Abstract
Human Ku70 is a well-known endogenous nuclear protein involved in the non-homologous end joining pathway to repair double-stranded breaks in DNA. However, Ku70 has been studied in multiple contexts and grown into a multifunctional protein. In addition to the extensive functional study of Ku70 in DNA repair process, many studies have emphasized the role of Ku70 in various other cellular processes, including apoptosis, aging, and HIV replication. In this review, we focus on discussing the role of Ku70 in inducing interferons and proinflammatory cytokines as a cytosolic DNA sensor. We explored the unique structure of Ku70 binding with DNA; illustrated, with evidence, how Ku70, as a nuclear protein, responds to extracellular DNA stimulation; and summarized the mechanisms of the Ku70-involved innate immune response pathway. Finally, we discussed several new strategies to modulate Ku70-mediated innate immune response and highlighted some potential physiological insights based on the role of Ku70 in innate immunity.
Keywords: Ku70, Ku heterodimer, cytosolic DNA sensing, innate immunity, interferons, HIV replication
Introduction
Innate immunity includes diverse areas of host defense response to pathogen invasion, such as bacterial or viral infection. In this system, pattern recognition receptors (PRRs) expressed in host cells recognize the conserved pathogen-associated molecular patterns (PAMPs) which are derived from microbes and then mediate innate immune responses (Medzhitov and Janeway, 1997; Medzhitov and Janeway, 2000; Akira et al., 2006; Pichlmair and Reis e Sousa, 2007; Mogensen, 2009; Takeuchi and Akira, 2009; Yoneyama and Fujita, 2010). Detection of pathogenic cytosolic nucleic acids: double-stranded (ds) or single-stranded (ss) DNA and RNA is essential to initiate innate immunity. PRR families include the retinoic acid-inducible gene I (RIG-I)-like receptors, toll-like receptors (TLRs), and a diverse member of cytosolic DNA sensors (Bowie and Haga, 2005; Kaisho and Akira, 2006; Yoneyama and Fujita, 2008; Beutler, 2009; Kawai and Akira, 2009; Yoneyama and Fujita, 2009; Barber, 2011; Keating et al., 2011; Thompson et al., 2011; Paludan and Bowie, 2013; Dempsey and Bowie, 2015). Once PAMPs are sensed by PRRs, the recognition subsequently mediates intracellular signaling pathways and activates transcription factors, interferon (IFN) regulatory factors (IRFs) or nuclear factor κB (NF-κB), which in turn leads to the increased production of antiviral interferons and proinflammatory cytokines (Lee and Kim, 2007; Mogensen, 2009).
DNA is a potent trigger of innate immune responses in host cells (Sharma and Fitzgerald, 2011). Many studies have emphasized the importance of cytosolic DNA sensing in the innate immune response against invading pathogens. The DNA-mediated innate immune response includes diverse signaling pathways leading to the production of IFN-α, IFN-β, interleukin (IL)-1β, or IL-18 (Christensen and Paludan, 2016). For instance, the DNA-dependent activator of IFN-regulatory factors (DAI) (Takaoka, 2007) is the first identified DNA sensor to recognize dsDNA and activate the STING-TBK1-IRF3 signaling pathway. After that, gamma-interferon-inducible protein (IFI16) (Unterholzner et al., 2010; Monroe et al., 2014; Thompson et al., 2014) and DEAD-box helicase 41 (DDX41) (Zhang et al., 2011c) were found as cytosolic DNA sensors in diverse cellular processes to recognize DNA. Leucine-rich repeat flightless-interacting protein 1 (LRRFIP1), another discovered cytosolic DNA sensor, binds dsDNA and activates β-catenin to induce downstream signaling (Yang et al., 2010). DEAH box protein 9 (DHX9) and DHX36 bind with dsDNA in dendritic cells and activate NF-κB through myeloid differentiation primary response 88 (MyD88) (Kim et al., 2010; Zhang et al., 2011b). More recently, cyclic GMP-AMP Synthase (cGAS) has been identified as a cytosolic DNA sensor (Gao et al., 2013; Sun et al., 2013; Cai et al., 2014; Zhang et al., 2014; Xia et al., 2016). Once cGAS detects dsDNA, it undergoes a conformational change to open the catalytic pocket followed by synthesis of cGAMP from ATP and GTP: a potent activator of the STING-TBK1-IRF3 signaling pathway. In addition to the induction pathway of IFNs, Absent in melanoma 2 (AIM2) has been found to associate with cytosolic DNA and activate inflammasomes by recruiting apoptosis-associated speck-like protein (ASC) and pro-caspase-1, and then produce mature forms of IL-1β and IL-18 (Burckstuummer, 2009; Fernandes-Alnemri et al., 2009; Hornung, 2009).
The DNA-mediated innate immune response is not restricted to the induction of type I IFNs and proinflammatory cytokines: cytosolic DNA also induces type III IFNs. Type III IFNs are new members of the IFN family (Ank et al., 2006; Ank and Paludan, 2009; Donnelly and Kotenko, 2010; Kotenko, 2011; Syedbasha and Egli, 2017). Type III IFNs are also called IFN-λs, which include IFN-λ1, IFN-λ2, IFN-λ3 (also known as IL-29, IL-28A, and IL-28B, respectively) and IFN-λ4 (Ank et al., 2006; Uzé and Monneron, 2007; Ank and Paludan, 2009; Kotenko, 2011; Booth and George, 2013; Lu et al., 2015). Compared with type I IFNs, they use a different heterodimeric receptor complex (IFN-λR1/IL-10R2) to get into the cells (Ank and Paludan, 2009; Kotenko, 2011). Similar to type I IFNs, stimulation by virus infection or TLR agonist induces type III IFNs (Kotenko et al., 2003; Coccia et al., 2004; Spann et al., 2004). Of note, Donnelly et al. found that the gene encoding the mouse ortholog of human IFNL1 contains a stop codon in the region of exon 1 and lacks the entire exon 2. Therefore, the gene Ifnl1 in mice does not encode a functional IFN-λ1 protein (Donnelly and Kotenko, 2010).
Ku70 and Ku80, proteins with molecular weight (MW) of 70 KDa and 80 KDa, respectively, are the essential components in the non-homologous end-joining (NHEJ) pathway. They are first identified in humans (Mimori et al., 1981). Ku70 is encoded by the X-ray repair cross-complementing protein (XRCC) 6 gene located on chromosome 22, and Ku80 is encoded by the XRCC5 gene on chromosome 2. Hetero dimerization of Ku70 and Ku80 is essential for the stability of each protein. The lacking of one subunit leads to dramatically decreased intracellular level of the other subunit, suggesting that most Ku70 and Ku80 exist as a heterodimer (Nussenzweig et al., 1996; Gu et al., 1997). Such functional homologs have been identified in some prokaryotic lineages and almost all eukaryotes, including vertebrates, insects, and fungi (Dynan and Yoo, 1998; Aravind and Koonin, 2001; Bowater and Doherty, 2006). The Ku70/Ku80 heterodimer (so-called Ku) and a catalytic kinase subunit (DNA-PKcs) are often referred to as the subunit of the DNA-dependent protein kinase (DNA-PK) complex, which assembles in response to DNA double-strand breaks to repair the damaged DNA via NHEJ pathway (Fell and Schild-Poulter, 2015). The region between residues 439–592 at Ku80 C-terminus interacts with DNA-PKcs (Gell and Jackson, 1999; Singleton et al., 1999; Davis et al., 2013) and promotes the autophosphorylation of DNA-PKcs at DNA double-stranded breaks.
Ku70 and Ku80 are predominantly observed in the nucleus (Koike, 2002). Following translation of each protein in the cytosol, the Ku subunits can translocate from the cytoplasm into the nucleus together (Koike, 2002), or independently (Koike et al., 1999a), since each subunit possesses its own nuclear localization signal (NLS) (Koike et al., 2000). However, further functional studies have reported that Ku70 is not only involved in nuclear activities like DNA repair, transcription, and replication but is also involved in multiple cytosolic activities. Bax, a cytoplasmic protein, has been discovered to interact with Ku70, and this Ku70-Bax binding is indicated to inhibit Bax-mediated apoptosis (Cohen et al., 2004). In addition, many studies have implicated that cytosolic Ku has been shown to serve as a PRR that recognizes viral DNA in human cells and then induces type I and type III interferons or proinflammatory cytokines (Zhang et al., 2011a; Ferguson et al., 2012; Li et al., 2016b; Sui et al., 2017; Wang et al., 2017; Burleigh et al., 2020; Sui et al., 2021; Wang et al., 2021). This article summarized and discussed the Ku70-mediated innate immune response in detail and then highlighted potential strategies to modulate the innate immune cascades. The homology modeling for Ku70 and Ku80 is illustrated in Figure 1 , the diverse functions of Ku70 are listed in Figure 2 . The studies for Ku70 related to innate immunity are summarized in Table 1 and illustrated in Figure 3 . Beyond the role of Ku70 in innate immunity, roles of Ku70 in viral life cycle of Human Immune deficiency virus (HIV) ( Figure 4 ) and in bacterial pathogen invasion are reviewed and discussed.
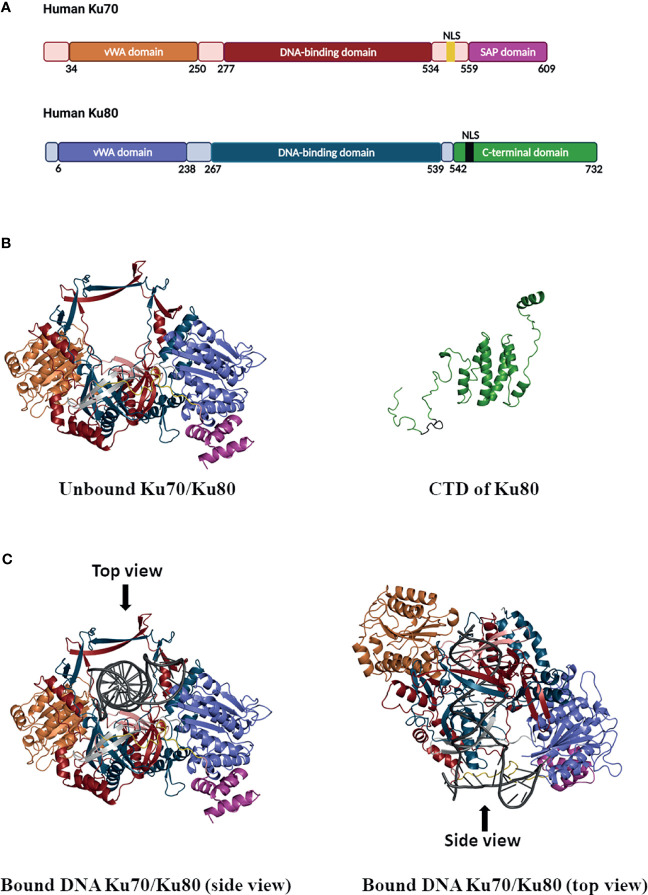
Figure 1.
Schematic of Ku70/80 heterodimer domains and ribbon diagrams. (A) Domains in Ku70 and Ku80. In Ku70, the vWA domain is colored in orange, the DNA-binding domain is colored in firebrick, the SAP domain is colored in pink, the nuclear localization sequence (NLS: 539–556) is colored in yellow, and other parts are colored in light pink. In Ku80, the vWA domain is colored in purple, the DNA-binding domain is colored in blue, the c-terminal domain of Ku80 is colored in green, the NLS (561-569) of Ku80 is colored in black, and other parts are colored in light grey. (B) Unbound Ku70/Ku80 heterodimer with a view of Ku70 NLS (yellow) in the front (left panel). The range in the Ku70 model is from 35–609 amino acids, where the first 34 residues in the N-terminal domain (NTD) are truncated. The range in the Ku80 model is from 6–541 amino acids, where the first 5 residues in the NTD and residues 542–732 in the C-terminal domain (CTD) are truncated. The CTD domain of Ku80 is colored in green, and the corresponding NLS (561–569) is colored in black (right panel). (C) The structure model of the Ku70/Ku80 heterodimer bound with DNA. The left and the right panel demonstrate the side and top view, respectively, and bound DNA is colored in dark grey.
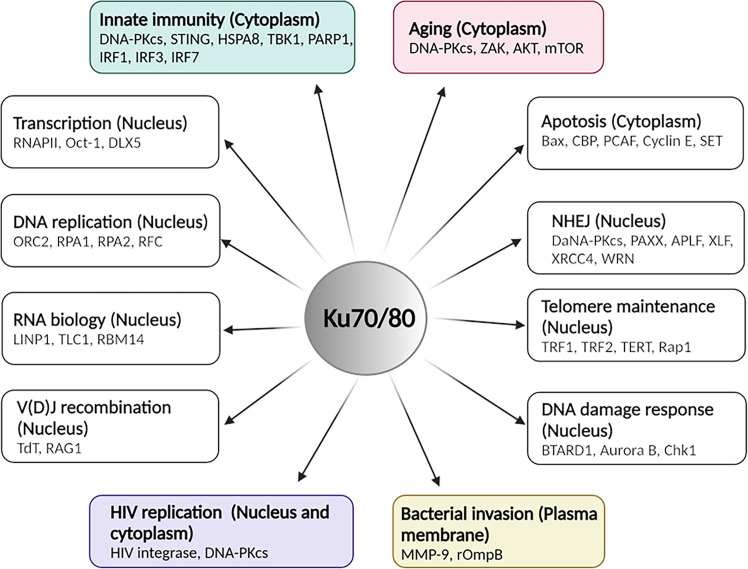
Figure 2.
The overview of functions of Ku70/80 heterodimer in various cellular contexts. Ku70/80’s involvement in multiple cellular activities with indicated cellular localization and critical mediators. This illustration was created by using BioRender. The figure was adapted from the 2021 review by Abbasi et al. (Abbasi et al., 2021).
Table 1.
List of studies about Ku70-involved innate immune response.
| Sensor proteins | The source of nucleotides | Host cells | Signaling pathway | Induced cytokines | In vivo | References |
|---|---|---|---|---|---|---|
| Ku70 | Plasmid DNA, bacterial DNA. HSV-2G, HSV-1 | HEK, RD, THP-1, macrophages | STING-TBK1-IRF3, IRF1, and IRF7 pathway | IFN-λ1 | N/A | (Zhang et al., 2011a; Sui et al., 2017; Sui et al., 2021) |
| DNA-PK | VACV, E. coli, ISD, HSV-1, MVA | Fibroblasts, MEF | STING-TBK1-IRF3 | IFN-β, CXCL10, IL-6 | Mice | (Ferguson et al., 2012; Peters et al., 2013; Scutts et al., 2018) |
| Ku70 | pAAV-HBV plasmid, HBV | Liver-derived cells: Sk-Hep-1, Hep G2, Huh7, primary HSECs | DNA-PKcs and PARP1-IRF1 | CCL3, CCL5 | HBV-infected human patients | (Li et al., 2016b) |
| Ku70 | HTLV-1 RTI ssDNA90 | HeLa cells, PMA-THP-1 | STING-TBK1-IRF3 | IFN-β, IFN-λ, and TNF-α | N/A | (Wang et al., 2017) |
| DNA-PK | CT DNA | Human U937 cells, primary human hepatocytes, human fibroblasts |
HSPA8-IRF3 (STING-independent sensing pathway) | IFN-β | N/A | (Burleigh et al., 2020) |
| Ku70/Ku80 | ISD | Jurkat T cells, aged CD4+ T cells | ZAK-AKT-mTOR | IL-2, IFN-γ, T-cell proliferation | Mice | (Wang et al., 2021) |
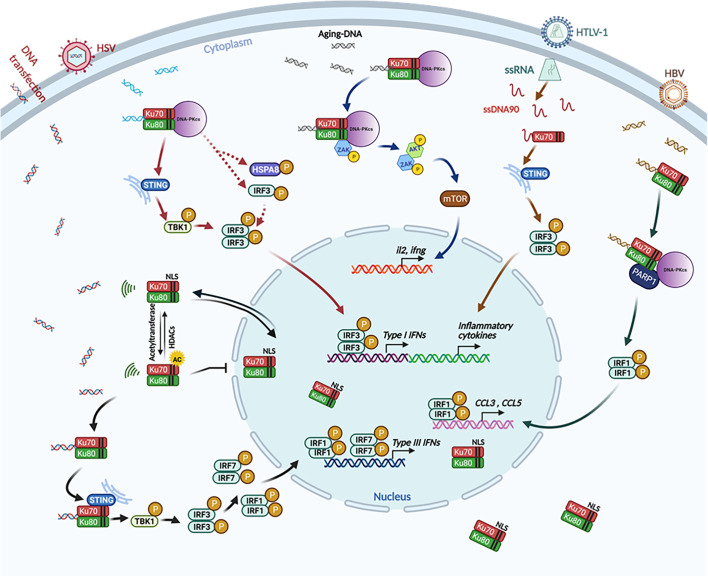
Figure 3.
The involvement of Ku70 as a cytosolic DNA sensor to activate the innate immune response. Ku70 is identified as a cytosolic DNA sensor that induces type III IFNs through a STING-TBK1-IRF3, IRF1, and IRF7 signaling pathway. In this signaling pathway, cytoplasmic translocation of Ku70 is an initial and essential step (black arrows) (Zhang et al., 2011a; Sui et al., 2017; Sui et al., 2021); Ku70 and Ku80, together with DNA-PKcs (DNA-PK), are also involved in a STING-dependent (Ferguson et al., 2012) (solid arrow) and STING-independent (Burleigh et al., 2020) (dashed arrow) pathway to induce type I IFNs (red arrows). Ku70 is reported to sense HTLV-1 transcription intermediate product ssDNA90 and interacts with STING to induce IFNs and inflammatory cytokines, thereby modulating HTLV-1 replication (Wang et al., 2017) (brown arrows). The Ku70/80 heterodimer recognizes HBV-infection-derived DNA, then activates DNA-PKcs and PARP1 to induce CCL3 and CCL5 inflammatory cytokines (Li et al., 2016b) (green arrows). DNA-PK (the complex of DNA-PKcs, Ku70, and Ku80) senses aging-related cytoplasmic DNA in CD4+ T cells. This DNA sensing then induces T-cell proliferation and activation, as well as autoimmunity through the ZAK-AKT-mTOR pathway (Wang et al., 2021) (blue arrows). This illustration was created by using BioRender.
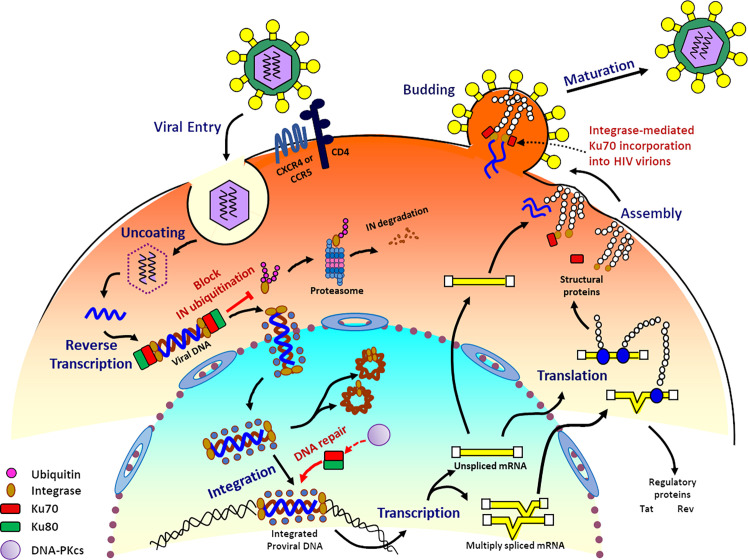
Figure 4.
Ku70 is an indispensable host cellular factor in the early and late stages of the HIV-1 replication cycle. The interaction of IN with Ku70 during HIV reverse transcription prevents IN from degradation by the K48-linked Ub proteasome pathway. The interaction between Ku70 and IN decreases the modification level of IN by Ub in the cells. During the integration step, the initial binding of Ku70 and HIV-1 IN facilitates the recruitment of other members of the DNA-PK complex to the post-integration site. Then Ku70 serves as a member of DNA-PK and participates in the DNA gaps repair process through the NHEJ pathway, thereby completing the integration of viral DNA into the cell genome and enabling the HIV-1 viral replication. Ku70 is also packaged into HIV particles as early as its assembly stage and becomes part of HIV virions, and this process is mediated by HIV IN.
Structure and Diverse Functions of Ku70 and Ku80
Ku70 forms a heterodimer with Ku80. Homology modeling for human Ku70 and Ku80, the proteins alone, and the complex with an oligo DNA substrate are shown in Figure 1 . The entire structure of Ku70/Ku80 was not crystallized (Walker et al., 2001) due to the difficult experimental conditions of solving regions such as the Ku70 NLS. The homology modeling technique was used here to predict such missing segments. The hetero-oligomeric modeling pipeline implemented in SWISS-MODEL (Biasini et al., 2014) was used to predict the Ku70/Ku80 dimer structure. The target sequences of Ku70 (XRCC6, UniProt: P12956) and Ku80 (XRCC5, UniProt: P13010) were used as the input, and the crystal structure (PDB ID: 1JEQ (Walker et al., 2001)) was retrieved as the template for the final modeling. The target modeling segments show high sequence identities with their respective templates when aligned with those templates (95% for Ku70 and 97% for Ku80). Finally, the Ku70 structure was modeled, including 575 amino acids with the first 34 amino acids truncated, while the Ku80 structure with amino acids 6–541 was modeled. To create the C-terminal domain of the Ku80 model, we used the PDB ID of 6ZHE (Chaplin et al., 2021) as the template due to its relative completeness in this domain ( Figure 1B ). For incorporating the DNA coordinate into the Ku70/Ku80 model ( Figure 1C ), the PDB structure of 1JEY (Walker et al., 2001) was used as the anchor for fitting the model. Both the Ku70 and Ku80 protein possess a three-domain topology, including an N-terminal vWA (von Willebrand factor A) domain, a DNA-binding domain, and a C-terminal arm (Walker et al., 2001) ( Figure 1A ). The homology modeling of Ku heterodimer suggested a quasi-symmetric basket-like molecule with a narrow-preformed ring, which facilitates the binding of DNA to Ku (Walker et al., 2001) ( Figures 1B, C ).
The N-terminal vWA domains of Ku70 or Ku80 are composed of a six-stranded β-sheet in a Rossman fold (Walker et al., 2001). Disrupting the vWA domains in yeast Ku70/80 has been found to impair the function of Ku in DNA repair and telomere regulation (Ribes-Zamora et al., 2007). Although the amino edge of the vWA domain locates close to the DNA-binding groove, the vWA domain is not required for DNA binding. However, the Ku vWA domains may facilitate protein-protein interactions. For instance, the vWA domain of Ku80 has been found to interact with APLF, an NHEJ repair protein important for recruiting other repair factors (Grundy et al., 2013). So, the N-terminal vWA domains have minimal contribution to heterodimerization or DNA binding but are potentially involved in protein-protein interactions.
Meanwhile, the middle domain consists of a seven-stranded anti-parallel β-barrel and plays an essential role in Ku DNA binding and heterodimerization (Walker et al., 2001). Heterodimerization leads to a positively charged DNA-binding ring that fits sterically around the minor and major DNA grooves. Ku threads inwards on DNA like a nut threaded onto a bolt, with Ku70 positioned close and Ku80 far away to the DNA end (Yoo et al., 1999; Doherty and Jackson, 2001; Abbasi et al., 2021). Ku binds to dsDNA ends, 5′ and 3′ overhangs, or blunt ends with a higher binding affinity (Kd = 10–9 M). And it has a much lower binding affinity with circular DNA or the ends of single-stranded DNA (ss DNA) (Fell and Schild-Poulter, 2015).
C terminal regions of Ku contain a flexible linker and a globular structural domain ( Figure 1 ). The C-terminal region of Ku70 contains a SAP (SAF-A/B, Acinus, and PIAS) domain encoded by residues 559–609 (Walker et al., 2001). Studies on other SAP domain proteins have implicated that SAP domains can bind DNA (Göhring et al., 1997; Suzuki et al., 2009). Using a pair of even shorter versions of Ku70, the Ku70_251-438 and Ku70_439-609 truncated mutants, Anisenko et al. have determined that the dsDNA is bound within the C-terminal part of the protein containing SAP domain (Anisenko et al., 2017b). While DNA binding to Ku, the SAP domain undergoes displacement, making itself close to the DNA-binding region of the Ku heterodimer (Rivera-Calzada et al., 2007; Makowski et al., 2016). Even the exact function of Ku70-SAP has not been completely investigated yet, the helical C-terminal arms of Ku contribute to heterodimerization and stabilize the interaction of Ku to DNA (Walker et al., 2001; Keijzers, 2018).
Notably, Ku70 and Ku80 per se possess an NLS in the molecule (as shown in Figure 1 with Ku70 NLS in yellow and Ku80 NLS in black). The various NLS are classified into two types based on the pattern of molecular sequences: (1) a single cluster of basic amino acids, such as the NLS of the SV40 T-antigen, and (2) a bipartite type in which two basic amino acid regions are separated by a stretch of approximately 10 non-basic amino acids (Görlich and Mattaj, 1996). The Ku70 NLS belongs to type 2, and the sequence of Ku70 NLS is highly conserved among human, mouse, rat, hamster, and chicken (Koike et al., 1999b). While importing into the nucleus of the cells, the Ku70 NLS is recognized by the nuclear targeting complex, PTAC58, and PTAC97 (Koike et al., 1999b). Given that Ku70 is an NLS-possessing protein, it has been found predominately in the nucleus of unstimulated cells, such as HeLa, HEK (human embryonic kidney cells), and rhabdomyosarcoma (RD) cells (Sui et al., 2021).
Consistent with the illustrated structure, many studies have suggested that Ku possesses unusual DNA-binding properties, binding potently to the ends of dsDNA molecules in a sequence-independent manner. The unusual end-binding properties are required for various nuclear processes, such as NHEJ DNA repair (Critchlow and Jackson, 1998; Dobbs et al., 2010; Radhakrishnan et al., 2014; Menon and Povirk, 2016; Scully et al., 2019), V(D)J recombination of immunoglobin (Jackson and Jeggo, 1995; Fugmann et al., 2000; Bassing et al., 2002), telomerase maintenance (Bertuch and Lundblad, 2003; Indiviglio and Bertuch, 2009; Wood et al., 2015; Shay and Wright, 2019; Sui et al., 2020), transcription (Li et al., 1995; Giffin et al., 1996; Ono et al., 1996; Giffin et al., 1997; Dynan and Yoo, 1998; Mo and Dynan, 2002; Bunch et al., 2015), DNA damage response (Wang et al., 2000; Zhou and Elledge, 2000; Rouse and Jackson, 2002; Harper and Elledge, 2007; Jackson and Bartek, 2009; Fell and Schild-Poulter, 2012; Nowsheen and Yang, 2012; Blackford and Jackson, 2017), RNA biology (Yoo and Dynan, 1998; Peterson et al., 2001; Stellwagen et al., 2003; Ting et al., 2005; Pfingsten et al., 2012; Lamaa et al., 2016; Zhang et al., 2016b; Dutertre and Vagner, 2017; Shao et al., 2020; Thapar et al., 2020), and DNA replication (Barnes and Rio, 1997; Shao et al., 1999; Novac et al., 2001; Cosgrove et al., 2002; Matheos et al., 2002; Schild-Poulter et al., 2003; Park et al., 2004; Rampakakis et al., 2008; Miyoshi et al., 2009; Foster et al., 2011; Abdelbaqi et al., 2013; Sánchez and Russell, 2015; Teixeira-Silva et al., 2017). Such unusual DNA-binding properties also facilitate Ku70’s activities in the cytoplasm of the cells. Those activities include participating in Bax-mediated apoptosis (Cohen et al., 2004; Gomez et al., 2007; Mazumder et al., 2007; Kim et al., 2014) and serving as a cytosolic DNA sensor to activate the DNA-mediated innate immune response. An overview of Ku70/80 heterodimer functions in the various cellular processes is illustrated in Figure 2 . In the following paragraphs, we will discuss roles of Ku70 in innate immunity, aging-related cytoplasmic DNA sensing, HIV replication and bacterial invasion in detail.
Ku70 is Identified as a Novel Cytosolic DNA Sensor That Mediates Innate Immune Responses
Our lab previously reported, for the first time, that Ku70 is a novel DNA sensor to induce expression of IFN-λ1 rather than that of type-I IFNs (Zhang et al., 2011a). Plasmid DNA transfection or DNA virus infection-mediated IFN-λ1 induction was detected in HEK cells, RD cells, monocyte-derived macrophages, immature dendric cells, and—with a much lesser level—HeLa cells (Zhang et al., 2011a). These results indicated that the Ku70-mediated IFN-λ1 induction is consistently presented in multiple cell types.
Different forms of DNA transfection (e.g., single-stranded DNA, fragmented human genomic DNA, and bacterial DNA) and infection of DNA virus induce IFN-λ1 (Zhang et al., 2011a); IFN-λ1 mRNA was induced by both supercoiled or linearized forms of DNA plasmids. However, the linearized plasmid DNA significantly enhanced activation. This result was consistently supported by the property of Ku, which detects the end structure of DNA. Zhang et al. confirmed that over 500 bp of DNA triggers IFN-λ1 induction with the dependency of DNA length. By contrast, the production of IFN-λ1 was not detected with the transfection of DNA that was only 50 bp in length (Zhang et al., 2011a). One study demonstrated that titration of Ku to a fixed amount of linear dsDNA fragments produced ladders of shifted bands, which are proportional to the length of DNA. This data implicated that many Ku heterodimers bind to multiple sites on one dsDNA in a sequence-independent pattern (Blier et al., 1993). Based on those Ku properties, it was apparent that Ku70 induces activation of IFN-λ1 and that Ku70 recognizes intracellular DNA by DNA transfection or infection with a DNA virus, such as herpes simplex virus (HSV) type 1 (HSV-1) or type 2 (HSV-2), without any restriction in structure or sequence (Zhang et al., 2011a).
In addition to the fact that Ku70 senses DNA to induce type III IFNs, subsequent other studies indicated that Ku70 perse, or Ku70 in Ku70/Ku80 heterodimer, or Ku70 in the DNA-PK complex involves in the induction of type I IFNs and other inflammatory cytokines directly or indirectly. For example, Ku70 has also been reported to detect human T-lymphotropic virus type 1 (HTLV-1) reverse intermediate product ssDNA90 and induce IFN-α, IFN-β, IFN-λ, and RANTES (Wang et al., 2017). Additionally, the Ku70/Ku80 heterodimer senses the in vitro adenovirus-delivered hepatitis B virus (HBV) DNA and induces CCL3 and CCL5, thereby implicating that Ku70 modulates HBV replication (Li et al., 2016b). More interestingly, a recent study suggested that the Ku70/80 complex senses cytoplasmic DNA in aged CD4+ T cells and that this detection potentiated T-cell activation and aging-related autoimmune responses (Wang et al., 2021). Furthermore, Ferguson et al. reported that DNA-PK, a heterotrimeric protein complex composed of Ku70, Ku80, and DNA-PKcs, is able to activate downstream STING-TBK1-IRF3 signaling pathway when it recognizes foreign DNA (Ferguson et al., 2012). It has been further demonstrated that DNA-PK co-localizes with vaccinia virus (VACV) DNA during VACV infection. Virus infection-mediated IFN response is aborted when the components of DNA-PK were knocked-out (Ferguson et al., 2012).
Ku70, Ku protein, or DNA-PK have been implicated in having a role in sensing a variety of DNA or DNA viruses without restrictions. More importantly, many other DNA sensors, such as cGAS, require binding of double-stranded DNA to activate the sensor protein: a conformation change, thereby activating downstream signaling (Cai et al., 2014; Dempsey and Bowie, 2015; Xia et al., 2016). However, the Ku protein or DNA-PK complex does not have such conformational restriction; therefore, the Ku/DNA-PK-mediated innate immune response may become a perfect complementary pathway in the host defense system when other DNA-sensing pathways are impaired.
The Downstream Signaling Pathway of Ku70
Many studies have indicated that Ku70, as a cytosolic DNA sensor, binds with DNA and mediates the downstream signaling pathway. However, “What is the adapter at the downstream signaling of Ku70?” was the next question. In Ku70-mediated type III IFN response, an investigation was initiated from the observation of IFN-λ1 induction in HEK and 293T (SV40-T antigen transformed HEK cell line) cells with GFP-encoding DNA plasmid transfection. With a similar green fluorescence signal observed between HEK 293 cells and 293T cells, DNA-induced IFN-λ1 induction was detected in HEK 293 cells but not in 293T cells. By comparing the expression level among different signal mediators associated with the cytosolic sensor, we found that the stimulator of interferon genes (STING) is not endogenously expressed in 293T cells. The gain-of-function and loss-of-function study confirmed the hypothesis that STING is the downstream adaptor of Ku70 to activate the IFN-λ1 signaling pathway. The co-immunoprecipitation assay further illustrated that Ku70 interacts with STING in the cytoplasm and forms a complex upon DNA stimulation (Sui et al., 2017). At this point, the activating pathway is quite similar to the DNA-PK-mediated STING-dependent pathway. DNA-PK was reported as a DNA cytosolic sensor to induce IFN-α or IFN-β (Ferguson et al., 2012). However, the interaction between DNA-PK and the downstream STING is in a transient pattern. After binding at three hours after DNA stimulation, STING dissociates from the complex, and this dissociation activates downstream signaling (Ferguson et al., 2012). STING is also the downstream target of Ku70 in sensing HTLV-1 intermediate product ssDNA90 and, therefore, induces type I interferons and inflammatory cytokines through phosphorylation of IRF3 (Wang et al., 2017).
In addition to STING as the downstream adaptor of Ku70 or DNA-PK, other proteins, namely DNA-PKcs and PARP1, are also reported as the adaptor proteins to Ku70/80 in sensing HBV DNA (Li et al., 2016b). While sensing aging-related DNA cytoplasmic accumulation, DNA-PK interacts with ZAK, AKT, and mTOR, inducing T-cell proliferation and aging-related autoimmunity (Wang et al., 2021). Another study recently claimed that DNA-PK is a potent sensor that activates the innate immune response with STING-independent signaling pathway. However, this pathway only exists in human cells and is not present in mouse cells (Burleigh et al., 2020). In this pathway, HSPA8/HSC70 is the adaptor protein for inducible phosphorylation and then activates downstream innate immune signaling (Burleigh et al., 2020). All those different Ku70-involved mechanisms determine the diverse patterns of innate immune response in a cell-type-dependent pattern. The coexistence of various molecular mechanisms is always an interesting topic in the research field of innate immunity.
Compared with the induction of type I IFNs, the kinetics of Ku70-mediated IFN-λ1 induction indicates a delayed induction profile. The IFN-λ1 mRNA expression is initiated at about 12 hours after DNA transfection. A profound protein level of IFN-λ1 can be detected at 48 hours after DNA transfection (Sui et al., 2017). As we know, cGAS- or IFI16-mediated innate immune response is usually induced as an earlier event after stimulation: for example, at six hours after stimulation (Unterholzner et al., 2010; Sun et al., 2013; Cai et al., 2014). The activation of downstream of cGAS or IFI16 is the STING-TBK1-IRF3 signaling pathway. IRF3 is endogenously expressed in most cells. The activation of IRF3 is detected at three hours after stimulation, indicating that IRF3 facilitates the induction as a faster and earlier event. By contrast, Ku70-mediated IFN-λ1 induction relies on activating the STING-TBK1-IRF3, IRF1, and IRF7 axis (Zhang et al., 2011a; Sui et al., 2017). IRF3 is activated first to produce a profound expression of IRF1 and IRF7, since IRF1 and IRF7 are not endogenously expressed in the cells. Once IRF1 and IRF7 are produced, IFN-λ1 and then can be significantly induced. In summary, similar to other DNA sensor-mediated innate immune responses, the kinetics of Ku70 involved innate immune response depends on the specific signaling pathway by which interferon or inflammatory cytokines are produced.
The Cytoplasmic Translocation of Ku70 Is The Initial Step For Ku70 as a Cytosolic DNA Sensor
Ku70 was initially characterized as a DNA repair protein; its primary function serves in the nucleus. However, more and more studies have identified Ku70 as a cytosolic DNA sensor that mediates innate immune response. The downstream adaptor STING and another protein, HSPA8, were all found in the cells’ cytoplasm (Ferguson et al., 2012; Sui et al., 2017; Burleigh et al., 2020). So how can a nuclear protein sense cytosolic DNA and thereby initiate a downstream signaling pathway? This question led to identify the molecular mechanism at an earlier time point. In the case of Ku70-mediated IFN-λ1 induction by DNA transfection, Ku70 was observed predominately located in the cytoplasm of the cells, thereby facilitating the interaction between STING and Ku70 (Sui et al., 2017). So, it was speculated that upon DNA stimulation, Ku70 translocates from the nucleus to the cytoplasm of the cells. And this process is closely correlated with the induction of IFN-λ1.
The subsequent study using confocal microscopy confirmed that the cytoplasmic translocation of Ku70 is observed in the cells in which IFN-λ1 is induced by DNA transfection, such as HEK and RD cells. And consistently, such cytoplasmic translocation of Ku70 is not observed in HeLa cells, and similarly, DNA transfection does not induce IFN-λ1 induction in HeLa cells (Sui et al., 2021). In addition to DNA plasmid transfection, HSV-1, a DNA virus infection, also triggered the cytoplasmic translocation in HEK cells with a virus-strain-dependent manner. Ku70 cytoplasmic translocation and IFN-λ1 induction only in HEK cells infected with the HSV-1 McKrae strain. Those results further emphasized that the cytoplasmic translocation of Ku70 is a required step for Ku70-mediated IFN-λ1 induction (Sui et al., 2021). A quantification analysis with Western blot using cytosolic fractions was adapted to characterize the accumulation kinetics of cytoplasmic Ku70. The data demonstrated that the cytoplasmic translocation of Ku70 was started right after DNA stimulation and obtained the highest level at six hours after transfection, and then the accumulation of cytoplasmic Ku70 returned to a similar level as that in unstimulated cells (Sui et al., 2021). These data testified two points. First, the cytoplasmic translocation of Ku70 is a kinetics process. Ku70 translocates freely from the nucleus to the cytoplasm or from the cytoplasm back to the nucleus. How DNA transfection triggers the translocation remains unclear, but we hypothesized it is due to a change in a dynamic balance between the accumulation level of Ku70 in the nuclear and the cytoplasm. When cytosolic Ku70 recognizes and associates with cytoplasmic DNA, such interaction may interrupt the equilibrium between the cytosolic and the nuclear Ku70 and then drive the translocation of Ku70 from the nucleus to the cytoplasm. Second, the kinetic study further demonstrated that the translocation of Ku70 from the nucleus to the cytoplasm is an initial and essential step in the DNA-mediated IFN-λ1 innate immune response. Compared with the time course of IFN-λ1 induction, the translocation of Ku70 occurred one hour right after DNA transfection and peaked at six hours. All this happened before IFN-λ1 induction. Consequently, it is reasonable to speculate that the translocation of Ku70 happened first and that IFN-λ1 induction occurred later since we further confirmed that recombinant IFN-λ1 does not induce the cytoplasmic translocation of Ku70 (Sui et al., 2021). Like Ku70, IFI16, another DNA sensor protein, has been reported to recognize and sense DNA not only in the cytoplasm but also in the nucleus of the cells, and its sensing capabilities depend on the distribution of IFI16 (Li et al., 2012; Dell’oste et al., 2014; Ansari et al., 2015). IFI16 detects and binds to herpes viral DNA in the nucleus of the cells. However, detection of transfected DNA or cytoplasmic viral DNA occurs in the cytoplasm. Ku70 ubiquitously expresses in the nucleus as a nuclear protein; however, there is no evidence to indicate that Ku70 can also serve as a nuclear DNA sensor protein.
In addition to our detailed study about the cytoplasmic translocation of Ku70, Li et al. also reported that the cytoplasmic-translocated Ku70/Ku80 complex senses HBV DNA and then induces hepatitis-associated chemokine secretion (Li et al., 2016b). This kind of nuclear-cytoplasmic (N-C) trafficking has become a conventional mechanism for these multifunctional DNA sensors. As we know, cGAS recognizes cytosolic DNA. This detection produces the second messenger 2’3’-cGAMP, and the cGAMP in turn initiates STING-dependent downstream signaling to induce type I IFNs. However, more recently, Sun et al. demonstrated that cGAS is located both in the cytoplasm and in the nucleus, and cGAS is required to export into the cytoplasm in response to DNA stimulation. (Sun et al., 2021). Therefore, the N-C trafficking is required for Ku70 and other multiple-functional proteins to conduct their cytosolic and nuclear activities.
The Cooperative Pattern of Ku70, Ku80, and DNA-PKcs In Mediating Innate Immune Response
Ku70 is a subunit of the heterotrimeric protein complex DNA-PK composing of Ku80 and the catalytic subunit DNA-PKcs. While we identified Ku70 as a novel cytosolic DNA sensor that induces IFN-λ1 innate immune response (Zhang et al., 2011a; Sui et al., 2017; Sui et al., 2021), we hope to determine whether Ku80 or DNA-PKcs are also involved in the cytosolic-DNA-sensing activity.
It has been reported that DNA-PK serves as a PRR, recognizing cytoplasmic DNA and inducing the production of type I IFNs (Ferguson et al., 2012; Burleigh et al., 2020). The Ku heterodimer (Walker et al., 2001) and DNA-PKcs (Hammarsten and Chu, 1998) can directly bind to DNA; however, in the absence of either Ku70 or Ku80, the binding affinity of DNA-PKcs with DNA is dramatically decreased (Yaneva et al., 1997). These findings implicated that each subunit of the DNA-PK complex plays an essential role. Consistent with this study, we also observed the existence of Ku80, but not DNA-PKcs, in the complex of Ku70-STING (Sui et al., 2017). Additionally, we observed the co-localization of Ku80 with Ku70 in the nucleus of unstimulated cells and the cytoplasm of DNA-transfection-stimulated cells. Those data suggest that Ku80 is translocated with Ku70 from the nucleus to the cytoplasm (Sui et al., 2021).
However, we previously reported that DNA-mediated IFN-λ1 induction substantially decreased, when Ku70 is transiently knocked down; in contrast, knocking down of Ku80 has no impact on the induction of IFN-λ1. To further validate the role of Ku70 and Ku80 in DNA-mediated innate immune response, the IFN‐λ1 promoter reporter assay by overexpressing each subunit was utilized in the study. The result from the assay consistently demonstrated that overexpression of Ku70 highly activates the IFN‐λ1 promoter. However, the overexpression of Ku80 had no impact on IFN‐λ1 promoter activation. Moreover, the results of the co-immunoprecipitation assay directly exclude the presence of DNA-PKcs in the complex of Ku70-STING. Therefore, all those studies suggested that Ku80 and DNA-PKcs may not be directly involved in DNA-mediated IFN-λ1 induction (Zhang et al., 2011a; Sui et al., 2017).
In studies about DNA-PK as the cytosolic DNA sensor to induce the innate immune response, it seems that DNA-PKcs is the key factor to mediate downstream signaling and that the involvement of Ku70 or Ku80 enhances the sensing capability of DNA-PKcs (Ferguson et al., 2012; Burleigh et al., 2020). In HBV infection, the Ku70/80 complex senses infected HBV DNA, and DNA-PKcs and PARP1 act as a downstream adaptor to activate hepatitis-associated chemokine secretion (Li et al., 2016b).
Ku70-involved innate immune response shows various patterns for the participation and function of Ku70, Ku80, and DNA-PKcs. In general, like their function in the DNA repair process, they work together as a whole complex (Ferguson et al., 2012; Burleigh et al., 2020; Wang et al., 2021), but in the case of type-III IFN response and Ku70 sensing HTLV-1, the functional element is Ku70 itself. However, we have become aware that when Ku70 or Ku80 is expressed individually, neither of them are stable (Satoh et al., 1995) and that the absence of one of the subunits leads to a remarkable reduction in the stable level of the other one (Errami et al., 1996; Gu et al., 1997; Singleton et al., 1997). Consequently, it is hard to precisely elucidate the function of Ku70 or Ku80 alone by completely knocking out one or the other. Further study will help to illuminate the detailed molecular mechanism of how Ku70, Ku80, or DNA-PKcs cooperate and facilitate DNA-sensing activity.
The Potential Regulation Factors Involved in Ku70-Mediated Innate Immune Response
Further studies have reported that Ku70, predominantly located in the nucleus of the cells, has a cytoplasmic translocation from the nucleus, then conducts its cytosolic activities, such as sensing invading cytosolic DNA to induce an innate immune response (Zhang et al., 2011a; Sui et al., 2017; Sui et al., 2021) or binding with invading viral elements/proteins to modulate virus replication (Li et al., 2016b). Therefore, abundant amounts of cytoplasmic protein accumulation seem to be essential for Ku70 to successfully recognize cytosolic DNA and activate the downstream IFN signaling pathway. Our observations, nuclear retention of Ku70, because of the treatment with leptomycin B, severely attenuates the IFN-λ1 response to DNA stimulation (Sui et al., 2021), indicating that cytoplasmic translocation is a critical factor for Ku70’s cytosolic DNA sensing.
Our group confirmed that acetylation at Ku70-NLS regulates the localization of Ku70 in the nucleus or in the cytoplasm, which is consistent with the finding from other groups (Fujimoto et al., 2018), and we first reported that acetylation modulates Ku70’s DNA‐sensing activity (Sui et al., 2021). While importing into the nucleus, Ku70 has to interact with the Impα/Impβ complex to facilitate nuclear translocation. With the acetylation at the region of Ku70-NLS, the interaction between acetylated Ku70 and the Impα/Impβ complex is severely decreased. Therefore, acetylated Ku70 is predominantly located in the cytoplasm of the cells (Fujimoto et al., 2018). In line with Fujimoto’s finding, we further demonstrated that acetylated Ku70 highly induces DNA-mediated IFN-λ1 induction (Sui et al., 2021).
The acetylation level of a protein depends on the dynamic balance between the activity of acetylation and deacetylation enzymes (Ansari et al., 2015; Gong et al., 2019). Multiple lysine residues have been identified as acetylation locations on Ku70 and Ku80 (Cohen et al., 2004; Subramanian et al., 2013; Al-Emam et al., 2014; Koike et al., 2017). Acetylation at Ku70 lysine residues, K539, K542 (Subramanian et al., 2005; Subramanian et al., 2013) and K317, K331, K338 (Al-Emam et al., 2014) impaired the function of Ku70 in NHEJ, since those lysine residues of Ku70 are required for Ku70 binding with dsDNA ends during NHEJ process. Two histone acetyltransferase enzymes, CBP and PCAF, are responsible for Ku acetylation (Cohen et al., 2004). Histone deacetylases (HDACs), a family of deacetylation enzymes, regulate the deacetylation of multiple non‐histone proteins and, therefore, impact functions by changing their activity, such as cellular localization and protein-protein interactions (Subramanian et al., 2005; Roger et al., 2011). More than 50 non‐histone proteins, including p53 and Ku70, have been defined as the substrates of HDACs (Chaudhary et al., 2014; Gong et al., 2019). Trichostatin A (TSA), an inhibitor sensitive to class I/II deacetylases, was utilized in our study to evaluate the impact of this deacetylase inhibitor on the Ku70 cytoplasmic accumulation and DNA-mediated IFN-λ1 induction. The data implicated that TSA treatment dose-dependently enhances the cytoplasmic accumulation of Ku70 and increases DNA-mediated IFN-λ1 induction. (Sui et al., 2021). As we demonstrated in our study, the relationship of the acetylation levels of Ku70 and DNA-mediated innate immune response may provide a simple and elegant strategy, modulating the acetylation levels of the target protein to regulate its localization-dependent activities.
Ku70 and Ku80 are generally believed to always form heterodimers. And it has been consistently confirmed in our previous study that Ku80 translocates from the nucleus to the cytoplasm together with Ku70. The confocal microscopy analysis indicated that Ku70 and Ku80 colocalized together in the nucleus of unstimulated cells, and then both translocate from the nucleus to the cytoplasm upon a DNA stimulation (Sui et al., 2021). However, Koik et al. demonstrated that the localization of Ku80 does not entirely coincide with that of Ku70, Ku80 protein was transported to the nucleus without heterodimerization with Ku70. The Ku80 NLS was demonstrated to be mediated to the nuclear rim by two components of PTAC58 and PTAC97. This findings support the idea that Ku80 can translocate to the nucleus using its own NLS independent of the translocation of Ku70 (Koike et al., 1999a). On the other hand, using the site-directed mutagenesis technique, the same group demonstrated that Ku70 can also translocate to the nucleus without heterodimerization with Ku80 or independent of DNA-PK autophosphorylation (Koike et al., 2000).
The N-C or C-N translocation of DNA-PKcs is rarely reported. We have confirmed DNA-PKcs is not involved in Ku70-mediated IFN-λ1 induction. Co-immunoprecipitation assay suggested DNA-PKcs is not present in the Ku70-STING complex. Therefore, implicating that DNA-PKcs does not translocate together with Ku70 or Ku80 from the nucleus to the cytoplasm upon a DNA transfection or DNA virus infection (Sui et al., 2017). Other factors may involve in facilitating the translocation of DNA-PKcs. Further study is required to help us understand the translocation of DNA-PKcs.
Regulation of the N-C translocation has been defined as an essential mechanism to control protein activities. Whether some other cellular factors may facilitate Ku70, Ku, or DNA-PK to respond to specific stimuli by regulating its nuclear or cytoplasmic localization remains to be identified. Better understanding the N-C translocation of Ku70, Ku80 and DNA-PKcs may provide unique insights into the multiple functions of Ku70 in the DNA repair process, Bax-mediated apoptosis, and innate immune response.
In addition to regulating the N-C transport, the detailed mechanisms regarding Ku70-mediated innate immunity offer multiple strategies to downregulate cytosolic DNA-induced autoimmunity or enhance innate immune response under the context of DNA vaccination. For example, Wang et al. found that the Ku70/Ku80 heterodimer recognizes aging-related DNA accumulation in the cytoplasm of human or mouse CD4+ T cells. The sensing by the Ku complex further recruits DNA-PKs on the site and triggers the phosphorylation of ZAK. Subsequently, it activates the AKT-mTOR signaling pathway, which enhances the proliferation of CD4+ T cells and accelerates the pathology progress of experimental autoimmune encephalomyelitis (EAE) in mice (Wang et al., 2021). Consequently, based on the discovered molecular mechanism, the group further developed an inhibitor specific against ZAK to dampen the pathology progress of EAE (Wang et al., 2021).
It is known that many viruses possess the system to escape from the innate immune response by host cells. The mechanism of the immune escape by a DNA virus, Vaccinia virus (VACV), has been investigated (Bowie and Unterholzner, 2008; Elde et al., 2012). The VACV C16 protein was reported as the first protein to inhibit DNA-PK-mediated signaling (Peters et al., 2013). It has been demonstrated that the C-terminal region of C16 binds directly to the Ku70/Ku80 complex, therefore blocking the sensing of Ku to DNA. The protein VACV C16 is not endogenously expressed on VACV strain Western Reserve. So the intranasal infection of this virus strain in mice leads to enhanced innate immune response and less symptoms of viral infection-related sickness (Fahy et al., 2008; Peters et al., 2013). Another protein of VACV, C4, is later identified targeting DNA-PK to inhibit DNA-PK-mediated signaling. VACV C4 possesses a similar sequence as C16, so it shares a similar mechanism to block DNA binding to DNA-PK by binding to the Ku complex. The absence of C4 promotes innate and adaptive immune responses (Scutts et al., 2018). Overall, these findings demonstrate that viral proteins help to evade the sensing of the viral genome by inhibiting the activity of PRR, therefore highlighting alternative strategies to regulate the innate immune response.
Similar to VACV, DNA virus HSV-1 has also shown the ability to evade innate immune responses in host cells (Su et al., 2016; Lum and Cristea, 2021). Studies from Zheng’s lab implicated that HSV-1 VP24, a serine protease, could also inhibit dsDNA-initiated IFN production by blocking the interaction between IRF3 and TBK1 and therefore dampening the phosphorylation of IRF3 (Zhang et al., 2016a). Another study demonstrated that HSV-1 VP16 could interrupt IRF3 recruiting the CREB-binding protein coactivator, thus inhibiting IRF3-mediated downstream signaling (Xing et al., 2013). Furthermore, US3 of HSV-1, another viral protein kinase, has been reported to prevent IRF3 activation and inhibit type-I IFN production by hyper phosphorylating IRF3 at Ser175 (Wang et al., 2013; Wang et al., 2014). It has been reported that HSV-1 ICP27 interacts with TBK1 and STING, which impairs the activation of downstream transcription factor IRF3. (Christensen et al., 2016). Our previous study also found that the Ku70-mediated type-III IFN response was induced in an HSV-1 strain-dependent manner: Infection with the HSV-1 McKrae strain triggers the cytoplasmic translocation of Ku70 and induces IFN-λ1 induction, while infection with the HSV-1 MacIntyre strain does not. Therefore, we speculated that the HSV-1 MacIntyre strain might encode specific viral proteins that may inhibit the signaling pathway of IFN induction. Further studies are needed to identify the specific molecular mechanism for HSV-1 immune evasion. As we listed above, all these observations implicated that HSV-1-encoded viral proteins to facilitate HSV-1 immune evasion could interrupt the downstream signaling of DNA-mediated signaling pathway, therefore providing potential strategies to regulate any signaling pathway with shared downstream signaling adaptors, such as STING, TBK1, and IRF3.
In summary, with an aim to highlight innate immune response mediated by DNA virus infection in a battle against viral infection, a better understanding of the interplay between host innate immune response and viral immune evasion would provide intriguing novel strategies to help develop diverse therapies to treat viral infection-related diseases.
Beyond the Role of Ku70 In Innate Immunity: A Role of Ku70 in HIV Replication Cycle and Bacterial Internalization
HIV needs many cellular factors to facilitate its replication (Emig-Agius et al., 2014). Ku70 and Ku80 are reported as host partners involving in HIV replication (Waninger et al., 2004; Studamire and Goff, 2008; Santos et al., 2012; Schweitzer et al., 2013; Emig-Agius et al., 2014; Hultquist et al., 2016; Li et al., 2016a). Several studies found that Ku70/Ku80 heterodimer binds with HIV genomic RNA or TAR RNA at the 5’ end of mRNA transcripts. Those data further implicated that the Ku complex may regulate the transcription process of HIV. (Kaczmarski and Khan, 1993; Yoo and Dynan, 1998); the interactions between Ku and HIV RNA may also impact the transcription level of HIV and the latency property of HIV (Manic et al., 2013). Additionally, Ku could also regulate transcriptional elongation by interacting with the RNA hairpin structure of 7SK snRNA, a scaffold protein for forming the 7SK snRNP complex (Shadrina et al., 2016; Shadrina et al., 2020). Several contradictory studies also show that Ku involves in retroviral DNA integration (Daniel et al., 1999; Baekelandt et al., 2000; Daniel et al., 2004; Knyazhanskaya et al., 2016) in the transcription of integrated provirus (Jeanson and Mouscadet, 2002; Tyagi et al., 2011; Manic et al., 2013; Shadrina et al., 2016), and in functions of HIV-1 matrix protein (Li et al., 2016a). Another evidence demonstrated that the DNA-PK complex involves in the induction of apoptosis in activated CD4+ T cells at the early stage of HIV infection (Cooper et al., 2013).
HIV-1 integrase (IN) is an essential viral enzyme involving in several viral replication steps. Meanwhile, IN is also an unstable protein and degraded by the N-end rule pathway through the host ubiquitin-proteasome machinery (Mulder and Muesing, 2000). However, it remains unknown how HIV-1 IN is protected from degradation during HIV replication. Zheng et al. demonstrated that Ku70 from host cells interacts with HIV-1 IN and prevents it from the Lys48-linked polyubiquitination proteasomal pathway. Additionally, Ku70 can decrease the overall protein polyubiquitination level and specifically deubiquitinate IN by binding with HIV-1 IN (Zheng et al., 2011). Mutagenic studies by Anisenko et al. showed that the amino acid residues 51-160 of HIV-1 IN interacts with 251-438 aa of Ku70. It is further reported that the N-terminal region (1-250 aa) of Ku70 interacts with the α6-helix region located at the 200-220 residues of IN, and the single mutations at E212A or L213A abrogate the interaction. Those findings highlighted the essential role of the 200-220 aa residues of IN in forming a complex with Ku70 (Anisenko et al., 2017a).
Additionally, knockdown of Ku70 significantly inhibits the HIV-1 virus replication in virus-producing cell lines or HIV-infected CD4+ T cells, and the copy number of two-long terminal repeat (LTR) circles and integrated proviral DNA cannot be detected. Those data implicated that Ku70 is an indispensable factor at the early and the late stages of HIV-1 replication (Zheng et al., 2011) (as illustrated in Figure 4 ). HIV-1 IN is an essential enzyme in HIV virions and integrates the proviral DNA into the host genomic DNA. Integration is a critical step during HIV-1 replication. (Cherepanov et al., 2003; Faure et al., 2005; Passos et al., 2017). In detail, IN binds viral DNA and then catalyzes the cleavage of dinucleotides from both 3’-ends of viral DNA. The complex of 3’-processed viral DNA and IN helps recruit some other viral and cellular proteins as cofactors. Subsequently, the whole complex imports into the nucleus. The second step of integration happens in the nucleus of the host cells. IN inserts the processed viral DNA into one strand of the genomic DNA of host cells (Lesbats et al., 2016). This insertion leads to 5-nucleotide gaps (Vincent et al., 1990; Vink et al., 1990; Lesbats et al., 2016). As a result, 3’-ends of viral DNA are then covalently associated with the cellular DNA. However, an overhang is formed at the 5′-ends because of an unpaired dinucleotide (Knyazhanskaya et al., 2019). In order to complete the integration process, the intermediate product has to be repaired (Knyazhanskaya et al., 2016). Knyazhanskaya et al. proposed that the direct binding between Ku70 and HIV-1 IN greatly facilitates the recruitment of Ku80 and DNA-PKcs to the integration site. And then, the whole DNA-PK complex sufficiently functions in initiating the DNA repair process by the NHEJ pathway and resumes efficient HIV-1 replication (Knyazhanskaya et al., 2019).
Interestingly, Zheng et al. found that Ku70 is incorporated into HIV viral particles (Zheng et al., 2011). Nascent HIV viruses contain Gag and GagPol polyproteins, and viral genomic RNAs (as illustrated in Figure 4 ). The polyproteins are composed of several HIV proteins and immature forms of IN, located at the C’- terminus end (Imamichi et al., 2021). Thus, Ku70 maybe incorporated in the virion during assembly via the immature IN. As IN regulates viral maturation (Engelman et al., 1995; Bukovsky and Göttlinger, 1996; Balakrishnan et al., 2013; Hoyte et al., 2017; Imamichi et al., 2021). Further study may reveal more roles of Ku70 during retrovirus infection and replication.
Overall, many studies have provided examples of how HIV-1 viruses commandeer host cellular machinery to protect themselves and facilitate viral replication (Zheng et al., 2011). Consequently, identifying the host cell factors that participate in these processes and determining their functions in HIV viral replication may lead to discovering novel therapeutic targets to fight HIV (Adamson and Freed, 2010; Tintori et al., 2014). Ku70 may become an ideal therapeutic target to treat patients infected with multi-drug-resistant HIV variants.
As we discussed in the current review, cytosolic Ku70, which is translocated from the nucleus to the cytoplasm, can sense cytosolic DNA to induce innate immune response (Zhang et al., 2011a; Ferguson et al., 2012; Li et al., 2016b; Sui et al., 2017; Wang et al., 2017; Burleigh et al., 2020; Sui et al., 2021; Wang et al., 2021), and can inhibit Bax-mediated apoptosis (Sawada et al., 2003; Mazumder et al., 2007). Additionally, Ku70 has also been found localized in the plasma membrane, where it can interact with metalloprotease 9 (MMP-9) (Monferran et al., 2004b), fibronectin (Monferran et al., 2004a) and participate in heterologous and homologous cell adhesion (Koike, 2002). It was also reported that the cell-surfaced Ku70 acts as a receptor for the infection of Rickettsia conorii (R. conorii), a negative gamma bacterium; the rickettsial protein, rOmpB, binds to Ku70 as a ligand. The interaction plays an important role in initiating infection signals, ultimately leading to bacterial entry (Martinez et al., 2005). The plasma membrane-associated Ku70 has also been identified in lipid rafts, and so it has been speculated that the existence of Ku70 within these domains may play an essential role in pathogen entry and signal transduction (Lucero et al., 2003).
Beyond the role of Ku70 in innate immunity, those studies about the involvement of Ku70 in pathogen invasion and HIV replication highlighted a further understanding of the interplay between the host protein Ku70 and pathogen. Further investigation could lead to the development of novel, efficacious therapies in the treatment and prevention of infectious diseases.
Conclusions and Perspectives
The study of Ku70/80 is expanding to encompass numerous research fields, including regulatory processes. More and more promising research emphasizes the role of Ku in innate immunity, the development of a small-molecule Ku inhibitor (Weterings et al., 2016), and the essential clinical relevance of Ku. Exploring the molecular mechanism by which the Ku- or DNA-PK-involved innate immune response confers various strategies to regulate innate immune cascade and could shed light on the role of Ku70 in autoimmune diseases, vaccine development, or aging-related abnormalities. Further investigation could lead to more discoveries at both the basic and translational research levels.
Delineation of the molecular mechanisms of Ku70-mediated innate immune response, especially the cytoplasmic translocation of Ku70, provides novel strategies to regulate innate immune cascades in response to the invasion of foreign microbe DNA or the accumulation of abnormal cellular DNA. Some autoimmune diseases are caused by the persistent induction of proinflammatory cytokines and IFNs. The emergence of mutations in some genes, including Trex1, RNase H, SAMHD1, and others (Crow et al., 2006a; Crow et al., 2006b; Rice et al., 2009; Crow et al., 2015) leads to the abnormal accumulation of cellular DNA. Those abnormal cytoplasmic DNAs serve as dangerous PAMPs and are recognized by potential PRRs in host cells, and then initiate continuous production of innate immune cytokines. Hypothetically, inhibition of the cytoplasmic translocation of DNA sensors, such as Ku70 and IFI16, with some small compounds is expected to abrogate the sensing of cytosolic DNA, therefore downregulating IFN response and providing effective interventions for these autoimmune diseases. Similar strategies may also be used to decrease the over-response of host cells to some viral infections (Sun et al., 2021).
Future research may reveal a more comprehensive understanding of the multiple roles of Ku70, especially in the field of Ku70-involved innate immune networks. These findings would help us solve some remaining questions: how Ku70 regulates its activities in the nucleus and the cytoplasm, and whether it is possible that Ku70 also serves as a nucleus DNA sensor like IFI16 (Kerur et al., 2011; Unterholzner and Bowie, 2011; Li et al., 2012; Dell’oste et al., 2014; Ansari et al., 2015; Roy et al., 2019). Overall, a better understanding of the multiple functions of Ku70 at both the cellular and organismal level would provide new insights into treatments of infectious diseases and autoimmune abnormalities.
Author Contributions
HS and TI conceptualized the work and contributed to writing the manuscript. MH and WC contributed to homology modeling for human Ku70 and Ku80. All authors contributed to editing the manuscript and approved the submitted version.
Funding
This research was supported (in part) by the National Institute of Allergy and Infectious Diseases. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E.
Author Disclaimer
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank Y. Sei for creating the illustration of the HIV life cycle.
Abbreviations
DNA-PK, DNA-dependent protein kinase; NHEJ, non-homologous end-joining; PRRs, pattern recognition receptors; PAMPs, pathogen-associated molecular patterns; TLRs, Toll-like receptors; RIG-I, retinoic acid-inducible gene I; NF-κB, nuclear factor κB; IFN, interferon; IRF3, Interferon regulatory factor 3; IL-1β, interleukin-1βb; DDX41, DEAD-box helicase 41; IFI16, gamma-interferon-inducible protein; DAI, DNA-dependent activator of IFN-regulatory factors; dsDNAs, double-stranded DNAs; LRRFIP1, Leucine-rich repeat flightless-interacting protein 1; DHX, DEAH box protein; AIM2, absent in melanoma 2; PYHIN, pyrin- and HIN200-domain-containing protein; MyD88, myeloid differentiation primary response 88; cGAS, cyclic GMP-AMP synthase; ASC, apoptosis-associated speck-like protein; XRCC, X-ray repair cross-complementing protein; NLS, nuclear localization signal; RD, rhabdomyosarcoma; VACV, vaccinia virus; HBV, hepatitis B virus; HSV, herpes simplex virus; HDACs, histone deacetylases; TSA, trichostatin A; N–C, nuclear-cytoplasmic; EAE, experimental autoimmune encephalomyelitis; IN, integrase; Ub, ubiquitination; CT DNA, calf thymus sonicated DNA; HTLV-1, human T-lymphotropic virus type 1; NTD, N-terminal domain; CTD, C-terminal domain; ISD, interferon stimulatory DNA.
References
- Abbasi S., Parmar G., Kelly R. D., Balasuriya N., Schild-Poulter C. (2021). The Ku Complex: Recent Advances and Emerging Roles Outside of Non-Homologous End-Joining. Cell Mol. Life Sci. 78 (10), 4589–4613. doi: 10.1007/s00018-021-03801-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelbaqi K., Di Paola D., Rampakakis E., Zannis-Hadjopoulos M. (2013). Ku Protein Levels, Localization and Association to Replication Origins in Different Stages of Breast Tumor Progression. J. Cancer 4, 358–370. doi: 10.7150/jca.6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson C. S., Freed E. O. (2010). Novel Approaches to Inhibiting HIV-1 Replication. Antiviral Res. 85, 119–141. doi: 10.1016/j.antiviral.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. (2006). Pathogen Recognition and Innate Immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Al-Emam A., Arbon D., Kysela B. (2014). Deacetylation of Ku70 Regulates Ionizing-Radiation Induced DNA Damage Responses in Human Cells. BMC Genomics 15, P24. doi: 10.1186/1471-2164-15-S2-P24 [DOI] [Google Scholar]
- Anisenko A. N., Knyazhanskaya E. S., Zalevsky A. O., Agapkina J. Y., Sizov A. I., Zatsepin T. S., et al. (2017. a). Characterization of HIV-1 Integrase Interaction With Human Ku70 Protein and Initial Implications for Drug Targeting. Sci. Rep. 7, 5649. doi: 10.1038/s41598-017-05659-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisenko A. N., Knyazhanskaya E. S., Zatsepin T. S., Gottikh M. B. (2017. b). Human Ku70 Protein Binds Hairpin RNA and Double Stranded DNA Through Two Different Sites. Biochimie 132, 85–93. doi: 10.1016/j.biochi.2016.11.001 [DOI] [PubMed] [Google Scholar]
- Ank N., Paludan S. R. (2009). Type III IFNs: New Layers of Complexity in Innate Antiviral Immunity. Biofactors 35, 82–87. doi: 10.1002/biof.19 [DOI] [PubMed] [Google Scholar]
- Ank N., West H., Bartholdy C., Eriksson K., Thomsen A. R., Paludan S. R. (2006). Lambda Interferon (IFN-λ), a Type III IFN, Is Induced by Viruses and IFNs and Displays Potent Antiviral Activity Against Select Virus Infections In Vivo . J. Virol. 80, 4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari M. A., Dutta S., Veettil M. V., Dutta D., Iqbal J., Kumar B., et al. (2015). Herpesvirus Genome Recognition Induced Acetylation of Nuclear IFI16 Is Essential for Its Cytoplasmic Translocation, Inflammasome and IFN-? Responses. PLoS Pathog. 11, e1005019. doi: 10.1371/journal.ppat.1005019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Koonin E. V. (2001). Prokaryotic Homologs of the Eukaryotic DNA-End-Binding Protein Ku, Novel Domains in the Ku Protein and Prediction of a Prokaryotic Double-Strand Break Repair System. Genome Res. 11 (8), 1365–1374. doi: 10.1101/gr.181001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baekelandt V., Claeys A., Cherepanov P., Clercq E. D., Strooper B. D., Nuttin B., et al. (2000). DNA-Dependent Protein Kinase Is Not Required for Efficient Lentivirus Integration. J. Virol. 74 (23), 11278–11285. doi: 10.1128/JVI.74.23.11278-11285.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan M., Yant S. R., Tsai L., O’sullivan C., Bam R. A., Tsai A., et al. (2013). Non-Catalytic Site HIV-1 Integrase Inhibitors Disrupt Core Maturation and Induce a Reverse Transcription Block in Target Cells. PLoS One 8, e74163. doi: 10.1371/journal.pone.0074163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber G. N. (2011). Cytoplasmic DNA Innate Immune Pathways. Immunol. Rev. 243, 99–108. doi: 10.1111/j.1600-065X.2011.01051.x [DOI] [PubMed] [Google Scholar]
- Barnes G., Rio D. (1997). DNA Double-Strand-Break Sensitivity, DNA Replication, and Cell Cycle Arrest Phenotypes of Ku-Deficient Saccharomyces Cerevisiae. Proc. Nat. Acad. Sci. 94 (3), 867–872. doi: 10.1073/pnas.94.3.867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing C. H., Swat W., Alt F. W. (2002). The Mechanism and Regulation of Chromosomal V(D)J Recombination. Cell 109, S45–S55. doi: 10.1016/S0092-8674(02)00675-X [DOI] [PubMed] [Google Scholar]
- Bertuch A. A., Lundblad V. (2003). The Ku Heterodimer Performs Separable Activities at Double-Strand Breaks and Chromosome Termini. Mol. Cell. Biol. 23 (22), 8202–8215. doi: 10.1128/MCB.23.22.8202-8215.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B. A. (2009). TLRs and Innate Immunity. Blood 113, 1399–1407. doi: 10.1182/blood-2008-07-019307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., et al. (2014). SWISS-MODEL: Modelling Protein Tertiary and Quaternary Structure Using Evolutionary Information. Nucleic Acids Res. 42, W252–W258. doi: 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford A. N., Jackson S. P. (2017). ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 66, 801–817. doi: 10.1016/j.molcel.2017.05.015 [DOI] [PubMed] [Google Scholar]
- Blier P. R., Griffith A. J., Craft J., Hardin J. A. (1993). Binding of Ku Protein to DNA. Measurement of Affinity for Ends and Demonstration of Binding to Nicks. J. Biol. Chem. 268, 7594–7601. doi: 10.1016/S0021-9258(18)53216-6 [DOI] [PubMed] [Google Scholar]
- Booth D., George J. (2013). Loss of Function of the New Interferon IFN-λ4 May Confer Protection From Hepatitis C. Nat. Genet. 45, 119–120. doi: 10.1038/ng.2537 [DOI] [PubMed] [Google Scholar]
- Bowater R., Doherty A. J. (2006). Making Ends Meet: Repairing Breaks in Bacterial DNA by Non-Homologous End-Joining. PLoS Genet. 2, e8. doi: 10.1371/journal.pgen.0020008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie A. G., Haga I. R. (2005). The Role of Toll-Like Receptors in the Host Response to Viruses. Mol. Immunol. 42, 859–867. doi: 10.1016/j.molimm.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Bowie A. G., Unterholzner L. (2008). Viral Evasion and Subversion of Pattern-Recognition Receptor Signalling. Nat. Rev. Immunol. 8, 911–922. doi: 10.1038/nri2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukovsky A., Göttlinger H. (1996). Lack of Integrase can Markedly Affect Human Immunodeficiency Virus Type 1 Particle Production in the Presence of an Active Viral Protease. J. Virol. 70, 6820–6825. doi: 10.1128/jvi.70.10.6820-6825.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch H., Lawney B. P., Lin Y.-F., Asaithamby A., Murshid A., Wang Y. E., et al. (2015). Transcriptional Elongation Requires DNA Break-Induced Signalling. Nat. Commun. 6, 10191. doi: 10.1038/ncomms10191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstuummer T. (2009). An Orthogonal Proteomic-Genomic Screen Identifies AIM2 as a Cytoplasmic DNA Sensor for the Inflammasome. Nat. Immunol. 10, 266–272. doi: 10.1038/ni.1702 [DOI] [PubMed] [Google Scholar]
- Burleigh K., Maltbaek J. H., Cambier S., Green R., Gale M., James R. C., et al. (2020). Human DNA-PK Activates a STING-Independent DNA Sensing Pathway. Sci. Immunol. 5, eaba4219. doi: 10.1126/sciimmunol.aba4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Chiu Y.-H., Chen ,. Z. J. (2014). The cGAS-cGAMP-STING Pathway of Cytosolic DNA Sensing and Signaling. Mol. Cell 54, 289–296. doi: 10.1016/j.molcel.2014.03.040 [DOI] [PubMed] [Google Scholar]
- Chaplin A. K., Hardwick S. W., Liang S., Kefala Stavridi A., Hnizda A., Cooper L. R., et al. (2021). Dimers of DNA-PK Create a Stage for DNA Double-Strand Break Repair. Nat. Struct. Mol. Biol. 28, 13–19. doi: 10.1038/s41594-020-00517-x [DOI] [PubMed] [Google Scholar]
- Chaudhary N., Nakka K. K., Chavali P. L., Bhat J., Chatterjee S., Chattopadhyay S. (2014). SMAR1 Coordinates HDAC6-Induced Deacetylation of Ku70 and Dictates Cell Fate Upon Irradiation. Cell Death Dis. 5, e1447–e1447. doi: 10.1038/cddis.2014.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., et al. (2003). HIV-1 Integrase Forms Stable Tetramers and Associates With LEDGF/p75 Protein in Human Cells. J. Biol. Chem. 278, 372–381. doi: 10.1074/jbc.M209278200 [DOI] [PubMed] [Google Scholar]
- Christensen M. H., Jensen S. B., Miettinen J. J., Luecke S., Prabakaran T., Reinert L. S., et al. (2016). HSV-1 ICP27 Targets the TBK1-Activated STING Signalsome to Inhibit Virus-Induced Type I IFN Expression. EMBO J. 35 (13), 1385–1399. doi: 10.15252/embj.201593458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen M. H., Paludan S. R. (2016). Viral Evasion of DNA-Stimulated Innate Immune Responses. Cell. Mol. Immunol. 14, 4. doi: 10.1038/cmi.2016.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia E. M., Severa M., Giacomini E., Monneron D., Remoli M. E., Julkunen I., et al. (2004). Viral Infection and Toll-Like Receptor Agonists Induce a Differential Expression of Type I and λ Interferons in Human Plasmacytoid and Monocyte-Derived Dendritic Cells. Eur. J. Immunol. 34, 796–805. doi: 10.1002/eji.200324610 [DOI] [PubMed] [Google Scholar]
- Cohen H. Y., Lavu S., Bitterman K. J., Hekking B., Imahiyerobo T. A., Miller C., et al. (2004). Acetylation of the C Terminus of Ku70 by CBP and PCAF Controls Bax-Mediated Apoptosis. Mol. Cell 13, 627–638. doi: 10.1016/S1097-2765(04)00094-2 [DOI] [PubMed] [Google Scholar]
- Cooper A., Garcia M., Petrovas C., Yamamoto T., Koup R. A., Nabel G. J. (2013). HIV-1 Causes CD4 Cell Death Through DNA-Dependent Protein Kinase During Viral Integration. Nature 498, 376–379. doi: 10.1038/nature12274 [DOI] [PubMed] [Google Scholar]
- Cosgrove A. J., Nieduszynski C. A., Donaldson A. D. (2002). Ku Complex Controls the Replication Time of DNA in Telomere Regions. Genes Dev. 16 (19), 2485–2490. doi: 10.1101/gad.231602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchlow S. E., Jackson S. P. (1998). DNA End-Joining: From Yeast to Man. Trends Biochem. Sci. 23, 394–398. doi: 10.1016/S0968-0004(98)01284-5 [DOI] [PubMed] [Google Scholar]
- Crow Y. J., Chase D. S., Lowenstein Schmidt J., Szynkiewicz M., Forte G. M. A., Gornall H. L., et al. (2015). Characterization of Human Disease Phenotypes Associated With Mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am. J. Med. Genet. Part A 167, 296–312. doi: 10.1002/ajmg.a.36887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow Y. J., Hayward B. E., Parmar R., Robins P., Leitch A., Ali M., et al. (2006. a). Mutations in the Gene Encoding the 3′-5′ DNA Exonuclease TREX1 Cause Aicardi-Goutières Syndrome at the AGS1 Locus. Nat. Genet. 38, 917–920. doi: 10.1038/ng1845 [DOI] [PubMed] [Google Scholar]
- Crow Y. J., Leitch A., Hayward B. E., Garner A., Parmar R., Griffith E., et al. (2006. b). Mutations in Genes Encoding Ribonuclease H2 Subunits Cause Aicardi-Goutières Syndrome and Mimic Congenital Viral Brain Infection. Nat. Genet. 38 (8), 910–916. doi: 10.1038/ng1842 [DOI] [PubMed] [Google Scholar]
- Daniel R., Greger J. G., Katz R. A., Taganov K. D., Wu X., Kappes J. C., et al. (2004). Evidence That Stable Retroviral Transduction and Cell Survival Following DNA Integration Depend on Components of the Nonhomologous End Joining Repair Pathway. J. Virol. 78 (16), 8573–8581. doi: 10.1128/JVI.78.16.8573-8581.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R., Katz R. A., Skalka A. M. (1999). A Role for DNA-PK in Retroviral DNA Integration. Science 284 (5414), 644–647. doi: 10.1126/science.284.5414.644 [DOI] [PubMed] [Google Scholar]
- Davis A. J., Lee K.-J., Chen D. J. (2013). The N-Terminal Region of the DNA-Dependent Protein Kinase Catalytic Subunit Is Required for Its DNA Double-Stranded Break-Mediated Activation*. J. Biol. Chem. 288, 7037–7046. doi: 10.1074/jbc.M112.434498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’oste V., Gatti D., Gugliesi F., De Andrea M., Bawadekar M., Lo Cigno I., et al. (2014). Innate Nuclear Sensor IFI16 Translocates Into the Cytoplasm During the Early Stage of In Vitro Human Cytomegalovirus Infection and Is Entrapped in the Egressing Virions During the Late Stage. J. Virol. 88, 6970–6982. doi: 10.1128/JVI.00384-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey A., Bowie A. G. (2015). Innate Immune Recognition of DNA: A Recent History. Virology 479-480, 146–152. doi: 10.1016/j.virol.2015.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs T. A., Tainer J. A., Lees-Miller S. P. (2010). A Structural Model for Regulation of NHEJ by DNA-PKcs Autophosphorylation. DNA Repair 9, 1307–1314. doi: 10.1016/j.dnarep.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty A. J., Jackson S. P. (2001). DNA Repair: How Ku Makes Ends Meet. Curr. Biol. 11, R920–R924. doi: 10.1016/S0960-9822(01)00555-3 [DOI] [PubMed] [Google Scholar]
- Donnelly R. P., Kotenko S. V. (2010). Interferon-Lambda: A New Addition to an Old Family. J. Interferon Cytokine Res. 30, 555–564. doi: 10.1089/jir.2010.0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre M., Vagner S. (2017). DNA-Damage Response RNA-Binding Proteins (DDRBPs): Perspectives From a New Class of Proteins and Their RNA Targets. J. Mol. Biol. 429, 3139–3145. doi: 10.1016/j.jmb.2016.09.019 [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Yoo S. (1998). Interaction of Ku Protein and DNA-Dependent Protein Kinase Catalytic Subunit With Nucleic Acids. Nucleic Acids Res. 26, 1551–1559. doi: 10.1093/nar/26.7.1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde N. C., Child S. J., Eickbush M. T., Kitzman J. O., Rogers K. S., Shendure J., et al. (2012). Poxviruses Deploy Genomic Accordions to Adapt Rapidly Against Host Antiviral Defenses. Cell 150, 831–841. doi: 10.1016/j.cell.2012.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emig-Agius D., Olivieri K., Pache L., Shih H. L., Pustovalova O., Bessarabova M., et al. (2014). An Integrated Map of HIV-Human Protein Complexes That Facilitate Viral Infection. PLoS One 9, e96687. doi: 10.1371/journal.pone.0096687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A., Englund G., Orenstein J. M., Martin M. A., Craigie R. (1995). Multiple Effects of Mutations in Human Immunodeficiency Virus Type 1 Integrase on Viral Replication. J. Virol. 69, 2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errami A., Smider V., Rathmell W. K., He D. M., Hendrickson E. A., Zdzienicka M. Z., et al. (1996). Ku86 Defines the Genetic Defect and Restores X-Ray Resistance and V(D)J Recombination to Complementation Group 5 Hamster Cell Mutants. Mol. Cell Biol. 16, 1519–1526. doi: 10.1128/MCB.16.4.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy A. S., Clark R. H., Glyde E. F., Smith G. L. (2008). Vaccinia Virus Protein C16 Acts Intracellularly to Modulate the Host Response and Promote Virulence. J. Gen. Virol. 89 (Pt 10), 2377–2387. doi: 10.1099/vir.0.2008/004895-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure A., Calmels C., Desjobert C., Castroviejo M., Caumont-Sarcos A., Tarrago-Litvak L., et al. (2005). HIV-1 Integrase Crosslinked Oligomers Are Active In Vitro . Nucleic Acids Res. 33, 977–986. doi: 10.1093/nar/gki241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell V. L., Schild-Poulter C. (2012). Ku Regulates Signaling to DNA Damage Response Pathways Through the Ku70 Von Willebrand A Domain. Mol. Cell. Biol. 32 (1), 76–87. doi: 10.1128/MCB.05661-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell V. L., Schild-Poulter C. (2015). The Ku Heterodimer: Function in DNA Repair and Beyond. Mutat. Res/Rev Mutat. Res. 763, 15–29. doi: 10.1016/j.mrrev.2014.06.002 [DOI] [PubMed] [Google Scholar]
- Ferguson B. J., Mansur D. S., Peters N. E., Ren H., Smith G. L. (2012). DNA-PK Is a DNA Sensor for IRF-3-Dependent Innate Immunity. Elife 1, e00047. doi: 10.7554/eLife.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T., Yu J. W., Datta P., Wu J., Alnemri E. S. (2009). AIM2 Activates the Inflammasome and Cell Death in Response to Cytoplasmic DNA. Nature 458, 509–513. doi: 10.1038/nature07710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster S. S., Balestrini A., Petrini J. H. J. (2011). Functional Interplay of the Mre11 Nuclease and Ku in the Response to Replication-Associated DNA Damage. Mol. Cell. Biol. 31 (21), 4379–4389. doi: 10.1128/MCB.05854-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann S. D., Lee A. I., Shockett P. E., Villey I. J., Schatz D. G. (2000). The RAG Proteins and V(D)J Recombination: Complexes, Ends, and Transposition. Annu. Rev. Immunol. 18, 495–527. doi: 10.1146/annurev.immunol.18.1.495 [DOI] [PubMed] [Google Scholar]
- Fujimoto H., Ikuta T., Koike A., Koike M. (2018). Acetylation of Nuclear Localization Signal Controls Importin-Mediated Nuclear Transport of Ku70. bioRxiv. doi: 10.1101/403485 [DOI] [Google Scholar]
- Gao D., Wu J., Wu Y.-T., Du F., Aroh C., Yan N., et al. (2013). Cyclic GMP-AMP Synthase Is an Innate Immune Sensor of HIV and Other Retroviruses. Science 341, 903–906. doi: 10.1126/science.1240933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell D., Jackson S. P. (1999). Mapping of Protein-Protein Interactions Within the DNA-Dependent Protein Kinase Complex. Nucleic Acids Res. 27, 3494–3502. doi: 10.1093/nar/27.17.3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffin W., Kwast-Welfeld J., Rodda D. J., Préfontaine G. G., Traykova-Andonova M., Zhang Y., et al. (1997). Sequence-Specific DNA Binding and Transcription Factor Phosphorylation by Ku Autoantigen/DNA-Dependent Protein Kinase: PHOSPHORYLATION OF Ser-527 OF THE RAT GLUCOCORTICOID RECEPTOR*. J. Biol. Chem. 272, 5647–5658. doi: 10.1074/jbc.272.9.5647 [DOI] [PubMed] [Google Scholar]
- Giffin W., Torrance H., Rodda D. J., Préfontaine G. G., Pope L., Haché R. J. G. (1996). Sequence-Specific DNA Binding by Ku Autoantigen and Its Effects on Transcription. Nature 380, 265–268. doi: 10.1038/380265a0 [DOI] [PubMed] [Google Scholar]
- Göhring F., Schwab B. L., Nicotera P., Leist M., Fackelmayer F. O. (1997). The Novel SAR-Binding Domain of Scaffold Attachment Factor A (SAF-A) Is a Target in Apoptotic Nuclear Breakdown. EMBO J. 16 (24), 7361–7371. doi: 10.1093/emboj/16.24.7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez J. A., Gama V., Yoshida T., Sun W., Hayes P., Leskov K., et al. (2007). Bax-Inhibiting Peptides Derived From Ku70 and Cell-Penetrating Pentapeptides. Biochem. Soc. Trans. 35, 797–801. doi: 10.1042/BST0350797 [DOI] [PubMed] [Google Scholar]
- Gong P., Wang Y., Jing Y. (2019). Apoptosis Induction Byhistone Deacetylase Inhibitors in Cancer Cells: Role of Ku70. Int. J. Mol. Sci. 20 (7), 1601. doi: 10.3390/ijms20071601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Mattaj I. W. (1996). Nucleocytoplasmic Transport. Science 271 (5255), 1513–1519. doi: 10.1126/science.271.5255.1513 [DOI] [PubMed] [Google Scholar]
- Grundy G. J., Rulten S. L., Zeng Z., Arribas-Bosacoma R., Iles N., Manley K., et al. (2013). APLF Promotes the Assembly and Activity of Non-Homologous End Joining Protein Complexes. Embo J. 32 (1), 112–125. doi: 10.1038/emboj.2012.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Jin S., Gao Y., Weaver D. T., Alt F. W. (1997). Ku70-Deficient Embryonic Stem Cells Have Increased Ionizing Radiosensitivity, Defective DNA End-Binding Activity, and Inability to Support V(D)J recombination. Proc. Natl. Acad. Sci. 94, 8076–8081. doi: 10.1073/pnas.94.15.8076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarsten O., Chu G. (1998). DNA-Dependent Protein Kinase: DNA Binding and Activation in the Absence of Ku. Proc. Natl. Acad. Sci. 95, 525–530. doi: 10.1073/pnas.95.2.525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. W., Elledge S. J. (2007). The DNA Damage Response: Ten Years After. Mol. Cell 28, 739–745. doi: 10.1016/j.molcel.2007.11.015 [DOI] [PubMed] [Google Scholar]
- Hornung V. (2009). AIM2 Recognizes Cytosolic dsDNA and Forms a Caspase-1-Activating Inflammasome With ASC. Nature 458, 514–518. doi: 10.1038/nature07725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyte A. C., Jamin A. V., Koneru P. C., Kobe M. J., Larue R. C., Fuchs J. R., et al. (2017). Resistance to Pyridine-Based Inhibitor KF116 Reveals an Unexpected Role of Integrase in HIV-1 Gag-Pol Polyprotein Proteolytic Processing. J. Biol. Chem. 292, 19814–19825. doi: 10.1074/jbc.M117.816645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist J. F., Schumann K., Woo J. M., Manganaro L., Mcgregor M. J., Doudna J., et al. (2016). A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells. Cell Rep. 17, 1438–1452. doi: 10.1016/j.celrep.2016.09.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamichi T., Bernbaum J. G., Laverdure S., Yang J., Chen Q., Highbarger H., et al. (2021). Natural Occurring Polymorphisms in HIV-1 Integrase and RNase H Regulate Viral Release and Autoprocessing. J. Virol. Jvi0132321. doi: 10.1128/jvi.01323-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiviglio S. M., Bertuch A. A. (2009). Ku’s Essential Role in Keeping Telomeres Intact. Proc. Natl. Acad. Sci. U. S. A. 106 (30), 12217–12218. doi: 10.1073/pnas.0906427106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. P., Bartek J. (2009). The DNA-Damage Response in Human Biology and Disease. Nature 461, 1071–1078. doi: 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. P., Jeggo P. A. (1995). DNA Double-Strand Break Repair and V(D)J Recombination: Involvement of DNA-PK. Trends Biochem. Sci. 20, 412–415. doi: 10.1016/S0968-0004(00)89090-8 [DOI] [PubMed] [Google Scholar]
- Jeanson L., Mouscadet J.-F. (2002). Ku Represses the HIV-1 Transcription: Identification of a Putative Ku Binding Site Homologous to the Mouse Mammary Tumor Virus Nre1 Sequence in the HIV-1 Long Terminal Repeat*. J. Biol. Chem. 277, 4918–4924. doi: 10.1074/jbc.M110830200 [DOI] [PubMed] [Google Scholar]
- Kaczmarski W., Khan S. A. (1993). Lupus Autoantigen Ku Protein Binds HIV-1 TAR RNA In Vitro . Biochem. Biophys. Res. Commun. 196, 935–942. doi: 10.1006/bbrc.1993.2339 [DOI] [PubMed] [Google Scholar]
- Kaisho T., Akira S. (2006). Toll-Like Receptor Function and Signaling. J. Allergy Clin. Immunol. 117, 979–987. doi: 10.1016/j.jaci.2006.02.023 [DOI] [PubMed] [Google Scholar]
- Kawai T., Akira S. (2009). The Roles of TLRs, RLRs and NLRs in Pathogen Recognitionint. Int. Immunol. 21 (4), 317–337. doi: 10.1093/intimm/dxp017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating S. E., Baran M., Bowie A. G. (2011). Cytosolic DNA Sensors Regulating Type I Interferon Induction. Trends Immunol. 32, 574–581. doi: 10.1016/j.it.2011.08.004 [DOI] [PubMed] [Google Scholar]
- Keijzers G. (2018). “Ku70 and Ku80,” in Encyclopedia of Signaling Molecules. Ed. Choi S. (Cham: Springer International Publishing; ), 2781–2786. [Google Scholar]
- Kerur N., Veettil M.V., Sharma-Walia N., Bottero V., Sadagopan S., Otageri P., et al. (2011). IFI16 Acts as a Nuclear Pathogen Sensor to Induce the Inflammasome in Response to Kaposi Sarcoma-Associated Herpesvirus Infection. Cell Host Microbe 9, 363–375. doi: 10.1016/j.chom.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.-B., Kim D.-W., Park J. W., Jeon Y.-J., Kim D., Rhee S., et al. (2014). Inhibition of Ku70 Acetylation by INHAT Subunit SET/TAF-Iβ Regulates Ku70-Mediated DNA Damage Response. Cell. Mol. Life Sci. 71, 2731–2745. doi: 10.1007/s00018-013-1525-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Pazhoor S., Bao M., Zhang Z., Hanabuchi S., Facchinetti V., et al. (2010). Aspartate-Glutamate-Alanine-Histidine Box Motif (DEAH)/RNA Helicase A Helicases Sense Microbial DNA in Human Plasmacytoid Dendritic Cells. Proc. Natl. Acad. Sci. 107, 15181–15186. doi: 10.1073/pnas.1006539107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knyazhanskaya E., Anisenko A., Shadrina O., Kalinina A., Zatsepin T., Zalevsky A., et al. (2019). NHEJ Pathway Is Involved in Post-Integrational DNA Repair Due to Ku70 Binding to HIV-1 Integrase. Retrovirology 16, 30. doi: 10.1186/s12977-019-0492-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knyazhanskaya E. S., Shadrina O. A., Anisenko A. N., Gottikh M. B. (2016). Role of DNA-Dependent Protein Kinase in the HIV-1 Replication Cycle. Mol. Biol. 50, 567–579. doi: 10.1134/S0026893316040075 [DOI] [PubMed] [Google Scholar]
- Koike M. (2002). Dimerization, Translocation and Localization of Ku70 and Ku80 Proteins. J. Radiat. Res. 43, 223–236. doi: 10.1269/jrr.43.223 [DOI] [PubMed] [Google Scholar]
- Koike M., Ikuta T., Miyasaka T., Shiomi T. (1999. a). Ku80 can Translocate to the Nucleus Independent of the Translocation of Ku70 Using Its Own Nuclear Localization Signal. Oncogene 18, 7495–7505. doi: 10.1038/sj.onc.1203247 [DOI] [PubMed] [Google Scholar]
- Koike M., Ikuta T., Miyasaka T., Shiomi T. (1999. b). The Nuclear Localization Signal of the Human Ku70 Is a Variant Bipartite Type Recognized by the Two Components of Nuclear Pore-Targeting Complex. Exp. Cell Res. 250, 401–413. doi: 10.1006/excr.1999.4507 [DOI] [PubMed] [Google Scholar]
- Koike M., Shiomi T., Koike A. (2000). Ku70 Can Translocate to the Nucleus Independent of Ku80 Translocation and DNA-PK Autophosphorylation. Biochem. Biophys. Res. Commun. 276, 1105–1111. doi: 10.1006/bbrc.2000.3567 [DOI] [PubMed] [Google Scholar]
- Koike M., Yutoku Y., Koike A. (2017). Cloning of Canine Ku80 and Its Localization and Accumulation at DNA Damage Sites. FEBS Open Bio 7, 1854–1863. doi: 10.1002/2211-5463.12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S. V. (2011). IFN-λs. Curr. Opin. Immunol. 23, 583–590. doi: 10.1016/j.coi.2011.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S. V., Gallagher G., Baurin V. V., Lewis-Antes A., Shen M., Shah N. K., et al. (2003). IFN-λs Mediate Antiviral Protection Through a Distinct Class II Cytokine Receptor Complex. Nat. Immunol. 4, 69–77. doi: 10.1038/ni875 [DOI] [PubMed] [Google Scholar]
- Lamaa A., Le Bras M., Skuli N., Britton S., Frit P., Calsou P., et al. (2016). A Novel Cytoprotective Function for the DNA Repair Protein Ku in Regulating P53 mRNA Translation and Function. EMBO Rep. 17 (4), 508–518. doi: 10.15252/embr.201541181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S., Kim Y.-J. (2007). Signaling Pathways Downstream of Pattern-Recognition Receptors and Their Cross Talk. Annu. Rev. Biochem. 76, 447–480. doi: 10.1146/annurev.biochem.76.060605.122847 [DOI] [PubMed] [Google Scholar]
- Lesbats P., Engelman A. N., Cherepanov P. (2016). Retroviral DNA Integration. Chem. Rev. 116, 12730–12757. doi: 10.1021/acs.chemrev.6b00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Diner B. A., Chen J., Cristea I. M. (2012). Acetylation Modulates Cellular Distribution and DNA Sensing Ability of Interferon-Inducible Protein IFI16. Proc. Natl. Acad. Sci. 109, 10558–10563. doi: 10.1073/pnas.1203447109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Frederick K. M., Haverland N. A., Ciborowski P., Belshan M. (2016. a). Investigation of the HIV-1 Matrix Interactome During Virus Replication. Prot. Clin. Appl. 10 (2), 156–163. doi: 10.1002/prca.201400189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wu Y., Zheng X., Cong J., Liu Y., Li J., et al. (2016. b). Cytoplasm-Translocated Ku70/80 Complex Sensing of HBV DNA Induces Hepatitis-Associated Chemokine Secretion. Front. Immunol. 7, 569. doi: 10.3389/fimmu.2016.00569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G. C., Yang S. H., Kim D., Nussenzweig A., Ouyang H., Wei J., et al. (1995). Suppression of Heat-Induced Hsp70 Expression by the 70-kDa Subunit of the Human Ku Autoantigen. Proc. Natl. Acad Sci. U. S. A. 92 (10), 4512–4516. doi: 10.1073/pnas.92.10.4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucero H., Gae D., Taccioli G. E. (2003). Novel Localization of the DNA-PK Complex in Lipid Rafts: A Putative Role in the Signal Transduction Pathway of the Ionizing Radiation Response. J. Biol. Chem. 278, 22136–22143. doi: 10.1074/jbc.M301579200 [DOI] [PubMed] [Google Scholar]
- Lu Y.-F., Goldstein D. B., Urban T. J., Bradrick S. S. (2015). Interferon-λ4 Is a Cell-Autonomous Type III Interferon Associated With Pre-Treatment Hepatitis C Virus Burden. Virology 476, 334–340. doi: 10.1016/j.virol.2014.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum K., Cristea I. (2021). Host Innate Immune Response and Viral Immune Evasion During Alphaherpesvirus Infection. Curr. Issues Mol. Biol. 42, 635–686. doi: 10.21775/cimb.042.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski M. M., Willems E., Jansen P. W. T. C., Vermeulen M. (2016). Cross-Linking Immunoprecipitation-MS (xIP-MS): Topological Analysis of Chromatin-Associated Protein Complexes Using Single Affinity Purification*. Mol. Cell. Proteomics 15, 854–865. doi: 10.1074/mcp.M115.053082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manic G., Maurin-Marlin A., Laurent F., Vitale I., Thierry S., Delelis O., et al. (2013). Impact of the Ku Complex on HIV-1 Expression and Latency. PLoS One 8, e69691. doi: 10.1371/journal.pone.0069691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. J., Seveau S., Veiga E., Matsuyama S., Cossart P. (2005). Ku70, a Component of DNA-Dependent Protein Kinase, Is a Mammalian Receptor for Rickettsia conorii. Cell 123, 1013–1023. doi: 10.1016/j.cell.2005.08.046 [DOI] [PubMed] [Google Scholar]
- Matheos D., Ruiz M. T., Price G. B., Zannis-Hadjopoulos M. (2002). Ku Antigen, an Origin-Specific Binding Protein That Associates With Replication Proteins, Is Required for Mammalian DNA Replication. Biochim. Biophys. Acta (BBA) Gene Structure Expression 1578, 59–72. doi: 10.1016/S0167-4781(02)00497-9 [DOI] [PubMed] [Google Scholar]
- Mazumder S., Plesca D., Kinter M., Almasan A. (2007). Interaction of a Cyclin E Fragment With Ku70 Regulates Bax-Mediated Apoptosis. Mol. Cell. Biol. 27 (9), 3511–3520. doi: 10.1128/MCB.01448-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Janeway C. (1997). Innate Immunity: The Virtues of a Nonclonal System of Recognition. Cell 91, 295–298. doi: 10.1016/S0092-8674(00)80412-2 [DOI] [PubMed] [Google Scholar]
- Medzhitov R., Janeway C. (2000). Innate Immunity. N. Engl. J. Med. 343, 338–344. doi: 10.1056/NEJM200008033430506 [DOI] [PubMed] [Google Scholar]
- Menon V., Povirk L. F. (2016). End-Processing Nucleases and Phosphodiesterases: An Elite Supporting Cast for the Non-Homologous End Joining Pathway of DNA Double-Strand Break Repair. DNA Repair 43, 57–68. doi: 10.1016/j.dnarep.2016.05.011 [DOI] [PubMed] [Google Scholar]
- Mimori T., Akizuki M., Yamagata H., Inada S., Yoshida S., Homma M. (1981). Characterization of a High Molecular Weight Acidic Nuclear Protein Recognized by Autoantibodies in Sera From Patients With Polymyositis-Scleroderma Overlap. J. Clin. Invest. 68, 611–620. doi: 10.1172/JCI110295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T., Kanoh J., Ishikawa F. (2009). Fission Yeast Ku Protein Is Required for Recovery From DNA Replication Stress. Genes Cells 14 (9), 1091–1103. doi: 10.1111/j.1365-2443.2009.01337.x [DOI] [PubMed] [Google Scholar]
- Mo X., Dynan W. S. (2002). Subnuclear Localization of Ku Protein: Functional Association With RNA Polymerase II Elongation Sites. Mol. Cell. Biol. 22 (22), 8088–8099. doi: 10.1128/MCB.22.22.8088-8099.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen T. H. (2009). Pathogen Recognition and Inflammatory Signaling in Innate Immune Defenses. Clin. Microbiol. Rev. 22, 240–273. doi: 10.1128/CMR.00046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monferran S., Muller C., Mourey L., Frit P., Salles B. (2004. a). The Membrane-Associated Form of the DNA Repair Protein Ku Is Involved in Cell Adhesion to Fibronectin. J. Mol. Biol. 337, 503–511. doi: 10.1016/j.jmb.2004.01.057 [DOI] [PubMed] [Google Scholar]
- Monferran S., Paupert J., Dauvillier S., Salles B., Muller C. (2004. b). The Membrane Form of the DNA Repair Protein Ku Interacts at the Cell Surface With Metalloproteinase 9. EMBO J. 23, 3758–3768. doi: 10.1038/sj.emboj.7600403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe K. M., Yang Z., Johnson J. R., Geng X., Doitsh G., Krogan N. J., et al. (2014). IFI16 DNA Sensor Is Required for Death of Lymphoid CD4 T Cells Abortively Infected With HIV. Science 343, 428–432. doi: 10.1126/science.1243640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder L. C. F., Muesing M. A. (2000). Degradation of HIV-1 Integrase by the N-End Rule Pathway *. J. Biol. Chem. 275, 29749–29753. doi: 10.1074/jbc.M004670200 [DOI] [PubMed] [Google Scholar]
- Novac O., Matheos D., Araujo F. D., Price G. B., Zannis-Hadjopoulos M. (2001). In Vivo Association of Ku With Mammalian Origins of DNA Replication. Mol. Biol. Cell. 12 (11), 3386–3401. doi: 10.1091/mbc.12.11.3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowsheen S., Yang E. S. (2012). The Intersection Between DNA Damage Response and Cell Death Pathways. Exp. Oncol. 34, 243–254. [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig A., Chen C., Da Costa Soares V., Sanchez M., Sokol K., Nussenzweig M. C., et al. (1996). Requirement for Ku80 in Growth and Immunoglobulin V(D)J Recombination. Nature 382, 551–555. doi: 10.1038/382551a0 [DOI] [PubMed] [Google Scholar]
- Ono M., Tucker P. W., Capra J. D. (1996). Ku Is a General Inhibitor of DNA-Protein Complex Formation and Transcription. Mol. Immunol. 33, 787–796. doi: 10.1016/0161-5890(96)00030-2 [DOI] [PubMed] [Google Scholar]
- Paludan S. R., Bowie A. G. (2013). Immune Sensing of DNA. Immunity 38, 870–880. doi: 10.1016/j.immuni.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-J., Ciccone S. L. M., Freie B., Kurimasa A., Chen D. J., Li G. C., et al. (2004). A Positive Role for the Ku Complex in DNA Replication Following Strand Break Damage in Mammals*. J. Biol. Chem. 279, 6046–6055. doi: 10.1074/jbc.M311054200 [DOI] [PubMed] [Google Scholar]
- Passos D. O., Li M., Yang R., Rebensburg S. V., Ghirlando R., Jeon Y., et al. (2017). Cryo-EM Structures and Atomic Model of the HIV-1 Strand Transfer Complex Intasome. Sci. (New York NY) 355, 89–92. doi: 10.1126/science.aah5163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters N. E., Ferguson B. J., Mazzon M., Fahy A. S., Krysztofinska E., Arribas-Bosacoma R., et al. (2013). A Mechanism for the Inhibition of DNA-PK-Mediated DNA Sensing by a Virus. PLoS Pathog. 9, e1003649. doi: 10.1371/journal.ppat.1003649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S. E., Stellwagen A. E., Diede S. J., Singer M. S., Haimberger Z. W., Johnson C. O., et al. (2001). The Function of a Stem-Loop in Telomerase RNA Is Linked to the DNA Repair Protein Ku. Nat. Genet. 27, 64–67. doi: 10.1038/83778 [DOI] [PubMed] [Google Scholar]
- Pfingsten J. S., Goodrich K. J., Taabazuing C., Ouenzar F., Chartrand P., Cech T. R. (2012). Mutually Exclusive Binding of Telomerase RNA and DNA by Ku Alters Telomerase Recruitment Model. Cell 148, 922–932. doi: 10.1016/j.cell.2012.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A., Reis e Sousa C. (2007). Innate Recognition of Viruses. Immunity 27, 370–383. doi: 10.1016/j.immuni.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Radhakrishnan S. K., Jette N., Lees-Miller S. P. (2014). Non-Homologous End Joining: Emerging Themes and Unanswered Questions. DNA Repair 17, 2–8. doi: 10.1016/j.dnarep.2014.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampakakis E., Di Paola D., Zannis-Hadjopoulos M. (2008). Ku Is Involved in Cell Growth, DNA Replication and G1-S Transition. J. Cell Sci. 121, 590–600. doi: 10.1242/jcs.021352 [DOI] [PubMed] [Google Scholar]
- Ribes-Zamora A., Mihalek I., Lichtarge O., Bertuch A. A. (2007). Distinct Faces of the Ku Heterodimer Mediate DNA Repair and Telomeric Functions. Nat. Struct. Mol. Biol. 14, 301–307. doi: 10.1038/nsmb1214 [DOI] [PubMed] [Google Scholar]
- Rice G. I., Bond J., Asipu A., Brunette R. L., Manfield I. W., Carr I. M., et al. (2009). Mutations Involved in Aicardi-Goutières Syndrome Implicate SAMHD1 as Regulator of the Innate Immune Response. Nat. Genet. 41, 829–832. doi: 10.1038/ng.373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Calzada A., Spagnolo L., Pearl L. H., Llorca O. (2007). Structural Model of Full-Length Human Ku70–Ku80 Heterodimer and Its Recognition of DNA and DNA-PKcs. EMBO Rep. 8, 56–62. doi: 10.1038/sj.embor.7400847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger T., Lugrin J., Le Roy D., Goy G., Mombelli M., Koessler T., et al. (2011). Histone Deacetylase Inhibitors Impair Innate Immune Responses to Toll-Like Receptor Agonists and to Infection. Blood 117, 1205–1217. doi: 10.1182/blood-2010-05-284711 [DOI] [PubMed] [Google Scholar]
- Rouse J., Jackson S. P. (2002). Interfaces Between the Detection, Signaling, and Repair of DNA Damage. Science 297 (5581), 547–551. doi: 10.1126/science.1074740 [DOI] [PubMed] [Google Scholar]
- Roy A., Ghosh A., Kumar B., Chandran B. (2019). IFI16, a Nuclear Innate Immune DNA Sensor, Mediates Epigenetic Silencing of Herpesvirus Genomes by Its Association With H3K9 Methyltransferases SUV39H1 and GLP. eLife 8, e49500. doi: 10.7554/eLife.49500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez A., Russell P. (2015). Ku Stabilizes Replication Forks in the Absence of Brc1. PLoS One 10 (5), e0126598. doi: 10.1371/journal.pone.0126598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos S., Obukhov Y., Nekhai S., Bukrinsky M., Iordanskiy S. (2012). Virus-Producing Cells Determine the Host Protein Profiles of HIV-1 Virion Cores. Retrovirology 9, 65. doi: 10.1186/1742-4690-9-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M., Wang J., Reeves W. H. (1995). Role of Free P70 (Ku) Subunit in Posttranslational Stabilization of Newly Synthesized P80 During DNA-Dependent Protein Kinase Assembly. Eur. J. Cell Biol. 66, 127–135. [PubMed] [Google Scholar]
- Sawada M., Sun W., Hayes P., Leskov K., Boothman D. A., Matsuyama S. (2003). Ku70 Suppresses the Apoptotic Translocation of Bax to Mitochondria. Nat. Cell Biol. 5, 320–329. doi: 10.1038/ncb950 [DOI] [PubMed] [Google Scholar]
- Schild-Poulter C., Matheos D., Novac O., Cui B., Giffin W., Ruiz M. T., et al. (2003). Differential DNA Binding of Ku Antigen Determines Its Involvement in DNA Replication. DNA Cell Biol. 22, 65–78. doi: 10.1089/104454903321515887 [DOI] [PubMed] [Google Scholar]
- Schweitzer C. J., Jagadish T., Haverland N., Ciborowski P., Belshan M. (2013). Proteomic Analysis of Early HIV-1 Nucleoprotein Complexes. J. Proteome Res. 12, 559–572. doi: 10.1021/pr300869h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Panday A., Elango R., Willis N. A. (2019). DNA Double-Strand Break Repair-Pathway Choice in Somatic Mammalian Cells. Nat. Rev. Mol. Cell Biol. 20, 698–714. doi: 10.1038/s41580-019-0152-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scutts S. R., Ember S. W., Ren H., Ye C., Lovejoy C. A., Mazzon M., et al. (2018). DNA-PK Is Targeted by Multiple Vaccinia Virus Proteins to Inhibit DNA Sensing. Cell Rep. 25, 1953–1965.e1954. doi: 10.1016/j.celrep.2018.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadrina O., Garanina I., Korolev S., Zatsepin T., Van Assche J., Daouad F., et al. (2020). Analysis of RNA Binding Properties of Human Ku Protein Reveals Its Interactions With 7SK snRNA and Protein Components of 7SK snRNP Complex. Biochimie 171-172, 110–123. doi: 10.1016/j.biochi.2020.02.016 [DOI] [PubMed] [Google Scholar]
- Shadrina O. A., Knyazhanskaya E. S., Korolev S. P., Gottikh M. B. (2016). Host Proteins Ku and HMGA1 As Participants of HIV-1 Transcription. Acta Naturae 8, 34–47. doi: 10.32607/20758251-2016-8-1-34-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao R.-G., Cao C.-X., Zhang H., Kohn K. W., Wold M. S., Pommier Y. (1999). Replication-Mediated DNA Damage by Camptothecin Induces Phosphorylation of RPA by DNA-Dependent Protein Kinase and Dissociates RPA: DNA-PK Complexes. Embo J. 18 (5), 1397–1406. doi: 10.1093/emboj/18.5.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z., Flynn R. A., Crowe J. L., Zhu Y., Liang J., Jiang W., et al. (2020). DNA-PKcs has KU-Dependent Function in rRNA Processing and Haematopoiesis. Nature 579, 291–296. doi: 10.1038/s41586-020-2041-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Fitzgerald K. A. (2011). Innate Immune Sensing of DNA. PLoS Pathog. 7, e1001310. doi: 10.1371/journal.ppat.1001310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay J. W., Wright W. E. (2019). Telomeres and Telomerase: Three Decades of Progress. Nat. Rev. Genet. 20, 299–309. doi: 10.1038/s41576-019-0099-1 [DOI] [PubMed] [Google Scholar]
- Singleton B. K., Priestley A., Steingrimsdottir H., Gell D., Blunt T., Jackson S. P., et al. (1997). Molecular and Biochemical Characterization of Xrs Mutants Defective in Ku80. Mol. Cell Biol. 17, 1264–1273. doi: 10.1128/MCB.17.3.1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton B. K., Torres-Arzayus M. I., Rottinghaus S. T., Taccioli G. E., Jeggo P. A. (1999). The C Terminus of Ku80 Activates the DNA-Dependent Protein Kinase Catalytic Subunit. Mol. Cell. Biol. 19 (5), 3267–3277. doi: 10.1128/MCB.19.5.3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann K. M., Tran K.-C., Chi B., Rabin R. L., Collins P. L. (2004). Suppression of the Induction of Alpha, Beta, and Gamma Interferons by the NS1 and NS2 Proteins of Human Respiratory Syncytial Virus in Human Epithelial Cells and Macrophages. J. Virol. 78, 4363–4369. doi: 10.1128/JVI.78.8.4363-4369.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen A. E., Haimberger Z. W., Veatch J. R., Gottschling D. E. (2003). Ku Interacts With Telomerase RNA to Promote Telomere Addition at Native and Broken Chromosome Ends. Genes Dev. 17 (19), 2384–2395. doi: 10.1101/gad.1125903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studamire B., Goff S. P. (2008). Host Proteins Interacting With the Moloney Murine Leukemia Virus Integrase: Multiple Transcriptional Regulators and Chromatin Binding Factors. Retrovirology 5, 48. doi: 10.1186/1742-4690-5-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C., Hada M., Opipari A. W., Castle V. P., Kwok R. P. S. (2013). CREB-Binding Protein Regulates Ku70 Acetylation in Response to Ionization Radiation in Neuroblastoma. Mol. Cancer Res. 11, 173. doi: 10.1158/1541-7786.MCR-12-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian C., Opipari A. W., Bian X., Castle V. P., Kwok R. P. S. (2005). Ku70 Acetylation Mediates Neuroblastoma Cell Death Induced by Histone Deacetylase Inhibitors. Proc. Natl. Acad. Sci. U. S. A. 102, 4842. doi: 10.1073/pnas.0408351102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H., Chen Q., Imamichi T. (2021). Cytoplasmic-Translocated Ku70 Senses Intracellular DNA and Mediates Interferon-Lambda1 Induction. Immunology 163, 323–337. doi: 10.1111/imm.13318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Zhang S., Chen B. P. C. (2020). DNA–dependent Protein Kinase in Telomere Maintenance and Protection. Cell. Mol. Biol. Lett. 25, 2. doi: 10.1186/s11658-020-0199-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H., Zhou M., Imamichi H., Jiao X., Sherman B. T., Lane H. C., et al. (2017). STING Is an Essential Mediator of the Ku70-Mediated Production of IFN-λ1 in Response to Exogenous DNA. Sci. Signal. 10 (488), 1–11. doi: 10.1126/scisignal.aah5054 [DOI] [PubMed] [Google Scholar]
- Sun H., Huang Y., Mei S., Xu F., Liu X., Zhao F., et al. (2021). A Nuclear Export Signal Is Required for cGAS to Sense Cytosolic DNA. Cell Rep. 34, 108586. doi: 10.1016/j.celrep.2020.108586 [DOI] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X. G., Chen Z. J. (2013). Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science 339, 786–791. doi: 10.1126/science.1232458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C., Zhan G., Zheng C. (2016). Evasion of Host Antiviral Innate Immunity by HSV-1, an Update. Virol. J. 13, 38. doi: 10.1186/s12985-016-0495-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R., Shindo H., Tase A., Kikuchi Y., Shimizu M., Yamazaki T. (2009). Solution Structures and DNA Binding Properties of the N-Terminal SAP Domains of SUMO E3 Ligases From Saccharomyces Cerevisiae and Oryza Sativa. Proteins 75 (2), 336–347. doi: 10.1002/prot.22243 [DOI] [PubMed] [Google Scholar]
- Syedbasha M., Egli A. (2017). Interferon Lambda: Modulating Immunity in Infectious Diseases. Front. Immunol. 8, 119. doi: 10.3389/fimmu.2017.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A. (2007). DAI (DLM-1/ZBP1) Is a Cytosolic DNA Sensor and an Activator of Innate Immune Response. Nature 448, 501–505. doi: 10.1038/nature06013 [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. (2009). Innate Immunity to Virus Infection. Immunol. Rev. 227, 75–86. doi: 10.1111/j.1600-065X.2008.00737.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira-Silva A., Ait Saada A., Hardy J., Iraqui I., Nocente M. C., Fréon K., et al. (2017). The End-Joining Factor Ku Acts in the End-Resection of Double Strand Break-Free Arrested Replication Forks. Nat. Commun. 8, 1982. doi: 10.1038/s41467-017-02144-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapar R., Wang J. L., Hammel M., Ye R., Liang K., Sun C., et al. (2020). Mechanism of Efficient Double-Strand Break Repair by a Long Non-Coding RNA. Nucleic Acids Res. 48, 10953–10972. doi: 10.1093/nar/gkaa784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. R., Kaminski J. J., Kurt-Jones E. A., Fitzgerald K. A. (2011). Pattern Recognition Receptors and the Innate Immune Response to Viral Infection. Viruses 3, 920–940. doi: 10.3390/v3060920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. R., Sharma S., Atianand M., Jensen S. B., Carpenter S., Knipe D. M., et al. (2014). Interferon γ-Inducible Protein (IFI) 16 Transcriptionally Regulates Type I Interferons and Other Interferon-Stimulated Genes and Controls the Interferon Response to Both DNA and RNA Viruses. J. Biol. Chem. 289, 23568–23581. doi: 10.1074/jbc.M114.554147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting N. S. Y., Yu Y., Pohorelic B., Lees-Miller S. P., Beattie T. L. (2005). Human Ku70/80 Interacts Directly With hTR, the RNA Component of Human Telomerase. Nucleic Acids Res. 33, 2090–2098. doi: 10.1093/nar/gki342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintori C., Brai A., Fallacara A. L., Fazi R., Schenone S., Botta M. (2014). Protein–protein Interactions and Human Cellular Cofactors as New Targets for HIV Therapy. Curr. Opin. Pharmacol. 18, 1–8. doi: 10.1016/j.coph.2014.06.005 [DOI] [PubMed] [Google Scholar]
- Tyagi S., Ochem A., Tyagi M. (2011). DNA-Dependent Protein Kinase Interacts Functionally With the RNA Polymerase II Complex Recruited at the Human Immunodeficiency Virus (HIV) Long Terminal Repeat and Plays an Important Role in HIV Gene Expression. J. Gen. Virol. 92 (Pt 7), 1710–1720. doi: 10.1099/vir.0.029587-0 [DOI] [PubMed] [Google Scholar]
- Unterholzner L., Bowie A. G. (2011). Innate DNA Sensing Moves to the Nucleus. Cell Host Microbe 9, 351–353. doi: 10.1016/j.chom.2011.05.001 [DOI] [PubMed] [Google Scholar]
- Unterholzner L., Keating S. E., Baran M., Horan K. A., Jensen S. B., Sharma S., et al. (2010). IFI16 Is an Innate Immune Sensor for Intracellular DNA. Nat. Immunol. 11, 997–1004. doi: 10.1038/ni.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzé G., Monneron D. (2007). IL-28 and IL-29: Newcomers to the Interferon Family. Biochimie 89, 729–734. doi: 10.1016/j.biochi.2007.01.008 [DOI] [PubMed] [Google Scholar]
- Vincent K. A., York-Higgins D., Quiroga M., Brown P. O. (1990). Host Sequences Flanking the HIV Provirus. Nucleic Acids Res. 18, 6045–6047. doi: 10.1093/nar/18.20.6045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C., Groenink M., Elgersma Y., Fouchier R. A., Tersmette M., Plasterk R. H. (1990). Analysis of the Junctions Between Human Immunodeficiency Virus Type 1 Proviral DNA and Human DNA. J. Virol. 64, 5626–5627. doi: 10.1128/jvi.64.11.5626-5627.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J. R., Corpina R. A., Goldberg J. (2001). Structure of the Ku Heterodimer Bound to DNA and Its Implications for Double-Strand Break Repair. Nature 412, 607–614. doi: 10.1038/35088000 [DOI] [PubMed] [Google Scholar]
- Wang Y., Cortez D., Yazdi P., Neff N., Elledge S. J., Qin J. (2000). BASC, a Super Complex of BRCA1-Associated Proteins Involved in the Recognition and Repair of Aberrant DNA Structures. Genes Dev. 14, 927–939. [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Fu Z., Li X., Liang Y., Pei S., Hao S., et al. (2021). Cytoplasmic DNA Sensing by KU Complex in Aged CD4+ T Cell Potentiates T Cell Activation and Aging-Related Autoimmune Inflammation. Immunity 54 (4), 632–647.e639. doi: 10.1016/j.immuni.2021.02.003 [DOI] [PubMed] [Google Scholar]
- Wang J., Kang L., Song D., Liu L., Yang S., Ma L., et al. (2017). Ku70 Senses HTLV-1 DNA and Modulates HTLV-1 Replication. J. Immunol. 199, 2475–2482. doi: 10.4049/jimmunol.1700111 [DOI] [PubMed] [Google Scholar]
- Wang K., Ni L., Wang S., Zheng C., Hutt-Fletcher L. (2014). Herpes Simplex Virus 1 Protein Kinase US3 Hyperphosphorylates P65/RelA and Dampens NF-κB Activation. J. Virol. 88 (14), 7941–7951. doi: 10.1128/JVI.03394-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang K., Lin R., Zheng C. (2013). Herpes Simplex Virus 1 Serine/Threonine Kinase US3 Hyperphosphorylates IRF3 and Inhibits Beta Interferon Production. J. Virol. 87 (23), 12814–12827. doi: 10.1128/JVI.02355-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waninger S., Kuhen K., Hu X., Chatterton J. E., Wong-Staal F., Tang H. (2004). Identification of Cellular Cofactors for Human Immunodeficiency Virus Replication via a Ribozyme-Based Genomics Approach. J. Virol. 78 (23), 12829–12837. doi: 10.1128/JVI.78.23.12829-12837.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weterings E., Gallegos A. C., Dominick L. N., Cooke L. S., Bartels T. N., Vagner J., et al. (2016). A Novel Small Molecule Inhibitor of the DNA Repair Protein Ku70/80. DNA Repair 43, 98–106. doi: 10.1016/j.dnarep.2016.03.014 [DOI] [PubMed] [Google Scholar]
- Wood A. M., Laster K., Rice E. L., Kosak S. T. (2015). A Beginning of the End: New Insights Into the Functional Organization of Telomeres. Nucleus 6, 172–178. doi: 10.1080/19491034.2015.1048407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Wang S., Gao P., Gao G., Fan Z. (2016). DNA Sensor cGAS-Mediated Immune Recognition. Protein Cell 7 (11), 777–791. doi: 10.1007/s13238-016-0320-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J., Ni L., Wang S., Wang K., Lin R., Zheng C. (2013). Herpes Simplex Virus 1-Encoded Tegument Protein VP16 Abrogates the Production of Beta Interferon (IFN) by Inhibiting NF-κB Activation and Blocking IFN Regulatory Factor 3 To Recruit Its Coactivator CBP. J. Virol. 87 (17), 9788–9801. doi: 10.1128/JVI.01440-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaneva M., Kowalewski T., Lieber M. R. (1997). Interaction of DNA-Dependent Protein Kinase With DNA and With Ku: Biochemical and Atomic-Force Microscopy Studies. EMBO J. 16, 5098–5112. doi: 10.1093/emboj/16.16.5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., An H., Liu X., Wen M., Zheng Y., Rui Y., et al. (2010). The Cytosolic Nucleic Acid Sensor LRRFIP1 Mediates the Production of Type I Interferon via a [Beta]-Catenin-Dependent Pathway. Nat. Immunol. 11, 487–494. doi: 10.1038/ni.1876 [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Fujita T. (2008). Structural Mechanism of RNA Recognition by the RIG-I-Like Receptors. Immunity 29, 178–181. doi: 10.1016/j.immuni.2008.07.009 [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Fujita T. (2009). RNA Recognition and Signal Transduction by RIG-I-Like Receptors. Immunol. Rev. 227, 54–65. doi: 10.1111/j.1600-065X.2008.00727.x [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Fujita T. (2010). Recognition of Viral Nucleic Acids in Innate Immunity. Rev. Med. Virol. 20 (1), 4–22. doi: 10.1002/rmv.633 [DOI] [PubMed] [Google Scholar]
- Yoo S., Dynan W. S. (1998). Characterization of the RNA Binding Properties of Ku Protein. Biochemistry 37, 1336–1343. doi: 10.1021/bi972100w [DOI] [PubMed] [Google Scholar]
- Yoo S., Kimzey A., Dynan W. S. (1999). Photocross-Linking of an Oriented DNA Repair Complex: Ku BOUND AT A SINGLE DNA END*. J. Biol. Chem. 274, 20034–20039. doi: 10.1074/jbc.274.28.20034 [DOI] [PubMed] [Google Scholar]
- Zhang X., Brann T. W., Zhou M., Yang J., Oguariri R. M., Lidie K. B., et al. (2011. a). Cutting Edge: Ku70 Is a Novel Cytosolic DNA Sensor That Induces Type III Rather Than Type I IFN. J. Immunol. 186, 4541–4545. doi: 10.4049/jimmunol.1003389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., He Q., Hu Z., Feng Y., Fan L., Tang Z., et al. (2016. b). Long Noncoding RNA LINP1 Regulates Repair of DNA Double-Strand Breaks in Triple-Negative Breast Cancer. Nat. Struct. Mol. Biol. 23, 522–530. doi: 10.1038/nsmb.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Kim T., Bao M., Facchinetti V., Jung S. Y., Ghaffari A. A., et al. (2011. b). DDX1, DDX21, and DHX36 Helicases Form a Complex With the Adaptor Molecule TRIF to Sense dsRNA in Dendritic Cells. Immunity 34, 866–878. doi: 10.1016/j.immuni.2011.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Su C., Zheng C., Sandri-Goldin R. M. (2016. a). Herpes Simplex Virus 1 Serine Protease VP24 Blocks the DNA-Sensing Signal Pathway by Abrogating Activation of Interferon Regulatory Factor 3. J. Virol. 90 (12), 5824–5829. doi: 10.1128/JVI.00186-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yeruva L., Marinov A., Prantner D., Wyrick P. B., Lupashin V., et al. (2014). The DNA Sensor, Cyclic GMP–AMP Synthase, Is Essential for Induction of IFN-β During Chlamydia Trachomatis Infection. J. Immunol. 193, 2394–2404. doi: 10.4049/jimmunol.1302718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y. (2011. c). The Helicase DDX41 Senses Intracellular DNA Mediated by the Adaptor STING in Dendritic Cells. Nat. Immunol. 12, 959–965. doi: 10.1038/ni.2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Ao Z., Wang B., Jayappa K. D., Yao X. (2011). Host Protein Ku70 Binds and Protects HIV-1 Integrase From Proteasomal Degradation and Is Required for HIV Replication. J. Biol. Chem. 286, 17722–17735. doi: 10.1074/jbc.M110.184739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.-B. S., Elledge S. J. (2000). The DNA Damage Response: Putting Checkpoints in Perspective. Nature 408, 433–439. doi: 10.1038/35044005 [DOI] [PubMed] [Google Scholar]