Abstract
To acquire sufficient mineral nutrients such as phosphate (Pi) from the soil, most plants engage in symbiosis with arbuscular mycorrhizal (AM) fungi. Attracted by plant-secreted strigolactones (SLs), the fungi colonize the roots and form highly branched hyphal structures called arbuscules inside inner cortex cells. The host plant must control the different steps of this interaction to maintain its symbiotic nature. However, how plants sense the amount of Pi obtained from the fungus, and how this determines the arbuscule lifespan, are far from understood. Here, we show that Medicago truncatula SPX-domain containing proteins SPX1 and SPX3 regulate root Pi starvation responses, in part by interacting with PHOSPHATE RESPONSE REGULATOR2, as well as fungal colonization and arbuscule degradation. SPX1 and SPX3 are induced upon Pi starvation but become more restricted to arbuscule-containing cells upon the establishment of symbiosis. This induction in arbuscule-containing cells is associated with the presence of cis-regulatory AW-boxes and transcriptional regulation by the WRINKLED1-like transcription factor WRI5a. Under Pi-limiting conditions, SPX1 and SPX3 facilitate the expression of the SL biosynthesis gene DWARF27, which could help explain the increased fungal branching in response to root exudates. Later, in arbuscule-containing cells, SPX1 and SPX3 redundantly control arbuscule degradation. Thus, SPX proteins play important roles as phosphate sensors to maintain a beneficial AM symbiosis.
In Medicago, two phosphate-sensing SPX proteins control phosphate homeostasis and the degradation of arbuscules to ensure a beneficial mycorrhizal symbiosis.
Introduction
In nature, plants usually face low mineral phosphate (Pi) availability in the soil, which limits their growth and development (Rouached et al., 2010). To deal with such Pi limitation, plants typically induce a set of Pi starvation-induced (PSI) genes to acquire more Pi from the soil and to increase Pi use efficiency (Bari et al., 2006; Zhou et al., 2008). In addition, most land plants engage in a symbiosis with arbuscular mycorrhizal (AM) fungi to increase their Pi acquisition efficiency (Smith and Read, 2008). Under Pi starvation conditions, plant roots release strigolactones (SLs), which enhance spore germination and hyphal branching to initiate a symbiotic association (Akiyama et al., 2005; Besserer et al., 2006). Subsequently, the fungus colonizes the roots and forms highly branched hyphal structures, called arbuscules, inside root cortex cells, and its hyphae continue to form extensive networks in the soil. The hyphae can efficiently reach the scarcely available Pi, which they deliver to the plant in return for carbon (fatty acids and sugars; Luginbuehl and Oldroyd, 2017).
The maintenance of proper Pi homeostasis is important for plant growth and development, as either too low or too high Pi concentrations in plant cells can be harmful to the plant (Wang et al., 2014). Therefore, plants continuously sense and signal the Pi status in response to their environment. Also during the symbiosis, the plant must integrate Pi status with fungal colonization and arbuscule development to keep the interaction beneficial. However, how a plant determines how much Pi it obtains locally at the arbuscules, and how it regulates Pi homeostasis in relation to arbuscule development, is still an open question (Ezawa and Saito, 2018; Müller and Harrison, 2019).
Arbuscules form a symbiotic interface where Pi is provided to the host plant (Ezawa and Saito, 2018). These relatively short-lived structures are degraded and removed from the cortical cells after 2–7 days (Kobae and Hata, 2010). This transient characteristic is thought to give the plant a means to locally abort arbuscules when they do not deliver sufficient nutrients due to their age (Lanfranco et al., 2018). In line with this, the loss of the arbuscule-containing cell-specific PHOSPHATE TRANSPORTER4 (PT4), responsible for transporting Pi across the peri-arbuscular membrane into the plant cell, leads to the premature degradation of arbuscules in the model legume Medicago truncatula (Medicago; Javot et al., 2007). This requires the activity of the MYB1 transcription factor, which induces the expression of hydrolytic enzymes such as cysteine proteases and chitinases to degrade the arbuscules (Floss et al., 2017). Intriguingly, Medicago can also adjust the amount of carbon that it delivers to the fungus depending on the amount of Pi obtained from the fungus (Kiers et al., 2011). AM fungal strains that deliver more Pi were shown to receive more carbon compared to less cooperative strains. This so-called reciprocal rewarding indicates that the plant is able to locally monitor the amount of Pi that it obtains from the fungus.
SPX domain-containing proteins have emerged as key sensors and regulators of Pi homeostasis and signaling (Jung et al., 2018). The SPX domain is named after the Suppressor of Yeast gpa1 (Syg1), the cyclin-dependent kinase inhibitor in the yeast PHO pathway (Pho81), and the human Xenotropic and Polytropic Retrovirus receptor 1 (Xpr1) (Barabote et al., 2006). This domain is able to sense the Pi status of a cell by binding with high affinity to inositol polyphosphates (PP-InsPs; Wild et al., 2016; Jung et al., 2018). Changes in PP-InsPs levels in response to Pi deficiency are thought to modulate the activity of SPX-containing proteins and their interactors. The mode of action of single SPX domain-containing proteins in the Pi starvation response (PSR) has been best studied in rice (Oryza sativa) and Arabidopsis thaliana. The nucleus-localized SPX1 and SPX2 proteins in Arabidopsis were shown to interact with PHOSPHATE STARVATION RESPONSE1 (AtPHR1), a MYB-like transcription factor belonging to the GARP family (Safi et al., 2017) that together with its homologs controls Pi starvation-induced gene expression (Puga et al., 2014; Sun et al., 2016). The binding of AtSPX1/2 to AtPHR1 occurs at high Pi concentrations and prevents the binding of AtPHR1 to the PHR1 binding site (P1BS) cis-regulatory element present in the promoters of many PSI genes. Similar regulation has been reported in rice, where OsSPX1 and OsSPX2 control the activity of OsPHR2 in a Pi-dependent manner (Wang et al., 2014). Other SPX members, such as OsSPX4 are localized to the cytoplasm where they control the cytoplasm-to-nucleus shuttling of OsPHR2 in a Pi-dependent manner (Hu et al., 2019). Furthermore, OsSPX4 regulates nitrate and Pi balance in rice by interacting with the nitrate transporter OsNRT1.1B, which triggers OsSPX4 degradation upon nitrate perception (Hu et al., 2019). OsSPX3 and OsSPX5 localize to both the cytoplasm and nucleus and redundantly modulate Pi homeostasis as functional repressors of OsPHR2 (Shi et al., 2014).
Given their key role in sensing and signaling of Pi status in cells, we studied the role of SPX proteins during AM symbiosis. We identified two SPX genes that are strongly upregulated upon AM symbiosis, most specifically in the arbuscule-containing cells of Medicago. We show that these genes regulate Pi homeostasis under nonmycorrhizal conditions and control the degradation of arbuscules during symbiosis.
Results
SPX1 and SPX3 are strongly induced upon Pi starvation and in arbuscule-containing cells
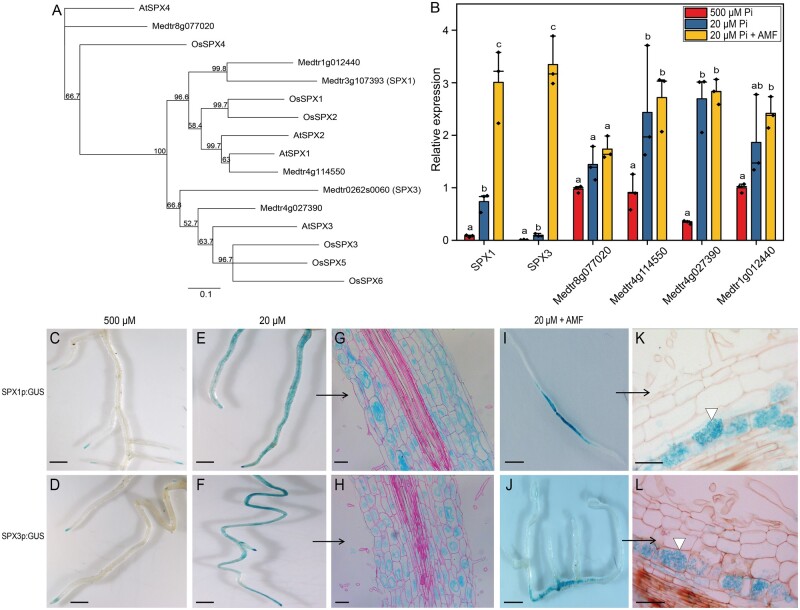
To identify SPX genes that might be important players during AM symbiosis, we first performed phylogenetic analysis of all single SPX domain proteins from Medicago, in relation to SPX proteins from Arabidopsis and rice. This analysis identified six SPX family members (Figure 1A). We analyzed their expression in the roots of plants grown under high (500 μM) Pi conditions, low (20 μM) Pi conditions, and in 20-μM Pi plus the AM fungus Rhizophagus irregularis for 3 weeks. Real-time quantitative polymerase chain reaction (qRT-PCR) analyses showed that two SPX genes, MtSPX1 (Medtr3g107393; hereafter SPX1) and MtSPX3 (Medtr0262s0060; hereafter SPX3) were strongly induced by Pi starvation as well as during AM symbiosis (Figure 1B). Previous transcriptome analyses of laser microdissected arbuscule-containing cells showed that these two SPX genes are predominantly expressed in arbuscule-containing cells (Supplemental Figure S1).
Figure 1.
Medicago SPX1 and SPX3 are induced by Pi starvation and arbuscular mycorrhizal fungal infection. A, Phylogenetic relationship of SPX proteins in Medicago, Arabidopsis, and rice. The unrooted tree was constructed using Geneious R11.0 by the neighbor-joining method, with bootstrap probabilities based on 500 replicates shown on the nodes. The identifiers of the Arabidopsis and rice SPX proteins are listed in Supplemental Table S4. B, qRT-PCR analysis of Medicago (Jemalong A17) SPX expression at high Pi (500 μM), low Pi (20 μM), and arbuscular mycorrhizal (20-μM Pi plus AM fungi [AMF]) conditions. SPX1 and SPX3 are induced under low Pi conditions, and even more strongly upon symbiosis with AMF. Medicago ELONGATION FACTOR1 (MtEF1) was used as an internal reference. Data shown are the individual values of three independent plants; bars indicate the maximum and median. Different letters indicate significant differences (P < 0.05) between treatments (ANOVA followed by Tukey’s honestly significant difference). C and D, SPX1 and SPX3 are expressed in root tips under 500-μM Pi conditions. Scale bar =1 cm. E–H, SPX1 and SPX3 at 20-μM Pi, (G) and (H) are longitudinal sections of (E) and (F). Sections were counterstained with 0.1% ruthenium red. Scale bar in (C and D), 1 cm, in (E and F), 100 μm. I–L, SPX1 and SPX3 are highly and specifically induced in arbuscule-containing cortical cells (3-weeks post-inoculation). K and L are longitudinal sections of (I) and (J), white triangle points to arbuscule-containing cell. Scale bar in (I–J), 1 cm, in (K–L), 100 μm.
To examine the spatial expression patterns of SPX1 and SPX3 in more detail, we analyzed the expression of promoter-GUS reporter constructs in transgenic Medicago roots grown for 3 weeks in 500-µM Pi, 20-µM Pi, and 20-μM Pi plus R. irregularis. Plants grown in 500-µM Pi showed only weak GUS signal in the root tip, while under low Pi conditions, strong GUS activity was detected throughout the root for both constructs (Figure 1, C–F). Sectioning of these roots showed that the SPX1 and SPX3 promoters were active in multiple cell types, including cortex and epidermis, in roots grown at low Pi, but not at high Pi (Figure 1, G and H). Upon AM symbiosis, the GUS signal became much more restricted to the arbuscule-containing cells (Figure 1, I–L; Supplemental Figure S2A). Very low GUS signals were observed in the non-colonized sections of the mycorrhizal roots. The predominant arbuscule-related expression was further confirmed by promoter:NLS-3×GFP analyses (Supplemental Figure S2B) and by examining available laser microdissection data (Supplemental Figure S1; Hogekamp and Küster, 2013; Zeng et al., 2018). The finding that both SPX genes had the same expression pattern suggested that they may have similar functions, playing dual roles in the PSR and AM symbiosis.
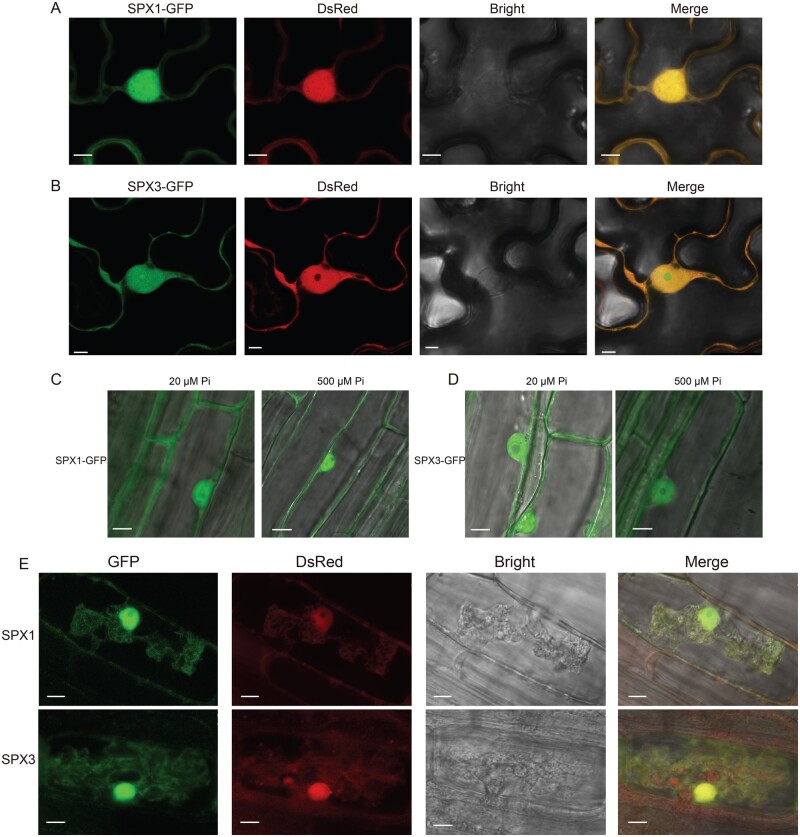
To study the subcellular localization of SPX1 and SPX3, we expressed C-terminal GFP-fusion constructs in Medicago roots and Nicotiana benthamiana leaves using either the constitutive Lotus japonicus Ubiquitin1 (LjUB1) promoter or their endogenous promoters. In all cases, including arbuscule-containing cells expressing SPX–GFP fusion proteins under the control of their endogenous promoters, both fusion proteins localized to the cytoplasm as well as the nucleus (Figure 2, A–E). The subcellular localization was not influenced by Pi levels (Figure 2, C and D).
Figure 2.
SPX1 and SPX3 localize to the nucleus and cytoplasm. A, Confocal images of SPX1-GFP expressed from a constitutive LjUbiquitin promoter in N. benthamiana leaves. A co-expressed UBp:DsRed marker localizes to the nucleus and cytoplasm. Different panels represent the following channels (from left to right): GFP, DsRED, Bright field, and all channels merged. Scale bar = 8 μm. B, Confocal images of LjUBp:SPX3-GFP in N. benthamiana leaves. Scale bar = 5 μm. C, Confocal images of LjUBp:SPX1-GFP in M. truncatula roots localizing to the nucleus and cytoplasm under low Pi and high Pi conditions. Scale bar = 10 μm. D, Confocal images of LjUBp:SPX3-GFP expressed in M. truncatula roots under low Pi and high Pi conditions. Scale bar = 8 μm. E, Confocal images of SPX1-GFP and SPX3-GFP expressed from their own promoter in M. truncatula roots in arbuscule-containing cells. Scale bar = 10 μm.
To explain the transcriptional regulation of SPX1 and SPX3, we examined their presumed promoter regions for known cis-regulatory elements involved in Pi starvation-induced expression and arbuscule-specific regulation. This identified a P1BS (GNATATNC) binding site for the MYB transcription factor PHR (Bustos et al., 2010), a key regulator of PSI gene expression (Figure 3A). The presence of this element suggests that both SPX genes are under the control of PHR-dependent transcriptional regulation upon Pi stress. Furthermore, we identified cis-regulatory AW-boxes (CG(N)7CNANG) and CTTC elements (CTTCTTGTTC) in the promoter regions of both genes, which are binding sites for WRINKLED1-like transcription factors that were recently found to regulate arbuscule-specific expression (Figure 3A). These elements can been found in the promoters of many arbuscule-enhanced genes, including genes involved in fatty acid synthesis and transport as well as genes required for Pi uptake from the arbuscules (Xue et al., 2018; Jiang et al., 2018; Pimprikar and Gutjahr, 2018). To study whether SPX1 and SPX3 are regulated by WRINKLED1-like transcription factors, we overexpressed WRI5a under the control of the LjUB1 promoter in Medicago roots under high Pi conditions. qPCR analyses showed that SPX1 and SPX3 expression was significantly induced upon WRI5a overexpression (Figure 3B). This result suggests that the arbuscule-specific expression of SPX1 and SPX3 may be controlled by WRI5a. The presence of both P1BS and AW-box cis-regulatory elements may explain the observed expression patterns under different conditions.
Figure 3.
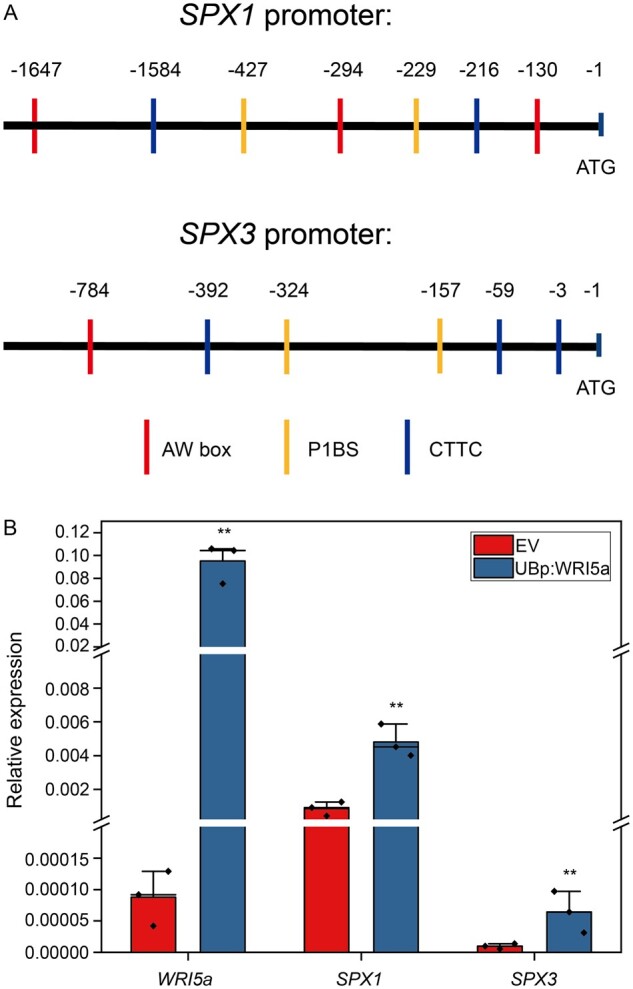
SPX1 and SPX3 expression is regulated by WRI5a. A, The SPX1 and SPX3 promoters contain P1BS (GXATATXC), AW box (CG(X)7CXAXG), and CTTC (CTTCTTGTTC) cis-regulatory elements. B, qPCR analyses showing that overexpression of WRI5a in roots grown under high Pi conditions induces SPX1 and SPX3 expression. Data shown are the individual values of three individually transformed roots. Data significantly different from the corresponding EV controls are indicated **P < 0.01 (Student’s t test).
SPX1 and SPX3 regulate Pi homeostasis
To study the functions of SPX1 and SPX3, we identified spx1 (NF13203_high_1) and spx3 (NF4752_high_18) Tnt1-retrotransposon insertion mutants (Supplemental Figure S3A). Genotyping by PCR confirmed the Tnt1 insertion. We generated the spx1 spx3 double mutant by crossing spx1 to spx3 (Supplemental Figure S3B). RT-PCR confirmed the impairment of SPX1 and/or SPX3 expression in the respective mutant lines (Supplemental Figure S3C).
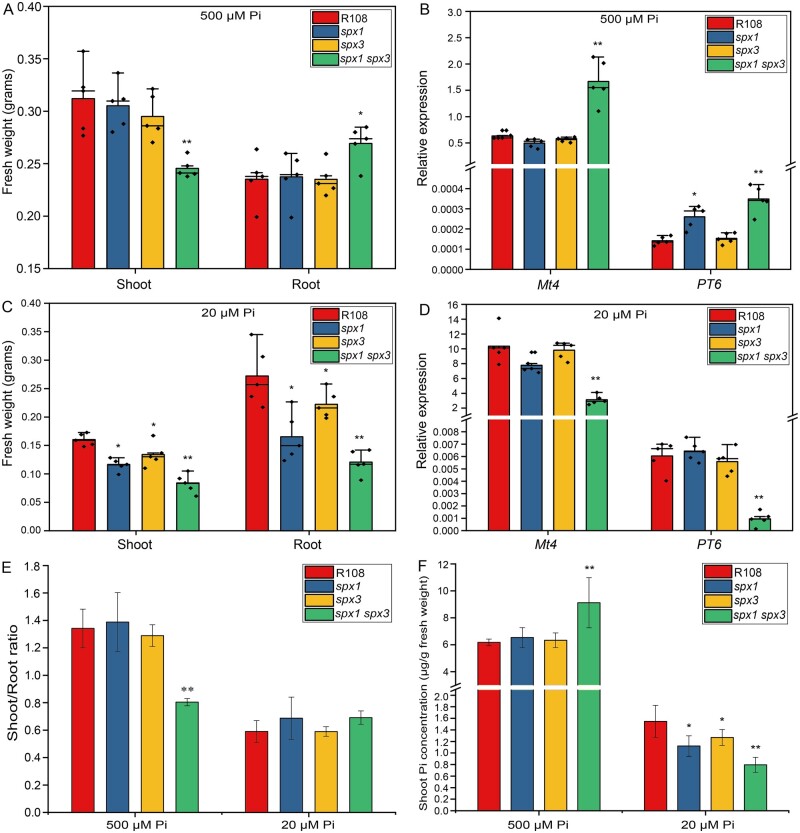
Since SPX proteins are thought to negatively regulate PHR activity under high Pi conditions to prevent the overaccumulation of Pi, we first analyzed the expression of PSI genes in the mutants and R108 wild-type under high and low Pi conditions by qRT-PCR. The PSI genes Mt4 (Burleigh and Harrison, 1999) and the Pi transporter encoding gene PT6 (Mbodj et al., 2018; Hu et al., 2019) were expressed at significantly higher levels in the spx1 spx3 double mutant under high Pi conditions compared to the wild-type (Figure 2B). This coincided with a decreased shoot:root fresh weight ratio indicative of a PSR in the double mutant (Figure 4, A and E). Furthermore, Pi levels were higher in the shoots of the double mutant compared to wild-type plants grown at 500-μM Pi (Figure 4F). Prolonged growth at 500-μM Pi led to typical Pi toxicity symptoms (yellow coloring of the leaf margins) in the leaves of the double mutant (Supplemental Figure S4A). The single mutant lines did not show obvious phenotypes when grown under 500-μM Pi conditions (Figure 4, A, E, and F). These results suggest that SPX1 and SPX3 play negative roles in the PSR when ample Pi is available.
Figure 4.
SPX1 and SPX3 regulate Pi homeostasis. A, Fresh weights of R108 wild-type, spx1, spx3, and spx1 spx3 plants grown for 3 weeks under 500-μM Pi conditions. B, Relative expression of Mt4 and PT6 in the root samples from (A), as determined by qPCR. MtEF1 was used as a reference gene. C, Fresh weights of R108 wild-type, spx1, spx3, and spx1 spx3 plants grown for 3 weeks under 20-μM Pi conditions. D, Relative expression of Mt4 and PT6 in the root samples shown in (C). Data shown are the individual values of five biological replicates. E, Shoot-to-root ratios of R108, spx1, spx3, and spx1 spx3 plants from (A and C). F, Shoot cellular Pi concentrations in R108, spx1, spx3, and spx1 spx3 plants from (A and C). Values in (E and F) represent mean ± se of five replicate plants. All data significantly different from the corresponding R108 wild-type controls are indicated *P < 0.05; **P < 0.01 (Student’s t test).
Under low (20 μM) Pi conditions, the fresh weights of the spx1 and spx3 single mutants were significantly reduced compared to R108 wild-type plants, and the spx1 spx3 double mutant showed an additive effect (Figure 4, C–F; Supplemental Figure S4B). Furthermore, the leaves of the double mutant showed anthocyanin accumulation indicative of Pi starvation stress (Supplemental Figure S4B). Consistently, the PSI genes Mt4 and PT6 were expressed at lower levels in the double mutant compared to the wild-type (Figure 4D), while the overexpression of SPX1/3 enhanced the expression of Mt4 and PT6 under low Pi conditions (Supplemental Figure S5). Shoot Pi concentrations in both single and double mutant plants were significantly lower compared to the wild-type (Figure 4F). These results suggest that SPX1 and SPX3 also play positive roles in the PSR under limiting (20 µM) Pi conditions.
Overall, these results indicate that SPX1 and SPX3 enhance the PSR under low Pi conditions and inhibit this response under high Pi conditions.
SPX1 and SPX3 interact with PHR2
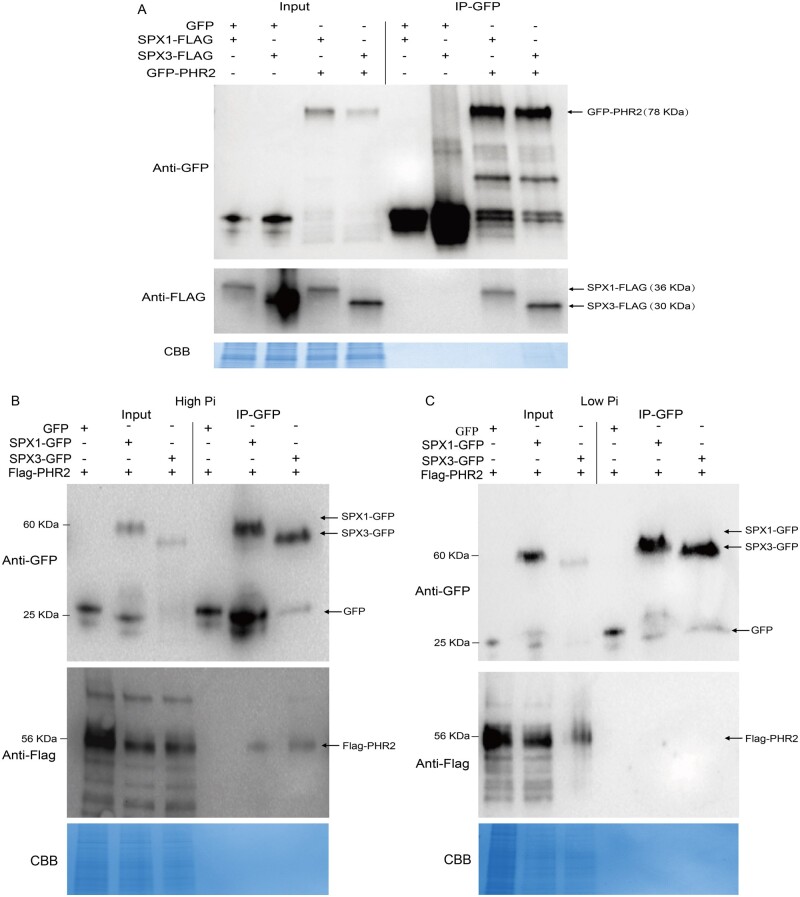
In Arabidopsis and rice, SPX proteins interact with PHR and inhibit its activity under high Pi conditions (Wang et al., 2014; Puga et al., 2014). Therefore, we checked whether SPX1 and SPX3 could also interact with Medicago PHR homologs. Phylogenetic analysis indicated the presence of three PHR-like proteins in Medicago (Supplemental Figure S6). Co-immunoprecipitation (Co-IP) analysis of GFP-tagged PHR with FLAG-tagged SPX1/3 proteins expressed in N. benthamiana leaves revealed a clear interaction of both SPX1 and SPX3 with MtPHR2 (Medtr1g080330; hereafter PHR2; Figure 5A). No significant interaction was found for the other two Medicago PHR-like proteins. To study the Pi dependency of the interaction, we co-expressed GFP-tagged SPX1/3 together with Flag-tagged PHR2 in Medicago roots under high and low Pi conditions. Co-IP analysis showed that SPX1 and SPX3 interacted with PHR2 under high Pi conditions, but no interaction was observed under low Pi conditions (Figure 5, B and C).
Figure 5.
SPX1 and SPX3 interact with PHR2 under high Pi conditions. A, Immunoblot of Co-IP samples showing the interaction of SPX1 and SPX3 with PHR2. FLAG-tagged SPX1 or SPX3 was co-expressed with free GFP or GFP-tagged PHR2 in N. benthamiana leaves. IP of GFP-tagged proteins shows Co-IP of the FLAG-tagged SPX proteins. B, Immunoblot of Co-IP samples showing an interaction of PHR2 with SPX1 and SPX3 in Medicago (Jemalong A17) roots grown under high Pi conditions. Free GFP, GFP-tagged SPX1, or SPX3 were co-expressed with Flag-tagged PHR2 in Medicago roots. IP of GFP-tagged proteins shows the Co-IP of the FLAG-tagged PHR2 proteins. C, Immunoblot of co-IP samples showing that SPX1 and SPX3 do not interact with PHR2 under low (20 μM) Pi conditions. Coomassie brilliant blue staining shows total protein levels as a loading control.
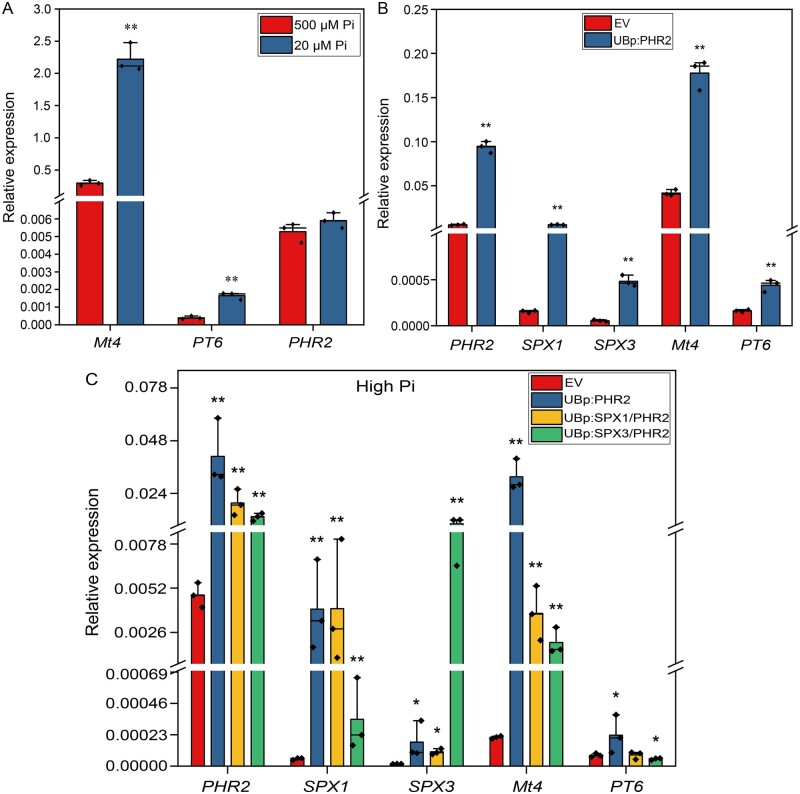
To study whether PHR2 is indeed involved in the PSR, we overexpressed PHR2 using the LjUB1 promoter in Medicago roots and analyzed its effect on the expression of PSI genes Mt4 and PT6 (Mbodj et al., 2018; Hu et al., 2019). Both Mt4 and PT6 were strongly induced under Pi limiting conditions and, like SPX1 and SPX3, were also induced upon PHR2 overexpression (Figure 6, A and B). PHR2 itself was not regulated in a Pi-dependent manner at the transcriptional level (Figure 6A), in analogy to its homologs AtPHR1 (Bustos et al., 2010) and OsPHR2 (Zhou et al., 2008).
Figure 6.
SPX1 and SPX3 regulate Pi homeostasis. A, qPCR analysis showing Pi starvation-induced expression of Mt4 and PT6, but not MtPHR2 in Medicago (Jemalong A17) roots. Data shown are the individual values of three plants. Data significantly different from 500-μM Pi conditions are indicated **P < 0.01 (Student’s t test). B, qPCR analysis showing that overexpression of MtPHR2 (LjUBp:PHR2) induced SPX1, SPX3, Mt4, and PT6 expression in Medicago A17 transgenic roots grown for 4 days under low Pi (20 µM) conditions. C, qPCR results showing that the overexpression (using the LjUB1 promoter) of SPX1 or SPX3 together with PHR2 under high Pi conditions induced Mt4 and PT6 expression less strongly compared to the overexpression of PHR2 alone in Medicago A17 transgenic roots. Data shown in (B and C) are the individual values of three independently transformed roots. Data significantly different from the corresponding EV transformed controls are indicated *P < 0.05; **P < 0.01 (Student’s t test). MtEF1 was used as a reference gene for normalization. Relative expression was calculated via the 2−ΔCt method.
To determine whether the observed Pi-related phenotypes were due to the negative regulation of PHR2 activity by SPX1 and SPX3, we overexpressed SPX1 or SPX3 together with PHR2 using the control of the LjUB1 promoter in Medicago roots. Overexpression of SPX1 or SPX3 indeed inhibited the induction of Mt4 and PT6 by PHR2 under high Pi conditions (Figure 6C). We noticed that the (over)expression level of PHR2 was ∼2× lower for the co-expression constructs containing SPX1 and SPX3 compared to overexpression of PHR2 alone. This is likely a result of the expression construct rather than an effect of SPX1/3 on the activity of the LjUB1 promoter. The lower induction of Mt4 upon co-expression of SPX1/3 supports the inhibitory effect of SPX1/3 on the activity of PHR2 when sufficient Pi levels are reached.
SPX1 and SPX3 regulate arbuscule degeneration
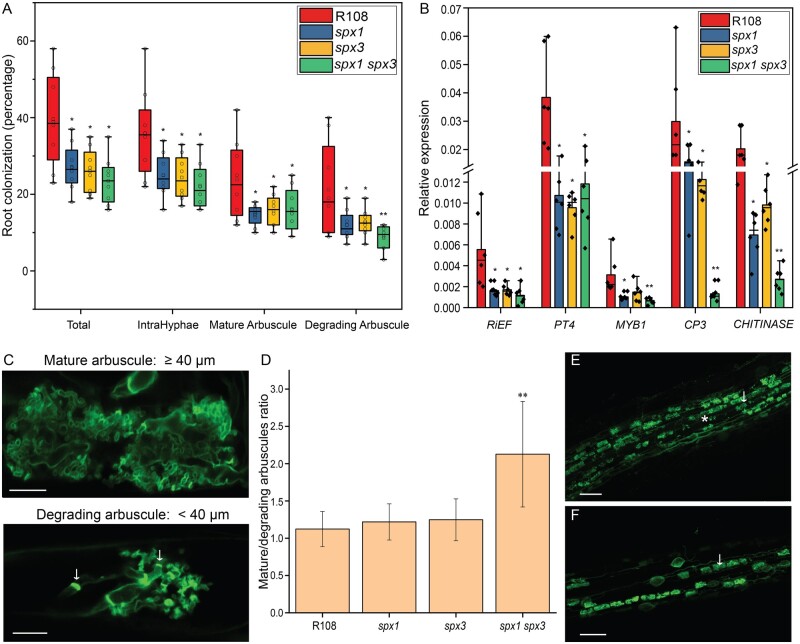
Next, we examined the roles of SPX1 and SPX3 in the interaction with AM fungi. Three weeks after inoculation with R. irregularis spores, we quantified mycorrhization in the spx1 and spx3 single mutants, the spx1 spx3 double mutant, and R108 wild-type controls using the magnified intersections method (McGonigle et al., 1990). Eight plants were used as replicates for each line. Compared to R108, single mutant and double mutant plants all showed significantly lower root colonization levels and arbuscule abundance (Figure 7A). The analysis of the transcript levels of RiEF and PT4, molecular markers for fungal colonization and arbuscule abundance, respectively, confirmed the lower colonization levels in the mutants (Figure 7B).
Figure 7.
SPX1 and SPX3 regulate AM colonization and arbuscule degeneration. A, Quantification of mycorrhization levels in R108 wild-type, spx1, spx3, and spx1 spx3 3 weeks post inoculation with R. irregularis. Eight independently transformed roots were used as replicates for each sample. Quantification was performed using the magnified intersections method (McGonigle et al., 1990). Mature arbuscule: larger or equal to 40 μm; Degrading arbuscules: smaller than 40 μm with septa (white arrow in C). Data significantly different from R108 wild-type controls are indicated *P < 0.05; **P < 0.01 (Student’s t test). B, Expression levels of RiEF, PT4, CP3, CHITINASE, and MYB1 in root samples from (A), as determined by the qPCR analysis. MtEF1 was used as a reference gene for normalization. Data significantly different from R108 wild-type controls are indicated *P < 0.05; **P < 0.01 (Student’s t test). C, Representative images of mature and degrading arbuscules in WGA-Alexa488 stained roots 3 weeks post-inoculation. Arrow points to septa. Scale bar = 10 μm. D, Ratio of mature-to-degrading arbuscules in R108, spx1, spx3, and spx1 spx3 mycorrhizal samples from (A). Values represent mean ± se of eight independently transformed roots. Data significantly different from R108 wild-type controls are indicated **P < 0.01 (Student’s t test). E, F, Representative images of WGA-Alexa488 stained R. irregularis in R108 and spx1 spx3. White arrow marks a mature arbuscule. Asterisk marks a degrading arbuscule. Scale bar = 100 μm.
Because SPX1 and SPX3 are most strongly expressed in arbuscule-containing cells in mycorrhized roots, we quantified arbuscule morphology in more detail. We defined arbuscules larger or equal to 40 μm as “mature” arbuscules, and arbuscules smaller than 40 μm with typical features of degradation, including visible septa, as “degrading” arbuscules (Figure 7C). Interestingly, there were significantly fewer degrading arbuscules in the spx1 spx3 double mutant compared to R108 wild-type plants, resulting in a much higher mature/degrading arbuscule ratio (Figure 7, A, E, and F). The spx1 and spx3 single mutants showed a similar mature/degrading arbuscule ratio to R108 wild-type (Figure 5A), suggesting that SPX1 and SPX3 play redundant roles in regulating arbuscule degradation. To further determine the effects on arbuscule morphology, we monitored PT4, CP3 and CHITINASE expression after normalization to fungal RiEF expression. Significantly higher PT4 expression and lower CP3 and CHITINASE expression were detected in the spx1 spx3 double mutant (Supplemental Figure S7A). In line with this finding, analysis of arbuscule size (length) distribution also showed that the number of large (50–80 μm) arbuscules was higher in spx1 spx3 compared to R108 wild-type, whereas fewer small (20–30 and 30–40 μm) arbuscules were observed in spx1 spx3.
Arbuscule degradation in Medicago is regulated by the transcription factor MYB1, which controls the expression of hydrolase genes such as CYSTEINE PROTEASE3 (CP3) and CHITINASE (Floss et al., 2017). qRT-PCR analysis revealed strongly impaired expression of these hydrolase genes in the spx1 spx3 double mutant (Figure 7B). Similar phenotypes were observed upon knockdown of both SPX1 and SPX3 by RNA interference (Supplemental Figure S8, A and B). To further confirm that the phenotype was indeed caused by a mutation in SPX1 and SPX3, we complemented the spx1 spx3 double mutant by driving SPX1 and SPX3 expression from by their native promoters in A. rhizogenes-transformed roots. This indeed complemented the mycorrhization levels, arbuscule abundance, and marker gene expression to wild-type levels (Supplemental Figure S8, C and D), showing that the phenotypes were not caused by background insertions/mutation in the mutant lines.
Overall, these results indicate that both SPX1 and SPX3 have positive effects on AM colonization levels and redundantly regulate arbuscule degradation.
SPX1 and SPX3 may control AM colonization by regulating SL levels
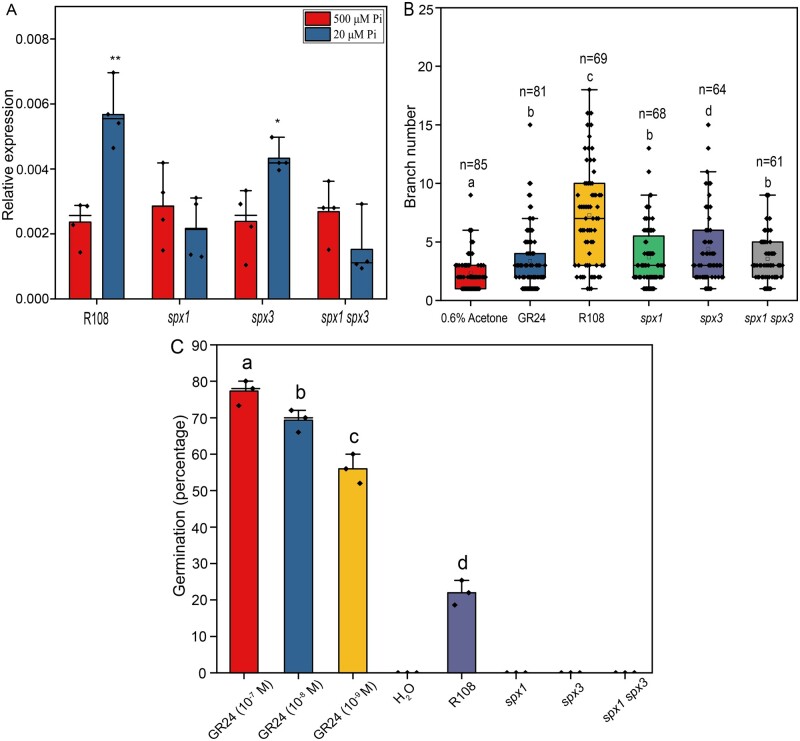
To explain the positive roles of SPX1 and SPX3 in mycorrhizal colonization, we measured the expression level of MtDWARF27 (D27), a key gene required for SL biosynthesis (Hao et al., 2009; Liu et al., 2011). This gene showed little or no induction upon Pi starvation in spx1 and spx3 single and double mutants compared to wild-type plants (Figure 8A). Under low Pi conditions, SL levels increase drastically in several species, and this induction in SL biosynthesis correlates with increased D27 expression under this condition (Liu et al., 2011; Figure 8A). The D27 expression was induced when SPX1 and SPX3 were overexpressed together under low Pi conditions using the LjUB1 promoter in A. rhizogenes-transformed roots (Supplemental Figure S9A). Under high Pi conditions, the D27 expression did not appear to be affected (Supplemental Figure S10B). This revealed a positive effect of SPX1 and SPX3 on the D27 expression under low Pi conditions, and thereby possibly on SL levels.
Figure 8.
SPX1 and SPX3 regulate the expression of the SL biosynthesis gene D27. A, The induction of D27 expression under low Pi conditions is impaired in spx1, spx3, and spx1 spx3 mutants. Data shown are the individual values of three plant replicates. Data significantly different from the corresponding controls are indicated *P < 0.05; **P < 0.01 (Student’s t test). B, Rhizophagus irregularis spores treated with root exudates of R108, spx1, spx3, and spx1 spx3 for eight days. About 0.01-μM GR24 and 0.6% acetone were, respectively, used as positive and negative controls. Hyphal branch numbers of germinated spores treated with root exudates of spx1, spx3, and spx1 spx3 mutants are significantly lower compared to spores treated with R108 root exudates. n indicates the number of spores counted. Data shown are the individual values of the replicates. C, Germination of P. ramosa seeds induced by treatment with root exudates and GR24. Data shown are individual values of three replicates of 50 seeds each. Different letters in (B and C) indicate significant differences (P < 0.05) between treatments (ANOVA followed by Tukey’s honestly significant difference).
Unfortunately, we were unable to measure SL levels in the R108 genetic background, perhaps because they were below the level of detection, or perhaps as yet unknown SL derivates are present in R108. As an alternative, we used an AM hyphal branching assay as a proxy for SL levels in root exudates (Besserer et al., 2006, 2008). Exudates collected from the spx1/3 mutant roots grown under Pi limiting conditions were much less able to induce R. irregularis branching compared to R108 wild-type root exudates (Figure 8B;Supplemental Figure S9C). As an additional assay, we applied the root exudates to the seeds of the parasitic plant Phelipanche ramosa and scored the germination rates after 12 days. Parasitic plants like P. ramosa sense plant SLs as host-detection signals that stimulate their germination (Cardoso et al., 2011). The exudates of R108 plants induced 21% germination, while no clear germination was induced upon the application of the exudates of spx1, spx3, or spx1 spx3 (Figure 8C;Supplemental Figure S9D). Together, these data suggest that SL levels are reduced in the spx mutants, although an additional effect on other root exudates cannot be ruled out.
Overexpression of SPX1 and SPX3 increases arbuscule degradation
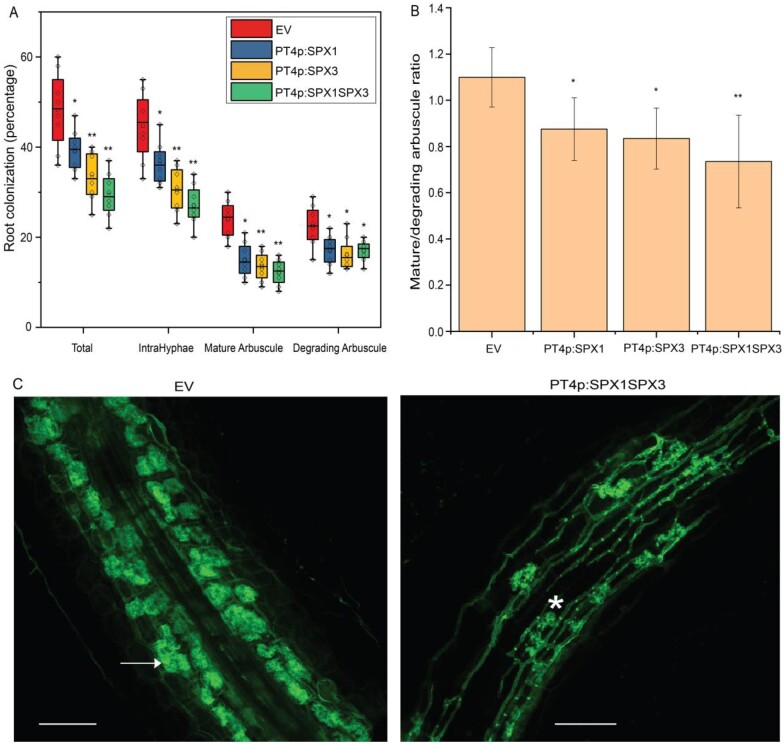
The mutant analyses showed that SPX1 and SPX3 redundantly regulate arbuscule degradation. To further study this, we overexpressed SPX1, SPX3, or both in M. truncatula A17 roots using the arbuscule-specific PT4 promoter. Compared to empty vector (EV)-transformed roots, decreased colonization levels were observed in all SPX overexpressing roots when expressed from the PT4 promoter (Figure 9A). This coincided with decreased arbuscule abundance and an increased ratio of degrading arbuscule compared to mature arbuscule classes (Figure 9, A–C). Transcript levels of the markers for fungal biomass (RiEF), healthy arbuscule abundance (PT4), and arbuscule degradation (MYB1, CP3, and CHITINASE) all confirmed the visual phenotyping results (Supplemental Figure S10, A and B). Because the colonization levels in SPX overexpressing roots were much lower than EV control roots and because SPX1 and SPX3 are normally highly induced in arbuscule-containing cells the overall SPX1/3 expression levels did not exceed those in the EV controls (Figures 1, B and 9, A;Supplemental Figure S10A). Overall, these results further strengthen the notion that SPX1/3 play a role in regulating arbuscule degradation.
Figure 9.
Overexpression of SPX1/3 under the control of the MtPT4 promoter induces arbuscule degradation. A, Quantification of mycorrhization levels in M. truncatula A17 roots expressing EV, PT4p:SPX1, PT4p:SPX3, and PT4p:SPX1-PT4p:SPX3 3-weeks post inoculation with R. irregularis. Eight independently transformed plants were used as replicates for each sample. Quantification was performed using the magnified intersections method (McGonigle et al., 1990). B, Ratio of mature-to-degrading arbuscules in EV, PT4p:SPX1, PT4p:SPX3, and PT4p:SPX1-PT4p:SPX3 mycorrhizal samples from (A). Values represent mean ± sd of eight independently transformed roots. C, Representative confocal images of WGA-Alexa488 stained R. irregularis in EV and PT4p:SPX1-PT4p:SPX3 roots. White arrow marks a mature arbuscule. Asterisk marks a degrading arbuscule. Scale bar = 100 μm. All data in (A and B) significantly different from the corresponding EV controls are indicated *P < 0.05; **P < 0.01 (Student’s t test).
Discussion
SPX proteins have emerged as key sensors and signaling regulators of cellular Pi status in plants (Wild et al., 2016; Wang et al., 2014; Puga et al., 2014; Shi et al., 2014; Hu et al., 2019). Here, we show that the Medicago single SPX-domain proteins SPX1 and SPX3 not only regulate Pi homeostasis under nonsymbiotic conditions, but also regulate root colonization and arbuscule degradation during AM symbiosis. This offers important insight into the Pi-dependent regulation of this agriculturally and ecologically important symbiosis.
Under nonsymbiotic conditions, SPX1 and SPX3 control Pi homeostasis in part through the regulation of the PHR2 activity. In analogy to the situation in Arabidopsis and rice, Pi starvation leads to the activation of the PHR activity to control transcriptional responses. Among the targets of PHR2 are the SPX1/3 genes. Both SPX1 and SPX3 bind to PHR2 under high Pi conditions and negatively affect the PSR to prevent the overaccumulation of Pi. We show that SPX1 and SPX3 also control the induction of the SL biosynthesis gene D27 (Liu et al., 2011) under Pi-limiting conditions. This suggests that SPX1 and SPX3 play an additional positive role in the transcription of Pi-starvation induced genes under low Pi conditions. This is further supported by the observation that SPX1 and SPX3 play a positive role in the PSR, increasing fresh weight and shoot Pi concentration when grown under low Pi conditions. Under high Pi conditions, they negatively regulate the PHR-controlled PSR to prevent Pi toxicity effects. PHR-independent roles have also been suggested for AtSPX4 in the regulation of shoot phosphorus status in Arabidopsis (Osorio et al., 2019).
Since D27 plays an essential role in the biosynthesis of SLs (Hao et al., 2009), reduced SL levels could explain the lower colonization levels observed in the spx mutants, as these key signal molecules induce the growth and branching of AM fungi (Besserer et al., 2006; Tsuzuki et al., 2016). This hypothesis is supported by the reduced stimulatory effect of root exudates from the spx1 and spx3 mutant on AM hyphal branching and P. ramosa germination compared to exudates from R108 wild-type. However, the current inability to measure SL levels in the R108 genetic background prevented us from confirming this directly. Therefore, it cannot be excluded that additional stimulatory components in the exudates may be affected in the spx mutants. Furthermore, impaired arbuscule development can also lead to reduced colonization levels, as it may reduce the amount of carbon transferred to the fungus. In the spx1 spx3 mutant, at least morphologically, mature arbuscules appeared to be more prominent, but still the expression level of PT4 (as a marker for functional arbuscules) seemed to be lower compared to the wild-type. This suggests that overall, fewer functional arbuscules may have been present, which could also have contributed to the reduced colonization levels in the mutant background.
The induction of SPX1 and SPX3 upon Pi starvation or PHR2 overexpression fits with the presence of the P1BS cis-regulatory element in the promoters of these genes. However, after the establishment of AM symbiosis, the expression of both SPX1 and SPX3 becomes restricted from a ubiquitous expression pattern to a more restricted high expression level in arbuscule-containing cells. This suggests that PHR activity in the noncolonized root cortical and epidermal cells is inhibited upon a functional AM symbiosis in a non-cell autonomous manner. It has been proposed that AM fungi may interfere with the direct Pi uptake of plants (Smith et al., 2004; Christophersen et al., 2009; Yang et al., 2012; Wang et al., 2020), although the mechanisms for this are still unknown. Another possibility is a more systemic regulation of the PSR through hormonal or peptide signaling as the plant is obtaining Pi from the fungus (Müller and Harrison, 2019). Although we cannot pinpoint the exact time after the initiation of symbiosis (or the level of colonization) at which the shift in expression occurs based on our analyses, we already detected relatively little expression in noncolonized root cells, or in non-transgenic roots in the same composite plants, after 3 weeks of inoculation, suggesting a rather rapid systemic regulation.
It is currently not known whether PHR2 is active in arbuscule-containing cells, as contrasting roles for the P1BS element in the expression of symbiotic Pi transporters have been reported (Chen et al., 2011; Lota et al., 2013). Low expression of PHR2 was detected in laser microdissected arbuscule-containing cells (Supplemental Figure S11); however, the presence of SPX1/3 would be expected to suppress PHR2 activity upon Pi supply by the arbuscules, as it does under nonsymbiotic conditions. Instead, the induction of SPX1 and SPX3 in arbuscule-containing cells is associated with the presence of multiple AW-boxes and CTTC elements in the promoters of both genes. These cis-regulatory elements are found in many genes that are induced in arbuscule-containing cells. These elements are bound by WRINKLED1-like transcription factors that are themselves activated by the key GRAS transcription factor RAM1, which controls arbuscule formation upon activation of the common symbiotic signaling pathway (Jiang et al., 2018; Xue et al., 2018; Limpens and Geurts, 2018; Pimprikar and Gutjahr, 2018). The control of SPX1/3 expression by this symbiotic pathway is supported by the observation that overexpressing WRI5a induced SPX1 and SPX3 expression (Figure 4B). A further link among RAM1, WRI5a, and SPX3 expression is suggested by the lack of SPX3 induction in the ram1 mutant (Supplemental Table S1; Luginbuehl et al., 2017).
Since Pi levels are most likely not limiting in cells containing active arbuscules, the impaired induction of CP3 and CHITINASE and the associated arbuscule degradation in the spx1 spx3 double mutant argues for the involvement of SPX1/3 interacting proteins other than PHR2. To understand how SPX1 and SPX3 control arbuscule degradation, efforts are underway to identify interactors of both SPX1 and SPX3 in arbuscule-containing cells. So far, we have detected interactions between SPX1/3 and the known regulator of arbuscule degradation MYB1 or its interactors NSP1 and DELLA1 (Floss et al., 2017), suggesting that additional regulators of arbuscule degradation remain to be identified.
It is tempting to speculate that SPX proteins may also be involved in the crosstalk with other nutrients supplied by the fungus, such as nitrogen. This suggestion is based on the involvement of OsSPX4 in nitrate signaling to control the PSR (Hu et al., 2019) and the role of nitrogen in controlling arbuscule development (Breuillin-Sessoms et al., 2015). For example, the premature arbuscule degradation in the pt4 mutant was also suppressed when the plants were grown under N-limiting conditions (Breuillin-Sessoms et al., 2015). This was shown to depend on the ammonium transceptor AMT2;3. It will therefore be interesting to test whether SPX1 and SPX3 contribute to this nutrient crosstalk in Medicago.
In conclusion, we revealed important roles for Pi sensing SPX proteins in the regulation of AM symbiosis to enhance the Pi acquisition efficiency of plants. SPX proteins play a role at both the pre-contact stage as well as in the termination of symbiosis by controlling the degradation of arbuscules. The latter role could be essential for maintaining the beneficial nature of the interaction. In nature, plants are most often colonized by multiple different AM strains that can differ in the amount of nutrients they supply (Kiers et al., 2011). Therefore, SPX proteins can provide a means to locally monitor whether a fungal partner provides sufficient nutritional benefits. Further unraveling how nutrient sensing and homeostasis are regulated during AM symbiosis will be pivotal to understanding the ecological workings of this key symbiosis and how to best exploit it for more sustainable agricultural practices.
Materials and methods
Plant and fungal materials
Medicago truncatula A17 and R108 seedlings were grown and transformed as described (Limpens et al., 2004). The spx1 (NF13203) and spx3 (NF4752) Tnt1-insertion lines were obtained from the Noble Research Institute (https://medicago-mutant.noble.org/mutant/index.php). Homozygous spx1 and spx3 mutants were identified by PCR and crossed to obtain the spx1 spx3 double mutant. Primers used are listed in Supplemental Table S2. Plants were grown in SC10 RayLeach containers (Stuewe and Sons, Canada) with a premixed sand:clay (1:1 v/v) mixture and watered with 10 mL 1/2 Hoagland medium with 20-µM (low Pi) or 500-µM H2PO4 (high Pi) twice a week in a 16-h daylight chamber at 21°C, as described previously (Zeng et al., 2018). Rhizophagus irregularis DAOM197198 spores (Agronutrion, France) were washed through three layers of filter mesh (220 µm, 120 µm, 38 µm) before inoculation. For inoculation, 200 spores were placed ∼2 cm below the seedling roots upon planting.
Constructs
Most constructs were produced using the Golden Gate cloning system (Engler et al., 2014) Constructs for RNAi were generated via Gateway cloning and constructs for Y2H experiments were generated using In-fusion cloning (Takara, Japan). Primers used are listed in Supplemental Table S2. Vectors used for cloning are listed in Supplemental Table S3. All newly made vectors were confirmed by Sanger sequencing.
Phylogenic analysis
Phylogenetic analysis was performed using the Geneious R11.0 software package (https://www.geneious.com). Medicago, Arabidopsis and rice SPX or MYB family protein sequences were collected from PLAZA (https://bioinformatics.psb.ugent.be/plaza/) or NCBI (https://www.ncbi.nlm.nih.gov/). Proteins were aligned using MAFFT in Geneious R11.0. An unrooted SPX phylogenetic tree was generated using the neighbor-joining tree builder with 500 bootstraps. The alignment and tree information are provided in Supplemental File S1. Identifiers of genes used are listed in Supplemental Table S4.
GUS histochemical analyses
To create the MtSPX1p-GUS and MtSPX3p-GUS constructs, a 1,824-bp promoter region upstream of the MtSPX1 (Medtr3g107393) ATG start codon and a 1,260-bp region upstream of the MtSPX3 (Medtr0262s0060) ATG start codon was amplified by PCR from M. truncatula A17 genomic DNA as the promoter, respectively. The MtSPX1p-GUS and MtSPX3p-GUS constructs were introduced into Medicago plants using Agrobacterium rhizogenes-mediated root transformation (Limpens et al., 2004). GUS staining was done as described (An et al., 2019). Briefly, transgenic roots were harvested based on DsRed fluorescence (red fluorescent marker present in the constructs) and washed twice with PBS buffer for 10 min. The roots were incubated in GUS reaction buffer (3% sucrose, 10-mM EDTA, 2-mM potassium-ferrocyanide, 2-mM potassium-ferricyanide, and 1 mg·mL-1 X-Gluc in 100-mM PBS, pH 7.0) for 30 min under a vacuum, followed by incubated at 37°C for 1 h. The stained roots were fixed in fixation buffer (5% glutaraldehyde in 100-mM Pi buffer, pH 7.2) for 2 h under a vacuum at room temperature, followed by dehydration in an ethanol series (20%, 30%, 50%, 70%, 90%, 100%) for 10 min each. Root segments were embedded in Technovit 7100 (Hereus-Kulzer, Germany), cut into 8-μm longitudinal sections with a microtome (Leica RM2255), and stained with 0.1% Ruthenium Red for 5 min. Images were taken under a Leica DM5500 B microscope.
Co-IP and immunoblotting
FLAG-tagged SPX1 and SPX3 and GFP-tagged PHR2 (Medtr1g080330) constructs were transiently expressed in N. benthamiana leaves as described (Zeng et al., 2018). Total proteins were isolated from the samples using Co-IP buffer (10% glycerol, 50-mM Tris–HCl pH = 8.0, 150-mM NaCl, 1% Igepal CA 630, 1-mM PMSF, 20-µM MG132, one tablet protease inhibitor cocktail). GFP-Trap agarose beads (Chromotek) were used to immunoprecipitate the GFP protein complexes. Immunoblotting was performed as described (Bungard et al., 2010), using 1:5,000 diluted anti-GFP-HRP and anti-FLAG-HRP antibodies (Miltenyi Biotec, USA) in combination with the ECL Western Blotting Substrate (Bio-Rad) for detection.
RNA interference
A SPX1 SPX3 hairpin construct was generated targeting both SPX1 and SPX3 mRNA using the Gateway system (Invitrogen, USA). Overlap PCR was used to combine 534 bp of the SPX1 cDNA sequence and 489 bp of the SPX3 cDNA sequence, and the resulting 1023 bp sequence was cloned into the pENTR/D-TOPO entry vector. Primers used are listed in Supplemental Table S2. Subsequently, the modified pK7GWIWG2(II)-AtEF1 RR vector (Zeng et al., 2020) was used for an LR reaction to obtain the final hairpin silencing construct.
Measurement of Pi concentration
The inorganic Pi concentration in shoots was measured as described (Zhou et al., 2008). Briefly, Medicago shoots were crushed into a fine powder in liquid nitrogen and rigorously vortexed for 1 min in 1 mL 10% (w/v) of perchloric acid. The homogenate was diluted ten-fold with 5% (w/v) perchloric acid and placed on ice for 30 min, followed by centrifugation at 10,000g for 10 min at 4°C to collect the supernatant. The molybdenum blue method was used to measure Pi content in the supernatant. To prepare the molybdenum blue solution, 6 mL solution A (0.4% ammonium molybdate dissolved in 0.5-M H2SO4) was mixed with 1-mL 10% ascorbic acid. About 2 mL of molybdenum blue solution was added to 1 mL of the sample supernatant and incubated in a water bath for 20 min at 42°C. The absorbance was measured at 820 nm, and Pi content was calculated by comparison to a standard curve. The Pi concentration was normalized to the shoot fresh weight.
RNA isolation and qRT-PCR
RNA was isolated from the samples using a Qiagen Plant RNA Mini kit according to the manufacturer’s instructions, including an on-column DNase treatment step. cDNA was generated using an iScript cDNA Synthesis kit (Bio-Rad) using 300-ng total RNA as template. qRT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad) in a Bio-Rad CFX connect real-time system; two-step program with 40 cycles (95°C 10 s, 60°C 1 min) followed by a heat dissociation step (from 65°C to 95°C). Primers used for qPCR are listed in Supplemental Table S2. Medicago ELONGATION FACTOR1 (EF1) was used as a reference for normalization. Relative expression levels were calculated as 2−△ct with three technical replicates for each sample.
AM quantification
Mycorrhizal roots were stained with WGA-Alexafluor 488 (Thermo Fisher Scientific, USA). AM quantification was performed using the magnified intersections method as described (McGonigle et al., 1990). In short, a Leica DM5500 B microscope equipped with an eyepiece crosshair was used to inspect the intersections between the crosshair and roots at 200× magnification. The following categories were noted in each intersection: root only, hyphopodium, extraradical hyphae, intracellular hyphae (Intrahyphae), mature arbuscule (equal to or larger than 40 µm), degrading arbuscule (<40 µm, and presence of septa). In cases where at one intersection more than one category was observed, each category was counted once at that position. One hundred intersections were inspected for each sample (containing 30 cm of root pieces), and the percentage of each category was calculated.
Root exudate collection and quantification of SLs
SL analysis was done as described (Liu et al., 2011; van Zeijl et al., 2015). For each genotype, six seedlings were grown in a X-stream 20 aeroponic system (Nutriculture) operating with 5 L of 1/2 Hoagland medium (Hoagland, 1950) containing 500-μM Pi in a greenhouse with natural light, 28°C, 60% relative humidity. After 4 weeks, Pi starvation was initiated by replacing the high Pi medium with 1/2 Hoagland medium containing 20-μM Pi for 1 week. The medium was refreshed with new low-Pi 1/2 Hoagland medium 24 h before exudate collection. The resulting 5-L sample containing 24-h exudate was purified and concentrated by loading onto a pre-equilibrated C18 column (Grace Pure C18-Fast 5,000 mg/20 mL). The column was washed with 50 mL of deionized water, followed by 50 mL of 30% acetone. SLs were eluted with 50 mL 60% acetone and measured using a Quadcore as described (Kohlen et al., 2011).
Rhizophagus irregularis branching assay
Rhizophagus irregularis spore germination and branching assays were performed as previously described (Besserer et al., 2006; Tsuzuki et al., 2016). One hundred spores were grown on Solid M medium (Bécard and Fortin, 1988) with 0.6% acetone, 0.01-μM GR24, or 100-fold diluted root exudates (as collected above) in the dark at 22°C. Hyphal branches were counted 8 days after inoculation under a Leica M165 FC microscope.
Phelipanche ramosa germination assay
Germination of P. ramosa seeds was performed as reported before (Kohlen et al., 2011). Briefly, cleaned P. ramose seeds were preconditioned on wet filter paper for 10 days at 21°C. For each replicate, 1-cm discs containing approximately 50 preconditioned seeds were treated with 50 μL of root exudates (described above). As positive controls, 50 μL of 10−7, 10−8, and 10−9 M of the synthetic SL GR24 were applied. Demineralized water was used as a negative control. After 12 days, the germination rates were counted under a binocular microscope.
Confocal microscopy
The subcellular localization of fluorescently tagged proteins and structure of the arbuscules using WGA-Alexafluor 488 stained roots were analyzed under a Leica SP8 confocal microscope (for GFP/Alexafluor 488: excitation 488 nm, emission 500–540 nm; for DsRED/mCherry: excitation 552, emission 580–650 nm).
Statistical analyses
For pairwise comparisons, data were analyzed using a t test built in EXCEL with tail 1, type 2. Analysis of variation (ANOVA) using the Origin 2018 software package was used to test differences across two groups of data with default settings. Replicates used per experiment are indicated in the corresponding figure legends. ANOVA tables are provided in Supplemental File S2.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: MtSPX1 (Medtr3g107393), MtSPX3 (Medtr0262s0060), Mt4 (U76742.1), PT6 (Medtr1g069935), MtPHR2 (Medtr1g080330), MtEF1 (Medtr6g021800), MtCP3 (Medtr4g107930), MtCHITINASE (Medtr6g079630), MtPT4 (Medtr1g028600), MtWRI5a (XM_013590339), MtDWARF27 (MTR_1g471050), RiEF1 (XM_025321412.1), LjUBIQUITIN1 (AB303069), AtSPX1 (At5g20150), AtSPX2 (At2g26660), AtSPX3 (At2g45130), AtSPX4 (At5g15330), OsSPX1 (LOC_Os06g40120), OsSPX2 (LOC_Os02g10780), OsSPX4 (LOC_Os10g25310), OsSPX4 (LOC_Os03g61200), OsSPX5 (LOC_Os03g29250), OsSPX6 (LOC9271158), AtPHR1 (At4g28610), MtMYB1 (Medtr7g068600).
Supplemental data
The following materials are available in the online version of this article. Supplemental Figure S1. SPX1 and SPX3 are highly expressed in arbuscule-containing cells.
Supplemental Figure S2. SPX1 and SPX3 are specifically expressed in arbuscule-containing cells.
Supplemental Figure S3. spx1, spx3, and spx1 spx3 Tnt1-retrotransposon insertion lines.
Supplemental Figure S4. Phenotypes of spx1, spx3, and the double-mutant spx1 spx3.
Supplemental Figure S5. qPCR results showing that overexpression of SPX1/3 under low Pi conditions induces Mt4 and PT6 expression in Medicago A17 transgenic roots.
Supplemental Figure S6. Phylogenetic tree of MYB family proteins from Medicago, Arabidopsis, and rice generated using the neighbor-joining tree builder in Geneious R11.0.
Supplemental Figure S7. SPX1 and SPX3 regulate arbuscule degradation.
Supplemental Figure S8. SPX1 and SPX3 control arbuscular mycorrhization.
Supplemental Figure S9. SPX1 and SPX3 likely regulate SL biosynthesis.
Supplemental Figure S10. qPCR analysis of Medicago A17 transgenic roots overexpressing SPX1/3 under the control of the PT4 promoter.
Supplemental Figure S11. PHR2 is (weakly) expressed in arbuscule-containing cells based on RNA-seq data of laser microdissected roots colonized by R. irregularis.
Supplemental Table S1. SPX1 and SPX3 expression fold changes in wild-type and ram1-1 roots during mycorrhization.
Supplemental Table S2. Primers used in this study.
Supplemental Table S3. Constructs generated using the Golden Gate cloning system.
Supplemental Table S4. Identifiers of genes used for phylogenetic analysis.
Supplemental File S1. Alignment files.
Supplemental File S2. ANOVA Tables.
Supplementary Material
Acknowledgments
We would like to thank Rene Geurts for critical reading and comments on the manuscript.
Funding
P.W. is supported by the China Scholarship Council (CSC) grant 201606310038 and by the Dutch research school Experimental Plant Sciences (EPS).
Conflict of interest statement. There are no conflicts of interest.
P.W. and E.L. conceived and designed the experiments. P.W., R.S., and W.K. performed the experiments and/or data analyses. J.L. performed crossing. P.W., T.B., and E.L. wrote the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plcell) is: Erik Limpens (erik.limpens@wur.nl).
References
- Akiyama K, Matsuzaki KI, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435:824–827 [DOI] [PubMed] [Google Scholar]
- An J, Zeng T, Ji C, de Graaf S, Zheng Z, Xiao TT, Deng X, Xiao S, Bisseling T, Limpens E, et al. (2019). A Medicago truncatula SWEET transporter implicated in arbuscule maintenance during arbuscular mycorrhizal symbiosis. New Phytol 224:396–408 [DOI] [PubMed] [Google Scholar]
- Barabote RD, Tamang DG, Abeywardena SN, Fallah NS, Fu JYC, Lio JK, Mirhosseini P, Pezeshk R, Podell S, Salampessy ML, et al. (2006) Extra domains in secondary transport carriers and channel proteins. Biochim Biophys Acta - Biomembr 1758:1557–1579 [DOI] [PubMed] [Google Scholar]
- Bari R, Pant BD, Stitt M, Scheible WR (2006) PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol 141:988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécard G, Fortin JA (1988) Early events of vesicular–arbuscular mycorrhiza formation on Ri T‐DNA transformed roots. New Phytol 108: 211–218 [DOI] [PubMed] [Google Scholar]
- Besserer A, Bécard G, Jauneau A, Roux C, Séjalon-Delmas N (2008) GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the arbuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol 148:402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Bécard G, Séjalon-Delmas N (2006) Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 4:1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuillin-Sessoms F, Floss DS, Karen Gomez S, Pumplin N, Ding Y, Levesque-Tremblay V, Noar RD, Daniels DA, Bravo A, Eaglesham JB,et al. (2015). Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter4 mutants is dependent on the ammonium transporter 2 family protein AMT2;3. Plant Cell 27: 1352–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL (2010) Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science 329:1201–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ (1999) The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol 119:241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso C, Ruyter-Spira C, Bouwmeester HJ (2011) Strigolactones and root infestation by plant-parasitic Striga, Orobanche and Phelipanche spp. Plant Sci 180: 414–420 [DOI] [PubMed] [Google Scholar]
- Chen A, Gu M, Sun S, Zhu L, Hong S, Xu G (2011) Identification of two conserved cis-acting elements, MYCS and P1BS, involved in the regulation of mycorrhiza-activated phosphate transporters in eudicot species. New Phytol 189:1157–1169 [DOI] [PubMed] [Google Scholar]
- Christophersen HM, Smith FA, Smith SE (2009) Arbuscular mycorrhizal colonization reduces arsenate uptake in barley via downregulation of transporters in the direct epidermal phosphate uptake pathway. New Phytol 184:962–974 [DOI] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert TM, Werner S, Jones JDG, Patron NJ, Marillonnet S (2014) A Golden Gate modular cloning toolbox for plants. ACS Synth Biol 3:839–843 [DOI] [PubMed] [Google Scholar]
- Ezawa T, Saito K (2018) How do arbuscular mycorrhizal fungi handle phosphate? New insight into fine-tuning of phosphate metabolism. New Phytol 220:1116–1121 [DOI] [PubMed] [Google Scholar]
- Floss DS, Gomez SK, Park HJ, MacLean AM, Müller LM, Bhattarai KK, Lévesque-Tremblay V, Maldonado-Mendoza IE, Harrison MJ (2017) A transcriptional program for arbuscule degeneration during AM symbiosis is regulated by MYB1. Curr Biol 27:1206–1212 [DOI] [PubMed] [Google Scholar]
- Hao L, Renxiao W, Qian Q, Meixian Y, Xiangbing M, Zhiming F, Cunyu Y, Biao J, Zhen S, Jiayang L, et al. (2009) DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21:1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogekamp C, Küster H (2013) A roadmap of cell-type specific gene expression during sequential stages of the arbuscular mycorrhiza symbiosis. BMC Genomics 14:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Jiang Z, Wang W, Qiu Y, Zhang Z, Liu Y, Li A, Gao X, Liu L, Qian Y, et al. (2019). Nitrate–NRT1.1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat Plants 5:401–413 [DOI] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104:1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Xie Q, Wang W, Yang J, Zhang X, Yu N, Zhou Y, Wang E (2018) Medicago AP2-domain transcription factor WRI5a is a master regulator of lipid biosynthesis and transfer during mycorrhizal symbiosis. Mol Plant 11:1344–1359 [DOI] [PubMed] [Google Scholar]
- Jung JY, Ried MK, Hothorn M, Poirier Y (2018) Control of plant phosphate homeostasis by inositol pyrophosphates and the SPX domain. Curr Opin Biotechnol 49:156–162 [DOI] [PubMed] [Google Scholar]
- Kiers ET, Duhamel M, Beesetty Y, Mensah JA, Franken O, Verbruggen E, Fellbaum CR, Kowalchuk GA, Hart MM, Bago A, et al. (2011) Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 333:880–882 [DOI] [PubMed] [Google Scholar]
- Kobae Y, Hata S (2010) Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol 51: 341–353 [DOI] [PubMed] [Google Scholar]
- Kohlen W, Charnikhova T, Liu Q, Bours R, Domagalska MA, Beguerie S, Verstappen F, Leyser O, Bouwmeester H, Ruyter-Spira C (2011) Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host arabidopsis. Plant Physiol 1 55:974–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfranco L, Fiorilli V, Gutjahr C (2018) Partner communication and role of nutrients in the arbuscular mycorrhizal symbiosis. New Phytol 220:1031–1046 [DOI] [PubMed] [Google Scholar]
- Limpens E, Geurts R (2018) Transcriptional regulation of nutrient exchange in arbuscular mycorrhizal symbiosis. Mol Plant 11:1421–1423 [DOI] [PubMed] [Google Scholar]
- Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55:983–992 [DOI] [PubMed] [Google Scholar]
- Liu W, Kohlen W, Lillo A, den Camp RO, Ivanov S, Hartog M, Limpens E, Jamil M, Smaczniak C, Kaufmann K, et al. (2011) Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23:3853–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lota F, Wegmüller S, Buer B, Sato S, Bräutigam A, Hanf B, Bucher M (2013) The cis-acting CTTC-P1BS module is indicative for gene function of LjVTI12, a Qb-SNARE protein gene that is required for arbuscule formation in Lotus japonicus. Plant J 74:280–293 [DOI] [PubMed] [Google Scholar]
- Luginbuehl LH, Menard GN, Kurup S, Van Erp H, Radhakrishnan GV, Breakspear A, Oldroyd GED, Eastmond PJ (2017) Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356:1175–1178 [DOI] [PubMed] [Google Scholar]
- Luginbuehl LH, Oldroyd GED (2017) Understanding the arbuscule at the heart of endomycorrhizal symbioses in plants. Curr Biol 27:R952–R963 [DOI] [PubMed] [Google Scholar]
- Mbodj D, Effa-Effa B, Kane A, Manneh B, Gantet P, Laplaze L, Diedhiou AG, Grondin A (2018) Arbuscular mycorrhizal symbiosis in rice: Establishment, environmental control and impact on plant growth and resistance to abiotic stresses. Rhizosphere 8:12–26 [Google Scholar]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA (1990) A new method which gives an objective measure of colonization of roots by vesicular—arbuscular mycorrhizal fungi. New Phytol 115:495–501 [DOI] [PubMed] [Google Scholar]
- Müller LM, Harrison MJ (2019) Phytohormones, miRNAs, and peptide signals integrate plant phosphorus status with arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol 50:132–139 [DOI] [PubMed] [Google Scholar]
- Osorio MB, Ng S, Berkowitz O, De Clercq I, Mao C, Shou H, Whelan J, Jost R (2019) SPX4 acts on PHR1-dependent and -independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiol 181:332–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimprikar P, Gutjahr C (2018) Transcriptional Regulation of Arbuscular Mycorrhiza Development. Plant Cell Physiol 59: 673–690 [DOI] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodriguez J, et al. (2014). SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci USA 111:14947–14952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H, Arpat AB, Poirier Y (2010) Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol Plant 3:288–299 [DOI] [PubMed] [Google Scholar]
- Safi A, Medici A, Szponarski W, Ruffel S, Lacombe B, Krouk G (2017) The world according to GARP transcription factors. Curr Opin Plant Biol 39:159–167 [DOI] [PubMed] [Google Scholar]
- Shi J, Hu H, Zhang K, Zhang W, Yu Y, Wu Z, Wu P (2014) The paralogous SPX3 and SPX5 genes redundantly modulate Pi homeostasis in rice. J Exp Bot 65:859–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Read D (2008) Mycorrhizal Symbiosis, Ed 3. Academic Press, Cambridge, UK [Google Scholar]
- Smith SE, Smith FA, Jakobsen I (2004) Functional diversity in arbuscular mycorrhizal (AM) symbioses: the contribution of the mycorrhizal P uptake pathway is not correlated with mycorrhizal responses in growth or total P uptake. New Phytol 162:511–524 [Google Scholar]
- Sun L, Song L, Zhang Y, Zheng Z, Liu D (2016) Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol 170:499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki S, Handa Y, Takeda N, Kawaguchi M (2016) Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mol Plant-Microbe Interact 29:277–286 [DOI] [PubMed] [Google Scholar]
- Wang S, Chen A, Xie K, Yang X, Luo Z, Chen J, Zeng D, Ren Y, Yang C, Wang L, et al. (2020). Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc Natl Acad Sci USA 117:16649–16659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wenyaun R, Shi J, Zhang L, Dan X, Yang C, Li C, Wu Z, Liu Y, Yanan Y, et al. (2014). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc Natl Acad Sci USA 111:14953–14958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild R, Gerasimaite R, Jung JY, Truffault V, Pavlovic I, Schmidt A, Saiardi A, Jacob Jessen H, Poirier Y, Hothorn M, et al. (2016). Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352:986–990 [DOI] [PubMed] [Google Scholar]
- Xue L, Klinnawee L, Zhou Y, Saridis G, Vijayakumar V, Brands M, Dörmann P, Gigolashvili T, Turck F, Bucher M (2018) AP2 transcription factor CBX1 with a specific function in symbiotic exchange of nutrients in mycorrhizal Lotus japonicus. Proc Natl Acad Sci USA 115:E9239–E9246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SY, Grønlund M, Jakobsen I, Grotemeyer MS, Rentsch D, Miyao A, Hirochika H, Santhosh Kumar C, Sundaresan V, Salamin N,et al. (2012). Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the PHOSPHATE TRANSPORTER1 gene family. Plant Cell 24: 4236–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeijl A, Liu W, Xiao TT, Kohlen W, Yang WC, Bisseling T, Geurts R (2015) The strigolactone biosynthesis gene DWARF27 is co-opted in rhizobium symbiosis. BMC Plant Biol 15:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng T, Holmer R, Hontelez J, te Lintel-Hekkert B, Marufu L, de Zeeuw T, Wu F, Schijlen E, Bisseling T, Limpens E (2018) Host- and stage-dependent secretome of the arbuscular mycorrhizal fungus Rhizophagus irregularis. Plant J 94:411–425 [DOI] [PubMed] [Google Scholar]
- Zeng T, Rodriguez-Moreno L, Mansurkhodzaev A, Wang P, van den Berg W, Gasciolli V, Cottaz S, Fort S, Thomma BPHJ, Bono JJ, et al. (2020). A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytol 225:448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao FC, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146:1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.