Abstract
This study aimed to determine the presence of cryptosporidiosis in immunosuppressed patients hospitalized at the Clinic of Haematology and Oncohaematology in a form of routine screening. Samples were collected from November 2019 to February 2020, when the first wave of the Coronavirus pandemic occurred in Slovakia. A total of 36 samples were collected from patients hospitalized at the Clinic of Haematology and Oncohaematology, both from the open ward and the intensive care unit. For the diagnosis of cryptosporidiosis, a nested PCR targeting the gp60 gene and the SSU rRNA locus was used. From the 36 samples, Cryptosporidium parvum subtype IIaA17G1R1 was diagnosed in 9 patients (7 from the open ward and 2 from the intensive care unit), all hospitalized at the clinic at the same time, in February 2020. The occurrence of the same species and subtype, Cryptosporidium parvum IIaA17G1R1, in 9 patients hospitalized at the same time, both at the open ward and the intensive care unit may suggest a possible transmission occurred at the clinic.
Keywords: Cryptosporidium parvum, Immunosuppression, Malignancies, Transmission, Slovakia
Introduction
Cryptosporidiosis is a significant infection, causing diarrhea among humans. A recent systematic meta-analysis including findings from 221 publications estimates the pooled global prevalence of cryptosporidiosis as high as 7.6%. The prevalence is highest in people living in low-income countries, people with gastrointestinal symptoms, and in children under 5 years of age (Dong et al. 2020). In developed countries, the prevalence ranges from 3.2% in the EU/EEA to 3.8% in the USA (ECDC 2019; CDC 2019). The infection is transmitted mainly via the fecal–oral route. Contaminated water and food are the most important transmission pathways, but Cryptosporidium spp. can also be transmitted by direct contact with an infected human or animal and rarely via air (Hunter and Nichols 2002; Sponseller et al. 2014).
Infections caused by Cryptosporidium spp. are one of the most frequent causes of mortality especially in children under 5 years of age (Lozano et al. 2012; Striepen 2013; Shoultz et al. 2016). To this day, 47 valid Cryptosporidium species and more than 100 distinguished genotypes have been recognized (Holubová et al. 2019; Bolland et al. 2020; Ježková et al. 2021; Zahedi et al. 2021). Human infections are most commonly caused by Cryptosporidium parvum and Cryptosporidium hominis, although other species and genotypes have been recognized, mainly in humans suffering from immunodeficiency, including C. meleagridis; C. felis; C. canis; C. cuniculus; C. ubiquitum; C. viatorum; C. muris; C. suis; C. fayeri; C. andersoni; C. bovis; C. scrofarum; C. tyzzeri; C. erinacei; and Cryptosporidium horse, skunk, and chipmunk I genotypes (Cacciò et al. 2002; Wolska-Kusnierz et al. 2007; Xiao 2010; Zahedi et al. 2016; Ryan et al. 2016).
In immunocompetent individuals, cryptosporidium gastroenteritis is typically self-limiting, and patients usually recover within 2 weeks. On the other hand, chronic diarrhea is present in immunosuppressed patients (Ryan et al. 2016). Immunosuppression, either impaired or constant, is often associated with oncological diseases or their treatment (Sulżyc-Bielicka et al. 2018). Immunosuppression and diarrhea are known side effects in patients undergoing cancer treatment, although few studies were focused on the occurrence of cryptosporidiosis in patients with malignancies (Hassanein et al. 2012).
In this study, we have focused on the occurrence and subtyping of Cryptosporidium spp. in patients with hemato-oncological diseases, hospitalized at the Clinic of Haematology and Oncohaematology. This study is also a follow-up to our previous studies about the occurrence of Cryptosporidium spp. in immunosuppressed and immunocompetent patients, animals, and in environmental samples to further expand the knowledge of individual species, genotypes, and subtypes circulating in Slovakia (Hasajová et al. 2014; Danišová et al. 2017; Kalinová et al. 2018; Hatalová et al. 2019).
Materials and methods
From November 2019 to February 2020 a total of 36 samples of feces were collected from diarrheic patients hospitalized at the Clinic of Haematology and Oncohaematology of the Louis Pasteur University Hospital (LPUH) in Kosice. Samples were obtained from patients from the open ward (n = 28) and the intensive care unit (ICU; n = 8). Samples were transferred to the Department of Epidemiology, Faculty of Medicine of the Pavol Jozef Safarik University in Kosice and stored at -20 °C until analysis. Characteristics of samples according to the date of sampling are summarized in Table 1. Nucleic acids were extracted using the DNA Sorb-B nucleic acid extraction kit (AmpliSense, Russia) according to the manufacturer’s guide. Isolated nucleic acids were stored at -20 °C until the use in PCR. Nested PCR was used for the amplification of the 60 kDa glycoprotein gene (gp60) of C. parvum and C. hominis since they are the most common species diagnosed in humans (Xiao 2010). After gp60 subtyping, nested PCR using primers targeting the SSU rRNA locus were used for all samples (Leetz et al. 2007) to diagnose other species of Cryptosporidium and to ensure that the contamination did not occur during subtyping. For all reactions, a 20 µl mix was prepared using 5 × HOT FIREPol Blend Master Mix Ready to Load (Solis BioDyne, Estonia), 0.1 µM of the respective primers, and 5 µl of DNA template. All reactions were carried out in Biometra T1 Thermocycler (AnalyticJenna). Final products of a length of 450 bp (for primers targeting the gp60 gene) or 250 bp (for primers targeting the SSU locus) were analyzed in 1.5% agarose gel dyed with GoodView™ Nucleic Acid Stain in TBE buffer. Positive samples were sent for sequencing. Final sequences were compared with homologous sequences stored in GenBank using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Subtypes of Cryptosporidium were assessed according to the rules of subtyping, using serine-coding trinucleotides TCA, TCT, and TCG at the 5′end of the gp60 gene and other repetitive and non-repetitive sequences following the tandem repeats (Xiao 2010).
Table 1.
Characteristics of patients’ samples according to the date of sampling
| Date of sampling | No. of samples | Ward | ICU | ||||
|---|---|---|---|---|---|---|---|
| Male | Female | Total | Male | Female | Total | ||
| 18 November 2019 | 8 | 4 | 4 | 8 | 0 | 0 | 0 |
| 27 November 2019 | 10 | 5 | 2 | 7 | 1 | 2 | 3 |
| 28 November 2019 | 3 | 1 | 1 | 2 | 0 | 1 | 1 |
| 23 January 2020 | 6 | 2 | 3 | 5 | 0 | 1 | 1 |
| 24 February 2020 | 9 | 3 | 4 | 7 | 1 | 1 | 2 |
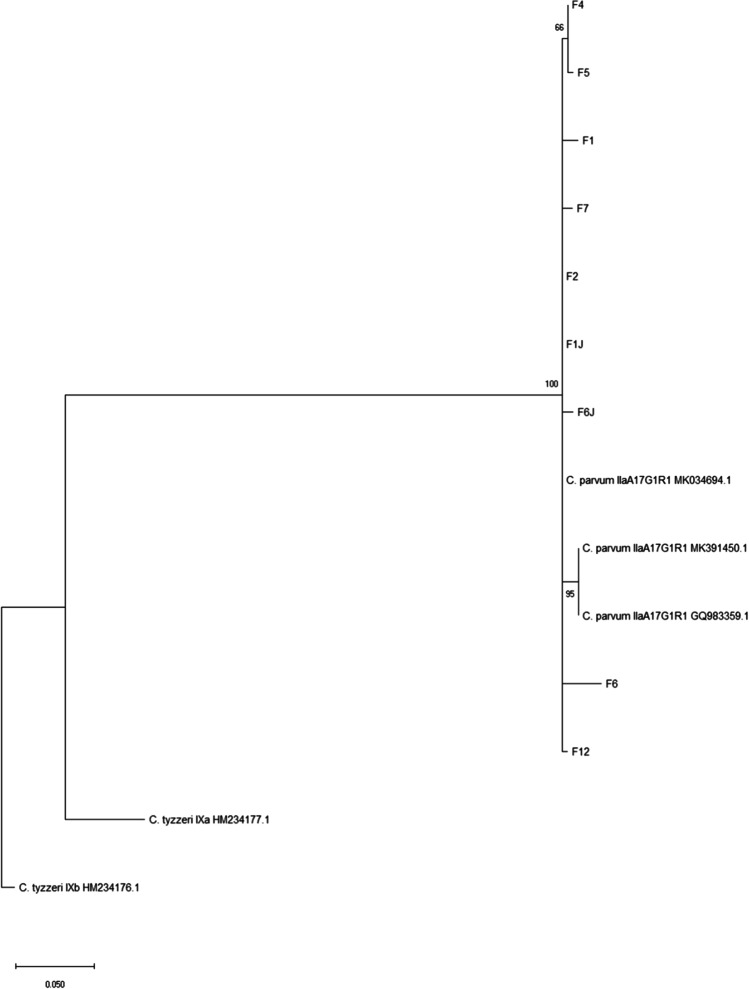
The consensus sequences were edited and assembled using BioEdit program. Then sequences were aligned using MUSCLE Multiple Sequence Alignment with reference sequences from GenBank. The phylogenetic tree was constructed by maximum likelihood method using the program MEGA-X (Molecular Evolutionary Genetics Analysis software). Bootstrap values were calculated from 1,000 replicates. C. tyzzeri was used as an out-group. The phylogenetic tree is represented in Fig. 1.
Fig. 1.
Phylogenetic tree constructed by maximum likelihood method
Parallel to PCR, all patients from this study were tested for common nosocomial strains of bacteria in the microbiological laboratory of the Louis Pasteur University Hospital.
Results and discussion
After PCR, from all 36 samples obtained from November to February, only 9 were positive for Cryptosporidium spp. using primers targeting the gp60 gene. All 9 samples where Cryptosporidium spp. was detected came from February 2020. In these 9 samples from patients hospitalized at the same time, C. parvum subtype IIaA17G1R1 was diagnosed in all cases, both in the open ward and the ICU. Primers targeting the SSU region showed positivity for C. parvum only in five samples and also obtained in February 2020. The lower rate of amplification by primers targeting the SSU region in comparison to gp60 primers may be explained by their lower specificity and/or low DNA yield in the isolates. All other samples collected from November 2019 were negative. Parallel microbiological examination by the LPUH laboratory showed that one patient positive for C. parvum IIaA17G1R1, hospitalized in the ICU during February 2020, was also suffering from Klebsiella pneumoniae type CRE (carbapenem-resistant Enterobacteriaceae). Association between PCR results, dates of sampling, and hemato-oncological diseases of the patients are summarized in Table 2.
Table 2.
Association between PCR results, date of sampling and hemato-oncological diagnoses
| Date of sampling | Sample number | Sex | Diagnosis | PCR results |
|---|---|---|---|---|
| 24 February 2020 | F1 | M | B-NHL DLBCL | C. parvum IIaA17G1R1 |
| 24 February 2020 | F2 | M | NS-HL, after allo-SCT | C. parvum IIaA17G1R1 |
| 24 February 2020 | F4 | F | PCNSL DLBCL | C. parvum IIaA17G1R1 |
| 24 February 2020 | F5 | F | TTP | C. parvum IIaA17G1R1 |
| 24 February 2020 | F6 | F✝ | B-NHL DLBCL | C. parvum IIaA17G1R1 |
| 24 February 2020 | F7 | F | AML | C. parvum IIaA17G1R1 |
| 24 February 2020 | F12 | M✝ | AML | C. parvum IIaA17G1R1 |
| 24 February 2020 | F1J | M✝ | AML, after MUD allo-SCT | C. parvum IIaA17G1R1 |
| 24 February 2020 | F6J | F✝ | AML | C. parvum IIaA17G1R1 |
Abbreviations: allo-SCT allogeneic stem cell transplantation, AML acute myeloid leukemia, B-NHL B cell non-Hodgkin lymphoma, DLBCL diffuse large B cell lymphoma, MUD matched unrelated donor, NS-HL nodular sclerosis Hodgkin lymphoma, PCNSL primary central nervous system lymphoma, TTP thrombotic thrombocytopenic purpura
Cryptosporidium and other opportunistic pathogens are common causes of life-threatening infections with severe or fatal outcomes, especially in immunosuppressed patients (Iqbal et al. 1999). Cryptosporidium spp. are important parasites in cancer patients, in which a severe form of cryptosporidiosis occurs most frequently (Hunter and Nichols 2002). In recent years, several studies investigated the relationship between cancer and parasites (Oliveira 2014; Samuel 2016). Most studies describe infections caused by Cryptosporidium as a consequence of immunosuppression caused by cancer and its therapy (Sulżyc-Bielicka et al. 2018). These studies were conducted using samples from patients undergoing chemotherapy, therefore fostering the development of opportunistic infections. In these studies, the prevalence of cryptosporidiosis varied from 1.3 to 80% (Hassan et al. 2002; Al-Qobati et al. 2012). A recent study shows that by 2040, the incidence of cancer is expected to rise to 29.5 million new cases per year (Perera et al. 2021). Due to this fact, the probability of the occurrence of opportunistic parasites in cancer patients will increase.
A study conducted in France between the years 2015 and 2017 declared 210 cases of cryptosporidiosis in immunosuppressed patients with solid organ transplantation (51%), HIV (31%), bone marrow transplantation (8%), and malignancy. From the total 210 cases, 80% of the patients were treated with corticosteroids, 19% were on chemotherapy, and 3% had anti-cytokine biotherapy. Patients on mycophenolate mofetil therapy and solid organ transplantation recipients appeared to be the most vulnerable group, especially during the first 6 months after the transplantation (Costa et al. 2018).
Since the 1980s, numerous outbreaks of cryptosporidiosis have been reported in healthcare facilities around the world, including the transmission of the parasites to healthcare workers from immunosuppressed and immunocompetent patients, and between patients (Baxby et al. 1983; Vandenberg et al. 2012; Feng et al. 2012; Goñi et al. 2015).
In our study, from the total 36 samples obtained from November 2019 to February 2020, none was diagnosed with Cryptosporidium spp., except samples obtained in February. In these 9 samples, the presence of the same species and subtype was detected, C. parvum IIaA17G1R1, both in patients from the open ward, and also in patients from the ICU. Before hospitalization, none of the patients showed any clinical signs of cryptosporidiosis, suggesting that a possible transmission of the pathogen occurred at the clinic.
C. parvum IIaA17G1R1 was also diagnosed in previous studies from Slovakia, in immunocompetent humans, and also immunosuppressed adults and children, and animals, suggesting it as a very common subtype in our country (Hatalová et al. 2019).
Our study came across limitations. On 11 March 2020, the World Health Organization declared the novel Coronavirus (COVID-19) outbreak a global pandemic (WHO Director-General’s opening remarks at the media briefing on COVID19 -March 2020). On the same date, the Government of Slovakia declared a state of emergency and enhanced restrictive measures countrywide, followed by a lockdown of the country for several months (Uznesenie vlády Slovenskej republiky 147/2020 Z.z.). The hospital and the Clinic of Haematology and Oncohaematology closed to visitors during this period, and sanitary regime was increased. For these reasons, new stool samples and samples of water and smears from the environment were not obtained. This fact led us to postpone the assessment of the source of the infection and the transmission mode until the epidemiological situation becomes more favorable. Repeated sampling could not have been conducted from four of the patients, because they are now deceased.
Conclusion
The detection of the same species and subtype of Cryptosporidium in 9 patients hospitalized at the Clinic of Haematology and Oncohaematology at the same time, both at the open ward and on the ICU, indicates that a possible transmission of the pathogen may have occurred. When the situation allows, we will continue sampling to further monitor the situation at the Clinic of Haematology and Oncohaematology. In case of repeated occurrences of cryptosporidiosis, we will conduct an epidemiological investigation to assess the source of the infection and transmission modes of Cryptosporidium spp. in patients suffering from hemato-oncological diseases. Along with the staff, these patients should be systematically screened for cryptosporidiosis before and during hospitalization to avoid possible transmission from healthcare personnel to patients and from patients to personnel, eliminating the possibility of an infection.
Acknowledgements
This article is dedicated to the memory of Dorota Hatalova, M.Ed. Special thanks to Mr. Fraser Kyle during the manuscript preparation and correction.
Funding
This study was supported by grants from the Slovak Research and Development Agency (grant no. APVV-0134) and Scientific Grant Agency VEGA (VEGA MŠVVaŠ a SAV) (grant no. VEGA 1/0084/18 and VEGA 1/0359/21).
Data availability
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Declarations
Ethics approval
Approval for the study was obtained from the Ethics Committee of the Faculty of Medicine at P. J. Safarik University in Kosice. The study was performed following the ethical standards as laid down in the Declaration of Helsinki of 1975 and revised in 2008.
Consent to participate
Participation in the study was voluntary and anonymous, and informed consent was obtained before the medical examination.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Elena Hatalova, Email: elena.hatalova@upjs.sk.
Tomas Guman, Email: tomas.guman@upjs.sk.
Veronika Bednarova, Email: veronika.bednarova@upjs.sk.
Vladimira Turcok Simova, Email: vladimira.turcok.simova@student.upjs.sk.
Mariia Logoida, Email: mariia.logoida@student.upjs.sk.
Monika Halanova, Email: monika.halanova@upjs.sk.
References
- Al-Qobati SA, Al-Maktari MT, Al-Zoa AMB, Derhim M (2012) Intestinal parasitosis among Yemeni patients with cancer, Sana’a, Yemen. J Egypt Soc Parasitol 42:727–734. 10.12816/0006356 [DOI] [PubMed]
- Baxby D, Hart CA, Taylor C. Human cryptosporidiosis: possible case of hospital cross infection. Br Med J. 1983;287:1760–1761. doi: 10.1136/bmj.287.6407.1760-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland SJ, Zahedi A, Oskam C, et al (2020) Cryptosporidium bollandi n. sp. (Apicomplexa: Cryptosporidiiae) from angelfish (Pterophyllum scalare) and Oscar fish (Astronotus ocellatus). Exp Parasitol 217:. 10.1016/j.exppara.2020.107956 [DOI] [PubMed]
- Cacciò S, Pinter E, Fantini R, et al. Human infection with Cryptosporidium felis: case report and literature review. Emerg Infect Dis. 2002;8:85–86. doi: 10.3201/eid0801.010269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC - Centers for Disease Control and Prevention. Cryptosporidiosis summary report — National Notifiable Diseases Surveillance System, United States, 2018. Atlanta, Georgia: U.S. Department of Health and Human Services, CDC, 2019. Centers for Disease Control MMWR Office. Retrieved from https://www.cdc.gov/healthywater/surveillance/cryptosporidium/cryptosporidium-2018.html
- Costa D, Razakandrainibe R, Sautour M, et al. Human cryptosporidiosis in immunodeficient patients in France (2015–2017) Exp Parasitol. 2018;192:108–112. doi: 10.1016/j.exppara.2018.08.001. [DOI] [PubMed] [Google Scholar]
- Danišová O, Valenčáková A, Stanko M, et al. Rodents as a reservoir of infection caused by multiple zoonotic species/genotypes of C. parvum, C. hominis, C. suis, C. scrofarum, and the first evidence of C. muskrat genotypes I and II of rodents in Europe. Acta Trop. 2017;172:29–35. doi: 10.1016/j.actatropica.2017.04.013. [DOI] [PubMed] [Google Scholar]
- Dong S, Yang Y, Wang Y, et al. Prevalence of cryptosporidium infection in the global population: a systematic review and meta-analysis. Acta Parasitol. 2020;65:882–889. doi: 10.2478/s11686-020-00230-1. [DOI] [PubMed] [Google Scholar]
- ECDC - European Centers for Disease Control and Prevention. Cryptosporidiosis annual epidemiological report for 2017. Surveillance report, 10 Oct 2019. Retrieved from https://www.ecdc.europa.eu/en/publications-data/cryptosporidiosis-annual-epidemiological-report-2017
- Feng Y, Wang L, Duan L, et al. Extended outbreak of cryptosporidiosis in a pediatric hospital, China. Emerg Infect Dis. 2012;18:312–314. doi: 10.3201/eid1802.110666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goñi P, Almagro-Nievas D, Cieloszyk J, et al. Cryptosporidiosis outbreak in a child day-care center caused by an unusual Cryptosporidium hominis subtype. Enferm Infecc Microbiol Clin. 2015;33:651–655. doi: 10.1016/j.eimc.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Hasajová A, Valenčáková A, Malčeková B, et al. Significantly higher occurrence of cryptosporidium infection in Roma children compared with non-Roma children in Slovakia. Eur J Clin Microbiol Infect Dis. 2014;33:1401–1406. doi: 10.1007/s10096-014-2082-2. [DOI] [PubMed] [Google Scholar]
- Hassan SI, Sabry H, Amer NM, Shalaby MA, Mohamed NA, Gaballah H. Incidence of cryptosporidiosis in immunodeficient cancer patients in Egypt. J Egypt Soc Parasitol. 2002;32:33–46. [PubMed] [Google Scholar]
- Hassanein SMA, Abd-El-Latif MMS, Hassanin OM, et al. Cryptosporidium gastroenteritis in Egyptian children with acute lymphoblastic leukemia: magnitude of the problem. Infection. 2012;40:279–284. doi: 10.1007/s15010-011-0230-5. [DOI] [PubMed] [Google Scholar]
- Hatalová E, Valenčáková A, Luptáková L, et al. The first report of animal genotypes of Cryptosporidium parvum in immunosuppressed and immunocompetent humans in Slovakia. Transbound Emerg Dis. 2019;66:243–249. doi: 10.1111/tbed.13009. [DOI] [PubMed] [Google Scholar]
- Holubová N, Zikmundová V, Limpouchová Z, et al. Cryptosporidium proventriculi sp. n. (Apicomplexa: Cryptosporidiidae) in Psittaciformes birds. Eur J Protistol. 2019;69:70–87. doi: 10.1016/j.ejop.2019.03.001. [DOI] [PubMed] [Google Scholar]
- Hunter PR, Nichols G. Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clin Microbiol Rev. 2002;15:145–154. doi: 10.1128/CMR.15.1.145-154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Munir MA, Khan MA. Cryptosporidium infection in young children with diarrhea in Rawalpindi, Pakistan. Am J Trop Med Hyg. 1999;60:868–870. doi: 10.4269/ajtmh.1999.60.868. [DOI] [PubMed] [Google Scholar]
- Ježková J, Limpouchová Z, Prediger J, et al (2021) Cryptosporidium myocastoris n. Sp. (apicomplexa: Cryptosporidiidae), the species adapted to the nutria (myocastor coypus). Microorganisms 9:. 10.3390/microorganisms9040813 [DOI] [PMC free article] [PubMed]
- Kalinová J, Valenčáková A, Hatalová E, et al. Occurrence of Cryptosporidium in the water basins of nitra region, slovakia. Acta Trop. 2018;179:36–38. doi: 10.1016/j.actatropica.2017.12.019. [DOI] [PubMed] [Google Scholar]
- Leetz AS, Sotiriadou I, Ongerth J, Karanis P. An evaluation of primers amplifying DNA targets for the detection of Cryptosporidium spp. using C. parvum HNJ-1 Japanese isolate in water samples. Parasitol Res. 2007;101:951–962. doi: 10.1007/s00436-007-0567-y. [DOI] [PubMed] [Google Scholar]
- Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira G. Cancer and parasitic infections: similarities and opportunities for the development of new control tools. Rev Soc Bras Med Trop. 2014;47:1–2. doi: 10.1590/0037-8682-0013-2014. [DOI] [PubMed] [Google Scholar]
- Perera SK, Jacob S, Wilson BE, et al. Global demand for cancer surgery and an estimate of the optimal surgical and anaesthesia workforce between 2018 and 2040: a population-based modelling study. Lancet Oncol. 2021;22:182–189. doi: 10.1016/S1470-2045(20)30675-6. [DOI] [PubMed] [Google Scholar]
- Ryan U, Zahedi A, Paparini A. Cryptosporidium in humans and animals—a one health approach to prophylaxis. Parasite Immunol. 2016;38:535–547. doi: 10.1111/pim.12350. [DOI] [PubMed] [Google Scholar]
- Samuel F (2016) Opportunistic parasitism: parasitic association with the host that has compromised immune system. J Bacteriol Parasitol 07: 10.4172/2155-9597.1000261
- Shoultz DA, de Hostos EL, Choy RKM (2016) Addressing cryptosporidium infection among young children in low-income settings: the crucial role of new and existing drugs for reducing morbidity and mortality. PLoS Negl Trop Dis. 10 [DOI] [PMC free article] [PubMed]
- Sponseller JK, Griffiths JK, Tzipori S. The evolution of respiratory cryptosporidiosis: evidence for transmission by inhalation. Clin Microbiol Rev. 2014;27:575–586. doi: 10.1128/CMR.00115-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striepen B. Time to tackle cryptosporidiosis. Nature. 2013;503:189–191. doi: 10.1038/503189a. [DOI] [PubMed] [Google Scholar]
- Sulżyc-Bielicka V, Kołodziejczyk L, Jaczewska S, et al (2018) Colorectal cancer and Cryptosporidium spp. infection. PLoS One 13:. 10.1371/journal.pone.0195834 [DOI] [PMC free article] [PubMed]
- UZNESENIE VLÁDY SLOVENSKEJ REPUBLIKY č. 366 (2020, June 10) . Retrieved from https://www.slov-lex.sk/pravne-predpisy/SK/ZZ/2020/147/20200610
- Vandenberg O, Robberecht F, Dauby N, et al. Management of a cryptosporidium hominis outbreak in a day-care center. Pediatr Infect Dis J. 2012;31:10–15. doi: 10.1097/INF.0b013e318235ab64. [DOI] [PubMed] [Google Scholar]
- WHO Director-General's opening remarks at the media briefing on COVID-19 - 11 March 2020. Retrieved from https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020
- Wolska-Kusnierz B, Bajer A, Caccio S, et al. Cryptosporidium infection in patients with primary immunodeficiencies. J Pediatr Gastroenterol Nutr. 2007;45:458–464. doi: 10.1097/MPG.0b013e318054b09b. [DOI] [PubMed] [Google Scholar]
- Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2010;124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- Zahedi A, Bolland SJ, Oskam CL, Ryan U (2021) Cryptosporidium abrahamseni n. sp. (Apicomplexa: Cryptosporidiiae) from red-eye tetra (Moenkhausia sanctaefilomenae). Exp Parasitol 223: 10.1016/j.exppara.2021.108089 [DOI] [PubMed]
- Zahedi A, Paparini A, Jian F, et al. Public health significance of zoonotic Cryptosporidium species in wildlife: critical insights into better drinking water management. Int J Parasitol Parasites Wildl. 2016;5:88–109. doi: 10.1016/j.ijppaw.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.