ABSTRACT
Soil viruses are abundant, but the influence of the environment and climate on soil viruses remains poorly understood. Here, we addressed this gap by comparing the diversity, abundance, lifestyle, and metabolic potential of DNA viruses in three grassland soils with historical differences in average annual precipitation, low in eastern Washington (WA), high in Iowa (IA), and intermediate in Kansas (KS). Bioinformatics analyses were applied to identify a total of 2,631 viral contigs, including 14 complete viral genomes from three deep metagenomes (1 terabase [Tb] each) that were sequenced from bulk soil DNA. An additional three replicate metagenomes (∼0.5 Tb each) were obtained from each location for statistical comparisons. Identified viruses were primarily bacteriophages targeting dominant bacterial taxa. Both viral and host diversity were higher in soil with lower precipitation. Viral abundance was also significantly higher in the arid WA location than in IA and KS. More lysogenic markers and fewer clustered regularly interspaced short palindromic repeats (CRISPR) spacer hits were found in WA, reflecting more lysogeny in historically drier soil. More putative auxiliary metabolic genes (AMGs) were also detected in WA than in the historically wetter locations. The AMGs occurring in 18 pathways could potentially contribute to carbon metabolism and energy acquisition in their hosts. Structural equation modeling (SEM) suggested that historical precipitation influenced viral life cycle and selection of AMGs. The observed and predicted relationships between soil viruses and various biotic and abiotic variables have value for predicting viral responses to environmental change.
KEYWORDS: lysogeny, auxiliary metabolic gene, grassland soil, metagenome, soil bacteriophage, soil virus
INTRODUCTION
Viruses are highly abundant in soil, with estimates of 107 to 1010 per gram of soil by microscopic enumeration (1, 2). Recent advances in metagenomic sequencing and bioinformatics have provided more details about the types of DNA viruses that inhabit different soil ecosystems (3–5), including thawing permafrost (3, 4), dry Antarctic soil (6), forest soil (7), and a range of soils from a global ecosystem comparison (5). These metagenomic analyses have suggested that the majority of soil DNA viruses are bacteriophages (3–5), although giant viruses that infect eukaryotes have been detected in forest soil (7) and permafrost (8). Additionally, soil viruses identified from metagenomes have been shown to have auxiliary metabolic genes (AMGs) that potentially encode an array of metabolic functions outside essential host infection and viral replication (3, 4). The high prevalence, host interactions, and intriguing auxiliary metabolic functions of DNA viruses highlight their potential ecological importance in soil.
Climate change is currently influencing the soil environment, with unknown consequences on soil viruses. For example, changes in precipitation patterns result in soil moisture shifts that influence soil biotic and abiotic properties (9). Field and laboratory studies have shown that changes in soil moisture impact the microbial composition and functional potential of the soil microbiome (10, 11). However, it is currently unknown how changes in precipitation shape the soil virosphere and influence viral types, abundances, activities, and lytic/lysogenic lifecycles (12, 13). During the lytic life cycle, bacteriophages replicate inside the host and are released over short intervals via lethal disruption of host cells (14). In contrast, during the lysogenic life cycle, a prophage does not directly result in virion production or release. Instead, the prophage is replicated along with the host cell genome during binary fission (14). Information about viral life strategies is key to predicting how soil viruses will regulate host dynamics in response to shifts in precipitation due to climate change (15, 16). By microscopy, abundance of virus-like particles in soil has been positively correlated with soil water content (17). This could be a result of increased bacterial numbers and enhanced diffusion/advection under high soil moisture conditions, which could increase the chances of virus-host encounters as well as prophage induction to a lytic life cycle. In contrast, bacteria extracted from dry Antarctic soils have been shown to contain high proportions of temperate viruses (18). These findings lead to a hypothesis that lysogeny is higher in dry soils than in wetter soils. This hypothesis is particularly important to test due to the roles that viruses play in the regulation of host abundances and other, yet unexplored, auxiliary metabolic functions.
Here, we aimed to contribute new information to this complex problem by directly comparing viral diversity and abundance, prevalence of lysogeny, and metabolic potential in three grassland soils across the continental United States with historical differences in annual precipitation. One extreme was an arid grassland soil from eastern Washington (WA), an area with hot dry summers and 180 mm average annual precipitation (19). The other extreme was a grassland soil from Iowa (IA), with large amounts of annual precipitation (934 mm) and tile drainage to prevent soil moisture saturation (20, 21). We also collected soil from Kansas (KS), a state with shifting precipitation due to climate change (9), having trends toward drier summers in the southwest and wetter summers in the northeast. The KS location was Konza native prairie with 835 mm annual rainfall (19). We used a variety of bioinformatics approaches to determine differences in viral lifestyles, abundances, and functional potentials from bulk soil metagenomes that were obtained across the locations. The results of this study indicate major differences in soil viruses along the precipitation gradient and provide context for predicting how future changes in climate will influence the soil virosphere.
RESULTS
Soil samples were collected in triplicate in the fall of 2017 across the three grassland locations having differences in historical annual precipitation (IA > KS > WA). For each location, one deeply sequenced composite metagenome (19) (>1 terabase [Tb]) was obtained to detect viral contigs using a bioinformatics workflow (Fig. 1). In addition, each of the three field soil samples were individually sequenced (∼0.5 Tb each) to provide three replicate metagenomes for a statistical comparison of the impact of historical annual precipitation on viral types, relative abundances, diversity, and AMGs. Therefore, there were four metagenomes per site (one large and three smaller) for a total of 12 metagenomes. The soil samples were also analyzed for soil biotic and abiotic properties, including soil moisture content at the time of sampling, chemical composition and microbial community biomass, composition, and diversity (Fig. S1; Fig. 2). Although KS had a historically lower annual precipitation than IA, at the time of sampling, KS had the highest average soil moisture content (30.32%), followed by IA (20.22%) and WA (5.03%). Unlike the other two sites, the IA soil was tile drained to prevent the soil from being saturated, which might be one explanation for its relatively lower soil moisture content at the time of sampling. Alternatively, the relatively lower soil moisture content in IA compared to KS at the time of sampling could be due to timing of recent precipitation events. The WA soil also had a much higher pH (8.67) compared to the other two soils (IA, pH 6.81; KS, pH 6.25). There were also significant differences in organic matter content (IA > WA, P < 0.01; IA > KS, P < 0.05) and iron concentrations (IA > WA, P < 0.01) across some of the locations (Fig. S1).
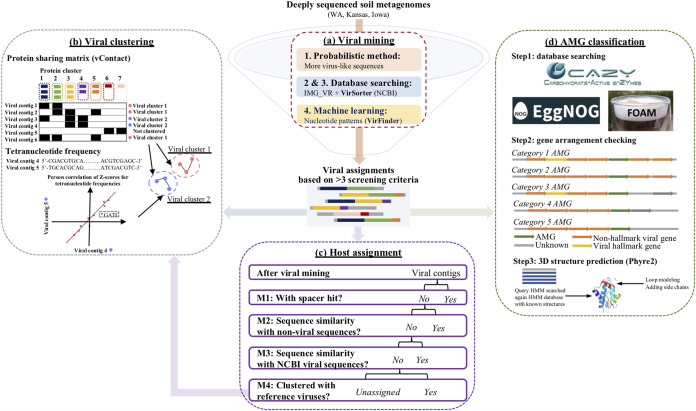
FIG 1.
Overview of bioinformatic workflow. The bioinformatic workflow comprises four modules. (a) Viral mining. Contigs with length greater than 2,500 bp (bp) were selected and screened for viral sequences by an integrated approach that combines a probabilistic approach (1) together with database searching (2) against IMG-VR (63) and (3) VirSorter (66) (NCBI), and machine learning (3). Four different criteria were then used to determine highly confident viral assignments. Only those that corroborated at least three out of the four criteria were included as confident viral sequences for further analyses. (b) Viral clustering. An integrated viral clustering network was constructed based on a protein sharing matrix by vContact (v2.0.9.10) (22). The remaining viral contigs were then linked to the clusters according to their tetranucleotide frequencies (pyani v0.2.9) (92). (c) Host assignments. The primary method (M1) used to assign the hosts of viral contigs was via matching CRISPR spacers in the nonviral contigs with the classified taxon. The additional host assignment methods (M2 to M4) were based on sequence similarity to either nonviral sequences (M2) or reference viruses (M3 based on local alignment; M4 based on protein sharing and tetranucleotide frequency). (d) Auxiliary metabolic gene (AMG) classification. Potential AMGs were first annotated by searching different functional gene databases (CAZY [84], EggNOG [61], and FOAM [85]). Potential AMGs were then classified into five categories based on their gene arrangements, and only those that contained viral genes both upstream and downstream and had a confirmed 3D structure reconstruction by Phyre2 (87) were considered AMG candidates.
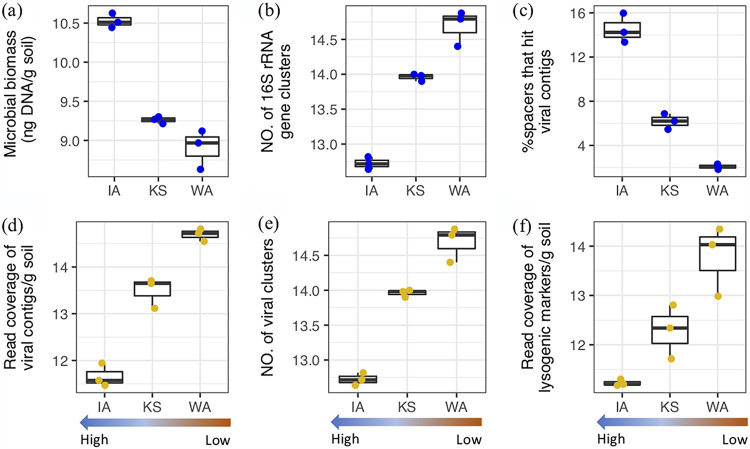
FIG 2.
Shifts in grassland soil viral and microbial communities along a gradient of historical precipitation (Iowa [IA] > Kansas [KS] > Washington [WA]). (a) Estimates of microbial biomass based on DNA yield per gram of source soil. The same color scheme was applied to all panels, with microbial data in blue and viral data in yellow. (b) Microbial diversity based on the number of 16S rRNA genes clustered at 97% identity. (c) Percentage of CRISPR spacers extracted from each soil metagenome that exactly matched viral contigs identified from the same site. (d) Estimates of viral abundances in the three soils. The sum of the average read coverage of the viral contigs identified in each soil (identity, >95%; coverage, >80%) was used to represent the viral abundance at each site. (e) Viral diversity based on clustering of detected viral contigs using a protein sharing matrix and tetranucleotide frequency (details in Materials and Methods). (f) Abundance of viral lysogenic markers. The total read coverage (identity, >95%; coverage, >80%) of viral genes encoding integrases and excisionases was used to assess the prevalence of lysogeny. All of the values shown in panels a, b, d, e, and f were first normalized to gram of soil and then log transformed. Statistical tests were performed via pairwise comparisons among the three grasslands (n = 3). Significant differences were determined using t tests in the R package (“rstatix”). In each boxplot (panels a, b, c, d, e, and f), the top and bottom of each box represent the 25th and 75th percentiles, respectively, and the center line indicates the median.
Soil chemical data for soils across a gradient of historical precipitation (Iowa [IA] > Kansas [KS] > Washington [WA]). Significant differences were determined by t test in the R package (“rstatix”; *, P < 0.05; **, P < 0.01). In each boxplot, the top and bottom of each box represent the 25th and 75th percentiles, respectively, and the central line shows the median. “OM” represents organic matter in the source soil. Download FIG S1, PDF file, 0.03 MB (35.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
A 16S rRNA gene-based analysis revealed that the grassland microbial communities were dominated by Actinobacteria and Proteobacteria. The microbial community composition was more similar in IA and KS compared to WA (Fig. S2). The microbial biomass was estimated by the DNA concentration extracted from the soil, with the highest estimates in IA compared to KS (P < 0.05) and WA (P < 0.01) (Fig. 2a). Microbial diversity, based on the number of 16S rRNA gene clusters (at 97% identity) enumerated in the metagenomes, exhibited the opposite trend, with the lowest diversity in IA and highest in WA (IA versus KS, P < 0.05; IA versus WA, P < 0.05; Fig. 2b).
Prokaryotic community compositions for soils collected across a gradient of historical precipitation (Iowa [IA] > Kansas [KS] > Washington [WA]). The community compositions of the prokaryotic species are shown in the heatmap with log transformed read coverages that are color-coded (the warmer the color, the higher the coverage). The prokaryotic communities detected from the three soils were clustered by composition similarity. Download FIG S2, PDF file, 0.05 MB (48KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Viral abundance, diversity, and potential activity across the precipitation gradient.
A total of 2,631 viral contigs (>2.5 Kb with an average length of 11 Kb) were identified from the three grassland soils, and 14 of these were complete and high-quality viral genomes (Table S1 and Fig. S3). The total number of viral contigs was highest in WA (1,577 contigs), followed by KS (756 contigs), and lowest in IA (298 contigs). This trend still held when mapping reads from three additional replicate metagenomes per site to the viral contigs and normalizing per gram of soil (IA versus KS, P < 0.05; IA versus WA, P < 0.05; KS versus WA, P < 0.05; Fig. 2d).
Visualization of 14 complete viral genomes assembled from soil metagenomes. The putative AMGs, hypothetical genes, and viral hallmark and nonhallmark genes are color-coded, and the arrangement of these genes is shown in linear and circular views of the 14 high-quality complete viral genomes. The genome length is illustrated by the scale of the backbone representing the viral genome in the linear view or specified in number of base pairs in the circular view. The order of the colored blocks represents the arrangement of the predicted genes with annotations. Download FIG S3, PDF file, 1.5 MB (1.5MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Viral host assignments and clustering information for each identified viral contig (summary table containing information on the following: assigned viral hosts and respective methods used for viral assignment, viral clusters and respective clustering method used, CRISPR spacer hit(s), length, and read coverage for each viral contig identified) Download Table S1, XLSX file, 0.8 MB (843.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
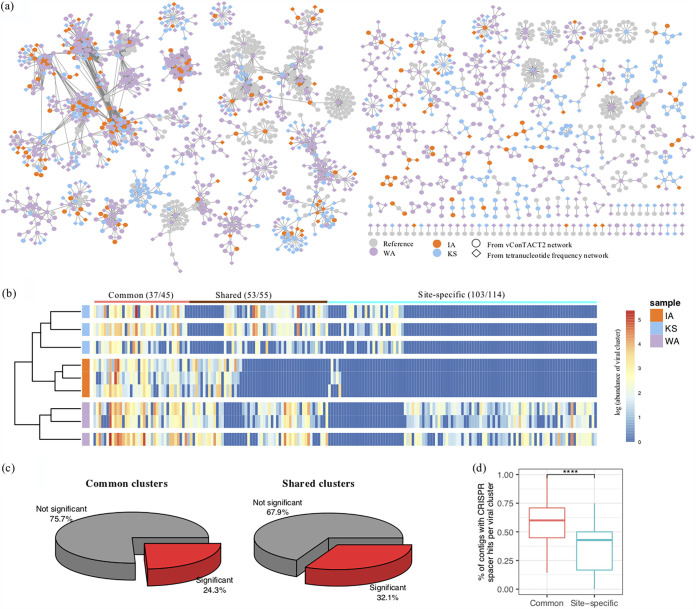
The number of viral clusters per sample (richness) was used to represent viral diversity and compared across the grasslands with a historical precipitation gradient. Only 63% of the viral contigs (1,666 out of 2,631 viral contigs) could be grouped into clusters. In total, 214 viral clusters were obtained. We first analyzed the contigs using vConTACT (22), which relates different contigs to each other based on a protein sharing matrix which clustered 58% of the viral contigs into 143 clusters (nodes in circular shape, Fig. 3a). Another 5% of the viral contigs (139 viral contigs; nodes in diamond shape, Fig. 3a) were added to the vConTACT clusters as extended edges based on correlation of their tetranucleotide frequency Z scores. An interactive network of viral clustering (Fig. S4) is available for a closer exploration of these clusters that can be filtered by the complexity of each cluster (number of edges). The majority of the clusters (144) were novel and did not cluster with known viruses in reference databases, and the rest were clustered with NCBI reference genomes representing viruses spanning four orders, Caudovirales, Kalamavirales, Petitvirales, and Tubulavirales. These results highlight that soil viruses are highly diverse and largely uncharacterized.
FIG 3.
Viral contig clusters identified from Washington, Kansas, and Iowa grassland soil metagenomes. (a) Viral contigs detected from Washington (WA, purple), Kansas (KS, blue), and Iowa (IA, orange) grassland metagenomic sequences were clustered together with NCBI reference viruses (gray). Viral contigs that primarily clustered based on their protein sharing matrices are shown as closed circles, and contigs that were added to the vConTACT network based on their Z score correlations by tetranucleotide frequencies are shown as diamonds. (b) The heatmap illustrates abundance estimates for each viral cluster detected in the three replicate metagenomes from each site. The estimated abundances were log transformed, and warmer colors represent higher abundances. The abundance profile was grouped according to the similarity of the viral cluster composition. Viral clusters that were common and detected in all soil locations are labeled “common” (red line). The clusters that were found in any two of the three soil locations are labeled “shared” (brown line). Clusters that were unique to each site are labeled “site-specific” (blue line). (c) The percentage of common and shared viral clusters that differed according to differences in historical precipitation. Viral clusters with significantly differential abundances across sites were colored in red (P < 0.05), with the remainder in gray. (d) Percentage of contigs with CRISPR spacer hits in common clusters (red) versus site-specific clusters (in blue). In each boxplot, the top and bottom of each box represent the 25th and 75th percentiles, respectively, and the center line shows the median.
An interactive network for viral clustering. An interactive and reader-friendly clustering network that enables close exploration of each individual viral cluster. The interactive network contains the detected soil viral contigs and NCBI viruses and can be filtered based on the complexity or relative representation of the clusters (number of the edges). Download FIG S4, HTML file, 3.6 MB (3.6MB, html) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Similar to differences in viral relative abundance and in microbial diversity across locations, the number of viral clusters was highest in WA (compared to IA and KS, P < 0.05; Fig. 2e) and lowest in IA (compared to KS and WA, P < 0.05; Fig. 2e). A total of 45 viral clusters were detected across all of the grassland soils, referred to as “common clusters.” Of these, 37 contained viral contigs with genome coverages of >50% in the replicate metagenomes (Fig. 3b). The estimated abundances of the common clusters accounted for an average of 67% of the IA virosphere, 55% of the KS virosphere, and 50% of the WA virosphere (Fig. 3b). An additional 55 clusters were shared between two of the three sites, and the majority of these clusters (23) were also detected in the replicate metagenomes (“shared clusters”; genome coverage, >50%; Fig. 3b). The remaining 114 clusters were unique to only one soil location (“site-specific clusters”). The majority of the site-specific clusters (103) were also detected in the replicate metagenomes (genome coverage, >50%; Fig. 3b). The remaining clusters that were not detected in the replicate metagenomes could represent contigs that were in low abundance.
Differential abundance of the common and shared viral clusters that were detected in the replicate genomes was calculated and compared across samples. Many of these viral clusters significantly responded to differences in historical precipitation (24% of common clusters and 32% of shared clusters; Fig. 3c). Common clusters contained significantly more viral contigs that showed signs of host interaction, as was evident by their significantly higher frequency of CRISPR spacer hits than site-specific clusters (P < 0.01; Fig. 3d).
Potential virus-host interactions across the precipitation gradient.
Potential virus-host interactions at the time of sampling were determined by sequence homology searches for exact matches of CRISPR spacers against the viral contigs (24). The CRISPR spacers were bioinformatically identified from the contigs representing cellular genomes. This method has been previously demonstrated for predicting potential interactions between viruses and prokaryotes (5, 25). A total of 3,708, 2,791, and 746 CRISPR spacers were matched to the viral contigs detected from IA, KS, and WA, respectively (average of absolute counts from three replicates for each soil). As prokaryotes keep an inventory of viral genome fragments from previous and current viral infections as arrays of CRISPR spacers (26), we used the percentage of spacers that were exact matches to the viral contigs identified at the time of sampling to inform the relative frequency of more recent virus-host interactions. These differences were significant across the precipitation gradient (IA versus WA, IA versus KS, and KS versus WA; P < 0.05; Fig. 2c), with a higher percentage in the historically wetter IA soil (15%), followed by KS (6%) and WA (2%). In addition, we assessed the coverage of lysogenic markers, such as genes encoding integrases and excisionases, that have previously been used as proxies for the prevalence of lysogeny (27). The relative abundances of these markers for lysogeny were higher in the historically drier WA soil (Fig. 2f). These findings suggest that temperate phages were more abundant in historically drier soil.
Four nested methods were applied to assign putative hosts to the viral contigs (M1 to M4, Fig. 1c). One percent of the nonviral contigs that are representative of possible viral hosts contained CRISPR arrays, yielding 12,892 spacers, with 8,481 of them being unique. These were used for CRISPR matching (3, 4) (M1, Fig. 1c) to assign 43% of the viral contigs to their hosts. Subsequently, to increase the number of host assignments, a second method was applied to assess the sequence similarities of viral contigs to nonviral sequences (M2, Fig. 1c). Using this approach, hosts for an additional 8% of the viral contigs were assigned. The third method (M3, Fig. 1c) assigned an additional 1% of the viral contigs to host taxa according to their sequence similarities to NCBI reference viral genomes with known hosts. Finally, the fourth method (M4, Fig. 1c) assigned potential hosts to another 10% of the viral contigs by clustering them with known reference viruses.
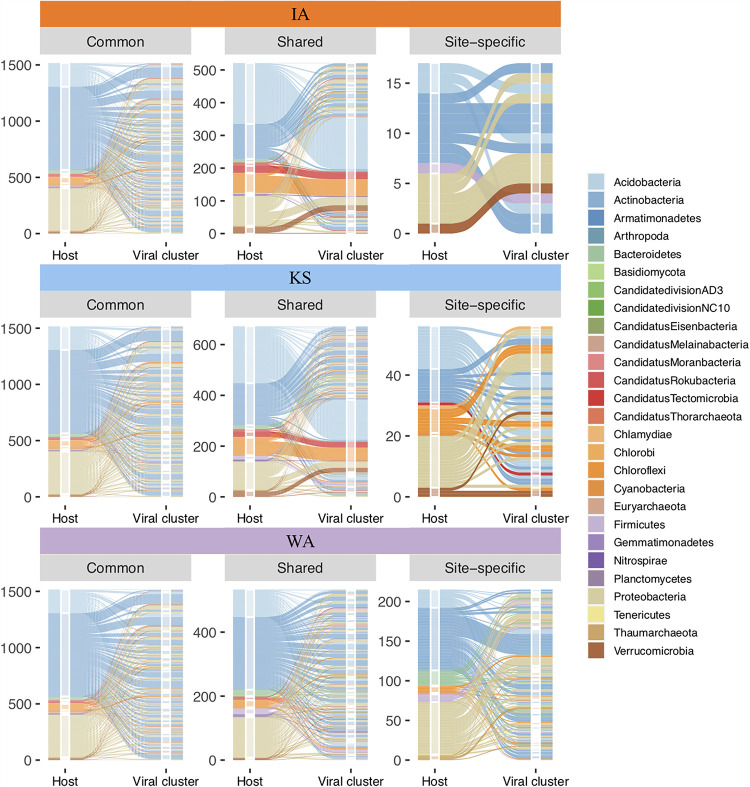
Virus-host pairings revealed that the detected soil viruses potentially targeted a broad range of host phyla across kingdoms, including archaea, bacteria, and fungi (Fig. 4). Although the viral communities recovered from the three grasslands differed in relative abundance and overall composition, the majority of the detected viral contigs were consistently assigned to Acidobacteria, Actinobacteria, and Proteobacteria as the hosts, which also represent the dominant grassland soil microorganisms (Fig. S2). These dominant bacterial phyla were also the primary hosts of the common viral clusters (Fig. 4). For the shared viral clusters, the host composition of KS and IA grasslands were more similar than those for WA (Fig. 4). In contrast, the site-specific clusters had different predicted hosts, thus illustrating unique assemblages of soil viruses and hosts across sites with differences in historical precipitation (Fig. 4). Interestingly, some of the host assignments that we found have not previously been reported for soil viruses in thawing permafrost (3, 4) and the Earth’s virome study (5) (e.g., Candidatus groups NC10 [28] and Rokubacteria [29]).
FIG 4.
Predicted virus-host pairing within viral clusters detected in soil metagenomes across a gradient of historical precipitation (Iowa [IA] > Kansas [KS] > Washington [WA]). Alluvial plots illustrate virus-host pairing using the integrated approach described in Fig. 1c (host assignment). The plots are grouped by the grassland soil location (IA, orange; KS, blue; WA, purple). The number of predicted virus-host pairs are shown on the y axis. Within each soil location, virus-host pairs were plotted for viral clusters that were common to all three locations (common), shared among two of three locations (shared), or only found in one location (site-specific). For each plot, the left stratum represents host assignment, colored by phylum, and the right stratum shows viral clusters separated by horizontal white lines. The flow of the pairing is colored by the host lineage assigned. The height of the colored strata demonstrates the relative dominance of each host phylum that the identified soil viruses were predicted to target.
Within VC_110, one of the most abundant common clusters, there were a total of 71 viral contigs, and 6 of these contigs were potential viral generalists (5). Each of the 6 viral contigs had more than 10 spacer hits and could be assigned with a broad host range spanning across several phyla, including Actinobacteria, Proteobacteria, Acidobacteria, Chloroflexi, Planctomycetes, and Verrucomicrobia (Table S1).
Viral auxiliary metabolic genes (AMGs) differed across the precipitation gradient.
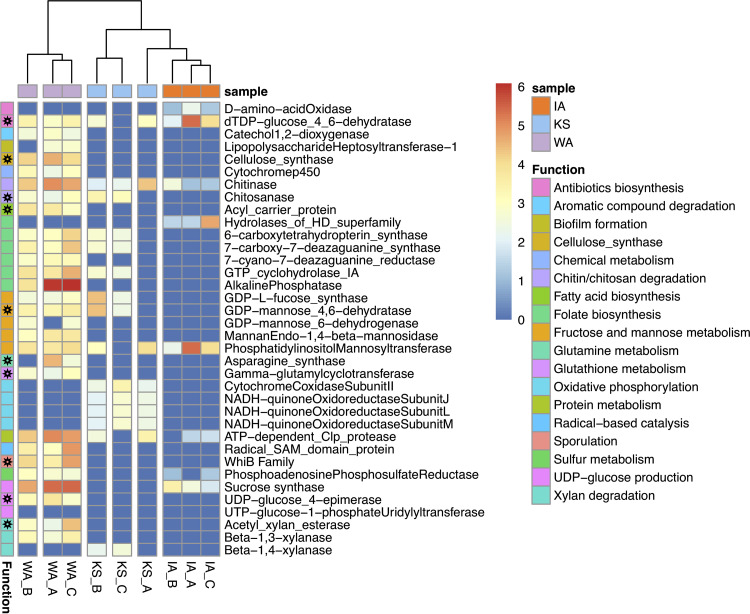
We used a three-step approach to classify auxiliary metabolic genes (AMGs) from the detected viral contigs (details in Fig. 1d) and to estimate their abundances across the three locations (Fig. 5). A total of 94 putative AMGs were found that were represented in 18 metabolic pathways, including energy acquisition and carbon metabolism (Table S2). The most confident AMG assignments (category 1, Fig. 1d) had viral hallmark genes on the same contig as the AMG. There were 65 AMGs that fulfilled category 1 criteria; 14 of these had viral hallmark genes both upstream and downstream of the AMG. The other 29 AMGs fulfilled category 2 criteria (Fig. 1d) without viral hallmark genes but with viral nonhallmark genes both upstream and downstream (the genomic context of category 1 and 2 AMGs is provided in Table S2). The viral contigs with AMGs are visualized in Fig. S5. Identified AMGs were dereplicated based on annotated functional categories, and AMG richness was used to represent the range of unique auxiliary metabolic potentials of the viral community of each site. The historically driest site (WA) had the highest AMG richness, with 28 that were involved in 17 pathways, followed by KS with 15 AMGs involved in 7 pathways and IA with 8 AMGs involved in 7 pathways (Fig. 5).
FIG 5.
Auxiliary metabolic genes (AMGs) detected in three grassland soil metagenomes across a gradient of historical precipitation (Iowa [IA] > Kansas [KS] > Washington [WA]). The heatmap illustrates abundance estimates for each putative AMG in three replicate metagenomes (IA, orange; KS, blue; WA, purple). The estimated abundances represent log transformed read coverages of the viral contigs detected with the AMGs, and warmer colors represent higher abundances. The abundance profile is clustered by sample according to the similarity of AMG composition. AMGs are further grouped by KEGG Orthology (“function,” details in Table S2). AMGs with a black star in the “function” cells are also detected in the complete viral genomes (Fig. S3).
Visualization of viral contigs that were identified to contain AMGs. Linear view of the 67 viral contigs detected with at least one of the category 1 or 2 putative AMGs that are listed in Table S2. The genes predicted from the viral contigs are color-coded, with categories 1 and 2 AMGs in dark purple, categories 3 to 5 AMGs in light purple, hypothetical genes in grey, viral hallmark genes in yellow, and viral nonhallmark genes in orange. Download FIG S5, PDF file, 1.8 MB (1.8MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
94 putative AMGs detected from large (>1 Tb) soil metagenomesaaA list of putative AMGs identified on viral contigs (category 1 and 2 AMGs; a detailed description of AMG categories in “AMG classification”). The table contains information of full and abbreviated functional annotations (“Annotation” and “Gene identifier used for visualization”), length of the nucleotide sequences (“Length”), positions of the AMGs that were detected on the viral contigs (“Start” and “End”), sites where the AMGs were detected (“Sample”), statistics from the sequence homology analysis, including “E-value,” “bit score/coverage%,” and the genomic contexts of viral contigs with AMGs detected (“Gene Arrangement,” “AMG_index,” “AMG_upstream,” “‘AMG_downstream”) Table S2, XLSX file, 0.02 MB (25.9KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
An unexpected finding was the discovery of bacteriophages carrying putative AMGs with homology to parts of the oxidative respiratory chain. These AMGs were found on viral contigs from the KS grassland; one contig contained a cytochrome c oxidase, and one contig contained three subunits for NADH-quinone oxidoreductase (Fig. 5 and Fig. S5). The putative AMGs annotated as three subunits for NADH-quinone oxidoreductase on one viral contig were classified as category 2 AMGs with viral genes both upstream and downstream after manual inspection (details in Materials and Methods, Table S2, and Fig. S5). AMGs encoding NADH-quinone oxidoreductase subunits were all located on the same contig; genes for subunits L and J were located next to each other, and the gene for subunit M was located 16 Kb downstream. Each of the three AMGs had at least one viral nonhallmark gene upstream and downstream, suggesting that they were carried on a virus (Table S2 and Fig. S5). To our knowledge, this is the first report of finding AMGs involved in oxidative respiration in soil viruses and could be analogous to the discovery of AMGs involved in photosynthetic production of ATP via proteorhodopsin in marine viruses (30). Examples of other intriguing auxiliary metabolic potentials in viral contigs included (i) an acyl carrier protein (category 2 AMG), a key enzyme for fatty acid biosynthesis in all domains of life, (ii) genes involved in the production of UDP-glucose (category 2 AMG), a substrate for cellulose biosynthesis, (iii) an alkaline phosphatase (category 2 AMG), a key enzyme involved in phosphate cycling, and (iv) genes regulating bacterial sporulation (Fig. 5).
Another set of putative AMGs were found to be involved in metabolizing a diverse range of carbon compounds with various complexities, including xylan, cellulose, chitin/chitosan, catechol, pectate, fructose, and mannose (Fig. 5). For example, we detected three AMGs predicted to be involved in xylan degradation—acetyl xylan esterase (category 1 AMG with viral hallmark genes both upstream and downstream), β-1,3-xylanase (category 1 AMG with viral hallmark genes both upstream and downstream), and β-1,4-xylanase (category 2 AMG) that converts xylan to xylose. Genes encoding viral chitinases were also identified in metagenomes from all 3 grassland soils. In contrast, catechol 1,2-dioxygenase (category 1 AMG with a viral hallmark gene downstream), an enzyme involved in the breakdown of aromatic compounds, was only found in the WA grassland soil. Five putative AMGs involved in mannose metabolism were also identified on 16 viral contigs; 15 were category 1 AMGs and 1 was a category 2 AMG. Among these, the mannan endo-1,4-beta-mannosidase AMG was previously confirmed to be active in cleaving β-1,4-linked mannose, a plant-derived polymer, in a soil virosphere from thawing permafrost (3). In addition, 6 enzymes involved in folate biosynthesis were detected as putative AMGs on 17 viral contigs (13 category 1 and 4 category 2). Three of the AMGs for folate biosynthesis (6-pyruvoyltetrahydropterin/6-carboxytetrahydropterin synthase, 7-carboxy-7-deazaguanine synthase, GTP cyclohydrolase) were located on the same viral contigs in WA and KS metagenomes.
We also found putative AMGs on completely closed viral genomes (Fig. S3), which enables connections of viral AMGs to the other viral genes. All 14 of the complete viral genomes were characterized as novel bacteriophages with no qualified matches to NCBI viral genomes (coverage, >50%; identity, >50%). Two of the complete viral genomes were detected from KS and 12 from WA. The AMGs annotated on these complete viral genomes included those involved in metabolism of xylan, cellulose, mannose, chitin/chitosan, and pectate. As these 14 soil viruses were assigned to the dominant bacterial taxa as putative hosts (i.e., Actinobacteria, Acidobacteria, and Proteobacteria), the identified auxiliary metabolic functions carried by the soil viruses could potentially impact the turnover of organic matter in soil by influencing the host communities.
Structural equation modeling suggested causal relationships between biotic and abiotic data.
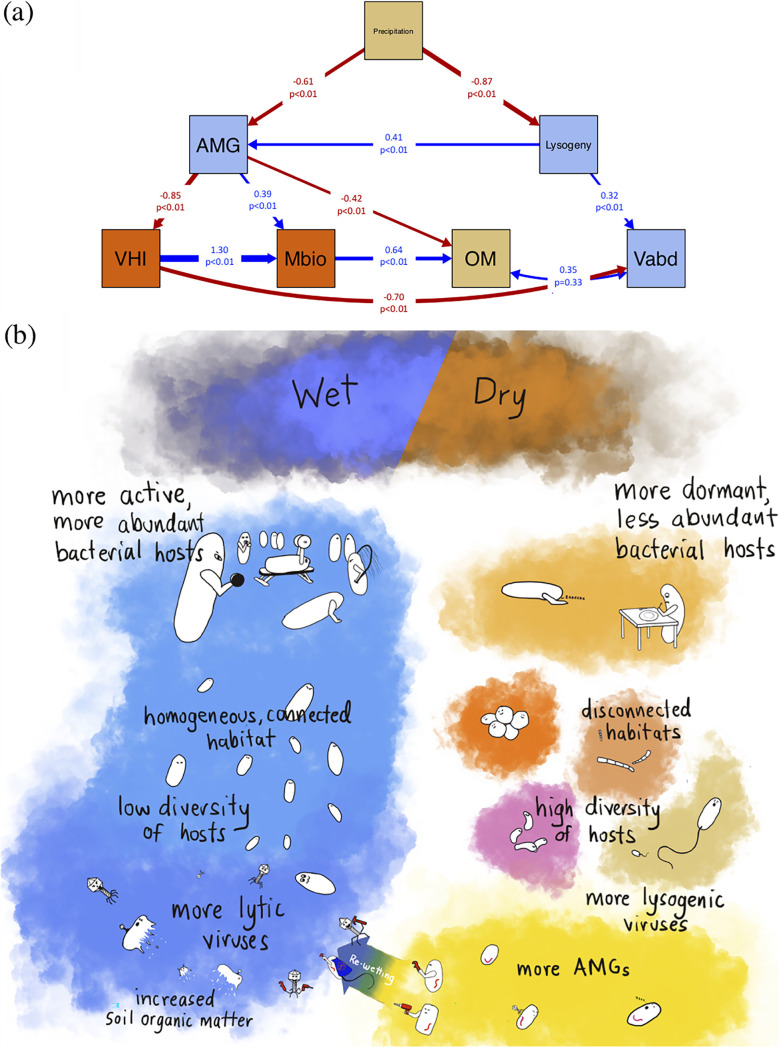
We applied structural equation modeling (SEM) to a constructed model based on the above-described findings to determine potential dependencies between environmental factors, soil viruses, and microbial communities across the historical precipitation gradient (31, 32). The resulting SEM provided a good fit as evaluated by a chi-squared (χ2) test (χ2 = 7.16, degree of freedom, or df, = 10, P = 0.71) and other commonly used indices (goodness of fit, or GFI, = 0.83, comparative fit index, or CFI, = 0.99, standardized root mean square residual, or SRMR, = 0.04) (33). Thus, the SEM provides a basis to inform possible causal relationships among the data for future evaluation (Fig. 6a). We recognize that this model could be strengthened with additional data, so here it is presented as a foundational framework for future validation.
FIG 6.
Structural equation model-supported predictions of the influence of biotic and abiotic factors on grassland soil viruses. (a) Structural equation modeling (SEM) was applied to test the conceptual relationships between abiotic properties (brown boxes), microbial abundance, and virus-host interactions (orange boxes) and the virosphere (blue boxes). The estimated abundances of viral integrases and excisionases were used to infer a lysogenic lifestyle (“lysogeny” in blue box). The total read coverages of the viral contigs with differential abundances (data in Fig. 3c) were used to represent the viral abundances in each location (“Vabd” in blue box). The DNA yield per gram of soil was used to estimate the microbial biomass (“Mbio” in orange box). The percentage of CRISPR spacers that were exact matches to the detected viral contigs was used to represent the degree of virus-host interactions (“VHI” in orange box, data shown in Fig. 2c). Organic matter (“OM” in brown box) represents the percentage of soil organic matter. The number of auxiliary metabolic categories detected from each grassland is noted as “AMG” (blue box). Blue and red arrows represent positive and negative pathways, respectively. Arrow width is proportional to the strength of the relationship, and numbers on the arrows are the path coefficients and the P value. The direction of arrows represents the direct impact of one variable on another supported by SEM. Parameters evaluating the model fitness were Chi-square (χ2) = 7.16, df = 10, P = 0.71, goodness of fit, or GFI, = 0.83, comparative fit index, or CFI, = 0.99, and standardized root mean square residual, or SRMR, = 0.04. (b) An illustration based on the SEM model to summarize viral responses to either wet or dry soil conditions and their associated impacts on the soil microbial community. In wet soil where the environment is more homogenous and microbes are higher in biomass and lower in diversity, viruses are more active and frequently interact with hosts, resulting in more host lysis. The carbon released due to host lysis may contribute to the organic matter pool in soil. In dry soil where the soil habitat is more disconnected, microbial diversity is higher and so is the associated virosphere. Instead of immediately lysing the hosts, temperate viruses carrying AMGs are selected. The hosts (lysogens) may in turn benefit from the auxiliary metabolic functions carried by the viruses, both to cope with the dry environment and when moisture becomes available.
The SEM-based analysis showed that historical precipitation influenced viral lysogeny (“Lysogeny,” path coefficient = –0.87, P < 0.01), with higher lysogeny in drier soils. In turn, lysogeny was positively correlated with viral population density (“Vabd,” path coefficient = 0.32, P < 0.01), and AMG richness (“AMG,” path coefficient = 0.41, P < 0.01). Historical precipitation was also predicted to have a significant impact on AMGs (“Precipitation” to “AMG,” path coefficient = –0.61, P < 0.01), as soils with lower precipitation had more diverse AMGs. Our SEM analysis linked higher AMG richness to reduced virus-host interactions through the CRISPR-Cas system (“VHI,” path coefficient = –0.85, P < 0.01), and as a result AMG richness indirectly affected viral abundance (“Vabd,” path coefficient = –0.70, P < 0.01). In addition, the presence of viral AMGs positively influenced microbial biomass (“Mabd,” path coefficient = 0.39, P < 0.01). Both AMGs and microbial biomass, rather than viral abundance, were associated with soil organic matter content (“AMG” to “OM,” path coefficient = –0.42, P < 0.01; “Mabd” to “OM,” path coefficient = 0.64, P < 0.01; “Vabd” and “OM,” path coefficient = 0.35, P = 0.33). As a result of the SEM analysis, we propose that the auxiliary metabolic potentials carried by soil viruses may increase host fitness via metabolic complementation in stressful soil environments (“AMG” to “Mbio,” path coefficient = 0.39, P < 0.01), similar to what has previously been proposed for marine systems (34).
DISCUSSION
Here, we aimed to determine how differences in historical precipitation influence viral ecology. We compared viral abundance, diversity, lysogenic markers, and AMGs in bulk soil metagenomes from three geographically distinct grassland/prairie soil locations in IA, KS, and WA. We acknowledge that the different soils varied in many other ways, in addition to differences in precipitation. For example, there were differences in soil chemistry between the locations that could confound interpretation of our results. Also, KS and IA were much more similar in many respects, including historical precipitation, compared to the arid soil in WA. We hypothesized that historically drier soil environments would select for temperate viruses, where the hosts are less abundant and less active than in historically wetter soil environments. This hypothesis was supported by the SEM-based analysis that revealed a strong influence of historical precipitation and associated biotic and abiotic factors on relative abundance and activity of soil viruses, with a potential impact on soil organic matter content. Interestingly, in the historically drier WA soil, where microbial biomass was lower, a more diverse and abundant DNA viral community was detected (Fig. 2). This apparent dichotomy could be explained by a preference for temperate viruses in the virosphere of historically dry soils. Indeed, we found several indications of lysogeny in the WA grassland, including significantly fewer virus-host interactions via the CRISPR-Cas system and more viral integrases and excisionases. A preference for lysogeny, therefore, may allow soil viruses to persist under dry conditions.
Because soil DNA viruses are primarily bacteriophages that infect dominant bacterial taxa such as Acidobacteria, Proteobacteria, and Verrucomicrobia as observed in this study, bacteriophage predation pressures may contribute to substantial bacterial cell death under certain conditions (3–5). For instance, cell lysis may be enhanced when soil conditions become favorable for the increase of microbial biomass (2) and/or for the transition from lysogenic to lytic viral life cycles. Our data suggested that temperate phages were more prevalent in the historically dry WA soil, as has been observed in other dry soils (18). Thus, we predict that the switch of temperate prophages to lytic life cycles, for example following a wetting event, could have profound impacts on bacterial host dynamics and organic matter cycling in traditionally dry soils.
We also found higher viral diversity in soil with lower historical precipitation. A possible explanation for this finding is that as soil dries, it experiences reduced hydrologic connectivity that can promote niche differentiation and lead to higher microbial and viral diversity within soil microenvironments (15). Other factors that could also be contributing toward the differences between the grassland soils include differences in pH. For example, WA had a much higher pH than the other two soils. Because pH is a strong driver of soil microbial community composition (35) and the dominant bacterial taxa in the studied grasslands were targeted by viruses, pH may also indirectly influence viral diversity. However, IA and KS had similar soil characteristics, including pH and viral host composition, but still exhibited significant differences in their viral communities that were aligned with differences in historical precipitation. Other work across the Stordalen permafrost thaw gradient has suggested that viral community composition was correlated with bacterial community composition, pH, and soil moisture content (3). Together with our data, these results highlight a dynamic viral response to host variation and environmental variables (36).
Our findings also indicate that despite differences in environmental variables and soil biogeochemistry across the sites, grassland soils harbor some common viral groups. The majority of the viral clusters were site specific and not shared between the three soil locations. However, 21% of the viral clusters were found across all three soils (Fig. 3). These common clusters accounted for more than half of the estimated viral load of each grassland soil. The common clusters were most abundant in IA, the site with the highest historical precipitation. In addition to the high abundance, these common clusters were enriched for viral contigs that actively interact with the hosts via the CRISPR-Cas system. Some of the common clusters contained viral contigs that represented putative generalists that recently interacted with more than one bacterial host. We propose that representative viruses in common viral clusters are well adapted to diverse ecological contexts in grassland soils, and importantly, are able to partner with many species that are prevalent in soil microbiomes. In contrast, viral representatives in site-specific clusters may reflect the impact of historical differences in precipitation and/or soil biogeochemical factors on viral type selection and diversity. Further studies of the patterns of common and site-specific viral clusters can fuel our knowledge of viral ecology in different soil matrices and how biotic/abiotic factors play a role in structuring viral communities.
Similarly, some of the common clusters contained potential viral generalists that infected more than one host. The concept of “viral generalists” with expanded host ranges has long been suspected (37), and recent studies have reported viruses with hosts across orders (38) or even phyla (5, 38, 39). Examples include a global virome survey that found several viral clusters associated with hosts from different phyla (5). In addition, four phages isolated from Lake Michigan were shown to infect different bacterial phyla (39). A broad host range facilitates the ability of the viruses to cope with stresses encountered in soil, including dispersal limitation in heterogeneous environments (40–43). We hypothesize that viruses with an expanded host range could be a beneficial strategy for more successful infections in soil environments. We acknowledge that the bioinformatic assignment of virus-host linkages only suggests possible virus-host pairings with chances of introducing false-positive assignments. More validation is therefore needed to determine the host range of soil viruses.
Soil bacteriophages with predominantly lysogenic lifecycles also have the potential to provide metabolic and evolutionary advantages (44, 45) to their hosts. Our SEM analysis supported this hypothesis and linked incidence of lysogeny to AMG richness. Furthermore, AMG richness was also connected to host interactions via CRISPR-Cas immunity, which indirectly influences both microbial and viral abundance (Fig. 6a). Finally, the historically driest soil had the highest number of AMGs, suggesting a selection for viruses with a wider range of auxiliary metabolic potentials that could be beneficial to hosts coping with environmental stresses. These results provide new perspectives on potential impacts of AMGs carried by temperate viruses on the soil microbial community that remain to be further tested with paired expression data.
We discovered putative AMGs with a range of metabolic functions on the viral contigs and genomes, including energy acquisition via oxidative respiration, fatty acid biosynthesis, phosphorus cycling, precursor processes for central metabolism (e.g., folate biosynthesis), host sporulation (e.g., whiB), and carbon cycling (Fig. 5). Some of these AMGs have previously been detected on viral contigs in other studies (4, 46, 47), but several were novel. For example, we found AMGs that could potentially encode three subunits of NADH oxidoreductase (all on one contig) and cytochrome c oxidase that are involved in the oxidative respiratory chain that generates ATP. We propose that this finding is analogous to the finding of ATP production via proteorhodopsin in marine viruses (48), although this remains to be experimentally validated. As viruses need to use hosts’ ATP for DNA packaging and capsid maturation (49), the oxidative phosphorylation-related AMGs could enhance host energy production that potentially benefits viral replication during infection. We were also able to detect AMGs on some of our high-quality, complete viral genomes, including a set of putative carbon metabolic genes (Fig. S3), e.g., for metabolizing xylan, cellulose, mannose, chitin/chitosan, and pectate, the common components of plant and fungal cell walls (50, 51). The AMG candidates that we detected here further contribute new knowledge of how soil viruses could potentially contribute to the metabolic functions of soil microbiomes.
In summary, this study provides new knowledge of soil viruses across three disparate grassland locations with historical differences in annual precipitation. SEM-based analysis inferred possible causal relationships between precipitation and the soil viral and microbial communities. Based on the SEM, we present a conceptual model of the soil viral response to either wet or dry soil conditions (Fig. 6b). In drier soil temperate viruses are more prevalent, perhaps because their microbial hosts are less abundant and less active than in wetter soils. In contrast, in wet soil where the host microbes are more active and abundant, viruses would also be more active with more frequent viral-host interactions (as observed via CRISPR-Cas), potentially resulting in more host lysis. The higher turnover of microbial biomass due to viral lysis could also be linked to higher organic matter content in wet soil than in dry soil. Furthermore, the higher prevalence of AMGs in dry soil may mirror the “batten down the hatches” strategy proposed for marine viruses, where the infecting viruses turn the hosts into lysogens, increasing host fitness, and therefore their own, via metabolic complementation under stress conditions (34). Although future work is needed to experimentally validate this conceptual model, it should serve as a valuable framework for hypothesis testing.
MATERIALS AND METHODS
Soil sampling and DNA extraction.
Three surface soil cores (top 5 to 15 cm) were collected from each of three randomly selected field block locations (∼10 m apart) from grassland soils in Iowa (41°55′14″N, 93°44′59″ W), Kansas (39°N 06′12″ N, 96°36′50″W), and Washington (46°15′04″N, 119°43′43″W) in the fall of 2017. WA and KS represented an unmanaged, native grassland and native prairie, respectively. Iowa was a reconstructed grassland/prairie and was tile drained. The individual soil cores were shipped in sterile foil packets on blue ice to the Pacific Northwest National Laboratory (PNNL) for processing. Upon receipt at PNNL, the three soil cores per field block location were immediately combined, sieved (4 mm), aliquoted into 50-ml Falcon tubes, flash-frozen, and stored at −80°C until processing. This resulted in a total of three field soil replicates per grassland soil location. The field soil replicate samples were sent to Kuo Lab testing (Pasco, WA) for soil chemical analysis (E1), following standard protocols (51).
Total community DNA was extracted from multiple 0.25-g samples for each of the three field replicates to achieve sufficiently high DNA yield for sequencing. Extraction was performed using a PowerSoil DNA extraction kit (Qiagen, Germantown, MD) following the manufacturer’s instructions with the addition of a 20 min, 55°C incubation prior to bead beating. The eluted DNA was quantified with the Qubit BR DNA quantification kit (Invitrogen, Waltham, MA). DNA yield from the soils was used for biomass estimates. The total amount of soil used for generating the respective metagenomes was used to normalize the data per gram of source soil.
Metagenome sequencing and assembly.
Metagenome libraries were prepared with the TruSeq PCR free kit (Illumina, San Diego, CA) using 1 μg DNA as starting material. For obtaining metagenomes with deep coverage of soil virosphere, the DNA extracted from the field replicates were combined to obtain enough DNA for sequencing, resulting in one large metagenome for each location (IA, KS, and WA). Each of the large metagenomes per grassland location was sequenced using seven lanes of an Illumina HiSeq X (Fulgent, California; noted as “large metagenome”), resulting in each having a size of ∼1.1 Tb with 7.3 to 7.7 billion paired-end reads (19). Assembly of the large metagenomes was performed using the metaHipMer pipeline (52) on the NERSC Cori platform (https://docs.nersc.gov/systems/cori/). The details of sequence processing and assembly parameters were described previously (19). Sequence information of the metagenomes with accompanying summary statistics is in Table S3.
Statistical information for soil metagenomes and assembled complete viral genomesaaThe table includes numbers of quality reads and bases, numbers of assembled contigs and N50 value for the “large” (>1 Tb) metagenome for each grassland soil. The download link and DOI are available for large and “replicate” metagenomes and the complete viral genomes that were assembled from the large metagenomes Table S3, XLSX file, 0.01 MB (12KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
In addition, the three field replicates per grassland location were sequenced separately (Illumina HiSeq X; Genewiz, New Jersey), yielding an average of 0.5 Tb sequence data per replicate (Table S3; noted as “replicate metagenome”). Each of the replicate metagenomes was used to calculate the depth of coverage of the contigs assembled from the respective large metagenome for statistical analyses. All of the reads of the replicate metagenomes were trimmed to remove the Illumina adaptors and to retain high-quality reads (score of >30 and length of >36 bases), as recommended by Trimmomatic (v0.33) (53).
To avoid double counting, the quality-filtered forward reads were used to align to the contigs that were assembled from the large metagenomes using BamM (v1.7.3, bamm make, https://github.com/Ecogenomics/BamM), and the reads that mapped were filtered using stringent cutoffs, i.e., percent identity higher than 0.95 and percent alignment greater than 0.80 (BamM v1.7.3, bamm filter). After filtering, the abundances of the assembled contigs in the large metagenomes were estimated based on the average base coverage mapped by the reads of the replicate metagenomes (SAMtools v1.9, SAMtools depth, http://www.htslib.org/doc/). The read coverage for each detected viral contig was then checked by the breadth of genome coverage using InStrain (v1.3.9) (23). Abundance estimates for the viral contigs with quality-filtered paired reads covering >50% of the contig length were included for the following analyses to minimize false-positive detection of contigs as previously described (23). The abundance estimates of the viral contigs with genome coverages of less than 50% in the replicate metagenomes were transformed to zero for the following analyses. Finally, the read coverage was normalized to the number of reads per gram of source soil that were used for DNA extraction to enable cross-site comparisons.
Bioinformatic identification of viral contigs.
Metagenome-assembled contigs with lengths longer than 2.5 Kb were included in our pipeline for bioinformatic assignment of viruses. Here, we also acknowledge that inclusion of contigs that are shorter than 10 Kb has been discouraged (54) due to a likelihood of introducing false positives. Because of our stringent downstream analyses to confirm viral contigs, we retained a more permissive length cutoff of 2.5 Kb. For confident viral assignment, we integrated both database-dependent and -independent assignment approaches and specified four criteria using the stringent cutoffs suggested previously (3, 54–56) (Fig. 1a). The first approach is a probabilistic method. Contigs passing screening using this probabilistic method were noted as passing criterion 1 (“1. Probabilistic method” in Fig. 1a). Genes were predicted and translated using Prodigal (57) and searched against several custom and existing viral and microbial genome databases using hmmsearch (Hmmer v3.1b2 [58], E value cutoff of 1.0e−05). The five databases searched included three viral databases: (i) a custom database curated previously (55) that comprises 25,218 viral protein hidden Markov models (HMMs) built upon viral protein-coding genes from NCBI viral genomes (55), (ii) a custom database comprising 1,147 curated viral protein families (Pfam [59], Table S4), and (iii) the existing Nucleo-Cytoplasmic Viruses Orthologous Groups (NCVOG) protein set (60). In addition, the EggNOG bacterial and archaeal databases (61) were screened to discriminate viral proteins from those known to be bacterial or archaeal. The bit scores obtained from the five databases were ranked, and taxon annotations were assigned to the highest bit scores, with a minimum bit score of 50 as the threshold (62). Contigs with more “viral-like” than “bacterial or archaeal-like” genes were considered likely viral candidates and satisfied criterion 1.
A list of Pfams that were used to annotate viral proteins. Download Table S4, TXT file, 0.4 MB (384.5KB, txt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
A second comprehensive contig-based search was also used to screen for soil viral contigs, thus satisfying our criteria 2 and 3 (“2 & 3. Database searching” in Fig. 1a). The contigs were searched against two virus-specific databases: (i) IMG/VR (released on 1 July 2018), the largest publicly available database that includes both cultured and uncultured viral sequences (63), and (ii) the curated RefseqVirus genomes database (NCBI RefseqVirus [64] v69, January 2014). The length-filtered assemblies (>2,500 bp) were compared to the viral sequences in IMG/VR using a basic local alignment search tool, BLASTN (65). Contigs having E value cutoffs equal to or lower than 1.0e−05 fulfilled our criterion (Fig. 1a) and were retained and checked by the other criteria. Additionally, for contig-based searching, we used the existing tool, VirSorter (66), which searches against the RefseqVirus genomes and evaluates the searches using a strategic scoring matrix. If the contigs passed screening by VirSorter (categories 1 to 3) (66), they satisfied our criterion 3 (Fig. 1a). We recognize that category 3 VirSorter contigs, characterized as “possible” viral contigs, are more likely to be false positives than categories 1 and 2. However, inclusion of category 3 contigs also provides room for detection of smaller lysogenic phages and novel viruses that cannot be annotated using the reference databases that are implanted in VirSorter (66). The contigs satisfying criterion 3 were thus passed through three additional screening criteria (criteria 1, 2, and 4) to constrain the searches. The third approach differentiated the viral nucleotide (8-mer) frequency patterns from nonviral ones by a machine learning approach using VirFinder (67). The contigs with P values less than 0.05 and scores greater than 0.9 passed criterion 4 (Fig. 1a). Contigs that satisfied at least three of the four criteria were assigned as identified viral contigs that were used for the following analyses.
The 14 complete viral genomes were identified as “high-quality” and closed by the detection of direct terminal repeats (DTRs) using CheckV (v0.7.0) (68).
Detection of lysogenic markers.
To assess viral lysogeny, reads for lysogenic markers, e.g., genes encoding viral integrase and excisionase (69, 70), were recruited from the three replicate metagenomes from each grassland location (IA, KS, and WA). Nucleotide sequences from a total of 20,712 unique viral integrases and 329 unique excisionases were collected from NCBI Virus and used as a reference database for lysogenic markers (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/, accessed on 16 December2020; sequence d-replication using VSEARCH v2.13.4, derep_fulllength). The same read mapping strategies described above were applied here as well. In brief, the quality-filtered forward reads were aligned to the curated lysogenic marker reference database using BamM (v1.7.3, bamm make, https://github.com/Ecogenomics/BamM), and the mapped reads with percent identity higher than 0.95 and percent alignment greater than 0.80 were retained (BamM v1.7.3, bamm filter). The relative abundances of the known lysogenic markers in each of the three replicate soils per site were estimated based on the average base coverage mapped by the reads of the replicate metagenomes (SAMtools v1.9, SAMtools depth, http://www.htslib.org/doc/).
Viral clustering.
The soil viral diversity was inferred by grouping highly confident viral contigs into clusters. vConTACT (v2.0.9.10) (71) was applied using the default parameters to first compute the protein similarities using Diamond (72), to generate the protein clusters using the Markov cluster algorithm (MCL) and to cluster the viral contigs according to the protein sharing matrix using ClusterONE (73) (for details refer to the published protocol at https://dx.doi.org/10.17504/protocols.io.x5xfq7n). The clusters generated by vConTACT (71) were visualized in a network with viral contigs as the nodes with the strength of node connectivity as the edges. Because of incomplete assemblies from soil metagenomes, contigs that were related and with multiple viral features without a significant number of shared proteins were unable to be clustered (71). Therefore, to further cluster more viral sequences from the assembled soil metagenomes, tetranucleotide frequency analysis (67, 74–76) was also implemented. The tetranucleotide frequency-based clustering method has been previously tested to group nonoverlapping but related viral contigs (74). To carefully benchmark the sensitivity and suitability of this method, Pearson correlations were calculated between Z score distributions for every tetranucleotide in a subset of viral contigs screened from grassland soils and in deposited NCBI viral genomes (64) (a total of 77 archaeal viruses, 1,675 bacteriophages, and 443 fungal viruses) after removing the dereplicated strings or substring using VSEARCH (77). When the same contig was assigned to multiple clusters, only the pairings with the highest correlation coefficients (greater than 0.6) were retained. The network in Fig. S6 shows that the tetranucleotide frequency-based method can cluster the related viral contigs according to the taxonomy annotations. We then applied this complementary method to the viral contigs that were initially unclustered by vConTACT (22). The additional viral contigs that were able to be clustered using tetranucleotide frequency analysis were merged into the vConTACT network as extended edges.
Network of viral clusters that was obtained using the tetranucleotide frequency method. This network was used as a proof-of-concept for applying tetranucleotide frequency as an approach to cluster related viral contigs. NCBI viruses and a subset of the viral contigs detected in this study were clustered using the tetranucleotide frequency-based method. The clustering of these viral sequences is visualized in the network with each node as a viral sequence and are colored by the types of the viruses or the sites where the viral sequences were detected. NCBI archaeal viruses are highlighted in red nodes, with NCBI bacterial viruses in green and fungal viruses in yellow. Viral sequences detected in Iowa, Kansas, and Washington grassland soils are colored in orange, blue, and purple, respectively. The viral clusters are generally grouped according to their taxonomic annotations, e.g., Sangervirae and Rudiviridae. Download FIG S6, PDF file, 2.9 MB (2.9MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We also created an interactive network display for more selective visualization of the viral clusters of interest, using TrelliscopeJS (https://github.com/hafen/trelliscopejs) together with R packages visNetwork (78) and networkD3 (79) (Fig. S4).
Host assignment.
We used a nested approach to assign host taxonomy to the screened viral contigs (host assignment module, Fig. 1c). The viral contigs were first searched for spacer hits from microbial clustered regularly interspaced short palindromic repeat (CRISPR) regions, a hallmark of virus-host interactions (M1, Fig. 1c). Spacers were identified from the assembled contigs of the three soil metagenomes using MinCED (https://github.com/ctSkennerton/minced) and searched against the screened viral contigs using “blastn-short task” with previously described parameters (25, 80). The taxonomy of nonviral contigs containing CRISPR regions was predicted using the Contig Annotation Tool (CAT, https://github.com/dutilh/CAT, default cutoffs). Putative host phylogeny was assigned based on the taxonomy annotation of the nonviral contigs carrying spacers that matched the screened viral contigs. The percentage of the detected CRISPR spacers that were exact matches to the identified viral contigs was used to estimate the relative frequency of virus-host interactions across the sites at the time of sampling.
As CRISPR arrays are not universally found in all prokaryotes, especially many soil bacteria (81), and assembling repetitive regions from metagenomes can be problematic (82), more complementary methods were needed to assign hosts. Thus, viral contigs with no matching CRISPR spacers were assigned to a potential host using a sequence homology screening approach based on assumptions that viruses will share genes with their hosts (83) and with relevant reference viruses. The viral contigs were searched against nonviral contigs from the same soil metagenomes that had taxonomy assignments (M2, Fig. 1c) and against NCBI viral genomes with known hosts (BLASTN [65]; thresholds of E value, 10−3; bit score, 50) (M3, Fig. 1c). Unassigned viral contigs inherited host assignments from clustered reference phages with known hosts (M4, Fig. 1c). Due to the largely uncharacterized viral and host diversity in soil, the rest of viral contigs remained unassigned.
AMG classification.
Potential auxiliary metabolic genes (AMGs) were first annotated with the following three databases: the KEGG orthology database by EggNOG (61), the carbohydrate-active enzyme database (84) (CAZy, dbCAN HMMdb release 7.0) with stringent searching parameters, E value of <1e−15, and coverage of >0.35, and the functional ontology assignments for metagenomes database (FOAM [85], hmmscan, –cut_tc) (AMG classification module, step 1, Fig. 1d). Potential AMGs were then determined by checking gene positions on the viral contigs with or without viral hallmark genes and classified into five categories (step 2, Fig. 1d). Category 1 AMGs were present on contigs with “viral-like” genes (as annotated by the viral database searching from module 1) both up- and downstream and contained at least one viral hallmark gene, such as structural genes (“capsid,” “portal protein,” “tail,” “coat”), terminases, or integrases. Category 2 AMGs were those with virus-like genes both up- and downstream but no viral hallmark genes. Category 3 AMGs were those with viral hallmark genes and virus-like genes only upstream or downstream. Category 4 AMGs were those with virus-like genes only upstream or downstream but no viral hallmark genes. Lastly, category 5 AMGs were those located at the edge of the viral contigs. We only selected categories 1 and 2 as AMG candidates for the following steps.
After search ranking and gene arrangement checking, the third checkpoint was to predict the 3D structure of the translated AMG and search against the Protein Data Bank (PDB [86]) through Phyre2 (87) (step 3, Fig. 1d). We only included viral metabolic genes in categories 1 and 2 that had highly confident Phyre2 predictions (100%) as putative AMGs. These stringent criteria were used to reduce the potential of host genome contamination and to identify AMGs with high confidence.
As chitosanases and chitinases are known to share structural similarities with viral lysozymes (88, 89), we further screened the putative chitosanase and chitinase AMGs against a profile of lysozyme HMMs to assess their sequence similarities. The profile of lysozyme HMMs includes five pfams (PF13702, PF00959, PF04965, PF18013, and PF00062) and a lysozyme HMM built upon the newly self-curated lysozyme sequences that were deposited at NCBI Virus (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/virus?SeqType_s=Protein, accessed on 16 November 2020). The putative chitosanase and chitinase AMGs were searched against the lysozyme HMMs using hmmsearch with default options (Hmmer v3.1b2 [58]). The putative chitosanase and chitinase AMGs without lysozyme HMM hits were retained.
Microbial diversity estimation.
The bacterial and archaeal 16S rRNA gene reads were first recruited from the replicate metagenomes of each grassland location (IA, KS, and WA) by mapping to the well-curated 16S RefSeq database at NCBI (https://www.ncbi.nlm.nih.gov/refseq/targetedloci/16S_process/) via BamM (v1.7.3, bamm make). The read mappings were filtered by percent identity higher than 0.95 and percent alignment greater than 0.80 (BamM v1.7.3, bamm filter). The mapped 16S rRNA gene RefSeq sequences were then clustered at 97% identity by CD-HIT (v4.8.1) (90) to inform the microbial composition and diversity of the site.
Structural equation modeling.
We applied structural equation modeling (SEM) (31, 32) to test the strengths of the following hypotheses: (i) historical precipitation has an impact on soil viral life cycles and auxiliary metabolic potentials; (ii) Changes in viral life cycles and abundances impact the soil microbial community; and (iii) Viral and microbial dynamics influence soil organic matter concentration. The code availability of the applied SEM can be found in the section describing “Data and Code Availability.”
The quantitative data for soil viruses that were coded into the model included estimated relative viral abundance, estimated relative abundance of potentially lysogenic viruses, and richness of putative AMGs. The total read coverage of the viral contigs with differential relative abundances (data in Fig. 3c) was used to represent the viral abundance at each site. The estimated abundances of viral integrases and excisionases (details in the section of “Detection of Lysogenic Markers”) were used to quantify viral lysogeny. The number of auxiliary metabolic categories detected from each grassland was used to estimate AMG richness. Microbial biomass was estimated by DNA yield per gram of soil (data shown in Fig. 2a). The degree of virus-host interactions was inferred by the percentage of CRISPR spacers that were exact matches to the detected viral contigs (data shown in Fig. 2c). The environmental data included historical precipitation for each site and organic matter content (%).
A Chi-square (χ2) test was performed, and multiple statistical parameters were calculated (i.e., goodness of fit, or GFI, comparative fit index, or CFI, standardized root mean square residual, or SRMR) to evaluate the model fit (cutoffs: χ2 P > 0.05, GFI > 0.8, CFI > 0.9, SRMR < 0.08). After selecting the model with the best fit to the data, R package semPaths (91) was used to create the SEM diagram with the result of path analysis in SEM with statistics. The connection and strength of the resolved paths in the SEM model were evaluated by path coefficients representing the change of dependent variable with a unit change in the explanatory variable and P values demonstrating the significance of the paths (P < 0.05 was considered significant in this study). The direct impact of one variable on another that was supported by the SEM was plotted as a directional arrow.
Data and code availability.
Raw reads and the assembled data of the large metagenomes (TmG.1.0) were packaged and previously published (19). Replicate metagenomes (WA-TmG.2.0, KS-TmG.2.0, IA-TmG.2.0) along with the 14 complete viral genomes assembled from the large metagenomes (Iso-VIG14.1.0) have been deposited in the PNNL Project DataHub repository with DOIs (https://data.pnl.gov/group/7/nodes/dataset/13708) and are available to download. The download link for each data set is included in Table S3. The detailed metadata are included in each data package. The R codes used in this study, including the details of graphical and statistical packages and the structural equation model, viral contigs screened from the metagenome assemblies and lysozyme HMMs, can be found at https://github.com/Ruonan0101/GrasslandVirome_PNNL.
ACKNOWLEDGMENTS
This work was supported by the Department of Energy (DOE) Office of Biological and Environmental Research (BER) and is a contribution of the scientific focus area “phenotypic response of the soil microbiome to environmental perturbations.” PNNL is operated for the DOE by Battelle Memorial Institute under contract DE-AC05-76RL01830.
We also thank Rob Egan and Kathy Yelick from the Joint Genome Institute and NERSC, respectively, at the Lawrence Berkeley National Laboratory, and their funding through the DOE-supported ExaBiome Project for supplying computational resources for the metagenome assemblies.
J.K.J. and K.S.H. obtained funding for the study. R.W. performed the majority of the bioinformatics analyses. W.C.N. performed the sequencing and assembly statistics. S.J.F. and S.L.B. conducted the soil experiments and sample collection. M.R.D. and Y.F. extracted DNA from the soil samples. I.G. constructed the interactive viral clustering network. J.E.M. plotted the conceptual model. J.E.M., K.S.H., E.B.G., and M.R.D. provided their expert insights to interpret the data. All authors contributed to writing the manuscript.
We declare no conflicts of interest.
Footnotes
This article is a direct contribution from Janet K. Jansson, a Fellow of the American Academy of Microbiology, who arranged for and secured reviews by Eric Wommack, University of Deleware, and Joanne Emerson, University of California, Davis.
Citation Wu R, Davison MR, Nelson WC, Graham EB, Fansler SJ, Farris Y, Bell SL, Godinez I, Mcdermott JE, Hofmockel KS, Jansson JK. 2021. DNA viral diversity, abundance, and functional potential vary across grassland soils with a range of historical moisture regimes. mBio 12:e02595-21. https://doi.org/10.1128/mBio.02595-21.
Contributor Information
Janet K. Jansson, Email: janet.jansson@pnnl.gov.
Frederick M. Ausubel, Mass General Hospital
REFERENCES
- 1.Trubl G, Solonenko N, Chittick L, Solonenko SA, Rich VI, Sullivan MB. 2016. Optimization of viral resuspension methods for carbon-rich soils along a permafrost thaw gradient. PeerJ 4:e1999. doi: 10.7717/peerj.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williamson KE, Fuhrmann JJ, Wommack KE, Radosevich M. 2017. Viruses in soil ecosystems: an unknown quantity within an unexplored territory. Annu Rev Virol 4:201–219. doi: 10.1146/annurev-virology-101416-041639. [DOI] [PubMed] [Google Scholar]
- 3.Emerson JB, Roux S, Brum JR, Bolduc B, Woodcroft BJ, Jang HB, Singleton CM, Solden LM, Naas AE, Boyd JA, Hodgkins SB, Wilson RM, Trubl G, Li C, Frolking S, Pope PB, Wrighton KC, Crill PM, Chanton JP, Saleska SR, Tyson GW, Rich VI, Sullivan MB. 2018. Host-linked soil viral ecology along a permafrost thaw gradient. Nat Microbiol 3:870–880. doi: 10.1038/s41564-018-0190-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trubl G, Jang HB, Roux S, Emerson JB, Solonenko N, Vik DR, Solden L, Ellenbogen J, Runyon AT, Bolduc B, Woodcroft BJ, Saleska SR, Tyson GW, Wrighton KC, Sullivan MB, Rich VI. 2018. Soil viruses are underexplored players in ecosystem carbon processing. mSystems 3:e00076-18. doi: 10.1128/mSystems.00076-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Rubin E, Ivanova NN, Kyrpides NC. 2016. Uncovering Earth’s virome. Nature 536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 6.Bezuidt OK, Lebre PH, Pierneef R, León-Sobrino C, Adriaenssens EM, Cowan DA, Van de Peer Y, Makhalanyane TP. 2020. Phages actively challenge niche communities in Antarctic soils. mSystems 5:e00234-20. doi: 10.1128/mSystems.00234-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz F, Alteio L, Goudeau D, Ryan EM, Feiqiao BY, Malmstrom RR, Blanchard J, Woyke T. 2018. Hidden diversity of soil giant viruses. Nat Commun 9:4881. doi: 10.1038/s41467-018-07335-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Legendre M, Lartigue A, Bertaux L, Jeudy S, Bartoli J, Lescot M, Alempic J-M, Ramus C, Bruley C, Labadie K, Shmakova L, Rivkina E, Couté Y, Abergel C, Claverie J-M. 2015. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc Natl Acad Sci USA 112:E5327–E5335. doi: 10.1073/pnas.1510795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook BI, Smerdon JE, Seager R, Coats S. 2014. Global warming and 21st century drying. Clim Dyn 43:2607–2627. doi: 10.1007/s00382-014-2075-y. [DOI] [Google Scholar]
- 10.Roy Chowdhury T, Lee J-Y, Bottos EM, Brislawn CJ, White RA, Bramer LM, Brown J, Zucker JD, Kim Y-M, Jumpponen A, Rice CW, Fansler SJ, Metz TO, McCue LA, Callister SJ, Song H-S, Jansson JK. 2019. Metaphenomic responses of a native prairie soil microbiome to moisture perturbations. mSystems 4:e00061-19. doi: 10.1128/mSystems.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnard RL, Osborne CA, Firestone MK. 2015. Changing precipitation pattern alters soil microbial community response to wet-up under a Mediterranean-type climate. ISME J 9:946–957. doi: 10.1038/ismej.2014.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerson JB. 2019. Soil viruses: a new hope. mSystems 4:e00120-19. doi: 10.1128/mSystems.00120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Goethem MW, Swenson TL, Trubl G, Roux S, Northen TR. 2019. Characteristics of wetting-induced bacteriophage blooms in biological soil crust. mBio 10:e02287-19. doi: 10.1128/mBio.02287-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobbs Z, Abedon ST. 2016. Diversity of phage infection types and associated terminology: the problem with ‘Lytic or lysogenic’. FEMS Microbiol Lett 363:fnw047. doi: 10.1093/femsle/fnw047. [DOI] [PubMed] [Google Scholar]
- 15.Jansson JK, Hofmockel KS. 2020. Soil microbiomes and climate change. Nat Rev Microbiol 18:35–46. doi: 10.1038/s41579-019-0265-7. [DOI] [PubMed] [Google Scholar]
- 16.Stocker T. 2014. Climate change 2013: the physical science basis: Working Group I contribution to the fifth assessment report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, UK. [Google Scholar]
- 17.Williamson KE, Radosevich M, Wommack KE. 2005. Abundance and diversity of viruses in six Delaware soils. Appl Environ Microbiol 71:3119–3125. doi: 10.1128/AEM.71.6.3119-3125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williamson KE, Radosevich M, Smith DW, Wommack KE. 2007. Incidence of lysogeny within temperate and extreme soil environments. Environ Microbiol 9:2563–2574. doi: 10.1111/j.1462-2920.2007.01374.x. [DOI] [PubMed] [Google Scholar]
- 19.Nelson WC, Anderson LN, Wu R, McDermott JE, Bell SL, Jumpponen A, Fansler SJ, Tyrrell KJ, Farris Y, Hofmockel KS, Jansson JK. 2020. Terabase metagenome sequencing of grassland soil microbiomes. Microbiol Resour Announc 9:e00718-20. doi: 10.1128/MRA.00718-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Upton RN, Bach EM, Hofmockel KS. 2019. Spatio-temporal microbial community dynamics within soil aggregates. Soil Biol Biochem 132:58–68. doi: 10.1016/j.soilbio.2019.01.016. [DOI] [Google Scholar]
- 21.Jarchow ME, Liebman M. 2013. Nitrogen fertilization increases diversity and productivity of prairie communities used for bioenergy. Gcb Bioenergy 5:281–289. doi: 10.1111/j.1757-1707.2012.01186.x. [DOI] [Google Scholar]
- 22.Bolduc B, Jang HB, Doulcier G, You Z-Q, Roux S, Sullivan MB. 2017. vConTACT: an iVirus tool to classify double-stranded DNA viruses that infect Archaea and Bacteria. PeerJ 5:e3243. doi: 10.7717/peerj.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olm MR, Crits-Christoph A, Bouma-Gregson K, Firek BA, Morowitz MJ, Banfield JF. 2021. inStrain profiles population microdiversity from metagenomic data and sensitively detects shared microbial strains. Nat Biotechnol 39:727–736. doi: 10.1038/s41587-020-00797-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puschnik AS, Majzoub K, Ooi YS, Carette JE. 2017. A CRISPR toolbox to study virus-host interactions. Nat Rev Microbiol 15:351–364. doi: 10.1038/nrmicro.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards RA, McNair K, Faust K, Raes J, Dutilh BE. 2016. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol Rev 40:258–272. doi: 10.1093/femsre/fuv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGinn J, Marraffini LA. 2019. Molecular mechanisms of CRISPR-Cas spacer acquisition. Nat Rev Microbiol 17:7–12. doi: 10.1038/s41579-018-0071-7. [DOI] [PubMed] [Google Scholar]
- 27.Luo E, Aylward FO, Mende DR, DeLong EF. 2017. Bacteriophage distributions and temporal variability in the ocean’s interior. mBio 8:e01903-17. doi: 10.1128/mBio.01903-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Z, Cai C, Wang J, Xu X, Zheng P, Jetten MS, Hu B. 2016. A novel denitrifying methanotroph of the NC10 phylum and its microcolony. Sci Rep 6:32241. doi: 10.1038/srep32241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anantharaman K, Hausmann B, Jungbluth SP, Kantor RS, Lavy A, Warren LA, Rappé MS, Pester M, Loy A, Thomas BC, Banfield JF. 2018. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J 12:1715–1728. doi: 10.1038/s41396-018-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Needham DM, Yoshizawa S, Hosaka T, Poirier C, Choi CJ, Hehenberger E, Irwin NAT, Wilken S, Yung C-M, Bachy C, Kurihara R, Nakajima Y, Kojima K, Kimura-Someya T, Leonard G, Malmstrom RR, Mende DR, Olson DK, Sudo Y, Sudek S, Richards TA, DeLong EF, Keeling PJ, Santoro AE, Shirouzu M, Iwasaki W, Worden AZ. 2019. A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators. Proc Natl Acad Sci USA 116:20574–20583. doi: 10.1073/pnas.1907517116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisenhauer N, Bowker MA, Grace JB, Powell JR. 2015. From patterns to causal understanding: structural equation modeling (SEM) in soil ecology. Pedobiologia 58:65–72. doi: 10.1016/j.pedobi.2015.03.002. [DOI] [Google Scholar]
- 32.Guo X, Gao Q, Yuan M, Wang G, Zhou X, Feng J, Shi Z, Hale L, Wu L, Zhou A, Tian R, Liu F, Wu B, Chen L, Jung CG, Niu S, Li D, Xu X, Jiang L, Escalas A, Wu L, He Z, Van Nostrand JD, Ning D, Liu X, Yang Y, Schuur EAG, Konstantinidis KT, Cole JR, Penton CR, Luo Y, Tiedje JM, Zhou J. 2020. Gene-informed decomposition model predicts lower soil carbon loss due to persistent microbial adaptation to warming. Nat Commun 11:1–12. doi: 10.1038/s41467-020-18706-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cangur S, Ercan I. 2015. Comparison of model fit indices used in structural equation modeling under multivariate normality. J Mod App Stat Meth 14:152–167. doi: 10.22237/jmasm/1430453580. [DOI] [Google Scholar]
- 34.Warwick-Dugdale J, Buchholz HH, Allen MJ, Temperton B. 2019. Host-hijacking and planktonic piracy: how phages command the microbial high seas. Virol J 16:1–13. doi: 10.1186/s12985-019-1120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rousk J, Bååth E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- 36.Santos-Medellin C, Zinke LA, Ter Horst AM, Gelardi DL, Parikh SJ, Emerson JB. 2021. Viromes outperform total metagenomes in revealing the spatiotemporal patterns of agricultural soil viral communities. ISME J:1–15. doi: 10.1038/s41396-021-00897-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elena SF, Agudelo-Romero P, Lalić J. 2009. The evolution of viruses in multi-host fitness landscapes. Open Virol J 3:1–6. doi: 10.2174/1874357900903010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peters DL, Lynch KH, Stothard P, Dennis JJ. 2015. The isolation and characterization of two Stenotrophomonas maltophilia bacteriophages capable of cross-taxonomic order infectivity. BMC Genomics 16:1–15. doi: 10.1186/s12864-015-1848-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malki K, Kula A, Bruder K, Sible E, Hatzopoulos T, Steidel S, Watkins SC, Putonti C. 2015. Bacteriophages isolated from Lake Michigan demonstrate broad host-range across several bacterial phyla. Virol J 12:164. doi: 10.1186/s12985-015-0395-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santiago FE. 2017. Local adaptation of plant viruses: lessons from experimental evolution. Mol Ecol 26:1711–1719. doi: 10.1111/mec.13836. [DOI] [PubMed] [Google Scholar]
- 41.Woolhouse ME, Gowtage-Sequeria S. 2005. Host range and emerging and reemerging pathogens. Emerg Infect Dis 11:1842–1847. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLeish M, Sacristán S, Fraile A, Garcia-Arenal F. 2017. Scale dependencies and generalism in host use shape virus prevalence. Proc R Soc B 284:20172066. doi: 10.1098/rspb.2017.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross A, Ward S, Hyman P. 2016. More is better: selecting for broad host range bacteriophages. Front Microbiol 7:1352. doi: 10.3389/fmicb.2016.01352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nasir A, Kim KM, Caetano‐Anollés G. 2017. Long‐term evolution of viruses: a Janus‐faced balance. Bioessays 39:1700026. doi: 10.1002/bies.201700026. [DOI] [PubMed] [Google Scholar]
- 45.Ramisetty BCM, Sudhakari PA. 2019. Bacterial ‘grounded’ prophages: hotspots for genetic renovation and innovation. Front Genet 10:65. doi: 10.3389/fgene.2019.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Smet J, Zimmermann M, Kogadeeva M, Ceyssens P-J, Vermaelen W, Blasdel B, Jang HB, Sauer U, Lavigne R. 2016. High coverage metabolomics analysis reveals phage-specific alterations to Pseudomonas aeruginosa physiology during infection. ISME J 10:1823–1835. doi: 10.1038/ismej.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson CL, Sullivan MB, Fernando SC. 2017. Dietary energy drives the dynamic response of bovine rumen viral communities. Microbiome 5:1–19. doi: 10.1186/s40168-017-0374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM. 2006. Community genomics among stratified microbial assemblages in the ocean’s interior. Science 311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 49.Serwer P, Wright ET. 2017. ATP-driven contraction of phage T3 capsids with DNA incompletely packaged in vivo. Viruses 9:119. doi: 10.3390/v9050119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettolino FA, Walsh C, Fincher GB, Bacic A. 2012. Determining the polysaccharide composition of plant cell walls. Nat Protoc 7:1590–1607. doi: 10.1038/nprot.2012.081. [DOI] [PubMed] [Google Scholar]
- 51.Lenardon MD, Munro CA, Gow NA. 2010. Chitin synthesis and fungal pathogenesis. Curr Opin Microbiol 13:416–423. doi: 10.1016/j.mib.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Georganas E, Egan R, Hofmeyr S, Goltsman E, Arndt B, Tritt A, Buluç A, Oliker L, Yelick K. 2018. Extreme scale de novo metagenome assembly, p 122–134. In SC18: International Conference for High Performance Computing, Networking, Storage and Analysis. IEEE, New York, NY. doi: 10.1109/SC.2018.00013. [DOI] [Google Scholar]
- 53.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roux S, Adriaenssens EM, Dutilh BE, Koonin EV, Kropinski AM, Krupovic M, Kuhn JH, Lavigne R, Brister JR, Varsani A, Amid C, Aziz RK, Bordenstein SR, Bork P, Breitbart M, Cochrane GR, Daly RA, Desnues C, Duhaime MB, Emerson JB, Enault F, Fuhrman JA, Hingamp P, Hugenholtz P, Hurwitz BL, Ivanova NN, Labonté JM, Lee K-B, Malmstrom RR, Martinez-Garcia M, Mizrachi IK, Ogata H, Páez-Espino D, Petit M-A, Putonti C, Rattei T, Reyes A, Rodriguez-Valera F, Rosario K, Schriml L, Schulz F, Steward GF, Sullivan MB, Sunagawa S, Suttle CA, Temperton B, Tringe SG, Thurber RV, Webster NS, Whiteson KL, et al. 2019. Minimum information about an uncultivated virus genome (MIUViG). Nat Biotechnol 37:29–37. doi: 10.1038/nbt.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paez-Espino D, Pavlopoulos GA, Ivanova NN, Kyrpides NC. 2017. Nontargeted virus sequence discovery pipeline and virus clustering for metagenomic data. Nat Protoc 12:1673–1682. doi: 10.1038/nprot.2017.063. [DOI] [PubMed] [Google Scholar]
- 56.Ahlgren NA, Fuchsman CA, Rocap G, Fuhrman JA. 2019. Discovery of several novel, widespread, and ecologically distinct marine Thaumarchaeota viruses that encode amoC nitrification genes. ISME J 13:618–631. doi: 10.1038/s41396-018-0289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser L. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yutin N, Wolf YI, Raoult D, Koonin E. 2009. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol J 6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pearson WR. 2013. An introduction to sequence similarity (“homology”) searching. Curr Protoc Bioinformatics 42:3.1. 1–3.1. 8. doi: 10.1002/0471250953.bi0301s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paez-Espino D, Roux S, Chen I-MA, Palaniappan K, Ratner A, Chu K, Huntemann M, Reddy TBK, Pons JC, Llabrés M, Eloe-Fadrosh EA, Ivanova NN, Kyrpides NC. 2019. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res 47:D678–D686. doi: 10.1093/nar/gky1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brister JR, Ako-Adjei D, Bao Y, Blinkova O. 2015. NCBI viral genomes resource. Nucleic Acids Res 43:D571–D577. doi: 10.1093/nar/gku1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 66.Roux S, Enault F, Hurwitz BL, Sullivan MBJP. 2015. VirSorter: mining viral signal from microbial genomic data. PeerJ 3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ren J, Ahlgren NA, Lu YY, Fuhrman JA, Sun FJM. 2017. VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome 5:69. doi: 10.1186/s40168-017-0283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nayfach S, Camargo AP, Eloe-Fadrosh E, Roux S, Kyrpides N. 2020. CheckV: assessing the quality of metagenome-assembled viral genomes. BioRxiv doi: 10.1101/2020.05.06.081778. [DOI] [PMC free article] [PubMed]
- 69.Knowles B, Silveira CB, Bailey BA, Barott K, Cantu VA, Cobián-Güemes AG, Coutinho FH, Dinsdale EA, Felts B, Furby KA, George EE, Green KT, Gregoracci GB, Haas AF, Haggerty JM, Hester ER, Hisakawa N, Kelly LW, Lim YW, Little M, Luque A, McDole-Somera T, McNair K, de Oliveira LS, Quistad SD, Robinett NL, Sala E, Salamon P, Sanchez SE, Sandin S, Silva GGZ, Smith J, Sullivan C, Thompson C, Vermeij MJA, Youle M, Young C, Zgliczynski B, Brainard R, Edwards RA, Nulton J, Thompson F, Rohwer F. 2016. Lytic to temperate switching of viral communities. Nature 531:466–470. doi: 10.1038/nature17193. [DOI] [PubMed] [Google Scholar]
- 70.Coutinho FH, Silveira CB, Gregoracci GB, Thompson CC, Edwards RA, Brussaard CP, Dutilh BE, Thompson FL. 2017. Marine viruses discovered via metagenomics shed light on viral strategies throughout the oceans. Nat Commun 8:15955–15912. doi: 10.1038/ncomms15955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bin Jang H, Bolduc B, Zablocki O, Kuhn JH, Roux S, Adriaenssens EM, Brister JR, Kropinski AM, Krupovic M, Lavigne R, Turner D, Sullivan MB. 2019. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat Biotechnol 37:632–639. doi: 10.1038/s41587-019-0100-8. [DOI] [PubMed] [Google Scholar]
- 72.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 73.Nepusz T, Yu H, Paccanaro A. 2012. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods 9:471–472. doi: 10.1038/nmeth.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trifonov V, Rabadan R. 2010. Frequency analysis techniques for identification of viral genetic data. mBio 1:e00156-10. doi: 10.1128/mBio.00156-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kang DD, Froula J, Egan R, Wang Z. 2015. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deaton J, Yu F, Quake S. 2017. PhaMers identifies novel bacteriophage sequences from thermophilic hot springs. bioRxiv doi: 10.1101/169672. [DOI]
- 77.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Almende B, Thieurmel B, Robert T. 2016. visNetwork: network visualization using “vis. js” Library. R package version 1.0.3. https://CRAN.R-project.org/package=visNetwork. Accessed 14 November 2019.
- 79.Allaire J, Ellis P, Gandrud C, Kuo K, Lewis B, Owen J, Russell K, Rogers J, Sese C, Yetman C. 2017. Package ‘networkD3’. https://github.com/eliztang/networkD3. Accessed 14 November 2019..
- 80.Biswas A, Gagnon JN, Brouns SJ, Fineran PC, Brown CM. 2013. CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol 10:817–827. doi: 10.4161/rna.24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 82.Kuśmirek W, Nowak R. 2018. De novo assembly of bacterial genomes with repetitive DNA regions by dnaasm application. BMC Bioinformatics 19:273. doi: 10.1186/s12859-018-2281-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiang YN, Penadés JR, Chen J. 2019. Genetic transduction by phages and chromosomal islands: the new and noncanonical. PLoS Pathog 15:e1007878. doi: 10.1371/journal.ppat.1007878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prestat E, David MM, Hultman J, Taş N, Lamendella R, Dvornik J, Mackelprang R, Myrold DD, Jumpponen A, Tringe SG, Holman E, Mavromatis K, Jansson JK. 2014. FOAM (functional ontology assignments for metagenomes): a hidden Markov model (HMM) database with environmental focus. Nucleic Acids Res 42:e145. doi: 10.1093/nar/gku702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Berman HM, Bourne PE, Westbrook J, Zardecki C. 2003. The Protein Data Bank, p 394–410. In Protein structure. CRC Press, Boca Raton FL. [Google Scholar]
- 87.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 Web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Monzingo AF, Marcotte EM, Hart PJ, Robertus JD. 1996. Chitinases, chitosanases, and lysozymes can be divided into procaryotic and eucaryotic families sharing a conserved core. Nat Struct Biol 3:133–140. doi: 10.1038/nsb0296-133. [DOI] [PubMed] [Google Scholar]
- 89.Holm L, Sander C. 1994. Structural similarity of plant chitinase and lysozymes from animals and phage: an evolutionary connection. FEBS Lett 340:129–132. doi: 10.1016/0014-5793(94)80187-8. [DOI] [PubMed] [Google Scholar]
- 90.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Epskamp S, Epskamp MS, MplusAutomation S. 2019. Package ‘semPlot’. Path diagrams and visual analysis of various SEM packages’ output. https://cran.r-project.org/package=semPlot. Accessed 15 February 2021.
- 92.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. doi: 10.1039/C5AY02550H. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Soil chemical data for soils across a gradient of historical precipitation (Iowa [IA] > Kansas [KS] > Washington [WA]). Significant differences were determined by t test in the R package (“rstatix”; *, P < 0.05; **, P < 0.01). In each boxplot, the top and bottom of each box represent the 25th and 75th percentiles, respectively, and the central line shows the median. “OM” represents organic matter in the source soil. Download FIG S1, PDF file, 0.03 MB (35.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Prokaryotic community compositions for soils collected across a gradient of historical precipitation (Iowa [IA] > Kansas [KS] > Washington [WA]). The community compositions of the prokaryotic species are shown in the heatmap with log transformed read coverages that are color-coded (the warmer the color, the higher the coverage). The prokaryotic communities detected from the three soils were clustered by composition similarity. Download FIG S2, PDF file, 0.05 MB (48KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Visualization of 14 complete viral genomes assembled from soil metagenomes. The putative AMGs, hypothetical genes, and viral hallmark and nonhallmark genes are color-coded, and the arrangement of these genes is shown in linear and circular views of the 14 high-quality complete viral genomes. The genome length is illustrated by the scale of the backbone representing the viral genome in the linear view or specified in number of base pairs in the circular view. The order of the colored blocks represents the arrangement of the predicted genes with annotations. Download FIG S3, PDF file, 1.5 MB (1.5MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Viral host assignments and clustering information for each identified viral contig (summary table containing information on the following: assigned viral hosts and respective methods used for viral assignment, viral clusters and respective clustering method used, CRISPR spacer hit(s), length, and read coverage for each viral contig identified) Download Table S1, XLSX file, 0.8 MB (843.6KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
An interactive network for viral clustering. An interactive and reader-friendly clustering network that enables close exploration of each individual viral cluster. The interactive network contains the detected soil viral contigs and NCBI viruses and can be filtered based on the complexity or relative representation of the clusters (number of the edges). Download FIG S4, HTML file, 3.6 MB (3.6MB, html) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Visualization of viral contigs that were identified to contain AMGs. Linear view of the 67 viral contigs detected with at least one of the category 1 or 2 putative AMGs that are listed in Table S2. The genes predicted from the viral contigs are color-coded, with categories 1 and 2 AMGs in dark purple, categories 3 to 5 AMGs in light purple, hypothetical genes in grey, viral hallmark genes in yellow, and viral nonhallmark genes in orange. Download FIG S5, PDF file, 1.8 MB (1.8MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
94 putative AMGs detected from large (>1 Tb) soil metagenomesaaA list of putative AMGs identified on viral contigs (category 1 and 2 AMGs; a detailed description of AMG categories in “AMG classification”). The table contains information of full and abbreviated functional annotations (“Annotation” and “Gene identifier used for visualization”), length of the nucleotide sequences (“Length”), positions of the AMGs that were detected on the viral contigs (“Start” and “End”), sites where the AMGs were detected (“Sample”), statistics from the sequence homology analysis, including “E-value,” “bit score/coverage%,” and the genomic contexts of viral contigs with AMGs detected (“Gene Arrangement,” “AMG_index,” “AMG_upstream,” “‘AMG_downstream”) Table S2, XLSX file, 0.02 MB (25.9KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Statistical information for soil metagenomes and assembled complete viral genomesaaThe table includes numbers of quality reads and bases, numbers of assembled contigs and N50 value for the “large” (>1 Tb) metagenome for each grassland soil. The download link and DOI are available for large and “replicate” metagenomes and the complete viral genomes that were assembled from the large metagenomes Table S3, XLSX file, 0.01 MB (12KB, xlsx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
A list of Pfams that were used to annotate viral proteins. Download Table S4, TXT file, 0.4 MB (384.5KB, txt) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Network of viral clusters that was obtained using the tetranucleotide frequency method. This network was used as a proof-of-concept for applying tetranucleotide frequency as an approach to cluster related viral contigs. NCBI viruses and a subset of the viral contigs detected in this study were clustered using the tetranucleotide frequency-based method. The clustering of these viral sequences is visualized in the network with each node as a viral sequence and are colored by the types of the viruses or the sites where the viral sequences were detected. NCBI archaeal viruses are highlighted in red nodes, with NCBI bacterial viruses in green and fungal viruses in yellow. Viral sequences detected in Iowa, Kansas, and Washington grassland soils are colored in orange, blue, and purple, respectively. The viral clusters are generally grouped according to their taxonomic annotations, e.g., Sangervirae and Rudiviridae. Download FIG S6, PDF file, 2.9 MB (2.9MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
Raw reads and the assembled data of the large metagenomes (TmG.1.0) were packaged and previously published (19). Replicate metagenomes (WA-TmG.2.0, KS-TmG.2.0, IA-TmG.2.0) along with the 14 complete viral genomes assembled from the large metagenomes (Iso-VIG14.1.0) have been deposited in the PNNL Project DataHub repository with DOIs (https://data.pnl.gov/group/7/nodes/dataset/13708) and are available to download. The download link for each data set is included in Table S3. The detailed metadata are included in each data package. The R codes used in this study, including the details of graphical and statistical packages and the structural equation model, viral contigs screened from the metagenome assemblies and lysozyme HMMs, can be found at https://github.com/Ruonan0101/GrasslandVirome_PNNL.