Abstract
Neurodegenerative diseases affect millions of people worldwide and the presence of various physiological barriers limits the accessibility to the brain and reduces the efficacy of various therapies. Moreover, new carriers having targeting properties to specific brain regions and cells are needed in order to improve therapies for the brain disorder treatment. In this study, for the first time, Myelin nanoVesicles (hereafter defined MyVes) from brain-extracted myelin were produced. The MyVes have an average diameter of 100–150 nm, negative zeta potential, spheroidal morphology, and contain lipids and the key proteins of the myelin sheath. Furthermore, they exhibit good cytocompatibility. The MyVes were able to target the white matter and interact mainly with the microglia cells. The preliminary results here presented allow us to suppose the employment of MyVes as potential carrier to target the white matter and microglia in order to counteract white matter microglia-related diseases.
Keywords: Nanovesicles, Myelin nanovesicles, Brain delivery, Withe matter, Microglia cells
Graphical abstract
Highlights
-
•
Bio-fabrication of brain tissue derived nanovesicles: myelin nanovesicles.
-
•
Myelin nanovesicles contain the main proteins of the myelin sheath (myelin basic protein and myelin proteolipid protein).
-
•
Myelin nanovesicles can lade a drug/molecule and cross a blood–brain barrier model.
-
•
Myelin nanovesicles target white matter and microglia cells.
1. Introduction
Central nervous system (CNS) disorders are a broad category of medical conditions (more than 600 human disorders) characterized by brain dysfunction that limits normal activities. They include cancer, degenerative nerve diseases, genetic brain disorders, stroke, traumatic brain injury (TBI), Parkinson’s disease, multiple sclerosis (MS), amyotrophic lateral sclerosis, Huntington’s disease, Prion diseases, Alzheimer’s disease (AD), and other less frequent dementias [1].
The treatment of CNS disorders and the administration routes remain, to date, a most interesting and attractive challenge for the researchers. Among the routes of administration to the brain, intracerebral and intrathecal injection is appropriate for therapeutic interventions but invasive approaches. Peripheral or systemic administration is a less invasive and safer route, offering the advantages of larger injection volumes and multiple dosages. However, the presence of physiological barriers, such as the blood–brain barrier (BBB) and blood–cerebrospinal fluid barrier limits the accessibility to the brain and reduces the efficacy of various therapies.
Recently, intranasal administration has been addressed as an alternative route for drug brain delivery [2,3]. In fact, intranasal administration bypasses the BBB and the first-pass hepatic clearance of the drugs [4]. However, factors such as a limited dosing volume, small absorption surface, presence of degrading enzymes, and other variables attributable to patient congestion and presence of mucus limit the efficacy of intranasal route. For these reasons, new nasal formulations and delivery devices, designed for nose–brain drug delivery, are now being studied in preclinical models and early clinical stages [5]. The successful claims are only limited to animal models [5].
Therefore, formulations selectively targeting specific regions and cells (neuron, microglia, or astrocytes), and able to bypass the physiological barriers are essential factors to develop an efficient therapy for the treatment of brain disorders. Nanotechnology delivery systems to the brain can be an exciting and promising platform for overcoming the problems mentioned above.
To improve brain targeting, many researches have focused on the development of new strategies based on colloidal drug delivery systems (DDSs), such as nanoparticles, liposomes, dendrimers, and nanogels [[6], [7], [8]]. To perform nanocarrier functions they must possess specific properties, such as small and narrow size distribution, drug loading capacity, ability to control the drug release, and to protect the drug from the surrounding environments. If opportunely formulated, a nanocarrier can allow the reduction of the dose frequency administration, the release of the drug to a specific cellular target, stability, ability to cross the barriers, and biocompatibility. Nanoparticles up to approximately 100 nm facilitate drug transport through the BBB and, due to the human cell average size (10–100 μm), they can be easily absorbed by the cells and release the drug inside [9]. Moreover, the presence of distinct receptors on the BBB surface, capable of binding specific ligands (transferrin, insulin, apolipoprotein E, leptin, glutathione), can facilitate the nanodevices internalization mechanism allowing the achievement of the target delivery site and their transit through the BBB [10].
Recently, extracellular vesicles (EVs) and artificial vesicles have been explored as important tools in biomedicine and pharmaceutics [11,12].
EVs are cell-derived membrane structures, which have been proposed as DDS for brain delivery and as therapeutic agents [13]. Among EVs, exosomes are emerging as a promising tool for therapeutic delivery strategies due to their characteristics, such as stability, biocompatibility, ability to invisibility when circulating in the bloodstream, ability to overcome natural barriers, and intrinsic targeting capabilities. In addition, the appropriate design of their membrane could generate vectors with unprecedented capabilities for targeting specific organs or tissues. However, it is urgent to improve the safety, the source (need in vitro culture of patient cells), the extraction protocols, the reproducibility, and the characterization processes. The content and composition of EVs released from producing cells depend on their physiological status. All this anticipates intensive research in the field over the next years and novel approaches to efficiently produce EVs. To date, concerns with yield and heterogeneity have hindered the clinical use of EVs [14].
Cells or cell membrane–based systems have received increasing interest as DDS [15]. Recently, whole cells such as red blood cells, stem cells [15], and immunological cells [16] have been proposed as drug carriers [17]. Moreover, studies have demonstrated the ability to form cell membrane vesicles [[18], [19], [20], [21]] using biotechnological approaches. In addition, the cell plasma membrane can be engineered to improve the performance and efficacy of the carrier. Recently, synaptosomes have been used as vesicles for delivery of the mitochondria (mitochondrial transplantation) in neuronal cells, against neurodegenerative diseases [22].
The inflammatory process is a common feature of several CNS pathologies. Microglial cells are the innate immune cells within the CNS and their activity is involved in various pathologies [23]. In particular, microglial cells could be activated in the early stages of the disease process or during chronic inflammation, participating in the destruction of tissues through the release of proinflammatory factors [23]. Additionally, clinical observations of white matter–related diseases, such as MS, AD, and TBI, commonly reported the activation of white matter resident microglia, underlining its pivotal role [24]. Different DDSs showed an increase in the drug uptake in microglia [[25], [26], [27], [28], [29], [30], [31]]. Several approaches have been investigated such as conjugation with specific microglia receptor ligands or peptides to the nanoparticles surface to allow uptake, taking advantage of the phagocytic properties of microglia [25]. However, up to date no carriers, having a specific ability in targeting the white matter microglia, have been identified. This property could be of primary importance to fight several diseases involving the resident microglia in this region.
In this study, we propose, unprecedented nanovesicles derived from brain tissue, as a new potential carrier with an enhanced tropism for the brain tissues (from brain-to-brain). The nanovesicles were produced from myelin brain, and hereafter they will be identified as Myelin nanoVesicles (MyVes). An attempt to produce similar vesicles, with applications for the structural and functional analysis of myelin, dates back to 1986 [32], but with little success [33]. The MyVes are designed to be produced with an easy, efficient, cost-effective, and reproducible production protocol. Furthermore, they have controlled dimensions, negative zeta potential, high stability, and cytocompatibility. The obtained results suggested that MyVes are able to cross a BBB in vitro model and could have a potential application as carrier for targeting specific brain regions, such as white matter and preferentially interacting with microglial cells. In this first study, the main aim is to propose a proof of concept on the potential applicability of these novel systems, for targeting microglia and white matter to counteract the active microglia in white matter–related diseases, such as MS, AD, and TBI.
2. Experimental section
2.1. Materials and instruments
Bidistilled water (Millopore, Merck Italy®), isopropanol, tert-butanol (tBuOH), N,N-dimethylformamide (DMF), and Alexa Fluor 488 succinimidyl ester were acquired from Sigma-Aldrich srl (Italy). Dispersions were vigorously mixed by using an Ultraturrax (T 25, Janke & Kunkel Ika-Labortechnik). The size and dimensional distribution of particles were measured using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK). Measurements were executed at a fixed angle of 173° and a temperature of 25°C. Z-potential values were obtained using the laser Doppler velocimetry and phase analysis light scattering. Ultracentrifugation was performed by Optima XPN-100 Ultracentrifuge (Beckman Coulter). DI-8-ANEPPS 4-(2-[6-(dioctylamino)-2-naphthalenyl] ethenyl)-1-(3-sulfopropyl) pyridinium (Thermo Fisher Scientific). NanoSight NS300 (Malvern Panalytical, UK). Talos Arctica TEM operating at 200 kV and equipped with a Falcon3 camera operated in linear mode (Thermo Fisher Scientific, Waltham, MA). Bruker Alpha FTIR spectrometer. Drugs detection was performed by using a HPLC Agilent instrument 1260 Infinity equipped with a Quaternary Pump VL G1311C and a DAD detector 1260 VL.
2.2. Isolation and extraction of myelin
Wistar rats were purchased from Charles River Laboratories (Calco-Lecco, Italy). Explanted rat brains were donated from Palermo University (Italy) in accordance with the authorization number 69636.N.JCO approved by Italian Ministry of Health (Rome, Italy). The cortex (60–80 mg) was quickly removed and homogenized in 180 μl of 0.32 M sucrose with 10% protease (Amersham Biosciences, Milan, Italy) and phosphatase cocktail inhibitors (cocktail II and III; Sigma-Aldrich, Milan, Italy) with a Dounce on ice. An aliquot (10 μl) was immediately flash-freeze in liquid nitrogen and saved as total homogenate. The homogenate (170 μl) was mixed with 720 μl of 2 M sucrose and 300 μl of 0.1 mM CaCl2, transferred to 5 ml ultracentrifuge tube, overlayed carefully with 1 M sucrose solution and ultracentrifuged at 127,000 Relative Centrifugal Force (RCF) for 3 h at 4°C by using TLA110 rotor (Beckman Coulter, Brea, CA) [34]. The floating myelin layer on top was collected and washed twice in deionized water by centrifugation at 14,000 rpm for 15 min. Subsequently, the pellet containing the myelin extract was cold lyophilized.
2.3. Total protein extraction and SDS page
Total proteins were extracted by dissolving total homogenate (10 μl) and the myelin power (1 mg) in radio-immunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5% Triton X-100, 2 mM phenylmethylsulphonyl fluoride, 1 mM dithiothreitol 0.1% sodium dodecyl sulfate with protease inhibitors (Amersham Biosciences) and phosphatase inhibitors (cocktail II and III; Sigma-Aldrich). Total protein concentrations were determined by Bradford protein assay (Bio-Rad). Protein samples (30 μg) were submitted to 10% Sodium Dodecyl Sulphate - PolyAcrylamide Gel Electrophoresis (SDS PAGE) and staining with Coomassie Brilliant Blue.
2.4. Production of nanovesicles
Solubility tests were carried out for myelin extract to detect water soluble solvents potentially suitable for the nanoprecipitation procedure. In particular, extract powder was dispersed (up to 8 mg/ml) in tert-butanol, DMF, isopropanol, ethanol, and methanol. Nanoprecipitation by solvent injection technique [35,36] was performed to produce nanovesicles. In particular, 1 ml of solutions of myelin in DMF or tert-butanol (0.1 mg/ml) was added with a microsyringe over a 20-fold excess of ultrapure water under Ultraturrax® agitation (8000 rpm for 15 min). After agitation, samples produced by using DMF were dialyzed (cut-off 3500 Da) in order to remove the solvent and finally freeze dried (FD). Instead, samples produced by using tert-butanol were directly FD. In comparison, 1 ml of aqueous dispersion of myelin (0.1 mg/ml) was directly injected in a 20-fold excess of ultrapure water, mixed under Ultraturrax® (8000 rpm for 15 min) and then FD. Size, dimensional distribution of polydispersity index (PDI) and zeta potential of particles was measured just after solvent injection procedure or on FD samples that were re-dispersed in water at 0.1 mg/ml and sonicated (10 min, 35 kHz).
2.5. Synaptosomal vesicles isolation
Synaptosomal vesicles were isolated using the method descripted in Picone et al. [22]. Briefly, the cortex (60 mg) was quickly removed and homogenized in 180 μl of 0.32 M sucrose with 10% protease (Amersham Biosciences) and phosphatase cocktail inhibitors (cocktail II and III; Sigma-Aldrich) with a Dounce on ice. The homogenate was mixed with 720 μl of 2 M sucrose and 300 μl of 0.1 mM CaCl2, transferred to 5 ml ultracentrifuge tube and ultracentrifuged at 127,000 RCF for 3 h at 4°C by using TLA110 rotor (Beckman Coulter). The synaptosomal band at the sucrose layer interface was collected. Then, the synaptosomal band was transferred into an ultracentrifuge tube and centrifuged at 18,200 RCF for 15 min at 4°C in TLA110 rotor (Beckman Coulter). The supernatant was discarded and the pellet, containing the synaptosomal vesicles, was resuspended in phosphate-buffered saline.
2.6. Labeling of fluorescence probes into vesicles and drug loading suitability
For fluorescence labeling 500 μl of Alexa Fluor 488 succinimidyl ester in DMF (2.5 mg/ml) were mixed with 5 ml of the re-dispersed MyVes or synaptosomal vesicles (Synapt-Ves) (0.1 mg/ml), and purified by repeated ultracentrifugation steps (15 min, 40,000 rpm) (MyVes-Alexa 488, Synapt-Ves-Alexa 488), then diluted to the proper concentration before use. We loaded MyVes with the Di-8-ANEPPS dye, which exhibits a high fluorescence in apolar environments, such as the lipid membrane. As previously reported [37], 5 × 1010 particles/ml are stained with 500 nM of DI-8-ANEPPS (Ex/Em: 467/631 nm; Thermo Fisher Scientific) previously filtered by 20 nm filters (Whatman Anotop filters). After 1 h at room temperature, to eliminate the excess probe, the MyVes loaded with DI-8-ANEPPS were dialyzed (dialytic membrane 1 kDa) in water for 24 h at room temperature. Fluorescence intensity was determined by the Wallac Victor 2 1420 Multilabel Counter (PerkinElmer, Inc.) microplate reader. The excitation and emission wavelengths were set at 488 and 530 nm, respectively. The drugs doxorubicin hydrochloride, zileuton, and syrolymus, selected as models, were loaded exploiting the fabrication procedure used for MyVes. Briefly, myelin (1 mg/ml) and drugs (0.5 mg/ml) were respectively dissolved in DMF. The dispersions were then added drop by drop to an excess of DI water under Ultraturrax mixing (8000 rpm, 15 min). The suspensions were then filtered with a 5 μm filter, then dialyzed (cut-off 3500 Da) over pure water, finally were collected and FD. For the analysis of dimensional distributions and zeta potential, FD samples were dispersed in water, sonicated, and analyzed. For loading measurements, weighted amounts of samples were dispersed in DMF and drugs were quantified by High-performance liquid chromatography (HPLC) methods using a reversed-phase C18 Luna column (5 μm of 250 mm x 4.6 mm internal diameter; Phenomenex). In particular, Dox HCl was quantified by eluting with a mobile phase composed of methanol and ammonium phosphate 10 mM, pH 4.5 (70:30 v/v) with a flow rate of 0.6 ml/min at 260 nm [38]. Instead, zileuton and sirolymus were quantified using a mobile phase of water/methanol (30/70, v/v) using a flow rate of 0.6 ml/min, at 260 nm [39,40]. The procedures were repeated in triplicate and drug loading was expressed as amount of drug loaded respect total amount of samples (w/w %).
2.7. Nanoparticle tracking analysis
Nanoparticle size distribution and concentration were measured using a NanoSight NS300 (Malvern Panalytical). The instrument was equipped with a 488 nm laser, a high-sensitivity sCMOS camera and a syringe pump. The MyVes particles were diluted in particle-free water (Water, HPLC grade, Sigma-Aldrich, filtered by 20 nm using Whatman Anotop filters) to generate a dilution in which 20–120 particles per frame were tracked to obtain a concentration. For each sample, 5 experiment videos of 60 s duration were analyzed using NTA 3.4 Build 3.4.003 (camera level 15–16) with syringe pump speed 60. A total of 1500 frames were examined per sample, which were captured and analyzed by applying instrument-optimized settings using a suitable detection threshold so that the observed particles are marked with a red cross and that no more than five blue crosses are seen. Further settings such as blur size and Max Jump Distance were set to “automatic” and viscosity was set to water (0.841–0.844 cP).
2.8. ATR-FTIR measurements
Attenuated total reflectance-fourier transform infrared (ATR-FTIR) spectra were collected using a Bruker Alpha FTIR spectrometer from 4000/cm to 400/cm. In total, 5 μl of sample was mounted on the diamond ATR crystal and a thin dry film was obtained by slow evaporation of the buffer solvent under ambient conditions. The measurements were performed at room temperature, immediately after drying the sample. Resolution was set at 2/cm. Each measurement is the result of the average of 64 scans.
2.9. Cryo-EM
Immediately after preparation 3.5 μl of MyVes at the final concentration of 1 mg/ml was applied on a copper 300-mesh Quantifoil R2/1 holey carbon grid, previously glow discharged for 30 s at 30 mA using a GloQube system (Quorum Technologies, Laughton, UK). After 60 s of incubation, the grid was plunge-frozen in liquid ethane using a Vitrobot Mk IV (Thermo Fisher Scientific, Waltham, MA) operating at 4°C and 100% room humidity. Images of the vitrified specimens were acquired using a Talos Arctica Transmission Electron Microscopy operating at 200 kV and equipped with a Falcon3 camera operated in a linear mode (all from Thermo Fisher Scientific). Images with an applied defocus of 3 μm were acquired with a total accumulated dose of 40 electrons per A2 at nominal magnifications of 28,000×.
2.10. Proteomic analysis
Sample preparation
Before performing in-solution enzymatic digestion, the pellet was delipidated through four washes in ether/ethanol (2:1) as reported elsewhere [41]. The pellet was washed twice in water, re-dissolved in 50 μl of 50 mM NH₄HCO₃ (pH 8.2) and 0.1% RapiGest SF surfactant (Waters, Milan, Italy), and kept 15 min at 55°C to promote sample denaturation. At this stage, the protein content was calculated performing a fluorimetric assay with the Qubit Protein Assay Kit (Qubit 1.0 Fluorometer;Thermo Fisher Scientific). Finally, an amount of sample equivalent to 10 μg of quantified proteins was reduced by adding 0.1 M, 1,4-dithiothreitol in 50 mM NH₄HCO₃ pH 8.2 (3 h, RT), alkylated with 0.2 M iodoacetamide in the same buffer solution (in the dark at RT for 1 h) and digested overnight at 37°C by a solution of porcine trypsin (Sequencing Grade Modified Trypsin, Porcine, Promega, USA) corresponding to an enzyme–substrate ratio of 1:50 [42]. The peptide mixture solutions were vacuum dried (Concentrator Plus; Eppendorf, Germany), dissolved in 100 μl of H2O + 5% formic acid (FA), and analyzed by Reversed Phase-nano-High-Performance Liquid Chromatography-nano-Electrospray Ionization-tandem mass spectrometry (RP-nHPLC/nESI-MS/MS). The reproducibility of MS data was assessed analyzing the sample in triplicate.
Mass spectrometric analysis
Mass spectrometry data were acquired on a Thermo Fisher Scientific Orbitrap Fusion Tribrid® (Q-OT-qIT) mass spectrometer (Thermo Fisher Scientific) as elsewhere reported [42]. Liquid chromatography was carried out with a Thermo Scientific Dionex UltiMate 3000 RSLC nanosystem (Sunnyvale, CA) first loading the peptide mixtures onto a trapping column (Acclaim® Nano-Trap C18 Column; 100 μm i.d. × 2 cm, 5 μm particle size, 100 Å) with washes of H2O + 0.1% FA for 3 min at a flow rate of 7 μl/min, and subsequently onto a PepMap® RSLC C18 EASY-Spray column (75 μm i.d. × 50 cm, 2 μm particle size, 100 Å). The peptides were eluted by a linear gradient of ACN + 0.1% FA in H2O + 0.1% FA, 5% for 3 min, followed by 5%–20% in 32 min, 20%–40% in 30 min, 40%–60% in 20 min, and 60%–98% in 15 min. The elution was completed with 98% of acetonitrile (ACN) + 0.1% FA for 5 min, 98%–5% in 1 min, and a subsequent re-equilibration at 5% of ACN + 0.1% FA for 20 min. The elution was achieved with a flow rate of 0.25 μl/min at 40°C. The eluting peptide cations were converted to gas-phase ions by electrospray ionization using the same instrumental parameters reported in [42].
Database search and protein identification
MS data were analyzed with PEAKS X Pro de novo sequencing software v. 10 (Bioinformatics Solutions Inc., Waterloo, ON). MS data were searched against the 8125 reviewed entries of Rattus Norvegicus downloaded from UniProt database (March 2021). The c-RAP contaminant database (common Repository of Adventitious Proteins) was included in the database search. Database search was carried out using the following fixed modification: (i) full tryptic peptides with a maximum of three missed cleavage sites and (ii) cysteine carbamidomethylation. The variable modifications selected were: (i) transformation of N-terminal glutamine and N-terminal glutamic acid residue to pyroglutamic acid form; (ii) oxidation of methionine; and (iii) deamidation of asparagine and glutamine. The mass tolerance threshold was set to 10 ppm for each precursor; the maximum mass error for each fragment was set to 0.6 Da. Validation of the peptide spectral matches (PSM) was performed with a Target Decoy PSM Validator node based on q-values at 1% false discovery rate.
Each protein was considered identified if detected at least twice out of three replicates, with a minimum of two peptides and at least one unique peptide in each sample. Proteins containing the same peptides which could not be differentiated according to (Mass specrometry/Mass spectrometry) MS/MS analysis were grouped to satisfy the principles of parsimony. The final list of identified proteins so obtained and reported in the Supplementary Materials (Table S1) was normalized for the average area of the most abundant protein in the sample (myelin basic protein, MBP).
The mass spectrometry proteomics data have been deposited in the Proteome Xchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [43] with the dataset identifier <PXD026282>.
2.11. Cell cultures
SH-SY5Y cells generously provided by Dr. Venera Cardile, University of Catania, Italy, were cultured in T25 tissue culture flasks. Complete Dulbecco’s Modified Eagle’s Medium and F12 (DMEM/F12; 1:1) was used, supplemented with 10% fetal bovine serum (FBS) (GIBCO Invitrogen, Milan, Italy), 100 U/ml penicillin and 100 U/ml streptomycin, and 2 mM l-glutamine.
BV2 microglial cells were cultured in D’ME’M (GIBCO, Carlsbad, CA) supplemented with 10% FBS (Gibco), 100 U/ml penicillin and 100 U/ml streptomycin, and 2 mM l-glutamine.
HuH7 human liver cancer and H292 human pulmonary mucoepidermoid carcinoma cells were cultured with RPMI 1640 medium (Celbio srl, Milan, Italy) supplemented with 10% FBS (GIBCO Invitrogen), 100 U/ml penicillin and 100 U/ml streptomycin, and 2 mM l-glutamine.
Human epithelial cell line Caco-2 was cultured with MEM/F12 supplemented with 10% (v/v) supplemented with 10% FBS (Gibco), 100 U/ml penicillin and 100 U/ml streptomycin, and 2 mM l-glutamine. All the cell lines were grown in a humidified atmosphere of 95% air and 5% CO2 at 37°C.
2.12. Determination of cell viability
Cell viability was measured by MTS assay (Promega). MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3carboxymethoxyphenyl)-2-(4-sulphophenyl)-2H-tetrazolium] was utilized according to the manufacturer’s instructions. Cells were seeded at a density of 2 × 104 cells/well on 96-well plates. After the treatment, with different doses (0.5, 1, and 1.5 μg/ml) of MyVes for 48 h, 20 μl of the MTS solution was added to each well, and incubated for 4 h at 37°C, 5% CO2. The absorbance was read at 490 nm on the Microplate reader Wallac Victor (PerkinElmer). Results were expressed as the percentage of MTS reduction relative to the control. The treated cultured cells and the controls were morphologically analyzed by microscopy inspection on an Axio Scope 2 microscope (Zeiss).
2.13. Permeability assays by BBB cell model
For permeability experiments, the Caco-2 cell monolayer was cultured on Transwell, polycarbonate filter inserts (0.4-mm pore size; 12 mm diameter, Costar) in 12-well plates. The 1 × 105 CaCo2 cells were seeded on the Transwell inserts. The monolayer integrity was evaluated by measuring transepithelial electrical resistance (TEER) at different time (0, 1, and 9 days). The apical chamber was filled with MyVes-Alexa 488 (5 μl of MyVes-Alexa 488 at 1.34e+11 ± 1.13e+10 particles/ml in 200 μl of cell medium) and NaFluo (negative control) [6] at different times (0, 1, 4, and 20 h) and 0.2 ml was picked up from the basal chamber. Fluorescence intensity was determined by the Wallac Victor 2 1420 Multilabel Counter (PerkinElmer, Inc.) microplate reader. The excitation and emission wavelengths were set at 485 and 530 nm, respectively.
2.14. Genotoxicity assay on BBB cellular model
At the final point of the BBB permeability experiments, the medium was removed and cells, in the apical chamber, were washed twice with phosphate-buffered saline (PBS) and subsequently stained with acridine orange/PBS solution with calcium and magnesium (Sigma) at 100 μg/ml for 10 s at room temperature. After three wash in PBS solution, cells are quickly examined by fluorescence microscopy (Leica). Acridine orange is a cell permeating nucleic acid binding dye that emits green fluorescence when bound to double-strand DNA and red fluorescence when bound to single-stranded DNA (ssDNA) or RNA. This staining technique allows discriminating between intact (green nuclei) and damaged DNA in cells (red nuclei).
2.15. Cerebellar slices
Brain slices (BS) were prepared from bovine brain or C57BL/6 mouse. Bovine brain cortex was cut into coronal sections of 400–500 μm and mouse brain serial coronal cryostat sections of 14 μm were prepared. The tissue slices were placed in a Petri dish containing PBS with 100 units/ml of penicillin and 100 μg/ml of streptomycin. The BS was incubated with MyVes-Alexa 488 (10 μg/ml) or Synapt-Ves-Alexa 488 (10 μg/ml) for 3 h or with NileRed for 10 min; the untreated BS was used as controls. Furthermore, some BS was incubated with NileRed dye for 10 min, washed twice with PBS, and subsequently stained with MyVes-Alexa 488 (10 μg/ml) for 3 h. Then, the BS was washed twice with PBS and observed by the Typhoon 8600 fluorescence scanner (GE Healthcare). Exc/Emi 488/530 for MyVes-Alexa 488 and Synapt-Ves-Alexa 488, Exc/Emi 530/630 for NileRed.
2.16. Myelin nanovesicles cellular uptake
For interaction studies, SH-SY5Y, BV2, H292, and A549 cells were sub-cultured once they reached 60% confluence in 96-well plates. The cells were incubated for 4 and 24 h with 5 μl of MyVes-Alexa 488 in 100 μl of cell medium (1.34e+11 ± 1.13e+10 particles/ml). After three wash in PBS solution, the cells are examined by fluorescence microscopy (Leica). The excitation and emission wavelengths were set at 488 and 530 nm, respectively.
2.17. Statistical analysis
All experiments were repeated three times, and in each experiment the samples were taken in triplicate. The results are presented as mean ± standard deviation. Statistical evaluation was conducted by analysis of variance, for analysis of significance. Results with a P value ≤ 0.05 were considered statistically significant, ∗P ≤ 0.05, ∗∗P ≤ 0.01.
3. Results and discussion
3.1. Bio-fabrication of the myelin nanovesicles
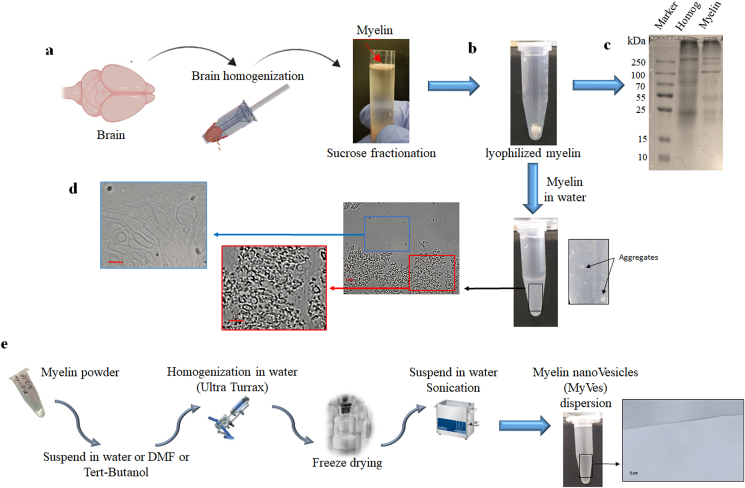
To produce MyVes, myelin was isolated from the rat brain cortex using the sucrose fractionation procedure [34]. Different technical approaches can be used to extract and purify myelin. The method used involve homogenization of the tissue in isotonic sucrose solution, followed by the isolation of myelin membranes by density gradient centrifugation [33,34]. We used 0.32 M sucrose to homogenize the tissue, during this process; the isotonic sucrose causes the myelin sheath to peel from the axon. Subsequently, we chose to subject the sample to an ultracentrifugation in high-density sucrose gradient (1 M); this process allows, compared to lower densities, to obtain a myelin fraction rich in fragments and bilayer structures [33]. After ultracentrifugation, the floating fraction containing myelin (Fig. 1a) was collected, washed and lyophilized (Fig. 1b).
Fig. 1.
Bio-fabrication of the myelin nanovesicles. (a) Schematic representation of the myelin extraction procedure starting from brain tissue by homogenization and fractionation of sucrose. (b) Representative picture of lyophilized myelin in a vial. (c) Protein electrophoresis and Coomassie blue staining of proteins extracted from total homogenate (Homog) and myelin powder (Myelin). (d) Visualization of the structure of myelin suspended in water, aggregate structures, and multiple micrometric vesicles are shown. (e) Schematic representation of the protocol for bio-fabrication of myelin nanovesicles dispersion; myelin powder was dispersed in water or DMF or tert-butanol at 0.1 mg/ml and precipitated in an excess of ultrapure water. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Myelin is composed of about 40% water, proteins, and a high percentage of lipids. The dry mass is characterized from 70% to 85% of lipids, from 15% to 30% of proteins [44], and its composition is highly conserved. To test for the presence of proteins, the myelin powder extracted (Fig. 1b) was analyzed by SDS-PAGE and stained with blue Coomassie (Fig. 1c). Moreover, the total protein fraction, present in extracted myelin and determined by Bradford assay, was about 22% of the total myelin; this value being in line with previous studies in literature [44].
Myelin extract resulted in soluble DMF and tert-butanol while formed aggregates in water (Fig. 1d), isopropanol, ethanol, and methanol. In particular, by microscopy inspection the myelin suspended in water presented aggregate structures and large size micrometric multiple vesicles (Fig. 1d). To obtain nanometric vesicles, nanoprecipitation by a solvent injection procedure was then performed by dripping of the solutions of myelin extracts in water under Ultraturrax® (U) mixing (Fig. 1e). The simple dispersion of myelin extracts in water, without the employment of organic solvent, generates, after Ultraturrax mixing (sample Water U) (Table 1), nanovesicles of about 144 nm. Re-dispersion of FD samples produced vesicles with a Z-average of 130 nm (sample Water U-FD) (Table 1). In fact, as shown in Fig. 1e, the absence of micrometric structures, revealed by microscopic inspection, were observed in the MyVe dispersion in water, after nanoprecipitation (Fig. 1e). Instead, nanoprecipitation performed by using DMF as organic solvent to solubilize myelin extracts generates, after Ultraturrax mixing (sample DMF U in Table 1), nanoparticles with a Z-average of 83 nm. Re-dispersion of FD sample achieved from aqueous nanoprecipitation of myelin dissolved in DMF (sample DMF U-FD) and tert-butanol (sample tert-Butanol U-FD) showed a Z-average of 103 and 71 nm, respectively. Data reported showed how nanoprecipitation allowed the fabrication of vesicles with a nanometric dimension, and vesicles stability was just minimally affected by freeze-drying procedure. Moreover, we have measured ζ-potential values to determine nanovesicle stability in the solution. Our results (Table 1), with the ζ-potential values, ranging from −37 to −42 mV, were consistent with an anionic nanovesicle surface with high stability in water [45]. PDI, used to estimate the average uniformity of a particle solution, has been measured and shown in Table 1.
Table 1.
Experimental conditions employed for MyVes production and physical characteristics: Z-average, ζ-potential, and PDI values.
| Fabrication procedure | Z-average (nm) | ζ-potential (mV) | PDI |
|---|---|---|---|
| Water U | 144 | −39 | 0.331 |
| Water U-FD | 130 | −37 | 0.243 |
| DMF U | 83 | −40 | 0.297 |
| DMF U-FD | 103 | −42 | 0.399 |
| Tert-butanol U-FD | 71 | −37 | 0.286 |
Therefore, here we have presented the bio-fabrication of myelin-derived nanovesicles with an easy, scalable, time-efficient (within 3 h), and cost-effective manufacturing method. Furthermore, neither cell cultures nor lab-intensive efforts were needed to isolate MyVes compared, for example, to the production of natural EVs [46].
3.2. Myelin nanovesicle characterization
For biological applications, we chose to characterize and use MyVes produced in water (water U-FD in Table 1). Besides the obvious advantage of being produced in water, these particles presented the lower PDI, average size of about 130 nm, and high stability assessed by ζ-potential. First, the MyVes produced in water were characterized by nanoparticle tracking analysis (NTA) (Fig. 2a). NTA exploits laser light to illuminate freely diffusing particles, tracks their Brownian motion by analysis of monochrome images, and estimates their size distribution through an algorithm based on Stokes–Einstein relation. Compared to dynamic light scattering (DLS; Table 1), where an average particle size is obtained (ensemble measurement) and the particle size distribution is polarized toward large particles, NTA, due to the particle-by-particle measurement, provides better resolution for mixtures of particles that vary in size [47]. The NTA size distribution indicates that most of the particles in solution have diameter mode of about 100 nm and a small fraction have diameter mode of about 160 nm (Fig. 2a). The other particles present in solution are numerically negligible. Moreover, 0.1 mg/ml of MyVes preparation correspond to a total concentration of 1.34e+11 ± 1.13e+10 particles/ml. Infrared spectroscopy (IR) has been used as a fast and simple method for EV characterization via measurements of absorption peaks of functional groups [48]. In particular, to verify the presence of lipids in our sample we performed ATR-FTIR analysis [49,50]. Specifically, we analyzed two different regions of the FTIR spectrum: 4000-2500/cm (Fig. 2b) and 1200-800/cm (Fig. 2c), where bands related to main functional groups such as –COOH, –NH, –OH, and –CH occur. The most intense absorption in the 4000–2500/cm region is due to different –OH stretching vibrations of water molecules indicating the presence of residual water trapped in the sample. Fig. 2b shows the presence of two prominent broad bands at 2930 and 2884/cm that can be attributed to the asymmetric and symmetric stretching vibrations of the carbon–hydrogen bonds (–CH2 and –CH3) [48,50]. In Fig. 2c, the presence of phospholipids is confirmed by the two main peaks at 1109 and 1041/cm due to stretching of PO−2 groups (Fig. 2b) [48,50]. To investigate the morphological characteristics typical of a vesicle-like structure, MyVes cryo-EM measurements were performed (Fig. 2d). In particular, in Fig. 2d representative examples of vesicles with single double membrane, spherical and with a diameter of 100–150 nm, comparable to the dynamic light scattering (Table 1) and NTA (Fig. 2a) analysis, without dense electron load in the lumen were shown. The near absence of multilayer vesicles is due to the high-density fractionation of sucrose used for myelin extraction, which contains mostly single-bilayer myelin vesicles and fragments [33].
Fig. 2.
Myelin nanovesicles characteristics of (a) MyVes size distribution graph by NTA tool. ATR-FTIR absorption spectrum (b) contributions from specific lipid groups are visible in the C–H stretching region (3000–2750/cm) and (c) in the asymmetric and symmetric stretching region of PO2− (1200–1000/cm). (d) Representative Cryo-EM images of MyVes; scale bar is 100 nm. (e) Pie graph of MyVes proteins identified by top-down proteomic approach. (f) Schematic representation of the hypothetical structure of MyVes. (g) Schematic illustration of the efficiency of the MyVes preparation.
Subsequently, in order to characterize the MyVes proteomic composition, here, we performed in-solution digestion of the MyVes, and peptides were analyzed by Reversed Phase- nano- Ultra-High-Performance Liquid Chromatography-nano-Electrospray Ionization-tandem mass spectrometry (RP-nUHPLC/nESI-MS/MS) with subsequent database search against the Rat UniProt database. A total of 442 proteins were identified. Interestingly, the main known myelin proteins, such as MBP, myelin proteolipid protein (PLP), 2′-3′-cyclic-nucleotide 3′-phosphodiesterase (CNP), myelin-associated glycoprotein (MAG), calmodulin-1, and myelin-oligodendrocyte glycoprotein (MOG), were present in the MyVes and represent about 70% of the total identified protein content (Fig. 2e). These results are in agreement with the literature data, where classical myelin proteins, with structural roles, such as MBP, PLP, CNP, MAG, MOG, and Oligodendrocyte specific protein (OSP), in both human and mouse myelin, are known to make up nearly all myelin proteins [51]. In particular, MBP and PLP are nearly 85% of the protein content in myelin [52]. Recently, main myelin proteins (MBP, PLP, MOG, CNP) were identified in brain tissue-derived EVs [53] and in oligodendrocyte-derived EVs [54].
PLP and MBP are found in compact internodal myelin. MBP through the positive charge interacts with the negatively charged internal part of the plasma membrane, bringing the layers of myelin closer together; therefore, it is essential for the formation of myelin sheath in the CNS [55]. PLP mainly plays a stabilizing role by bringing together the extracellular flaps of the myelin membrane [56]. Other myelin proteins (475 proteins) have also been identified although with a different experimental proteomic approach [51], similar to the 442 proteins identified here. The complete list of the remaining proteins that make up a small minority in percentage terms is provided in Tables S1 and S2. Functional enrichments in the MyVes-identified proteins were highlighted by using the Fun Rich tool (Fig. S3). As expected, when the cellular component was taken into account, the enrichment affects the neuron projections, axons, and synapses (Fig. S3). Biological processes and pathways (Fig. S3) in which the identified proteins are involved included the synaptic vesicle endocytosis and transport, brain development, and metabolic processes, suggesting that the MyVes-identified proteins, quantitatively present as “minor” proteins of myelin may play important roles in oligodendrocytes and myelin biology [51]. A MyVes hypothetical structure containing lipids and proteins is represented by the image shown in Fig. 2f. Furthermore, to demonstrate the efficiency of the MyVes preparation method, Fig. 2g shows the schematic illustration of the preparation steps, starting from the amount of brain tissue up to the high concentration of particles (particles/ml) obtained.
3.3. Myelin nanovesicles cytocompatibility
To determine the potential translational relevance of MyVes as a brain delivery system, we evaluated, as a first step, the effect on the cell viability of the MyVes in different cell lines: neuronal (SH-SY5Y), microglia (BV2), epithelial (H292), and hepatocyte (HuH7) cells. The cells were treated for 48 h with MyVes at increasing concentrations. The results reported in Fig. 3 showed that the percentage of viability of the treated cells is similar to the control, indicating a good MyVes cytocompatibility. Moreover, no proliferative effect was observed (Fig. 3). The cytocompatibility was also confirmed by microscopic inspection, where no morphological change was detected in all treated cell lines (Fig. 3). The main factors affecting cytotoxicity, for nanosystems, are material, size, shape, composition, surface charge, and surface hydrophobicity [57]. Positive surface charge, hydrophobic surfaces, fiber shape, and small size (<100 nm) correlate with increased nanoparticle cytotoxicity [57]. Moreover, literature data indicated a cellular administration of myelin at doses ranging from 1 to 100 μg/ml [58,59]. Therefore, possibly due to their nature (myelin) (Fig. 1b) and their properties such as negative surface charge, dimensions (Table 1 and Fig. 2a), and spherical shape (Fig. 2d), these vesicles, at the administered doses, do not induce any alterations in cellular viability.
Fig. 3.
Myelin nanovesicles cytocompatibility. Cell viability and morphological analysis of (a) microglia (BV2), (b) epithelial lung cells (H292), (c) hepatocytes (HuH7) and (d) neurons (SH-SY5Y) treated for 48 h with MyVes at different doses.
3.4. MyVes labeling with fluorescent probes and drug loading suitability
As reported in the previous section, both proteins and lipids are constituent elements of these new nanovesicles. For this reason, in order to track their fate it is possible to label the protein component using the succinimidyl ester Alexa Fluor 488 (MyVes-Alexa 488) (Fig. 4a). The small reduction of the ζ-potential (Fig. 4a), compared to the empty nanovesicle (Table 1) suggest that binding of the Alexa Fluor 488 succinimidyl ester to the proteins was performed. Moreover, the results of the fluorescence intensity (Fig. 4b) showed that the procedure was suitable for staining the MyVes and, therefore, as vesicle tracing. We used the fluorophore Di-8-ANEPPS, as a lipophilic mimetic compound, to detect tethering suitability of MyVes membrane. Di-8-ANEPPS is a no fluorescent molecule, which emits an intense fluorescence when intercalated in the lipophilic environment (Fig. 4c). The high fluorescence intensity shown in Fig. 4d conceivably suggest that the lipophilic nature of the MyVes membrane could be exploited for tethering (o labeling) purposes.
Fig. 4.
Labeling and loading of fluorescent probes in MyVes. (a) Schematic representation and ζ-potential of the MyVes labeled with succinimidyl ester Alexa Fluor 488 (MyVes-Alexa 488). (b) Histogram of the fluorescence intensity of the vehicle (PBS), MyVes and NyVes-Alexa 488. (c) Schematic representation of MyVes labeled with Di-8-ANEPPS (NyVes-Di-8). (d) Histogram of the fluorescence intensity the vehicle (PBS), MyVes and NyVes-Di-8. ∗∗P ≤ 0.01.
In order to have a preliminary assessment of potential suitability of MyVes as drug carrier, three different model drugs have been loaded, simply exploiting for drug encapsulation of the solvent injection procedure described for MyVes fabrication. Doxorubicin hydrochloride (Dox HCl), zileuton, and syrolymus were considered as examples of drugs with increasing logP (1.3, 1.6, and 6 respectively). Table S4 reports physical properties of vesicles produced, and drug loading values.
Data show a detectable amount of drugs loaded, reaching values of 2.98, 2.01, and 1.97 w/w %, for Dox HCl, syrolymus, and zileuton, respectively (Fig. S4). Just a slight increase in loading of Dox HCl compared to zileuton and syrolymus was registered, with a significant reduction in zeta potential and increased Z-average, which could be attributed to a superficial ionic interaction of the drug with the phospholipidic double layer and a possible vesicle destabilization. Even if the low loading efficiency (less than 10% for all three drugs) evidenced that parameters of solvent injection procedure should be more specifically tuned or fabrication procedure opportunely changed in order to improve drugs loaded amount, the data observed suggest the potential suitability of MyVes as drug carrier [60,61].
3.5. In vitro BBB model permeation study
The impermeability characteristic of the BBB is considered the main reason for the failure of the attempts to achieve therapeutic concentrations of the drug in the brain [7]. In order to evaluate the potential permeability efficiency of MyVes through the BBB, we used Transwell-cultured human epithelial cells (CaCo2), a suitable model for studying solute and particle transport through the BBB [[62], [63], [64]]. To characterize the BBB model, the CaCo2 cells plated in Lab-Tek chambers were subjected to TEER [65] measurements. The data shown in Fig. 5a reported that at 9 days, the TEER value was approximately 500 Ohm∙cm2 and the occurred formation of the cell monolayer as revealed by nuclear fluorescence staining (Hoechst) (Fig. 5a). These results indicated that the cellular monolayer mimics the BBB.
Fig. 5.
Crossing a BBB model. (a) Schematic representation of human epithelial cells (CaCo2) cultured in Transwell insert, the nuclei of the cell monolayer were stained with Hoechst 3342 fluorescence dye (blue), and the TEER values were reported in the histogram. (b) Histogram of the fluorescence intensity of the MyVes and NaFluo (negative control) detected in the acceptor chamber at different times (0, 1, 4, and 20 h). (c) Fluorescence image of the cell monolayer stained with acridine dye at the end of 20 h. ∗P ≤ 0.05, ∗∗P ≤ 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
MyVes-Alexa 488 and NaFluo (negative control) were loaded into the apical chamber of the Transwell insert (Fig. 5a). Subsequently, small quantities of the samples were taken from the basolateral chamber at different times (0, 1, 4, and 20 h) and the fluorescence signal was measured (Fig. 5b). The intensity of the fluorescence signal increased in a time-dependent manner (Fig. 5b), indicating the ability of MyVes to cross the BBB in vitro model.
Furthermore, to explore whether the MyVes crossing the BBB model could induce cellular stress and/or alteration of the monolayer, inducing breakdown of the BBB model, the cellular monolayer was stained with acridine dye at the end of the permeability experiments, using non-labeled MyVes. Acridine is a nucleic acid-binding dye that permeates cells and emits a green fluorescence when bound to double-stranded DNA and a red fluorescence when bound to single-stranded DNA (ssDNA), being ssDNA a typical marker of apoptosis. By fluorescence microscopic analysis, the cells showed green fluorescence only (Fig. 5c) indicating the absence of toxic effect of MyVes during the BBB crossing process. Several studies have reported that nanoparticles between 1 and 100 nm facilitate drug transport across the BBB due to their smaller size [9]. In addition, brain-targeted drug success and overcoming BBB have been reported in mice by using nanovesicles with a mean diameter of 100 nm [66,67]. The ability of MyVes to cross the BBB cellular model could be due to some proteins present in MyVes, such as transferrin, apolipoprotein E, and glutathione (Table S1), which are known to promote the crossing of nanosystems through the BBB [10].
3.6. MyVes interact with the brain white matter and microglia cells
The brain is composed of an outer layer of grey matter, consisting of cell bodies, dendrites, and unmyelinated axons, and an inner white matter, consisting mainly of myelinated axons (Fig. 6a). Organotypic cerebellar slice cultures appear to be the best alternative to in vivo experiments and the most used model to study the etiology of new therapeutic strategies of brain disease [68]. Moreover, it is very difficult to obtain the myelin sheath in vitro by co-culture techniques. As a first test, we used 400–500 μm thick slices of bovine brain, where the white and grey matter, brain-specific areas, are visible and clearly recognizable thanks to the brain bovine size (Fig. 6a).
Fig. 6.
Ability of MyVes to interact with white matter. (a) Image of the coronal section of the bovine brain, where the gray matter formed by the cell body of neurons and the white matter formed by myelinated axons were visible. (b) Bovine BS 400–500 μm thick, incubated or not (Control) with NileRed dye, MyVes-Alexa 488 (MyVes), and MyVes-Alexa 488 with NileRed dye (MyVes-NileRed), analyzed by fluorescence scanner. The high magnification of fluorescent portions of the brain sections and their merged are shown in the squares. (c) Coronal brain mouse slides stained with NileRed dye and MyVes-Alexa 488 (MyVes) visualized by fluorescence scanner; cortex (CCx), corpus callosum (CC), striatum (ST), external capsule (EC), and lateral olfactory tract (LOT). The high magnification of fluorescent portions of the brain sections and their merged, are shown in the squares.
Bovine BS was incubated with MyVes-Alexa 488 to test the affinity for gray or white matter. The myelin sheath in BS was stained with NileRed fluorescence dye (red) [69] (Fig. 6b). By fluorescence scanner analysis, MyVes-Alexa 488 (green) was detected in the same region of the NileRed (Red) indicating the capacity of MyVes to interact with myelin sheath in white matter (Fig. 6b). Furthermore, to confirm this interaction we used coronal mouse brain sections (Fig. 6c), where the typical global distribution of myelin in the whole brain [70] was shown by NileRed staining (Fig. 6c). The brain slide incubated with MyVes-Alexa 488 showed that the MyVes colocalized with NileRed stain, confirming the ability of MyVes to interact with myelin (Fig. 6c). In order to determine if the localization of MyVes to myelin sheath is specific, we used a vesicle of different nature, obtained from the synaptic terminals, called synaptosomal vesicle [22]. Results indicated that these vesicles showed a different localization with respect to MyVes, demonstrating the specificity of MyVes for white matter (Fig. S5).
The ability of MyVes to interact with the myelin sheath possibly depends on the nature of the nanovesicles. In fact, the myelin sheath is a tightly compact multilayer membrane that has a specific set of proteins important for the structure and formation of myelin sheath such as MBP and PLP [56].
Studies on the interaction of MBP with myelinated lipids indicated the electrostatic nature of the attraction between MBP and lipids, showing that there are approximately 20 anionic lipid molecules per MBP molecule in myelin [70]. Furthermore, the evidence indicates that the hydrophobic segments of MBP interact with the hydrophobic lipid chains by partially penetrating the lipid bilayers [71,72]. Furthermore, the 116–150 PLP fragment plays an important role in the interaction of PPL with the lipid bilayer in the myelin sheath [73]. It is possible that MyVes due to the presence of MBP and/or PLP, which represent the 53% and 9% of the total MyVes protein, respectively (Fig. 2e), interact with the myelin sheath in a similar way to the formation of the myelin sheath.
Alterations in white matter can result in various neuronal diseases, such as MS, AD, TBI, and vascular cognitive impairment and dementia including subcortical ischemic vascular dementia [24]. White matter–related diseases have commonly reported microglia activation, indicating that it plays a pivotal role in these diseases [24]. Moreover, in human and rat brains, the white matter contains a significantly higher number of microglia and macrophages than the gray matter [74,75]. Recent studies reported that potential therapeutics associated with modulation of microglia could improve white matter integrity in various injury models [23]. To evaluate whether interaction of MyVes depends on the cell type, nervous cell lines (neurons SH-SY5Y and microglia cells BV2) or not (epithelial cells H292 and hepatocytes HuH7) were incubated with MyVes-Alexa 488 for 4 and 24 h. The relative fluorescence images showed that the highest levels of fluorescence were detected in the microglia (BV2) cells (Fig. 7).
Fig. 7.
Interaction of MyVes to microglia cells. Representative fluorescence images of microglia (BV2), epithelial (H292), hepatocyte (HuH7), and neuron (SH-SY5Y) cells incubated with MyVes-Alexa 488 for 4 and 24 h. High-magnification images of the cells are highlighted in the circles.
These results indicate that MyVes have a preferential targeting ability for microglial cells. This effect is in agreement with the literature; in fact, microglia are the key effector cells that remove myelin debris [59]. Several studies showed that myelin phagocytosis by microglia and macrophages triggers the release of proinflammatory cytokines and nitric oxide [[76], [77], [78]], suggesting that phagocytosis of myelin could enhance neuroinflammation. In contrast, other studies reported an anti-inflammatory effect of microglia myelin phagocytosis [58,59,79]. Liu et al. [79] proposed that phagocytosis of myelin modulates activation of microglial cells with a biphasic temporal pattern: (i) enhanced production of proinflammatory mediators during the first phase and (ii) a pronounced suppression in the second one.
Our system is different from the studies indicated, and the nanostructuring of myelin leads to the production of nanovesicles that could acquire different biological properties with respect to the unstructured myelin. In fact, the physical–chemical properties of nanostructures such as size, surface chemistry, surface charge, and shape are important factors influencing the binding and absorption by microglia and the biological response [25]. Studies are needed to evaluate the microglia MyVes potential effect. The liver acts as a biological filtration system sequestering 30%–99% of the nanoparticles administered through the bloodstream [80]; the reduced uptake shown by hepatocytes (HuH7) could suggest the potential ability of MyVes to escape to the hepatic clearance.
The CNS resident immune cells, microglia, are emerging as a promising cellular target for neurodegeneration considering their prominent role in neuroinflammation diseases [25]. Different studies reported that nanoparticle DDSs increase the uptake of drugs in microglia cells [[25], [26], [27], [28], [29], [30], [31]]. Different approaches such as conjugation of microglia receptor-specific ligands or peptides to the nanoparticle surface to achieve targeted delivery, exploiting the phagocytic properties of microglia were investigated [25]. Different researches indicate that the drugs encapsulated in the nanosystems, after microglia phagocytose, are able to modulate their activation and related neuroinflammation [25]. Moreover, expression of phagocytic receptors on microglia is increased under neuroinflammatory conditions and this condition is particularly beneficial for targeted delivery of nanotherapeutics to these cells [25].
However, a specific DDS potentially able to target white matter and microglia has not yet been identified. The preliminary biological data here reported, obtained by ex vivo BS and in vitro cells, suggest the potential role of MyVes as suitable carrier for this targeting. However, further in vivo studies are needed to confirm these interactions.
Therefore, the MyVes are vesicles produced with an easy, scalable, efficient, cost-effective, and reproducible protocol. They have an average diameter of 100–150 nm, negative zeta potential, and key proteins of the myelin sheath (MBP and PLP). In addition, they can target a specific region of the brain, white matter, and interact preferentially with microglia cells, actors involved in white matter-related disease, such as MS (Fig. 8).
Fig. 8.
Schematic representation of myelin nanovesicle production from brain tissue by myelin extraction and nanostructuring. Biophysical characteristics and biological applications are summarized.
Thanks to the characteristics reported in this first study, MyVes could be further considered to evaluate their potential as carriers to load and deliver an immunomodulatory drug to the active microglia residing in the white matter. Furthermore, we cannot exclude that the interaction with myelinated fibers may also find application in the repair of damaged myelin sheath. However, future in vivo investigations are needed to evaluate their effective potential.
4. Conclusions
In this study, we propose, for the first time, the bio-fabrication of myelin-based nanovesicles with an easy, scalable, efficient, economical, and reproducible production protocol. The MyVes produced have an average diameter of 100–150 nm, negative zeta potential (−37mv), spheroidal morphology, lipids, and main proteins of the myelin sheath and show good cytocompatibility. Furthermore, we provide proof of concept of the potential suitability for brain targeting of these novel carriers. In fact, due to their biophysical and biological properties, MyVes cross an in vitro BBB model, reach a specific brain region, the white matter, and interact with microglia cells. For these reasons, we propose MyVes as a potential tool to counteract white matter-related microglial diseases, such as MS. However, this work proposes the first basis and future investigations and optimizations, through in vivo studies, are needed to evaluate their effective potential.
Authorship statement
Conception and design of study: P. Picone; D. Nuzzo; F.S. Palumbo; V. Vetri; A. Bongiovanni; V. Muccilli; P. Cancemi.
Acquisition of data: P. Picone; D. Nuzzo; F.S Palumbo; S. Federico; G. Adamo; V. Muccilli; V. Giglio; A. Chaves; S. Anselmo, G. Sancataldo; V. Di Liberto.
Analysis and/or interpretation of data: P. Picone; D. Nuzzo; F.S. Palumbo; P. Cancemi; V. Vetri; G. Pitarresi.
Drafting the manuscript: P. Picone; D. Nuzzo; F.S. Palumbo.
Revising the manuscript critically for important intellectual content: S. Federico; G. Pitarresi; G. Adamo; A. Bongiovanni; A. Chaves; P. Cancemi; V. Muccilli; V. Giglio; V. Vetri; S. Anselmo; G. Sancataldo; V. Di Liberto.
Approval of the version of the manuscript to be published: P. Picone; D. Nuzzo; F.S. Palumbo; S. Federico; G. Pitarresi; G. Adamo; A. Bongiovanni; A. Chaves; P. Cancemi; V. Muccilli; V. Giglio; V. Vetri; S. Anselmo; G. Sancataldo; V. Di Liberto.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Istituto per la Ricerca e l’Innovazione Biomedica (IRIB) of CNR. The authors gratefully acknowledge the Bio-Nanotech Research and Innovation Tower of the University of Catania (BRIT; PON project financed by the Italian Ministry for Education University and Research MIUR) for the availability of the Orbitrap Fusion mass spectrometer. The authors wish to thank Dr. Daniela Giacomazza for the English revision of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2021.100146.
Contributor Information
Pasquale Picone, Email: pasquale.picone@irib.cnr.it.
Domenico Nuzzo, Email: domenico.nuzzo@cnr.it.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Matilla-Dueñas A., Corral-Juan M., Rodríguez-Palmero Seuma A., Vilas D., Ispierto Lourdes, Morais S., Sequeiros J., Alonso I., Volpini V., Serrano-Munuera C., Pintos-Morell G., Álvarez R., Sánchez I., editors. Inc Advances in Experimental Medicine and Biology. Vol. 1031. Springer; Cham: 2017. Rare neurodegenerative diseases: clinical and genetic update; pp. 443–496. [DOI] [PubMed] [Google Scholar]
- 2.Veronesi M.C., Alhamami M., Miedema S.B., Yun Y., Ruiz-Cardozo M., Vannier M.W. Imaging of intranasal drug delivery to the brain. Am J Nucl Med Mol Imag. 2020;10:1–31. [PMC free article] [PubMed] [Google Scholar]
- 3.Picone P., Sabatino M.A., Ditta L.A., Amato A., San Biagio P.L., Mulè F., a Giacomazza D., Dispenza C., Di Carlo M. Nose-to-brain delivery of insulin enhanced by a nanogel carrier. J. Contr. Release. 2018;28:23–36. doi: 10.1016/j.jconrel.2017.11.040. [DOI] [PubMed] [Google Scholar]
- 4.Badhe R.V., Nipate S.S. Book Wiley Online Library; 2020. Nasal Bioadhesive Drug Delivery Systems and Their Applications. [DOI] [Google Scholar]
- 5.Islam S.U., Shehzad A., Ahmed M.B., Lee Y.S. Intranasal delivery of nanoformulations: a potential way of treatment for neurological disorders. Molecules. 2020;21:1929. doi: 10.3390/molecules25081929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picone P., Ditta L.A., Sabatino M.A., Militello V., San Biagio P.L., Di Giacinto M.L., Cristaldi L., Nuzzo D., Dispenza C., Giacomazza D., Di Carlo M. Ionizing radiation-engineered nanogels as insulin nanocarriers for the development of a new strategy for the treatment of Alzheimer's disease. Biomaterials. 2016;80:179–194. doi: 10.1016/j.biomaterials.2015.11.057. [DOI] [PubMed] [Google Scholar]
- 7.Sharma G., R Sharma A., Lee S., Bhattacharya M., Nam J., Chakraborty C. Advances in nanocarriers enabled brain targeted drug delivery across blood brain barrier. Int. J. Pharm. 2019;559:360–372. doi: 10.1016/j.ijpharm.2019.01.056. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y., Peng Z., Seven E.S., Leblanc R.M. Crossing the blood-brain barrier with nanoparticles. J. Contr. Release. 2018;28:290–303. doi: 10.1016/j.jconrel.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 9.Amit A., Mukta A., Ajaz U., Sabahuddin S., Ahmed M.S., Mahmoud A.S., Syed A.U.R., Mohi I.M.A., Mohamed A.S. Recent expansions of novel strategies towards the drug targeting into the brain. Int. J. Nanomed. 2019;14:5895–5909. doi: 10.2147/IJN.S210876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgieva J.V., Hoekstra D., Zuhorn I.S. Smuggling drugs into the brain: an overview of ligands targeting transcytosis for drug delivery across the blood-brain barrier. Pharmaceutics. 2014;17:557–583. doi: 10.3390/pharmaceutics6040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leggio L., Arrabito G., Ferrara V., Vivarelli S., Paternò G., Marchetti B., Pignataro B., Iraci N. Mastering the tools: natural versus artificial vesicles in nanomedicine. Adv. Healthc. Mater. 2020;9 doi: 10.1002/adhm.202000731. [DOI] [PubMed] [Google Scholar]
- 12.Nuzzo D., Picone P. Multiple sclerosis: focus on extracellular and artificial vesicles, nanoparticles as potential therapeutic approaches. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22168866. 8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haney M.J., Klyachko N.L., Zhao Y., Gupta R., Plotnikova E.G., He Z., Patel T., Piroyan A., Sokolsky M., Kabanov A.V., Batrakova E.V. Exosomes as drug delivery vehicles for Parkinson's disease therapy. J. Contr. Release. 2015;10:18–30. doi: 10.1016/j.jconrel.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong X. Current strategies for brain drug delivery. Theranostics. 2018;8:1481–1493. doi: 10.7150/thno.21254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan S., Wu T., Zhang D., Zhang Z. Cell or cell membrane-based drug delivery systems. Theranostics. 2015;5:863–881. doi: 10.7150/thno.11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu D., Dong X., Shi X., Zhang C., Wang Z. Neutrophil-based drug delivery systems. Adv. Mater. 2018;30:1706245. doi: 10.1002/adma.201706245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y., Sua J., Liua G., Chen J., Zhang X., Jiang R. Zhang M., Qiu M. Advances of blood cell-based drug delivery systems. Eur. J. Pharmaceut. Sci. 2017;96:115–128. doi: 10.1016/j.ejps.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Zhang P., Liua Gang, Chend Xiaoyuan. Nanobiotechnology: cell membrane-based delivery systems. Nano Today. 2017;13:7–9. doi: 10.1016/j.nantod.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J., Chu D.F., Wang Z.J. Cell membrane-formed nanovesicles for disease-targeted delivery. J. Contr. Release. 2016;224:208–216. doi: 10.1016/j.jconrel.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J., Wang S.H., Wang Z.J. High yield, scalable and remotely drug-loaded neutrophil-derived extracellular vesicles (evs) for anti-inflammation therapy. Biomaterials. 2017;135:62–73. doi: 10.1016/j.biomaterials.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong X., Gao J., Zhang C.Y., Hayworth C., Frank M., Wang Z. Neutrophil membrane-derived nanovesicles alleviate inflammation to protect mouse brain injury from ischemic stroke. ACS Nano. 2019;26:1272–1283. doi: 10.1021/acsnano.8b06572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Picone P., Porcelli G., Bavisotto C.C., Nuzzo D., Galizzi G., San Biagio P.L., Bulone D., Di Carlo M., Synaptosomes New vesicles for neuronal mitochondrial transplantation. J. Nanobiotechnol. 2021;19 doi: 10.1186/s12951-020-00748-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subhramanyam C.S., Wang C., Hu Q., Dheen S.T. Microglia-mediated neuroinflammation in neurodegenerative diseases. Semin. Cell Dev. Biol. 2019;94:112–120. doi: 10.1016/j.semcdb.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Lee J., Hamanaka G., Lo E.H., Arai K. Heterogeneity of microglia and their differential roles in white matter pathology. CNS Neurosci. Ther. 2019;25:1290–1298. doi: 10.1111/cns.13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao N., Francis N.L., Calvelli H.R., Moghe P.V. Microglia-targeting nanotherapeutics for neurodegenerative diseases. APL Bioeng. 2020;4 doi: 10.1063/5.0013178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cahalane C., Bonezzi J., Shelestak J., Clements R., Boika A., Yun Y.H., Shriver L.P. Targeted delivery of anti-inflammatory and imaging agents to microglial cells with polymeric nanoparticles. Mol. Pharm. 2020;17:1816–1826. doi: 10.1021/acs.molpharmaceut.9b00489. [DOI] [PubMed] [Google Scholar]
- 27.Nance E., Kambhampati S.P., Smith E.S., Zhang Z., Zhang F., Singh S., Johnston M.V., Kannan R.M., Blue M.E., Kannan S. Dendrimer-mediated delivery of N-acetyl cysteine to microglia in a mouse model of Rett syndrome. J. Neuroinflammation. 2017;14:252. doi: 10.1186/s12974-017-1004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma R., Kim S.Y., Sharma A., Zhang Z., Kambhampati S.P., Kannan S., Kannan R.M. Activated microglia targeting dendrimer–minocycline conjugate as therapeutics for neuroinflammation. Bioconjugate Chem. 2017;28:2874–2886. doi: 10.1021/acs.bioconjchem.7b00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papa S., Rossi F., Ferrari R., Mariani A., De Paola M., Caron I., Fiordaliso F., Bisighini C., Sammali E., Colombo C., Gobbi M., Canovi M., Lucchetti J., Peviani M., Morbidelli M., Forloni G., Perale G., Moscatelli D., Veglianese P. Selective nanovector mediated treatment of activated proinflammatory microglia/macrophages in spinal cord injury. ACS Nano. 2013;26:9881–9895. doi: 10.1021/nn4036014. [DOI] [PubMed] [Google Scholar]
- 30.Papa S., Ferrari R., De Paola M., Rossi F., Mariani A., Caron I., Sammali E., Peviani M., Dell'Oro V., Colombo C., Morbidelli M., Forloni G., Perale G., Moscatelli D., Veglianese P. Polymeric nanoparticle system to target activated microglia/macrophages in spinal cord injury. J. Contr. Release. 2014;174:15–26. doi: 10.1016/j.jconrel.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 31.Peviania M., Palmiero U.C., Cecere F., Milazzo R., Moscatelli D., Biffi A. Biodegradable polymeric nanoparticles administered in the cerebrospinal fluid: brain biodistribution, preferential internalization in microglia and implications for cell-selective drug release. Biomaterials. 2019;209:25–40. doi: 10.1016/j.biomaterials.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Lin L.F., Bartlett C., Lees M.B. Preparation and characterization of unilamellar myelin vesicles. J. Biol. Chem. 1986;261:16241–16246. [PubMed] [Google Scholar]
- 33.Sedzik J., Blaurock A.E. Myelin vesicles: what we know and what we do not know. J. Neurosci. Res. 1995;41:145–152. doi: 10.1002/jnr.490410202. [DOI] [PubMed] [Google Scholar]
- 34.Frankly W., Taglialatela G. A method to determine insulin responsiveness in synaptosomes isolated from frozen brain tissue. J. Neurosci. Methods. 2016;261:128–134. doi: 10.1016/j.jneumeth.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D'Addio S.M., Prud'homme R.K. Controlling drug nanoparticle formation by rapid precipitation. Adv. Drug Deliv. Rev. 2011;63:417–426. doi: 10.1016/j.addr.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 36.Guimarães D., Cavaco-Paulo A., Nogueira E. Design of liposomes as drug delivery system for therapeutic applications. Int. J. Pharm. 2021;601:120571. doi: 10.1016/j.ijpharm.2021.120571. [DOI] [PubMed] [Google Scholar]
- 37.Adamo G., Fierli D., Romancino D.P., Picciotto S., Barone M.E., Aranyos A., Božič D., Morsbach S., Raccosta S., Stanly C., Paganini C., Gai M., Cusimano A., Martorana V., Noto R., Carrotta R., Librizzi F., Randazzo L., Parkes R., Capasso U.P., Rao E., Paterna A., Santonicola P., Iglič A., Corcuera L., Kisslinger A., Schiavi E.D., Liguori G.L., Landfester K., Kralj-Iglič V., Arosio P., Pocsfalvi G., Touzet N., Manno M., Bongiovanni A. Nanoalgosomes: introducing extracellular vesicles produced by microalgae. J. Extracell. Vesicles. 2021;10 doi: 10.1002/jev2.12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Craparo E.F., Cabibbo M., Conigliaro A., Barreca M.M., Musumeci T., Giammona G., Cavallaro G. Rapamycin-loaded polymeric nanoparticles as an advanced formulation for macrophage targeting in atherosclerosis. Pharmaceutics. 2021;7:503. doi: 10.3390/pharmaceutics13040503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greenfield R.S., Kaneko T., Daues A., Edson M.A., Fitzgerald K.A., Olech L.J., Grattan J.A., Spitalny G.L., Braslawsky G.R. Evaluation in vitro of adriamycin immunoconjugates synthesized using an acid-sensitive hydrazone linker. Cancer Res. 1990;50:6600–6607. [PubMed] [Google Scholar]
- 40.Craparo E.F., Luxenhofer R., Cavallaro G. Development of polymer-based nanoparticles for zileuton delivery to the lung: PMeOx and PMeOzi surface chemistry reduces interactions with mucins SE Drago. Nanomedicine. 2021;26:102451. doi: 10.1016/j.nano.2021.102451. [DOI] [PubMed] [Google Scholar]
- 41.Greenfield S., Norton W.T., Morell P. Quaking mouse: isolation and characterization of myelin protein. J. Neurochem. 1971;18:2119–2128. doi: 10.1111/j.1471-4159.1971.tb05070.x. [DOI] [PubMed] [Google Scholar]
- 42.Di Francesco A., Cunsolo V., Saletti R., Svensson B., Muccilli V., De Vita P., Foti S. Quantitative label-free comparison of the metabolic protein fraction in old and modern Italian wheat genotypes by a shotgun approach. Molecules. 2021;26:2596. doi: 10.3390/molecules26092596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A.F., Ternent T., Brazma A., Vizcaíno J.A. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morell P., H Quarles R. In: Characteristic Composition of Myelin. Basic Neurochemistry: Molecular, Cellular and Medical Aspects. sixth ed. Siegel G.J., Agranoff B.W., Albers R.W., editors. Lippincott-Raven; Philadelphia: 1999. [Google Scholar]
- 45.Samimi S., Maghsoudnia N., Baradaran R., Dorkoosh E.F. Book: Characterization and Biology of Nanomaterials for Drug Delivery- Nanoscience and Nanotechnology in Drug Delivery Micro and Nano Technologies. 2019. Chapter 3 - lipid-based nanoparticles for drug delivery systems; pp. 47–76. [DOI] [Google Scholar]
- 46.Ilahibaks N.F., Lei Z., Mol E.A., Deshantri A.K., Jiang L., Schiffelers R.M., Vader P., Sluijter J.P.G. Biofabrication of cell-derived nanovesicles: a potential alternative to extracellular vesicles for regenerative medicine. Cells. 2019;25:1509. doi: 10.3390/cells8121509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gandham S., Su X., Wood J., Nocera A.L., Alli S.C., Milane L., Zimmerman A., Amiji M., Ivanov A.R. Technologies and standardization in research on extracellular vesicles. Trends Biotechnol. 2020;38:1066–1098. doi: 10.1016/j.tibtech.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mihály J., Deák R., Szigyártó I.C., Bóta A., Beke-Somfai T., Varga Z. Characterization of extracellular vesicles by IR spectroscopy: fast and simple classification based on amide and C-H stretching vibrations. Biochim. Biophys. Acta. 2017;1859:459–466. doi: 10.1016/j.bbamem.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 49.Nzai J.M., Proctor A. Determination of phospholipids in vegetable oil by Fourier transform infrared spectroscopy. J. Am. Oil Chem. Soc. 1998;75:1281–1289. doi: 10.1007/s11746-998-0173-x. [DOI] [Google Scholar]
- 50.Casal H.L., Mantsch H.H. Polymorphic phase Behaviour of phospholipid membranes studied by infrared spectroscopy. Biochim. Biophys. Acta. 1984;779:381–401. doi: 10.1016/0304-4157(84)90017-0. [DOI] [PubMed] [Google Scholar]
- 51.Ishii A., Dutta R., Wark G.M., Hwang S.-Il, Han D.K., Trapp B.D., Pfeiffer S.E., Bansal R. Human myelin proteome and comparative analysis with mouse myelin. Proc. Natl. Acad. Sci. U. S. A. 2009;25:14605–14610. doi: 10.1073/pnas.0905936106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanrobaeys F., Van C.R., Dhondt G., Devreese B., Van B.J. Profiling of myelin proteins by 2D-gel electrophoresis and multidimensional liquid chromatography coupled to MALDI TOF-TOF mass spectrometry. J. Proteome Res. 2005;4:2283–2293. doi: 10.1021/pr050205c. [DOI] [PubMed] [Google Scholar]
- 53.Huang Y., Cheng L., Turchinovich A., Mahairaki V., Troncoso J.C., Pletniková O., Haughey N.J., Vella L.J., Hill A.F., Zheng L., Witwer K.W. Influence of species and processing parameters on recovery and content of brain tissue-derived extracellular vesicles. J. Extracell. Vesicles. 2020;30:1785746. doi: 10.1080/20013078.2020.1785746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Casella G., Rasouli J., Boehm A., Zhang W., Xiao D., Ishikawa L.L.W., Thome R., Li X., Hwang D., Porazzi P., Molugu S., Tang H.Y., Zhang G.X., Ciric B., Rostami A. Oligodendrocyte-derived extracellular vesicles as antigen-specific therapy for autoimmune neuroinflammation in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aba0599. eaba0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moscarello M.A. Cell Biology and Pathology of Myelin: Evolving Biological Concepts and Therapeutic Approaches. 1997. Myelin basic protein, the ‘executive’ molecule of the myelin membrane; pp. 13–25. [Google Scholar]
- 56.Ozgen H., Baron W., Hoekstra D., Kahya N. Oligodendroglial membrane dynamics in relation to myelin biogenesis. Cell. Mol. Life Sci. 2016;73:3291–3310. doi: 10.1007/s00018-016-2228-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fröhlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012;7:5577–5591. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boven L.A., Van Meurs M., Van Zwam M., Wierenga-Wolf A., Hintzen R.Q., Boot R.G., Aerts J.M., Amor S., Nieuwenhuis E.E., Laman J.D. Myelin-laden macrophages are anti-inflammatory, consistent with foam cells in multiple sclerosis. Brain. 2006;129:517–526. doi: 10.1093/brain/awh707. [DOI] [PubMed] [Google Scholar]
- 59.Grajchen E., Wouters E., van de Haterd B., Haidar M., Hardonnière K., Dierckx T., Van Broeckhoven J., Erens C., Hendrix S., Kerdine-Römer S., Hendriks J.J.A., Bogie J.F.J. CD36-mediated uptake of myelin debris by macrophages and microglia reduces neuroinflammation. J. Neuroinflammation. 2020;17:224. doi: 10.1186/s12974-020-01899-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaafar-Maalej C., Diab R., Andrieu V., Elaissari A., Fessi H. Ethanol injection method for hydrophilic and lipophilic drug-loaded liposome preparation. J. Liposome Res. 2010:20 228–243. doi: 10.3109/08982100903347923. [DOI] [PubMed] [Google Scholar]
- 61.Pons M., Foradada M., Estelrich J. Liposomes obtained by the ethanol injection method. Int. J. Pharm. 1993;95:51–56. doi: 10.1016/0378-5173(93)90389-W. [DOI] [Google Scholar]
- 62.Garberg P., Ball M., Borg N., Cecchelli R., Fenart L., Hurst R.D., Lindmark T., Mabondzo A., Nilsson J.E., Raub T.J., Stanimirovic D., Terasaki T., Oberg J.O., Osterberg T. In vitro models for the blood-brain barrier. Toxicol. Vitro. 2005;19:299–334. doi: 10.1016/j.tiv.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 63.García-Salvador A., Domínguez-Monedero A., Gómez-Fernández P., García-Bilbao A., Carregal-Romero S., Castilla J., Goñi-de-Cerio F. Evaluation of the influence of astrocytes on in vitro blood-brain barrier models. Altern. Lab Anim. 2020;48:184–200. doi: 10.1177/0261192920966954. [DOI] [PubMed] [Google Scholar]
- 64.Hellinger E., Veszelka S., Tóth A.E., Walter F., Kittel A., Bakk M.L., Tihanyi K., Háda V., Nakagawa S., Duy T.D., Niwa M., Deli M.A., Vastag M. Comparison of brain capillary endothelial cell-based and epithelial (MDCK-MDR1, Caco-2, and VB-Caco-2) cell-based surrogate blood-brain barrier penetration models. Eur. J. Pharm. Biopharm. 2012;82:340–351. doi: 10.1016/j.ejpb.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 65.Lee J.B., Zgair A., Taha D.A., Zang X., Kagan L., Kim T.H., Kim M.G., Yun H.Y., Fischer P.M., Gershkovich P. Quantitative analysis of lab-to-lab variability in Caco-2 permeability assays. Eur. J. Pharm. Biopharm. 2017;114:38–42. doi: 10.1016/j.ejpb.2016.12.027. [DOI] [PubMed] [Google Scholar]
- 66.Liu C., Liu X.N., Wang G.L., Hei Y., Meng S., Yang L.F., Yuan L., Xie Y. A dual-mediated liposomal drug delivery system targeting the brain: rational construction, integrity evaluation across the blood–brain barrier, and the transporting mechanism to glioma cells. Int. J. Nanomed. 2017;12:2407–2425. doi: 10.2147/IJN.S131367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Danaei M., Dehghankhold M., Ataei S., Hasanzadeh Davarani F., Javanmard R., Dokhani A., Khorasani S., Mozafari M.R. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics. 2018;10:57. doi: 10.3390/pharmaceutics10020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frédéric D., Jean-Luc D., Neel D., Schneider A., Poulain B., Bossu J.L. Organotypic cultures of cerebellar slices as a model to investigate demyelinating disorders. Expet Opin. Drug Discov. 2017;12:1011–1022. doi: 10.1080/17460441.2017.1356285. [DOI] [PubMed] [Google Scholar]
- 69.Grochmal J., Teo W., Gambhir H., Kumar R., Stratton J.A., Dhaliwal R., Brideau C., Biernaskie J., Stys P.K., Midha R. A novel approach to 32-channel peripheral nervous system myelin imaging in vivo, with single axon resolution. J. Neurosurg. 2018;130:163–171. doi: 10.3171/2017.6.JNS17239. [DOI] [PubMed] [Google Scholar]
- 70.Mei F., Christin Chong S.Y., Chan J.R. Myelin-based inhibitors of oligodendrocyte myelination: clues from axonal growth and regeneration. Neurosci. Bull. 2013;29:177–188. doi: 10.1007/Fs12264-013-1319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boggs J.M., Moscarello M.A., Papahadjopoulos D. In: Lipid–Protein Interactions. Jost P.C., Griffith O.H., editors. Wiley; New York: 1982. pp. 1–51. [Google Scholar]
- 72.Bates I.R., Feix J.B., Boggs J.M., Harauz G. An immunodominant epitope of myelin basic protein is an amphipathic alpha-helix. J. Biol. Chem. 2004;279:5757–5764. doi: 10.1074/jbc.M311504200. [DOI] [PubMed] [Google Scholar]
- 73.Houbre D., Schindler P., Trifilieff E., Luu B., Duportail G. Selectivity of lipid-protein interaction with myelin proteolipids PLP and DM-20. A fluorescence anisotropy study. Biochim. Biophys. Acta. 1990;1029:136–142. doi: 10.1016/0005-2736(90)90446-u. [DOI] [PubMed] [Google Scholar]
- 74.Carlson S.L., Parrish M.E., Springer J.E. Acute inflammatory response in spinal cord following impact injury. Exp. Neurol. 1998;151:77–88. doi: 10.1006/exnr.1998.6785. [DOI] [PubMed] [Google Scholar]
- 75.Savchenko V.L., McKanna J.A., Nikonenko I.R. Microglia and astrocytes in the adult rat brain: comparative immunocytochemical analysis demonstrates the efficacy of lipocortin 1 immunoreactivity. Neuroscience. 2000;96:195–203. doi: 10.1016/s0306-4522(99)00538-2. [DOI] [PubMed] [Google Scholar]
- 76.Williams K., Ulvestad E., Waage A., Antel J.P., McLaurin J. Activation of adult human derived microglia by myelin phagocytosis in vitro. J. Neurosci. Res. 1994;38:433–443. doi: 10.1002/jnr.490380409. [DOI] [PubMed] [Google Scholar]
- 77.Mosley K., Cuzner M.L. Receptor-mediated phagocytosis of myelin by macrophages and microglia: effect of opsonization and receptor blocking agents. Neurochem. Res. 1996;21:481–487. doi: 10.1007/BF02527713. [DOI] [PubMed] [Google Scholar]
- 78.Van der Laan L.J., Ruuls S.R., Weber K.S., Lodder I.J., Dopp E.A., Dijkstra C.D. Macrophage phagocytosis of myelin in vitro determined by flow cytometry: phagocytosis is mediated by CR3 and induces production of tumor necrosis. J. Neuroimmunol. 1996;70:145–152. doi: 10.1016/s0165-5728(96)00110-5. [DOI] [PubMed] [Google Scholar]
- 79.Liu Y., Hao W., Letiembre M., Walter S., Kulanga M., Neumann H., Fassbender K. Suppression of microglial inflammatory activity by myelin phagocytosis: role of p47-PHOX-mediated generation of reactive oxygen species. J. Neurosci. 2006;13:12904–12913. doi: 10.1523/JNEUROSCI.2531-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Y.N., Poon W., Tavares A.J., McGilvray I.D., Chan W.C.W. Nanoparticle-liver interactions: cellular uptake and hepatobiliary elimination. J. Contr. Release. 2016;28:332–348. doi: 10.1016/j.jconrel.2016.01.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.