Abstract
Sequencing of large cohorts offers an unprecedented opportunity to identify rare genetic variants and to find novel contributors to human disease. We used gene-based collapsing tests to identify genes associated with glucose, HbA1c and type 2 diabetes (T2D) diagnosis in 379,066 exome-sequenced participants in the UK Biobank. We identified associations for variants in GCK, HNF1A and PDX1, which are known to be involved in Mendelian forms of diabetes. Notably, we uncovered novel associations for GIGYF1, a gene not previously implicated by human genetics in diabetes. GIGYF1 predicted loss of function (pLOF) variants associated with increased levels of glucose (0.77 mmol/L increase, p = 4.42 × 10–12) and HbA1c (4.33 mmol/mol, p = 1.28 × 10–14) as well as T2D diagnosis (OR = 4.15, p = 6.14 × 10–11). Multiple rare variants contributed to these associations, including singleton variants. GIGYF1 pLOF also associated with decreased cholesterol levels as well as an increased risk of hypothyroidism. The association of GIGYF1 pLOF with T2D diagnosis replicated in an independent cohort from the Geisinger Health System. In addition, a common variant association for glucose and T2D was identified at the GIGYF1 locus. Our results highlight the role of GIGYF1 in regulating insulin signaling and protecting from diabetes.
Subject terms: Genetic association study, Type 2 diabetes, Genetic predisposition to disease, Quantitative trait
Introduction
Human genetics provides powerful methods for understanding the roles of genes and proteins in disease and can lead to new therapeutic hypotheses and drug targets. Genetic evidence based on sequence variants within coding regions of the genome is better at predicting the efficacy and safety of novel therapeutics than evidence from genome-wide association studies (GWAS), which tend to involve common noncoding variants1–3. Among coding variants, predicted loss of function (pLOF) variants are particularly informative in association studies because they establish a direct causal link between reduction in gene function and biological outcomes. Additionally, rare missense variants predicted to be deleterious can provide valuable biological insights4,5. However, interrogation of the effects of such variants is hampered by the rarity of these variants and the cohort sizes needed to identify associations6. Exome or whole-genome sequencing of large biobanks coupled with gene-level aggregation of rare high impact variants can help to circumvent these challenges4. Biobanks offer a considerable advantage over case–control cohorts as they contain richer phenotyping data which often includes biomarker measurements as well as disease diagnoses. This allows a more complete understanding of the biological consequences of damaging variants in particular genes7,8.
Type 2 diabetes (T2D) is a disease that has been extensively studied in traditional array-based GWAS with hundreds of associations identified to date9–12. Although these studies have given insight into some of the biological mechanisms contributing to T2D, most of the reported associations are with variants in non-coding regions, making identification of the causal gene challenging. More recently, exome sequencing has been applied to discover protein-coding variants that alter the risk of developing T2D. Sequencing of 20,791 T2D cases followed by the use of gene-based collapsing tests (to aggregate predicted damaging variants) identified associations of SLC30A8, MC4R and PAM with T2D diagnosis5.
Using whole exome sequences from the UK Biobank (UKBB) we performed gene-level collapsing tests to examine the association of pLOF and damaging missense variants in ~ 17,000 genes with biomarkers of glycemic control, glucose, and glycated hemoglobin (HbA1c), as well as T2D diagnosis.
Results
Gene-level associations with glucose, HbA1c and T2D
We used 454,787 whole exome sequences from the UK Biobank (UKBB) to identify rare variants with a minor allele frequency (MAF) ≤ 1% likely to have functional impact; pLOF variants (i.e. frameshift, stop gain, splice donor or splice acceptor variants) called as high confidence by LOFTEE13 or missense variants predicted to be damaging (Combined Annotation Dependent Depletion [CADD] score ≥ 25). We identified 726,422 rare pLOF variants affecting 16,477 genes, 58.5% of which were singletons (carried by a single individual), and 2.14 million damaging missense variants in 17,312 genes, 49.6% of which were singletons (Supplementary Table 1).
Given the large proportion of variants present in just a single individual, we used gene-based collapsing tests to look for associations with biomarkers of glycemic control and T2D diagnosis. We used two variant aggregation strategies; (1) pLOF variants with MAF ≤ 1% and (2) damaging missense variants with MAF ≤ 1% and performed burden testing in the unrelated European ancestry population (n = 363,977) adjusting for age, sex and genetic ancestry via 12 principal components.
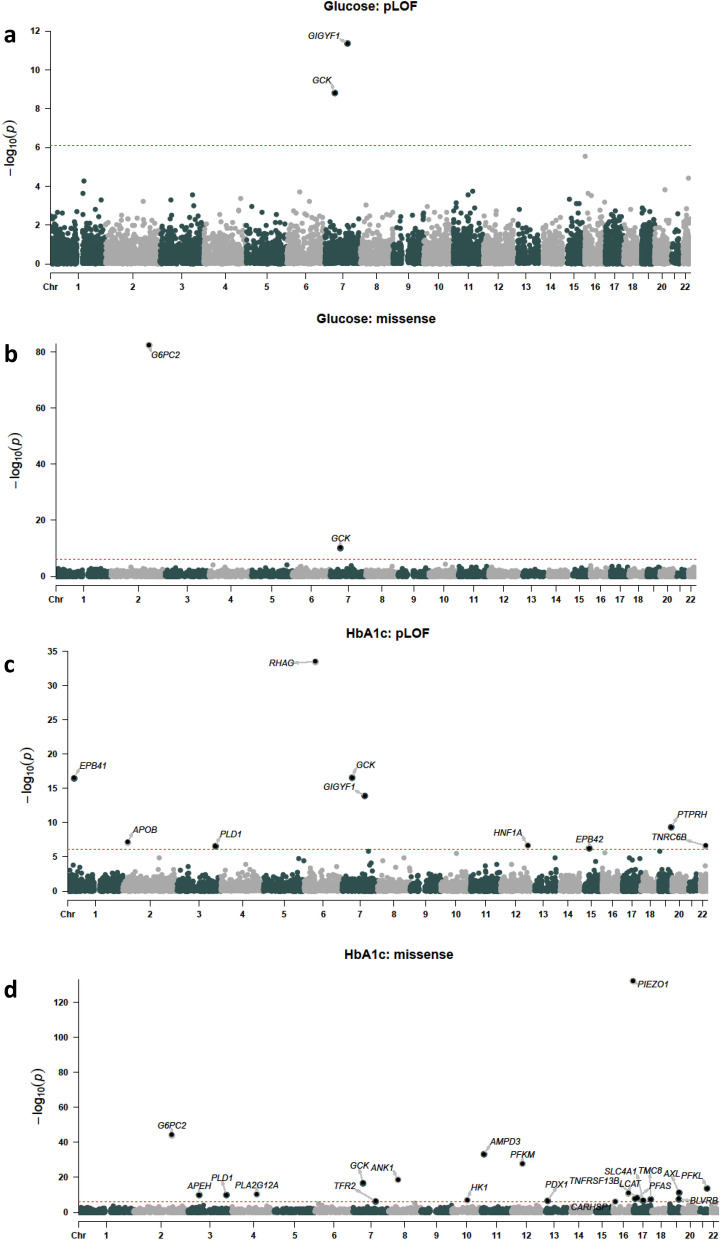
First, we tested genes for association with glucose and HbA1c levels. We required at least ten variant carriers per gene to have measurements based on an examination of genomic inflation at different carrier thresholds (Supplementary Figure 1). Using a p-value threshold adjusted for the number of variant sets and phenotypes tested (p ≤ 7.82 × 10–7), four variant sets in three genes significantly associated with glucose levels: GCK pLOF (p = 1.56 × 10–9, 1.24 mmol/L increase), GCK damaging missense (p = 6.15 × 10–11, 0.61 mmol/L increase), GIGYF1 pLOF (p = 4.42 × 10–12, 0.77 mmol/L increase) and G6PC2 damaging missense variants (p = 4.62 × 10–83, 0.33 mmol/L decrease) (Fig. 1, Table 1). The same variant sets also associated with HbA1c levels along with 27 additional sets including HNF1A pLOF (p = 2.14 × 10–7, 4.01 mmol/mol increase), TNRC6B pLOF (p = 2.36 × 10–7, 3.94 mmol/mol increase) and PDX1 damaging missense variants (p = 2.54 × 10–7, 0.41 mmol/mol increase) (Fig. 1, Table 1 and Supplementary Table 2).
Figure 1.
Gene-level associations with glucose and HbA1c levels. (A) pLOF associations with glucose levels. (B) Damaging missense variant (CADD score ≥ 25) associations with glucose levels. (C) pLOF associations with HbA1c. (D) Damaging missense variant associations with HbA1c levels. The red line indicates the threshold for significance, genes with significant associations are labeled.
Table 1.
Gene-level associations with glucose and HbA1c levels.
| Gene | Variant set | Title | p-value | Effect (SD) | Effect (IFCC units) | 95% CI− (IFCC units) | 95% CI+ (IFCC units) | IFCC units | n carrier measured |
|---|---|---|---|---|---|---|---|---|---|
| GCK | pLOF | Glucose | 1.56 × 10–9 | 1.00 | 1.24 | 0.84 | 1.65 | mmol/L | 35 |
| GIGYF1 | pLOF | Glucose | 4.42 × 10–12 | 0.62 | 0.77 | 0.55 | 0.98 | mmol/L | 121 |
| GCK | missense | Glucose | 6.15 × 10–11 | 0.49 | 0.61 | 0.42 | 0.79 | mmol/L | 173 |
| G6PC2 | missense | Glucose | 4.62 × 10–83 | − 0.27 | − 0.33 | − 0.36 | − 0.3 | mmol/L | 5128 |
| GCK | pLOF | HbA1c | 2.64 × 10–17 | 1.29 | 8.75 | 6.73 | 10.78 | mmol/mol | 38 |
| GIGYF1 | pLOF | HbA1c | 1.28 × 10–14 | 0.64 | 4.33 | 3.23 | 5.43 | mmol/mol | 129 |
| HNF1A | pLOF | HbA1c | 2.14 × 10–7 | 0.59 | 4.01 | 2.50 | 5.53 | mmol/mol | 68 |
| TNRC6B | pLOF | HbA1c | 2.36 × 10–7 | 0.58 | 3.94 | 2.45 | 5.43 | mmol/mol | 70 |
| RHAG | pLOF | HbA1c | 3.31 × 10–34 | − 0.86 | − 5.81 | − 6.75 | − 4.88 | mmol/mol | 179 |
| EPB41 | pLOF | HbA1c | 3.14 × 10–17 | − 0.53 | − 3.58 | − 4.41 | − 2.75 | mmol/mol | 226 |
| PTPRH | pLOF | HbA1c | 4.39 × 10–10 | 0.11 | 0.74 | 0.51 | 0.97 | mmol/mol | 2924 |
| APOB | pLOF | HbA1c | 6.94 × 10–8 | 0.23 | 1.57 | 1.00 | 2.15 | mmol/mol | 478 |
| PLD1 | pLOF | HbA1c | 2.99 × 10–7 | 0.23 | 1.56 | 0.96 | 2.16 | mmol/mol | 438 |
| EPB42 | pLOF | HbA1c | 6.11 × 10–7 | − 0.31 | − 2.08 | − 2.90 | − 1.26 | mmol/mol | 234 |
| GCK | missense | HbA1c | 1.86 × 10–17 | 0.56 | 3.83 | 2.94 | 4.71 | mmol/mol | 201 |
| G6PC2 | missense | HbA1c | 6.71 × 10–45 | − 0.18 | − 1.21 | − 1.38 | − 1.04 | mmol/mol | 5574 |
| PFAS | missense | HbA1c | 2.09 × 10–8 | − 0.05 | − 0.32 | − 0.44 | − 0.21 | mmol/mol | 12,621 |
| PDX1 | missense | HbA1c | 2.54 × 10–7 | 0.06 | 0.41 | 0.25 | 0.56 | mmol/mol | 6694 |
| PIEZO1 | missense | HbA1c | 1.0 × 10–132 | − 0.15 | − 1.00 | − 1.07 | − 0.92 | mmol/mol | 26,726 |
| AMPD3 | missense | HbA1c | 7.28 × 10–34 | 0.13 | 0.86 | 0.72 | 1.00 | mmol/mol | 8258 |
| PFKM | missense | HbA1c | 2.16 × 10–28 | − 0.28 | − 1.92 | − 2.26 | − 1.58 | mmol/mol | 1353 |
| ANK1 | missense | HbA1c | 3.12 × 10–19 | − 0.13 | − 0.87 | − 1.06 | − 0.68 | mmol/mol | 4342 |
| PFKL | missense | HbA1c | 2.69 × 10–14 | 0.10 | 0.68 | 0.50 | 0.85 | mmol/mol | 5245 |
| AXL | missense | HbA1c | 4.11 × 10–12 | − 0.08 | − 0.54 | − 0.69 | − 0.39 | mmol/mol | 6827 |
| LCAT | missense | HbA1c | 1.29 × 10–11 | − 0.27 | − 1.82 | − 2.34 | − 1.29 | mmol/mol | 565 |
| PLA2G12A | missense | HbA1c | 5.74 × 10–11 | − 0.08 | − 0.51 | − 0.67 | − 0.36 | mmol/mol | 6761 |
| PLD1 | missense | HbA1c | 1.51 × 10–10 | 0.08 | 0.57 | 0.40 | 0.75 | mmol/mol | 5180 |
| APEH | missense | HbA1c | 1.86 × 10–10 | 0.29 | 1.96 | 1.36 | 2.57 | mmol/mol | 429 |
| TNFRSF13B | missense | HbA1c | 4.63 × 10–9 | − 0.08 | − 0.51 | − 0.68 | − 0.34 | mmol/mol | 5470 |
| BLVRB | missense | HbA1c | 2.53 × 10–8 | − 0.07 | − 0.48 | − 0.65 | − 0.31 | mmol/mol | 5533 |
| TMC8 | missense | HbA1c | 3.01 × 10–8 | 0.11 | 0.77 | 0.50 | 1.05 | mmol/mol | 2095 |
| HK1 | missense | HbA1c | 1.08 × 10–7 | − 0.16 | − 1.08 | − 1.48 | − 0.68 | mmol/mol | 988 |
| SLC4A1 | missense | HbA1c | 1.76 × 10–7 | − 0.15 | − 1.04 | − 1.43 | − 0.65 | mmol/mol | 1025 |
| TFR2 | missense | HbA1c | 4.24 × 10–7 | − 0.12 | − 0.84 | − 1.16 | − 0.51 | mmol/mol | 1491 |
| CARHSP1 | missense | HbA1c | 6.78 × 10–7 | − 0.13 | − 0.86 | − 1.20 | − 0.52 | mmol/mol | 1360 |
pLOF or damaging missense variants (CADD score ≥ 25) were aggregated per gene. The effect is shown in standard deviations (SD) of transformed values as well as in International Federation of Clinical Chemistry (IFCC) units.
CI confidence interval.
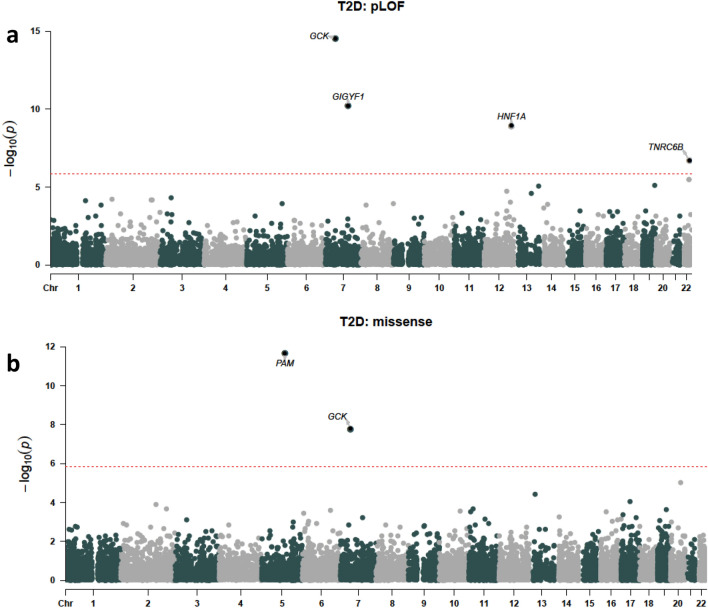
We then tested aggregated pLOF and damaging missense variants in all genes for association with T2D diagnosis (n = 24,695 cases). Six variant sets significantly associated with T2D (p ≤ 1.46 × 10–6); pLOF variants in GIGYF1, GCK, HNF1A and TNRC6B and damaging missense variants in GCK and PAM (Fig. 2, Table 2 and Supplementary Table 2). As the time of available follow-up differs between England, Scotland, and Wales, we controlled for country of recruitment in the regression. In addition, we confirmed that significant hits did not associate with country of recruitment (all p > 0.035) and that hits remained significant when only data from England were considered (Supplementary Table 3). Our results also remained significant when we adjusted for follow-up time in the burden test (Supplementary Table 4).
Figure 2.
Gene-level associations with T2D. (A) pLOF associations. (B) Damaging missense variant (CADD score ≥ 25) associations. The red line indicates the threshold for significance, genes with significant associations are labeled.
Table 2.
Gene-level associations with T2D diagnosis.
| Gene | Variant set | Title | p-value | OR | 95% CI− | 95% CI+ | N cases | N carrier | N carrier cases | N expected |
|---|---|---|---|---|---|---|---|---|---|---|
| GCK | pLOF | T2D | 2.96 × 10–15 | 14.16 | 7.33 | 27.34 | 24,695 | 40 | 19 | 2.71 |
| GIGYF1 | pLOF | T2D | 6.14 × 10–11 | 4.15 | 2.71 | 6.37 | 24,695 | 131 | 29 | 8.89 |
| HNF1A | pLOF | T2D | 1.23 × 10–9 | 5.27 | 3.08 | 9 | 24,695 | 73 | 20 | 4.95 |
| TNRC6B | pLOF | T2D | 2.00 × 10–7 | 4.44 | 2.53 | 7.79 | 24,695 | 71 | 17 | 4.82 |
| PAM | missense | T2D | 2.26 × 10–12 | 1.31 | 1.21 | 1.41 | 24,695 | 9357 | 801 | 634.85 |
| GCK | missense | T2D | 1.70 × 10–8 | 2.96 | 2.03 | 4.32 | 24,695 | 202 | 34 | 13.71 |
pLOF or damaging missense variants (CADD score ≥ 25) were aggregated per gene.
OR odds ratio, CI confidence interval.
Sex-specific analyses of glucose, HbA1c and T2D were performed but did not identify associations for additional genes (Supplementary Tables 5–7). Meta-analysis of results for males and females gave similar effect estimates to the primary analysis, with one additional association detected, CREB3L3 pLOF and HbA1c levels. We did not observe significant heterogeneity in effects between sexes (Supplementary Tables 8, 9, 10).
We also tested genes for association with glucose, HbA1c and T2D in the African ancestry (n = 6221), South Asian ancestry (n = 7553) and East Asian ancestry (n = 1315) populations of UKBB. The only statistically significant association was between MYH6 damaging missense variants and HbA1c levels in the East Asian ancestry population (p = 1.11 × 10–6, 5.10 mmol/mol decrease) (Supplementary Table 11). Several of the variant sets significant in our primary analysis were nominally significant in the other populations. These included G6PC2 damaging missense variants which associated with decreased glucose levels in the African and South Asian ancestry populations (p = 0.01 and p = 8.31 × 10–4 respectively, Supplementary Tables 11 and 12). Carriers of GIGYF1 pLOF variants were too rare to test associations of these variants in the other populations.
Identification of genes with a biological role in diabetes
Variants in two genes, GCK and GIGYF1, significantly associated with glucose, HbA1c and T2D diagnosis, strongly suggesting a biological role in diabetes; GCK is involved in Mendelian forms of diabetes while GIGYF1 has not previously been implicated by genetics in the disease. Both GCK and GIGYF1 are located on chromosome 7 but are 56 Mb apart, strongly suggesting that these signals are independent; this independence was confirmed by conditional analysis (Supplementary Table 13). Two additional variant sets, HNF1A pLOF and TNRC6B pLOF, had genome-wide associations with both T2D diagnosis and HbA1c levels while G6PC2 damaging missense variants associated with decreased levels of both glucose and HbA1c but not T2D diagnosis (Table 3).
Table 3.
Genes and variant sets associated with multiple diabetes-related traits.
| Gene | Variant set | p-value glucose | Effect glucose | p-value HbA1c | Effect Hba1c | p-value T2D | OR T2D |
|---|---|---|---|---|---|---|---|
| GCK | pLOF | 1.56 × 10–9 | 0.999 | 2.64 × 10–17 | 1.292 | 2.96 × 10–15 | 14.16 |
| HNF1A | pLOF | 0.01 | 0.317 | 2.14 × 10–7 | 0.592 | 1.23 × 10–9 | 5.27 |
| GIGYF1 | pLOF | 4.42 × 10–12 | 0.616 | 1.28 × 10–14 | 0.639 | 6.14 × 10–11 | 4.15 |
| GCK | missense | 6.15 × 10–11 | 0.487 | 1.86 × 10–17 | 0.565 | 1.70 × 10–8 | 2.96 |
| PAM | missense | 0.92 | 0.001 | 0.009 | 0.026 | 2.26 × 10–12 | 1.31 |
| TNRC6B | pLOF | 4.01 × 10–5 | 0.507 | 2.36 × 10–7 | 0.582 | 2.00 × 10–7 | 4.44 |
| PDX1 | missense | 0.02 | 0.029 | 2.54 × 10–7 | 0.060 | 3.99 × 10–5 | 1.21 |
| PFAS | missense | 0.32 | 0.009 | 2.09 × 10–8 | − 0.048 | 4.43 × 10–4 | 0.88 |
| G6PC2 | missense | 4.62 × 10–83 | − 0.266 | 6.71 × 10–45 | − 0.179 | 0.97 | 1.00 |
Variant sets significant for at least one trait in our primary analysis that are also associated with additional diabetes traits (p ≤ 0.0016, 32 sets tested) are shown. Effect is shown in SD of transformed values or as an odds ratio (OR).
To see which other significant genes were likely to have a role in diabetes we looked at all variant sets with a significant glucose, HbA1c, or T2D association and examined whether they had associations with additional diabetes traits (p ≤ 0.0016, correcting for 32 sets tested). Damaging missense variants in PDX1 and PFAS, which significantly associated with HbA1c levels in our primary analysis, associated with T2D diagnosis using this threshold (Table 3 and Supplementary Table 14).
Many HbA1c associations appeared to be secondary to effects on red blood cells. 22 out of 31 variant sets associated with HbA1c did not show effects on glucose levels or T2D diagnosis (Supplementary Table 14) and were not implicated in Mendelian forms of diabetes. Out of these 22 variant sets, 12 were in genes implicated in Mendelian disorders affecting red blood cells (for example EPB42 and TFR2; see Supplementary Table 15) and an additional five had highly significant associations with red blood cell traits in our data (p ≤ 7.82 × 10–7; Supplementary Table 16).
We focused on the variant sets associated with multiple diabetes traits as these are strong candidates for regulating glucose homeostasis and/or causally contributing to T2D risk. The genes fall into three main groups; known MODY (maturity-onset diabetes of the young) genes (GCK, HNF1A and PDX1)14, known genes reported in previous exome-wide analyses of glucose levels or T2D (G6PC2 and PAM)5,15, and novel genes not previously implicated by genetics in diabetes (GIGYF1, TNRC6B and PFAS).
Because obesity is linked to the development of T2D, we adjusted for body mass index (BMI) in the regression and found that the association of these genes with diabetes-related traits remained significant (Supplementary Tables 17 and 18).
We used the generalized linear mixed model implemented by SAIGE-Gene which accounts for relatedness and adjusts for unbalanced case–control ratios16 to verify association of our variant sets of interest with glucose, HbA1c, and T2D diagnosis. SAIGE-Gene was run in the European ancestry population including related individuals (n = 398,574). Using the p-value thresholds previously employed, all associations were statistically significant using this method apart from the associations of TNRC6B pLOF with HbA1c (p = 6.85 × 10–6) and T2D diagnosis (p = 4.77 × 10–5) which were less significant (Supplementary Table 19).
To maximize power to detect associations for rare variants, our original analysis of glucose and HbA1c included individuals with a diabetes diagnosis. Associations for all variant sets of interest were at least nominally significant when such individuals were excluded from the analysis (Supplementary Table 20). For GIGYF1 pLOF, there was still a substantial effect on glucose (p = 2.95 × 10–8, effect = 0.53 SD) and HbA1c (p = 8.29 × 10–7, effect = 0.43 SD) levels in carriers without a formal diabetes diagnosis.
For the MODY genes, GCK, HNF1A and PDX1, we also tested pathogenic or likely pathogenic variants annotated in ClinVar17 for association with glucose, HbA1c and T2D—either alone or combined with the variants used in our primary analysis. Pathogenic and likely pathogenic variants in GCK and HNF1A associated with T2D diagnosis and markers of glycemic control and added signal beyond what was seen in the primary analysis (Supplementary Tables 21 and 22).
Validation of GIGYF1 pLOF associations using independent datasets
We sought to use independent measurements of glucose and HbA1c to perform sensitivity analysis and verify the associations of interest seen in our primary analysis which used measurements taken as part of the UKBB assessment. To do this we extracted lab test values for glucose and HbA1c from primary care data, which is available for approximately half of the cohort. In gene-based burden tests all variant sets showed a direction of effect consistent with that seen in the primary analysis and 10 out of 12 of these were significant when correcting for the number of tests performed (p ≤ 0.004). This included the association of GIGYF1 pLOF with glucose (p = 2.10 × 10–6, effect = 0.65 SD) and HbA1c (p = 1.19 × 10–5, effect = 0.74 SD) levels (Supplementary Figure 2 and Supplementary Table 23).
We then assessed whether rare variants in GIGYF1 and the other novel genes associated with T2D replicated in a completely independent exome-sequencing cohort. Gene-based tests in European ancestry individuals from the Geisinger Health System (GHS; 25,846 T2D cases and 63,749 controls) confirmed the association of GIGYF1 pLOF with T2D (p = 0.01, OR = 1.8). We did not replicate the association of TNRC6B pLOF with T2D. We tested an expanded PFAS variant set (pLOF + deleterious missense) and did not detect an association with T2D (Supplementary Table 24). Notably variant set definitions varied somewhat from those used in our primary analysis (see “Methods”).
Multiple variants contribute to associations with diabetes diagnosis and biomarkers
To examine whether specific variants were driving the associations with diabetes traits we conducted “leave-one-out” burden tests. The association of PAM missense variants with T2D diagnosis was driven entirely by a previously reported variant Ser539Trp (rs78408340; p = 0.43 when Ser539Trp is excluded). For all other variant sets, multiple variants contributed to the associations observed (Supplementary Figure 3). Notably, when singleton variants were excluded, half of the associations no longer reached significance including those for GCK pLOF and glucose (p = 0.0015 without singletons versus p = 1.56 × 10–9) and GIGYF1 pLOF and T2D (p = 2.9 × 10–5 without singletons versus p = 6.14 × 10–11) (Supplementary Table 25), demonstrating the power of including singletons in gene-based tests.
For the variants contributing to our novel discovered associations, GIGYF1 pLOF, TNRC6B pLOF and PFAS damaging missense variants, we examined the quality scores, sequencing depth, transcripts affected and presence of contributing variants in the gnomAD database. We found that for GIGYF1 and PFAS the variants contributing most to the associations had good quality scores and depth and were present in gnomAD. In contrast, TNRC6B is a highly constrained gene and the most common pLOF variant is not present in gnomAD (see “Supplementary Note” and Supplementary Figure 4). This observation along with the fact that the association of TNRC6B pLOF with T2D did not replicate in GHS leads us to view this association with suspicion.
Replication of published gene-level associations with T2D and associations for T2D drug target genes
The association between predicted damaging variants in PAM and T2D diagnosis was previously reported in an exome-sequencing study performed by Flannick et al.5. We examined whether the other two significant genes in the study, SLC30A8 and MC4R, associated with diabetes traits in our analysis. Both pLOF and damaging missense variants in SLC30A8 associated with reduced levels of HbA1c and glucose and suggestively associated with decreased incidence of T2D diagnosis (Supplementary Table 26). Combining SLC30A8 pLOF and missense variants resulted in more significant associations with glucose (p = 2.71 × 10–6), HbA1c (p = 8.64 × 10–10) and T2D diagnosis (p = 0.005) (Supplementary Table 26). There were no MC4R high confidence pLOF variants in our dataset and MC4R predicted damaging missense variants did not associate with diabetes-related traits in our study (all p > 0.19). We note that the MC4R Ile269Asn variant driving the association in Flannick and colleagues’ analysis is absent from our dataset, consistent with the fact that it is absent from European populations in gnomAD.
We also examined whether we detect associations for the 8 genes encoding T2D drug targets (GLP1R, IGF1R, PPARG, INSR, SLC5A2, DPP4, KCNJ11, ABCC8). Variant sets in three of these genes, DPP4, GLP1R and KCNJ11 significantly associated with either T2D diagnosis or HbA1c levels (p ≤ 0.003 correcting for 15 variant sets tested) and an additional 4 genes had a nominally significant association with T2D and/or HbA1c (Supplementary Figure 5 and Supplementary Table 27).
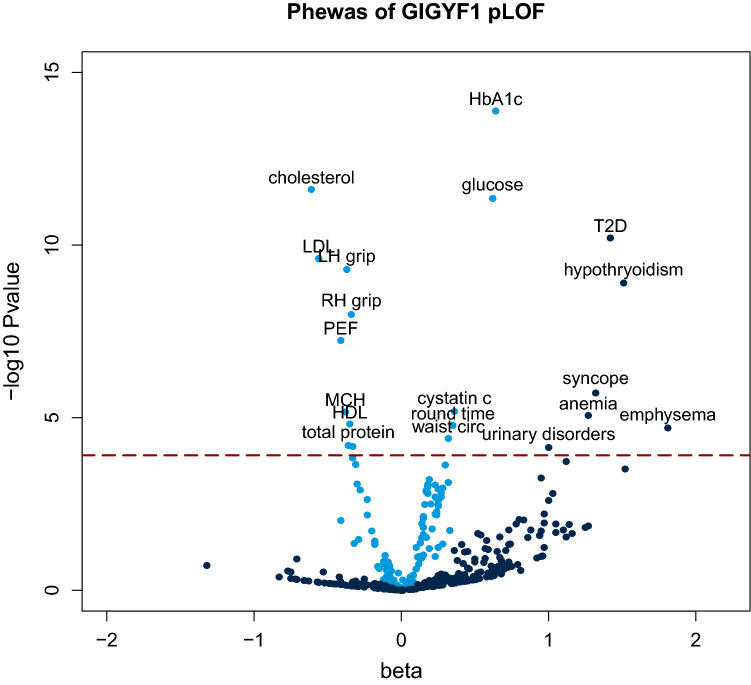
PheWAS of GIGYF1 pLOF reveals associations with cholesterol levels, hypothyroidism and complications of diabetes
The most significant novel associations were seen for GIGYF1 pLOF which associated with increased glucose and HbA1c levels as well as increased incidence of T2D diagnosis. To give additional insight into the biological roles of GIGYF1 we performed a phenome-wide association study (PheWAS) testing GIGYF1 pLOF for association with 142 quantitative traits and 262 ICD10-coded diagnoses (Fig. 3).
Figure 3.
PheWAS of GIGYF1 pLOF. The x-axis is the beta (effect size in standard deviations) for the association and the y-axis is − log10 (p-value). Quantitative traits are colored light blue and ICD10 diagnoses colored dark blue. Phenome-wide significant associations are labeled. The dashed line indicates the p-value threshold for phenome-wide significance. Protein; total protein, RH grip; right hand grip strength, round time: time to complete round (cognitive test), LH grip; left hand grip strength, PEF; peak expiratory flow.
GIGYF1 pLOF strongly associated with decreased levels of total cholesterol (p = 2.44 × 10–12, effect = − 0.61 SD) which was, in large part, driven by LDL cholesterol (p = 2.40 × 10–10, effect = − 0.56 SD) although an effect on HDL cholesterol was also observed (Table 4). To understand the extent to which this is influenced by the use of cholesterol-lowering medication in diabetics, we adjusted for medication use in the regression and also performed a separate analysis excluding those on cholesterol-lowering medication. The association between GIGYF1 pLOF and LDL cholesterol levels was significant in both analyses (Supplementary Table 28). GIGYF1 pLOF also associated with decreased grip strength and decreased peak expiratory flow. Notably, GIGYF1 pLOF also associated with increased levels of the kidney injury biomarker cystatin c (p = 6.65 × 10–6, effect = 0.36 SD) and increased diagnosis of urinary system disorders (p = 7.32 × 10–5, OR = 2.71) (Tables 4 and 5).
Table 4.
PheWAS of GIGYF1 pLOF – quantitative traits.
| Gene | Variant set | Title | p-value | Effect (SD) | 95% CI− | 95% CI+ | n carrier measured |
|---|---|---|---|---|---|---|---|
| GIGYF1 | pLOF | HbA1c | 1.28 × 10–14 | 0.64 | 0.48 | 0.80 | 129 |
| GIGYF1 | pLOF | Cholesterol | 2.44 × 10–12 | − 0.61 | − 0.78 | − 0.44 | 128 |
| GIGYF1 | pLOF | Glucose | 4.42 × 10–12 | 0.62 | 0.44 | 0.79 | 121 |
| GIGYF1 | pLOF | LDL cholesterol | 2.40 × 10–10 | − 0.56 | − 0.73 | − 0.38 | 128 |
| GIGYF1 | pLOF | Apolipoprotein b | 2.52 × 10–10 | − 0.56 | − 0.73 | − 0.39 | 127 |
| GIGYF1 | pLOF | LH grip strength | 5.11 × 10–10 | − 0.37 | − 0.49 | − 0.25 | 131 |
| GIGYF1 | pLOF | RH grip strength | 1.01 × 10–8 | − 0.34 | − 0.46 | − 0.23 | 131 |
| GIGYF1 | pLOF | Peak expiratory flow | 5.73 × 10–8 | − 0.41 | − 0.56 | − 0.26 | 114 |
| GIGYF1 | pLOF | Cystatin c | 6.65 × 10–6 | 0.36 | 0.20 | 0.51 | 128 |
| GIGYF1 | pLOF | Mean corpuscular hemoglobin | 6.80 × 10–6 | − 0.38 | − 0.55 | − 0.22 | 128 |
| GIGYF1 | pLOF | HDL cholesterol | 1.53 × 10–5 | − 0.35 | − 0.52 | − 0.19 | 121 |
| GIGYF1 | pLOF | Time to complete round (cognitive test) | 1.67 × 10–5 | 0.35 | 0.19 | 0.51 | 129 |
| GIGYF1 | pLOF | Waist circumference | 3.98 × 10–5 | 0.32 | 0.16 | 0.47 | 130 |
| GIGYF1 | pLOF | Total protein | 6.45 × 10–5 | − 0.36 | − 0.53 | − 0.18 | 121 |
| GIGYF1 | pLOF | Apolipoprotein a | 6.88 × 10–5 | − 0.33 | − 0.49 | − 0.17 | 121 |
Significant associations with quantitative traits are shown (p ≤ 1.22 × 10–4). Effect is shown in standard deviations (SD) of transformed values.
RH right hand, LH left hand, CI confidence interval.
Table 5.
PheWAS of GIGYF1 pLOF—ICD10-coded diagnoses.
| Gene | Variant set | Title | p-value | OR | 95% CI− | 95% CI+ | N cases | N carrier cases | N expected |
|---|---|---|---|---|---|---|---|---|---|
| GIGYF1 | pLOF | E11 T2D | 6.14 × 10–11 | 4.15 | 2.71 | 6.37 | 24,695 | 29 | 8.89 |
| GIGYF1 | pLOF | E03 other hypothyroidism | 1.25 × 10–9 | 4.53 | 2.78 | 7.38 | 19,417 | 21 | 6.99 |
| GIGYF1 | pLOF | R55 syncope and collapse | 1.90 × 10–6 | 3.75 | 2.18 | 6.47 | 12,706 | 15 | 4.57 |
| GIGYF1 | pLOF | D50 iron deficiency anemia | 8.52 × 10–6 | 3.56 | 2.04 | 6.23 | 12,886 | 14 | 4.64 |
| GIGYF1 | pLOF | J43 emphysema | 1.99 × 10–5 | 6.13 | 2.67 | 14.10 | 3015 | 6 | 1.09 |
| GIGYF1 | pLOF | N39 other disorders of urinary system | 7.32 × 10–5 | 2.71 | 1.66 | 4.45 | 24,581 | 19 | 8.85 |
Significant associations with ICD10-coded diagnoses are shown (p ≤ 1.22 × 10–4).
OR odds ratio, CI confidence interval.
After diabetes, the most significant disease association of GIGYF1 pLOF was with increased risk of hypothyroidism (p = 1.25 × 10–9, OR = 4.53). 21 out of the 131 GIGYF1 pLOF carriers had a diagnosis of unspecified hypothyroidism and seven of these also had a diagnosis of T2D. Given the autoimmune component in hypothyroidism and type 1 diabetes (T1D), we examined the association of GIGYF1 pLOF with T1D diagnoses but did not detect a significant association (p = 0.1). GIGYF1 pLOF also significantly associated with increased risk of syncope and collapse (p = 1.92 × 10–6, OR = 3.75) (Table 5).
Other phenome-wide significant associations with quantitative traits included waist circumference, total protein and mean corpuscular hemoglobin as well increased time to complete a cognitive test (Table 4). To ensure that the association of GIGYF1 pLOF with HbA1c was independent of effects on hemoglobin we adjusted for mean corpuscular hemoglobin level and verified that the association remained highly significant (p = 4.10 × 10–12). GIGYF1 pLOF also associated with increased diagnosis of emphysema and anemia (Table 5).
Common variants at GIGYF1 associate with glucose, T2D and GIGYF1 expression
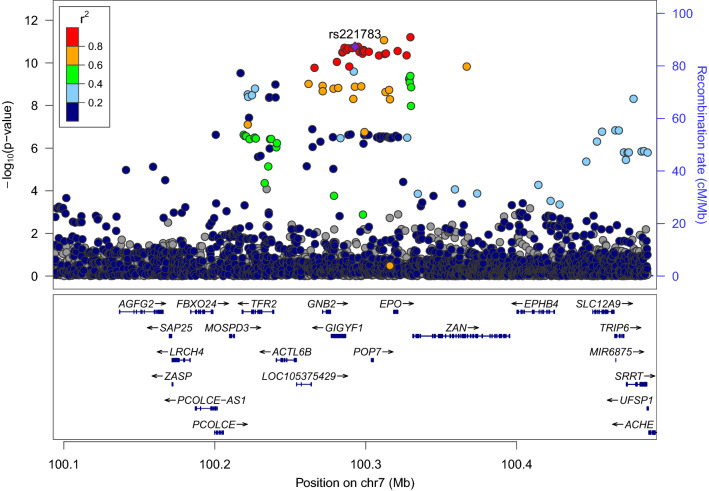
We looked for more common variants that could further implicate the GIGYF1 locus in diabetes. We tested array genotyped and imputed variants at the GIGYF1 locus for association with glucose levels in 294,042 unrelated European ancestry individuals with measurements available. We found a cluster of variants in a linkage disequilibrium block covering GIGYF1 and EPO significantly associating with glucose levels (Fig. 4). This signal is represented by rs221783, an intergenic variant whose minor T allele associated with decreased glucose (p = 1.8 × 10–11, effect = − 0.03 SD) and HbA1c (p = 3.6 × 10–7, effect = − 0.02 SD) levels as well as increased cholesterol (p = 7.0 × 10–12, effect = 0.03 SD). This variant also associated with a decreased risk of T2D (p = 0.005, OR = 0.96) and hypothyroidism (p = 6.95 × 10–7, OR = 0.92) (Table 6). rs221783 is the best eQTL (R2 > 0.8) for GIGYF1 in several tissues including pancreas, adipose and thyroid18 (Supplementary Table 29). In all tissues, the T allele associating with decreased glucose and decreased T2D risk associated with increased GIGYF1 expression. Conditional analysis showed that the glucose and HbA1c associations of GIGYF1 pLOF and rs221783 are independent of each other (Supplementary Table 30).
Figure 4.
Locus plot of glucose associations at the GIGYF1 locus. Association results for array genotyped and imputed variants are shown. The purple diamond represents the lead variant rs221783. Other variants are colored according to correlation (R2) with this marker (legend at top-left). The region displayed is chr7: 100092914–100492914. Genomic coordinates are for hg19.
Table 6.
Common variant associations at the GIGYF1 locus.
| Phenotype | Chrom | Pos (hg19/hg38) | Ref (effect allele) | Alt | rsid | MAF (%) | p-value | Effect (beta/OR) | 95% CI− | 95% CI+ |
|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | 7 | 100292914/100695291 | T | C | rs221783 | 11 | 1.82 × 10–11 | − 0.03 | − 0.03 | − 0.02 |
| HbA1c | 7 | 100292914/100695291 | T | C | rs221783 | 11 | 3.58 × 10–7 | − 0.02 | − 0.03 | − 0.01 |
| Cholesterol | 7 | 100292914/100695291 | T | C | rs221783 | 11 | 7.00 × 10–12 | 0.03 | 0.02 | 0.03 |
| LDL | 7 | 100292914/100695291 | T | C | rs221783 | 11 | 6.25 × 10–10 | 0.02 | 0.02 | 0.03 |
| T2D | 7 | 100292914/100695291 | T | C | rs221783 | 11 | 0.005 | 0.96 | 0.93 | 0.99 |
| Hypothyroidism | 7 | 100292914/100695291 | T | C | rs221783 | 11 | 6.95 × 10–7 | 0.92 | 0.88 | 0.95 |
Associations for the array-typed variant rs221783. For quantitative traits the effect is shown in standard deviations (beta) and for diagnoses as an odds ratio (OR).
MAF minor allele frequency.
The association of rs221783 with glucose levels replicated in Biobank Japan (p = 1.7 × 10–4, effect = − 0.05 SD for T allele)19 whilst in FinnGen, rs221783 showed a nominal association with T2D diagnosis (p = 0.02, OR = 0.96 for T allele) (Supplementary Table 31). The association with thyroid disease has been replicated elsewhere20.
The independent glucose and T2D associations at the GIGYF1 locus and their replication in other datasets further support the hypothesis that decreasing GIGYF1 levels predisposes to diabetes while increasing GIGYF1 levels may protect from diabetes.
Identification of causal genes at GWAS loci
Given the fact that the GIGYF1 locus harbors both rare and common variants associated with T2D we examined whether our study points to the causal gene at additional GWAS loci. For 558 variants associated with T2D in a recent study by Vujkovic et al.9 we tested whether either of the two closest genes associated with T2D or HbA1c levels in our study. Just nine genes close to these 558 variants significantly associated with T2D or HbA1c (p ≤ 2.41 × 10–5 adjusting for 2071 variant sets tested)—ANK1, GCK, HNF1A, TNRC6B, SLC30A8, NF1, IRS2, CFTR and HNF4A (Supplementary Figure 6 and Supplementary Table 32). Most of these genes are already known to be causal for T2D including GCK, HNF1A, SLC30A8, IRS2 and HNF4A. Given that there is a common variant association with T2D at TNRC6B but conflicting results for TNRC6B pLOF in UKBB and GHS, further study of this locus may be warranted.
Discussion
Our results highlight the power of whole exome sequencing to make novel discoveries relevant to human disease and to detect known associations of Mendelian disease genes. Gene-level aggregation and burden testing of rare pLOF and predicted damaging missense variants identified genes associating with T2D and biomarkers of glycemic control. These included several genes not previously implicated in diabetes, GIGYF1, TNRC6B and PFAS, as well as GCK, HNF1A and PDX1, known MODY genes14,21–23. We also identified PAM and G6PC2, genes highlighted by other rare-variant studies of T2D and glucose levels5,15. Gene-level tests were needed to detect the majority of these associations owing to the rarity of the variants. For example, out of 363,977 individuals, just 40 carried a pLOF variant in GCK and 131 carried a pLOF variant in GIGYF1. In general, singleton variants contributed a large part of the signal arguing strongly, as others have done4, for including such variants in gene-based collapsing tests. One limitation of this study is its focus on individuals of European ancestry. Although we also performed gene-level tests in the other sub-populations of UKBB we were underpowered to detect associations as they represent a small proportion of the cohort. The only significant association in non-European ancestry populations was between MYH6 damaging missense variants and HbA1c levels in the East Asian ancestry population. Given the small number of carriers and known function of MYH6 in cardiac function24,25 this may be a false positive and replication is required. An important area for further study will be assessing the impact of rare variants on T2D risk in larger multi-ethnic cohorts.
Our finding that rare variants in the MODY genes, GCK, HNF1A and PDX1 associate with common T2D are consistent with results reported by Bonneford et al.26. Bonneford et al. defined pathogenic variants in MODY genes according to the American Society of Human Genetics criteria27. In contrast, we used bioinformatic methods to select variants likely to have functional impact in all genes in the genome. This allowed us to identify associations for genes not previously implicated in diabetes such as GIGYF1. The fact that we detected associations for MODY genes strongly suggests that we are enriching for variants with functional impact. However, for GCK and HNF1A, adding known pathogenic variants did increase the significance of detected associations.
Test statistic inflation can be a challenge when testing rare variants as statistical assumptions break down when the number of carriers expected to have the disease of interest is low4,28. To avoid false positives in our analysis of diabetes, we initially examined associations with glucose and HbA1c because quantitative traits are less susceptible to inflation. All the variant sets that associated with T2D also affected HbA1c and/or glucose levels giving us confidence in these associations. A targeted analysis of the genes encoding T2D drug targets revealed HbA1c and/or T2D associations for variants in several of these genes. The lack of association for variants in some of these drug target genes may in part be due to a lack of statistical power for genes with small numbers of rare variant carriers. However, for some of these genes such as SLC5A2 (encoding SGLT2) we do not detect associations with diabetes traits despite having a reasonable number of variant carriers.
We uncovered novel associations with T2D and biomarkers of glycemic control for aggregated variants in GIGYF1, TNRC6B and PFAS and attempted replication of these associations in exome-sequenced individuals from GHS. The association of GIGYF1 pLOF with T2D replicated in GHS but we did not replicate associations for TNRC6B and PFAS. There are differences between these two cohorts; UKBB is a population-based cohort with T2D diagnoses obtained from inpatient records while GHS is a health system-based cohort and includes both inpatient and outpatient diagnoses. There is a larger effect size for GIGYF1 pLOF in UKBB compared to GHS which may be due to these differences in ascertainment. Differences in the definition of the variant sets tested or the frequency of the relevant variants may have contributed to the failure to replicate the TNRC6B and PFAS associations. Alternatively, this may suggest that the TNRC6B and PFAS associations are false positives.
We focused our analysis on understanding the consequences of GIGYF1 pLOF as it strongly associated with glucose, HbA1c and T2D and the T2D association replicated in GHS. GIGYF1 encodes a protein that was initially identified for its binding to the adapter protein GRB10 (GRB10 interacting GYF protein 1) which negatively regulates both the insulin and IGF-1 receptors29,30. Transfection of cells with GRB10-binding fragments of GIGYF1 led to greater activation of both the insulin and IGF-1 receptors30. This supports a hypothesis whereby GIGYF1 enhances insulin signaling by reducing the negative regulation of the insulin receptor by GRB10. When GIGYF1 is reduced, as is the case in individuals carrying pLOF variants, GRB10 presumably inhibits insulin signaling to a greater degree thereby reducing the action of insulin in its target tissues and leading to increased risk of T2D. However, the exact mechanistic details of these interactions remain to be determined. GRB10 variants have also been reported to associate with T2D and glycemic traits although interpretation of these results is complicated by imprinting31,32. GIGYF1 is broadly expressed with high levels observed in endocrine tissues, pancreas and brain18,33. GIGYF1 and the related protein GIGYF2 have also been implicated in translational repression34 and translation-coupled mRNA decay35 suggesting biological roles beyond regulation of insulin and IGF-1 receptor signaling.
PheWAS of GIGYF1 pLOF revealed a strong association with decreased cholesterol levels reflecting altered energy homeostasis in carriers. An inverse relationship between glucose and cholesterol levels has been observed for variants in other genes36. We also observed several associations that could reflect complications of diabetes in GIGYF1 pLOF carriers including increased cystatin c levels and increased diagnosis of urinary disorders, suggesting renal complications, as well as syncope and collapse which may be a side-effect of hyperglycemia and/or hypoglycemia in diabetics. Other associations may reflect poor health in carriers including decreased grip strength and decreased peak expiratory flow.
GIGYF1 pLOF associated with a 4.5-fold increased risk of hypothyroidism and GIGYF1 is highly expressed in thyroid18,33 consistent with a biological function in this tissue. IGF-1 and insulin have been implicated in the proliferation of thyroid cells which may, in part, explain the association with thyroid dysfunction37–39. An alternative possibility is that GIGYF1 contributes to thyroid function by affecting secretion of thyroid stimulating hormone in the anterior pituitary gland. Another explanation is that shared autoimmune mechanisms contribute to thyroid dysfunction and diabetes in pLOF carriers and that some of the carriers diagnosed with T2D have features of latent autoimmune diabetes in adults40. Damaging variants in GIGYF1 have recently been implicated in conferring risk for developmental delay and autism spectrum disorders41. Consistent with this, we see an association of GIGYF1 pLOF with increased time to complete a cognitive test. It may be that metabolic aberrations in carriers affect cognitive performance, that brain development is altered due to perturbation of IGF-1 signaling, or that other functions of GIGYF1 such as regulation of mRNA expression and decay are responsible for cognitive phenotypes.
In addition to replicating the association of GIGYF1 pLOF with T2D in an independent cohort we also used common genetic variants to further investigate the role of the GIGYF1 locus in diabetes. A non-coding variant at the GIGYF1 locus associated with glucose levels and T2D, and this replicated in independent datasets. This variant associated with increased GIGYF1 expression but a lower risk of T2D. This direction of effect is consistent with what we see for the pLOF variants—reduced levels of GIGYF1 increases diabetes risk but increased levels of GIGYF1 are protective.
We observed an intersection of rare and common variant associations at GIGYF1 as well as at MODY genes such as GCK, HNF1A and HNF4A. However, in general, our gene-level analysis of rare variants did not identify many additional causal genes at GWAS loci; out of 558 variants associated with T2D9 just nine had rare variant associations at a nearby gene.
We assessed the impact of pLOF and predicted damaging missense variants on glycemic traits and uncovered a hitherto unappreciated role for GIGYF1 in regulating blood sugar and protecting from T2D. By highlighting the importance of GIGYF1 and GRB adapter proteins in modulating insulin signaling this finding may lead to new therapeutic approaches for the treatment of diabetes. Discoveries such as this are only possible by combining health-related data with the sequencing of rare variants on a biobank scale.
Methods
The UK Biobank resource and data access
The UK Biobank (UKBB) recruited ~ 500,000 participants in England, Wales, and Scotland between 2006 and 201042. Written informed consent was obtained from all participants. Phenotypic data available includes age, sex, biomarker data and self-reported diseases collected at the time of baseline assessment as well as disease diagnoses from inpatient hospital stays, the cancer registry and death records obtained through the NHS. Approximately half of the participants also have diagnoses from primary care available. Array genotypes are available for nearly all participants and exome sequencing data is available for 454,787 participants. The data used in this study were obtained from the UKBB through application 26041.
Population definition and PC calculation for subjects with exome data
Subject quality control was performed by Regeneron Genetics Center (RGC) and removed subjects with evidence of contamination, unresolved duplications, sex discrepancies and discordance between exome sequencing and genotyping data. Genetic relationships between participants were determined by RGC using the PRIMUS program43. For the unrelated subset all first- and second-degrees relatives and some third-degree relatives were excluded.
Populations were defined through a combination of self-reported ethnicity and genetic principal components. We selected the unrelated individuals who identify as White (European), Black (African), Asian (South Asian) or Chinese (East Asian) (Field 21000) and ran an initial principal component analysis (PCA) on high quality common variants using eigenstrat44. SNPs were filtered for missingness across individuals < 2%, MAF > 1%, regions of known long range LD45, and pruned to independent markers with pairwise LD < 0.1. We then projected the principal components (PCs) onto related individuals and removed all individuals ± 3 standard deviations from the mean of PCs 1–6. A final PC estimation was performed in eigenstrat44 using unrelated subjects. We then projected related individuals onto the PCs.
Exome sequencing and variant calling
DNA was extracted from whole blood and was sequenced by the RGC as described elsewhere46. Briefly, the xGen exome capture was used and reads were sequenced using the Illumina NovaSeq 6000 platform. Reads were aligned to the GRCh38 reference genome using BWA-mem47. Duplicate reads were identified and excluded using the Picard MarkDuplicates tool (Broad Institute). Variant calling of SNVs and indels was done using the WeCall variant caller (Genomics Plc.) to produce a GVCF for each subject. GVCFs were combined to using the GLnexus joint calling tool48. Post-variant calling filtering was applied using the Goldilocks pipeline46. Variants were annotated using the Ensembl Variant Effect Predictor v9549 which includes a LOFTEE plug-in to identify high confidence (HC) pLOF variants13. Combined Annotation Dependent Depletion (CADD) scores were generated using the Whole Genome Sequence Annotator (WGSA) AMI version 0.8.
Phenotype definitions
Blood biochemistry values were obtained for glucose (Field 30740) and HbA1c (Field 30750) from UKBB and inverse rank normalized using the RNOmni R package50, resulting in an approximately normal distribution. For sex-stratified analyses, glucose and HbA1c values were inverse rank normalized separately in males and females.
For disease diagnoses, ICD10 codes were obtained from inpatient hospital diagnoses (Field 41270), causes of death (Field 40001 and 40002) and the cancer registry (Field 40,006) from UKBB. Diagnoses also included additional hospital episode statistics (HESIN) and death registry data made available by UKBB in July 2020. T2D was defined as ICD10 E11. For the purposes of excluding diagnosed diabetics from the glucose and HbA1c analysis we defined diabetes as ICD10 codes E10–E14 which includes both T1D and T2D diagnoses.
For phenome-wide analyses, a selection of quantitative traits was obtained from other fields, encompassing anthropometric measurements, blood counts, as well as blood and urine biochemistry. Beyond these measurements, we selected additional quantitative traits found to be heritable (h2 significance flagged as at least “nominal” with a confidence level flagged as “medium” or “high”) by the Neale lab28, using PHESANT to transform values to quantitative traits when necessary as they describe. These included the results of cognitive tests. All quantitative traits were inverse rank normalized using the RNOmni R package50. For burden testing, we required at least ten carriers to have measurements. We also tested associations with ICD10-coded diagnoses (using three character codes) that had more than 500 cases in the European ancestry participants with exome data and at least one expected case carrier based on variant frequency and disease prevalence.
Glucose and HbA1c values were also extracted from primary care data available for about half of the cohort using the following read codes. Glucose: read 2 codes 44U..,44g.., 44g1.,44TJ.,44f..,44TK.,44f1.,44g0.,44f0. and read 3 codes XM0ly, X772z, XE2mq; HbA1c: read 2 codes 42W5., 44TB., 66Ae0, 44TC., 42W4. and read 3 codes XaPbt, X772q, XaWP9, XaBLm, XaERp. Values were converted to IFCC units where necessary. Aberrantly high (≥ 45 mmol/L for glucose, ≥ 300 mmol/mol for HbA1c) and extremely low values (≤ 0.6 mmol/L for glucose, ≤ 10 mmol/mol for HbA1c) were excluded. The mean measurement per individual was then taken and inverse rank normalized prior to association testing. The mean age at measurement was also extracted and used as a covariate in the regression.
Individuals taking cholesterol-lowering medication were identified using self-reported medications recorded at their UKBB interview (Field 20003) and whether cholesterol-lowering medications were recorded using the touchscreen questionnaire (Fields 6177 and 6153).
Gene-based association testing
For gene-based tests, autosomal rare pLOF variants were identified as follows; LOFTEE high confidence LOFs, MAF ≤ 1%, missingness across individuals ≤ 2%, HWE p-value ≥ 10–10. Predicted damaging missense variants were defined as missense variants with a CADD PHRED-scaled score ≥ 25, MAF ≤ 1%, missingness across individuals ≤ 2%, HWE p-value ≥ 10–10. Only genes with more than one pLOF variant or damaging missense variant were tested.
Burden testing was performed using glm in R, using a gaussian model for quantitative traits and a binomial model for case–control analyses. Genotype was coded according to a dominant model; 0 (no variant) or 1 (any number of variants). We adjusted for age, sex and the first 12 PCs of genetic ancestry in the regression. Additionally, when testing for association with disease diagnoses, we included country of recruitment as a covariate as the time of available follow-up differs between England, Scotland and Wales. Recruitment country was defined using the location of the relevant UKBB recruitment center (Field 54). Associations were later confirmed using just participants recruited in England. Follow-up duration was defined as the time from recruitment to UKBB until censoring, death, or the last date on which inpatient diagnoses were obtained. For case–control analyses we only ran tests where there was at least one expected case carrier based on variant frequency and disease prevalence. For quantitative traits we required at least ten carriers to have measurements.
For glucose and HbA1c, to convert effect sizes from normalized values back to measured units, the estimates from the regression were multiplied by the standard deviation of these traits in the entire cohort.
For sex-stratified analyses, males and females were defined using a combination of recorded sex (Field 31) and genetic sex (Field 22001). Meta-analysis of males and females was performed using the standard error weighted method in METAL and heterogeneity was tested using Cochrane’s Q test51.
SAIGE-Gene was run using the SAIGE R package (v0.36.5)52 using settings recommend by the developers and an additive model, related individuals were included.
T2D drug targets were defined according to Flannick et al.5.
Manhattan plots were created using the R Package CMplot (https://github.com/YinLiLin/R-CMplot).
Array association testing
Genotypes were obtained through array genotyping and imputation as described previously53. Population definition and PC estimation for individuals with array data was performed as previously described54. We tested all variants with imputation quality score (info) ≥ 0.8 and minor allele frequency (MAF) ≥ 0.1% in a 200 Mb region around GIGYF1 for association with glucose, HbA1c, T2D and hypothyroidism. Association analyses were performed using an additive model in PLINK adjusting for age at recruitment to UKBB, sex and the first 12 PCs of genetic ancestry. We also adjusted for country of recruitment where appropriate. The most significant variant with info > 0.95 was selected as the lead variant at the locus.
We replicated the association of rs221783 with glucose using available summary statistics for Biobank Japan for the trait “blood sugar” (http://jenger.riken.jp/en/result)19. We replicated the association of this variant with T2D diagnosis using summary statistics from FinnGen release 3 for the phenotype “E4_DM2” (https://www.finngen.fi/en/access_results). The effect allele in these datasets was the alternate allele “C”. For consistency with the UKBB associations we have shown the effect for the “T” allele.
Meta-analysis of the UKBB and replication dataset association results was performed with the METAL software package using the standard error weighted method51.
Region plots were created using LocusZoom55. LD calculations were performed in the European ancestry population for array variants in a 500 kb sliding window as follows; we extracted genotypes with info > 0.9, rounded them to whole numbers, mean-imputed missing genotypes and used the R “cor” function to compute R which was then squared to get an R2 value.
Gene expression and eQTL analysis
The expression of GIGYF1 in various tissues was assessed using the GTEx portal (accessed 08/04/2020)18 and Human Protein Atlas (http://www.proteinatlas.org)33. eQTL data for rs221783 was obtained from GTEx v8. For each tissue of interest, the best eQTL for GIGYF1 was identified (GTEx v8 “eGene”). R2 for rs221783 and the best GIGYF1 eQTL was calculated as described above.
Replication analysis in GHS
The GHS MyCode Community Health Initiative study is a health system-based cohort and has been described previously56. A subset of participants sequenced as part of the GHS-Regeneron Genetics Center DiscovEHR partnership were included in this study. T2D status was defined based on meeting at least one of the following criteria: (1) clinical encounters due to or problem-lists diagnosis code for type 2 diabetes (ICD-10 code E11), or (2) HbA1c greater than 6.5%, or (3) use of diabetic oral hypoglycemic medicine. Controls were participants who did not meet any of the criteria for case definition. Individuals were excluded from the analysis if they had clinical encounters due to or problem-lists diagnosis code for type 1 diabetes (ICD-10 code E10), or if they were treated with insulin but not with oral hypoglycemic medicines.
Exome-sequencing, variant calling, quality control and gene-based tests were performed as previously described57. Variant sets tested were pLOF variants (GIGYF1 and TNRC6B) or pLOF plus missense variants predicted to be deleterious by 5/5 algorithms (PFAS) with MAF < 1%. The following variants were classified as pLOF variants: frameshift-causing indels, variants affecting splice acceptor and donor sites, variants leading to stop gain, stop loss and start loss. The five missense deleterious algorithms used were SIFT58, PolyPhen2 (HDIV), PolyPhen2 (HVAR)59, LRT60, and MutationTaster61. Association testing was performed in the European ancestry population using the Firth logistic regression test implemented in REGENIE62.
Identification of potential causal genes at GWAS loci
For 558 variants identified as associating with T2D9 we mapped the two closest protein coding genes using bedtools. This resulted in 1118 genes for which we had tested 2071 variant sets (pLOF and/or damaging missense) in our primary analysis. Genes with p < 2.41 × 10–5 (correcting for 2071 variant sets tested) for HbA1c or T2D were considered significant.
Ethics statement
The UK Biobank study was approved by the National Health Service National Research Ethics Service and all participants provided written informed consent to participate in the study. The UK Biobank resource is an approved Research Tissue Bank and is registered with the Human Tissue Authority, which means that researchers who wish to use it do not need to seek separate ethics approval (unless re-contact of participants is required). Information about ethics oversight in the UK Biobank can be found at https://www.ukbiobank.ac.uk/ethics/. This research has been conducted using the UK Biobank resource, application 26041. Research in GHS was approved by the GHS IRB, approval number 2006-0258. Written informed consent was obtained from all participants in GHS.
Supplementary Information
Acknowledgements
This research has been conducted using the UK Biobank Resource (Project 26041). We would like to thank the participants and researchers of UK Biobank for creating an open-access resource. We thank the UK Biobank Exome Sequencing Consortium and UK Biobank for facilitating exome sequencing of participants. We also thank the participants of the GHS MyCode initiative as well as participants and investigators of the FinnGen study and Biobank Japan. We thank Mark McCarthy and Anna Gloyn for comments on the manuscript. Data management and analytics were performed using the REVEAL/SciDB translational analytics platform from Paradigm4.
Author contributions
A.D, L.W., M.P., A.F.C. and P.N. performed computational analyses; P.A., L.L. and A.B. performed replication analysis in GHS; A.D. wrote the manuscript. All authors interpreted results and edited the manuscript.
Funding
This study was funded by Alnylam Pharmaceuticals and Regeneron Pharmaceuticals.
Data availability
For the primary analysis in UKBB, all phenotypic data and array genotypes used in this study are accessible through application to UKBB. Currently, exome sequencing data for ~ 200,000 participants is available63; the remainder of the exome data used is scheduled for release by UKBB in 2021. Full summary statistics for gene-level tests performed in UKBB will be made available upon publication. For the replication analysis in Geisinger Health System, the full summary statistics for all replication analyses performed are supplied in Supplemental Table 24. Summary statistics for Biobank Japan are available at http://jenger.riken.jp/en/result and summary statistics for FinnGen are available at https://www.finngen.fi/en/access_results.
Competing interests
A.D, L.W., M.P., A.F.C., L.B., G.H. and P.N. are employees and stockholders of Alnylam Pharmaceuticals. P.A., L.L. and A.B. are employees and stockholders of Regeneron Pharmaceuticals.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Aimee M. Deaton, Email: adeaton@alnylam.com
Regeneron Genetics Center:
Goncalo Abecasis, Aris Baras, Michael Cantor, Giovanni Coppola, Aris Economides, Luca A. Lotta, John D. Overton, Jeffrey G. Reid, Alan Shuldiner, Katia Karalis, Andrew Deubler, Katherine Siminovitch, Christina Beechert, Caitlin Forsythe, Erin D. Fuller, Zhenhua Gu, Michael Lattari, Alexander Lopez, John D. Overton, Thomas D. Schleicher, Maria Sotiropoulos Padilla, Louis Widom, Sarah E. Wolf, Manasi Pradhan, Kia Manoochehri, Ricardo H. Ulloa, Xiaodong Bai, Suganthi Balasubramanian, Andrew Blumenfeld, Boris Boutkov, Gisu Eom, Lukas Habegger, Alicia Hawes, Shareef Khalid, Olga Krasheninina, Rouel Lanche, Adam J. Mansfield, Evan K. Maxwell, Mona Nafde, Sean O’Keeffe, Max Orelus, Razvan Panea, Tommy Polanco, Ayesha Rasool, Jeffrey G. Reid, William Salerno, Jeffrey C. Staples, Michael Cantor, Dadong Li, Adam Locke, Niek Verweij, Jonas Nielsen, Jonas Bovijn, Tanima De, Mary Haas, Parsa Akbari, Olukayode Sosina, Marcus B. Jones, Jason Mighty, Michelle G. LeBlanc, and Lyndon J. Mitnaul
DiscovEHR Collaboration:
Goncalo Abecasis, Aris Baras, Michael Cantor, Giovanni Coppola, Aris Economides, Luca A. Lotta, John D. Overton, Jeffrey G. Reid, Alan Shuldiner, Andrew Deubler, Katia Karalis, Christina Beechert, Caitlin Forsythe, Erin D. Fuller, Zhenhua Gu, Michael Lattari, Alexander Lopez, Thomas D. Schleicher, Maria Sotiropoulos Padilla, Karina Toledo, Louis Widom, Sarah E. Wolf, Manasi Pradhan, Kia Manoochehri, Ricardo H. Ulloa, Xiaodong Bai, Suganthi Balasubramanian, Leland Barnard, Andrew Blumenfeld, Gisu Eom, Lukas Habegger, Alicia Hawes, Shareef Khalid, Evan K. Maxwell, William Salerno, Jeffrey C. Staples, Ashish Yadav, Dadong Li, Marcus B. Jones, Lyndon J. Mitnaul, Jason Mighty, Andrew Deubler, Katia Karalis, Katherine Siminovitch, Lance J. Adams, Jackie Blank, Dale Bodian, Derek Boris, Adam Buchanan, David J. Carey, Ryan D. Colonie, F. Daniel Davis, Dustin N. Hartzel, Melissa Kelly, H. Lester Kirchner, Joseph B. Leader, David H. Ledbetter, J. Neil Manus, Christa L. Martin, Raghu P. Metpally, Michelle Meyer, Tooraj Mirshahi, Matthew Oetjens, Thomas Nate Person, Christopher Still, Natasha Strande, Amy Sturm, Jen Wagner, and Marc Williams
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-99091-5.
References
- 1.King EA, Davis JW, Degner JF. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 2019;15:e1008489. doi: 10.1371/journal.pgen.1008489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen PA, Born DA, Deaton AM, Nioi P, Ward LD. Phenotypes associated with genes encoding drug targets are predictive of clinical trial side effects. Nat. Commun. 2019;10:1579. doi: 10.1038/s41467-019-09407-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cirulli ET, et al. Genome-wide rare variant analysis for thousands of phenotypes in over 70,000 exomes from two cohorts. Nat Commun. 2020;11:542. doi: 10.1038/s41467-020-14288-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flannick J, et al. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature. 2019;570:71–76. doi: 10.1038/s41586-019-1231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moutsianas L, et al. The power of gene-based rare variant methods to detect disease-associated variation and test hypotheses about complex disease. PLoS Genet. 2015;11:e1005165. doi: 10.1371/journal.pgen.1005165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bush WS, Oetjens MT, Crawford DC. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat. Rev. Genet. 2016;17:129–145. doi: 10.1038/nrg.2015.36. [DOI] [PubMed] [Google Scholar]
- 8.Diogo D, et al. Phenome-wide association studies across large population cohorts support drug target validation. Nat. Commun. 2018;9:4285. doi: 10.1038/s41467-018-06540-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vujkovic M, et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 2020;52:680–691. doi: 10.1038/s41588-020-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue A, et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat. Commun. 2018;9:2941. doi: 10.1038/s41467-018-04951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Ellinghaus D, Franke A, Howie B, Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur. J. Hum. Genet. 2012;20:801–805. doi: 10.1038/ejhg.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott RA, et al. An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes. 2017;66:2888–2902. doi: 10.2337/db16-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N. Engl. J. Med. 2001;345:971–980. doi: 10.1056/NEJMra002168. [DOI] [PubMed] [Google Scholar]
- 15.Wessel J, et al. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nat. Commun. 2015;6:5897. doi: 10.1038/ncomms6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou W, et al. Scalable generalized linear mixed model for region-based association tests in large biobanks and cohorts. Nat. Genet. 2020;52:634–639. doi: 10.1038/s41588-020-0621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landrum MJ, et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguet F, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanai M, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018;50:390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 20.Saevarsdottir S, et al. FLT3 stop mutation increases FLT3 ligand level and risk of autoimmune thyroid disease. Nature. 2020;584:619–623. doi: 10.1038/s41586-020-2436-0. [DOI] [PubMed] [Google Scholar]
- 21.Froguel P, et al. Close linkage of glucokinase locus on chromosome 7p to early-onset non-insulin-dependent diabetes mellitus. Nature. 1992;356:162–164. doi: 10.1038/356162a0. [DOI] [PubMed] [Google Scholar]
- 22.Ellard S. Hepatocyte nuclear factor 1 alpha (HNF-1 alpha) mutations in maturity-onset diabetes of the young. Hum. Mutat. 2000;16:377–385. doi: 10.1002/1098-1004(200011)16:5<377::AID-HUMU1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat. Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 24.Holm H, et al. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat. Genet. 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ching YH, et al. Mutation in myosin heavy chain 6 causes atrial septal defect. Nat. Genet. 2005;37:423–428. doi: 10.1038/ng1526. [DOI] [PubMed] [Google Scholar]
- 26.Bonnefond A, et al. Pathogenic variants in actionable MODY genes are associated with type 2 diabetes. Nat. Metab. 2020;2:1126–1134. doi: 10.1038/s42255-020-00294-3. [DOI] [PubMed] [Google Scholar]
- 27.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Churchhouse, C. Details and Considerations of the UK Biobank GWAS. Neale lab.http://www.nealelab.is/blog/2017/9/11/details-and-considerations-of-the-uk-biobank-gwas (2017).
- 29.Holt LJ, Siddle K. Grb10 and Grb14: enigmatic regulators of insulin action–and more? Biochem. J. 2005;388:393–406. doi: 10.1042/BJ20050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giovannone B, et al. Two novel proteins that are linked to insulin-like growth factor (IGF-I) receptors by the Grb10 adapter and modulate IGF-I signaling. J. Biol. Chem. 2003;278:31564–31573. doi: 10.1074/jbc.M211572200. [DOI] [PubMed] [Google Scholar]
- 31.Prokopenko I, et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 2014;10:e1004235. doi: 10.1371/journal.pgen.1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rampersaud E, et al. Identification of novel candidate genes for type 2 diabetes from a genome-wide association scan in the Old Order Amish: Evidence for replication from diabetes-related quantitative traits and from independent populations. Diabetes. 2007;56:3053–3062. doi: 10.2337/db07-0457. [DOI] [PubMed] [Google Scholar]
- 33.Uhlen M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 34.Peter D, et al. GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression. Genes Dev. 2017;31:1147–1161. doi: 10.1101/gad.299420.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weber R, et al. 4EHP and GIGYF1/2 mediate translation-coupled messenger RNA decay. Cell Rep. 2020;33:108262. doi: 10.1016/j.celrep.2020.108262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klimentidis YC, et al. Type-2 diabetes with low LDL-C: Genetic insights into a unique phenotype. bioRxiv. 2019 doi: 10.1101/837013. [DOI] [Google Scholar]
- 37.Clement S, Refetoff S, Robaye B, Dumont JE, Schurmans S. Low TSH requirement and goiter in transgenic mice overexpressing IGF-I and IGF-Ir receptor in the thyroid gland. Endocrinology. 2001;142:5131–5139. doi: 10.1210/endo.142.12.8534. [DOI] [PubMed] [Google Scholar]
- 38.Kimura T, et al. Regulation of thyroid cell proliferation by TSH and other factors: A critical evaluation of in vitro models. Endocr. Rev. 2001;22:631–656. doi: 10.1210/edrv.22.5.0444. [DOI] [PubMed] [Google Scholar]
- 39.Zaballos MA, Santisteban P. FOXO1 controls thyroid cell proliferation in response to TSH and IGF-I and is involved in thyroid tumorigenesis. Mol. Endocrinol. 2013;27:50–62. doi: 10.1210/me.2012-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mishra R, Hodge KM, Cousminer DL, Leslie RD, Grant SFA. A global perspective of latent autoimmune diabetes in adults. Trends Endocrinol. Metab. 2018;29:638–650. doi: 10.1016/j.tem.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Satterstrom FK, et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell. 2020;180:568–584.e523. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allen N, et al. UK Biobank: Current status and what it means for epidemiology. Health Policy Technol. 2012;1:123–126. doi: 10.1016/j.hlpt.2012.07.003. [DOI] [Google Scholar]
- 43.Staples J, et al. PRIMUS: rapid reconstruction of pedigrees from genome-wide estimates of identity by descent. Am. J. Hum. Genet. 2014;95:553–564. doi: 10.1016/j.ajhg.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, L., Zhang, W. & Li, Q. AssocTests: An R package for genetic association studies. J. Stat. Softw. 94, 1–26. 10.18637/jss.v094.i05 (2020).
- 45.Price AL, et al. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 2008;83:132–135. doi: 10.1016/j.ajhg.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Hout CV, et al. Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature. 2020;586:749–756. doi: 10.1038/s41586-020-2853-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin MF, et al. GLnexus: Joint variant calling for large cohort sequencing. bioRxiv. 2018;343:970. doi: 10.1101/343970. [DOI] [Google Scholar]
- 49.McLaren W, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCaw ZR, Lane JM, Saxena R, Redline S, Lin X. Operating characteristics of the rank-based inverse normal transformation for quantitative trait analysis in genome-wide association studies. bioRxiv. 2019 doi: 10.1101/635706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou W, et al. Efficiently controlling for case–control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bycroft C, et al. Genome-wide genetic data on ~ 500,000 UK Biobank participants. bioRxiv. 2017 doi: 10.1101/166298. [DOI] [Google Scholar]
- 54.Ward LD, et al. Genome-wide association study of circulating liver enzymes reveals an expanded role for manganese transporter SLC30A10 in liver health. J. bioRxiv. 2020 doi: 10.1101/2020.05.19.104570. [DOI] [Google Scholar]
- 55.Pruim RJ, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dewey FE, et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016 doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 57.Akbari P, et al. Sequencing of 640,000 exomes identifies GPR75 variants associated with protection from obesity. Science. 2021 doi: 10.1126/science.abf8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat. Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 59.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013 doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 62.Mbatchou J, et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 2021;53:1097–1103. doi: 10.1038/s41588-021-00870-7. [DOI] [PubMed] [Google Scholar]
- 63.Szustakowski JD, et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat. Genet. 2021;53:942–948. doi: 10.1038/s41588-021-00885-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For the primary analysis in UKBB, all phenotypic data and array genotypes used in this study are accessible through application to UKBB. Currently, exome sequencing data for ~ 200,000 participants is available63; the remainder of the exome data used is scheduled for release by UKBB in 2021. Full summary statistics for gene-level tests performed in UKBB will be made available upon publication. For the replication analysis in Geisinger Health System, the full summary statistics for all replication analyses performed are supplied in Supplemental Table 24. Summary statistics for Biobank Japan are available at http://jenger.riken.jp/en/result and summary statistics for FinnGen are available at https://www.finngen.fi/en/access_results.