ABSTRACT
Introduction
Human gut microbiota plays a crucial role in providing protective responses against pathogens, particularly by regulating immune system homeostasis. There is a reciprocal interaction between the gut and lung microbiota, called the gut-lung axis (GLA). Any alteration in the gut microbiota or their metabolites can cause immune dysregulation, which can impair the antiviral activity of the immune system against respiratory viruses such as severe acute respiratory syndrome coronavirus (SARS-CoV) and SARS-CoV-2.
Areas covered
This narrative review mainly outlines emerging data on the mechanisms underlying the interactions between the immune system and intestinal microbial dysbiosis, which is caused by an imbalance in the levels of essential metabolites. The authors will also discuss the role of probiotics in restoring the balance of the gut microbiota and modulation of cytokine storm.
Expert opinion
Microbiota-derived signals regulate the immune system and protect different tissues during severe viral respiratory infections. The GLA’s equilibration could help manage the mortality and morbidity rates associated with SARS-CoV-2 infection.
KEYWORDS: Gut microbiota, gut microbial-host-immune axis, gut-lung axis, coronavirus, Covid-19, respiratory complications
1. Introduction
Influenza and other respiratory virus infections are one of the leading causes of morbidity and mortality worldwide [1]. A competent immune system reduces mortality by protecting the host against viral infections and reducing susceptibility to secondary bacterial infections [2]. The World Health Organization (WHO) has stated that severe coronavirus disease 2019 (COVID-19) is a pandemic challenge [3]. COVID-19 is a lethal condition in patients with comorbidity, particularly those with immune system disorders. A healthy immune system can help people overcome clinical complications and return to work faster [4]. COVID-19 seems to cause a wide range of extrapulmonary difficulties affecting multiple organ systems. The pooled prevalence of all gastrointestinal (GI) problems was 17.6% in an early meta-analysis of 60 trials involving 4243 patients, most of whom were from China. The most prevalent presenting symptom was anorexia (26.8%), followed by diarrhea (12.5%), nausea/vomiting (10.2%), and abdominal pain (9.2%) [5]. Critically ill COVID-19 patients often develop GI complications during hospitalization, such as bowel ischemia, pancreatitis, GI bleeding, elevated transaminases, ogilvie syndrome, and severe ileus [6,7].
Gut microbiota is essential for the development of immune responses [8]. There are approximately 38 trillion microorganisms in our body, including bacteria, fungi, archaea, viruses, and other eukarya. Bacteria are more common among them, with at least 2,000 different species [9]. The gastrointestinal tract (GIT) is the main habitat of microbiota, since it provides the largest interface surface (250–400 m2) of the body and is nutrient-rich. The GIT comprises over 2,000 bacteria species from 12 distinct phyla. The microbiome contains 3 million genes that are 150 to 500 times more than the number of genes in the human genome [10]. The gut microbiota can affect food digestion, production of energy and vitamins (B1, B5, B12, K, and folic acid), biliary acids deconjugation, and other vital biochemical aspects of our life [11,12]. On the other hand, microbiota has a remarkable impact on the activation and function of immune cells. Accordingly, a persistent microbial community imbalance in the gut is known as dysbiosis [13], increasing the risk of infections and autoimmune diseases [14–19].
Many studies have recently shown a regulatory association between the gut microbiota and other organs, such as the gut–lung axis, gut–brain axis [20], gut–liver axis [21], gut-host hormonal axis [22], and gut–skin axis [23]. Noticeably, cross-talk between the healthy gut microbiota and lungs can significantly prevent respiratory tract infection in both influenza A virus and COVID-19 [24,25]. Based on these interactions, any alterations in the gut microbiota may directly affect the function of other organs. Furthermore, more than 50% of patients with inflammatory bowel disease (IBD) and 33% of patients with irritable bowel syndrome (IBS) are susceptible to respiratory complications as a result of dysbiosis without a history of acute or chronic respiratory disease [26,27].
The interaction of the gut microbiome and immune system [10] plays a protective role in preventing severe infection and controlling the viral load. The immune system is naïve in newborns and inefficient in older adults. Both groups are susceptible to viral infections and have a high mortality rate [28,29]. Recent molecular studies have also revealed differences in gut microbiota composition among infants, toddlers, adults, and the elderly [30]. It appears that an altered gut microbiota composition can make an infant more susceptible to infections and allergic disease [31]. Dysbiosis, or disruptions in the hemostasis of gut microbiota, may cause an uncontrolled and intense immune response to viral infections. Dysbiosis occurs when there is disparity in the number of normal bacteria and a rise in commensal, and/or opportunistic gut bacteria. Moreover, eubiosis is characterized by a species diversity dominated by members of only four bacterial phyla, including Firmicutes (e.g. Lactobacillus and Clostridium), Bacteroidetes (e.g. Bacteroides), Proteobacteria (e.g. Escherichia), and Actinobacteria (e.g. Bifidobacterium) [32,33]. Antibiotics, high-fat diet, stress, and air pollution have all been linked to dysbiosis, which has been shown to affect other body systems [17,34]. During COVID-19 infection, microbiota dysbiosis such as Klebsiella oxytoca, Lactic Acid Bacteria, and Tobacco mosaic virus were discovered [35]. In addition, a study found a link between the severity of COVID-19 infection and the prevalence of Clostridium ramosum, Coprobacillus, Clostridium hathewayi, Faecalibacterium prausnitzii, and Alistipes onderdonkii [36].
The present narrative review discusses the interaction between the healthy microbiome and the immune system during the initiation and progression of COVID-19. Finally, we overview the protective role of probiotics in the current COVID-19 pandemic.
2. Gut microbiota regulates the immune system
The number of microorganisms in the GI tract is reported to be more than 1014. The total number of microbes encompasses about 10 times more than human cells [37]. Bacteria that inhabit the microbiota of the human gut are classified into phyla, classes, orders, families, genera, and species. Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia are the significant gut microbial phyla [38,39]. Recent research has discovered that species and strains of microorganisms can control the host defense against different pathogens. According to the rodent studies, Bacteroides fragilis strain No. 9343 protects against colitis, while strain 86–5443-2-2 induces colitis [17,40]. Clostridial cluster IV and XIVa have also been shown to cause colonic Treg cells [41].
On the other hand, the density of DNA and RNA viruses that comprise the intestinal virome is equal to that of bacterial cells. At the same time, they may exceed bacterial cells by a 20:1 ratio on gut mucosal surfaces and within mucus layers [42]. Through inducing immune system tolerance, the gut virome maintains and develops diversity in the human gut microbiota in a dynamic equilibrium with the host. The regulation of human metabolism, inflammation, and carcinogenesis are all influenced by gut virome. Recent evidence showed that Bacteriophages play a role in human liver metabolism and immune response modulation [43]. Developing an immunoregulatory network that protects against induction of mucosal IgE, which is linked to allergy susceptibility, is also dependent on intestinal microbial diversity during early-life colonization [44].
The gut microbiota, including bacteria, archaea, and eukarya, colonize within the GI tract and co-evolve with the host over thousands of years to develop an intricate and mutually beneficial interaction. The gut microbiota influences the host immune system’s induction, education, and function [45,46]. On the other hand, the immune system has largely evolved to maintain the host’s symbiotic relationship with these diverse and evolving microorganisms. This immune system-microbiota alliance interweaves the innate and adaptive arms of immunity in a dialog that selects, calibrates, and terminates responses. This interaction also enables the maintenance of regulatory pathways to maintain tolerance to innocuous antigens [45–48]. Gut microbiota may modulate systemic immunity by regulating innate and adaptive immune homeostasis by developing metabolites and antimicrobial peptides against various pathogens [10,49,50]. Several studies showed a cross-talk between the gut microbiota and antigen-presenting cells (APCs) and neutrophil regulation, besides forming CD4 + T cells both within and outside the intestine [41,44,51–54]. A shift in the gut microbiota’s composition can have either a pathological or beneficial effects, mediated by the gut microbiota’s regulation of specific CD4 + T cell subtypes [55]. Dysbiosis is linked to dysimmunity, commonly described as T-helper 2 (Th2)-overactived and Treg-deficient state [19]. The gut microbiome’s influence on the T cell compartment of the adaptive immune system is the best-understood mechanism by which the gut microbiota influences the systemic immunological response [46]. The GIT microbiota has been found to influence T cell development into Th1, Th2, and Th17 cells, as well as regulatory T cells [47]. It has been reported that segmented filamentous bacteria (SFBs) directly regulate the secretion of various antibacterial peptides leading to altered secretion of IL-17 by Th17 cells [56,57]. The number of Th17 cells in the lamina propria and the production of IgA in the intestinal mucosa were found to be reduced by SFBs [58,59].
In response to the microbiota signal, secretory IgA (sIgA), IL-12, IL-22, and IFN-α are secreted. sIgA regulates local immunity, viral infection colonization, and proliferation [60]. Short-chain fatty acids (SCFAs) are one of the common metabolites produced by the gut microbiota and rapidly absorbed by intestinal epithelial cells (IECs) [61]. SCFAs regulate immune cell bioactivity, such as glucose homeostasis, gene expression profile, anticancer activity, and inflammatory response. SCFAs can also serve as a source of energy for T cell differentiation (Th1, Th2, Th17, and T regulatory (Treg) cells). Interestingly, SCFAs can induce tolerance in dendritic cells (DCs) which contributes to the differentiation of naïve CD4 + T cells into Treg cells [62]. Common SCFAs include propionate (form Bacteroidetes), butyrate (form Firmicutes), and acetate (from most gut anaerobes) [60,63]. Butyrate enhances the differentiation of Treg cells and prevents the development of systemic inflammation [48]. The potential of microbiota-released signaling molecules to enter the circulation also enables resident bacteria in the gut to affect the immune system during immune cell formation in hematopoiesis, and hence alter infection response [64]. Moreover, the SCFA butyrate induces bone marrow monocyte differentiation from an inflammatory to a more tolerogenic phenotype [48]. A decrease in butyrate-producing bacteria and gut dysbiosis, in particular, resulted in reduced IL-22 production, which is necessary for gut and lung epithelial barrier integrity [65]. Acetate is the most abundant SCFA produced in high quantities by Bifidobacteria, and is found in the gut lumen and peripheral circulation [66]. Acetate modulates intestinal inflammation by activating G protein-coupled receptor 3 (GPR43) [67], thereby contributing to the maintenance of gut epithelial barrier function [68]. Acetate also has anti-inflammatory characteristics in neutrophils, decreasing NF-kB activation by suppressing the levels of pro-inflammatory mediators such as lipopolysaccharide-induced TNF-α [66].
The mucus in the intestines and lungs is considered the first defense line against pathogen colonization [69]. The luminal layer of the gut mucosa is a suitable site for the colonization of commensal bacteria. Furthermore, the gut microbiota regulates the integrity of the mucosal barrier, provides bacterial metabolites [37,70], and regulates the immunoregulatory functions of IECs by modulating the expression of antimicrobial factors [71,72]. As one of the critical bacterial metabolites, butyrate regulates transepithelial fluid transport, reduces mucosal inflammation and oxidative stress, improves the epithelial defense barrier, and controls visceral sensitivity and intestinal motility [73]. Moreover, butyrate provides a critical line of immunologic defense in the intestine via induction of DCs tolerance in the epithelium, which is associated with mucosal IgA production. Butyrate also suppresses mast cell degranulation in the intestinal mucosa and limits circulating inflammatory mediators’ production [74]. So, the absence of the gut mucosa reduces the commensal bacteria adhesion and IECs integrity and result in direct contact with bacteria which eventually causes inflammation in the gut [75]. On the other hand, SCFAs induce the expression of pattern recognition receptors (PPRs) on DCs and macrophages, which regulate cytokine secretion and antibody synthesis (sIgA and IgM) [76].
The balanced production of antiviral cytokines is associated with specific GIT bacteria, including Faecalibacterium, Oscillibacter, Pseudoflavonifractor, Anaerotruncus, and Bifidobacterium. It has been reported that antibiotic administration in chickens resulted in higher susceptibility to the avian influenza virus (AIV) and reduced expression of IFN-α, IFN-β, and IL-22 levels [77]. The synthesis of microbial peptides in the gut is regulated by the key active components of innate immunity such as α-defensin (DEFA) and C-type lectins (REG3g and REG3b) [60]. Menendez et al. showed that germ-free mice had a significant decrease in the expression of DEFA [78]. Toll-like receptors (TLRs) and NOD-like receptors (NLRs) on DCs and macrophages help the immune system to distinguish between useful and pathogenic bacteria [79]. IECs also express these transmembrane receptors, which are involved in the recognition of different microbe-associated molecular patterns (MAMPs) e.g. peptidoglycan, capsular polysaccharides, lipopolysaccharides (LPS), and flagellin and bacterial DNA CpG motifs [80].
Gut-associated lymphoid tissue (GALT), composed of lamina propria, intraepithelial lymphocytes, mesenteric lymph nodes, Peyer patches, and isolated lymphoid follicles, helps defend against external factors that penetrate the luminal mechanical barrier [38]. Experimental evidence has established the role and function of gut microbiota in GALT regulation [81,82]. Accordingly, germ-free mice developed hypoplastic Peyer’s patches/mesenteric lymph nodes in the small intestine, but no isolated lymphoid follicles (ILFs) [83]. The first colonization of gut microorganisms in full-term infants depends on GALT maturation and GI-Blood Barrier critical closure. These interactions result in the formation of symbiotic conditions, which are defined as a balance between immunity and infections [84].
During early life, critical host immune-microbiota interactions occur, which may have long-term effects on multiple immune arms that contribute to immune homeostasis and sensitivity to infectious and inflammatory diseases later. However, the mechanisms underlying these interactions are remaining unclear. In this regard, future studies are needed to determine the long-term effects of mild dysbiosis states during the neonatal period on adult immunity and the risk of immune-mediated diseases [44].
3. Gut microbiota–lung axis
A healthy lung contains 10–100 bacteria per 1,000 cells [85]. Bacteroidetes, Firmicutes, and Proteobacteria are the most common phyla colonizing the healthy lung [86]. There is a constant cross-talk between the gut and lung mucosal compartments through the mesenteric lymphatic system and lung lymph nodes. Physical interaction, quorum-sensing molecules, and synthesis of antimicrobial agents could all be involved in these interactions, which could modulate the lung’s immune response [87] .Changes in the microbial composition of the lung can influence the community of gut microbiota and vice versa. Due to possible barrier dysfunction, bacterial translocation from the gut to the lungs has been documented in sepsis and acute respiratory distress syndrome [88]. The gut and respiratory tract have been linked to control immunological responses, and dysbiosis in the gut microbiota leads to development of respiratory disease [89]. Mechanisms involved in microbiota-lung-gut-axis alteration in COVID-19 include direct lung damage, ACE2 expression, gut microbiota as lungs’ defense against SARS-CoV-2, and immune response [90]. In 2012, researchers demonstrated that colonization of the influenza virus in the respiratory tract of mice causes intestinal dysbiosis by increasing Enterobacteriaceae, while reducing Lactobacilli and Lactococci [91]. In consistent with these findings, IBD and IBS patients with many lung disorders, including asthma, chronic obstructive pulmonary disease (COPD), influenza virus, and coronavirus, exhibited dysbiosis in their airway microbiota [92–94]. As illustrated in Figure 1,Figure 2 the commensal intestinal microbiota plays an essential role in regulating the immune response against respiratory virus infections such as influenza and coronavirus [8,95].
Figure 1.
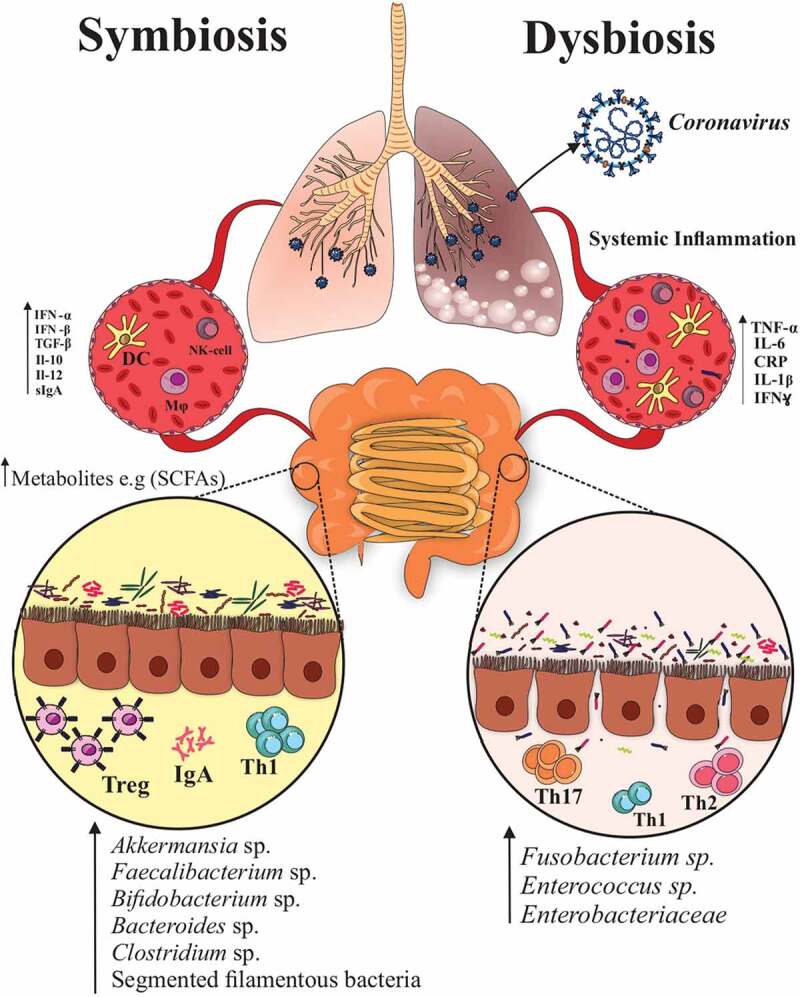
The gut-lung axis plays a critical role in the control of SARS-CoV-2 virulence. The gut microbiota regulates the innate and adaptive immunity by producing bacterial metabolites (SCFAs) and antimicrobial peptides against different pathogens. In addition, they regulate the integrity of the mucosal barrier and immune homeostasis. Dysbiosis can make negative impact on the balance and recruitment of immune cells in the lungs and increase inflammatory cytokines such as IL-6, TNF-α, and IL-1β, which could be the most important predisposing factor for sever COVID-19 infection
Figure 2.
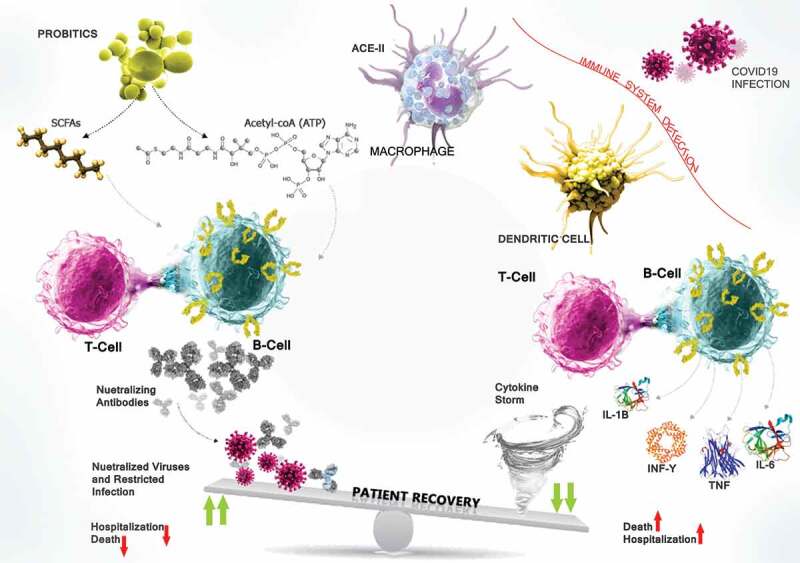
The interaction between the SARS-CoV-2 spike protein and ACE2 on the DCs and MQs contributes to the pathogenicity of COVID-19. Most critically ill patients in ICU, who are suffering from ARDS, have high levels of inflammatory cytokines owing to the complex immune dysregulation. Thus, the NAbs can potentially block the interaction between the SARS-CoV-2 spike protein and ACE2 on the cell membrane and thus prevent the entry of the virus and can control viral load. There are supporting evidence suggesting that probiotics affect pulmonary health through gut-lung cross-talk. MAMPs are the microbiota-derived products/metabolites, such as SCFAs that have increased cellular levels of acetyl-CoA, ATP, and lipid biogenesis to drive plasma B cell’s differentiation and induce IgAs and NAbs secration against viral respiratory infections particularly SARS-CoV-2 and modulate immune reactions
Moreover, alterations of the lung microbiota following infection can cause intestinal complications. Accordingly, P. aeruginosa pneumonia can reduce IECs proliferation and improve mucosal repair [96,97]. According to a meta-analysis, GI symptoms such as nausea, vomiting, diarrhea, and abdominal pain were seen in 17.6% of SARS-CoV-2 infected patients, and they were more common in severe cases [5]. These GI signs could be caused by SARS-CoV-2 directly infecting enterocytes through the GLA, or by immunoregulatory mechanisms [98]. On the other hand, dysbiosis in the gut microbiota negatively impacts on the balance and recruitment of immune cells in the lungs, which could be the most important predisposing factor in the development of respiratory tract infections [99]. In addition, this immune cell imbalance in the lungs reduces a load of certain beneficial bacteria (Bifidobacteria, Akkermansia, Faecalibacterium, Lachnospira, and Veillonella), which has been linked to an increased risk of asthma in neonates [100,101]. Table 1 summarizes the association between chronic respiratory diseases and changes in the microbial composition of the gut or respiratory tract.
Table 1.
The association between respiratory chronic diseases and alterations of the intestinal and respiratory tract microbial composition
| Type of Disease/Infection | Alteration of gut bacteria |
Subject | Reference | ||
|---|---|---|---|---|---|
| Decreased | Increased | ||||
| Asthma (in children) |
Akkermansia muciniphila, Faecalibacterium prausnitzii Bifidobacteria |
Clostridia | Human | [102] | |
| [103] | |||||
| Asthma (in adults) |
Bacteroidetes (specifically), Prevotella spp. |
Proteobacteria (Haemophilus, Moraxella, Neisseria spp.) Firmicutes (Lactobacillus spp.) | Human | [85,104] | |
| Chronic Obstructive Pulmonary Disease (COPD) | Enterobacter cloacae, Citrobacter, Eggerthella, Pseudomonas, Anaerococcus, Proteus, Clostridium difficile, Salmonella | Human | [105] | ||
| Cystic fibrosis (in children) |
Bacteroides vulgatus Bacteroides uniformis Firmicutes Faecalibacterium prausnitzii, Bifidobacterium adolescentis Bifidobacterium catenulatum, Eubacterium rectale | Streptococcus, Staphylococcus, Veillonella dispar, Clostridium difficile, Pseudomonas aeruginosa, Escherichia coli | Human | [106] | |
| Cystic fibrosis (in adults) |
Bacteroidetes, Faecalibacterium Prausnitzii | Firmicutes, Ruminococcus gnavus, Enterobacteriaceae, Clostridia species Proteobacteria phylum (Pseudomonas, Haemophilus, Burkholderia), Actinobacteria phylum | Human | [107–111] | |
| Lung cancer | Bifidobacterium sp., Actinobacteria sp. Dialister, Enterobacter, Escherichia–Shigella, Fecalibacterium, and Kluyvera | Enterococcus sp. Veillonella, Bacteroides, Fusobacterium | Human | [112,124] | |
| Type of Disease/Infection | Alteration of respiratory tract bacteria | Subject | Reference | ||
| Decreased | Increased | ||||
| Coronaviruses and Adenoviruses infection | M. catarrhalis | Human | [113] | ||
| COPD and Rhinovirus infection | Proteobacteria phylum | Human | [130] | ||
| Rhinovirus infection | H.parainfluenzae, Neisseria subflava, S. aureus | Human | [114] | ||
| Rhinovirus infection (in children) |
S. pneumoniae | Human | [115] | ||
| COPD and hRV infection | H. influenza | Human | [116] | ||
| hRV infection | Haemophilus and Neisseria spp. |
Propionibacterium S. pneumonia H. influenza |
Human | [113,117] | |
| Enteroviruses | S. pneumoniae | Human | [113] | ||
| Influenza (in children) |
S. aureus | Prevotella, Streptobacillus, Porphyromonas, Granulicatella, Veillonella, Fusobacterium, Haemophilus. | Human | [118] | |
| Influenza A H1N1 infection | Firmicutes (Staphylococcus and Streptococcus spp.), Proteobacteria (Pseudomonas amygdali, P. fluorescens, Pseudomonas sp. UK4, Acinetobacter baumanii and A. junii) Moraxella and Enterobacter spp. | Human | [119] | ||
| Haemophilus influenza infection | Neisseria | Human | [120] | ||
| Influenza, Parainfluenza, Rhinovirus, Respiratory Syncytial Virus (RSV), Coronavirus, Adenovirus, Metapneumovirus |
Haemophilus and Moraxella |
Human | [120] | ||
| Pseudomonas aeruginosa infection | Prevotella and Flavobacterium | Human | [120] | ||
| H7N9 virus infection | Faecalibacterium | Human | [121] | ||
| Respiratory Syncytial Virus (RSV) | H.influenzae | Human | [113] | ||
| Tuberculosis infection | Roseburia, Faecalibacterium, Phascolarctobacterium, Eubacterium | Human | [122] | ||
| Recurrent tuberculosis | Bacteroidetes, Prevotella and Lachnospira | Proteobacteria, Actinobacteria | Human | [123] | |
Metabolites of gut bacteria can reach other organs via the bloodstream and regulate immune responses at distal mucosal sites such as the lungs. This control can be seen in the induction of antibody synthesis and anti-inflammatory responses [99,124]. Microbiota and viruses interact in both direct and indirect ways [125]. Microbiota-produced lipopolysaccharides (LPS) and peptidoglycans compete with viruses to bind to cell surface receptors, such as TLRs.COVID-19 infection symptoms are found to be more severe in the absence of LPS. TLR4 is one of the leading players of the immune system against COVID-19 infection. Indirectly, microbiota can regulate the secretion of IFN-α and antimicrobial peptides, which are essential in preventing virus-host cell fusion. Therefore, they can prevent viral colonization using the S protein [126–128]. According to the experimental studies in a mouse model, dysbiosis in the lung impacts the immune system, reducing the recruitment of APCs to the lungs and phagocytic activity. In addition, less intensive recruitment of immune cells results in an increase in viral load in the lungs [129,130] and a decrease in IFN-α and -β production which can negatively impact T cell priming [130]. It was shown that active resident memory B cells in the lungs also required encountering lung microbiota [131]. This interaction is reciprocal, and any changes in the gut microbiota directly affect the host’s defense against acute respiratory pathogens as well as the risk of death [87].
4. Gut microbiota and immune system in COVID-19
Changes in the gut microbiota increase the risk of infectious, inflammatory, and endocrine diseases. Since there is no approved treatment or vaccine for many respiratory viral infections [132], a healthy immune system can play a key role in reducing the severity of symptoms and mortality. SARS-CoV belongs to the subfamily of Orthocoronavirinae in the family of Coronaviridae. The enveloped and crown-like SARS-CoV virus is the second-largest genome size (27–32 kb) and single-strand positive-sense RNA (+ssRNA) of all RNA viruses (27–32 kb). Lv et al. reported an imbalance of fungal and bacterial flora in COVID-19 patients [133]. A study of the diverse array of fungal species, which refers to the diverse array of fungal species the gut mycobiota [134] showed that the most common symptom of fungal mycobiota dysbiosis in COVID-19-infected patients was the depletion of Aspergillus and Penicillium. Interestingly, the gut mycobiota profiles of COVID-19 patients with mild and severe symptoms were similar. In COVID-19 patients, the gut virome, especially in those with GI infections, can be very significant [135]. COVID-19 patients had significant differences in their fecal mycobiome at the time of hospitalization compared to controls, including Candida albicans enrichment and a highly heterogeneous mycobiome composition that lasted up to 12 days after nasopharyngeal clearance of SARS-CoV-2, according to a recent report [136]. Fecal metabolomic analysis in COVID-19 patients has revealed possible amino acid-related pathways that relate gut microbiota to inflammation [137]. In patients with COVID-19, Zuo et al. studied the temporal transcriptional activity of SARS-CoV-2 and its relationship with longitudinal fecal microbiota modifications. They showed that SARS-CoV-2 RNA was found in 46.7% of stool samples of patients without GI symptoms. In fecal samples with high SARS-CoV-2 active viral transcription and replication in vitro (infectivity), the numbers of particular opportunistic pathogens, such as Collinsella aerofaciens and Morganella morganii spp. were higher than in fecal samples with low or no SARS-CoV-2 infectivity [138]. In a previous investigation, qRT-PCR revealed that 14 of the 15 patients with COVID-19 (93%) tested positive for SARS-CoV-2 in fecal samples. By contrast, stool viral RNA metagenomics revealed that seven (47%) of them were positive for SARS-CoV-2 [136]. Fecal viral RNA metagenomics only detected positive samples with an abundance of >3.2 × 104 copies per mL inoculum, as determined by qRT-PCR [138]. In COVID-19 patients with moderate fever, Zhou et al. evaluated the correlation between gut microbiota dysbiosis and abnormal immunological responses. Patients with fever had a significant different gut microbiota composition than those who did not have a fever. Patients with fever had higher opportunistic infections, including Enterococcus faecalis and Saccharomyces cerevisiae [139]. In the cross-sectional study conducted by Gu et al., it was discovered that the microbiological profile of COVID-19 patients differed from that of patients with influenza A and healthy controls. They reported that COVID-19 patients had a lower microbial diversity, a higher concentration of opportunistic bacteria (Rothia, Streptococcus, Actinomyces, and Veillonella), and a lower proportion of beneficial microbes [140].COVID-19 severity is thought to be caused by a cytokine storm [36], which is linked to the gut bacterial population. Several immunomodulatory gut commensals, including Faecalibacterium prausnitzii, Eubacterium rectale, and bifidobacteria were reduced in COVID-19 patients [141]. Moreover, butyrate-producing bacteria including Faecalibacterium prausnitzii, Clostridium butyricum, Clostridium leptum, and Eubacterium were found to be in lower abundance [142]. Beneficial commensals including Eubacterium ventriosum, Faecalibacterium prausnitzii, Lachnospiraceae taxa, Roseburia, and Bacteriodes spp. like B. dorei, B. massiliensis, B. ovatus, and B. thetaiotaomicron, as well as Bacteriodes spp. like B. dorei, B. massiliensis, B. ovatus were diminished in COVID-19 cases, which correlated with the severity of the disease [143]. The previous studies have indicated that microbiota-derived SCFAs play an important role in promoting beneficial IL-18 and IL-22 secretion [144]. Moreover, Proteobacteria regulate the strong natural antiviral activity of IFN-α secretion [145]. The major phyla in the healthy gut and lungs are Bacteroidetes and Firmicutes. According to Yasui et al. feeding Lactobacillus casei to BALB/c mouse pups before inoculation with influenza virus decreases viral titers in the nasal lavage fluid and increases the activity of pulmonary natural killer (NK) cells. They demonstrated that the survival rate was nearly tripled under this type of regime [146]. Nasal administration of Lactobacillus rhamnosus also induced protection against respiratory syncytial virus infection in mice [147]. The SARS-CoV entry by TLRs (TLR3 or TLR7) results in airway inflammation [76]. Similarly, oral administration of immunobiotic L.rhamnosus in BALB/c mice was found to modulate antiviral immunity in the respiratory system. Activation of TLR3 and NK cells stimulate DCs to produce IL-12 as a critical factor in the development of effector CD4 Th1 and cytotoxic TCD8 lymphocyte responses [148,149].
Through the immediate response of the innate immune system to the coronavirus infection, macrophages and DCs regulate the virulence and disease outcomes. Furthermore, macrophages and DCs upregulate IFN-α production [76,150]. Some studies have shown that SARS-CoV inhibits the production of IFN-α and IFN-β [76]. Cinatl et al. showed that SARS-CoV could suppress the secretion of IFN-α, IFN-β, IL-18, and macrophage inhibitory factor (MIF), and downregulate the expression of antiviral genes (MIF). In addition, SARS-CoV could upregulate chemoattractant cytokines (CXC chemokines), IL-8, and oligoadenylate synthetase 2 (OAS2) in two intestinal cell lines, Caco2 and CL-14 [151]. In another study, it has been demonstrated that SARS-CoV infection downregulates the expression of IFN-α, IFN-β, as well as NFkβ, and also reduces the p56 level in Caco2 and 293 cell lines [152]. If the first line of immune response fails in eliminating the viral pathogens, the adaptive immune response is activated seven days following the SARS-CoV infection and this gap is a golden time for virus replication. The Angiotensin II converting enzyme (ACE2) is an essential receptor for infecting human cells by coronaviruses [153]. Fischer and colleagues have shown that gut microbiota and a healthy diet directly regulate the expression of ACE2 and the activation of the adaptive immune response following viral infection [154]. In this regard, the gut microbiota via its metabolite signals can regulate the immune response, reduce viral replication, and increase the survival rate during an epidemic and pandemic of viral infection [124].
5. Probiotics and a diet, modulation of severe complications in COVID-19
Although the evidence suggests that the gut contains a remarkable number of immune cells [155], the gut microbiome plays a crucial role in protecting the host against pathogens. Dysbiosis leads to break tolerance to harmless bacteria, inefficient immune response, and susceptibility to viral infections. Probiotics, known as beneficial live microorganisms, regulate dysbiosis and restore eubiosis [156]. Lactic acid bacteria (LAB) (Lactobacillus and Bifidobacterium spp) are the main probiotics species [157] that are well-known to the immune system and can regulate immune responses [158]. Lactobacillus spp acting via different pathways has antiviral features and could be considered a potent inducer of IFN-α and IFN-β production. Mullish et al. showed evidence for probiotics’ possible effect in reducing viral upper respiratory tract infections (URTI) and maybe COVID-19, especially in overweight/obese adults. They discovered that probiotic supplementation could reduce URTI symptoms by 27%, with adults ≥ 45 years old and/or with a BMI ≥ 30 kg/m2, obtaining the significant benefit of risk reduction [159]. In one early trial, probiotic supplementation to 28 hospitalized COVID-19 patients resulted in an eightfold decreased rate of respiratory failure compared to 42 patients receiving only medical treatment [160]. Probiotics treatment (Bacillus subtilis, Enterococcus faecalis, and Lactobacillus rhamnosus GG) was also found to lower ventilator dependency in COVID-19 patients who were severely ill compared to placebo. The mechanisms of probiotics’ antiviral effects are still unclear [98]. Prebiotics are substrates that host certain microorganisms and are used to provide a health benefit such as fructans, galactomannan, oligosaccharides, arabinooligosaccharides, lactosucrose, acid lactobionic, and polyunsaturated fatty acids [161]. Prebiotics can modulate the composition and function of the human microbiota, inducing immune system activation and pathogen inhibition in the GIT. To date, however, there is no evidence that prebiotics are associated with COVID-19 infections in any way [162].
Fecal microbiota L. gasseri SBT2055 (LG2055) is resistant to bile acid and can be colonized in the human gut [163]. The most current data support the regulatory role of LG2055 in the IgA secretion, which inhibits the influenza A virus infection in mice [164–166]. Eguchi and colleagues showed the antiviral activity of L. gasseri SBT2055 in both in vivo and in vitro models. These bacteria reduced viral load in the lungs and suppressed pro-inflammatory cytokine production [167]. Recently, it has been demonstrated that the cell walls of L. acidophilus NCFM and live Lactobacillus spp enhanced the immune system’s response to viral respiratory infection. Heat-killed L. casei [168], L. pentosus [169], L. plantarum [170], L. reuteri [170], and L. rhamnosus CRL1505 [171] participated in the regulation of the immune response in the respiratory tract and reduced lung damage after influenza and respiratory syncytial virus infections (RSV). The direct competition in binding to TLR3 between viral particles and local bacteria present in the lung resulted in decreased virus colonization in mice [172]. Bifidobacterium, the other group of LAB, regulates lung inflammation and infection. Supplementation with B. breve and B. longum (BL) improved the survival rate, restored lung injury, and eventually restored intestinal homeostasis [173–175]. Treatment with a Bifidobacterium probiotic can modulate pro-inflammatory cytokine levels and increase IL-10 and Foxp3+ Treg expression in the lungs. Animal studies have demonstrated that oral administration of B. longum BB536 reduces the incidence of influenza and fever, and can also regulate proliferation of the influenza virus in the lungs [176].
In comparison with oral treatment, nasal administration of L. rhamnosus CRL1505 tends to be a more effective way to significantly activate the Th1 response and CD103+c DCs [177]. Likewise, the nasal administration of Lactobacillus leads to faster colonization on the respiratory epithelium than oral way. The nasal administration is highly effective in suppressing the virus-induced inflammation and also diminishes the expression of inflammatory cytokines and chemokines (CXCL10, CXCL1, CCL2, TNF, IL-6) that damage lung tissue [170]. The effects of probiotics on the regulation of the immune system in infectious respiratory diseases have been summarized in Table 2.
Table 2.
The effects of probiotics on the immune system regulation in infectious respiratory diseases
| Probiotic | Efficacy | Subjects | Administration Rout |
Reference |
|---|---|---|---|---|
| L. acidophilus | Induced secretion of IFN‐α, IFN-β, and IL-12 from DCs via TLR2 and TLR-3 | Mice | Oral | [146] |
| L. rhamnosus CRL1506 | Production of IFN-α and IL-6 | Mice | Oral | [51] |
| L. casei Shirota (LcS) | Activated Th1 immunity, phagocytic activity, NK cells activity, and production of mucosal IgA | Mice/Elderly people | Oral/ Intranasal | [178,179] |
| [168] | ||||
| L.delbrueckii ssp. bulgaricus OLL1073R-1 | Increased NK cells activity | Elderly people | Oral | [180] |
| L. gasseri SBT2055 (LG2055) | Induced IFN-α & IFN-β production | Mice | Oral | [150] |
| Heat-killed Lactobacillus plantarum L-137 | Increased phagocytic activity and NK cells activity, acquired immunity, proliferative response of T cells, and increased number of T cells | mice/ Healthy adults | Oral | [181,182] |
| Heat-killed L. pentosus | Modulation of Th1/Th2, enhanced NK cells activity, induced production of both IL-12 and IL-10 | Mice | Oral | [169] |
| Heat-killed L. plantarum | Stimulation of IFN-β and IL-10 production | Mice | Intranasal | [170] |
| L. reuteri | Stimulation of IFN-α production | Mice | Nasal | [171] |
| L. rhamnosus CRL1505 | Th1 response significantly activated CD103+c DCs | Mice | Oral | [154] |
|
L. Casei Shirota (LcS) |
Activation of Th1 immunity | Mice | Intranasal | [168] |
| L. fermentum | Activation of macrophages | Mice | Intranasal | [113] |
| L. pentosus S-PT84 | Enhanced splenic NK cells, modulating the Th1/Th2 balance | Mice | Intranasal | [183] |
| L. johnsonii | Reduction in the total number and proportion of activated CD11c+/CD11b+ CD11c+/CD8+ cells, reduced expression of airway Th2 cytokine | Mice | Intranasal | [184] |
| B. breve YIT4064 | Enhanced IgG against IFV | Mice | Oral | [185] |
| B. longum BB536 | Reduced the incidence of IFV and fever effect on host cellular immunity, enhanced production of Th1 cytokines, declined plasma IFN-Ƴ levels, enhanced NK cell activity and neutrophil bactericidal activity. | Mice | Oral | [171] |
| B. longum MM-2 | Anti-viral effect, enhanced activity of NK cells in the lungs and spleen, Increased expression of IFN-α and Th1-related cytokines, | Mice/healthy adults | Oral | [171] |
Absorptive trapping and the development of lipopolysaccharides, which binds to and destabilizes the viral structure, are two mechanisms by which gut commensal bacteria (including those often found in probiotics) suppress viruses that enter the host via the upper respiratory tract [186]. The gut-lymph hypothesis, which describes gut bacteria within the draining chyle from the lower GIT entering the lymphatic system, is also one possible route by which probiotic bacteria may translocate from the gut to the lung [187]. Additional indirect pathways include modulating interferon responses in the lung stromal cells [188], promotion of APCs migration, and T cell antiviral responses mediated by TLRs [129].
Modifying nutrient availability via diet may be another effective microbiome-modulating strategy. A healthy diet is even more critical due to its effect on the microbial metabolome and richness of the microbiota diversity [189]. Therefore, alteration of the gut microbiota can increase the production of SCFAs via diet modification. Interestingly, adding indigestible carbohydrates increased butyrate production, which is linked to improved lung function. Also, fiber-rich diets can modulate innate immunity and reduce the GI signs and mortality rate from respiratory diseases [190,191]. Collectively, probiotics and Mediterranean lifestyle are protective, increase survival rate, and protect lung tissue against the destructive effects of respiratory viruses (SARS-COV2, IFV, RSV, etc.) and could be considered an alternative therapy for improving immune response.
6. Fecal microbiota transplantation in COVID-19 patients
Fecal microbiota transplantation (FMT) is a novel treatment that has shown to be helpful in the treatment of recurrent Clostridioides difficile infections (rCDI). FMT has the potential to help with a variety of dysbiosis-related disorders. It refers to transferring a healthy person’s distal gut microbia communities to another patient’s intestinal tract. This treatment has been proven to restore a disrupted microbial diversity and related microbial functional networks, with a success rate of 90% for rCDI patients [192]. According to Bradley et al., decreased intestinal microbiota following antibiotic administration could cause a shift in the interferon signature triggered by commensals in the lung epithelium, promoting early influenza virus proliferation in the respiratory tract [188]. By monitoring COVID-19 patients, Liu et al. revealed persistent alteration in the fecal microbiome composition after recovery and discharge from hospital [193]. They reported that Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria had relative abundances of 41.0%, 42.9%, 9.2%, and 4.0%, respectively, which differed from the general population. In healthy people, the dominant phyla are Firmicutes (60%) and Bacteroidetes (20%) [194]. Multiple research has shown that the FMT can promote immune function and consequently have an indirect protective effect against influenza virus infection [188]. Tiffany et al., used FMT on rhesus monkeys infected with chronic SIV who were receiving antiretroviral medication. After antibiotic therapy, microbiota shift was observed, whereas Th17 and Th22 frequencies in peripheral blood increased and CD4 T cell activation in the intestinal tract reduced following FMT [195]. Liu et al. investigated the effects of FMT on GI symptoms, gut dysbiosis, and immune status in 11 discharged COVID-19 patients. After FMT, the COVID-19’s peripheral lymphocyte subset was altered. The gut microbiota was restored, and GI disorders were relieved, suggesting that FMT could be used as a therapeutic and rehabilitative intervention [193]. Further research is needed to understand whether FMT could be a viable technique for altering the gut microbiome to improve COVID-19’s residual effect.
7. Conclusion
The link between human microbiota and COVID-19 is still unknown. This narrative review aimed to assess and summarize available evidence on the relationship between the microbiome and COVID-19 in patients in the pandemic era. The current study uncovered several major issues that call for more investigation on microbiota in COVID-19 patients. There is a crucial cross-talk between the gut microbiota and the lungs through the GLA. The protection of the gut microbiota hemostasis, especially in the absence of treatment for a viral respiratory infection, could be more practicable to regulate the immune system response and reduce the GI disorder after COVID-19 infection. Healthy gut microbiota might lead to asymptomatic or mild COVID-19 infection without any severe clinical complications. Probiotics have been shown to modulate the occurrence and severity of diseases, implying that they may be used to treat or prevent COVID-19. By preserving the human GI or lung, probiotics could help prevent COVID-19. More than ever, this pandemic has highlighted the importance of the gut microbial–host-immune axis and the impact of the fecal metabolome on inflammation control and regulation of the immune system. To investigate the possible preventive and therapeutic effects of probiotics against SARS-CoV-2 infection, in vitro and clinical studies are necessary.
8. Expert opinion
During hospitalization, gut microbiota composition may influence the immune response and change the severity and prognosis of COVID-19 [196]. Thus, a microbiome-oriented risk-assessment profile could be used to identify those at risk [197]. Additionally, since the human microbiome is adaptive and may be changed through dietary modifications [198], we believe that further research is urgently needed to assess the impact of microbiota and diet on COVID-19 [198]. However, the detection of enriched inflammatory-associated gut microorganisms in COVID-19 is controversial. The question is if they enriched or this was happened due to the reduction of the other microorganisms. It seems that longer follow-up is needed for COVID-19 patients, such as three months to 1 year after virus clearance. This will enable us to find a correlation of gut microbiota composition to long-term persisting symptoms. In addition, this can assess the possible alteration of the gut microbiota dysbiosis post-recovery. Also, it can help to determine whether enrichment or depletion of specific gut microorganisms can contribute to future health problems [141].
On the other hand, the described gut dysbiosis may just be a response to patients’ health and defensive statuses rather than playing a central role in disease severity. Accordingly, it is not clear how much clinical practice during hospitalization can affect the gut microbiota composition in COVID-19 patients. Of note, it is anticipated that 50% to 75% of COVID-19 patients received antibiotics, while just about 7% of them displayed bacterial infections [199]. Moreover, according to Chinese studies, antibiotics were given to 58.71% of COVID-19 patients, while 2–36% suffering from diarrhea. Even though probiotics have only a little efficacy in treating antibiotic-induced diarrhea, the use of probiotics has been indicated to reduce susceptibility to recurrent infection [98]. Previously, it has been found that polypharmacy significantly impacts microbiota composition and that more alterations could be observed if the number of co-administered drugs increases [200].
In conclusion, the implication of sufficient, safe, and cost-benefit pre- and probiotics can support the microbiota. Their prescription can be used as an adjuvant therapy to modulate COVID-19 progression or as a preventive strategy for non-infected individuals who are at risk during the COVID-19 pandemic.
Funding Statement
This paper was not funded.
Article highlights
Gut microbiota is essential for host immune system’s induction, education, function, development of immune responses, and regulates the integrity of the mucosal barrier, provides bacterial metabolites, and regulates the immunoregulatory functions of intestinal epithelial cells by modulating the expression of antimicrobial factors.
Dysbiosis is linked to dysimmunity, commonly described as a T-helper 2 (Th2)-overactive, and Treg-deficient state.
Cross-talk between the healthy gut microbiota and lungs can significantly prevent respiratory tract infection in both influenza A virus and COVID-19 through reciprocal interaction, quorum-sensing molecules, and synthesis of antimicrobial agents.
In COVID-19 patients, probiotic treatment could result in an eight-fold lower rate of respiratory failure.
It seems that fecal microbiota transplantation could be used as a therapeutic and rehabilitative intervention in the COVID-19 patients.
Declaration of interests
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or conflict with the subject matter or materials discussed in this manuscript apart from those disclosed.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Mathew S, Smatti MK, Al Ansari K, et al. Mixed viral-bacterial infections and their effects on gut microbiota and clinical illnesses in children. Sci Rep. 2019;9(1):865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanada S, Pirzadeh M, Carver KY, et al. Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Front Immunol. 2018;9:2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu H, Wei L, Niu P.. The novel coronavirus outbreak in Wuhan, China. Glob Health Res Policy. 2020;5:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cully M. Immune status could determine efficacy of COVID-19 therapies. Nat Rev Drug Discov. 2020;19(7):431–434. [DOI] [PubMed] [Google Scholar]
- 5.Cheung KS, Hung IFN, Chan PPY, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology. 2020;159(1):81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Moheb M, Naar L, Christensen MA, et al. Gastrointestinal complications in critically ill patients with and without COVID-19. JAMA. 2020;324(18):1899–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaafarani HM, El Moheb M, Hwabejire JO, et al. Gastrointestinal complications in critically ill patients with COVID-19. Ann Surg. 2020;272(2):e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budden KF, Gellatly SL, Wood DLA, et al. Emerging pathogenic links between microbiota and the gut–lung axis. Nature Rev Microbiol. 2017;15(1):55. [DOI] [PubMed] [Google Scholar]
- 9.Hugon P, Dufour J-C, Colson P, et al. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect Dis. 2015;15(10):1211–19. [DOI] [PubMed] [Google Scholar]
- 10.Cianci R, Pagliari D, Piccirillo CA, et al. The microbiota and immune system crosstalk in health and disease. Mediators of Inflammation. 2018;2018:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu G, Jiang Y, Yao Y, et al. Ovotransferrin ameliorates the dysbiosis of immunomodulatory function and intestinal microbiota induced by cyclophosphamide. Food Funct. 2019;10(2):1109–1122. [DOI] [PubMed] [Google Scholar]
- 12.Xie Q, Pan M, Huang R, et al. Modulation of the small intestinal microbial community composition over short-term or long-term administration with Lactobacillus plantarum ZDY2013. J Dairy Sci. 2016;99(9):6913–21. [DOI] [PubMed] [Google Scholar]
- 13.Belizário JE, Faintuch J. Microbiome and gut dysbiosis. Exp Suppl. 2018;109:459–476. [DOI] [PubMed] [Google Scholar]
- 14.Round JL, Mazmanian SKJNRI. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamada N, Seo S-U, Chen GY, et al. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–35. [DOI] [PubMed] [Google Scholar]
- 16.Flint HJ, Scott KP, Louis P, et al. The role of the gut microbiota in nutrition and health. Nat Clin Pract Gastroenterol Hepatol. 2012;9(10):577. [DOI] [PubMed] [Google Scholar]
- 17.Mazmanian SK, Round JL, Kasper DLJN. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453(7195):620–5. [DOI] [PubMed] [Google Scholar]
- 18.Gérard PJP. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens. 2014;3(1):14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mezouar S, Chantran Y, Michel J, et al. Microbiome and the immune system: from a healthy steady-state to allergy associated disruption. Hum Microbiome J. 2018;10:11–20. [Google Scholar]
- 20.Fleck A-K, Schuppan D, Wiendl H,et al. Gut–CNS-axis as possibility to modulate inflammatory disease activity—Implications for multiple sclerosis. Int J Mol Sci. 2017;18(7):1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiest R, Albillos A, and Trauner, M et al. Targeting the gut-liver axis in liver disease. J Hepatol . 2017;67(5):1084–1103. [DOI] [PubMed] [Google Scholar]
- 22.Yurkovetskiy L, Burrows M, Khan A, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39(2):400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salem I, Ramser, A, and Isham, N, et al. The gut microbiome as a major regulator of the gut-skin axis Front Microbiol . 2018;9:1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sencio V, Barthelemy A, Tavares LP, et al. Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short-chain fatty acid production. Cell Rep. 2020;30(9):2934–2947. [DOI] [PubMed] [Google Scholar]
- 25.Schuijt TJ, Lankelma JM, Scicluna BP, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. 2016;65(4):575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keely S, Talley NJ, Hansbro PMJMI. Pulmonary-intestinal cross-talk in mucosal inflammatory disease. Mucosal Immunol. 2012;5(1):7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yazar A, Atis S, Konca K, et al. Respiratory symptoms and pulmonary functional changes in patients with irritable bowel syndrome. Am J Gastroenterol. 2001;96(5):1511–1516. [DOI] [PubMed] [Google Scholar]
- 28.Lotz MT, Peebles RS. Mechanisms of respiratory syncytial virus modulation of airway immune responses. Curr Allergy Asthma Rep. 2012;12(5):380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brodin P, Davis MM. Human immune system variation. Nat Rev Immunol. 2017;17(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Odamaki T, Kato K, Sugahara H, et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fouhy F, Ross RP, Fitzgerald GF, et al. Composition of the early intestinal microbiota: knowledge, knowledge gaps and the use of high-throughput sequencing to address these gaps. Gut Microbes. 2012;3(3):203–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine KM, Priedigkeit N, Basudan A, et al. FGFR4 overexpression and hotspot mutations in metastatic ER+ breast cancer are enriched in the lobular subtype. NPJ Breast Cancer. 2019;5(1):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell SL, Gold MJ, Reynolds LA, et al. Perinatal antibiotic-induced shifts in gut microbiota have differential effects on inflammatory lung diseases. J Allergy Clin Immunol. 2015;135(1):100–9. e5. [DOI] [PubMed] [Google Scholar]
- 35.Han Y, Jia, Z, and Shi, J, et al. The active lung microbiota landscape of COVID-19 patients. medRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 36.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J. 2017;474(11):1823–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scarpellini E, Fagoonee S, Rinninella E, et al. Gut microbiota and liver interaction through immune system cross-talk: a comprehensive review at the time of the SARS-COV-2 pandemic. J Clin Med. 2020;9(8):2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rinninella E, Raoul P, Cintoni M, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rabizadeh S, Rhee K-J, Wu S, et al. Enterotoxigenic Bacteroides fragilis: a potential instigator of colitis. Inflamm Bowel Dis. 2007;13(12):1475–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atarashi K, Tanoue T, Shima T, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukhopadhya I, Segal, JP, and Carding, SR, et al. The gut virome: the ‘missing link’between gut bacteria and host immunity? Therap Adv Gastroenterol. 2019;12:1756284819836620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scarpellini E, Ianiro G, Attili F, et al. The human gut microbiota and virome: potential therapeutic implications. Digestive Liver Dis. 2015;47(12):1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535(7610):75–84. [DOI] [PubMed] [Google Scholar]
- 47.Owaga E, Hsieh R-H, Mugendi B, et al. Th17 cells as potential probiotic therapeutic targets in inflammatory bowel diseases. Int J Mol Sci. 2015;16(9):20841–20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng H-Y, Ning, MX, and Chen, DK, et al. Interactions between the gut microbiota and the host innate immune response against pathogens. Front Immunol. 2019;10. DOI: 10.3389/fimmu.2019.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pagliari D, Piccirillo CA, Larbi A, et al. The interactions between innate immunity and microbiota in gastrointestinal diseases. J Immunol Res. 2015;2015:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivanov II, Atarashi K, Manel N, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaudino SJ, Kumar P. Cross-Talk between antigen presenting cells and T cells impacts intestinal homeostasis, bacterial infections, and tumorigenesis. Front Immunol. 2019;10:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bene K, Varga Z, Petrov VO, et al. Gut microbiota species can provoke both inflammatory and tolerogenic immune responses in human dendritic cells mediated by retinoic acid receptor alpha ligation. Front Immunol. 2017;8:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu H-J, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3(1):4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flannigan KL, Denning TL. Segmented filamentous bacteria‐induced immune responses: a balancing act between host protection and autoimmunity. Immunology. 2018;154(4):537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lécuyer E, Rakotobe S, Lengliné-Garnier H, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40(4):608–620. [DOI] [PubMed] [Google Scholar]
- 58.Salzman NHJG. m., Paneth cell defensins and the regulation of the microbiome: detente at mucosal surfaces. Gut Microbes. 2010;1(6):401–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–689. [DOI] [PubMed] [Google Scholar]
- 60.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19(1):29–41. [DOI] [PubMed] [Google Scholar]
- 61.Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol (Lausanne). 2020;11:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corrêa‐Oliveira R, Fachi JL, Vieira A, et al. Regulation of immune cell function by short‐chain fatty acids. Clin Transl Immunology. 2016;5(4):e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosshart SP, Vassallo BG, Angeletti D, et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. 2017;171(5):1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khosravi A, Yáñez, A, and Price, JG, et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe. 2014;15(3):374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lo BC, Shin, SB, and Canals Hernaez, D, et al. IL-22 preserves gut epithelial integrity and promotes disease remission during chronic Salmonella infection. J Immunol. 2019;202(3):956–965. [DOI] [PubMed] [Google Scholar]
- 66.Chen GY, Liu, M, and Wang, F, et al. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J Immunol. 2011;186(12):7187–7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lukasova M, Malaval, C, and Gille, A, et al. Nicotinic acid inhibits progression of atherosclerosis in mice through its receptor GPR109A expressed by immune cells. J Clin Investig. 2011;121(3):1163–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gustafsson JK, Ermund A, Johansson MEV, et al. An ex vivo method for studying mucus formation, properties, and thickness in human colonic biopsies and mouse small and large intestinal explants. Am J Physiol Gastrointest Liver Physiol. 2012;302(4):G430–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jung T-H, Park JH, Jeon W-M, et al. Butyrate modulates bacterial adherence on LS174T human colorectal cells by stimulating mucin secretion and MAPK signaling pathway. Nutr Res Pract. 2015;9(4):343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Diamond G, Beckloff N, Weinberg A, et al. The roles of antimicrobial peptides in innate host defense. Curr Pharm Des. 2009;15(21):2377–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim D, Zeng MY, Núñez G, et al. The interplay between host immune cells and gut microbiota in chronic inflammatory diseases. Exp Mol Med. 2017;49(5):e339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Canani RB, Costanzo, MD, and Leone L, et al. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol. 2011;17(12):1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salvi PS, Cowles RA. Butyrate and the intestinal epithelium: modulation of proliferation and inflammation in homeostasis and disease. Cells. 2021;10(7):1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiminez JA, Uwiera, TC, and Abbott, DW, et al. Butyrate supplementation at high concentrations alters enteric bacterial communities and reduces intestinal in flammation in mice infected with Citrobacter rodentium. MSphere. 2017;2(4);e00243–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frieman M, Heise M, Baric RJVR. SARS coronavirus and innate immunity. Virus Res. 2008;133(1):101–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yitbarek A, Taha-Abdelaziz, K, and Hodgins, DC, et al. Gut microbiota-mediated protection against influenza virus subtype H9N2 in chickens is associated with modulation of the innate responses. Sci Rep. 2018;8(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Menendez A, Willing, BP, and Montero, M, et al. Bacterial stimulation of the TLR-MyD88 pathway modulates the homeostatic expression of ileal Paneth cell α-defensins. J Innate Immun. 2013;5(1):39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cianci R, et al. Tissue infiltrating lymphocytes: the role of cytokines in their growth and differentiation. J Biol Regul Homeostat Agents. 2010;24(3):239. [PubMed] [Google Scholar]
- 80.Francino MPJP. Early development of the gut microbiota and immune health. Pathogens. 2014;3(3):769–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arrazuria R, Pérez V, Molina E, et al. Diet induced changes in the microbiota and cell composition of rabbit gut associated lymphoid tissue (GALT). Sci Rep. 2018;8(1):14103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ahluwalia B, Magnusson MK, Öhman L. Mucosal immune system of the gastrointestinal tract: maintaining balance between the good and the bad. Scand J Gastroenterol. 2017;52(11):1185–1193. [DOI] [PubMed] [Google Scholar]
- 83.Fagarasan S, Muramatsu M, Suzuki K, et al. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298(5597):1424–1427. [DOI] [PubMed] [Google Scholar]
- 84.Kasarello K, Sajdel-Sulkowska EM. Developmental significance of early gut-associated lymphoid tissue (GALT)-microbiota interactions in health and disease: creating balance between tolerance and inflammation in children. Open J Pediatr Child Health. 2019;4(1):040–6. doi: 10.17352/ojpch.000019. [DOI] [Google Scholar]
- 85.Sze MA, Dimitriu PA, Hayashi S, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;185(10):1073–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marsland BJ, Trompette A, Gollwitzer ESJAOTATS. The gut–lung axis in respiratory disease. Ann Am Thorac Soc. 2015;12(Supplement 2):S150–S156. [DOI] [PubMed] [Google Scholar]
- 87.Enaud R, Prevel R, Ciarlo E, et al. The gut-lung axis in health and respiratory diseases: a place for inter-organ and inter-kingdom crosstalks. Front Cell Infect Microbiol. 2020;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dickson RP, Singer BH, Newstead MW, et al. Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat Microbiol. 2016;1(10):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fanos V, Pintus, MC, and Pintus, R, et al. Lung microbiota in the acute respiratory disease: from coronavirus to metabolomics. J Pediatr Neonatal Individ Med(JPNIM). 2020;9(1): e090139 [Google Scholar]
- 90.Battaglini D, Robba C, Fedele A, et al. The role of dysbiosis in critically ill patients with COVID-19 and acute respiratory distress syndrome. Front Med. 2021;8:826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Looft T, Allen HKJGM. Collateral effects of antibiotics on mammalian gut microbiomes. Gut Microbes. 2012;3(5):463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fujimura KE, Lynch SVJCH. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe. 2015;17(5):592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Young RP, Hopkins RJ, Marsland B, et al. The gut–liver–lung axis. Modulation of the innate immune response and its possible role in chronic obstructive pulmonary disease. Am J Resp Cell Mol Biol. 2016;54(2):161–169. [DOI] [PubMed] [Google Scholar]
- 94.Baradaran Ghavami SH, Sharokh, SH, and Hossein-Khannazer, N, et al. IBD patients could be silent carriers for novel coronavirus and less prone to its severe adverse events: true or false? Cell J. 2020;22(Suppl 1):151–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Denny JE, Powell WL, Schmidt NW, et al. Local and long-distance calling: conversations between the gut microbiota and intra-and extra-gastrointestinal tract infections. Front Cell Infect Microbiol. 2016;6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coopersmith CM, Stromberg PE, Davis CG, et al. Sepsis from Pseudomonas aeruginosa pneumonia decreases intestinal proliferation and induces gut epithelial cell cycle arrest. Crit Care Med. 2003;31(6):1630–1637. [DOI] [PubMed] [Google Scholar]
- 97.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rajput S, Paliwal, D, and Naithani, M, et al. COVID-19 and gut microbiota: a potential connection. Indian J Clin Biochem. 2021;36(3) ;1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Anand S, Mande SS. Diet, microbiota and gut-lung connection. Front Microbiol. 2018;9:2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Arrieta M-C, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. [DOI] [PubMed] [Google Scholar]
- 101.Lynch S, Sitarik, AR, and Havstad, S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation Nat Med . 2016;22(10):1187–1191 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Demirci M, Tokman HB, Uysal HK, et al. Reduced Akkermansia muciniphila and Faecalibacterium prausnitzii levels in the gut microbiota of children with allergic asthma. Allergol Immunopathol (Madr). 2019;47(4):365–371. [DOI] [PubMed] [Google Scholar]
- 103.Kalliomäki M, Kirjavainen P, Eerola E, et al. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107(1):129–134. [DOI] [PubMed] [Google Scholar]
- 104.Hilty M, Burke, C, and Pedro, H, et al. Disordered microbial communities in asthmatic airways. PloS One. 2010;5(1):e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Charlson ES, Bittinger K, Haas AR, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184(8):957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Antosca KM, Chernikova DA, Price CE, et al. Altered stool microbiota of infants with cystic fibrosis shows a reduction in genera associated with immune programming from birth. J Bacteriol. 2019;201(16):e00274–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Burke DG, Fouhy, F, and Harrison, MJ, et al. The altered gut microbiota in adults with cystic fibrosis. BMC Microbiol. 2017;17(1):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fouhy F, Ronan NJ, O’Sullivan O, et al. A pilot study demonstrating the altered gut microbiota functionality in stable adults with cystic fibrosis. Sci Rep. 2017;7(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Debyser G, Mesuere B, Clement L, et al. Faecal proteomics: a tool to investigate dysbiosis and inflammation in patients with cystic fibrosis. J Cyst Fibros. 2016;15(2):242–250. [DOI] [PubMed] [Google Scholar]
- 110.Blainey PC, Milla, CE, and Cornfield, DN, et al. Quantitative analysis of the human airway microbial ecology reveals a pervasive signature for cystic fibrosis. Sci Transl Med. 2012;4(153):153ra130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kolak M, Karpati, F, and Monstein, H-J, et al. Molecular typing of the bacterial flora in sputum of cystic fibrosis patients. Int J Med Microbiol. 2003;293(4):309–317. [DOI] [PubMed] [Google Scholar]
- 112.Zhuang H, Cheng L, Wang Y, et al. Dysbiosis of the gut microbiome in lung cancer. Front Cell Infect Microbiol. 2019;9:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rosas-Salazar C, Shilts MH, Tovchigrechko A, et al. Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J Infect Dis. 2016;214(12):1924–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hofstra JJ, Matamoros, S, and van de Pol, MA, et al. Changes in microbiota during experimental human Rhinovirus infection. BMC Infect Dis. 2015;15(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Karppinen S, Teräsjärvi, J, and Auranen, K, et al. Acquisition and transmission of Streptococcus pneumoniae are facilitated during rhinovirus infection in families with children. Am J Respir Crit Care Med. 2017;196(9):1172–1180. [DOI] [PubMed] [Google Scholar]
- 116.Molyneaux PL, Mallia P, Cox MJ, et al. Outgrowth of the bacterial airway microbiome after rhinovirus exacerbation of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;188(10):1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Allen EK, Koeppel, AF, and Hendley, JO, et al. Characterization of the nasopharyngeal microbiota in health and during rhinovirus challenge. Microbiome. 2014;2(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lu H-F, Li, A, and Zhang, T, et al. Disordered oropharyngeal microbial communities in H7N9 patients with or without secondary bacterial lung infection. Emerg Microbes Infect. 2017;6(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Leung R-K, Zhou J-W, Guan W, et al. Modulation of potential respiratory pathogens by pH1N1 viral infection. Clin Microbiol Infect. 2013;19(10):930–935. [DOI] [PubMed] [Google Scholar]
- 120.Rogers GB, Shaw D, Marsh RL, et al. Respiratory microbiota: addressing clinical questions, informing clinical practice. Thorax. 2015;70(1):74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Qin N, Zheng, B, and Yao, J, et al. Influence of H7N9 virus infection and associated treatment on human gut microbiota. Sci Rep. 2015;5:14771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Saitou M, Nemoto D, Utano K, et al. Identification of intestinal abnormalities in patients with active pulmonary tuberculosis using small bowel capsule endoscopy. Endosc Int Open. 2018;6(9):E1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Luo M, Liu Y, Wu P, et al. Alternation of gut microbiota in patients with pulmonary tuberculosis. Front Physiol. 2017;8:822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang D, Li, S, and Wang, N, et al. The cross-talk between gut microbiota and lungs in common lung diseases. Front Microbiol. 2020;11: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Neu U, Mainou BA, Evans MJ. Virus interactions with bacteria: partners in the infectious dance. PLoS Pathog. 2020;16(2):e1008234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mustafa S, Balkhy H, Gabere MN. Current treatment options and the role of peptides as potential therapeutic components for Middle East Respiratory Syndrome (MERS): a review. J Infect Public Health. 2018;11(1):9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Du L, He Y, Zhou Y, et al. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7(3):226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zeppa SD, Agostini, D, and Piccoli, G, et al. Gut microbiota status in COVID-19: an unrecognized player? Front Cell Infect Microbiol. 2020;10 576551 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ichinohe T, Pang IK, Kumamoto Y, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci. 2011;108(13):5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Abt MC, Osborne L, Monticelli L, et al. Commensal bacteria calibrate the activation threshold of innate antiviral. immunity. 2012;37(1):158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Allie SR, Bradley JE, Mudunuru U, et al. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat Immunol. 2019;20(1):97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov Ther. 2020;14(1):58–60. [DOI] [PubMed] [Google Scholar]
- 133.Lv L, Gu S, Jiang H, et al. Gut mycobiota alterations in patients with COVID-19 and H1N1 infections and their associations with clinical features. Commun Biol. 2021;4(1):480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Forbes JD, Bernstein CN, Tremlett H, et al. A fungal world: could the gut mycobiome be involved in neurological disease? Front Microbiol. 2019;9:3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lv L, Gu, S, and Jiang, H, et al. Gut mycobiota alterations in patients with COVID-19 and H1N1 and their associations with clinical features Commun Biol . 2021;4(1): 480 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159(3):944–955. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gou W, Fu, Y, and Yue, L, et al., Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. MedRxiv, 2020.
- 138.Zuo T, Liu, Q, and Zhang, F, et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut. 2021;70(2):276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhou Y, Shi X, Fu W, et al. Gut microbiota dysbiosis correlates with abnormal immune response in moderate COVID-19 patients with fever. J Inflamm Res. 2021;14:2619–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gu S, Chen Y, Wu Z, et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 Influenza. Clin Infect Dis. 2020;71(10):2669–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yeoh YK, Zuo T, Lui GC-Y, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70(4):698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Tang L, Gu S, Gong Y, et al. Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID-19 severity. Engineering. 2020;6(10):1178–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Khatiwada S, Subedi A. Lung microbiome and coronavirus disease 2019 (COVID-19): possible link and implications. Hum Microbiome J. 2020;17:100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kalina U, Koyama N, Hosoda T, et al. Enhanced production of IL-18 in butyrate-treated intestinal epithelium by stimulation of the proximal promoter region. Eur J Immunol. 2002;32(9):2635–2643. [DOI] [PubMed] [Google Scholar]
- 145.Domínguez-Cherit G, Lapinsky, SE, and Macias, AE, et al. Critically ill patients with 2009 influenza A (H1N1) in Mexico. JAMA. 2009;302(17):1880–1887. [DOI] [PubMed] [Google Scholar]
- 146.Yasui H, Kiyoshima J, Hori TJCDLI. Reduction of influenza virus titer and protection against influenza virus infection in infant mice fed Lactobacillus casei Shirota. Clin Vaccin Immunol. 2004;11(4):675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tomosada Y, Chiba E, Zelaya H, et al. Nasally administered Lactobacillus rhamnosus strains differentially modulate respiratory antiviral immune responses and induce protection against respiratory syncytial virus infection. BMC Immunol. 2013;14(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mailliard RB, Son Y-I, Redlinger R, et al. Dendritic cells mediate NK cell help for Th1 and CTL responses: two-signal requirement for the induction of NK cell helper function. J Immunol. 2003;171(5):2366–2373. [DOI] [PubMed] [Google Scholar]
- 149.Kitazawa H, Villena JJFII. Modulation of respiratory TLR3-anti-viral response by probiotic microorganisms: lessons learned from Lactobacillus rhamnosus CRL1505. Front Immunol. 2014;5:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yilla M, Harcourt BH, Hickman CJ, et al. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res. 2005;107(1):93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cinatl J Jr., Hoever G, Morgenstern B, et al. Infection of cultured intestinal epithelial cells with severe acute respiratory syndrome coronavirus. Cell Mol Life Sci. 2004;61(16):2100–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Spiegel M, Schneider K, Weber F, et al. Interaction of severe acute respiratory syndrome-associated coronavirus with dendritic cells. J Gen Virol. 2006;87(7):1953–60. [DOI] [PubMed] [Google Scholar]
- 153.Simmons G, Gosalia DN, Rennekamp AJ, et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci U S A. 2005;102(33):11876–11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fischer DD, Kandasamy S, Paim FC, et al. Protein malnutrition alters tryptophan and angiotensin-converting enzyme 2 homeostasis and adaptive immune responses in human rotavirus-infected gnotobiotic pigs with human infant fecal microbiota transplant. Clin Vaccin Immunol. 2017;24(8):e00172–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Vighi G, Marcucci F, Sensi L, et al. Allergy and the gastrointestinal system. Clin Exp Immunol. 2008;153(Suppl 1):3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mikelsaar M, Lazar V, Onderdonk A, et al. Do probiotic preparations for humans really have efficacy? Microb Ecol Health Dis. 2011;22(1):10128. [Google Scholar]
- 157.Kong YH, Shi, Q, and Han, N, et al. Structural modulation of gut microbiota in rats with allergic bronchial asthma treated with recuperating lung decoction. Biomed Environ Sci. 2016;29(8):574–583. [DOI] [PubMed] [Google Scholar]
- 158.Villena J, Salva S, Barbieri N, et al. Immunobiotics for the Prevention of Bacterial and Viral Respiratory Infections Probiotics 1. Boca Raton: CRC Press. 2013:128–168. 9780429090684 . [Google Scholar]
- 159.Mullish BH, Marchesi JR, McDonald JAK, et al. Probiotics reduce self-reported symptoms of upper respiratory tract infection in overweight and obese adults: should we be considering probiotics during viral pandemics? Gut Microbes. 2021;13(1):1900997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bron PA, Kleerebezem M, Brummer R-J, et al. Can probiotics modulate human disease by impacting intestinal barrier function? Br J Nutr. 2017;117(1):93–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Davani-Davari D, Negahdaripour M, Karimzadeh I, et al. Prebiotics: definition, types, sources, mechanisms, and clinical applications. Foods. 2019;8(3):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Olaimat AN, Aolymat I, Al-Holy M, et al. The potential application of probiotics and prebiotics for the prevention and treatment of COVID-19. NPJ Science Food. 2020;4(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fujiwara S, Seto Y, Kimura A, et al. Establishment of orally‐administered Lactobacillus gasseri SBT2055SR in the gastrointestinal tract of humans and its influence on intestinal microflora and metabolism. J Appl Microbiol. 2001;90(3):343–352. [DOI] [PubMed] [Google Scholar]
- 164.Takahashi H, Fujita T, Suzuki Y, et al. Monitoring and survival of Lactobacillus gasseri SBT2055 in the human intestinal tract. Microbiol Immunol. 2006;50(11):867–870. [DOI] [PubMed] [Google Scholar]
- 165.Sakai F, Hosoya T, Ono-Ohmachi A, et al. Lactobacillus gasseri SBT2055 induces TGF-β expression in dendritic cells and activates TLR2 signal to produce IgA in the small intestine. PloS One. 2014;9(8):e105370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nakayama Y, Moriya T, Sakai F, et al. Oral administration of Lactobacillus gasseri SBT2055 is effective for preventing influenza in mice. Sci Rep. 2014;4:4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Eguchi K, Fujitani N, Nakagawa H, et al. Prevention of respiratory syncytial virus infection with probiotic lactic acid bacterium Lactobacillus gasseri SBT2055. Sci Rep. 2019;9(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hori T, Kiyoshima J, Shida K, et al. Effect of intranasal administration oflactobacillus casei Shirota on influenza virus infection of upper respiratory tract in mice. Clin Diagn Lab Immunol. 2001;8(3):593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vickers NJJCB. Animal communication: when i’m calling you, will you answer too? Curr Biol. 2017;27(14):R713–15. [DOI] [PubMed] [Google Scholar]
- 170.Gabryszewski SJ, Bachar O, Dyer KD, et al. Lactobacillus-mediated priming of the respiratory mucosa protects against lethal pneumovirus infection. J Immunol. 2011;186(2):1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Villena J, Barbieri N, Salva S, et al. Enhanced immune response to pneumococcal infection in malnourished mice nasally treated with heat‐killed Lactobacillus casei. Microbiol Immunol. 2009;53(11):636–46. [DOI] [PubMed] [Google Scholar]
- 172.Weiss G, Rasmussen S, Zeuthen LH, et al. Lactobacillus acidophilus induces virus immune defence genes in murine dendritic cells by a Toll‐like receptor‐2‐dependent mechanism. Immunology. 2010;131(2):268–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Khailova L, Petrie B, Baird CH, et al. Lactobacillus rhamnosus GG and Bifidobacterium longum attenuate lung injury and inflammatory response in experimental sepsis. PLoS One. 2014;9(5):e97861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Iwabuchi N, Xiao J-Z, Yaeshima T, et al. Oral administration of Bifidobacterium longum ameliorates influenza virus infection in mice. Biol Pharm Bull. 2011;34(8):1352–1355. [DOI] [PubMed] [Google Scholar]
- 175.Kawahara T, Takahashi T, Oishi K, et al. Consecutive oral administration of Bifidobacterium longum MM‐2 improves the defense system against influenza virus infection by enhancing natural killer cell activity in a murine model. Microbiol Immunol. 2015;59(1):1–12. [DOI] [PubMed] [Google Scholar]
- 176.Namba K, Hatano M, Yaeshima T, et al. Effects of Bifidobacterium longum BB536 administration on influenza infection, influenza vaccine antibody titer, and cell-mediated immunity in the elderly. Biosci Biotechnol Biochem. 2010;74(5):939–945. [DOI] [PubMed] [Google Scholar]
- 177.Furuhashi K, Suda T, Hasegawa H, et al. Mouse lung CD103+ and CD11bhigh dendritic cells preferentially induce distinct CD4+ T-cell responses. Am J Respir Cell Mol Biol. 2012;46(2):165–172. [DOI] [PubMed] [Google Scholar]
- 178.Gleeson M, Pyne DB. Respiratory inflammation and infections in high‐performance athletes. Immunol Cell Biol. 2016;94(2):124–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Van Puyenbroeck K, Hens N, Coenen S, et al. Efficacy of daily intake of Lactobacillus casei Shirota on respiratory symptoms and influenza vaccination immune response: a randomized, double-blind, placebo-controlled trial in healthy elderly nursing home residents. Am J Clin Nutr. 2012;95(5):1165–1171. [DOI] [PubMed] [Google Scholar]
- 180.Sindhu KN, Sowmyanarayanan TV, Paul A, et al. Immune response and intestinal permeability in children with acute gastroenteritis treated with Lactobacillus rhamnosus GG: a randomized, double-blind, placebo-controlled trial. Clinl Infect Dis. 2014;58(8):1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hirose Y, Murosaki, S, and Yamamoto, Y, et al. Daily intake of heat-killed Lactobacillus plantarum L-137 augments acquired immunity in healthy adults. J Nutr. 2006;136(12):3069–3073. [DOI] [PubMed] [Google Scholar]
- 182.Maeda N, Nakamura, R, and Hirose, Y, et al. Oral administration of heat-killed Lactobacillus plantarum L-137 enhances protection against influenza virus infection by stimulation of type I interferon production in mice. Int Immunopharmacol. 2009;9(9):1122–1125. [DOI] [PubMed] [Google Scholar]
- 183.Izumo T, Maekawa, T, and Ida, M, et al. Effect of intranasal administration of Lactobacillus pentosus S-PT84 on influenza virus infection in mice. Int Immunopharmacol. 2010;10(9):1101–1106. [DOI] [PubMed] [Google Scholar]
- 184.Fujimura KE, Demoor T, Rauch M, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A. 2014;111(2):805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yasui H, Kiyoshima J, Hori T, et al. Protection against influenza virus infection of mice fed Bifidobacterium breve YIT4064. Clin Diagn Lab Immunol. 1999;6(2):186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bandoro C, Runstadler JA, Lowen AC. Bacterial lipopolysaccharide destabilizes influenza viruses. MSphere. 2017;2(5):e00267–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Deitch EA. Gut-origin sepsis: evolution of a concept. Surgeon. 2012;10(6):350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Bradley KC, Finsterbusch K, Schnepf D, et al. Microbiota-driven tonic interferon signals in lung stromal cells protect from influenza virus infection. Cell Rep. 2019;28(1):245–256. e4. [DOI] [PubMed] [Google Scholar]
- 189.Albenberg LG, Wu GDJG. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology. 2014;146(6):1564–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Varraso R, Chiuve SE, Fung TT, et al. Alternate Healthy Eating Index 2010 and risk of chronic obstructive pulmonary disease among US women and men: prospective study. BMJ. 2015;350:h286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.King DE, Egan BM, Woolson RF, et al. Effect of a high-fiber diet vs a fiber-supplemented diet on C-reactive protein level. Arch Intern Med. 2007;167(5):502–506. [DOI] [PubMed] [Google Scholar]
- 192.Kazemian N, Kao D, Pakpour S. Fecal microbiota transplantation during and post-COVID-19 pandemic. Int J Mol Sci. 2021;22(6):3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Liu F, Ye S, Zhu X, et al. Gastrointestinal disturbance and effect of fecal microbiota transplantation in discharged COVID-19 patients. J Med Case Rep. 2021;15(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Stewart CJ, Ajami NJ, O’Brien JL, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562(7728):583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mutlu EA, Keshavarzian A, Losurdo J, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10(2):e1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Vabret N, Britton GJ, Gruber C, et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910–941 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Verdoni L, Mazza A, Gervasoni A, et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395(10239):1771–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chhibber-Goel J, Gopinathan S, Sharma A. Interplay between severities of COVID-19 and the gut microbiome: implications of bacterial co-infections? Gut Pathog. 2021;13(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Goncalves Mendes Neto A, Lo KB, Wattoo A, et al. Bacterial infections and patterns of antibiotic use in patients with COVID‐19. J Med Virol. 2021;93(3):1489–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Vila AV, Collij, V, and Sanna, S, et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat Commun. 2020;11(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]


