Abstract
Genomic tumor profiling by next-generation sequencing (NGS) allows for large-scale tumor testing to inform targeted cancer therapies and immunotherapies, and to identify patients for clinical trials. These tests are often underutilized in patients with late-stage solid tumors and are typically performed in centralized specialty laboratories, thereby limiting access to these complex tests. Personal Genome Diagnostics Inc., elio tissue complete NGS solution is a comprehensive DNA-to-report kitted assay and bioinformatics solution. Comparison of 147 unique specimens from >20 tumor types was performed using the elio tissue complete solution and Foundation Medicine's FoundationOne test, which is of similar size and gene content. The analytical performance of all genomic variant types was evaluated. In general, the overall mutational profile is highly concordant between the two assays, with agreement in sequence variants reported between panels demonstrating >95% positive percentage agreement for single-nucleotide variants and insertions/deletions in clinically actionable genes. Both copy number alterations and gene translocations showed 80% to 83% positive percentage agreement, whereas tumor mutation burden and microsatellite status showed a high level of concordance across a range of mutation loads and tumor types. The Personal Genome Diagnostics Inc., elio tissue complete assay is comparable to the FoundationOne test and will allow more laboratories to offer a diagnostic NGS assay in house, which will ultimately reduce time to result and increase the number of patients receiving molecular genomic profiling and personalized treatment.
Over the last decade, improvements in next-generation sequencing (NGS) have allowed the technology to bridge from the research and translational settings into clinical care pathways. Through increased accuracy and efficiency of massively parallel sequencing and reduced costs and turnaround times, many obstacles for its routine clinical use have been mitigated, thus increasing acceptance and uptake from the medical community. Although NGS has been applied to many disease settings, it has emerged as an impactful technology in cancer care by allowing for rapid detection of multiple genomic alterations for personalized tumor profiling. In this regard, not only has NGS increased the research and medical community's knowledge of cancer biology and the mechanisms of disease progression, but it also provides a window into unique molecular profiles of patient populations for whom a targeted therapy may be appropriate.1 The National Comprehensive Cancer Network and the Association for Molecular Pathology guidelines currently recommend molecular profiling in advanced non–small-cell lung cancer and list NGS as an efficient option, especially when testing for multiple genomic alterations.2,3 Guidelines for other indications similarly recommend biomarker testing, but do not always specify a technique or approach. However, a multitude of clinical trials utilizing NGS are underway in nearly all indications, contributing to a better understanding of the genomic landscape of cancer and providing a wealth of potential therapeutic targets. The results of such trials will help to shape future clinical guidelines and move NGS and precision medicine to the forefront of clinical oncology.
As clinical evidence evolves and more actionable biomarkers are identified, more patients are benefiting from a precision medicine strategy. For example, a large-scale tumor sequencing study has reported actionable mutations in driver genes for 40% to 50% of patients.4 Furthermore, other studies have shown that 30% to 40% of patients treated with a biomarker-matched therapy experienced a rate of progression-free survival that was >30% better than those not treated with targeted therapies.5 As a result of this increased clinical utility, a patient's molecular profile is predicted to become standard of care in oncology.6 In addition, comprehensive NGS assays allow for detection of multiple genomic alterations simultaneously from a single patient sample; therefore, clinicians can obtain more information from a limited sample.7 This personalized approach to cancer care is achieved only through highly sensitive detection of all genomic variant types [sequence mutations, insertions/deletions (indels), translocations, copy number alterations, and microsatellite instability]. The comprehensive nature of NGS has also facilitated the generation of novel cancer signatures, previously unfeasible with conventional techniques, such as tumor mutation burden (TMB). TMB acts as a composite genomic score, often reported as mutations per exome or mutations per megabase (Muts/Mb) of DNA, which highlights the overall mutation load of a tumor. Recent clinical observations show that tumors with high mutation burden are more responsive to immune checkpoint inhibition than tumors with fewer mutations.8, 9, 10, 11, 12, 13, 14, 15
Despite the value of precision oncology, <20% of late-stage patients receive genomic testing to help determine the most effective treatment path for their particular cancer.16 In addition, most testing occurs in a limited number of commercial send-out laboratories independent of the local institutional care pathway. This lack of access is driven by several factors, many of which are related to the need to send samples to specialty laboratories, where close to 80% of comprehensive genomic profiling tests are performed today.16 Alternatively, distributed commercial kits provide a local testing solution for any molecular diagnostic laboratory with next-generation sequencing capabilities. Personal Genome Diagnostics, Inc. (PGDx; Baltimore, MD), kitted NGS solution for use in local laboratories includes both library chemistries as well as automated bioinformatics that have been rigorously developed in parallel under design control to produce highly accurate and robust clinical results. In addition to retaining samples and data in the local network and knowledgebase, institutions benefit from a 4- to 5-day turnaround time to result when onboarding clinical NGS in house. To compare performance results between a centralized laboratory and the PGDx distributable kit, this study provides the concordance of analytical results between the Foundation Medicine, Inc. (Cambridge, MA), FoundationOne test and the PGDx elio tissue complete NGS solution.
Materials and Methods
Under Duke University Institutional Review Board–approved protocol Pro00091621, formalin-fixed, paraffin-embedded tissue blocks for 147 unique specimens were used for the evaluation. All tissue blocks had been previously sent to Foundation Medicine for the FoundationOne test as part of standard of care at Duke University Health System. For this comparative genomic research, additional unstained slides from the same block were prepared, de-identified, and sent to PGDx for the research and development team to run the PGDx elio tissue complete assay in house before commercial availability of the decentralized kitted solution for customer laboratories. Genomic results from FoundationOne testing were de-identified and analyzed in comparison to the PGDx results.
For this comparator study, the research use only version of the PGDx next-generation sequencing elio tissue complete assay was utilized to detect somatic tumor alterations across 505 genes, including 23 and 28 genes for translocations and copy number alternations, respectively, as well as genomic signatures (microsatellite status and TMB) from genomic DNA isolated from formalin-fixed, paraffin-embedded tumor tissue. The PGDx elio tissue complete laboratory workflow consists of a targeted hybrid capture–based chemistry and genome sequencing using the Illumina (San Diego, CA) NextSeq, with batching of up to 15 samples and 1 external control per sequencing run. Following sequencing, an automated pipeline of software version 3.2.2 for bioinformatic analysis evaluates each case for sample quality and then identifies and reports genomic alterations.17 PGDx elio tissue complete uses a comprehensive set of quality control metrics to ensure robust high confidence variant calls (Supplemental Table S1). At the sequencing run level, elio tissue complete requires even lane fraction and that a large percentage of the reads are high quality. For each individual sample, a minimum coverage across the exonic regions on the panel is required. Samples are also checked for possible contamination in silico through a bioinformatic analysis of genome haplotypes.
For elio tissue complete TMB determination, following alignment to the reference genome, candidate single-nucleotide variants (SNVs) and indels are assessed by the automated pipeline, and variants that do not meet the acceptance criteria are excluded from the TMB score. Only exonic, high-quality variants, based on the PGDx machine learning model, are assessed; and most common germline variants (as annotated by dbSNP and Exome Aggregation Consortium databases) are removed. Synonymous and nonsynonymous sequence variants at >5% variant allele frequency are included in the variants assessed for TMB determination, as they may reflect the rate of mutational processes in the tumor. Common drivers [designated by Catalogue of Somatic Mutations in Cancer, US Food and Drug Administration (FDA), or National Comprehensive Cancer Network], however, are removed from consideration for the TMB score. TMB was compared on the basis of the quantitative TMB score, reported as Muts/Mb, determined from the regions sequenced in both assays. The sequenced TMB value for the PGDx elio tissue complete assay represents the numbers of candidate variants identified in the regions of interest divided by the number of bp sequenced, and it is reported as the number of mutations identified per megabase of genomic sequence. A linear regression was used to derive a comparability map between scores for the two assays. PGDx's TMB algorithm was validated against whole exome sequencing using 118 samples across eight tumor types [data presented in the FDA 510(k) Premarket Notification page, Food and Drug Administration website, Next Generation Sequencing Based Tumor Profiling Test, K192063, PGDx elio tissue complete; https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/pmn.cfm?ID=K192063, last accessed May 10, 2021). PGDx is also involved in an effort to harmonize TMB reporting results with Friends of Cancer Research.18
PGDx elio tissue complete determines microsatellite status by evaluating the observed length of >60 homopolymer tracts across regions of interest in combination with mutational signatures of synonymous and nonsynonymous mutations to derive a genomic signature score. The algorithm was trained using 170 samples during feasibility studies and validated using 283 samples across 18 tumor types [data presented in the FDA 510(k) Premarket Notification page, Food and Drug Administration website, Next Generation Sequencing Based Tumor Profiling Test, K192063, PGDx elio tissue complete; https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/pmn.cfm?ID=K192063, last accessed May 10, 2021). Comparison to the FoundationOne test was performed by comparing each sample's status as microsatellite instability (MSI) or microsatellite stable. In some cases, the FoundationOne test results did not have microsatellite status available. Discrepancies were investigated by assessing the presence of MSI-high (MSI-H)–associated variants present in the sample and confirmed by PCR-based testing. Confirmatory PCR testing was done by targeting five mononucleotide short tandem repeat and two pentanucleotide short tandem repeats and performed on tumor and normal DNA dissected from adjacent unstained formalin-fixed, paraffin-embedded sections. The size of each short tandem repeat is resolved using capillary electrophoresis on the Genetic Analyzer and analyzed using GeneMapper software version 5.0 (ThermoFisher, Waltham, MA). A sample is interpreted as MSI-H if two or more short tandem repeat alleles contain novel repeat lengths in the tumor tissue when compared with normal.
For calculating positive percentage agreement (PPA) and positive predictive value, each PGDx elio call status was compared with the orthogonal assay status reported by FoundationOne. Sequence mutations (SNVs and indels) were evaluated in genes and genomic regions determined to be present in both FoundationOne and PGDx elio tissue complete (Supplemental Tables S2 and S3). Characterization of variants as somatic hotspots was based on prevalence in Catalogue of Somatic Mutations in Cancer version 72 with >25 exact hits.19 Germline variants were annotated on the basis of prevalence in ExAC version 0.3.1, Genome Aggregation Database (gnomAD) version 2.02, and dbSNP version 150 databases. Copy number alterations were evaluated for focal amplifications only based on the gene reported amplified and considered in agreement if the reported amplified gene matched between assays. A total of 28 genes were evaluated in the comparison of copy number alterations. Gene translocations were compared on the basis of the reported gene partners and considered in agreement if both reported gene partners matched between assays. Two cases with discordant translocation results (one NTRK1 and one EGFR) were subsequently tested with a third orthogonal assay, Archer FusionPlex (Invitae, Boulder, CO), which assesses translocation positivity in RNA. In both cases, additional tissue was sectioned (5 × 10-μm sections) from the same formalin-fixed, paraffin-embedded tissue block that was assessed by the PGDx and FoundationOne assays and sent for confirmation testing.
Results
Analytical performance was evaluated across 147 samples representing >20 tumor types that were tested with both PGDx elio tissue complete and FoundationOne (Table 1). Both assays rely on targeted sequencing of selected genomic regions and are similar in size, at 2.2 Mb for elio tissue complete and 1.8 Mb for FoundationOne. Most of the gene content that is targeted by these tests is shared between panels, with 505 genes reported by PGDx elio tissue complete compared with 324 reported by FoundationOne (Figure 1 and Supplemental Tables S2 and S3). A comprehensive set of genomic alterations that are reported by both panels were compared, including tumor mutation burden, microsatellite status, SNVs, indels, copy number alterations, and translocations.
Table 1.
The Number of Samples Evaluated for Each Tumor Type
| Tumor types | Samples, n |
|---|---|
| Colorectal | 20 |
| CNS | 20 |
| Ovarian | 19 |
| Prostate | 17 |
| Uterine | 14 |
| Lung | 13 |
| Breast | 10 |
| Other | 34 (including gastric, bladder, and thyroid) |
There were 147 unique samples across >20 tumor types. Other tumor type category encompasses the tumor types with three or fewer samples represented in the cohort.
CNS, central nervous system.
Figure 1.

Overlap in gene content between Personal Genome Diagnostics, Inc. (PGDx), elio tissue complete and FoundationOne is shown with the number of genes in each assay. The reported size in megabases for the regions of interest (ROIs) targeted by the panel is also provided, including the ROI utilized for the evaluation of tumor mutation burden (TMB).
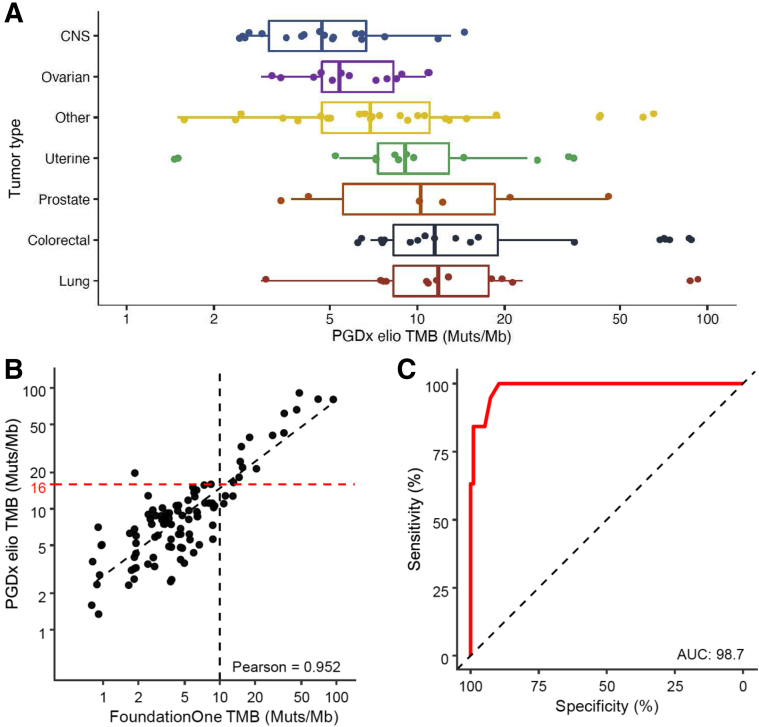
TMB was evaluated in samples where FoundationOne TMB results were available across a range of mutation loads and tumor types (n = 99) (Figure 2A). Both assays report TMB scores as Muts/Mb sequenced in terms of their respective panel sizes and content. TMB scores were highly concordant with a Pearson correlation coefficient of >0.95 in a quantitative comparison of the TMB values reported by both assays (Figure 2B). To illustrate the comparability between assays for TMB, a mapping between TMB scores was obtained on the basis of a linear regression to provide comparable scores between assays at any cutoff. Concordance between TMB scores reported in both assays was high, with an area under the curve of 0.98 across all TMB scores (Figure 2C). Using this model, a FoundationOne TMB score of 10 Muts/Mb corresponds to a PGDx elio TMB score of 16 Muts/Mb (Figure 2B).
Figure 2.
Concordance for reported values for tumor mutation burden (TMB). A: The range of Personal Genome Diagnostics, Inc. (PGDx), elio tissue complete TMB scores for the predominant tumor types evaluated. B: Comparison between FoundationOne and PGDx elio tissue complete TMB scores, as measured by mutations per megabase (Muts/Mb) sequenced. A linear correlation between the values is provided; Pearson correlation coefficient >0.95. A correspondence between the FoundationOne TMB score of 10 Muts/Mb (black) and the PGDx elio tissue complete TMB score of 16 Muts/Mb (red) is shown by dashed lines at the axis. C: The sensitivity, specificity, and area under the curve (AUC) for various corresponding TMB score cutoffs for PGDx elio tissue complete compared with FoundationOne. n = 99. CNS, central nervous system.
Classification of samples as microsatellite stable and instable was highly concordant, with 96% overall agreement (90/93 cases in agreement) for the cases evaluated (Table 2). Ninety-three samples from the cohort were available with microsatellite status, and the remaining 54 samples had PGDx elio tissue complete microsatellite status information (52 microsatellite stable and 2 MSI-H) but did not have reported results from the FoundationOne test.
Table 2.
Concordance of 93 Samples with Reported Microsatellite Status
| Microsatellite status (n = 93 cases∗) | ||||
|---|---|---|---|---|
| 96% Overall agreement | elio Tissue complete |
|||
| MSI-H | MSS | Indeterminate | ||
| FoundationOne | MSI-H | 7 | 0 | 0 |
| MSS | 1 | 83 | 1 | |
| Ambiguous | 1 | 0 | 0 | |
MSI-H, microsatellite instability–high; MSS, microsatellite stable.
A total of 54 samples did not have FoundationOne microsatellite reporting available. Of those, 52 were MSS and 2 were MSI-H by elio tissue complete.
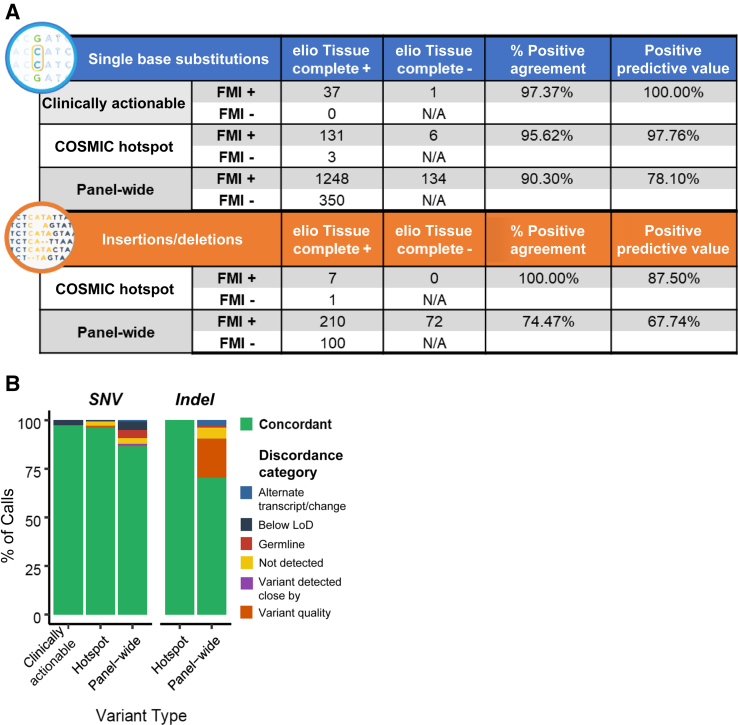
Agreement in sequence mutations reported between panels was high, with >95% PPA for SNVs and indels in clinically actionable genes and known somatic hotspots. Agreement of >90% PPA was observed for SNVs across all shared gene content, including variants of unknown significance (Figure 3A). Remaining differences in calls for SNVs were primarily in variants of unknown significance at low variant allele frequency <5% variant allele frequency or germline variants (Figure 3B). Agreement in reported indel variants was high for somatic hotspot mutations, with seven concordant calls of eight indel calls evaluated (Figure 3A). In variants of unknown significance, agreement for indel variants was lower, with differences primarily associated with lower-quality sequences, such as homopolymer or repetitive genomic regions (Figure 3B). These genomic regions tend to have higher limits of detection and would be expected to have reduced call rates between observations.
Figure 3.
Concordance for single-nucleotide variant (SNV) and insertion/deletions (Indel) sequence mutations. A: Agreement is shown between somatic calls for clinically actionable mutations, Catalogue of Somatic Mutations in Cancer (COSMIC) hotspots, and the remainder of variants reported by both panels (panel-wide). B: Fraction of calls in agreement between FoundationOne and elio tissue complete (green) and discordance (FMI+/elio−) categories identified after raw data investigation. Variants listed as discordant due to an alternate transcript/change were reported at the same genomic position but annotated with a different transcript. Discordant variants below the limit of detection (LoD) were identified in elio tissue complete at a variant allele frequency below the limit of detection and were filtered. Germline variants were annotated as germline based on the dbSNP and Exome Aggregation Consortium databases and removed, whereas variants noted as detected close by were detected within three amino acids of the FoundationOne reported position and could be discordant because of alignment differences. Variants filtered because of quality were identified but failed the Personal Genome Diagnostics, Inc., bioinformatic machine learning score threshold and were filtered. Those listed as not detected were not detected by elio tissue complete following visual inspection of the raw data. FMA, Foundation Medicine; N/A, not applicable.
Gene translocations were evaluated in 23 genes with an observed agreement of 82.8% PPA (Figure 4A). Differences in calls were attributed to low-level events or alternative fusion genes and secondary events in the reports for two of the five discordant calls. Copy number alterations reported by both assays were evaluated in 28 genes, with an observed agreement of 80.6% PPA (Figure 4A). Differences in calls were primarily attributed to low-level copy gains (Figure 4B). Copy number calls were concordant between assays at higher levels and in cases where all exons across a gene indicated a copy number amplification (Figure 4C).
Figure 4.
Concordance for structural alterations. A: Agreement is shown for genes evaluated for translocations and copy number alterations. B: Fold sequence change observed in Personal Genome Diagnostics, Inc. (PGDx), elio tissue complete for each of the genes evaluated for copy number alterations. The various data point colors indicate agreement or disagreement for the copy number alteration call reported by the assays. C: The relationship between the observed fold sequence change and the fraction of genomic segments observed to be amplified in PGDx elio tissue complete. The various data point colors indicate agreement or disagreement for the copy number alteration call reported by the assays. FMI, Foundation Medicine; NPA, negative percentage agreement; NPV, negative predictive value; PPA, positive percentage agreement; PPV, positive predictive value.
Discussion
In this study, analysis of 147 samples and from 20 tumor types showed that overall, the agreement between the PGDx assay and the FoundationOne test is excellent (>95% PPA) for SNVs and indels in clinically actionable genes and known somatic hotspots. The same tissue block that was used for testing with FoundationOne during standard of care was recut for this project and tested with PGDx elio tissue complete. However, it is standard practice for a pathologist to limit sample size and enrich for tumor content by circling tumor for use. It is possible that different regions of the tumor were selected by the Foundation Medicine pathologist and the Duke pathology team. Because of this, differences in the region of tumor selected by the pathologists could result in differences in the mutational calls due to tumor heterogeneity, even within the same block. When evaluating discordant calls (Supplemental Table S4), most of the discordant findings were variants of unknown significance with either low sequencing quality or low variant allele frequency (<5% variant allele frequency) or were likely germline variants. For germline calls, both panels report variants in tumor samples without a matched normal and rely on population databases and filtering rules to identify and exclude germline variants from reports. Differences in these databases and rules can result in reporting of germline calls, especially in cases of more rare private germline variants. Furthermore, because of variations in the bioinformatics filtering criteria, including variant allele frequency thresholds, germline filtering, and sequence coverage and quality, and in the specificity and sensitivity, for each assay, some differences are expected (Supplemental Table S1 provides a summary of PGDx elio tissue complete quality metrics and passing criteria). For the PGDx elio tissue complete assay, variants are scored for quality using a machine learning approach that incorporates experimental data with in silico training, which has been shown to decrease false-positive calls and improve sensitivity.17
Agreement for copy number alterations and gene translocations was 80.6% and 82.8% PPA, respectively. For copy number alterations, differences in calls were attributed to low-level gains. The reporting of low-level copy gains is especially impacted by differences in cutoffs and rules for reporting copy number, such as the predicted tumor purity of the sample. There were three low-level ERBB2 amplification events identified by FoundationOne that did not meet criteria for the reporting of focal amplification by PGDx elio tissue complete. The observed fold changes in these discordant cases were low, at 2.2-fold and 2.4-fold. Worth noting, the criteria for reporting of ERBB2 focal amplifications by PGDx elio tissue complete has been demonstrated to be highly concordant with HER2 fluorescence in situ hybridization [40/46 (87.0%) PPA, excluding borderline fluorescence in situ hybridization ratios ranging from 1.5 to 2.5]. There were four samples that showed signs of a MET amplification in PGDx elio tissue complete but were not reported as amplified by FoundationOne. Although exact reporting rules were not available to compare between both assays, differences in thresholds for reporting of copy number alternations as well as the identification and reporting of polysomy events can lead to differences in reported results (Supplemental Tables S1 and S4).
For the gene translocations, two discordant cases were investigated with a third assay, RNA-based Archer FusionPlex. In one case, FoundationOne called an NTRK1 translocation positive, whereas PGDx elio tissue complete was negative. The Archer analysis was also negative for this case, agreeing with PGDx elio tissue complete. In a second case, an EGFR translocation was reported by FoundationOne, which was negative by PGDx elio tissue complete. This case was positive by Archer for EGFR translocation, agreeing with FoundationOne. These disagreements could reflect differences in genomic regions of interest (ROIs) or thresholds of the assays but could also be a result of multiple clones in the sample being assessed (Supplemental Tables S1 and S4).
Agreement was excellent in the evaluation of TMB (Pearson correlation coefficient >0.95) across the samples evaluated, where TMB data from FoundationOne were available (n = 99). There was a strong linear relationship between the TMB scores reported by the assays, although the reported values in terms of Muts/Mb are specific to each assay. The TMB calculation is panel dependent because of differences in gene content and size, composition, reporting rules for mutations, and strategies for filtering rare germline calls. Efforts to streamline the reporting of TMB have been purposed to better harmonize with whole-exome sequencing results, including the use of a TMB calibration tool by the Friends of Cancer Research.18 With the recent approval of drugs, such as pembrolizumab, in patients with high TMB, an understanding of the correspondence between assay cutoffs for TMB is important. Although there is no standard approach to TMB calculation, a value of 10 Muts/Mb has been widely viewed as a tissue-agnostic cutoff for high TMB based on the FoundationOne test. According to a linear regression model, there is a correspondence between the FoundationOne TMB score of 10 Muts/Mb and the PGDx elio tissue complete TMB score of 16 Muts/Mb. The difference in score thresholds is, again, because of a combination of unique characteristics, including panel assay content and sequence mutation filters, such as germline filters. Any comparison of assays that contain large cancer gene panels has expected variability because of these factors.18 Although FoundationOne has historically provided qualitative labeling of low, indeterminate, or high, this tiered designation has now been removed from newer reports.
Agreement was also excellent in the evaluation of microsatellite stability/instability (96% agreement from a total of 93 cases). This is critical with the importance of MSI-H as a biomarker for immune checkpoint inhibitor therapy, such as pembrolizumab, in tumors determined to have mismatch repair deficiency, resulting in microsatellite instability regardless of tumor type.20 Differences in microsatellite status may be observed because of differences in mononucleotide tract assessed and different methods utilized to evaluate microsatellites. The criteria for reporting borderline status can also lead to differences in call status as FoundationOne reporting includes an MSI-ambiguous status, whereas PGDx elio tissue complete includes an MSI-indeterminate status. There were only three samples that showed disagreement in MSI-H call status, two of which exhibited characteristics of being borderline. In one non–colorectal cancer sample, the genomic signature score of the PGDx assay and an MSH3 deletion supported MSI-H status, although FoundationOne identified the sample as MSI-ambiguous. Although the sample exceeded the overall genomic score threshold for the PGDx assay for MSI-H status, only 16% of homopolymer tracts were observed to be shortened, consistent with being a borderline case. PCR-based orthogonal testing revealed four unstable mononucleotide markers and one pentanucleotide marker, thus confirming PGDx designation of MSI-H. An additional sample was identified by the PGDx assay as MSI-H but microsatellite stable in FoundationOne. This colorectal cancer sample exhibited strong indications of MSI-H status, including a shortening in 31% of microsatellite tracts evaluated by the PGDx assay, as well as a strong genomic signature for instability, including two MSH3 deletions at 19% and 22% mutant allele fraction. However, this sample also contained two co-occurring APC insertions at 20% and 24%, which have been shown to be mutually exclusive with MSI-H status.21 With the high level of support for MSI-H status but co-occurring APC mutations, it is possible this patient's tumor has a high degree of heterogeneity and that FoundationOne and PGDx sequenced different subclones of the tumor. Ultimately, PCR-based orthogonal testing revealed five unstable mononucleotide markers, thus confirming PGDx designation of MSI-H. Last, another non–colorectal cancer sample was designated as microsatellite stable by FoundationOne and indeterminate by PGDx elio tissue complete. PGDx microsatellite status is determined using a proprietary combination of two scoring systems. The first scoring method analyzes >60 microsatellite tracts, including four of the tracts used in the Bethesda panel, and assesses the lengths of these tracts to generate a score. The second method uses SNVs that are indicative of microsatellite instability. The two scores are used together to determine whether the sample has microsatellite instability. Indeterminate status by PGDx is reported for MSI when there is a significant disagreement in status between the two scoring systems. Investigation into quality control metrics of this indeterminate sample revealed no coverage or quality issues; therefore, confirmatory testing was ordered. Although matched normal was not available, PCR testing showed low amplification, and yielded indeterminate results.
Next-generation sequencing is revolutionizing the practice of oncology by allowing rapid detection of multiple genomic alterations that could suggest response to targeted therapies in the clinical or clinical research setting. However, only a small fraction of late-stage cancer patients receives genomic testing at the present time, with most of these tests being performed by large specialty laboratories using samples sent in from the patient's local institution. Small- to medium-sized clinical laboratories may not have sufficient personnel, time, or financial means required for achieving FDA clearance for a laboratory-developed test to offer locally. The FDA has become increasingly involved in the regulation of laboratory-developed tests, especially those based on NGS technology. In addition, FDA-approved companion diagnostics may be required for a patient to have access to drugs that target certain genomic biomarkers or pathways, although studies have shown that laboratory-developed tests for these Clinical Laboratory Improvement Amendments–approved laboratories perform as well as or better than the FDA-approved companion diagnostics assays.22 Furthermore, in many cases, laboratories must submit technical assessments, including details of the assay validation, to Medicare administration contractors before being provided with coverage determination.
In April 2020, PGDx received FDA clearance for the elio tissue complete in vitro diagnostic assay, consisting of 505 full-coding genes covering SNVs, indels, ERBB2 copy number alternations, and four translocations (ALK, RET, NTRK2, and NTRK3) as well as microsatellite status and TMB [FDA 510(k) Premarket Notification page, Food and Drug Administration website, Next Generation Sequencing Based Tumor Profiling Test, K192063, PGDx elio tissue complete; https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/pmn.cfm?ID=K192063, last accessed May 10, 2021). The comprehensive, pan-cancer kitted assay is the first of its kind to be cleared for clinical use by health professions to guide clinical management and identify opportunities for clinical trial participation. Having access to an FDA-cleared comprehensive genomic profiling off-the-shelf kit eliminates several key challenges in running a multidisciplinary, expansive cancer care program based on comprehensive tumor profiling. Faster turnaround times and integration of NGS into the existing diagnostics care pathway for cancer patients at their treating institution improve the timeliness of care. Immediate access to all data also allows for more thoughtful and innovative integration into the electronic medical record, aiding in electronic decision support. Testing of hospitalized patients or during a hospital encounter becomes more feasible without the compliance concerns of diagnosis-related group-based billing. Although reimbursement has historically been a hurdle for this type of testing with patients being burdened to cover the cost, the Centers for Medicare & Medicaid Services has become a reliable payer for comprehensive NGS testing of the Duke cancer patient population. In fact, PGDx recently announced that MolDx (Columbia, SC) and Novitas (Mechanicsburg, PA) issued local coverage determinations for the FDA-cleared elio tissue complete test, establishing reimbursement for laboratory facilities across 40 states, thus enabling broader access to comprehensive genomic profiling for cancer patients. Although this covers a large minority of patients with advanced cancer at Duke, many others rely on private insurers that do not typically provide coverage for comprehensive tumor profiling, and this gap must be addressed to ensure access to precision medicine for all.
Local institutions vary in the ability to perform NGS assays in house, but even institutions with relevant expertise and equipment may lack the resources to develop and validate a broad NGS panel with complex biomarkers, such as MSI-H and TMB. In addition, it will be necessary to keep such tests up to date with current literature and clinical needs as the practice of precision oncology evolves. The PGDx elio tissue complete is an FDA-cleared kitted assay that now allows local institutions with sequencing capabilities to provide high-quality NGS testing in house to improve turnaround times and the ability to address sample quality issues. In addition, laboratories and health systems can maintain local access to granular data, which can be used in the support of translational research. The PGDx elio tissue complete is a viable alternative to send-out testing for local institutions seeking a path to provide rapid and accurate tumor profiling to patients and may increase the number of patients who have access to tumor-based genomic testing.
Acknowledgments
We thank the Duke University BioRepository and Precision Pathology Center (BRPC), a shared resource of the Duke University School of Medicine and Duke Cancer Institute, for tissue histology services performed for this project. The BRPC receives support from the National Cancer Institute's Cancer Center Support grant 2P30-CA014236. We also thank the Personal Genome Diagnostics, Inc., Product Development Teams for technical assistance and data gathering for this study.
Footnotes
Supported through joint collaboration between Duke University Medical Center, Department of Pathology, and Personal Genome Diagnostics Inc. (PGDx). Study samples were provided by the Duke Biorepository and Precision Pathology Center, and library preparation, sequencing, and data analysis were provided by PGDx.
Disclosures: J.B.J., K.C.V., L.A.K., K.M.R.G., and S.V.A. are employed by Personal Genome Diagnostics, Inc.; S.J.M. is a consultant for AstraZeneca.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2021.07.004.
Contributor Information
Kristen L. Deak, Email: kristen.deak@duke.edu.
Jennifer B. Jackson, Email: jjackson@pgdx.com.
Author Contributions
In collaboration, Duke and Personal Genome Diagnostics, Inc., designed the study and wrote the manuscript, and all authors approved submission and publication of the manuscript.
Supplemental Data
References
- 1.Mardis E.R. Genome sequencing and cancer. Curr Opin Genet Dev. 2012;22:245–250. doi: 10.1016/j.gde.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindeman N.I., Cagle P.T., Aisner D.L., Arcila M.E., Beasley M.B., Bernicker E.H., Colasacco C., Dacic S., Hirsch F.R., Kerr K., Kwiatkowski D.J., Ladanyi M., Nowak J.A., Sholl L., Temple-Smolkin R., Solomon B., Souter L.H., Thunnissen E., Tsao M.S., Ventura C.B., Wynes M.W., Yatabe Y. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol. 2018;13:323–358. doi: 10.1016/j.jtho.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 3.National Comprehensive Cancer Network . 2019. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®): Non-Small Cell Lung Cancer. Version 3. Plymouth Meeting, PA: National Comprehensive Cancer Network. [Google Scholar]
- 4.Massard C., Michiels S., Ferté C., Le Deley M.-C., Lacroix L., Hollebecque A., Verlingue L., Ileana E., Rosellini S., Ammari S., Ngo-Camus M., Bahleda R., Gazzah A., Varga A., Postel-Vinay S., Loriot Y., Even C., Breuskin I., Auger N., Job B., De Baere T., Deschamps F., Vielh P., Scoazec J.-Y., Lazar V., Richon C., Ribrag V., Deutsch E., Angevin E., Vassal G., Eggermont A., André F., Soria J.-C. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7:586–595. doi: 10.1158/2159-8290.CD-16-1396. [DOI] [PubMed] [Google Scholar]
- 5.Marquart J., Chen E.Y., Prasad V. Estimation of the percentage of US patients with cancer who benefit from genome-driven oncology. JAMA Oncol. 2018;4:1093–1098. doi: 10.1001/jamaoncol.2018.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartmaier R.J., Albacker L.A., Chmielecki J., Bailey M., He J., Goldberg M.E., Ramkissoon S., Suh J., Elvin J.A., Chiacchia S., Frampton G.M., Ross J.S., Miller V., Stephens P.J., Lipson D. High-throughput genomic profiling of adult solid tumors reveals novel insights into cancer pathogenesis. Cancer Res. 2017;77:2464. doi: 10.1158/0008-5472.CAN-16-2479. [DOI] [PubMed] [Google Scholar]
- 7.Thomas R.K., Baker A.C., Debiasi R.M., Winckler W., Laframboise T., Lin W.M. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 8.Alexandrov L.B., Nik-Zanial S., Wedge D.C., Aparicio A.J.R., Behjati S. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan T.A., Yarchoan M., Jaffee E., Swanton C., Quezada S.A., Stenzinger A., Peters S. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cristescu R., Mogg R., Ayers M., Albright A., Murphy E., Yearley J., Sher X., Liu X.Q., Lu H., Nebozhyn M., Zhang C., Lunceford J.K., Joe A., Cheng J., Webber A.L., Ibrahim N., Plimack E.R., Ott P.A., Seiwert T.Y., Ribas A., McClanahan T.K., Tomassini J.E., Loboda A., Kaufman D. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362:eaar3593. doi: 10.1126/science.aar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hellmann M.D., Ciuleanu T.-E., Pluzanski A., Lee J.S., Otterson G.A., Audigier-Valette C., Minenza E., Linardou H., Burgers S., Salman P., Borghaei H., Ramalingam S.S., Brahmer J., Reck M., O’Byrne K.J., Geese W.J., Green G., Chang H., Szustakowski J., Bhagavatheeswaran P., Healey D., Fu Y., Nathan F., Paz-Ares L. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378:2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J., Lee W., Yuan J., Wong P., Ho T.S., Miller M.L., Rekhtman N., Moreira A.L., Ibrahim F., Bruggeman C., Gasmi B., Zappasodi R., Maeda Y., Sander C., Garon E.B., Merghoub T., Wolchok J.D., Schumacher T.N., Chan T.A. Cancer immunology: mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Snyder A., Makarov V., Merghoub T., Yuan J., Zaretsky J.M., Desrichard A., Walsh L.A., Postow M.A., Wong P., Ho T.S., Hollmann T.J., Bruggeman C., Kannan K., Li Y., Elipenahli C., Liu C., Harbison C.T., Wang L., Ribas A., Wolchok J.D., Chan T.A. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Topalian S.L., Drake C.G., Pardoll D.M. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yarchoan M., Hopkins A., Jaffee E.M. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vadas A. Immuno-oncology is making pharma step up its diagnostics game. In Vivo. 2018;36:2–8. [Google Scholar]
- 17.Wood D.E., White J.R., Georgiadis A., Van Emburgh B., Parpart-Li S., Mitchell J., Anagnostou V., Niknafs N., Karchin R., Papp E., McCord C., LoVerso P., Riley D., Diaz L.A., Jones S., Sausen M., Velculescu V.E., Angiuoli S.V. A machine learning approach for somatic mutation discovery. Sci Transl Med. 2018;10:eaar7939. doi: 10.1126/scitranslmed.aar7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merino D.M., McShane L.M., Fabrizio D., Funari V., Chen S.J., White J.R., Wenz P., Baden J., Barrett J.C., Chaudhary R., Chen L., Chen W.S., Cheng J.H., Cyanam D., Dickey J.S., Gupta V., Hellmann M., Helman E., Li Y., Maas J., Papin A., Patidar R., Quinn K.J., Rizvi N., Tae H., Ward C., Xie M., Zehir A., Zhao C., Dietel M., Stenzinger A., Stewart M., Allen J. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. J Immunother Cancer. 2020;8:e000147. doi: 10.1136/jitc-2019-000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E., Fish P., Harsha B., Hathaway C., Jupe S.C., Kok C.Y., Noble K., Ponting L., Ramshaw C.C., Rye C.E., Speedy H.E., Stefancsik R., Thompson S.L., Wang S., Ward S., Campbell P.J., Forbes S.A. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang L., Chang M., Chang H.M., Chang F. Microsatellite instability: a predictive biomarker for cancer immunotherapy. Appl Immunohistochem Mol Morphol. 2018;26:e15–e21. doi: 10.1097/PAI.0000000000000575. [DOI] [PubMed] [Google Scholar]
- 21.Schell M.J., Yang M., Teer J.K., Lo F.Y., Madan A., Coppola D., Monteiro A.N.A., Nebozhyn M.V., Yue B., Loboda A., Bien-Willner G.A., Greenawalt D.M., Yeatman T.J. A multigene mutation classification of 468 colorectal cancers reveals a prognostic role for APC. Nat Commun. 2016;7:11743. doi: 10.1038/ncomms11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim A.S., Bartley A.N., Bridge J.A., Kamel-Reid S., Lazar A.J., Lindeman N.I., Long T.A., Merker J.D., Rai A.J., Rimm D.L., Rothberg P.G., Vasalos P., Moncur J.T. Comparison of laboratory-developed tests and FDA-approved assays for BRAF, EGFR, and KRAS testing. JAMA Oncol. 2018;4:838–841. doi: 10.1001/jamaoncol.2017.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.