blue light intensity, whereas there was little change in the cry2-1 mutant (Figure 2, E and F; Supplemental Figure S1G). These data suggest that the CRY2-mediated blue-light signaling indeed mimics cold acclimation and thus regulate freezing tolerance.
Low temperature enhances the stability of phosphorylated CRY2
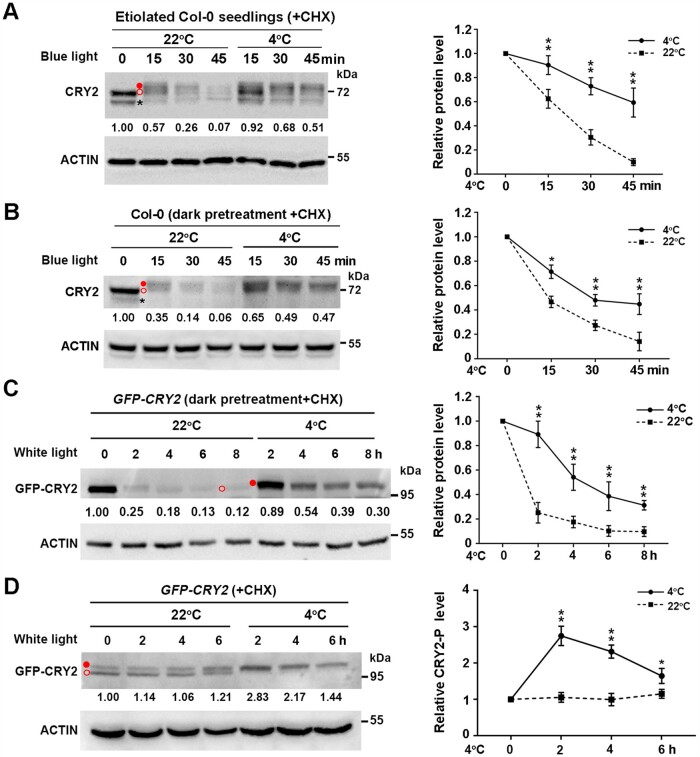
CRY2 is phosphorylated and activated by blue light, but phosphorylated CRY2 is also rapidly degraded (Shalitin et al., 2002; Wang and Lin, 2020). This prompted us to investigate whether cold affects the activation and/or stability of CRY2. We pretreated 10-day-old etiolated Col-0 seedlings grown at 22°C with the protein biosynthesis inhibitor cycloheximide (CHX) for 0.5 h and kept the plants at 22°C or shifted them to 4°C in the dark. Immunoblot analysis revealed only a single band of CRY2 protein in etiolated seedlings, which remained unchanged even after exposure to cold (Supplemental Figure S2A). However, after CHX-treated etiolated seedlings were shifted to constant blue light (20 µmol m−2 s−1), we readily detected the upper migration of phosphorylated CRY2 before its rapid degradation at 22°C (Figure 3A;Supplemental Figure S2A), which is consistent with previous findings (Shalitin et al., 2002). In contrast, more phosphorylated CRY2 was detected after 15 min, 30 min, and 45 min of cold treatment (Figure 3A). Thus, blue light-induced degradation of phosphorylated CRY2 is suppressed by cold stress.
Figure 3.
Cold promotes the stability of phosphorylated CRY2 induced by blue light. A, Effects of cold exposure on CRY2 protein levels in 10-day-old etiolated wild-type Col-0 seedlings transferred to blue light (20 µmol m−2 s−1) for the indicated time. Seedlings were treated with 300 µM CHX for 0.5 h, followed by transfer to 4°C or back to 22°C in the dark or in blue light in the presence of CHX. B and C, Effects of cold exposure on CRY2 protein levels in Col-0 seedlings and GFP-CRY2 transgenic lines. A 13-day-old LD-grown Col-0 and GFP-CRY2 transgenic seedlings were pretreated in the dark at 22°C for 24 h, treated with 300 µM CHX for 0.5 h, followed by transfer to 4°C or back to 22°C for the indicated times in blue light (B) or white light (C) (20 µmol m−2 s−1). D, Effects of cold exposure on GFP-CRY2-P protein levels in GFP-CRY2 transgenic seedlings in constant white light (20 µmol m−2 s−1). A 13-day-old LD-grown GFP-CRY2 seedlings were treated with 300 µM CHX for 0.5 h at ZT2 (Zeitgeber 2; 2 h after dawn), followed by treatment at 22°C or 4°C for the indicated times in constant white light (20 µmol m−2 s−1). In (A–D), total proteins were extracted and subjected to immunoblot analysis with anti-CRY2 (A and B) or anti-GFP (C and D) antibodies to detect CRY2 protein. Actin served as a loading control. Immunoblot results were quantified using ImageJ software. Relative protein levels are shown to the right of the blots, with relative protein levels at 0 h set to 1.00. Quantitative data are means of three independent experiments ±sd. Asterisks indicate significant differences compared with 22°C under the same light conditions (*P < 0.05, **P < 0.01, Student’s t test). Open red circle, nonphosphorylated CRY2; filled red circle, phosphorylated CRY2; asterisk, nonspecific band.
To further examine the regulation of phosphorylated CRY2 by low temperature, we transferred 13-day-old wild-type seedlings grown in LD conditions to the dark at 22°C for 24 h, followed by a return to 22°C or exposure to 4°C in constant blue light (20 µmol m−2 s−1). The degradation of CRY2 after this pretreatment was similar to that seen in etiolated seedlings upon blue light exposure, and under both conditions, the degradation of phosphorylated CRY2 was dramatically inhibited by cold stress (Figure 3B). We obtained similar results with transgenic seedlings overexpressing GFP-CRY2 that were transferred to the dark at 22°C for 24 h, followed by white light (20 µmol m−2 s−1) or blue light (5 µmol m−2 s−1) illumination (Figure 3C;Supplemental Figure S2B). In addition, we detected both phosphorylated and nonphosphorylated forms of CRY2 at 22°C in GFP-CRY2 transgenic seedlings under constant white light, whereas cold treatment significantly promoted the conversion of nonphosphorylated CRY2 to phosphorylated CRY2 (Figure 3D). Together with the finding that blue light-dependent phosphorylation of CRY2 enhances its activity (Wang and Lin, 2020), these data suggest that low temperature enhances the stability of CRY2, thereby promoting its activity and positively modulating cold acclimation.
CRY2 attenuates the interaction of COP1 and HY5
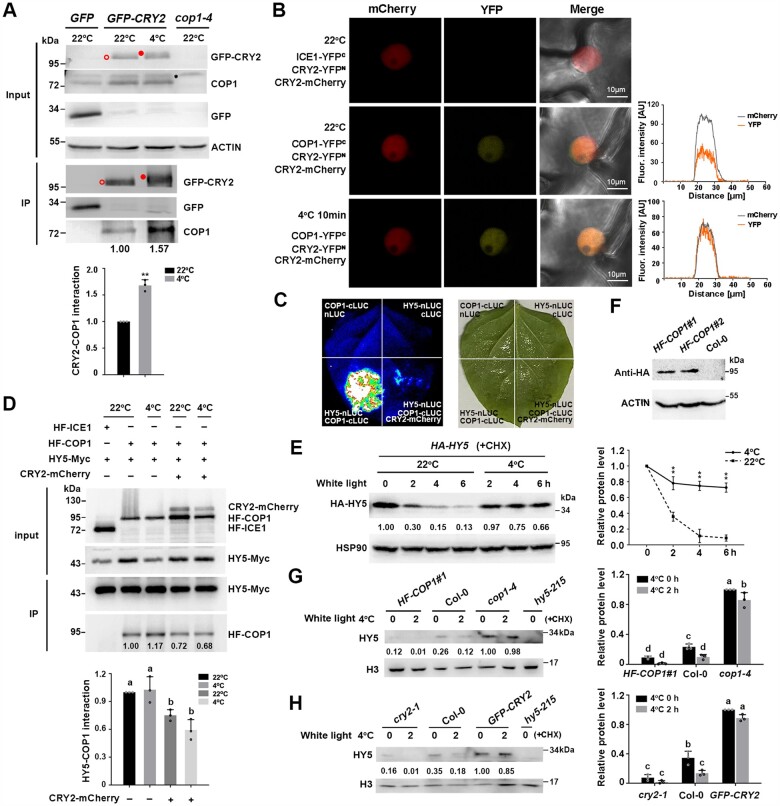
The COP1–HY5 regulatory module is crucial for regulating cold acclimation in plants (Catala et al., 2011). Interactions among CRY2, COP1, and SPA inhibit the E3 ubiquitin ligase activity of CULLIN4 (CUL4)COP1-SPAs (Wang and Lin, 2020). Therefore, we wondered whether CRY2 regulates plant cold responses via the COP1–HY5 complex. To this end, we performed a co-immunoprecipitation (co-IP) assay to examine the interaction between CRY2 and COP1 using transgenic lines overexpressing GFP-CRY2 exposed to 4°C treatment for 1 h, with seedlings maintained at 22°C used as controls. We immunoprecipitated the proteins with GFP agarose beads and detected them with anti-COP1 antibody, which was verified using the cop1-4 mutant (Figure 4A). More COP1 co-immunoprecipitated with GFP-CRY2 in cold-treated seedlings than in control seedlings (Figure 4A), indicating that the interaction between CRY2 and COP1 is enhanced by cold treatment. Consistent with this result, bimolecular fluorescence complementation (BiFC) assays showed that the CRY2–COP1 interaction in the nucleus was promoted by exposure to 4°C compared with 22°C, as reflected by the optical density map across nuclei (Figure 4B).
Figure 4.
CRY2 attenuates the interaction between COP1 and HY5 under cold stress. A, co-IP assays showing the interaction between CRY2 and COP1 in vivo. 13-d-old GFP-CRY2 transgenic seedlings were exposed to cold treatment (4°C, 1 h) or maintained at 22°C under white light. Transgenic seedlings transformed with 35S:GFP empty vector were used as a negative control. Total protein extracts were immunoprecipitated with GFP agarose beads. GFP, GFP-CRY2 and COP1 were detected with anti-GFP or anti-COP1 antibodies. cop1-4 was used to verify the specificity of anti-COP1 antibody. Open red circle, nonphosphorylated CRY2; filled red circle, phosphorylated CRY2; asterisk, nonspecific band. Each bar represents the means of three independent experiments ± sd (**P < 0.01, Student’s t test). B, BiFC showing the interaction between COP1 and CRY2 in N. benthamiana epidermal cells. COP1-YFPN or ICE1- YFPN (as a control), CRY2-YFPC and CRY2-mCherry were co-infiltrated in N. benthamiana leaves. The same cell was imaged under a fluorescence microscope at 22°C and cooled with ice for 10 min. Fluorescence intensity was measured by ZEN 3.2 software. Scale bars, 10 μm. A representative experiment from three independent experiments is shown. C, Firefly LUC complementation imaging assay showing that CRY2 attenuates the COP1–HY5 interaction in N. benthamiana leaves. A representative experiment from three independent experiments is shown. D, Co-IP assays show that CRY2 attenuates the interaction between COP1 and HY5 proteins in vivo. Arabidopsis mesophyll protoplasts co-transfected with CRY2-mCherry, HF-COP1, and/or HY5-Myc were incubated at 22°C for 16 h in the dark and exposed to 4°C for 1 h in white light or maintained at 22°C in white light. Total protein extracts were immunoprecipitated with HA Sepharose beads. Proteins (input) and IP proteins were detected using anti-HA, anti-CRY2, and anti-Myc antibodies. E, Immunoblot analysis of HY5 protein levels in HA-HY5 seedlings. The 13-day-old LDgrown seedlings were treated with 300 µM CHX for 0.5 h, followed by treatment at 4°C or maintained at 22°C in constant white light (20 µmol m−2 s−1). Total proteins (E) were extracted from the samples, and HY5 was detected with anti-HA antibody. HSP90 was used as a loading control. Immunoblot results were quantified using ImageJ software. F, COP1 protein levels of Col-0 and COP1 overexpression transgenic lines (HF-COP1#1, HF-COP1#2). COP1 protein was detected with anti-HA antibody. G and H, Immunoblot analysis of HY5 protein levels in cop1-4, HF-COP1#1 (G), cry2-1, and GFP-CRY2 (H) seedlings. The 13-day-old LD-grown seedlings were treated as described in (E). Nuclear proteins were extracted and HY5 was detected with anti-HY5 antibodies. hy5-215 mutant was used to verify the specificity of anti-HY5 antibody. Histone was used as a loading control. Immunoblot results were quantified using ImageJ software. In (E, G, and H), the relative protein levels of HA-HY5, cop1-4, GFP-CRY2 at 0 h were set to 1.00. In (D, E, G, and H), data are means of three independent experiments ± sd. In (D, G, and H), different letters represent significant differences at P < 0.05 (one-way ANOVA and Tukey’s multiple comparison tests). In (E), asterisks indicate significant differences compared with 22°C under the same light conditions (**P < 0.01, Student’s t test).
We then examined whether CRY2 affects the interaction of COP1 with HY5 by performing firefly luciferase (LUC) complementation imaging. When COP1-cLUC and HY5-nLUC were co-infiltrated in Nicotiana benthamiana leaves, we visualized very strong luminescence signals, reflecting the interaction between COP1 and HY5. However, when CRY2-mCherry was co-infiltrated with COP1-cLUC and HY5-nLUC, LUC activity dramatically decreased (Figure 4C). Co-IP assays further indicated that the co-transfection of CRY2-mCherry with HA-FLAG-COP1 (HF-COP1) and HY5-Myc in Arabidopsis protoplasts compromised the interaction between COP1 and HY5 at both 22°C and 4°C (Figure 4D). Therefore, the interaction of COP1 and HY5 interaction is attenuated by CRY2.
We next asked whether the stability of HY5 is mediated by CRY2 at low temperatures. In the presence of CHX, HY5 protein levels in transgenic plants overexpressing HA-HY5 dramatically decreased at 22°C under white light; however, HY5 became stabilized after transfer to 4°C (Figure 4E). We then extracted nuclear proteins and assessed HY5 protein levels with an anti-HY5 antibody in wild-type Col-0, the cry2-1 mutant, and transgenic plants overexpressing GFP-CRY2 before and after cold treatment. The HF-COP1#1 overexpression line (Figure 4F) and cop1-4 mutant were used as controls, and the anti-HY5 antibody was verified using hy5-215 (Figure 4G). As expected, before cold treatment, HY5 protein levels were significantly lower in wild-type Col-0 than in cop1-4 but higher than in the HF-COP1#1 overexpression line. After cold treatment, HY5 protein levels were significantly downregulated in Col-0 and HF-COP1#1 plants but largely unchanged in cop1-4 (Figure 4G), supporting the notion that the stability of HY5 is enhanced under cold stress when COP1 is inactivated (Catala et al., 2011). In contrast, before cold treatment, HY5 protein levels were significantly higher in Col-0 than in cry2-1 but lower than in GFP-CRY2 overexpression seedlings. After cold treatment, HY5 protein levels significantly decreased in Col-0 and cry2-1 but remained stable in GFP-CRY2 seedlings (Figure 4H). These results indicate that the stability of HY5 is positively regulated by CRY2. Therefore, cold-stabilized CRY2 interacts with COP1 and competes with HY5 for binding to COP1, thereby enhancing the stability of HY5 under cold stress.
CRYs act upstream of HY5 and COP1 to regulate freezing tolerance
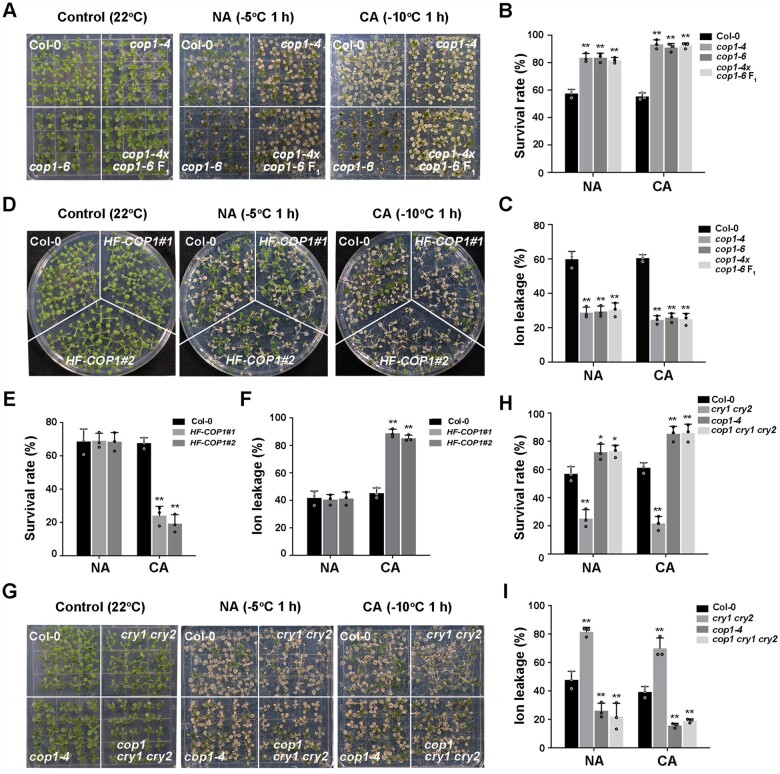
To explore the genetic interactions between CRYs and COP1 in regulating plant responses to cold stress, we examined the freezing tolerance of weak alleles of cop1 (cop1-4 and cop1-6), HF-COP1 overexpression lines and cop1-4 cry1 cry2 (cop1 cry1 cry2) mutants (Mao et al., 2005). Both cop1-4 and cop1-6 mutants showed constitutive freezing tolerance without cold acclimation, with more significant freezing tolerance after cold acclimation (Figure 5, A–C). An allelism test indicated that the F1 progeny of a cross between cop1-4 and cop1-6 exhibited freezing tolerance that was comparable with that of each cop1 mutant (Figure 5, A–C), validating the notion that the observed freezing tolerance phenotype is a consequence of partial inactivation of COP1. Moreover, compared with the wild-type, two independent HF-COP1 overexpression transgenic lines displayed significantly increased freezing sensitivity only after cold acclimation (Figure 5, D–F). The triple mutant cop1 cry1 cry2 had a freezing tolerance phenotype similar to that of cop1-4 (Figure 5, G–I). These data suggest that COP1 acts downstream of CRYs to negatively regulate freezing tolerance.
Figure 5.
COP1 acts genetically downstream of CRYs to negatively regulate plant freezing tolerance. A–C, Freezing phenotypes (A), survival rates (B), and ion leakage (C) of Col-0, cop1-4, cop1-6, and cop1-4 × cop1-6 F1 seedlings. D–F, Freezing phenotypes (D), survival rates (E), and ion leakage (F) of Col-0 and COP1 overexpression transgenic lines (HF-COP1#1, HF-COP1#2). G–I, Freezing phenotypes (G), survival rates (H), and ion leakage (I) of Col-0, cry1 cry2, cop1-4, and cop1 cry1 cry2 seedlings. In (B–I), data are means of three independent experiments ± SEM; each experiment was repeated three times (n = 30). Asterisks indicate significant differences compared with Col-0 under the same treatment (*P < 0.05, **P < 0.01, Student’s t test).
The null mutant hy5-215 displayed a freezing-sensitive phenotype, which is consistent with previous findings (Catala et al., 2011), whereas UBQ10:3×HA-HY5 overexpression (HA-HY5) lines (Li et al., 2020) showed enhanced acquired freezing tolerance compared with the wild-type after cold acclimation (Supplemental Figure S3, A–C). Therefore, we generated cry2-1 HA-HY5 plants by genetic crossing and discovered that cry2-1 HA-HY5 seedlings phenocopied HA-HY5 seedlings in terms of their freezing tolerance specifically after cold acclimation (Supplemental Figure S3, D–F). These results demonstrate that CRY2 acts genetically upstream of HY5 to positively regulate cold acclimation.
As CBF genes are critical for cold acclimation in plants (Jia et al., 2016; Zhao et al., 2016), we investigated whether the CRY2–COP1–HY5 module regulates cold acclimation via the CBF pathway. The expression of CBFs and their target genes (such as COR15B, galactinol synthase3 [GOLS3], and responsive to desiccation 29A [RD29A]) was indistinguishable between the wild-type, cry2, hy5-215, and GFP-CRY2 and HA-HY5 overexpression transgenic lines before or after cold treatment (Supplemental Figure S4, A and B). Moreover, the expression levels of CBFs and their target genes were significantly lower in cop1-4 and higher in HF-COP1#1 overexpression lines than in the wild-type (Supplemental Figure S4C), which are the opposite to their freezing phenotypes. These results suggest that the function of CRY2, COP1, and HY5 in regulating freezing tolerance is mainly independent of the CBF signaling pathway.
Cold-induced expression of BBX7 and BBX8 is regulated by CRY2, COP1, and HY5
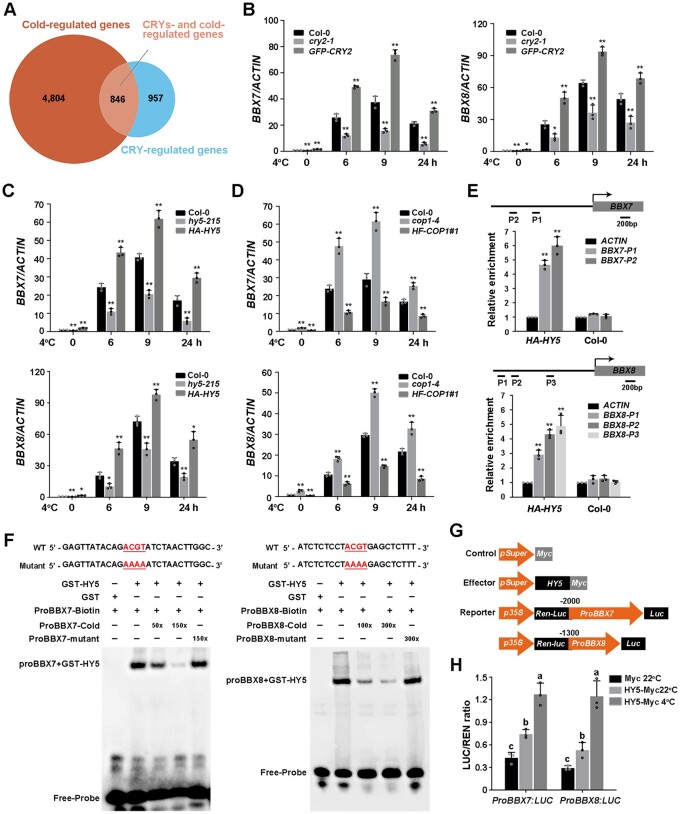
To further define the mechanism underlying CRY2-mediated cold signaling, we analyzed published RNA-Seq data of blue light-responsive genes regulated by CRYs (Wang et al., 2016) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA318638; |log2| ≥ 1, adjusted P < 0.05) and COR genes identified in previous studies (Song et al., 2021; http://www.ncbi.nlm.nih.gov/sra/PRJNA732005; |log2| ≥ 1, adjusted P < 0.05). Interestingly, among the 1,803 CRY-regulated genes, 846 are COR genes, representing 46.9% of CRY-regulated genes (Figure 6A;Supplemental Data Set S1). These data highlight the important role of CRYs in regulating COR gene expression.
Figure 6.
Cold-induced expression of BBX7 and BBX8 is regulated by CRY2–COP1–HY5. A, Venn diagram indicating the number of CRY-regulated COR genes by comparing CRY-regulated genes (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA318638; |log2| ≥ 1, adjusted P < 0.05) with COR genes (http://www.ncbi.nlm.nih.gov/sra/PRJNA732005; |log2| ≥ 1, adjusted P < 0.05) identified in previous studies. B–D, Relative expression levels of BBX7 and BBX8 under cold treatment in Col-0, cry2-1 and GFP-CRY2 (B), hy5-215 and HA-HY5 (C), and cop1-4 and HF-COP1#1 (D). Total RNA was extracted from 10-day-old LD-grown seedlings treated at 4°C for the indicated times. ACTIN2/8 was used as a normalization control. Expression in untreated Col-0 was set to 1.00. E, ChIP assays showing that HY5 binds to the BBX7 and BBX8 promoters in vivo. The 13-day-old HA-HY5 or WT (Col-0) seedlings grown under LD conditions were harvested for ChIP analysis using HA beads, and the amounts of the indicated DNA in the immune complex were analyzed by qRT-PCR. The ACTIN fragment was amplified as a control. Relative enrichment was calculated as Input% (indicated DNA)/Input% (control). F, EMSAs showing that HY5 binds to the promoters of BBX7 (P1) and BBX8 (P3) in vitro. Each biotin-labeled DNA fragment was incubated with recombinant GST-HY5 or GST proteins. Competition assays for labeled promoter sequences were performed by adding an excess of unlabeled wild-type or mutated probes. G, Schematic representation of the control, effector, and reporter constructs used in the dual-LUC assays. The LUC reporter constructs harboring BBX7 and BBX8 promoters were used as reporters. Effector constructs harboring the HY5 coding sequences were driven by the Super promoter. The Myc tag sequence driven by the Super promoter was used as a negative control. H, Transcriptional activation experiments show that HY5 activates the expression of BBX7 and BBX8 in vivo. Construct combinations were co-transfected in Arabidopsis protoplasts. Following incubation in the dark for 16 h, the protoplasts were treated in the light at 22°C (control) or 4°C for 1 h, and the samples were collected for dual-LUC analysis. LUC activity was normalized to Renilla (REN) LUC activity. In (B–E, and H), data are means of three independent experiments ± sd. In (B–D), asterisks indicate significant differences compared with Col-0 under the same treatment (*P < 0.05, **P < 0.01, Student’s t test). In (E), asterisks indicate significant differences compared with ACTIN fragment in HA-HY5 or WT (**P < 0.01, Student’s t test). In (H), different letters represent significant differences at P < 0.05 (one-way ANOVA and Tukey’s multiple comparison tests).
Next, we aimed to identify COR genes that regulate by CRYs–COP1–HY5 module. Previous studies established a regulatory network consisting of BBXs and HY5 that controls the expression of a large number of light-responsive genes (Xu, 2020). Interestingly, in our previous transcriptome, deep sequencing (RNA-seq) data of wild-type Col-0 plants exposed to cold, two BBX-containing transcription factor genes, BBX7 and BBX8, were among the genes most significantly upregulated by low temperature (Jia et al., 2016; Song et al., 2021), prompting us to ask whether BBXs are regulated by the CRY2–COP1–HY5 module in response to cold. Quantitative RT-PCR (Real-time polymerase chain reaction) analysis revealed that the cold-induced expression levels of BBX7 and BBX8 were lower in cry2-1 and hy5-215 but higher in GFP-CRY2 and HA-HY5 overexpression lines than in the wild-type (Figure 6, B and C). Conversely, the induction of BBX7 and BBX8 expression by cold was stronger in the cop1-4 mutant but weaker in HF-COP1#1 overexpression plants than in the wild-type (Figure 6D). These results demonstrate that the cold-induced expression of BBX7 and BBX8 is positively regulated by CRY2 and HY5, but repressed by COP1.
HY5 directly binds to the promoters of BBX7 and BBX8
HY5 positively regulates target gene expression by binding to the Z-box and other cis-acting elements such as the ACGT-containing element (ACE) in their promoters (Catala et al., 2011; Heng et al., 2019). The BBX7 and BBX8 promoters contain two and three ACE elements, respectively. Chromatin immunoprecipitation (ChIP)-qRT-PCR assays showed that HY5 was associated with the regions of the BBX7 and BBX8 promoters containing ACEs in vivo (Figure 6E). We then performed electrophoretic mobility shift assays (EMSAs) to test whether HY5 directly binds to the promoters of BBX7 and BBX8 in vitro. Recombinant GST-HY5 bound directly to the ACE-containing fragments of the BBX7 (P1) and BBX8 promoters (P3) (Figure 6F). Moreover, increasing amounts of unlabeled wild-type probes markedly reduced HY5 binding to biotin-labeled probes, whereas unlabeled mutant probes were unable to compete for HY5 binding (Figure 6F), indicating that HY5 directly binds to the ACE motifs in the BBX7 and BBX8 promoters in vitro.
In addition, we performed a dual-LUC assay in Arabidopsis protoplasts transfected with Super:HY5-Myc and ProBBX7/8:LUC to determine whether the expression of BBX7 and BBX8 is regulated by HY5 (Figure 6G). HY5 protein levels were much higher at 4°C than at 22°C in Arabidopsis protoplasts transfected with Super:HY5-Myc and ProBBX7/8:LUC (Supplemental Figure S5), which is consistent with earlier results (Figure 4E). Accordingly, the ProBBX7:LUC and ProBBX8:LUC reporters were activated by HY5 at 22°C, and this effect was more prominent at 4°C (Figure 6H). These data indicate that HY5 activates BBX7 and BBX8 expression by directly binding to their promoters.
BBX7 and BBX8 redundantly function downstream of HY5 to positively regulate plant freezing tolerance
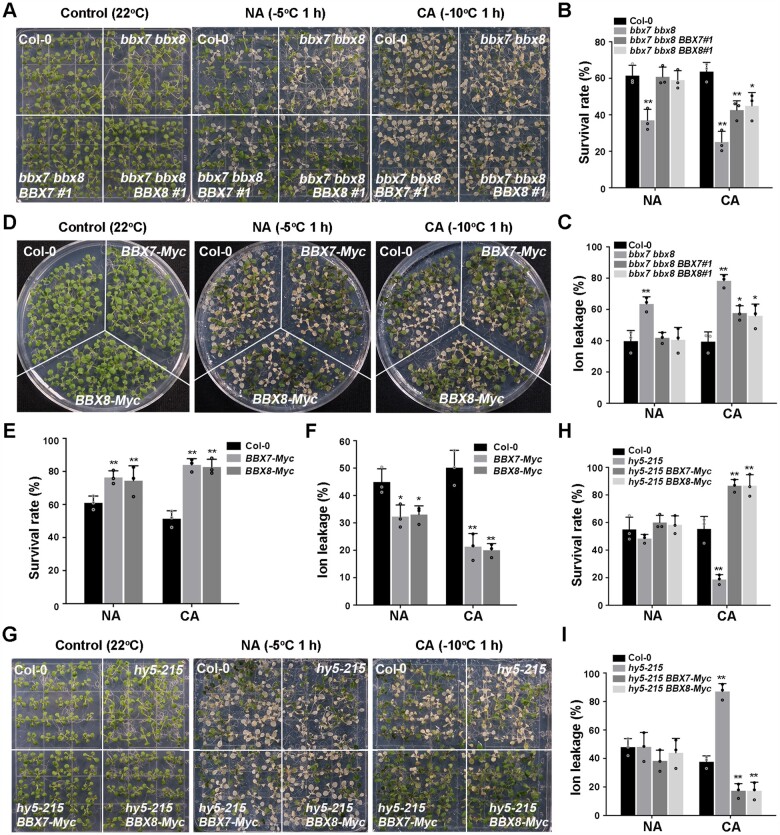
To determine the exact role of BBX7 and BBX8 in cold stress responses, we generated two independent mutant lines (bbx7-1 and bbx7-2) by CRISPR (clustered regularly interspaced short palindromic repeats)/CRISPR-associated protein 9 (Cas9)-mediated genome editing and produced the bbx7 bbx8 double mutant by crossing the genome-edited bbx7-1 allele with bbx8-1 (SALK_061961) (Supplemental Figure S6, A–C). The bbx7-1 and bbx7-2 mutants contained 86-bp and 71-bp deletions from –3 relative to the ATG (with the A set to +1), respectively, resulting in a complete knockout of BBX7 (Supplemental Figure S6, A–E). Freezing tolerance assays revealed that the survival rate and ion leakage of nonacclimated bbx7-1, bbx7-2 and bbx8-1 single mutants were not significantly different from those of the wild-type (Supplemental Figure S7, A–F). Nonacclimated bbx7 bbx8 double mutants displayed mild freezing sensitivity (Figure 7, A–C; Supplemental Figure S7, D–F). After cold acclimation, the bbx7-1, bbx7-2, and bbx8-1 single mutants exhibited freezing sensitivity, and the bbx7 bbx8 double mutant was more sensitive than the single mutants (Figure 7, A–C; Supplemental Figure S7, A–F). Transformation with genomic fragments spanning the BBX7 or BBX8 locus fully rescued the freezing tolerance seen in nonacclimated bbx7 bbx8 double mutant seedlings and partially rescued the freezing tolerance in acclimated bbx7 bbx8 double mutant seedlings (Figure 7, A–C; Supplemental Figure S6, D–G).
Figure 7.
BBX7 and BBX8 redundantly act downstream of HY5 to positively regulate plant freezing tolerance. A–C, Freezing phenotypes (A), survival rates (B), and ion leakage (C) of Col-0, the bbx7 bbx8 double mutant, and bbx7 bbx8 BBX7#1, bbx7 bbx8 BBX8#1 complementation lines. D–F, Freezing phenotypes (D), survival rates (E), and ion leakage (F) of Col-0, BBX7-Myc and BBX8-Myc overexpression lines. G–I, Freezing phenotypes (G), survival rates (H), and ion leakage (I) of Col-0, hy5-215, hy5-215 BBX7-Myc, and hy5-215 BBX8-Myc transgenic lines. In (B, C, E, F, H, and I), data are means of three independent experiments ± sem (n = 30). Asterisks indicate significant differences compared with Col-0 under the same treatment (*P < 0.05, **P < 0.01, Student’s t test).
We also generated BBX7-Myc and BBX8-Myc overexpression lines (Supplemental Figure S6, D, E, and G); they showed significantly enhanced freezing tolerance compared with the wild-type with or without cold acclimation (Figure 7, D–F). Moreover, overexpression of BBX7 or BBX8 in the hy5-215 mutant fully suppressed the freezing sensitivity of the hy5-215 mutant (Figure 7, G–I; Supplemental Figure S6, D, E, and G). These results demonstrate that BBX7 and BBX8 act downstream of HY5 to redundantly promote freezing tolerance in plants.
Identification of COR genes regulated by HY5 and BBX7/8
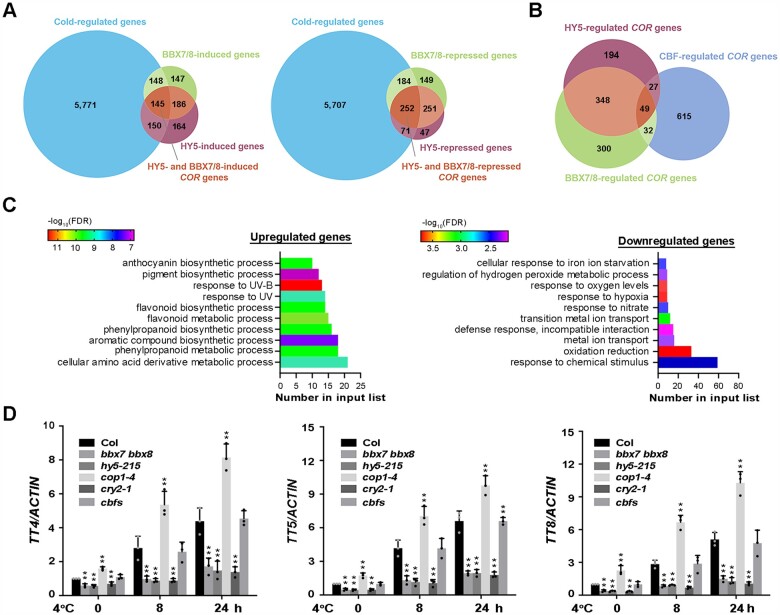
To further explore the mechanism underlying the roles of HY5 and BBX7/8 in regulating plant freezing tolerance, we performed RNA-sequencing (RNA-seq) to analyze the transcriptomes of 10-day-old wild-type Col-0, hy5-215, and bbx7 bbx8 seedlings treated with 4°C for 0 h, 8 h, and 24 h to identify HY5- and BBX7/8-regulated COR genes. HY5- and BBX7/8-regulated COR genes were selected based on the following criteria: genes are up- or downregulated by cold stress (|log2| ≥ 1, adjusted P < 0.05) and are up- or downregulated in the hy5-215 or bbx7 bbx8 mutant compared with wild-type Col-0 under cold stress. A total of 6,214 genes were identified in wild-type seedlings after cold treatment (Supplemental Data Set S2), which were defined as COR genes. Among these, 729 COR genes (293 upregulated and 436 downregulated) were regulated by BBX7/8, representing 11.7% of all COR genes (Figure 8A;Supplemental Data Set S3). Meanwhile, 618 COR genes (295 upregulated and 323 downregulated) were regulated by HY5, representing 9.9% of all COR genes (Figure 8A;Supplemental Data Set S4).
Figure 8.
Transcriptome profiling analyses identify cold-regulated BBX7/8- and HY5-dependent genes. A, Venn diagrams indicating the numbers of BBX7/8- and HY5-regulated COR genes. B, Venn diagram indicating the numbers of COR genes regulated by BBX7/8, HY5, and CBFs. C, GO enrichment analysis of BBX7/8- and HY5-regulated COR genes described in (A). Left panel indicates BBX7/8- and HY5-coactivated COR genes (145), and right panel indicates BBX7/8- and HY5-corepressed COR genes (252). D, Expression of BBX7/8- and HY5-induced COR genes in Col-0, bbx7 bbx8, hy5-215, cry2-1, cop1-4, and cbfs under cold treatment. Expression in Col-0 at 22°C was set to 1.00. ACTIN2/8 was used as a normalization control. Data are the means of three independent experiments ± sd. Asterisks indicate significant differences compared with Col-0 under the same treatment (*P < 0.05, **P < 0.01, Student’s t test).
Subsequently, we analyzed COR genes that are regulated by both BBX7/8 and HY5; 397 COR genes (145 upregulated, 252 downregulated) were identified (Figure 8B;Supplemental Data Set S5). These data demonstrate that BBX7/8 and HY5 share a large set of common downstream genes. As expected, when we compared these genes with CBF-dependent COR genes (http://www.ncbi.nlm.nih.gov/sra/PRJNA732005; |log2| ≥ 1, adjusted P < 0.05), only 49 genes were found to be commonly regulated by CBFs and the HY5-BBX7/8 module (Figure 8B), represent only ∼6.8% of CBF-regulated COR genes. These data suggest that HY5-BBX7/8 regulate COR genes, primarily in a CBF-independent manner.
Gene Ontology (GO) enrichment analysis revealed that the COR genes that were upregulated by HY5-BBX7/8 were mainly involved in the biosynthesis and metabolism of anthocyanins, flavonoids, aromatic compounds, and amino acid derivatives and the response to ultraviolet signals. In contrast, HY5-BBX7/8-downregulated COR genes were mainly enriched in oxidation–reduction, response to hypoxia, nitrate, and ion stress and ion transport (Figure 8C). To confirm the changes in expression of HY5-BBX7/8-downregulated COR genes in wild-type, bbx7 bbx8, and hy5-215 plants, we performed qRT-PCR in plants under a time-course of cold treatment. As expected, several anthocyanin biosynthesis-related genes (such as TRANSPARENT TESTA4 [TT4], TT5, and TT8) showed constitutively reduced expression in bbx7 bbx8, hy5-215, and cry2-1 compared with the wild-type before cold treatment. Moreover, their cold-induced expression in these mutants was significantly suppressed compared with the wild-type (Figure 8D;Supplemental Data Set S6–S8), suggesting that these genes are indeed positively regulated by BBX7/8, HY5, and CRY2 under both normal conditions and cold stress and that this regulation is more prominent under cold stress. The expression patterns of these genes in cop1 were opposite to those in bbx7 bbx8, hy5-215, and cry2-1 mutants, whereas there were no significant differences in the expression of these genes between the cbfs mutant and wild-type (Figure 8D). Taken together, these results suggest that the CRY2–COP1–HY5–BBX7/8 module regulates cold acclimation by modulating the expression of a set of COR genes, primarily independently of the CBF signaling pathway.
Discussion
Many components that participate in light signaling pathways contribute to plant cold tolerance (Franklin and Whitelam, 2007; Catala et al., 2011; Lee and Thomashow, 2012; Jiang et al., 2017, 2020). However, whether CRY-mediated blue-light signaling is involved in plant cold responses remains unclear. In this study, we provide evidence for the important roles of the blue-light receptors CRYs in regulating plant cold acclimation and freezing tolerance. First, phenotypic analyses revealed that CRY1 and CRY2 positively regulate freezing tolerance, and blue-light mimics cold acclimation to enhance plant freezing tolerance. Second, biochemistry analyses demonstrated that cold promotes the stability of blue light-induced phosphorylated CRY2, thereby enhancing its activity. Third, at low temperatures, the interaction of phosphorylated CRY2 with COP1 is enhanced, which releases HY5 from COP1, thereby preventing COP1-mediated HY5 degradation. Fourth, RNA-seq demonstrated that approximately half of CRY-regulated genes are COR genes. In addition, BBX7 and BBX8 redundantly function downstream of HY5 to positively regulate plant freezing tolerance by modulating the expression of a set of COR genes, which occurs largely independently of the CBF pathway. These findings thus uncover how CRY2 governs plant cold acclimation through a CBF-independent COP1–HY5–BBX7/8–CORs cold signaling pathway (Figure 9).
Figure 9.
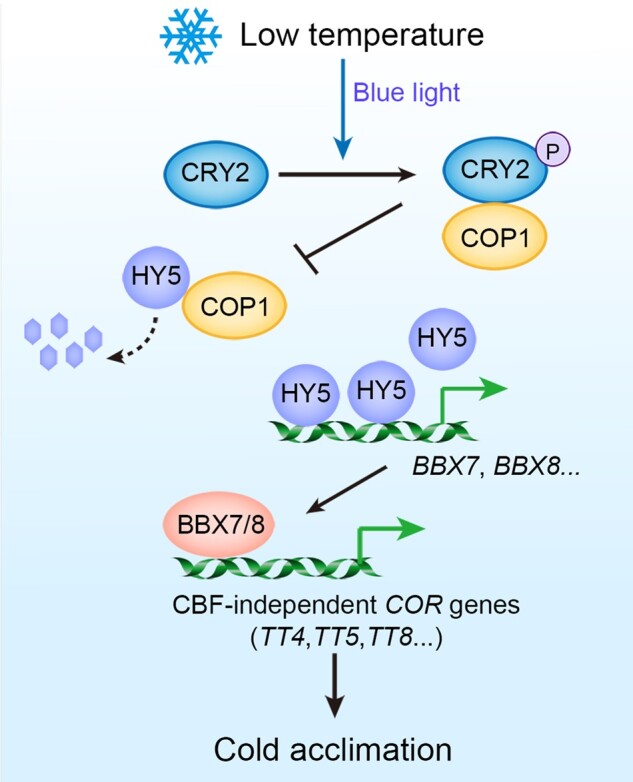
A working model for the CRY2–COP–HY5–BBX7/8-CORs cold signaling pathway. Under cold stress, phosphorylated CRY2 (induced by blue light) is stabilized and interacts with COP1 to compete with HY5, thereby inhibiting the degradation of HY5. BBX7 and BBX8 function as direct HY5 targets to positively regulate freezing tolerance by modulating the expression of a set of COR genes, including anthocyanin biosynthetic genes, mainly independently of the CBF pathway.
The red light photoreceptor phyB functions as a thermosensor that perceives ambient temperature changes (10–30°C) both during the daytime and at night (Jung et al., 2016; Legris et al., 2016; Qiu et al., 2019). Moreover, we recently showed that phyB is stabilized by cold, which promotes the degradation of PIF1, PIF4, and PIF5, thus leading to increased CBF protein accumulation, COR expression, and freezing tolerance in Arabidopsis (Jiang et al., 2020). In this study, we uncovered the important role of CRY2 in promoting acquired freezing tolerance in a CBF-independent manner. Notably, the enhanced stabilization of phosphorylated CRY2 upon cold stress only occurs in the presence of blue light during the daytime. These findings indicate that both phyB and CRY2 are necessary for the full acquisition of cold acclimation: phyB enhances plant cold tolerance in both the light and dark, at least in part through the well-characterized CBF pathway, whereas CRY2 positively regulates plant cold responses only in the light independently of the CBF pathway. Consistent with these observations, a recent study showed that the dark reversion rate of the cryptochrome flavin from the active state (FADH°) to the inactive state (FADOX) was lower at 15°C than at 25°C (Pooam et al., 2021). A similar scenario was observed in liverwort (M. polymorpha), showing that the conversion of phototropin from its phosphorylated active form to its nonphosphorylated inactive form was delayed at low temperature in a blue light-dependent manner (Fujii et al., 2017). Further study will be needed to dissect whether CRY2 acts as a cold sensor to perceive cold signals directly.
Two E3 ligases, the CUL4-based COP1–SPA1 E3 ligase complex and CUL3-based Bric-a-Brac/Tramtrack/Broad (LRB 1–3) E3 ligases, are known to mediate the blue light-dependent degradation of CRY2 (Liu et al., 2016; Chen et al., 2021). CUL3LRBs target photoactivated and phosphorylated CRY2 for rapid ubiquitination and degradation, whereas CUL4COP1/SPA is responsible for sustained ubiquitination and degradation of CRY2 in plants under prolonged exposure to light (Liu et al., 2016; Chen et al., 2021). The increased stability of phosphorylated CRY2 at low temperatures may be partly due to the reduced activity of proteasomes at low temperatures (Herbel et al., 2013). Consistently, the blue light-dependent degradation of CRY2 mediated by COP1 was dramatically suppressed by cold stress (Supplemental Figure S8). It is also possible that the function of LRBs in degrading CRY2 is compromised at low temperatures, which awaits further investigation.
CRY1 and CRY2 compete with COP1 substrates for interaction with COP1, repressing the ubiquitin ligase activity of COP1 and thereby stabilizing COP1 substrates during photomorphogenesis (Ponnu et al., 2019). Here, we demonstrated that cold-stabilized phosphorylated CRY2 interferes with the association of COP1 and HY5, promoting HY5 accumulation in the light. In agreement with this finding, HY5 stability decreased in the cry2-1 mutant but increased in GFP-CRY2 overexpressing lines compared with the wild-type. The nucleocytoplasmic partitioning of COP1 is regulated by light at 22°C, and multiple photoreceptors, including both Phys and CRYs, mediate the light-induced depletion of COP1 from the nucleus (von Arnim and Deng, 1994; Osterlund and Deng, 1998). A previous study showed that a YFP–COP1 fusion protein was barely detectable in the nucleus in the dark at 4°C (Catala et al., 2011). We propose that cold induces the nuclear depletion of COP1 in the dark, while cold-stabilized CRY2 inhibits the interaction of COP1 with HY5 in blue light; cold thus represses COP1 activity both in the dark and in blue light.
CBF transcription factors are classified as “first wave” transcription factors, as they are rapidly and transiently induced by cold, reaching their expression peak after 2–3 h of cold treatment before quickly returning to basal levels (Park et al., 2015). In this study, BBX7 and BBX8 expression reached its peak after 9 h of cold treatment before decreasing, although these genes remained more highly expressed 24 h into cold treatment than before exposure to cold. These data suggest that BBX7 and BBX8 may belong to a “second wave” of transcription factors (Park et al., 2015). Our genetic and biochemical evidence demonstrates that BBX7/8 act downstream of HY5 and commonly regulate a set of COR genes; among these, only 6.8% are governed by CBFs. Therefore, HY5-BBX7/8 mediate a cold signaling pathway that is largely independent of the CBFs. Our RNA-seq data reveal that a set of HY5-BBX7/8-co-regulated COR genes are involved in anthocyanin biosynthesis, which is consistent with the finding that HY5 regulates anthocyanin biosynthesis during cold stress (Catala et al., 2011). Considering that anthocyanins act as antioxidants to remove reactive oxygen species (Winkel-Shirley, 2002; Pourcel et al., 2007) and thus contribute to plant freezing tolerance (Li et al., 2017b), the HY5–BBX7/8 module might promote freezing tolerance (at least partially) by positively regulating the expression of anthocyanin biosynthetic genes.
Materials and methods
Plant materials and growth conditions
The Arabidopsis thaliana Columbia (Col-0) accession was used as a wild-type. The cry2-1, cop1-4, cop1-6, cry1-104, cry1 cry2, cop1-4 cry1 cry2 (cop1 cry1 cry2) (Mao et al., 2005), cbfs (Jia et al., 2016), and hy5-215 (Gangappa et al., 2013) mutants; 35S:GFP-CRY1 (Ma et al., 2016), 35S:GFP-CRY2 (Yu et al., 2009), and UBQ10:3×HA-HY5 (Li et al., 2020) transgenic plants used in this study were described previously. The T-DNA insertion mutant bbx8-1 (SALK_061961) was obtained from Arabidopsis Biological Resource Center (Columbus, OH).
The CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated 9) technique was performed as previously described (Xing et al., 2014) to generate the bbx7-1 and bbx7-2 single mutants. We searched for and identified 23-bp specific target sites (5′-N20NGG-3′) within exons of the BBX7 genomic sequence on the website http://cbi.hzau.edu.cn/cgi-bin/CRISPR. The targets were cloned into the pHSE401 vector, generating BBX7-pHSE401. The resulting vectors were transformed into wild-type Col-0 plants by the floral dip method (Clough and Bent, 1998). Vector construction and mutant identification were performed as described (Xing et al., 2014).
The hy5-215 BBX8-Myc, hy5-215 BBX7-Myc, cry2 HA-HY5, bbx7 bbx8, and cop1-4 x cop1-6 F1 plants were generated by crossing, and homozygous lines were genotyped and kept for further study. Unless otherwise indicated, Arabidopsis plants were grown at 22°C on 1/2 Murashige and Skoog (MS) medium (Sigma-Aldrich) containing 0.8% agar and 1.5% sucrose under LD conditions (16-h light/8-h dark). Plants at 22°C were grown under 80–100 μmol·m−2·s−1 cool-white fluorescent illumination and transferred to 4°C (for cold acclimation) under 20–25 μmol·m−2·s−1 cool-white fluorescent illumination. Both the blue and red light sources used in this study were LED cold light sources with a light intensity of 5–20 μmol·m−2·s−1.
Plasmid construction and plant transformation
Genomic fragments containing 1.5-, 2.0-, and 1.3-kb DNA sequences upstream of CRY2, BBX7, and BBX8 were amplified and cloned into pCAMBIA1300 (containing a GFP tag) to generate the CRY2:CRY2-GFP, BBX7:BBX7-GFP, and BBX8:BBX8-GFP constructs, respectively.
The CRY2 coding region was fused with GFP tag in the pSuper1300 vector (pCAMBIA1300 vector containing a Super promoter, which consists of three copies of the octopine synthase upstream-activating sequence in front of the manopine synthase promoter) (Ni et al., 1995) to generate Super:CRY2-GFP. The COP1 coding region was fused with HA and FLAG tag in the pCM1307 to generate the 35S:HF-COP1 construct. The HY5, BBX7, and BBX8 coding regions were fused with MYC tag in the pSuper1300 vector to generate the Super:HY5-Myc, Super:BBX7-Myc, and Super:BBX8-Myc constructs, respectively. The CRY2 coding region was fused with mCherry in the pSuper1300 vector to generate the Super:CRY2-mCherry construct.
All of the resulting vectors were transformed into Arabidopsis plants via the floral dip method (Clough and Bent, 1998). 35S:GFP, Super:CRY2-GFP, Super:BBX7-Myc, Super:BBX8-Myc, and 35S:HF-COP1 were transformed to wild-type Col-0. CRY2:CRY2-GFP was transformed into the cry2-1 mutant. BBX7:BBX7-GFP or BBX8:BBX8-GFP was transformed into the bbx7 bbx8 mutant. T3 homozygous transgenic plants were used for analysis. The specific primers used in this study are listed in Supplemental Data Set S9.
Freezing tolerance and ion leakage assays
Freezing tolerance and ion leakage assays were performed as described previously (Ding et al., 2015). Arabidopsis seedlings were grown at 22°C for 13 days on 1/2 MS plates containing 0.8% agar and subjected to the freezing assay conducted in a freezing chamber (Percival). Compared with noncold acclimation (NA), cold acclimation (CA)-treated seedlings were grown at 4°C under continuous white light for 2 days before the freezing assay. The freezing assays for both NA and CA seedlings were conducted as follows: seedlings were maintained in the dark at 0°C for 1 h, and the temperature was then dropped by 1°C/h until the temperature described in the figure legends was reached. After freezing treatment, the seedlings were shifted to 4°C and kept in the dark for 12 h before being transferred to LD conditions at 22°C for 3 days. The survival rate was then calculated based on the ability of green growth points to continue to generate new leaves.
After the freezing treatment and recovery for 3 days, the seedlings were collected in 15-mL tubes containing 10 mL of deionized water, and the electrical conductivity (EC) was measured as S0. The samples were gently shaken at 22°C for 30 min, and the resulting EC was measured as S1. The samples were boiled for 1 h and shaken at 22°C for 30 min, and the resulting EC was measured as S2. Electrolyte leakage was calculated using the formula: (S1-S0)/(S2-S0).
RNA extraction and qRT-PCR
Total RNA was extracted from 10-day-old seedlings with TRIzol Reagent (Invitrogen) and reverse transcribed by M-MLV reverse transcriptase (Promega). qRT-PCR was performed with a SYBR Green PCR Master Mix kit (Takara, Kusatsu, Japan). Analysis was performed using the Applied Biosystems StepOnePlus real-time PCR system. Whole plant seedlings from wild-type and different genotypes on the same petri dish were collected separately at the same time. Three independent experiments were conducted. Relative transcript levels were normalized to ACTIN2. The reaction and the calculation of relative expression levels were performed as described previously (Miura et al., 2007). The specific primers used in this study are listed in Supplemental Data Set S9.
Protein extraction and immunoblot analysis
Total proteins were extracted from the samples in immunoprecipitation (IP) buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM PMSF, 1 mM EDTA, 20 mM NaF, and 1× Protease inhibitor cocktail). The sample was centrifuged at 12,000g for 15 min. Total proteins were quantified, separated on 10% SDS/PAGE (Sodium Dodecyl Sulfate/Polyacrylamide Gel Electropheresis) gels, and transferred to PVDF membranes (Bio-Rad). Immunoblotting was performed using anti-CRY2 antibody (polyclonal antibody produced by Shanghai Youke Biotechnology, using Arabidopsis CRY2 protein as the antigen) at 1:1,000 (v/v) dilution, anti-HA antibody (Abmart, Cat. No: H3663) at 1:5,000 (v/v) dilution, anti-Actin antibody (EASYBIO, BE0027-100) at 1: 5,000 (v/v) dilution, anti-GFP antibody (Abmart, Cat. No: M20004) at 1:5,000 (v/v) dilution, and anti-HY5 antibody (Li et al., 2010) at 1:500 (v/v) dilution.
Bimolecular fluorescence complementation assays
BiFC assays were performed as described (Waadt and Kudla, 2008). The full-length coding sequence of COP1 was cloned into the pSPYCE vector (CE), resulting in COP1-YCE, while the full-length coding sequence of CRY2 was cloned into the pSPYNE vector (NE), resulting in CRY2-YNE. ICE1-YCE was used as a negative control. The COP1-YCE, CRY2-YNE, and CRY2-mCherry constructs were co-transformed into N. benthamiana leaves. Three days after infiltration, YFP fluorescence signals were observed under a confocal laser-scanning microscope (LSM710, Carl Zeiss).
Split LUC complementation assay
The split LUC complementation assay was performed as previously described (Chen et al., 2008). Vectors harboring COP1-cLUC, CRY2-mCherry, and HY5-nLUC were co-transformed into N. benthamiana leaves and expressed for 72 h, and the LUC signals were captured by a charge-coupled device camera (1300B; Roper) at −110°C within 15 minutes of exposure.
Co-immunoprecipitation assays
For co-IP experiments using 35S:CRY2-GFP and 35S:GFP (as a control), total proteins were extracted from plants in IP buffer (as mentioned above) after treatment at 22°C or 4°C for 1 h. The protein extracts were incubated with GFP-trap agarose beads (Chromotek, Cat. No: GTA-20) at 4°C for 2 h. The samples were washed four times with ice-cold IP buffer (without protease inhibitors), and 5× loading buffer was added before separating the samples on 8% SDS/PAGE gels. Immunoblotting was performed using anti-GFP antibody at 1:5,000 (v/v) dilution or anti-COP1 antibody (Saijo et al., 2003) at 1:1,000 (v/v) dilution.
For co-IP experiments, the 35S:HF-COP1, 35S:HF-ICE1 (Ding et al., 2015), Super:CRY2-mCherry, and Super:HY5-Myc plasmids were purified and transformed into Arabidopsis mesophyll protoplasts. Total proteins were extracted with IP buffer, and the protein extracts were incubated with anti-HA beads (Sigma-Aldrich, Cat. No: A7470) at 4°C for 2 h. The samples were washed four times with IP buffer and used for immunoblotting with anti-HA, anti-Myc (Sigma-Aldrich, Cat. No: M4439), and anti-CRY2 antibodies.
Electrophoretic mobility shift assay
EMSA was performed using biotin-labeled probes and a Light Shift Chemiluminescent EMSA Kit (Thermo Fisher) with minor modifications. Purified GST-HY5 protein was added to the binding reaction; GST protein was used as a negative control. The binding reactions were incubated at 25°C for 30 min and separated on 8% native polyacrylamide gels in 0.5×TBE (Tris-Borate-EDTA) buffer. The biotin-labeled and mutant probes used in this study are shown in Supplemental Data Set S9.
Chromatin immunoprecipitation assays
ChIP assays were performed as described previously (Li et al., 2017a) with minor modifications. HA-HY5 or wild-type (Col-0) seedlings were grown at 22°C on 1/2 MS medium for 2 weeks under LD conditions. A 2 g plant tissue sample was used in the ChIP experiment. Chromatin was isolated from the samples and sonicated, and DNA fragments associated with HA-HY5 protein were coimmunoprecipitated using anti-HA beads (Sigma-Aldrich). The enrichment of DNA fragments was quantified by qRT-PCR using the primers listed in Supplemental Data Set S9.
Dual-LUC assay
For the dual-LUC assay, the BBX7 and BBX8 promoters were fused to the firefly LUC gene in the pGreenII 0800-LUC plasmid (Hellens et al., 2005). The plasmids were purified and transformed into Arabidopsis mesophyll protoplasts. Following incubation in the dark for 16 h and treatment in the light at 22°C or 4°C for 1 h, the protoplasts were collected for the dual-LUC assay. The Promega dual-LUC reporter assay system was used for the dual-LUC assays according to the manufacturer’s instructions. The specific primers used in this study are listed in Supplemental Data Set S9.
RNA-seq analysis
Ten-day-old Col-0, hy5-215 and bbx7 bbx8 seedlings grown under LD conditions were treated at 4°C for 0, 8, or 24 h. Total RNA was extracted from the plants with TRIzol Reagent (Invitrogen) for subsequent sequencing analysis. The libraries were sequenced on the Illumina NovaSeq 6000 platform and 150-bp paired-end reads were generated. Raw data (raw reads) in fastq format were subjected to quality control using FastQC (v.0.11.9) (Davis et al., 2013). The reads were mapped to the Arabidopsis genome (TAIR10) using HISAT2 (v.2.2.0) (Kim et al., 2019) with default parameters. The read counts of each gene were obtained by FeatureCounts (v.2.0.1) (Liao et al., 2014), and the TPM of each gene was calculated using an in-house R script. Differential-expression analysis was performed using DESeq2 (v.1.30.0 R package) (Love et al., 2014). P-values were adjusted using the Benjamini–Hochberg procedure (Benjamini and Hochberg, 1995). Differentially expressed genes were defined based on the following criteria: a DESeq2 adjusted P <0.05 and a twofold-change cut-off in expression compared with the control samples. Three independent replicates were performed in this experiment.
Statistical analysis
For statistical tests related to RNA-seq, see the RNA-seq analysis section. All statistical tests not related to RNA-seq analysis were conducted in GraphPad Prism™ version 9. One-way analysis of variance and Tukey’s multiple comparison tests or two-tailed Student’s t-test were performed. The statistical results are shown in Supplemental File S1.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under accession numbers CRY2 (AT1G04400), CRY1 (AT4G08920), COP1 (AT2G32950), HY5 (AT5G11260), CBF1 (AT4G25490), CBF2 (AT4G25470), CBF3 (AT4G25480), COR15B (AT2G42530), GOLS3 (AT1G09350), RD29A (AT5G52310), BBX7 (AT3G07650), BBX8 (AT5G48250), TT4 (AT5G13930), TT5 (AT3G55120), TT8 (AT4G09820).
RNA-seq data were submitted to the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under accession number SRP327295.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Gene and protein levels of CRYs in the corresponding transgenic plants, and ion leakage of cry mutant and CRY-overexpression plants after freezing treatment.
Supplemental Figure S2. Cold promotes the stability of blue light-dependent phosphorylation of CRY2.
Supplemental Figure S3. HY5 positively regulates acquired freezing tolerance in Arabidopsis.
Supplemental Figure S4. Expression of CBF genes and their target genes in CRY2-COP1-HY5-related mutants.
Supplemental Figure S5. Immunoblot analysis of the HY5 proteins used in the transcriptional activation experiments described in Figure 6I.
Supplemental Figure S6. BBX7 and BBX8 positively regulate plant freezing tolerance.
Supplemental Figure S7. Gene and protein levels of BBX7 and BBX8 in the corresponding transgenic plants.
Supplemental Figure S8. The degradation of CRY2 by COP1 is attenuated under cold stress.
Supplemental Data Set S1. Cold-responsive (COR) genes in wild-type plants (Col-8 h/Col-0 h and Col-24 h/Col-0 h).
Supplemental Data Set S2. BBX7/8-regulated COR genes (Col-8 h/bbx7 bbx8-8 h and Col-24 h/bbx7 bbx8-24 h).
Supplemental Data Set S3. HY5-regulated COR genes (Col-8 h/hy5-215-8 h and Col-24 h/hy5-215-24 h).
Supplemental Data Set S4. BBX7/8 and HY5 co-regulated COR gene expression.
Supplemental Data Set S5. BBX7/8-regulated genes at 22°C (Col-0 h/bbx7 bbx8-0 h).
Supplemental Data Set S6. HY5-regulated genes at 22°C (Col-0 h/hy5-215-0 h).
Supplemental Data Set S7. Differences in COR genes co-regulated by HY5 and BBX7/8 at 22°C (hy5-215-0 h/Col-0 h and bbx7 bbx8-0 h/Col-0 h).
Supplemental Data Set S8. Overlap of blue-light-responsive genes regulated by CRYs and COR genes.
Supplemental Data Set S9. Primers used for qRT-PCR.
Supplemental File S1. Statistical analyses.
Supplementary Material
Acknowledgments
We thank Drs Hongquan Yang and Rongcheng Lin for providing the seeds of mutants and transgenic plants.
Funding
This work was supported by the National Key Research and Development Project (2020YFA0509902) and the National Natural Science Foundation of China (31872658, 32022008 and 31921001).
Conflict of interest statement. The authors declare no conflict of interests.
S.Y. supervised the project. Y.L. performed the experiments. D.F. and S.W. analyzed the RNA-seq data. H.L. and J.L. provides resources. All authors discussed and interpreted the data. Y.L. and S.Y. wrote the article with the help of Y.S. and J.L.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Shuhua Yang (yangshuhua@cau.edu.cn).
References
- Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate—a practical and powerful approach to multiple testing. J Royal Stat Soc B Stat Methodol 57: 289–300 [Google Scholar]
- Blazquez MA, Ahn JH, Weigel D (2003) A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet 33: 168–171 [DOI] [PubMed] [Google Scholar]
- Bursch K, Toledo-Ortiz G, Pireyre M, Lohr M, Braatz C, Johansson H (2020) Identification of BBX proteins as rate-limiting cofactors of HY5. Nat Plants 6: 921–928 [DOI] [PubMed] [Google Scholar]
- Catala R, Medina J, Salinas J (2011) Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc Natl Acad Sci USA 108: 16475–16480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Hu X, Liu S, Su T, Huang H, Ren H, Gao Z, Wang X, Lin D, Wohlschlegel JA, et al. (2021) Regulation of Arabidopsis photoreceptor CRY2 by two distinct E3 ubiquitin ligases. Nat Commun 12: 2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M (2008) LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-Box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MPA, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ (2013) Kraken: A set of tools for quality control and analysis of high-throughput sequence data. Methods 63: 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Shi Y, Yang S (2020) Molecular regulation of plant responses to environmental temperatures. Mol Plant 13: 544–564 [DOI] [PubMed] [Google Scholar]
- Ding Y, Li H, Zhang X, Xie Q, Gong Z, Yang S (2015) OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell 32: 278–289 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC (2007) Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat Genet 39: 1410–1413 [DOI] [PubMed] [Google Scholar]
- Fujii Y, Tanaka H, Konno N, Ogasawara Y, Hamashima N, Tamura S, Hasegawa S, Hayasaki Y, Okajima K, Kodama Y (2017) Phototropin perceives temperature based on the lifetime of its photoactivated state. Proc Natl Acad Sci USA 114: 9206–9211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2014) The BBX family of plant transcription factors. Trends Plant Sci 19: 460–470 [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Crocco CD, Johansson H, Datta S, Hettiarachchi C, Holm M, Botto JF (2013) The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 25: 1243–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad M, Albaqami M, Pooam M, Kernevez E, Witczak J, Ritz T, Martino C, Ahmad M (2020) Cryptochrome mediated magnetic sensitivity in Arabidopsis occurs independently of light-induced electron transfer to the flavin. Photochem Photobiol Sci 19: 341–352 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng Y, Lin F, Jiang Y, Ding M, Yan T, Lan H, Zhou H, Zhao X, Xu D, Deng XW (2019) B-Box containing proteins BBX30 and BBX31, acting downstream of HY5, negatively regulate photomorphogenesis in Arabidopsis. Plant Physiol 180: 497–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hense A, Herman E, Oldemeyer S, Kottke T (2015) Proton transfer to flavin stabilizes the signaling state of the blue light receptor plant cryptochrome. J Biol Chem 290: 1743–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbel V, Orth C, Wenzel R, Ahmad M, Bittl R, Batschauer A (2013) Lifetimes of Arabidopsis cryptochrome signaling states in vivo. Plant J 74: 583–592 [DOI] [PubMed] [Google Scholar]
- Jia Y, Ding Y, Shi Y, Zhang X, Gong Z, Yang S (2016) The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol 212: 345–353 [DOI] [PubMed] [Google Scholar]
- Jiang B, Shi Y, Zhang X, Xin X, Qi L, Guo H, Li J, Yang S (2017) PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc Natl Acad Sci USA 114: E6695–E6702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Shi Y, Peng Y, Jia Y, Yan Y, Dong X, Li H, Dong J, Li J, Gong Z, et al. (2020) Cold-induced CBF-PIF3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in Arabidopsis. Mol Plant 13: 894–906 [DOI] [PubMed] [Google Scholar]
- Job N, Yadukrishnan P, Bursch K, Datta S, Johansson H (2018) Two B-Box proteins regulate photomorphogenesis by oppositely modulating HY5 through their diverse C-terminal domains. Plant Physiol 176: 2963–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao MJ, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. (2016) Phytochromes function as thermosensors in Arabidopsis. Science 354: 886–889 [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C, Viczian A, Kircher S, Schafer E, Nagy F (2015) Molecular mechanisms for mediating light-dependent nucleo/cytoplasmic partitioning of phytochrome photoreceptors. New Phytol 206: 965–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Thomashow MF (2012) Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc Natl Acad Sci U S A 109: 15054–15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Ince YC, Fankhauser C (2019) Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat Commun 10: 5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CC, Neme M, Hiltbrunner A, Wigge PA, Schafer E, Vierstra RD, Casal JJ (2016) Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354: 897–900 [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ye K, Shi Y, Cheng J, Zhang X, Yang S (2017a) BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol Plant 10: 545–559 [DOI] [PubMed] [Google Scholar]
- Li J, Li G, Wang H, Wang Deng X (2011) Phytochrome signaling mechanisms. Arabidopsis Book 9: e0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Terzaghi W, Gong Y, Li C, Ling JJ, Fan Y, Qin N, Gong X, Zhu D, Deng XW (2020) Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat Commun 11: 1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee JH, Zhang H, Shen Y, Wang H, et al. (2010) Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Li YJ, Zhang FJ, Zhang GZ, Jiang XY, Yu HM, Hou BK (2017b) The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J 89: 85–103 [DOI] [PubMed] [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Lin F, Jiang Y, Li J, Yan T, Fan L, Liang J, Chen ZJ, Xu D, Deng XW (2018) B-BOX DOMAIN PROTEIN28 negatively regulates photomorphogenesis by repressing the activity of transcription factor HY5 and undergoes COP1-mediated degradation. Plant Cell 30: 2006–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang Q, Liu B, Wang W, Wang X, Park J, Yang Z, Du X, Bian M, Lin C (2016) The blue light-dependent polyubiquitination and degradation of Arabidopsis cryptochrome2 requires multiple E3 ubiquitin ligases. Plant Cell Physiol 57: 2175–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Gen Biol 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu XD, Zhou CM, Xu PB, Luo Q, Lian HL, Yang HQ (2015) Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant 8: 467–478 [DOI] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H (2016) Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc Natl Acad Sci U S A 113: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Zhang YC, Sang Y, Li QH, Yang HQ (2005) From The Cover: A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci U S A 102: 12270–12275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM (2007) SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ni M, Cui D, Einstein J, Narasimhulu S, Vergara CE, Gelvin SB (1995) Strength and tissue-specificity of chimeric promoters derived from the octopine and mannopine synthase Genes. Plant J 7: 661–676 [Google Scholar]
- Osterlund MT, Deng XW (1998) Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J 16: 201–208 [DOI] [PubMed] [Google Scholar]
- Park E, Park J, Kim J, Nagatani A, Lagarias JC, Choi G (2012) Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J 72: 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Lee CM, Doherty CJ, Gilmour SJ, Kim Y, Thomashow MF (2015) Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J 82: 193–207 [DOI] [PubMed] [Google Scholar]
- Pedmale UV, Huang SC, Zander M, Cole BJ, Hetzel J, Ljung K, Reis PAB, Sridevi P, Nito K, Nery JR, et al. (2016) Cryptochromes Interact Directly with PIFs to Control Plant Growth in Limiting Blue Light. Cell 164: 233–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnu J, Riedel T, Penner E, Schrader A, Hoecker U (2019) Cryptochrome 2 competes with COP1 substrates to repress COP1 ubiquitin ligase activity during Arabidopsis photomorphogenesis. Proc Natl Acad Sci U S A [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooam M, Dixon N, Hilvert M, Misko P, Waters K, Jourdan N, Drahy S, Mills S, Engle D, Link J, et al. (2021) Effect of temperature on the Arabidopsis cryptochrome photocycle. Physiol Plant 172: 1653–1661 [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Cheynier V, Lepiniec L, Debeaujon I (2007) Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci 12: 29–36 [DOI] [PubMed] [Google Scholar]
- Qiu YJ, Li MN, Kim RJA, Moore CM, Chen M (2019) Daytime temperature is sensed by phytochrome B in Arabidopsis through a transcriptional activator HEMERA. Nat Commun 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeber VM, Bajaj I, Rohde M, Schmulling T, Cortleven A (2021) Light acts as a stressor and influences abiotic and biotic stress responses in plants. Plant Cell Environ 44: 645–664 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C (2002) Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763–767 [DOI] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, Oven-Krockhaus SZ, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Ding Y, Yang S (2018) Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci 23: 623–637 [DOI] [PubMed] [Google Scholar]
- Song Y, Zhang X, Li M, Yang H, Fu D, Lv J, Ding Y, Gong Z, Shi Y, Yang S (2021) The direct targets of CBFs: in cold stress response and beyond. J Integr Plant Biol doi: 10.1111/jipb.13161 [DOI] [PubMed] [Google Scholar]
- Song Z, Bian Y, Liu J, Sun Y, Xu D (2020) B-box proteins: Pivotal players in light-mediated development in plants. J Integr Plant Biol 62: 1293–1309 [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- von Arnim AG, Deng XW (1994) Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Waadt R, Kudla J (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc 2008: pdb prot4995. [DOI] [PubMed] [Google Scholar]
- Wang Q, Lin CT (2020) Mechanisms of cryptochrome-mediated photoresponses in plants. Annu Rev Plant Biol 71: 103–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zuo ZC, Wang X, Gu LF, Yoshizumi T, Yang ZH, Yang L, Liu Q, Liu W, Han YJ, Kim JI, Liu B, Wohlschlegel JA, Matsui M, Oka Y, Lin CT (2016) Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Xing H, Dong L, Wang Z, Zhang H, Han C, Liu B, Wang X, Chen Q (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DQ (2020) COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol 228: 1748–1753 [DOI] [PubMed] [Google Scholar]
- Xu DQ, Jiang Y, Li JG, Lin F, Holm M, Deng XW (2016) BBX21, an Arabidopsis B-box protein, directly activates HY5 and is targeted by COP1 for 26S proteasome-mediated degradation. Proc Natl Acad Sci USA 113: 7655–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A, Bakshi S, Yadukrishnan P, Lingwan M, Dolde U, Wenkel S, Masakapalli SK, Datta S (2019) The B-Box-containing microprotein miP1a/BBX31 regulates photomorphogenesis and UV-B protection. Plant Physiol 179: 1876–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XH, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang HY, Huang J, Lee J, Kaufman L, Lin CT (2009) Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21: 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Huai JL, Shang FF, Xu G, Tang WJ, Jing YJ, Lin R (2017) A PIF1/PIF3-HY5-BBX23 transcription factor cascade affects photomorphogenesis. Plant Physiol 174: 2487–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Zhang Z, Xie S, Si T, Li Y, Zhu JK (2016) Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol 171: 2744–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.