Abstract
We developed a fully-automated dicentric chromosome assay (DCA) in multiwell plates. All operations, from sample loading to chromosome scoring, are performed, without human intervention, by the second-generation Rapid Automated Biodosimetry Tool II (RABiT-II) robotic system, a plate imager and custom software, FluorQuantDic. The system requires small volumes of blood (30 μl per individual) to determine radiation dose received as a result of a radiation accident or terrorist attack. To visualize dicentrics in multiwell plates, we implemented a non-classical protocol for centromere FISH staining at 37°C. The RABiT-II performs rapid analysis of chromosomes after extracting them from metaphase cells. With the use of multiwell plates, many samples can be screened at the same time. Thus, the RABiT-II DCA provides an advantage during triage when risk-based stratification and medical management are required for a large population exposed to unknown levels of ionizing radiation.
INTRODUCTION
After a large-scale radiological or nuclear event, significant numbers of people may be exposed to ionizing radiation. These individuals will require quick and reliable dose assessment for risk-based stratification of the radiation-exposed population. Automated biodosimetry assays, based on scoring of cytogenetic end points in multiwell plates (1), have the potential to provide dose assessments for thousands of individuals per day.
Previously, at the Center for Radiological Research we developed an automated platform for biological dosimetry, the second-generation Rapid Automated Biodosimetry Tool II (RABiT-II) (2). The RABiT-II uses a commercially-available high-throughput screening (HTS) system, consisting of robotics, liquid handling devices and imagers. HTS systems are widely used to perform chemical, genetic or pharmacological tests in universities and in the pharmaceutical industry. Implementing biodosimetry assays on these platforms will enable their wide deployment, greatly increasing the capacity for running these assays in an emergency. Practically, any HTS system that meets certain minimal criteria (e.g., the availability of an automated incubator) can be assembled as a RABiT-II system.
The dicentric chromosome assay (DCA) is based on scoring dicentric chromosomes in irradiated samples, which are aberrant chromosomes with two active centromeres. This assay achieves highly reproducible results and is recognized as the gold standard [International Atomic Energy Agency (IAEA), International Organization for Standardization (ISO)], which is recommended as a reference method for validating other biodosimetry assays (3). The main disadvantage of this method is that it is labor-intensive, time-consuming and requires experienced scorers. Even large cytogenetic laboratory networks, using multiple satellite scoring locations, can only analyze some hundreds of samples per day (4, 5). Use of the RABiT-II system can increase throughput of the DCA because many samples can be run at the same time.
In general, centromeres can be visualized using the fluorescence in situ hybridization (FISH) technique with peptide nucleic acid (PNA) or bridged nucleic acid (BNA) probes (Fig. 1) (6). In the RABiT-II system, images of the chromosomes labeled with PNA or BNA can be acquired using a 96-well plate imager, making automated recognition of the dicentric chromosomes straightforward. However, there are two technical issues that potentially limit the application of the DCA with FISH probes on the RABiT-II system.
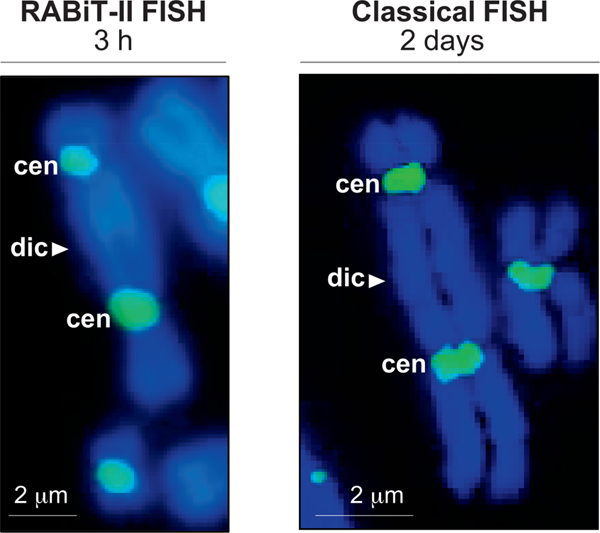
FIG. 1.
Comparison of the RABiT-II and the classical FISH staining with centromere probes. Panel A: Representative image of a dicentric stained with centromere probe at 37°C (without rehydration of samples in PBS, incubation in formaldehyde, pepsin treatment, dehydration in ethanol series and washes). Image captured at 20× magnification (BioTek Cytation Cell 1). Scale bar 2 μm. Panel B: Representative image of a dicentric after staining following classical FISH protocol at 80°C (including rehydration of samples in PBS, incubation in formaldehyde, pepsin treatment, dehydration in ethanol series and washes). Image captured at 63× magnification (Metafer 4 Master Station). Scale bar = 2 μm; dic = dicentric chromosome; cen = centromere.
The first issue is that for fluorescent staining in classical FISH protocols, the chromosomes are denatured at high temperatures (approximately 80°C) to allow access of the fluorescent probes to the centromeres. For our implementation of the RABiT-II, a plate heater cannot be utilized due to the risk that the plastic multiwell plates will be deformed by the high temperatures. The second issue is that traditionally, the dicentric analysis requires observation of the whole nuclei to identify the dicentrics (7). Considering the large number of various elements in the mitotic figure (chromosomes, fragments, etc.), the parallel analysis of multiple samples is a difficult task that requires significant computational resources (8).
To overcome these issues and to allow automated DCA using the RABiT-II system, we modified the procedure for staining centromeres as well as the general approach of scoring chromosomes. Specifically, we developed a protocol for centromere FISH staining at low temperatures (not higher than 37°C) that can be realized in plastic plates of the RABiT-II system. Additionally, to simplify the automated image processing, instead of analysis of whole mitotic cells, we performed a disruption of mitotic cells using mild acidic treatment that releases all chromosomes from metaphase nuclei (Fig. 2A). After this, recognition of dicentrics becomes a relatively easy task for an automated identifier: the software needs to detect only individual chromosomes and count the number of centromeres in each, an easier alternative for automated DCA (Fig. 2B).
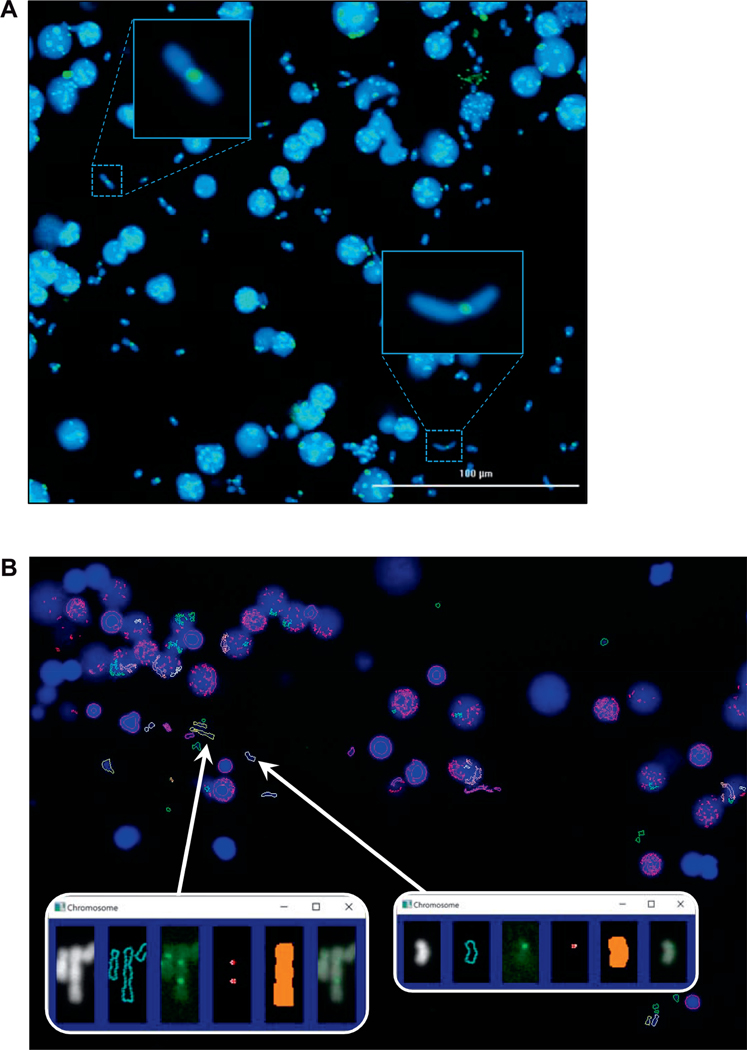
FIG. 2.
Automated detection of dicentrics in the RABiT-II system. Panel A: Chromosomes released from mitotic cells (30 μl of whole blood), 0 Gy sample. Panel B: Output from the image analysis software FluorQuantDic version 3.3. Large blue objects are intact nuclei. White outlines correspond to identified monocentric chromosomes. Red outlines are objects which are either too big or too small to be scored as chromosomes. Green objects are too round to be chromosomes. Yellow objects are identified dicentric chromosomes. Inserts are identified dicentric (left) and monocentric (right) chromosomes. Panels from left to right: chromosome image (DAPI); the identified chromosome outline; centromere signal (FITC); identified centromeres; the mask for selecting which centromeres are associated with the chromosome; a merged DAPI/FITC image.
Here, we introduce the robotic-compatible DCA and describe its implementation on the RABiT-II system. We perform fundamental comparisons between the results achieved using the RABiT-II DCA and a conventional DCA and discuss the application of our system to actual triage situations.
MATERIALS AND METHODS
Ethics Statement
All studies were performed in accordance with the International Ethical Guidelines for Epidemiological Studies (9) and approved by an Institutional Review Board at Columbia University (Protocol approval number AAAR0643). Unless otherwise noted, all reagents and plasticware were purchased through Thermo Fisher Scientific™, Inc. (Waltham, MA).
Elements and Programs of the RABiT-II System
In this work we used a cell::explorer HTS system (PerkinElmer® Inc., Waltham, MA) housed at the Columbia Genome Center (New York, NY). This system consists of sub-stations surrounding an anthropomorphic robotic arm (Denso Inc., Long Beach, CA). A list and descriptions of relevant elements can be found elsewhere (2). A human operator defines the protocol and coordinate sub-stations using PerkinElmer’s plate::works automation control and scheduling software. A BioTek® Cytation™ 1 Cell Imaging Multi-Mode Reader (Thermo Fisher Scientific) was used as a plate imager; alternatively, other microplate imagers (e.g., the GE InCell Analyzer (Pittsburgh, PA), or the Perkin Elmer Operetta) can be used. MetaSystems Group, Inc. (Waltham, MA) in principle, also offers an imager for inverted 96-well plates.3 However, none of these imagers were tested at the time of writing this manuscript.
Blood Sampling and Irradiation
Whole blood from healthy human donors was collected after informed consent (IRB approval no. AAAR0643) into heparinized vacutainer tubes. Aliquots (1 ml) were pipetted into 2D-barcoded tubes (Matrix Storage Tubes) and transported to a Gammacell®−40 cesium-137 (137Cs) irradiator (Atomic Energy of Canada, Ltd., Chalk River, Canada). Samples were γ-ray exposed to 0, 2, 4, 6, 8 and 10 Gy at a dose rate of 73.0 cGy/min (0.73 Gy/min) at a height of 2 mm in the chamber. The Gammacell-40 calibration is verified annually by the chief medical physicist at Columbia University Irving Medical Center’s Department of Radiation Oncology using thermoluminescent dosimeters (TLDs) placed at several locations on the floor and inside of the irradiator
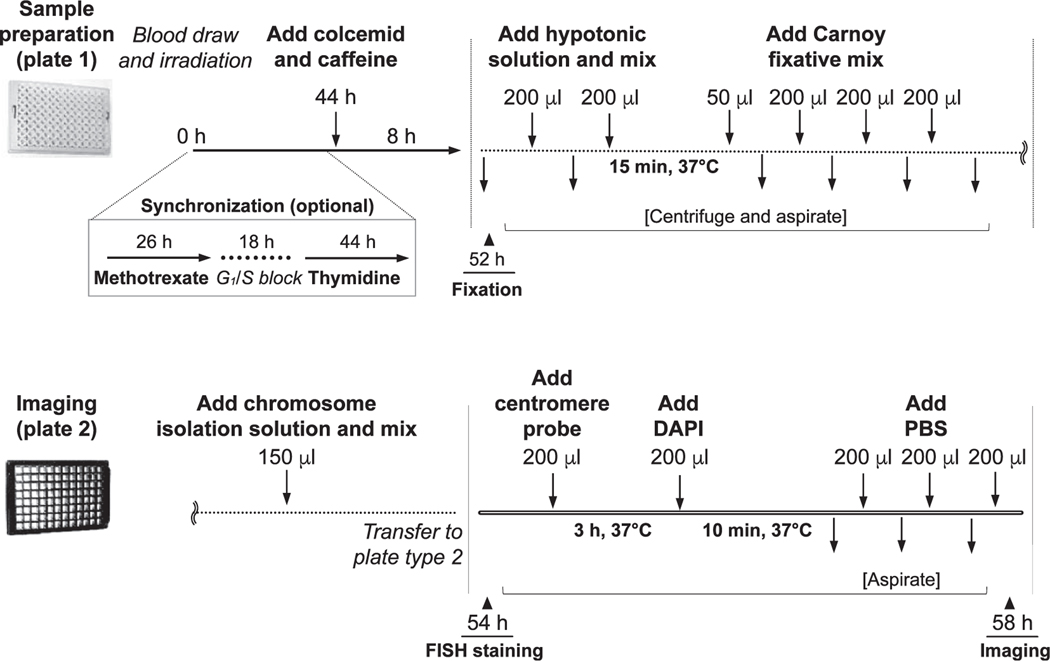
RABiT-II Blood Culturing and Fixation
After γ-ray irradiation, 30-μl blood aliquots were loaded into a 96-well plate (Corning 96-well Expanded Volume Polypropylene Not Treated Microplate, Standard Height, V-Bottom; Corning Inc, Corning, NY) preloaded with 270 μl of PB-MAX karyotyping media (final volume 300 μl/well). Plates were placed into the system incubator at 37°C with 5% CO2 humidified air. After 44 h, 40 μl of the “old” PB-MAX media was replaced automatically with 40 μl of fresh media containing 30 μl colcemid and 10 μl caffeine (0.1 μg/ml and 0.3 mg/ml final concentrations, respectively). Cells were returned to the incubator and cultured for an additional 8 h (total incubation time of 52 h), then harvested, treated in 150 μl 0.075 M potassium chloride for 15 min at 37°C to swell the cells, and fixed three times with 3:1 methanol:acetic acid (200 μl each). After fixation, the plates containing cell pellets (50 μl) were either immediately processed by FISH staining and acquisition, or sealed with aluminum microplate seals and stored at 4°C for future use.
Quality Control of the RABiT-II DCA Protocol
Whole blood cell incubation time in the RABiT-II DCA protocol is 52 h, which is longer than the standard 48 h. To assess cell cycle kinetics, the blood cells were cultured in a moderately toxic concentration of 5-bromo-2′-deoxyuridine (BrdU, final concentration 7 μg/ml; Sigma Aldrich®, St. Louis, MO) as described elsewhere (10). For 48-h and 52-h cultures, BrdU was added at the beginning of the cultures. As a positive control we included longer cultures (72 h) where BrdU was added after 24 h of culture initiation. The substitution of BrdU was confirmed using staining with Hoechst 33258 (final concentration 0.5 μg/ml) as described elsewhere (11). For analysis, a total 1,000 metaphases selected at random were checked for presence first-, second- and third-division metaphases.
RABiT-II Cell Synchronization
As will be shown below, chromosome morphology plays an important role in the ability of the RABiT-II system to detect dicentric chromosomes. Synchronization increases the fraction of long chromosomes (12) suitable for the RABiT-II chromosome identifier software. To accomplish this, the RABiT-II system can be re-programmed to arrest cells at the G1/S border by a temporary block of DNA replication with methotrexate. To initiate the block, 30 μl of old PB-MAX should be replaced by 30 μl of fresh media containing methotrexate (0.1 μM final concentration; Sigma Aldrich) at 26 h after initiation of cell culture. To release cells from block and accumulate mitotic nuclei, a similar replacement should be performed 18 h later (44 h of incubation) with 70 μl of fresh PB-MAX media containing thymidine, colcemid and caffeine (10 μM, 0.1 μ/ml and 0.3 mg/ml final concentrations, respectively). Synchronization is an optional procedure and can be omitted.
RABiT-II Extraction of Chromosomes
To release chromosomes from mitotic nuclei, the fixed cells (50 μl) in multiwell plates were mixed with 150 μl of 1:1 acetic acid:-deionized water (total 200 μl) and pipetted 10 times. Then samples were transferred by dispenser from plastic plates to glass-bottom 96-well plates (Brooks Automation Inc., Chelmsford, MA) preloaded with 200 μl of 3:1 methanol:acetic acid. After centrifugation of the plates for 4 min at 250g, the intact nuclei and chromosomes were attached to the surface of glass-bottom multiwell plates. The remaining liquid was aspirated and plates were allowed to dry for 10 min at room temperature.
RABiT-II Centromere FISH Staining without Heat Denaturation
After complete evaporation of fixative, 200 μl of the centromere staining solution, consisting of 90% formamide, 2× saline sodium citrate buffer [0.33 M sodium chloride in a 0.03-M sodium citrate buffer (pH 7.0)], and 0.025 μg/ml–1 of centromere probe was directly added to each well without pre-hybridization and denaturing steps. In this study we tested several probes for low-temperature centromere FISH:
PNA (CENP-B): 5′ ATTCGTTGGAAACGGGA [FAM] 3′ (PNA Bio, Newbury Park, CA);
BNA with 7 bases: 5′ +A+TT+CG+TTGGAA+AC+GG+GA [FAM] 3′ (Bio-Synthesis, Lewisville, TX); and
BNA with 9 bases: 5′ +A+TT+CG+TT+GG+AA+AC+GG+GA [FAM] 3′ (Bio-Synthesis),
where +N is an incorporated BNA base.
The probes were allowed to hybridize for 3 h at 37°C. Then 200 μl of DAPI (1.5 μg/ml) was dispensed into plates and mixed with the centromere staining solution. After 10 min of incubation, the liquid (400 μl) was aspirated from wells, plates were rinsed two times with 200 μl of PBS, and 200 μl of fresh PBS was loaded for imaging.
RABiT-II Image Acquisition and Chromosomal Analysis
After extraction of chromosomes and centromere staining, color image acquisition was performed on the DAPI and FITC channels using a low-magnification objective lens (20×). Automated processing of one well (60 mm2) takes approximately 1 h. A total of 450 paired DAPI and FITC images (size 380 × 280 μm, 1,152 × 832 pixels, 16 bit) were obtained and stored in the image gallery. Upon completion of acquisition, all captured images were automatically screened for normal and aberrant chromosomes using a custom software program (written in-house), FluorQuantDic version 3.3. This software written in Visual C++ (Visual Studio 2013; Microsoft® Corp., Redmond, WA), using the OpenCV version 3.2 open source computer vision libraries (www.opencv.org). The software identifies the chromosomes extracted from mitotic nuclei (DAPI images) and then looks for bright foci (centromeres) overlapping the chromosomes (FITC images). The DAPI images were binarized using an adaptive thresholding segmentation algorithm, which assigns each pixel a value of 1 (if it is significantly brighter than pixels in a 15 × 15-pixel neighborhood) or zero otherwise. Detected chromosomes are then located as the binary large objects (BLOBs) using the algorithm described elsewhere (13). Only chromosomes between 10 and 200 pixels (0.35–7 μm2) were chosen for analysis. Clustered chromosomes (>5 within a 100 × 100-pixel square) and chromosomes touching the image edges were rejected. Centromeres were identified from the FITC channel by binarizing a small section of the image, corresponding to the identified area of a single chromosome, using Otsu’s algorithm (14) with a threshold multiplier of 1.2, and BLOBs smaller than 20 pixels (0.7 μm2). Validated objects were classified as acentric, monocentric, dicentric or multicentric chromosomes based on the number of detected spots. Where possible, multithreading was used to allow several chromosomes on the same image to be analyzed simultaneously on a multi-core processor. Image analysis for one well took approximately 10 min on a Dell Precision desktop with a quad-core Xeon E5–1620 version 3 processor (B&H Foto & Electronics Corp., New York, NY).
Classical DCA and Chromosomal Analysis
Classical DCA was performed in parallel with the RABiT-II DCA using aliquots from the same blood samples. After irradiation, whole blood (0.5 ml) was cultured in cell culture flasks containing 4.5 ml of PB-MAX media in a humidified 5% CO2 incubator at 37°C. Metaphase preparations and centromere staining were performed manually using standard protocols, as described elsewhere (15). Briefly, the samples were spread onto glass microscope slides and air-dried overnight. On the next day, slides were rehydrated in PBS, fixed in 4% formaldehyde, washed with PBS, digested with HCl-pepsin solution, washed with PBS and dehydrated in a 70%–85%–100% ethanol series. Chromosomes fixed on glass slides were denatured 3 min by heat (80°C) in the presence of 20 μl of FITC-labeled PNA probe and kept in the dark for 3 h. After hybridization, the slides were washed two times in 70% formamide and TBS, with 0.05% Tween™−20 solutions at room temperature. Samples were counterstained with Vectashield® Antifade Mounting Medium containing DAPI. Metaphase cell images were acquired on a Metafer 4 Master Station (MetaSystems) consisting of an automated acquisition module AutoCapt version 3.9.1, a Zeiss Plan-Apochromat 63x/1.40 oil immersion objective and a CoolCube 1 Digital High Resolution CCD Camera. Chromosomal analysis was performed using Isis software (MetaSystems). Scoring of dicentric chromosomes was performed by an experienced scorer according to the IAEA manual (3). Analysis was performed in complete metaphases containing 46 centromeres; each dicentric was accompanied by an acentric fragment. A total 50 cells [sufficient for triage (16)] per dose point were scored. All samples were blinded to the scorer and decoded after analysis. Dicentric chromosome yields were adjusted by the number of multicentric configurations according to IAEA recommendations (tricentric chromosome is equal to two dicentrics, etc.) (3).
Reference Dose-Response Curves and Statistical Analysis
Generation of preliminary dose-response calibration curves for both assays, their validation and statistical analysis was performed using dose estimate software version 5.2 (17). The dicentric yield for each dose point was calculated by dividing the total number of dicentrics by the entire number of metaphases or chromosomes (depending on the assay type) counted per dose point. The coefficients of the fitted curves were derived for collectively pooled individuals. The dose-effect relationship was determined by the coefficient of correlation (r). The 95% confidence intervals (CI), goodness-of-fit parameters, chi-squared test, dispersion index (σ2/y) and its normalized unit (u) were evaluated to indicate consistency of the data with the linear-quadratic model and its conformity with the Poisson distribution. Due to an inability to access individual cell statistics in the RABiT-II DCA, the dispersion index and u value were calculated only for the classical DCA.
Validation of the Curves and Intercomparison of the Assays
Validation of the dose-response curves utilized whole blood from two donors (male and female) that was not used to generate the calibration curves. Blood aliquots (1 ml) were irradiated to 1, 3, 5 and 8 Gy in 2D-barcoded tubes. After irradiation, one half of the blood (0.5 ml) was used for classical DCA, and the rest for RABiT-II DCA. Both assays were performed side-by-side as described in previous sections. After centromere staining, the multiwell plates were analyzed automatically using the FluorQuantDic software, while the microscope slides with metaphase spreads were examined manually by an investigator blinded to the doses and mode of exposure. In total, one well per individual (RABiT-II DCA) or 50 metaphase cells per sample (classical DCA) were examined. After scoring, the doses from each blinded sample were calculated from generated dose-response curves using the Dose Estimate software. For both assay types, the mean of the calculated dose estimations (8 replicates per dose point) and the corresponding 95% CI were determined to achieve accuracy of the dose prediction.
RESULTS
FISH Probes Bind to Centromeres without Heat
Since the RABiT-II system can only provide temperatures of 25°C (room temperature) or 37°C (in the automated incubator), it cannot perform the standard centromere FISH staining protocol, which requires heating to 80°C. Therefore, instead of thermal denaturation, we used a chemical denaturation to allow the probes to access the target sequences on the centromeres. Various probes (based on BNA and PNA chemistries; see Methods) were tested for their ability to bind centromeres at a low temperature. Considering the short length (17 nucleotides) and G/C content (47.1%) of our probes, we calculated that hybridization at 37°C in a standard 2× SSC buffer would be more favorable at higher concentrations of formamide (more than 80%). The strength of the fluorescent signal was assessed for different hybridization times and variable probe cocktail compositions (i.e., formamide, SSC and probe concentration). The BNA probe with 7 bases did not hybridize well under any conditions. The BNA probe with 9 bases and the PNA probe worked in a range of formamide concentrations from 80% to 90% and gave a strong signal with low background after 3 h of incubation at 37°C when the concentration of formamide was equal to 90%. After 3 h of staining under the same conditions (0.025 μg/ml–1 of probe), BNA probes gave a cleaner and brighter (1.78 fold) peak intensity signal on chromosomes than PNA probe (data not shown). Extension of the hybridization time (overnight at 37°C) increased the signal. Additionally, to accelerate staining, we excluded from our FISH protocol all pre-treatment, post-treatment and washing steps that are traditionally used in conventional protocols. Correspondence between our staining method (no pepsin, no ethanol treatment, no washes, centromere labeling at 37°C) and classical PNA-FISH staining (including pepsin and ethanol treatment, washes and centromere labeling at 80°C) is demonstrated in Fig. 1. In both cases we observed bright fluorescence of centromere regions with no lacking regions or background fluorescence over the chromosome length.
Dicentric Analysis in Multiwell Plates
The initial time course for the RABiT-II DCA has been considered, which follows the IAEA recommendations (3). Preliminary experiments showed insufficient yield of chromosomes from severely damaged cells (at or above 6 Gy). To enable analysis at high doses, we treated the cells with caffeine (18), which is known to abrogate the ATM-mediated G2/M checkpoint and increase the number of damaged cells that can progress to mitosis (19). The effect of caffeine was determined at the end of cell culturing by scoring mitotic figures without disruption of metaphases. Compared to control samples (without caffeine), the mitotic index of blood cells irradiated to 10 Gy increased approximately threefold in the presence of caffeine (Fig. 3A). Additionally, to increase the yield of chromosomes we extended colcemid treatment time from 4 h (standard 48 h protocol) to 8 h (termination of culture at 52 h). Prolonged treatment resulted in an approximately 15.5× higher number of metaphases available for analysis (data not shown). At the moment of termination of cultures (52 h), we observed 93.8% and 6.2% metaphases in the first and second cell divisions, respectively (average of two donors, one male and one female) (Fig. 3B, Supplementary Table S1; http://dx.doi.org/10.1667/RR15266.1.S1). Preliminary microscopic examination of samples without disruption of mitotic cells revealed considerable metaphase-to-metaphase variation in chromosomal shapes in the same well (Supplementary Fig. S1). Frequently observed morphologies were classified into several groups (Fig. 4A), roughly corresponding to the sub-phases of mitosis previously described by Rieder et al. (20) (Supplementary Fig. S1). We attributed the presence of different chromosomal morphologies in the same sample to the asynchronous growth conditions of blood cultures. While most of the “linear” chromosomes were recognized by our software effectively (Fig. 2B), the X-shaped chromosomes caused complications in the automated recognition (Fig. 4B). Thus, to collect chromosomes with relatively uniform shapes (long, mildly condensed, with closed arms) that are in the early stages of mitosis, we synchronized cell population with methotrexate and thymidine (12) in a 96-well plate. In general, the proportions of mitotic cells in synchronized and asynchronized cultures were comparable to each other (Fig. 4C). Overall, synchronization resulted in several notable effects. First, the number of chromosomes with uniform shape increased (Fig. 4D). Specifically, the number of chromosomes detectable by the software (long with closed arms) was increased from 30% to 87%; in contrast, the number of chromosomes with unpaired arms (not detectable) decreased from 8% to only 1% (Fig. 4E and Supplementary Table S2). Second, the average length of chromosomes (based on length of largest human chromosome 1) was approximately 2× longer after synchronization than before (7.1 μm vs. 3.6 μm; data not shown). In multiple technical repeats, we observed that a drop of whole blood (30 μl) from a healthy individual contains thousands of chromosomes suitable for analysis (Fig. 2A). An established outline of the RABiT-II DCA protocol is shown in Fig. 5.
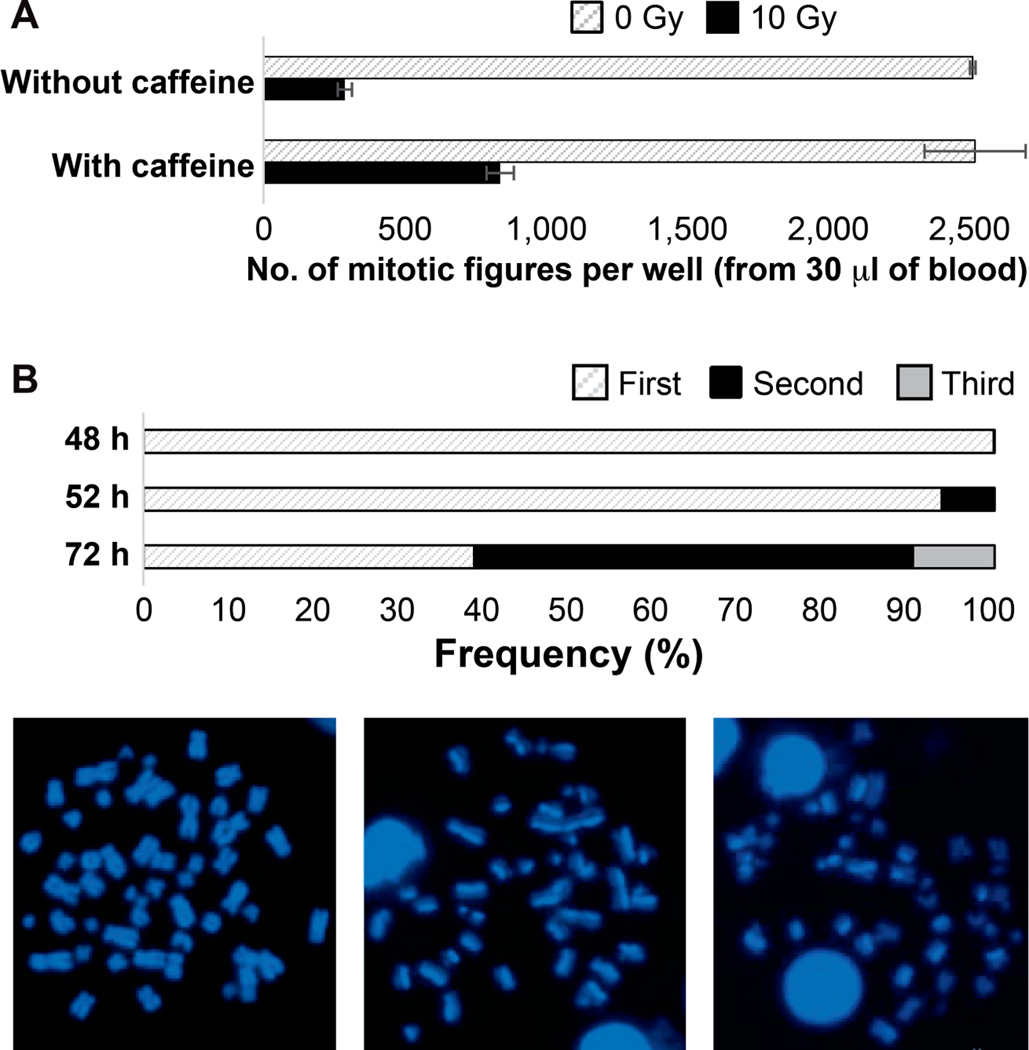
FIG. 3.
Assay development and quality control. Panel A: Caffeine allows G2/M arrested lymphocytes to enter mitosis after severe DNA damage. Quantification data represents total number of mitotic figures scored in one well after 10 Gy irradiation without (286 mitotic cell spreads per well) or with (834 mitotic cell spreads per well) caffeine. Data and standard errors were calculated based on three independent experimental trials. Panel B: Frequency of second division metaphases in the RABiT-II DCA protocol after 52 h of culture. Images represent differentiation of sister chromatids by 33258 Hoechst fluorescence after different cycles of BrdU incorporation: first (bifiliar BrdU incorporation, on the left), second (unifiliar BrdU incorporation, in the middle), and third (one fourth BrdU incorporation, on the right) cell cycle metaphase spreads.
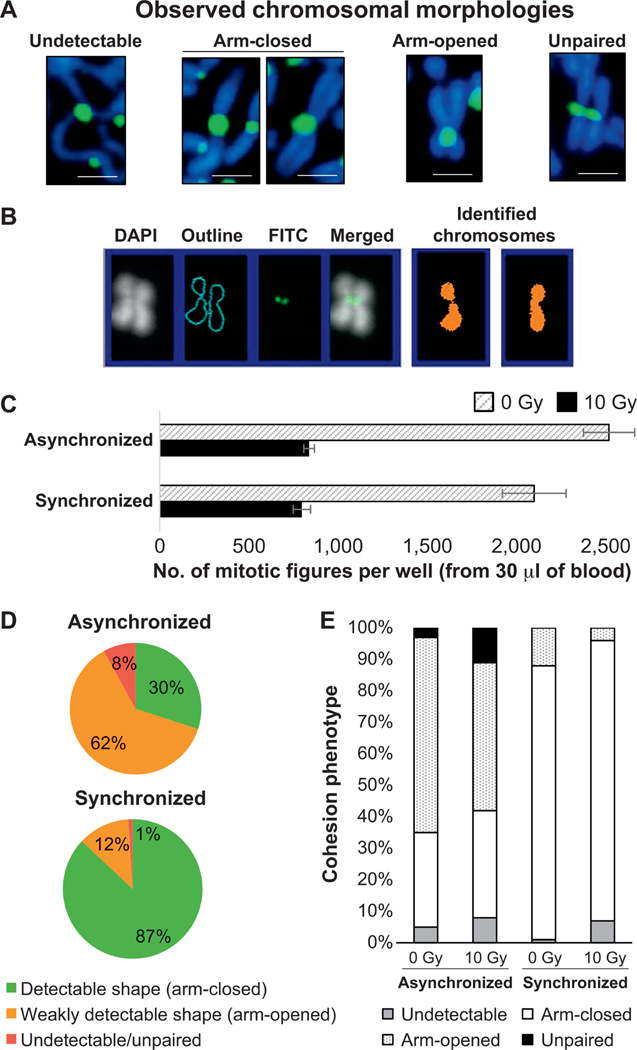
FIG. 4.
Uniform shapes of chromosomes after synchronization. Panel A: Common chromosomal morphologies frequently observed in the same well (from drop of blood from healthy volunteer, no irradiation, asynchronously cultured, 8 h incubation with colcemid). Images representing chromosome 1 were cropped and magnified from Supplementary Fig. S1 (http://dx.doi.org/10.1667/RR15266.1.S1). Panel B: FluorQuantDic classifier misrecognized the chromosome with opened arms as two chromosomes with closed arms. Panel C: Quantification data of mitotic figures before and after synchronization. Standard errors were calculated based on three independent experimental trials. Panel D: Proportion of mitotic cells with variable shapes of chromosomes (that can or cannot be detected using FluorQuantDic software) before and after synchronization. Panel E: Quantification results for distribution of chromosomal shapes before and after synchronization calculated for 1,000 prometaphase cells (0 Gy and 10 Gy irradiated samples). White and dashed populations represent cells with closed and open arms, respectively. Dark gray and black bars refer to undetectable or unpaired populations, respectively.
FIG. 5.
Schematic of the end-to-end automated RABiT-II DCA. The operating parameters are divided into several subprograms, which allows the user to perform various tasks including routine controls of the cell culture, synchronization, harvesting, fixation, staining and sample imaging.
Intercomparison with Conventional Method
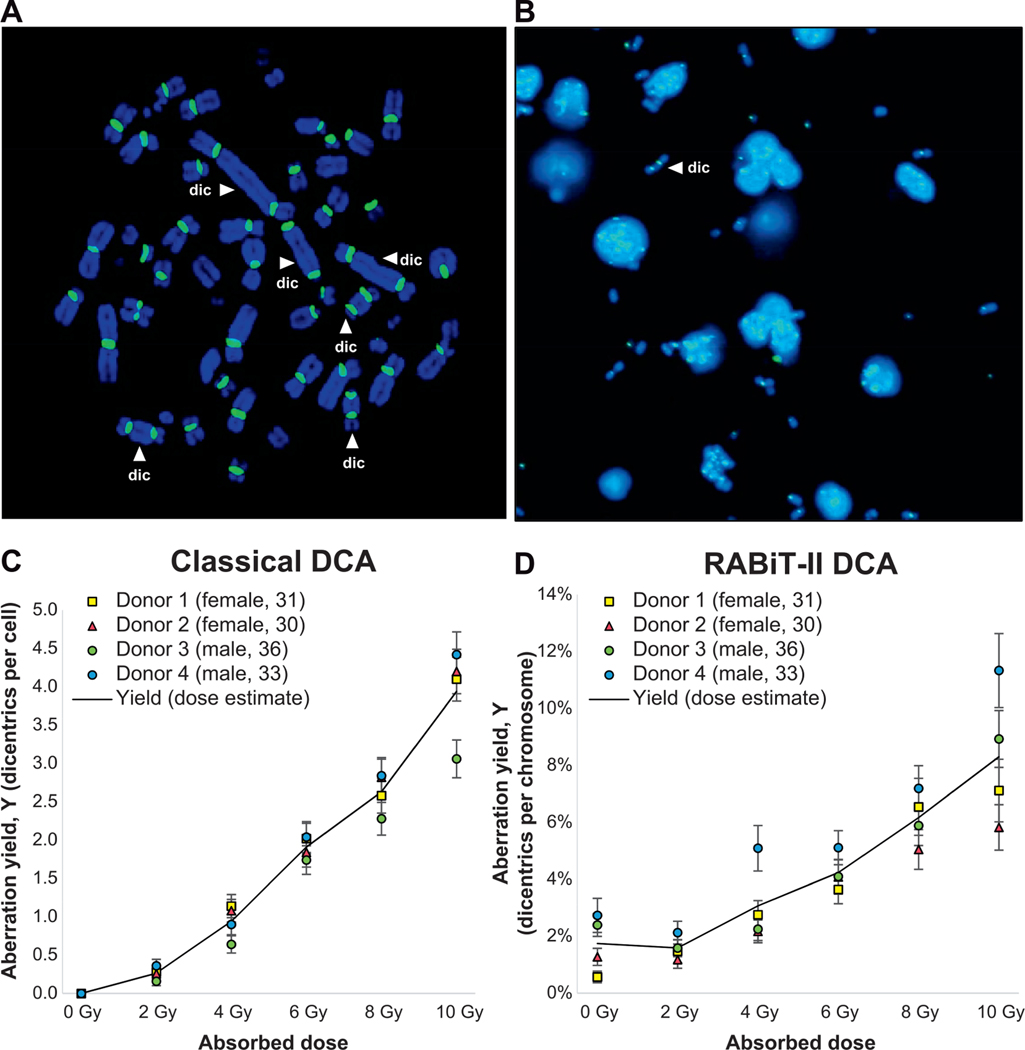
After establishing the RABiT-II DCA, we analyzed the agreement between our assay and the classical DCA. Both assays were performed side-by-side, using aliquots from the same irradiated blood from four individuals. Scoring in the classical DCA was performed manually (Fig. 6A), while in the RABiT-II DCA scoring was fully automated (Fig. 6B). The raw data for the RABiT-II DCA and classical DCA are provided in the Supplementary Tables S3 and S4, respectively (http://dx.doi.org/10.1667/RR15266.1.S1). The results of pooled scoring for the four individuals indicate an increase of dicentric yields with dose up to at least 8 Gy (Tables 1 and 2). For the RABiT-II DCA we assessed the number of chromosomes scored and the frequency and yields of dicentric chromosomes among them (Table 1). For the classical DCA we were able to assess additional parameters, such as intercellular distribution of dicentrics and Pearson’s correlation coefficients (Table 2). For the majority of data points, the dispersion index values were close to 1 and u values were between −1.96 and 1.96, confirming good agreement with a Poisson distribution. Deviations from the Poisson distribution were observed only at 10 Gy (u value of −2.81), which is not uncommon for high radiation doses (21). Dose-response calibration curves for the classical and the RABiT-II DCA are shown in Fig. 6C and D, respectively. Reproducibility between technical repeats is characterized by small uncertainty at each dose point. The resultant linear and quadratic yield parameters of the fitted curves and goodness-of-fit test results were calculated using dose estimate software (Table 3). For both assays, the data fitted to linear-quadratic equation, Y = C + αD + βD2, and the yield of dicentric chromosomes increased with the dose (r = 0.9973 and r = 0.9977).
FIG. 6.
Dose-response calibration curves for the RABiT-II and classical DCA. Panel A: Dicentric scoring strategy in the classical DCA. Representative example of a metaphase cell (10 Gy irradiated sample) suitable for analysis (complete set of centromeres, dicentrics are accompanied by acentric fragments, etc.). For each sample, analysis was performed in 50 metaphases using Isis software. Distribution of identified dicentrics (indicated by white arrows) was recorded; multicentrics are converted to the dicentric chromosome equivalents. Panel B: Dicentric scoring strategy in the RABiT-II DCA. Representative image (4 Gy sample) used for analysis. Scoring was performed automatically by the FluorQuantDic software (450 images per sample). A dicentric is indicated by the white arrow. Panel C: Preliminary dose-response curve for 137Cs γ rays for classical DCA. The left axis shows the yield of DCs as a fraction of 50 cells scored. Panel D: Preliminary dose-response curve for 137Cs γ rays performed using the RABiT-II system and scored without human intervention. The left axis shows the yield of dicentrics as a fraction of normal chromosomes.
TABLE 1.
Frequencies of Dicentric Chromosomes, Scored Using RABiT-II DCA for Different Gamma-Ray Doses Detected with FluorQuantDic Version 3.3
| Dose (Gy) | Chromosomes | Dicentric chromosomes | Yield ± standard error |
|---|---|---|---|
| 0 | 4,997 | 77 | 0.015 ± 0.002 |
| 2.0 | 5,693 | 93 | 0.016 ± 0.002 |
| 4.0 | 4,879 | 142 | 0.029 ± 0.002 |
| 6.0 | 5,279 | 227 | 0.043 ± 0.003 |
| 8.0 | 4,126 | 255 | 0.062 ± 0.004 |
| 10.0 | 3,062 | 251 | 0.082 ± 0.005 |
Note. Data pooled from four donors: two females (donors 1 and 2), ages 31 and 30, respectively; and two males (donors 3 and 4), ages 36 and 33, respectively.
TABLE 2.
Classical DCA Frequencies and Intercellular Distribution of Dicentrics for Different Gamma-Ray Doses Scored Manually
| Distribution of dicentrics |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dose (Gy) | Cells | DC | D0 | D1 | D2 | D3 | D4 | D5 | D6 | D7 | D8 | D9 | Y ± SE | σ2/y | u test |
| 0 | 200 | 0 | 200 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.000 ± 0.000 | 1.00 | 0.00 |
| 2.0 | 200 | 53 | 150 | 47 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.265 ± 0.036 | 0.85 | –1.49 |
| 4.0 | 200 | 188 | 81 | 70 | 32 | 14 | 3 | 0 | 0 | 0 | 0 | 0 | 0.940 ± 0.069 | 1.04 | 0.44 |
| 6.0 | 200 | 382 | 19 | 60 | 70 | 35 | 7 | 6 | 2 | 1 | 0 | 0 | 1.910 ± 0.098 | 0.81 | –1.88 |
| 8.0 | 200 | 526 | 18 | 30 | 48 | 52 | 28 | 15 | 7 | 1 | 1 | 0 | 2.630 ± 0.115 | 0.95 | –0.55 |
| 10.0 | 200 | 789 | 2 | 7 | 33 | 45 | 38 | 42 | 18 | 10 | 4 | 1 | 3.940 ± 0.140 | 0.72 | –2.81 |
Notes. Data pooled from four donors: two females (donors 1 and 2), ages 31 and 30, respectively; and two males (donors 3 and 4), ages 36 and 33, respectively. DC=dicentric chromosomes; Y ± SE: yield ± standard error; σ2/y=dispersion index (σ=variance, y=mean); u=normalized unit of the dispersion index.
TABLE 3.
Comparison of the RABiT-II DCA and Classical DCA Linear and Quadratic Yield Coefficients and Goodness-of-Fit Parameters Calculated with the Dose Estimate Software
| Assay type | c ± SE | α ± SE | β ± SE | χ2 | DF | P value | r |
|---|---|---|---|---|---|---|---|
| Classical DCA | 0.0000 ± 0.0000 | 0.1062 ± 0.0338 | 0.0295 ± 0.0047 | 11.2400 | 3 | 0.0105 | 0.9973 |
| RABiT-II DCA | 0.0145 ± 0.0016 | 0.0008 ± 0.0010 | 0.0006 ± 0.0001 | 2.7720 | 3 | 0.4282 | 0.9977 |
Note. c = Background frequency of dicentrics; SE = standard error; χ = weighted chi squared; DF = degrees of freedom; r = correlation coefficient.
Validation
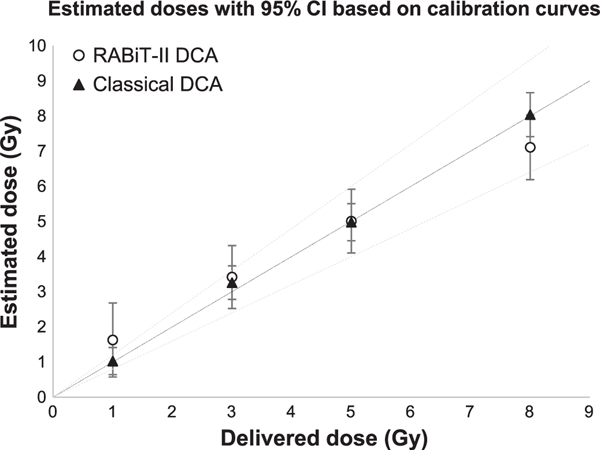
To assess reproducibility of dose-response calculations and compare prediction accuracy for RABiT-II DCA and classical DCA, we irradiated blood samples from two additional individuals and performed a side-by-side blind test using both assays. Estimated doses were calculated manually based on 50 cells per data point (classical DCA) or automatically based on the fraction of dicentric chromosomes identified among 450 images (one well) per data point (RABiT-II DCA). Table 4 summarizes whether the actual delivered dose fell within the 95% CI of the dose, as estimated using the Dose Estimate software. While for classical DCA estimated doses showed good agreement with delivered doses, reliable dose confirmation for the RABiT-II DCA was possible at doses higher than 3 Gy (Fig. 7).
TABLE 4.
Dose Estimation of the Blind Test Based on Gamma-Ray-Induced Dicentric Yields for Validation of the Reference Rabit-II and Classical Dose-Response Curves
| Assay | Delivered dose (Gy) | Chromosomes/cells | DC | Estimated dose (Gy) | 95% CI |
|
|---|---|---|---|---|---|---|
| Lower | Upper | |||||
| RABiT-II DCA |
1.00 | 4,654 | 81 | 1.63 | 0.67 | 2.85 |
| 3.00 | 2,885 | 70 | 3.42 | 2.13 | 4.57 | |
| 5.00 | 2,266 | 76 | 5.01 | 3.84 | 6.13 | |
| 8.00 | 3,264 | 165 | 7.11 | 6.27 | 7.96 | |
| Classical DCA |
1.00 | 50 | 7 | 1.03 | 0.47 | 1.81 |
| 3.00 | 50 | 33 | 3.26 | 2.52 | 4.09 | |
| 5.00 | 50 | 63 | 4.98 | 4.21 | 5.81 | |
| 8.00 | 50 | 138 | 8.04 | 7.25 | 8.87 | |
Note. DC = dicentric chromosomes; CI = confidence interval.
FIG. 7.
The reliability of individual dose estimations by RABiT-II DCA and classical DCA. Four dose estimations of samples from two different donors (male and female, 29 and 30 years old, respectively) and the corresponding 95% CI were determined at each dose. The RABiT-II DCA and classical DCA calibration curves were used for dose-prediction calculations. Reconstructed doses are based on eight replicates for each donor; black line is an ideal reconstruction (reconstructed dose = physical dose) and dashed lines are ±20%.
DISCUSSION
In this work we developed a high-throughput DCA for rapid triage in the event of a radiation mass casualty scenario. Our goal in developing this assay was to identify those individuals at risk for acute radiation syndrome, having received an absorbed dose of more than 2 Gy. The RABiT-II system screens radiation-induced dicentric chromosomes in a small amount of blood (30 μl) sampled from individuals who may been exposed to an unknown dose of ionizing radiation (96 people per plate). The entire DCA procedure, including staining of centromeres with FISH probes, was miniaturized to the volume of a single well in a 96-well plate (rather than using tubes or flasks and imaging on microscope slides). The RABiT-II DCA has several principal differences from classical DCA which were implemented for ease of automation (Table 5).
TABLE 5.
Key Differences between Classical DCA and the RABiT-II DCA
| Assay characteristics | Conventional DCA | RABiT-II DCA |
|---|---|---|
| Culturing step | ||
| Cultivation | Tubes/flasks | Multiwell plate (96 samples) |
| Starting blood volume | 500 μl | 30 μl |
| Working volume | 5 ml per tube or flask | 300 μl per well |
| Lymphocyte isolation | Optional | No |
| FISH staining | ||
| Sampling | Glass slide | Multiwell plate |
| Pre-/post-treatment | Yes | No |
| Washes | Yes | No |
| Hybridization temperature | 75–80°C | 37°C |
| Probe composition | With blocking reagents | Formamide only |
| Max time | 1–2 days | 4 h |
| Chromosome analysis | ||
| Magnification | 63× | 20× |
| Target objects | Whole metaphases | Individual chromosomes |
| Parameter | Dicentrics per metaphase | Dicentrics per chromosomes |
| Triage scoring | ||
| Operation | Manual/semi-automated (human) | Automated (RABiT-II) |
| Throughput | Less than 1 h/sample; with QuickScan 20 min/sample | 10 min/sample |
With classical FISH staining, high-temperature (80°C) denaturation is required prior to centromere labeling. We have developed a FISH-staining protocol that does not require temperatures higher than 37°C and can thus be performed in plastic multiwell plates. This technique, based on the non-denaturing FISH-labeling protocol (22–25), allows the probes to bind to centromeres at low temperatures in a simple buffer consisting of only formamide and SSC. In addition, we eliminated time-consuming washes and chromosome treatment (with ethanol and pepsin) from the staining protocol. This reduces the time for centromere labeling: the samples can be processed for image acquisition and dicentric analysis within approximately 3 h after harvesting and fixation, instead of the approximately 2 days required for the conventional centromere FISH protocol.
Chromosomes in the early stages of mitosis (long, with unseparated chromatids) were more easily recognized by our software than those that were more condensed with a higher degree of chromatid separation. The morphology of chromosomes is precisely regulated throughout cell cycle by cohesins, which keep chromosomal arms closed or opened, and condensins, which condense chromosomal DNA (26, 27). Thus, to increase yield of analyzable chromosomes, we synchronized blood cells and harvested them in the early stage of mitosis. This method is normal for routine blood cultures and results in longer chromosomes and more mitotic cells (12).
Traditionally, dicentrics are scored in mitotic cells. This complicates automated analysis and requires manual or semi-automated scoring, whereby a trained human scorer locates dicentric chromosomes within the cell or re-checks candidate dicentrics selected by the software. Rather than analyze whole mitotic figures, our method of dicentric screening focuses on individual chromosomes, extracted from mitotic cells. This approach does not require identification of metaphases suitable for scoring (with a complete set of centromeres, without overlapping chromosomes, etc.), increases the total number of chromosomes participating in analysis and simplifies the software for chromosomal identification. The FluorQuantDic software we have written counts spots (centromeres) instead of analyzing the morphologic configuration of the chromosomes; this method does not require high-resolution imaging. Acquisition at a magnification of 20× has several benefits: a reduced number of images required to cover a single well; shortened imaging time per frame; relaxing the need for focusing due to the higher depth of field of the objective; and eliminating the need for oil immersion, which is complicated in an automated imager.
To determine how well our system compares to classic measurements of radiation-induced dicentrics, we generated dose-response calibration curves for both assays and performed a side-by-side blind comparison of its capability and accuracy to assess radiation dose. We analyzed four dose points (two individuals, two blind doses each) using both assays. The classical DCA determined the doses very accurately. While the RABiT-II DCA partially sacrificed this accuracy, the system was able to complete centromere FISH staining and the measurements in a shorter period of time. In the mass-casualty scenario, the medical planners allow for reduced sensitivity of the DCA to improve throughput of the assay (16). The estimated time to complete screening of four individuals (calibration curve donors) using centromere FISH staining was 60 h and 88 h for the RABiT-II and the classical DCA, respectively (including cell culturing, fixation, staining with centromere probes, acquisition and dicentric analysis). Comparison was performed using centromere FISH staining, whereas classical dicentric analysis of four individuals with Giemsa staining and QuickScan could be completed in less than 60 h, which is much shorter than automated RABiT-II DCA with fluorescent probes. In that regard our system should be more beneficial after a large-scale radiation accident (mass casualty event) when a significantly larger number of samples can be processed at the same time to accelerate sample throughput in triage.
Analysis of metaphase cells in the classical DCA can provide very important information on uniformity of the radiation field during exposure, distribution of ionizing events between cells, possibility of partial-body exposure (28) and LET (29). Since the RABiT-II DCA is not based on analysis of individual cells, the assay cannot provide information on inhomogeneity of radiation dose distribution, and therefore cannot determine partial-body doses for nonuniform exposures. We expect the RABiT-II DCA to provide information on the average whole-body dose as a laboratory or hospital-based high-throughput bioassay (30) for risk-based stratification of exposed individuals. If necessary, the partial-body injuries can be determined by scoring 500 cells manually using conventional DCA. Use of our robotic system during triage will help to distinguish between those who are and who are not at risk for developing acute radiation syndrome, who will require immediate hospitalization for advanced medical attention (≥2Gy). Hospital bioassays can be used to re-confirm results obtained by the RABiT-II.
Currently, a limitation of this system is that the low-dose dicentric yields are overestimated due to poorly resolved, touching chromosomes or clusters (false dicentrics) misrecognized by the FluorQuantDic software (Supplementary Fig. S2; http://dx.doi.org/10.1667/RR15266.1.S1). In addition to continued optimization of the scoring algorithms, we are working to improve detection with the addition of telomere probes in the RABiT-II DCA. Having a positive identification of telomeres would allow easy rejection of chromosome clusters and more precise identification of dicentric chromosomes.
The main advantage of the RABiT-II DCA is that it can be performed on already-existing and deployed robotic platforms. Thus, it potentially allows for surge capacity for dicentric analysis orders of magnitude beyond that available through conventional techniques, without the use of experienced cytogenetic scorers. We have used a Perkin-Elmer cell::explorer system and BioTek Cytation imager, although many other HTS robotic systems with the same capacities are available at universities and pharmaceutical companies.
Supplementary Material
Table S1. Distribution of first-, second- and third-division metaphases among 1,000 cells in 48-, 52- and 72-h cultures from two donors.
Table S2. Observed frequency of different arm cohesion phenotypes before and after synchronization.
Table S3. RABiT-II DCA yields after in vitro γ-ray irradiation of blood samples derived from four healthy individuals (scored automatically using FluorQuantDic version 3.3).
Table S4. Classical DCA yields and intercellular distribution of dicentric aberrations after in vitro γ-ray irradiation of blood samples derived from four healthy individuals (scored manually).
Fig. S1. Representative images of chromosome spreads obtained from asynchronously growing blood cells arrested in metaphase by colcemid. A variety of chromosomal configurations are observed in different metaphases. Panel A: DNA strands are weakly condensed. Panel B: Progressively condensed chromosomes with long length; chromosome arms keep firm connection. Panel C: Arms begin to open due to cleavage and release of cohesin. Panel D: Chromosomes begin to shorten in length by condensing. Panel E: Chromosomes become very short, arms are completely opened and firm connection remains only at the centromere region (“X-shape”). Panel F: Arms and centromeres are split. Panel G: Other phases. (Inserts are published and modified, with permission, from: Rieder CL, Palazzo RE. Colcemid and the mitotic cycle. J Cell Sci 1992; 102 Pt 3:387–92). White dashed boxes are cropped chromosomes used for Fig. 6A.
Fig. S2. Representative examples of chromosome clusters that lead to errors at low doses.
ACKNOWLEDGMENTS
We thank Maria Taveras for blood collection and Dr. Mònica Pujol for help with classical centromere FISH staining protocol. This work was supported by National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) (contract no. HHSN272201600040C).
Footnotes
MetaSystems (personal communication).
Editor’s note. The online version of this article (DOI: 10.1667/RR15266.1) contains supplementary information that is available to all authorized users.
REFERENCES
- 1.Repin M, Turner HC, Garty G, Brenner DJ. Next generation platforms for high-throughput biodosimetry. Radiat Prot Dosimetry 2014; 159:105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Repin M, Pampou S, Karan C, Brenner DJ, Garty G. RABiT-II: Implementation of a high-throughput micronucleus biodosimetry assay on commercial biotech robotic systems. Radiat Res 2017; 187:492–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cytogenetic dosimetry: Applications in preparedness for and response to radiation emergencies. EPR-Biodosimetry 2011; Vienna: International Atomic Energy Agency; 2011. [Google Scholar]
- 4.Sullivan JM, Prasanna PGS, Grace MB, Wathen LK, Wallace RL, Koerner JF, et al. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys 2013; 105:540–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flood AB, Boyle HK, Du G, Demidenko E, Nicolalde RJ, Williams BB, et al. Advances in a framework to compare biodosimetry methods for triage in large-scale radiation events. Radiat Prot Dosimetry 2014; 159:77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karachristou I, Karakosta M, Pantelias A, Hatzi VI, Karaiskos P, Dimitriou P, et al. Triage biodosimetry using centromeric/telomeric PNA probes and Giemsa staining to score dicentrics or excess fragments in non-stimulated lymphocyte prematurely condensed chromosomes. Mutat Res Toxicol Environ Mutagen 2015; 793:107–14. [DOI] [PubMed] [Google Scholar]
- 7.M’kacher R, Maalouf EEL, Ricoul M, Heidingsfelder L, Laplagne E, Cuceu C, et al. New tool for biological dosimetry: Reevaluation and automation of the gold standard method following telomere and centromere staining. Mutat Res Mol Mech Mutagen 2014. December;770:45–53. [DOI] [PubMed] [Google Scholar]
- 8.Shirley B, Li Y, Knoll JHM, Rogan PK. Expedited radiation biodosimetry by automated dicentric chromosome identification (ADCI) and dose estimation. J Vis Exp 2017; 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International guidelines for ethical review of epidemiological studies. Law Med Health Care 1991; 19:247–58. [PubMed] [Google Scholar]
- 10.Crossen PE, Morgan WF. Analysis of human lymphocyte cell cycle time in culture measured by sister chromatid differential staining. Exp Cell Res 1977; 104:453–7. [DOI] [PubMed] [Google Scholar]
- 11.Latt SA, Stetten G, Juergens LA, Willard HF, Scher CD. Recent developments in the detection of deoxyribonucleic acid synthesis by 33258 Hoechst fluorescence. J Histochem Cytochem 1975; 23:493–505. [DOI] [PubMed] [Google Scholar]
- 12.Bangs CD, Donlon TA. Metaphase chromosome preparation from cultured peripheral blood cells. Curr Protoc Hum Genet 2005; Chapter 4:Unit 4.1. [DOI] [PubMed]
- 13.Suzuki S, Abe K. Topological structural analysis of digitized binary images by border following. Comput Vision Graph Image Process 1985; 30:32–46. [Google Scholar]
- 14.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 1979; 9:62–6. [Google Scholar]
- 15.Karachristou I, Karakosta M, Pantelias A, Hatzi V, Pantelias G, Thanassoulas A, et al. Biodosimetry for high-dose exposures based on dicentric analysis in lymphocytes released from the G2-block by caffeine. Radiat Prot Dosimetry 2016; 172:230–7. [DOI] [PubMed] [Google Scholar]
- 16.Wilkins RC, Romm H, Oestreicher U, Marro L, Yoshida MA, Suto Y, et al. Biological dosimetry by the triage dicentric chromosome assay - Further validation of international networking. Radiat Meas 2011; 46:923–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ainsbury EA, Lloyd DC. Dose estimation software for radiation biodosimetry. Health Phys 2010; 98: 290–5. [DOI] [PubMed] [Google Scholar]
- 18.Pujol M, Barquinero J-F, Puig P, Puig R, Caballín MR, Barrios L. A new model of biodosimetry to integrate low and high doses. PLoS One 2014; 9:e114137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou BB, Chaturvedi P, Spring K, Scott SP, Johanson RA, Mishra R, et al. Caffeine abolishes the mammalian G(2)/M DNA damage checkpoint by inhibiting ataxia-telangiectasia-mutated kinase activity. J Biol Chem 2000; 275:10342–8. [DOI] [PubMed] [Google Scholar]
- 20.Rieder CL, Palazzo RE. Colcemid and the mitotic cycle. J Cell Sci 1992; 102 Pt 3:387–92. [DOI] [PubMed] [Google Scholar]
- 21.Lemos-Pinto MMP, Cadena M, Santos N, Fernandes TS, Borges E, Amaral A. A dose-response curve for biodosimetry from a 6 MV electron linear accelerator. Brazilian J Med Biol Res 2015; 48:908–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuadrado A, Golczyk H, Jouve N. A novel, simple and rapid nondenaturing FISH (ND-FISH) technique for the detection of plant telomeres. Potential used and possible target structures detected. Chromosome Res 2009; 17:755–62. [DOI] [PubMed] [Google Scholar]
- 23.Fu S, Chen L, Wang Y, Li M, Yang Z, Qiu L, et al. Oligonucleotide probes for ND-FISH analysis to identify rye and wheat chromosomes. Sci Rep 2015; 5:10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Durm M, Haar FM, Hausmann M, Ludwig H, Cremer C. Non-enzymatic, low temperature fluorescence in situ hybridization of human chromosomes with a repetitive alpha-satellite probe. Z Naturforsch C 1997; 52:82–8. [PubMed] [Google Scholar]
- 25.Genet MD, Cartwright IM, Kato TA. Direct DNA and PNA probe binding to telomeric regions without classical in situ hybridization. Mol Cytogenet 2013; 6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haarhuis JHI, Elbatsh AMO, Rowland BD. Cohesin and its regulation: on the logic of x-shaped chromosomes. Dev Cell 2014; 31:7–18. [DOI] [PubMed] [Google Scholar]
- 27.Peters J-M, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev 2008; 22:3089–114. [DOI] [PubMed] [Google Scholar]
- 28.Prasanna PGS, Coleman CN. Cytogenetic biodosimetry. In: Lenhart MK, editor. Medical consequences of radiological and nuclear medicine. Fort Sam Houston, TX: Office of the Surgeon General, Dept. of the Army, USA and US Army Medical Department Center and School; 2012. p. 267–79. (https://bit.ly/2HWfkLn) [Google Scholar]
- 29.Mestres M, Caballin MR, Schmid E, Stephan G, Sachs R, Barrios L, et al. Analysis of alpha-particle induced chromosome aberrations in human lymphocytes, using pan-centromeric and pan-telomeric probes. Int J Radiat Biol 2004; 80:737–44. [DOI] [PubMed] [Google Scholar]
- 30.Grace MB, Moyer BR, Prasher J, Cliffer KD, Ramakrishnan N, Kaminski J, et al. Rapid radiation dose assessment for radiological public health emergencies: roles of NIAID and BARDA. Health Phys 2010; 98:172–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Distribution of first-, second- and third-division metaphases among 1,000 cells in 48-, 52- and 72-h cultures from two donors.
Table S2. Observed frequency of different arm cohesion phenotypes before and after synchronization.
Table S3. RABiT-II DCA yields after in vitro γ-ray irradiation of blood samples derived from four healthy individuals (scored automatically using FluorQuantDic version 3.3).
Table S4. Classical DCA yields and intercellular distribution of dicentric aberrations after in vitro γ-ray irradiation of blood samples derived from four healthy individuals (scored manually).
Fig. S1. Representative images of chromosome spreads obtained from asynchronously growing blood cells arrested in metaphase by colcemid. A variety of chromosomal configurations are observed in different metaphases. Panel A: DNA strands are weakly condensed. Panel B: Progressively condensed chromosomes with long length; chromosome arms keep firm connection. Panel C: Arms begin to open due to cleavage and release of cohesin. Panel D: Chromosomes begin to shorten in length by condensing. Panel E: Chromosomes become very short, arms are completely opened and firm connection remains only at the centromere region (“X-shape”). Panel F: Arms and centromeres are split. Panel G: Other phases. (Inserts are published and modified, with permission, from: Rieder CL, Palazzo RE. Colcemid and the mitotic cycle. J Cell Sci 1992; 102 Pt 3:387–92). White dashed boxes are cropped chromosomes used for Fig. 6A.
Fig. S2. Representative examples of chromosome clusters that lead to errors at low doses.