ABSTRACT
Rural communities often rely on groundwater for potable water supply. In this study, untreated groundwater samples from 28 shallow groundwater wells in Finland (<10 m deep and mostly supplying untreated groundwater to <200 users in rural areas) were assessed for physicochemical water quality, stable water isotopes, microbial water quality indicators, host-specific microbial source tracking (MST) markers, and bacterial community composition, activity, and diversity (using amplicon sequencing of the 16S rRNA gene and 16S rRNA). Indications of surface water intrusion were identified in five wells, and these indications were found to be negatively correlated, overall, with bacterial alpha diversity (based on amplicon sequencing of the 16S rRNA gene). High levels of turbidity, heterotrophs, and iron compromised water quality in two wells, with values up to 2.98 nephelometric turbidity units (NTU), 16,000 CFU/ml, and 2,300 μg/liter, respectively. Coliform bacteria and general fecal indicator Bacteroidales bacteria (GenBac3) were detected in 14 and 10 wells, respectively (albeit mostly at low levels), and correlations were identified between microbial, physicochemical, and environmental parameters, which may indicate impacts from nearby land use (e.g., agriculture, surface water, road salt used for deicing). Our results show that although water quality was generally adequate in most of the studied wells, the continued safe use of these wells should not be taken for granted.
IMPORTANCE Standard physicochemical water quality analyses and microbial indicator analyses leave much of the (largely uncultured) complexity of groundwater microbial communities unexplored. This study combined these standard methods with additional analyses of stable water isotopes, bacterial community data, and environmental data about the surrounding areas to investigate the associations between physicochemical and microbial properties of 28 shallow groundwater wells in Finland. We detected impaired groundwater quality in some wells, identified potential land use impacts, and revealed indications of surface water intrusion which were negatively correlated with bacterial alpha diversity. The potential influence of surface water intrusion on groundwater wells and their bacterial communities is of particular interest and warrants further investigation because surface water intrusion has previously been linked to groundwater contamination, which is the primary cause of waterborne outbreaks in the Nordic region and one of the major causes in the United States and Canada.
KEYWORDS: groundwater, bacteria, drinking water, microbial diversity, potable water, 16S rRNA, wells, rural, water supply, water quality, isotopes, groundwater bacteria
INTRODUCTION
Groundwater is the world’s largest freshwater resource and is estimated to provide potable water for up to half of the global population, supplying many major cities and towns, as well as most rural areas (1–4). Shallow groundwater resources (e.g., <10 m below the land surface) are widespread globally (5) and are commonly exploited throughout the developing and developed world because they can provide reliable supplies of water in a technically and economically feasible manner (2, 6–8). However, shallow groundwater resources are particularly vulnerable to contamination, not only because of their proximity to the ground surface but also because of shortcomings in the management and maintenance of groundwater wells (9–13). Groundwater contamination is widely recognized as an important public health issue (7), but more work is needed to understand and mitigate the threats to groundwater systems in rural areas. These smaller systems often suffer from a lack of attention and resources and generally have more problems than larger systems (14–20).
Threats to the safe use of shallow groundwater wells for potable water supply can arise through contamination events that often correlate with changes in the physicochemical and/or microbial parameters of the water. Physicochemical changes may be caused by, for example, the influence of nearby ditches, sand or gravel pits, or salted roads in wintertime (21–24), whereas microbial contamination is typically caused by the introduction of pathogenic microbes from animal waste and/or human sewage into the water supply from nearby agricultural activities, livestock, wild animals, septic tanks, sewage systems, or surface water sources (13, 25–35). Microbes known to be associated with groundwater contamination include (i) fecal indicator bacteria, such as Escherichia coli, intestinal enterococci, Clostridium, and Bacteroides, (ii) pathogenic bacteria, such as pathogenic strains of E. coli and some species of Salmonella, Shigella, and Campylobacter, (iii) pathogenic viruses, such as enterovirus, norovirus, rotavirus, hepatovirus A, and adenovirus, and (iv) protozoa, such as Cryptosporidium and Giardia (7, 36, 37).
The microbial contamination of groundwater is a widespread occurrence globally and continues to cause outbreaks of gastrointestinal illness in both developing and developed countries (11, 37, 38). It is currently the primary cause of waterborne outbreaks in the Nordic region (36, 39), as well as one of the major causes in the United States and Canada (7, 40–42). Many of these outbreaks have been associated with private or community groundwater wells in rural areas (7, 40–42). Such wells are often operated by untrained personnel (43), and in many cases the water is pumped to users without treatment, which means that good groundwater quality and an intact well structure are essential to enable safe water use (13, 30, 35, 44–46). Unfortunately, these conditions are not always guaranteed, and outbreaks can arise due to poor well construction, insufficient depth of protective layer above the water table, floods and surface runoffs, fissures in bedrock, or the leakage and blockage of nearby wastewater pipes (13, 33, 34, 44–48).
Many studies of groundwater microbiology focus largely on the detection of indicator microbes such as E. coli and coliform bacteria and how these might indicate potential risks to human health (13, 31, 49–51). Recently, however, the composition, activity, and diversity of microbial communities in groundwater are being more thoroughly investigated and understood (52–66). As a result, it is becoming clear that groundwater and groundwater wells should not be treated as inert systems but rather as complex ecosystems containing a wide variety of (often uncultured) microbes that interact with each other and their environment in ways that are not yet fully known. These interactions may have implications for the management of these wells and for ensuring good drinking water quality for the people who rely on them.
The aim of our study was to examine the physicochemical, microbial, and environmental differences between shallow groundwater wells in rural areas, with the goal of discovering associations which may have implications for detecting and mitigating contamination. Our principal hypothesis was that nearby land use and/or nearby hydrology and hydrogeology (e.g., streams, lakes, bogs, and fens) would be the major factors influencing the water quality and diversity of bacterial communities in the groundwater wells. We explored this hypothesis by analyzing physicochemical and microbial data from untreated groundwater samples, stable water isotope data from nearby surface water sources, and site-specific environmental data gathered via maps and on-site evaluations.
RESULTS AND DISCUSSION
Variation in physicochemical parameters between wells indicates potential links with environmental data.
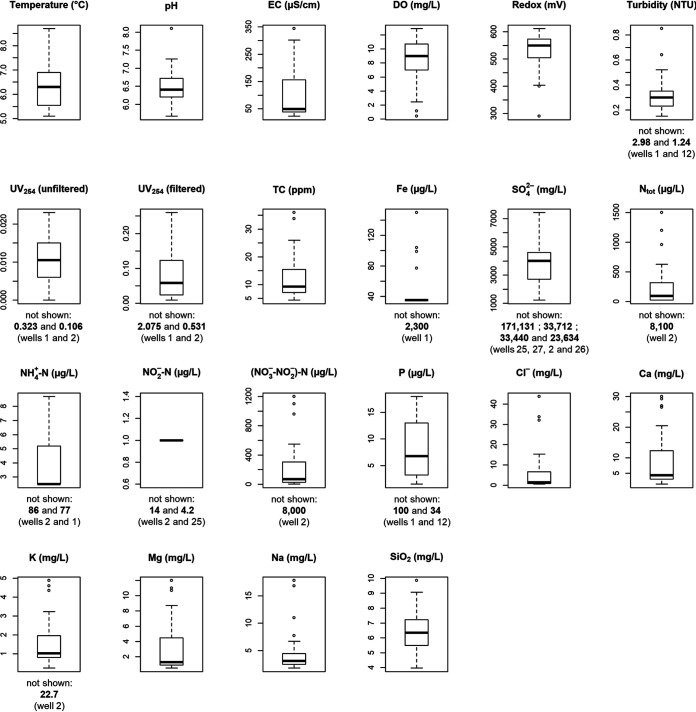
The groundwater from most wells was oligotrophic (low in nutrients such as N and P), with high levels of dissolved oxygen (DO; median: 9 mg/liter) and slightly acidic pH (median: 6.4) (Fig. 1). Some wells had high pH (up to 8.1) because of alkalinization material in the wells (e.g., well 21) or because of naturally high levels of Ca and Mg in the water (e.g., well 23). Well 1 stood out as having the highest overall temperature (8.7°C) and turbidity (2.98 nephelometric turbidity units [NTU]), the highest concentrations of Fe (2,300 μg/liter) and P (100 μg/liter), and the lowest redox potential (71.5 mV) and DO (0.42 mg/liter) levels. Reddish-brown staining and slime were observed on pipes at this site, suggesting that the high Fe levels were causing high water turbidity and growth of iron-oxidizing bacteria such as Gallionella (29.9% of all reads in the cDNA-derived 16S amplicons from this site were attributed to this genus). Also, this well was located in a clay-rich coastal area of a kind that, in Finland, is often associated with acid sulfate soils that can leach metals like Fe (67). High P levels in this well might be explained by the fact that low DO levels can cause Fe oxides in soils and aquifers to dissolve and release adsorbed P into the water (68). Well 2 had the highest overall concentrations of total nitrogen (Ntot; 8,100 μg/liter), ammonium nitrogen (NH4+-N; 86 μg/liter), nitrite nitrogen (NO2−-N; 14 μg/liter), combined nitrate and nitrite nitrogen [(NO3−+NO2−)-N; 8,000 μg/liter], and K (22.7 mg/liter), as well as the largest nearby field area (72 ha within 1 km2 of the well), suggesting that high input of these nutrients may be coming from nearby agriculture (69). Well 25 had the highest concentrations of both sulfate (SO42−; 171 mg/liter) and silica (SiO2) (9.86 mg/liter). The SO42− may have come from sulfur-containing fertilizers used in nearby agriculture (the well had the fourth largest nearby field area) or from the weathering of rocks and minerals, which are also potential sources of SiO2 (70, 71). Several wells also showed comparatively high concentrations of Na and chloride (Cl−), potentially indicating the infiltration of surface waters carrying road salt components into wells near major roads (23), as sodium chloride (NaCl) is the main road salt used in Finland. The strongest example of this was well 26, which had the highest concentrations of both Na (17.8 mg/liter) and Cl− (43.7 mg/liter), as well as the largest total nearby road length (8,074 m within 1 km2 of the well).
FIG 1.
Boxplots showing physicochemical data for the 28 shallow groundwater wells. Some relatively extreme values were removed to improve plot readability. These values are indicated below the plots from which they were removed, with the corresponding well numbers given in respective order. Medians were not significantly affected by removal of these values. Boxplots were generated in R.
Indications of surface water intrusion identified in five wells.
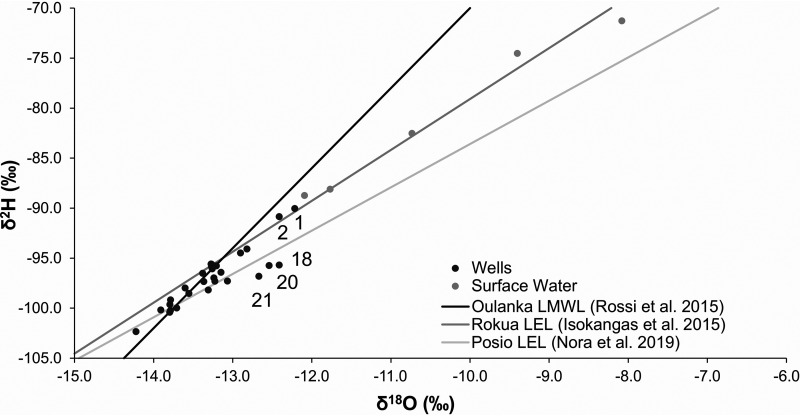
Most groundwater samples taken from the wells had stable water isotopes in the vicinity of the local rainfall line (i.e., the Oulanka local meteoric water line [LMWL]) (72), with δ18O values varying between −14.2 and −12.8, δ2H values varying between −102.3 and −94.1, and d-excess values varying between 7.3 and 11.4 (Fig. 2). This indicated that the source of the water in these wells was local precipitation and especially snowmelt. These are the main expected sources of groundwater recharge, and therefore the isotope signals of most wells were typical of groundwater. Contrastingly, the collected surface water samples mainly followed the Rokua local evaporation line (LEL) (73). Wells 1 and 2, which were previously identified as exceptional based on their physicochemical data, were exceptional here too. Groundwater samples taken from these wells deviated from the bulk of the samples by having stable water isotopes that followed the Rokua LEL rather than the Oulanka LMWL, with δ2H values of −90 and −90.8 and d-excess values of 7.7 and 8.5, respectively. Groundwater samples from wells 18, 20, and 21 were also exceptional, following the Posio LEL rather than the Oulanka LMWL, with d-excess values varying between 3.6 and 4.6 (a range different from that of the other wells). The surface water evaporation signal, the LEL, can vary regionally, depending on local conditions. For this reason, wells 18, 20, and 21 were closer to Posio LEL than to Rokua LEL. Wells 18 and 20 were relatively unremarkable based on physicochemical data, but well 21 had above average pH, Ca, electrical conductivity (EC), and total carbon (TC) levels. Overall, wells 1, 2, 18, 20, and 21 appear to have indications of surface water intrusion based on the evaporation signal in stable water isotopes. Although well 18 had the largest nearby surface water area of all wells (29 ha within 1 km2 of the well), “total nearby surface water area” (median: 3.54 ha within 1 km2 of the well) alone did not seem to be an important predictor of surface water indication in stable water isotopes. Neither did “distance to nearest surface water” (median: 80 m), with only one of the five identified wells (well 18) having a below-median value (55 m). Risk for surface water intrusion into these wells may therefore depend more on the surrounding hydrogeological conditions and precipitation patterns and/or factors relating to well construction and maintenance. Stable water isotopes have previously been used to identify surface water intrusion into wells (74), but given that the groundwater samples analyzed here represent only a single time point, repeated or continuous sampling campaigns would be needed to better understand the dynamics of the surface and well waters at these sites.
FIG 2.
Stable water isotope results from the wells compared to surface water samples and to rainfall. Data for Oulanka local meteoric water line (LMWL) and Rokua and Posio local evaporation lines (LEL) were taken from previous studies (72, 73, 99). Wells with various d-excess values and/or alignment with LEL lines are marked with well numbers.
Microbiological water quality was impaired in some wells.
Low microbial loads were detected in most wells (Table 1). The median total heterotrophic plate count was 125 CFU/ml, although some wells, such as well 2 and well 21, had comparatively high counts (1,200 CFU/ml and 16,000 CFU/ml, respectively), possibly related to surface water indications observed in stable water isotopes. E. coli was not detected in any of the wells, and virtually no coliphages or spores of sulfite-reducing clostridia were detected either, except for a very low level of F-specific coliphages (0.04 PFU/liter) in well 2 and an observation of clostridia (1 CFU/100 ml) in well 1. Coliform bacteria were detected in half of the wells, but mostly at low levels (<20 CFU/liter), with the highest levels being in well 17 (260 CFU/liter) and well 2 (80 CFU/liter). General fecal indicator Bacteroidales bacteria (GenBac3) were detected in DNA extracts from 3 wells (wells 1, 2, and 15) and in cDNA extracts from 10 wells (wells 1, 2, 7, 8, 15, 16, 20, 21, 24, and 26), but no human-specific fecal indicator Bacteroides bacteria (HF183) were detected. Wells 1, 2, and 15 also had above-median levels of NH4+-N, P, coliform bacteria, and buildings within 200 m distance from the well. Gene copies of Gram-negative bacteria were prevalent in all wells, although the RNA copy numbers of Gram-negative bacteria remained below the relatively high limit of detection in eight wells. Well 2, which exhibited relatively high levels of nutrients in the physicochemical analyses and a surface water signal in stable water isotopes, was also the most exceptional well here in terms of microbial findings, exhibiting the highest levels of 16S rRNA gene copies (0.08 genome copies [GC]/ml) and rRNA copies (2.6 GC/ml) of Bacteroidales, the highest levels of 16S rRNA gene copies (2,500 GC/ml) and rRNA copies (370,000 GC/ml) of Gram-negative bacteria, and the second-highest level of heterotrophic plate counts (1,200 CFU/ml).
TABLE 1.
Summary of microbial indicators in the 28 studied groundwater wellsa
| Microbial indicator | Min | Median | Max | Well no. (relatively extreme values)b |
|---|---|---|---|---|
| E. coli (CFU/100 ml) | 0 | 0 | 0 | Indicator not detected |
| Coliform bacteriac (CFU/liter) | 0 | 0 | 260 | Well 17 (260), well 2 (80) |
| Coliform bacteriad (CFU/liter) | 0 | 0 | 210 | Well 17 (210), well 2 (80) |
| SSRC (CFU/100 ml) | 0 | 0 | 1 | Well 1 (1) |
| Heterotrophic bacteria (CFU/ml) | 5 | 125 | 16,000 | Well 21 (16,000), well 2 (1,200) |
| Somatic coliphages (PFU/liter) | 0 | 0 | 0 | Indicator not detected |
| F-specific coliphages (PFU/liter) | 0 | 0 | 0.04 | Well 2 (0.04) |
| Bacteroidales rRNA gene (GenBac3) (GC/100 ml) | 0 | 0 | 8 | Well 2 (8), well 1 (7), well 15 (4) |
| Bacteroidales rRNA (GenBac3) (GC/100 ml) | 0 | 0 | 260 | Well 2 (260), well 15 (183), well 14 (81), well 8 (67) |
| Bacteroides rRNA gene (HF183) (GC/100 ml) | 0 | 0 | 0 | Indicator not detected |
| Bacteroides rRNA (HF183) (GC/100 ml) | 0 | 0 | 0 | Indicator not detected |
| Gram-negative bacteria (rRNA gene) (GC/100 ml) | 1,400 | 13,000 | 250,000 | Well 2 (250,000), well 14 (100,000), well 1 (96,000) |
| Gram-negative bacteria (rRNA) (GC/100 ml) | 0 | 450,000 | 37,000,000 | Well 2 (37,000,000), well 21 (9,700,000) |
SSRC, spores of sulphite-reducing Clostridia; GC, genome copies.
Sites with values greater than one standard deviation above the median.
SFS 3016 method.
ISO 9308-1 method.
Differences observed in alpha diversity metrics of bacterial DNA- and cDNA-derived 16S amplicons.
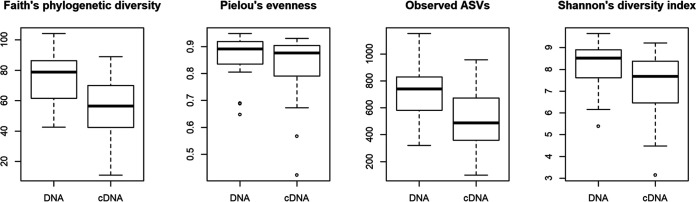
Bacterial 16S rRNA amplicons were sequenced from DNA and cDNA. On average, approximately 35,400 quality-filtered sequences were obtained per library. The median number of observed amplicon sequence variants (ASVs) and median values for Faith’s phylogenetic diversity and Shannon diversity were lower for the cDNA libraries than for the DNA libraries (Fig. 3 and Table 2). The lower diversity in cDNA libraries may indicate a selective activation of bacterial taxa (i.e., not all present taxa are equally active and a small number of active taxa dominate), or it may simply be a reflection of the fact that DNA persists longer than RNA in natural waters, such that DNA-based diversity is better preserved.
FIG 3.
Boxplots showing differences between alpha diversity metrics in DNA- and cDNA-derived 16S amplicons. Boxplots were generated in R.
TABLE 2.
Summary of alpha diversity metrics
| Alpha diversity metric | Min | Median | Max | Well no. (relatively extreme values)a |
|
|---|---|---|---|---|---|
| High | Low | ||||
| Faith’s PD (DNA) | 42.6 | 78.81 | 104.2 | Well 9 (104.24), well 23 (94.56) | Well 2 (42.63), well 21 (48.23), well 27 (55.03), well 16 (57.22), well 18 (59.87), well 7 (60.23), well 15 (60.39), well 22 (62.57) |
| Faith’s PD (cDNA) | 11.1 | 56.54 | 89.04 | Well 23 (89.04), well 26 (88.00), well 5 (79.86), well 20 (79.29), well 10 (77.86) | Well 21 (11.05), well 12 (21.11), well 2 (22.18), well 14 (23.35), well 1 (24.64), well 15 (33.27) |
| Pielou’s evenness (DNA) | 0.65 | 0.89 | 0.95 | Well 2 (0.65), well 22 (0.69), well 15 (0.69), well 21 (0.81) | |
| Pielou’s evenness (cDNA) | 0.42 | 0.88 | 0.93 | Well 12 (0.42), well 15 (0.57), well 2 (0.67), well 21 (0.67), well 28 (0.71) | |
| Observed ASVs (DNA) | 321 | 742 | 1153 | Well 9 (1153), well 8 (971), well 17 (942) | Well 2 (321), well 21 (404), well 27 (451), well 22 (492), well 16 (500), well 18 (538) |
| Observed ASVs (cDNA) | 100 | 489 | 957 | Well 26 (957), well 20 (831), well 23 (819), well 10 (809), well 5 (807), well 3 (718) | Well 21 (100), well 12 (174), well 14 (174), well 2 (184), well 1 (227) |
| Shannon’s diversity (DNA) | 5.39 | 8.52 | 9.65 | Well 9 (9.65) | Well 2 (5.39), well 22 (6.15), well 15 (6.33), well 21 (6.98), well 27 (7.40), well 18 (7.41), well 16 (7.44) |
| Shannon’s diversity (cDNA) | 3.15 | 1.53 | 9.21 | Well 26 (9.21) | Well 12 (3.15), well 21 (4.48), well 15 (4.63), well 2 (5.06), well 14 (5.92), well 28 (5.98), well 1 (6.1) |
Sites with values greater than one standard deviation above or below the median.
Wells 2 and 21 were exceptional here again, being characterized by relatively low values of all four alpha diversity metrics in both DNA and cDNA libraries. This is possibly related to surface water influence noted by the signal in stable water isotopes. The relatively high levels of heterotrophic bacteria and gene copy numbers of Gram-negative bacteria in wells 2 and 21, together with low alpha diversity values, suggest that, at the time of sampling, the bacterial communities in these wells were dominated by a limited group of active bacteria. Wells 9, 23, and 26 had relatively high levels of at least three alpha diversity metrics.
Different dominant taxa in DNA and cDNA libraries and some exceptional wells.
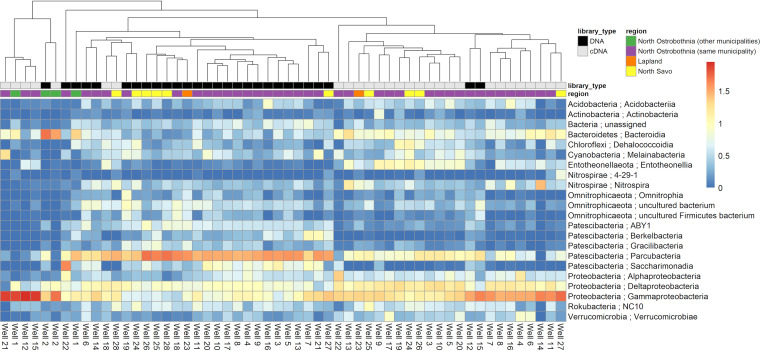
Taxonomic classification of 16S sequences revealed differences between DNA and cDNA libraries (Fig. 4). The most commonly identified bacterial taxa in the DNA-derived 16S amplicons—based on mean relative abundance values of phyla, with Proteobacteria split to the class level—were Patescibacteria (43.5%), Gammaproteobacteria (11.5%), Omnitrophicaeota (7.5%), Deltaproteobacteria (6.6%), and Bacteroidetes (4.5%). Members of the Patescibacteria superphylum have previously been shown to dominate DNA-derived 16S rRNA gene amplicons in groundwater environments (58, 59, 61, 75). These bacteria have particularly small cell sizes and are not easily cultivated (56). The most commonly identified taxa in the cDNA libraries were Gammaproteobacteria (31.9%), Deltaproteobacteria (11.8%), Patescibacteria (8.4%), Bacteroidetes (8.0%), and Entotheonellaeota (4.4%). The most commonly identified taxa in the control libraries were Firmicutes (32.1%), Gammaproteobacteria (28.8%), Actinobacteria (15.8%), Bacteroidetes (11.8%), and Cyanobacteria (2.3%). In 20 of the wells, Patescibacteria were dominant in DNA libraries and Gammaproteobacteria were dominant in cDNA libraries. However, three wells (wells 2, 12, and 15) had DNA libraries with >10% higher relative abundances of Gammaproteobacteria than of Patescibacteria, combined with above-median numbers of coliform bacteria, heterotrophic plate counts, and P concentrations. Wells 2 and 15 also had above-median concentrations of NH4+-N and both DNA-derived and RNA-derived copy numbers of a general fecal Bacteroidales marker (GenBac3). Well 2 also had an indication of surface water intrusion in stable water isotopes. Similarly, two wells (wells 18 and 28) had cDNA libraries with >15% higher relative abundances of Patescibacteria than of Gammaproteobacteria. Both of these wells had above-median values for EC, Ntot, (NO3-+NO2−)-N, SO42−, Cl-, Ca, K, Mg, Na, “nearby field area,” and “number of nearby buildings” and below-median values for “distance to surface water.” Well 18 also had an indication of surface water intrusion in stable water isotopes. Based on these observations, it seems as though a Gammaproteobacteria-Patescibacteria ratio of >1 in DNA libraries or <1 in cDNA libraries could perhaps serve as some kind of water quality indicator in these wells, although further work would be needed to verify this. Escherichia coli was not detected in any of the DNA or cDNA libraries, nor were any of the other bacterial pathogens which were screened in this study.
FIG 4.
Heatmap showing differences in bacterial communities based on taxonomic classifications of DNA- and cDNA-derived 16S amplicons generated in QIIME 2 using the SSU SILVA 132 majority taxonomy. The heatmap was generated in R (using the pheatmap package) (113) from the log-transformed relative abundance values of bacterial classes which had a relative abundance of 5% or more in at least one library. Columns were clustered using average linkage hierarchical clustering based on the Bray–Curtis dissimilarity matrix of the data set (using the vegan package) (116).
There was also some evidence that bacterial taxa with certain metabolic lifestyles are found at higher relative abundance in wells with suitable physicochemical properties. For example, well 1, which had the highest levels of Fe, had one of the highest relative abundances of the Gallionella genus of iron-oxidizing bacteria in DNA (29.9%) and cDNA (6%) libraries. Well 25, which had the highest levels of SO42−, had the highest relative abundances of Beggiatoaceae DNA (1.14%) and cDNA (11.14%), a family of sulfur-oxidizing bacteria. In addition, some wells with high levels of Bacteroidales in quantitative PCR (qPCR) and reverse transcriptase quantitative PCR (RT-qPCR) also had high relative abundances of Bacteroidetes in 16S rRNA gene and rRNA libraries. For example, well 2 had the highest levels of Bacteroidales DNA (80 GC/liter) and cDNA (2,600 GC/liter) in (RT)-qPCR and the highest relative abundance of Bacteroidetes, the phylum containing the order Bacteroidales, in DNA (51.2%) and cDNA (35.3%) libraries. It is worth noting, however, that due to the compositional nature of 16S rRNA amplicon sequencing data, the absolute abundances of the bacteria identified by 16S rRNA amplicon sequencing remain unknown (76).
DNA- and cDNA-derived 16S amplicons largely clustered apart, indicating general differences between “present” (DNA) and metabolically “active” (cDNA) bacterial communities. Differences between present and active microbial communities are often observed in other studies (61, 64, 77). However, use of amplicon sequencing of the 16S rRNA to approximate the active fraction of a bacterial community has its limitations (78).
Some of the cDNA libraries formed a small outlier group during clustering (far left of Fig. 4), with noticeably higher relative abundances of Gammaproteobacteria and lower relative abundances of Parcubacteria than those of the other cDNA libraries. Some of these exceptional libraries were from wells previously identified as exceptional based on physicochemical data, microbiological analysis results, stable water isotopes, or Gammaproteobacteria-Patescibacteria ratio: wells 1, 2, 12, 15, and 21, for example. Such abnormalities may warrant further investigation of potential risks to the continued use of these wells for potable water supply.
Physicochemical and microbial correlations and potential land use impacts.
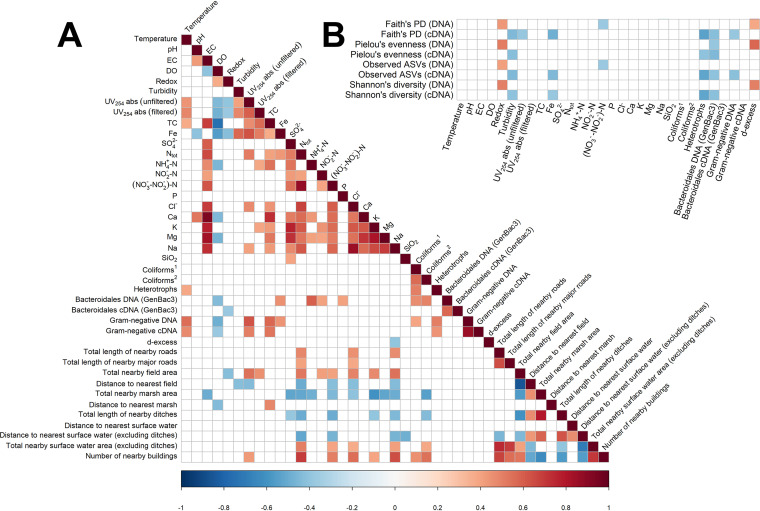
Many statistically significant correlations (P < 0.05) were identified among and between physicochemical and microbial parameters (Fig. 5A). Fe had a strong positive correlation with UV254 absorbance of unfiltered water (0.64) and a moderate positive correlation with turbidity (0.58). Turbidity can be caused by clay, silt, nonliving organic particulates, plankton, microbes, or suspended organic or inorganic matter. Turbidity caused by suspended inorganic matter is particularly common in groundwater, and precipitated iron oxides/hydroxides are one source (as was visibly observed at well 1) (43, 79, 80). Turbidity is known to influence absorbance throughout the UV spectrum (81, 82), and high positive correlation coefficients have previously been reported between turbidity and UV254 absorbance (83).
FIG 5.
Correlograms showing (A) correlations between physicochemical data, microbiological data, and environmental data and (B) correlations between physicochemical data and alpha diversity metrics. Both correlograms were constructed using a Spearman rank-based correlation coefficient matrix and associated P values. Only the statistically significant correlations (P < 0.05) are shown. Red colors are positive correlations. Blue colors are negative correlations. In each case, the intensity of the color indicates the strength of the correlation. Spearman rank-based correlation coefficients were calculated using the rcorr function from the Hmisc R package (114), and correlograms were produced using the corrplot function from the corrplot R package (115). 1SFS 3016 method, 2ISO 9308-1 method.
There was a strong negative correlation between DO and TC (−0.74), which may be related to the fact that heterotrophic microbes typically consume organic carbon most efficiently via aerobic respiration. There were positive correlations between TC and Gram-negative DNA (0.57) and cDNA (0.54) and between UV254 absorbance of unfiltered water and Gram-negative DNA (0.63) and cDNA (0.51). TC and UV254 absorbance of unfiltered water were also positively correlated with each other (0.57). This is unsurprising, perhaps, as UV254 absorbance is an indicator for total organic carbon (TOC) and dissolved organic carbon (DOC), higher levels of which enable better growth of bacteria in water (84–86). Positive correlation coefficients have been previously reported in groundwater for bacterial colony counts and UV254 absorbance (87) and for DOC and Gram-negative bacteria such as coliforms (88).
“Total length of nearby roads” was positively correlated with Ntot, Cl−, and Na, the first of which may originate from vehicle emissions and the latter two from road salt (23, 89). “Total nearby field area” was positively correlated with Na (0.54), UV254 absorbance of unfiltered water (0.54), Ntot (0.52), Cl− (0.50), K (0.48), NO2−-N (0.42), and (NO3−+NO2−)-N (0.40), many of which may be linked to fertilizer use (90–92). Roads and fields were not correlated with microbial data.
“Distance to nearest surface water (excluding ditches)” was negatively correlated with Ntot (−0.52), (NO3−+NO2−)-N (−0.47), and Na (−0.47), and “total nearby surface water area” was positively correlated with the same chemicals (0.50, 0.42, 0.40), suggesting that surface waters may be a potential source of these chemicals in the groundwater wells. “Number of nearby buildings” was positively correlated with Ntot (0.69), Na (0.65), Cl− (0.55), coliforms (0.55 and 0.46), (NO3−+NO2−)-N (0.50), and UV254 absorbance of unfiltered water (0.43). Ntot, Na, and Cl− might appear here, partly because buildings and roads are positively correlated (0.67 and 0.55). The limitations of correlation analysis are quite visible here, as roads, surface water area, buildings, and fields are mostly all positively correlated with one another, making it difficult to determine the exact sources of various chemical and microbial parameters. No statistically significant correlations were identified between environmental data and alpha diversity metrics, suggesting that the environmental data collected in this study were not sufficient to explain differences in bacterial alpha diversity between wells (Table S1).
Bacterial alpha diversity correlated negatively with surface water intrusion and positively with redox potential.
Several statistically significant correlations (P < 0.05) were identified between the physicochemical data and alpha diversity metrics (Fig. 5B). For the DNA-derived 16S rRNA gene amplicons, redox potential had positive correlations with all four alpha diversity metrics (range: 0.42 to 0.52), and d-excess had positive correlations with Pielou’s evenness (0.57), Shannon’s diversity (0.51), and Faith’s phylogenetic diversity (PD; 0.38). Low d-excess values are indicative of surface water intrusion, so intrusion appears to be associated with relatively lower bacterial diversity in the groundwater wells. Further work would be needed to test causation, but perhaps intrusion can cause reduced diversity by introducing generalist bacteria or fresh surface materials from surface water and soil into the naturally occurring, largely oligotrophic groundwater community (93). Microbial communities in groundwater have been shown to react sensitively to surface water intrusion in the context of riverbank filtration, with losses of resident taxa indicated by declining alpha diversity (66), similar to the low diversity found here. However, by contrast, a recent study of a fractured bedrock aquifer found that proximity to the recharge area gave prominence to high bacterial diversity, with the authors proposing that this high diversity was largely due to episodic input of surface soil-derived bacteria (65). Thus, the influence of surface water intrusion on bacterial diversity may be site dependent, or otherwise variable, and warrants further investigation. Yan et al. studied a series of wells at different points on a single hillslope above a fractured bedrock aquifer in the temperate broadleaf biome (65), whereas the wells studied here are largely from sand and gravel aquifers at different locations within the boreal biome, mostly with relatively flat surrounding topographies. These and other differences (e.g., precipitation rates, exact aquifer type and structure, recharge rates, well structure and maintenance) may give rise to different associations between surface water intrusion and bacterial diversity. Microbial communities catalyze important biogeochemical processes in groundwater, such as the turnover of carbon and other nutrients, as well as pollutant attenuation (57, 94), so disturbances and changes to their community composition via surface water intrusion may have important implications for drinking water quality and safety, and thereby for the proper management of groundwater wells, given that surface water intrusion has previously been identified as a direct risk factor for waterborne outbreaks (13, 39, 47).
For the cDNA libraries, turbidity, heterotrophs, and Bacteroidales DNA (GenBac3) had moderate negative correlations with all four alpha diversity metrics (ranges: −0.44 to −0.40, −0.40 to −0.54, and −0.45 to −0.41), Fe had weak to moderate negative correlations with Faith’s phylogenetic diversity, observed ASVs, and Shannon’s diversity (range: −0.48 to −0.38), Gram-negative DNA had weak to moderate negative correlations with Faith’s phylogenetic diversity and observed ASVs, and UV254 absorbance of unfiltered water had a moderate negative correlation with Faith’s phylogenetic diversity (−0.39).
Nonmetric multidimensional scaling.
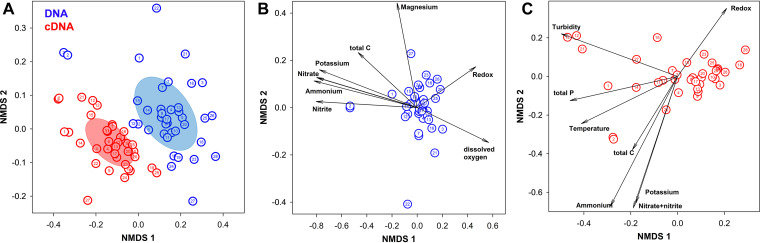
Nonmetric multidimensional scaling (NMDS) based on sequence analysis of bacterial 16S rRNA on the DNA and cDNA level revealed overall differences between DNA- and cDNA-based bacterial communities (Fig. 6A). In addition, fitting of environmental parameters to the NMDS plots for DNA- and cDNA-based bacterial communities revealed that several environmental parameters correlated significantly with bacterial communities (Fig. 6B and C). The compositions of DNA- and cDNA-based communities are influenced by redox potential, total carbon, potassium, ammonium nitrogen (NH4+-N), and combined nitrate and nitrite nitrogen [(NO3−+NO2−)-N]. DNA-based communities were additionally influenced by magnesium, dissolved oxygen, and nitrite nitrogen (NO2−-N), whereas cDNA-based communities were additionally influenced by turbidity, total phosphorus, and temperature. Nutrients such as N, P, and C, and factors such as temperature, redox potential, and dissolved oxygen, are well known to influence the ability of specific bacterial taxa to survive and propagate in various ecosystems (95). As groundwater is often considered an oligotrophic (nutrient-poor) environment (93), the idea that nutrients such as N and P could influence bacterial community composition is not entirely surprising, but the possibility that nutrient inputs from agriculture or surface waters could alter bacterial community composition in these shallow groundwater wells is still worth considering in case it may have implications for maintaining a safe potable water supply.
FIG 6.
Nonmetric multidimensional scaling (NMDS) based on sequence analysis of bacterial 16S rRNA on the DNA (blue) and cDNA (red) levels. Comparison of bacterial communities on the DNA and cDNA levels (A) and effect of environmental parameters on bacterial community composition on the DNA (B) and cDNA (C) levels. In panel A, dispersion ellipses indicate centroids of microbial communities on the DNA and cDNA levels. In panels B and C, selected environmental parameters (P ≤ 0.05) fitted to the ordinations are indicated by arrows. Well numbers are indicated inside the data points.
Revised EU Drinking Water Directive and water safety planning.
The recently revised EU Drinking Water Directive 2020/2184 promotes risk-based approaches and better transparency for drinking water consumers throughout the European Union. However, it remains to be seen how the new directive will affect the smallest water suppliers, because it does not require EU member states to carry out risk assessments on water suppliers supplying 10 to 100 m3 per day or serving 50 to 500 people, and water quality sampling for these supplies need be conducted only once or twice a year. Finnish national legislation requires risk assessments at even the smallest supplies, but other countries may choose to exempt these sites to reduce potential administrative burden.
Given that shallow groundwater resources are often particularly vulnerable to contamination (9–13), and that investments in interventions aimed at improving rural community water supplies are highly cost beneficial in the developed world (96), we propose that risk assessments should be more carefully considered for shallow groundwater wells, especially in cases where surface water intrusion or other risks are indicated. Such risk assessments would involve thorough sanitary surveys (water safety planning) accompanied by, if possible, detailed microbial and physicochemical investigations which include the use of novel analytical methods (e.g., analyses of stable water isotopes, biomarkers, and microbial communities). These assessments would lead to better understanding and predicting of contamination events and better aversion of potential negative health consequences through remedial actions, such as applying water treatment and decontamination and/or eliminating the contamination source(s). In the event that such detailed site-specific analyses are not possible, due to time and resource constraints, findings from this study and similar studies can serve as a starting point for interpreting potential risks to water quality in shallow groundwater wells.
Conclusions, limitations, and outlook.
Our findings provide further evidence that groundwater wells should not be treated as inert structures but rather as complex ecosystems influenced by many factors that are not yet fully known. We pinpointed several potentially problematic wells on the basis of combined physicochemical, microbial, or environmental parameters that may be linked to various nearby water quality risk factors arising from the impacts of land use such as agriculture, roads, surface water, and other human activity. Future work will consider seasonal variation in physicochemical, microbial, and environmental parameters—something which was not assessed here—and further explore the question of surface water intrusion to better assess its risk to water quality and safety and its association with, and potential influence on, groundwater microbial communities, as well as metagenomic analysis of selected groundwater samples to investigate the functional capabilities of microorganisms in these wells.
MATERIALS AND METHODS
Groundwater wells and sampling methods.
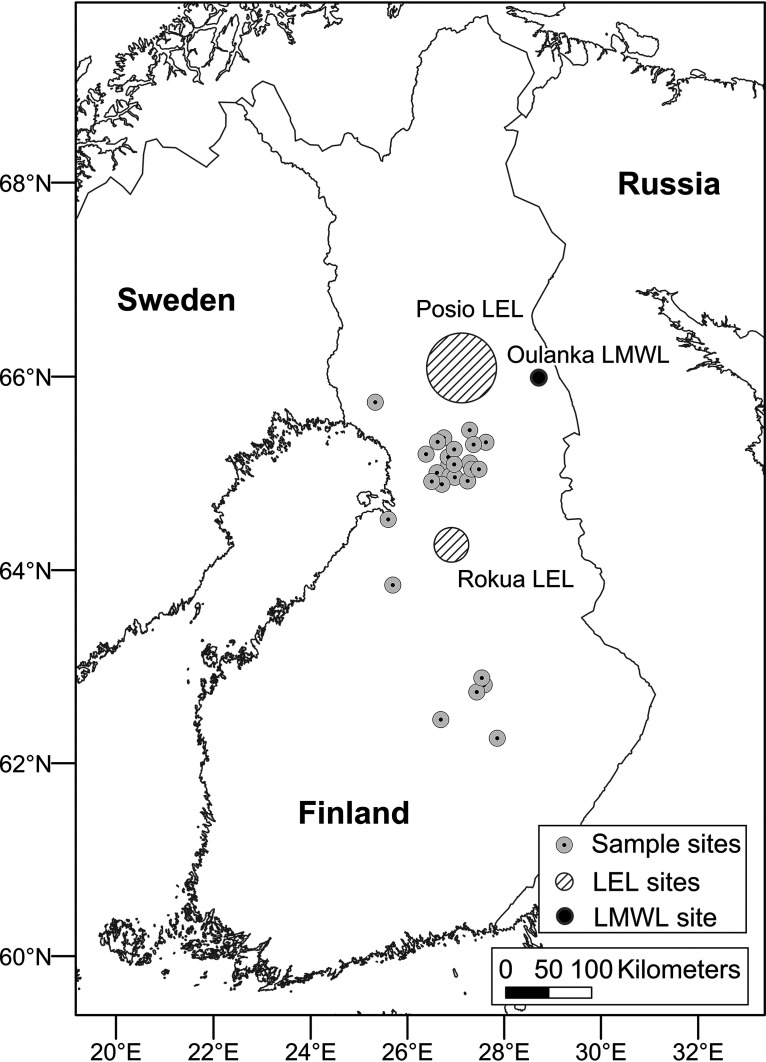
Water sampling for this study was carried out during October/November 2018 at 28 shallow groundwater wells used as sources of potable water in the North Ostrobothnia, North Savo, and Lapland regions of Finland (between 66°15′ and 62°15′N, and 24°30′ and 28°30′E) (Fig. 7). The studied sites are virtually all in rural locations and have mostly flat surrounding topographies, estimated annual precipitation of 600 to 700 mm, and shallow groundwater tables (about 3 m below the land surface, on average). Twenty of the wells (wells 3 to 22) are in a single, sparsely populated municipality in North Ostrobothnia (between 65°45′ and 65°0′N, and 26°15′ and 27°45′E). Wells 1 and 2 are in other municipalities of North Ostrobothnia. Well 23 is in Lapland, and wells 24 to 28 are in North Savo. Relevant characteristics of all 28 shallow groundwater wells are shown in Table 3. Raw water samples, before any treatment processes, were collected from each well aseptically from a sampling tap into sample containers. In the case of wells 11, 13, and 21, the raw water sample was alkalized due to presence of alkalization material in the well. Prior to stable water isotope analysis, the samples were stored at 4°C. At each well, a large volume of groundwater (200 liters) was filtered by a dead-end ultrafiltration method (DEUF; ASAHI Rexeed-25A, Asahi Kasei Medical Co., Ltd., Tokyo, Japan) as described earlier by reference 77 to concentrate the otherwise highly diluted microbes for further analysis. The flow rate during DEUF sampling was adjusted to around 1 liter/18 s (3.33 liter/min), which enabled 200 liters of water to be filtered in about 1 h. Sample containers and DEUF capsules were transported in cool boxes immediately (within 24 h after sample collection) to the laboratories for physicochemical and microbiological analysis.
FIG 7.
Map of the well locations and the sampling sites and regions for additional stable water isotope samples for rain (black point; Oulanka LMWL, reference 72) and surface water evaporation (lined areas; Rokua LEL, reference 73, and Posio LEL, reference 99). LMWL, local meteoric water line; LEL, local evaporation line.
TABLE 3.
Characteristics of the 28 shallow groundwater wellsa
| Well no. | Treatment status | Users | Water intake (m3/day) | Year changes to well structure last made | Well type | Well depth (m) | GW depth near well (m) | Potential nearby risk factors (within 1 km2) |
|---|---|---|---|---|---|---|---|---|
| 1 | UV, ALK, CH | 7,000b | NA | 1993 | Tube | ≥8 | 1.5 | A, SW, R, RA, R, S, C |
| 2 | UV, ALK | 6,400b | 250 | 1978 | Dug | 6 | 3 | A, SL |
| 3 | None | 190 | 32.9 | 1984 | Dug | 6 | 2.5 | M |
| 4 | None | 10 | 1 | 1992 | Dug | ∼3 | 1.5 | SG, M |
| 5 | ALK | 40 | 24 | 1974 | Dug | 3 | 1.5 | A |
| 6 | None | 150 | 16 | 1986 | Tube | 8 | 4 | D |
| 7 | None | <50 | <5.5 | NA | NA | NA | NA | M, SW, R |
| 8 | None | <100 | 13.7 | 1980s | Dug | 6 | 2 | M, A, SGP |
| 9 | None | 100–200 | 11 | 1979 | Dug | 5 | 2 | R, MW, SW |
| 10 | None | 105 | 8.9 | 1984 | Dug | 5.5 | 2 | M, D, P |
| 11 | ALKc | 100 | <11 | 1987 | Dug | 6 | 4.2 | B, R, SG |
| 12 | None | ∼170 | 12 | 1984 | Dug | 7 | 2 | SG |
| 13 | ALKc | ∼280 | 11 | 1979 | Dug | 5 | 2.5 | M, P |
| 14 | None | 50 | 8.3 | 1979 | Dug | NA | 2 | M, D |
| 15 | None | 150 | 125 | 1983 | Dug | 7 | 3 | SW, B, SR |
| 16 | ALK | 150 | 65 | 1983 | Tube | 11 | 3 | WW |
| 17 | None | 50–60 | 16.4 | 1984 | Dug | 4 | 1 | SG, SW |
| 18 | None | 155 | 71.2 | 1983 | Dug | 6 | 3.5 | R, SW, SG |
| 19 | UV, ALK | 4,200b | 650 | 1961 | Dug | 7.5 | 3 | SG, SA, S, R, SW |
| 20 | None | 80 | 27.4 | 1984 | Dug | 4 | ∼2 | M, D |
| 21 | ALKc | <50 | 10 | 1980s | Dug | 3.5 | 1 | SW, SG |
| 22 | None | 28 | 5 | 1989 | Tube | 7 | >0.5 (artesian spring) | M |
| 23 | UV, ALK | 20,000b | NA | 2007 | Tube | 7 | 4 | M, D, SW |
| 24 | UV, ALK | 1,000 | 170 | 2014 | Tube | 9.2 | 2 | P, SG |
| 25 | ALK | >200 | 38 | 1990 | Tube | 7.5 | 3 | A, SW, SG, SL, C |
| 26 | UV, ALK | 20,000b | 600–1,000 | 1969 | Dug | 9 | 5–10 | B, R, SW, T |
| 27 | ALK | 500 | 120 | 1987 | Tube | NA | NA | A, SL |
| 28 | ALK | 2,000 | 400 | 1988 | Tube | NA | NA | R, A, SL, SG |
aALK, alkalization; UV, UV disinfection; CH, chemical purification; GW depth, groundwater depth; R, roads; SW, surface water; M, marsh; D, ditches; A, agriculture; B, buildings; P, peat production; RA, recreational area; C, cemetery; S, school; SL, slurry storage tank; SG, sand or gravel pit; MW, meltwater; WW, wastewater; SR, ski resort; SA, swimming area; T, town; NA, not applicable.
Water served from several wells to the same network.
Raw water samples have been alkalized.
Physicochemical water quality analyses.
Temperature, pH, dissolved oxygen (DO), redox potential, and electrical conductivity (EC) of groundwater samples were measured on site using portable field meters, according to the manufacturer’s instructions (SenTix 940, FDO 925, SenTix ORP 900, TetraCon 325; WTW, Weilheim, Germany). Field redox potential values were converted to standard redox potential values by temperature-based adjustment. Analyses of total nitrogen (Ntot), ammonium nitrogen (NH4+-N), nitrite nitrogen (NO2−-N), phosphorus (P), combined nitrate and nitrite nitrogen (NO3−+NO2−)-N, chloride (Cl−), calcium (Ca), potassium (K), magnesium (Mg), sodium (Na), and silica (SiO2) were performed in an accredited commercial laboratory according to international standards for chemical water quality. Iron (Fe) and sulfate (SO42−) concentrations were determined colorimetrically via the phenanthroline method and the barium gelatin method, respectively (97, 98). Total carbon (TC) values were determined using a Sievers 900 portable TOC analyzer. Turbidity (EN 27027:1994) was determined using a Hach Ratio XR turbidity meter. UV absorbance values of unfiltered and 0.45-μm-filtered water samples were determined at 254 nm (UV254) using a UV-1800 spectrophotometer (Shimadzu, Japan) according to the manufacturer’s instructions.
Stable water isotope analyses.
To identify anomalies which may indicate surface water intrusion into the groundwater wells, stable water isotope (δ18O, δ2H) analyses were conducted on untreated groundwater samples from each of the wells and on water samples taken from nearby surface water sources, such as rivers and lakes. (These well and surface water samples were collected in 15-ml high-density polyethylene tubes, which were rinsed with the sampled water before filling.) The isotope ratios 2H/1H and 18O/16O were determined using cavity ring-down spectroscopy with a Picarro L2130-i analyzer. All isotope ratios are expressed in δ notation relative to Vienna Standard Mean Ocean Water 2 (VSMOW2) with precision for δ18O and δ2H values of δ0.025‰ and δ0.1‰, respectively. The stable water isotope samples were compared to regional results for rainwater and surface water signals. A local meteoric water line (LMWL) based on data collected from the Oulanka region was used as the rainwater reference (72). For the surface water local evaporation line (LEL), the references were from the Rokua region (73) and from the Posio municipality, collected in a parallel project by the Geological Survey of Finland in 2018 (99). Surface water is prone to evaporation, which can cause deuterium isotope values to differentiate from oxygen isotope values. This deuterium excess (d-excess = δ2H − 8 δ18O) (100) was determined for the well water samples in order to study the effect of evaporative fractionation on the samples potentially resulting from surface water intrusion.
DEUF capsule elution and coliphage analyses.
In the laboratory, DEUF capsules were eluted as described earlier (77) except that the secondary concentration of DEUF eluates of 35 to 250 ml was performed by filtration through 0.22-μm Millipore Express PLUS membrane filters (Merck KGaA, Darmstadt, Germany). Polyethylene glycol (PEG) precipitation of filtrate (200 to 500 ml) after Millipore Express PLUS membrane filtration was performed as described earlier (101), and analyses of somatic coliphages and F-specific coliphages were performed immediately from PEG precipitates using a double agar layer (DAL) procedure (USEPA Method 1601; with excess precipitate being stored at −75°C or lower). Millipore Express PLUS membranes were stored at −75°C or lower prior to nucleic acid extraction.
Enumeration of microbial indicators.
Escherichia coli and coliform bacteria were analyzed from untreated groundwater samples according to standard methods using membrane filtration with LES Endo agar medium and Chromocult coliform agar medium (SFS 3016 and ISO 9308-1). Spores of sulfite-reducing clostridia were enumerated from water samples after heat treatment of membranes for 15 min at 75°C and incubation for 2 days on tryptose sulfite cycloserine (TSC) agar (ISO 6461-2). Heterotrophic bacteria were enumerated from water samples by the spread-plate technique on Reasoner’s 2 agar (R2A) medium (Difco, USA) and incubated at 22 ± 2°C for 7 days (102, 103).
Analysis of host-specific MST markers and high-throughput 16S rRNA gene amplicon sequencing.
Total nucleic acids were extracted from DEUF concentrates on membrane filters as described previously in reference 104 using Chemagic DNA plant kit (Perkin Elmer, Waltham, MA, USA). Total RNA was further purified using Ambion Turbo DNA-free DNase kit (Life Technologies, Carlsbad, CA, USA). cDNA was synthesized using Invitrogen Superscript IV VILO system (Thermo Fisher Scientific, Waltham, MA, USA) and used in the 16S rRNA analysis. The total RNA was stored at −75°C or lower, while the cDNA and the DNA extracts were stored at −20°C until use. The gene copy numbers of general fecal indicator Bacteroidales bacteria (GenBac3), human-specific fecal indicator Bacteroides bacteria (HF183), and Gram-negative bacteria (105) in the samples (including extraction and filtration blanks) were measured from cDNA and DNA extracts as described previously (106). The qPCR assays were performed as described previously (107), by processing 16 μl of RNA in a cDNA synthesis (reverse transcription [RT]). The qPCRs were performed using a QuantStudio 6 real-time PCR system (Applied Biosystems) in 20 μl volume using the TaqMan Environmental PCR master mix (Life Technologies), all with primers and probes at final concentrations 0.2 μM (IDT Technologies, Inc.). The cycling conditions included 95°C for 10 min of enzyme activation and predenaturation followed by 40 cycles at 95°C for 15s of denaturation and at 60°C for 60s of annealing. Standard curves were generated using artificial gene fragments (gBlocks, IDT Technologies, Inc.) containing the sequences for each of the targeted genes. In qPCR, undiluted and 10-fold diluted cDNA and DNA samples were used as qPCR templates to detect PCR polymerase inhibition. For samples in which PCR inhibition was detected, qPCR data were generated using the results from diluted samples. Background signals, if detected in the filtration blanks, were subtracted from all the results to generate the final qPCR and RT-qPCR data per assay. The limit of detection (LOD) was set as 3 copies per reaction. The final qPCR, equivalent LOD (eLOD), and equivalent limit of quantification (eLOQ) values were calculated after taking into account the volume/mass of the processed sample, factors associated with the different processing steps of the RNA and DNA manipulations, and the dilutions used for each sample analyzed.
Subsamples of the nucleic acids were sent to Macrogen Inc. (Seoul, South Korea) for amplicon generation and subsequent sequencing. Bacterial 16S rRNA genes were amplified from DNA (targeting all bacteria present) and cDNA (traditionally considered to target only metabolically active bacteria) using the primers Bakt_341F (5′-CCTACGGGNGGCWGCAG-3′) and Bakt_805R (5′-GACTACHVGGGTATCTAATCC-3′), which target the V3–V4 variable region of the 16S rRNA gene (108). Amplicons were sequenced as 300 bp pair-end reads using the Illumina MiSeq platform. Some samples were sequenced in duplicate to check for reproducibility. Negative controls from different sample processing steps were included in the qPCR and high-throughput amplicon sequencing analysis (tube control for sampling/elution, elution solution, membrane filtration, nucleic acid extraction).
Sequencing data preprocessing and taxonomic classification.
The 16S rRNA amplicon data for DNA and cDNA libraries were processed and analyzed via the QIIME 2 pipeline (version 2018.11) (109). The DADA2 denoise-paired QIIME 2 plugin was used, with the parameters --p-trim-left-f 9, --p-trim-left-r 9, --p-trunc-len-f 290, and --p-trunc-len-r 250, to trim sequences (to remove bad quality reads with quality score of <20) and to denoise and merge trimmed reads to produce a table of amplicon sequence variants (ASVs) (110). The ASV table was rarefied to a sampling depth of 2,504, which excluded four samples with sequence counts below this threshold (three of these were controls, and one was a duplicate DNA sample). Remaining duplicates were checked for consistency and merged. Taxonomic classification of the ASVs was performed via the q2-feature-classifier plugin in QIIME 2 (111) using a naive Bayes classifier trained on the V3–V4 variable region of representative 16S rRNA sequences. These representative 16S rRNA sequences were derived by clustering 16S rRNA sequences from the SILVA rRNA database (release 132) into operational taxonomic units (OTUs) based on 99% sequence identity (112). The default confidence cutoff of 70% was used in assigning taxonomic labels, as this is designed to balance precision and recall in classifying 16S rRNA sequences (111). Nontarget sequences such as archaeal, mitochondrial, and chloroplastic sequences were filtered out, so that only bacterial sequences remained.
Assessment of bacterial diversity and bacterial indicator analysis.
Taxonomic classifications of bacterial DNA- and cDNA-based communities were screened for taxa relevant to water safety, e.g., species such as Escherichia coli, Clostridium perfringens, and Pseudomonas aeruginosa and genera such as Klebsiella, Aeromonas, Arcobacter, Enterococcus, Legionella, Mycobacterium, Yersinia, and Listeria. Alpha diversity metrics for bacterial DNA- and cDNA-based communities, namely, Faith’s phylogenetic diversity, Pielou’s evenness, observed ASVs, and Shannon diversity, were calculated based on the rarefied ASV table using the QIIME2 diversity plugin. Relative abundance values for the bacterial DNA- and cDNA-based communities were calculated in R, and a heatmap showing relative abundance values of selected taxa was generated using the pheatmap function from the pheatmap R package (113).
Correlation analysis.
Environmental land use data were extracted from maps produced by the National Land Survey of Finland (Maanmittauslaitos) using their online map-viewing tool, MapSite. For each of the wells, measurements were made of distances to, and lengths or areas of, nearby roads, fields, marshes, surface water sources, and buildings. “Nearby” land use was defined as land use occurring within 1 km2 of the well. In addition to the map-based analyses, site inspections were conducted during sampling visits to examine the immediate surroundings for potential risk factors, and well operators were probed for information on nearby land use and details of any past or suspected problems (e.g., high turbidity, surface water intrusion, fecal indicators). Correlations were sought between physicochemical data, microbial indicator data, environmental land use data, and bacterial alpha diversity data via calculation of Spearman rank-based correlation coefficients using the rcorr function from the Hmisc R package (114). Spearman was used here because Shapiro-Wilk tests carried out in R showed that many parameters were not normally distributed. Correlograms were produced using the corrplot function from the corrplot R package (115). Only statistically significant Spearman correlation coefficients (P < 0.05) are reported here, with the following ranges being used for discussion of correlations: very strong (>0.8 or <−0.8), strong (between 0.6 and 0.8, or between −0.6 and −0.8), moderate (between 0.4 and 0.6, or between −0.4 and −0.6), weak (between 0.3 and 0.4).
Nonmetric multidimensional scaling.
Nonmetric multidimensional scaling (NMDS) plots using weighted UniFrac distance matrices (calculated from the rarefied ASV table in QIIME 2) were created in R using the metaMDS function in the vegan package (116). Environmental variables (e.g., turbidity, dissolved oxygen, nitrate, ammonium) were fitted to the NMDS plots using the envfit function. Only environmental variables that were significantly correlated with community composition (as identified in envfit with a P value of ≤0.05) were considered for the figures.
Data availability.
The 16S rRNA amplicon sequencing data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under primary accession number PRJEB41020 (https://www.ebi.ac.uk/ena/browser/view/PRJEB41020).
ACKNOWLEDGMENTS
We acknowledge the well operators for their cooperation and participation in the study, Tuomo Reinikka, Jussi Keränen, Elisangela Heiderscheidt, Tiina Heiskanen, and Tarja Rahkonen for assistance with field and laboratory work, and the CSC: IT Centre for Science (CSC: tietotekniikan keskus) for computational resources. Support for this study was provided by the Finnish Ministry of Agriculture and Forestry (Maa- ja metsätalousministeriö), the Land and Water Technology Support Association (Maa- ja vesitekniikan tuki ry), the Academy of Finland, and the KAUTE and Erkki Paasikivi Foundations (Kaupallisten ja teknillisten tieteiden tukisäätiö KAUTE ja Erkki Paasikiven säätiö).
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Katharina Kujala, Email: katharina.kujala@oulu.fi.
Vincent J. Denef, University of Michigan-Ann Arbor
REFERENCES
- 1.Foster S, Chilton J, Moench M, Cardy F, Schiffler M. 2008. Groundwater in rural development: facing the challenges of supply and resource sustainability. The World Bank, Washington, DC. [Google Scholar]
- 2.Morris BL, Lawrence ARL, Chilton PJC, Adams B, Calow RC, Klinck BA. 2003. Groundwater and its susceptibility to degradation: a global assessment of the problem and options for management. division of early warning and assessment, United Nations Environment Program, Nairobi, Kenya. [Google Scholar]
- 3.UN World Water Assessment Program (WWAP). 2003. The United Nations world water development report: water for people, water for life (executive summary). UNESCO Publishing, Paris, France. [Google Scholar]
- 4.Giordano M. 2009. Global groundwater? Issues and solutions. Annu Rev Environ Resour 34:153–178. doi: 10.1146/annurev.environ.030308.100251. [DOI] [Google Scholar]
- 5.Fan Y, Li H, Miguez-Macho G. 2013. Global patterns of groundwater table depth. Science 339:940–943. doi: 10.1126/science.1229881. [DOI] [PubMed] [Google Scholar]
- 6.Close M, Dann R, Ball A, Pirie R, Savill M, Smith Z. 2008. Microbial groundwater quality and its health implications for a border-strip irrigated dairy farm catchment, South Island, New Zealand. J Water Health 6:83–98. doi: 10.2166/wh.2007.020. [DOI] [PubMed] [Google Scholar]
- 7.Hynds PD, Thomas MK, Pintar KDM. 2014. Contamination of groundwater systems in the US and Canada by enteric pathogens, 1990–2013: a review and pooled-analysis. PLoS One 9:e93301. doi: 10.1371/journal.pone.0093301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nadeau S, Rosa E, Cloutier V, Daigneault R-A, Veillette J. 2015. A GIS-based approach for supporting groundwater protection in eskers: application to sand and gravel extraction activities in Abitibi-Témiscamingue, Quebec, Canada. J Hydrol Reg Stud 4:535–549. doi: 10.1016/j.ejrh.2015.05.015. [DOI] [Google Scholar]
- 9.Flanagan SM, Montgomery DL, Ayotte JD. 2001. Shallow ground-water quality in the Boston, Massachusetts metropolitan area. USGS water-resources investigations report 2001–4042. United States Geological Survey (USGS). [Google Scholar]
- 10.Röling WFM, van Breukelen BM, Braster M, Lin B, van Verseveld HW. 2001. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl Environ Microbiol 67:4619–4629. doi: 10.1128/AEM.67.10.4619-4629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard G, Pedley S, Barrett M, Nalubega M, Johal K. 2003. Risk factors contributing to microbiological contamination of shallow groundwater in Kampala, Uganda. Water Res 37:3421–3429. doi: 10.1016/S0043-1354(03)00235-5. [DOI] [PubMed] [Google Scholar]
- 12.Tiquia SM, Schleibak M, Schlaff J, Floyd C, Benipal B, Zakhem E, Murray KS. 2008. Microbial community profiling and characterization of some heterotrophic bacterial isolates from river waters and shallow groundwater wells along the Rouge River, Southeast Michigan. Environ Technol 29:651–663. doi: 10.1080/09593330801986998. [DOI] [PubMed] [Google Scholar]
- 13.Pitkänen T, Karinen P, Miettinen IT, Lettojärvi H, Heikkilä A, Maunula R, Aula V, Kuronen H, Vepsäläinen A, Nousiainen L-L, Pelkonen S, Heinonen-Tanski H. 2011. Microbial contamination of groundwater at small community water supplies in Finland. Ambio 40:377–390. doi: 10.1007/s13280-010-0102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulsmann A. 2005. Small systems large problems: a European inventory of small water systems and associated problems. WeKnow (Web-based European Knowledge Network on Water)/Endware Report. [Google Scholar]
- 15.Daniels B, Weinthal E, Hudson B. 2008. Is an exemption from US groundwater regulations a loophole or a noose? Policy Sci 41:205–220. doi: 10.1007/s11077-008-9064-0. [DOI] [Google Scholar]
- 16.Hulsmann A, Smeets P. 2011. Towards a guidance document for the implementation of a risk assessment for small water supplies in the European Union: overview of best practices. KWR Watercycle Research Institute Report, Nieuwegein, Netherlands. [Google Scholar]
- 17.Hendry S, Akoumianaki I. 2016. Governance and management of small rural water supplies: a comparative study. CREW - Scotland’s Centre of Expertise for Waters, Aberdeen, Scotland, UK. [Google Scholar]
- 18.Gunnarsdottir MJ, Persson KM, Andradottir HO, Gardarsson SM. 2017. Status of small water supplies in the Nordic countries: characteristics, water quality and challenges. Int J Hyg Environ Health 220:1309–1317. doi: 10.1016/j.ijheh.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Gunnarsdottir MJ, Gardarsson SM, Schultz AC, Albrechtsen H-J, Hansen LT, Gerlach Bergkvist KS, Rossi PM, Klöve B, Myrmel M, Persson KM, Eriksson M, Bartram J. 2020. Status of risk-based approach and national framework for safe drinking water in small water supplies of the Nordic water sector. Int J Hyg Environ Health 230:113627. doi: 10.1016/j.ijheh.2020.113627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McFarlane K, Harris LM. 2018. Small systems, big challenges: review of small drinking water system governance. Environ Rev 26:378–395. doi: 10.1139/er-2018-0033. [DOI] [Google Scholar]
- 21.Hatva T. 1994. Effect of gravel extraction on groundwater, p 427–434. In Future groundwater resources at risk. International Association of Hydrological Sciences (IAHS), IAHS Helsinki Conference, June 1994, Helsinki, Finland. [Google Scholar]
- 22.Vadas PA, Srinivasan MS. 2007. Hydrology and groundwater nutrient concentrations in a ditch-drained agroecosystem. J Soil Water Conserv 62:178–188. [Google Scholar]
- 23.Salminen JM, Nystén TH, Tuominen SM. 2011. Review of approaches to reducing adverse impacts of road deicing on groundwater in Finland. Water Qual Res J 46:166–173. doi: 10.2166/wqrjc.2011.002. [DOI] [Google Scholar]
- 24.Smerdon BD, Mendoza CA, Devito KJ. 2012. The impact of gravel extraction on groundwater dependent wetlands and lakes in the Boreal Plains, Canada. Environ Earth Sci 67:1249–1259. doi: 10.1007/s12665-012-1568-4. [DOI] [Google Scholar]
- 25.Sworobuk JE, Law CB, Bissonnette GK. 1987. Assessment of the bacteriological quality of rural groundwater supplies in Northern West Virginia. Water Air Soil Pollut 36:163–170. doi: 10.1007/BF00450627. [DOI] [Google Scholar]
- 26.McKeon DM, Calabrese JP, Bissonnette GK. 1995. Antibiotic resistant gram-negative bacteria in rural groundwater supplies. Water Res 29:1902–1908. doi: 10.1016/0043-1354(95)00013-B. [DOI] [Google Scholar]
- 27.Cho J-C, Kim S-J. 2000. Increase in bacterial community diversity in subsurface aquifers receiving livestock wastewater input. Appl Environ Microbiol 66:956–965. doi: 10.1128/AEM.66.3.956-965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valenzuela M, Lagos B, Claret M, Mondaca MA, Pérez C, Parra O. 2009. Fecal contamination of groundwater in a small rural dryland watershed in central Chile. Chilean J Agric Res 69:235–243. [Google Scholar]
- 29.Cool G, Rodriguez MJ, Bouchard C, Levallois P, Joerin F. 2010. Evaluation of the vulnerability to contamination of drinking water systems for rural regions in Québec, Canada. J Environ Plan Manag 53:615–638. doi: 10.1080/09640561003727128. [DOI] [Google Scholar]
- 30.Pitkänen T, Juselius T, Isomäki E, Miettinen I, Valve M, Kivimäki A-L, Lahti K, Hänninen M-L. 2015. Drinking water quality and occurrence of Giardia in Finnish small groundwater supplies. Resources 4:637–654. doi: 10.3390/resources4030637. [DOI] [Google Scholar]
- 31.Ferguson AS, Layton AC, Mailloux BJ, Culligan PJ, Williams DE, Smartt AE, Sayler GS, Feighery J, McKay LD, Knappett PSK, Alexandrova E, Arbit T, Emch M, Escamilla V, Ahmed KM, Alam MJ, Streatfield PK, Yunus M, van Geen A. 2012. Comparison of fecal indicators with pathogenic bacteria and rotavirus in groundwater. Sci Total Environ 431:314–322. doi: 10.1016/j.scitotenv.2012.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gunnarsdottir MJ, Gardarsson SM, Andradottir HO. 2013. Microbial contamination in groundwater supply in a cold climate and coarse soil: case study of norovirus outbreak at Lake Mývatn, Iceland. Hydrol Res 44:1114–1128. doi: 10.2166/nh.2013.076. [DOI] [Google Scholar]
- 33.Wallender EK, Ailes EC, Yoder JS, Roberts VA, Brunkard JM. 2014. Contributing factors to disease outbreaks associated with untreated groundwater. Ground Water 52:886–897. doi: 10.1111/gwat.12121. [DOI] [PubMed] [Google Scholar]
- 34.Moreira NA, Bondelind M. 2017. Safe drinking water and waterborne outbreaks. J Water Health 15:83–96. doi: 10.2166/wh.2016.103. [DOI] [PubMed] [Google Scholar]
- 35.Kauppinen A, Pitkänen T, Miettinen IT. 2018. Persistent norovirus contamination of groundwater supplies in two waterborne outbreaks. Food Environ Virol 10:39–50. doi: 10.1007/s12560-017-9320-6. [DOI] [PubMed] [Google Scholar]
- 36.Guzman-Herrador B, Carlander A, Ethelberg S, Freiesleben de Blasio B, Kuusi M, Lund V, Löfdahl M, MacDonald E, Nichols G, Schönning C, Sudre B, Trönnberg L, Vold L, Semenza J, Nygård K. 2015. Waterborne outbreaks in the Nordic countries, 1998 to 2012. Eurosurveillance 20:21160. doi: 10.2807/1560-7917.ES2015.20.24.21160. [DOI] [PubMed] [Google Scholar]
- 37.Murphy HM, Prioleau MD, Borchardt MA, Hynds PD. 2017. Review: epidemiological evidence of groundwater contribution to global enteric disease, 1948–2015. Hydrogeol J 25:981–1001. doi: 10.1007/s10040-017-1543-y. [DOI] [Google Scholar]
- 38.Hrudey SE, Hrudey EJ. 2007. Published case studies of waterborne disease outbreaks: evidence of a recurrent threat. Water Environ Res 79:233–245. doi: 10.2175/106143006x95483. [DOI] [PubMed] [Google Scholar]
- 39.Kløve B, Kvitsand HML, Pitkänen T, Gunnarsdottir MJ, Gaut S, Gardarsson SM, Rossi PM, Miettinen I. 2017. Overview of groundwater sources and water-supply systems, and associated microbial pollution, in Finland, Norway and Iceland. Hydrogeol J 25:1033–1044. doi: 10.1007/s10040-017-1552-x. [DOI] [Google Scholar]
- 40.Moffatt H, Struck S. 2011. Water-borne disease outbreaks in Canadian small drinking water systems. National Collaborating Centres for Public Health, Canada. [Google Scholar]
- 41.Craun GF, Brunkard JM, Yoder JS, Roberts VA, Carpenter J, Wade T, Calderon RL, Roberts JM, Beach MJ, Roy SL. 2010. Causes of outbreaks associated with drinking water in the United States from 1971 to 2006. Clin Microbiol Rev 23:507–528. doi: 10.1128/CMR.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pons W, Young I, Truong J, Jones-Bitton A, McEwen S, Pintar K, Papadopoulos A. 2015. A systematic review of waterborne disease outbreaks associated with small non-community drinking water systems in Canada and the United States. PLoS One 10:e0141646. doi: 10.1371/journal.pone.0141646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.WHO. 2011. Guidelines for drinking-water quality, 4th ed. World Health Organization, Geneva. [Google Scholar]
- 44.Lahti K, Hiisvirta L. 1995. Causes of waterborne outbreaks in community water systems in Finland: 1980–1992. Wat Sci Tech 31:33–36. doi: 10.2166/wst.1995.0552. [DOI] [Google Scholar]
- 45.Isomäki E. 2006. Pienet pohjavesilaitokset Suomessa. Vesitalous 47:11–15. [Google Scholar]
- 46.Isomäki E, Valve M, Kivimäki A-L. 2006. Small waterworks in Finland, p 91–95. In 5th Nordic drinking water conference, Reykjavík, Iceland. [Google Scholar]
- 47.Miettinen IT, Zacheus O, von Bonsdorff C-H, Vartiainen T. 2001. Waterborne epidemics in Finland in 1998–1999. Water Sci Technol 43:67–71. doi: 10.2166/wst.2001.0713. [DOI] [PubMed] [Google Scholar]
- 48.Hynds PD, Misstear BD, Gill LW. 2012. Development of a microbial contamination susceptibility model for private domestic groundwater sources. Water Resour Res 48:W12504. [Google Scholar]
- 49.Powell KL, Cronin AA, Pedley S, Barrett MH. 2001. Microbiological quality of groundwater in UK urban aquifers: do we know enough?, p 91–96. In Groundwater quality: natural and enhanced restoration of groundwater pollution. International Association of Hydrological Sciences (IAHS), Groundwater Quality 2001 Conference, Sheffield, UK. [Google Scholar]
- 50.Llopis-González A, Sánchez A, Requena P, Suárez-Varela M. 2014. Assessment of the microbiological quality of groundwater in three regions of the Valencian Community (Spain). Int J Environ Res Public Health 11:5527–5540. doi: 10.3390/ijerph110505527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keesari T, Ramakumar KL, Prasad MBK, Chidambaram S, Perumal P, Prakash D, Nawani N. 2015. Microbial evaluation of groundwater and its implications on redox condition of a multi-layer sedimentary aquifer system. Environ Process 2:331–346. doi: 10.1007/s40710-015-0067-5. [DOI] [Google Scholar]
- 52.Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, Thomas BC, Banfield JF. 2013. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. mBio 4:e00708-13. doi: 10.1128/mBio.00708-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523:208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 54.Hug LA, Thomas BC, Brown CT, Frischkorn KR, Williams KH, Tringe SG, Banfield JF. 2015. Aquifer environment selects for microbial species cohorts in sediment and groundwater. ISME J 9:1846–1856. doi: 10.1038/ismej.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hug LA, Thomas BC, Sharon I, Brown CT, Sharma R, Hettich RL, Wilkins MJ, Williams KH, Singh A, Banfield JF. 2016. Critical biogeochemical functions in the subsurface are associated with bacteria from new phyla and little studied lineages. Environ Microbiol 18:159–173. doi: 10.1111/1462-2920.12930. [DOI] [PubMed] [Google Scholar]
- 56.Luef B, Frischkorn KR, Wrighton KC, Holman H-YN, Birarda G, Thomas BC, Singh A, Williams KH, Siegerist CE, Tringe SG, Downing KH, Comolli LR, Banfield JF. 2015. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun 6:6372. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 57.Long PE, Williams KH, Hubbard SS, Banfield JF. 2016. Microbial metagenomics reveals climate-relevant subsurface biogeochemical processes. Trends Microbiol 24:600–610. doi: 10.1016/j.tim.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 58.Bruno A, Sandionigi A, Rizzi E, Bernasconi M, Vicario S, Galimberti A, Cocuzza C, Labra M, Casiraghi M. 2017. Exploring the under-investigated “microbial dark matter” of drinking water treatment plants. Sci Rep 7:44350. doi: 10.1038/srep44350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar S, Herrmann M, Thamdrup B, Schwab VF, Geesink P, Trumbore SE, Totsche K-U, Küsel K. 2017. Nitrogen loss from pristine carbonate-rock aquifers of the Hainich Critical Zone Exploratory (Germany) is primarily driven by chemolithoautotrophic anammox processes. Front Microbiol 8:1951. doi: 10.3389/fmicb.2017.01951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mauffret A, Baran N, Joulian C. 2017. Effect of pesticides and metabolites on groundwater bacterial community. Sci Total Environ 576:879–887. doi: 10.1016/j.scitotenv.2016.10.108. [DOI] [PubMed] [Google Scholar]
- 61.Schwab VF, Herrmann M, Roth V-N, Gleixner G, Lehmann R, Pohnert G, Trumbore S, Küsel K, Totsche KU. 2017. Functional diversity of microbial communities in pristine aquifers inferred by PLFA- and sequencing-based approaches. Biogeosciences 14:2697–2714. doi: 10.5194/bg-14-2697-2017. [DOI] [Google Scholar]
- 62.Geesink P, Tyc O, Küsel K, Taubert M, van de Velde C, Kumar S, Garbeva P. 2018. Growth promotion and inhibition induced by interactions of groundwater bacteria. FEMS Microbiol Ecol 94:fiy164. [DOI] [PubMed] [Google Scholar]
- 63.Lazar CS, Lehmann R, Stoll W, Rosenberger J, Totsche KU, Küsel K. 2019. The endolithic bacterial diversity of shallow bedrock ecosystems. Sci Total Environ 679:35–44. doi: 10.1016/j.scitotenv.2019.04.281. [DOI] [PubMed] [Google Scholar]
- 64.Herrmann M, Geesink P, Yan L, Lehmann R, Totsche KU, Küsel K. 2020. Complex food webs coincide with high genetic potential for chemolithoautotrophy in fractured bedrock groundwater. Water Res 170:115306. doi: 10.1016/j.watres.2019.115306. [DOI] [PubMed] [Google Scholar]
- 65.Yan L, Herrmann M, Kampe B, Lehmann R, Totsche KU, Küsel K. 2020. Environmental selection shapes the formation of near-surface groundwater microbiomes. Water Res 170:115341. doi: 10.1016/j.watres.2019.115341. [DOI] [PubMed] [Google Scholar]
- 66.Fillinger L, Hug K, Griebler C. 2021. Aquifer recharge viewed through the lens of microbial community ecology: initial disturbance response, and impacts of species sorting versus mass effects on microbial community assembly in groundwater during riverbank filtration. Water Res 189:116631. doi: 10.1016/j.watres.2020.116631. [DOI] [PubMed] [Google Scholar]
- 67.Fältmarsch RM, Åström ME, Vuori K-M. 2008. Environmental risks of metals mobilised from acid sulphate soils in Finland: a literature review. Boreal Environ Res 13:444–456. [Google Scholar]
- 68.Domagalski JL, Johnson H. 2012. Phosphorus and groundwater: establishing links between agricultural use and transport to streams. U.S. Geological Survey Fact Sheet 2012–3004, United States Geological Survey (USGS). [Google Scholar]
- 69.Burkartaus D MR, Stoner JD. 2008. Nitrogen in groundwater associated with agricultural systems, p 177–202. In Nitrogen in the environment: sources, problems, and management. Hatfield JL, Follett RF (ed). Elsevier B.V., Amsterdam, Netherlands. [Google Scholar]
- 70.Piispanen R, Nykyri T. 1997. Acidification of groundwater in water-filled gravel pits – a new environmental and geomedical threat. Environ Geochem Health 19:111–126. doi: 10.1023/A:1018454622669. [DOI] [Google Scholar]
- 71.Porowski A, Porowska D, Halas S. 2019. Identification of sulfate sources and biogeochemical processes in an aquifer affected by peatland: insights from monitoring the isotopic composition of groundwater sulfate in Kampinos National Park, Poland. Water 11:1388. doi: 10.3390/w11071388. [DOI] [Google Scholar]
- 72.Rossi PM, Marttila H, Jyväsjärvi J, Ala-Aho P, Isokangas E, Muotka T, Kløve B. 2015. Environmental conditions of boreal springs explained by capture zone characteristics. J Hydrol 531:992–1002. doi: 10.1016/j.jhydrol.2015.11.009. [DOI] [Google Scholar]
- 73.Isokangas E, Rozanski K, Rossi PM, Ronkanen A-K, Kløve B. 2015. Quantifying groundwater dependence of a sub-polar lake cluster in Finland using an isotope mass balance approach. Hydrol Earth Syst Sci 19:1247–1262. doi: 10.5194/hess-19-1247-2015. [DOI] [Google Scholar]
- 74.Hunt RJ, Coplen TB, Haas NL, Saad DA, Borchardt MA. 2005. Investigating surface water–well interaction using stable isotope ratios of water. J Hydrol 302:154–172. doi: 10.1016/j.jhydrol.2004.07.010. [DOI] [Google Scholar]
- 75.Herrmann M, Wegner C-E, Taubert M, Geesink P, Lehmann K, Yan L, Lehmann R, Totsche KU, Küsel K. 2019. Predominance of Cand. Patescibacteria in groundwater is caused by their preferential mobilization from soils and flourishing under oligotrophic conditions. Front Microbiol 10:1407. doi: 10.3389/fmicb.2019.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gloor GB, Macklaim JM, Pawlowsky-Glahn V, Egozcue JJ. 2017. Microbiome datasets are compositional: and this is not optional. Front Microbiol 8:2224. doi: 10.3389/fmicb.2017.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Inkinen J, Jayaprakash B, Siponen S, Hokajärvi A-M, Pursiainen A, Ikonen J, Ryzhikov I, Täubel M, Kauppinen A, Paananen J, Miettinen IT, Torvinen E, Kolehmainen M, Pitkänen T. 2019. Active eukaryotes in drinking water distribution systems of ground and surface waterworks. Microbiome 7:99. doi: 10.1186/s40168-019-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. 2013. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J 7:2061–2068. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Filip A, Vuskovic B, Strundjalic P. 1974. Correlation between turbidity and iron content of the filter effluent of well origin. J Am Water Works Assoc 66:166–168. doi: 10.1002/j.1551-8833.1974.tb01995.x. [DOI] [Google Scholar]
- 80.DeZuane J. 1997. Handbook of drinking water quality, 2nd ed. John Wiley & Sons, Inc, New York, NY. [Google Scholar]
- 81.Hu Y, Wen Y, Wang X. 2016. Novel method of turbidity compensation for chemical oxygen demand measurements by using UV–vis spectrometry. Sens Actuators B Chem 227:393–398. doi: 10.1016/j.snb.2015.12.078. [DOI] [Google Scholar]
- 82.Li J, Tong Y, Guan L, Wu S, Li D. 2019. A turbidity compensation method for COD measurements by UV–vis spectroscopy. Optik 186:129–136. doi: 10.1016/j.ijleo.2019.04.096. [DOI] [Google Scholar]
- 83.Karanfil T, Erdogan I, Schlautman M. 2005. The impact of filtrate turbidity on UV254 and SUVA254 determinations. J Am Water Works Assoc 97:125–136. doi: 10.1002/j.1551-8833.2005.tb10893.x. [DOI] [Google Scholar]
- 84.Dobbs RA, Wise RH, Dean RB. 1972. The use of ultra-violet absorbance for monitoring the total organic carbon content of water and wastewater. Water Res 6:1173–1180. doi: 10.1016/0043-1354(72)90017-6. [DOI] [Google Scholar]
- 85.Reid J, Cresser M, Macleod D. 1980. Observations on the estimation of total organic carbon from u.v. absorbance for an unpolluted stream. Water Res 14:525–529. doi: 10.1016/0043-1354(80)90220-1. [DOI] [Google Scholar]
- 86.Peacock M, Evans CD, Fenner N, Freeman C, Gough R, Jones TG, Lebron I. 2014. UV-visible absorbance spectroscopy as a proxy for peatland dissolved organic carbon (DOC) quantity and quality: considerations on wavelength and absorbance degradation. Environ Sci Process Impacts 16:1445–1461. doi: 10.1039/c4em00108g. [DOI] [PubMed] [Google Scholar]
- 87.Ikonen J, Pitkänen T, Miettinen I. 2013. Suitability of optical, physical and chemical measurements for detection of changes in bacterial drinking water quality. Int J Environ Res Public Health 10:5349–5363. doi: 10.3390/ijerph10115349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Francy DS, Helsel DR, Nally RA. 2000. Occurrence and distribution of microbiological indicators in groundwater and stream water. Water Environ Res 72:152–161. doi: 10.2175/106143000X137220. [DOI] [Google Scholar]
- 89.Bettez ND, Marino R, Howarth RW, Davidson EA. 2013. Roads as nitrogen deposition hot spots. Biogeochemistry 114:149–163. doi: 10.1007/s10533-013-9847-z. [DOI] [Google Scholar]
- 90.Huhtanen P, Ahvenjärvi S, Heikkilä T. 2000. Effects of sodium sulphate and potassium chloride fertilizers on the nutritive value of timothy grown on different soils. AFSci 9:105–120. doi: 10.23986/afsci.5653. [DOI] [Google Scholar]
- 91.Chiy PC, Phillips CJ. 2002. Effects of sodium and sulphur fertilisers on the concentrations of nitrate and potentially toxic elements (PTEs) in water leached from permanent pasture. J Sci Food Agric 82:816–822. doi: 10.1002/jsfa.1113. [DOI] [Google Scholar]
- 92.Scherer HW. 2005. Fertilizers and fertilization, p 20–26. In Encyclopedia of Soils in the Environment. Elsevier Ltd, Amsterdam, Netherlands. [Google Scholar]
- 93.Griebler C, Lueders T. 2009. Microbial biodiversity in groundwater ecosystems. Freshw Biol 54:649–677. doi: 10.1111/j.1365-2427.2008.02013.x. [DOI] [Google Scholar]
- 94.Griebler C, Avramov M. 2015. Groundwater ecosystem services: a review. Freshw Sci 34:355–367. doi: 10.1086/679903. [DOI] [Google Scholar]
- 95.Pepper IL, Gerba CP, Gentry TJ. 2015. Environmental microbiology, 3rd ed. Elsevier Inc, New York, NY. [Google Scholar]
- 96.Hunter PR, Pond K, Jagals P, Cameron J. 2009. An assessment of the costs and benefits of interventions aimed at improving rural community water supplies in developed countries. Sci Total Environ 407:3681–3685. doi: 10.1016/j.scitotenv.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 97.Fortune WB, Mellon MG. 1938. Determination of iron with o-phenanthroline: a spectrophotometric study. Ind Eng Chem Anal Ed 10:60–64. doi: 10.1021/ac50118a004. [DOI] [Google Scholar]
- 98.Tabatabai MA. 1974. A rapid method for determination of sulfate in water samples. Environ Lett 7:237–243. doi: 10.1080/00139307409437403. [DOI] [Google Scholar]
- 99.Nora J, Rossi P, Sanaksenaho R, Lindholm A. 2019. Lapin POSKI2 – hankkeen erillisselvitys: Isotooppitutkimukset (POSKI2 project, Lapland – separate study on water isotopes). Geological Survey of Finland, Groundwater Unit, Rovaniemi, Finland. [Google Scholar]
- 100.Dansgaard W. 1964. Stable isotopes in precipitation. Tellus 16:436–468. doi: 10.3402/tellusa.v16i4.8993. [DOI] [Google Scholar]
- 101.Kauppinen A, Pitkänen T, Al-Hello H, Maunula L, Hokajärvi A-M, Rimhanen-Finne R, Miettinen IT. 2019. Two drinking water outbreaks caused by wastewater intrusion including sapovirus in Finland. Int J Environ Res Public Health 16:4376. doi: 10.3390/ijerph16224376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reasoner DJ, Geldreich EE. 1985. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Greenberg AE, Clesceri LS, Eaton ED. 1992. Standard methods for the examination of water and wastewater, 18th ed. American Public Health Association, Washington, DC. [Google Scholar]
- 104.Brester C, Ryzhikov I, Siponen S, Jayaprakash B, Ikonen J, Pitkänen T, Miettinen IT, Torvinen E, Kolehmainen M. 2020. Potential and limitations of a pilot-scale drinking water distribution system for bacterial community predictive modelling. Sci Total Environ 717:137249. doi: 10.1016/j.scitotenv.2020.137249. [DOI] [PubMed] [Google Scholar]
- 105.Kärkkäinen PM, Valkonen M, Hyvärinen A, Nevalainen A, Rintala H. 2010. Determination of bacterial load in house dust using qPCR, chemical markers and culture. J Environ Monit 12:759–768. doi: 10.1039/b917937b. [DOI] [PubMed] [Google Scholar]
- 106.Tiwari A, Kauppinen A, Pitkänen T. 2019. Decay of Enterococcus faecalis, Vibrio cholerae and MS2 coliphage in a laboratory mesocosm under brackish beach conditions. Front Public Health 7:269. doi: 10.3389/fpubh.2019.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pitkänen T, Ryu H, Elk M, Hokajärvi A-M, Siponen S, Vepsäläinen A, Räsänen P, Santo Domingo JW. 2013. Detection of fecal bacteria and source tracking identifiers in environmental waters using rRNA-based RT-qPCR and rDNA-based qPCR assays. Environ Sci Technol 47:13611–13620. doi: 10.1021/es403489b. [DOI] [PubMed] [Google Scholar]
- 108.Herlemann DP, Labrenz M, Jürgens K, Bertilsson S, Waniek JJ, Andersson AF. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5:1571–1579. doi: 10.1038/ismej.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Gregory Caporaso J. 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kolde R. 2019. pheatmap: pretty heatmaps. R package. https://CRAN.R-project.org/package=pheatmap.
- 114.Harrell FE. 2020. Hmisc: Harrell miscellaneous. R package. https://CRAN.R-project.org/package=Hmisc.
- 115.Wei T. 2017. corrplot: visualization of a correlation matrix. R package. https://github.com/taiyun/corrplot.
- 116.Oksanen J. 2020. vegan: community ecology package. R package. https://CRAN.R-project.org/package=vegan.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00179-21_Supp_1_seq14.pdf, PDF file, 0.1 MB (133.2KB, pdf)
Data Availability Statement
The 16S rRNA amplicon sequencing data for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under primary accession number PRJEB41020 (https://www.ebi.ac.uk/ena/browser/view/PRJEB41020).