Abstract
One conserved feature among angiosperms is the development of flat thin leaves. This developmental pattern optimizes light capture and gas exchange. The blue light (BL) receptors phototropins are required for leaf flattening, with the null phot1phot2 mutant showing curled leaves in Arabidopsis (Arabidopsis thaliana). However, key aspects of their function in leaf development remain unknown. Here, we performed a detailed spatiotemporal characterization of phototropin function in Arabidopsis leaves. We found that phototropins perceive light direction in the blade, and, similar to their role in hypocotyls, they control the spatial pattern of auxin signaling, possibly modulating auxin transport, to ultimately regulate cell expansion. Phototropin signaling components in the leaf partially differ from hypocotyls. Moreover, the light response on the upper and lower sides of the leaf blade suggests a partially distinct requirement of phototropin signaling components on each side. In particular, NON PHOTOTROPIC HYPOCOTYL 3 showed an adaxial-specific function. In addition, we show a prominent role of PHYTOCHROME KINASE SUBSTRATE 3 in leaf flattening. Among auxin transporters, PIN-FORMED 3,4,7 and AUXIN RESISTANT 1 (AUX1)/LIKE AUXIN RESISTANT 1 (LAX1) are required for the response while ABCB19 has a regulatory role. Overall, our results show that directional BL perception by phototropins is a key aspect of leaf development, integrating endogenous and exogenous signals.
Phototropins perceive light direction in the leaf and control the auxin signaling pattern to regulate blade flattening.
Introduction
Plants are photoautotrophic organisms, and as such optimization of light capture is key for their success. In most land plants, leaves are the major photosynthetic organ (Chitwood and Sinha, 2016). Leaves start developing in the shoot apical meristem, where primordia differentiate. Primordia are patterned early with different gene expression domains in the portion closer to the meristem (adaxial side) versus distally from the meristem (abaxial side) and in the medio-lateral and proximo-distal axes. In the juxtaposition between the adaxial and abaxial side, the marginal domain will promote leaf expansion. Early in development, cells divide, and in Arabidopsis (Arabidopsis thaliana) an arrest front in cell division moves from the tip to the base of the leaf. Subsequently, cells expand and a growth arrest front follows the same pattern from tip to base (Xiong and Jiao, 2019; Heisler and Byrne, 2020). In summary, leaf shape depends on the early patterning of the primordia, cell division rates, and coordinated expansion of cell layers.
One conserved feature of angiosperm leaves is their flat shape, with wide thin blades. This shape optimizes light capture, as well as gas exchange and temperature regulation (Inoue et al., 2008; de Carbonnel et al., 2010). In addition to endogenous cues, environmental signals, in particular light, also control leaf flattening. The red light (RL) and the far-red light receptor phytochrome B (phyB) promote leaf curling, and in shade conditions phyB inactivation triggers blade flattening (Kozuka et al., 2012; Johansson and Hughes, 2014). Blue light (BL) perceived by phototropins (phot1 and phot2 in Arabidopsis) promotes leaf expansion and flattening (Sakai et al., 2001; Takemiya et al., 2005). However, whether phototropins act as sensor of light direction to control blade flattening is currently unknown.
Phototropins are membrane-associated serine–threonine kinases containing two light, oxygen, and voltage domains allowing them to perceive BL and ultraviolet light (Legris and Boccaccini, 2020). Their role in stem phototropism is well understood. A directional light stimulus establishes a light gradient within the stem creating a phototropin activation gradient. Activated phototropins are autophosphorylated and interact with members of the NON-PHOTOTROPIC HYPOCOTYL 3 (NPH3)/ROOT PHOTOTROPISM 2 (RPT2)-Like (NRL) and Phytochrome Kinase Substrate (PKS) families. NPH3 and RPT2 have a role in stem phototropism and the control of leaf flattening (Inoue et al., 2008; de Carbonnel et al., 2010; Harada et al., 2012; Christie et al., 2018). NPH3 is phosphorylated by phot1 as an early step of the signaling pathway (Sullivan et al., 2021). Another direct phototropin phosphorylation target is PKS4 (Demarsy et al., 2012). Among PKS proteins, PKS1 and PKS4 have a role in stem phototropism, while PKS2 has a role in leaf flattening (de Carbonnel et al., 2010; Kami et al., 2014). While the molecular function of these proteins remains unknown, they have been related to auxin transport or signaling (de Carbonnel et al., 2010; Kami et al., 2014). In the stem, unilateral irradiation leads to an auxin gradient across the organ. Higher auxin concentrations on the shaded side of the stem promote cell expansion, while growth on the lit side is inhibited causing bending toward the light (Legris and Boccaccini, 2020). Auxin transporters of the PIN-formed (PIN) family are required for hypocotyl bending (Willige et al., 2013). In particular, in response to unilateral BL, PIN3 in the endodermis re-localizes to the outer membrane, allowing auxin transport from the vasculature to the epidermis to control growth (Ding et al., 2011). Phot1-mediated phosphorylation of the ABCB19 auxin transporter also regulates hypocotyl phototropism by promoting basipetal auxin fluxes (Christie et al., 2011).
The plant hormone auxin has a strong role during leaf development (Xiong and Jiao, 2019; Heisler and Byrne, 2020). In the meristem, primordia differentiate following an auxin response maxima. Auxin has also been implicated in the establishment and/or maintenance of the adaxial–abaxial polarity with lower auxin in the adaxial side of the blade promoting the adaxial fate, and allowing flattening (Guan et al., 2017). Auxin signaling in the middle domain is required for leaf expansion. Auxin is synthesized in the margins of expanded leaves and transported toward the stem. In accordance with these roles, several mutants in auxin synthesis, signaling or transport genes fail to grow flat leaves, and exogenous application of auxin or auxin signaling inhibitors promote blade curling (Watahiki and Yamamoto, 1997; de Carbonnel et al., 2010; Jenness et al., 2019; Xiong and Jiao, 2019; Heisler and Byrne, 2020; Jenness et al., 2020).
Provided that leaf development relies on internal cues, auxin being one of them, and that phototropins regulate leaf shape and can regulate auxin signaling, we set out to evaluate the mechanisms underlying the interaction between light perception and leaf development to regulate blade flattening. Using specific light treatments, we performed a spatiotemporal analysis to determine the role of phototropin activation on leaf shape. We show that throughout leaf development light direction perceived by phototropins regulates cell expansion to regulate blade morphology.
Results
Leaf shape depends not only on the perception of BL, but also on its site of perception
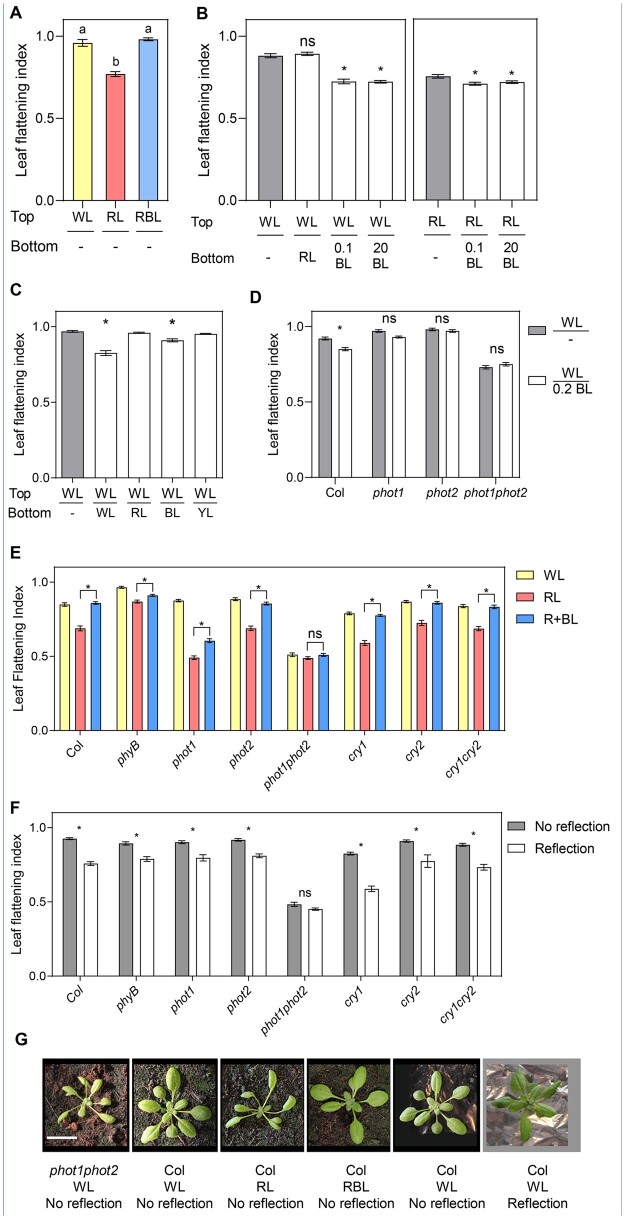
BL perceived by phototropins is required to grow flat leaves (Takemiya et al., 2005; de Carbonnel et al., 2010; Kozuka et al., 2012; Jenness et al., 2020). However, whether this depends on phototropins’ ability to sense light direction remains unclear. In most plants, but in particular in Arabidopsis which is a rosette, leaves intercept light with the adaxial side, and most of it is absorbed by the photosynthetic pigments, creating a steep BL gradient throughout the leaf (Vogelmann et al., 1989; Paradiso et al., 2020). Given that, a similar signal perceived by phototropins triggers stem phototropism, we evaluated whether phototropins also perceive light direction in the leaf. We measured the leaf flattening index (LFI), as the ratio between the leaf blade area before and after artificially flattening it (de Carbonnel et al., 2010), in plants grown in RL with addition of 0.1 µmol m−2 s−1 of BL either from the top or from below (Figure 1, A and B). While plants grown with 0.1 µmol m−2 s−1 BL from the top grew flat leaves (Figure 1A), those grown in RL plus BL from below had curled leaves with an even stronger flattening defect than those grown in the absence of BL (Figure 1B). Since plants grown on plates had a different morphology than soil-grown plants (Supplemental Figure S1B), we also performed similar analyses in the latter conditions. We covered the soil surface with colored aluminum foil, which reflects ambient light, irradiating the abaxial side of leaves (Supplemental Figure S1C). Dark aluminum foil did not affect leaf development compared to uncovered soil (Supplemental Figure S1, A and B). Reflection of White light (WL) triggered downward bending of the leaf blade (Figure 1, C and G and Supplemental Figure S1C). As observed on plates, reflection of BL was enough to interfere with normal leaf flattening, while reflection of RL or yellow light (YL) did not affect flattening (Figure 1C and Supplemental Figure S1C). These results confirm that BL is required to grow flat leaves and show that the site of BL perception in the leaf affects its shape. In addition to changing leaf shape, in response to reflected light plants also accumulated more biomass (Supplemental Figure S1D). This suggests that in a more heterogeneous light environment changing leaf shape may improve light interception to optimize photosynthesis.
Figure 1.
Leaf shape responds to light direction in a phototropin-dependent manner. A, LFI in Col plants grown in plates for three weeks in WL, RL, or RBL. Bars represent mean ± se of 16–30 leaves. Different letters represent significant differences among means (P < 0.05) in an ANOVA followed by Tukey’s test. B, LFI in Col plants grown in plates for two weeks and transferred to WL or RL from the top supplemented with RL, 0.1 µmol m−2 s−1 blue light (0.1 BL) or 20 µmol m−2 s−1 BL (20 BL) from the bottom or dark bottom (−) for three days. Bars represent mean ± se of 56–60 leaves. *P < 0.05 in ANOVA followed by Dunnett’s multiple comparison test. In each case, treatments involving light from below were compared to the same light from the top (WL or RL) without light from below (gray bars). C, LFI in Col plants grown in WL for three weeks in soil covered with aluminum foil, which reflects various colors (see spectra of the reflected light in Supplemental Figure S1B). YL: yellow light. Bars represent mean ± se of 30–88 leaves. *P < 0.05 in ANOVA followed by Dunnett’s multiple comparison test. Treatments involving light from below were compared to the treatment without light from below (−, gray bar). D, LFI in Col and phototropin mutants grown in plates in WL for 2 weeks and transferred to WL from the top and darkness (−) or 0.2 µmol m−2 s−1 BL (0.2 BL) from the bottom for three days. Bars represent mean ± se of 34–92 leaves. *P < 0.05 in ANOVA followed by Sidak’s multiple comparison test between the two treatments for each genotype. E, LFI in Col and photoreceptor mutant plants grown in soil for 2 weeks and transferred to WL, RL, or RBL for 1 week. Bars represent mean ± se of 36–50 leaves. *P < 0.05 in ANOVA followed by Tukey’s multiple comparison test. The complete ANOVA results including comparisons with WL can be found in Supplemental Table S3. F, LFI in Col and photoreceptor mutant plants grown in WL for 3 weeks in soil covered with dark (No reflection) or clear (Reflection) aluminum foil. Bars represent mean ± se of 16–29 plants. *P < 0.05 in ANOVA followed by Sidak’s multiple comparison test. G, Representative pictures of phot1phot2 and Col plants grown in selected light conditions. Scale bar: 2 cm.
Phototropins perceive light direction in the leaf
In agreement with previous reports (Takemiya et al., 2005; de Carbonnel et al., 2010; Kozuka et al., 2012; Jenness et al., 2020), phototropins were the main photoreceptors perceiving BL and promoting flattening when BL reaches the adaxial side of the leaf (Figure 1, E and G). This was also the case when the abaxial side of the leaf was irradiated (Figure 1F). When plants were grown on plates, both phot1 and phot2 were required to respond to very low BL intensities (Figure 1D). When plants were grown on soil, both in response to light from the top or below phot1 and phot2 triggered responses to low BL intensities and only the double mutant phot1phot2 showed no BL response (Figure 1, E and F).
phyB influences flattening in an opposite way to the phototropins (Kozuka et al., 2012). In our conditions, phyB mutants were not very different from Col in WL, although, consistent with previous findings, phyB leaves were flatter than Col particularly in RL (Figure 1E and Supplemental Figure S1E; Kozuka et al., 2012). However, phyB did not compromise significantly the response to light from below, showing that phyB does not contribute to the perception of light direction (Figure 1F). Some leaf development defects in phyB-9 are caused by a second mutation in VENOSA4 present in the phyB-9 allele (ven4-bnen; Yoshida et al., 2018), but this was not the case for leaf flattening (Supplemental Figure S1E). Interestingly, cry1 had an effect on leaf flattening, with the cry1-304 mutant showing slightly curled leaves when light was perceived on the adaxial side and responding more to light from below (Figure 1, E and F). However, the double cry1cry2 mutant responded similarly to the wild-type and in all cases cry mutants responded to the light stimulus (Figure 1, E and F). Hence, cryptochromes may have a regulatory role but not directly control leaf shape by directional light. In summary, similar to what is known in hypocotyls and inflorescence stems, phototropins perceive light direction in leaves to control blade curvature.
The perception of light direction in the leaf involves differential phototropin activation in the adaxial and abaxial layers
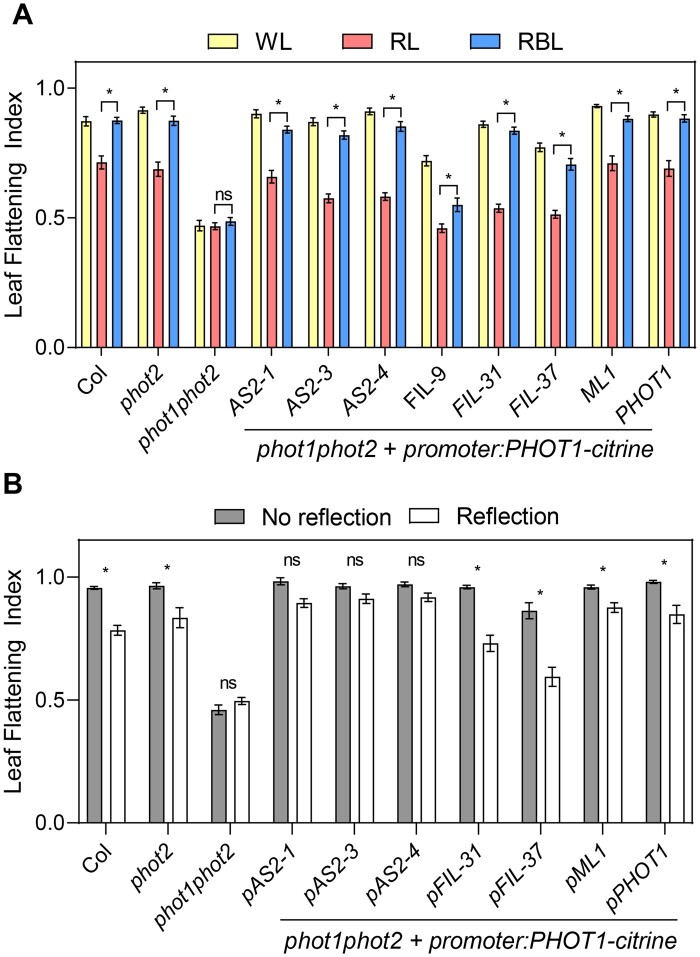
In contrast to the radial symmetry of the hypocotyl, Arabidopsis leaves have a bilateral polarity with well-defined adaxial and abaxial domains (Xiong and Jiao, 2019; Heisler and Byrne, 2020). Taking advantage of this, we used promoters to express PHOT1 on the adaxial or the abaxial side of the leaf. If leaf flattening is controlled by differential phototropin activation, changing the expression domain of phototropins is expected to influence the response to light direction. phot1phot2 mutants were complemented with PHOT1-citrine expressed under the promoter of ASYMMETRIC LEAVES 2 (pAS2) or FILAMENTOUS FLOWER (pFIL), inducing its expression predominantly in the adaxial or abaxial domains, respectively (Supplemental Figure S2B; Guan et al., 2017; Xiong and Jiao, 2019). With light from the top most transgenic lines showed complementation of the phot1phot2 flattening defect (Figure 2A). However, lines expressing PHOT1 on the adaxial side of the leaf showed better complementation, and among the three lines expressing PHOT1 on the abaxial side of the leaf one complemented poorly and another one partially (pFIL-9 and pFIL-37, respectively; Supplemental Table S4). The site of PHOT1 expression affected leaf flattening in response to light from the abaxial side more clearly. Lines expressing PHOT1 predominantly in the adaxial leaf side (pAS2 lines) did not respond to the abaxial light stimulus, while those expressing PHOT1 predominantly in the abaxial side (pFIL lines) robustly responded to the abaxial light stimulus (Figure 2B). Expressing PHOT1 in the epidermal layers under the MERISTEM LAYER 1 promoter (pML1; Preuten et al., 2013) had a similar effect as expressing it in the whole blade under the pPHOT1 promoter (Preuten et al., 2015), suggesting that, a phot1 activation gradient throughout the leaf blade was not required, but differential activation on each epidermal layer was sufficient to regulate blade shape. The response to light corresponded with the PHOT1 expression pattern rather than with PHOT1 levels, since lines expressing similar levels showed contrasting responses (pAS2-1 and pFIL-31) and lines expressing different levels showed the same trend (pFIL-9 and pFIL-37 or pML1 and pAS2-4, Supplemental Figure S2A). However, we note that among the pFIL lines expression level and phot1phot2 complementation with light coming from the top corresponded (Figure 2A and Supplemental Figure S2A). Collectively, these results indicate that in the leaf a directional light stimulus leads to differential phototropin activation on the adaxial versus abaxial layers, which controls blade flattening.
Figure 2.
The PHOT1 expression domain regulates leaf shape. A, LFI in plants grown in soil for two weeks and transferred to WL, RL, or RBL for 1 week. Bars represent mean ± se of 17–28 leaves. *P < 0.05 in ANOVA followed by Tukey’s multiple comparison test. The complete ANOVA results including comparisons with WL can be found in Supplemental Table S4. B, LFI in plants grown in soil covered with dark (No reflection) or clear (Reflection) aluminum foil for 3 weeks. pAS2-1, pAS2-3, pAS2-4, pFIL-9, pFIL-31, and pFIL-37 are independent transgenic lines expressing PHOT1-citrine under the promoters of ASYMMETRIC LEAVES2 (AS2) or FILAMENTOUS FLOWER (FIL), respectively. pML1 and pPHOT1 lines express PHOT1-citrine under the control of the MERISTEM LAYER 1 and PHOTROPIN 1 promoters, respectively. pAS2 drives PHOT1 expression in the adaxial domain, and pFIL in the abaxial domain. Bars represent mean ± se of 6–28 plants. *P < 0.05 in ANOVA followed by Sidak’s multiple comparison test.
Asymmetric requirement for some phototropin signaling elements in the control of leaf flattening
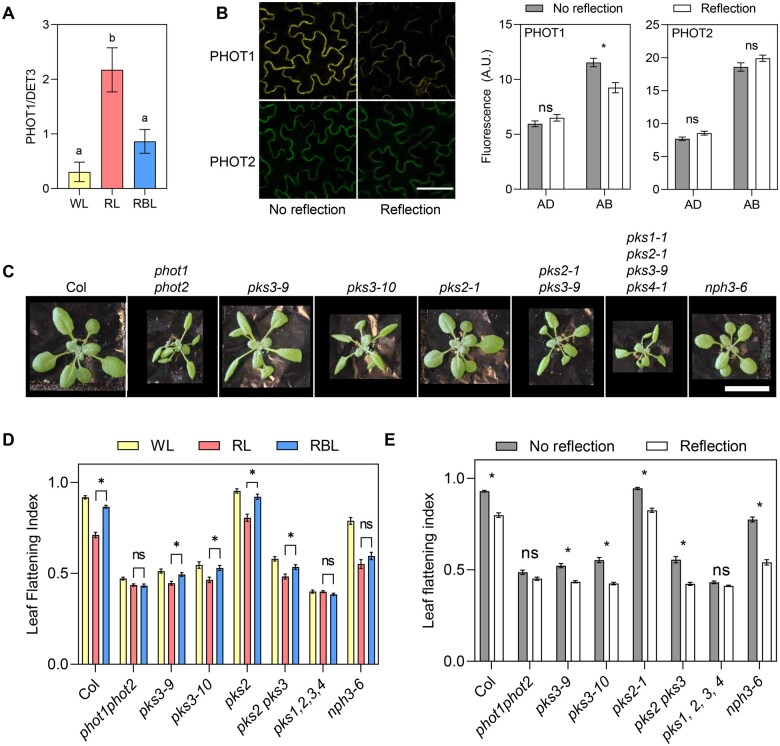
Early phototropin signaling during hypocotyl phototropism involves autophosphorylation of the receptor, in some cases followed by its degradation, and interaction with signaling members of the NRL and PKS families (Sakamoto and Briggs, 2002; Kong et al., 2006; Roberts et al., 2011; Christie et al., 2018; Legris and Boccaccini, 2020; Sullivan et al., 2021). We evaluated whether these mechanisms were conserved in leaf responses to BL. Consistent with observations in hypocotyls and in young leaves, in our conditions endogenous PHOT1 levels decreased in response to BL from the top, with a stronger effect of higher BL intensities (Figure 3A and Supplemental Figure S3A; Sakamoto and Briggs, 2002; Kozuka et al., 2011). In response to light irradiating the abaxial side a reduction in the fluorescent signal could be observed in the abaxial epidermis for pML1:PHOT1-citrine, but not for pCER6:PHOT2-GFP (Figure 3B). The same response was found in total protein levels in the same transgenic lines evaluated by western blotting (Supplemental Figure S3B). This is consistent with higher stability of phot2 than phot1 in response to BL in hypocotyls and leaves (Sakamoto and Briggs, 2002; Kong et al., 2006; Kozuka et al., 2012).
Figure 3.
Early phototropin signaling mechanisms in leaves. A, PHOT1 levels in leaf blades of 2-week-old Col plants transferred to WL, RL, or RBL for 1 d. Protein blots were probed with anti-PHOT1 antibody, and DET3 was used as a loading control. Quantification of western blots shown in Supplemental Figure S3A. Each bar represents mean ± se of eight replicates coming from two independent experiments. Different letters indicate significant differences among means in ANOVA followed by Tukey’s multiple comparisons test (P < 0.05). B, PHOT1 levels decrease in the abaxial epidermis in response to reflected light. Plants grown for 3 weeks on soil covered with dark (No reflection) or clear (Reflection) aluminum foil. Left: Confocal images of the abaxial epidermis of plants expressing pML1:PHOT1-citrine or pCER6:PHOT2-GFP. Scale bar 50 µm. Right: quantification of microscopy images. Each bar represents mean ± se of four replicates. *P < 0.05 in ANOVA followed by Sidak’s multiple comparisons test. C, Pictures of Col and phototropin, pks, and nph3 mutant plants grown for three weeks on soil with no reflection. Scale bar: 2 cm. D, LFI in plants grown on soil for two weeks and transferred to WL, RL, or RBL for one week. Bars represent mean ± se of 10–20 plants. *P < 0.05 in ANOVA followed by Tukey’s multiple comparison test. The complete ANOVA results including comparisons with WL can be found in Supplemental Table S5. E, LFI in plants grown on soil covered with dark (No reflection) or clear (Reflection) aluminum foil for 3 weeks. Bars represent mean ± se of 20–45 leaves. *P < 0.05 in ANOVA followed by Sidak’s multiple comparison test.
NRLs and PKS proteins regulate phototropin-mediated flattening in response to light from the top (Inoue et al., 2008; de Carbonnel et al., 2010; Harada et al., 2012). To compare the phototropin signaling network in the adaxial and the abaxial sides of the leaf, we measured LFI in response to light from the top and from below in nph3 and pks mutants (Figure 3, C–E). nph3 mutants showed partially impaired leaf flattening when grown in WL (Figure 3, C–E) and NPH3 was required to respond to low BL intensities coming from the top, as reported previously (Figure 3D;Inoue et al., 2008). However, when light was reflected and reached the abaxial side of the leaf, nph3 mutants responded strongly (Figure 3E), suggesting that the early light signaling mechanisms controlling leaf flattening differ between the adaxial and the abaxial sides of the blade. The PKS family has four members in Arabidopsis, but so far only pks1, pks2, and pks4 null mutants were available. To evaluate the role of PKS3, we used CRISPR to create null alleles of PKS3, namely pks3-9 and pks3-10. These mutants showed a striking flattening defect, comparable to phot1phot2 mutants when grown in WL, suggesting that PKS3 is a key phototropin signaling component in the leaf blade (Figure 3C). However, single mutant alleles maintained a small response to light (Figure 3, D and E). PKS2, which was implicated in phototropin signaling in the leaf (de Carbonnel et al., 2010), did not show an effect in our conditions when mutated alone or in combination with PKS3 (Figure 3, D and E). Nevertheless, the quadruple pks1pks2pks3pks4 mutant showed an extreme flattening defect in all conditions tested, and did not respond to any light stimuli, showing that, while PKS3 has a major role in leaf flattening, there is functional redundancy among PKS family members.
Phototropins regulate leaf flattening reversibly and during the expansion phase
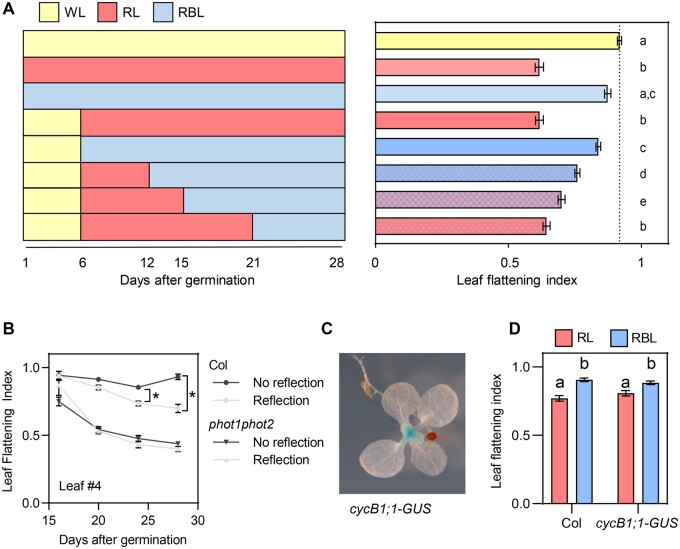
To change organ shape plants can regulate cell number, size, or shape. During the first steps of leaf development the primordia undergoes cell division, followed by a front of cell expansion moving from the tip to the base of the blade (Xiong and Jiao, 2019). To determine whether phototropins control cell division or cell expansion in the leaf, we activated or inactivated phototropins at different times during development by transferring plants to WL, RL, or red + blue light (RBL). Plants grown in RL or RBL since Day 0 after germination showed the same LFI as those transferred 6 or 9 d after germination, when leaves 1 and 2 were already more than 4-mm long and cell division should be finished (Marrocco et al., 2009), suggesting that phototropins regulate leaf shape during the expansion phase (Figure 4; Supplemental Figure S4). Consistent with this, LFI was comparable between Col and phot1phot2 leaves during early development, and gradually phot1phot2 curled downward while Col leaves remained flat (Figure 4B). In addition, the response to reflected light in Col plants occurred late in development and increased as leaves expanded (Figure 4B). These results indicate that phototropins regulate leaf shape in large part by controlling cell expansion. To test this further, we used plants expressing the cell division marker pCYCB1;1::NterCYCB1-GUS (cycB1;1-GUS; Marrocco et al., 2009) and stained them after 14 d of growth, prior to transfer to light treatments (Figure 4, C and D). We observed no GUS staining in leaves 1 and 2, which later responded to phototropin inactivation by a RL treatment, showing that leaf flattening was controlled during cell expansion (Figure 4, C and D). Finally, the response to light signals was reversible as long as the treatments occurred while the leaf was still expanding. Transferring plants from RL to RBL at Day 12 or 15 after germination resulted in a higher LFI than keeping them in RL, but if the transfer was done at Day 21 after germination BL could not promote leaf flattening (Figure 4A and Supplemental Figure S4). We conclude that phototropin-mediated leaf flattening occurs at least in part through light-controlled cellular expansion.
Figure 4.
Phototropins control leaf shape reversibly during the expansion phase. A, LFI of Col plants grown on soil and transferred to WL, RL, or RBL at different time points. Measurements were done 28 d after germination. Bars represent mean ± se of 40–48 leaves. Different letters represent significant differences among means (P < 0.05) in an ANOVA followed by Tukey’s test. Left: scheme of the treatments. B, LFI in Col and phot1phot2 plants grown on soil covered with dark (No reflection) or clear (Reflection) aluminum. Results correspond to leaf 4 during development since leaves are big enough to be measured. Each point represents mean ± se 8 to 11 plants. *P < 0.05 in ANOVA followed by Sidak’s multiple comparison test. Within each genotype, the effect of light was tested at each time point, and only the significant effects are shown. C, GUS staining of plants expressing cycB1;1-GUS grown on soil covered with clear (Reflection) or dark (No reflection) aluminum foil. Staining was performed 2 weeks after germination, when leaves 1 and 2 were more than 3-mm long, but before they respond to the light stimulus. D, LFI in plants expressing cycB1;1-GUS grown in WL for 2 weeks and transferred to RL or RBL for 1 week. Bars represent mean ± se of 16–22 leaves. Different letters represent significant differences among means (P < 0.05) in an ANOVA followed by Tukey’s test.
Phototropins regulate the auxin signaling pattern in the medio-lateral and abaxial–adaxial axes of the leaf blade
In stem phototropism, higher auxin levels on the shaded than on the lit side of the hypocotyl leads to asymmetric growth (Ding et al., 2011; Willige et al., 2013; Legris and Boccaccini, 2020). To evaluate the role of auxin signaling during leaf phototropism, we determined the distribution pattern of the auxin output reporter pDR5rev::3XVENUS-N7 (DR5:VENUS; Heisler et al., 2005) in the blade of expanding leaves using epifluorescence microscopy. In WL, the DR5:VENUS signal was higher on the abaxial side of the blade than on the adaxial side, where it was restricted to the margins (Figure 5). In conditions where leaves curled downward, i.e. reflected light from below or the absence of BL (RL), the VENUS signal increased on the adaxial side (Figure 5, A and B). Adding low intensities of BL, which was enough to promote flattening (Figure 1), partially reduced the VENUS signal on the adaxial side of the blade and increased VENUS signal in the abaxial side, restoring the distribution pattern found in WL (Figure 5A). This response depended on phototropins, since the double phot1phot2 mutant had an altered DR5:VENUS expression pattern in WL and did not respond to the light treatments (Figure 5, C and D). These results are consistent with a model where phototropins create an auxin response gradient in the leaf. However, in transverse cuts of fixed leaves expressing DR5:VENUS, we only detected VENUS signal in the epidermis (Supplemental Figure S5). Taken together with our data indicating that pML1:PHOT1 complemented phot1phot2 (Figure 2), these results suggest a particularly important role of phototropin activation and auxin signaling on both epidermal layers to control leaf flattening. In particular, BL-activated phototropins in the adaxial epidermis inhibit auxin signaling in the center of the blade and increase auxin signaling in the abaxial epidermis to promote flattening.
Figure 5.
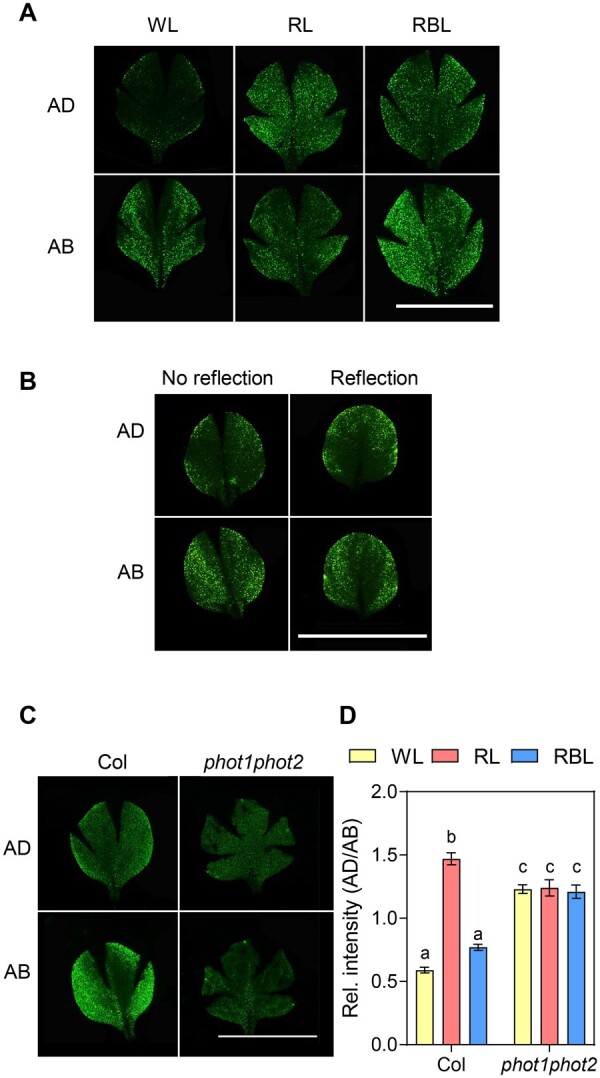
Leaf shape correlates with the auxin signaling pattern in the blade. A, Epifluorescence images of leaf blades of 2-week-old plants expressing DR5:VENUS transferred for 1 week to WL, RL, and RBL. For each leaf, images were taken on the adaxial (AD) and abaxial (AB) side. Scale bar 1 cm. B, Epifluorescence images of leaf blades of plants expressing DR5:VENUS grown on soil covered with dark (No reflection) or clear (Reflection) aluminum foil for 3 weeks. For each leaf, images were taken on the AD and AB side. Scale bar 1 cm. C, Epifluorescence images of leaf blades of Col and phot1phot2 plants expressing DR5:VENUS grown for 2 weeks and transferred for 1 week to WL, RL, and RBL. For each leaf, images were taken on the AD and AB side. Scale bar 1 cm. D, Quantification the VENUS fluorescence in epifluorescence images of whole blades. For each leaf, images were taken on the AD and AB side, DR5:VENUS fluorescence was quantified and the ratio between fluorescence in each side was calculated. Each bar represents mean ± se of 10–16 leaves. Different letters represent significant differences among means (P < 0.05) in an ANOVA followed by Tukey’s test.
Light-regulated leaf flattening involves auxin transport
Changes in the DR5 promoter activity could be due to changes in auxin synthesis, transport, or sensitivity. The strong role of auxin transport in hypocotyl phototropism (Christie et al., 2011; Ding et al., 2011; Willige et al., 2013) prompted us to focus on auxin transport during leaf flattening. First, we used the auxin transport inhibitor N-1-naphthylphthalamic acid (NPA) to block auxin transport during the leaf expansion phase. Treating plants with this compound had a similar effect on leaf flattening as inactivating phototropins (Figure 6A). This phenotypic response corresponded with changes in the DR5:VENUS pattern. In accordance to what we observed in response to light and phototropin activity, NPA treatment increased the adaxial/abaxial ratio of the DR5 signal (Figure 6B). However, the difference between the margins and the center of the blade was still apparent in this condition. These results suggest that while high RL intensity (as in RL and RBL) increases auxin signaling in the adaxial side, BL enhances auxin signaling in the abaxial side in a process requiring auxin transport to promote flattening.
Figure 6.
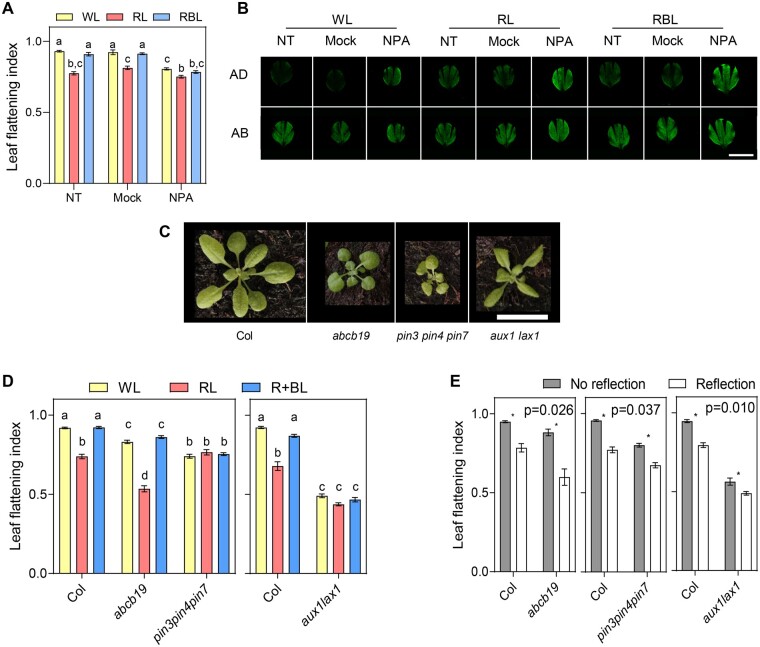
Auxin transporters are required for light-regulated leaf flattening. A, LFI of Col plants grown for 2 weeks and transferred to WL, RL, or RBL for 1 week. During this week, plants were sprayed with NPA, mock solution (Mock), or not treated (NT). Bars represent mean ± se of 30 plants. Different letters represent significant differences among means (P < 0.05) in an ANOVA followed by Tukey’s test. B, Epifluorescence images of leaf blades of Col plants expressing DR5:VENUS grown for 2 weeks and transferred for 1 week to WL, RL, or RBL for 1 week while being treated with NPA, mock solution (Mock), or NT. For each leaf, images were taken on the AD and AB side. Scale bar: 0.5 cm. C, Pictures of 3-week-old Col and various auxin transporters mutants. Scale bar: 2 cm. D, LFI in plants of various genotypes grown on soil for 2 weeks and transferred to WL, RL, or RBL for 1 week. Bars represent mean ± se of 18–56 plants. Different letters represent significant differences among means (P < 0.05) in an ANOVA followed by Tukey’s test. E, LFI in plants of various genotypes grown on soil covered with dark (No reflection) or clear (Reflection) aluminum foil for 3 weeks. Bars represent mean ± se of 14–441 plants. *P < 0.05 in ANOVA followed by Sidak’s multiple comparison test. Significance of the interaction term (Genotype × Treatment) in the ANOVA is shown in the top right corner in each case.
To determine which transporters play a role in this response, we grew mutants affected in the major classes of auxin transporters. All tested auxin transporters families played a role in leaf flattening when plants were grown in standard conditions (Figure 6, C–E). To evaluate whether this defect was related to light-regulated leaf development, we measured LFI in response to light signals in these mutants. abcb19 mutants responded to light irradiated from the top or from below (Figure 6, D and E), indicating that this auxin transporter is not essential to promote leaf flattening by BL. However, it showed a regulatory role in the response, since this mutant responded more to light from below as shown by the significant interaction term in the analysis of variance (ANOVA) (Figure 6E). In agreement with the observation that NPA inhibits BL-regulated leaf flattening, pin3pin4pin7 triple mutants did not respond to light from the top and showed a reduced response to reflected light (Figure 6, D and E). Interestingly, among the mutants tested, aux1lax1 was the only one showing a striking resemblance to the phot1phot2 rosette morphology (Figure 6C). Also, this mutant showed a very similar flattening defect as phot1phot2, with no response to BL from above, and a reduced response to reflected light (Figure 6, D and E). Overall, these results indicate that phototropins regulate auxin transport to control leaf flattening and uncover a potential mechanism of phototropin signaling involving AUX1/LAX auxin transporters.
Discussion
Light direction perceived by phototropins is a key aspect regulating leaf blade flattening
Leaves are the main light harvesting organs in plants, and their shape influences photosynthesis through light interception and gas exchange. Leaf flattening is strongly controlled by the developmental program from early patterning through expansion (Chitwood and Sinha, 2016; Xiong and Jiao, 2019; Heisler and Byrne, 2020). Given the tight relationship between leaf shape and light interception, we focused on how BL signals control leaf flattening, and found that phototropins perceive light direction to control leaf shape (Figures 1 and 2). This effect, combined with the role of phototropins on leaf positioning and anatomy, indicates strong interactions between BL signaling and endogenous cues in controlling leaf development according to the environment (Inoue et al., 2008; de Carbonnel et al., 2010; Kozuka et al., 2011). Indeed, phototropin mutants show reduced biomass accumulation, which can at least in part be explained by their leaf morphology (Takemiya et al., 2005; Inoue et al., 2008; de Carbonnel et al., 2010). In response to reflected light, we found that Arabidopsis changes leaf shape and also accumulate more fresh weight (Supplemental Figure S1D). This suggests that in a heterogeneous light environment leaf curling may contribute to light interception and photosynthesis. However, more experiments are needed to directly link this change in biomass accumulation to leaf morphology. Interestingly, the response to light direction in the leaf is conserved in lettuce, where leaves irradiated on the abaxial side showed a curled phenotype (Wang et al., 2021).
Phototropins control leaf flattening reversibly, late in development, in expanding leaves when cell division has ended (Figure 4). This role of light on blade expansion presumably occurs in the epidermis, which has major effects on growth regulation (Savaldi-Goldstein et al., 2007). This is consistent with complementation of phot1phot2 leaf curling by epidermal expression of PHOT1-citrine (Figure 2). Moreover, in lettuce light from below affects epidermal cell shape and expansion to regulate leaf curvature (Wang et al., 2021). phot2 regulates palisade mesophyll development in a light-dependent manner (Kozuka et al., 2011). However, we observed changes in leaf flattening in response to low BL and in phot2 mutants (Figure 1). In addition, plants expressing phot2 in the epidermis respond to light to control leaf flattening but not to regulate palisade mesophyll development (Kozuka et al., 2011). We conclude that phototropins play a primary role in the epidermis to control blade flattening.
Leaf flattening corresponds with the pattern of auxin signaling reporter DR5:VENUS. In conditions where leaves were flat, the DR5:VENUS signal was high in the margins and in the abaxial epidermis, while downward curling corresponded with an increase in the DR5:VENUS signal in the center of the adaxial epidermis (Figure 5). The adaxial/abaxial ratio of auxin signaling corresponded with leaf flattening and depended on BL, phototropins, and auxin transport (Figures 5 and 6). We cannot exclude the possibility that an auxin pattern in the adaxial epidermis (margins versus center) also explains leaf flattening, since both aspects were affected in phot1phot2. The role of abaxial auxin concentration on leaf flattening has been studied in earlier stages of leaf development (Guan et al., 2017; Xiong and Jiao, 2019; Heisler and Byrne, 2020). Although some controversy exists on whether higher auxin in the abaxial side of the primordium is an early signal creating the adaxial–abaxial patterning, it is possible that this signal helps maintain tissue patterning (Heisler and Byrne, 2020). It would be interesting to know whether phototropins affect auxin distribution early in development. However, we note that the phot1phot2 leaf flattening phenotype is most obvious late in development highlighting their importance during leaf expansion (Figure 4B). Whether changes in the DR5 expression pattern reflect changes in auxin levels or downstream signaling requires further investigations. NPH4/ARF7, which is required for hypocotyl phototropism, is developmentally regulated showing expression predominantly in the adaxial side of the primordia, as most activator auxin response factors (ARFs) (Guan et al., 2017). Moreover, arf7 mutants show deficient leaf flattening, with a phenotype similar to phot1phot2 suggesting a role for auxin signaling in light-controlled leaf flattening (Watahiki and Yamamoto, 1997).
All auxin transporter mutants tested showed defects in leaf flattening and in response to light (Figure 6). However, the only mutant resembling the phot1phot2 phenotype was aux1lax1 (Figure 6). AUX/LAX proteins control phyllotaxis, venation, and margin development (Reinhardt et al., 2003; Swarup and Bhosale, 2019; Xiong and Jiao, 2019). Interestingly, these genes also control leaf flattening in tomato (Pulungan et al., 2018). PINs have many roles in leaf development, including primordia establishment, phyllotaxis, and early patterning (Xiong and Jiao, 2019; Heisler and Byrne, 2020). The triple pin3pin4pin7 mutant did not respond to light signals, suggesting that PINs are essential to regulate leaf flattening (Figure 6). Nevertheless, the leaf and rosette morphology of pin3pin4pin7 differs strongly from phot1phot2 (Figure 6C). Given the many roles of PINs in development, it is possible that earlier defects in leaf patterning render the mutants unable to respond to light signals. However, inhibiting PINs with NPA late in development caused similar flattening defects as inactivating phototropins, indicating that during blade expansion polar auxin transport is required for leaf flattening (Figure 6A). ABCB19 is involved in phototropin-dependent leaf positioning and flattening (Jenness et al., 2020). In our experiments, and consistent with previous reports, the abcb19 mutant had a flattening defect in WL and RBL (Figure 6). However, abcb19 responded to light signals from the top (Figure 6D) and it responded to light from below more than Col (Figure 6E). Hence, in the conditions analyzed here, ABCB19 is not required for phototropin-mediated leaf flattening, but it has a regulatory role. We conclude that multiple auxin transporters regulate leaf flattening but based on the mutant phenotype AUX1 and LAX1 might be more directly involved in phototropin signaling. This contrasts with the minor role of AUX/LAX in hypocotyl phototropism (Christie et al., 2011; Hohm et al., 2014), suggesting that partially different mechanisms control phot-mediated auxin gradients formation in hypocotyls versus leaves.
Conserved aspects and differences of phototropin signaling between leaf blades and hypocotyls
Our study reveals that general principles of phototropin action are conserved between stem phototropism and leaf flattening, in particular following light interception from the adaxial leaf side. In both cases, phototropins perceive light direction and regulate auxin signaling to change cell expansion and ultimately organ curvature (Figures 1, 4, and and 5; Legris and Boccaccini, 2020). In hypocotyl phototropism, phot1 responds to a broad BL intensity range and phot2 only responds to higher light intensities. In leaves phot1 and phot2 respond to low BL intensities (below 0.4 µmol m−2 s−1) on both leaf sides, although phot1 has a more prominent role in response to low light intensities coming from the top (Figure 1). The current model for hypocotyl phototropism is that directional light establishes a light gradient across the organ leading to a phototropin activation gradient to determine growth orientation. The situation in leaves is different because the abaxial and adaxial sides respond differently to BL (Figure 1). The lower side is more sensitive to light than the upper side, since low BL applied to the abaxial side triggers bending despite high BL reaching the adaxial side (Figure 1B). phot1-mediated sensing on both sides of the leaf was further established using PHOT1-citrine under the control of pAS2 and pFIL promoters (Figure 2). Moreover, we could determine that in response to BL phot1 is downregulated (Figure 3), which is linked to desensitization in hypocotyls (Roberts et al., 2011). Higher light exposure of the adaxial side may trigger asymmetric phot1 degradation, thereby explaining the higher light sensitivity of the lower side of the leaf.
Differential light sensitivity of both leaf sides may also be caused by different signaling mechanisms on these developmentally distinct leaf parts. In line with this idea, the nph3 mutant has distinct phenotypes depending on the side of leaf irradiation (Figure 3). Consistent with previous data, NPH3 is required to promote leaf flattening in response to LB irradiation from the top (Figure 3;Inoue et al., 2008). In contrast, NPH3 may inhibit the BL response on the abaxial side (Figure 3, D and E). NPH3 is essential for hypocotyl phototropism (Christie et al., 2018). It is therefore surprising that NPH3 may counteract phototropin signaling depending on the tissue type. One alternative interpretation of the nph3 leaf phenotype is that blade curvature is a balance between light responses on both sides of the leaf. In the absence of NPH3 phototropin signaling in the adaxial side of the leaf could be impaired, but response to light from below could be normal, causing an imbalance and explaining the nph3 phenotype. Irrespective of the mechanism, our results show that phototropins can perceive and respond to light in the abaxial epidermis in an NPH3-independent manner possibly involving other members of the NRL family such as RPT2, which also regulates leaf flattening (Harada et al., 2012).
In the hypocotyl PKS1, PKS2, and PKS4 have a major role regulating phototropism (Kami et al., 2014). Here, we found that PKS3 is the main PKS protein involved in leaf flattening (Figure 3). pks3 mutants showed a flattening defect comparable to phot1phot2 and reduced responses to light signals. However, as observed for hypocotyl phototropism, there is functional redundancy among the members of the PKS family to control leaf BL responses (Figure 5). This is consistent with the reported roles in leaf flattening and positioning of PKS1 and PKS2, and the flattening defect found in pks1pks2pks4 triple mutant (de Carbonnel et al., 2010). PKS2 functions in phot2 signaling controlling flattening (de Carbonnel et al., 2010). In our experiments, pks3 mutants had a reduced but significant response to low BL, suggesting that the phot1 pathway was still partially active in this mutant (Figure 3). Interestingly, PKS proteins were previously shown to act at the interface between internal and external cues. During hypocotyl phototropism in etiolated seedlings the response of pks mutants depends on whether light reaches the cotyledons or the hook (Kami et al., 2014). Results presented here for PKS3 in leaf flattening further show the central role of PKS proteins in coordinating plant development with the environment.
Materials and methods
Plant material
The following Arabidopsis (Arabidopsis thaliana; Col-0) mutants were used previously: phot1-5, phot2-1, phot1-5 phot2-1, nph3-6 (de Carbonnel et al., 2010); phyB-9, phyB-9 ven4-bnen (Yoshida et al., 2018), cry1-304, cry2-1, cry1-304 cry2-1 (Boccaccini et al., 2020); aux1-21 lax2-1 (Hohm et al., 2014), pin3-3 pin4-101 pin7-101 (Willige et al., 2013), abcb19-101 (Jenness et al., 2019).
The following transgenic lines were described before: pML1:PHOT1-citrine, pPHOT1:PHOT1-citrine (Preuten et al., 2013), pCER6:PHOT2-GFP (Kozuka et al., 2011), pCYCB1;1::NterCYCB1-GUS (Marrocco et al., 2009), pDR5rev::3XVENUS-N7 (Heisler et al., 2005).
pDR5rev::3XVENUS-N7 in phot1-5 phot2-1, as well as multiple pks mutants containing pks3-9, were obtained by crossing. The methods used to genotype the mutations appear in Supplemental Tables S1 and S2. The presence of the pDR5rev::3XVENUS-N7 transgene was selected by resistance to BASTA.
Generation of CRISPR mutant alleles of PKS3
To create pks3-9 and pks3-10 mutants the protocol by Hyun et al. (2015) was followed. pYB196 and pRG-ext-CCR5 were provided by George Coupland. The sequence of the guide used was 5′-AGATCATGTTGATTCCACGG-3′.
The selected mutant alleles were named pks3-9 and pks3-10. pks3-9 has a 2-bp deletion and a mutation in position 192 (Col sequence 5′-TGATTCCA-3′, mutant allele 5′-CGTTTT-3′), which creates an early stop codon resulting in a 73-aminoacid protein. pks3-10 has a base insertion (A) in position 199, which creates an early stop codon and a truncated 74-aminoacid protein.
Generation of transgenic lines expressing PHOT1 under pAS2 and pFIL promoters
For pAS2::PHOT1-citrine, the backbone and coding sequence (CDS) PHOT1-citrine were obtained from pPHOT1::PHOT1-citrine (Preuten et al., 2013). The pAS promoter was amplified from pCRII-TOPO pAS2 (Wu et al., 2008). For pFIL::PHOT1-citrine, the backbone and pFIL promoter were obtained from pGWB-NB1-pFIL (Tameshige et al., 2013). The PHOT1-citrine CDS was obtained from pPHOT1::PHOT1-citrine (Preuten et al., 2013). Transgenic plants were generated by introduction of the plant expression constructs into pSOUP-containing Agrobacterium tumefaciens strain GV3101. Transformation of phot1-5phot2-1 plants was done by floral dipping. Based on segregation of Basta-resistance, homozygous T3 lines with a single transgene were selected.
Reverse Transcription quantitative PCR (RT-qPCR)
Plants were grown on plates for 12 d in continuous light. For RNA extraction, only the aerial part containing hypocotyls, the shoot apical meristem, and small leaves of ∼1-mm long were grinded in liquid nitrogen. RNA extraction was performed with the RNAeasy plant mini kit (Quiagen). Each sample consists of six plants, except for pFIL-9 (4 plants) and pFIL-37 (8 plants), and three replicates per line were harvested. The reverse transcription was done using SuperScript II (Invitrogen) from 1 µg of RNA. Transgene expression levels were measured quantifying the citrine transcript with primers LAP038 and LAP045. Three housekeeping genes were used: GAPDH, UBC, YLS8. Primers used for this analysis are listed in Supplemental Table S2.
Growth conditions
Seeds were sowed on soil (CL TON KOKOS, CLASSIC Profisubstrat, Einheits erde) in square plastic pots (8 cm × 8 cm × 7 cm, width × depth × height) watered with Solbac solution 1:400 (Andemat Biocontrol). After stratifying for 3–5 d plants were grown in 16:8-light/dark cycles in a walk-in incubator equipped with white and red LEDs, photosynthetic photon flux density (PPFD) = 100 µmol m−2 s−1, at 22°C.
For experiments involving light reflection seeds were sowed on 0.8% (w/v) water-agar plates, stratified for 3–5 d and grown for 4 d in light until they were de-etiolated. A square of aluminum foil was placed over the soil, and four small holes (2–3 mm diameter) were done on each corner. The holes were filled with moistened soil, and seedlings were transplanted inside.
In Figure 1, A, B, and D, seeds were surface-sterilized and sowed in square transparent plates (10 × 10 × 2 cm3) with half MS media (Duchefa) plus 0.8% agar (Roth).
Light treatments
WL, RL and RBL treatments were performed in a Percival incubator, model AR36-L3, set to 16:8-light/dark cycles, 22°C, relative humidity 70%. WL was obtained with fluorescent tubes (OSRAM Lumilux cool white L18W/840), RL was obtained with red LEDs and RB was obtained with red LEDs combined with fluorescent tubes. PPFD was set to 100 µmol m−2 s−1 in all cases. The red, blue, and far-red values (µmol m−2 s−1) were WL = 20; 7; 1.45; RL = 60; 0.003; 0.1; RBL = 60; 0.4; 0.1.
In Figure 1, A, B, and D, plants were grown on plates in Percival incubators model AR22-LX, set to 16:8-light/dark cycles, 22°C. WL was obtained with fluorescent tubes (OSRAM Lumilux cool white L18W/840), RL was obtained with red LEDs. Light from below was obtained with LEDs (FloraLED, CLF plant climatics).
Light intensities were measured with a radiometer IL 1400A (International Light, USA) using a W filter (#9540) and PAR (#21777), TBLU (#21853), TRED (#22237), and TFR (#22238) filters.
LFI measurement
Leaf blades were placed on top of an agar plate with the adaxial face up and imaged. Then they were flattened performing cuts in the margins and sticking the adaxial side on transparent tape. The flattened leaves were imaged again. Image analysis was performed with Image J. Pictures were converted to 8-bit and a manual threshold was applied to segment the whole blade. The whole blade was selected using the Wand (tracing) tool. LFI was calculated as the ratio Area (before flattening)/Area (after flattening) for each leaf. Unless stated otherwise, all measurements were done on leaves 1 and 2.
Fresh weight measurement
Plants were grown on soil covered with dark or clear aluminum foil for three weeks. Rosettes were harvested cutting in the upper part of the hypocotyl and weighed immediately after.
Fluorescence microscopy
Confocal microscopy images were taken with an LSM 710 confocal microscope (Zeiss). For imaging DR5:VENUS samples were excited with an Argon laser (514 nm, 100%), and detection was done between 520 and 560 nm, gain 607.8. For pAS2:PHOT1-citrine and pFIL:PHOT1-citrine samples were excited with an Argon laser (514 nm, 30%) and detection was done between 520 and 560 nm, gain 1063.6. In addition, a channel was set to detect chlorophyll, exciting with an Argon laser (514 nm, 30%) and, detecting between 600 and 750 nm, gain 572.7. For pML1:PHOT1-citrine samples were excited with an Argon laser (514 nm, 100%) and detection was done between 520 and 560 nm, gain 1051.7. For pCER6:PHOT2-GFP samples were excited with an Argon laser (488 nm, 50%) and detection was done between 505 nm and 530 nm, gain 1051.7. For the calcofluor white stain samples were excited with a 405 nm laser, 1.2% and detection was done between 420 and 470 nm, gain 491.8.
In Supplemental Figure S5, leaves were fixed in 4% PFA for 2 h, washed with 1× phosphate-buffered saline (PBS) and stained with calcofluor white (0.1%) in PBS applying vacuum and staining overnight at room temperature. Stained leaves were embedded in 1.5% agarose, cut with a razor blade, and mounted on a glass bottom dish with PBS.
In Figures 5 and 6, B, leaves were harvested and immediately mounted between two coverslips with water, doing cuts in the margins to allow flattening. For each leaf, images were taken on the adaxial and abaxial side using a Leica M205 FCA stereomicroscope equipped with a GFP filterset (excitation 470/40, emission 525/50).
In Figure 5D, DR5:VENUS fluorescence from whole blades was quantified using ImageJ, applying a threshold to segment the whole blade and selecting it with the wand (tracing) tool.
In Figure 3B, images were taken from the adaxial and abaxial epidermis of plants expressing pML1:PHOT1-citrine or pCER6:PHOT2-GFP. From each leaf, five images were taken from each epidermis, and in each image five region of interests (ROIs) of 3.4 µm2 were quantified using Image J.
GUS staining
Leaves were harvested and fixed in 90% acetone for 4 h and rinsed twice with 50 mM NaPO4 before vacuum infiltrating with the staining solution (four times 15 min each). The staining solution contained: EDTA pH 8.5 10 mM, NaHPO4 50 mM, Triton C-100 0.1%, X-Gluc 0.5 mg mL−1 in water. Samples were incubated at 37°C overnight and cleared with 70% EtOH at 4°C, over various days changing EtOH regularly. Images were taken with a Leica M205 FCA stereomicroscope.
Western blot
Plants were grown for two weeks and transferred to WL, RL, or RBL at CT8 for 24 h. Leaf blades were harvested on liquid nitrogen, each sample consisting of leaves 1 and 2 from one plant.
Total proteins were extracted by grinding the seedlings in 50 μL 2× Laemmli buffer with 10% β-mercaptoethanol. Samples were heated 5 min at 95°C and centrifuged for 1 min; 10 μL per sample were loaded in a pre-cast 4%–15% gradient agarose gel (Mini-PROTEAN TGX, BioRad). Electrophoresis was performed at 100 mV during 75 min. Transfer was performed using the TransBlot Turbo system (BioRad). Detection of PHOT1-citrine was done using Living Colors anti-GFP antibody JL-8 (632381, Clontech), 1:4,000 in PBS 5% milk 0.1% tween (PBSTM). Detection of endogenous PHOT1 was done using anti-PHOT1 1:5,000 in PBSTM (Christie et al., 1998; Cho et al., 2007). DET3 and Histone 3 were used as a loading controls, and detected using anti-DET3 1:20,000 in PBSTM (Schumacher et al., 1999), or anti-Histone H3 (ab1791, abcam) 1:3,000 in PBSTM. Membranes were blocked for 1 h at room temperature with PBSTM and incubated overnight with the corresponding primary antibody at 4°C. Anti-PHOT1, anti-DET3, and anti-Histone H3 were detected with anti-rabbit IgG, HRP conjugate (W4011, Promega) 1:2,500 in PBSTM. Anti-GFP was detected with anti-mouse IgG, HRP conjugate (W4021, Promega) 1:5,000 in PBSTM. Membranes were revealed using Immobilion Western Chemiluminiscent HRP substrate (WBKLS0500, Millipore). Chemiluminiscence was detected using an ImageQuant LAS4000 mini (GE Healthcare). Image quantification was performed using ImageJ.
NPA treatment
A stock solution of NPA (Prod No. N0926.0250, Duchefa) was prepared to a concentration of 10 mM in DMSO and kept at −20°C. On the day of the treatment, a new dilution was performed in water plus 0.15% Tween-20 to a final concentration of 10 μM. The mock solution consisted of 1:1,000 DMSO in 0.15% Tween-20. NPA was sprayed from above the day plants were transferred to the light treatment and every 2 d. LFI and DR5:VENUS signal were measured after 7 d.
Accession numbers
The Arabidopsis Genome Initiative numbers for the genes mentioned in this article are as follows: AT2G18790 (PHYB), AT3G45780 (PHOT1), AT5G58140 (PHOT2), AT4G08920 (CRY1), AT1G04400 (CRY2),AT2G02950 (PKS1), AT1G14280 (PKS2), AT1G18810 (PKS3), AT5G04190 (PKS4), AT5G64330 (NPH3), AT3G28860 (ABCB19), AT1G70940 (PIN3), AT2G01420 (PIN4), AT1G23080 (PIN7), AT2G38120 (AUX1), AT5G01240 (LAX1), AT4G21750 (ML1), AT1G68530 (CER6), AT1G65620 (AS2), AT1G68530 (FIL), AT1G13440 (GAPDH), AT5G25760 (UBC), AT5G08290 (YLS8).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Light treatments and growth conditions.
Supplemental Figure S2 . Expression levels and distribution pattern of pAS2 and pFIL lines.
Supplemental Figure S3 . PHOT1 levels decrease with BL in the leaf blade.
Supplemental Figure S4 . Phototropins control flattening reversibly and late in development.
Supplemental Figure S5 . DR5:VENUS signal is detected in the epidermis.
Supplemental Table S1. Genotyping conditions to select mutations in crosses.
Supplemental Table S2. Primers used in this study.
Supplemental Table S3. Full ANOVA results for Figure 1E.
Supplemental Table S4. Full ANOVA results for Figure 2A.
Supplemental Table S5. Full ANOVA results for Figure 3D.
Supplementary Material
Acknowledgments
We thank Prof. George Coupland for sharing the plasmids used for CRISPR, Prof. John Christie for the anti-PHOT1 antibody, and Prof. Akira Nagatani for sharing seeds carrying pCER6:PHOT2-GFP. The plasmid pCRII-TOPO pAS2 was provided by Prof. Patricia Springer, and the plasmid pGWB-NB1-pFIL by Prof. Kiyoshi Tatematsu. We thank Dr Mark Jennes for his comment on the phenotype of abcb mutants and for sending seeds. The CIF facility at the University of Lausanne assisted us with confocal microscopy.
Funding
This work was supported by the University of Lausanne, the Swiss National Science Foundation (grant no. 310030B_179558 to C.F.), European Molecular Biology Organization (ALTF 46-2017 to M.L.), Human Frontier Science Program (LT000829/2018-L to M.L.), and European Commission Marie Curie fellowship (grant no. H2020–MSCA–IF–2017–796443 to M.L.).
Conflict of interest statement. None declared.
M.L., B.B.S-E., and C.F. conceived the original research plans; M.L., B.B.S.-E., M.T., and L.A.P. performed the experiments and analyzed the data; M.L. and C.F. wrote the article with contributions of all the authors; C.F. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/General-Instructions) is Christian Fankhauser (Christian.fankhauser@unil.ch).
References
- Boccaccini A, Legris M, Krahmer J, Allenbach-Petrolati L, Goyal A, Galvan-Ampudia C, Vernoux T, Karayekov E, Casal JJ, Fankhauser C (2020) Low blue light enhances phototropism by releasing cryptochrome1-mediated inhibition of PIF4 expression. Plant Physiol 183: 1780–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood DH, Sinha NR (2016) Evolutionary and environmental forces sculpting leaf development. Curr Biol 26: R297–R306 [DOI] [PubMed] [Google Scholar]
- Cho H-Y, Tseng T-S, Kaiserli E, Sullivan S, Christie JM, Briggs WR (2007) Physiological roles of the light, oxygen, or voltage domains of phototropin 1 and phototropin 2 in Arabidopsis. Plant Physiol 143: 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR (1998) Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science 282: 1698–1701 [DOI] [PubMed] [Google Scholar]
- Christie JM, Suetsugu N, Sullivan S, Wada M (2018) Shining light on the function of NPH3/RPT2-like proteins in phototropin signaling. Plant Physiol 176: 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie JM, Yang H, Richter GL, Sullivan S, Thomson CE, Lin J, Titapiwatanakun B, Ennis M, Kaiserli E, Lee OR, et al. (2011) phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol 9: e1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carbonnel M, Davis P, Roelfsema MRG, Inoue SI, Schepens I, Lariguet P, Geisler M, Shimazaki KI, Hangarter R, Fankhauser C (2010) The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol 152: 1391–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarsy E, Schepens I, Okajima K, Hersch M, Bergmann S, Christie J, Shimazaki Ki, Tokutomi S, Fankhauser C (2012) Phytochrome kinase substrate 4 is phosphorylated by the phototropin 1 photoreceptor. EMBO J 31: 3457–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Galvan-Ampudia CS, Demarsy E, Langowski L, Kleine-Vehn J, Fan Y, Morita MT, Tasaka M, Fankhauser C, Offringa R, et al. (2011) Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol 13: 447–452 [DOI] [PubMed] [Google Scholar]
- Guan C, Wu B, Yu T, Wang Q, Krogan NT, Liu X, Jiao Y (2017) Spatial auxin signaling controls leaf flattening in Arabidopsis. Curr Biol 27: 2940–2950.e2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Takemiya A, Inoue SI, Sakai T, Shimazaki KI (2012) Role of RPT2 in Leaf Positioning and Flattening and a Possible Inhibition of phot2 Signaling by phot1. Plant Cell Physiol 54: 36–47 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Byrne ME (2020) Progress in understanding the role of auxin in lateral organ development in plants. Curr Opin Plant Biol 53: 73–79 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hohm T, Demarsy E, Quan C, Allenbach Petrolati L, Preuten T, Vernoux T, Bergmann S, Fankhauser C (2014) Plasma membrane H+‐ATPase regulation is required for auxin gradient formation preceding phototropic growth. Mol Syst Biol 10: 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun Y, Kim J, Cho SW, Choi Y, Kim J-S, Coupland G (2015) Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 241: 271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue SI, Kinoshita T, Takemiya A, Doi M, Shimazaki KI (2008) Leaf Positioning of Arabidopsis in Response to Blue Light. Mol Plant 1: 15–26 [DOI] [PubMed] [Google Scholar]
- Jenness MK, Carraro N, Pritchard CA, Murphy AS (2019) The Arabidopsis ATP-BINDING CASSETTE transporter ABCB21 regulates auxin levels in cotyledons, the root pericycle, and leaves. Front Plant Sci 10: 806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenness MK, Tayengwa R, Murphy AS (2020) An ATP-binding cassette transporter, ABCB19, regulates leaf position and morphology during phototropin1-mediated blue light responses. Plant Physiol 184: 1601–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson H, Hughes J (2014) Nuclear Phytochrome B regulates leaf flattening through phytochrome interacting factors. Mol Plant 7: 1693–1696 [DOI] [PubMed] [Google Scholar]
- Kami C, Allenbach L, Zourelidou M, Ljung K, Schütz F, Isono E, Watahiki MK, Yamamoto KT, Schwechheimer C, Fankhauser C (2014) Reduced phototropism in pks mutants may be due to altered auxin-regulated gene expression or reduced lateral auxin transport. Plant J 77: 393–403 [DOI] [PubMed] [Google Scholar]
- Kong SG, Suzuki T, Tamura K, Mochizuki N, Hara‐Nishimura I, Nagatani A (2006) Blue light‐induced association of phototropin 2 with the Golgi apparatus. Plant J 45: 994–1005 [DOI] [PubMed] [Google Scholar]
- Kozuka T, Kong SG, Doi M, Shimazaki KI, Nagatani A (2011) Tissue-autonomous promotion of palisade cell development by Phototropin 2 in Arabidopsis. Plant Cell 23: 3684–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozuka T, Suetsugu N, Wada M, Nagatani A (2012) Antagonistic regulation of leaf flattening by Phytochrome B and Phototropin in Arabidopsis thaliana. Plant Cell Physiol 54: 69–79 [DOI] [PubMed] [Google Scholar]
- Legris M, Boccaccini A (2020) Stem phototropism toward blue and ultraviolet light. Physiol Plant 169: 357–368 [DOI] [PubMed] [Google Scholar]
- Marrocco K, Thomann A, Parmentier Y, Genschik P, Criqui MC (2009) The APC/C E3 ligase remains active in most post-mitotic Arabidopsis cells and is required for proper vasculature development and organization. Development 136: 1475–1485 [DOI] [PubMed] [Google Scholar]
- Paradiso R, De Visser PH, Arena C, Marcelis LF (2020) Light response of photosynthesis and stomatal conductance of rose leaves in the canopy profile: the effect of lighting on the adaxial and the abaxial sides. Funct Plant Biol 47: 639–650 [DOI] [PubMed] [Google Scholar]
- Preuten T, Blackwood L, Christie JM, Fankhauser C (2015) Lipid anchoring of Arabidopsis phototropin 1 to assess the functional significance of receptor internalization: should I stay or should I go? New Phytol 206: 1038–1050 [DOI] [PubMed] [Google Scholar]
- Preuten T, Hohm T, Bergmann S, Fankhauser C (2013) Defining the site of light perception and initiation of phototropism in Arabidopsis. Curr Biol 23: 1934–1938 [DOI] [PubMed] [Google Scholar]
- Pulungan SI, Yano R, Okabe Y, Ichino T, Kojima M, Takebayashi Y, Sakakibara H, Ariizumi T, Ezura H (2018) SlLAX1 is required for normal leaf development mediated by balanced adaxial and abaxial pavement cell growth in tomato. Plant Cell Physiol 59: 1170–1186 [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Pesce E-R, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Roberts D, Pedmale UV, Morrow J, Sachdev S, Lechner E, Tang X, Zheng N, Hannink M, Genschik P, Liscum E (2011) Modulation of phototropic responsiveness in Arabidopsis through ubiquitination of phototropin 1 by the CUL3-Ring E3 ubiquitin ligase CRL3(NPH3). Plant Cell 23: 3627–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Kagawa T, Kasahara M, Swartz TE, Christie JM, Briggs WR, Wada M, Okada K (2001) Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci U S A 98: 6969–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Briggs WR (2002) Cellular and subcellular localization of phototropin 1. Plant Cell 14: 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Peto C, Chory J (2007) The epidermis both drives and restricts plant shoot growth. Nature 446: 199–202 [DOI] [PubMed] [Google Scholar]
- Schumacher K, Vafeados D, McCarthy M, Sze H, Wilkins T, Chory J (1999) The Arabidopsis det3 mutant reveals a central role for the vacuolar H+–ATPase in plant growth and development. Genes Dev 13: 3259–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S, Waksman T, Henderson L, Paliogianni D, Lütkemeyer M, Suetsugu N, Christie JMJB (2021) Regulation of Plant Phototropic Growth by NPH3/RPT2-like Substrate Phosphorylation and 14-3-3 Binding. bioRxiv 439135 [DOI] [PMC free article] [PubMed]
- Swarup R, Bhosale R (2019) Developmental roles of AUX1/LAX auxin influx carriers in plants. Front Plant Sci 10: 1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemiya A, Inoue S-I, Doi M, Kinoshita T, Shimazaki K-I (2005) Phototropins promote plant growth in response to blue light in low light environments. Plant Cell 17: 1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameshige T, Fujita H, Watanabe K, Toyokura K, Kondo M, Tatematsu K, Matsumoto N, Tsugeki R, Kawaguchi M, Nishimura M (2013) Pattern dynamics in adaxial-abaxial specific gene expression are modulated by a plastid retrograde signal during Arabidopsis thaliana leaf development. PLoS Genet 9: e1003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelmann T, Bornman J, Josserand S (1989) Photosynthetic light gradients and spectral regime within leaves of Medicago sativa. Philos Trans R Soc London Ser B Biol Sci 323: 411–421 [Google Scholar]
- Wang M, Wei H, Jeong BR (2021) Lighting direction affects leaf morphology, stomatal characteristics, and physiology of head lettuce (Lactuca sativa L.). Int J Mol Sci 22: 3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watahiki MK, Yamamoto KT (1997) The massugu1 mutation of Arabidopsis identified with failure of auxin-induced growth curvature of hypocotyl confers auxin insensitivity to hypocotyl and leaf. Plant Physiol 115: 419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Ahlers S, Zourelidou M, Barbosa IC, Demarsy E, Trevisan M, Davis PA, Roelfsema MR, Hangarter R, Fankhauser C, et al. (2013) D6PK AGCVIII kinases are required for auxin transport and phototropic hypocotyl bending in Arabidopsis. Plant Cell 25: 1674–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lin W-C, Huang T, Poethig RS, Springer PS, Kerstetter RA (2008) KANADI1 regulates adaxial–abaxial polarity in Arabidopsis by directly repressing the transcription of ASYMMETRIC LEAVES2. Proc Natl Acad Sci U S A 105: 16392–16397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Jiao Y (2019) The diverse roles of auxin in regulating leaf development. Plants (Basel) 8: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Sarmiento-Manus R, Yamori W, Ponce MR, Micol JL, Tsukaya H (2018) The Arabidopsis phyB-9 mutant has a second-site mutation in the VENOSA4 gene that alters chloroplast size, photosynthetic traits, and leaf growth. Plant Physiol 178: 3–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.