Abstract
Purpose
To evaluate radiomic features extracted from standard static images (20–40 min p.i.), early summation images (5–15 min p.i.), and dynamic [18F]FET PET images for the prediction of TERTp-mutation status in patients with IDH-wildtype high-grade glioma.
Methods
A total of 159 patients (median age 60.2 years, range 19–82 years) with newly diagnosed IDH-wildtype diffuse astrocytic glioma (WHO grade III or IV) and dynamic [18F]FET PET prior to surgical intervention were enrolled and divided into a training (n = 112) and a testing cohort (n = 47) randomly. First-order, shape, and texture radiomic features were extracted from standard static (20–40 min summation images; TBR20–40), early static (5–15 min summation images; TBR5–15), and dynamic (time-to-peak; TTP) images, respectively. Recursive feature elimination was used for feature selection by 10-fold cross-validation in the training cohort after normalization, and logistic regression models were generated using the radiomic features extracted from each image to differentiate TERTp-mutation status. The areas under the ROC curve (AUC), accuracy, sensitivity, specificity, and positive and negative predictive value were calculated to illustrate diagnostic power in both the training and testing cohort.
Results
The TTP model comprised nine selected features and achieved highest predictability of TERTp-mutation with an AUC of 0.82 (95% confidence interval 0.71–0.92) and sensitivity of 92.1% in the independent testing cohort. Weak predictive capability was obtained in the TBR5–15 model, with an AUC of 0.61 (95% CI 0.42–0.80) in the testing cohort, while no predictive power was observed in the TBR20–40 model.
Conclusions
Radiomics based on TTP images extracted from dynamic [18F]FET PET can predict the TERTp-mutation status of IDH-wildtype diffuse astrocytic high-grade gliomas with high accuracy preoperatively.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00259-021-05526-6.
Keywords: Radiomics, [18F]FET PET, TERTp-mutation, Glioma
Introduction
Mutations in the telomerase reverse transcriptase promoter (TERTp), leading to telomerase activation and lengthened telomeres, play an important role in the formation of brain cancer and individual prognosis [1–3]. In diffuse astrocytic high-grade gliomas without mutation of the isocitrate dehydrogenase gene (IDH-wildtype), TERTp mutations are reported to be associated with poor overall survival [4–6]. Molecular genetic analysis of the TERTp-mutation status has therefore gained increasing attention in the clinical routine diagnosis of IDH-wildtype diffuse astrocytic gliomas and will be included in the upcoming glioma WHO classification [7–9].
Molecular imaging using positron emission tomography (PET) with radiolabelled amino acids such as O-(2-[18F]-fluoroethyl)-L-tyrosine ([18F]FET) is a useful tool for the characterization and evaluation of primary brain neoplasms [10–12], and its application in the clinical management of brain tumour patients has been recommended by the Response Assessment in Neuro-Oncology (RANO) Working Group [13–17]. While static image data (standard 20–40 min summation images) are particularly used for the delineation of the tumour extent, the assessment of dynamic [18F]FET PET data has been shown to provide additional information about tumour biology [18]. More aggressive gliomas (i.e. high-grade gliomas and/or IDH-wildtype gliomas) were shown to be characterized by a high tracer uptake within the first 5–15 min post injection (p.i.) with subsequent curve decrease, while less aggressive gliomas (i.e. low grade gliomas and/or IDH-mutant gliomas) typically show a slowly increasing [18F]FET uptake with highest values in the later time frames [12, 19, 20]. As the early peak uptake in aggressive gliomas is missed in the standard 20–40 min p.i. summation images, it does not surprise that the maximal tumour-to-background ratio (TBRmax) evaluation obtained in early summation images (5–15 min p.i.) was reported to perform better than the standard static TBRmax values (20–40 min p.i.) for the differentiation between low-grade and high-grade gliomas [17], which led to the suggestion to include these early summation images for a better glioma characterization. Another interesting parameter derived from dynamic [18F]FET PET is the minimal time-to-peak (TTPmin), which is extracted from the time-activity-curves and was reported to provide prognostic information [21]. Interestingly, an early TTPmin was associated with an aggressive disease course in newly diagnosed gliomas and was able to predict an IDH-wildtype status [22, 23]. Yet, in our recently published study investigating [18F]FET uptake characteristics in TERTp mutant and TERTp wildtype glioblastomas, neither the standard TBRmax as static parameter nor TTPmin as dynamic parameter were associated with the TERTp-mutation status [24].
In recent years, radiomics have been increasingly investigated as a promising non-invasive tool for accurate diagnosis and prognosis assessment by converting medical images into high-dimensional quantitative image features and establishing predictive models [25–32]. However, radiomics have not been applied for the detection of TERTp mutations on [18F]FET PET images so far. Therefore, the aim of this study was to evaluate radiomic features extracted from standard static images (20–40 min p.i.), early summation images (5–15 min p.i.) as well as dynamic [18F]FET PET images for the prediction of the TERTp-mutation status in patients with newly diagnosed IDH-wildtype diffuse astrocytic high-grade glioma.
Materials and methods
Patients
Patients with primary diagnosis of a glioma who had received a pre-treatment dynamic [18F]FET PET scan at the Department of Nuclear Medicine of the LMU Munich between December 2005 and June 2016 were screened for this retrospective study. Inclusion criteria were (1) neuropathologically confirmed IDH-wildtype diffuse astrocytic gliomas (WHO grade III or IV) according to the updated 2016 WHO classification [33], (2) availability of the TERTp-mutation status, and (3) pre-treatment dynamic [18F]FET PET scan (ECAT EXACT HR + , Siemens Healthineers, Inc., Erlangen, Germany Siemens Medical Systems, Inc., Erlangen, Germany). [18F]FET-negative gliomas (tumour-to-background ratio, TBR < 1.6) were excluded. All patients had given written informed consent prior to the PET scan as part of the clinical routine. The retrospective analysis of PET imaging data was approved by the institutional ethics committee (604–16). A total of 61% of the investigated patients (97/159) have been evaluated in a previous study [24].
Histopathology and molecular genetic analysis
Histopathology and molecular genetic analyses were performed at the Institute of Neuropathology, LMU Munich, Germany. All patients initially classified according to the 2007 WHO brain tumour classification [34] were re-classified according to the 2016 WHO classification [33]. The IDH-mutation status and TERTp-mutation status were evaluated according to clinical standard protocols [35, 36].
[18F]FET PET imaging
[18F]FET PET scans were performed at the Department of Nuclear Medicine, LMU Munich, Germany. Images were acquired by using an ECAT EXACT HR + PET scanner (Siemens Healthineers, Inc., Erlangen, Germany) with the standard protocol [11, 37]. Exactly 180 MBq of [18F]FET were injected after a 15-min transmission scan with a 68Ge rotating rod source. After tracer injection up to 40 min post injection in 3-D mode consisting of 16 frames (7 × 10 s, 3 × 30 s, 1 × 2 min, 3 × 5 min, and 2 × 10 min) with a reconstructed voxel size of 2.03 × 2.03 × 2.43 mm3 and matrix size of 128 × 128 × 63, dynamic emission recording was finished. Two-dimensional filtered back-projection reconstruction algorithm using a 4.9-mm Hann Filter was applied for image reconstruction, then corrected for attenuation, decay, dead time, and random and scattered coincidences. When relevant motion was visible in dynamic PET data, a frame-wise correction was performed by using PMOD fusion tool (version 3.5, PMOD Technologies, Zurich, Switzerland) after frame-wise checking for motion.
Segmentation of tumour volumes and brain background
First, a background activity was extracted from a large crescent-shaped volume of interest (VOI) in the contralateral healthy hemisphere as published previously [38]. For tumour segmentation, a VOI was drawn using a TBR-threshold of 1.6 in static 20–40 min p.i. summation images as suggested by Pauleit et al. [39]. All segmentations were processed within the PMOD View tool (version 3.5, PMOD Technologies, Zurich, Switzerland).
Image normalization and TTP image generation
We used the in-house developed software described previously by Kaiser et al. [40] (C + + with integration of the ROOT data analysis framework, version 6.22/08, Cern, Switzerland and ITK segmentation and registration toolkit 4.13.3, National Library of Medicine) to generate voxel-wise parametric images. Then we normalized the image values with the mean background value derived from each image by using the VOI of background to generate early 5–15 min p.i. (TBR5–15) and late 20–40 min p.i. (TBR20–40) TBR images. For TTP images, time-activity curves (TAC) were extracted from each voxel, which were then classified according to the time frame reaching the peak uptake (i.e. (1) < 5 min, (2) 5–10 min, (3) 10–15 min, (4) 15–20 min, (5) 20–30 min, and (6) 30–40 min). To avoid influence from early blood flush, TTP analyses did not include the first 2.7 min p.i. [40]. In case of a positive late slope (15–40 min p.i.), the TTP was always assigned to group 6.
Radiomic feature extraction
Radiomic features from parametric images were extracted with PyRadiomics (version 3.0.1) [41] as introduced previously by Kaiser et al. [42], and complied with the Imaging Biomarker Standardization Initiative (IBSI) guidelines [43]. Before extraction, images were resampled to isotropic voxels using linear interpolation in PyRadiomics (size 2.03 × 2.03 × 2.03 mm 3). Classes of features extracted from TBR5–15, TBR20–40, and TTP images included first-order features, shape features, and texture features. No image filters were used. The chosen fixed intensity bin size was set to the average interquartile range divided by 4, which led to 0.18 for TBR5–15 images and 0.13 for TBR20–40 images [42, 44]. As the smallest time frame duration considered in the TTP categories was 5 min, this was used as the fixed bin width for radiomics calculation of TTP images.
Feature selection
Before feature extraction, a stratified random split was used to assign 70% of the patients to the training cohort (n = 112) and the remaining 30% to the testing cohort (n = 47), with a balanced distribution of TERTp-wildtype and TERTp-mutation.
Features were standardized as follows: for each feature, we calculated the mean value and the standard deviation. The mean value was subtracted from each individual value, which was then divided by the standard deviation. Feature normalization was computed only in the training cohort and then applied on the testing cohort. Since the number of features was large, we compared the similarity of each feature pair. If the Pearson correlation coefficient (PCC) value of the feature pair was larger than 0.99, we removed one of them. After this process, the number of the features was reduced and each feature was independent to each other. The recursive feature elimination (RFE) based on logistic regression classifier was performed to reduce redundant features and select potential TERTp-mutation related features [45]. Considering the imbalance of comparison groups, we performed the weighted logistic regression in the ‘balanced’ mode, which gives higher weight to the minority class and lower weight to the majority class and therefore automatically adjusts weights inversely proportional to class frequencies in the input data [46]. Each iteration removes a feature which is considered least important. After stratified split-based 10-fold cross-validation, the area under the receiver operating characteristic curve (AUC) of the model in the training cohort was used to determine the optimal number of features.
Model construction and testing
Logistic regression (LR) models were built to predict the TERTp-mutation status by fitting the selected radiomic features. Each model was generated by using only the radiomic features extracted from each image (i.e. TBR5–15, TBR20–40, and TTP images) separately. According to the coefficients of selected features generated by the LR models [47], the risk probability of TERTp-mutation was calculated by the following formula:
is the value of selected features, is the coefficient of selected features, and represents the intercept. In case of , TERTp-mutation status was considered as positive by the LR model.
Model testing was applied to the independent testing cohort, which was not involved in the process of model training. The workflow of the process is presented in Fig. 1.
Fig. 1.
The workflow of process. TBR tumour-to-background ratio, TTP time-to-peak, RFE recursive feature elimination, LR logistic regression, AUC area under the receiver operating characteristic curve, PPV positive predictive value, NPV negative predictive value
Statistical analysis
To evaluate the model performance, receiver operating characteristic curve (ROC) analysis was performed in the training and testing cohort. The AUC was calculated as quantitative measure to illustrate diagnostic power. The accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated. 95% confidence intervals (CI) were calculated by using a non-parametric bootstrap method, which was repeated 1000 times to get a bootstrap distribution of the results.
Categorical variables or continuous variables were reported as numbers and percentages or as mean and standard deviation. Categorical variables were compared by the χ2 test, and continuous variables were compared by the Mann–Whitney U test. P < 0.05 were considered statistically significant. Statistical analyses were programmed in Python (v. 3.8.5; https://www.python.org/).
Results
Patient characteristics
A total of 159 patients (median age, 60.2 years; range, 19–82 years) were enrolled in this study. Exactly 31 patients (19.50%) were diagnosed with TERTp-wildtype, and 128 patients had TERTp mutation. The clinical characteristics are presented in Table 1. There were no significant differences between the training and testing cohorts with regard to age, sex, WHO grade, and TERTp mutation status, with TERTp-wildtype rates of 19.64% and 19.15%, respectively.
Table 1.
Clinical characteristics of the patients
| Training cohort (n = 112) | Testing cohort (n = 47) | ||||
|---|---|---|---|---|---|
| TERTp-mutation | TERTp-wildtype | TERTp-mutation | TERTp-wildtype | P | |
| Characteristic | (n = 90) | (n = 22) | (n = 38) | (n = 9) | 0.8958 |
| Age, years | 58.1 ± 12.3 | 59.2 ± 11.2 | 0.3699 | ||
| Sex | |||||
| Female | 45 (40.2%) | 17 (54.8%) | 0.1449 | ||
| Male | 67 (59.8%) | 14 (45.2%) | |||
| WHO grade | |||||
| III | 39 (34.8%) | 14 (29.8%) | 0.5389 | ||
| IV | 73 (65.2%) | 33 (70.2%) | |||
Data are means ± standard deviations or numbers of patients with percentages in parentheses. P value was derived from the univariate association analyses between each clinical parameter. Calculated by using the independent sample t test for continuous variables and the χ2 test for categoric variables
Radiomic feature extraction and selection
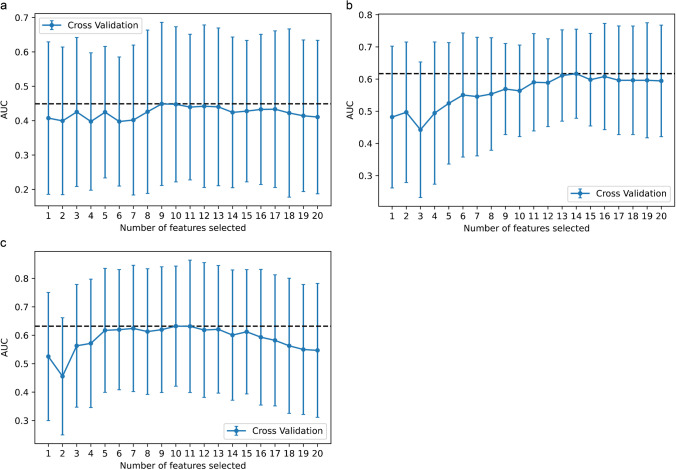
In this study, 107 radiomic features of candidates were generated from standard static images (20–40 min p.i.), early summation images (5–15 min p.i.), and dynamic [18F]FET PET images respectively, including first-order statistics, shape-based features, and texture features. After PCC process, 80 TBR20–40 features, 83 TBR5–15 features, and 91 TTP features were retained. For the TBR20–40 model, based on the AUC of the 10-fold cross-validation on the training cohort, 14 features were finally selected to fit the LR model after performing the RFE method. For the TBR5–15 model and the TTP model, 9 features and 10 features were selected for inclusion in the LR model, respectively (Fig. 2).
Fig. 2.
The feature selection process of the RFE method. Each iteration removes a feature that is considered least important and corresponds to a 10-fold cross-validation. After 10-fold cross-validation, the AUC of the model in the training cohort was used to determine the optimal number of features. The minimum AUC of feature number was selected. a TBR5–15 model, b TBR20–40, and c TTP model; 9, 14, and 10 features were selected respectively. RFE recursive feature elimination, AUC area under the receiver operating characteristic curve
Diagnostic Validation of the TBR20–40 model, TBR5–15 model, and TTP model
According to the above-mentioned formula, the risk probabilities of TERTp-mutation were calculated. The coefficients of selected features in the TBR20–40 model and TBR5–15 model are shown in Table S1. The coefficients of selected features in the TTP model are shown in Table 2.
Table 2.
Coefficients of selected features in the TTP model
| Features | Coefficients |
|---|---|
| SmallDependenceLowGreyLevelEmphasis | 1.508 |
| Energy | 1.404 |
| SmallDependenceHighGreyLevelEmphasis | − 1.283 |
| GreyLevelNonUniformityNormalized | − 1.235 |
| LeastAxisLength | − 1.219 |
| Busyness | − 0.916 |
| ShortRunHighGreyLevelEmphasis | − 0.699 |
| Maximum2DDiameterColumn | 0.654 |
| LowGreyLevelZoneEmphasis | − 0.626 |
| LargeDependenceHighGreyLevelEmphasis | 0.606 |
Intercept is 0.599 in the TTP model. Details of features were shown in Supplementary Information
No predictive power was observed in the TBR20–40 model with an AUC of only 0.49 (95% CI 0.30–0.69) in the testing cohort (AUC of 0.90 in the training cohort (95% CI 0.85–0.95); see Fig. S1). The TBR5–15 model demonstrated weak predictive capability to predict a TERTp-mutation (Fig. 3a, b), with an AUC of 0.61 (95% CI 0.42–0.80) in the testing cohort and an AUC of 0.80 (95% CI 0.71–0.89) in the training cohort. The TTP model showed the strongest predictive power and achieved an AUC of 0.82 (95% CI 0.71–0.92) and 0.90 (95% CI 0.84–0.95) in the testing cohort and training cohort, respectively (Fig. 3c, d).
Fig. 3.
a TBR5–15 model reached an AUC of 0.80 in the training cohort, and b an AUC of 0.61 in the testing cohort. c TTP model reached an AUC of 0.90 in the training cohort, and d an AUC of 0.82 in the testing cohort. AUC area under the receiver operating characteristic curve
Detailed information about the performance of each model is shown in Table 3.
Table 3.
Performance of each model
| TBR5–15 | TBR20–40 | TTP | ||||
|---|---|---|---|---|---|---|
| Training cohort | Testing cohort | Training cohort | Testing cohort | Training cohort | Testing cohort | |
| AUC | 0.80 | 0.61 | 0.90 | 0.49 | 0.90 | 0.82 |
| AUC 95%CI | (0.71–0.89) | (0.42–0.80) | (0.85–0.95) | (0.30–0.69) | (0.84–0.95) | (0.71–0.92) |
| Accuracy | 0.75.0% | 66.0% | 83.0% | 66.0% | 78.6% | 83.0% |
| Sensitivity | 73.3% | 73.7% | 81.1% | 73.7% | 77.8% | 92.1% |
| Specificity | 81.8% | 33.3% | 90.9% | 33.3% | 81.8% | 44.4% |
| PPV | 94.3% | 82.4% | 97.3% | 82.4% | 94.6% | 87.5% |
| NPV | 42.9% | 23.1% | 54.1% | 23.1% | 47.4% | 57.1% |
CI confidence interval
Discussion
Our study showed that radiomics based on dynamic [18F]FET PET data can reliably predict the TERTp-mutation status of IDH-wildtype diffuse astrocytic high-grade gliomas. Best predictability was reached using the TTP model derived from dynamic PET, and weak predictive capability was obtained with radiomics based on early summation images (5–15 min p.i.), while no reliable information about the TERTp-mutation status was possible based on the standard summation images (20–40 min p.i.).
Previous studies have shown that patients with IDH-wildtype TERTp-mutant glioblastoma have a significantly shorter progression free and overall survival compared to those with TERT-wildtype status. Therefore, TERTp-mutation status is now considered to be an important diagnostic and prognostic factor in primary glioblastomas and especially in patients with IDH-wildtype glioma [3, 5, 8, 9, 48]. TERTp-mutations indicate tumours that require aggressive and immediate treatments [3]. Hence, a preoperative tool for the prediction of a TERTp-mutation would be useful for early decision making and clinical management of patients with suspected glioma.
Several studies have analyzed the value of MRI based radiomics to predict the TERTp-mutation status in brain tumour patients [49–51]. Although these studies reported to achieve high accuracy values in the range of 79.88–93.80%, only WHO grade II or/and III gliomas have been considered and a limited number of patients has been investigated [49–51]. Besides, Tian et al. established a multiparameter MRI based radiomics model for the prediction of the TERTp-mutation status in patients with high-grade glioma [52], but ignored that TERTp-mutations play different roles in different IDH phenotypes [48].
Compared with conventional MRI, amino acid PET has been shown to be more sensitive in the definition of brain tumour extent [39], and dynamic [18F]FET uptake parameters extracted from the TAC have shown to be an independent biomarker for prognosis [53, 54]. Several studies have reported the informative value of [18F]FET PET-based radiomics in personalized clinical decisions and individualized treatment selection [27–29, 55]. Lohmann et al. found textural feature analysis in combination with TBRs to better differentiate brain metastasis recurrence from radiation injury than TBRs alone, and [18F]FET PET radiomics achieved a higher accuracy than the best standard FET PET parameter (TBRmax) to diagnose patients with pseudoprogression [27, 55]. Haubold et al. utilized multiparametric [18F]FET PET/MRI and MR fingerprinting to decode and phenotype cerebral gliomas, which may serve as an alternative to invasive tissue characterization [28]. In addition, Carles et al. evaluated the prognostic value of [18F]FET PET radiomics after re-irradiation, and found it could contribute to the selection of recurrent glioblastoma patients benefiting from re-irradiation [29]. However, all studies included radiomics based on standard static images (20–40 min p.i.) only and did not extract radiomic features derived from dynamic [18F]FET PET as well as early summation images (5–15 min p.i.) even though two studies have shown the impact of dynamic parameters on radiomics [32, 56]. Furthermore, no study has evaluated the potential to predict the TERTp-mutation status by [18F]FET PET radiomics so far.
This study included standard static images (20–40 min p.i.), early summation images (5–15 min p.i.), and dynamic [18F]FET PET images to develop the radiomic models. A total of 107 features were extracted from each image. Our TTP model, built from ten dynamic [18F]FET PET features selected by RFE, achieved the highest AUC of 0.82 in the independent testing cohort, indicating that the TERTp-mutation status can be predicted by using [18F]FET PET based radiomics. Notably, our former study did neither find an association between the TERTp-mutation status and traditional static [18F]FET PET parameters (TBRmax and TBRmean in static 20–40 min summation images) nor the standard dynamic parameter TTPmin [24].
Interestingly, radiomics based on the standard TBR20–40 model showed a low performance for the prediction of the TERTp-mutation status, and even the TBR5–15 model, generated from nine early summation [18F]FET PET features, had an accuracy of only 66% and an AUC of 0.61 in the testing cohort. With a high prediction accuracy of 83% in the TTP model, our study demonstrates that radiomic features extracted from dynamic PET data can achieve a higher performance level than models based on static PET data. Remarkably, the sensitivity of the TTP model reached 92.1% in the testing cohort, so that patients with aggressive TERTp-mutant glioma can be identified non-invasively with high probability [3]. With the generated multivariate LR-based formula, health practitioners will be able to calculate the patient individual risk probability of bearing a TERTp-mutation before neurosurgical intervention. Our study shows that even sophisticated radiomic analysis of static [18F]FET PET imaging cannot replace dynamic acquisitions, at least with regard to the prediction of the TERTp-mutation status.
Traditional dynamic [18F]FET PET parameters such as the classification of the time-activity curve (increasing vs. decreasing or increasing vs. plateau vs. decreasing), the slope or the TTPmin were most frequently calculated from a mean VOI-TAC of the tumour or from the hot-spot of the tumour with a 90% isocontour [10, 12, 19]. Considering the heterogeneity of gliomas, it may happen that the hot-spot in standard summation images does not correspond to the most suspicious tumour aggressiveness when only considering TTPmin and TAC and that, therefore, the most aggressive areas are inadvertently not evaluated. In contrast, we extracted the dynamic [18F]FET uptake information in every voxel within the tumour VOI and generated TTP images. This approach, which was first introduced by Kaiser et al. [40, 42], ensures that the dynamic information including the heterogeneity of uptake kinetics is extracted and that radiomics can be performed on the prognostically valuable dynamic data. The correlation between tumour heterogeneity and TERTp-mutation status can be considered in GreyLevelNonUniformityNormalized (GLNN) feature, which was used in the TTP model (see Table 2). GLNN belongs to Gray Level Dependence Matrix (GLDM), which is mathematically equal to first order–uniformity and is a measure of the homogeneity of the image array. A low value implies a greater heterogeneity, which was correlated with the TERTp-mutation, indicating that tumours with more heterogeneous TTP images are more likely to be classified as TERTp-mutant glioma.
Several limitations of this study should be discussed. First, the number of investigated patients is relatively small. However, it needs to be considered that we analyzed a very homogeneous group of patients with newly diagnosed and untreated IDH-wildtype diffuse astrocytic high-grade glioma. To exclude any influence by scanner type, all images in this study were derived from the same PET scanner, which limited the number of patients as well. In order to increase the number of patients, multi-centre validation studies are needed which, however, require phantom studies and harmonization of reconstruction parameters to make images from different PET scanners comparable. Another approach to directly harmonize features extracted from different devices may be to use the ComBat method [57]. In addition, our results are difficult to extrapolate to other centres, as the PET images analyzed in this study were acquired with our old PET scanner with fixed time frames, resulting in relatively long time frames (predominantly 5 and 10 min) in the dynamic analysis which could not be changed afterwards, and were reconstructed using filtered back-projection, while most PET centres now use other reconstruction methods such as ordered subset expectation maximization (OSEM). Furthermore, radiomic features were only extracted from the [18F]FET-positive tumour VOI to construct the model. Besides the tumour VOI, the remaining image (with normal seeming tissue) may still contain invisible but useful information. To analyze the entire images, deep learning methods will be necessary. Furthermore, our study focused on PET-based radiomics only. A combination with MRI may improve the performance of the prediction model and should be evaluated in future studies.
Conclusion
While conventional [18F]FET PET parameters assessed by standard analyses have previously shown no association with the TERTp-mutation status, radiomic models can predict the TERTp-mutation status of IDH-wildtype diffuse astrocytic high-grade gliomas with high accuracy preoperatively. Notably, this is only the case for radiomics based on dynamic image data (TTP model) instead of standard summation images (20–40 min). Further external validation in multi-centre studies with a larger number of patients is needed to evaluate the potential for clinical applications.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
Conceptualization: Zhicong Li and Nathalie L. Albert; Data acquisition: Adrien Holzgreve, Viktoria C. Ruf, Bogdana Suchorska, Vera Wenter, Stefanie Quach, Jochen Herms and Marcus Unterrainer; Image processing: Lena Kaiser; Implementation and application of the software for generation of parametric maps and for radiomic feature extraction using Pyradiomics: Lena Kaiser; Feature selection and machine learning implementation and application: Zhicong Li; Formal analysis: Adrien Holzgreve; Writing—original draft preparation: Zhicong Li, Lena Kaiser and Nathalie L. Albert; Writing—review and editing: All; Funding acquisition, Zhicong Li and Nathalie L. Albert; Supervision: Peter Bartenstein, Jörg-Christian Tonn and Nathalie L. Albert. All authors have read and agreed to the published version of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the Collaborative Research Centre SFB-824 of the Deutsche Forschungsgemeinschaft (DFG) and by the Else Kröner-Fresenius-Stiftung. The China Scholarship Council (CSC) funded this study for Zhicong Li.
Data availability
The data that support the findings of this study are available on request from the corresponding author, N.L.A. The data are not publicly available due to the privacy of research participants.
Code availability
The codes that support the findings of this study are available from the Z.L. and L.K., upon reasonable request.
Declarations
Ethics approval
Ethical approval of the retrospective data analysis was given by the institutional review board of the LMU (# 606–16) in accordance with the ICH Guideline for Good Clinical Practice (GCP) and the Declaration of Helsinki.
Consent to participate
All patients have given written informed consent prior to the PET examination.
Consent for publication
Not applicable.
Conflict of interest
N.L.A. is a member of the Neuroimaging Committee of the EANM. All other authors declare that they have no relationships or interests that could have direct or potential influence or impart bias on the work.
Footnotes
This article is part of the Topical Collection on Oncology - Brain
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
2/24/2022
The original version of this paper was updated to add the missing compact agreement Open Access funding note.
References
- 1.Arita H, Narita Y, Takami H, Fukushima S, Matsushita Y, Yoshida A, et al. TERT promoter mutations rather than methylation are the main mechanism for TERT upregulation in adult gliomas. Acta Neuropathol. 2013;126:939–941. doi: 10.1007/s00401-013-1203-9. [DOI] [PubMed] [Google Scholar]
- 2.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simon M, Hosen I, Gousias K, Rachakonda S, Heidenreich B, Gessi M, et al. TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro Oncol. 2015;17:45–52. doi: 10.1093/neuonc/nou158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Labussière M, Di Stefano AL, Gleize V, Boisselier B, Giry M, Mangesius S, et al. TERT promoter mutations in gliomas, genetic associations and clinico-pathological correlations. Br J Cancer. 2014;111:2024–2032. doi: 10.1038/bjc.2014.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinagre J, Almeida A, Pópulo H, Batista R, Lyra J, Pinto V, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4:2185. doi: 10.1038/ncomms3185. [DOI] [PubMed] [Google Scholar]
- 7.Rushing EJ. WHO classification of tumors of the nervous system: preview of the upcoming 5th edition. memo - Magazine of European Medical Oncology. 2021. 10.1007/s12254-021-00680-x.
- 8.Brat DJ, Aldape K, Colman H, Holland EC, Louis DN, Jenkins RB, et al. cIMPACT-NOW update 3: recommended diagnostic criteria for “Diffuse astrocytic glioma, IDH-wildtype, with molecular features of glioblastoma, WHO grade IV”. Acta Neuropathol. 2018;136:805–810. doi: 10.1007/s00401-018-1913-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louis DN, Wesseling P, Aldape K, Brat DJ, Capper D, Cree IA, et al. cIMPACT-NOW update 6: new entity and diagnostic principle recommendations of the cIMPACT-Utrecht meeting on future CNS tumor classification and grading. Brain Pathol. 2020;30:844–856. doi: 10.1111/bpa.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jansen NL, Suchorska B, Wenter V, Schmid-Tannwald C, Todica A, Eigenbrod S, et al. Prognostic significance of dynamic 18F-FET PET in newly diagnosed astrocytic high-grade glioma. J Nucl Med. 2015;56:9–15. doi: 10.2967/jnumed.114.144675. [DOI] [PubMed] [Google Scholar]
- 11.Jansen NL, Suchorska B, Wenter V, Eigenbrod S, Schmid-Tannwald C, Zwergal A, et al. Dynamic 18F-FET PET in newly diagnosed astrocytic low-grade glioma identifies high-risk patients. J Nucl Med. 2014;55:198–203. doi: 10.2967/jnumed.113.122333. [DOI] [PubMed] [Google Scholar]
- 12.Popperl G, Kreth FW, Mehrkens JH, Herms J, Seelos K, Koch W, et al. FET PET for the evaluation of untreated gliomas: correlation of FET uptake and uptake kinetics with tumour grading. Eur J Nucl Med Mol Imaging. 2007;34:1933–1942. doi: 10.1007/s00259-007-0534-y. [DOI] [PubMed] [Google Scholar]
- 13.Albert NL, Weller M, Suchorska B, Galldiks N, Soffietti R, Kim MM, et al. Response assessment in neuro-oncology working group and European Association for Neuro-Oncology recommendations for the clinical use of PET imaging in gliomas. Neuro Oncol. 2016;18:1199–1208. doi: 10.1093/neuonc/now058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langen KJ, Watts C. Neuro-oncology: amino acid PET for brain tumours—ready for the clinic? Nat Rev Neurol. 2016;12:375–376. doi: 10.1038/nrneurol.2016.80. [DOI] [PubMed] [Google Scholar]
- 15.Galldiks N, Langen KJ, Albert NL, Chamberlain M, Soffietti R, Kim MM, et al. PET imaging in patients with brain metastasis-report of the RANO/PET group. Neuro Oncol. 2019;21:585–595. doi: 10.1093/neuonc/noz003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Law I, Albert NL, Arbizu J, Boellaard R, Drzezga A, Galldiks N, et al. Joint EANM/EANO/RANO practice guidelines/SNMMI procedure standards for imaging of gliomas using PET with radiolabelled amino acids and [(18)F]FDG: version 1.0. Eur J Nucl Med Mol Imaging. 2019;46:540–557. doi: 10.1007/s00259-018-4207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albert NL, Winkelmann I, Suchorska B, Wenter V, Schmid-Tannwald C, Mille E, et al. Early static (18)F-FET-PET scans have a higher accuracy for glioma grading than the standard 20–40 min scans. Eur J Nucl Med Mol Imaging. 2016;43:1105–1114. doi: 10.1007/s00259-015-3276-2. [DOI] [PubMed] [Google Scholar]
- 18.Kunz M, Albert NL, Unterrainer M, la Fougere C, Egensperger R, Schüller U, et al. Dynamic 18F-FET PET is a powerful imaging biomarker in gadolinium-negative gliomas. Neuro Oncol. 2019;21:274–284. doi: 10.1093/neuonc/noy098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansen NL, Graute V, Armbruster L, Suchorska B, Lutz J, Eigenbrod S, et al. MRI-suspected low-grade glioma: is there a need to perform dynamic FET PET? Eur J Nucl Med Mol Imaging. 2012;39:1021–1029. doi: 10.1007/s00259-012-2109-9. [DOI] [PubMed] [Google Scholar]
- 20.Jansen NL, Suchorska B, Wenter V, Eigenbrod S, Schmid-Tannwald C, Zwergal A, et al. Dynamic 18F-FET PET in newly diagnosed astrocytic low-grade glioma identifies high-risk patients. J Nucl Med. 2014;55:198–203. doi: 10.2967/jnumed.113.122333. [DOI] [PubMed] [Google Scholar]
- 21.Fleischmann DF, Unterrainer M, Bartenstein P, Belka C, Albert NL, Niyazi M. 18F-FET PET prior to recurrent high-grade glioma re-irradiation—additional prognostic value of dynamic time-to-peak analysis and early static summation images? J Neurooncol. 2017;132:277–286. doi: 10.1007/s11060-016-2366-8. [DOI] [PubMed] [Google Scholar]
- 22.Vettermann F, Suchorska B, Unterrainer M, Nelwan D, Forbrig R, Ruf V, et al. Non-invasive prediction of IDH-wildtype genotype in gliomas using dynamic 18F-FET PET. Eur J Nucl Med Mol Imaging. 2019;46:2581–2589. doi: 10.1007/s00259-019-04477-3. [DOI] [PubMed] [Google Scholar]
- 23.Suchorska B, Giese A, Biczok A, Unterrainer M, Weller M, Drexler M, et al. Identification of time-to-peak on dynamic 18F-FET-PET as a prognostic marker specifically in IDH1/2 mutant diffuse astrocytoma. Neuro Oncol. 2018;20:279–288. doi: 10.1093/neuonc/nox153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unterrainer M, Ruf V, von Rohr K, Suchorska B, Mittlmeier LM, Beyer L, et al. TERT-promoter mutational status in glioblastoma—is there an association with amino acid uptake on dynamic (18)F-FET PET? Front Oncol. 2021;11:645316. doi: 10.3389/fonc.2021.645316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohmann P, Elahmadawy MA, Gutsche R, Werner JM, Bauer EK, Ceccon G, et al. FET PET radiomics for differentiating pseudoprogression from early tumor progression in glioma patients post-chemoradiation. Cancers (Basel). 2020;12. 10.3390/cancers12123835. [DOI] [PMC free article] [PubMed]
- 28.Haubold J, Demircioglu A, Gratz M, Glas M, Wrede K, Sure U, et al. Non-invasive tumor decoding and phenotyping of cerebral gliomas utilizing multiparametric (18)F-FET PET-MRI and MR Fingerprinting. Eur J Nucl Med Mol Imaging. 2020;47:1435–1445. doi: 10.1007/s00259-019-04602-2. [DOI] [PubMed] [Google Scholar]
- 29.Carles M, Popp I, Starke MM, Mix M, Urbach H, Schimek-Jasch T, et al. FET-PET radiomics in recurrent glioblastoma: prognostic value for outcome after re-irradiation? Radiat Oncol. 2021;16:46. doi: 10.1186/s13014-020-01744-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian J, Herman MG, Brinkmann DH, Laack NN, Kemp BJ, Hunt CH, et al. Prediction of MGMT status for glioblastoma patients using radiomics feature extraction from (18)F-DOPA-PET imaging. Int J Radiat Oncol Biol Phys. 2020;108:1339–1346. doi: 10.1016/j.ijrobp.2020.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohmann P, Meißner AK, Kocher M, Bauer EK, Fink GR, et al. Feature-based PET/MRI radiomics in patients with brain tumors. Neurooncol Adv. 2020;2:iv15–iv21. doi: 10.1093/noajnl/vdaa118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaragori T, Oster J, Roch V, Hossu G, Chawki MB, Grignon R, et al. (18)F-FDOPA PET for the non-invasive prediction of glioma molecular parameters: a radiomics study. J Nucl Med. 2021 doi: 10.2967/jnumed.120.261545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 34.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thon N, Eigenbrod S, Kreth S, Lutz J, Tonn JC, Kretzschmar H, et al. IDH1 mutations in grade II astrocytomas are associated with unfavorable progression-free survival and prolonged postrecurrence survival. Cancer. 2012;118:452–460. doi: 10.1002/cncr.26298. [DOI] [PubMed] [Google Scholar]
- 36.Biczok A, Kraus T, Suchorska B, Terpolilli NA, Thorsteinsdottir J, Giese A, et al. TERT promoter mutation is associated with worse prognosis in WHO grade II and III meningiomas. J Neurooncol. 2018;139:671–678. doi: 10.1007/s11060-018-2912-7. [DOI] [PubMed] [Google Scholar]
- 37.Langen KJ, Bartenstein P, Boecker H, Brust P, Coenen HH, Drzezga A, et al. German guidelines for brain tumour imaging by PET and SPECT using labelled amino acids. Nuklearmedizin. 2011;50:167–173. doi: 10.3413/nuk-2011041. [DOI] [PubMed] [Google Scholar]
- 38.Unterrainer M, Vettermann F, Brendel M, Holzgreve A, Lifschitz M, Zahringer M, et al. Towards standardization of 18F-FET PET imaging: do we need a consistent method of background activity assessment? EJNMMI Res. 2017;7:48. doi: 10.1186/s13550-017-0295-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pauleit D, Floeth F, Hamacher K, Riemenschneider MJ, Reifenberger G, Muller HW, et al. O-(2-[18F]fluoroethyl)-L-tyrosine PET combined with MRI improves the diagnostic assessment of cerebral gliomas. Brain. 2005;128:678–687. doi: 10.1093/brain/awh399. [DOI] [PubMed] [Google Scholar]
- 40.Vomacka L, Unterrainer M, Holzgreve A, Mille E, Gosewisch A, Brosch J, et al. Voxel-wise analysis of dynamic 18F-FET PET: a novel approach for non-invasive glioma characterisation. EJNMMI Res. 2018;8:91. doi: 10.1186/s13550-018-0444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Can Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.can-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaiser LGM, Ahmadi SA, Unterrainer M, Holzgreve A, Mille E, Gosewisch A, Brosch J, Suchorska B, Navab N, Tonn JC, Ziegler S, Bartenstein P, Albert NL, Böning G. Annual Congress of the European Association of Nuclear Medicine October 12–16, 2019 Barcelona, Spain. Eur J Nucl Med Mol Imaging. 2019;46:1–952. doi: 10.1007/s00259-019-04486-2. [DOI] [Google Scholar]
- 43.Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. arXiv preprint arXiv:161207003. 2016.
- 44.Leijenaar RT, Nalbantov G, Carvalho S, van Elmpt WJ, Troost EG, Boellaard R, et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep. 2015;5:11075. doi: 10.1038/srep11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Granitto PM, Furlanello C, Biasioli F, Gasperi F. Recursive feature elimination with random forest for PTR-MS analysis of agroindustrial products. Chemometr Intell Lab. 2006;83:83–90. doi: 10.1016/j.chemolab.2006.01.007. [DOI] [Google Scholar]
- 46.Tomz M, King G, Zeng L. ReLogit: rare events logistic regression. 2003. 2003;8:27. 10.18637/jss.v008.i02.
- 47.Makhoul J. Linear prediction: a tutorial review. Proc IEEE. 1975;63:561–580. doi: 10.1109/PROC.1975.9792. [DOI] [Google Scholar]
- 48.Vuong HG, Altibi AMA, Duong UNP, Ngo HTT, Pham TQ, Chan AK, et al. TERT promoter mutation and its interaction with IDH mutations in glioma: combined TERT promoter and IDH mutations stratifies lower-grade glioma into distinct survival subgroups-A meta-analysis of aggregate data. Crit Rev Oncol Hematol. 2017;120:1–9. doi: 10.1016/j.critrevonc.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 49.Jiang C, Kong Z, Zhang Y, Liu S, Liu Z, Chen W, et al. Conventional magnetic resonance imaging-based radiomic signature predicts telomerase reverse transcriptase promoter mutation status in grade II and III gliomas. Neuroradiology. 2020;62:803–813. doi: 10.1007/s00234-020-02392-1. [DOI] [PubMed] [Google Scholar]
- 50.Park CJ, Han K, Kim H, Ahn SS, Choi D, Park YW, et al. MRI features may predict molecular features of glioblastoma in isocitrate dehydrogenase wild-type lower-grade gliomas. AJNR Am J Neuroradiol. 2021;42:448–456. doi: 10.3174/ajnr.A6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang S, Fan Z, Sun Z, Li Y, Liu X, Liang Y, et al. Radiomics features predict telomerase reverse transcriptase promoter mutations in world health organization grade II gliomas via a machine-learning approach. Front Oncol. 2020;10:606741. doi: 10.3389/fonc.2020.606741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tian H, Wu H, Wu G, Xu G. Noninvasive prediction of TERT promoter mutations in high-grade glioma by radiomics analysis based on multiparameter MRI. Biomed Res Int. 2020;2020:3872314. doi: 10.1155/2020/3872314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kunz M, Thon N, Eigenbrod S, Hartmann C, Egensperger R, Herms J, et al. Hot spots in dynamic (18)FET-PET delineate malignant tumor parts within suspected WHO grade II gliomas. Neuro Oncol. 2011;13:307–316. doi: 10.1093/neuonc/noq196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bauer EK, Stoffels G, Blau T, Reifenberger G, Felsberg J, Werner JM, et al. Prediction of survival in patients with IDH-wildtype astrocytic gliomas using dynamic O-(2-[(18)F]-fluoroethyl)-L-tyrosine PET. Eur J Nucl Med Mol Imaging. 2020;47:1486–1495. doi: 10.1007/s00259-020-04695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lohmann P, Stoffels G, Ceccon G, Rapp M, Sabel M, Filss CP, et al. Radiation injury vs. recurrent brain metastasis: combining textural feature radiomics analysis and standard parameters may increase (18)F-FET PET accuracy without dynamic scans. Eur Radiol. 2017;27:2916–2927. doi: 10.1007/s00330-016-4638-2. [DOI] [PubMed] [Google Scholar]
- 56.Lohmann P, Lerche C, Bauer EK, Steger J, Stoffels G, Blau T, et al. Predicting IDH genotype in gliomas using FET PET radiomics. Sci Rep. 2018;8:13328. doi: 10.1038/s41598-018-31806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orlhac F, Boughdad S, Philippe C, Stalla-Bourdillon H, Nioche C, Champion L, et al. A postreconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med. 2018;59:1321–1328. doi: 10.2967/jnumed.117.199935. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author, N.L.A. The data are not publicly available due to the privacy of research participants.
The codes that support the findings of this study are available from the Z.L. and L.K., upon reasonable request.