Abstract
Background
Despite that machine learning (ML)-based MRI has been evaluated for diagnosis of axillary lymph node metastasis (ALNM) in breast cancer patients, diagnostic values they showed have been variable. In this study, we aimed to assess the use of ML to classify ALNM on MRI and to identify potential covariates that might influence the diagnostic performance of ML.
Methods
A systematic research of PubMed, Embase, Web of Science, and the Cochrane Library was conducted until 27 December 2020 to collect the included articles. Subgroup analysis was also performed.
Findings
Fourteen studies assessing a total of 2247 breast cancer patients were included in the analysis. The overall AUC for ML in the validation set was 0.80 (95% confidence interval [CI] 0.76–0.83) with a negative predictive value of 0.83. The pooled sensitivity and specificity were 0.79 (95% CI 0.74–0.84) and 0.77 (95% CI 0.73–0.81), respectively. In the subgroup analysis of the validation set, T1-weighted contrast-enhanced (T1CE) imaging with ML yielded a higher sensitivity (0.80 vs. 0.67 vs. 0.76) than the T2-weighted fat-suppressed (T2-FS) imaging and diffusion-weighted imaging (DWI). Support vector machines (SVMs) had a higher specificity than linear regression (LR) and linear discriminant analysis (LDA) (0.79 vs. 0.78 vs. 0.75), whereas LDA showed a higher sensitivity than LR and SVM (0.83 vs. 0.70 vs. 0.77).
Interpretation
MRI sequences and algorithms were the main factors that affect the diagnostic performance of ML. Although its results were encouraging with the pooled sensitivity of around 0.80, it meant that 1 in 5 women that would go with undetected metastases, which may have a detrimental effect on the overall survival for 20% of patients with positive SLN status. Despite that a high NPV of 0.83 meant that ML could potentially benefit those with negative SLN, it might also translate to 1 in 5 tests being false negative. We would like to suggest that ML may not be yet usable in clinical routine especially when patient survival is used as a primary measurement of its outcome.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13244-021-01034-1.
Keywords: Artificial intelligence, Axillary lymph node metastasis, Machine learning, Magnetic resonance imaging
Key points
Sentinel lymph node biopsy (SLNB) with machine learning (ML) might be more helpful to breast cancer patients because ML might prevent over-treatment due to its sensitivity of around 0.80 to classify ALNM.
T1CE with ML is more sensitive than T2-FS and DWI (0.80 vs. 0.67 vs. 0.76, respectively).
Support vector machine (SVM) is more specific than linear regression (LR) and linear discriminant analysis (LDA) (0.79 vs. 0.78 vs. 0.75, respectively), whereas LDA is more sensitive than LR and SVM (0.83 vs. 0.70 vs.0.77, respectively).
Introduction
Breast cancer is one of the most common malignancies worldwide, accounting for 30% of all new cancer diagnoses in 2018 among American women [1]. As axillary lymph node status in breast cancer patients is crucial for pathologic staging, it is also used as a prognostic indicator and for clinical patient management, therapeutic guidance, and survival predictions [2, 3]. Although axillary lymph node dissection (ALND) is the gold standard for evaluating axillary lymph node metastasis (ALNM), ALND might not confer a survival advantage [4]. Sentinel lymph node biopsies (SLNBs) are used widely and can reduce ALND complications [5]. However, SLNBs are invasive procedures that could be associated with fewer disadvantages such as lymphoedema and sensory loss (the risks of 5% and 11%, respectively) [6]. One way of SLNBs is to surgically remove of one or a few axillary lymph nodes, whereas over 70% of SLNBs are negative, thus questioning the generic use of this invasive procedure [7]. In addition, another way of SLNBs is to inject a radiotracer or ultrasound with fine-needle aspiration, which, however, is difficult to perform in primary hospitals due to lack of practical experience and nuclear medicine or other relevant facilities. Therefore, it would be more than advantageous to research and develop some noninvasive approaches to predict ALNMs preoperatively.
Ultrasound, mammogram, PET/CT, and MRI have been used to diagnose ALNMs during breast cancer staging. Ultrasonography showed a sensitivity and a specificity of 33–86.2% and 40.5–96.2%, respectively [8–13]. The sensitivity and specificity of mammogram procedures were 21% and 99.5%, respectively [11]. The overall sensitivities and specificities of PET/CT were reported to be 20–80% and 88.6–97%, respectively [8–10, 13, 14]. In addition, ultrasonography is convenient but is also dependent on operator experience. Mammogram and PET-CT can result in unnecessary exposure to harmful ionising radiation. Conversely, due to its low inter-observer variability, hardly any radiation, and improved diagnostic contrast, MRI has become a routine noninvasive diagnostic tool.
Machine learning (ML) is a branch of artificial intelligence that includes algorithms that could enhance diagnosis, treatments, and follow-up neuro-oncology visits by analysing enormous complex datasets [15, 16]. In recent years, there have been some studies on the use of ML to predict ALNM in breast cancer patients. The use of ML in predicting ALNM is not dependent on operator experience levels and is more objective with good repeatability. In addition, the diagnostic performance of ML might be further improved. To avoid overfitting and to adequately assess ML performance, proper training should involve k-fold cross-validation or external testing. However, the results so far are far from being consistent even among themselves. What’s more, no meta-analysis has previously been done to assess the use of ML for predicting ALNM. To address this problem, the present meta-analysis pooled all the published studies concerning the diagnostic performance of ML-based MRI in the prediction of ALNM in breast cancer patients.
Materials and methods
Literature search and study selection
A search in PubMed, Embase, Web of Science, and the Cochrane Library was performed until 27 December 2020. It used almost all Medical Subject Heading (MeSH) terms available and free keywords for “Machine Learning”, “Transfer Learning”, “Breast Neoplasms”, “Breast Tumor”, “Breast Cancer”, “Lymphatic Metastasis”, and “Lymph Node Metastasis”. A search of the reference lists from included studies was also performed.
Two reviewers selected potentially relevant studies independently based on the title and abstract, and disagreements were resolved by a third reviewer to reach a consensus.
Studies were included if (1) the research subject was limited to human subjects in English; (2) the diagnostic performance pertaining to sensitivity and specificity was reported; (3) a histopathologically confirmed ALNM was present in breast cancer patients; and (4) ML was applied to predict ALNM without defined limit for age or sample size.
Studies were excluded if (1) the publication was on animal research, a conference abstract, or a review article, and (2) the study reported on overlapping patient cohorts.
Data extraction
Data from the included studies were collected by two investigators independently, and discrepancies between were resolved with the help of a third investigator. Each study was initially identified by identifying the author’s name and the year of publication. A spreadsheet was used to extract total patient populations, numbers of abnormal and normal lymph nodes, and sensitivity and specificity of ML detection. Other information included the study design, algorithms, data sources, MRI sequences, image segmentations, magnet field strengths, and manufacturers.
Quality assessment
The quality of studies and likelihood of bias were conducted according to the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) [17], which has two main areas, viz. risk of bias and concerns regarding applicability. The tool consists of four domains, including patient selection, index test, reference standard, and flow and timing. The first three domains were also assessed in terms of applicability concerns using high, low, or unclear ratings. For individual studies, each domain was considered at a high, low, or unclear risk of bias. If the answers to all signalling questions for a domain were “yes”, the risk of bias would be judged as low. If answers to any signalling questions were “no or unclear”, the risk of bias would be judged as high or unclear. Two reviewers performed quality assessments independently and discrepancies between them were resolved by a third reviewer to reach a consensus. Review Manager 5.3 (RevMan 5.3) software (Cochrane Collaboration, Oxford, UK) was used.
Statistical analysis
Numerical values for sensitivity and specificity were extracted and their true positive (TP), false positive (FP), false negative (FN), and true negative (TN) values were recalculated. Threshold analysis was performed, and a Spearman correlation coefficient and p value were obtained. Symmetry or asymmetry summary receiver operating characteristic (SROC) curves were used to evaluate threshold effects according to the p value of the b coefficient using measures of effectiveness (MOEs) modelling. Cochran-Q tests and the inconsistency indices (I2) of the sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), negative predictive value(NPV), and diagnostic odds ratio (DOR) were used to explore heterogeneity. If I2 < 50% and p > 0.05, the fixed-effects model was used; otherwise, the random-effects model was used to pool these five effect sizes. The criteria of heterogeneity for the I2 values were 0–25% (very low), 25–50% (low), 50–75% (medium), and > 75% (high), respectively. Subgroup analysis was further performed to explore the sources of heterogeneity that were performed based on MRI sequences, magnet field strengths, image segmentation methods, and ML algorithms. Sensitivity analysis was used to assess the robustness of the meta-analysis by verifying if the size of a research study can affect the pooled results. Deeks funnel plot was used to assess publication bias. Data analysis was performed using Stata14.0 (StataCorp LP, College Station, TX) and MetaDisc1.4 (http://www.hrc.es/investigacion/metadisc_en.htm) software. For each of the parameters (1.5 T; 3.0 T; T2-FS; T1CE; DWI; SVM; LR; LDA; 2D and 3D), we constructed forest plots for pooled sensitivities, specificities, PLR, NLR, and DOR. Others such as field strength, sequence, algorithm, and segmentation were compared by using a Student t test, Mann–Whitney U test, or a one-way analysis of variance. For studies describing different results in a classifier due to multiple kernels in ML, performance of these studies was selected with the highest one.
Results
Literature search and data extraction
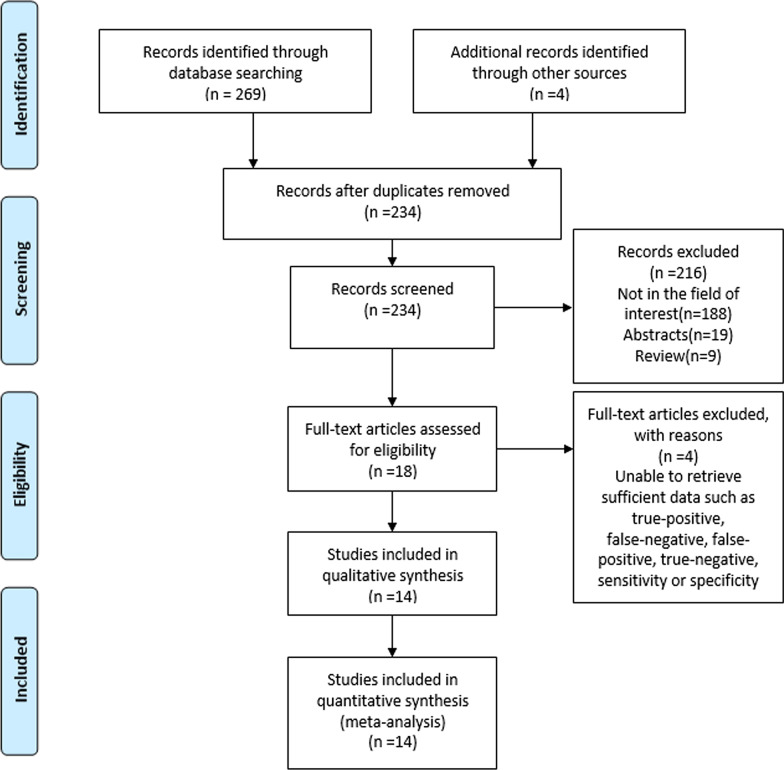
A detailed study selection process is presented in Fig. 1. There were 273 potentially eligible citations. After removing 39 duplicate records, 234 records were screened. With 216 citations based on the title and abstract excluded, 18 full-text articles were assessed for eligibility. After revision, a total of 14 original articles [18–31] that included 2247 breast cancer patients (954 abnormal lymph nodes and 1508 normal lymph nodes) were eventually included in the study. The patient and study characteristics are described in Table 1, and the baseline characteristics are shown in Table 2.
Fig. 1.
A flow diagram of the literature review and study selection
Table 1.
Study and patient characteristics
| Author (year of publication) | Data source | # Abnormal/normal nodes | # Patients | Magnet field strength, manufacturer | Study design |
|---|---|---|---|---|---|
| Hongna Tan (2020) | Single institution | 119/210 | 329 | 3.0 T GE | Retro |
| Thomas Ren (2020) | Single institution | 66/193 | 99 | 1.5 T GE | Retro |
| Xiao Zhang (2019) | Single institution | 55/91 | 146 | 1.5 T Philips | Retro |
| Jia Liu (2019) | Single institution | 35/27 | 62 | 3.0 T GE | Retro |
| Karl D Spuhler (2019) | Single institution | 55/108 | 163 | 1.5 T GE | Retro |
| Lu Han (2019) | Single institution | 148/263 | 411 | 1.5 T GE | Retro |
| Jiaxiu Luo (2018) | Single institution | 74/98 | 172 | 1.5 T Philips | Retro |
| Chunling Liu (2019) | Single institution | 55/108 | 163 | 1.5 T GE | Retro |
| Yuhao Dong (2018) | Single institution | 55/91 | 146 | 1.5 T Philips | Retro |
| Meijie Liu (2020) | Single institution | 78/86 | 164 | 3.0 T GE | Pros |
| Xiaoyu Cui (2019) | Single institution | 52/63 | 102 | 3.0 T Siemens | Retro |
| Demircioglu (2020) | Single institution | 34/50 | 84 | 1.5 T Siemens | Retro |
| Arefan (2020) | Single institution | 80/74 | 154 | 3.0 T Siemens | Retro |
| Fusco (2018) | Single institution | 48/46 | 52 | 1.5 T Aurora | Retro |
Retro, retrospective; Pros, prospective
Table 2.
Baseline characteristics of the included studies
| Author (year of publication) | Algorithm | Sequences | Segmentation | Dataset | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| Hongna Tan (2020) | SVM | T2-FS | 3D | Training set | 84.38% | 72.25% |
| Validation set | 65.22% | 81.08% | ||||
| Thomas Ren (2020) | CNN | T1CE | 2D | Validation set | 92.10 ± 2.90% | 79.30 ± 5.10% |
| Xiao Zhang (2019) | RF | DWI | 3D | Validation set | 83.30% | 74.20% |
| T2-FS | 77.80% | 87.10% | ||||
| Jia Liu (2019) | SVM | T1CE | 3D | Training set | 75.00% | 76.00% |
| Validation set | 71.00% | 100.00% | ||||
| Xgboost | Training set | 89.00% | 76.00% | |||
| Validation set | 86.00% | 83.00% | ||||
| LR | Training set | 71.00% | 71.00% | |||
| Validation set | 71.00% | 83.00% | ||||
| Karl D Spuhler (2019) | CNN | T1CE | 3D | Testing set | 72.20% | 88.90% |
| Lu Han (2019) | SVM | T1CE | 2D | Training set | 89.00% | 57.00% |
| Validation set | 78.00% | 72.00% | ||||
| Jiaxiu Luo (2018) | SVM | DWI | 3D | Testing set | 86.70% | 83.30% |
| Chunling Liu (2019) | LR | T1CE | 3D | Training set | 76.10% | 66.70% |
| Validation set | 81.90% | 77.80% | ||||
| Yuhao Dong (2018) | LR | T2-FS | 3D | Training set | 66.30 ± 0.30% | 81.60 ± 0.20% |
| Validation set | 60.00 ± 0.60% | 74.70 ± 0.40% | ||||
| DWI | Training set | 74.00 ± 0.20% | 80.80 ± 0.20% | |||
| Validation set | 69.50 ± 0.50% | 75.70 ± 0.40% | ||||
| T2-FS and DWI | Training set | 66.30 ± 0.30% | 81.60 ± 0.20% | |||
| Validation set | 70.00 ± 0.80% | 74.70 ± 0.50% | ||||
| Meijie Liu (2020) | LR | T1CE | 2D | Validation set | 64.00% | 79.00% |
| Xiaoyu Cui (2019) | SVM | T1CE | 3D | Validation set | 94.90% | 77.96% |
| KNN | 89.39% | 87.18% | ||||
| LDA | 80.31% | 67.78% | ||||
| Demircioglu (2020) | LR | T1CE and T2-FS | 3D | Validation set | 71.00% | 74.00% |
| Arefan (2020) | LDA | T1CE | 2D | Testing set | 60.00% | 87.00% |
| RF | 60.00% | 86.00% | ||||
| NB | 81.00% | 67.00% | ||||
| KNN | 74.00% | 54.00% | ||||
| SVM | 70.00% | 71.00% | ||||
| LDA | 3D | 63.00% | 92.00% | |||
| RF | 64.00% | 90.00% | ||||
| NB | 85.00% | 62.00% | ||||
| KNN | 67.00% | 58.00% | ||||
| SVM | 81.00% | 50.00% | ||||
| LDA | 2D | Validation set | 82.00% | 76.00% | ||
| RF | 89.00% | 68.00% | ||||
| NB | 73.00% | 82.00% | ||||
| KNN | 79.00% | 75.00% | ||||
| SVM | 65.00% | 88.00% | ||||
| LDA | 3D | 82.00% | 78.00% | |||
| RF | 89.00% | 70.00% | ||||
| NB | 69.00% | 76.00% | ||||
| KNN | 75.00% | 75.00% | ||||
| SVM | 73.00% | 79.00% | ||||
| Fusco (2018) | LDA | T1CE | 3D | Validation set | 88.50% | 77.80% |
CNN, convolutional neural networks; SVM, support vector machine; LR, linear regression; KNN, k-nearest neighbor; LDA, linear discriminant analysis; NB, naive Bayes; RF, random forest; T2-FS, fat-suppressed T2; T1CE, contrast-enhanced T1. The algorithm pooled was chosen with the highest performance
Data quality assessment
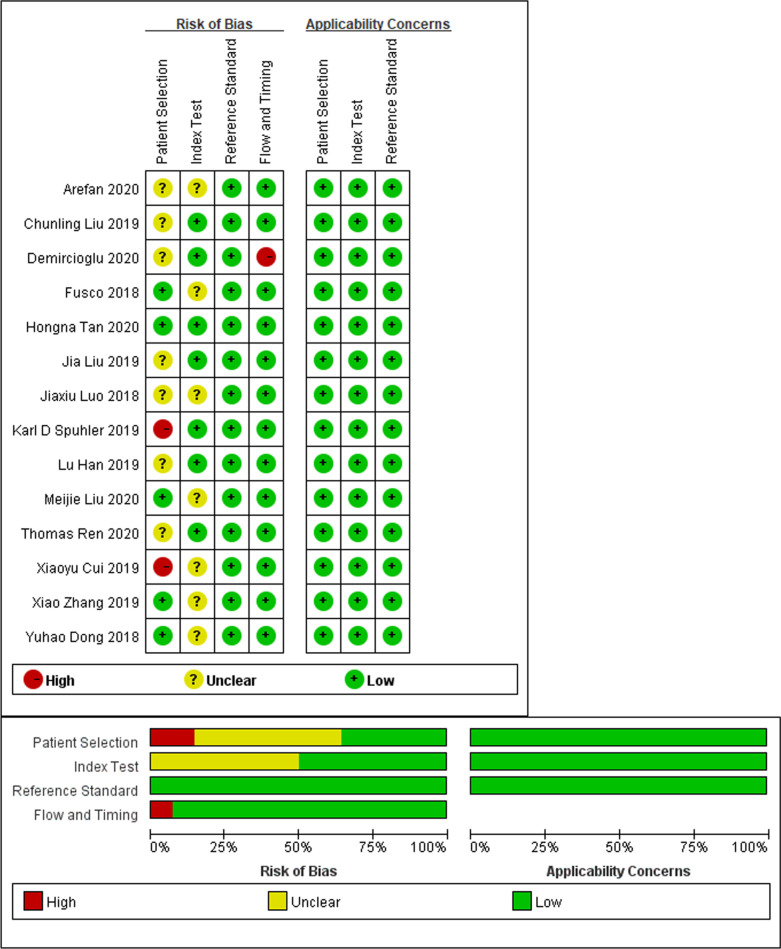
Results of the QUADAS-2 assessments are shown in Fig. 2 and the Additional file 1: Supplementary Materials. Most included studies were regarded as having a low to moderate risk of bias with low concerns over their applicability. In particular, only one study scored a low risk of bias in all domains [19]. For the patient selection domain, two studies were considered to have a high risk of bias due to their non-consecutive or random patient enrolment [24, 27]. Seven studies were considered to have a risk that is uncertain because they did not explain how patients were enrolled [20–23, 26, 28, 30]. For the index test domain, seven studies [18, 22, 25, 27, 29–31] were considered to have an unclear risk because they lacked pre-specified thresholds. Further, for the reference standard domain, all studies were classified as having a low risk, and all of the selected studies used biopsy and/or histopathology as the reference standard. Lastly, for the domain of flow and timing, one study [21] was considered to be at high risk because it did not explain whether all of its patients were included in the study.
Fig. 2.
Methodologic quality of the included studies assessed according to the Quality Assessment of Diagnostic Accuracy Study 2 tool for risk of bias and applicability concerns. Green represents low, yellow unclear, and red high risk of bias
Data analysis
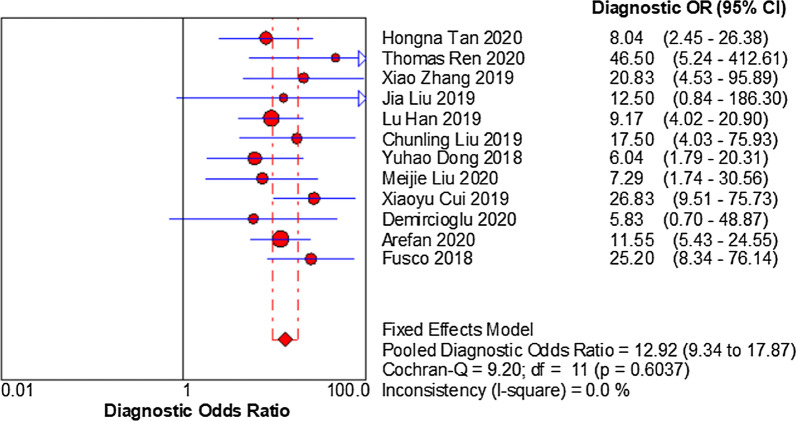
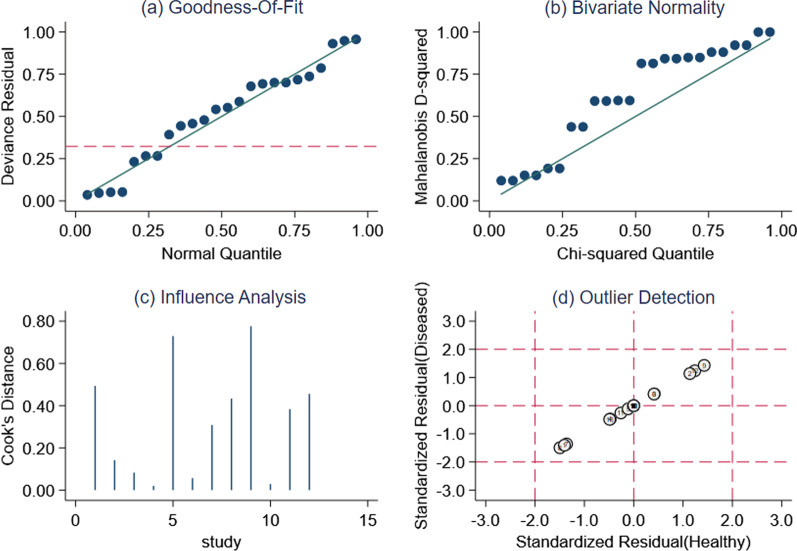
There are different numbers for training, testing, and validation studies because there were eight studies which only showed the validation or test results [18, 20, 21, 24, 25, 27, 29, 30], whereas six others only showed the results of training, testing/validation [19, 22, 23, 26, 28, 31]. In the test set, the sensitivity and specificity of the pooled three studies were 76% and 82%, respectively. In the validation set, the sensitivity and specificity of the pooled twelve studies were 79% and 77%, respectively. The Spearman correlation coefficient in the validation set was − 0.083 (p = 0.799), indicating no threshold effect discovered. Next, no “shoulder arm” plot was observed in the SROC curve. Since I2 of sensitivity, specificity, PLR, NLR, DOR = 21.60%, 0.00%, 0.00%, 24.20%, 0.00% < 50.00%, p values were 0.174, 0.994, 0.969, 0.206, 0.604, respectively, a model of fixed-effects to pool effect sizes was chosen (Fig. 3). The pooled sensitivity, specificity, PLR, NLR, and DOR were 0.79 (95%Cl 0.74–0.84), 0.77 (95%Cl 0.73–0.81), 3.47 (95%Cl 2.91–4.14), 0.27 (95%Cl 0.22–0.33), 12.92 (95%Cl 9.34–17.87), respectively (Fig. 4). The AUC was 0.80(95%Cl 0.76–0.81). More interestingly, for those patients predicted to have negative SLN, they achieved a high NPV of 0.83. Sensitivity analysis that removed studies with potential bias showed results consistent with the primary meta-analysis, which were conducted to assess robustness of the synthesised results. Sensitivity analysis showed that twelve original articles had better stability and reliability and relatively high quality (Fig. 5).
Fig. 3.
A forest plot of single studies for the pooled diagnostic odds ratio and 95% confidence interval (CI) in the validation group. Horizontal lines represent the 95% CI of the point estimates. Each red circle represents the area under the curve (AUC) of the individual studies, and the box size indicates the study size. The red diamond indicates the pooled AUC of all twelve studies
Fig. 4.
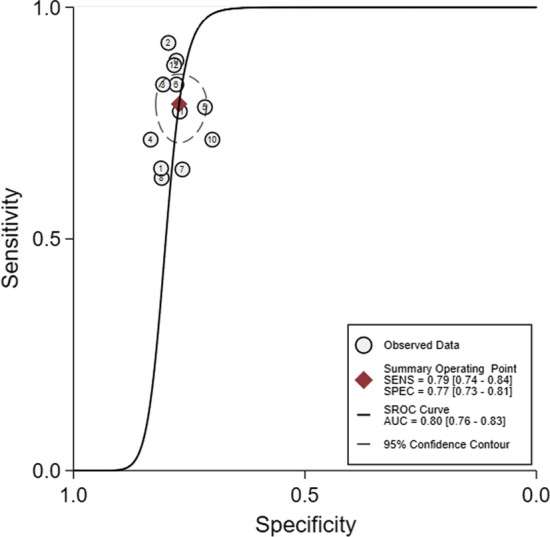
Summary receiver operating characteristics (SROC) curve regarding the diagnostic performance of machine learning in predicting axillary lymph node metastasis in breast cancer patients. SROC curve (solid line) and summary point (red square). Every circle represents the sensitivity and specificity estimate from one study, and the size of the circle reflects the sample size. The pooled area under the curve was 0.80
Fig. 5.
Sensitivity analysis. a Goodness of fit; b bivariate normality; c influence analysis; and d outlier detection. All original articles had high stability and reliability and relatively high quality in the validation set
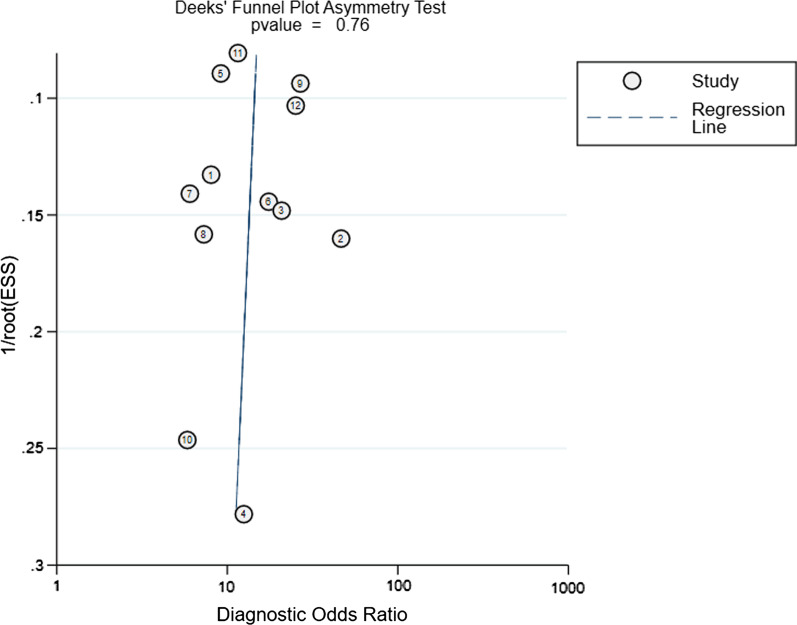
Since the studies of the test set were too small to draw any reliable conclusions, subgroup analysis was performed only in the validation set (Table 3). T1CE with ML yielded a higher sensitivity (0.80 vs. 0.67 vs. 0.768) than T2-FS and DWI (p < 0.05). The number of combining T2-FS and DWI or T2-FS and T1CE was only one study, which, nonetheless, was too small to draw any reliable conclusions. In addition, MRI magnet field strength also affected the diagnostic performance of ML. In comparison with 1.5 T, studies using 3.0 T had a better sensitivity (0.86 vs. 0.81) (p > 0.05). In algorithm, SVM demonstrated a higher specificity than LR and LDA (0.79 vs. 0.78 vs. 0.75), whereas LDA showed a higher sensitivity than LR and SVM (0.83 vs. 0.70 vs.0.77) (p < 0.05). ML performed better for 3D than 2D, in which the pooled sensitivities were 0.80 versus 0.77 and specificities were 0.78 versus 0.76 (p > 0.05). In addition, the Deeks funnel plot revealed that there was no obvious publication bias. (p = 0.870, Fig. 6).
Table 3.
The performance evaluation of the datasets and subgroups
| Dataset | No. of studies | Sensitivity | Specificity | PLR | NLR | DOR |
|---|---|---|---|---|---|---|
| Testing set | 3 | 0.76 (0.64–0.86)# | 0.82 (0.72–0.89)## | 3.90 (2.49–6.11)## | 0.29 (0.19–0.46)# | 12.88 (5.99–27.71)## |
| Validation set | 12 | 0.79 (0.74–0.84)# | 0.77 (0.73–0.81)# | 3.47 (2.91–4.14)# | 0.27 (0.22–0.33)# | 12.92 (9.34–17.87)# |
| Overall validation group | ||||||
| Magnetic field strength (T) | ||||||
| 1.5 T | 7 | 0.81 (0.75–0.87)# | 0.76 (0.71–0.81)# | 3.38 (2.70–4.24)# | 0.25 (0.18–0.34)# | 13.15 (8.37–20.66)# |
| 3.0 T | 5 | 0.86 (0.78–0.89)** | 0.76 (0.70–0.82)# | 3.46 (2.69–4.46)# | 0.23 (0.11–0.48)** | 17.78 (10.54–30.00)## |
| Sequence | ||||||
| T2-FS | 3 | 0.67 (0.54–0.79)# | 0.80 (0.71–0.88)# | 3.43 (2.22–5.29)## | 0.41 (0.28–0.59)# | 8.01 (4.01–16.00)## |
| T1CE | 8 | 0.80 (0.75–0.84)## | 0.80 (0.75–0.84)# | 4.04 (3.25–5.02)# | 0.25 (0.19–0.31)## | 16.27 (11.08–23.89)# |
| DWI | 2 | 0.76 (0.60–0.89)# | 0.75 (0.63–0.85)# | 3.10 (1.96–4.91)# | 0.31 (0.17–0.56)# | 10.00 (3.97–25.17)# |
| Algorithm | ||||||
| SVM | 5 | 0.77 (0.71–0.82)** | 0.79 (0.73–0.83)## | 3.67 (2.87–4.70)# | 0.30 (0.19–0.47)* | 13.43 (8.62–20.93)## |
| LR | 4 | 0.70 (0.58–0.81)# | 0.78 (0.68–0.85)# | 3.20 (2.17–4.71)# | 0.37 (0.26–0.55)# | 8.60 (4.44–16.66)# |
| LDA | 3 | 0.83 (0.77–0.88)# | 0.75 (0.68–0.81)# | 3.35 (2.58–4.35)## | 0.22 (0.16–0.31)# | 14.78 (9.01–24.26)# |
| Segmentation | ||||||
| 2D | 4 | 0.77 (0.70–0.83)## | 0.76 (0.70–0.82)# | 3.30 (2.55–4.28)# | 0.29 (0.21–0.39)# | 11.46 (7.05–18.62)# |
| 3D | 9 | 0.80 (0.75–0.84)## | 0.78 (0.73–0.82)# | 3.56 (2.88–4.39)# | 0.26 (0.21–0.34)## | 13.52 (9.27–19.72)# |
PLR, positive likelihood ratio; NLR, negative likelihood ratio; DOR, diagnostic odds ratio; SVM, support vector machine; LR, linear regression; LDA, linear discriminant analysis; T2-FS, fat-suppressed T2; T1CE, contrast-enhanced T1; #, 0–25% of I2 values; ##, 25–50%; *, 50–75%; **, > 75%; # and ##, fixed-effects model; * and **, random-effects model
Fig. 6.
The Deeks funnel plot asymmetry test to determine publication bias in the literature evaluation (p = 0.76) indicated there was no obvious publication bias. Each dot represents an individual study
Discussion
ALNM can determine the prognosis and treatment of breast cancer patients. Thus, it is imperative to find an accurate and reproducible way to detect ALNM. To address this issue, this meta-analysis was intended to assess the applicability of ML to the classification of ALNM on MRI and to the identification of potential covariates that influence the diagnostic performance of ML.
DWI is a commonly performed sequence with promising results for the evaluation of breast lesions because their images can be obtained in a short time without contrast agents [32]. However, most lesions show images with a relatively inferior quality and increase blurrings and distortions on DWI, which can make it difficult to accurately segment the lesions. In contrast, T2-weighted imaging (T2WI) can clearly depict edema, hemorrhage, mucus, and cystic fluid, which can be valuable for evaluation of breast masses [33]. T2-FS can also show the lesion boundaries clearly. Dynamic contrast-enhanced (DCE)-MRI with numerous scanning phases is sensitive to the change in tissue vessel perfusion and permeability [34]. However, compared with T2-FS, DCE-MRI had a better sensitivity. Demircioglu et al. [21] reported that ALNM classifications that combined T2-FS with T1CE were moderate with an AUC of 0.710, but this meta-analysis discovered that the number of combined sequences was not big enough to draw any reliable conclusions from them. Ren et al. [20], Han et al. [26] and Liu et al. [28] used the T1CE first axial phase images to predict ALNM and achieved AUCs of 0.91, 0.78, and 0.81 in the validation sets, respectively. Liu et al. [23], Liu et al. [18], Arefan et al. [22], Fusco et al. [29], and Cui et al. [27] use the peak enhanced phase images achieved AUCs of 0.85, 0.74, 0.82, 0.81, and 0.77 in the validation sets, respectively. Theoretically, 3 T imaging has been shown to be able to improve image quality due to its better performance resulting from higher spatial resolution [35]. This meta-analysis revealed that, in comparison with 1.5 T, studies using 3.0 T had a better sensitivity (0.86 vs. 0.81), albeit without significant differences. This should be further validated with a larger size of samples in the future.
Different as they are, algorithms have advantages of their own. Linear discriminant analysis (LDA) models can distinguish and identify a linear decision boundary between classes [36]. Support vector machine (SVM), as one of the most popular classifying techniques, is an excellent algorithm that can be utilised to model misspecifications and can effectively handle high-dimensional data [37]. Linear regression (LR) is also suitable for the regression of high-dimensional data. In this meta-analysis, SVM demonstrated a higher specificity than LR and LDA, whereas LDA showed a higher sensitivity than LR and SVM. Yu et al. [38] used LR to identify ALNM in the development and validation set with AUCs of 0.88 and 0.85, respectively. Takada et al. [39] used a decision tree to predict ALNM (AUCs of 0.770 and 0.772 in the training and validation sets, respectively). Schacht et al. [40] and Ha et al. [41] used neural net classifiers to predict ALNM, achieving an AUC of 0.880 and an accuracy of 84.30%. As probabilistic classifiers, naive Bayes models are constructed using the Bayes theorem of conditional probabilities [42]. XGBoost is an optimised distributed gradient boosting library designed to be highly efficient, flexible, and portable. A random forest is a multitude of trees, but they are differentiated by a random selection of the variables to reduce correlations between the fitted trees [43]. The k-nearest neighbor algorithm is a tool used to exploit the local information in classification and regression problems [44]. Convolutional neural networks (CNNs) can obtain global and local image information directly from the convolution kernels [45, 46]. However, the data of these algorithms were insufficient. Multiple ML models should be used in clinical routine in order that the ensemble of models has a better diagnostic performance than individual one.
The 2D image segmentation method, using a single tumour slice, can make 2D analysis susceptible to the choices of the single representative slice chosen by human experts, whereas 3D analysis is not susceptible to this variation because they cover all tumour volumes. Although it is exceedingly time-consuming to perform manual segmentation using 3D imaging, most tumour characteristics can be captured with 3D tumour volumes. Thus, this may indicate why 3D performs better than 2D, albeit we did not find any significant differences.
The results of this meta-analysis were encouraging owing to its pooled sensitivity of around 0.80, which, however, means that 1 in 5 women that would go with undetected metastases and this may have a detrimental effect on overall survival for 20% of patients with positive SLN status. Although a high NPV of 0.83 means that ML may potentially benefit those with negative SLN, who account for over 70% of all breast cancer patients [7], by helping them eliminate the unnecessary invasive lymph node removal and avoid the overtreatment of axillary fossa accompanied by the associated serious complications, this would translate to 1 in 5 tests being false negative. In view of this, we would like to admit that ML may not be yet usable in routine clinical checkups especially when using patient survival as a primary measurement of outcome.
Several limitations to our meta-analysis are noticeable. First, only fourteen original research articles met the selection criteria as there were not many studies about ALNM in breast cancer patients. We were also unable to retrieve sufficient data for some studies. Second, the algorithm classification performances varied from feature selection to alterations in the linear, quadratic, cubic, and Gaussian kernel functions; therefore, we chose the highest performers that might have affected the performance results. Finally, in all cases, the use of data was from only one single institution, which may not be sufficient to demonstrate the replicability of our findings.
In conclusion, MRI sequences and algorithms constitute the main factors that affect the diagnostic performance of machine learning. Combining SLNB with ML, it would be more helpful to breast cancer patients. In future research, larger, well-designed, conducted, and reported trials are needed for better performances.
Supplementary Information
Additional file 1. Detailed data quality assessment.
Abbreviations
- ALNM
Axillary lymph node metastasis
- AUC
Area under the receiver operating characteristic curve
- CI
Confidence interval
- CNN
Convolutional neural network
- DOR
Diagnostic odds ratio
- LDA
Linear discriminant analysis
- LR
Linear regression
- ML
Machine learning
- NLR
Negative likelihood ratio
- PLR
Positive likelihood ratio
- QUADAS-2
Quality Assessment of Diagnostic Accuracy Studies-2
- SROC
Summary receiver operating characteristic
- SVM
Support vector machines
- T1CE
Contrast-enhanced T1
- T2-FS
Fat-suppressed T2
Authors' contributions
CC and YQ contributed equally in the collection and analysis of data, and in the preparation of the manuscript. FG was responsible for administrative support. XZ was in charge of language editing. HC and DZ were responsible for thoroughly proof reading. All authors read and approved the final manuscript.
Funding
This study has received funding by the National Natural Science Foundation of China (No. 8193000682).
Availability of data and materials
All data generated or analysed during this study are included in this article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not required.
Competing interests
One of the authors of this manuscript (Xiaoyue Zhou) is an employee of Siemens Healthineers. The remaining authors declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Cianfrocca M, Goldstein LJ. Prognostic and predictive factors in early-stage breast cancer. Oncologist. 2004;9(6):606–616. doi: 10.1634/theoncologist.9-6-606. [DOI] [PubMed] [Google Scholar]
- 3.Fitzgibbons PL, Page DL, Weaver D, et al. Prognostic factors in breast cancer. College of American Pathologists Consensus Statement 1999. Arch Pathol Lab Med. 2000;124(7):966–978. doi: 10.5858/2000-124-0966-PFIBC. [DOI] [PubMed] [Google Scholar]
- 4.Benson JR, Della RG. Management of the axilla in women with breast cancer. Lancet Oncol. 2007;8(4):331–348. doi: 10.1016/S1470-2045(07)70103-1. [DOI] [PubMed] [Google Scholar]
- 5.Yip CH, Taib NA, Tan GH, et al. Predictors of axillary lymph node metastases in breast cancer: is there a role for minimal axillary surgery? World J Surg. 2009;33(1):54–57. doi: 10.1007/s00268-008-9782-7. [DOI] [PubMed] [Google Scholar]
- 6.Mansel RE, Fallowfield L, Kissin M, et al. Randomized multicenter trial of sentinel node biopsy versus standard axillary treatment in operable breast cancer: the ALMANAC Trial. J Natl Cancer Inst. 2006;98(9):599–609. doi: 10.1093/jnci/djj158. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed M, Usiskin SI, Hall-Craggs MA, et al. Is imaging the future of axillary staging in breast cancer? Eur Radiol. 2014;24:288–293. doi: 10.1007/s00330-013-3009-5. [DOI] [PubMed] [Google Scholar]
- 8.He X, Sun L, Huo Y, et al. A comparative study of 18F-FDG PET/CT and ultrasonography in the diagnosis of breast cancer and axillary lymph node metastasis. Q J Nucl Med Mol Imaging. 2017;61(4):429–437. doi: 10.23736/S1824-4785.17.02785-6. [DOI] [PubMed] [Google Scholar]
- 9.An YS, Lee DH, Yoon JK, et al. Diagnostic performance of 18F-FDG PET/CT, ultrasonography and MRI. Detection of axillary lymph node metastasis in breast cancer patients. Nuklearmedizin. 2014;53(3):89–94. doi: 10.3413/Nukmed-0605-13-06. [DOI] [PubMed] [Google Scholar]
- 10.Hwang SO, Lee SW, Kim HJ, et al. The comparative study of ultrasonography, contrast-enhanced MRI, and (18)F-FDG PET/CT for detecting axillary lymph node metastasis in T1 breast cancer. J Breast Cancer. 2013;16(3):315–321. doi: 10.4048/jbc.2013.16.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valente SA, Levine GM, Silverstein MJ, et al. Accuracy of predicting axillary lymph node positivity by physical examination, mammography, ultrasonography, and magnetic resonance imaging. Ann Surg Oncol. 2012;19(6):1825–1830. doi: 10.1245/s10434-011-2200-7. [DOI] [PubMed] [Google Scholar]
- 12.Lee MC, Eatrides J, Chau A, et al. Consequences of axillary ultrasound in patients with T2 or greater invasive breast cancers. Ann Surg Oncol. 2011;18(1):72–77. doi: 10.1245/s10434-010-1171-4. [DOI] [PubMed] [Google Scholar]
- 13.Monzawa S, Adachi S, Suzuki K, et al. Diagnostic performance of fluorodeoxyglucose-positron emission tomography/computed tomography of breast cancer in detecting axillary lymph node metastasis: comparison with ultrasonography and contrast-enhanced CT. Ann Nucl Med. 2009;23(10):855–861. doi: 10.1007/s12149-009-0314-9. [DOI] [PubMed] [Google Scholar]
- 14.Guney IB, Dalci K, Teke ZT, et al. A prospective comparative study of ultrasonography, contrast-enhanced MRI and 18F-FDG PET/CT for preoperative detection of axillary lymph node metastasis in breast cancer patients. Ann Ital Chir. 2020;91:458–464. [PubMed] [Google Scholar]
- 15.Villanueva-Meyer JE, Chang P, Lupo JM, et al. Machine learning in neurooncology imaging: from study request to diagnosis and treatment. AJR Am J Roentgenol. 2019;212(1):52–56. doi: 10.2214/AJR.18.20328. [DOI] [PubMed] [Google Scholar]
- 16.Senders JT, Zaki MM, Karhade AV, et al. An introduction and overview of machine learning in neurosurgical care. Acta Neurochir (Wien) 2018;160(1):29–38. doi: 10.1007/s00701-017-3385-8. [DOI] [PubMed] [Google Scholar]
- 17.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 18.Liu M, Mao N, Ma H, et al. Pharmacokinetic parameters and radiomics model based on dynamic contrast enhanced MRI for the preoperative prediction of sentinel lymph node metastasis in breast cancer. Cancer Imaging. 2020;20(1):65. doi: 10.1186/s40644-020-00342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan H, Gan F, Wu Y, et al. Preoperative prediction of axillary lymph node metastasis in breast carcinoma using radiomics features based on the fat-suppressed T2 sequence. Acad Radiol. 2020;27(9):1217–1225. doi: 10.1016/j.acra.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Ren T, Cattell R, Duanmu H, et al. Convolutional neural network detection of axillary lymph node metastasis using standard clinical breast MRI. Clin Breast Cancer. 2020;20(3):e301–e308. doi: 10.1016/j.clbc.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 21.Demircioglu A, Grueneisen J, Ingenwerth M, et al. A rapid volume of interest-based approach of radiomics analysis of breast MRI for tumor decoding and phenotyping of breast cancer. PLoS One. 2020;15(6):e234871. doi: 10.1371/journal.pone.0234871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arefan D, Chai R, Sun M, et al. Machine learning prediction of axillary lymph node metastasis in breast cancer: 2D versus 3D radiomic features. Med Phys. 2020;47:6334–6342. doi: 10.1002/mp.14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Sun D, Chen L, et al. Radiomics analysis of dynamic contrast-enhanced magnetic resonance imaging for the prediction of sentinel lymph node metastasis in breast cancer. Front Oncol. 2019;9:980. doi: 10.3389/fonc.2019.00980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spuhler KD, Ding J, Liu C, et al. Task-based assessment of a convolutional neural network for segmenting breast lesions for radiomic analysis. Magn Reson Med. 2019;82(2):786–795. doi: 10.1002/mrm.27758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Zhong L, Zhang B, et al. The effects of volume of interest delineation on MRI-based radiomics analysis: evaluation with two disease groups. Cancer Imaging. 2019;19(1):89. doi: 10.1186/s40644-019-0276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han L, Zhu Y, Liu Z, et al. Radiomic nomogram for prediction of axillary lymph node metastasis in breast cancer. Eur Radiol. 2019;29(7):3820–3829. doi: 10.1007/s00330-018-5981-2. [DOI] [PubMed] [Google Scholar]
- 27.Cui X, Wang N, Zhao Y, et al. Preoperative prediction of axillary lymph node metastasis in breast cancer using radiomics features of DCE-MRI. Sci Rep. 2019;9(1):2240. doi: 10.1038/s41598-019-38502-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C, Ding J, Spuhler K, et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer by radiomic signatures from dynamic contrast-enhanced MRI. J Magn Reson Imaging. 2019;49(1):131–140. doi: 10.1002/jmri.26224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fusco R, Sansone M, Granata V, et al. Use of quantitative morphological and functional features for assessment of axillary lymph node in breast dynamic contrast-enhanced magnetic resonance imaging. Biomed Res Int. 2018;2018:2610801. doi: 10.1155/2018/2610801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo J, Ning Z, Zhang S, et al. Bag of deep features for preoperative prediction of sentinel lymph node metastasis in breast cancer. Phys Med Biol. 2018;63(24):245014. doi: 10.1088/1361-6560/aaf241. [DOI] [PubMed] [Google Scholar]
- 31.Dong Y, Feng Q, Yang W, et al. Preoperative prediction of sentinel lymph node metastasis in breast cancer based on radiomics of T2-weighted fat-suppression and diffusion-weighted MRI. Eur Radiol. 2018;28(2):582–591. doi: 10.1007/s00330-017-5005-7. [DOI] [PubMed] [Google Scholar]
- 32.Choi BH, Baek HJ, Ha JY, et al. Feasibility study of synthetic diffusion-weighted MRI in patients with breast cancer in comparison with conventional diffusion-weighted MRI. Korean J Radiol. 2020;21(9):1036–1044. doi: 10.3348/kjr.2019.0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westra C, Dialani V, Mehta TS, et al. Using T2-weighted sequences to more accurately characterize breast masses seen on MRI. AJR Am J Roentgenol. 2014;202(3):W183–W190. doi: 10.2214/AJR.13.11266. [DOI] [PubMed] [Google Scholar]
- 34.Liu Z, Li Z, Qu J, et al. Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin Cancer Res. 2019;25(12):3538–3547. doi: 10.1158/1078-0432.CCR-18-3190. [DOI] [PubMed] [Google Scholar]
- 35.Chatterji M, Mercado CL, Moy L. Optimizing 1.5-Tesla and 3-Tesla dynamic contrast-enhanced magnetic resonance imaging of the breasts. Magn Reson Imaging Clin N Am. 2010;18(2):207–224. doi: 10.1016/j.mric.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 36.Stark GF, Hart GR, Nartowt BJ, et al. Predicting breast cancer risk using personal health data and machine learning models. PLoS One. 2019;14(12):e226765. doi: 10.1371/journal.pone.0226765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luckett DJ, Laber EB, El-Kamary SS et al (2020) Receiver operating characteristic curves and confidence bands for support vector machines. Biometrics [DOI] [PMC free article] [PubMed]
- 38.Yu Y, Tan Y, Xie C, et al. Development and validation of a preoperative magnetic resonance imaging radiomics-based signature to predict axillary lymph node metastasis and disease-free survival in patients with early-stage breast cancer. JAMA Netw Open. 2020;3(12):e2028086. doi: 10.1001/jamanetworkopen.2020.28086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takada M, Sugimoto M, Naito Y, et al. Prediction of axillary lymph node metastasis in primary breast cancer patients using a decision tree-based model. BMC Med Inform Decis Mak. 2012;12:54. doi: 10.1186/1472-6947-12-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schacht DV, Drukker K, Pak I, Abe H, Giger ML, et al. Using quantitative image analysis to classify axillary lymph nodes on breast MRI: a new application for the Z 0011 Era. Eur J Radiol. 2015;84(3):392–397. doi: 10.1016/j.ejrad.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ha R, Chang P, Karcich J, et al. Axillary lymph node evaluation utilizing convolutional neural networks using MRI dataset. J Digit Imaging. 2018;31(6):851–856. doi: 10.1007/s10278-018-0086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lorena AC, Jacintho LFO, Siqueira MF, et al. Comparing machine learning classifiers in potential distribution modelling. Exp Syst Appl. 2011;38(5):5268–5275. doi: 10.1016/j.eswa.2010.10.031. [DOI] [Google Scholar]
- 43.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 44.Pan X, Luo Y, Xu Y. K-nearest neighbor based structural twin support vector machine. Knowl Based Syst. 2015;88:34–44. doi: 10.1016/j.knosys.2015.08.009. [DOI] [Google Scholar]
- 45.Wang F, Liu X, Yuan N, et al. Study on automatic detection and classification of breast nodule using deep convolutional neural network system. J Thorac Dis. 2020;12(9):4690–4701. doi: 10.21037/jtd-19-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin HC, Roth HR, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Detailed data quality assessment.
Data Availability Statement
All data generated or analysed during this study are included in this article.