Abstract
Human cells lacking functional p53 exhibit a partial deficiency in nucleotide excision repair (NER), the pathway for repair of UV-induced DNA damage. The global genomic repair (GGR) subpathway of NER, but not transcription-coupled repair (TCR), is mainly affected by p53 loss or inactivation. We have utilized mouse embryo fibroblasts (MEFs) lacking p53 genes or downstream effector genes of the p53 pathway, gadd45 (Gadd45a) or p21 (Cdkn1a), as well as MEFs lacking both gadd45 and p21 genes to address the potential contribution of these downstream effectors to p53-associated DNA repair. Loss of p53 or gadd45 had a pronounced effect on GGR, while p21 loss had only a marginal effect, determined by measurements of repair synthesis (unscheduled DNA synthesis), by immunoassays to detect removal of UV photoproducts from genomic DNA, and by assays determining strand-specific removal of CPDs from the mouse dhfr gene. Taken together, the evidence suggests a role for Gadd45, but relatively little role for p21, in DNA repair responses to UV radiation. Recent evidence suggests that Gadd45 binds to UV-damaged chromatin and may affect lesion accessibility. MEFs lacking p53 or gadd45 genes exhibited decreased colony-forming ability after UV radiation and cisplatin compared to wild-type MEFs, indicating their sensitivity to DNA damage. We provide evidence that Gadd45 affects chromatin remodelling of templates concurrent with DNA repair, thus indicating that Gadd45 may participate in the coupling between chromatin assembly and DNA repair.
The tumor suppressor p53 is an important mediator of cellular responses to DNA damage in mammalian cells. In some cell types, p53 activation triggers apoptosis (30), while in other cell types, p53 serves a protective function, attributable not only to the activation of cell cycle checkpoints, but also enhancement of DNA repair (12; reviewed recently in reference 41). P53 plays a role in nucleotide excision repair (NER), the pathway for repair of UV-induced DNA damage, bulky carcinogen adducts, and DNA damage caused by cancer chemotherapy agents such as cis-dichloro-diammine-platinum (cisplatin) (see references 11–13 and 38–41 and references therein). Loss of p53 function, as occurs frequently in human cancer cells, leads to decreased DNA repair of these types of lesions, and in some cell types, this is reflected by increased cellular sensitivity to these agents (7, 9, 13, 16, 20, 39–41).
An important question concerning the connection between p53 and NER is to what extent the p53 protein participates directly in DNA repair (28, 48), versus p53 transcriptionally regulated gene products that contribute to the NER response. For example, p53 has been shown to directly associate with TFIIH, an NER component (28, 48), while genes implicated in repair, such as DDB2 (23, 24) and gadd45, are p53 regulated (24, 25). In addition, gadd45 is UV responsive, even in p53-deficient cells (51). In the case of gadd45, its protein product is known to associate with proliferating cell nuclear antigen (PCNA), core histones, p21, and MTK1 (6, 27, 38, 45, 47), and reduced repair, as measured by host cell reactivation of UV-damaged plasmid reporter, was observed in RKO cells expressing antisense Gadd45 (40). However, the conclusion that Gadd45 contributes to NER is complicated by several issues. First, the antisense approach only suppressed Gadd45 expression, but did not ablate it. Second, the study was carried out with a human tumor line that contained additional genetic changes. Third, one such mutation resulted in the mismatch repair-deficient phenotype, and mismatch repair has been implicated in damage recognition (32). Moreover, RKO cells express higher levels of Gadd45 than are observed for most human cell lines (40). Here we have used mouse embryo fibroblasts (MEFs) carrying homozygous deletions of the p53 gene or deletions of known component genes (downstream effectors) of the p53 pathway p21 (Cdkn1a, also known as cip1, waf1, or sdi1) and gadd45 (Gadd45a, also known as Gadd45α) or MEFs lacking both gadd45 and p21 (gadd45/p21-null cells). The present study is the first to explore the components of the p53 pathway that contribute to DNA repair in an isogenic, primary cell system carrying only the defined genetic alterations. Rodent cells do, however, exhibit intrinsically slower global genomic repair (GGR) of cyclobutane pyrimidine dimers (CPDs) than human cells (14).
Recent studies using gadd45-null cells showed that Gadd45 contributes to maintenance of genomic stability, inasmuch as cells lacking gadd45 genes exhibited multiple chromosome abnormalities, and gadd45-deficient mice showed increased radiation carcinogenesis (22). While perturbation in the control of G2 cell cycle progression was also observed, control of G1 checkpoints after either ionizing radiation (IR) or UV radiation as well as radiation-induced apoptosis was equivalent in gadd45−/− and wild-type cells (22). Although the components of the p53 pathway involved in apoptosis and cell cycle checkpoint control are well known, much less is known about p53 and the roles of its downstream effectors in the maintenance of genomic stability, including DNA repair. For example, p53 mutants have been isolated from human cancers that retain the apoptotic and cell cycle arrest properties of wild-type p53, but are nonetheless associated with genomic instability and carcinogenesis (43).
Two approaches were used to assess acute responses to UV irradiation in the MEF lines, measurements of DNA repair synthesis as unscheduled DNA synthesis (UDS) after UV irradiation and quantification of major photoproducts remaining at various times after UV irradiation. Long-term cytotoxicity responses in these cell lines after treatment with UV radiation or cisplatin suggest that p53-downstream effectors—Gadd45 in particular—contribute to NER responses and may influence cellular sensitivity to certain DNA-damaging agents.
A potential mechanism by which Gadd45 might interact with cellular NER was suggested by a recent study showing that Gadd45 interacts with chromatin (6). In this study, Gadd45 was found to have some similarity to other acidic chromatin-interacting proteins and was found to bind to UV-irradiated nucleosomes. Gadd45 also disrupted histone DNA associations in vitro (6). We now present evidence that Gadd45, in the presence of other nuclear factors, affects chromatin remodelling of damaged plasmid templates concurrent with DNA repair in vitro.
MATERIALS AND METHODS
MEFs.
Mouse embryos at 13.5 days of gestation were isolated in utero and cells were dispersed by using a razor blade. Resultant cells were cultured overnight, and were frozen as soon as was practical. MEFs from matched littermates were screened for the presence or absence of the appropriate gene sequences by Southern blotting and/or by PCR for the presence of diagnostic restriction fragments. MEFs lacking p53 genes were from previously published studies (25). The generation of mice lacking p21 genes has been described previously (8). Mice hemizygous for the presence or absence of p21 genes were bred to generate matched sets of p21+/+ and p21−/− MEFs. The construction of gadd45−/− and gadd45/p21-null mice has been reported elsewhere (22). The gadd45 gene disruption ablates all but 37 carboxy-terminal residues of the expected protein product and all of the essential promoter region. As controls in the UDS studies, we used the rb−/− MEFs (kindly provided by Allan Bradley, Baylor College of Medicine or p16ink4a−/− MEFs (kindly provided by Ronald DePinho, Dana-Farber Cancer Center). Cells were stored in liquid nitrogen, and upon thawing, were used within one to three passages. Results were obtained from two p53−/− MEF lines, three gadd45−/− lines, and two p21−/− lines. Only one MEF line was available carrying the gadd45/p21-null genotype. MEFs from matched littermates were used whenever possible.
UDS technique.
UDS assays were carried out essentially as described previously (40). Primary MEFs grown on glass slides were transferred to 150-mm2 dishes, and each respective nullizygous MEF line was irradiated side-by-side with corresponding wild-type (+/+) controls. Cells were irradiated with 20 J of UV radiation m−2 (254 nm) and were incubated for 3 h in serum-free medium containing [3H]thymidine (10 μCi per ml). Experiments with lower doses yielded similar results, but fewer tritium grains. Alternatively, cells were exposed to 100 μM cisplatin for 5 h concurrent with [3H]thymidine uptake. Cells were processed for autoradiography, and nuclei were photographed by using a ×100 objective (Olympus model AX70) under oil-immersion optics. The number of tritium grains per nucleus was determined from the photomicrographs by using a manual colony counter. Negative controls consisted of unirradiated MEFs, and the human xeroderma pigmentosum XP-A cell line XP12BE (40).
In experiments using serum-starved MEFs arrested in G0, cells were incubated in serum-free RPMI 1640 lacking isoleucine for 48 h, and UDS assays were performed as described above. Alternatively, cells were arrested in G1 and G2 by staurosporine treatment (50 nM, 24 h [35]). UDS assays were used as described above.
PCNA immunostaining.
Cells were grown on glass slides and irradiated with 20 J of UV radiation m−2 as for UDS studies. After 1.5, 3, or 6 h, slides were incubated in the presence of 1% Triton X-100 and methanol fixed. PCNA was detected with antibody PC10 (Oncogene Science, Inc.) followed by fluorescein-conjugated goat anti-mouse immunoglobulin G (Sigma).
Global genomic NER assay.
The relative number of UV-induced photoproducts, and 6-4 pyrimidine-pyrimidone photoproducts (6-4 pps) in total unreplicated genomic DNA from cells collected at various times following UV irradiation was determined using an immunoblot assay, as previously described (12). Briefly, exponentially growing cells were labeled with [3H]thymidine, washed with phosphate-buffered saline, and irradiated with 10 J of UV m−2 using a 15-W germicidal UV lamp delivering predominantly 254-nm light. Cells were either lysed immediately for an initial sample or were incubated in growth medium containing 5-bromodeoxyuridine (BrdU) to density label newly replicated DNA and then lysed at various times. Density labelling was performed during repair periods to allow unreplicated DNA to be isolated by cesium chloride isopycnic density gradient sedimentation. Equal amounts from each DNA sample were fixed to a Hybond N+ nylon membrane (Amersham) in triplicate by using a slot-blot apparatus. The membrane was incubated with mouse monoclonal antibodies specific for either CPDs or 6-4 pps (34) and horseradish peroxidase-conjugated secondary antibody. Enhanced chemiluminescence (Amersham) and phosphorimager analysis (Bio-Rad model GS-363) were employed for detecting the primary antibodies. Following antibody detection, equal DNA loading to each slot of the membrane was confirmed by scintillation spectrophotometry of the 3H-DNA on individual pieces cut from the membrane.
Strand-specific repair assays.
The rates of photoproduct removal were determined within the transcribed (TS) and nontranscribed (NTS) strand of a 14-kbp BamHI restriction fragment spanning the central region of the mouse dhfr gene, as described previously (36). Cells were irradiated with 10 Jm−2 of UV radiation, lysed immediately for an initial sample, or incubated for the times indicated to allow photoproduct repair. The frequency of induction and rate of removal of CPDs from TS and NTS of the dhfr gene was measured by treating purified BamHI-digested DNA with T4 endonuclease V (TEV), and then quantifying the reappearance of the full-length restriction fragments in DNA from cells allowed various times to remove the lesions. BamHI-treated samples from each time point were treated or mock treated with TEV, electrophoresed in parallel under denaturing conditions, Southern-transferred to a membrane, and then hybridized to strand-specific RNA probes generated by in vitro transcription. The ratio of full-length restriction fragments in the TEV-treated and untreated samples was determined by phosphorimager analysis and was used to calculate the average number of TEV-sensitive sites (unrepaired lesions) per fragment by using the Poisson distribution (12).
Survival responses to UV irradiation or cisplatin.
MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assays were carried out as described previously (10), but were terminated at 7 days. For cisplatin, cells were treated in microtiter wells for 72 h (40). For UV radiation experiments, cells were irradiated in 60-cm2 dishes and were dispensed in microtiter plates for quantitation on day 7. Assays were quantified by the use of a microtiter plate reader (E-max; Molecular Devices, Inc.). All survival data are expressed relative to untreated cells in the same experiment, as previously described (10).
Clonogenic survival experiments were conducted as described previously (38–40), except that irradiated feeder layers were used to alleviate the poor plating efficiency of MEFs. p53-null MEFs were used as feeder cells and were irradiated with 100 Gy of ionizing radiation. Approximately 5 × 105 feeder cells were applied per 10-cm2 dish. Each respective MEF line was then plated on the feeder layer and treated with DNA-damaging agents (UV radiation or cisplatin). Some experiments were also performed without feeder layers. While irradiated feeder cells did not grow, colonies (>100 cells/colony) of each respective MEF line were visible after 12 days and were counted by a manual colony counter as described previously (38–40).
Cell cycle analysis.
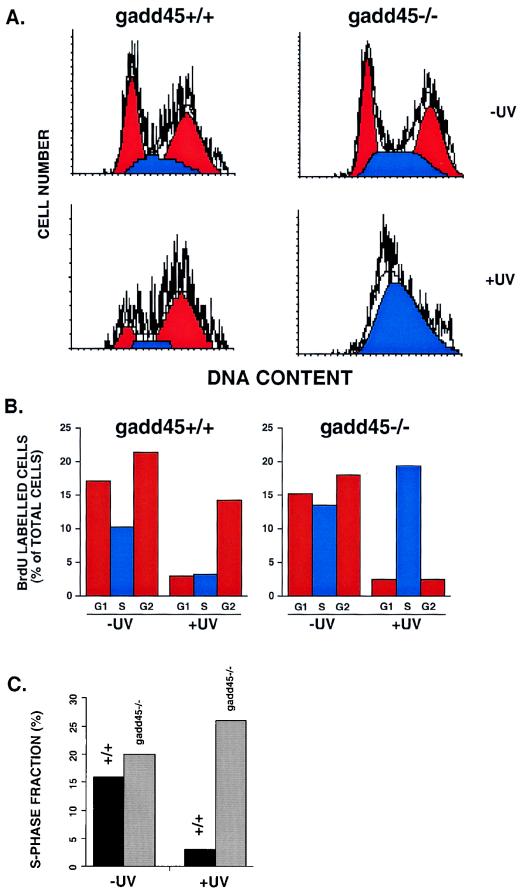
Wild-type or gadd45−/− MEFs were prepared for fluorescence-activated cell sorter (FACS) analysis as described previously (9, 10). Cells were arrested in G0 by serum deprivation and irradiated with 10 J of UV radiation m−2 (254 nm) and were incubated in complete medium containing 10 μM BrdU for 24 h. BrdU incorporation was detected by an anti-BrdU fluoroscein-conjugated antibody (Becton-Dickinson) (10). Propidium iodide (PI) staining for DNA content was as previously described (9, 10). Cell cycle analysis was performed using a Becton-Dickinson FACScan flow cytometer. The Cell-Quest software package (Becton-Dickinson) was used to analyze the data in which 15,000 BrdU-positive cells were analyzed in each individual sample. All available evidence from other studies showed a lack of effect of gadd45 deficiency on the first G1 (nor the second G1) arrest after DNA damage (29, 49). Attention was therefore focused on S and G2/M phases by the gating of BrdU-labeled cells. Only actively cycling cells are counted by this technique.
The S-phase fraction was also measured by [3H]thymidine pulse labelling 15 h after UV irradiation. Asynchronous wild-type or gadd45−/− MEFs were irradiated with 10 J of UV radiation m−2, incubated for 15 h in complete medium, and then labeled for 3 h with [3H]thymidine. Autoradiography was conducted as for UDS experiments. Although UDS was not detected after 15 h (i.e., most of the UDS occurs within the first few hours), the percentage of S-phase cells 15 h after UV irradiation was determined from photographic fields similar to those shown in Fig. 1A.
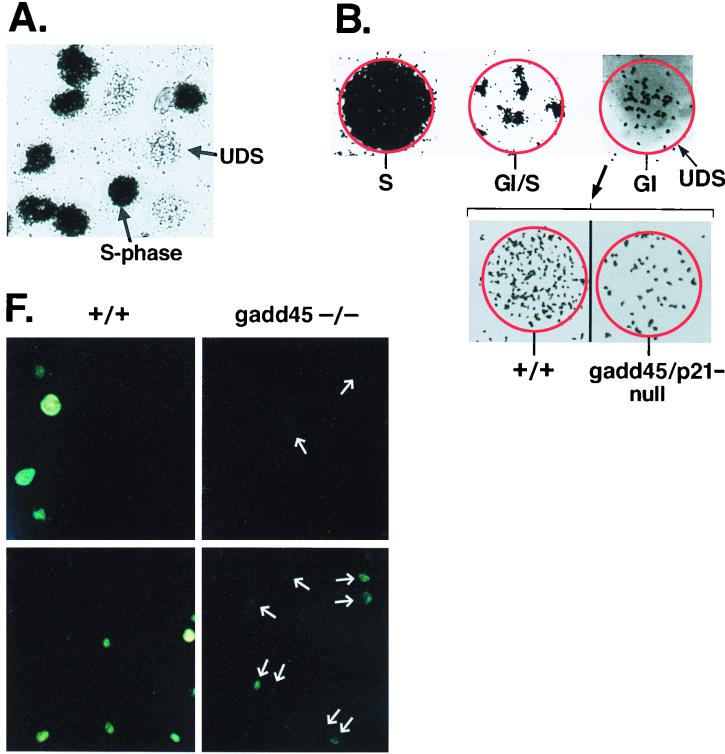
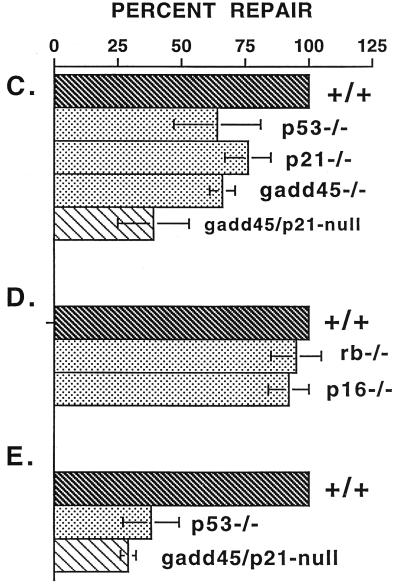
FIG. 1.
DNA repair deficiency in cells lacking p53 or p53 effector genes, gadd45 and p21, measured by UDS. (A) Illustration of UDS technique. A low-magnification (×20) field of cells irradiated with 20 J of UV radiation m−2 is shown. Nuclei were made visible by the incorporation of [3H]thymidine during either replicative or repair DNA synthesis. S-phase nuclei appear black upon the photographic emulsion, while non-S-phase cells exhibit UDS. The p21−/− and gadd45/p21-null lines did exhibit a higher S-phase fraction (50%) compared to wild-type or gadd45−/− lines (15 to 20%). (B) Distinctions between replicative DNA synthesis and repair synthesis (UDS). S-phase or G1/S transitional nuclei exhibited a different pattern of tritium incorporation from that observed in G1 or G2 nuclei (×100 oil immersion). Because nuclear membranes were not always visible on photomicrographs, the nucleus is outlined in red. Note the visualization of replicon clusters in G1/S transitional nuclei, while a G1 nucleus exhibits only UDS. The number of UDS grains per nucleus was quantitated, providing a measure of the number of sites of repair synthesis per nucleus. In comparing wild-type and (+/+) mutant MEFs, differences were observed in numbers of UDS grains per nucleus. UDS experiments were terminated 3 h after UV radiation (compare UDS results to the 4-h time point shown in Fig. 2). (C to E) Compiled data from several experiments such as those shown in panel B. Values obtained for wild-type cells were defined as 100%. (C) Compiled data for UDS after UV irradiation. The following numbers of nuclei were counted: wild-type, 652; p53−/−, 88; p21−/−, 129; gadd45−/−, 177; gadd45/p21-null, 157. The values shown are means ± standard deviations. (D) Compiled data, control MEF lines, UDS after UV irradiation. The following numbers of nuclei were counted: wild-type, 51; rb−/−, 22; p16−/−, 20. The values shown are means ± standard deviations. (E) Compiled data for UDS after cisplatin treatment. The following numbers of nuclei were counted: wild-type, 116; p53−/−, 93; gadd45/p21, 81. The values shown are means ± standard deviations. (F) PCNA immunostaining (Triton resistant) in wild-type and gadd45−/− MEFs 1.5 h after UV radiation (top) or 3 h after UV radiation (bottom). While all wild-type nuclei were clearly visible, gadd45−/− nuclei were much less visible; their positions are marked by arrows. The PCNA staining defect was most evident at 1.5 h and recovered to near normal after 6 h (not shown).
In vitro NER-chromatin assembly assay.
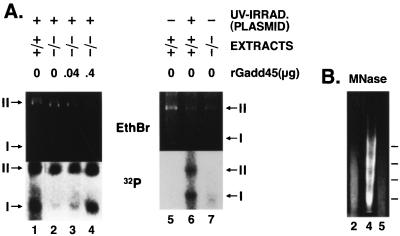
Whole-cell extracts (WCE) and nuclear (NUC) extracts were prepared from stimulated, growing wild-type or gadd45−/− mouse lymphoblasts (22). Assays were conducted in the presence of 100 μg of WCE and 50 μg of NUC extracts, utilizing a UV-damaged plasmid template (>20 lesions per plasmid molecule [38, 39]). WCE are fully competent for NER, while NUC extracts provide chromatin assembly factors and DNA topoisomerases as described previously (15). Assays were carried out in 50-μl reaction mixtures as described previously (38, 39). Approximately 150 fmol of radioactive nucleotide was incorporated during repair synthesis and subsequent steps, as determined by phosphorimager quantitation. Recombinant Gadd45 (human; rGadd45) was added to gadd45-null extracts to result in increased recovery of repaired (labeled by 32P) and subsequently remodeled (supercoiled and assembled into nucleosomes) plasmid template. rGadd45 was highly purified by high-pressure liquid chromatography (6) (a gift from F. Carrier, University of Maryland). For micrococcal nuclease (MNase) studies, in vitro reactions were conducted in parallel to those above, except that 32P-dCTP was omitted. MNase (0.001 U) (Sigma) was added directly to the reaction mixtures, incubated for an additional 5 min, and terminated by phenol-chloroform extraction, and ethanol precipitation.
The “uncoupling” of the repair reaction from the chromatin remodelling step was attempted. Damaged plasmid templates were first incubated with WCE in the presence of [32P]dCTP (3 h). Reactions were terminated by phenol-chloroform extraction and ethanol precipitation. Templates were then incubated with NUC extracts in the absence of radiolabel (3 h). In accord with the results of Gaillard et al. (15), the two processes could not be dissociated.
RESULTS
Measurements of DNA repair synthesis in situ by UDS technique.
DNA repair synthesis in UV-irradiated MEFs was quantified by unscheduled DNA synthesis (UDS; see Materials and Methods). UDS relies on the ability to distinquish replicative DNA synthesis from repair synthesis. Cells in S phase at the time of irradiation exhibit dense nuclear labelling (Fig. 1A). Cells that traverse the G1/S boundary during the 3-h labelling period exhibit a characteristic pattern of “replicon initiation” synthesis (Fig. 1B). The G1 phase of the cell cycle is 15 to 16 h in MEFs (21), while the duration of the UDS experiments is 3 h. Therefore, although MEFs spend a majority of the time in G1, few cells would traverse the G1/S boundary during this type of experiment. Repair synthesis (UDS) was detected in all non-S phase, and non-G1/S transitional cells, irrespective of whether cells were in G1 or in G2 (Fig. 1B). NER-defective XP12BE cells showed little or no UDS (results not shown and reference 40). In NER-competent cells, the number of grains per nucleus is a direct measure of the number of sites of DNA repair synthesis per nucleus and is linear with UV dose (2, 5, 14, 40). When the wild-type and mutant MEFs were compared, there was a clear difference in the number of grains per nucleus (Fig. 1B). We assayed only G1 nuclei in the current study for purposes of consistency; i.e., G2 nuclei were assayed in separate experiments but yielded results similar to those shown for G1 nuclei (G2 results not shown). A number of independent experiments were conducted for each respective pair of wild-type and mutant MEF lines (in total, eight experiments were conducted for p53−/−; four experiments for p21−/−, five experiments for gadd45−/−, and eight experiments for gadd45/p21-null MEFs). As negative controls, two experiments each were conducted in rb−/− and p16−/− MEFs. The data from all of these experiments (average relative number of grains per nucleus) are summarized in Fig. 1C to E.
Compared to the wild-type, p53−/− MEFs exhibited 35 to 70% of the normal UDS in response to UV irradiation, in agreement with other experimental approaches in which NER was found to be decreased in cells lacking functional p53 (11–13, 39, 40). Importantly, gadd45−/− MEFs exhibited UDS that was 61 to 71% of the wild-type level (P < 0.0001 by Student t test), while the UV-induced UDS response of gadd45/p21-null MEFs was 23 to 55% of the wild-type level (P < 0.0001 by Student t test) (Fig. 1C). An additional series of experiments utilized cisplatin as a DNA-damaging agent (Fig. 1E). In these experiments, the UDS response to cisplatin treatment in p53−/− MEFs was 25 to 50% of wild-type, while gadd45/p21-null MEFs showed 25 to 35% UDS compared to the wild type (P < 0.0001 by Student t test) (Fig. 1E).
Included in some studies, as controls, were MEFs lacking rb or p16ink4a tumor suppressor genes (see Materials and Methods for details). Neither rb−/− nor p16ink4a−/− MEFs exhibited an NER defect as measured by UDS (Fig. 1D), although they did exhibit deregulated cell cycle regulation. Therefore, reduced (UDS) labelling in G1 (observed in p53−/− and gadd45−/− MEF lines) is not due to accelerated S-phase entry. Indeed, p16−/− MEFs exhibited a very high S-phase fraction (approximately 50%; results not shown), yet showed normal repair measured as UDS (Fig. 1D). UDS studies with serum-starved (G0) wild-type or p53−/− MEFs yielded similar results (not shown), which indicated that the NER defect associated with p53 occurs even in noncycling cells. Because replicative DNA synthesis and repair synthesis are two distinct processes (Fig. 1B), the S-phase fraction of a given cell line does not appear to be relevant to UDS. Analysis of UDS data was confined to 2N (G1) nuclei merely for purposes of consistency. There is evidence that NER does not differ between G1 and G2 cells (33 [discussed in reference 41]). Moreover, p53−/− MEFs that were synchronized or arrested in G2 also exhibited reduced UDS (results not shown).
PCNA is recruited to sites of DNA damage, and PCNA immunostaining may be used to reflect DNA repair responses (2). PCNA immunostaining was reduced in gadd45-null MEFs, which is consistent with the UDS results (Fig. 1F). In the absence of DNA damage, there appeared to be no differences in PCNA expression between the two cell lines, as measured by Western blotting (results not shown).
Repair of CPDs and 6-4 pps in global genomic DNA.
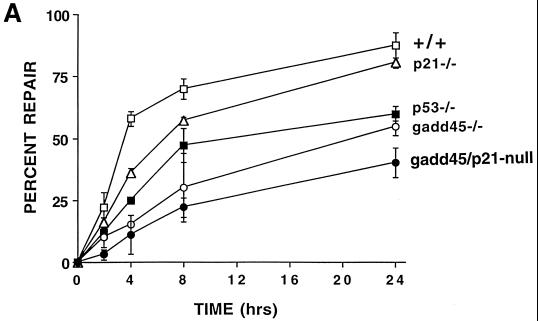
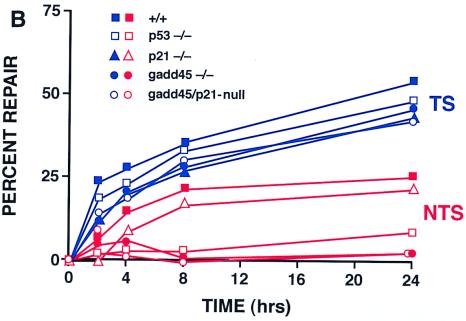
Experiments were performed to measure specifically the removal of the major UV-induced photoproducts CPDs or 6-4 pyrimidine-pyrimidone photoproducts (6-4 pps) from global genomic DNA by using an immunoblot assay with monoclonal antibodies to each of these photoproducts. The repair of 6-4 pps from global genomic DNA showed substantial differences between the different MEF lines, as shown in Fig. 2A. Wild-type MEFs demonstrated efficient removal of 6-4 pps, with 63% of the lesions repaired by 4 h and 75% of the lesions repaired by 8 h after UV irradiation. (The level of photoproducts in unirradiated cells is designated as 100% removal or repair.) p53-deficient MEFs exhibited a defect in repair of 6-4 pps, as well as CPDs (36), in line with previous studies conducted with human cells (11–13). Note that gadd45−/− and gadd45/p21-null MEFs exhibited the greatest defect, with only 18 to 27% of the lesions being repaired even after 24 h while p21−/− MEFs showed essentially normal repair of 6-4 pps after 24 h (Fig. 2A).
FIG. 2.
Deficient UV photoproduct repair in genomic DNA isolated from p53- or gadd45-null MEF lines. (A) Kinetics of 6-4 pp removal in MEFs determined by immunoassays following 10 J of UV irradiation m−2. Data were from two independent experiments conducted in triplicate. The values shown are means ± standard deviations. Note that 100% repair was defined relative to unirradiated cells. The bulk of the damage is repaired in wild-type cells within the first 3 to 4 h (compare the 4-h time point with the UDS data in Fig. 1). (B) Strand-specific repair of CPDs within the dhfr gene measured by TEV assays following 10 J of UV irradiation m−2. Repair of the NTS (red) was markedly reduced in gadd45−/− or gadd45/p21-null MEFs. Each of the MEF lines exhibited near normal TCR repair of the TS (blue).
Strand-specific repair assays.
Quantitative Southern blotting of TEV-treated DNA from UV-irradiated cells was used to examine CPD removal from TS and NTS strands of the dhfr gene. Previous studies of p53-deficient human cells showed that p53 deficiency did not significantly affect repair of the dhfr TS (repaired by TCR), but did markedly affect repair of the NTS (11–13) (repaired by GGR). Similar to p53-deficient human cells, MEFs lacking p53 or gadd45 genes exhibited nearly normal TCR of the transcribed strand of the dhfr gene, but defective GGR of the NTS of the dhfr gene (Fig. 2B). Importantly, p21−/− cells exhibited nearly normal levels of repair of either strand (Fig. 2B) (J. M. Ford, unpublished data).
Cellular sensitivity to UV irradiation or cisplatin.
In some cell types, loss of p53 function and the corresponding decrease in NER capacity sensitize cells to agents that produce DNA damage that is repaired by NER (7, 9, 10, 13, 16, 20, 39, 40). These agents include, in addition to UV irradiation, many chemical cross-linking agents, such as cisplatin or nitrogen mustards. We tested p53−/−, p21−/−, gadd45−/−, and gadd45/p21-null MEFs for sensitivity to some of these agents by using 7-day MTT (thiazolyl blue) cell survival assays. Each of the mutant MEF lines displayed enhanced sensitivity to UV radiation or cisplatin compared to wild-type MEFs (Table 1). Similar results were obtained using melphalan (a nitrogen mustard; results not shown). Table 1 shows 50% inhibitory concentration (IC50) data, which is the dose or concentration of a given agent required to give 50% survival, for each MEF line.
TABLE 1.
Sensitivity of MEFs to UV irradiation or cisplatina
| MEF line | IC50
|
|
|---|---|---|
| UV radiation (J m−2) | Cisplatin (μM) | |
| +/+ | 12.0 ± 2.0 | 9.0 ± 0.5 |
| gadd45−/− | 7.0 ± 2.0 | 4.0 ± 1.0 |
| p21−/− | 8.0 ± 2.5 | 4.0 ± 1.0 |
| gadd45/p21-null | 2.0 ± 1.0 | 1.5 ± 0.5 |
| p53−/− | 1.0 ± 0.5 | 1.0 ± 0.5 |
Shown are IC50s of the indicated agents (mean of five or more independent experiments ± standard deviations). Determinations were by 7-day MTT assays.
We also conducted clonogenic survival experiments (results not shown), as in previous studies (39, 40). MEFs exhibited poor plating efficiencies at low density, a problem alleviated through the use of feeder layers. Clonogenic survival after 10 of J UV radiation m−2 demonstrated a mean surviving fraction of 65% for wild-type cells and only 25% for gadd45/p21-null cells. After 20 J of UV radiation m−2, wild-type cells exhibited 51% mean survival, compared to only 9% for gadd45/p21-null cells. Cisplatin yielded similar results (not shown). Thus, the mutant MEFs were more sensitive to UV radiation and cisplatin than the wild type, as determined by both types of assays, consistent with other reports (16, 20). There were, however, differences in the actual doses of DNA-damaging agents required, between the two types of survival assays, owing perhaps to differences in cell density, e.g., the presence or absence of feeder cells. (Higher doses of DNA damage were required to achieve equivalent cell killing in the presence of feeder layers, probably because feeder cells secrete cytokines that promote cell survival.)
Reduced S-phase progression in gadd45-null cells after UV radiation.
The results shown in Fig. 1 and 2 provided strong evidence that Gadd45 is one component of the p53 pathway that contributes to DNA repair. In the present study, absence of Gadd45 resulted in the persistence of photolesions (Fig. 2), lesions that are known to interfere with replicative DNA synthesis. While gadd45−/− cells have already been shown to have normal p53-mediated G1 checkpoints after IR or UV radiation (22), we focused on S-phase progression by employing FACS analyses of MEFs in the presence or absence of UV damage. The data are summarized in Fig. 3A and B. Cell cycle profiles of wild-type and gadd45−/− cells were similar in the absence of DNA damage and indicated that the cells were actively growing. However, after UV irradiation, a pronounced S-phase accumulation of gadd45−/− cells was observed. This result is similar to those reported by other groups regarding UV-irradiated p53-deficient cells (7, 10, 20) and is consistent with defective NER and subsequent replicative arrest. We also determined the S-phase fractions 15 h after UV irradiation by [3H]thymidine labeling. Consistent with the FACS analysis, gadd45−/− MEFs exhibited a pronounced S-phase fraction 15 h after UV (Fig. 3C). Thus, gadd45−/− MEFs exhibit an S-phase delay after UV irradiation, due perhaps to persistence of UV photoproducts (Fig. 2), consistent with a slow DNA repair phenotype resulting in inhibition of DNA replication at damaged sites (Fig. 3).
FIG. 3.
Pronounced S-phase delay following UV-damage in gadd45−/− MEFs. (A) Flow cytometric profiles shown by PI staining. In the absence of UV irradiation, cell cycle profiles were similar (50 to 60% G1, 10 to 25% S, 20 to 35% G2/M). G1 and G2 peaks are in shown in red; the S-phase fraction is shown in blue. After UV irradiation, gadd45−/− MEFs exhibited a marked S-phase delay. The results shown correspond to 24 h after 10 J of UV irradiation m−2. Only BrdU-positive cells that were actively cycling are shown (10). The gating of BrdU-positive cells was designed to exclude cells arrested in the first G1, which as cited in other studies, was not affected by the presence or absence of Gadd45. The technique further distinquishes the p53/gadd45-mediated response to UV radiation from p53/p21-mediated G1 arrest, as discussed in the text. (B) Summary of cell cycle profiles in MEFs in the absence or presence of UV radiation. The results shown in panel A are summarized by bar graphs. Again, gadd45−/− cells are delayed in S-phase progression. Results shown correspond to 24 h after 10 J of UV irradiation m−2. Only BrdU-positive cells that were actively cycling are shown (10). (C) Determination of S-phase fractions 15 h after UV irradiation by [3H]thymidine pulse-labelling. UV-irradiated gadd45−/− MEFs exhibit a pronounced S-phase fraction 15 h after UV damage. The results were obtained from two or more independent experiments.
Biochemical NER assays to explore Gadd45 function(s).
The core NER reaction can be carried out in vitro in the presence of WCE (1, 37). However, WCE extracts may be relatively depleted of a number of nuclear proteins including DNA topoisomerases and chromatin remodeling factors, that while not required, may contribute to NER (14, 15, 50). The contributions of nuclear proteins to in vitro NER reactions, can be assessed by the addition of nuclear fractions (NUC) to WCE-NER reaction mixtures (15). By this approach, chromatin assembly and disassembly have been identified as processes closely linked to NER (15, 18, 23, 26, 50). WCE and NUC extracts were prepared from mouse lymphoblasts derived from either wild-type or gadd45−/− animals. The presence of Gadd45, either as an endogenous component of NUC extracts from wild-type mouse cells or added exogenously to gadd45−/− extracts, promoted chromatin assembly on plasmids undergoing or having undergone NER in vitro, although the amount of recombinant Gadd45 (400 ng) exceeded the amount endogenous to wild-type extracts (20 to 50 ng). Ethidium bromide (EtBr) staining was used to show approximate equal loading of the lanes, while repair synthesis was measured by 32P incorporation (panels marked 32P) into the damaged plasmids. Note that while very little form I DNA was recovered from the reactions (total plasmid DNA shown by EtBr staining), an appreciable amount of radiolabeled (repaired) plasmid DNA was recovered as form I DNA; i.e., repaired plasmids were preferentially assembled into nucleosome ladders, which showed a characteristic MNase digestion pattern (Fig. 4B). Recovery of (repaired) form I DNA was enhanced in the presence of Gadd45 (Fig. 4A). The chromatin remodeling reaction could not be uncoupled from the NER reaction (reference 15 and results not shown).
FIG. 4.
(A) Gadd45 affects in vitro NER-chromatin assembly in experiments with mouse cell extracts. WCE and NUC extracts prepared from wild-type (+/+) or gadd45−/− (−/−) mouse lymphoblasts were incubated with UV-damaged plasmid template (left panel). While little form I DNA was recovered from the reactions, evidenced by EtBr staining (total DNA), a greater fraction of repaired (radiolabeled) DNA was recovered as form I. Gadd45 affects the recovery of form I (repaired, radiolabeled) DNA, which is ordered into nucleosome ladders, evidenced by MNase digestion (shown in panel B). Gadd45 was either endogenous to wild-type extracts, or recombinant Gadd45 (rGadd45) was added to gadd45−/− extracts in the amounts indicated (micrograms). (Right panel) Plasmid templates lacking UV damage exhibited low levels of 32P labelling. Lanes show approximate 32P incorporation as follows: 1 to 4, 150 fmol; 6, 190 fmol; 5 and 7, 11 fmol. Form II DNA was predominantly nicked, while form I DNA was predominantly supercoiled. (B) MNase digestion of plasmids recovered from in vitro NER-chromatin assembly assays, corresponding to lanes 2, 4, and 5 in panel A. In the absence of Gadd45 (lane 2), the predominantly form II plasmid DNA did not exhibit nucleosome ladders after MNase digestion. In the presence of Gadd45 (lane 4), laddering was observed, indicative of chromatin assembly. Undamaged plasmids were not assembled into chromatin (lane 5).
DISCUSSION
Effect of p53 pathway on the GGR subpathway of NER.
NER can be classified into two broad subpathways: TCR and GGR. Recent studies show that p53 affects primarily GGR (11–13), although one study suggested a contribution of p53 to TCR in addition to GGR (46). In the present study, we examined both GGR and TCR, and as shown previously in human cells lacking p53 function, GGR was markedly affected. Defective GGR repair of 6-4 pps was more pronounced in the gadd45-deficient MEFs than in the p53-deficient MEFs (Fig. 2A), suggesting that Gadd45, like p48-XPE (DDB2, the p48 DNA-damage-binding protein that is defective in a subset of xeroderma pigmentosum group E patients [24]) contributes considerably to the p53-mediated NER response. The contributions of p53-regulated genes Gadd45 and p48-XPE to DNA repair responses define a new paradigm of p53 function, separable from other known p53 functions in cell cycle arrest and apoptosis. While p21 is a major mediator of p53-mediated G1 cell cycle arrest, p21 contributes relatively little to DNA repair at early times (3- to 4-h time points in Fig. 1 and 2), and not at all at later time points (24 h; Fig. 2A). On the other hand, approximately 50% of the 6-4 lesions persist in the gadd45-deficient cells even after 24 h (Fig. 2A). We also assayed strand-specific repair of TS and NTS strands of the dhfr gene by using TEV assays. MEFs lacking p53 or gadd45 genes exhibited defective CPD repair of the NTS, but near normal TCR of the TS (Fig. 2B). Thus, a pronounced GGR defect associated with p53 or gadd45 loss is clearly shown by both types of assays.
Relationship of p53-mediated NER to G1 checkpoint and p21.
The p53-mediated NER response appears to be distinct from the G1 cell cycle checkpoint, as indicated by several lines of evidence: Li-Fraumeni fibroblasts heterozygous for mutant or wild-type p53 genes retain the G1 checkpoint, but nonetheless exhibit an NER defect (11); HPV16-E7 oncoprotein expression, which blocks Rb, and hence blocks G1 cell cycle arrest, had no effect on NER (40). In the present study, rb- or p16-null MEFs exhibited altered G1 cell cycle control, but not altered NER, while gadd45-null MEFs showed no G1 checkpoint abnormality (22), but deficient NER (Fig. 1). Although the UDS experiments measured NER primarily in G1 nuclei, these could be readily distinguished from G1/S transitional nuclei (Fig. 1B), and, moreover, p53-null cells synchronized or arrested in G1 or G2 also showed an NER defect (results not shown). Although p21 is a major mediator of p53-induced G1 arrest, p21-null MEFs exhibited only a very slight NER defect (discussed below), contrary to what one would predict were the NER defect dependent on or coincident with the G1 checkpoint.
The potential contribution of p21 to DNA repair has been unclear. Some studies have shown reduced capacity for NER and UV sensitivity in p21-null HCT116 cells (31). In the present study, a modest but statistically significant decrease in UDS (75% of the wild-type level) was observed in p21−/− MEFs 3 h after UV irradiation, and gadd45/p21-null MEFs exhibited a greater defect than either of the single nullizygous mutants, which was again statistically significant. Indeed, p21 and gadd45 appear to be “additive” in their effects on NER measurements taken 3 h after irradiation (Fig. 1C), an interesting and novel observation that could possibly be due to subtle cell cycle differences, with due consideration of the caveats mentioned in the preceding paragraph. The 6-4 pp removal experiments are consistent with the UDS results (3 to 4 h after UV irradiation), but these experiments also showed that p21−/− cells recovered normal levels of NER after 24 h (Fig. 2A). TEV assays likewise showed essentially no effect of p21 deficiency on NER 24 h after UV irradiation (Fig. 2B). Thus, the effect of p21 on NER appears to be minimal, and Gadd45 contributes considerably to the p53-mediated NER response. Note that at the 3-h time point, where greater than 50% of the 6-4 pps lesions are removed from genomic DNA of wild-type cells, only 15% of the lesions were removed in gadd45−/− cells. After 24 h, wild-type and p21−/− cells were essentially identical in 6-4 lesion repair, while gadd45−/− cells still retained about 50% of the 6-4 lesions (Fig. 2A).
If p21 does contribute, albeit minimally, to NER, as suggested by Fig. 1, it is likely that this function is unrelated to p21 activity as a cyclin-dependent kinase inhibitor, because, again, neither HPV16-E7 expression, nor rb or p16 deficiency recapitulates the effect of p53 or gadd45 deficiency on NER. Moreover, it is conceivable that any effect of p21 on NER may require the presence of additional genetic alterations, such as mutS deficiency in HCT116 cells or gadd45 deficiency in the gadd45/p21-null MEFs. The complex relationship of p53-mediated DNA repair to the activation of cell cycle checkpoint(s) will be the subject of future studies.
Cell cycle checkpoint responses to UV radiation may differ from the well-known G1 checkpoint response elicited by IR (and also G1 arrest by UV radiation to some extent [25, 51]) and mediated by cyclin-dependent kinase inhibitor p21. In contrast, p53-mediated responses to UV radiation can involve the S phase and G2/M delays, which may reflect the greater lesion frequency of UV photoproducts compared to IR-induced damages. The cell cycle analyses in Fig. 3 further dissociate the p53/gadd45-mediated response to UV radiation from the p53/p21-mediated response to IR, in that the presence or absence of Gadd45 showed no effect on G1 arrest in the present and previous experiments (reference 22 and results not shown), and in fact, the experiments in Fig. 3A and B, employing BrdU incorporation, were specifically designed to exclude cells arrested in the first G1 phase from the analysis.
Gadd45-deficient cells exhibit slow repair.
It should be noted that p53 protein and a number of downstream effector proteins are present at appreciable basal levels in many cell types, including normal human and mouse fibroblasts (7). This has also been observed for the p48-XPE gene product. p48-XPE is expressed at higher basal levels in p53 wild-type cells than p53 mutant cells, but is induced after DNA damage only in p53 wild-type cells (24). This may explain why DNA repair functions are significantly enhanced in p53-wild-type cells at early times (3 to 4 h after UV irradiation; Fig. 1 and 2). On the other hand, MEFs nullizygous for gadd45 genes did not achieve normal levels of 6-4 pp repair even after 24 h (Fig. 2A). TEV assays likewise showed a pronounced NER deficiency in gadd45-null cells (Fig. 2B). The persistence of lesions in gadd45-deficient cells led to S-phase delay 24 h after UV irradiation (Fig. 3). S-phase delay may represent an active checkpoint response or may reflect a blockage of replication fork progression at UV-induced lesions, as observed in other NER-defective cells (17). The cell cycle results shown in Fig. 3 are consistent with an NER defect causing replicon stalling at damaged nucleotide bases (17). Persistence of lesions (e.g., after 24 h as shown in Fig. 2) may continue to trigger cell cycle checkpoint responses, including p53, although gadd45 deficiency produced no overt changes in p53 or p21 mRNA or protein expression at early times (3 to 12 h) following DNA damage (results not shown).
Cytotoxic responses to DNA-damaging agents.
In this study and others, cells lacking p53 or components of the p53 pathway were sensitized to UV- or cisplatin-induced DNA damage (previously cited). These findings provide a counterpoint to the prevalent view that cells lacking functional p53 are often desensitized to DNA damage (attributed to escape from apoptosis). There are a number of implications that follow. (i) Many epithelial cell types may die by mechanisms other than apoptosis, e.g., cytotoxicity of DNA damage (3). (ii) In these cell types, loss of functional p53 either has no effect on cell death responses, or in the case of UV or cisplatin damage, can result in enhanced cell death. (iii) Downstream effectors of the p53 pathway can contribute to cell survival responses, although DNA repair is only one of multiple parameters that influence cytotoxic responses to DNA damage. For example, p21−/− cells also were more sensitive to UV radiation, due probably to G1 checkpoint loss. It will be of interest to explore further the role of p53 in cell death or survival responses in epithelial and other cell types in which p53 activation is associated with DNA repair (survival) responses rather than apoptosis.
Coupling of later stages of NER to chromatin assembly.
The mechanism of NER consists of several steps including incision of the DNA strand carrying the damage, displacement of an approximately 30-nucleotide oligomer containing the damaged base(s), resynthesis of the correct sequence using the complementary strand as a template, and ligation. Because mammalian cells package their genomes into chromatin, one may also consider that destabilization of the preexisting chromatin structure may affect early phases of NER, inasmuch as nucleosomes may impede NER (14). One may also consider that restoration of chromatin structure, i.e., repositioning of nucleosomes along newly repaired DNA, would be required to maintain proper regulation of genomic functions. In fact, other studies have shown that the NER process is tightly linked to chromatin assembly (15). Specifically, the passage of the DNA polymerase associated with repair synthesis was found to promote chromatin formation. This was demonstrated by the finding that plasmid templates undergoing NER in vitro were preferentially assembled into nucleosomes (over those that were not repaired, and therefore retain DNA damage). Stated another way, this means that the repair process promotes nucleosome assembly when all factors are available. The chromatin assembly factor CAF1 was implicated as a mediator of the chromatin remodelling process, because nuclear extracts containing CAF1 could carry out the nucleosome assembly step, while CAF1-deficient extracts could not (15). Moreover, the chromatin assembly step could not be uncoupled from NER, because plasmids that were repaired in WCE extracts, recovered by extraction and ethanol precipitation, and then subsequently incubated with nuclear extracts did not exhibit nucleosome assembly (15). These findings implicated CAF1 in a late step of NER, in which repaired DNA is then repackaged into a native configuration. Supercoiled plasmids were consequently recovered from in vitro repair reactions (15). We show evidence that Gadd45 participates in late-stage NER steps involving chromatin assembly. As mentioned above, nucleosome assembly is inhibited by DNA damage, particularly in the form of bulky lesions or UV damage which may cause helical distortions. It is clear from Fig. 4 that Gadd45 promotes one or more activities associated with this process.
p53-regulated gene products involved in chromatin accessibility.
A recent study showed that Gadd45 binds to UV-damaged chromatin and may affect accessibility to sites of DNA damage (6). A number of reports in the literature suggest that chromatin accessibility proteins such as yeast and human CAF-1 (mentioned above and in references 15 and 26), and Saccharomyces RAD7 and RAD16 (50), while not components of the core NER complex, can specifically contribute to the GGR subpathway of NER. XPC, which is also required for normal GGR (44), has also been implicated in damage recognition which could involve either DNA damage or chromatin damage (3, 44). Such a role for Gadd45 would be consistent with the present results, wherein MEFs lacking gadd45 exhibit defective GGR. An additional link between p53 and the GGR subpathway of NER is suggested by the finding that the xeroderma pigmentosum group E (XP-E) p48 gene, like gadd45, is transcriptionally regulated by p53 (24). The NER defect in XP-E likewise affects the GGR subpathway (23, 24). Interestingly, the p48-XPE protein shares sequence homology with CAF-1 (23), while Gadd45 shares some homology to other chromatin accessibility proteins (6).
One means by which chromatin accessibility factors may contribute to NER is by facilitating the binding of damage recognition proteins and/or other proteins involved in DNA damage processing, to sites of DNA damage. One such protein, PCNA, is known to bind strongly to damaged chromatin after UV irradiation. The binding of PCNA to damaged chromatin is defective in XP cells, strongly suggesting that the “recruitment” of PCNA to damaged chromatin reflects its involvement in NER (determined by immunostaining of Triton-resistant PCNA [2]). In the present study, PCNA immunostaining after UV radiation was defective in Gadd45-deficient cells (Fig. 1F). This finding suggests that PCNA may be one such protein whose interaction with damage sites is affected by the presence or absence of Gadd45 (2), which interestingly has been shown to associate with PCNA (19, 38, 47).
Since naked plasmid DNA was introduced to the in vitro DNA repair reactions (Fig. 4), the assay may not address early events involving recognition of damaged DNA in chromatin. However, the present results clearly illustrate a role for Gadd45 in chromatin assembly. A possible role for Gadd45 in chromatin accessibility early in the reaction cannot be excluded. Indeed, the RAD7 and RAD16 proteins, previously shown to be chromatin accessibility factors, that enhanced NER, but were not required for NER (50), have more recently been shown to act as a part of a DNA damage sensor mechanism, by binding to UV-damaged chromatin as a component of the damage recognition step (18). One implication is that Gadd45 signalling may participate in DNA damage responses, at least in some cases, upstream of p53 in the damage-response pathway (49), as would be predicted were Gadd45 to play a role in damage recognition. Such a postulated role for Gadd45 early in the repair reaction would be consistent with the UDS experiments, in which gadd45-deficient cells exhibited a defect in the repair synthesis step of NER (Fig. 1), but could also be due to the temporal coupling of repair synthesis with latter-stage chromatin remodeling (Fig. 4) (15).
In summary, we have used a genetic approach to dissect components of the p53 pathway that contribute to DNA repair responses. In particular, Gadd45 contributes appreciably to DNA repair, while p21 contributes relatively little. One interpretation is that p53-associated NER may be independent of the G1 cell cycle checkpoint mediated by p21 (25). Recent studies showed that Gadd45 binds to UV-damaged chromatin, perhaps facilitating access to regions of DNA damage (6). The p53-associated NER response may therefore be mediated at the level of chromatin accessibility to sites of DNA damage.
ACKNOWLEDGMENTS
This work was supported in part by Clinical Investigator Award KO8-CA64330 (J.M.F.) and Outstanding Investigator grant CA44349 (P.C.H.), both from the National Cancer Institute; and by grant 1RG84-002-15 from the American Cancer Society (M.L.S.). We thank Jay Robbins for helpful advice on UDS experiments and Rodney S. Nairn, Maureen A. Harrington, Rick Bockrath, and members of the Fornace and Hanawalt laboratories for critical comments on the manuscript.
REFERENCES
- 1.Aboussekhra A, Biggerstaff M, Shivji M, Vilpo J, Moncollin V, Podust V, Protic M, Hubscher U, Egly J-M, Wood R D. Mammalian DNA excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 2.Aboussekhra A, Wood R D. Detection of nucleotide excision repair incisions in human fibroblasts by immunostaining for PCNA. Exp Cell Res. 1995;221:326–332. doi: 10.1006/excr.1995.1382. [DOI] [PubMed] [Google Scholar]
- 3.Baxter B K, Smerdon M J. Nucleosome unfolding during DNA repair in normal and xeroderma pigmentosum (group C) human cells. J Biol Chem. 1998;273:17517–17524. doi: 10.1074/jbc.273.28.17517. [DOI] [PubMed] [Google Scholar]
- 4.Brown J M, Wouters B G. Apoptosis, p53, and tumor cell sensitivity to anticancer agents. Cancer Res. 1999;59:1391–1399. [PubMed] [Google Scholar]
- 5.Burk P G, Lutzner M A, Clarke D D, Robbins J H. Ultraviolet-stimulated thymidine incorporation in xeroderma pigmentosum lymphoblasts. J Lab Clin Med. 1971;77:759–767. [PubMed] [Google Scholar]
- 6.Carrier F, Georgel P T, Pourquier P, Blake M, Kontny H U, Antinore M J, Gariboldi M, Myers T G, Weinstein J N, Pommier Y, Fornace A J., Jr Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol Cell Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cistulli C A, Kaufman W K. P53-dependent signalling sustains DNA replication and enhances clonogenic survival in 254 nm ultraviolet-irradiated human fibroblasts. Cancer Res. 1998;58:1993–2002. [PubMed] [Google Scholar]
- 8.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 9.Fan S, Smith M L, Rivet D J, Duba D, Zhan Q, Kohn K W, Fornace A J, Jr, O'Connor P M. Disruption of p53 function sensitizes breast cancer MCF7 cells to cisplatin and pentoxifylline. Cancer Res. 1995;55:1649–1654. [PubMed] [Google Scholar]
- 10.Fan S, Chang J K, Smith M L, Duba D, Fornace A J, Jr, O'Connor P M. Cells lacking CIP1/WAF1 genes exhibit preferential sensitivity to cisplatin and nitrogen mustard. Oncogene. 1997;14:2127–2136. doi: 10.1038/sj.onc.1201052. [DOI] [PubMed] [Google Scholar]
- 11.Ford J M, Hanawalt P C. Li-Fraumeni syndrome fibroblasts homozygous for p53 mutations are deficient in global DNA repair but exhibit normal transcription-coupled repair and enhanced UV-resistance. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ford J M, Hanawalt P C. Expression of wild-type p53 is required for efficient global genomic nucleotide excision repair in UV-irradiated human fibroblasts. J Biol Chem. 1997;272:28073–28080. doi: 10.1074/jbc.272.44.28073. [DOI] [PubMed] [Google Scholar]
- 13.Ford J M, Baron E L, Hanawalt P C. Human fibroblasts expressing the human papillomavirus E6 gene are deficient in global genomic nucleotide excision repair and sensitive to ultraviolet irradiation. Cancer Res. 1998;58:599–603. [PubMed] [Google Scholar]
- 14.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: American Society for Microbiology Press; 1995. pp. 283–310. [Google Scholar]
- 15.Gaillard P-H, Martini E M D, Kaufman P D, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor-1. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 16.Gorospe M, Wang X, Holbrook N J. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol Cell Biol. 1998;18:1400–1407. doi: 10.1128/mcb.18.3.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths T D, Ling S Y. Effect of UV light on DNA chain growth and replicon initiation in xeroderma pigmentosum variant cells. Mutagenesis. 1991;6:247–251. doi: 10.1093/mutage/6.4.247. [DOI] [PubMed] [Google Scholar]
- 18.Guzder S N, Sung P, Prakash L, Prakash S. Yeast RAD7-RAD16 complex, specific for nucleotide excision repair of the nontranscribed DNA strand, is an ATP-dependent DNA damage sensor. J Biol Chem. 1997;272:21665–21668. doi: 10.1074/jbc.272.35.21665. [DOI] [PubMed] [Google Scholar]
- 19.Hall P A, Kearsey J M, Coates P J, Norman D G, Warbrick E, Cox L S. Characterization of the interaction between PCNA and Gadd45. Oncogene. 1995;10:2427–2433. [PubMed] [Google Scholar]
- 20.Hawkins D S, Demers G W, Galloway D A. Inactivation of p53 enhances sensitivity to multiple chemotherapeutic agents. Cancer Res. 1996;56:892–898. [PubMed] [Google Scholar]
- 21.Herrera R E, Sah V P, Williams B O, Mäkelä T P, Weinberg R A, Jacks T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol Cell Biol. 1996;16:2402–2407. doi: 10.1128/mcb.16.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollander M C, Sheikh M S, Bulavin D, Lundgren K, Augeri-Henmueller L, Shehee R, Molinaro T, Kim K, Tolosa E, Ashwell J, Rosenberg M, Zhan Q, Fernandez-Salguero P, Morgan W F, Deng C, Fornace A J., Jr Genomic instability in gadd45-deficient mice. Nat Genet. 1999;23:176–184. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- 23.Hwang B J, Toering S, Francke U, Chu G. p48 activates a UV-damaged-DNA binding factor and is defective in xeroderma pigmentosum group E cells that lack binding activity. Mol Cell Biol. 1998;18:4391–4399. doi: 10.1128/mcb.18.7.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang B J, Ford J M, Hanawalt P C, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kastan M B, Zhan Q, El-Deiry W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 26.Kaufman P D, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-1. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 27.Kearsey J, Coates P J, Prescott A R, Warbrick E, Hall P A. Gadd45 is a nuclear cell cycle regulated protein which interacts with p21Cip1. Oncogene. 1995;11:1675–1683. [PubMed] [Google Scholar]
- 28.Leveillard T, Andera L, Bissonette N, Schaeffer L, Bracco L, Egly J-M, Waslyk B. Functional interactions between p53 and the TFIIH complex are affected by tumour-associated mutations. EMBO J. 1996;15:1615–1624. [PMC free article] [PubMed] [Google Scholar]
- 29.Little J B, Nagasawa H, Keng P C, Yu Y, Li C Y. Absence of radiation-induced G1 arrest in two closely-related human lymphoblast cell lines that differ in p53 status. J Biol Chem. 1995;270:11033–11036. doi: 10.1074/jbc.270.19.11033. [DOI] [PubMed] [Google Scholar]
- 30.Lowe S W, Ruley H E, Jacks T, Housman D E. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 31.McDonald E R, Wu G S, Waldman T, El-Deiry W S. Repair defect in WAF1/CIP1−/− human cancer cells. Cancer Res. 1996;56:2250–2255. [PubMed] [Google Scholar]
- 32.Mellon I, Champe G N. Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide excision repair of the lactose operon in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:1292–1297. doi: 10.1073/pnas.93.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell D L, Cleaver J E, Lowery M P, Hewitt R R. Induction and repair of (6-4) photoproducts in normal human and xeroderma pigmentosum variant cells during the cell cycle. Mutat Res. 1995;337:161–167. doi: 10.1016/0921-8777(95)00020-k. [DOI] [PubMed] [Google Scholar]
- 34.Mori T, Nakane M, Hattori T, Matsunaga T, Ihara M, Nikaido O. Simultaneous establishment of monoclonal antibodies specific for either cyclobutane pyrimidine dimer or (6-4) photoproduct from the same mouse immunized with ultraviolet-irradiated DNA. Photochem Photobiol. 1991;54:225–232. doi: 10.1111/j.1751-1097.1991.tb02010.x. [DOI] [PubMed] [Google Scholar]
- 35.Orr M S, Reinhold W, Yu L, Schreiber-Agus N, O'Connor P M. An important role for the retinoblastoma protein in staurosporine-induced G1 arrest in murine embryo fibroblasts. J Biol Chem. 1998;273:3803–3807. doi: 10.1074/jbc.273.7.3803. [DOI] [PubMed] [Google Scholar]
- 36.Prost S, Ford J M, Taylor C, Doig J, Harrison D J. Hepatitis B-x protein inhibits p53-dependent DNA repair in primary mouse hepatocytes. J Biol Chem. 1998;273:33327–33332. doi: 10.1074/jbc.273.50.33327. [DOI] [PubMed] [Google Scholar]
- 37.Sancar A. Mechanisms of DNA excision repair. Science. 1994;266:1954–1956. doi: 10.1126/science.7801120. [DOI] [PubMed] [Google Scholar]
- 38.Smith M L, Chen I-T, Zhan Q, Bae I, Chen C-Y, Gilmer T M, Kastan M B, O'Connor P M, Fornace A J., Jr Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 39.Smith M L, Chen I-T, Zhan Q, O'Connor P M, Fornace A J., Jr Involvement of the p53 tumor suppressor in repair of UV-type DNA damage. Oncogene. 1995;10:1053–1059. [PubMed] [Google Scholar]
- 40.Smith M L, Kontny H U, Zhan Q, Sreenath A, O'Connor P M, Fornace A J., Jr Antisense GADD45 expression results in decreased DNA repair and sensitizes cells to UV-irradiation or cisplatin. Oncogene. 1996;13:2255–2263. [PubMed] [Google Scholar]
- 41.Smith M L, Fornace A J., Jr P53-mediated protective responses to UV-irradiation. Proc Natl Acad Sci USA. 1997;94:12255–12257. doi: 10.1073/pnas.94.23.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith M L, Bortnick R A, Sheikh M S, Fornace A J., Jr Chromatin relaxation by overexpression of mutant p53, HPV16-E6, or cyclin G transgenes. Exp Cell Res. 1998;242:235–243. doi: 10.1006/excr.1998.4078. [DOI] [PubMed] [Google Scholar]
- 43.Smith P D, Crossland S, Parker G, Osin P, Brooks L, Waller J, Philp E, Crompton M, Gusterson B, Allday M, Crook T. Novel p53 mutants selected in BRCA-associated tumors which dissociate transformation suppression from other wild-type p53 functions. Oncogene. 1999;18:2451–2459. doi: 10.1038/sj.onc.1202565. [DOI] [PubMed] [Google Scholar]
- 44.Sugasawa K, Ng J M, Masutani C, Iwai S, van der Spek P J, Eker A P, Hanaoka F, Bootsma D, Hoeijmakers J H J. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 45.Takakawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-reponsive MTK1/MEKK4/MAPKKK pathway. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 46.Therrian J P, Drouin R, Baril C, Drobetsky E A. Human cells compromised for p53 function exhibit defective global and transcription-coupled nucleotide excision repair, whereas cells compromised for pRb function are defective only in global repair. Proc Natl Acad Sci USA. 1999;96:15038–15043. doi: 10.1073/pnas.96.26.15038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vairapandi M, Balliet AG, Fornace A J, Jr, Hoffman B, Liebermann D A. The differentiation primary response gene MyD118, related to GADD45, encodes for a nuclear protein which interacts with PCNA and p21Waf1/Cip1. Oncogene. 1996;12:2579–2594. [PubMed] [Google Scholar]
- 48.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J-M, Wang Z, Friedberg E C, Evans M K, Taffe B G, Bohr V A, Weeda G, Hoeijmakers J H J, Forrester K, Harris C C. p53 modulation of TFIIH-associated nucleotide excision repair activity. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 49.Wang X-W, Zhan Q, Coursen J D, Khan M, Kontny H U, Yu L, Hollander M C, O'Connor P M, Fornace A J, Jr, Harris C C. Gadd45 induction of a G2-M cell cycle checkpoint. Proc Natl Acad Sci USA. 1999;80:875–879. doi: 10.1073/pnas.96.7.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Wei S, Reed S H, Wu X, Svejstrup J Q, Feaver W J, Kornberg R D, Friedberg E C. The RAD7, RAD16, and RAD23 genes of Saccharomyces cerevisiae: requirement for transcription-independent nucleotide excision repair in vitro and interactions between the gene products. Mol Cell Biol. 1997;17:635–643. doi: 10.1128/mcb.17.2.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhan Q, Fan S, Smith M L, Bae I, Yu K, Alamo I, O'Connor P M, Fornace A J., Jr Abrogation of p53 function affects gadd gene responses to DNA base-damaging agents and starvation. DNA Cell Biol. 1996;15:805–815. doi: 10.1089/dna.1996.15.805. [DOI] [PubMed] [Google Scholar]