Abstract
Adjuvants are critical for improving the quality and magnitude of adaptive immune responses to vaccination. Lipid nanoparticle (LNP)-encapsulated nucleoside-modified mRNA vaccines have shown great efficacy against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), but the mechanism of action of this vaccine platform is not well-characterized. Using influenza virus and SARS-CoV-2 mRNA and protein subunit vaccines, we demonstrated that our LNP formulation has intrinsic adjuvant activity that promotes induction of strong T follicular helper cell, germinal center B cell, long-lived plasma cell, and memory B cell responses that are associated with durable and protective antibodies in mice. Comparative experiments demonstrated that this LNP formulation outperformed a widely used MF59-like adjuvant, AddaVax. The adjuvant activity of the LNP relies on the ionizable lipid component and on IL-6 cytokine induction but not on MyD88- or MAVS-dependent sensing of LNPs. Our study identified LNPs as a versatile adjuvant that enhances the efficacy of traditional and next-generation vaccine platforms.
Keywords: adjuvant, vaccine, lipid nanoparticle, Tfh cell, SARS-CoV-2, influenza virus, germinal centers, IL-6
Graphical abstract
The mechanism of action of nucleoside-modified mRNA-LNP vaccines is unknown. Alameh et al. demonstrate that LNPs can possess adjuvant activity and promote robust induction of Tfh cell, B cell, and humoral responses when utilized in mRNA and protein subunit vaccines in mice. IL-6 induction and the ionizable lipid component are critical for the adjuvant activity of LNPs.
Introduction
Vaccines are essential to prevent or control infectious disease outbreaks, often through induction of protective antibody (Ab) responses (Iwasaki, 2016). The process leading to generation of high-affinity Abs occurs in germinal centers (GCs) of secondary lymphoid organs (Victora and Nussenzweig, 2012). In GCs, B cells undergo somatic hypermutation (SHM) of B cell receptor genes, which is followed by positive selection of high-affinity GC B cells. Ultimately, GC B cells differentiate into high-affinity memory B cells (MBCs) and Ab-secreting long-lived plasma cells (LLPCs) (Griffiths et al., 1984; McKean et al., 1984; Weigert et al., 1970). T follicular helper (Tfh) cells are a subset of CD4+ T cells specialized in regulating affinity maturation of B cells in GCs (Crotty, 2019; Vinuesa et al., 2016). Induction of Tfh cells is critical for durable, protective Ab responses (Crotty, 2019).
Recombinant protein subunit vaccines are often poorly immunogenic and require addition of adjuvants to enhance the induction of durable protective immune responses (Del Giudice et al., 2018). Despite ongoing efforts to identify safe adjuvants or novel vaccine platforms that induce strong Tfh cell responses, limited success has been achieved (Linterman and Hill, 2016). Recent studies have reported negligible or modest Tfh cell generation after immunization of mice and non-human primates with protein antigens formulated with US Food and Drug Administration (FDA)-approved adjuvants such as aluminum hydroxide and MF59 (Liang et al., 2017; Mastelic Gavillet et al., 2015; Olafsdottir et al., 2016). These findings may partially explain why currently licensed adjuvants do not support the generation of durable, broadly protective Abs against difficult pathogens, such as human immunodeficiency virus 1 (HIV-1), hepatitis C virus, influenza virus, Plasmodium species, and others. This underscores the critical need for new, more effective Tfh cell-promoting adjuvants (Havenar-Daughton et al., 2017; Linterman and Hill, 2016).
mRNA-based vaccines have recently proven highly effective against infectious diseases (Alameh et al., 2020; Bettini and Locci, 2021; Pardi et al., 2018b). One of the most promising vaccine platforms comprises nucleoside-modified mRNA encapsulated in lipid nanoparticles (mRNA-LNPs) (Pardi et al., 2015). Importantly, nucleoside-modified mRNA-LNP vaccines developed against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by Moderna and Pfizer/BioNTech have received approval for human use in multiple countries around the world. In previous studies, we demonstrated that a single dose of nucleoside-modified mRNA-LNP vaccines elicits potent Tfh cell and GC B cell responses as well as sustained and protective Ab responses against influenza virus infection in mice (Pardi et al., 2018a). Additionally, mRNA-LNPs induced superior Tfh cell responses compared with an adjuvanted protein subunit vaccine in rhesus macaques (Pardi et al., 2018a). The mechanism of Tfh cell induction by mRNA-LNP vaccines is not known. Several studies have demonstrated that nucleoside-modified mRNAs do not induce strong inflammatory responses (Karikó et al., 2008, 2011). Although the effects of LNPs on immune system cell activation have been investigated minimally, a number of studies have indicated that some LNPs could have intrinsic adjuvant activity (Awasthi et al., 2019b; Shirai et al., 2020; Swaminathan et al., 2016a, 2016b).
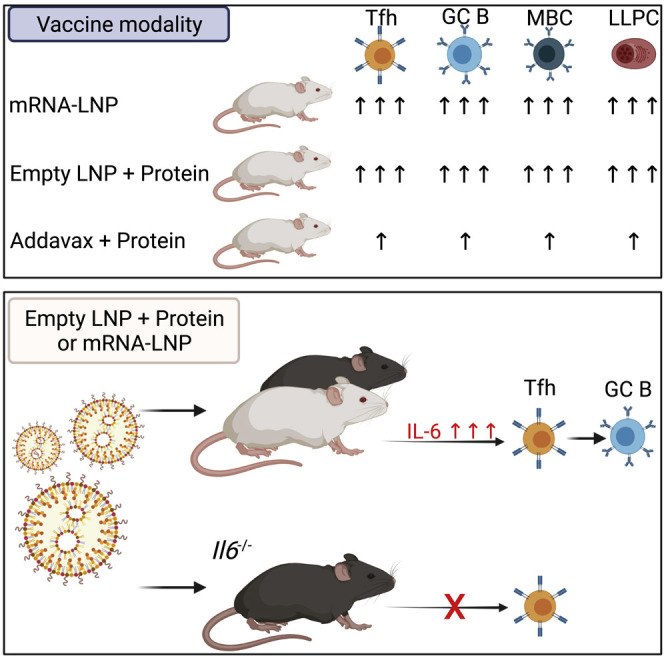
In this report, we demonstrated that the LNP formulation used in previous studies to deliver mRNA (Awasthi et al., 2019a; Freyn et al., 2020, 2021; Pardi et al., 2017, 2018a, 2018c, 2019; Weissman et al., 2021) is an effective Tfh cell-inducing adjuvant that can be utilized in mRNA and protein subunit vaccines. The induction of antigen-specific Tfh cells by LNP-containing protein vaccines was superior than that induced by AddaVax (an MF59-like adjuvant)-formulated vaccines and was coupled with generation of antigen-specific GC B cells, LLPCs, MBCs, and durable, protective Ab responses. Mechanistically, the capacity of this LNP formulation to elicit robust Tfh and GC B cell responses in mice depended on the presence of the ionizable lipid component and induction of the pro-Tfh cytokine interleukin-6 (IL-6). This conclusion was supported by a measurable wave of IL-6 production following LNP injection and by the deeply blunted Tfh and GC B cell responses in IL-6-deficient mice immunized with LNP-formulated protein and mRNA vaccines.
This study is an important advancement in the field of vaccine development because it identifies LNPs as a potent immunostimulatory component of mRNA vaccines and sheds light on the mechanism of Tfh cell induction of this recently licensed vaccine platform. Furthermore, our findings indicated that LNP formulations could be exploited as a potent adjuvant not only for mRNA vaccination but also for improving the efficacy of the FDA-approved protein subunit vaccine format.
Results
LNPs possess strong adjuvant activity and enhance the efficacy of protein subunit vaccines
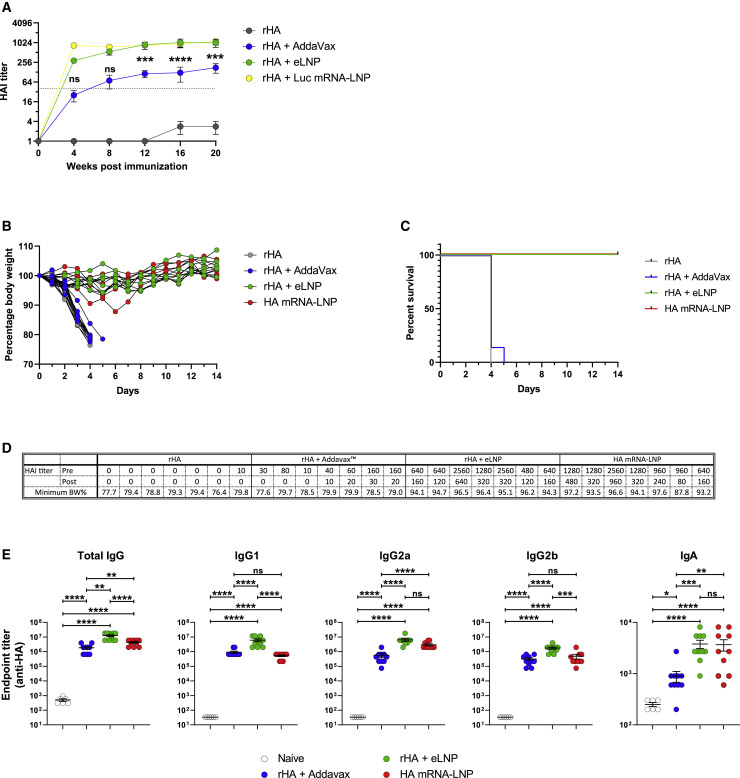
The nucleoside-modified mRNA-LNP vaccine platform is one of the most promising vaccine modalities and has the ability to induce robust cellular and humoral immune responses, as demonstrated by multiple preclinical (Alameh et al., 2020) and clinical studies (Jackson et al., 2020; Mulligan et al., 2020; Sahin et al., 2020; Walsh et al., 2020). The mechanism of action of this vaccine type has not been investigated in detail. The adjuvant activity of nucleoside-modified mRNA-LNPs can stem from the mRNA, from the LNP, or from both components. To address this, we prepared LNPs containing nucleoside-modified mRNA or no payload (empty LNP [eLNP]). We mixed A/Puerto Rico/8/1934 (PR8, H1N1) influenza virus hemagglutinin recombinant protein (rHA) with mRNA-LNP containing firefly luciferase (Luc)-encoding mRNA or with eLNP equivalent to the same amount of lipid and administered them intramuscularly (IM) to mice. Animals were bled 4, 8, 12, 16 and 20 weeks after immunization and serum hemagglutination inhibition (HAI) titers were measured as a simple readout of the quality and magnitude of humoral responses induced by vaccination (Figure 1 A). We found that rHA + eLNP and rHA + Luc mRNA-LNP induced similarly high PR8 HAI titers at most time points examined, demonstrating that this LNP formulation possesses adjuvant activity. Our study also included a group of mice that were injected with rHA + AddaVax as a benchmark. The HAI titers induced by LNP-containing vaccines were an order of magnitude higher compared with those elicited by the AddaVax-adjuvanted protein vaccine. Luc mRNA did not significantly influence the HAI titers induced by the LNP adjuvant. Non-adjuvanted rHA induced negligible HAI titers, as expected. Of note, LNP-adjuvanted vaccination-induced HAI titers remained stable for at least 20 weeks (Figure 1A). Most licensed vaccines are delivered IM, but some vaccines are administered intradermally (ID) (i.e., Fluzone ID); thus, we evaluated the various vaccine formats by ID inoculation (Figure S1). Similar to the IM route, the rHA + eLNP vaccine induced long-lasting high PR8 HAI titers after a single immunization, whereas the rHA + AddaVax vaccine induced lower HAI titers. In the ID study, we immunized a group of mice with PR8 HA mRNA-LNP and found that the mRNA-LNP vaccine induced PR8 HAI titers comparable with the rHA + eLNP vaccine.
Figure 1.
LNP-adjuvanted HA mRNA and protein subunit vaccines induce durable, protective humoral responses
(A) BALB/c mice received a single IM immunization with 10 μg of rHA mixed with eLNP (an equal amount of LNP as in 30 μg mRNA-LNP), 30 μg Luc mRNA-LNP, or AddaVax. PR8 HAI titers were followed for 20 weeks. Data are pooled from two independent experiments. n = 10 mice/group. The horizontal dotted line represents the protective HAI value (1:40). Data are shown as mean + SEM.
(B–D) BALB/c mice were immunized ID with 10 μg of rHA, rHA + Addavax, or rHA + eLNP or 10 μg HA mRNA-LNP. Animals were terminally bled 9 months after injection, and immune sera were transferred into naive mice. Mice were challenged with PR8 virus, and survival and weight loss were followed daily. n = 7 mice/group in two independent experiments.
(B) Weight of serum-transferred mice as percentage of starting weight.
(C) Survival as percentage of total number of live animals per group.
(D) HAI titers of pre-transfer immune sera and sera of injected mice at the time of infection (2 h after transfer). The lowest body weight percentage reached during 14 days after infection is shown.
(E) Titers of HA-binding IgG, IgG1, IgG2a, IgG2b, and IgA were determined by endpoint dilution ELISA using sera of immunized mice collected 4 weeks after IM immunization. n = 10 animals per treatment group, n = 6 naive animals in two independent experiments.
Statistical analysis: In (A), two-way ANOVA with Bonferroni’s multiple comparisons test; comparisons of AddaVax and eLNP groups are shown for each time point; ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. In (E), one-way ANOVA with Bonferroni’s multiple comparisons test was performed on log transformed values; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S1.
To unequivocally demonstrate that this LNP formulation, in combination with rHA, is a superior adjuvant compared with AddaVax at inducing protective Ab responses, we tested these vaccine formats in a challenge experiment. Mice were immunized with rHA alone or supplemented with AddaVax or eLNP, and one group was injected with HA mRNA-LNP. Mice were terminally bled 9 months after immunization, and sera from vaccinated animals were transferred into naive mice, which were subsequently infected with PR8 influenza virus. HAI titers of sera of immunized mice and serum-transferred mice were determined, and the morbidity and mortality of the serum recipient animals were followed daily for 2 weeks. Mice that received sera from rHA- or rHA + AddaVax-immunized animals started losing weight rapidly (as early as day 2) and were euthanized around day 4, when they reached the humane endpoint of 20% weight loss. However, no or only minimal weight loss could be observed in mice injected with rHA + eLNP or mRNA-LNP vaccine immune sera (Figures 1B–1D).
To further examine the quality of Ab responses, we performed ELISA assays and determined the endpoint dilution titers of immunoglobulin G (IgG), IgG1, IgG2a, IgG2b, and IgA induced in the serum 4 weeks after a single immunization with these vaccines. The rHA + eLNP vaccine induced significantly higher titers of total IgG and all tested IgG subclasses in comparison with rHA + AddaVax (Figure 1E). Of note, we were also able to detect low levels of IgA, and LNPs outperformed AddaVax in IgA induction too. These data indicate that our LNP formulation is capable of inducing functionally variegated Ab responses.
These experiments provided proof of concept that the LNP formulation used in this study has intrinsic adjuvant property, is at least partially responsible for the potency of nucleoside-modified mRNA-LNP vaccines, and might be repurposed as a standalone adjuvant for the protein subunit vaccine platform.
LNP-adjuvanted vaccines efficiently support Tfh cell differentiation
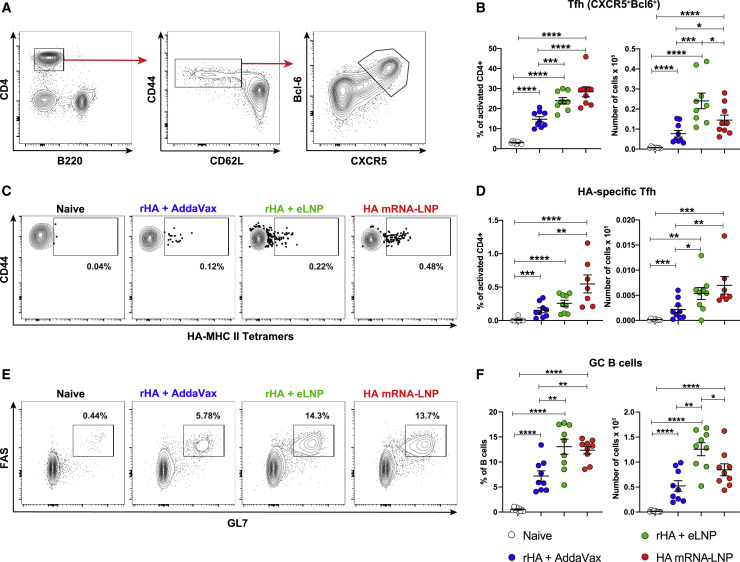
Because our LNP formulation induced high PR8 HAI titers (Figure 1), and Tfh cells play a fundamental role in regulating high-quality Ab responses, we hypothesized that LNP-adjuvanted protein vaccines have the capacity to efficiently induce Tfh cell differentiation. In line with our hypothesis, potent Tfh cell generation has been reported previously for nucleoside-modified mRNA-LNP vaccines (Lederer et al., 2020; Lindgren et al., 2017; Pardi et al., 2018a). To investigate Tfh cell responses, mice were immunized IM or ID with a single dose of rHA + eLNP or rHA + AddaVax. As a positive control, a group of mice was immunized with PR8 HA mRNA-LNP. Tfh cell responses were examined in the draining lymph nodes (dLNs) by flow cytometry 12 days after vaccination (Figures 2 and S2). Animals immunized with rHA + eLNP or HA mRNA-LNP presented higher numbers of CXCR5+Bcl6+ (Figures 2A, 2B, and S2A) and CXCR5+PD-1+ (Figures S2B–S2D) Tfh cells compared with rHA + AddaVax-immunized and naive animals, regardless of the route of vaccine administration. Analysis of PR8 HA-specific Tfh cell numbers, determined using PR8 HA-specific major histocompatibility complex (MHC) class II tetramers, yielded similar findings (Figures 2C, 2D, and S2E–S2G).
Figure 2.
LNP-adjuvanted HA mRNA and protein subunit vaccines induce robust antigen-specific Tfh cell and GC B cell responses
BALB/c mice received a single IM immunization with PR8 HA mRNA-LNP or rHA mixed with eLNP or AddaVax as described in Figure 1. Tfh and GC B cell responses were examined at day 12 in dLNs by flow cytometry.
(A and B) Tfh cell (CD4+B220−CD44hiCD62L−Bcl6+CXCR5+) representative gating strategy (A) and percentages and absolute numbers (B).
(C and D) HA-specific Tfh cell (HA-MHC class II tetramer+ Tfh cells) representative flow cytometry (C) and percentages and absolute numbers (D).
(E and F) GC B cell (CD4/CD8−CD19+FAS+GL7+) representative flow cytometry (E) and percentages and absolute numbers (F).
n = 7–9 mice/group. Data were combined from three independent experiments. Symbols represent individual animals. Data shown are mean + SEM. Statistical analysis: unpaired two-tailed Mann-Whitney U test was conducted. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figures S2 and S3.
T follicular regulatory (Tfr) cells are important modulators of Tfh and B cells in response to foreign Th2 antigens and autoantigens (Clement et al., 2019; Crotty, 2019; Fonseca et al., 2019). To investigate the balance of Tfh to Tfr cells after immunization with an LNP-adjuvanted vaccine, mice were immunized IM with a single dose of rHA + eLNP or rHA + AddaVax. Tfh and Tfr cell responses were measured in dLNs 7, 14, 21, and 28 days after immunization (Figure S3). In agreement with our previous study with nucleoside-modified mRNA-LNP vaccines (Lederer et al., 2020), we found that (Foxp3−) Tfh cells peaked around day 7 after immunization and were largely resolved by day 28 after immunization, as confirmed by a near-baseline Tfh/Tfr cell ratio by day 28 after immunization (Figures S3A and S3C). Additionally, neither vaccine substantially changed the absolute number of Tfr cells post vaccination at the time points analyzed (Figure S3B).
Because Tfh cells provide help to B cells in GCs, we hypothesized that LNP-adjuvanted vaccine formats can induce strong GC B cell responses in dLNs. Indeed, the rHA + eLNP and HA mRNA-LNP vaccines elicited potent differentiation of GC B cells (FAS+GL7+), as measured by flow cytometry (Figures 2E, 2F, and S2H). These data demonstrate that this LNP formulation efficiently induces Tfh and GC B cells after a single IM or ID immunization in mice.
LNP-adjuvanted vaccines induce strong LLPC and MBC responses
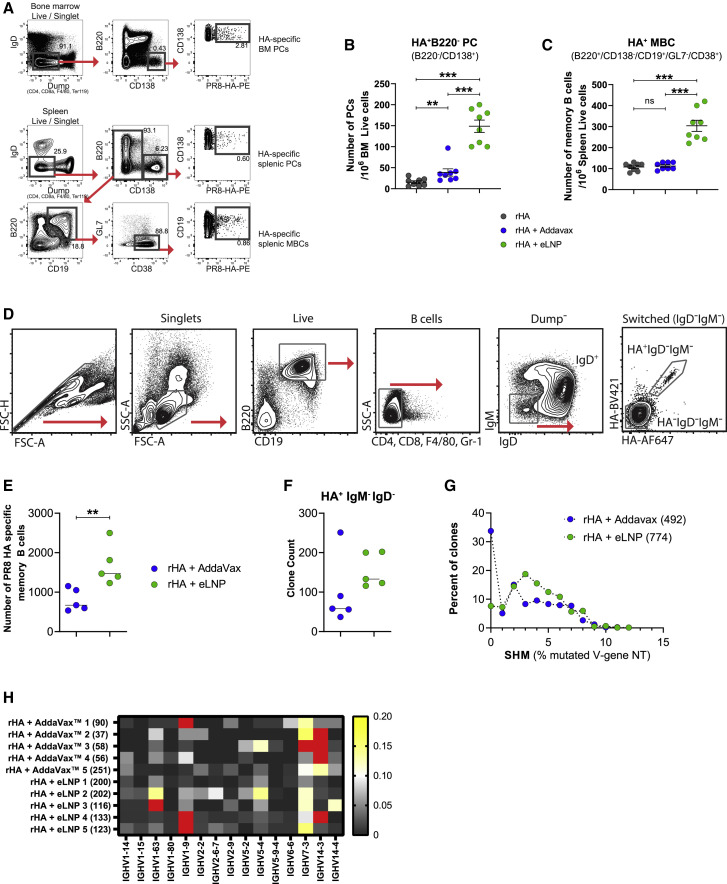
Durable, affinity-matured Ab responses are associated with the induction of GC reactions, giving rise to LLPCs and MBCs (Sallusto et al., 2010). Because we were able to measure strong Tfh cell, GC B cell, and durable Ab responses after a single immunization with LNP-adjuvanted vaccines (Figures 1, 2, and S1–S3), we hypothesized that this LNP formulation may also efficiently induce LLPC and MBC responses. To directly evaluate this hypothesis, mice were immunized IM or ID with a single dose of rHA + eLNP or rHA + AddaVax. Antigen-specific LLPC (IgD−/DUMP−/CD138+/B220−/PR8 HA+) and MBC (IgD−/DUMP−/CD138−/B220+/CD19+/CD38+/GL7−/PR8 HA+) absolute numbers were determined 20 weeks after immunization in the bone marrow and spleen, respectively (Figures 3 A–3C and S4). In agreement with its capacity to efficiently promote generation of Tfh and GC B cells, the rHA + eLNP vaccine induced stronger PR8 HA-specific LLPC and MBC responses than rHA + AddaVax.
Figure 3.
The LNP-adjuvanted protein vaccine induces strong antigen-specific LLPC and class-switched, somatically mutated MBC responses
(A–C) BALB/c mice received a single IM immunization with rHA or rHA mixed with eLNP or AddaVax as described in Figure 1. PR8 HA-specific LLPC and MBC responses were examined in bone marrow and spleen, respectively, 20 weeks after immunization. n = 8 mice/group. Symbols represent individual animals. Data shown are mean + SEM and are pooled from two independent experiments.
(A) Gating strategy for measuring HA-specific LLPC and MBC responses.
(B and C) PR8 HA-specific LLPC (IgD−Dump−B220−CD138+PR8 HA+, B) and MBC (IgD−Dump−CD138−B220+CD19+CD38+GL7−PR8 HA+, C) responses.
(D–H) Immune repertoire profiling of PR8 HA-binding MBCs. Mice received a single ID immunization with rHA formulated with eLNP or AddaVax. PR8 HA-specific MBCs were sorted from spleens 80 days after immunization.
(D) Sorting strategy.
(E) Number of PR8 HA-specific MBCs recovered from the sort.
(F) Clone counts of IgM−IgD− HA-binding cells. Symbols represent individual animals.
(G) HA-binding clones binned by their somatic hypermutation (SHM) percentage in the IGHV gene (compared with the nearest germline gene). Each clone counts only once.
(H) IGHV gene usage in sorted HA+ MBCs (the heatmap indicates the fraction of clones). Red cells denote values that are between 0.21–0.59.
In (G) and (H), IGHV genes present in at least 6 mice were included. Values in parentheses denote clone numbers. In (D)–(H), data were obtained from one experiment; n = 5 per group. Statistical analysis: in (B), (C), (E), and (F), unpaired two-tailed Mann-Whitney U test was conducted. ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001. See also Figure S4.
The LNP-adjuvanted protein subunit vaccine elicits SHM in MBCs
Ab diversity is critical to recognize a large number of foreign antigens and can be enhanced by SHM in the immunoglobulin genes. The process of SHM, coupled with selection for antigen binding, leads to affinity maturation (Griffiths et al., 1984; McKean et al., 1984; Weigert et al., 1970). Potent neutralizing Abs against certain pathogens (for example, HIV and influenza virus) are often highly somatically mutated (Sok et al., 2013; Victora and Wilson, 2015). Given the critical role of Tfh cells in driving Ab affinity maturation and our studies demonstrating strong Tfh and GC B cell differentiation after immunization with LNP-adjuvanted vaccines, we hypothesized that our LNP formulation can elicit Abs with high levels of SHM.
Mice were immunized with eLNP- or AddaVax-adjuvanted rHA vaccines as described above. Splenic IgD−IgM− HA-binding B cells were sorted (Figure 3D) at day 80 after immunization and subjected to bulk sequencing of Ab heavy-chain gene rearrangements, as we have described previously (Johnson et al., 2020). As additional controls, we also sorted and sequenced B cells that were IgD+ (naive) or HA−IgD−IgM− (class switched). Confirming our previous data (Figure 3C), there was a trend toward higher numbers of HA-binding class-switched cells (Figure 3E) and clones (Figure 3F) among the cells sorted from animals that received the rHA + eLNP vaccine compared with the rHA + AddaVax vaccine. Because of the relatively low number of HA-binding B cells, we pooled the data from each vaccine group to evaluate SHM. Mice that received the rHA + eLNP vaccine had fewer unmutated clones than mice that received the rHA + AddaVax vaccine (7.6% versus 33.7%) (Figure 3G). Furthermore, among clones with mutations, those derived from mice that received the rHA + eLNP vaccine had more of them, as evidenced by a right-shifted distribution of SHM that also presented a significantly higher median value (two-tailed Mann-Whitney test, p < 0.0001) (Figure 3G). Among HA binders, VH1-9, VH7-3, and VH14-3 were used most frequently in both vaccinated groups of mice (Figure 3H), in line with previous data from immunized C57BL/6 animals (Johnson et al., 2020). These data suggest that the rHA + eLNP vaccine is more effective than the rHA + AddaVax vaccine at generating larger numbers of diverse HA-binding MBC clones with a higher degree of somatic mutation.
LNPs can be utilized to generate potent mRNA and protein subunit vaccines against SARS-CoV-2
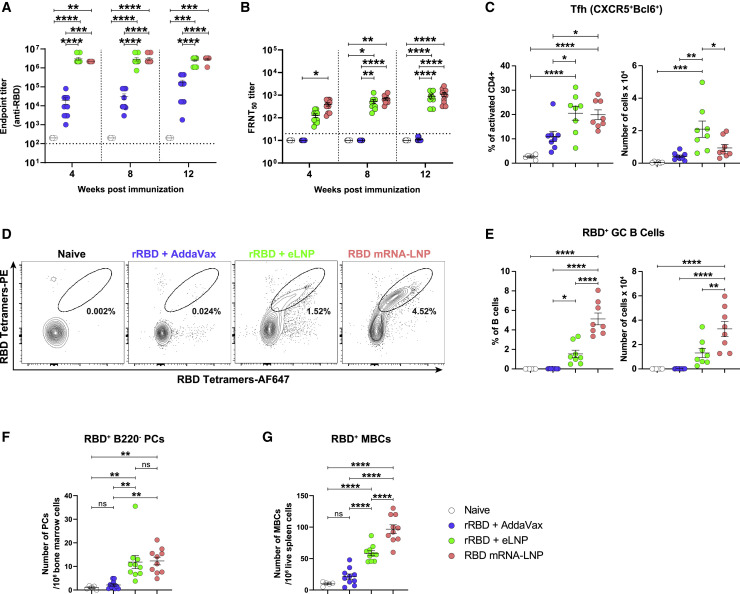
SARS-CoV-2 infection is a serious ongoing pandemic, and vaccines eliciting protective immune responses against the virus will be critical for resolving the current public health crisis (Krammer, 2020). We and others have demonstrated that SARS-CoV-2 nucleoside-modified mRNA-LNP vaccines induced robust, protective immune responses in animals as well as in humans (Bettini and Locci, 2021; Corbett et al., 2020a, 2020b; Jackson et al., 2020; Laczkó et al., 2020; Lederer et al., 2020; Mulligan et al., 2020; Sahin et al., 2020; Walsh et al., 2020; Weissman et al., 2021). To demonstrate that this LNP formulation can provide adjuvant activity for other antigens, we tested eLNP- and AddaVax-adjuvanted SARS-CoV-2 spike protein subunit vaccines in mice. Animals were immunized with a single dose of recombinant SARS-CoV-2 spike protein receptor binding domain (rRBD) adjuvanted with eLNP or AddaVax. As a positive control, a group of mice was immunized with nucleoside-modified RBD mRNA-LNP (Laczkó et al., 2020; Lederer et al., 2020). In line with our previous findings (Laczkó et al., 2020; Lederer et al., 2020), LNP-adjuvanted vaccines elicited durable RBD-specific IgG titers that were significantly higher than the AddaVax-elicited responses (Figure 4 A). Using a vesicular stomatitis virus (VSV)-based pseudovirus neutralization assay, we observed high and sustained neutralization titers with the LNP-adjuvanted vaccines and no measurable titers with the AddaVax-formulated rRBD vaccine, showing the superiority of this LNP formulation over AddaVax (Figure 4B).
Figure 4.
LNP-adjuvanted SARS-CoV-2 mRNA and protein subunit vaccines induce robust cellular and humoral immune responses
BALB/c mice received a single IM immunization with 10 μg of RBD mRNA-LNP or recombinant SARS-CoV-2 RBD (rRBD) formulated with eLNP or AddaVax.
(A) RBD-specific IgG titers were determined by endpoint dilution ELISA. Horizontal dotted line represents the limit of detection.
(B) Neutralizing Ab titers were measured using a VSV-based pseudovirus neutralization assay. Horizontal dotted line represents the limit of detection. Focus reduction neutralization titer 50% (FRNT50) titers below the limit of detection are reported as half of the limit of detection.
(C–E) Tfh and GC B cell responses were determined in dLNs 12 days after immunization. n = 8 mice/group.
(C) Percentages and absolute numbers of Tfh cells (CD4+B220−CD44hiCD62L−Bcl6+CXCR5+).
(D) Representative frequencies of RBD-specific GC B cells (CD4/CD8−CD19+FAS+GL7+RBD+).
(E) Frequencies and absolute numbers of RBD-specific GC B cells (defined as in D).
(F and G) Antigen-specific (F) LLPCs (IgD−Dump−B220−CD138+RBD+) in bone marrow and (G) MBCs (IgD−Dump−CD138−B220+CD19+CD38+GL7−RBD+) in the spleen 12 weeks after immunization.
In (A) (B), (F), and (G), n = 10 mice/group. In (A)–(C) and (E)–(G), symbols represent individual animals. Data are pooled from two independent experiments. Data shown are mean + SEM. Statistical analysis: in (A) and (B), two-way ANOVA with Bonferroni’s multiple comparisons test; differences are only shown within time points. In (C) and (E)–(G), unpaired two-tailed Mann-Whitney U test; ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figures S5 and S6.
Similar to the PR8 HA studies, we demonstrated that the rRBD + eLNP and RBD mRNA-LNP vaccines induced robust Tfh cell responses and antigen-specific GC B cell responses (Figures 4C–4E and S5). Moreover, LNP-adjuvanted vaccines induced robust antigen-specific LLPC and MBC responses 12 weeks after a single vaccine administration in the bone marrow and spleen, respectively (Figures 4F, 4G, and S6).
We provide evidence that the eLNP-adjuvanted SARS-CoV-2 rRBD vaccine induced potent cellular and humoral immune responses in mice after a single immunization, confirming our findings with rHA + eLNP vaccination.
The ionizable lipid is responsible for the adjuvant activity of the LNP formulation
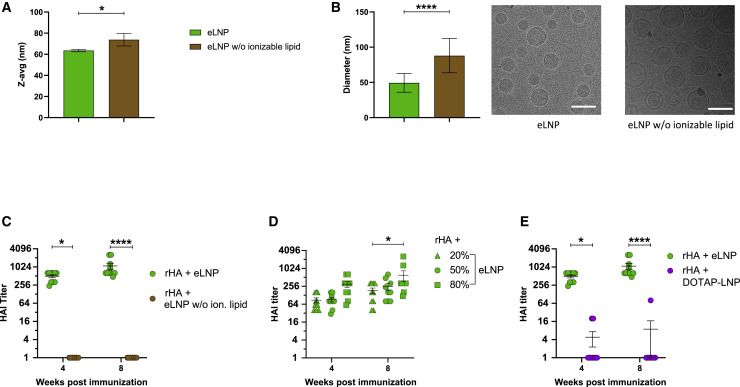
LNPs are prepared by mixing an aqueous solution with an organic solution comprised of an ionizable lipid, cholesterol, helper lipids, and a polyethylene glycol (PEG)-lipid at different molar ratios (Kulkarni et al., 2019). To investigate whether the adjuvant effect is due to the composition or physicochemical characteristics of the LNP, we generated eLNPs with or without the ionizable lipid. Using dynamic light scattering (DLS), we demonstrated that the average sizes (Z-average) of the two formulations were similar, with slightly larger particles formed in the absence of the ionizable lipid (Figure 5 A). eLNPs with or without the ionizable lipid formed similar, spherical structures, as seen by cryoelectron microscopy (cryo-EM) (Figure 5B). In line with the DLS data, omission of the ionizable lipid had the tendency to create slightly larger particles with typical unilamellar/liposome type structures. In contrast, eLNP with ionizable lipid showed typical electron-dense structures.
Figure 5.
The ionizable lipid component is critical for the adjuvant activity of LNPs
(A and B) Physicochemical characterization of the eLNPs with or without ionizable lipids determined by (A) DLS or (B) cryo-EM. The scale bars represent 100 nm.
(C) BALB/c mice received a single IM immunization with rHA mixed with eLNPs with or without the ionizable lipid, and PR8 HAI titers were determined 4 and 8 weeks after vaccine administration.
(D) BALB/c mice received a single IM immunization with 10 μg of rHA mixed with eLNPs with various amounts of ionizable lipid, and PR8 HAI titers were determined 4 and 8 weeks after vaccine administration.
(E) BALB/c mice received a single IM immunization with 10 μg of rHA mixed with eLNP or DOTAP-LNP, and PR8 HAI titers were determined 4 and 8 weeks after vaccine administration.
In (C)–(E), n = 10 mice/group. Symbols represent individual animals. Data are pooled from two independent experiments. In (A)–(E), data are shown as mean + SEM. Statistical analysis: in (A) and (B), unpaired two-tailed Mann-Whitney U test. In (C)–(E), two-way ANOVA with Bonferroni’s multiple comparisons test; only significant differences within the same time point are shown. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗∗p ≤ 0.0001.
To evaluate the adjuvant properties of LNPs with and without the ionizable lipid, mice were immunized with rHA mixed with the two different formats of eLNP, and serum from immunized mice was collected for PR8 HAI assays. In line with our previous studies (Figure 1), eLNP with the ionizable lipid showed high HAI titers 4 and 8 weeks after vaccine administration (Figure 5C). Strikingly, eLNP not containing the ionizable lipid did not possess adjuvant activity, as demonstrated by the absence of HAI activity in sera from mice immunized with these reagents (Figure 5C). As physicochemical characterization of eLNPs with and without the ionizable lipid showed similar size distributions by DLS and spherical structures by cryo-EM, our data suggest that the presence of the ionizable lipid is a critical parameter for providing adjuvant activity to LNPs.
Next we generated particles with an increasing molar ratio of the ionizable lipid and immunized animals with rHA mixed with these formulations. We found that the molar ratio of the ionizable lipid in the eLNPs slightly increased their adjuvant activity at the highest lipid ratio used, as measured by PR8 HAI titers (Figure 5D). Next we compared the adjuvanticity of this proprietary ionizable lipid with a frequently used (not ionizable) cationic lipid, 1,2-dioleoyloxy-3-(trimethylammonium) propane (DOTAP). DOTAP was formulated into LNP instead of the ionizable lipid with the same ratios of any additional components as the eLNP. In contrast to the ionizable lipid used in eLNP, DOTAP-LNP-adjuvanted rHA failed to induce PR8 HAI titers, suggesting that not all cationic lipids used for nucleic acid delivery are potent adjuvants in an LNP vaccine format (Figure 5E).
MyD88-based signaling enhances mRNA-LNP vaccine potency but is not required for LNP adjuvanticity
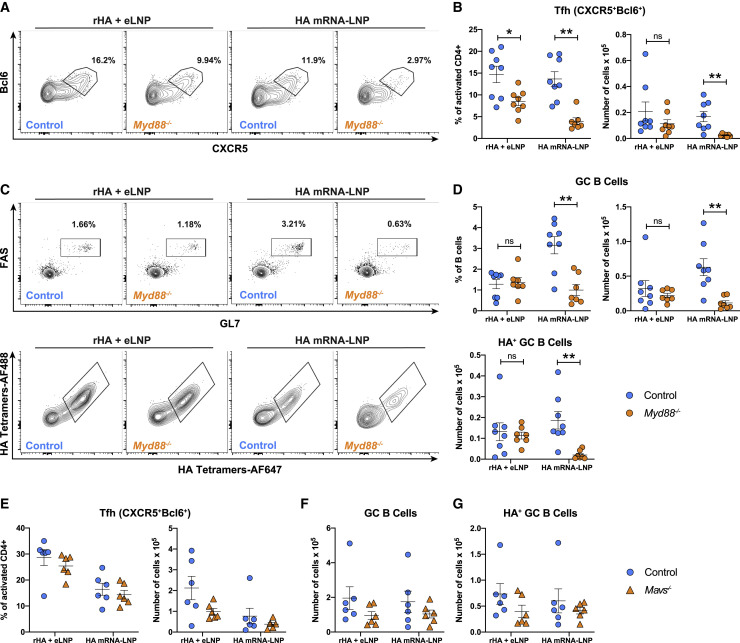
mRNA-LNP vaccines have inherent adjuvant activity, but it is not known how they are sensed and promote strong immune reactions. To uncouple the possible role of 1-methylpseudouridine-modified mRNA as an adjuvant from the LNP itself, we investigated the involvement of pathogen-associated molecular pattern (PAMP)-sensing receptors such as Toll-like receptors (TLRs) (Akira and Takeda, 2004; O’Neill et al., 2013), retinoic acid-inducible gene I (RIG-I) (Rehwinkel and Gack, 2020), and melanoma differentiation-associated protein 5 (MDA5) (Dias Junior et al., 2019) in sensing of the HA mRNA-LNP and rHA + eLNP vaccines. Since multiple TLRs could recognize RNA and its degradative products or the LNP itself, we immunized MyD88-deficient mice (Myd88 −/−) because most TLRs signal through this adaptor protein. Myd88 −/− (Akira and Takeda, 2004; Hou et al., 2008) and strain-matched control mice were immunized with HA mRNA-LNP or rHA + eLNP. Higher antigen doses (recombinant protein and mRNA) were used in these experiments than in previous studies to compensate for the lower GC responses to mRNA-LNP vaccines observed previously in C57BL/6 mice in comparison with BALB/c mice (Lederer et al., 2020). Tfh cell and GC B cell responses were analyzed by flow cytometry in dLNs 12 days after immunization. Myd88 −/− mice immunized with HA mRNA-LNP had a significant decrease in Tfh cell frequencies and absolute numbers compared with control mice as well as a noticeable drop in total and HA-specific GC B cell numbers (Figures 6 A–6D). However, when combined with rHA, the adjuvanticity of this LNP formulation seemed to be less reliant on MyD88-mediated mechanisms, as evidenced by only a minor decrease in Tfh cell frequency but not in Tfh, GC B cell, and HA+ GC B cell numbers when Myd88 −/− mice were immunized with the rHA + eLNP vaccine (Figures 6A–6D).
Figure 6.
Sensing through MyD88 enhances the potency of mRNA-LNP vaccines but is not required for LNP adjuvanticity
Myd88−/−, Mavs−/− and control mice received a single IM immunization with 30 μg PR8 HA mRNA-LNP or 30 μg rHA + eLNP. Tfh and GC B cell responses were examined 12 days after immunization in dLNs by flow cytometry.
(A and B) Tfh cell (CD4+B220−CD44hiCD62L−Bcl6+CXCR5+) representative flow cytometry (A) and percentages and absolute numbers (B) in Myd88−/− and control mice.
(C and D) GC B cell (CD3−CD19+FAS+GL7+) and HA-specific GC B cell (RBD−HA+) representative flow cytometry (C) and percentages and absolute numbers (D).
(E and F) Percentages and absolute numbers of Tfh cells (E) and absolute numbers of GC B cells and HA-specific GC B cells (F) in Mavs−/− and control mice.
In (A)–(D), n = 7–8 mice/group. In (E) and (F), n = 6 mice/group. Data were combined from three independent experiments in (A)–(D) and two independent experiments in (E) and (F). In (B) and (D)–(F), symbols represent individual animals. Data shown are mean + SEM. Statistical analysis: unpaired two-tailed Mann-Whitney U test was conducted. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001.
Next we investigated whether the cytosolic RNA sensors RIG-I/MDA5 could sense LNP-adjuvanted vaccines. We immunized mitochondrial anti-viral signaling (MAVS)-deficient mice (Mavs −/−) (Loo and Gale, 2011; Seth et al., 2005; Sun et al., 2006) with HA mRNA-LNP or rHA + eLNP. Mavs −/− mice lack MAVS protein, a common adaptor molecule of RIG-I and MDA5 (Loo and Gale, 2011; Seth et al., 2005). In contrast to Myd88 −/−, Mavs −/− mice showed no significant differences in Tfh cell or GC B cell frequencies and numbers compared with control mice (Figures 6E and 6F).
These data indicate that the 1-methylpseudouridine-modified, cellulose-purified mRNA used in our mRNA-LNP vaccines is sensed by receptors signaling through pathways relying on MyD88 but not MAVS and suggest a complementary effect of mRNA sensing in the adjuvanticity of mRNA-LNP vaccines. In contrast, our data ruled out a major role of MyD88- and MAVS-based signaling in the immunostimulatory activity of the LNP component.
LNPs induce robust IL-6 production, which is required for efficient induction of Tfh cells
To gain further insights into the mechanism of immune activation by LNP-based vaccines, mice were injected with eLNPs formulated with or without the ionizable lipid, Luc mRNA-LNP, AddaVax, or PBS. The immunostimulatory profile of the various reagents was examined by Luminex assay. Lipopolysaccharide (LPS) was used as positive control for cytokine induction. dLNs from immunized animals were collected 4 or 24 h after injection and analyzed for cytokine expression (Figure S7). We found that the eLNP containing the ionizable lipid (eLNP) and Luc mRNA-LNP induced large amounts of proinflammatory cytokines and chemokines 4 and 24 h after injection in comparison with PBS, whereas the LNP lacking the ionizable lipid (eLNP w/o ion. lip.) and AddaVax elicited lower cytokine and chemokine concentrations (Figure S7). The differences between AddaVax and the ionizable lipid-containing eLNP were particularly noticeable for GM-CSF (34×), IL-1β (22×), IL-5 (17×), IL-6 (≥12×), IP-10 (≥18×), KC (≥20×), LIF (23×), Lix (214×), and MIP-2 (≥49×) 4 h after injection. The signal intensity for several cytokines in the eLNP sample group was above the quantitative range of the plate reader (indicated in Figure S7); thus, the fold changes for these cytokines may be greater than noted above.
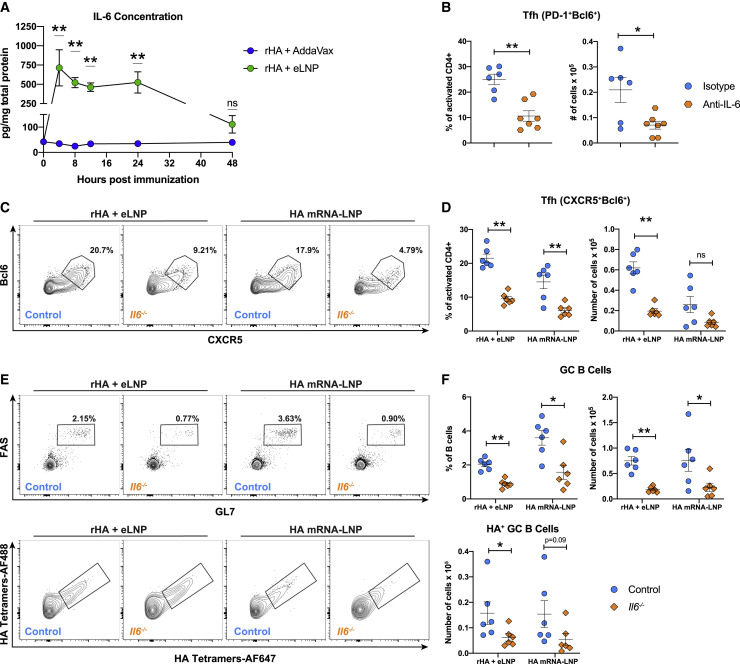
Importantly, we found that our eLNP formulation induced elevated production of IL-6 (Figure S7), a cytokine critical for early Tfh cell differentiation in mice (Crotty, 2019; Vinuesa et al., 2016). To investigate the role of IL-6 in the adjuvanticity of this LNP formulation, we sought to confirm that IL-6 is induced in dLNs when LNP is injected via the IM route. Mice were immunized with rHA adjuvanted with AddaVax or eLNP, and dLNs were collected 4, 8, 12, 24, and 48 h after immunization. In agreement with the Luminex data, we found that IL-6 production peaked 4 h after immunization and remained elevated for at least 24 h before beginning to decline by 48 h (Figure 7 A).
Figure 7.
IL-6 is crucial for LNP-induced GC reactions
(A) BALB/c mice received a single IM immunization with 10 μg rHA adjuvanted with eLNP or AddaVax. IL-6 levels were determined by ELISA in dLN lysates at various times after immunization.
(B) BALB/c mice received anti-IL-6 or isotype mAb 1 day prior to and every other day after a single IM immunization with 10 μg rHA + eLNP. Tfh cell (CD4+B220−CD44hiCD62L−PD-1+CXCR5+) percentages and absolute numbers were analyzed in dLNs 5 days after immunization.
(C–F) Il6−/− and control mice received a single IM immunization with 30 μg PR8 HA mRNA-LNP or 30 μg rHA + eLNP. Twelve days later, Tfh and GC B cell responses were examined in dLNs by flow cytometry.
(C and D) Tfh cell (CD4+B220−CD44hiCD62L−Bcl6+CXCR5+) representative flow cytometry (C) and percentages and absolute numbers (D).
(E and F) GC B cell (CD3−CD19+FAS+GL7+, top panel) and antigen-specific GC B cell (RBD−HA+, bottom panel) representative flow cytometry (E) and percentages and absolute numbers (F).
In (A)–(F), n = 6–7 mice/group. Data were combined from two independent experiments. In (B), (D), and (F), symbols represent individual animals. Data are shown as mean + SEM. Statistical analysis: unpaired two-tailed Mann-Whitney U test was conducted. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001. See also Figure S7.
Because IL-6 has been shown to be an early regulator of Tfh cell differentiation in mice (Choi et al., 2013), we first investigated whether LNP-induced IL-6 is indeed a relevant driver of Tfh cell induction 5 days after immunization. Mice received an IL-6-blocking monoclonal antibody (mAb; anti-IL-6 mAb) or an isotype control mAb 1 day prior to immunization with rHA + eLNP and then every other day after immunization for the duration of the study. Frequencies and absolute numbers of Tfh cells were decreased in mice treated with anti-IL-6 mAb in comparison with isotype mAb-treated animals (Figure 7B). To confirm the role of IL-6 in Tfh cell differentiation and function, we immunized IL-6-deficient mice (Il6 −/−) or strain-matched control mice with HA mRNA-LNP or rHA + eLNP and analyzed Tfh and GC B cell responses 12 days after immunization. We observed that, with HA mRNA-LNP or rHA + eLNP, IL-6 was critical for induction of Tfh and GC responses, as evidenced by a noticeable decrease in Tfh frequency in Il6 −/− mice (Figures 7C and 7D). The decrease in Tfh cell absolute numbers between control and Il6 −/− mice in the HA mRNA-LNP group did not show a significant difference (Figures 7C and 7D). We assume that the limited size of our test group may have been the reason for this. In addition to a decrease in Tfh cells, total and antigen-specific GC B cell frequencies and absolute numbers were also reduced (Figures 7E and 7F) in Il6 −/− mice in the HA mRNA-LNP and rHA + eLNP groups. These data indicate impairment of the GC response supported by this LNP formulation in the absence of IL-6.
These data show that the ionizable lipid-containing LNP drives the production of IL-6 in dLNs of mice and that LNP-mediated IL-6 production is critical for the induction of Tfh cells in response to mRNA and recombinant protein vaccines adjuvanted with LNPs.
Discussion
Vaccination is one of the most efficient and cost-effective means of preventing and containing pathogenic infections in human and animal populations. Protection from infection can be achieved by eliciting neutralizing Abs (Iwasaki, 2016), LLPCs, and MBCs (Sallusto et al., 2010) through the induction of Tfh cell responses in GCs (Havenar-Daughton et al., 2017). Thus, identification of vaccine platforms and adjuvants that strongly promote Tfh cell and GC responses is critically important for developing effective vaccines for current and emerging infectious diseases.
In this study, we describe a potent and versatile adjuvant, an ionizable LNP formulation that elicits robust Tfh cell responses and durable protective Ab titers when combined with a variety of vaccine antigens. LNPs have been used widely in preclinical and clinical evaluation of mRNA-based vaccines (Alameh et al., 2020), and, importantly, two SARS-CoV-2 nucleoside-modified mRNA vaccines have received approval for vaccination of humans in countries around the world as the first vaccines against the ongoing coronavirus disease 2019 (COVID-19) pandemic. These vaccines utilize proprietary LNPs that protect mRNA from degradation and facilitate delivery into the cytoplasm of host cells for subsequent expression and presentation to the immune system. Here we found that the LNP formulation investigated in this study can be utilized as an adjuvant for recombinant protein vaccines, paving the way for the development of next-generation, highly effective protein-based vaccines. Our LNP formulation potently induced SHM in antigen-specific MBCs. This is a relevant finding because development of broadly neutralizing (and often highly somatically hypermutated) Abs against certain rapidly mutating pathogens, such as HIV-1 and influenza virus, is particularly challenging (Doria-Rose and Joyce, 2015).
Although LNPs have been used in various mRNA vaccine formulations for years (Alameh et al., 2020), the source of adjuvant activity and the range of applicability of this type of adjuvant are not known. A number of vaccine studies by us and others have suggested the possible adjuvant effect of LNPs. We found that non-antigen mRNA-LNP successfully adjuvants an HA protein vaccine to induce high HAI titers (Pardi et al., 2018a). Shirai et al. (2020) investigated an LNP formulation as a possible adjuvant for influenza vaccines. Although their LNP provided increased responses with a split virus vaccine format, no adjuvant effect was found when the LNPs were mixed with HA protein. Additionally, they reported no induction of proinflammatory cytokine (IL-6 and G-CSF) production or recruitment of inflammatory immune cells after administration of their LNPs to mice. Another group investigated the adjuvant effect of a Merck proprietary LNP formulation in two closely related studies. They showed that protein subunit vaccines of hepatitis B surface antigen and ovalbumin mixed with this LNP induced similar responses as the aluminum-based adjuvant IMO or the TLR4 agonist 3-O-deacetytaled monophosphoryl lipid A (Swaminathan et al., 2016a). In a related report, the same LNP was examined for an adjuvant effect formulated into a tetravalent subunit dengue vaccine in mice, guinea pigs, and non-human primates, and high titers of neutralizing Abs were observed (Swaminathan et al., 2016b). Because of the difference in the antigens used, the dosing/immunization regimen, established adjuvants used as comparators, and experiments performed, it is difficult to directly compare the results with our findings. Of note, Tfh cell, LLPC, and MBC induction, the protective efficacy of various vaccine formulations, and the mechanism of action of LNPs were not examined in these studies. This points out the importance of development and screening/testing of LNP formulations as adjuvants for favorable immunostimulatory profiles in studies such as these.
As nucleoside-modified mRNA-LNP vaccines are being deployed in humans at a large scale for the first time ever, studies examining the mechanism of action become critical to improve and fine-tune the efficacy of such vaccines. Both licensed COVID-19 mRNA-LNP vaccines contain LNPs with ionizable cationic lipids. Our study suggests that the high efficacy of these vaccines might at least partially stem from the potent adjuvant properties of the ionizable lipid component of LNPs. Importantly, we demonstrate here that the eLNP formulation used in our study is endowed with an intrinsic ability to promote secretion of IL-6 in mice. IL-6 levels were elevated 4 h after LNP injection and waned by 48 h. LNP-induced IL-6 appeared to be largely responsible for the induction of Tfh cells, as supported by the severe reduction of Tfh cell generation after rHA + eLNP immunization in IL-6-deficient mice or in animals treated with an IL-6-blocking mAb in comparison with wild-type or isotype mAb-treated controls, respectively. IL-6 is a well-established pro-Tfh cell-inducing factor (Crotty, 2014) and an important regulator of early Tfh cell differentiation. By signaling through the IL-6 receptor complex, this cytokine promotes expression of B cell lymphoma 6 (Bcl6), the Tfh cell lineage-defining transcription factor in activated CD4 T cells (Nurieva et al., 2009). Furthermore, IL-6 is the most potent inducer of the Tfh cell-signature cytokine IL-21 in mice (Crotty, 2014). IL-6 is not a universally required regulator of Tfh cell biology; it appears to be only marginally important for murine Tfh cell differentiation in certain viral infections or protein immunizations (Eddahri et al., 2009; Eto et al., 2011; Poholek et al., 2010) and relevant in others (De Giovanni et al., 2020; Riteau et al., 2016). A key determinant for IL-6 involvement in Tfh cell differentiation in diverse infection/immunization models is the timing of IL-6 induction (De Giovanni et al., 2020). VSV infection has been shown to cause an early peak of IL-6 that, in turn, promotes Tfh cell differentiation. Conversely, lymphocytic choriomeningitis virus (LCMV) drives more modest and delayed IL-6 production that does not have any major role in shaping Tfh cell responses. Because we observed noticeable IL-6 production as early as 4 h after injection of eLNP and reduced Tfh cell differentiation 5 days after immunization with rHA + eLNP in mice where IL-6 was blocked, it is likely that, similar to what was described by De Giovanni et al. (2020) during VSV infection, “early” IL-6 induction by LNPs also accounts for robust Tfh cell differentiation in our study.
An aspect that still remains to be determined is how this LNP triggers IL-6 production in a prompt and efficient fashion. Our work indicates that this LNP formulation is not sensed by most TLRs or RIG-I/MDA5. Indeed, Tfh cell and GC B cell responses to rHA + eLNP immunization were mostly unchanged in MyD88- and MAVS-deficient mice. Future studies will determine how LNPs are sensed and regulate IL-6 production. On the other hand, our study unveils a role of MyD88-based signaling in response to nucleoside-modified (1-methylpseudouridine-containing) mRNA-LNP vaccines. We found that the Tfh cell and GC B cell promotion capacity of mRNA-LNP vaccines was greatly diminished in MyD88-deficient mice. Similar to the licensed COVID-19 vaccines, the mRNA vaccines used in this study are 1-methylpseudouridine modified to reduce inflammation caused by PAMP-sensing mechanisms and enhance protein translation from the mRNA. Additionally, they are cellulose purified (Baiersdörfer et al., 2019) to remove double-stranded RNAs that are generated during mRNA synthesis, which could also trigger PAMP-sensing mechanisms and contribute to excessive inflammation (Hur, 2019). Our data indicate that the combination of nucleoside modification and cellulose purification might not completely abolish recognition of mRNA or its degradative products by one or multiple TLRs and that this residual capacity to promote TLR signaling is an active component of the ability of mRNA-LNP to foster GC responses. An alternative explanation for the observed phenotype could also be that, instead of (or besides) being sensed by TLRs, the 1-methylpseudouridine-modified mRNA and/or its degradative products trigger alternative sensing mechanisms that ultimately support generation of Tfh cells via production of cytokines binding to the IL-1R family, which also signals through the adaptor protein MyD88 (Martin and Wesche, 2002).
We believe that our study is a major advancement in the field of infectious disease vaccines because it provides insights into the mechanism of action of nucleoside-modified mRNA-LNP vaccines and suggest that currently approved protein subunit vaccines may be improved by using LNPs as adjuvants.
Limitations of the study
It is worth noting that the potential role of IL-6 in human Tfh cell biology is less evident than in mice. In humans, IL-21 production by CD4+ T cells is regulated predominantly by IL-12 (Locci et al., 2016; Schmitt et al., 2009), and addition of IL-6 to human CD4+ T cells in vitro does not seem to induce expression of Tfh signature markers such as CXCR5 or BCL6 (Schmitt et al., 2014). On the other hand, inborn errors of immunity leading to reduced STAT3 signaling capacity in humans are associated with reduced frequencies of circulating Tfh cells (Ma et al., 2012, 2015), suggesting a possibly unappreciated role of IL-6 in human Tfh cell differentiation (because IL-6 is one of the cytokines signaling via STAT3). Because SARS-CoV-2 nucleoside-modified mRNA-LNP vaccines promote robust GC B cell and Tfh cell responses in humans (Lederer et al., 2021; Mudd et al., 2021; Turner et al., 2021), we argue that a similar pro-Tfh cell-inducing activity is also present in humans and that the mechanism could also likely rely on LNP-induced IL-6 activity, as supported by a recently described STAT3 signature in T cells of SARS-CoV-2-vaccinated individuals (Arunachalam et al., 2021). Alternatively, LNPs might be capable of inducing additional cytokines that have been described to be potent regulators of human Tfh cell differentiation, such as IL-12, activin A, or transforming growth factor β (TGF-β) (Locci et al., 2016; Schmitt et al., 2009, 2014). Future studies will address these important questions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-biotin MACS microbeads | Miltenyi biotec | Cat#130-097-046 |
| CD16/CD32, clone 2.4G2 | BioXCell | Cat#BE0307 |
| anti-VSV Indiana G | Absolute Antibody | Cat#Ab01401-2.0 |
| B220, clone RA3-6B2, APC | Tonbo Biosciences | Cat#20-0452-U100 |
| B220, clone RA3-6B2, BUV496 | BD Biosciences | Cat#612950 |
| B220, clone RA3-6B2, BV650 | Biolegend | Cat#103241 |
| B220, clone RA3-6B2, PE | Biolegend | Cat#103207 |
| Bcl6, clone K112-91, Ax647 | BD Biosciences | Cat#624024 |
| CD138, clone 281-2, BV421 | Biolegend | Cat#142508 |
| CD138, clone 281-2, BV650 | BD Biosciences | Cat#564068 |
| CD19, clone 6D5, BV711 | Biolegend | Cat#115555 |
| CD19, clone 6D5, BV605 | Biolegend | Cat#115539 |
| CD19, clone 6D5, BV785 | Biolegend | Cat#115543 |
| CD25, clone PC61.5, AF488 | eBioScience | Cat#53-0251-82 |
| CD3, clone 145-2C11, BUV395 | BD Biosciences | Cat#565992 |
| CD3, clone 17A2, APC-Cy7 | Biolegend | Cat#100222 |
| CD38, clone 90, AF700 | Invitrogen | Cat#56-0381-82 |
| CD4, clone GK1.5, PE-Cy7 | Biolegend | Cat#100409 |
| CD4, clone H29.19, PE-Cy5 | Biolegend | Cat#130312 |
| CD4, clone RM4-5, Biotin | eBioScience | Cat#13-0042-85 |
| CD4, clone RM4-5, PerCP-Cy5.5 | Biolegend | Cat#100540 |
| CD44, clone IM7, BV605 | Biolegend | Cat#103047 |
| CD44, clone IM7, FITC | Biolegend | Cat#103005 |
| CD62L, clone MEL-14, BUV395 | BD Biosciences | Cat#740218 |
| CD8a, clone 53-6.7, Biotin | eBioScience | Cat#13-0081-85 |
| CD8a, clone 53-6.7, PerCP-Cy5.5 | Biolegend | Cat#100734 |
| CD8a, clone 53-6.7, PE-Cy7 | Biolegend | Cat#100722 |
| CD8a, clone 53-6.7, PE-Cy5 | BD Biosciences | Cat#553034 |
| CD8a, clone 53-6.7, PerCP-Cy5.5 | Biolegend | Cat#100734 |
| CXCR5, clone SPRCL5, Biotin | eBioScience | Cat#13-7185-82 |
| F4/80, clone BM8, Biotin | eBioScience | Cat#13-4801-82 |
| F4/80, clone BM8, PE-Cy5 | eBioScience | Cat#15-4801-82 |
| F4/80, clone BM8, PE-Cy7 | Biolegend | Cat#123114 |
| FAS, clone JO2, BV510 | BD Biosciences | Cat#563646 |
| FoxP3, clone FJK-16 s, PE-Cy7 | eBioScience | Cat#25-5773-82 |
| GL7, clone GL7, e660 | eBioScience | Cat#50-5902-82 |
| GL7, clone GL7, PerCP-Cy5.5 | Biolegend | Cat#144610 |
| GL7, clone GL7, AF488 | Biolegend | Cat#144612 |
| Gr-1, clone RB6-8C5, PE-Cy5 | Biolegend | Cat#108410 |
| IgD, clone 11-26c, e450 | eBioScience | Cat#48-5993-82 |
| IgD, clone 11-26c.2a, PE-Cy7 | Biolegend | Cat#405720 |
| IgD, clone 11-26c.2a, APC-Cy7 | Biolegend | Cat#405716 |
| IgD, clone 11-26c.2a, BV605 | Biolegend | Cat#405727 |
| IgG1 isotype control | Bio X Cell | Cat#BE0088 |
| IgM, clone II/41, PerCP-eF710 | Invitrogen | Cat#46-5790-82 |
| IgM clone R6-60.2, PE-CF594 | BD Biosciences | Cat#562565 |
| IL-6, clone MP5-20F3 | Bio X Cell | Cat#BE0046 |
| Live/Dead Aqua | Biolegend | Cat#423101 |
| Mouse IL-6 Uncoated ELISA kit | Invitrogen | Cat#88-7064 |
| PD-1, clone RMP1-30, PE-Cy7 | Biolegend | Cat#109110 |
| Ter-199, clone Ter-199, Biotin | Biolegend | Cat#116204 |
| Ter-199, clone Ter-199, PE-Cy7 | Biolegend | Cat#116222 |
| Streptavidin-conjugated AF488 | Biolegend | Cat#405235 |
| Streptavidin-conjugated AF647 | Biolegend | Cat#405237 |
| Streptavidin-conjugated BV421 | Biolegend | Cat#405225 |
| HRP conjugated anti-mouse secondary | Jackson Immunoresearch | Cat#715-035-150 |
| HRP conjugated anti-mouse IgG1 | Abcam | Cat#98693 |
| HRP conjugated anti-mouse IgG2a | Abcam | Cat#98698 |
| HRP conjugated anti-mouse IgG2b | Abcam | Cat#98703 |
| HRP conjugated anti-mouse IgA | Abcam | Cat#97235 |
| Chemicals and recombinant proteins | ||
| Addavax | Invivogen | Cat#vac-adx-10 |
| BD Brilliant Buffer | BD Biosciences | Cat#563794 |
| ACK Lysing buffer | Lonza | Cat#10-548E |
| Cellulose | Sigma-Aldrich | Cat#11363-250G |
| CleanCap | Trilink | Cat#N-7413 |
| CoMplete protease inhibitor cocktail | Sigma-Aldrich | Cat#11697498001 |
| EZ-Link Micro Sulfo-NHS-Biotinylation Kit | Thermo Fisher Scientific | Cat#21925 |
| Fixable Viability Dye eFluor 780 | eBioScience | Cat#65-0865-14 |
| FoxP3/Transcription Factor Staining Buffer Set | eBioScience | Cat#00-5523-00 |
| Illumina MiSeq Reagent kit | Illumina | Cat#MS-102-3003 |
| Lightning-Link® Rapid Alexa Fluor 647 | Novus Biologicals | Cat#336-0005 |
| Lightning-Link® R-Phycoerythrin (R-PE) | Novus Biologicals | Cat#703-0010 |
| Luminex 32-Plex | Millipore-Sigma | Cat#MCYTOMAG-70K |
| N1-methylpseudouridine-triphosphate | Trilink | Cat#N-1081 |
| Miltenyi LD columns | Miltenyi biotec | Cat#130-042-901 |
| M-Per lysis buffer | Thermo Fisher Scientific | Cat#78501 |
| Pierce BCA protein assay kit | Thermo Fisher Scientific | Cat#23227 |
| Pierce microBCA protein assay kit | Thermo Fisher Scientific | Cat#23235 |
| PR8 HA recombinant protein | Sino Biological | Cat#11684-V08H1 |
| QIAGEN Gentra DNA purification kit | QIAGEN | Cat#158689 |
| Recombinant RBD | Florian Krammer | N/A |
| SARS-CoV-2 RBD | Sino Biological | Cat#40592-VNAH |
| SARS-CoV-2 RBD | GenScript | Cat#Z03501 |
| KPL TMB 2-Component Microwell Peroxidase Substrate | Seracare | Cat#5120-0050 |
| Whole turkey blood | Lampire Biological Products | Cat# 7209401 |
| Deposited data | ||
| BCR sequencing data | Sequence Read Archive | BioProject: PRJNA669143 |
| Experimental models: Organisms/strains | ||
| Myd88tm1.1Defr (Myd88−/−) mouse | Jackson Lab | Cat#009088 |
| Mavsstm1zjc (Mavs−/−) mouse | Jackson Lab | Cat#008634 |
| Il6tm1Kopf (Il6−/−) mouse | Jackson Lab | Cat#002650 |
| C57BL/6 mouse | Jackson Lab | Cat#000664 |
| B6129SF2 mouse | Jackson Lab | Cat#101045 |
| BALB/c mouse | Jackson Lab | Cat#000651 |
| Experimental models: Cell lines | ||
| HEK293T/17 cell | ATCC | CRL11268 |
| Vero E6 cell stably expressing TMPRSS2 | Dr. Stefan Pohlmann | N/A |
| Recombinant DNA | ||
| SARS-CoV-2 receptor binding domain (RBD) | GenBank | MN908947.3 |
| Influenza A/Puerto Rico/8-MC/1934(H1N1) HA | GenBank | CY083950.1 |
| Softwares and algorithms | ||
| S6 FluoroSpot Analyzer CTL | Immunospot | N/A |
| FlowJo v10.3 | FlowJo LLC | N/A |
| FACSDIVA | BD Biosciences | N/A |
| GraphPad Prism | GraphPad | N/A |
| pRESTO version 0.5.10 | Vander Heiden et al., 2014 | https://presto.readthedocs.io |
| ImmuneDB v0.29.9 | Rosenfeld et al., 2018 | https://immunedb.readthedocs.io |
Resource availability
Lead contact
Further information and requests for supporting data, resources, and reagents should be directed to and will be fulfilled upon request by the Lead Contact, Norbert Pardi (pnorbert@pennmedicine.upenn.edu).
Materials availability
Reagents from this study are available upon request.
Experimental model and subject details
Mice
Eight-week-old BALB/c mice were purchased from Charles River Laboratories and The Jackson Laboratory and were housed either in a conventional or a Specific Pathogen Free (SPF) animal facility. Eight- to twelve-week-old Myd88 tm1.1Defr (Myd88 −/−), Mavs stm1zjc (Mavs −/−), Il6 tm1Kopf (Il6 −/−) and aged-matched control mice were purchased from The Jackson Laboratory and were housed in an SPF animal facility. C57BL/6 mice were the strain-matched controls for Myd88 −/− and Il6 −/− mice. B6129SF2 mice were the strain-matched controls for Mavs −/− mice. Female mice were used in all experiments of this study. Animals were randomly assigned to experimental groups. The investigators faithfully adhered to the “Guide for the Care and Use of Laboratory Animals” by the Committee on Care of Laboratory Animal Resources Commission on Life Sciences, National Research Council. Mouse studies were conducted under protocols approved by the Institutional Animal Care and Use Committees (IACUC) of the University of Pennsylvania. All animals were housed and cared for according to local, state and federal policies in an Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC)-accredited facility.
Cell lines
HEK293T/17 (originally from female fetus) cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Mediatech #MT10-013-CM) containing 10% fetal calf serum (FCS). Vero E6 cells stably expressing TMPRSS2 were a gift from Stefan Pohlman and were cultured in DMEM +10% FCS.
Method details
Production of empty LNPs and mRNA-LNP vaccines
Codon-optimized coding sequences of hemagglutinin (HA) of A/Puerto Rico/8/1934 (PR8), receptor binding domain (RBD, amino acids 1-14 fused with amino acids 319-541) of SARS-CoV-2 (Wuhan-Hu-1, GenBank: MN908947.3) and firefly luciferase (Luc) were synthesized and cloned into an mRNA production plasmid as described (Freyn et al., 2020). mRNA production was performed as described (Freyn et al., 2020). Briefly, mRNAs were produced to contain 101 nucleotide-long poly(A) tails. m1Ψ-5′-triphosphate instead of UTP was used to generate modified nucleoside-containing mRNA. Capping of the in vitro transcribed mRNAs was performed co-transcriptionally using the trinucleotide cap1 analog, CleanCap. mRNA was purified by cellulose purification, as described (Baiersdörfer et al., 2019). All mRNAs were analyzed by agarose gel electrophoresis and were stored frozen at −20°C. Cellulose-purified m1Ψ-containing RNAs were encapsulated in LNPs using a self-assembly process as previously described wherein an ethanolic lipid mixture of ionizable cationic lipid, phosphatidylcholine, cholesterol and polyethylene glycol-lipid was rapidly mixed with an aqueous solution containing mRNA at acidic pH (Maier et al., 2013). The LNP formulation used in this study is proprietary to Acuitas Therapeutics; the proprietary lipid and LNP composition are described in US patent US10,221,127. As the DOTAP-containing LNP could not be easily concentrated after the self-assembly process, the DOTAP-LNP was made using an extrusion process (Mui et al., 2003). The lipids (DOTAP, cholesterol, DSPC, PEG-lipid) were first solubilized in chloroform. The chloroform was removed using a stream of nitrogen gas and the lipid film was put under vacuum for 2 h to remove residual solvent. The lipid film was hydrated in PBS, freeze-thawed 5 times using liquid nitrogen and a 60°C water bath, and then extruded 10 times through 80 nm pore-sized filters. The RNA-loaded and empty particles were characterized and subsequently stored at −80°C at an RNA concentration of 1 μg μl-1 (in the case of loaded particles) and total lipid concentration of 30 μg μl-1 (both loaded and empty particles). The mean hydrodynamic diameter of mRNA-LNPs was ∼80 nm with a polydispersity index of 0.02-0.06 and an encapsulation efficiency of ∼95%. Two or three batches from each mRNA-LNP formulations were used in these studies and we did not observe variability in vaccine efficacy.
Recombinant proteins
The PR8 HA recombinant protein and the SARS-CoV-2 RBD protein used for vaccine studies were purchased from Sino Biological.
The PR8 HA and RBD proteins used for generating fluorescently-labeled probes for B cell analyses and PR8 HA ELISA studies were produced as described previously (Amanat et al., 2020; Margine et al., 2013; Stadlbauer et al., 2020). The RBD protein used for ELISA was purchased from GenScript.
Production of fluorescently-labeled proteins
Fluorescently-labeled recombinant RBD protein was prepared as previously described (Laczkó et al., 2020; Lederer et al., 2020). Briefly, rRBD was independently conjugated to either PE or AlexaFluor 647 using the Lightning-Link® R-Phycoerythrin (R-PE) and Lightning-Link (R) Rapid Alexa Fluor 647 according to manufacturer’s instructions. To create fluorescently labeled RBD and HA tetramers, recombinant RBD or HA was biotinylated using the EZ-Link Micro Sulfo-NHS-Biotinylation Kit. Streptavidin-conjugated Alexa Fluor 647, Alexa Fluor 488, or Brilliant Violet 421 were then added at a 6:1 molar ratio (biotinylated-protein to streptavidin-conjugate). Specifically, after the volume of fluorochrome needed to achieve a 6:1 molar ratio was determined, the total volume of fluorochrome was split into 10 subaliquots. These subaliquots were then added, on ice, to the biotinylated protein and mixed by pipetting every 10 minutes (for a total of 10 additions).
Dynamic light scattering (DLS)
The hydrodynamic diameter (Z-average size) of LNPs (with and without mRNA) was measured at 25°C by DLS using a Zetasizer Nano ZS instrument (Malvern Instruments Ltd) equipped with a solid-state HeNe 633nm laser at a scattering angle of 173°. LNP samples formulated at 1 mg/ml mRNA (or an equivalent of 1 mg/ml for empty LNP) were diluted 1:500 using sterile phosphate-buffered saline before determination of size/PDI. The Z-average is represented as diameter in nm ± width of distribution.
Cryo-electron microscopy imaging
eLNPs with and without ionizable lipid were vitrified using Vitrobot Mark IV System (FEI/Thermo Scientific), and frozen grids were imaged with 300 kV Titan Krios Cryo-TEM with a Falcon III camera (FEI/Thermo Scientific). Images were analyzed using ImageJ. Between 200 to 300 particles were manually measured to determine the diameter of the LNPs. The size of the LNPs are represented as diameter in nm ± standard deviation.
Mouse immunizations
BALB/c mouse immunizations
Vaccines were administered via the intramuscular (IM) route into the gastrocnemius muscle, or the intradermal (ID) route across two sites localized toward the base of the tail using a 3/10cc 29½G insulin syringe (Covidien). Mice were immunized with one of the following combinations: 10 μg recombinant PR8 HA (rHA), 10 μg rHA + 30 μg Luc mRNA-LNP (containing 30 μg mRNA and 900 μg total lipid), 10 μg rHA or rRBD + Addavax according to manufacturer’s instructions (equal volumes of protein and AddaVax™ were mixed), 10 μg rHA or rRBD + eLNP (total lipid content: 900 μg; equivalent to lipid content of 30 μg mRNA-LNP), or 10 μg PR8 HA or RBD mRNA-LNP.
In the IL-6 Ab blockade experiments, mice were given 0.5 mg of anti-IL6 antibody or isotype antibody intraperitoneally (IP) one day prior to immunization and 0.25 mg every other day following immunization.
Targeted mutant mouse immunizations
Mice were immunized IM in the gastrocnemius muscle. Mice were immunized with either 30 μg PR8 HA mRNA-LNP or 30 μg rHA + eLNP (total lipid content equivalent to 30 μg mRNA-containing mRNA-LNP).
Blood collection
Mice were isoflurane-anesthetized and blood was collected through the retro-orbital route. Serum was separated from blood following an incubation period of 30 minutes at room temperature, and samples were centrifuged at 10 000 g for 5 minutes in a non-refrigerated Eppendorf 5424 centrifuge. Separated serum was stored at −20°C.
Influenza virus challenge studies
8-week-old BALB/c mice were immunized ID with 10 μg rHA, 10 μg rHA + Addavax, 10 μg rHA + eLNP (30 μg mRNA-LNP equivalent) or 10 μg HA mRNA-LNP. Animals were terminally bled 9 months post-immunization, and HAI titers were determined. 400 μL of each individual immune sera was injected IP into naive recipient mice. Two hours after serum transfer, recipient mice were put under isoflurane anesthesia, and blood was collected for post transfer HAI titer determination. Then, mice were infected with 5000 focus forming units (FFU) of mouse-adapted A/Puerto Rico/8/1934 influenza virus (see IAV LD in Tapia et al., 2013) diluted in a final volume of 40 μL PBS. Viral solution was applied dropwise to nasal orifice, and was inhaled by serum-transferred mice. Weight loss was followed daily for 14 days. Animals were euthanized when they lost 20% of their starting bodyweight, as per IACUC protocol.
Hemagglutination inhibition assay
Sera were heat-inactivated (55°C) for 30 minutes, spun down at 11000 rpm for 2 minutes, and diluted 1:20 in PBS, then serially diluted 1:2 in 50 μl in 96-well U-bottom plates (lowest concentration: 1:2560) using a multichannel pipette. Then, four hemagglutinating doses of A/Puerto Rico/8/1934 virus was added in the same volume as diluted sera. Finally, 12.5 μL of turkey erythrocyte solution - washed twice in phosphate buffered saline and diluted to a final concentration of 2% (v/v) - was added, and gently mixed with the sera-virus solution (final volume of 125 μl). Samples were incubated for 45 minutes at room temperature, after which the plates were turned on the side for one minute, then scanned on a regular office scanner. HAI titers were determined as the highest dilution of the sample that inhibited four agglutinating doses of the influenza virus. Inhibition of agglutination was observed as the blood forming a “tear drop” shape.
Protein extraction from lymph nodes
Lymph nodes were collected at specific time points and snap-frozen in a mixture of isopropanol and dry ice and stored at −80°C until use. Frozen tissues were cut on dry ice, weighed (∼20 mg), and disrupted using the TissueLyzer® II system (QIAGEN). Tissues were disrupted using 5 mm steel beads (QIAGEN) under the following conditions: 2 × 30 Hz, 20 s per cycle. Homogenized tissues were resuspended in 750 μl of M-Per lysis buffer, in the presence of CoMplete protease inhibitor cocktail and incubated on ice for 30 minutes with inversions every 10 minutes. For the IL-6 ELISA, whole lymph nodes were resuspended in lysis buffer (as above) prior to homogenization (as above). Lysates were cleared by centrifugation (2270 g, 30 minutes, 4°C), transferred to new tubes, and stored at −80°C until use. An aliquot was diluted 1:100 in PBS and total protein content of the lysate sample was determined using the Pierce microBCA protein assay kit or measured undiluted using the Pierce BCA protein assay kit for the IL-6 ELISA.
Determination of cytokine levels (Luminex)
Tissue lysate collected at 4 and 24 hours post administration of test articles were assayed for the induction of a 32 pro-inflammatory cytokine panel (MCYTOMAG-70K; including TNF-α, IL-1β, IL-6, KC, and IFN-γ) using the Luminex® technology. Plates were designed using the Milliplex® assay builder (Millipore-Sigma) and subjected to the manufacturer’s quality control. For each plate, a standard curve was prepared by diluting the Milliplex® Pro Mouse Cytokine Standard 32-Plex in the Milliplex® in standard diluent followed by 4-fold serial dilutions from 1:4 to 1:65536 in the same diluent. Samples were thawed on ice, cleared by centrifugation (10,000 g, 10 minutes, 4°C), diluted 1:2 using the Milliplex® Sample diluent, and a volume of 25 μl transferred to assay plates prefilled with pooled capture Abs. The plates were incubated for 30 minutes under orbital shaking (800 rpm, room temperature), washed as per manufacturer recommendation, incubated with biotinylated detection Abs (30 minutes, 800 rpm, room temperature), washed and revealed post-incubation for 10 minutes with streptavidin-phycoerythrin (800 rpm, room temperature). Data was acquired on a BioPlex2100® system using RP1 PMT setting with a minimum of 50 beads per region analyzed. For each cytokine, a 5-parameter regression algorithm (5-PL) was used to fit the data and interpolate cytokine values in tissue lysate samples. In order to account for inter-plate variability, two samples (PBS and LPS) were used as inter-plate calibrators.
Enzyme linked immunosorbent assays (ELISAs)
Anti-HA Ab isotype/subclass and anti-RBD Ab ELISA
High Bind Stripwell Corning 96 Well Clear Polystyrene Microplates were coated overnight with 1 μg/ml purified recombinant PR8 HA or SARS-CoV-2 RBD. Plates were washed once with wash buffer (0.05% Tween-20 in PBS), and blocked for two hours at room temperature using a solution of heat inactivated, IgG depleted, protease free bovine serum albumin (2% w/v BSA in PBS). After blocking, plates were washed three times, and mouse sera was serially diluted in the blocking solution and incubated for 2 hours at room temperature. Plates were washed three times before the addition of horse radish peroxidase (HRP)-conjugated anti-mouse secondary Ab specific to total IgG or subclasses in blocking buffer (total IgG, IgG2a, IgG2b: 1:10 000, IgG1: 1:15000, IgA: 1:7500). Plates were incubated for 1.5 hours, washed three times before the addition of 100 μl per well of KPL TMB substrate for 8 minutes. The reaction was stopped with 50 μl 2N sulfuric acid, and the absorbance was measured at 450nm using a SpectraMax 190 microplate reader. Endpoint dilution titer was defined as the highest dilution of serum to give a value 0.01 OD greater than the background (no serum) cut-off OD value. All samples were run in technical duplicates.
IL-6 ELISA
The Mouse IL-6 Uncoated ELISA kit was used according to the manufacturer’s instructions. In brief, 96-well MaxiSorp plates were coated with IL-6 capture Ab overnight, blocked and washed according to the manufacturer’s instructions. For each plate, an IL-6 standard curve was prepared by dissolving lyophilized IL-6 in lysis buffer at 500 pg/ml and then performing a 2-fold serial dilution to ∼4 pg/ml. Tissue lysate samples (described in “Protein extraction from lymph nodes”) collected at 4, 8, 12, 24, and 48 hours post immunization were thawed on ice and cleared by centrifugation. 100 μl of each sample was added to the plates undiluted and diluted at a 1:1 ratio with ELISA diluent. Plates were incubated on an orbital shaker at room temperature for 2 hours, before detection Ab and HRP were added according to the manufacturer’s instructions. TMB was added to plates for 10 minutes before the addition of a 2N sulfuric acid stop solution. Absorbance was measured at 450 nm using a BioTek Synergy HT plate reader. A third order polynomial regression was used to fit the data and interpolate a standard curve in Prism v9.0.0. IL-6 concentrations were then normalized on a per sample basis by dividing the IL-6 concentration (pg/ml) generated from the ELISA by the total protein content of the lysate as determined by BCA (mg/ml) to yield pg IL-6 per mg of total protein. Finally, the normalized IL-6 concentrations from the undiluted and the diluted (1:1) samples were averaged prior to plotting. All samples were run in technical duplicates.
Pseudovirus neutralization assay
Production of VSV pseudotyped with SARS-CoV-2 S D614G
293T cells plated 24 hours previously at 5 X 106 cells per 10 cm dish were transfected using calcium phosphate with 35 μg of pCG1 SARS-CoV-2 S D614G delta18 expression plasmid encoding a codon optimized SARS-CoV S gene with an 18 residue truncation in the cytoplasmic tail (kindly provided by Stefan Pohlmann) with a single amino acid substitution (D614G) found in recent circulating variants. Twelve hours post transfection the cells were fed with fresh media containing 5mM sodium butyrate to increase expression of the transfected DNA. Thirty hours after transfection, the SARS-CoV-2 spike expressing cells were infected for 2-4 hours with VSV-G pseudotyped VSVΔG-RFP at an MOI of ∼1-3. After infection, the cells were washed twice with media to remove unbound virus. Media containing the VSVΔG-RFP SARS-CoV-2 pseudotypes was harvested 28-30 hours after infection and clarified by centrifugation twice at 6000 g then aliquoted and stored at −80°C until used for Ab neutralization analysis.
Ab neutralization assay using VSVΔG-RFP SARS-CoV-2
Vero E6 cells stably expressing TMPRSS2 were seeded in 100 μL at 2.5x104 cells/well in a 96 well collagen coated plate. The next day, 2-fold serially diluted serum samples were mixed with VSVΔG-RFP SARS-CoV-2 pseudotype virus (50-200 focus forming units/well) and incubated for 1 hour at 37°C. Also included in this mixture to neutralize any potential VSV-G carryover virus was 8G5F11, a mouse anti-VSV Indiana G, at a concentration of 100 ng/ml. The Ab-virus mixture was then used to replace the media on VeroE6 TMPRSS2 cells. 20 hours post infection, the cells were washed and fixed with 4% paraformaldehyde before visualization on an S6 FluoroSpot Analyzer (CTL). Individual infected foci were enumerated and the values compared to control wells without Ab. The focus reduction neutralization titer 50% (FRNT50) was measured as the greatest serum dilution at which focus count was reduced by at least 50% relative to control cells that were infected with pseudotype virus in the absence of mouse serum. FRNT50 titers for each sample were measured in two technical replicates performed on separate days.
Flow cytometry and cell sorting
Draining inguinal and/or popliteal lymph nodes were harvested after immunization, homogenized with a syringe plunger and filtered through a 40 μm cell strainer on ice. All staining steps were carried out at 4°C in FACS buffer (PBS with 2% heat inactivated FBS). Single cell suspensions were Fc blocked with anti-CD16/CD32 monoclonal Ab (mAb) prior to staining. Splenocytes were harvested from spleens by mechanical disruption between the frosted ends of microscope slides and filtered through 70 μm cell strainer. Bone marrow was flushed from femurs and tibia from each mouse using a 23 g X ¾” needle and syringe into RPMI-1640 media supplemented with 10% heat inactivated FBS and filtered through 70 μm cell strainer. Splenocytes and bone marrow cells were then subjected to red blood cell lysis for 5 min in 2 mL ACK buffer on ice and resuspended in 1 mL RPMI-1640 media.
Tfh and Tfr cell staining
Cells were incubated with biotinylated CXCR5 Ab for 1 hour, washed, followed by incubation with streptavidin-conjugated BV421 for 30 minutes. After washing, cells were incubated with 20 μg/ml I-A(d) Influenza A HA PE-conjugated tetramer (HNTNGVTAACSHE) in RPMI for 2 hours, then washed. This last step was only performed for the analysis of HA-specific Tfh cells. Cells were next incubated for 30 minutes with a cocktail of Fixable Viability dye eFluor780 and all other surface Abs. Cells were washed with FACS buffer, then fixed and permeabilized in FoxP3/Transcription Factor Staining Buffer Set according to manufacturer’s instructions before intranuclear staining with Bcl-6 (and Foxp3 for the Tfr staining) for 30 minutes. All incubations were performed at 4°C. Staining panel details in Tables S3–S5.
GC B cell staining
Cells were incubated for 30 minutes at 4°C with a cocktail containing Fixable Viability dye eFluor780 and a cocktail of surface Abs. For antigen-specific GC B cells: HA AF488 tetramer and HA AF647 tetramer, or RBD BV421 tetramer were also added during this step. Excess Abs were washed away, and cells were fixed with 1% paraformaldehyde (PFA) for 30 minutes prior to acquisition. Staining panel details in Tables S6 and S7.
All samples were acquired on a 5 laser Cytoflex LX (Beckman Coulter) or a 5 laser Aurora (Cytek), and data analyzed in FlowJo v10.
Antigen-specific LLPC and MBC
5 million cells were stained with fixable live dead aqua (Zombie Aqua) for 15 minutes at room temperature, washed with FACS buffer and stained with labeled HA or RBD probes and a cocktail of surface Abs (see below) in BD Brilliant Buffer for 15 minutes at 4°C. Cells were then washed and resuspended in FACS buffer (PBS + 0.2% bovine serum albumin). ∼2 million events per sample were acquired on a LSRII (PR8 HA experiments), or on BD Symphony A3 Lite (SARS-CoV-2 experiments) and analyzed with FlowJo 10.x software. Staining panel details in Table S1.
Cell sorting
Prior to sorting, splenic B cells were enriched by negative selection using Miltenyi LD columns, Miltenyi anti-biotin MACS microbeads, and biotin-conjugated CD4, CD8, F4/80 and Ter119 Abs, according to the manufacturer’s protocol. Enriched cells were stained with a cocktail of surface Abs and with recombinant HA conjugated to PE and AF647. IgD+ follicular B cells, HA+IgD-IgM- memory, and HA-IgD-IgM- B cells were sorted using a BD FACS Aria II sorter (see Figure 3D for gating strategy). Staining panel details in Table S2.
B cell receptor Sequencing
Sample processing, amplification, and library preparation
mouse spleens were harvested at day 80 post immunization, and single-cell suspension was prepared in RPMI-1640 media following physical tissue homogenization. Genomic DNA was extracted from sorted cells using the QIAGEN Gentra DNA purification kit following the manufacturer’s recommendations. Amplification and sequencing of Ab heavy chain gene rearrangements were performed as described previously (Johnson et al., 2020). Libraries were pooled at equimolar ratios and loaded onto an Illumina MiSeq in the Human Immunology Core Facility at the University of Pennsylvania, and subjected to pair-end sequencing (2x300 bp) using the Illumina MiSeq Reagent kit.
Processing and analysis of sequencing data
raw sequencing data in FASTQ format were processed with pRESTO version 0.5.10 (Vander Heiden et al., 2014). Paired reads were aligned and sequences were subsequently subjected to quality filtering as described previously (Johnson et al., 2020). IgBLAST was used for gene identification and ImmuneDB v0.29.9 was used for clonal inference (Rosenfeld et al., 2018). After gene identification, sequences were trimmed to IMGT position 20 to remove 5′ primer sequences. Sequences sharing the same VH gene, JH gene, and 85% CDR3 amino-acid similarity were then grouped into clones and clones averaging less than 85% IGHV nucleotide identity were excluded from further analysis to minimize sequencing artifacts.
Quantification and statistical analysis
Data were collected and expressed as average ± standard error of the mean (SEM). Statistical analysis was conducted using GraphPad Prism® v9.0.0 (GraphPad Software) software package.
Acknowledgments
N.P. was supported by the National Institute of Allergy and Infectious Diseases (NIAID; R01AI146101 and R01AI153064). M.L. was supported by NIAID (R01AI123738 and R01AI153064). D.W. is supported by NIH/NIAID (UM1-AI-144371, 1U19AI135902, U19-AI142596, R01-AI124429, and HHS-NIH-NIAID-BAA2018) and an SRA from BioNTech. E.L.P., W.M., J.K.K., and A.M.R. were supported by P01 AI106697. F.K. was supported by NIAID Collaborative Influenza Vaccine Innovation Centers (CIVIC) contract 75N93019C00051. D.A. was supported by NIAID (R01AI139123 and R01AI154932). The Igyártó lab is supported by NIAID (R01AI146420 and R01AI146101). We thank the NIH Tetramer Core Facility for producing the I-A(d) Influenza A HA PE-conjugated tetramer (HNTNGVTAACSHE) used in this study and the Human Immunology Core facility for assistance with immunoglobulin gene rearrangement sequencing (with infrastructure support from P30 CA016520 and P30-AI 0450080). We thank Dr. Florin Tuluc and Jennifer Murray of the Children’s Hospital of Philadelphia (CHOP) Flow Cytometry core facility for technical assistance. PR8 HA probes for initial LLPC and MBC studies were provided by Scott E. Hensley (University of Pennsylvania). Mouse-adapted A/Puerto Rico/8/1934 influenza virus for challenge studies was provided by Michael J. Hogan and Laurence C. Eisenlohr (CHOP). We thank Michael J. Hogan for critical reading of the manuscript. The graphical abstract was created with BioRender.
Author contributions
N.P. M.L., and B.Z.I. conceptualized the study. M.L., D.A., and M.P.C. designed the B cell studies. N.P., M.L., M.-G.A., I.T., and E.B. wrote the paper with help from co-authors. I.T., E.B., and K.L. graphed data and produced figures. H.M. produced mRNA vaccine antigens. Y.K.T., B.L.M., and P.J.C.L. produced empty LNP and encapsulated mRNAs into LNP and performed the physicochemical characterization of LNP. N.P., M.-G.A., I.T., K.L., S.N., C.S., E.B., and A.J. performed immunizations. S.N. and B.Z.I. contributed to the design of studies investigating the inflammatory properties of lipid nanoparticles. K.K. designed the untranslated region of the mRNA. D.L., M.-G.A., M.P., B.D., and I.T. performed HAI assays. M.-G.A., E.B., O.Y.S., and I.T. performed ELISAs. M.L., K.L., D.A., J.R.W., M.-G.A., E.B., and B.T.G. performed Tfh and B cell flow cytometry analyses. M.-G.A. designed and performed and M.-G.A. and I.T. analyzed the Luminex assay. E.T.L.P., W.M., A.M.R., M.P.C., J.J.K., and J.L.J. performed B cell sorting and B cell receptor sequencing experiments and analyzed the immune repertoire profiling data. P.B., P.H., and T.B.M. performed pseudovirus neutralization assays. F.K. and S.S. produced recombinant PR8 HA and RBD proteins used to produce fluorescently labeled probes. M.L., M.-G.A., E.B., and K.L. produced fluorescently labeled probes. I.T. and N.P. performed challenge studies.
Declaration of interests
In accordance with the University of Pennsylvania policies and procedures and our ethical obligations as researchers, we report that D.W. and N.P. are named on a patent describing the use of nucleoside-modified mRNA in lipid nanoparticles as a vaccine platform. We have disclosed those interests fully to the University of Pennsylvania, and we have in place an approved plan for managing any potential conflicts arising from licensing of our patents. K.K. is an employee of BioNTech. P.J.C.L., B.L.M., and Y.K.T. are employees of Acuitas Therapeutics, a company involved in the development of mRNA-LNP therapeutics. Y.K.T., D.W., and M.G.A. are named on patents that describe lipid nanoparticles for delivery of nucleic acid therapeutics, including mRNA and the use of modified mRNA in lipid nanoparticles as a vaccine platform. The Icahn School of Medicine at Mount Sinai has filed patent applications regarding SARS-CoV-2 and influenza virus vaccines that name F.K. as co-inventor.
Published: November 4, 2021; corrected online: May 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.11.001.
Supplemental information
Data and code availability
-
•
Raw BCR sequencing data for all mice and subsets are available on Sequence Read Archive (SRA) under BioProject: PRJNA669143. Processed AIRR-seq data are available on the AIRR Data Commons via the iReceptor portal (Corrie et al., 2018).
-
•
This paper does not report original code.
-
•
Raw data are available for reanalysis from the lead contact upon request.
References
- Akira S., Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Alameh M.G., Weissman D., Pardi N. Messenger RNA-Based Vaccines Against Infectious Diseases. Curr. Top. Microbiol. Immunol. 2020 doi: 10.1007/82_2020_202. Published online April 17, 2020. [DOI] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Scott M.K.D., Hagan T., Li C., Feng Y., Wimmers F., Grigoryan L., Trisal M., Edara V.V., Lai L., et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–416. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi S., Hook L.M., Pardi N., Wang F., Myles A., Cancro M.P., Cohen G.H., Weissman D., Friedman H.M. Nucleoside-modified mRNA encoding HSV-2 glycoproteins C, D, and E prevents clinical and subclinical genital herpes. Sci. Immunol. 2019;4:eaaw7083. doi: 10.1126/sciimmunol.aaw7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi S., Hook L.M., Swaminathan G., Cairns T.M., Brooks B., Smith J.S., Ditto N.T., Gindy M.E., Bett A.J., Espeseth A.S., et al. Antibody responses to crucial functional epitopes as a novel approach to assess immunogenicity of vaccine adjuvants. Vaccine. 2019;37:3770–3778. doi: 10.1016/j.vaccine.2019.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baiersdörfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Karikó K. A Facile Method for the Removal of dsRNA Contaminant from In Vitro-Transcribed mRNA. Mol. Ther. Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettini E., Locci M. SARS-CoV-2 mRNA Vaccines: Immunological Mechanism and Beyond. Vaccines (Basel) 2021;9:147. doi: 10.3390/vaccines9020147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.S., Eto D., Yang J.A., Lao C., Crotty S. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J. Immunol. 2013;190:3049–3053. doi: 10.4049/jimmunol.1203032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement R.L., Daccache J., Mohammed M.T., Diallo A., Blazar B.R., Kuchroo V.K., Lovitch S.B., Sharpe A.H., Sage P.T. Follicular regulatory T cells control humoral and allergic immunity by restraining early B cell responses. Nat. Immunol. 2019;20:1360–1371. doi: 10.1038/s41590-019-0472-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrie B.D., Marthandan N., Zimonja B., Jaglale J., Zhou Y., Barr E., Knoetze N., Breden F.M.W., Christley S., Scott J.K., et al. iReceptor: A platform for querying and analyzing antibody/B-cell and T-cell receptor repertoire data across federated repositories. Immunol. Rev. 2018;284:24–41. doi: 10.1111/imr.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity. 2019;50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Giovanni M., Cutillo V., Giladi A., Sala E., Maganuco C.G., Medaglia C., Di Lucia P., Bono E., Cristofani C., Consolo E., et al. Spatiotemporal regulation of type I interferon expression determines the antiviral polarization of CD4+ T cells. Nat. Immunol. 2020;21:321–330. doi: 10.1038/s41590-020-0596-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Giudice G., Rappuoli R., Didierlaurent A.M. Correlates of adjuvanticity: A review on adjuvants in licensed vaccines. Semin. Immunol. 2018;39:14–21. doi: 10.1016/j.smim.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Dias Junior A.G., Sampaio N.G., Rehwinkel J. A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation. Trends Microbiol. 2019;27:75–85. doi: 10.1016/j.tim.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N.A., Joyce M.G. Strategies to guide the antibody affinity maturation process. Curr. Opin. Virol. 2015;11:137–147. doi: 10.1016/j.coviro.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddahri F., Denanglaire S., Bureau F., Spolski R., Leonard W.J., Leo O., Andris F. Interleukin-6/STAT3 signaling regulates the ability of naive T cells to acquire B-cell help capacities. Blood. 2009;113:2426–2433. doi: 10.1182/blood-2008-04-154682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto D., Lao C., DiToro D., Barnett B., Escobar T.C., Kageyama R., Yusuf I., Crotty S. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS ONE. 2011;6:e17739. doi: 10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca V.R., Ribeiro F., Graca L. T follicular regulatory (Tfr) cells: Dissecting the complexity of Tfr-cell compartments. Immunol. Rev. 2019;288:112–127. doi: 10.1111/imr.12739. [DOI] [PubMed] [Google Scholar]
- Freyn A.W., Pine M., Rosado V.C., Benz M., Muramatsu H., Beattie M., Tam Y.K., Krammer F., Palese P., Nachbagauer R., et al. Antigen modifications improve nucleoside-modified mRNA-based influenza virus vaccines in mice. Mol. Ther. Methods Clin. Dev. 2021;22:84–95. doi: 10.1016/j.omtm.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyn A.W., Ramos da Silva J., Rosado V.C., Bliss C.M., Pine M., Mui B.L., Tam Y.K., Madden T.D., de Souza Ferreira L.C., Weissman D., et al. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020;28:1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G.M., Berek C., Kaartinen M., Milstein C. Somatic mutation and the maturation of immune response to 2-phenyl oxazolone. Nature. 1984;312:271–275. doi: 10.1038/312271a0. [DOI] [PubMed] [Google Scholar]
- Havenar-Daughton C., Lee J.H., Crotty S. Tfh cells and HIV bnAbs, an immunodominance model of the HIV neutralizing antibody generation problem. Immunol. Rev. 2017;275:49–61. doi: 10.1111/imr.12512. [DOI] [PubMed] [Google Scholar]
- Hou B., Reizis B., DeFranco A.L. Toll-like receptors activate innate and adaptive immunity by using dendritic cell-intrinsic and -extrinsic mechanisms. Immunity. 2008;29:272–282. doi: 10.1016/j.immuni.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S. Double-Stranded RNA Sensors and Modulators in Innate Immunity. Annu. Rev. Immunol. 2019;37:349–375. doi: 10.1146/annurev-immunol-042718-041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A. Exploiting Mucosal Immunity for Antiviral Vaccines. Annu. Rev. Immunol. 2016;34:575–608. doi: 10.1146/annurev-immunol-032414-112315. [DOI] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., et al. mRNA-1273 Study Group An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.L., Rosenthal R.L., Knox J.J., Myles A., Naradikian M.S., Madej J., Kostiv M., Rosenfeld A.M., Meng W., Christensen S.R., et al. The Transcription Factor T-bet Resolves Memory B Cell Subsets with Distinct Tissue Distributions and Antibody Specificities in Mice and Humans. Immunity. 2020;52:842–855.e6. doi: 10.1016/j.immuni.2020.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Kulkarni J.A., Witzigmann D., Chen S., Cullis P.R., van der Meel R. Lipid Nanoparticle Technology for Clinical Translation of siRNA Therapeutics. Acc. Chem. Res. 2019;52:2435–2444. doi: 10.1021/acs.accounts.9b00368. [DOI] [PubMed] [Google Scholar]
- Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H., 3rd, et al. A Single Immunization with Nucleoside-Modified mRNA Vaccines Elicits Strong Cellular and Humoral Immune Responses against SARS-CoV-2 in Mice. Immunity. 2020;53:724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer K., Castaño D., Gómez Atria D., Oguin T.H., 3rd, Wang S., Manzoni T.B., Muramatsu H., Hogan M.J., Amanat F., Cherubin P., et al. SARS-CoV-2 mRNA Vaccines Foster Potent Antigen-Specific Germinal Center Responses Associated with Neutralizing Antibody Generation. Immunity. 2020;53:1281–1295.e5. doi: 10.1016/j.immuni.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederer K., Parvathaneni K., Painter M.M., Bettini E., Agarwal D., Lundgreen K.A., Weirick M., Goel R.R., Xu X., Drapeau E.M., et al. Germinal center responses to SARS-CoV-2 mRNA vaccines in healthy and immunocompromised individuals. medRxiv. 2021 doi: 10.1016/j.cell.2022.01.027. 2021.09.16.21263686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Lindgren G., Sandgren K.J., Thompson E.A., Francica J.R., Seubert A., De Gregorio E., Barnett S., O’Hagan D.T., Sullivan N.J., et al. Vaccine priming is restricted to draining lymph nodes and controlled by adjuvant-mediated antigen uptake. Sci. Transl. Med. 2017;9:eaal2094. doi: 10.1126/scitranslmed.aal2094. [DOI] [PubMed] [Google Scholar]
- Lindgren G., Ols S., Liang F., Thompson E.A., Lin A., Hellgren F., Bahl K., John S., Yuzhakov O., Hassett K.J., et al. Induction of Robust B Cell Responses after Influenza mRNA Vaccination Is Accompanied by Circulating Hemagglutinin-Specific ICOS+ PD-1+ CXCR3+ T Follicular Helper Cells. Front. Immunol. 2017;8:1539. doi: 10.3389/fimmu.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman M.A., Hill D.L. Can follicular helper T cells be targeted to improve vaccine efficacy? F1000Res. 2016;5 doi: 10.12688/f1000research.7388.1. F1000 Faculty Rev-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M., Wu J.E., Arumemi F., Mikulski Z., Dahlberg C., Miller A.T., Crotty S. Activin A programs the differentiation of human TFH cells. Nat. Immunol. 2016;17:976–984. doi: 10.1038/ni.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y.M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.S., Avery D.T., Chan A., Batten M., Bustamante J., Boisson-Dupuis S., Arkwright P.D., Kreins A.Y., Averbuch D., Engelhard D., et al. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood. 2012;119:3997–4008. doi: 10.1182/blood-2011-11-392985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.S., Wong N., Rao G., Avery D.T., Torpy J., Hambridge T., Bustamante J., Okada S., Stoddard J.L., Deenick E.K., et al. Monogenic mutations differentially affect the quantity and quality of T follicular helper cells in patients with human primary immunodeficiencies. J. Allergy Clin. Immunol. 2015;136:993–1006.e1. doi: 10.1016/j.jaci.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margine I., Palese P., Krammer F. Expression of functional recombinant hemagglutinin and neuraminidase proteins from the novel H7N9 influenza virus using the baculovirus expression system. J. Vis. Exp. 2013:e51112. doi: 10.3791/51112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M.U., Wesche H. Summary and comparison of the signaling mechanisms of the Toll/interleukin-1 receptor family. Biochim. Biophys. Acta. 2002;1592:265–280. doi: 10.1016/s0167-4889(02)00320-8. [DOI] [PubMed] [Google Scholar]
- Mastelic Gavillet B., Eberhardt C.S., Auderset F., Castellino F., Seubert A., Tregoning J.S., Lambert P.H., de Gregorio E., Del Giudice G., Siegrist C.A. MF59 Mediates Its B Cell Adjuvanticity by Promoting T Follicular Helper Cells and Thus Germinal Center Responses in Adult and Early Life. J. Immunol. 2015;194:4836–4845. doi: 10.4049/jimmunol.1402071. [DOI] [PubMed] [Google Scholar]
- McKean D., Huppi K., Bell M., Staudt L., Gerhard W., Weigert M. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA. 1984;81:3180–3184. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd P.A., Minervina A.A., Pogorelyy M.V., Turner J.S., Kim W., Kalaidina E., Petersen J., Schmitz A.J., Lei T., Haile A., et al. SARS-CoV-2 mRNA vaccination elicits robust and persistent T follicular helper cell response in humans. bioRxiv. 2021 doi: 10.1101/2021.09.08.459485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mui B., Chow L., Hope M.J. Extrusion technique to generate liposomes of defined size. Methods Enzymol. 2003;367:3–14. doi: 10.1016/S0076-6879(03)67001-1. [DOI] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586:589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.H., Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill L.A., Golenbock D., Bowie A.G. The history of Toll-like receptors - redefining innate immunity. Nat. Rev. Immunol. 2013;13:453–460. doi: 10.1038/nri3446. [DOI] [PubMed] [Google Scholar]
- Olafsdottir T.A., Lindqvist M., Nookaew I., Andersen P., Maertzdorf J., Persson J., Christensen D., Zhang Y., Anderson J., Khoomrung S., et al. Comparative Systems Analyses Reveal Molecular Signatures of Clinically tested Vaccine Adjuvants. Sci. Rep. 2016;6:39097. doi: 10.1038/srep39097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E., et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R., et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., LaBranche C.C., Ferrari G., Cain D.W., Tombácz I., Parks R.J., Muramatsu H., Mui B.L., Tam Y.K., Karikó K., et al. Characterization of HIV-1 Nucleoside-Modified mRNA Vaccines in Rabbits and Rhesus Macaques. Mol. Ther. Nucleic Acids. 2019;15:36–47. doi: 10.1016/j.omtn.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S.J., Mui B.L., Tam Y.K., Karikó K., Barbosa C.J., Madden T.D., et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poholek A.C., Hansen K., Hernandez S.G., Eto D., Chandele A., Weinstein J.S., Dong X., Odegard J.M., Kaech S.M., Dent A.L., et al. In vivo regulation of Bcl6 and T follicular helper cell development. J. Immunol. 2010;185:313–326. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020;20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riteau N., Radtke A.J., Shenderov K., Mittereder L., Oland S.D., Hieny S., Jankovic D., Sher A. Water-in-Oil-Only Adjuvants Selectively Promote T Follicular Helper Cell Polarization through a Type I IFN and IL-6-Dependent Pathway. J. Immunol. 2016;197:3884–3893. doi: 10.4049/jimmunol.1600883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld A.M., Meng W., Luning Prak E.T., Hershberg U. ImmuneDB, a Novel Tool for the Analysis, Storage, and Dissemination of Immune Repertoire Sequencing Data. Front. Immunol. 2018;9:2107. doi: 10.3389/fimmu.2018.02107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A., Araki K., Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N., Liu Y., Bentebibel S.E., Munagala I., Bourdery L., Venuprasad K., Banchereau J., Ueno H. The cytokine TGF-β co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nat. Immunol. 2014;15:856–865. doi: 10.1038/ni.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N., Morita R., Bourdery L., Bentebibel S.E., Zurawski S.M., Banchereau J., Ueno H. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. 2009;31:158–169. doi: 10.1016/j.immuni.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shirai S., Kawai A., Shibuya M., Munakata L., Omata D., Suzuki R., Yoshioka Y. Lipid Nanoparticle Acts as a Potential Adjuvant for Influenza Split Vaccine without Inducing Inflammatory Responses. Vaccines (Basel) 2020;8:433. doi: 10.3390/vaccines8030433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D., Laserson U., Laserson J., Liu Y., Vigneault F., Julien J.P., Briney B., Ramos A., Saye K.F., Le K., et al. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog. 2013;9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlbauer D., Amanat F., Chromikova V., Jiang K., Strohmeier S., Arunkumar G.A., Tan J., Bhavsar D., Capuano C., Kirkpatrick E., et al. SARS-CoV-2 Seroconversion in Humans: A Detailed Protocol for a Serological Assay, Antigen Production, and Test Setup. Curr. Protoc. Microbiol. 2020;57:e100. doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Sun L., Liu H.H., Chen X., Seth R.B., Forman J., Chen Z.J. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Swaminathan G., Thoryk E.A., Cox K.S., Meschino S., Dubey S.A., Vora K.A., Celano R., Gindy M., Casimiro D.R., Bett A.J. A novel lipid nanoparticle adjuvant significantly enhances B cell and T cell responses to sub-unit vaccine antigens. Vaccine. 2016;34:110–119. doi: 10.1016/j.vaccine.2015.10.132. [DOI] [PubMed] [Google Scholar]
- Swaminathan G., Thoryk E.A., Cox K.S., Smith J.S., Wolf J.J., Gindy M.E., Casimiro D.R., Bett A.J. A Tetravalent Sub-unit Dengue Vaccine Formulated with Ionizable Cationic Lipid Nanoparticle induces Significant Immune Responses in Rodents and Non-Human Primates. Sci. Rep. 2016;6:34215. doi: 10.1038/srep34215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia K., Kim W.K., Sun Y., Mercado-López X., Dunay E., Wise M., Adu M., López C.B. Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity. PLoS Pathog. 2013;9:e1003703. doi: 10.1371/journal.ppat.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.S., O’Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden J.A., Yaari G., Uduman M., Stern J.N., O’Connor K.C., Hafler D.A., Vigneault F., Kleinstein S.H. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 2014;30:1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora G.D., Nussenzweig M.C. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Victora G.D., Wilson P.C. Germinal center selection and the antibody response to influenza. Cell. 2015;163:545–548. doi: 10.1016/j.cell.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa C.G., Linterman M.A., Yu D., MacLennan I.C. Follicular Helper T Cells. Annu. Rev. Immunol. 2016;34:335–368. doi: 10.1146/annurev-immunol-041015-055605. [DOI] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigert M.G., Cesari I.M., Yonkovich S.J., Cohn M. Variability in the lambda light chain sequences of mouse antibody. Nature. 1970;228:1045–1047. doi: 10.1038/2281045a0. [DOI] [PubMed] [Google Scholar]
- Weissman D., Alameh M.G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., et al. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe. 2021;29:23–31.e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw BCR sequencing data for all mice and subsets are available on Sequence Read Archive (SRA) under BioProject: PRJNA669143. Processed AIRR-seq data are available on the AIRR Data Commons via the iReceptor portal (Corrie et al., 2018).
-
•
This paper does not report original code.
-
•
Raw data are available for reanalysis from the lead contact upon request.