Abstract
Vaccination with the adenoviral-vector-based AstraZeneca ChAdOx1 nCov-19 (Vaxzevria) vaccine is efficient and safe. However, in rare cases vaccinated individuals developed life-threatening thrombotic complications, including thrombosis in cerebral sinus and splanchnic veins. Monitoring of the applied vector in vivo represents an important precondition to study the molecular mechanisms underlying vaccine-driven adverse effects now referred to as vaccine-induced immune thrombotic thrombocytopenia (VITT). We previously have shown that digital PCR (dPCR) is an excellent tool to quantify transgene copies in vivo. Here, we present a highly sensitive dPCR for in situ quantification of ChAdOx1 nCoV-19 copies. Using this method, we quantified vector copies in human plasma 24, 72, and 168 h post vaccination and in a variety of murine tissues in an experimental vaccination model 30 min post injection. We describe a method for high-sensitivity quantitative detection of ChAdOx1 nCoV-19 with possible implications to elucidate the mechanisms of severe ChAdOx1 nCov-19 vaccine complications.
Keywords: SARS-Cov2, ChAdOx1 nCoV-19, vaccination, digital PCR, adenoviral vector, vaccine-induced immune thrombotic thrombocytopenia (VITT), AZD1222, Vaxzevria
Graphical abstract

Badbaran et al. introduce a digital PCR technique for quantification of ChAdOx1 nCov-19 vaccine vector copies. Based on its excellent specificity and sensitivity, the new assay facilitates ChAdOx1 nCov-19 monitoring in different biological specimens and might thus help to elucidate the mechanisms underlying the rare but potentially severe vaccine-caused complications.
Introduction
Vaccination has been shown to be effective against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2).1 Adenoviral-vector-based vaccines represent one of the cornerstones of the ongoing vaccination programs worldwide. Unfortunately, in contrast to their good safety profile, single cases of severe thromboembolism, often combined with thrombocytopenia, were observed for the adenoviral-vector-based vaccines from Astra Zeneca (ChAdOx1 nCov-19, Vaxzevria)2, 3, 4 and, even more rarely, Johnson and Johnson (Ad26.COV2.S).5 By the beginning of April 2021, EudraVigilance reported a total of 169 cases of cerebral venous sinus thrombosis (CVST) and 53 cases of splanchnic vein thrombosis after vaccination with ChAdOx1 nCov-19.6 This severe complication, now referred to as vaccine-induced immune thrombotic thrombocytopenia (VITT, synonym TTS), to some extent resembles atypical heparin-induced thrombocytopenia (HIT) involving platelet-activating antibodies against platelet factor (PF) 4.3,4
Vector spread to distinct tissues might contribute to acute but potentially also long-term side effects (e.g., caused by spontaneous insertion events).7 Therefore, it is critical to develop methods that facilitate sensitive analysis of ChAdOx1 nCov-19 vector distribution. A better understanding might help to develop screening approaches for the progression of severe and serious adverse events and thus support prevention and early intervention measures.
We reasoned that digital PCR8 (dPCR) should be well suited to detect and quantify vector copies after vaccination in humans and also in experimental animal models. We previously showed applicability of dPCR to the detection of gene-modified cells in humans.9
Results
Establishment and testing of the dPCR assay
In order to identify an appropriate primer-probe combination, we first deciphered parts of the primary DNA sequence of the ChAdOx1 nCov-19 vector. To do so, we designed a nested PCR located in a suitable region of the spike-protein coding sequence10 in the vector (Figure S1). After nested PCR on genomic DNA (gDNA) from blood cells sampled 45 min post vaccination, we obtained a weak but distinct signal of the expected size (360 bp) visualized by gel electrophoresis (not shown). Sanger sequencing of the purified amplicon followed by in silico translation confirmed complete identity of the amino acid sequence encoded by the PCR fragment with the respective part (amino acids 1,105–1,210) of the published SARS-CoV-2 spike protein sequence (Figure 1).10 Meanwhile, the complete DNA sequence of the ChAdOx1 nCov-19 vector has been published, and full DNA sequence identity could be confirmed for the cloned fragment (Figure S1).11
Figure 1.
Primary sequence and in silico translation of the identified DNA of the ChAdOx1 nCov-19 vector
The shown amino acid sequence is identical with amino acids 1,105–1,210 of the published spike protein sequence.10 Target sequences of the primers (AZ-FP and AZ-RP) and the probe (AZ-P) used for the digital PCR (dPCR) are shaded in green and blue, respectively.
We next designed two PCR primers and an internal dual-labeled hydrolysis probe for dPCR (Figure 1; Materials and methods). To verify its sensitivity and specificity, we tested the new assay on negative control samples and a series of dilutions of the first PCR product. We performed duplex-dPCR with a well-established REF gene assay detecting single copies of the human RPP30 gene. As shown in Figure 2A, we observed excellent separation of positive and negative signals for both the RPP30- and the ChAdOx1 nCov-19-vector-specific PCRs. Based on the used dilutions, the highest amount of gDNA tested in our dilution series (approximately 5 ng) corresponds to approximately 750 cells or 1,500 haploid genomes. This corresponds very well to the maximal number of RPP30 signals in the dilution series (Figure 2B). As evident, the number of spike-amplicons (generated in the first PCR round) was approximately 6 times higher in the undiluted sample than the number of RPP30 amplicons. Notably, this ratio remained constant for all 16 dilutions tested, resulting in a high correlation of the two measured parameters (R2 = 0.9991, p < 0.0001; Figure 2C). Moreover, single copies of the targeted spike-amplicon were readily detected in the assay (Figure 2C), whereas all control samples (gDNA from ten non-vaccinated donors) were negative (not shown). This indicated high sensitivity and specificity of the dPCR assay on the DNA level.
Figure 2.
The new dPCR assay facilitates sensitive detection of ChAdOx1 nCov-19 vector DNA
(A) The depicted two-dimensional (2D) plot illustrates results of duplex dPCR simultaneously quantifying ChAdOx1 nCov-19 and RPP30 (REF gene) copies. As evident, positive and negative signals are clearly separated. (B) Quantification of signals of individual dilutions for the presence of ChAdOx1 nCov-19 (upper panel) and REF (lower panel) copies. Indicated numbers are Poisson corrected. (C) Highly significant correlation of ChAdOx1 nCov-19 and REF copy numbers in the dilution series. 16 different dilutions were tested. For dilutions containing fewer than 10 REF copies, dPCRs were performed in three independent replicates. Indicated are the Pearson coefficient r and the corresponding p value.
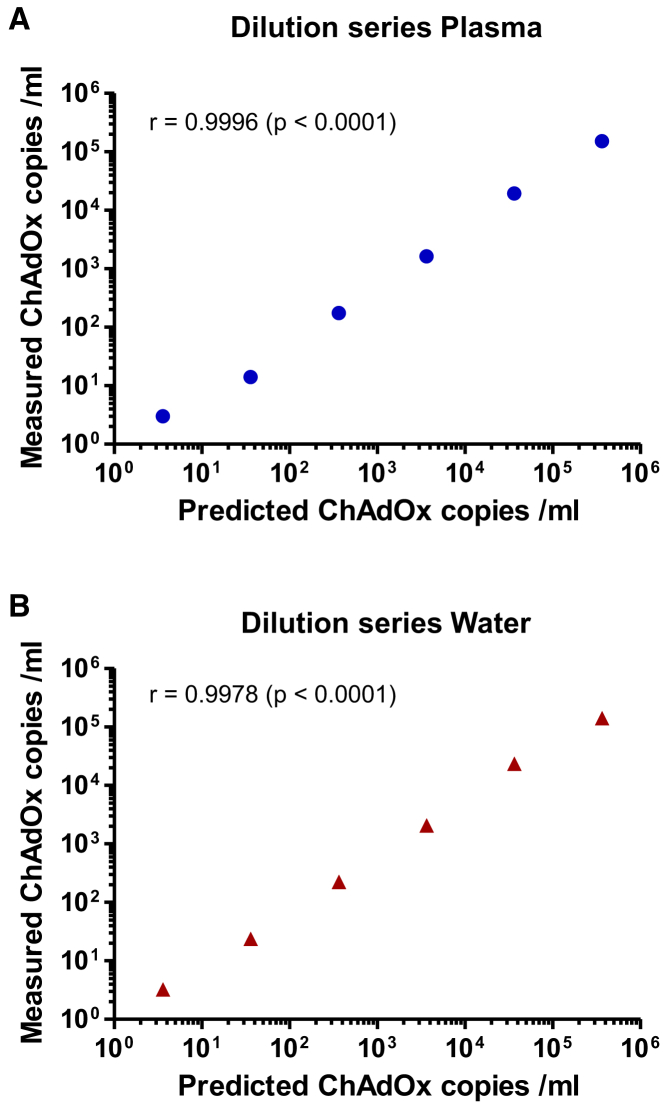
To assess practical utility of the assay, we next analyzed its sensitivity on vector particles from ChAdOx1 nCov-19 vaccine. We made serial tenfold dilutions of the vector in water and human plasma and isolated DNA and performed dPCR. Initial data indicated the (expected) saturation of all droplets precluding quantification, when more than 106 particles were present. Consequently, we focused on the last six dilution steps corresponding to expected amounts of 363,636 to 3.6 vector particles in the analyzed PCR samples. As evident, excellent correlation was observed between expected and measured vector copy numbers for dilutions in both plasma (Figure 3A) and water (Figure 3B). Moreover, single copies present in the last dilution could readily be detected and precisely quantified in water as well as in plasma. Interestingly, for both water and plasma, the copy numbers empirically determined by dPCR were slightly (by factors of 1.5–2) lower than the predicted ones. It seems most likely that this divergence reflects differences between the techniques used to measure copy numbers (qPCR versus dPCR), including diverse amplicons. Together, the obtained data confirm that the dPCR assay facilitates precise quantification of ChAdOx1 nCov-19 vector copies over a large range of particles.
Figure 3.
Dilution series of ChAdOx1 nCov-19 vector in plasma and water
ChAdOx1 nCov-19 vector particles were spiked into and serially diluted in (A) human plasma, and (B) water. Predicted copy numbers (x axes) based on the vector concentration in the vaccine (10 × 1010 per mL) specified by AstraZeneca are plotted against empirical numbers measured by dPCR (y axes). The latter represent mean values of at least four independent replicates (at least ten replicates for the last dilution). Values determined by dPCR were slightly (1.5–2 times) lower than predicted numbers (based on qPCR) but showed excellent correlation over five orders of magnitude. Pearson coefficients (r) and the corresponding p values are indicated.
High-sensitivity detection of ChAdOx1 nCov-19 vector DNA in biological specimens
After proving applicability of the method, we tested plasma samples from nine individuals obtained 24, 72, and 168 h post vaccination. We isolated cell-free DNA (cfDNA) from blood plasma for each time point. dPCRs were performed in duplex reactions using the RPP30 gene as reference to verify presence of cfDNA. At both 24 and 72 h post vaccination, we detected vector copies in all nine plasma samples, which contained variable amounts of cfDNA (Table 1). Interestingly, in six of the nine individuals, vector copy numbers were higher 72 h as compared to 24 h post vaccination (Figure 4A). These higher amounts did not correlate with the amounts of cfDNA or with the copy numbers of the reference gene RPP30 determined in parallel (Table 1). Expectedly, no ChAdOx1 nCov-19 vector DNA was found in any of the plasma samples 168 h post vaccination. All analyses were repeated three times in independent experiments (technical triplicates); mean values are shown in Figure 4A and Table 1. Taken together, mean copy numbers were more than two times higher at 72 h as compared to 24 h post vaccination. In summary, these data prove the usefulness of the presented dPCR assay to monitor the presence of ChAdOx1 nCov-19-vector-derived DNA in human blood after regular vaccination.
Table 1.
dPCR analyses of cfDNA in human plasma samples
| ID | Day | DNA (ng/μL) | Mean ChAdOx1 nCov-19 copies/mL | SD | Mean RPP30 copies/mL | SD |
|---|---|---|---|---|---|---|
| #01 | 1 | 44.4 | 178 | 20 | 3,649 | 170 |
| 3 | 43.9 | 134 | 47 | 1,640 | 314 | |
| 7 | 36.9 | 0 | 0 | 2,831 | 357 | |
| #02 | 1 | 35.4 | 18 | 18 | 5,920 | 2,056 |
| 3 | 34.3 | 31 | 16 | 2,084 | 154 | |
| 7 | 32.9 | 0 | 0 | 4,000 | 223 | |
| #03 | 1 | 47.6 | 62 | 34 | 3,098 | 553 |
| 3 | 38.3 | 20 | 8 | 1,115 | 134 | |
| 7 | 25.4 | 7 | 12 | 4,333 | 440 | |
| #04 | 1 | 47.2 | 126 | 41 | 2,862 | 466 |
| 3 | 42 | 476 | 47 | 3,685 | 641 | |
| 7 | 41 | 0 | 0 | 1,365 | 154 | |
| #05 | 1 | 38.9 | 33 | 22 | 7,702 | 130 |
| 3 | 41.1 | 224 | 60 | 3,667 | 630 | |
| 7 | 35.2 | 0 | 0 | 2,862 | 240 | |
| #06 | 1 | 36.5 | 83 | 36 | 1,631 | 329 |
| 3 | 44.3 | 119 | 86 | 1,418 | 235 | |
| 7 | 61.7 | 0 | 0 | 1,351 | 301 | |
| #07 | 1 | 67.5 | 26 | 23 | 2,311 | 255 |
| 3 | 60.4 | 320 | 113 | 2,733 | 792 | |
| 7 | 31 | 0 | 0 | 1,947 | 101 | |
| #08 | 1 | 53 | 41 | 30 | 3,200 | 335 |
| 3 | 43 | 20 | 17 | 1,640 | 148 | |
| 7 | 54 | 0 | 0 | 2,689 | 573 | |
| #09 | 1 | 20.4 | 18 | 18 | 13,847 | 2,519 |
| 3 | 27.6 | 67 | 23 | 1,649 | 309 | |
| 7 | 27.5 | 0 | 0 | 6,360 | 509 |
ID, identification number of volunteer; SD, standard deviation for technical triplicates.
Figure 4.
The novel dPCR assay quantifies ChAdOx1 nCov-19 DNA in human post-vaccination plasma samples and murine tissues in an experimental vaccination model
(A) ChAdOx1 nCov-19 copies were determined in human plasma from nine volunteers 24, 72, and 168 h post vaccination. Copy numbers were higher at 72 h in six out of the nine individuals. No copies were found in the blood 168 h post vaccination. Mean values and standard deviations (SDs) are shown for triplicate analyses. (B) Vector distribution in different mouse tissues 30 min after intradermal injection using an artificial vaccination model.12 Data represent mean values and SDs from four animals. All values for individual animals were measured in independent replicate (2–3) analyses. REF, copies of the reference gene EpoR.
Furthermore, we evaluated whether dPCR might also be applied to detect ChAdOx1 nCov-19 vector copies in different tissues after experimental vaccination in a dedicated mouse model. Particularly, the Miles mouse edema model was used to study proinflammatory reactions induced by the ChAdOx1 nCoV-19 vaccine. To this end, 50 μL of ChAdOx1 nCov-19 vaccine (corresponding to 5 × 109 vector particles) were intradermally injected into the dorsal skin of C57BL/6 mice.12 As reported elsewhere, intradermally injected vaccine triggered leakage in dermal vessels, a hallmark of inflammation.12 We aimed to analyze biodistribution of injected ChAdOx1 nCov-19 vaccine using our dPCR technique. To do so, challenged animals were sacrificed 30 min later, organs were excised, and gDNA was isolated from distinct organs/tissues as indicated in Figure 4B. Duplex dPCR was performed using the murine epoR gene as reference. As expected, huge numbers of ChAdOx1 nCov-19 vector copies were found at the injection site. Detection of ChAdOx1 nCov-19 vector copies in all tissues analyzed provided evidence for imminent vector spread into the bloodstream and different organ tissues (Figure 4B), which may have implications for adverse effects, such as VITT.12
Discussion
After vaccination with the adenoviral-vector-based ChAdOx1 nCov-19 vaccine, single cases of severe thromboembolism, often combined with thrombocytopenia, were reported.2, 3, 4 We reasoned that sensitive assessment of ChAdOx1 nCov-19 vector copy numbers and their kinetics in various tissues might contribute to a better understanding of mechanisms underlying these severe side effects.
To this end, we have developed a dPCR assay for the quantification of ChAdOx1 nCov-19 DNA copies. We show here that the new technique combines excellent sensitivity with high specificity. As discussed below, in order to prove its practical applicability, we also tested whether the assay facilitates quantification of ChAdOx1 nCov-19 DNA copies in vivo. Importantly, the obtained experimental data support the usefulness of our dPCR to quantitatively assess the presence of ChAdOx1 nCov-19 DNA in biological specimens, but they are too preliminary to draw biological conclusions.
The assay was successfully applied to quantify ChAdOx1 nCov-19 copies in plasma of vaccinated humans and also in a variety of tissues in experimentally vaccinated mice. In six of nine individuals we observed somewhat unexpected kinetics, with higher ChAdOx1 nCov-19 copy numbers 72 h post vaccination as compared to the 24-h value. This finding might reflect delayed release of ChAdOx1 nCov-19 vectors from the injection site to the bloodstream. In an alternative scenario, the observed long presence of ChAdOx1 nCov-19 vector particles in the blood might be caused by their binding to erythrocytes. Indeed, human (but not mouse) erythrocytes were previously shown to express the Coxsackie virus-adenovirus receptor (CAR) and bind adenoviral particles resulting in prolonged circulation.13 Finally, cell-free vector DNA in the plasma might, in principle, also result from the death of transduced, spike-overexpressing cells. Relevance as well as origin of the secondary increase in ChAdOx1 nCov-19 DNA copy numbers in the plasma deserve further research.
In the mouse leakage model,12 intradermal injection of hyper-physiological doses of ChAdOx1 nCov-19 resulted in rapid distribution of the vector to multiple tissues, including the brain. Determination of the actual (intra- versus extravasal, intra- versus extracellular) localization of the vector in different cell types will require future exploration. Potential contribution of vessel leakage to development of VITT has been discussed in detail elsewhere.12
In conclusion, the dPCR assay established here might be very useful to study the impact of ChAdOx1 nCov-19 vector distribution in vivo with implications for development and kinetics of side effects, both in vaccinated humans and experimental animal models. Notably, the presented dPCR principle might be readily adapted to other adenoviral-vector-based vaccines against SARS-CoV2 (e.g., Ad26.COV2.S, Sputnik), but also DNA-based vaccines against other viral infections.
Materials and methods
Decryption of parts of the codon-optimized spike sequence
To decipher a part of the primary DNA sequence of the spike coding sequence, we designed suitable primers for nested PCR based on the known amino acid sequence. 200 ng of gDNA isolated from blood cells drawn 45 min after vaccination with ChAdOx1 nCov-19 was used as template for the first reaction and 5 μL of a 1:50 dilution for the second. Both first and nested PCRs were performed in 100 μL final volume using Platinum PCR Supermix (Thermo Fisher, Kandel, Germany) following a 40-cycle protocol with the following conditions: initial denaturation: 94°C for 2 min; first 10 cycles: 94°C for 30 s, 50°C for 45 s, 72°C for 30 s; next 30 cycles: annealing temperature changed to 60°C; final extension, 7 min. Both primary and nested PCR products were visualized on an agarose gel. A single band of the expected size (360 bp) was found after nested PCR and Sanger sequenced (Eurofins, Ebersberg, Germany). The DNA sequence located between the primers and the corresponding amino acid sequence obtained by in silico translation are depicted in Figure 1. As evident, the amino acid sequence showed 100% homology to the respective part of the published spike sequence10 (Figure 1).
Design of primers and probes for dPCR
PCR primers and probes (Table 2) were designed with Primer Express version 3.0.1 (Thermo Fisher). All primers and probes were purchased from Eurofins.
Table 2.
Non-commercial dPCR primers and probes used in this study
| Primer/probe | Sequence | Final concentration |
|---|---|---|
| AZ-FP | 5′-TGGACCTGGGCGATATCAG-3′ | 900 nM |
| AZ-RP | 5′-TTCAGCCGGTCGATCTCTTT-3′ | 900 nM |
| AZ-P | 5′-FAM CAATGCCAGCGTCGT GAACATCCA-3′BHQ1 |
250 nM |
| mEpoR-FP | 5′-CGCCTGTGCAGATCCGATAA-3′ | 900 nM |
| mEpoR-RP | 5′-GCAGGCGGGGTCGCTACTC-3′ | 900 nM |
| mEpoR-P | 5′-HEX TTCTGAGGCGCCACT TTTGCAAGACC-3′BHQ1 |
250 nM |
Isolation of gDNA and cfDNA
gDNA from human blood cells was isolated from 200 μL peripheral blood using the QIAamp Blood kit (QIAGEN, Hilden, Germany) as previously described.9 To isolate gDNA from different mouse tissues, we made use of the blackPREP Rodent Tail DNA Kit (Analytik Jena, Jena, Germany) in accord with the manufacturer’s protocol.
cfDNA (both human and mouse) was isolated with QIAamp DSP Circulation NA Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. In short, 1 to 1.5 mL of human plasma and 0.5 mL of murine plasma were used for cfDNA isolation, which was eluted in 50 μL. Measured ChAdOx1 nCov-19 copy numbers were individually corrected to copies per 1 mL plasma.
dPCR protocol
dPCR was carried out as duplex reactions and analyzed with the QX100 Droplet Digital PCR System (Bio-Rad, Hercules, CA, USA).9,14 For human samples, an assay detecting the human ribonuclease P/MRP subunit p30 RPP30 gene (Bio-Rad Assay ID dHsaCP2500350) was used as reference (probe: HEX-BHQ1); for murine cells we included our in-house established reference assay detecting the murine EpoR gene (see above, based on Cornils et al.15). Per individual reaction, 250 nM of each probe (one FAM-, one HEX-labeled) and 900 nM of each primer, 11 μL 2× ddPCR Supermix (Bio-Rad), 0.5 μL (5 U) FastDigest restriction enzymes (MseI for human and ApaI for mouse gDNA, both Thermo Fisher), 8 μL gDNA (routinely at approximately 15 ng/μL), and water to a final volume of 22 μL were mixed and incubated for 10 min at 21°C for DNA digestion. The restriction enzymes were added to facilitate investigation of gDNA amounts >60 ng. In accordance with the manufacturer’s protocol, PCR mixes were transferred to the QX100 Droplet Generator (Bio-Rad), which generates approximately 20,000 droplets per single reaction. Droplet-containing water-in-oil emulsion was transferred to a 96-well, semi-skirted plate (Bio-Rad). The following protocol was used in a standard thermal cycler (Eppendorf, Hamburg, Germany): denaturation (95°C for 10 min), amplification cycles (94°C for 30 s, 60°C for 1 min; 40 times), a ramp rate of 1.5°C/s, and a final 10 min inactivation step at 98°C. After PCR, individual wells were analyzed simultaneously for FAM and HEX fluorescence using the QX100 droplet reader (Bio-Rad). Data were analyzed with QuantaSoft v.1.7 software (Bio-Rad) including automatic Poisson correction.9,14
Validation and application of the dPCR assay
In order to assess sensitivity and specificity of the dPCR, we used a dilution series of the PCR product of the first amplification round of the nested PCR; 16 different concentrations were tested. Those samples with expected copy numbers below 10 were tested in triplicate, and mean values were used to build the dilution curve. To test specificity, ten gDNA samples from healthy, non-vaccinated donors were tested in the duplex dPCR; all were highly positive for the REF gene but negative for ChAdOx1 nCov-19.
In order to assess recovery and detectability of dissolved ChAdOx1 nCov-19 vector, we performed serial dilutions of the original ChAdOx1 nCov-19 (AstraZeneca, LOT ABV3374) in water and plasma (first dilution 1:20, and further tenfold). DNA was isolated from plasma with QIAamp DSP Circulation NA Kit (QIAGEN), from water with the QIAamp Blood micro kit (QIAGEN) following the manufacturer’s instructions. The last six dilution steps were applied to dPCR. Based on the declared amount of ChAdOx1 nCov-19 vector particles (10 × 1010 per mL) and the dilution/pipetting schemes used, the calculated particle numbers in the analyzed dPCR samples ranged from 363,636 to 3.6 vector particles. For each but the last dilution, four independent dPCRs were performed; the last dilution was assayed at least ten times.
Volunteer samples
Blood samples from nine volunteers were collected at the University Medical Center Hamburg-Eppendorf after their prime vaccination. Donors provided written informed consent (ethical approval: PV4780). Volunteers received the vaccine ChAdOx1 nCov-19. At days 1, 3, and 7 post vaccination, EDTA-blood was drawn and centrifuged at 200 × g for 10 min at room temperature (RT). Afterward, plasma was transferred and was again centrifuged at 1,000 × g for 15 min at RT. Supernatant was stored at −80°C.
Mouse model
C57BL/6 mice of both sexes were anesthetized by intraperitoneal injection of ketamine (120 μg/g body weight [BW]) and xylazine (16 μg/g BW in saline, 10 μL/g BW), and 50 μL ChAdOx1 nCov-19 (5 × 109 vector particles; one-tenth of the human dose) of the AstraZeneca vaccine (LOT ABV3374) was intradermally injected into the dorsal region. The applied vaccine dose for a mouse (with 25 g body weight) is thus approximately 300× higher than an intramuscularly vaccinated 75-kg human being. After 30 min, mice were sacrificed by cervical dislocation and blood was drawn via cardiac puncture. To perfuse organs, the right atrium was opened, and 10 mL of ice-cold PBS was injected into the left ventricle of the heart before tissue samples were collected. Citrated blood samples were centrifuged at 1,000 × g for 10 min at 4°C, and gDNA was separately isolated from centrifuged cells (including thrombocytes) and plasma. Data are shown for blood cells. Mice were treated according to national guidelines for animal care at the animal facilities of University Medical Center Hamburg-Eppendorf and approved by local authorities (#56/18). All procedures were conducted in accordance with 3Rs (Replace, Reduce, Refine) principles.
Statistics
All dPCR analyses on mouse and human samples were performed in triplicate. Data shown represent mean values ± standard deviations (SDs). The Pearson correlation coefficient was determined using linear regression with a confidence interval of 95%. Statistical analysis was performed with GraphPad Prism (San Diego, CA, USA).
Acknowledgments
We thank Hanna Thode for expert technical assistance and the laboratory team at the UKE, especially My Linh Ly, Monika Friedrich, and Niclas Reneviér for collecting and processing the blood from volunteers as well as Amelie Sophie Alberti for study coordination. We acknowledge support by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) grants 125440785-SFB877 (T.R.), P6-KFO306 (T.R.), and 80750187-SFB841 (T.R. and B.F.) and the German Center for Infection Research (DZIF) SARS-CoV-2 Fast track fund TTU 01.921 (M.M.A.). We also are indebted to Stefan Kochanek for helpful information. Last but not least, the authors also wish to thank all volunteers for their willingness to support our research. The graphical abstract was created with BioRender.
Author contributions
B.F. conceived the study, designed primers, and wrote the manuscript; A.B. isolated DNAs and designed, performed, and analyzed dPCRs; R.K.M. and T.R. designed and performed the mouse study; C.D., J.W., A.F., S.C.M., M.M.A., and K.R. designed human studies and contributed human samples; all authors reviewed and edited the manuscript.
Declaration of interests
The authors declare no conflict of interest.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2021.10.002.
Supplemental information
References
- 1.Rossman H., Shilo S., Meir T., Gorfine M., Shalit U., Segal E. COVID-19 dynamics after a national immunization program in Israel. Nat. Med. 2021;27:1055–1061. doi: 10.1038/s41591-021-01337-2. [DOI] [PubMed] [Google Scholar]
- 2.Schultz N.H., Sørvoll I.H., Michelsen A.E., Munthe L.A., Lund-Johansen F., Ahlen M.T., Wiedmann M., Aamodt A.H., Skattør T.H., Tjønnfjord G.E., Holme P.A. Thrombosis and Thrombocytopenia after ChAdOx1 nCoV-19 Vaccination. N. Engl. J. Med. 2021;384:2124–2130. doi: 10.1056/NEJMoa2104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greinacher A., Thiele T., Warkentin T.E., Weisser K., Kyrle P.A., Eichinger S. Thrombotic Thrombocytopenia after ChAdOx1 nCov-19 Vaccination. N. Engl. J. Med. 2021;384:2092–2101. doi: 10.1056/NEJMoa2104840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scully M., Singh D., Lown R., Poles A., Solomon T., Levi M., Goldblatt D., Kotoucek P., Thomas W., Lester W. Pathologic Antibodies to Platelet Factor 4 after ChAdOx1 nCoV-19 Vaccination. N. Engl. J. Med. 2021;384:2202–2211. doi: 10.1056/NEJMoa2105385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muir K.L., Kallam A., Koepsell S.A., Gundabolu K. Thrombotic Thrombocytopenia after Ad26.COV2.S Vaccination. N. Engl. J. Med. 2021;384:1964–1965. doi: 10.1056/NEJMc2105869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.European Medicines Agency . 2021. AstraZeneca’s COVID-19 vaccine: EMA finds possible link to very rare cases of unusual blood clots with low blood platelets.https://www.ema.europa.eu/en/news/astrazenecas-covid-19-vaccine-ema-finds-possible-link-very-rare-cases-unusual-blood-clots-low-blood [Google Scholar]
- 7.Stephen S.L., Montini E., Sivanandam V.G., Al-Dhalimy M., Kestler H.A., Finegold M., Grompe M., Kochanek S. Chromosomal integration of adenoviral vector DNA in vivo. J. Virol. 2010;84:9987–9994. doi: 10.1128/JVI.00751-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sykes P.J., Neoh S.H., Brisco M.J., Hughes E., Condon J., Morley A.A. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- 9.Fehse B., Badbaran A., Berger C., Sonntag T., Riecken K., Geffken M., Kröger N., Ayuk F.A. Digital PCR Assays for Precise Quantification of CD19-CAR-T Cells after Treatment with Axicabtagene Ciloleucel. Mol. Ther. Methods Clin. Dev. 2020;16:172–178. doi: 10.1016/j.omtm.2019.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radukic M.T., Le D.T., Müller K.M. 2021. Nucleic acid sequence composition of the Oxford – AstraZeneca vaccine ChAdOx1 nCoV-19 (AZD1222, Vaxzevria). ResearchSquare.https://www.researchsquare.com/article/rs-799338/v1 [Google Scholar]
- 12.Greinacher A., Selleng K., Palankar R., Wesche J., Handtke S., Wolff M., Aurich K., Lalk M., Methling K., Völker U. Insights in ChAdOx1 nCov-19 Vaccine-induced Immune Thrombotic Thrombocytopenia (VITT) Blood. 2021 doi: 10.1182/blood.2021013231. Published online September 29, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carlisle R.C., Di Y., Cerny A.M., Sonnen A.F., Sim R.B., Green N.K., Subr V., Ulbrich K., Gilbert R.J., Fisher K.D. Human erythrocytes bind and inactivate type 5 adenovirus by presenting Coxsackie virus-adenovirus receptor and complement receptor 1. Blood. 2009;113:1909–1918. doi: 10.1182/blood-2008-09-178459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stahl T., Böhme M.U., Kröger N., Fehse B. Digital PCR to assess hematopoietic chimerism after allogeneic stem cell transplantation. Exp. Hematol. 2015;43:462–468.e1. doi: 10.1016/j.exphem.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Cornils K., Bartholomae C.C., Thielecke L., Lange C., Arens A., Glauche I., Mock U., Riecken K., Gerdes S., von Kalle C. Comparative clonal analysis of reconstitution kinetics after transplantation of hematopoietic stem cells gene marked with a lentiviral SIN or a γ-retroviral LTR vector. Exp. Hematol. 2013;41:28–38.e3. doi: 10.1016/j.exphem.2012.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.