Abstract
The ubiquitin system is an important part of the host cellular defense program during bacterial infection. This is in particular evident for a number of bacteria including Salmonella Typhimurium and Mycobacterium tuberculosis which—inventively as part of their invasion strategy or accidentally upon rupture of seized host endomembranes—become exposed to the host cytosol. Ubiquitylation is involved in the detection and clearance of these bacteria as well as in the activation of innate immune and inflammatory signaling. Remarkably, all these defense responses seem to emanate from a dense layer of ubiquitin which coats the invading pathogens. In this review, we focus on the diverse group of host cell E3 ubiquitin ligases that help to tailor this ubiquitin coat. In particular, we address how the divergent ubiquitin conjugation mechanisms of these ligases contribute to the complexity of the anti‐bacterial coating and the recruitment of different ubiquitin‐binding effectors. We also discuss the activation and coordination of the different E3 ligases and which strategies bacteria evolved to evade the activities of the host ubiquitin system.
Keywords: E3 ligase, innate immunity, Salmonella, ubiquitin, xenophagy
Subject Categories: Autophagy & Cell Death; Microbiology, Virology & Host Pathogen Interaction; Post-translational Modifications & Proteolysis
The ubiquitin system is an important part of the cellular defense against invading bacterial pathogens. This review discusses the function and interplay of the different E3 ligases that act in this tug of war between bacteria and the host cell.

Glossary
- ARIH1
ARIadne 1 homolog
- ATG
Autophagy‐related
- BMDM
Bone marrow‐derived macrophages
- BMP
Bone morphogenic protein
- CALCOCO
Calcium‐binding and coiled‐coil domain‐containing protein
- CC1
Coiled‐coil 1
- CRL
Cullin RING ligase
- DUB
Deubiquitinating enzyme
- GAS
Group A streptococcus
- GIR
Galectin interacting region
- GWAS
Genomewide association study
- HECT
Homologous to E6AP carboxyl terminus
- HHAR1
Homologues to human ARIadne 1
- HOIL‐IL
Heme‐oxidized IRP2 Ub ligase 1L
- HOIP
HOIL‐1‐interacting protein
- IBR
In between RING
- IKK
IκB kinase
- LAP
LC3‐associated phagocytosis
- LDD
Linear Ub‐determining domain
- LIR
LC3‐interacting region
- LPS
Lipopolysaccharide
- LRR
leucine‐rich repeat
- LRSAM1
Leucine‐rich repeat and sterile alpha motif‐containing protein 1
- LUBAC
Linear Ub assembly complex
- MAP1LC3B
Microtubule‐associated proteins 1A/1B light‐chain 3B
- NBR1
Neighbor of BRCA1
- NDP52
Nuclear dot protein 52
- NEL
Novel E3 ligase
- NEMO
NF‐kB essential modifier
- NF‐kB
Nuclear factor kappa‐B
- NOD1
Nucleotide‐binding oligomerization domain‐containing protein 1
- OMP
Outer membrane protein
- OPTN
Optineurin
- OTULIN
OTU domain DUB with LINear linkage specificity
- PAMP
Pathogen‐associated molecular pattern
- PE
Phosphatidylethanolamine
- PINK1
PTEN‐induced kinase 1
- RBR
RING‐between‐RING
- RING
Really interesting new gene
- RNF213
Ring finger protein 213
- SAM
Sterile alpha motif‐containing domain
- SCV
Salmonella‐containing vacuole
- SHARPIN
Shank‐associated RH domain interacting protein
- SIM
Structured illumination microscopy
- SLO
Streptolysin O
- Smurf1
SMAD Ub regulator factor 1
- SQSTM1
Sequestosome‐1
- TAK1
Transforming growth factor‐β‐activated kinase 1
- TBK1
TANK‐binding kinase 1
- TGF‐β
transforming growth factor beta
- TLR
Toll‐like receptor
- TRAF2
TNF receptor‐associated factor 2
- TRIM21
Tripartite motif‐containing protein 21
- TXN
Thioredoxin
- Ub
Ubiquitin
- UBA
Ubiquitin‐associated
- UBL
Ubiquitin‐like domain
- UPD
Unique Parkin domain
- UVRAG
UV radiation resistance associated
- ZF
Zinc finger
Introduction
Most mammalian cell types have the capacity to defend and protect themselves against pathogens, thereby contributing to cell‐autonomous innate immunity. Over the last two decades, macroautophagy has emerged as a critical innate immunity pathway that detects a variety of cytosolic pathogens and initiates their disposal.
Originally, macroautophagy has been described as a catabolic process that sequesters unwanted intracellular components, including misfolded proteins and dysfunctional organelles, and eliminates them through the lysosomal degradation machinery (Fig. 1A). A hallmark of macroautophagy is the de novo formation of a double‐membrane vesicle called autophagosome (Nakatogawa, 2020). Autophagosome biogenesis proceeds from an isolated crescent‐shaped membrane structure (called phagophore or isolation membrane) that is derived from preexisting endomembranes including the ER and that grows around, and ultimately encloses cytosolic components. To complete macroautophagy, autophagosome fuses with the lysosome, forming an autolysosome to allow disintegration of the inner membrane and the degradation of the sequestered material by diverse lysosomal hydrolases, such as lipases, proteases, glycosylases, and nucleases (Glick et al, 2010; Rubinsztein et al, 2012). To date, an increasing number of autophagy‐related (ATG) proteins have been identified that are essential for the formation of the autophagosome and its fusion with the lysosome. Their molecular details and mechanistic interactions have been worked out over the last decades, their description, however, would exceed the scope of this review and the interested reader is referred to some excellent recent reviews (Noda & Inagaki, 2015; Nakatogawa, 2020).
Figure 1. Selective macroautophagy.
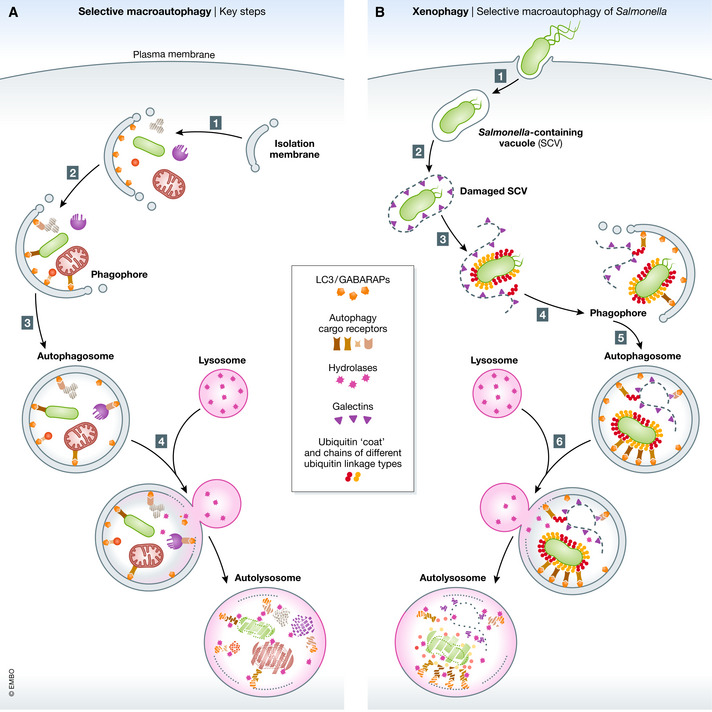
(A) Key steps in selective macroautophagy. (1) The process of macroautophagy is initiated by isolation membranes and vesicles that gradually expand and mature to phagophores decorated with membrane‐anchored LC3 and GABARAPs (orange hexagons). (2) These act as a binding site for autophagy cargo receptors (brown) allowing direct delivery and accumulation of cellular cargo. Finally, the phagophore closes forming a double‐membraned vesicle called autophagosome (3), which subsequently fuses with the lysosome forming an autolysosome (4), where the content is degraded by lysosomal enzymes. (B) Xenophagy, selective macroautophagy of Salmonella. (1) Salmonella enters host cells by forming a Salmonella‐containing vacuole (SCV) that protects it from the host surveillance system and serves as a replicative niche. (2) SCVs can rupture allowing access of host cytosolic proteins. (3) The exposed glycans, that are normally present on the outer side of the cell membrane, serve as a danger signals that are recognized by galectins (purple triangles). Besides, Salmonella is detected by E3 ligases that generate a dense Ub coat, consistent of different linkage‐type Ub chains (red and orange circles), around Salmonella. (4) Both, galectins and Ub recruit a variety of autophagy receptors (brown) that mediate Salmonella capture by LC3‐conjugated phagophores and Salmonella‐containing autophagosomes (5) are targeted for lysosomal degradation (6).
Macroautophagy comes in two flavors, depending on how degradation targets are sequestered. During bulk macroautophagy, the forming autophagosome engulfs randomly cytoplasmic components that are close by. This form of macroautophagy is active at a basal level in nutrient‐rich conditions, but can be upregulated in response to several stress conditions, such as starvation, thereby playing an integral part in recycling nutrients to maintaining energy homeostasis. By contrast, during selective macroautophagy, certain molecules, structures, and organelles are recognized via eat‐me signals by target‐specific proteins named autophagy receptors, which in turn mediate a stepwise recruitment of different macroautophagy machinery components to initiate and scaffold autophagosome formation exclusively around a distinct cargo (Reggiori et al, 2012; Stolz et al, 2014; Zaffagnini & Martens, 2016). Selective macroautophagy can be classified according to its cargo, including mitophagy (mitochondria), ribophagy (ribosomes), ER‐phagy (ER), lipophagy (lipid droplets), lysophagy (lysosomes), and pexophagy (peroxisomes) (Gatica et al, 2018).
However, cells utilize selective macroautophagy not only to clear and recycle their own cellular material, but can also direct the macroautophagy machinery to eliminate invaded cellular pathogens, a process commonly known as antimicrobial macroautophagy or xenophagy—from Greek for ‘foreign/strange’ and ‘eating’ (Hu et al, 2020). Non‐canonical forms of autophagy that do not involve the formation of double‐membrane autophagosomes such as LC3‐associated phagocytosis (LAP) also contribute to eliminate pathogens (Sanjuan et al, 2007; Mehta et al, 2014; Heckmann et al, 2017; Martinez, 2018). However, in this review, we will focus on our current mechanistic understanding of xenophagy and discuss recent advances on the role of the ubiquitin (Ub) machinery in targeting pathogenic bacteria for their clearance and how bacteria developed ways to fend off this process for survival or to even hijack it for their own benefit.
Mechanism of ubiquitin‐mediated xenophagy
Intracellular pathogens, such as the bacteria Shigella flexneri (S. flexneri), Mycobacterium tuberculosis (M. tuberculosis), Salmonella enterica serovar Typhimurium (S. Typhimurium), and Listeria monocytogenes (L. monocytogenes), are able to reside and reproduce inside the host cell and are responsible for many severe human diseases. Besides passive engulfment by macrophages and neutrophils via phagocytosis, bacteria evolved different strategies to actively invade endothelial and epithelial host cells (Cossart & Sansonetti, 2004). Following their entry, pathogens can either reside inside membranous compartments (vacuoles) or escape to the cytosol. Nevertheless, xenophagy can target these bacteria to restrict their growth and replication. Since S. Typhimurium is one of the best‐studied examples of bacteria that are targeted and eliminated by xenophagy (Herhaus & Dikic, 2018), we will use this model substrate to describe the different steps and key features of antimicrobial macroautophagy (Fig. 1B). After internalization, S. Typhimurium resides in an acidic compartment called the Salmonella‐containing vacuole (SCV). SCVs are used by S. Typhimurium as a permissive niche for survival and growth (LaRock et al, 2015). Numerous bacterial genes, including the effector SifA, are required for the extension of the membranous surface of the SCV to accommodate the replicating bacteria. However, in the early phase of infection (usually within the first hour), a subset of SCV membranes is damaged and specific signature molecules become exposed on the bacterial cell wall as well as on the luminal surface of the SCV. Specifically, β‐galactosides—which normally modify the extracellular side of the plasma membrane and the luminal surface of endosomes—are recognized on the damaged SCV membranes by the fast recruitment of the β‐galactoside‐binding lectin, galectin‐8 which acts as an essential damage and/or pattern‐recognition receptor and directly associates with the autophagy receptor NDP52 (also known as CALCOCO2) via its the galectin‐interacting region (GIR) motif (Thurston et al, 2012).
Besides β‐galactosides/galectin‐8‐initiated autophagy, damaged SCVs and exposed Salmonella activate the cellular Ub machinery and trigger the formation of a dense Ub coat dressing the whole Salmonella (Perrin et al, 2004). Ubiquitylation, the covalent attachment of Ub to substrate proteins, requires the orchestrated action of at least three enzymes working in a relay, an Ub‐activating enzyme (E1), an Ub‐conjugating enzyme (E2), and an Ub ligase (E3) (Hershko et al, 2000; Buetow & Huang, 2016). The E3s play a central part in substrate specificity and can be classified into three main families depending on the presence of characteristic domains and on the mechanism of Ub transfer to usually lysine residues of substrate proteins: really interesting new gene (RING) E3 ligases catalyze via allosteric activation of the E2˜Ub conjugate (˜ indicating a thioester bond) the direct Ub transfer from the E2 to the substrate, whereas homologous to the E6AP carboxyl terminus (HECT) E3 ligases and Ring‐Between‐Ring (RBR) E3 ligase utilize a catalytic cysteine forming a transient E3˜Ub thioester before mediating Ub transfer onto substrates (Zheng & Shabek, 2017). Substrates can be modified by single Ub molecules on one or multiple lysins (mono‐ or multi‐monoubiquitylation) and/or polyubiquitin chains (polyubiquitylation) can be formed by linking Ub molecules via one of their seven lysine residues (K6, K11, K27, K29, K33, K48, K63) or N‐terminal methionine (M1). These Ub chains are structurally distinct and can come in different variations, homotypic (single linkage type) or heterotypic (multiple, mixed linkage types) including branched polyubiquitin chains, resulting in a complex Ub code (Komander & Rape, 2012). Identity and origin of ubiquitylation targets forming the Ub coat were assessed by quantitative proteomics of S. Typhimurium‐infected epithelial cells and revealed ubiquitylation sites on several outer membrane‐associated and integral outer membrane proteins of Salmonella (Fiskin et al, 2016). Moreover, several chain types are associated with the Ub coat of S. Typhimurium including M1‐ (also known as linear Ub chains), K48‐, and K63‐polyubiquitin chain linkages, suggesting a complex structure and signaling platform of the Ub coat (van Wijk et al, 2012; Manzanillo et al, 2013). Over the last two decades, several E3 ligases like Smurf1, Parkin, ARIH1, LRSAM1, RNF213, and LUBAC, were identified to ubiquitylate more than one type of bacteria, and in some cases, the bacteria are targeted simultaneously by multiple E3 ligases. For instance, ARIH1, LRSAM1, RNF213, and LUBAC are known to colocalize with damaged SCVs and promote cytosolic S. Typhimurium Ub coat formation. There is also a group of E3 ligases like RNF166 (Heath et al, 2016), MARCH8 (Jin et al, 2017), TRIM21 (Hos et al, 2020), TRIM22 (Lou et al, 2018), and TRIM16 (Chauhan et al, 2016) that do not recognize the pathogen itself but recognize other aspects of infection like exposed glycans (Chauhan et al, 2016).
Similar to Galectin‐8, the bacterial Ub coat functions as a xenophagic eat‐me signal (Birmingham et al, 2006) and triggers the binding of autophagy receptors such as p62 (alias SQSTM1), NDP52 (alias CALCOCO2), and optineurin (OPTN) which are essential for the engulfment of bacteria by autophagosomes (Zheng et al, 2009; Wild et al, 2011; Thurston et al, 2012). These receptors are multifunctional proteins harboring distinct ubiquitin‐binding domains (UBD) with inherent specificities toward different ubiquitin chain types, as well as protein–protein interaction motifs that mediate the recruitment of autophagy machinery components to induce the formation and elongation of phagophore membranes proximal to bacteria (Kirkin & Rogov, 2019). The modular nature of these receptors is best understood in the case of NDP52 which employs a GIR and an Ub‐binding zinc finger (ZF) to associate with bacteria, a SKICH domain to recruit the autophagy promoting ULK1 and TBK1 kinases via their respective adaptors FIP200 and SINBAD/NAP1 as well as an LC3‐interacting region (LIR) to guide the engulfment of bacteria by the nascent autophagosome via LC3C (von Muhlinen et al, 2012; Thurston et al, 2012; Ravenhill et al, 2019). While NDP52’s ZF seems to promiscuously bind mono‐ and polyubiquitin (Xie et al, 2015), the UBAN domain of OPTN specifically recognizes M1‐ and K63‐linked ubiquitin chains of the Salmonella Ub coat (Wild et al, 2011). TBK1‐mediated phosphorylation of OPTN fosters its interaction with LC3, thereby supporting phagosome/autophagosome formation and restriction of Salmonella growth (Wild et al, 2011). Notably, the actions of multifunctional Ub‐binding autophagy receptors are not limited to xenophagy, but shared among other selective autophagy processes including aggrephagy and mitophagy (Turco et al, 2019; Vargas et al, 2019; Shi et al, 2020; Yamano et al, 2020). There are also some special cases, like xenophagy of Mycobacterium tuberculosis, where cytosolic Ub is directly binding to the UBA domain of the M. tuberculosis surface protein Rv1468c leading to the recruitment of p62 and LC3‐mediated delivery of the bacteria to lysosomes (Chai et al, 2019). Hence, Ub plays a central role as danger signal and sensor of cytosolic bacteria, highlighting the importance of the Ub machinery in xenophagy.
E3 ubiquitin ligases associated with xenophagy
The Ub coat on the surface of cytosolic bacteria including S. Typhimurium and the nonmotile ΔactA mutant of L. monocytogenes was first described by Perrin and colleagues in 2004 (Perrin et al, 2004). This seminal discovery initiated extensive research efforts in the past two decades to identify the Ub‐targeting machinery that recognizes and targets cytosol bacteria for Ub‐mediated xenophagy and innate immune responses. In this chapter, we will describe in a historic order the identification of xenophagy‐associated E3 ligases. We will discuss our current knowledge of their E3‐typical features and mechanisms in context of (i) sensing/recognition of cytosolic bacteria (ii) E3 ligase activation and (iii) type of Ub substrate modification.
LRSAM1
In 2011, the first E3 ligase, leucine‐rich repeat and sterile alpha motif‐containing protein 1 (LRSAM1), was functionally linked to Ub coat synthesis and xenophagy (Ng et al, 2011). LRSAM1, first shown to play a role in the endosomal sorting machinery (Amit et al, 2004; McDonald & Martin‐Serrano, 2008), belongs to the RING‐type E3 ligases that share the presence of a RING domain (Fig. 2). Canonical RING domains comprise conserved cysteines and histidines that allow coordinate binding of two zinc ions thereby stabilizing an overall globular three‐dimensional domain structure (Deshaies & Joazeiro, 2009). RING domains themselves lack Ub transfer activity, but accommodate the charged E2˜Ub in a distinct architecture known as the closed conformation that allosterically activates E2˜Ub for the nucleophilic attack by a suitable placed substrate lysine to form an isopeptide bond (Dou et al, 2012, 2013; Plechanovova et al, 2012; Pruneda et al, 2012; Branigan et al, 2020). Apart from the C‐terminal RING domain, LRSAM1 contains a leucine‐rich repeat (LRR) domain at the N terminus, a sterile alpha motif‐containing domain (SAM domain), and two coiled‐coil domains (CC1 and CC2), which are known to regulate the activity of E3 ligases by forming self‐associated oligomers (Bian et al, 2017).
Figure 2. The RING‐type E3 ligase LRSAM1 in Ub coat formation of cytosolic Salmonella .
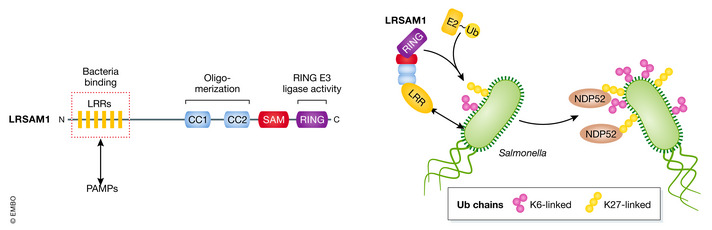
The domain architecture of LRSAM1 showing the leucine‐rich repeats (LRR) domains (red dotted box) that detects PAMPs of Salmonella and mediates bacterial binding, coiled‐coil (CC) domains, a sterile alpha motif (SAM) containing domain, and the C terminus RING domain with E3 ligase activity. LRSAM1‐mediated ubiquitylation of cytosolic Salmonella, preferentially K6‐ and K27‐linked chains, leads to the accumulation of autophagy cargo receptor NDP52 mediating the recruitment of the autophagy machinery.
LRRs are typically involved in the detection of pathogen‐associated molecular patterns (PAMPs) making LRSAM1 a suitably designed E3 for recognizing and ubiquitylating cytosolic bacteria. Notably, an LRR‐mediated recognition mechanism by E3 ligases was already proposed at the time when the Ub coat was first observed (Perrin et al, 2004). LRSAM1 colocalizes with xenophagy‐susceptible pathogens including S. Typhimurium, L. monocytogenes EGD‐e, S. flexneri, and invasive Escherichia coli, and colocalization is mediated by the LRR domain (Huett et al, 2012). The nature of PAMPs that are recognized by LRR is not characterized yet (Fig. 2). The RING domain of LRSAM1 is dispensable for bacterial association; however, it is essential to promote bacteria‐associated ubiquitylation with preference for K6‐ and K27‐linked chain formation in vitro. Whether the Ub coat is formed by bacteria‐bound autoubiquitylated LRSAM1 and/or by LRSAM1‐mediated ubiquitylation of bacterial surface proteins is not entirely clear. LRSAM1 accumulation with cytosolic Salmonella is peaking 40 min post‐infection coinciding with transient galectin‐8/NDP52 colocalization. However, LRSAM1 and NDP52 mark spatially different subdomains around Salmonella and are recruited independently (Huett et al, 2012; Thurston et al, 2012). Ub detection follows LRSAM1 occurring an hour post‐infection and is thought to foster NDP52 accumulation necessary for the recruitment of the autophagy machinery (Thurston et al, 2012).
The functional association of LRSAM1 with xenophagy is essential to control bacterial infection. Depletion of LRSAM1 in epithelial cells promotes cytoplasmic growth of S. Typhimurium. Lymphoblasts derived from Charcot–Marie–Tooth disease patients, which lack expression of LRSAM1 due to a frameshift that truncates the protein including the entire RING domain, are less efficient in controlling bacterial replication of the less virulent S. Typhimurium strain (i.e. NTCC12023; (Huett et al, 2012)). Conversely, the bactericide Biochanin A, a plant isoflavone derivate, was shown to enhance killing of Salmonella in infected HeLa cells and macrophages by reinforced LRSAM1/NDP52 accumulation and autophagosome formation (Zhao et al, 2018). The underlying mechanism of Biochanin A’s bactericidal activity is not known, though. LRSAM1, as a pattern‐recognition E3, may play a central role in the early step of anti‐bacterial defense by the establishment of the initial Ub coat on S. Typhimurium; however, it became clear that the Ub coat is further modified and re‐modeled by other xenophagy‐associated E3s to create an even more complex signaling platform on cytosolic bacteria.
PARKIN
Genomewide association studies (GWAS) identified some genetic polymorphism in the parkin gene (PRKN) that are also associated with increased susceptibility to intracellular pathogens, such as S. Typhimurium and Mycobacterium leprae, suggesting a potential connection between mitochondrial homeostasis and xenophagy via Parkin (Mira et al, 2004; Ali et al, 2006). However, Parkin is primarily known for its well‐established role in mitophagy and got much attention as it is frequently mutated in autosomal recessive juvenile Parkinsonism, a neurological disorder that leads to the progressive loss of dopaminergic neurons.
Parkin belongs to the RBR family of E3 ligases that are defined by common domain organization and catalytic mechanism (reviewed in (Cotton & Lechtenberg, 2020)). The catalytic RBR module consists of three zinc‐binding motifs: a RING1 domain with a canonical RING fold mediating E2 binding, followed by an In‐Between‐RING (IBR) domain, and finally a Rcat (also known as RING2) domain containing the catalytic cysteine (Fig. 3A). Specific for Parkin is the presence of an N‐terminal Ub‐like (UBL) domain that shares a structural fold similar to Ub and an additional zinc‐coordinating unique parkin domain (UPD) (also known as RING0) (Trempe et al, 2013). Both, UBL and UPD play a central role in regulating Parkin’s E3 ligase activity. The apo‐form of Parkin adopts a multilayered autoinhibited conformation, whereby the UBL and a repressor element (REP) occlude the E2 binding site in RING1, and the UPD domain masks the catalytic cysteine 431 (C431) in Rcat preventing the formation of the E3˜Ub thioester intermediate (Chaugule et al, 2011; Trempe et al, 2013; Wauer & Komander, 2013). Several elegant studies revealed a striking activation mechanism of Parkin in mitophagy. Parkin activation depends on PINK1 kinase, which itself accumulates and is activated on the outer membrane of damaged mitochondria (Matsuda et al, 2010; Narendra et al, 2010; Vives‐Bauza et al, 2010; Kondapalli et al, 2012). PINK1 phosphorylates Ub on serine 65 (pS65‐Ub) which increases Ub’s affinity for an allosteric site—a pS65‐Ub‐binding pocket—in Parkin. Once bound, pS65‐Ub induces a conformational change in the IBR domain resulting in the release of the inhibitory UBL from the RBR module, overall destabilizing the autoinhibitory conformation and making the Rcat accessible for transthiolation. Subsequently, freed UBL becomes a substrate of PINK1 and pS65‐UBL further fosters an open, active conformation (Ordureau et al, 2014; Kazlauskaite et al, 2015; Kumar et al, 2015; Sauve et al, 2015; Wauer et al, 2015; Yamano et al, 2015; Aguirre et al, 2017; Gladkova et al, 2018).
Figure 3. RBR‐type E3 ligases involved in coating cytosolic bacteria with ubiquitin.
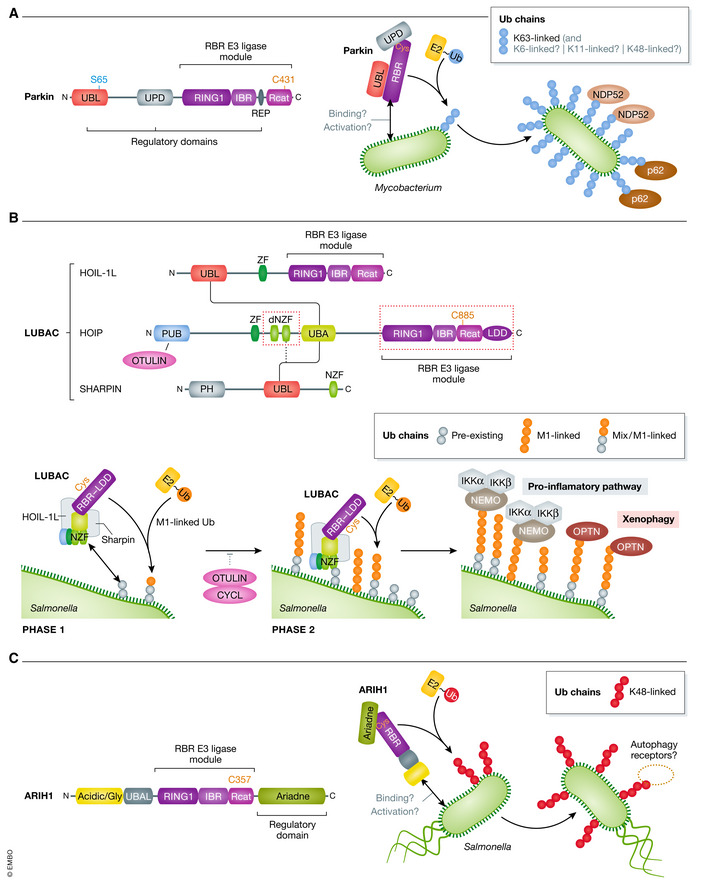
(A) Diagram of Parkin showing the Ub‐like (UBL) domain, the unique parkin domain (UPD), and the repressor element (REP), which form the regulatory part of Parkin, and the catalytic part comprising the RBR module at the C terminus. The mechanism for activation of Parkin in response to bacterial infection remains to be addressed but upon Mycobacterium infection, activated Parkin decorates the cytosolic bacteria with an Ub coat (predominantly K63 chains) that recruits p62 and NDP52 autophagy adaptors triggering xenophagy. (B) Domain organization of the three subunits of LUBAC: HOIP, HOIL‐IL, and SHARPIN. HOIL‐IL and SHARPIN interact via their respective UBL domain with the UBA domain of HOIP (solid lines), with potential contribution of the double NZF (dNZF) domain (dotted line). The dNZF and RBR domains of HOIP (red dotted boxes) are required for binding Salmonella in a two‐phase mechanism: in the first phase, LUBAC binds to preexisting Ub chains on Salmonella, in the second phase, enhanced HOIP activity mediates M1‐ and mixed M1‐linked Ub chains synthesis on these preexisting chains and triggers further LUBAC binding. The M1‐Ub chains recruit adapter proteins OPTN and NEMO which initiate xenophagy and pro‐inflammatory response, respectively. DUB activity of OTULIN and CYLD may counterbalance LUBAC activity. (C) Schematic view of ARIH1 domains showing the acidic/glycine region, UBA‐like domain (UBAL), the RBR module, and the regulatory Ariadne domain at the C terminus. ARIH1 decorates the Salmonella with K48‐Ub chains. The mechanism of its activation, binding to the cytosolic Salmonella, and the downstream action of ARIH1‐mediated K48‐Ub chains are not understood.
The PINK1/Parkin pathway is tightly linked with damaged mitochondria, however, and rather surprising, it was shown that Parkin is required for M. tuberculosis xenophagy in vitro. M. tuberculosis is a vacuolar pathogen that resides inside a phagosomal compartment after entering the cell. Due to the activity of mycobacterial ESX‐1 secretion system, the phagosomal membrane becomes permeable allowing components of the Ub‐xenophagy pathway to access M. tuberculosis (Watson et al, 2012). In infected murine bone marrow‐derived macrophages (BMDMs), Parkin colocalizes with M. tuberculosis and mediates a predominantly K63‐linked Ub coat surrounding M. tuberculosis phagosomes. This contrasts findings by Ordureau et al (2014) indicating that Parkin does not exhibit specificity for single chain types (Manzanillo et al, 2013; Ordureau et al, 2014). Parkin‐mediated ubiquitylation subsequently leads to the recruitment of autophagy receptors p62 and NDP52 via UBA and UBZ Ub‐binding domains, respectively, that facilitate the delivery of mycobacteria to autophagosomes (Ichimura et al, 2008; Watson et al, 2012; Manzanillo et al, 2013). In vivo data further support an important role of Parkin against a range of intracellular bacterial infections. Park2 −/− mice have an extreme susceptibility to M. tuberculosis and L. monocytogenes infections. Moreover, D. melanogaster and C. elegans strains deficient for Parc2 homologues are highly susceptible to infection by pathogens including L. monocytogenes, S. Typhimurium, and Mycobacterium marinum, suggesting a conserved role of Parkin in innate immunity across metazoans (Manzanillo et al, 2013). Importantly, while studies in cultured cells show involvement of many autophagy factors including ATG5, ATG12, ATG16L, and LC3 in restricting M. tuberculosis replication (Gutierrez et al, 2004; Dutta et al, 2012; Watson et al, 2012; Seto et al, 2013; Sakowski et al, 2015), comprehensive genetic investigations in mice revealed a unique and autophagy‐independent role of ATG5 in controlling resistance to M. tuberculosis infections (Kimmey et al, 2015). Given the lack of a clear correlation between in vitro and in vivo findings, Parkin’s precise in vivo role in killing of M. tuberculosis, with a possible functional link to ATG5 in early events of innate immunity, needs further investigation.
The gram‐positive Group A streptococcus (GAS) was the first bacterium with definitive evidence to be degraded by autophagy (Nakagawa et al, 2004). Generally, GAS is extracellular but has the ability to trigger its internalization into non‐immune cells (endothelial and epithelial cells) to evade immune surveillance (Molinari et al, 2000; Nakagawa et al, 2001). Intracellular GAS colocalizes to a large extent with LC3‐positive compartments which is dependent on the pore‐forming streptolysin O (SLO) (Nakagawa et al, 2004). SLO promotes the escape of GAS from the phagophore, which gives access to galectin‐8‐dependent Parkin‐mediated ubiquitylation of the GAS/phagophore compartment (Cheng et al, 2017). The autophagy receptors p62, NDP52, and NBR1—a p62 homolog, containing a PB1 domain to interact with p62, a UBA domain to interact with Ub, and a LIR motif (Kirkin et al, 2009; Lamark et al, 2009)—are essential for the recognition of Ub decorated GAS and initiation of LC3‐dependent xenophagic degradation (Thurston et al, 2009; Barnett et al, 2013). Consistent with the key role of galectin‐8 in this process, the efficiency of Parkin recruitment and subsequent GAS ubiquitylation is highly dependent on the abundance of galectin‐8. While exposed glycans can be sensed by other galectins in cells with low amounts of galectin‐8, the compensatory recognition of GAS by for example galectin‐3 does not lead to GAS elimination (Cheng et al, 2017). However, the differential roles of these two galectins in response to GAS remain poorly understood.
Although there are striking parallels between xenophagy and mitophagy pathways downstream of Parkin/ubiquitylation, tempting questions regarding how Parkin is recruited to cytosolic pathogens and what relieves Parkin’s autoinhibition, processes that might be mechanistically interconnected, remain unanswered with room for speculations. Recently, similar to findings for PRKN (Mira et al, 2004; Ali et al, 2006), common variants in PINK1 gene were associated with increased risk of leprosy, a chronic infectious, and neurological disease caused by Mycobacterium leprae (Wang et al, 2016). Hence, PINK1, in conjunction with Parkin, might have a role in M. leprae innate immune response via xenophagy. However, no study addressed yet, whether PINK1 accumulates on M. leprae/phagophores and initiates pS65‐Ub and/or pS65‐UBL‐mediated Parkin activation. At least, no pS65‐Ub was found to colocalize with S. Typhimurium (Thurston et al, 2012). Alternatively, Parkin activation could be PINK1‐independent in the context of xenophagy, and other sensor and activating molecules exist leading to recruitment as well as activation of Parkin (Singh et al, 2018). Addressing Parkin activation and sensing mechanism of cytosolic bacteria will be instrumental to further our understanding of Parkin’s role in xenophagy.
SMURF1
SMAD Ub regulator factor (Smurf1) was originally identified and characterized as negative regulator of bone morphogenic protein (BMP) and transforming growth factor beta (TGF‐β) signaling pathways, which play an essential role in embryogenesis and adult tissue homeostasis (Cao & Zhang, 2013). However, it is now evident that Smurf1 regulates a plethora of different cellular and developmentally important signaling pathways and networks impacting on a wide spectrum of biological roles including cell proliferation, DNA damage response, chromatin organization, and selective autophagy (recently reviewed in Koganti et al (2018)). The first hint linking Smurf1 to xenophagy came from Orvedahl et al (2011) who identified Smurf1 in a genomewide small interfering RNA screen for genes essential for autophagy of viruses and damaged mitochondria (Orvedahl et al, 2011). Drawing parallels between mitophagy and xenophagy, Franco et al (2017) subsequently showed that Smurf1 colocalizes with M. tuberculosis‐containing compartments mediating xenophagic degradation and suppression of M. tuberculosis replication in macrophages (Franco et al, 2017).
Smurf1 is a member of the HECT E3 Ub ligase family (Huibregtse et al, 1995; Scheffner & Staub, 2007; Scheffner & Kumar, 2014; Lorenz, 2018). HECT E3s have a modular architecture characterized by the C‐terminal HECT domain that consists of 2 lobes: the larger N‐lobe associates with the E2 enzyme (Huang et al, 1999) and the C‐lobe contains the catalytic cysteine and interacts with the Ub during the transfer reaction from the E2 (Scheffner et al, 1995; Kamadurai et al, 2009; Lorenz et al, 2013). While the HECT domain represents the catalytic module, substrate recruitment and regulation of the catalytic activity of HECT E3s are determined by their respective N‐terminal extensions (Sluimer & Distel, 2018). Specifically, Smurf1 belongs to the C2‐WW‐HECT (also known as NEDD4‐like) subfamily (Fig. 4) with two central WW domains which usually control protein–protein interaction by recruiting substrates and adaptor proteins containing proline‐rich PY motifs (sequence L/P‐P‐x‐Y) and phosphorylated Ser/Thr‐Pro sites (Chen & Sudol, 1995; Lu et al, 1999; Leon & Haguenauer‐Tsapis, 2009), and an N‐terminal C2 domain mediating membrane association via interactions with phospholipids and inositol phosphates (Plant et al, 1997; Angers et al, 2004; Dunn et al, 2004; Scott et al, 2020). In agreement with a role of C2 in regulating subcellular localization, the C2 domain is essential for the accumulation of Smurf1 at sites of M. tuberculosis, whereas the E3 ligase activity of Smurf1 is dispensable. Importantly, the C2 domain also participates in Smurf1 homo‐dimerization and intramolecular autoinhibition, whereby the C2 domain of one Smurf1 interacts in trans with the HECT domain of another Smurf1 (Fig. 4). It is tempting to speculate that C2 binding to M. tuberculosis and/or phagophore relieves the autoinhibitory conformation by liberating the HECT domain. Hence, coupling of recruitment and activation mechanisms might ensure spaciotemporal control of Smurf1‐mediated ubiquitylation. Similar to reports for Parkin (Watson et al, 2012), Smurf1 colocalization with a M. tuberculosis strain, which lacks the virulence factor and type VII secretion system ESX‐1, is reduced, suggesting that only damaged and permeabilized phagosomes promote the recruitment and activation of xenophagic Ub machineries.
Figure 4. The HECT‐type E3 ligase Smurf1 in Ub coat formation of cytosolic Mycobacterium .
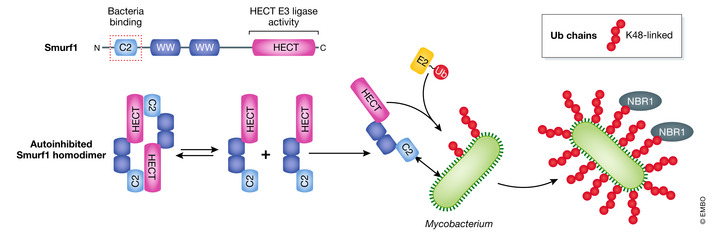
Domain schematic of Smurf1 highlighting the C2 domain required for bacterial binding (red dotted box) and C‐terminal HECT domain with E3 ligase activity. In its apo state, Smurf1 forms inactive homodimers. Interaction between the C2 domain and cytosolic Mycobacterium might relieve Smurf1’s autoinhibition and trigger K48‐linked Ub chain synthesis. Autophagy adaptor protein NBR1 recruits phagophores and initiates xenophagy.
Smurf1 mediates preferentially K48‐linked Ub chains on M. tuberculosis as well as L. monocytogenes that lead to binding of the autophagy receptor NBR1 (Franco et al, 2017). In addition, Smurf1 was reported to promote autophagosome maturation by K29/K33 ubiquitylation of UV radiation resistance associated (UVRAG), which is a key component of the phosphatidylinositol 3‐phosphate kinase complex localized in the endosomal compartment. Whether this Smurf1 activity contributes to xenophagy remains to be tested (Feng et al, 2019). Using a murine model of tuberculosis, it was shown that Smurf1 deficiency during the chronic, but not in the acute phase of M. tuberculosis infection, increased bacterial growth in lung and spleen causing increased pulmonary inflammation (Franco et al, 2017). Moreover, Smurf1 is essential to limit mycobacterial replication in human macrophages and associates with bacteria in lungs of patients with pulmonary tuberculosis. Together with the aforementioned E3s LRSAM1 and Parkin, Smurf1‐mediated K48‐linked Ub chains and the recruitment of NBR1 contributes further to the pattern of the signaling platform by the Ub coat mediating xenophagic elimination of cytosolic bacteria.
LUBAC
Super‐resolution microscopy (by structured illumination microscopy, SIM) of the Ub coat on cytosolic S. Typhimurium revealed microdomains/clusters of M1‐linked Ub chains that occur later than other linkage types (Noad et al, 2017; van Wijk et al, 2017). The concept of microdomains on bacterial surfaces was originally proposed for autophagy receptor proteins. The Brumell laboratory described that p62 and NDP52 are recruited independently, marking non‐overlapping microdomains on bacteria (Cemma et al, 2011). The discovery of Ub linkage‐specific microdomains suggests thus that this Ub code defines the discrete receptor pattern by recruiting Ub linkage‐specific receptors.
The only enzyme know so far to be specific for the synthesis of M1 linkages is the linear Ub assembly complex (LUBAC), a multimeric E3 Ub ligase composed of heme‐oxidized IRP2 Ub ligase 1L (HOIL‐1L), HOIL‐1 interacting protein (HOIP), and Shank‐associated RH domain interacting protein (SHARPIN) (reviewed in (Rittinger & Ikeda, 2017)) (Fig. 3B). Assembly of LUBAC is mediated by domain‐defined protein–protein interactions. HOIP’s UBA domain is required to associate with the respective UBL domains of HOIL‐1L and SHARPIN. The NZF1 domain of HOIP might additionally mediate HOIP‐SHARPIN binding. Moreover, HOIP was shown to associate via the PUB domain with the deubiquitinating (DUB) enzyme OTU domain DUB with LINear linkage specificity (OTULIN), and via the adapter protein SPARTA to the M1‐ and K63‐chain cleaving DUB CYLD (Keusekotten et al, 2013; Rivkin et al, 2013; Elliott et al, 2016; Hrdinka et al, 2016; Kupka et al, 2016). Both, HOIP and HOIL‐1L subunits contain the RBR domain cluster; however, HOIL‐1L’s activity is non‐essential for M1‐Ub chains synthesis by LUBAC (Kirisako et al, 2006). In fact, the RBR domain of HOIP plus a C‐terminal extension, known as the linear Ub‐determining domain (LDD), contains the minimal catalytic machinery required for the highly specific synthesis of M1 linkages (Smit et al, 2012; Stieglitz et al, 2012, 2013). However, apo full‐length HOIP is inactive. Detailed structural and mechanistic studies of inactive HOIP are still missing, but it is thought that the catalytic C885 is occluded involving the UBA domain. The autoinhibitory conformation is relieved in LUBAC assembly whereby HOIL‐1L via its UBL associates with the UBA domain of HOIP (Kirisako et al, 2006). Structural insight into HOIP’s active form was recently provided by the crystal structure of the RBR‐LDD fragment in complex with the charged E2, UBE2D˜Ub (Lechtenberg et al, 2016). This structure uncovered how UBE2D˜Ub is tightly embraced by the RBR, orientating the E2˜Ub thioester bond in close proximity to C885 primed to promote trans‐thiolyation. Furthermore, an allosteric Ub‐binding site was revealed. Ub or M1‐linked di‐Ub binding to this site apparently promotes binding efficiency of UBE2D˜Ub donor Ub, HOIP˜Ub thioester, and polyubiquitin chain formation (Lechtenberg et al, 2016).
Recruitment of LUBAC to cytosolic S. Typhimurium was dissected in detail by the Randow laboratory proposing a two‐phase mechanism (Noad et al, 2017) (Fig. 3B): First, LUBAC binds on preexisting Ub chains on S. Typhimurium mediated by the Ub‐binding dNFZ domain of HOIP. Second, efficient LUBAC accumulation depends on a feed forward loop requiring HOIP’s synthesis of M1‐linked Ub chains (likely on the preexisting chains), that in turn take part in the aforementioned allosteric activation of HOIP as well as the accelerated recruitment of yet more HOIP/LUBAC. This raises the tempting question whether the feed forward loop is counterbalanced by the activity of LUBAC‐associated DUBs, OTULIN and/or CYLD. Recent findings suggest that HOIP and OTULIN cooperate in regulating autophagy, whereby HOIP is required for autophagy initiation, whereas OTULIN is necessary for autophagy maturation steps (Chu et al, 2021; Weinelt & van Wijk, 2021). Curiously, OTULIN‐depleted cells had fewer LC3B‐positive autophagosome‐associated Salmonella, suggesting that OTULIN is not simply competing with HOIP and M1‐linked Ub chain synthesis to control xenophagy.
Originally, it was suggested that LUBAC recognizes the substrate target and attaches the first Ub on N‐terminal M1. Subsequently, this priming ubiquitin becomes substrate for M1 chain extension (Tokunaga et al, 2009; Smit et al, 2013; Fujita et al, 2014). However, an alternative model has been proposed by Emmerich et al (2013) whereby HOIP recognizes preexisting K63‐linked chains that serve as substrates for HOIP‐mediated M1‐linkage extensions that result in mixed (heterotypic) K63/M1 chains (Emmerich et al, 2013). Hence, the Ub coat surrounding cytosolic bacteria might not be limited to homotypic Ub chains, but also heterotypic chains of K63/M1 type. The M1‐linked Ub chain deposits on S. Typhimurium recruit the Ub‐binding effector proteins OPTN and NF‐kB Essential MOdifier (NEMO) and thereby transform the bacterial surface into a signaling platform coordinating two cellular defense pathways: xenophagy and the NF‐κB pro‐inflammatory pathway, respectively (Noad et al, 2017) (Fig. 3B). OPTN serves as an autophagy receptor protein that requires phosphorylation‐mediated activation by kinase TBK1 and recruits LC3‐coated phagophores to eliminate cytosolic S. Typhimurium (Wild et al, 2011; Rogov et al, 2013). Independent of OPTN, NEMO restricts together with the other subunits of the IKK complex (IKKα and IKKβ) proliferation of cytosolic S. Typhimurium via activation of the pro‐inflammatory NF‐κB pathway. This signaling axis has striking similarities to Toll‐like receptor (TLR)‐mediated signaling and might converge with other innate immune response pathways (Fiil & Gyrd‐Hansen, 2021).
ARIH1
The fifth E3 ligase entering the stage of Ub‐mediated xenophagy was ARIH1 (also known as Homologues to Human ARIadne 1 (HHAR)) (Fig. 3C). We isolated ARIH1 in a focused image‐based RNAi screen comprised of 14 known RBR E3 ligases that aimed to identify E3 ligases controlling replication of cytosolic S. Typhimurium and Ub coat formation (Polajnar et al, 2017). The recruitment of ARIH1 to cytosolic Salmonella is a fast response post‐infection, with kinetics comparable to galectin‐8 binding to damaged SCVs (Thurston et al, 2012; Polajnar et al, 2017). The molecular mechanism explaining of how ARIH1 recognizes Salmonella has not been determined, but ARIH1 is detected by super‐resolution microscopy in a patch‐like pattern around Salmonella cells even before the occurrence of the pattern‐recognition E3 LRSAM1 (see above), and mediates K48‐linked chain formation. ARIH1 together with LRSAM1 likely constitutes the first phase of E3 action and ubiquitylation on damaged SCVs (Polajnar et al, 2017). The observation that ARIH1 is also recruited to depolarized mitochondria might point to common molecular cues exposed on damaged membranes of these two compartments (Villa et al, 2017).
As discussed for the RBR E3s Parkin and HOIP, isolated ARIH1 is also autoinhibited, raising the question of how ARIH1 is activated during Salmonella infection. Structural studies of ARIH1 revealed that the C‐terminal Ariadne domain occludes the catalytic site cysteine C357 on the Rcat domain (Duda et al, 2013). To relieve the autoinhibition and to gain full Ub transfer activity, large‐scale conformational changes of ARIH1 are needed. Recently, an intriguing strategy/mechanism of ARIH1 activation was proposed. An analysis of the ARIH1 interactome revealed that ARHI1 binds directly to neddylated cullins (cullin‐1, ‐2, ‐3, 4a, and ‐4b) and to numerous neddylated cullin RING ligase (NEDD8‐CRLs) complexes (Kelsall et al, 2013; Scott et al, 2016). Recent cryo‐EM structural work and elegant biochemical studies suggest that binding of NEDD8‐CRL complexes repositions the inhibitory Ariadne domain and opens the structure making the catalytic C357 accessible to react with E2˜Ub (Kelsall et al, 2013; Scott et al, 2016; Horn‐Ghetko et al, 2021). Further, ARIH1 in complex with NEDD8‐CRLs complexes promotes fast and efficient monoubiquitylation of CRL clients, thus priming them for subsequent CRL‐mediated Ub chain elongation (Scott et al, 2016; Kelsall et al, 2019). Polajnar et al (2017) addressed whether NEDD8‐CRL‐dependent activation of ARIH1 plays a role in Salmonella xenophagy. Neither cullin depletion via RNAi nor treatment of cells with the NEDD8‐activating E1 enzyme inhibitor MLN4924, which abolishes all neddylation conjugations in the cell, did impact on Ub coat formation or did increase Salmonella replication, suggesting another activator and/or alternative activation mechanism of ARIH1 in the context of xenophagy. Despite the elusive activation mechanism, ARIH1‐mediated K48‐type Ub chains synthesis adds to the aforementioned, K63 and M1 Ub chain types to an unprecedented complexity of the Ub code on cytosolic bacteria. Notably, while K63 and M1 chains have apparently overlapping functions in the recruitment of autophagy receptors, ARIH1 is dispensable for p62‐, OPTN‐, and NDP52‐mediated xenophagy (Polajnar et al, 2017). The identification of downstream effectors of the K48‐type Ub chains on Salmonella is exciting goals for future studies.
RNF213
Besides the growing number of E3 enzymes and their contribution to the formation of the Ub coat that dress cytosolic bacteria, the identity of bacterial molecules that are conjugated with Ub was previously restricted to Salmonella outer membrane proteins (Fiskin et al, 2016). Otten and colleagues extended the landscape of ubiquitylation targets to lipopolysaccharide (LPS) of the outer surface of S. Typhimurium (Otten et al, 2021). LPS consists of Lipid A, which is anchored in the bacterial outer membrane, and oligosaccharides termed as inner‐, outer‐core, and O‐antigen. Using a set of Salmonella mutants that synthesize truncated versions of LPS in combination with chemical approaches that allowed discrimination between protein and non‐protein ubiquitin ligation, it was shown that lipid A is the minimal subunit to form an ester‐linked Ub conjugate with its hydroxy and/or phosphate groups (Fig. 5). To identify the enzyme that ubiquitylates LPS, Otten and colleagues fractionated HeLa cell lysates by applying sequential biochemical purification steps and isolated RNF213 as the cognate LPS‐active E3 ligase (Otten et al, 2021). With a mass of nearly 600 kDa, RNF213 is the largest identified E3 ligase in mammalian proteomes. A recent cryo‐EM structure of mouse RNF213 revealed distinct domain architectures of a dynein‐like AAA+ motor domain and a RING‐type domain‐containing E3 ligase module (Ahel et al, 2020) (Fig. 5). Interestingly, the dynein‐like AAA+ motor domain is required for LPS ubiquitylation, whereas—unexpectedly—no such requirement was found for the RING domain (Ahel et al, 2020; Otten et al, 2021). However, Otten and colleagues mapped a molecular motif within the E3 module, named RZ‐finger, that is typical for zinc‐binding domains, and showed that it is required for LPS ubiquitylation. Recent biochemical studies pinpointed an active site cysteine within the RZ finger that accepts Ub from the E2 enzyme UBCH7, suggesting that RNF213 represents an undescribed type of transthiolation E3 enzyme (preprint: Ahel et al, 2021). RNF213 knockout mouse embryonic fibroblasts (MEFs) or cells expressing RNF213 mutations corresponding to the RZ‐finger are severely defective in Ub coat formation around cytosolic Salmonella and also failed to recruit LUBAC and lacked LUBAC‐mediated M1 Ub chain synthesis. In agreement with the aforementioned dependency of LUBAC activity on pre‐formed Ub chains on Salmonella (Fig. 3B), RNF213 is a likely candidate for the first‐line E3 ligase activity, priming Salmonella with ubiquitylated LPS for xenophagy and innate immune response pathways. However, RNF213‐mediated immune response might not be limited to the gram‐negative bacterium Salmonella. A recent study suggested that RNF213 exerts a much broader antimicrobial activity, counteracting infections with the gram‐positive bacterium L. monocytogenes as well as several viral pathogens in vitro (preprint: Thery et al, 2021). Importantly, this finding would imply that LPS is unlikely the only target of RNF213 and other substrates might exist.
Figure 5. The atypical E3 ligase RNF213 ubiquitylates LPS on Salmonella .
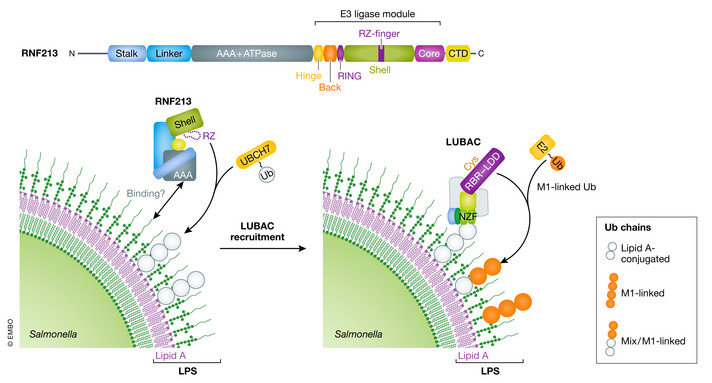
Schematic representation of the domain organization of RNF213 consistent of N‐terminal stalk, the central AAA+ ATPase, and the E3 ligase module. The AAA+ and RZ‐finger domains are required for ubiquitylation of lipid A/LPS on Salmonella’s outer surface. These Ub chains recruit LUBAC‐mediated xenophagy and pro‐inflammatory response pathways (see also Fig 3B).
Despite compelling evidence of a central role of RNF213’s E3 ligase in cell‐autonomous immunity studying cell systems, there are only limited data from in vivo studies. RNF213‐deficient mice have an increased susceptibility to L. monocytogenes infections (reprint: Thery et al, 2021); however, the function of RNF213 in innate immunity remains poorly understood in vivo. Several studies describe RNF213 as a major susceptibility gene for Moyamoya disease, a cerebrovascular disorder characterized by arterial stenosis and abnormal development of collateral blood vessels that frequently leads to brain strokes (Kamada et al, 2011; Liu et al, 2011). The physiological and pathophysiological role of RNF213 is little understood, but RNF213 was shown to be involved in lipid metabolism, lipid droplet formation, and signaling associated with cell death and immune‐system function (Banh et al, 2016; Piccolis et al, 2019; Sugihara et al, 2019). The majority of RNF213 mutations found in Moyamoya patients cluster in the composite E3 module; however, none of the tested mutations affected LPS ubiquitylation (Otten et al, 2021), suggesting that defective bacterial ubiquitylation might not play a role in Moyamoya disease. Still, to address whether bacterial infections are implemented in the development of the disease is worthy of future investigations. Moreover, it is tempting to speculate that RNF213 ligase activity for lipid substrates might regulate other types of selective macroautophagy, such as lipophagy. RNF213 as a potential immune sensor adds a new exciting player to the xenophagy machinery with interesting questions regarding precise enzymatic mechanism and substrate recognition/specificity for the future.
Interplay of xenophagic E3 ligases
Whether the different pathogen‐associated E3 ligases act in a hierarchically coordinated or independent random manner is not fully understood since systematic, time‐resolved analyses with comparable infection conditions across different bacteria are lacking. So far, studies have only examined pairs (Smurf1/Parkin, LRSAM1/Parkin, and RNF213/HOIP) and trios (ARIH1/LRSAM1/HOIP and Parkin/LRSAM1/HOIP) of these ligases in response to infection with M. tuberculosis and S. Typhimurium, respectively. Therefore, the emerging picture is sketchy at best. ARIH1 seems to translocate to Salmonella as soon as they evade into the cytosol followed by LRSAM1. Conversely, LRSAM1 tends to persist longer on these bacteria compared to ARIH1 (Polajnar et al, 2017). Given that ARIH1 is not required for the bacterial recruitment of LRSAM1 and that both ligases do not interact (Polajnar et al, 2017), it seems plausible that both ligases translocate to cytosolic Salmonella independent from each other as part of a first line of defense. While the kinetics of Smurf1, Parkin, HOIP, and RNF213 colocalization to cytosolic bacteria in relation to ARIH1 and LRSAM1 have not been monitored in detail, HOIP is only recruited to bacteria that already carry an initial Ub coat (Noad et al, 2017). Notably, binding to ubiquitylated bacteria is mediated by HOIP’s dNZF domain which has an in vitro preference for K63 chains (Noad et al, 2017). However, Parkin and LRSAM1 turned out to be dispensable for HOIP recruitment (Noad et al, 2017), whereas ARIH1 and Smurf1 have not been tested due to their preference to generate K48 chains. In the search of a ligase whose activity is responsible for the downstream recruitment of HOIP, the Randow laboratory recently provided compelling evidence that ubiquitylation of bacterial LPS by RNF213 is the priming event of this process (Otten et al, 2021). In the absence of RNF213, the recruitment of HOIP, M1‐Ub, NEMO, OPTN, and LC3 to bacteria was dramatically reduced and bacterial replication increased at 4 h after infection with S. Typhimurium. The observation that loss of RNF213‐mediated ubiquitylation also ablated the translocation of p62 and NDP52 to Salmonella suggests that RNF213 is a major driver of xenophagy at this time point of infection. Consistent with a rather late arrival of RNF213 at cytosolic bacteria 2‐3 h post‐infection, cells lacking RNF213 were still able to initially coat Salmonella with ubiquitin at 1 h post‐infection (Otten et al, 2021). This indicates that RNF213 constitutes a second but massive response wave to fend off cytosolic bacteria. Nevertheless, it remains to be clarified whether RNF213—like HOIP—requires a priming ubiquitylation event for its recruitment to Salmonella or depends on other cellular or bacterial factors for its translocation to bacteria. Conversely, it is fully possible that Ub moieties generated by RNF213 on bacterial LPS are extended by the other xenophagic ligases.
While the chain type preference of RNF213 is not known (Otten et al, 2021) and ablation of ARIH1, Smurf1, Parkin or HOIP reduces the bacterial ubiquitin coat by the chain type that is specifically generated by these E3 ligases (Manzanillo et al, 2013; Franco et al, 2017; Noad et al, 2017; Polajnar et al, 2017) their impact on other chain topologies has not been assessed comprehensively. However, the analysis of M1 chain decorated cytosolic bacteria in cells depleted of ARIH1 unveiled a first glance on an unexpected interplay between the different anti‐bacterial ligases. Surprisingly, HOIP’s activity compensated for a reduction of ARIH1 (Polajnar et al, 2017). While it remains to be seen whether loss of LRSAM1, Smurf1, or Parkin likewise triggers a robust increase in M1 chains on Salmonella and whether RNF213 plays a role in this compensatory ubiquitylation axis, the finding from ARIH1 depleted cells suggests that different E3 ligases may be competent for available Ub acceptor sites within the bacteria coat. Even more surprising, ARIH1 recruitment to Salmonella was found to be increased in the absence of LRSAM1 or HOIP which both tend to follow ARIH1 on cytosolic bacteria (Polajnar et al, 2017). This observation raises the question about the existence of feedback loops or hand‐over mechanisms between ARIH1, LRSAM1, and HOIP. A systematic mutual analysis of recruitment and activity dependencies would be highly informative to understand the coordinated action of the different anti‐bacterial ligases. However, such a study is currently lacking.
With the advantages in super‐resolution microscopy in the last years, it became increasingly clear that the Ub coat is a heterogenic assembly composed of at least K48, K63, and M1 chain types (van Wijk et al, 2012, 2017; Noad et al, 2017), which serve as specific direct binding sites on cytosolic bacteria for distinct cellular factors. These include selective autophagy receptors such as NDP52, p62, OPTN, and NBR1, innate immunity components such as NEMO and LUBAC as well as proteasomal subunits (Thurston et al, 2009; Cemma et al, 2011; Wild et al, 2011; Manzanillo et al, 2013; Franco et al, 2017; Noad et al, 2017; van Wijk et al, 2017). From this array of Ub‐binding partners, it is evident that the bacterial Ub coat integrates a number of different anti‐bacterial functions ranging from xenophagy to NF‐κB signaling which cooperatively ensures an efficient defense against cytosolic pathogens. Given that the pan‐Ub antibody FK2 recognizes almost all chains except K6 (Noad et al, 2017) and that the bacterial Ub coat has not been comprehensively sampled with chain type‐specific antibodies, we are still far away from grasping the full repertoire of host processes triggered by this assembly.
Hijacking the ubiquitin machinery by pathogenic bacteria
Bacterial pathogens, that invade and reside in host cells, have evolved various strategies to divert the host pathways for their advantage. They achieve these feats by secreting effector proteins or toxins that modify or interact with the host protein. Secretion of these effector proteins differs between species of bacteria, the diversity of the strain within species, and the exposure to different cytosolic proteins in different host cell types. Mediated by these proteins, Salmonella directs macroautophagy to facilitate its replication (Yu et al, 2014), Listeria modulates host actin protein to escape LC3‐positive membranes in macrophages (Cheng et al, 2018), Shigella modifies histone to inhibit activation of NF‐κB signaling (Arbibe et al, 2007), and many more. In this section of the review, we will only focus on the strategies employed by the cytosolic bacteria to evade or hijack the Ub machinery.
To prevent modification by E3 ligases and subsequent host responses, cytosol‐dwelling bacteria have evolved a number of different strategies: First, some bacteria actively avoid leaving their protective vacuoles. For example, at an early stage of infection when Salmonella resides in endosomes the bacteria divert several autophagy pathway components to repair damage on their surrounding membranes. This prevents the exposure of bacteria to the cytosol and allows a stepwise maturation of SCVs via recruitment of early and late endosomal markers as well as acidification (Kreibich et al, 2015). Likewise, Coxiella decorates its vacuole with LC3 to delay fusion with lysosomes (Beron et al, 2002; Gutierrez et al, 2005).
Second, other bacteria prevent ubiquitylation of their surfaces by molecular disguises (Table 1). Francisella tularensis modifies its surface with polysaccharide O‐antigen (Case et al, 2014), whereas Shigella secretes the effector protein IscB to camouflage its own outer membrane protein VirG (Ogawa et al, 2005) and Listeria evades detection by recruiting actin via its secreted effector ActA (Yoshikawa et al, 2009). Rickettsia uses its two protein–lysine methyltransferases (PKMTs) to methylate lysine residues of outer membrane proteins (Engstrom et al, 2021) whereas its outer membrane protein B (OmpB) physically shields other surface proteins in macrophages (Engstrom et al, 2019), thereby preventing their recognition and subsequent ubiquitylation.
Table 1.
Effector proteins employed by bacterial pathogens to escape or hijack components of the host ubiquitin machinery
| Pathogen | Effector | Protein type | Mechanism of action | Reference |
|---|---|---|---|---|
| Salmonella Typhimurium | SopA | HECT‐like | Ubiquitylates host E3 ligases TRIM65 and TRIM56 thereby blocking the expression of interferon‐γ and nucleic acid sensing receptor STING | Fiskin et al, (2017) |
| SspH1 | Novel E3 ligase family | Ubiquitylates serine/threonine kinase PKN1 resulting in its proteasomal degradation and suppression of NF‐kB pathway | Keszei et al, (2014) | |
| SspH2 | Novel E3 ligase family | Ubiquitylates and activates NOD1 signaling | Bhavsar et al, (2013) | |
| SlrP | Novel E3 ligase family | Ubiquitylates host thioredoxin (TXN) inducing host cell death | Bernal‐Bayard and Ramos‐Morales (2009) | |
| SseL | Deubiquitinase | Removes Ub aggregates on Salmonella and SCVs | Mesquita et al, (2012) | |
| Legionella pneumophila | RavZ | Cysteine protease | Inhibits recruitment of LC3 to LCV by irreversibly deconjugating LC3 from PE; Inhibits recruitment of Ub to LCV via unknown mechanism | Choy et al, (2012); Kubori et al, (2017) |
| RavD | Deubiquitinase | RavD specifically hydrolyzes Met1‐linked Ub chains directly antagonizing LUBAC activity and thus activation of NF‐kB | Wan et al, (2019) | |
| Shigella flexneri | IpaH 1.4 | Novel E3 ligase family | Interacts with HOIL and conjugate K48 chains to the catalytic domain of HOIP, thereby triggering its proteasomal degradation | de Jong et al, (2016) |
| IpaH 2.5 | Novel E3 ligase family | Interacts with HOIL and conjugate K48 chains to the catalytic domain of HOIP, thereby triggering its proteasomal degradation | de Jong et al, (2016) | |
| IpaH 4.5 | Novel E3 ligase family | Binds to p65 subunit of NF‐κB and modulates inflammatory response; Promotes K48 polyubiquitylation mediated degradation of TBK1, thereby shutting down the host anti‐bacterial response | Wang et al, (2013) and Zheng et al, (2016) | |
| IpaH 9.8 | Novel E3 ligase family | Ubiquitylates NEMO leading to its proteasomal degradation, thereby inhibiting the NF‐κB signaling pathway | Ashida et al, (2010) | |
| IpaH 0722 | Novel E3 ligase family | Inhibits PKC‐mediated activation of NF‐κB by ubiquitylating TRAF2 for proteasomal degradation | Ashida et al, (2013) | |
| IcsB | Fatty acyltransferase | Represses the recruitment of LC3 and NDP52 to Shigella; Escapes host autophagy indirectly by mediating fatty acylation and inactivation of CHMP5 protein | Baxt and Goldberg (2014); Liu et al, (2018) | |
| Listeria monocytogenes | ActA | Actin assembly‐inducing protein | Recruits Arp2/3 complex and Ena/VASP to disguise the bacterial surface from autophagic recognition | Yoshikawa et al, (2009) |
| Mycobacterium tuberculosis | SapM | Acid phosphatase | Prevents fusion of lysosomes with mycobacterium containing phagosomes | Zulauf et al, (2018) |
| PknG | Ser‐Thr protein kinase | Ubiquitylation of TRAF2 and TAB1 resulting in their proteasomal degradation | Wang et al, (2021) | |
| Group A Streptococcus (GAS) | SpeB | Cysteine protease | Degrades p62, NDP52, and NBR1 in the host cytosol | Barnett et al, (2013) |
| Rickettsia parkeri | OmpB | Outer membrane protein | Acts as a protective shield obstructing recognition by the autophagy machinery | Engstrom et al, (2019) |
| PKMT1/2 | Lysine methlytransferase | Modifies the lysine of outer membrane proteins with methyl groups and prevents their recognition by the host ubiquitin machinery | Engstrom et al, (2021) |
Third, still other bacteria interfere with the various components of the xenophagy machinery. M. tuberculosis employs the effector SapM to hydrolyze phagosomal PI3P and bind Rab7 to inhibit maturation of phagosomes and autophagosomes to phagolysosomes and autolysosomes, respectively (Vergne et al, 2005; Hu et al, 2015). In contrast, GAS serotype M1T1 secretes the cysteine protease SpeB that degrades p62, NBR1, and NDP52 (Barnett et al, 2013) and Legionella’s RavZ effector protein irreversibly deconjugates LC3 from phosphatidylethanolamine (PE), thereby preventing engulfment by autophagosomes (Choy et al, 2012).
Fourth, inflammatory signaling is commonly targeted by various pathogens since multiple innate immunity sensing pathways including detection of bacterial or damaged host membranes converge on this response. Bacterial survival strategies often involve the concerted action of several different effectors. Intriguingly, a number of bacteria including Salmonella, Shigella, and Legionella have developed E3‐like enzymes which specifically utilize the host ubiquitin system in their favor. This is remarkable since these pathogens do not possess genes encoding for ubiquitin themselves. An emerging theme of these mimicry effectors is to interact with host E3 ligases and modulate their functions in different ways to inhibit innate immune signaling. The Salmonella HECT‐like E3 ligase SopA engages two tripartite motif‐containing (TRIM) E3 ligases TRIM56 and TRIM65 which are involved in type I interferon expression (Kamanova et al, 2016). SopA inhibits both ligases via binding to their respective RING domains and triggering the proteasomal degradation of TRIM56 and TRIM65 by mediating their K48 and K11 polyubiquitylation (Fiskin et al, 2017). Moreover, Salmonella SspH1, SspH2, and SlrP represent so‐called novel E3 ligases (NELs) which do not share structural homology with any member of the three eukaryotic ligases families. NELs bind to host E2 enzymes, transfer the charged ubiquitin to a catalytic cysteine residue within their conserved ligase domain and use LRRs of varying lengths to interact with host substrates (Hicks & Galan, 2010; Maculins et al, 2016). By SspH1‐, SspH2‐ and SlrP‐mediated ubiquitylation of serine/threonine‐protein kinase N1 (PKN1), nucleotide‐binding oligomerization domain‐containing protein 1 (NOD1) and thioredoxin (TXN), respectively, Salmonella counteracts NF‐κB signaling and promotes host cell death (Haraga & Miller, 2006; Bernal‐Bayard & Ramos‐Morales, 2009; Bhavsar et al, 2013; Keszei et al, 2014). In contrast, Shigella secretes the NEL effector IpaH1.4 and IpaH2.5 that directly interact with HOIL‐1L and attach K48 chains to HOIP causing its proteasomal degradation and inhibition of NF‐κB pathway activation (de Jong et al, 2016). In parallel, ubiquitylation and degradation of the NF‐κB signaling components TRAF2, NEMO, and p65 are mediated by the Shigella NELs IpaH0722, IpaH9.8, and IpaH4.5, respectively (Ashida et al, 2013; Ashida & Sasakawa, 2015). In another notable example, M. tuberculosis protein kinase G (PknG) effector protein, which is a eukaryotic‐type serine/threonine kinase, inhibits NF‐κB signaling by acting as both E1 and E3 enzyme to trigger ubiquitylation and degradation of TRAF2 and TAK1 (Wang et al, 2021).
Lastly, Legionella RavD has deubiquitinase activity, which exclusively cleaves M1‐linked Ub chains in a similar manner than the host DUB OTULIN, and hence directly antagonizes the activity of LUBAC. Secreted RavD removes M1 chains from Legionella‐containing vacuoles and thereby suppresses NF‐κB signaling (Wan et al, 2019). Given the importance of Ub modifications, in particular M1 chains, for the defense against pathogens it is somewhat surprising that RavD is currently the only bacterial DUB known to hydrolyze M1 chains. However, effectors with deubiquitinase activity from other intracellular pathogens might have gone unrecognized since bacterial DUBs are very divergent from eukaryotic DUBs (Hermanns & Hofmann, 2019).
Concluding remarks and outlook
Since the discovery of the first bacteria‐targeting Ub E3 ligase, our knowledge about the role of Ub in host cell responses against cytosol intruding pathogens increased substantially—both at the level of the involved conjugation machinery and at the level of generated Ub chains. Since the in vitro and in vivo activities of the identified ubiquitin E3 ligases (ARIH1, LRSAM1, LUBAC, Smurf1, Parkin, and RNF213) do not exactly match the spectrum of chain types decorating cytosolic bacteria, the emerging picture is likely to become even more complex. A challenging task will be to systematically decipher the interdependence of these different E3 ligases with regard to their recruitment to bacteria and activation of ubiquitylation (Box 1). With the recent work from the Randow and Dikic laboratories (Noad et al, 2017; van Wijk et al, 2017), it becomes increasingly clear that Ub is not only attached to mark bacteria for clearance by xenophagy. Therefore, another important task will be to identify the full array of Ub chain type‐specific binding effector proteins and their anti‐bacterial activities. Lastly, it will be crucial to test these Ub‐mediated host cell detection and response cascades in (patho)physiological relevant settings across a number of different cytosol‐dwelling pathogens.
Box 1. In need of answers.
What determines the recognition and association of the E3 ligase machineries with cytosolic bacteria? What constitutes the specificity of xenophagic E3 ligases?
What is the full substrate spectrum on cytosolic bacteria that is conjugated with ubiquitin? Which bacterial outer membrane proteins are ubiquitylated by which E3 ligase? Does RNF213 ubiquitylate other non‐protein targets than LPS which is only present in gram‐negative bacteria?
What is the precise Ub chain type composition and architecture of the Ub coat on cytosolic bacteria? Is the Ub coat bacteria‐specific?
How are Ub and autophagy receptor microdomains on bacterial surfaces established and what are their roles in orchestrating xenophagic and innate immune response pathways?
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported Deutsche Forschungsgemeinschaft (FundRef DOI: 10.13039/501100001659) through the project grants BE 4685/6‐1 (CB) and AL 2389/1‐1 (AA). Open Access funding enabled and organized by Projekt DEAL.
EMBO reports (2021) 22: e52864.
See the Glossary for abbreviations used in this article.
Contributor Information
Christian Behrends, Email: Christian.Behrends@mail03.med.uni-muenchen.de.
Arno F Alpi, Email: aalpi@biochem.mpg.de.
References
- Aguirre JD, Dunkerley KM, Mercier P, Shaw GS (2017) Structure of phosphorylated UBL domain and insights into PINK1‐orchestrated parkin activation. Proc Natl Acad Sci U S A 114: 298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel J, Fletcher A, Grabarczyk DB, Roitinger E, Deszcz L, Lehner A, Virdee A, Clausen T (2021) E3 ubiquitin ligase RNF213 employs a non‐canonical zinc finger active site and is allosterically regulated by ATP. bioRxiv 10.1101/2021.05.10.443411 [PREPRINT] [DOI] [Google Scholar]
- Ahel J, Lehner A, Vogel A, Schleiffer A, Meinhart A, Haselbach D, Clausen T (2020) Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein‐like core and a distinct ubiquitin‐transfer mechanism. Elife 9: e56185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Vollaard AM, Widjaja S, Surjadi C, van de Vosse E, van Dissel JT (2006) PARK2/PACRG polymorphisms and susceptibility to typhoid and paratyphoid fever. Clin Exp Immunol 144: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Yakir L, Katz M, Zwang Y, Marmor MD, Citri A, Shtiegman K, Alroy I, Tuvia S, Reiss Y et al (2004) Tal, a Tsg101‐specific E3 ubiquitin ligase, regulates receptor endocytosis and retrovirus budding. Genes Dev 18: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers A, Ramjaun AR, McPherson PS (2004) The HECT domain ligase itch ubiquitinates endophilin and localizes to the trans‐Golgi network and endosomal system. J Biol Chem 279: 11471–11479 [DOI] [PubMed] [Google Scholar]
- Arbibe L, Kim DW, Batsche E, Pedron T, Mateescu B, Muchardt C, Parsot C, Sansonetti PJ (2007) An injected bacterial effector targets chromatin access for transcription factor NF‐kappaB to alter transcription of host genes involved in immune responses. Nat Immunol 8: 47–56 [DOI] [PubMed] [Google Scholar]
- Ashida H, Kim M, Schmidt‐Supprian M, Ma A, Ogawa M, Sasakawa C (2010) A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF‐kappaB‐mediated inflammatory response. Nat Cell Biol 12: 66–73; sup pp 61‐69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Nakano H, Sasakawa C (2013) Shigella IpaH0722 E3 ubiquitin ligase effector targets TRAF2 to inhibit PKC‐NF‐kappaB activity in invaded epithelial cells. PLoS Pathog 9: e1003409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Sasakawa C (2015) Shigella IpaH family effectors as a versatile model for studying pathogenic bacteria. Front Cell Infect Microbiol 5: 100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banh RS, Iorio C, Marcotte R, Xu Y, Cojocari D, Rahman AA, Pawling J, Zhang W, Sinha A, Rose CM et al (2016) PTP1B controls non‐mitochondrial oxygen consumption by regulating RNF213 to promote tumour survival during hypoxia. Nat Cell Biol 18: 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett T, Liebl D, Seymour L, Gillen C, Lim J, LaRock C, Davies M, Schulz B, Nizet V, Teasdale R et al (2013) The globally disseminated M1T1 clone of group A Streptococcus evades autophagy for intracellular replication. Cell Host Microbe 14: 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxt LA, Goldberg MB (2014) Host and bacterial proteins that repress recruitment of LC3 to Shigella early during infection. PLoS One 9: e94653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal‐Bayard J, Ramos‐Morales F (2009) Salmonella type III secretion effector SlrP is an E3 ubiquitin ligase for mammalian thioredoxin. J Biol Chem 284: 27587–27595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beron W, Gutierrez MG, Rabinovitch M, Colombo MI (2002) Coxiella burnetii localizes in a Rab7‐labeled compartment with autophagic characteristics. Infect Immun 70: 5816–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar AP, Brown NF, Stoepel J, Wiermer M, Martin DDO, Hsu KJ, Imami K, Ross CJ, Hayden MR, Foster LJ et al (2013) The Salmonella type III effector SspH2 specifically exploits the NLR co‐chaperone activity of SGT1 to subvert immunity. PLoS Pathog 9: e1003518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian W, Guo Y, Zhang Y, Li H (2017) The self‐association and activity regulation of LRSAM1 E3 ligase. Biochem Biophys Res Commun 485: 95–101 [DOI] [PubMed] [Google Scholar]
- Branigan E, Carlos Penedo J, Hay RT (2020) Ubiquitin transfer by a RING E3 ligase occurs from a closed E2˜ubiquitin conformation. Nat Commun 11: 2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH (2006) Autophagy controls Salmonella infection in response to damage to the Salmonella‐containing vacuole. J Biol Chem 281: 11374–11383 [DOI] [PubMed] [Google Scholar]
- Buetow L, Huang DT (2016) Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol 17: 626–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Zhang L (2013) A Smurf1 tale: function and regulation of an ubiquitin ligase in multiple cellular networks. Cell Mol Life Sci 70: 2305–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case ED, Chong A, Wehrly TD, Hansen B, Child R, Hwang S, Virgin HW, Celli J (2014) The Francisella O‐antigen mediates survival in the macrophage cytosol via autophagy avoidance. Cell Microbiol 16: 862–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cemma M, Kim PK, Brumell JH (2011) The ubiquitin‐binding adaptor proteins p62/SQSTM1 and NDP52 are recruited independently to bacteria‐associated microdomains to target Salmonella to the autophagy pathway. Autophagy 7: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Q, Wang X, Qiang L, Zhang Y, Ge P, Lu Z, Zhong Y, Li B, Wang J, Zhang L et al (2019) A Mycobacterium tuberculosis surface protein recruits ubiquitin to trigger host xenophagy. Nat Commun 10: 1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS, Walden H (2011) Autoregulation of Parkin activity through its ubiquitin‐like domain. EMBO J 30: 2853–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd M, Kimura T, Choi S, Peters R, Mandell M, Bruun J‐A et al (2016) TRIMs and galectins globally cooperate and TRIM16 and galectin‐3 Co‐direct autophagy in endomembrane damage homeostasis. Dev Cell 39: 13–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HI, Sudol M (1995) The WW domain of Yes‐associated protein binds a proline‐rich ligand that differs from the consensus established for Src homology 3‐binding modules. Proc Natl Acad Sci U S A 92: 7819–7823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MI, Chen C, Engstrom P, Portnoy DA, Mitchell G (2018) Actin‐based motility allows Listeria monocytogenes to avoid autophagy in the macrophage cytosol. Cell Microbiol 20: e12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YL, Wu YW, Kuo CF, Lu SL, Liu FT, Anderson R, Lin CF, Liu YL, Wang WY, Chen YD et al (2017) Galectin‐3 inhibits Galectin‐8/Parkin‐mediated ubiquitination of group A Streptococcus. MBio 8: e00899‐17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, Roy CR (2012) The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science 338: 1072–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Kang Y, Yan C, Yang C, Zhang T, Huo H, Liu Y (2021) LUBAC and OTULIN regulate autophagy initiation and maturation by mediating the linear ubiquitination and the stabilization of ATG13. Autophagy 17: 1684–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P, Sansonetti PJ (2004) Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304: 242–248 [DOI] [PubMed] [Google Scholar]
- Cotton TR, Lechtenberg BC (2020) Chain reactions: molecular mechanisms of RBR ubiquitin ligases. Biochem Soc Trans 48: 1737–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT (2012) BIRC7‐E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol 19: 876–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT (2013) Essentiality of a non‐RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat Struct Mol Biol 20: 982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Olszewski JL, Schuermann JP, Kurinov I, Miller DJ, Nourse A, Alpi AF, Schulman BA (2013) Structure of HHARI, a RING‐IBR‐RING ubiquitin ligase: autoinhibition of an Ariadne‐family E3 and insights into ligation mechanism. Structure 21: 1030–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R, Klos DA, Adler AS, Hicke L (2004) The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J Cell Biol 165: 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta RK, Kathania M, Raje M, Majumdar S (2012) IL‐6 inhibits IFN‐gamma induced autophagy in Mycobacterium tuberculosis H37Rv infected macrophages. Int J Biochem Cell Biol 44: 942–954 [DOI] [PubMed] [Google Scholar]
- Elliott P, Leske D, Hrdinka M, Bagola K, Fiil B, McLaughlin S, Wagstaff J, Volkmar N, Christianson J, Kessler B et al (2016) SPATA2 links CYLD to LUBAC, activates CYLD, and controls LUBAC signaling. Mol Cell 63: 990–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich CH, Ordureau A, Strickson S, Arthur JS, Pedrioli PG, Komander D, Cohen P (2013) Activation of the canonical IKK complex by K63/M1‐linked hybrid ubiquitin chains. Proc Natl Acad Sci U S A 110: 15247–15252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom P, Burke TP, Mitchell G, Ingabire N, Mark KG, Golovkine G, Iavarone AT, Rape M, Cox JS, Welch MD (2019) Evasion of autophagy mediated by Rickettsia surface protein OmpB is critical for virulence. Nat Microbiol 4: 2538–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom P, Burke TP, Tran CJ, Iavarone AT, Welch MD (2021) Lysine methylation shields an intracellular pathogen from ubiquitylation and autophagy. Sci Adv 7: eabg2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X, Jia Y, Zhang Y, Ma F, Zhu Y, Hong X, Zhou Q, He R, Zhang H, Jin J et al (2019) Ubiquitination of UVRAG by SMURF1 promotes autophagosome maturation and inhibits hepatocellular carcinoma growth. Autophagy 15: 1130–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiil BK, Gyrd‐Hansen M (2021) The Met1‐linked ubiquitin machinery in inflammation and infection. Cell Death Differ 28: 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskin E, Bhogaraju S, Herhaus L, Kalayil S, Hahn M, Dikic I (2017) Structural basis for the recognition and degradation of host TRIM proteins by Salmonella effector SopA. Nat Commun 8: 14004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskin E, Bionda T, Dikic I, Behrends C (2016) Global analysis of host and bacterial ubiquitinome in response to Salmonella Typhimurium infection. Mol Cell 62: 967–981 [DOI] [PubMed] [Google Scholar]
- Franco LH, Nair VR, Scharn CR, Xavier RJ, Torrealba JR, Shiloh MU, Levine B (2017) The ubiquitin ligase Smurf1 functions in selective autophagy of Mycobacterium tuberculosis and anti‐tuberculous host defense. Cell Host Microbe 21: 59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H, Rahighi S, Akita M, Kato R, Sasaki Y, Wakatsuki S, Iwai K (2014) Mechanism underlying IkappaB kinase activation mediated by the linear ubiquitin chain assembly complex. Mol Cell Biol 34: 1322–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica D, Lahiri V, Klionsky DJ (2018) Cargo recognition and degradation by selective autophagy. Nat Cell Biol 20: 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladkova C, Maslen SL, Skehel JM, Komander D (2018) Mechanism of parkin activation by PINK1. Nature 559: 410–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick D, Barth S, Macleod KF (2010) Autophagy: cellular and molecular mechanisms. J Pathol 221: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V (2004) Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119: 753–766 [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, Colombo MI (2005) Autophagy induction favours the generation and maturation of the Coxiella‐replicative vacuoles. Cell Microbiol 7: 981–993 [DOI] [PubMed] [Google Scholar]
- Haraga A, Miller SI (2006) A Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1. Cell Microbiol 8: 837–846 [DOI] [PubMed] [Google Scholar]
- Heath RJ, Goel G, Baxt LA, Rush JS, Mohanan V, Paulus GLC, Jani V, Lassen KG, Xavier RJ (2016) RNF166 determines recruitment of adaptor proteins during antibacterial autophagy. Cell Rep 17: 2183–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann BL, Boada‐Romero E, Cunha LD, Magne J, Green DR (2017) LC3‐associated phagocytosis and inflammation. J Mol Biol 429: 3561–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herhaus L, Dikic I (2018) Regulation of Salmonella‐host cell interactions via the ubiquitin system. Int J Med Microbiol 308: 176–184 [DOI] [PubMed] [Google Scholar]
- Hermanns T, Hofmann K (2019) Bacterial DUBs: deubiquitination beyond the seven classes. Biochem Soc Trans 47: 1857–1866 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A, Varshavsky A (2000) Basic Medical Research Award. The ubiquitin system. Nat Med 6: 1073–1081 [DOI] [PubMed] [Google Scholar]
- Hicks SW, Galan JE (2010) Hijacking the host ubiquitin pathway: structural strategies of bacterial E3 ubiquitin ligases. Curr Opin Microbiol 13: 41–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn‐Ghetko D, Krist DT, Prabu JR, Baek K, Mulder MPC, Klugel M, Scott DC, Ovaa H, Kleiger G, Schulman BA (2021) Ubiquitin ligation to F‐box protein targets by SCF‐RBR E3–E3 super‐assembly. Nature 590: 671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hos NJ, Fischer J, Hos D, Hejazi Z, Calabrese C, Ganesan R, Murthy AMV, Rybniker J, Kumar S, Kronke M et al (2020) TRIM21 is targeted for chaperone‐mediated autophagy during Salmonella typhimurium infection. J Immunol 205: 2456–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdinka M, Fiil B, Zucca M, Leske D, Bagola K, Yabal M, Elliott P, Damgaard R, Komander D, Jost P et al (2016) CYLD limits Lys63‐ and Met1‐linked ubiquitin at receptor complexes to regulate innate immune signaling. Cell Rep 14: 2846–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Wu J, Wang W, Mu M, Zhao R, Xu X, Chen Z, Xiao J, Hu F, Yang Y et al (2015) Autophagy regulation revealed by SapM‐induced block of autophagosome‐lysosome fusion via binding RAB7. Biochem Biophys Res Commun 461: 401–407 [DOI] [PubMed] [Google Scholar]
- Hu W, Chan H, Lu L, Wong KT, Wong SH, Li MX, Xiao ZG, Cho CH, Gin T, Chan MTV et al (2020) Autophagy in intracellular bacterial infection. Semin Cell Dev Biol 101: 41–50 [DOI] [PubMed] [Google Scholar]
- Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP (1999) Structure of an E6AP‐UbcH7 complex: insights into ubiquitination by the E2–E3 enzyme cascade. Science 286: 1321–1326 [DOI] [PubMed] [Google Scholar]
- Huett A, Heath RJ, Begun J, Sassi SO, Baxt LA, Vyas JM, Goldberg MB, Xavier RJ (2012) The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin‐dependent autophagy of intracellular Salmonella Typhimurium. Cell Host Microbe 12: 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Beaudenon S, Howley PM (1995) A family of proteins structurally and functionally related to the E6‐AP ubiquitin‐protein ligase. Proc Natl Acad Sci U S A 92: 2563–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kumanomidou T, Sou YS, Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K, Komatsu M (2008) Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem 283: 22847–22857 [DOI] [PubMed] [Google Scholar]
- Jin S, Tian S, Luo M, Xie W, Liu T, Duan T, Wu Y, Cui J (2017) Tetherin suppresses type I interferon signaling by targeting MAVS for NDP52‐mediated selective autophagic degradation in human cells. Mol Cell 68: 308–322.e304 [DOI] [PubMed] [Google Scholar]
- de Jong MF, Liu Z, Chen D, Alto NM (2016) Shigella flexneri suppresses NF‐kappaB activation by inhibiting linear ubiquitin chain ligation. Nat Microbiol 1: 16084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada F, Aoki Y, Narisawa A, Abe YU, Komatsuzaki S, Kikuchi A, Kanno J, Niihori T, Ono M, Ishii N et al (2011) A genome‐wide association study identifies RNF213 as the first Moyamoya disease gene. J Hum Genet 56: 34–40 [DOI] [PubMed] [Google Scholar]
- Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA (2009) Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin‐HECT(NEDD4L) complex. Mol Cell 36: 1095–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamanova J, Sun H, Lara‐Tejero M, Galan JE (2016) The Salmonella Effector Protein SopA modulates innate immune responses by targeting TRIM E3 ligase family members. PLoS Pathog 12: e1005552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A, Martinez‐Torres RJ, Wilkie S, Kumar A, Peltier J, Gonzalez A, Johnson C, Zhang J, Hope AG, Peggie M et al (2015) Binding to serine 65‐phosphorylated ubiquitin primes Parkin for optimal PINK1‐dependent phosphorylation and activation. EMBO Rep 16: 939–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsall IR, Duda DM, Olszewski JL, Hofmann K, Knebel A, Langevin F, Wood N, Wightman M, Schulman BA, Alpi AF (2013) TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin‐RING ligase complexes. EMBO J 32: 2848–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsall IR, Kristariyanto YA, Knebel A, Wood NT, Kulathu Y, Alpi AF (2019) Coupled monoubiquitylation of the co‐E3 ligase DCNL1 by Ariadne‐RBR E3 ubiquitin ligases promotes cullin‐RING ligase complex remodeling. J Biol Chem 294: 2651–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keszei AF, Tang X, McCormick C, Zeqiraj E, Rohde JR, Tyers M, Sicheri F (2014) Structure of an SspH1‐PKN1 complex reveals the basis for host substrate recognition and mechanism of activation for a bacterial E3 ubiquitin ligase. Mol Cell Biol 34: 362–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keusekotten K, Elliott P, Glockner L, Fiil B, Damgaard R, Kulathu Y, Wauer T, Hospenthal M, Gyrd‐Hansen M, Krappmann D et al (2013) OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1‐linked polyubiquitin. Cell 153: 1312–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, Debnath J, Virgin HW, Stallings CL (2015) Unique role for ATG5 in neutrophil‐mediated immunopathology during M. tuberculosis infection. Nature 528: 565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25: 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou Y‐S, Bjørkøy G, Nunn JL, Bruun J‐A, Shvets E, McEwan DG, Clausen TH, Wild P et al (2009) A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 33: 505–516 [DOI] [PubMed] [Google Scholar]
- Kirkin V, Rogov VV (2019) A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol Cell 76: 268–285 [DOI] [PubMed] [Google Scholar]
- Koganti P, Levy‐Cohen G, Blank M (2018) Smurfs in protein homeostasis, signaling, and cancer. Front Oncol 8: 295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81: 203–229 [DOI] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M et al (2012) PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol 2: 120080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreibich S, Emmenlauer M, Fredlund J, Ramo P, Munz C, Dehio C, Enninga J, Hardt WD (2015) Autophagy proteins promote repair of endosomal membranes damaged by the Salmonella type three secretion system 1. Cell Host Microbe 18: 527–537 [DOI] [PubMed] [Google Scholar]
- Kubori T, Bui XT, Hubber A, Nagai H (2017) Legionella RavZ plays a role in preventing ubiquitin recruitment to bacteria‐containing vacuoles. Front Cell Infect Microbiol 7: 384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Aguirre JD, Condos TEC, Martinez‐Torres RJ, Chaugule VK, Toth R, Sundaramoorthy R, Mercier P, Knebel A, Spratt DE et al (2015) Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. EMBO J 34: 2506–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupka S, De Miguel D, Draber P, Martino L, Surinova S, Rittinger K, Walczak H (2016) SPATA2‐mediated binding of CYLD to HOIP enables CYLD recruitment to signaling complexes. Cell Rep 16: 2271–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamark T, Kirkin V, Dikic I, Johansen T (2009) NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle 8: 1986–1990 [DOI] [PubMed] [Google Scholar]
- LaRock DL, Chaudhary A, Miller SI (2015) Salmonellae interactions with host processes. Nat Rev Microbiol 13: 191–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtenberg BC, Rajput A, Sanishvili R, Dobaczewska MK, Ware CF, Mace PD, Riedl SJ (2016) Structure of a HOIP/E2˜ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature 529: 546–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon S, Haguenauer‐Tsapis R (2009) Ubiquitin ligase adaptors: regulators of ubiquitylation and endocytosis of plasma membrane proteins. Exp Cell Res 315: 1574–1583 [DOI] [PubMed] [Google Scholar]
- Liu W, Morito D, Takashima S, Mineharu Y, Kobayashi H, Hitomi T, Hashikata H, Matsuura N, Yamazaki S, Toyoda A et al (2011) Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS One 6: e22542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Zhou Y, Peng T, Zhou P, Ding X, Li Z, Zhong H, Xu Y, Chen S, Hang HC et al (2018) N(epsilon)‐fatty acylation of multiple membrane‐associated proteins by Shigella IcsB effector to modulate host function. Nat Microbiol 3: 996–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz S (2018) Structural mechanisms of HECT‐type ubiquitin ligases. Biol Chem 399: 127–145 [DOI] [PubMed] [Google Scholar]
- Lorenz S, Cantor AJ, Rape M, Kuriyan J (2013) Macromolecular juggling by ubiquitylation enzymes. BMC Biol 11: 65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou J, Wang Y, Zheng X, Qiu W (2018) TRIM22 regulates macrophage autophagy and enhances Mycobacterium tuberculosis clearance by targeting the nuclear factor‐multiplicity kappaB/beclin 1 pathway. J Cell Biochem 119: 8971–8980 [DOI] [PubMed] [Google Scholar]
- Lu PJ, Zhou XZ, Shen M, Lu KP (1999) Function of WW domains as phosphoserine‐ or phosphothreonine‐binding modules. Science 283: 1325–1328 [DOI] [PubMed] [Google Scholar]
- Maculins T, Fiskin E, Bhogaraju S, Dikic I (2016) Bacteria‐host relationship: ubiquitin ligases as weapons of invasion. Cell Res 26: 499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU, Cox JS (2013) The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 501: 512–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J (2018) LAP it up, fuzz ball: a short history of LC3‐associated phagocytosis. Curr Opin Immunol 55: 54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou Y‐S, Saiki S, Kawajiri S, Sato F et al (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 189: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B, Martin‐Serrano J (2008) Regulation of Tsg101 expression by the steadiness box: a role of Tsg101‐associated ligase. Mol Biol Cell 19: 754–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P, Henault J, Kolbeck R, Sanjuan MA (2014) Noncanonical autophagy: one small step for LC3, one giant leap for immunity. Curr Opin Immunol 26: 69–75 [DOI] [PubMed] [Google Scholar]
- Mesquita FS, Thomas M, Sachse M, Santos AJ, Figueira R, Holden DW (2012) The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLoS Pathog 8: e1002743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira MT, Alcaïs A, Van Thuc N, Moraes MO, Di Flumeri C, Hong Thai VU, Chi Phuong M, Thu Huong N, Ngoc Ba N, Xuan Khoa P et al (2004) Susceptibility to leprosy is associated with PARK2 and PACRG. Nature 427: 636–640 [DOI] [PubMed] [Google Scholar]
- Molinari G, Rohde M, Guzman CA, Chhatwal GS (2000) Two distinct pathways for the invasion of Streptococcus pyogenes in non‐phagocytic cells. Cell Microbiol 2: 145–154 [DOI] [PubMed] [Google Scholar]
- von Muhlinen N, Akutsu M, Ravenhill BJ, Foeglein A, Bloor S, Rutherford TJ, Freund SM, Komander D, Randow F (2012) LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell 48: 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K et al (2004) Autophagy defends cells against invading group A Streptococcus. Science 306: 1037–1040 [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Nakata M, Kawabata S, Hamada S (2001) Cytochrome c‐mediated caspase‐9 activation triggers apoptosis in Streptococcus pyogenes‐infected epithelial cells. Cell Microbiol 3: 395–405 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H (2020) Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol 21: 439–458 [DOI] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng AC, Eisenberg JM, Heath RJ, Huett A, Robinson CM, Nau GJ, Xavier RJ (2011) Human leucine‐rich repeat proteins: a genome‐wide bioinformatic categorization and functional analysis in innate immunity. Proc Natl Acad Sci U S A 108(Suppl 1): 4631–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noad J, von der Malsburg A, Pathe C, Michel MA, Komander D, Randow F (2017) LUBAC‐synthesized linear ubiquitin chains restrict cytosol‐invading bacteria by activating autophagy and NF‐kappaB. Nat Microbiol 2: 17063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN, Inagaki F (2015) Mechanisms of autophagy. Annu Rev Biophys 44: 101–122 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C (2005) Escape of intracellular Shigella from autophagy. Science 307: 727–731 [DOI] [PubMed] [Google Scholar]
- Ordureau A, Sarraf S, Duda D, Heo J‐M, Jedrychowski M, Sviderskiy V, Olszewski J, Koerber J, Xie T, Beausoleil S et al (2014) Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell 56: 360–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orvedahl A, Sumpter R, Xiao G, Ng A, Zou Z, Tang YI, Narimatsu M, Gilpin C, Sun Q, Roth M et al (2011) Image‐based genome‐wide siRNA screen identifies selective autophagy factors. Nature 480: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten EG, Werner E, Crespillo‐Casado A, Boyle KB, Dharamdasani V, Pathe C, Santhanam B, Randow F (2021) Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. Nature 594: 111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin AJ, Jiang X, Birmingham CL, So NS, Brumell JH (2004) Recognition of bacteria in the cytosol of Mammalian cells by the ubiquitin system. Curr Biol 14: 806–811 [DOI] [PubMed] [Google Scholar]
- Piccolis M, Bond LM, Kampmann M, Pulimeno P, Chitraju C, Jayson CBK, Vaites LP, Boland S, Lai ZW, Gabriel KR et al (2019) Probing the global cellular responses to lipotoxicity caused by saturated fatty acids. Mol Cell 74: 32–44.e38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant PJ, Yeger H, Staub O, Howard P, Rotin D (1997) The C2 domain of the ubiquitin protein ligase Nedd4 mediates Ca2+‐dependent plasma membrane localization. J Biol Chem 272: 32329–32336 [DOI] [PubMed] [Google Scholar]
- Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT (2012) Structure of a RING E3 ligase and ubiquitin‐loaded E2 primed for catalysis. Nature 489: 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polajnar M, Dietz MS, Heilemann M, Behrends C (2017) Expanding the host cell ubiquitylation machinery targeting cytosolic Salmonella . EMBO Rep 18: 1572–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE (2012) Structure of an E3:E2˜Ub complex reveals an allosteric mechanism shared among RING/U‐box ligases. Mol Cell 47: 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenhill BJ, Boyle KB, von Muhlinen N, Ellison CJ, Masson GR, Otten EG, Foeglein A, Williams R, Randow F (2019) The cargo receptor NDP52 initiates selective autophagy by recruiting the ULK complex to cytosol‐invading bacteria. Mol Cell 74: 320–329 e326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Komatsu M, Finley K, Simonsen A (2012) Autophagy: more than a nonselective pathway. Int J Cell Biol 2012: 219625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittinger K, Ikeda F (2017) Linear ubiquitin chains: enzymes, mechanisms and biology. Open Biol 7: 170026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkin E, Almeida SM, Ceccarelli DF, Juang Y‐C, MacLean TA, Srikumar T, Huang H, Dunham WH, Fukumura R, Xie G et al (2013) The linear ubiquitin‐specific deubiquitinase gumby regulates angiogenesis. Nature 498: 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V, Suzuki H, Fiskin E, Wild P, Kniss A, Rozenknop A, Kato R, Kawasaki M, McEwan D, Löhr F et al (2013) Structural basis for phosphorylation‐triggered autophagic clearance of Salmonella . Biochem J 454: 459–466 [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Codogno P, Levine B (2012) Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov 11: 709–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakowski ET, Koster S, Portal Celhay C, Park HS, Shrestha E, Hetzenecker SE, Maurer K, Cadwell K, Philips JA (2015) Ubiquilin 1 promotes IFN‐gamma‐induced xenophagy of Mycobacterium tuberculosis . PLoS Pathog 11: e1005076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan MA, Dillon CP, Tait SWG, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S et al (2007) Toll‐like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450: 1253–1257 [DOI] [PubMed] [Google Scholar]
- Sauve V, Lilov A, Seirafi M, Vranas M, Rasool S, Kozlov G, Sprules T, Wang J, Trempe JF, Gehring K (2015) A Ubl/ubiquitin switch in the activation of Parkin. EMBO J 34: 2492–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Kumar S (2014) Mammalian HECT ubiquitin‐protein ligases: biological and pathophysiological aspects. Biochim Biophys Acta 1843: 61–74 [DOI] [PubMed] [Google Scholar]
- Scheffner M, Nuber U, Huibregtse JM (1995) Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nature 373: 81–83 [DOI] [PubMed] [Google Scholar]
- Scheffner M, Staub O (2007) HECT E3s and human disease. BMC Biochem 8(Suppl 1): S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Rhee DY, Duda DM, Kelsall IR, Olszewski JL, Paulo JA, de Jong A, Ovaa H, Alpi AF, Harper JW et al (2016) Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell 166: 1198–1214.e1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JL, Frick CT, Johnson KA, Liu H, Yong SS, Varney AG, Wiest O, Stahelin RV (2020) Molecular analysis of membrane targeting by the C2 domain of the E3 ubiquitin ligase Smurf1. Biomolecules 10: 229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto S, Tsujimura K, Horii T, Koide Y (2013) Autophagy adaptor protein p62/SQSTM1 and autophagy‐related gene Atg5 mediate autophagosome formation in response to Mycobacterium tuberculosis infection in dendritic cells. PLoS One 8: e86017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Chang C, Yokom AL, Jensen LE, Hurley JH (2020) The autophagy adaptor NDP52 and the FIP200 coiled‐coil allosterically activate ULK1 complex membrane recruitment. Elife 9: e59099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Kendall SL, Campanella M (2018) Common traits spark the mitophagy/xenophagy interplay. Front Physiol 9: 1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluimer J, Distel B (2018) Regulating the human HECT E3 ligases. Cell Mol Life Sci 75: 3121–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit JJ, van Dijk WJ, El Atmioui D, Merkx R, Ovaa H, Sixma TK (2013) Target specificity of the E3 ligase LUBAC for ubiquitin and NEMO relies on different minimal requirements. J Biol Chem 288: 31728–31737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit JJ, Monteferrario D, Noordermeer SM, van Dijk WJ, van der Reijden BA, Sixma TK (2012) The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING‐IBR‐RING domain and the unique LDD extension. EMBO J 31: 3833–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz B, Morris‐Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K (2012) LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep 13: 840–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz B, Rana RR, Koliopoulos MG, Morris‐Davies AC, Schaeffer V, Christodoulou E, Howell S, Brown NR, Dikic I, Rittinger K (2013) Structural basis for ligase‐specific conjugation of linear ubiquitin chains by HOIP. Nature 503: 422–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Ernst A, Dikic I (2014) Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 16: 495–501 [DOI] [PubMed] [Google Scholar]
- Sugihara M, Morito D, Ainuki S, Hirano Y, Ogino K, Kitamura A, Hirata H, Nagata K (2019) The AAA+ ATPase/ubiquitin ligase mysterin stabilizes cytoplasmic lipid droplets. J Cell Biol 218: 949–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery F, Martina L, Asselman C, Repo H, Zhang Y, Sedeyn K, Moschonas GD, Bredow C, Teo QW, Zhang J et al (2021) Ring Finger Protein 213 assembles into a sensor for ISGylated proteins with antimicrobial activity. bioRxiv 10.1101/2021.06.03.446796 [PREPRINT] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F (2009) The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin‐coated bacteria. Nat Immunol 10: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F (2012) Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482: 414–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S et al (2009) Involvement of linear polyubiquitylation of NEMO in NF‐kappaB activation. Nat Cell Biol 11: 123–132 [DOI] [PubMed] [Google Scholar]
- Trempe J‐F, Sauve V, Grenier K, Seirafi M, Tang MY, Menade M, Al‐Abdul‐Wahid S, Krett J, Wong K, Kozlov G et al (2013) Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340: 1451–1455 [DOI] [PubMed] [Google Scholar]
- Turco E, Witt M, Abert C, Bock‐Bierbaum T, Su MY, Trapannone R, Sztacho M, Danieli A, Shi X, Zaffagnini G et al (2019) FIP200 claw domain binding to p62 promotes autophagosome formation at ubiquitin condensates. Mol Cell 74: 330–346 e311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JNS, Wang C, Bunker E, Hao L, Maric D, Schiavo G, Randow F, Youle RJ (2019) Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol Cell 74: 347–362 e346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergne I, Chua J, Lee HH, Lucas M, Belisle J, Deretic V (2005) Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis . Proc Natl Acad Sci U S A 102: 4033–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa E, Proics E, Rubio‐Patino C, Obba S, Zunino B, Bossowski JP, Rozier RM, Chiche J, Mondragon L, Riley JS et al (2017) Parkin‐independent mitophagy controls chemotherapeutic response in cancer cells. Cell Rep 20: 2846–2859 [DOI] [PubMed] [Google Scholar]
- Vives‐Bauza C, Zhou C, Huang Y, Cui M, de Vries RLA, Kim J, May J, Tocilescu MA, Liu W, Ko HS et al (2010) PINK1‐dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A 107: 378–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Wang X, Huang C, Xu D, Wang Z, Zhou Y, Zhu Y (2019) A bacterial effector deubiquitinase specifically hydrolyses linear ubiquitin chains to inhibit host inflammatory signalling. Nat Microbiol 4: 1282–1293 [DOI] [PubMed] [Google Scholar]
- Wang D, Zhang DF, Feng JQ, Li GD, Li XA, Yu XF, Long H, Li YY, Yao YG (2016) Common variants in the PARL and PINK1 genes increase the risk to leprosy in Han Chinese from South China. Sci Rep 6: 37086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Jiang Z, Li Y, He X, Zhao J, Yang X, Zhu L, Yin Z, Li X, Wang X et al (2013) Shigella flexneri T3SS effector IpaH4.5 modulates the host inflammatory response via interaction with NF‐kappaB p65 protein. Cell Microbiol 15: 474–485 [DOI] [PubMed] [Google Scholar]
- Wang J, Ge P, Lei Z, Lu Z, Qiang L, Chai Q, Zhang Y, Zhao D, Li B, Su J et al (2021) Mycobacterium tuberculosis protein kinase G acts as an unusual ubiquitinating enzyme to impair host immunity. EMBO Rep 22: e52175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Manzanillo PS, Cox JS (2012) Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA‐sensing pathway. Cell 150: 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T, Komander D (2013) Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J 32: 2099–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T, Simicek M, Schubert A, Komander D (2015) Mechanism of phospho‐ubiquitin‐induced PARKIN activation. Nature 524: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinelt N, van Wijk SJL (2021) Ubiquitin‐dependent and ‐independent functions of OTULIN in cell fate control and beyond. Cell Death Differ 28: 493–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk SJ, Fiskin E, Putyrski M, Pampaloni F, Hou J, Wild P, Kensche T, Grecco HE, Bastiaens P, Dikic I (2012) Fluorescence‐based sensors to monitor localization and functions of linear and K63‐linked ubiquitin chains in cells. Mol Cell 47: 797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk SJL, Fricke F, Herhaus L, Gupta J, Hotte K, Pampaloni F, Grumati P, Kaulich M, Sou YS, Komatsu M et al (2017) Linear ubiquitination of cytosolic Salmonella Typhimurium activates NF‐kappaB and restricts bacterial proliferation. Nat Microbiol 2: 17066 [DOI] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C et al (2011) Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333: 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Li F, Wang Y, Wang Y, Lin Z, Cheng X, Liu J, Chen C, Pan L (2015) Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2. Autophagy 11: 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Kikuchi R, Kojima W, Hayashida R, Koyano F, Kawawaki J, Shoda T, Demizu Y, Naito M, Tanaka K et al (2020) Critical role of mitochondrial ubiquitination and the OPTN‐ATG9A axis in mitophagy. J Cell Biol 219: e201912144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K, Queliconi BB, Koyano F, Saeki Y, Hirokawa T, Tanaka K, Matsuda N (2015) Site‐specific interaction mapping of phosphorylated ubiquitin to uncover parkin activation. J Biol Chem 290: 25199–25211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T et al (2009) Listeria monocytogenes ActA‐mediated escape from autophagic recognition. Nat Cell Biol 11: 1233–1240 [DOI] [PubMed] [Google Scholar]
- Yu HB, Croxen MA, Marchiando AM, Ferreira RB, Cadwell K, Foster LJ, Finlay BB (2014) Autophagy facilitates Salmonella replication in HeLa cells MBio 5: e00865‐00814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini G, Martens S (2016) Mechanisms of Selective Autophagy. J Mol Biol 428: 1714–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Tang X, Guo N, An Y, Chen X, Shi C, Wang C, Li Y, Li S, Xu H et al (2018) Biochanin a enhances the defense against Salmonella enterica infection through AMPK/ULK1/mTOR‐mediated autophagy and extracellular traps and reversing SPI‐1‐dependent Macrophage (MPhi) M2 polarization. Front Cell Infect Microbiol 8: 318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Shabek N (2017) Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem 86: 129–157 [DOI] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH (2009) The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Immunol 183: 5909–5916 [DOI] [PubMed] [Google Scholar]
- Zheng Z, Wei C, Guan K, Yuan Y, Zhang Y, Ma S, Cao Y, Wang F, Zhong H, He X (2016) Bacterial E3 ubiquitin ligase IpaH4.5 of Shigella flexneri targets TBK1 To dampen the host antibacterial response. J Immunol 196: 1199–1208 [DOI] [PubMed] [Google Scholar]
- Zulauf KE, Sullivan JT, Braunstein M (2018) The SecA2 pathway of Mycobacterium tuberculosis exports effectors that work in concert to arrest phagosome and autophagosome maturation. PLoS Pathog 14: e1007011 [DOI] [PMC free article] [PubMed] [Google Scholar]


