Abstract
GATA transcription factors (TFs) are type IV zinc-finger proteins that have roles in plant development and growth. The 27 GATA TFs identified in the Brachypodium distachyon genome in this study were unevenly distributed across all five chromosomes and classified into four subgroups. Phylogenesis-related GATAs shared similar gene structures and conserved motifs. Expression profiles showed that all BdGATA genes were expressed in leaves and most were induced by PEG treatment. BdGATA13 was predominantly expressed in leaf tissue and phylogenetically close to OsSNFL1, AtGNC, and AtGNL. Its protein was detected in the nucleus by subcellular localization analysis. Overexpression of BdGATA13 in transgenic Arabidopsis resulted in darker green leaves, later flowering, and more importantly, enhanced drought tolerance compared to the wild type. BdGATA13 also promoted primary root development under GA treatment. These results lay a foundation for better understanding the function of GATA genes in B. distachyon and other plants.
Keywords: BdGATA13, Brachypodium distachyon, drought stress, gene expression, transgenesis
Introduction
GATA transcription factors (TFs), which have the consensus sequence W-G-A-T-A-R (W = T or A, R = G or A), are a class of regulators that exist in plants, fungi, and metazoans (Block and Shapira, 2015; Kobayashi and Masuda, 2016). The DNA-binding ligand of GATAs consists of a type IV zinc-finger motif with the consensus sequence CX2CX17–20CX2C followed by a basic region (Gupta et al., 2017). The first GATA gene NTL1 containing a zinc finger motif with a C-X2-C-X17-C-X2-C sequence was identified in tobacco (Nicotiana tabacum L.) (Daniel-Vedele and Caboche, 1993).
Most studies have found that plant GATA TFs play an important role in plant development and growth. GATA12 in Arabidopsis contributes to primary seed dormancy (Ravindran et al., 2017). An Arabidopsis B-GATA TF with an LLM domain in the C-terminus controlling leaf greening was characterized as a regulator of vegetative growth and development (Behringer et al., 2014; Bastakis et al., 2018); GNC regulates seed germination, stem elongation, and flowering time (Richter et al., 2010, 2013a; Chiang et al., 2012); and HAN is required to locate the proembryo boundary in early Arabidopsis embryos (Nawy et al., 2010). HAN is considered a boundary gene that regulates the development of shoot apical meristems and flower organs (Zhang et al., 2013). Arabidopsis GNC and CGA1, and rice (Oryza sativa L.) Cga1 regulate chloroplast development (Chiang et al., 2012; Hudson et al., 2013). Overexpression of OsGATA12 in rice causes increased leaf greenness, reduced leaf and tiller numbers, and ultimately affects yield-related traits (Lu et al., 2017). OsGATA7 in rice modulates BR (brassinosteroid)-mediated regulation of plant architecture and grain shape (Zhang et al., 2018); NL1 (NECK LEAF 1) regulates organogenesis during reproductive development in rice (Wang et al., 2009); and overexpression of Cga1 caused semi-dwarf height and reduced tillering (Hudson et al., 2013). Arabidopsis GATA12 is regulated by GA in a DELLA-dependent manner (Ravindran et al., 2017); GNC is implicated in regulation of carbon and nitrogen metabolism, and represses gibberellin signaling downstream of the DELLA proteins (Richter et al., 2010, 2013b; Chiang et al., 2012).
GATA TFs also respond to abiotic and biotic stresses. Expression profiles analysis show that rice, Brassica juncea, Cucumis sativus, and pepper GATA genes are in response to different abiotic stresses, including high temperature, salinity, cold, and drought treatments (Bhardwaj et al., 2015; Gupta et al., 2017; Yu et al., 2021; Zhang K. et al., 2021). Arabidopsis GNC and GNL participate in cold stress response (Richter et al., 2013a); overexpression of OsGATA16 in rice improves cold tolerance (Zhang H. et al., 2021); overexpression of OsGATA8 in rice improves drought tolerance (Nutan et al., 2020); and overexpression of SlGATA17 improves drought tolerance in tomato (Zhao et al., 2021). Studies also show that the expression patterns of AtGATA21, AtGATA22, OsGATA11, GmGATA44, and GmGATA58 are all inducible by nitrate (Scheible et al., 2004; Hudson et al., 2013; Zhang et al., 2015). And overexpression of wheat TaGATA1 enhanced resistance to Rhizoctonia cerealis (Liu et al., 2020). The above reports indicate that a full assessment of GATA TFs in plants is needed due to their importance in development and growth as well as stress response.
Genome-wide analyses of GATA TFs in plant species identified 29 genes in Arabidopsis, 28 in rice, 35 in apple (Malus × domestica Borkh.), and 64 in soybean (Glycine max (L.) Merill.) (Reyes et al., 2004; Zhang et al., 2015; Chen et al., 2017). However, a systematic analysis of GATA TFs in the model grass Brachpodium distachyon has not occurred. In this study, 27 GATA TFs were identified in a genome-wide search in B. distachyon and the functions of BdGATA13 in plant growth and in response to drought and GA treatments were investigated. The study sets a foundation for further studies on GATA genes.
Materials and Methods
Identification of Brachpodium distachyon GATA Genes
To identify GATA TFs genome-wide in B. distachyon we downloaded PF00320, a characteristic GATA domain, from the Pfam database (Mistry et al., 2021) and searched against the B. distachyon genome protein sequence. Twenty-nine Arabidopsis and 28 rice GATA protein sequences (Reyes et al., 2004) were used to BLAST (Basic Local Alignment Search Tool) against the genome protein sequence of B. distachyon with a threshold of <e–5 and identity of 50%. Redundant genes were manually removed before the NCBI-CDD and SMART programs were used to confirm that GATA TFs without a GATA domain were completely removed.
We used the ExPASy ProtParam1 to predict physicochemical properties of GATA TFs, and subcellular locations were predicted using CELLO v2.52. The sequences of cDNAs, coding sequence (CDS), proteins, and DNA genes were extracted from the Ensembl Plants database3.
Chromosome Location, Gene Duplication, and Phylogenetic Analyses
Gene location information was obtained from Ensembl Plants, and tandem and segmental duplication events were obtained from the PGDD (Plant Genome Duplication Database) (Lee et al., 2017) and visualized using TBtools (Chen et al., 2020). Un-rooted neighbour joining (NJ) and maximum likelihood (ML) trees were constructed using MEGA 7 with 1,000 bootstrap replications and the Poisson model based on the full-length protein sequences (Kumar et al., 2016), and visualized by Evolview v3 (Subramanian et al., 2019).
Gene Structure and Conserved Motif Analyses
Gene structures of BdGATA genes were displayed by GSDS (Gene Structure Display Server) (Hu et al., 2015) after submitting the CDS and DNA sequences (Hu et al., 2015). MEME Suite (Bailey et al., 2009) was used to predict conserved motifs with the following parameters: number of motifs set at six, and width of motifs set from 6 to 50. The structures were visualized using Evolview v3 (Subramanian et al., 2019).
Plant Growth and Stress Treatment, RNA Extraction, and cDNA Synthesis
Ten-day-old B. distachyon seedlings were planted in a growth chamber at 26/24°C (day/night) with a 14/10 h day/night photoperiod. Roots, stems, leaves, and spikes collected after heading were used for tissue expression analysis. To apply abiotic stresses 10-day-old seedlings were treated with salt (200 mM NaCl), drought (20% PEG), heat (45°C), cold (4°C), ABA (200 μM), and GA (10 μM) for 2 h in hydroponic culture to obtain whole plants for analysis. Materials were frozen in liquid nitrogen and stored at −80°C for further use. RNA extraction and cDNA synthesis were performed using the RNA Easy Fast Plant Tissue and FastKing gDNA Dispelling RT SuperMix (Tiangen Biotech, Beijing) Kits, respectively.
Real-Time Quantitative-PCR
Real-time quantitative (qRT)-PCR was performed in triplicate using SuperReal PreMix Plus SYBR Green (Tiangen Biotech). Data collection and analyses were conducted using an ABI7900 system (Applied Biosystems, Germany). Data were normalized to BdGAPDH and Atactin 8 as described previously (Hong et al., 2008; Reichel et al., 2016) and calculated using the 2–ΔΔCt analysis method (Livak and Schmittgen, 2001). Primers used for PCR are listed in Supplementary Table 1.
Vector Construction, Plant Transformation, and Subcellular Localization Assay
The full-length coding sequence of BdGATA13 was amplified by PCR and cloned into the pCambia-1301 vector harboring the CaMV35S promoter. The recombinant vector was transformed into Arabidopsis strain Col-0 using the GV3101-mediated floral dip method (Clough and Bent, 1998). Transgenic lines were screened using a 0.1% hygromycin B solution and further confirmed by PCR. The full-length BdGATA13 coding sequence without a stop codon was inserted into the pCambia-1301-GFP vector to produce construct 35S: BdGATA13-GFP. For subcellular localization assays this construct and the vector pCAMBIA1301-GFP were co-transformed into tobacco leaves. Subcellular localization in tobacco leaves using GFP and DAPI staining was expedited by confocal microscopy (Olympus IX83-FV1200, Japan).
Tolerance Assays Under Stress Conditions
Seeds of wild type Arabidopsis and transgenic lines were surface-sterilized and sown on 1/2 MS plates and incubated in darkness at 4°C for 48 h before germination. Chlorophyll content was measured according to a previous study (Zhang et al., 2011). For phenotypic assessment under drought stress, 5-day-old seedlings were transplanted to 1/2 MS plates containing 0, 100 and 200 mM mannitol and cultured at 23°C in a 16/8 h (light/darkness) photoperiod for 10 days. Five-day-old seedlings for GA treatment were transplanted to 1/2 MS plates containing 0, 0.25 and 0.5 μmol GA3 and cultured under the above conditions for 5 days. There were six replicates of each treatment; root lengths for each sample were measured by ImageJ (Rueden et al., 2017); and data were analyzed using Microsoft Excel 2010. Error bars in all graphs represent means ± S.D and “∗” (P < 0.05) or “∗∗” (P < 0.01) were used to indicate significant differences found by Student’s t-tests.
Results
Identification of Brachypodium distachyon GATAs
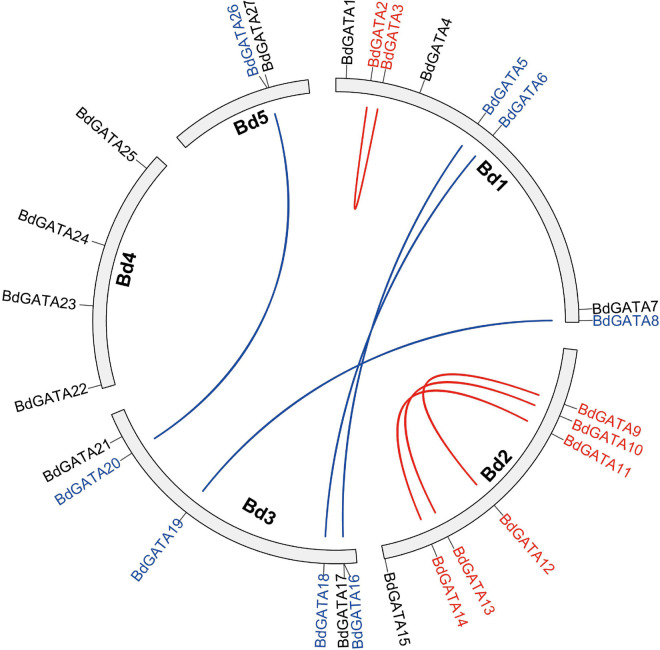
Twenty-seven GATAs were identified in the B. distachyon genome. To facilitate subsequent analysis, these GATA genes were named BdGATA1 to BdGATA27 according to their chromosomal location. The BdGATA loci were unevenly distributed and there were 2 to 8 genes on each chromosome (Figure 1). Eight BdGATA genes were present as four tandem duplications; and eight were in four segmental duplications.
FIGURE 1.
Chromosome locati on and analysis of duplicated BdGATAs. Red lines connect tandemly duplicated genes; blue lines connect segmentally duplicated genes.
BLAST of the CDS of each gene against the B. distachyon EST database in NCBI4 revealed that seven BdGATAs had no EST validation. Characteristic features of each gene were further examined. Average molecular weights, isoelectric points, and grand average hydropathicities were 35.478 KDa, 7.68, and −0.63, respectively. Detailed information for each gene is provided in Supplementary Table 2.
Phylogenetic Analysis of GATAs
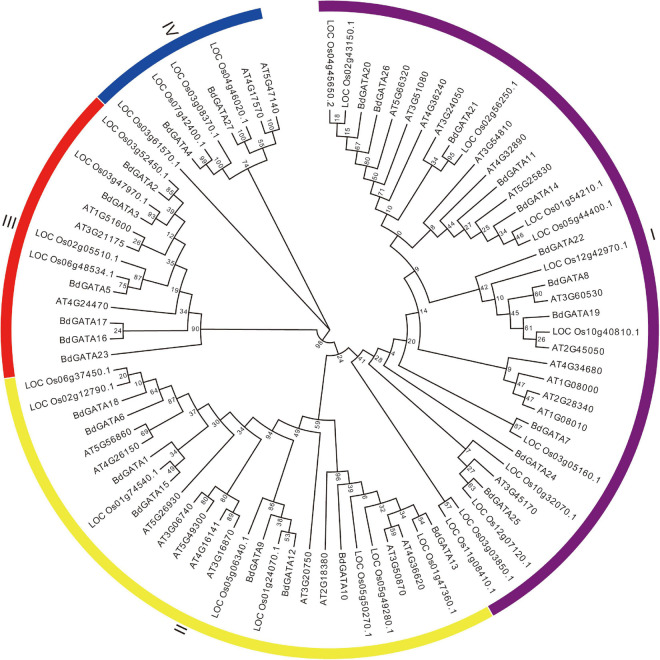
To elucidate the phylogenetic relationships among plant GATAs the 27 full-length GATA protein sequences in B. distachyon, 29 in Arabidopsis, and 28 in rice were extracted to build an un-rooted NJ tree. As shown in Figure 2 they were divided into four subgroups designated I, II, III, and IV based on bootstrap support. To further validate the reliability of the NJ tree the ML tree was also generated and formed the same subgroups (Supplementary Figure 1). The phylogenetically related genes were functionally conserved. For example, subgroup II members AT5G56860 (GNC), AT4G26150 (GNL), and LOC_Os06g37450 (OsGATA16) were found to participate in response to cold stress (Richter et al., 2013a; Zhang H. et al., 2021); and LOC_Os05g50270 (SNFL1) and LOC_Os02g12790 (Cga1) regulated plant architecture (Hudson et al., 2013; He et al., 2018); Overexpression of subgroup II members AT3G06740, AT3G16870, AT5G56860 (GNC), AT4G26150 (GNL), BdGATA15, and BdGATA18 in Arabidopsis showed increased chlorophyll accumulation and delayed flowering (Behringer et al., 2014). An unrooted NJ phylogenetic tree for B. distachyon placed 11, 8, 6, and 2 BdGATAs into subgroups I, II, III, and IV, respectively (Figure 3A).
FIGURE 2.
NJ tree of GATAs in plants. The tree included 27 GATAs from Brachypodium, 29 from Arabidopsis and 28 from rice, and construction was based on the full-length protein sequences. Four subgroups of GATAs were classified as I, II, III, and IV.
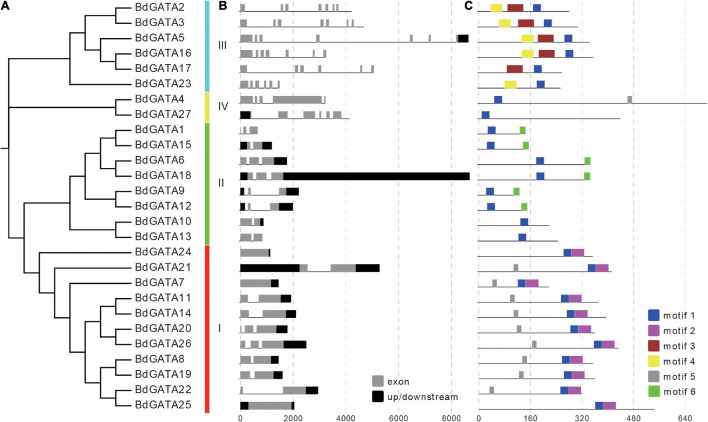
FIGURE 3.
Phylogenetic, gene structure, and conserved motif analysis of BdGATAs. (A) The phylogenetic tree divided GATAs into four subgroups. (B) Structures of BdGATA genes. Black rectangles indicate up/down stream sequences; gray rectangles indicate exons; gray lines indicate introns. (C) Conserved motif analysis of BdGATAs. Different motifs are identified and displayed in different colors.
Gene Structure and Conserved Motif Analysis
Analysis of the gene structures and conserved motifs in BdGATAs (Figure 3B) showed that the numbers of exons in BdGATA genes ranged from one to eight; exon numbers in subgroups I and II varied from three to eight whereas subgroups III and IV had one to three. Six types of motifs were identified in BdGATA proteins (Figure 3C); motif one was present in all BdGATAs whereas motifs two, three, and six were specific to subgroups IV, I, and III, respectively. Motif one formed the GATA domain.
Expression Pattern of GATA Genes
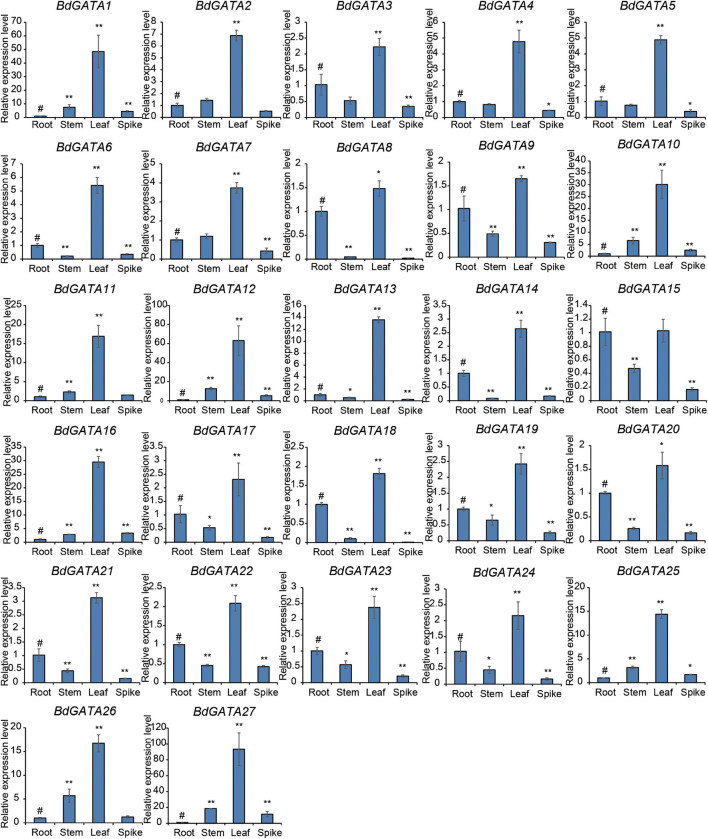
Analysis of the expression patterns of the GATA genes in root, stem, leaf, and spike tissues at different stages of plant development (Figure 4) showed that all BdGATA genes except BdGATA15 had much higher expression levels in the leaves than in other tissues. There were much lower differences among roots, stems, and spikes with some genes showing higher expression levels in roots but others with higher expression levels in stems or spikes.
FIGURE 4.
Expression patterns of BdGATAs in different tissues. The ordinate indicates the relative expression level, and the abscissa represents different tissues. The results were normalized against the expression of BdGAPDH as an internal control. Values are means ± SD (n = 3). Asterisks represent statistically significant differences between the indicated samples. Root (#) is used as control for each tissue. Student’s t-test: *P ≤ 0.05 and **P ≤ 0.01.
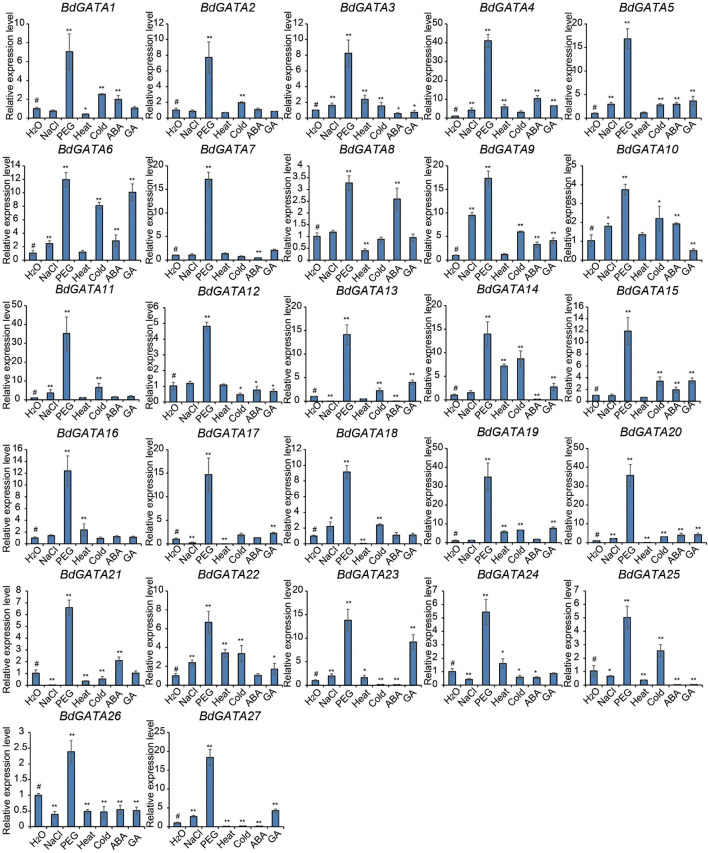
Gene expression under different abiotic stresses was also evaluated using RT-PCR (Figure 5). BdGATA genes also participated in abiotic stress responses. PEG treatment showed the greatest impact on expression of all the BdGATA genes. Other treatments caused changes in expression levels of some genes. For example, the expression levels of BdGATA6, BdGATA9, BdGATA13, BdGATA14, BdGATA15, BdGATA19, BdGATA23, and BdGATA27 were significantly upregulated by GA treatment; BdGATA6 and BdGATA15 were significantly upregulated and downregulated by cold treatment, respectively; and genes 12 and 6 were significantly upregulated and downregulated by salt treatment, respectively. Some genes, such as BdGATA26, showed consistent down-regulation under most treatments while others, such as BdGATA5 and BdGATA9, showed consistent up-regulation under most treatments.
FIGURE 5.
Expression patterns of BdGATAs under abiotic stresses. The ordinate indicates the relative expression level; and the horizontal axis represents different abiotic stresses, including salt (200 mM NaCl), drought (20% PEG), heat (45°C), cold (4°C), ABA (200 μM), and GA (10 μM) for 2 h in hydroponic culture. The results were normalized against the expression of BdGAPDH as an internal control. Values are means ± SD (n = 3). Asterisks represent statistically significant differences between the indicated samples. H2O (#) is used as control for each treatment. Student’s t-test: *P ≤ 0.05 and **P ≤ 0.01.
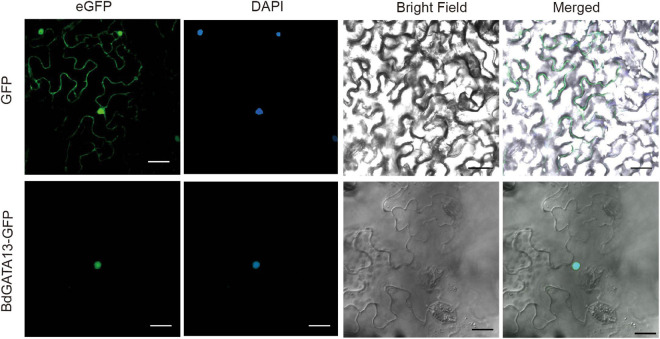
BdGATA13 Is Located in the Nucleus
A BdGATA13-eGFP fusion driven by the 35S promoter was transformed into tobacco leaves to investigate subcellular localization. The 35S:BdGATA13-eGFP fusion protein was detected in the nucleus (Figure 6), consistent with its predicted function as a TF.
FIGURE 6.
Subcellular localization of BdGATA13 protein. 35S: BdGATA13-GFP was transferred into tobacco leaves, and fluorescence signals of GFP were detected in tobacco leaf epidermal cells. Left panel, GFP image; middle panel, bright field; and right panel, merge of GFP and bright field. Bar, 10 μm.
Overexpression of BdGATA13 in Arabidopsis Increases Chlorophyll Content and Delays Flowering Time
All BdGATA genes were highly expressed in leaves (Figure 4), indicating that BdGATA genes have an important role in leaf growth and development. As one example, we investigated the function of BdGATA13, a gene predominantly expressed in leaf tissue but not previously studied. This gene is phylogenetically close to subgroup II genes LOC_Os05g50270 (SNFL1), GNC (AT5G56860), and GNL (AT4G26150). To further analyze whether their functions were conserved in the phylogeny, the CDS of BdGATA13 driven by the 35S promoter was transformed into Arabidopsis, and two transgenic lines (ox-5 and ox-15) showing different gene expression levels (Figure 7A) were generated and used for phenotypic analyses. Overexpression of BdGATA13 produced dark green seedling leaves (Figures 7B,C) by accumulation of chlorophyll (Figure 7D) under both dark and light conditions. Flowering time of the transgenic lines was also delayed (Figures 7E,F).
FIGURE 7.
Effects of the ectopic expression of BdGATA13 on greening and flowering in transgenic Arabidopsis. (A) Relative expression levels of two transgenic lines by qRT-PCR, N.D., not detected. (B) Phenotype of the WT and the two transgenic lines in 5-day-old seedlings grown under long-day conditions, bar = 1 mm. (C) Phenotypes of 5-day-old WT and two transgenic lines grown in darkness, bar = 1 mm. (D) Average and standard deviations of chlorophyll levels of WT and transgenic plants grown for 5 days in light or darkness (three biological replications). (E) Flowering time comparison of WT and transgenic lines grown in long-day conditions. (F) Comparison of days to flowering for WT and transgenic plants. Values are means ± SD (n = 3). Asterisks represent statistically significant differences between the indicated samples. Student’s t-test: *P ≤ 0.05, **P ≤ 0.01.
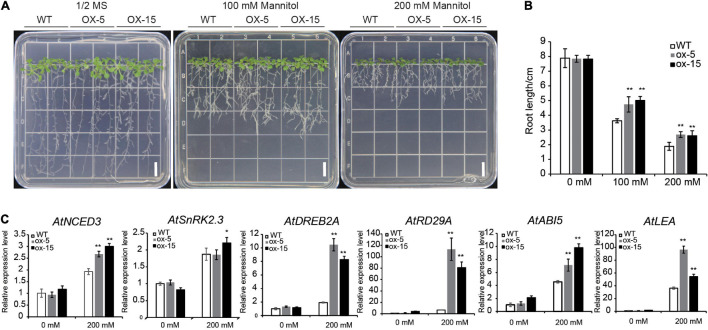
BdGATA13 Enhances Drought Tolerance in Transgenic Arabidopsis
The expression level of BdGATA13 was increased by PEG and GA treatments (Figure 5). Root growth of the transgenic lines on 1/2 MS medium was similar to wild type plants but was clearly increased relative to the WT under drought treatment (P < 0.01) (Figure 8A). When grown in 100 mM mannitol solution, the root lengths of the transgenic lines were increased by 30.70 and 38.08% compared to wild type plants (P < 0.01) (Figure 8B). When grown in 200 mM mannitol, the root lengths of the transgenic lines were increased by 42.41 and 38.19%, respectively (P < 0.01) (Figure 8B). These results clearly demonstrated that BdGATA13 enhanced drought tolerance in Arabidopsis.
FIGURE 8.
Overexpression of BdGATA13 enhanced tolerance to drought stress in Arabidopsis seedlings. (A) 5-day-old of seedlings of Arabidopsis were grown in 0, 100, and 200 mM mannitol for 10 day; bars = 1 cm. (B) Primary root lengths of WT and transgenic lines. (C) qRT-PCR results for six drought-related genes under 0 and 200 mM mannitol. Values are means ± SD (n = 3). Asterisks represent statistically significant differences between the indicated samples. Student’s t-tests: *P ≤ 0.05, **P ≤ 0.01.
The expression levels of several drought-responsive genes, including RD29A, ABI5, LEA, NCED3, SnRK2.3, and DREB2A, were also determined. Except for SnRK2.3, expression levels of these genes were induced by drought treatment (Figure 8C). These results indicated potential links between BdGATA13 and drought-related genes in Arabidopsis.
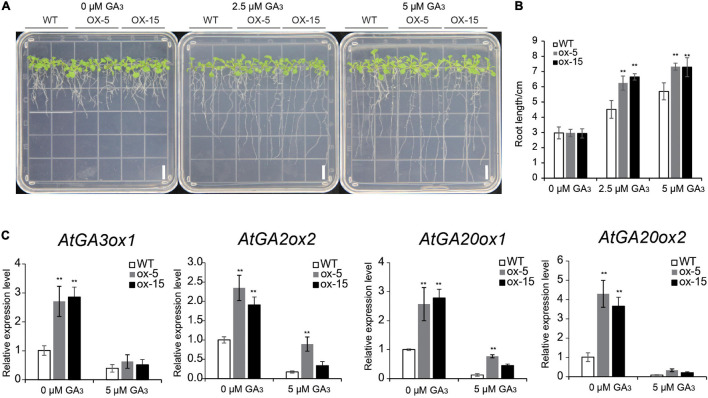
BdGATA13 Affects Plant Growth by Negative Regulation of GA Signaling
The expression level of BdGATA13 was upregulated by GA3 treatment. As shown in Figure 9A, the root lengths of the two transgenic lines did not differ from that of wild type plants under control conditions. However, under 2.5 μM GA3 treatment, the root lengths of the transgenic lines were 38.32–47.26% higher than the wild type (P < 0.01) and under 5 μM GA3 treatment, the root lengths of transgenic lines were 28.69% higher than the wild type (P < 0.01) (Figure 9B).
FIGURE 9.
Overexpression of BdGATA13 increased root length in Arabidopsis seedlings under GA3 treatment. (A) 5-day-old Arabidopsis seedlings were subjected to 0, 0.25, and 0.5 μmol GA3 treatment for 5 day; bars = 1 cm. (B) Comparison of primary roots in WT and transgenic lines. (C) qRT-PCR results for four gibberellin-related genes under 0 and 0.5 μmol GA3. Values are means ± SD (n = 3). Asterisks represent statistically significant differences between the indicated samples. Student’s t-tests: *P ≤ 0.05, **P ≤ 0.01.
The expression levels of gibberellin-related genes GA3ox1, GA2ox, GA20ox1, and GA20ox2 were also assayed. Expression levels of all four genes in the transgenic lines were higher than those in the WT without GA3 treatment (P < 0.01) (Figure 9C) and under 5 μM GA3 treatment expression levels of the four genes in transgenic plants were similar to the wild type (Figure 9C). These results suggested that BdGATA13 affects plant growth by negative regulation of GA signaling.
Discussion
Characteristics of GATA in Brachypodium distachyon
We identified 27 GATA TFs in the B. distachyon genome, a similar number to Arabidopsis (29) (Reyes et al., 2004), rice (28) (Gupta et al., 2017), pepper (Capsicum tetragonum L.) (28) (Yu et al., 2021), but less than in Brassica napus (96) (Zhu et al., 2020), apple (Melus pumila L.) (35) (Chen et al., 2017), and soybean [Glycine max (L.) Merill] (64) (Zhang et al., 2015). Among the 27 BdGATA genes, 16 were duplicated, including eight tandemly duplicated and eight segmently duplicated. The expression levels of the segmently duplicated genes were similar in all tissues. For example, BdGATA8 and BdGATA19 were segmently duplicated and showed low expression in stem and splike tissues but high expression in leaf tissue. These results indicated that gene duplication contributed to expansion of the GATA gene family in B. distachyon. This corresponds with findings in apple (Chen et al., 2017), Gossypium hirsutum L. (Zhang et al., 2019), B. napus (Zhu et al., 2020), and pepper (Yu et al., 2021).
Phylogenetic analysis of rice, Arabidopsis, and B. distachyon GATAs identified four subgroups (Figure 2). In the case of B. distachyon there were 11, 8, 6, and 2 members in each subgroup and subgroups shared similar gene structures and conserved motifs, and implying conserved functions among members within each subgroup (Figure 3). Among conserved motifs, motif one was present in all BdGATA members and formed a GATA domain. Expression pattern analysis showed that all BdGATA genes were highly expressed in leaf tissues. Members in the same subgroup had similar functions. For example, subgroup II members GNC (AT5G56860) and GNL (AT4G26150) contribute to chlorophyll biosynthesis as evidensed by chlorophyll accumulation in grown A. thaliana seedlings grown in light (Bastakis et al., 2018). Overexpression of Arabidopsis subgroup II members AT3G06740, AT3G16870, AT5G56860 (GNC), AT4G26150 (GNL), BdGATA15, and BdGATA18 showed increased chlorophyll accumulation and delayed flowering (Behringer et al., 2014). Subgroup II member BdGATA13 also accumulated chlorophyll when grown in light, confirming that BdGATA genes in the same subgroups have similar functions.
BdGATA13 Regulates Plant Development and Responds to Stress
All BdGATA genes had relatively high expression levels in leaf tissues indicating a significant role of GATA genes in leaf development. Overexpression of BdGATA13 in Arabidopsis caused plants to be greener, due to higher chlorophyll content than in wild type controls (Figures 4, 6). It was shown previously that overexpression of BdGATA4 (named BdGATA15 in this study), BdGATA6 (named BdGATA18 in this study), and SlGATA4, SlGATA5, and SlGATA7 from S. lycopersicon in Arabidopsis produced dark green leaves and accumulated high levels of chlorophyll when grown in light (Behringer et al., 2014). These results indicate that BdGATA genes have essential roles in chlorophyll biosynthesis.
In addition to regulating chlorophyll biosynthesis and chloroplast development GATA genes also function in seed germination, flowering time, and response to abiotic stress (Richter et al., 2010; Zhang et al., 2013). For example, the Arabidopsis gnc mutant flowered earlier than the wild type and overexpression of GNC showed a late-flowering phenotype (Richter et al., 2010); whereas in wheat, overexpression of TaZIM-A1 caused delayed flowering under long-day conditions (Liu et al., 2019). Our results also showed that overexpression of BdGATA13 caused delayed flowering in transgenic overexpression lines.
Real-time quantitative-PCR results showed that all BdGAGA genes were upregulated by PEG treatment, suggesting they function in response to drought. Overexpression of BdGATA13 promoted drought tolerance in transgenic plants by regulating the expression of drought-related genes such as RD29A, ABI5, LEA, NCED3, and DREB2A. Consistent with BdGATA13 results, overexpression of OsGATA8 in rice increased tolerance to drought stress (Nutan et al., 2020), and overexpression of SlGATA17 improved drought tolerance in tomato (Zhao et al., 2021).
Previous studies showed that Arabidopsis GNC is a transcriptional target downstream of GA (Richter et al., 2010). Some plant GATAs were induced by GA3. For example, the expression of GmGATA58 was promoted by GA3 treatment (Zhang et al., 2020). In the present study, the expression levels of four GA-related genes in transgenic lines grown under normal conditions, namely GA3ox1, GA2ox, GA20ox1, and GA20ox2, were higher than in the WT. With GA3 treatment, expression levels of these genes in BdGATA13 transgenic lines returned to normal levels, indicating that BdGATA13 plays a role in plant growth by negatively regulating GA signaling. The overall results suggest that BdGATA13 transcribes a TF that regulates various functions involved in abiotic stress and development in plants.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JG analyzed the data and wrote the manuscript. XB, KD, and XY helped to carry out the experiments. PG and MZ contributed to writing the manuscript. CH and WS contributed to the experimental design, provided the advice for data analysis, and assisted in writing the manuscript. All authors have read and approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully acknowledge help from Robert A McIntosh, University of Sydney, for English editing.
Footnotes
Funding
This work was supported by grants from the National Natural Science Foundation of China (31901541) and Natural Science Foundation of Shanxi Province (201901D211361). The funding body did not exert influence on the design of the study, and collection, analysis, and interpretation of data or in writing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.763665/full#supplementary-material
Maximum likelihood (ML) tree of GATAs in plants. The numbers of GATAs were 27 in Brachypodium, 29 in Arabidopsis, and 28 in rice, and construction was based on the full-length protein sequences. Four subgroups of GATAs were classified as I, II, III, and IV.
References
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37 W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastakis E., Hedtke B., Klermund C., Grimm B., Schwechheimer C. (2018). LLM-domain B-GATA transcription factors play multifaceted roles in controlling greening in Arabidopsis. Plant Cell 30 582–599. 10.1105/tpc.17.00947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer C., Bastakis E., Ranftl Q. L., Mayer K. F., Schwechheimer C. (2014). Functional diversification within the family of B-GATA transcription factors through the leucine-leucine-methionine domain. Plant Physiol. 166 293–305. 10.1104/pp.114.246660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj A. R., Joshi G., Kukreja B., Malik V., Arora P., Pandey R., et al. (2015). Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea. BMC Plant Biol. 15:9. 10.1186/s12870-014-0405-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block D. H., Shapira M. (2015). GATA transcription factors as tissue-specific master regulators for induced responses. Worm 4:e1118607. 10.1080/21624054.2015.1118607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. J., Chen H., Zhang Y., Thomas H. R., Frank M. H., He Y. H., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 1194–1202. 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Chen H. F., Shao H. X., Li K., Zhang D., Fan S., Li Y. M., et al. (2017). Genome-wide identification, evolution, and expression analysis of GATA transcription factors in apple (Malus× domestica Borkh.). Gene 627 460–472. 10.1016/j.gene.2017.06.049 [DOI] [PubMed] [Google Scholar]
- Chiang Y. H., Zubo Y. O., Tapken W., Kim H. J., Lavanway A. M., Howard L., et al. (2012). Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 160 332–348. 10.1104/pp.112.198705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Daniel-Vedele F., Caboche M. (1993). A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol. Gen. Genet. 240 365–373. 10.1007/BF00280388 [DOI] [PubMed] [Google Scholar]
- Gupta P., Nutan K. K., Singla-Pareek S. L., Pareek A. (2017). Abiotic stresses cause differential regulation of alternative splice forms of GATA transcription factor in rice. Front. Plant Sci. 8:1944. 10.3389/fpls.2017.01944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P., Wang X., Zhang X., Jiang Y., Tian W., Zhang X., et al. (2018). Short and narrow flag leaf1, a GATA zinc finger domain-containing protein, regulates flag leaf size in rice (Oryza sativa). BMC Plant Biol. 18:273. 10.1186/s12870-018-1452-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S. Y., Seo P. J., Yang M. S., Xiang F., Park C. M. (2008). Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 8:112. 10.1186/1471-2229-8-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Jin J., Guo A. Y., Zhang H., Luo J., Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31 1296–1297. 10.1093/bioinformatics/btu817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson D., Guevara D. R., Hand A. J., Xu Z., Hao L., Chen X., et al. (2013). Rice cytokinin GATA transcription Factor1 regulates chloroplast development and plant architecture. Plant Physiol. 162 132–144. 10.1104/pp.113.217265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Masuda T. (2016). Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 7:1811. 10.3389/fpls.2016.01811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. H., Kim J., Robertson J. S., Paterson A. H. (2017). Plant genome duplication database. Methods Mol. Biol. 1533 267–277. 10.1007/978-1-4939-6658-5_16 [DOI] [PubMed] [Google Scholar]
- Liu H., Li T., Wang Y. M., Zheng J., Li H. F., Hao C. Y., et al. (2019). TaZIM-A1 negatively regulates flowering time in common wheat (Triticum aestivum L.). J. Integr. Plant Biol. 61 359–376. 10.1111/jipb.12720 [DOI] [PubMed] [Google Scholar]
- Liu X., Zhu X., Wei X., Lu C., Shen F., Zhang X., et al. (2020). The wheat LLM-domain-containing transcription factor TaGATA1 positively modulates host immune response to Rhizoctonia cerealis. J. Exp. Bot. 71 344–355. 10.1093/jxb/erz409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu G., Casaretto J. A., Ying S., Mahmood K., Liu F., Bi Y. M., et al. (2017). Overexpression of OsGATA12 regulates chlorophyll content, delays plant senescence and improves rice yield under high density planting. Plant Mol. Biol. 94 215–227. 10.1007/s11103-017-0604-x [DOI] [PubMed] [Google Scholar]
- Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G. A., Sonnhammer E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49 D412–D419. 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T., Bayer M., Mravec J., Friml J., Birnbaum K. D., Lukowitz W. (2010). The GATA factor HANABA TARANU is required to position the proembryo boundary in the early Arabidopsis embryo. Dev. Cell 19 103–113. 10.1016/j.devcel.2010.06.004 [DOI] [PubMed] [Google Scholar]
- Nutan K. K., Singla-Pareek S. L., Pareek A. (2020). The Saltol QTL-localized transcription factor OsGATA8 plays an important role in stress tolerance and seed development in Arabidopsis and rice. J. Exp. Bot. 71 684–698. 10.1093/jxb/erz368 [DOI] [PubMed] [Google Scholar]
- Ravindran P., Verma V., Stamm P., Kumar P. P. (2017). A novel RGL2-DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol. Plant 10 1307–1320. 10.1016/j.molp.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Reichel M., Liao Y., Rettel M., Ragan C., Evers M., Alleaume A. M., et al. (2016). In planta determination of the mRNA-binding proteome of Arabidopsis etiolated seedlings. Plant Cell 28 2435–2452. 10.1105/tpc.16.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes J. C., Muro-Pastor M. I., Florencio F. J. (2004). The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 134 1718–1732. 10.1104/pp.103.037788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R., Bastakis E., Schwechheimer C. (2013a). Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiol. 162 1992–2004. 10.1104/pp.113.219238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R., Behringer C., Muller I. K., Schwechheimer C. (2010). The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and phytochrome-interacting factors. Genes Dev. 24 2093–2104. 10.1101/gad.594910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R., Behringer C., Zourelidou M., Schwechheimer C. (2013b). Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 110 13192–13197. 10.1073/pnas.1304250110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden C. T., Schindelin J., Hiner M. C., Dezonia B. E., Walter A. E., Arena E. T., et al. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 18:529. 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible W. R., Morcuende R., Czechowski T., Fritz C., Osuna D., Palacios-Rojas N., et al. (2004). Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 136 2483–2499. 10.1104/pp.104.047019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian B., Gao S., Lercher M. J., Hu S., Chen W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47 W270–W275. 10.1093/nar/gkz357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. P., Yin H. F., Qian Q., Yang J., Huang C. F., Hu X. H., et al. (2009). NECK LEAF 1, a GATA type transcription factor, modulates organogenesis by regulating the expression of multiple regulatory genes during reproductive development in rice. Cell Res. 19 598–611. 10.1038/cr.2009.36 [DOI] [PubMed] [Google Scholar]
- Yu C. Y., Li N., Yin Y. X., Wang F., Gao S. H., Jiao C. H., et al. (2021). Genome-wide identification and function characterization of GATA transcription factors during development and in response to abiotic stresses and hormone treatments in pepper. J. Appl. Genet. 62 265–280. 10.1007/s13353-021-00618-3 [DOI] [PubMed] [Google Scholar]
- Zhang C. J., Hou Y. Q., Hao Q. N., Chen H. F., Chen L. M., Yuan S. L., et al. (2015). Genome-wide survey of the soybean GATA transcription factor gene family and expression analysis under low nitrogen stress. PLoS One 10:e0125174. 10.1371/journal.pone.0125174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. J., Huang Y., Xiao Z. Y., Yang H. L., Hao Q. N., Yuan S. L., et al. (2020). A GATA transcription factor from soybean (Glycine max) regulates chlorophyll biosynthesis and suppresses growth in the transgenic Arabidopsis thaliana. Plants (Basel) 9:1036. 10.3390/plants9081036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. J., Liu J. X., Zhang Y. Y., Cai X. F., Gong P. J., Zhang J. H., et al. (2011). Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep. 30 389–398. 10.1007/s00299-010-0939-0 [DOI] [PubMed] [Google Scholar]
- Zhang H., Wu T., Li Z., Huang K., Kim N. E., Ma Z., et al. (2021). OsGATA16, a GATA Transcription Factor, confers cold tolerance by repressing OsWRKY45-1 at the seedling stage in rice. Rice 14:42. 10.1186/s12284-021-00485-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Jia L., Yang D., Hu Y., Njogu M. K., Wang P., et al. (2021). Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in cucumber (Cucumis sativus L.). Plants (Basel) 10:1626. 10.3390/plants10081626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. L., Zhou Y., Ding L., Wu Z. G., Liu R. Y., Meyerowitz E. M. (2013). Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25 83–101. 10.1105/tpc.112.107854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. J., Zhang Y., Zhang L. L., Huang H. Y., Yang B. J., Luan S., et al. (2018). OsGATA7 modulates brassinosteroids-mediated growth regulation and influences architecture and grain shape. Plant Biotechnol. J. 16 1261–1264. 10.1111/pbi.12887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zou X. Y., Huang Z., Fan S. M., Qun G., Liu A. Y., et al. (2019). Genome-wide identification and analysis of the evolution and expression patterns of the GATA transcription factors in three species of Gossypium genus. Gene 680 72–83. 10.1016/j.gene.2018.09.039 [DOI] [PubMed] [Google Scholar]
- Zhao T. T., Wu T. R., Pei T., Wang Z. Y., Yang H. H., Jiang J. B., et al. (2021). Overexpression of SlGATA17 promotes drought tolerance in transgenic tomato plants by enhancing activation of the phenylpropanoid biosynthetic pathway. Front. Plant Sci. 12:634888. 10.3389/fpls.2021.634888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W. G., Guo Y. Y., Chen Y. K., Wu D. Z., Jiang L. X. (2020). Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in Brassica napus. BMC Plant Biol. 20:543. 10.1186/s12870-020-02752-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Maximum likelihood (ML) tree of GATAs in plants. The numbers of GATAs were 27 in Brachypodium, 29 in Arabidopsis, and 28 in rice, and construction was based on the full-length protein sequences. Four subgroups of GATAs were classified as I, II, III, and IV.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.