Abstract
Streptococcal pyrogenic exotoxin A (SPE-A) and SPE-B have been implicated in the pathogenesis of severe group A streptococcal (GAS) disease. We studied 31 invasive GAS strains including 18 isolates from patients with toxic shock syndrome and 22 noninvasive strains isolated in The Netherlands between 1994 and 1998. These strains were associated with the different allelic variants of the gene encoding SPE-A. We selected endemic strains with speA-positive M and T serotypes: speA2-associated M1T1 and M22-60T12 strains, speA3-associated M3T3 strains, and speA4-associated M6T6 strains. Since speA1-positive isolates were not frequently encountered, we included speA1 strains of different serotypes. The GAS strains were compared genotypically by pulsed-field gel electrophoresis and phenotypically by the in vitro production of SPE-A and SPE-B. All strains within one M and T type appeared to be of clonal origin. Most strains produced SPE-A and SPE-B, but only a minority of the speA4-positive isolates did so. Among our isolates, speA1- and speA3-positive strains produced significantly more SPE-A than speA2- and speA4-carrying strains, while SPE-B production was most pronounced among speA1- and speA2-containing strains. There was a marked degree of variability in the amounts of exotoxins produced in vitro by strains that shared the same genetic profile. We conclude that the differences in the in vitro production of SPE-A and SPE-B between our selected strains with identical M and T types were not related to either genetic heterogeneity or the clinical course of GAS disease in the patient from whom they were isolated.
Although group A streptococcal (GAS) isolates are generally considered noninvasive pathogens that cause localized infections of nasopharyngeal mucosal surfaces and the skin, increasing numbers of reports have described invasive infections worldwide that result in necrotizing fasciitis, a toxic shock-like syndrome (TSS), and death (7, 15, 29, 38, 40, 44). Despite significant study, the cause of episodic changes in the severity of GAS disease has not yet been elucidated. Several factors have been implicated in the pathogenesis of severe GAS infections, including a decrease in herd immunity and the introduction of highly virulent mutant strains. The emergence of certain strain types, like M1T1 and M3T3, among invasive GAS isolates symbolizes the epidemiology of GAS pathogenicity (10, 14, 15, 29, 40, 41, 45). After a long period of absence, these strains, which are associated with streptococcal pyrogenic exotoxin A (SPE-A), started reappearing in several countries. SPEs belong to the major virulence factors in the pathogenesis of severe GAS infections. They show a remarkable degree of homology with staphylococcal enterotoxins B and C and are considered superantigens, exerting a series of important biological effects on the host, including the massive release of cytokines and the consequent induction of fever, erythematous skin reactions, and polyclonal T-cell activation (1, 3, 5, 6, 24, 39).
Different streptococcal exotoxins are known, and these have been designated SPE-A, SPE-B, SPE-C, SPE-F, and streptococcal superantigen (17, 27, 35, 44, 45). The highly conservative chromosomal gene encoding SPE-B (speB) is present in practically 100% of the GAS strains (4). It has been documented that SPE-B is identical to or is an allelic variant of streptococcal proteinase precursor (12, 13); it cleaves the interleukin 1β precursor and extracellular matrix proteins such as fibronectin (17, 18). Moreover, SPE-B appears to be involved in tissue invasion and destruction (22, 23). In addition, several reports have suggested an important role for SPE-B in the pathogenesis of serious GAS disease (15, 22, 23, 34). SPE-A and SPE-C are phage encoded, and their presence is restricted to a limited number of strains (29, 46). Far more streptococcal isolates from patients with TSS, other invasive GAS disease, or scarlet fever have been reported to produce SPE-A or at least to possess the gene that encodes SPE-A (speA) than GAS isolates in general (14, 20, 21, 29, 40, 43, 44). Four naturally occurring isotypes of SPE-A have been described: SPE-A1, SPE-A2, SPE-A3, and SPE-A4. The newer variants, SPE-A2 and SPE-A3, differ from the ancient SPE-A1 by one amino acid, whereas the most recently described isotype, SPE-A4, shows only 91% homology with the other allelic variants (32).
In this investigation, we studied speA-positive GAS isolates with certain M types which were isolated frequently in The Netherlands from 1994 to 1998 for the estimation of SPE-A and SPE-B production in vitro by comparing isolates that carry different speA alleles. For this purpose, we developed a sensitive technique for the detection of SPE-A and SPE-B produced by GAS isolates. Using this assay, we collected quantitative information on the production of SPE-A and SPE-B and related our data to the presence of speA1, speA2, speA3, or speA4 alleles in speA-positive GAS strains. In this study, we made a distinction between invasive isolates from patients with and without TSS and noninvasive throat isolates. Genetic heterogeneity among strains of the same M type was determined by pulsed-field gel electrophoresis (PFGE).
MATERIALS AND METHODS
Microorganisms.
A total of 53 clinical GAS strains were collected from Dutch patients during the period from 1994 to 1998 within the framework of a nationwide GAS surveillance program. Thirty-one invasive GAS strains were recovered from normally sterile sites. Twenty-two noninvasive strains were obtained from pharyngitis patients, who showed no signs of invasive infection. All GAS strains originated from different patients and were stored at −70°C upon receipt. Laboratory strains NY-5 (SPE-A positive), 279 (SPE-A negative but SPE-B positive), and A95/216 (SPE-B negative) were used as assay controls.
Strain typing.
For the characterization of isolates, T serotyping and M genotyping were performed as described by Kaufhold et al. (19). In addition, PCR technology was used to detect the speA, speB, and speC genes. Biotin-labeled oligonucleotide probes specific for speA1 (AACGTTGATATTTATGGT), speA2 (GATAAAAACATTGATATT), speA3 (GATATTTATAGTGTAGAA), and speA4 (GAAGAGCGTGTATTTATGGAGGGG) were used in a Southern blot hybridization of the PCR products to differentiate between the exotoxin variants as described by Schouls (42). Invasive and superficial GAS strains positive for speA1, speA2, speA3, or speA4 were selected, subjected to molecular typing, and tested for their production of SPE-A and SPE-B.
PFGE.
PFGE was performed with the Genepath group 1 reagent kit (Bio-Rad, Hercules, Calif.) as described in the instruction manual. Briefly, in situ cell lysis was carried out by overnight incubation at 37°C in Lysis Buffer I and lysozyme-lysostaphin. Proteolysis was achieved by overnight incubation at 50°C with proteinase K in proteinase K buffer. Then, the plugs were washed thoroughly and stored at 4°C. Next, DNA inserts were digested overnight at room temperature with 25 U of the SmaI enzyme in SmaI buffer per plug, followed by separation of the fragments at 180 V with a CHEF-DR II apparatus (Bio-Rad) with pulse times ranging from 5 to 50 s over 24 h. Polymerized bacteriophage lambda DNA standards (Bio-Rad) were used as molecular size markers. Gels were stained with ethidium bromide and were photographed under UV light, after which the PFGE patterns were compared visually. Restriction analysis of strains belonging to the same M type was performed simultaneously in the same run.
Culture and production of SPE-A and SPE-B.
Aliquots of freshly thawed bacteria were grown on fresh blood agar plates and were subsequently cultured in Todd-Hewitt broth (Difco, Detroit, Mich.) supplemented with 0.3% (wt/vol) glucose, 0.2% (wt/vol) NaHCO3, 0.2% (wt/vol) NaCl, 0.08% (wt/vol) Na2HPO4, and 0.02% (wt/vol) l-glutamine. The strains were sequentially grown twice at 37°C for 3 h to reach the logarithmic phase, followed by overnight culture to attain the stationary phase. The bacteria were spun down, and supernatants from the final culture were concentrated six times by ethanol precipitation. Subsequently, exotoxin-enriched preparations were subjected to inhibition enzyme-linked immunosorbent assay (ELISA) for the estimation of SPE-A and SPE-B production.
Antigens.
SPE-A was isolated from GAS strain NY-5 as described previously (26). SPE-B was kindly provided by J. M. Musser (Baylor College of Medicine, Houston, Tex.). Both antigens were ≥99% pure, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis combined with silver staining (26).
Antibodies against SPE-A and SPE-B.
High-titer serum from a selected healthy individual was used as the source of polyclonal antibodies against SPE-A and SPE-B.
Detection of SPE-A and SPE-B produced by GAS strains.
SPE production by the GAS strains to be tested was determined by competitive ELISAs. Ninety-six-well flat-bottom microtiter ELISA plates (3590; Costar, Cambridge, Mass.) were coated for 1 h with 100 μl of SPE-A or SPE-B (0.5 μg/ml in saline). Unless otherwise mentioned, all incubation steps were performed at 37°C. Phosphate-buffered saline supplemented with 0.1% Tween 20 was used in the washing procedures. Blocking was performed with 150 μl of 0.1% gelatin in twice-distilled water. Concentrated supernatants were then serially diluted in 96-well, flat-bottom microtiter plates (Greiner GmbH, Frickenhausen, Germany), mixed 1:1 with our high-titer reference serum (final concentration, 1:2,000), and incubated at 37°C for 1 h until equilibrium was reached. Aliquots of 100 μl were pipetted into the coated plates, and the plates were incubated for 1 h. Next, the plates were incubated with peroxidase-labeled sheep anti-human immunoglobulin G (IgG), and the ELISAs were developed with a hydrogen peroxide–3,3′,5,5′-tetramethylbenzidine mixture as the substrate. The optical densities (ODs) in the wells were determined with an ELISA microplate reader (Bio-Rad) operated at 450 nm. The OD values measured in the absence of the supernatant were taken as 100% reference values. Supernatants from strains NY-5 and 279 were used as SPE-A-positive and SPE-A-negative controls, respectively, while supernatants from strains 279 and 95/126 were used as SPE-B-positive and SPE-B-negative controls, respectively. All experiments were performed in triplicate. The inhibition of ELISA reactivity was used as a measure for SPE-A or SPE-B. Supernatant preparations from the strains which were included in this study were tested at five dilutions in washing buffer: as such and at 1:3, 1:10, 1:30, and 1:100. We used a 30% reduction of the positive control OD at 450 nm (OD450) as the threshold for exotoxin production. The highest dilution of concentrated supernatant which induced significant inhibition was used for the quantitation of toxin production. SPE production was depicted in arbitrary units, which represent the reciprocal value of the highest dilution of supernatant that induced ≥30% inhibition of ELISA reactivity. In our procedures, the lower detection limits for SPE-A and SPE-B were 5 and 1 ng/ml, respectively.
Statistics.
The nonparametric Kruskal-Wallis test was used to compare the production of SPE-A or SPE-B by GAS strains of different M/T types. Differences with P values below 0.05 were considered significant.
RESULTS
Typing of bacterial isolates.
After M and T typing and the determination of spe profiles, 53 clinical GAS isolates, of which 22 were noninvasive and 31 were invasive (18 isolates from patients with TSS and 13 isolates from patients with less severe infections), were included in the study. All isolates harbored speA and speB genes. The bacterial isolates comprised 17 M1T1 strains, 17 M3T3 strains, 5 M6T6 strains, and 9 M22-60T12 strains. Strains that possessed the speA1 allele were sporadically identified among both invasive and noninvasive isolates and were not found in association with any particular M or T strain type; five speA1-positive isolates of different types were included. The speA alleles and the clinical sources of the isolates are shown in Table 1.
TABLE 1.
M and T types, speA profiles, and sources of 53 speA-positive clinical GAS strainsa
| M and T type | Clinical status of source patient (no. of isolates) | speA allele |
|---|---|---|
| M49T8 | TSS (1) | speA1 |
| M49T14 | TSS (1) | speA1 |
| MntT? | Sepsis (1) | speA1 |
| MntT14 | Pharyngitis (1) | speA1 |
| MntT23 | Pharyngitis (1) | speA1 |
| M1T1 | TSS (10) | speA2 |
| M1T1 | Sepsis (1) | speA2 |
| M1T1 | Arthritis (1) | speA2 |
| M1T1 | Pharyngitis (5) | speA2 |
| M22-60T12 | TSS (1) | speA2 |
| M22-60T12 | Meningitis (1) | speA2 |
| M22-60T12 | Pharyngitis (7) | speA2 |
| M3T3 | TSS (5) | speA3 |
| M3T3 | Arthritis (3) | speA3 |
| M3T3 | Pneumonia (2) | speA3 |
| M3T3 | Sepsis (2) | speA3 |
| M3T3 | Pharyngitis (5) | speA3 |
| M6T6 | Meningitis (1) | speA4 |
| M6T6 | Necrotizing fasciitis (1) | speA4 |
| M6T6 | Pharyngitis (3) | speA4 |
Of the 53 strains, 22 were noninvasive strains and 31 were invasive strains.
A 100% correlation was observed between the following combinations: M1T1 strains and the speA2 gene, M22-60T12 strains and the speA2 gene, M3T3 strains and the speA3 gene, and M6T6 strains and the speA4 gene. All M and T types included were recovered from both invasive and noninvasive sites; M22-60T12 strains were predominantly isolated from pharyngeal swabs and were only sporadically isolated from normally sterile body compartments.
PFGE.
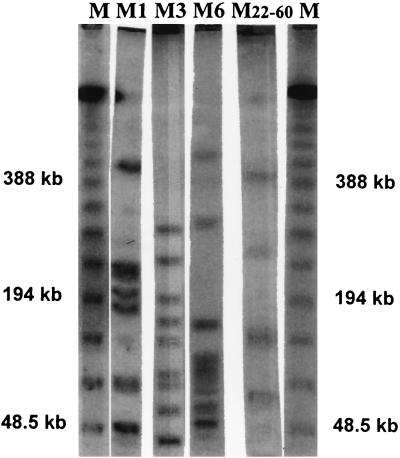
All isolates were subjected to PFGE, and most banding patterns between isolates that carried the same M protein were indistinguishable when restriction was done with SmaI (Fig. 1). For none of the strains were more than two fragment differences observed between strains of similar M and T types. Thus, all of the strains of a particular M and T type were considered to be of similar clonal origin. No major differences in PFGE patterns were observed between isolates of identical M and T types recovered from patients with TSS, otherwise invasive GAS isolates, or isolates from patients with pharyngitis. The five speA1-positive isolates each belonged to different serotypes, and all had different PFGE profiles (data not shown).
FIG. 1.
PFGE banding patterns of GAS strains specific for different M serotypes. Lanes: M, marker; M1, M1T1; M3, M3T3; M6, M6T6; M22-60, M22-60T12.
Production of SPE-A and SPE-B.
The production of SPE-A and SPE-B by these GAS strains was quantitated by competitive ELISA. Exotoxin-enriched supernatants were first incubated in solution with high-titer human serum at fixed concentrations as a source of anti-SPE-A and anti-SPE-B antibodies until equilibrium was reached. The concentration of free antibodies was then determined by ELISA. In order to guarantee the specificity of the assay, inhibition of ELISA reactivity was determined with supernatant preparations for negative control strains (strain 279 for SPE-A production and strain A95/126 for SPE-B production). OD values for the plates coated with SPE-A or SPE-B were identical to the values observed for the plates with Todd-Hewitt broth growth medium, indicating that cross-reactivity between the coating and other GAS products excreted into the supernatant did not occur. The supernatant preparation for strain NY-5, our source for the purification of SPE-A, was used as a positive control for the production of SPE-A (Fig. 2).
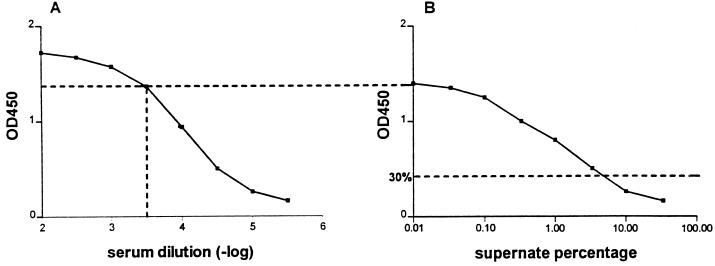
FIG. 2.
Principles of competitive ELISA for SPE-A. (A) The serum concentration appropriate for the competitive ELISA was deduced from the linear part of the dose-response curve obtained in the ELISA for SPE-A. (B) Supernatant preparations of the strains were incubated at several concentrations to equilibrium with a fixed serum concentration as determined from panel A. SPE-A-coated ELISA plates were incubated with this equilibrated mixture, and unbound antibody levels were subsequently determined. A 30% inhibition of the OD450 was taken as the lower threshold to designate a exotoxin-producing strain.
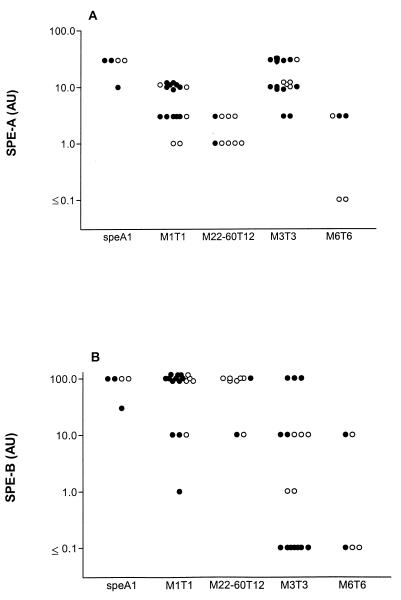
Similarly, the supernatant preparation for strain 279 was tested as a positive control for the production of SPE-B. The production of SPE-A and SPE-B by individual GAS strains is presented in Fig. 3. The majority of isolates were found to produce SPE-A as well as SPE-B in vitro. Significant differences in the amounts of SPE-A and SPE-B produced were observed between groups of strains with different speA allelic variants (P < 0.0005). Of the clinical isolates tested, SPE-A was detected in supernatant preparations for all speA1-, speA2-, and speA3-positive strains. speA4-positive strains did not always cause inhibition. The speA1- and speA3-positive strain types did not produce significantly different concentrations of SPE-A. Furthermore, no significant differences in SPE-A production were demonstrated between M22-60T12 and M6T6 strains. speA1 and speA3 strains produced significantly larger amounts of SPE-A than speA2 and speA4 strains (P < 0.005). The level of SPE-A production by M1T1 strains was lower than that by speA1 strains and speA3 M3T3 strains (P < 0.001) but was higher than that by M22-60T12 and M6T6 strains (P = 0.04).
FIG. 3.
Production of SPE-A (A) and SPE-B (B) by clinical GAS isolates as determined by competitive ELISA. SPE production is depicted in arbitrary units (AU), which represent the reciprocal value of the lowest dilution of supernatant which induced ≥30% inhibition of ELISA reactivity. Invasive isolates are represented by closed dots; noninvasive isolates are represented by open dots.
Overall, SPE-B was detected at higher concentrations than SPE-A. No significant differences in the levels of SPE-B were observed between speA1 and speA2 strains. In addition, no significant differences in the levels of SPE-B were found between speA3 and speA4 isolates. The level of production of SPE-B by speA1 isolates and speA2-positive strains of the M1T1 or M22-60T12 serotypes was significantly higher than that by M3T3 and M6T6 strains, some of which did not synthesize measurable amounts of SPE-B (P < 0.0005). The levels of exotoxin production in broth medium by strains of identical M types varied widely from none to strong, but no significant differences in SPE production by strains from patients with TSS, otherwise invasive strains, or noninvasive strains were observed.
DISCUSSION
In this study, we investigated the production of SPE-A and SPE-B by 53 endemic GAS strains and compared the levels of production by strains carrying different speA allelic variants. For this purpose, we included serotypes M1T1, M22-60T12, M3T3, and M6T6, which were recovered from patients with severe or mild GAS disease in The Netherlands during the period from 1994 to 1998 and which corresponded to the different speA allelic variants. Consequently, the isolates that were analyzed did not represent the wide variety of GAS types present in the community. In accordance with other Dutch M1T1, M3T3, and M6T6 types, all our strains hybridized with a probe specific for speA (40). M1T1 and M3T3 types accounted for about 50% of the Dutch isolates from patients with TSS, but the same types were also, now and then, encountered as causes of noninvasive infections (40, 41). Conspicuously, M22-60T12 isolates were often recognized among strains from patients with pharyngitis. Since the speA4 allele was detected only in M6T6 isolates, M6T6 types were included in the present study as well. Since speA1-positive strains only rarely occurred, we included five speA1-positive isolates which were of different M and T types for the detection of the production of exotoxins A1 and B. All M1T1 and M22-60T12 types contained speA2, all M3T3 types contained speA3, and all M6T6 types contained speA4. These data are in line with those from Swedish and Finnish studies that showed that all M1 strains possess the speA gene (15, 28, 33). In striking contrast, the connection between M1 and M3 strains and the speA gene among U.S. isolates was considerably lower (8). These large discrepancies with regard to the occurrence of speA may be related to the geographic region of where the strain is isolated.
PFGE was performed to discover whether toxin expression could be related to the genetic differences within strains of similar M and T types. The value of PFGE with SmaI in the molecular epidemiological typing of Streptococcus pyogenes, has been well established, yielding suitable and discriminatory macrorestriction patterns (2, 43, 47). Among the restricted numbers of isolates in our study, we could not detect significant differences in banding patterns between strains that shared identical M and T types. Moreover, regardless of the sources of the clinical isolates, all GAS isolates with similar M and T types appeared to be derived from the same genetic lineage, even though they may have originated from different regions of the country. These results are in line with those of others who did not find a correlation between the genomic type and the severity or outcome of disease among M1T1 isolates (28, 33). Accordingly, several other previous reports have demonstrated a strong clonality of M1T1 strains from different parts of the world (10, 25, 29–31, 43). While substantial genetic diversity among M1-expressing organisms has, however, been described, a single subclone that carries the speA gene and that has been recovered worldwide, including The Netherlands, was involved in most invasive episodes (25, 28, 31, 33). It seems obvious that our M1 isolates belong to this globally spread clone, which has been designated restriction fragment length polymorphism type 1a. Furthermore, Upton et al. (47) recently identified two subclones within the M3 type, and both of them contain the speA gene.
Some investigators reported a direct correlation between logarithmic growth and SPE-A production (16), while others mentioned that SPE-A is produced mainly during the short stationary interphases which interrupt growth (36). In contrast, SPE-B production was detected only when cultures entered the stationary phase (9). In our study, we tested exotoxin production in bacterial supernatant preparations at several time points in 1- to 24-h cultures starting from fresh mid-logarithmic-phase cultures. From these experiments, exotoxin production appeared to be most pronounced with two sequential incubations to the mid-logarithmic phase, followed by an overnight culture. The results indicated that the majority of our endemic strains containing the speA gene are indeed producers of exotoxins A and B in vitro. Purified SPE-A from laboratory strain NY-5 and human serum containing polyspecific IgG were used for the estimation of SPE-A production; considerable inhibition of ELISA reactivity was reached by all allelic SPE-A variants. Therefore, we conclude that anti-SPE-A antibodies are cross-reactive with all four allelic variants of SPE-A. As far as levels of SPE-A production were concerned, speA1 and speA3 strains induced significantly stronger inhibition than speA2 and speA4 strains. We considered the possibility that the anti-SPE-A antibodies in our reagent serum present a lower affinity or neutralizing capacity toward SPE-A2, SPE-A3, and especially, SPE-A4 than toward SPE-A1; SPE-A1 differs from SPE-A2 and SPE-A3 by only one amino acid, while the degree of homology with SPE-A4 is considerably lower. These differences may have influenced the results, since our ELISA plates were coated with NY-5-derived SPE-A1. However, subjection of GAS supernatant preparations to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining yielded results which were in line with the data obtained by inhibition ELISA (data not shown).
Our high-titer serum also appeared to be appropriate for the recognition of SPE-B. Our isolates generally produced SPE-B in larger amounts than SPE-A. Strong exotoxin B production was common in speA1 and speA2 strains, while nonproducers were found among the speA3 and speA4 strains. Interestingly, an enormous variation in the amounts of both SPE-A and SPE-B produced by strains of identical M and T types was noticed. Since these strains were indistinguishable by PFGE and were very likely genetically related, we conclude that toxin production is regulated at the transcription level. It is important to bear in mind that toxin production in vitro may not accurately reflect the regulation of protein expression in the host. In addition, it is not clear whether SPE-A production by invasive strains occurs in response to environmental conditions encountered in the host during tissue invasion and/or whether the toxin contributes to the invasiveness of the strain. The systemic effects in necrotizing fasciitis and TSS may be due to synergistic effects involving multiple streptococcal products. It is possible that speA is genetically linked to one or more other genes involved in TSS. Recently, Cleary et al. (11) reported that SPE-A expression is genetically unstable, suggesting the existence of a common regulatory circuit linking intracellular invasion, the M protein, the hyaluronic acid capsule, and SPE-A expression and providing a reversible genetic switch mechanism by which SPE-A and M1 expression could both vary (11).
Surprisingly, we observed higher percentages of SPE-A-producing strains than several other investigators: Talkington et al. (45) reported that 50% of M1 and M3 strains expressed SPE-A, Hauser et al. (14) observed 100% SPE-A production by M3 strains versus 20% SPE-A production by M1 strains, and Chaussee et al. (8) mentioned 43% SPE-A production by invasive speA-positive isolates. Holm and coworkers (15, 33) concluded that Scandinavian M1 strains produce very little or no SPE-A but large amounts of SPE-B, regardless of the clinical condition. We ascribe the discrepancies between our results and those from other groups to differences in procedures. Relative to other techniques that have been used (14–16, 21), the inhibition ELISA that we used is a very sensitive technique for the estimation of SPE-A and SPE-B, with both exotoxins being detected in nanogram amounts. The high-titer human serum that we used proved to be an excellent source of polyclonal antibodies against SPE-A and SPE-B. Furthermore, hyaluronic acid, which at high levels might interfere with the detection of SPE-A, did not disturb our assays, because the concentration factor of the bacterial supernatant preparations did not exceed sixfold.
Our data do not support the hypothesis that the clinical source of the GAS isolate determines the in vitro production of either exotoxin. With regard to this particular point, our data are in line with those of others who suggest an absence of a correlation between SPE-B production in vitro and the severity of GAS disease or any particular M type (8, 15, 33, 37, 45). A strong association between SPE-A production and the isolation of SPE-A-producing strains from patients with TSS has, however, been observed (14, 21, 29, 45), although the speA-positive TSS strains in those studies were compared with speA-negative strains isolated from other sources as well. Interestingly, a study with isolates with identical restriction fragment length polymorphism patterns within families showed that while the speA gene was maintained in all isolates, only invasive isolates expressed SPE-A in vitro (8).
The increase in the incidence of invasive GAS disease is accompanied by a shift in the appearance of speA alleles. Remarkably, the speA1 allele was already observed in GAS strains isolated in the first half of the century. Nowadays, however, speA1 is rarely found in current GAS strains, while speA2 and speA3 are frequently encountered (30, 32). The allelic variants of SPE-A may display qualitative or quantitative heterogeneity in one or more of the functions ascribed to SPE-A. Thus, it might be speculated that the allelic change from speA1 into speA2 or speA3 contributes to the progressive virulence of the bacteria. Bradford Kline and Collins (6) demonstrated that SPE-A3 has significantly enhanced mitogenic activity and affinity for class II MHC molecules those of SPE-A1. In contrast, SPE-A2 has slightly higher affinity for class II molecules than SPE-A1 but no increased mitogenic activity (6). Minor increases in production combined with the enhanced activities of exotoxin A could, however, result in disproportionate increases in toxicity and thus increased virulence in the host. This might be the result of both higher-level toxin production by SPE-A-positive strains compared with the for strains that do not synthesize SPE-A and a reflection of the greater toxicity of the current SPE-A alleles compared with that of the ancient SPE-A or other streptococcal toxins (20, 21).
In conclusion, the majority of speA-positive GAS strains endemic in The Netherlands synthesized SPE-A and SPE-B in vitro. In general, the selected M and T serotypes in combination with the speA allelic variants appeared to be correlated with the expression of exotoxins, with SPE-A production being most pronounced in our speA1 and speA3 strains and SPE-B production being most significant in our speA1 and speA2 strains. Nevertheless, genetically related isolates of identical M and T serotypes showed wide variations in levels of SPE-A and SPE-B excretion in vitro, but these variations were not decisive for the clinical course of disease: there were no significant differences in the levels of in vitro production of SPE-A and SPE-B by strains from patients with TSS, otherwise invasive strains, or noninvasive strains.
ACKNOWLEDGMENTS
We thank J. M. Musser for supplying SPE-B.
This work was supported by the Dutch Praeventie Fonds (project 002824180).
REFERENCES
- 1.Abe J, Forrester J, Nakahara T, Laferty J A, Kotzin B L, Leung D Y M. Selective stimulation of human T cells with streptococcal erythrogenic toxins A and B. J Immunol. 1991;146:3747–3750. [PubMed] [Google Scholar]
- 2.Bert F, Branger C, Lambert-Zechovsky N. Pulsed-field gel electrophoresis is more discriminating than multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for typing pyogenic streptococci. Curr Microbiol. 1997;34:226–229. doi: 10.1007/s002849900173. [DOI] [PubMed] [Google Scholar]
- 3.Betley M J, Borst D W, Regassa L B. Staphylococcal enterotoxins, toxic shock syndrome toxin and streptococcal pyrogenic exotoxins: a comparative study of their molecular biology. Chem Immunol. 1992;55:1–35. [PubMed] [Google Scholar]
- 4.Black C M, Talkington D F, Messmer T O, Facklam R R, Hornes E, Olsvik O. Detection of streptococcal pyrogenic exotoxin genes by a nested polymerase chain reaction. Mol Cell Probes. 1993;7:255–259. doi: 10.1006/mcpr.1993.1038. [DOI] [PubMed] [Google Scholar]
- 5.Bohach G A, Fast D J, Nelson R D, Schlievert P M. Staphylococcal and streptococcal pyrogenic toxins involved in toxic shock syndrome and related illnesses. Crit Rev Microbiol. 1990;17:251–272. doi: 10.3109/10408419009105728. [DOI] [PubMed] [Google Scholar]
- 6.Bradford Kline J, Collins C M. Analysis of the superantigenic activity of mutant and allelic forms of streptococcal pyrogenic exotoxin A. Infect Immun. 1996;64:861–869. doi: 10.1128/iai.64.3.861-869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carapetis C, Robins-Browne R, Martin D, Shelby-James T, Hogg G. Increasing severity of invasive group A streptococcal disease in Australia: clinical and molecular epidemiological features and identification of a new virulent M-nontypable clone. Clin Infect Dis. 1995;21:1220–1227. doi: 10.1093/clinids/21.5.1220. [DOI] [PubMed] [Google Scholar]
- 8.Chaussee M S, Liu J, Stevens D L, Ferretti J J. Genetic and phenotypic diversity among isolates of Streptococcus pyogenes from invasive infections. J Infect Dis. 1996;173:901–908. doi: 10.1093/infdis/173.4.901. [DOI] [PubMed] [Google Scholar]
- 9.Chaussee M S, Phillips E R, Feretti J J. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect Immun. 1997;65:1956–1959. doi: 10.1128/iai.65.5.1956-1959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleary P P, Kaplan E L, Handley J P, et al. Clonal basis for resurgence of serious Streptococcus pyogenes disease in the 1980s. Lancet. 1992;339:518–521. doi: 10.1016/0140-6736(92)90339-5. [DOI] [PubMed] [Google Scholar]
- 11.Cleary P P, McLandsborough L, Ikeda L, Cue D, Krawczak J, Lam H. High-frequency intracellular infection and erythrogenic toxin A expression undergo phase variation in M1 group A streptococci. Mol Microbiol. 1998;28:157–167. doi: 10.1046/j.1365-2958.1998.00786.x. [DOI] [PubMed] [Google Scholar]
- 12.Gerlach D, Knöll H, Köhler W, Ozegowski J H, Hribalova V. Isolation and characterization of erythrogenic toxins. V. Identity of erythrogenic toxin type B and streptococcal proteinase precursor. Zentol Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig. 1983;255:221–233. [PubMed] [Google Scholar]
- 13.Hauser A R, Schlievert P M. Nucleotide sequence of the streptococcal pyrogenic exotoxin type B gene and relationship between the toxin and the streptococcal proteinase precursor. J Bacteriol. 1990;172:4536–4542. doi: 10.1128/jb.172.8.4536-4542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hauser A R, Stevens D L, Kaplan E L, Schlievert P M. Molecular analysis of pyrogenic exotoxins from Streptococcus pyogenes associated with toxic shock-like syndrome. J Clin Microbiol. 1991;29:1562–1567. doi: 10.1128/jcm.29.8.1562-1567.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm S E, Norrby A, Bergholm A, Norgren M. Aspects of pathogenesis of serious group A streptococcal infections in Sweden, 1988–1989. J Infect Dis. 1992;166:31–37. doi: 10.1093/infdis/166.1.31. [DOI] [PubMed] [Google Scholar]
- 16.Houston C W, Feretti J J. Enzyme-linked immunosorbent assay for detection of type A streptococcal exotoxin: kinetics and regulation during growth of Streptococcus pyogenes. Infect Immun. 1981;33:862–869. doi: 10.1128/iai.33.3.862-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapur V, Topouzis S, Majesky M W, et al. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb Pathog. 1993;15:327–346. doi: 10.1006/mpat.1993.1083. [DOI] [PubMed] [Google Scholar]
- 18.Kapur V, Majewski M W, Li L-L, Black R A, Musser J M. Cleavage of interleukin 1β precursor to produce active IL-1β by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc Natl Acad Sci USA. 1993;90:7676–7680. doi: 10.1073/pnas.90.16.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufhold A, Podbielski A, Baumgarten G, Blokpoel M, Top J, Schouls L. Rapid typing of group A streptococci by the use of DNA amplification and non-radioactive allele-specific oligonucleotide probes. FEMS Microbiol Lett. 1994;119:19–26. doi: 10.1111/j.1574-6968.1994.tb06861.x. [DOI] [PubMed] [Google Scholar]
- 20.Köhler W, Gerlach D, Knöll H. Streptococcal outbreaks and erythrogenic toxin type A. Zentbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe A. 1987;266:104–115. doi: 10.1016/s0176-6724(87)80024-x. [DOI] [PubMed] [Google Scholar]
- 21.Lee, P. K., and P. M. Schlievert. Quantification and toxicity of group A streptococcal pyrogenic exotoxins in an animal model of toxic shock syndrome-like illness. J. Clin. Microbiol. 27:1890–1892. [DOI] [PMC free article] [PubMed]
- 22.Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser J M. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decrease mouse lethality of serotype M3 and M49 strains. J Clin Invest. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukomski S, Burns E H, Wyde P R, Podbielski A, Rurangirwa J, Moore-Poveda D K, Musser J M. Genetic inactivation of an extracellular cysteine protease (SpeB) expressed by Streptococcus pyogenes decreases resistance to phagocytosis and dissemination to organs. Infect Immun. 1998;66:771–776. doi: 10.1128/iai.66.2.771-776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marrack P, Kappler J. The staphylococcal enterotoxins and their relatives. Science. 1990;248:705–711. doi: 10.1126/science.2185544. [DOI] [PubMed] [Google Scholar]
- 25.Martin D R, Single L A. Molecular epidemiology of group A streptococcus M type 1 infections. J Infect Dis. 1993;167:1112–1117. doi: 10.1093/infdis/167.5.1112. [DOI] [PubMed] [Google Scholar]
- 26.Mascini E M, Hazenberg M A J, Verhage E A E, Holm S E, Verhoef J, Van Dijk H. A new procedure for the purification of streptococcal pyrogenic exotoxin A from Streptococcus pyogenes supernatant. Clin Diagn Lab Immunol. 1996;3:779–781. doi: 10.1128/cdli.3.6.779-781.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mollick J A, Miller G G, Musser J M, et al. A novel superantigen isolated from pathogenic strains of Streptococcus pyogenes with amino-terminal homology to staphylococcal enterotoxins B and C. J Clin Invest. 1993;92:710–719. doi: 10.1172/JCI116641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muotiala A, Seppälä H, Huovinen P, Vuopio-Varkila J. Molecular comparison of group A streptococci of T1M1 serotype from invasive and noninvasive infections in Finland. J Infect Dis. 1997;175:392–399. doi: 10.1093/infdis/175.2.392. [DOI] [PubMed] [Google Scholar]
- 29.Musser J M, Hauser A R, Kim M H, Schlievert P M, Nelson K, Selander R K. Streptococcus pyogenes causing toxic-shock-like syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc Natl Acad Sci USA. 1991;88:2668–2672. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musser J M, Kapur V, Kanjilal S, Shah U, Musher D M, Barg N L, Johnston K H, Schlievert P M, Henrichsen J, Gerlach D, Rakita RM, Tanna A, Cookson B D, Chang J C. Geographic and temporal distribution and molecular characterization of two highly pathogenic clones of Streptococcus pyogenes expressing allelic variants of pyrogenic exotoxin A (scarlet fever toxin) J Infect Dis. 1993;167:337–346. doi: 10.1093/infdis/167.2.337. [DOI] [PubMed] [Google Scholar]
- 31.Musser J M, Kapur V, Szeto J, Pan X, Swanson D S, Martin D R. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelson K, Schlievert P M, Selander R K, Musser J M. Characterization and clonal distribution of four alleles of the spe-A gene encoding pyrogenic exotoxin A (scarlet fever toxin) in Streptococcus pyogenes. J Exp Med. 1991;174:1271–1274. doi: 10.1084/jem.174.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norgren M, Norrby A, Holm S E. Genetic diversity in T1M1 group A streptococci in relation to clinical outcome of infection. J Infect Dis. 1992;166:1014–1020. doi: 10.1093/infdis/166.5.1014. [DOI] [PubMed] [Google Scholar]
- 34.Norrby-Teglund A, Pauksens P, Holm S E, Norgren M. Relation between low capacity of human sera to inhibit streptococcal mitogens and serious manifestations of disease. J Infect Dis. 1994;170:585–591. doi: 10.1093/infdis/170.3.585. [DOI] [PubMed] [Google Scholar]
- 35.Norrby-Teglund A, Newton D, Kotb M, Holm S E, Norgren M. Superantigenic properties of the group A streptococcal exotoxin SpeF (MF) Infect Immun. 1994;62:5227–5233. doi: 10.1128/iai.62.12.5227-5233.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozegowski J-H, Wollweber L, Vettermann S, Mueller P-J, Guenther E, Koehler W. Kinetics and regulation of erythrogenic toxins type A and C during growth of Streptococcus pyogenes. Zentbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig. 1996;283:271–285. doi: 10.1016/s0934-8840(96)80061-2. [DOI] [PubMed] [Google Scholar]
- 37.Reichardt W, Müller-Alouf H, Alouf J E, Köhler W. Erythrogenic toxins A, B and C: occurrence of the genes and exotoxin formation from clinical Streptococcus pyogenes strains associated with streptococcal toxic shock-like syndrome. FEMS Microbiol Lett. 1992;100:313–322. doi: 10.1111/j.1574-6968.1992.tb14058.x. [DOI] [PubMed] [Google Scholar]
- 38.Roggiani M, Schlievert P M. Streptococcal toxic shock syndrome, including necrotizing fasciitis and myositis. Curr Opin Infect Dis. 1994;7:423–426. [Google Scholar]
- 39.Roggiani M, Stoehr J A, Leonard B A B, Schlievert P M. Analysis of toxicity of streptococcal pyrogenic exotoxin A mutants. Infect Immun. 1997;65:2868–2875. doi: 10.1128/iai.65.7.2868-2875.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schellekens J F P, Schouls L, van Silfhout A, Elzenaar K, Brunings H, ten Broek H, Top J, van Leeuwen W J. The resurgence of group A streptococcal disease: characteristics of invasive infections in the Netherlands, 1993–1995. Ned Tijdschr Med Microbiol. 1995;3:78–83. [Google Scholar]
- 41.Schellekens J F P, Schouls L, van Pelt W, Esveld M, van Leeuwen W J. Group A streptococci: a change in virulence? Netherlands J Med. 1998;52:209–217. doi: 10.1016/s0300-2977(98)00034-5. [DOI] [PubMed] [Google Scholar]
- 42.Schouls L M, Blokpoel M C J, Elzenaar K P, Schellekens J F P, van Embden J D A, van Leeuwen W J. Genotyping of M- and exotoxin genes in the surveillance of group A streptococci infections in The Netherlands. RIVM Ann Sci Rep. 1992;119:155–158. [Google Scholar]
- 43.Stanley J, Linton D, Desai M, Efstratiou A, George R. Molecular subtyping of prevalent M serotypes of Streptococcus pyogenes causing invasive disease. J Clin Microbiol. 1995;33:2850–2855. doi: 10.1128/jcm.33.11.2850-2855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens D L, Tanner M H, Winship J, Swarts R, Ries K M, Schlievert P M, Kaplan E L. Severe group A streptococcal infections associated with a toxic shock-like syndrome and scarlet fever toxin A. N Engl J Med. 1989;321:1–7. doi: 10.1056/NEJM198907063210101. [DOI] [PubMed] [Google Scholar]
- 45.Talkington D F, Schwartz B, Black C M, Todd J K, Elliott J, Breimann R F, Facklam R R. Association of phenotypic and genotypic characteristics of invasive Streptococcus pyogenes isolates with clinical components of streptococcal toxic shock syndrome. Infect Immun. 1993;61:3369–3374. doi: 10.1128/iai.61.8.3369-3374.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tyler S D, Johnson W M, Huang J C, Ashton F E, Wang G, Low D E, Rozee K R. Streptococcal erythrogenic toxin genes: detection by polymerase chain reaction and association with disease in strains isolated in Canada from 1940 to 1991. J Clin Microbiol. 1992;30:3127–3131. doi: 10.1128/jcm.30.12.3127-3131.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Upton M, Carter P E, Orange G, Pennington G H. Genetic heterogeneity of M type 3 group A streptococci causing severe infections in Tayside, Scotland. J Clin Microbiol. 1996;34:196–198. doi: 10.1128/jcm.34.1.196-198.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weeks C A, Feretti J J. Nucleotide sequence of the type A streptococcal exotoxin (erythrogenic toxin) gene from Streptococcus pyogenes bacteriophage T12. Infect Immun. 1986;52:144–150. doi: 10.1128/iai.52.1.144-150.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]