Abstract
T cells provide critical immune surveillance to the central nervous system (CNS), and the cerebrospinal fluid (CSF) is thought to be a main route for their entry. Further characterization of the state of T cells in the CSF in healthy individuals is important for understanding how T cells provide protective immune surveillance without damaging the delicate environment of the CNS and providing tissue-specific context for understanding immune dysfunction in neuroinflammatory disease. Here, we have profiled T cells in the CSF of healthy human donors and have identified signatures related to cytotoxic capacity and tissue adaptation that are further exemplified in clonally expanded CSF T cells. By comparing profiles of clonally expanded T cells obtained from the CSF of patients with multiple sclerosis (MS) and healthy donors, we report that clonally expanded T cells from the CSF of patients with MS have heightened expression of genes related to T cell activation and cytotoxicity.
INTRODUCTION
The central nervous system (CNS) contains a unique immunological state, as the blood-brain and blood–cerebrospinal fluid (CSF) barriers exclude most peripheral cells to prevent damaging inflammation (1). However, pathogens are able to enter and damage the CNS, necessitating persistent immune surveillance. A major route for this surveillance is the CSF, which is predominantly composed of T cells whose trafficking involves the α4β1 integrin (1) and the T helper 1 (TH1)–associated chemokine receptor CXCR3 (2). T cells in the CNS are critical for the control of a range of pathogens in the CNS (3, 4). Disrupting T cell entry to the CNS can predispose individuals to the development of lethal infections, for example, by the JC polyomavirus after treatment with the α4 integrin binding antibody natalizumab (5). T cells have been shown to persist in the brain of mice after viral infection and adopt characteristics of tissue-resident memory T (TRM) cells, which may provide a way for T cells to be poised to respond to reinfection at the tissue site (6).
However, in addition to their critical role in protecting the CNS from pathogens, peripheral immune cells that enter the CNS can also drive neuroinflammation (7). The CSF provides insight into the state of the human CNS, as cellular, metabolic, and protein content can be reflective of a range of neurological diseases (8–11). In multiple sclerosis (MS), evidence of immune activation in the CSF, including antibodies made from clonally expanded B cells, is common metrics for diagnosis (12), and there is sharing of T and B cell populations between the brain parenchyma and CSF in MS (13, 14). These characteristics make the CSF a valuable way to assess immune function in the CNS in health and disease. Because strict trafficking requirements to the CNS paired with transcriptional changes T cells undergo within tissues (15) may result in distinct T cell profiles in the CSF, it is important to characterize CSF T cells in healthy individuals to provide tissue-specific context to T cell profiles in neuroinflammation.
Here, we used paired single-cell RNA sequencing (scRNA-seq) and T cell receptor (TCR) sequencing to characterize the continuum of transcriptional profiles of T cells from the blood and CSF of healthy individuals, including clonally expanded T cells that have been found in the CSF (16, 17). We then compared T cells from patients with MS to healthy individuals to define cluster-specific differences present in neuroinflammation, some of which we found were exemplified in clonally expanded T cells in the CSF from patients with MS. Last, we characterized T cells within the noninflamed brain parenchyma to identify potential ways that brain parenchymal cells and T cells may communicate.
RESULTS
Phenotypic continuum of T cell states between the blood and CSF
To define the composition and phenotypic states of cells within the CNS at steady state, we profiled the peripheral blood and CSF of healthy individuals (aged 27 to 38 years old), with no history of neurologic or immunologic disease and without symptoms of infection, using 5′ V(D)J 10x Genomics (n = 6 healthy donors, 50,483 cells after quality control) for scRNA-seq (fig. S1 and table S1). Single-cell characterization of total peripheral blood mononuclear cells (PBMCs) and CSF allowed for overall profiling of the composition of cell types involved in immune surveillance and more detailed characterization of how cell states are distinct between physiological compartments. We found as previously reported (18–20) that the CSF is dominated by T cells with a smaller number of monocytes (21) and dendritic cells, including recently described CSF-specific monocytes (22, 23), whereas B cells, classical monocytes, and natural killer (NK) cells are largely excluded (fig. S1).
T cells have previously been shown to have distinct characteristics within tissues due to the recruitment of specific cell types, development of tissue-resident phenotypes, and the influence of tissue-specific factors (24, 25). Visualization of conventional naive and memory CD4 and CD8 T cells (excluding γδ T cells, mucosal-associated invariant T cells, NK-like/effector CD8 T cells, and FOXP3-expressing regulatory T cells; fig. S1) from the blood and CSF of healthy donors (n = 6, 30,795 T cells; fig. S2) with the progression-preserving visualization tool potential of heat diffusion for affinity-based transition embedding (PHATE) (26) showed a phenotypic continuum of T cells between the blood and CSF (Fig. 1B). Clustering of these cells resulted in largely distinct clusters between the blood and CSF. Two clusters predominantly present in the blood were naïve CD4 (CD4 naïve blood) and naïve CD8 (CD8 naïve blood) that were defined by expression of genes including CCR7, SELL, and LEF1. The blood also contained a cluster of CD4 T cells (CD4 memory blood) marked by expression of molecules associated with memory T cells (S100A4) and markers of various T cell subsets including the TH2 transcription factor GATA3 and the TH17 marker KLRB1 (CD161) (27, 28). This cluster likely represents the heterogeneous population of memory T cells comprising multiple T cell subsets that is present in the peripheral blood (29). The last cluster predominantly found in the blood was a cluster of CD8 T cells (CD8 memory blood) that expressed markers of memory (S100A4) along with molecules of CD8 function including CCL5, GZMK, and NKG7.
Fig. 1. Phenotypic continuum of T cell states between the blood and CSF.
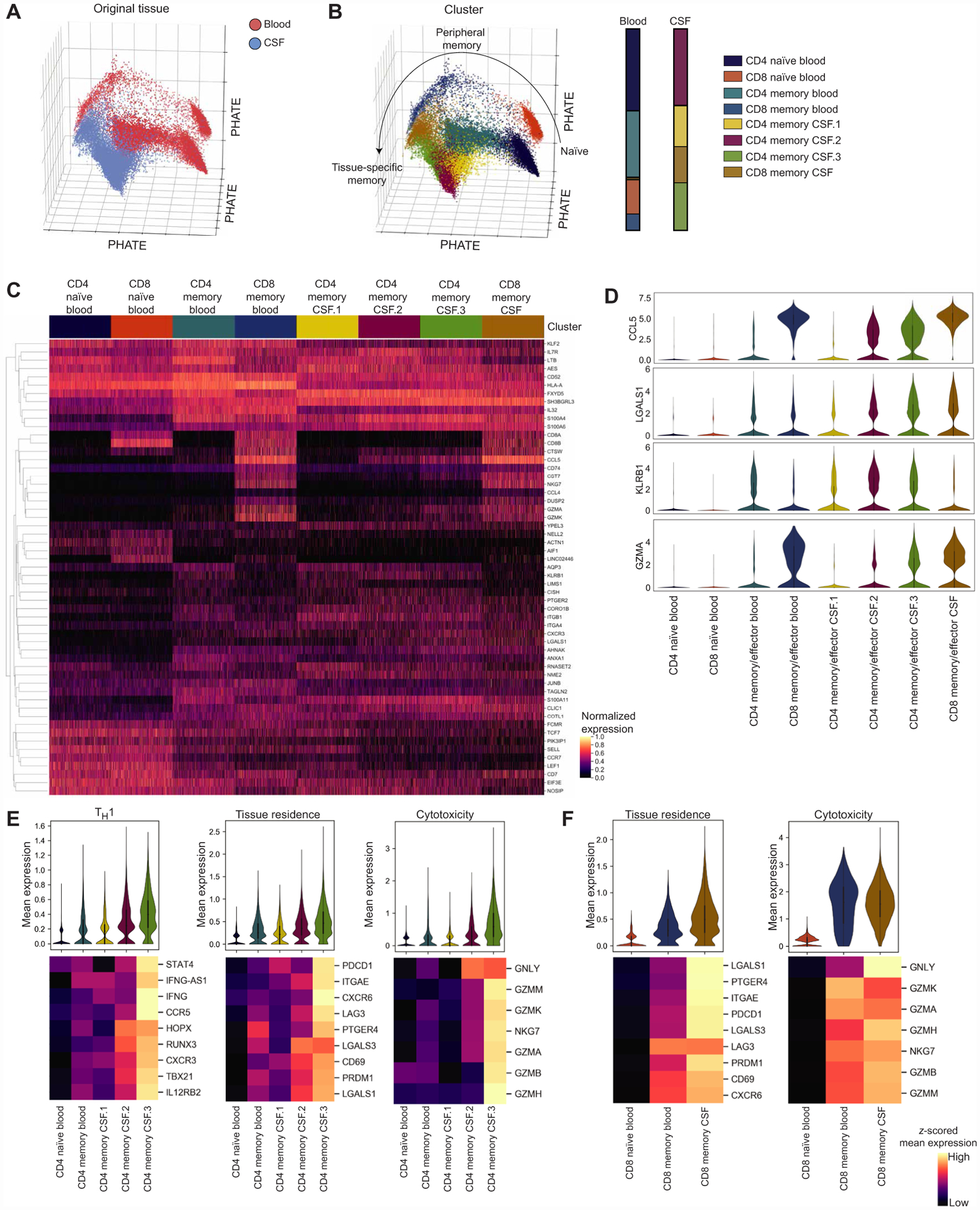
(A) Progression-preserving PHATE visualization of 16,752 T cells from the blood and 14,043 T cells from the CSF of healthy individuals (n = 6 healthy donors) colored by the tissue of origin. (B) PHATE visualization of healthy T cells colored by PhenoGraph clusters with bar graphs showing the composition of T cell clusters in the blood and CSF. Clusters are named if it contains CD4 or CD8 and naïve or memory (antigen-experienced) cells and the tissue where the cluster was predominantly present. (C) Heatmap of standard-scaled expression of the top 10 differentially expressed genes in each cluster in 1000 randomly sampled cells per cluster. Each column is a cell, and genes were hierarchically clustered. (D) Violin plots of expression for select genes presented in (C). (E) Violin plots of mean expression of genes related to TH1 function, tissue residence, and cytotoxicity for each cell in CD4 T cell clusters. Heatmaps show z-scored mean for each gene included in the gene module for each cluster, and genes were hierarchically clustered. CD4 memory CSF.2 and CD4 memory CSF.3 were compared with each other and all other CD4 clusters using a Kruskal-Wallis test with Dunn’s multiple comparisons correction. Full statistics are shown in table S2. (F) Violin plots of mean expression of genes related to tissue residence and cytotoxicity for each cell in CD8 T cell clusters. Heatmaps show z-scored mean expression for each gene included in the gene module for each cluster, and genes were hierarchically clustered. CD8 memory CSF was compared with both other CD8 clusters using a Kruskal-Wallis test with Dunn’s multiple comparisons correction. Full statistics are shown in table S2.
The CSF was largely composed of three clusters of memory CD4 T cells and one memory CD8 cluster, in line with previous findings that naïve T cells are largely excluded from the CSF (30). The CD4 memory CSF.1 cluster was marked by expression of the subunits of the alpha 4 (ITGA4) beta 1 (ITGB1) integrin, which has been shown to be important for T cell entry into the CNS (1), KLRB1, and S1PR1 that promotes T cell egress from tissues (31). The other two CD4 clusters in the CSF (CD4 memory CSF.2 and CD4 memory CSF.3) have phenotypes similar to each other characterized by the chemokine receptor CXCR3 associated with TH1 cells and CSF T cell trafficking (2, 32) and markers that have been found in tissue-resident T cells and T cells residing within nonlymphoid tissues, including CCL5 (24, 33, 34), LGALS1 (24, 25, 34), PTGER2 (35), and CD69 (36). However, CD4 memory CSF.3 was also defined by the cytotoxicity-related genes GZMA, GZMK, GZMM, and CST7 (37), whereas CD4 memory CSF.2 was not. Because these clusters were marked by CXCR3, which is associated with TH1 cells, and KLRB1 that is related to TH17 and nonclassic TH1 cells, it is possible that these cells are nonclassic TH1 cells (38). Staining for the chemokine receptors CXCR3 that marks TH1 cells and CCR6 that marks TH17 cells, we find that CXCR3+CCR6− and CXCR3+CCR6+ populations are the predominant memory CD4 T cell populations in the CSF, as the CSF contains significantly lower fractions of CXCR3−CCR6+ and CXCR3−CCR6− cells than the blood (fig. S3). This could support that nonclassic TH1 cells are present in the healthy CSF, because they have been show to coexpress CXCR3 and CCR6 (39) and previously IFNγ+CCR6+ T cells have been found in the CSF of patients with MS and donors with noninflammatory neurological diseases (40). In addition, KLRB1 may be associated with tissue-resident T cells (35). To further define and summarize the cell states of CD4 T cell clusters in the CSF, we examined the mean expression of genes known to be related to TH1 function, tissue residence, and cytotoxicity (Fig. 1E). We find that overall in CD4 CSF clusters, CD4 memory CSF.3 has the highest expression of genes related to all three signatures, followed by CD4 memory CSF.2, which had higher expression of all three signatures compared with CD4 clusters in the blood and CD4 memory CSF.1. This supports that, compared with the peripheral blood, the healthy CSF is enriched for CD4 T cells with TH1 function, including a CD4 subtype with cytotoxic capacity, and that these cells may adapt to the tissue environment in ways similar to what has been described in other tissues (24, 25, 34).
Last, the CSF contained a cluster of memory CD8 T cells (CD8 memory CSF) that was marked by cytotoxic genes (CCL5, NKG7, GZMK, GZMA, GZMM, and CCL4) and genes related to tissue residence [CD69, LGALS1, LGALS3 (24, 34, 36), PDCD1, PRDM1 (41, 42), and CXCR6 (43)]. CD8 memory CSF also expressed KLRG1 and EOMES, which suggests that these may represent a less differentiated tissue T cell population than canonical tissue-resident CD8 T cells, for example, similar to recently described tissue-resident effector cells (44). Cells in the CD8 memory CSF cluster were found to have increased mean expression of tissue residence and cytotoxicity-related genes compared with cells from CD8 clusters in the blood (Fig. 1F).
Together, these results suggest that CSF T cells are largely distinct from the blood, and three of the four clusters predominantly found in the CSF (CD4 memory CSF.2, CD4 memory CSF.3, CD8 memory CSF) share characteristics described in other nonlymphoid tissues, including previous descriptions of TRM cells in the brain (17, 45–47). Along with this profile, CD4 memory CSF.2 and CD4 memory CSF.3 T cells in the CSF express genes related to TH1 and nonclassic TH1 cells, whereas CD4 memory CSF.3 and CD8 memory CSF also express cytotoxicity-related genes. The final CSF cluster, CD4 memory CSF.1, expressed high levels of the integrin subunits necessary for CSF trafficking but did not share components of the transcriptional signature of nonlymphoid tissue T cells or tissue residence.
Gene expression dynamics across the blood-CSF T cell axis
To further characterize the phenotypic continuum of T cell states, we identified trends of gene expression as cells progress from a blood-like to CSF-like phenotype. To characterize the progression of naïve T cells through T cell differentiation in the periphery and tissue adaptation in the CSF, we used manifold enhancement of latent dimensions (MELD) to generate a continuous tissue score (see Materials and Methods) (48). Each cell therefore has a value reflecting how blood-like or CSF-like it is, providing an axis from the most prototypical blood to CSF T cell state (Fig. 2A). As expected, we saw that tissue score distributions of clusters demonstrate a progression from naïve to memory T cells clusters in the periphery, followed by clusters that predominate in the CSF (Fig. 2B). To further validate that this score is capturing known biology, we confirmed that when cells were ordered by tissue score, they mirrored trends of trafficking markers with established patterns between the blood and CSF (Fig. 2C) (49, 50).
Fig. 2. Gene expression dynamics across the blood-CSF T cell axis.
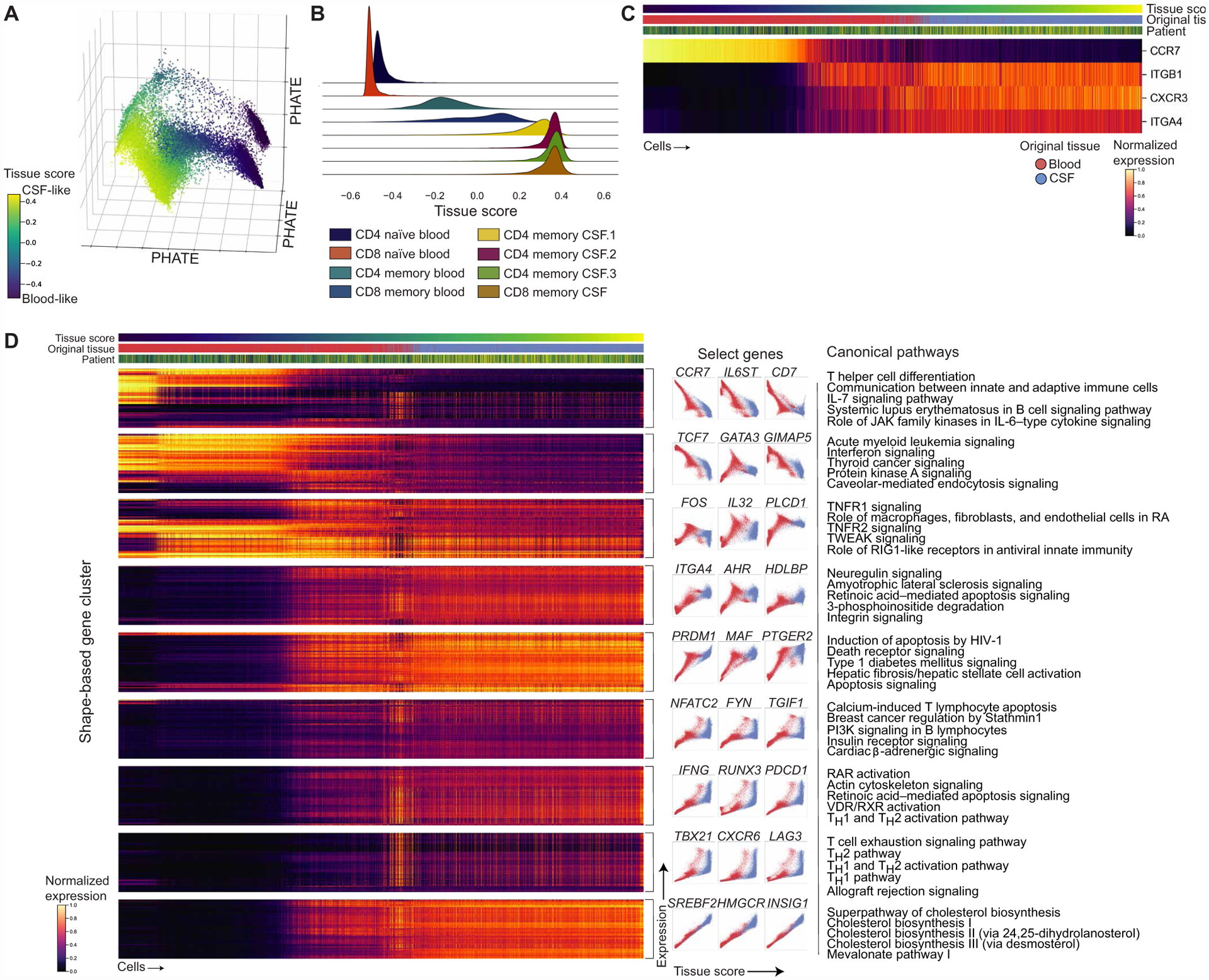
(A) PHATE visualization (Fig. 1) colored by the continuous tissue score representing the most blood-like to CSF-like state (see Materials and Methods). (B) Joy plot of the distribution of tissue scores for each T cell cluster. (C) Heatmap of chemokine receptors and integrin subunits with reported expression patterns between the blood and CSF with cells ordered by tissue score after gene imputation with MAGIC (see Materials and Methods). Bars above the heatmap show the tissue score, tissue of origin, and donor for each cell. Colored by standard-scaled imputed expression for each gene. (D) Heatmap of shape-based gene clusters (see Materials and Methods) based on gene dynamics with relation to the tissue score clustered by PhenoGraph after MAGIC (left). Bars above the heatmap show the tissue score, tissue of origin, and donor for each cell. Colored by standard-scaled imputed expression for each gene. Example plots depicting the gene shape in each cluster are shown (center) with the top five canonical pathways by P value for each gene cluster (right) (table S3). Genes within clusters were hierarchically clustered, and clusters were manually ordered. JAK, Janus kinase; TNFR1, tumor necrosis factor receptor 1; PI3K, phosphatidylinositol 3-kinase. TWEAK, TNF-related weak inducer of apoptosis; RIG1, retinoic acid-inducible gene I; RAR, retinoic acid receptor; VDR/RXR, vitamin D receptor/retinoid X receptor; RA, Rheumatoid Arthritis.
We then characterized gene expression dynamics by clustering genes based on their pattern in relation to the tissue score, or shape-based gene clustering, that resulted in clusters of genes with similar shapes or trends across the blood-CSF continuum (see Fig. 2D and Materials and Methods). To summarize genes present in these clusters, we used canonical pathway analysis that showed gene clusters decreasing in relation to the CSF state, as measured by tissue score, include pathways related to TH cell differentiation and interleukin-7 (IL-7) signaling, whereas gene clusters that increased with CSF tissue state include pathways related to T cell exhaustion and TH1 function (Fig. 2D and table S3). In addition, a cluster with genes related to cholesterol biosynthesis increased in relation to the tissue score, which may reflect TCR engagement driving a lipid biosynthesis program that includes genes found in this cluster, including HMGCR, HMGCS1, and SQLE (51). Although TH2 pathways were present in pathway analysis of genes increasing in CSF, we found that these pathways reflect more general T cell activation as GATA3, the master transcription factor driving TH2 responses, decreased in the CSF. Gene clusters that show an increase in expression in cells with high CSF-like tissue scores could be of particular interest because they may be candidates for pathways induced within the CSF by the tissue environment.
Because TH1- and exhaustion-related genes had similar patterns of gene expression in relation to the tissue score, for example, IFNG clustering with PDCD1 and TBX21 clustering with LAG3, as well as CD4 memory CSF.2 and CD4 memory CSF.3 having characteristics of TH1 T cells, we examined molecules representative of these functions by flow cytometry on the blood and CSF from a different cohort of healthy individuals (n = 3 healthy donors). We found that in the unstimulated state, because almost all stimulated T cells up-regulated CD69, there was a higher frequency of CD69+PD-1+ total (CD3+) T cells in the CSF, which are markers of activation and tissue residence, compared with the blood (Fig. 3A). Upon stimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin, the CSF had a higher frequency of total T cells expressing both interferon-γ (IFNγ) and programmed cell death protein-1 (PD-1) compared with T cells in the blood (Fig. 3B and fig. S4). This suggests that in healthy individuals, T cells in the CSF, including those expressing the coinhibitory receptor PD-1, retain their ability to mount effector TH1 responses. This aligns with previous literature finding that TRM cells express coinhibitory molecules but are able to produce robust cytokine responses (52, 53) and the ability of PD-1+ effector T cells in the peripheral blood producing heightened levels of IFNγ compared with PD-1−CD4+ T effector T cells (54).
Fig. 3. CSF T cells retain effector function capacity.
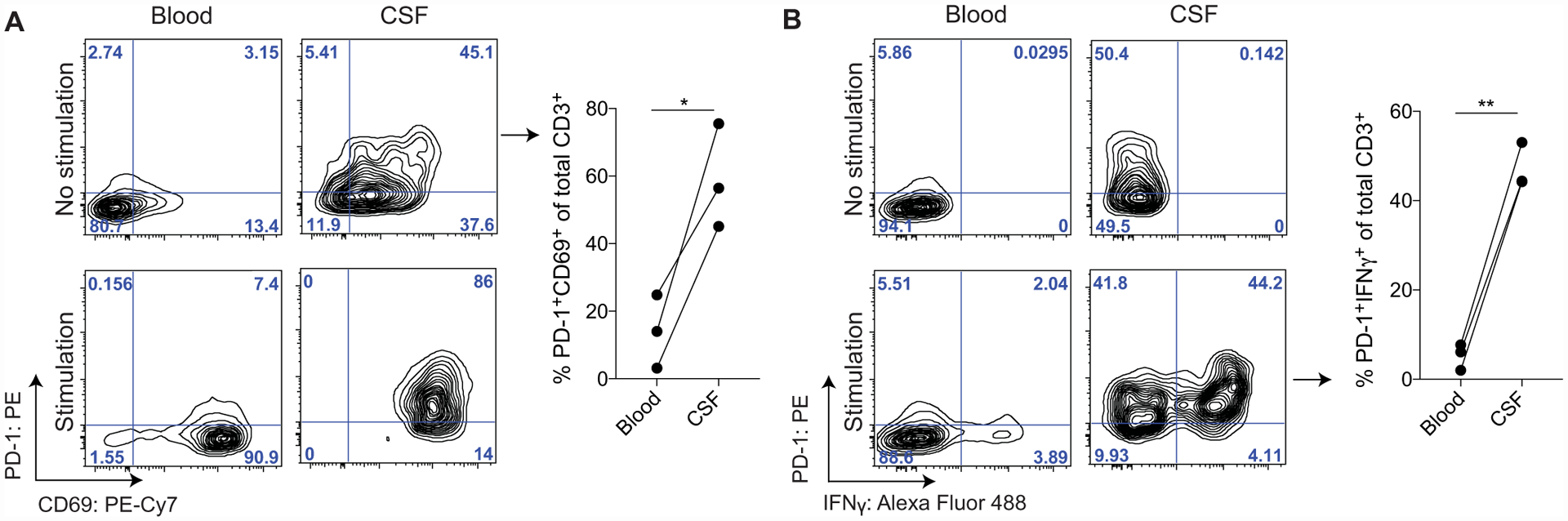
(A) Representative staining of total T cells (CD3+) from one healthy donor PBMCs and CSF after 4 hours in the presence of GolgiStop with or without stimulation (+PMA/ionomycin, 4 hours). Summary plots from the blood and CSF of three healthy individuals show the percentage of the total CD3+ fraction that is PD-1+CD69+ without stimulation. The blood and CSF were compared using a paired two-tailed t test, *P = 0.0359 (fig. S4 and table S10). (B) Representative staining of total T cells (CD3+) from one healthy donor PBMCs and CSF after 4 hours in the presence of GolgiStop with or without stimulation (+PMA/ionomycin, 4 hours). Summary plots from the blood and CSF of three healthy individuals show the percentage of the total CD3+ fraction that is PD-1+IFNγ+ with stimulation (right). The blood and CSF were compared using a paired two-tailed t test, **P = 0.0023 (fig. S4 and table S10).
Characterization of clonally expanded T cells in healthy CSF
We were next interested in integrating clonal information into our transcriptional analysis of conventional T cells using paired single-cell TCR sequencing using the 5′ V(D)J 10x Genomics platform (23,210 healthy blood and CSF T cells with TCRs). Clonally expanded T cells, or T cells with the same TCR, are evidence that a T cell has encountered and expanded in response to recognizing its cognate antigen and can have distinct signatures (55). All six donors had clonal groups, defined as groups of cells with the same CDR3α and CDR3β amino acid sequences, with cells shared between the blood and CSF, and five of six healthy controls had sufficient cell numbers to detect clonally expanded T cells within the CSF. We did not identify any shared CDR3s from conventional T cells between healthy donors. We leveraged clonal groups where we detected the same CDR3α/CDR3β in both the blood and CSF (n = 6 donors, 1010 cells, 284 groups of cells; fig. S5) to resolve whether T cells sharing a TCR but existing in different tissues have more divergent phenotypes than clonal groups detected only within the blood or CSF. We found that cells from the same clonal group but present in different tissues had greater average distances between principal components (PCs) as compared with clonal groups from the same compartment (Fig. 4, A and B), suggesting that cell states may diverge on the basis of their tissue environment. Differential expression analysis between the CSF and blood cells with clonal groups present in both tissues (n = 5 donors, 1008 cells) revealed that CSF cells within these clonal groups had increased expression of genes related to CNS trafficking (ITGB1), tissue residence (LGALS1 and CD69), and cholesterol homeostasis (LDLR, INSIG1, and SQLE) compared with the blood (fig. S5D and table S4).
Fig. 4. Characterization of clonally expanded T cells in healthy CSF.
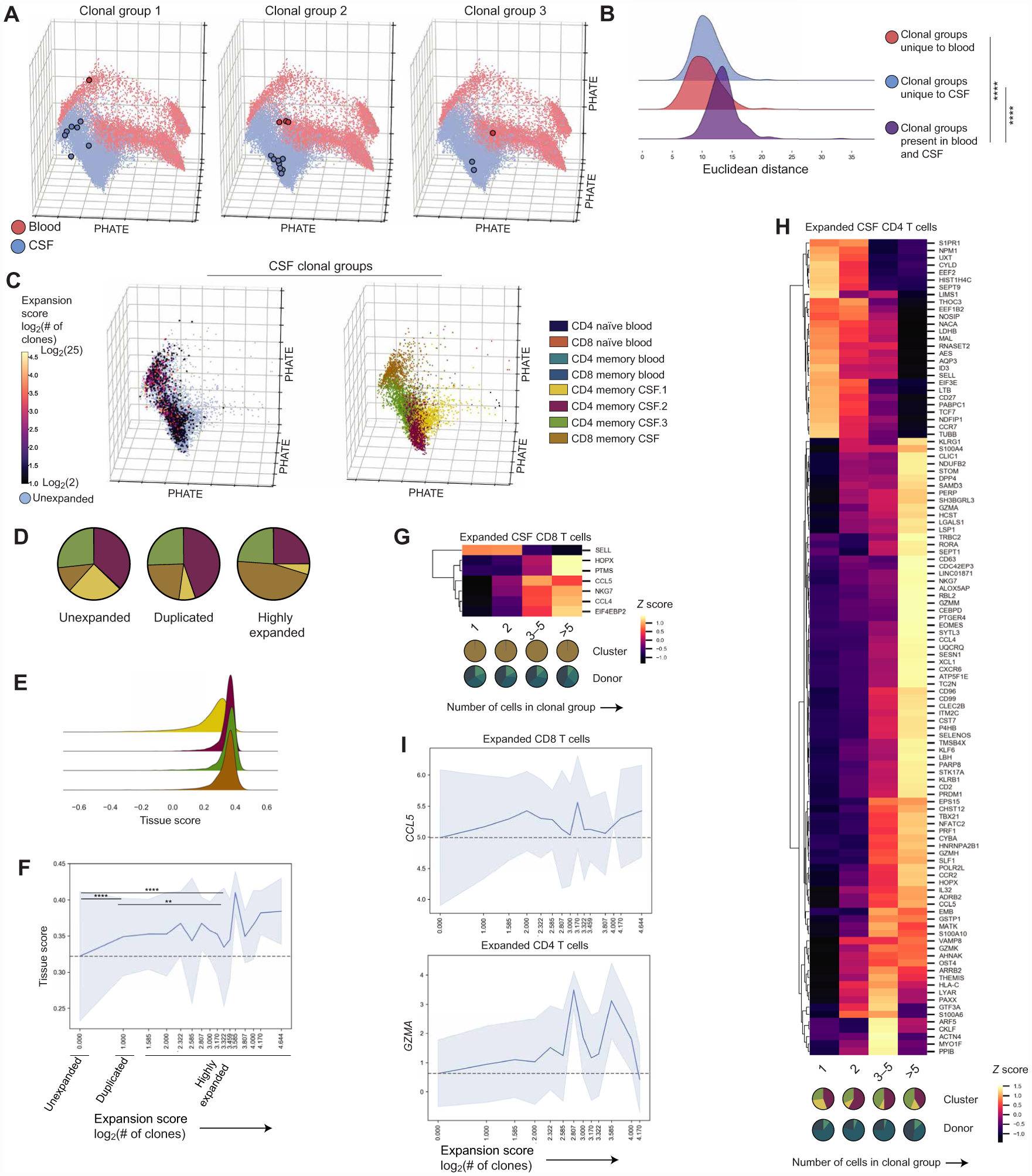
(A) PHATE visualization with three representative groups from three different donors of clonal groups with cells present in both the blood and CSF (fig. S5). (B) Joy plot showing Euclidean distances between the first 20 PCs of cells in the same clonal group but present in different tissues (purple; 284 clonal groups, n = 6 donors) or cells from clonal groups only in the blood (red; 129 clonal groups, n = 6 donors) or CSF (blue; 448 clonal groups, n = 5 donors). Distances compared by the Kruskal-Wallis test with Dunn’s multiple comparisons correction. ****P < 0.0001. (C) PHATE visualization showing CSF T cells where a TCR (CDR3α/CDR3β) was recovered, including unexpanded cells and expanded cells colored by the expansion score or log2(number of clones) in each group (left). PHATE visualization showing CSF T cells where a TCR was recovered colored by cluster (right). (D) Cluster distribution of unexpanded (7199 CSF T cells), duplicated (2 cells per clonal group, 792 CSF T cells), and highly expanded (>2 cells per clonal group, 708 CSF T cells) CSF T cells in healthy individuals where clonally expanded cells were present (n = 5 donors; fig. S5). Two-way analysis of variance (ANOVA) with Tukey’s multiple comparison correction (adjusted P values): CD4 memory CSF.1 (unexpanded > duplicated, P = 0.0021; unexpanded > highly expanded, P = 0.0007), CD4 memoryCSF.2 (unexpanded > highly expanded, P = 0.0021; duplicated > highly expanded, P = 0.0028), CD8 memory CSF (unexpanded < duplicated, P = 0.0051; unexpanded < highly expanded, P < 0.0001; duplicated < highly expanded, P < 0.0001). (E) Subset of the joy plot shown in Fig. 2B of tissue scores for T cell clusters predominantly in the CSF. Kruskal-Wallis test with Dunn’s multiple comparisons correction between the tissue score for CD4 memory CSF.1 and three other clusters predominantly in the CSF. P < 0.0001 for all comparisons. (F) Line plot of the mean tissue score and SD for each expansion score (n = 5 healthy donors). Gray dashed line shows the mean for unexpanded T cells. Tissue scores of unexpanded, duplicated, and highly expanded cells were compared by the Kruskal-Wallis test with Dunn’s multiple comparisons correction. Unexpanded versus duplicated and unexpanded versus highly expanded, ****P < 0.0001; duplicated versus highly expanded, **P = 0.0014. (G) Heatmap of differentially expressed genes in highly expanded (>2 clones per clonal group, n = 4 donors, 312 cells) compared with unexpanded (n = 4 donors, 751 cells) CSF CD8 T cells in healthy individuals with a MAST-estimated log fold change over log2(1.1) (10% change) and an FDR of <0.05 (see Materials and Methods and table S4) excluding specific TCR genes. Colored by z-scored values of mean expression for each gene. Pie charts below the graph reflect the cluster and patient composition for each expansion group. (H) Heatmap of differentially expressed genes in highly expanded (>2 clones per clonal group, 344 cells) compared with unexpanded (4883 cells) CSF CD4 T cells in healthy individuals (n = 3 donors) with a MAST-estimated log fold change over log2(1.1) (10% change) and an FDR of <0.05 (see Materials and Methods and table S4) excluding specific TCR genes. Colored by z-scored values of mean expression for each gene. Pie charts below the graph reflect the cluster and patient composition for each expansion group. (I) Line plots of the mean expression and SD of GZMA in CD4 CSF T cells (n = 3 donors) and CCL5 in CD8 CSF T cells (n = 4 donors) in relation to the expansion score. Gray dashed line shows the mean expression for unexpanded cells.
We then examined the nature of clonally expanded CSF T cells by calculating an expansion score, which we define as the log2 of the number of clones in the clonal group within the CSF. This score estimates the number of cell divisions each clonal group has undergone (Fig. 4C). The distribution of clones across previously defined clusters showed that compared with unexpanded cells [CDR3α/CDR3β recovered but did not match other TCRs (see Materials and Methods), 7199 CSF T cells from five healthy donors], both duplicated (two cells per clonal group detected, 792 cells from five healthy donors) and highly expanded (>2 cells per clonal group detected, 708 cells from five healthy donors) cells had a higher proportion of CD8 memory CSF T cells. In addition, highly expanded T cells had a lower proportion of CD4 memory CSF.2 cells. Both duplicated and highly expanded cells had a lower frequency of CD4 memory CSF.1 than unexpanded cells. That is, clonally expanded cells were largely distributed among the three clusters with characteristics of nonlymphoid or tissue-resident T cells (CD4 memory CSF.2, CD4 memory CSF.3, and CD8 memory CSF). The CD4 memory CSF.1 cluster also corresponds to the cluster with the lowest tissue score, or most blood-like, of the four clusters that dominate the CSF (Fig. 4E), supporting that this cluster may represent cells that recently entered the tissue and have not undergone tissue-specific changes.
Because clonally expanded T cells have been used as a readout of the relevance of T cells in the immune response (17) and the presence of clonally expanded TRM in tissues (56), we were interested in further interrogating the transcriptional state of expanded cells in healthy CSF. First, we found that highly expanded T cells had more CSF-like tissue scores, suggesting a more CSF-distinct state than duplicated T cells that also had more CSF-like scores compared with unexpanded cells (Fig. 4F). We then compared highly expanded CD8 (n = 4 healthy donors, 312 cells) and CD4 T cells (n = 3 healthy donors, 344 cells) with unexpanded CD8 (n = 4 donors, 751 cells) and CD4 T cells (n = 3 donors, 4,883 cells). In CD8 T cells, similar to what has recently been described in clonally expanded CD8 T cells in CSF from patients with Alzheimer’s disease (16), we found that clonally expanded CD8 T cells expressed higher levels of CD8 effector function–related molecules including NKG7, CCL5, and CCL4 and express lower levels of SELL or L-selectin that is associated with a naïve phenotype compared with unexpanded cells (Fig. 4, G and I, and table S4). In addition to reduced expression of genes related to tissue egress including S1PR1 and CCR7, clonally expanded CD4 T cells had increased expression of genes related to cytotoxicity (GZMK, GZMA, GZMM, GZMH, CST7, PRF1, CCL4, and CCL5), tissue residence [CXCR6, PRDM1, LGALS1, and PTGER4 (35, 57)], and TH1/nonclassic TH1 T cells (TBX21, RORA, and KLRB1) (Fig. 4, H and I, and table S4). Together, these finding suggest that clonally expanded T cells in healthy CSF represent a more extreme CSF-distinct state that is transcriptionally poised for effector responses in both CD4 and CD8 T cells. In CD4 T cells particularly, clonally expanded cells also express molecules associated with tissue residence.
Examining T cells during neuroinflammation in MS
After identifying the overall and clonally expanded T cell profiles in healthy individuals, we were interested in contrasting this cell state with T cells in the CSF during neuroinflammation. Using 5′ 10x Genomics with paired single-cell TCR information, we profiled the blood and CSF of patients with newly diagnosed relapsing-remitting MS (n = 5 patients, 54,676 total blood and CSF cells; table S1). As previously reported, patients with MS had an increased proportion of B cells present in the CSF compared with healthy controls and an increased proportion of a CD4 T cell cluster containing regulatory T cells (fig. S1) (23). However, the distribution of conventional naive and memory T cell clusters in either the blood or CSF from patients with MS did not notably differ from that of healthy controls (34,734 T cells; Fig. 5A, fig. S2, and table S5). Whereas these data suggested conventional T cell types between patients with MS and healthy controls are similar, differentially expressed genes were present within clusters, which may reflect potential functional changes present during neuroinflammation (Fig. 5B and table S6). Such genes included PCPB1 that has been shown to drive T cell pathogenicity in experimental autoimmune encephalomyelitis (58), which was increased in cells from patients with MS in all CSF but not in blood T cell clusters. Genes related to TCR engagement and T cell activation were more highly expressed by cells from patients with MS compared with healthy donors, including CD69 (59) that was up-regulated in all CSF clusters and IL32 (60) that was increased in all CSF clusters as well as naive T cells, which may suggest that some of these T cells may have more recently encountered antigen.
Fig. 5. Examining T cells during neuroinflammation in MS.
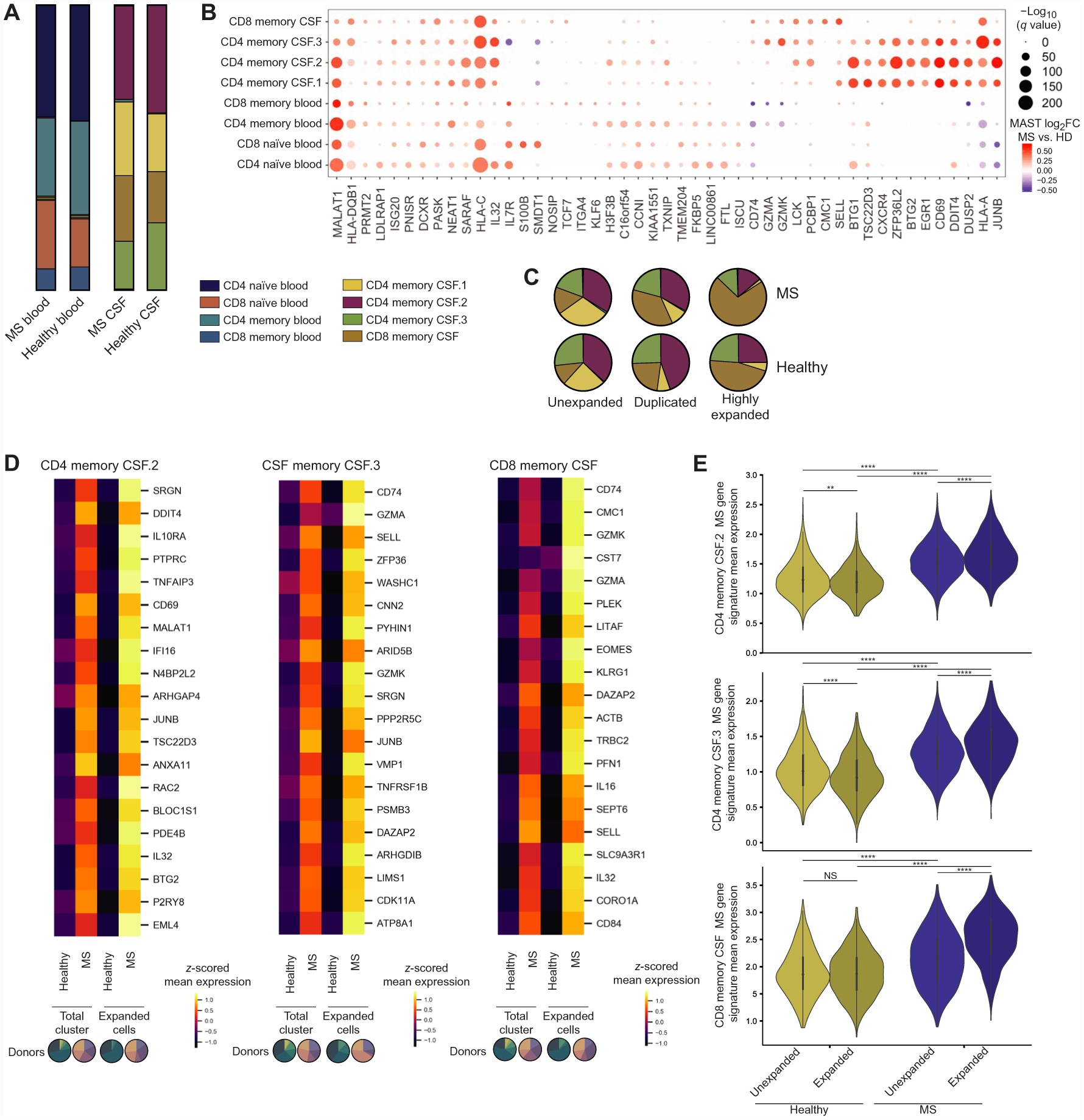
(A) T cell composition in the blood (15,482 cells) and CSF (19,252 cells) of patients with MS (n = donors) and healthy donors (Fig. 1). Composition in patients with MS and healthy individuals was compared by multiple t tests with FDR correction (desired FDR = 1%): no significant differences (table S5). (B) Dot plot of the top 10 differentially expressed genes by MAST-estimated log fold change (FC) between cells from patients with MS (n = 5) and healthy controls (n = 6) for each T cell cluster (table S6). Values are shown for all clusters where the gene was significantly differentially expressed (FDR < 0.05). Genes were hierarchically clustered. Dot size relates to −log10(q value), and color relates to MAST-estimated log fold change. (C) Cluster distribution of unexpanded (n = 5 patients with MS, 12,382 CSF T cells), duplicated (n = 5 patients with MS, 1,038 CSF T cells), and highly expanded CSF T cells (n = 5 patients with MS, 1170 CSF T cells) in MS. Compared with distribution in healthy individuals by multiple t tests with FDR correction (desired FDR = 1%): no significant differences (table S5). (D) Heat-maps of the top 20 genes differentially expressed genes between expanded cells in patients with MS (n = 5; CD4 memory CSF.2, 505 cells; CD4 memory CSF.3, 358 cells; CD8 memory CSF, 1207 cells) compared with healthy controls (n = 4; CD4 memory CSF.2, 528 cells; CD4 memory CSF.3, 363 cells; CD8 memory CSF, 496 cells) with the largest increase in MAST-estimated log fold change in expanded cells compared with patients with MS compared with healthy controls in the total cluster (n = 6 healthy donors; n = 5 patients with MS). Genes are ordered in descending order of difference in log fold change between MS versus healthy in expanded cells compared with total cluster. MAST-estimated log2 fold changes for the MS versus healthy donor comparison in expanded cells are all greater than a 15% [log2(1.15)] change. Analysis was performed for the three clusters that contained the majority of clonally expanded CSF T cells. Colored by z-scored values of mean expression for each gene. Pie charts below the graph reflect the patient composition for each expansion group. (E) Violin plots of mean expression for genes in the MS gene signature for each cell in each cluster for unexpanded and expanded CSF T cells in patients with MS (n = 5) and healthy donors (n = 4). Genes included in the gene score were the top 20 genes with the largest increase in MAST-estimated log2 fold change between patients with MS versus healthy individuals in expanded CSF T cells compared with the total cluster shown in Fig. 6D (fig. S5). Comparisons within each cluster were made by the Kruskal-Wallis test with Dunn’s multiple comparisons correction. **P = 0.0065, ****P < 0.0001, and NS = P > 0.9999.
We were then interested in comparing the state of clonally expanded T cells in the CSF between healthy controls and patients with MS, as MS is an organ-specific autoimmune disease with autoreactive T cells thought to be key initiators of inflammation (61). Similar to the overall T cell state, there was no significant difference in cluster distribution of unexpanded (12,382 CSF T cells from five patients with MS), duplicated (1038 CSF T cells from five patients with MS), or highly expanded T cells (1170 CSF T cells from five patients with MS) between healthy controls and patients with MS. Similarly, the proportions of unexpanded, duplicated, and highly expanded cells in the CSF were not significantly different between patients and healthy controls (table S5). One explanation for why we did not find an overall difference in clonally expanded conventional T cells in the CSF is that T cells are known to infiltrate the brain parenchyma in MS and are present in demyelinated lesions (14, 62), which may result in less overall clonal expansion in the CSF. We did not identify shared CDR3s between patients with MS, although we did identify three CDR3s that overlapped between healthy donors or patients with MS in either the blood or CSF (table S5). We found that when we compared clonally expanded CSF T cells from patients with MS (n = 5 patients with MS) to healthy donors (n = 4 healthy donors) within the three CSF clusters that contained the majority of clonally expanded CSF T cells, many of the genes overlapped with differentially expressed genes resulting from the comparison within the total cluster, which included all levels of expansion and cells with or without a recovered TCR. However, we found that the magnitude of differences between MS and healthy controls differed between the total cluster and solely expanded cells (Fig. 5D). From genes that were increased in MS in both the total cluster and expanded cells, we examined the top 20 genes with the largest increase in log2 fold change in expanded cells compared with the total cluster. That is, we looked at genes with a larger difference between cells from patients with MS and healthy controls in clonally expanded T cells compared with all T cells. Overall, many of these genes were related to T cell activation [CD4 memory CSF.2: CD69, RAC2 (63), and IL32 (64)], cytotoxicity (CSF memory CSF.3: GZMA, GZMK, and SRGN; CSF memory CD8: GZMK, CST7, PLEK, LITAF, and PFN1), and an effector phenotype (CSF memory CD8: CD74, EOMES, and KLRG1).
To further resolve the gene expression trends that underscored the difference in log2 fold change, we used the 20 genes from Fig. 5D as an MS gene signature for each cluster because they overlapped in both total and clonally expanded comparisons to healthy individuals. Examining the mean expression of those genes specifically in unexpanded and expanded cells in healthy individuals and patients with MS, we see that unexpanded cells from patients with MS have higher mean expression of these genes than unexpanded cells from healthy controls (Fig. 5E). Expression of the MS gene signature was then higher in clonally expanded cells compared with unexpanded cells from patients with MS, whereas the expression was decreased or was unchanged in healthy individuals (Fig. 5E). Although CD4 T cells are thought to be critical in MS immunopathology, it is notable that cells from the expanded CD8 memory CSF cluster showed a large increase in their MS gene score signature that includes various molecules related to cytotoxicity (GZMK, CST7, GZMA, and PFN1). Overall, this suggests that T cells in MS CSF broadly have signatures related to increased activation and cytotoxicity, which are further exemplified in clonally expanded T cells in MS but not healthy CSF.
Potential interactions between T cells and glia and neurons
To determine whether the CSF itself, containing factors derived from parenchymal cells (65), is able to affect T cell state, CXCR3+CD45RO+ T cells (fig. S7) were stimulated with anti-CD3/28 for 48 hours with or without the presence of healthy CSF supernatant. We found that the presence of CSF increased the expression of PRDM1, related to tissue residence, in CD4 and CD8 T cells and IFNG, related to TH1 function (Fig. 6A), in CD4 cells. This suggests that there may be factors within the CNS environment that can affect T cell function. We were then interested in characterizing T cells within the brain parenchyma that have been described in recent studies and identifying potential ways that brain parenchymal cells and T cells may communicate (46, 66). Thus, we examined T cells in the brain parenchyma from two different datasets. First, we generated a data-set by performing single-nucleus RNA sequencing (snRNA-seq) on the brain parenchyma from frozen post-mortem (n = 2 donors) and normal-appearing tissue resected during epilepsy surgery (n = 1 donor; table S1 and figs. S8 and S9) due to the inability to dissociate frozen brain samples into single cells. To increase T cell retrieval for each donor, we performed both snRNA-seq of total isolated brain nuclei, which includes glia and neurons, and selecting non-neuronal cells by sorting for NeuN− cells. This process enriched the fraction of recovered T cells (fig. S8). Combining these data provided a paired, high-resolution characterization of neurons and glia from the total nuclei and T cells from the enriched fraction. In addition, we identified T cells in an existing scRNA-seq dataset (67) from freshly processed normal-appearing brain tissue obtained during epilepsy surgery (n = 3 donors; table S1 and fig. S9). We found that in both snRNA-seq (n = 3; 38, 41, and 60 years old, 233 T cells) and scRNA-seq datasets spanning a range of donor ages (n = 3; 2, 2, and 62 years old, 319 T cells), T cells expressed genes related to tissue persistence, in agreement with previous reports (46), and cytotoxic and effector function (Fig. 6B and table S8). Although we could not directly compare brain parenchyma T cells with CSF and blood T cells because these datasets did not contain paired donors and were performed with different processing and technologies, this suggests that there may be overlap in these T cell states.
Fig. 6. Potential interactions between T cells and glia and neurons.
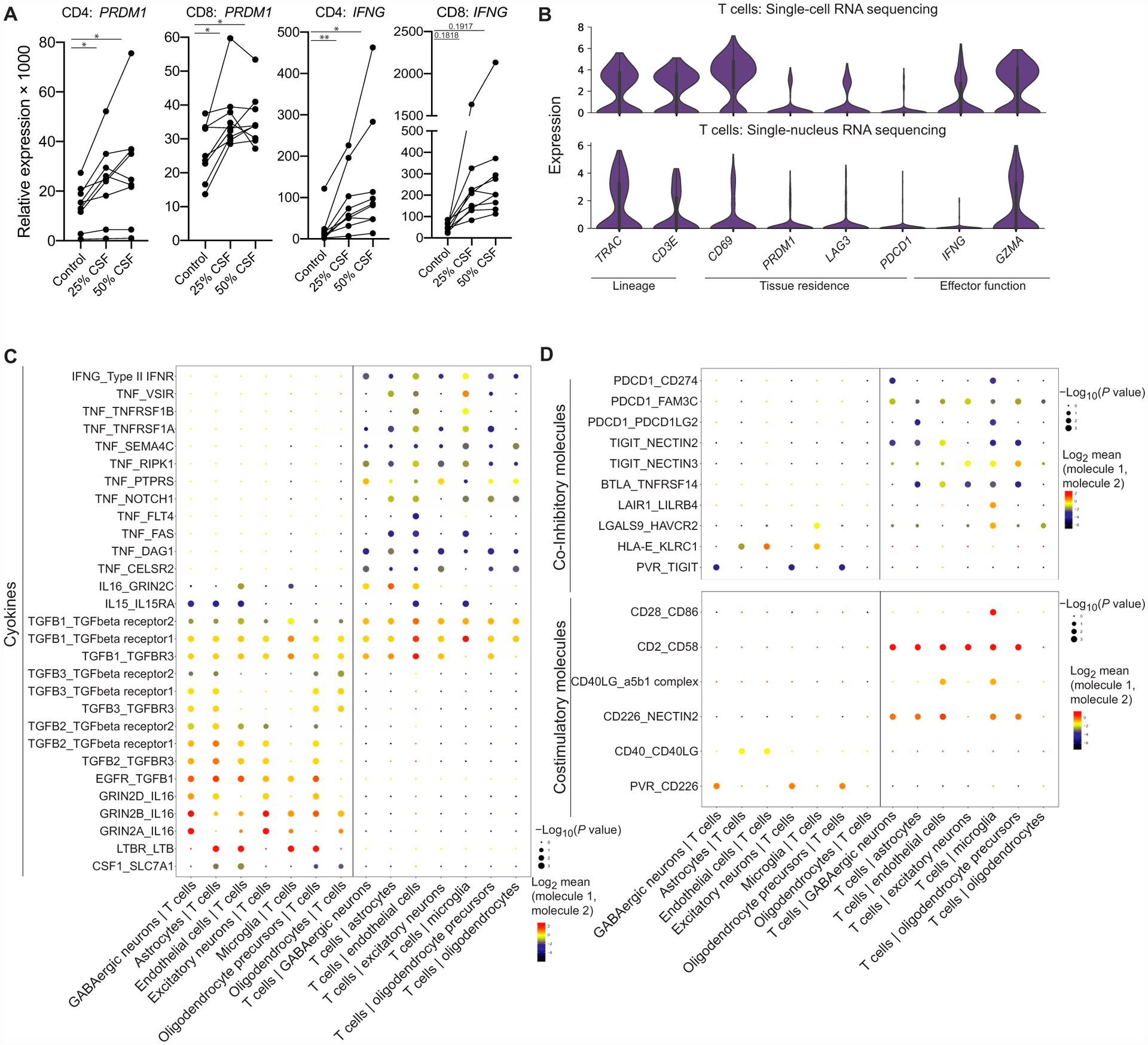
(A) qPCR results of CD4 or CD8 CD45RO+CXCR3+ T cells from the peripheral blood after they were stimulated with anti-CD3/anti-CD28 in XVIVO15 with or without the presence of healthy CSF supernatant for 48 hours (n = 9 healthy donors). Paired one-way ANOVA with Dunnett’s multiple comparisons test. PRDM1: *P = 0.0128 (CD4 25% CSF), *P = 0.0297 (CD4 50% CSF), *P = 0.0151 (CD8 25% CSF), and *P = 0.0351 (CD8 50% CSF); IFNG: **P = 0.0090 (CD4 25% CSF), **P = 0.0270 (CD4 50% CSF), P = 0.1818 (CD8 25% CSF), P = 0.1917 (CD8 50% CSF). (B) Violin plots of selected genes related to lineage, tissue residence, and functional gene expression in T cells within the brain parenchyma in scRNA-seq (top) (n = 3 donors, 319 T cells) and snRNA-seq (bottom) (n = 3 donors, 233 T cells) (table S8). (C) Dot plot of cell-cell interactions between T cells and glia/neurons from snRNA-seq data (n = 3, 233 T cells, 16,604 parenchymal cells) that involve cytokines calculated by CellPhoneDB (table S9). (D) Dot plot of cell-cell interactions between T cells and glia/neurons from snRNA-seq data (n = 3 donors, 233 T cells, 16,604 parenchymal cells) that involve coinhibitory and costimulatory molecules calculated by CellPhoneDB (table S9).
Because the snRNA-seq data were more representative of the cell types in the parenchyma, including astrocytes and neurons, we leveraged this dataset to identify candidate pathways of communication between T cells and parenchymal cells within the brain. We identified soluble and surface-surface receptor-ligand pairs between T cells and excitatory neurons, GABAergic neurons, oligodendrocytes, oligodendrocyte precursors, astrocytes, microglia, and endothelial cells (table S9). Soluble interactions may be specifically relevant for T cells within the brain parenchyma and the CSF, as the CSF contains factors produced from cells within the brain and could provide a pathway for T cell–derived factors to reach the parenchyma. We observed that transforming growth factor B cytokines, which are important factors in brain homeostasis (68) and can also drive features of TRM cells (69), provide a potential avenue for T cell and glia/neuron communication (Fig. 6C). Moreover, brain parenchymal cells expressed IL15, which can promote survival, homeostatic proliferation, and cytotoxicity in T cells (70). T cells also expressed cytokines, including IFNG and tumor necrosis factor (TNF), which may act on parenchymal cells (Fig. 6C). In addition to soluble interactions, we identified potential surface-surface interactions that may be relevant for T cells residing within the brain. These interactions include the expression of a balance of coinhibitory ligands by cells in the brain that may ligate receptors including PD-1 (PDCD1), T cell immunoreceptor with Ig and ITIM domains (TIGIT), and T cell immunoglobulin and mucin domain-containing protein 3 (TIM3, HAVCR2) on T cells (Fig. 6D), which could be a mechanism of restraining T cell function at steady state and potentially stimulatory interactions between parenchymal cells and T cells (Fig. 6D). Overall, these results generate hypotheses regarding potential ways that the brain milieu may be able to affect T cell state.
DISCUSSION
T cells are increasingly appreciated for their plasticity in adapting to tissue environments, allowing them to persist and orchestrate coordinated tissue-specific immune surveillance and other homeostatic functions (15). Here, we use scRNA-seq, single-cell TCR sequencing, and snRNA-seq to characterize T cell phenotypes across the healthy CSF, inflamed CSF, and noninflamed brain. We find that in the absence of inflammation, the CSF is largely characterized by T cells, including clonally expanded T cells, dominated by four T cell subtypes. We find that three of the four subtypes of T cells in the CSF have features described in T cells present in other nonlymphoid tissues and that clonally expanded T cells were present in these three clusters. TH1 function is likely enriched in the CSF through the importance of CXCR3 expression for T cell entry. Because cells are likely further shaped by signals they receive within the tissue, the plasticity of CD4 T cells within the clusters we found remains to be determined. For example, the cluster CD4 memory CSF.1 may reflect a population that has more recently entered the CSF because it does not share genes related to T cells in nonlymphoid sites, contains fewer clonally expanded T cells, and had the most blood-like tissue score of the CSF clusters. It is possible that this subset could differentiate into one of the other CSF CD4 phenotypes depending on the signals that the cells receive in the tissue. The similar profiles of CD4 memory CSF.2 and CD4 memory CSF.3 could also suggest that these may not represent completely distinct cell types but rather may reflect differences in length of time spent or the signals that they received within the tissue.
Because the immune system is increasingly recognized as existing across a continuum of phenotypic states, we translated the blood-CSF axis into a continuous tissue score. This allowed us to understand cellular states, gene expression trends, and T cell expansion in relation to a continuum capturing specific recruitment and adaptation of T cells in the CSF. We found that gene expression trends of molecules related to TH1 function and T cell exhaustion were similar, which we validated by showing that PD-1+ T cells in the CSF retain their ability to produce IFNγ. Integrating clonal information into our analysis, we found clonally expanded CSF T cells in healthy individuals represented a more extreme CSF T cell state compared with unexpanded CSF T cells, in line with transcriptional changes reflecting that both expanded CD4 and CD8 T cells may be poised for effector function, whereas CD4 cells also showed transcriptional changes in line with further tissue adaptation.
Although the CSF of newly diagnosed patients with MS and healthy donors consisted of similar conventional T cell types, we were able to define differences related to activation and cytotoxic gene expression in T cells from patients with MS that were further embodied by clonally expanded T cells. Although CD4 T cells are thought to be critical in MS immunopathology, CD8 T cells outnumber CD4 T cells in MS lesions and are able to damage parenchymal cells in the CNS (62, 71). It is therefore of interest that the clonally expanded T cells in the CD8 memory CSF cluster were enriched in a signature related to cytotoxic and effector function compared with unexpanded cells and expanded cells from healthy individuals. The specificity of clonally expanded cells in the CSF and the site and context of their expansion during homeostatic immune surveillance compared with during neuroinflammation remains of interest.
Although this study adds to the understanding of the T cell states involved in CNS immune surveillance and neuroinflammation in MS, there are limitations to this work. First, we characterize the T cell states in the CSF transcriptionally, which does not fully elucidate the effector potential of different T cell subtypes in the CSF in healthy individuals and patients with MS, especially upon antigen encounter and necessitates future validation at the protein and functional levels. We also examine newly diagnosed relapsing-remitting patients with MS, which does not necessarily represent the T cell state in progressive MS or other long-term neuroinflammation that may, for example, be further influenced by responses from parenchymal cells (72, 73). In line with this, although we cultured T cells with total CSF supernatant from healthy individuals, further characterization of components within the CSF that may influence T cell function in health and disease will be important for elucidating drivers of CSF T cell phenotypes. In addition, although we detect changes in clonally expanded T cells between patients with MS and healthy donors, we cannot determine whether this is due to their role in the ongoing autoimmune process or whether these cells may be influenced by a more inflammatory environment. Last, our analysis of potential cellular interactions between T cells and cells in the brain parenchyma is hypothesis-generating and does not preserve the spatial organization of the brain or the cellular protein content needed to verify cell-cell interactions.
Overall, our work has generated a single-cell RNA and TCR data-set that has allowed the integration of transcriptional and clonal information to further understand T cell surveillance of the CNS in healthy individuals and patients with MS. Defining the T cell states in the CNS, which has long been considered immune privileged, may shed further light on the mechanisms that allow for necessary immune surveillance without damaging inflammation, including the potential role of coinhibitory receptors on T cells in the CSF and parenchyma (74). Because T cells and the TH1 cytokine IFNγ have also been found to influence neurological functions including social behavior (75, 76), defining the profiles of CNS T cells may further elucidate their roles in homeostatic neurologic function in addition to contextualizing autoimmune, neurodegenerative, antitumor, and antipathogen immune responses within the immunologically unique CNS.
MATERIALS AND METHODS
Study design
The goal of this study was using scRNA-seq and single-cell TCR sequencing to define the state of T cell surveillance in the healthy CSF and comparing this with the CSF T cell state in neuroinflammation in newly diagnosed (diagnosis within 12 months of lumbar puncture) relapsing-remitting patients with MS. Healthy donors for paired blood draw and lumbar puncture (LP) or blood draw alone for in vitro experiments were recruited by International Review Board (IRB)–approved fliers or word of mouth. Patients with neuroinflammatory disease who were receiving an LP for diagnostic purposes were recruited through Yale New Haven Hospital and Yale Multiple Sclerosis Center. Patients and healthy controls were recruited as part of an IRB-approved study, and written consent was obtained. LPs were performed under sterile conditions. Post-mortem unfixed frozen human brain tissues were obtained from the Department of Pathology, Yale School of Medicine. Flash-frozen human brain tissue was obtained from epilepsy surgery of temporal lobectomy with written consent in the Department of Neurosurgery, Yale School of Medicine. Clinical and demographic characteristics of donors are shown in table S1. Sample sizes were determined on the basis of comparable studies, and the number of donors and cells used in analysis is specified for each figure. There was no randomization or blinding in our study.
Sample processing
PBMCs were isolated from fresh whole blood via density gradient by layering blood diluted 1:1 in phosphate-buffered saline (PBS) over Ficoll-Paque. Red blood cells were lysed using ACK lysing buffer (Gibco) per the manufacturer’s protocol and filtered through a 40-μm filter. Cells were resuspended in 1× Dulbecco’s PBS with 0.04% bovine serum albumin for loading into 10x Genomics at suspensions ranging from 800 to 1300 cells/μl. CSF was kept on ice until processing. Total CSF was spun for 10 min at 1500 rpm, and supernatant was aspirated. The pellet was then transferred to a 1.5-ml tube, spun for 10 min at 1500 rpm with supernatant, and then aspirated to leave 30 to 50 μl for cell counting and loading into 10x Genomics. If a small amount of red blood cell contamination was present in the CSF, then red blood cells were lysed in parallel with blood using ACK lysing buffer that was used as per the manufacturer’s protocol to lyse red blood cells immediately before loading cells (table S1). CSF from traumatic LPs that contain substantial contamination of peripheral blood cells was not included in this study. Blood and CSF samples were counted using a hemocytometer. For scRNA-seq, samples were run, and libraries were prepared using the 10x Genomics Chromium Single Cell V(D)J Reagent Kit v1 chemistry as per the manufacturer’s protocol. A target cell number of 6000 was used for the blood, and on the basis of cell number, CSF was loaded in total or was loaded for recovery of 6000 cells. Samples were sequenced on Illumina HiSeq 4000 or NovaSeq S4.
Human brain nuclei isolation and snRNA-seq
Nuclei were isolated from post-mortem frozen or flash-frozen human brain tissue as previously described (77) with modifications. Frozen tissue (50 to 100 mg) was homogenized in 15 ml of ice-cold nuclei homogenization buffer [2 M sucrose, 10 mM Hepes (pH 7.9), 25 mM KCl, 1 mM EDTA (pH 8.0), 10% glycerol, and ribonuclease (RNase) inhibitors freshly added (80 U/ml)] with Dounce tissue grinder (10 strokes with loose pestle and 10 strokes with tight pestle). The homogenate was transferred into an ultracentrifuge tube on top of 10 ml of fresh nuclei homogenization buffer and centrifuged at 24,000 rpm for 60 min at 4°C on high-speed centrifuge. The supernatant was removed, and the pellet was resuspended in 1 ml of nuclei resuspension buffer [15 mM Hepes (pH 7.4), 15 mM NaCl, 60 mM KCl, 2 mM MgCl2, 3 mM CaCl2, and RNase inhibitors freshly added (80 U/ml)] and counted on a hemocytometer. The nuclei were centrifuged at 5000 rpm for 10 min at 4°C and resuspended at a concentration of 500 to 1000 nuclei/μl for 10x Genomics Chromium loading. The snRNA-seq libraries were prepared by the Chromium Single Cell 3′ Reagent Kit v3 chemistry according to the manufacturer’s instructions (10x Genomics) and sequenced using Illumina HiSeq 4000. Methods and additional information for scRNA-seq of the brain parenchyma can be found in (67).
Non-neuronal nuclei enrichment and sequencing
The isolated nuclei were resuspended in blocking buffer [2% fetal calf serum (FCS) and nuclei resuspension buffer] and incubated at 4°C for 20 min, and NeuN antibody clone A60 conjugated with Alexa Fluor 488 (Sigma-Aldrich, MAB377X) was added at 1:1000 dilution and incubated for 30 min at 4°C. After 1× washing with blocking buffer, the nuclei were filtered through a 35-μm filter and subjected to sorting on a FACSAria for negative selection of NeuN− nuclei for 10x Genomics Chromium loading. The non-neuronal snRNA-seq libraries were prepared by the Chromium Single Cell 3′ Reagent Kit v3 chemistry according to the manufacturer’s instructions (10x Genomics) and sequenced using Illumina HiSeq 4000.
scRNA-seq analysis
Data alignment
For scRNA-seq, samples run on 10x Genomics were aligned to the human GRCh38 genome using CellRanger version 3.0.2. For TCR sequencing, reads from the V(D)J libraries were mapped to the human reference of vdj_GRCh38_alts_ensemble (available from 10x Genomics). For snRNA-seq of brain parenchyma, a custom pre-mRNA human genome reference was generated with GRCh38 that included pre-mRNA sequences, and snRNA-seq data were aligned to this GRCh38–pre-mRNA reference to map both unspliced pre-mRNA and mature mRNA using CellRanger version 3.0.2.
In vitro experiments
Peripheral blood was obtained from healthy individuals. PBMCs were isolated as previously described, and total T cells were magnetically enriched using the EasySep Human T Cell Enrichment Kit (STEMCELL Technologies) as per the manufacturer’s protocol. T cells were stained in PBS with 2% FCS for CD4 (1:45; allophycocyanin (APC)–Cy7, BioLegend, clone RPA-T4), CD8 [1:45; phycoerythrin (PE)–Texas Red, Abcam, clone RFT-8], CD45RO (1:12; PE-Cy7, Thermo Fisher Scientific, clone UCHL1), and CXCR3 (1:45; BV421, BD Biosciences, clone 1C6) for 30 min at 4°C. CD4+CD45RO+CXCR3+ and CD8+CD45RO+CXCR3+ cells were sorted into XVIVO15 + 5% FCS + penicillin (100 U/ml) and streptomycin (100 μg/ml). Round-bottom 96-well plates (Corning) were coated with anti-CD3 (1 μg/ml; UCHT1, BD Biosciences) for 4 hours and washed with PBS. CSF supernatant from healthy individuals was stored at the time of donation at −80°C and thawed on ice for in vitro experiments. A total of 20,000 T cells were plated per well in XVIVO15 (no added serum) with penicillin and streptomycin with soluble anti-CD28 (1 μg/ml; 28.2) (BD Biosciences) with or without 50 μl (25% total culture volume) or 100 μl (50% total culture volume) of healthy CSF supernatant. CSF supernatant from nine healthy controls was used, and in vitro T cell experiments were performed on three autologous and six nonautologous healthy blood donors. T cells were harvested and lysed for quantitative polymerase chain reaction (qPCR) analysis after 48 hours in culture.
Quantitative polymerase chain reaction
Total RNA was extracted with a ZR-96 Quick-RNA kit (Zymo Research) according to the manufacturer’s protocol. RNA was reverse-transcribed with TaqMan reverse transcription reagents (Applied Biosystems). Target genes were amplified using TaqMan Gene Expression Assay (B2M, HS00187842; IFNG, HS00989291; PRDM1, HS00153357) with TaqMan Fast Advanced Master Mix and read on a StepOne Real-Time PCR System (Applied Biosystems). Target gene relative expression was measured relative to B2M. Relative expression to B2M for each sample was calculated by (2−ΔCt) × 103.
Flow cytometry
For flow cytometry and intracellular staining of the healthy blood and CSF, PBMCs were isolated as previously described, CSF was spun at 1500 rpm for 10 min, and supernatant was aspirated. Cells (10 × 103 to 0.2 × 106) were incubated for 4 hours with or without PMA (250 μg/ml) and ionomycin (50 μg/ml) in the presence of GolgiStop (BD Biosciences). Cells were then incubated with Live/Dead Fixable Viability Dye (Invitrogen, L23102) for 15 min, washed with staining buffer (BD Biosciences, 554657), and fixed with fix/perm buffer (eBioscience, 00-5523-00). Last, cells were washed twice with staining buffer and resuspended in PBS. Blocking was performed with 5 μl of Fc Block (BioLegend, 422302) before adding the antibody mixture. Cell surface and intracellular staining were performed using CD4 (PerCP Cy5, BioLegend, clone SK3), CD69 (PE-Cy7, BioLegend, clone FN50), CD3 (APC-H7, BD Biosciences, clone SKS7), CD8 (Alexa Fluor 700, BioLegend, clone SK1), IFNG (Alexa Fluor 488, BD Biosciences, clone B27), CD45RA (Brilliant Violet 421, BioLegend, clone HI100), and PD-1 (PE, BioLegend, clone FH12.2H7). Data were acquired in a BD LSRII Fortessa flow cytometer and analyzed by FlowJo software v9.
For surface staining of the healthy blood and CSF, PBMCs were isolated as previously described, and T cells were isolated using the EasySep Human T Cell Enrichment Kit (STEMCELL Technologies) as per the manufacturer’s protocol. CSF was spun down at 1500 rpm for 5 min, and the supernatant was aspirated. For surface staining, cells were stained for 30 min at 4°C in PBS with 2% FCS. Cells for chemokine receptor analysis were stained with CD4 (1:40; APC-Cy7, BD Biosciences, clone RPA-T4), CD8 (1:40; PE–Texas Red, Abcam, clone RFT-8), CD45RO (1:10; fluorescein isothiocyanate, BD Biosciences, clone UCHL1), and CXCR3 (1:40; BV-421, BD Biosciences, clone 1C6). After staining, samples were washed and acquired on a BD FACSAria and analyzed by FlowJo software v10. Representative staining for all flow cytometry and sorting are shown in supplementary figures.
Statistical analysis
Statistical analyses were performed in GraphPad Prism 7 or R for differential expression, which was calculated using MAST, and the p.adjust function was used for FDR correction. For MAST results, an FDR of <0.05 was considered significant, and for CellPhoneDB, the P value cutoff was set to 0.01. For other statistics, a value of <0.05 for P value or adjusted P value was considered significant unless otherwise noted. Statistical analysis for cellular interactions was performed within the CellPhoneDB software. Sample sizes, statistical tests, and multiple comparison corrections are described in figure legends.
Supplementary Material
Fig. S1. Cell quality and lineage identification of healthy and MS PBMCs and CSF.
Fig. S2. PHATE representation of T cells.
Fig. S3. Chemokine receptor flow cytometry of healthy PBMCs and CSF.
Fig. S4. Gating strategy for total T cell selection for surface and intracellular staining.
Fig. S5. Analysis of healthy TCRs.
Fig. S6. Analysis of MS TCRs.
Fig. S7. Sample sort strategy for in vitro experiments.
Fig. S8. Sample sort strategy for non-neuronal nuclei enrichment.
Fig. S9. Cell quality and lineage identification in the brain parenchyma in snRNA-seq and scRNA-seq datasets.
Table S1. Donor information.
Table S2. Healthy T cell clusters.
Table S3. Gene clusters and canonical pathways.
Table S4. Healthy clonal group differentially expressed genes.
Table S5. T cell cluster distribution statistics.
Table S6. MS versus healthy T cell clusters differentially expressed genes.
Table S7. MS versus healthy expanded cells differentially expressed genes.
Table S8. T cells in brain parenchyma differentially expressed genes.
Table S9. Cell-cell interactions using CellPhoneDB.
Table S10. Raw data.
Acknowledgements:
We thank L. Devine and C. Wang for assistance with flow cytometry and sorting; G. Wang at Yale Center for Genome Analysis for support with 10x Genomics library preparation and sequencing; B. Goods, A. Tu, J. C. Love, and A. K. Shalek for assistance generating and analyzing preliminary data; E. Longbrake, S. Florin, and the staff at the Yale New Haven Hospital and Multiple Sclerosis Center for patient recruitment; S. Strittmatter and D. Spencer for providing brain parenchyma samples; S. Farhadian, P. Stathopolous, S. Spudich, and J. Chiarella for collection of patient CSF samples; and M. Askenase and A. Kitz for useful discussions and input on the manuscript.
Funding: This work was supported by grants to D.A.H. from the NIH (U19 AI089992, R25 NS079193, P01 AI073748, U24 AI11867, R01 AI22220, UM 1HG009390, P01 AI039671, P50 CA121974, and R01 CA227473), the National Multiple Sclerosis Society (NMSS) (CA 1061- A-18 and RG-1802–30153), the Nancy Taylor Foundation for Chronic Diseases, and Erase MS. This work was supported by the National Science Foundation Graduate Research Fellowship Program under grant DGE1752134 (to J.L.P) and Gruber Science Fellowship (to J.L.P.).
Footnotes
SUPPLEMENTARY MATERIALS
immunology.sciencemag.org/cgi/content/full/5/51/eabb8786/DC1
Supplementary Materials and Methods
Competing interests: D.A.H. has received funding for his lab from Bristol Myers Squibb, Sanofi, and Genentech. All other authors declared that they have no competing interests.
Data and materials availability: Data for all CSF scRNA-seq and single-cell TCR sequencing will be available through NCBI’s dbGaP at accession number phs002222.v1.p1, and brain parenchyma snRNA-seq data will be available through GEO at accession number GSE157277. scRNA-seq data of the brain parenchyma will be available in (67) at GEO accession number GSE155488. Additional data related to this paper may be requested from the authors. Code for scprep, PHATE, MAGIC, MELD, and graphtools is available at github.com/KrishnaswamyLab. Additional code is available upon request.
REFERENCES AND NOTES
- 1.Wilson EH, Weninger W, Hunter CA, Trafficking of immune cells in the central nervous system. J. Clin. Invest 120, 1368–1379 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen JE, Nansen A, Moos T, Lu B, Gerard C, Christensen JP, Thomsen AR, Efficient T-cell surveillance of the CNS requires expression of the CXC chemokine receptor 3. J. Neurosci 24, 4849–4858 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landrith TA, Harris TH, Wilson EH, Characteristics and critical function of CD8+ T cells in the Toxoplasma-infected brain. Semin. Immunopathol 37, 261–270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein RS, Hunter CA, Protective and pathological immunity during central nervous system infections. Immunity 46, 891–909 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Warnke C, Menge T, Hartung HP, Racke MK, Cravens PD, Bennett JL, Frohman EM, Greenberg BM, Zamvil SS, Gold R, Hemmer B, Kieseier BC, Stüve O, Natalizumab and progressive multifocal leukoencephalopathy: What are the causal factors and can it be avoided? Arch. Neurol 67, 923–930 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wakim LM, Woodward-Davis A, Bevan MJ, Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc. Natl. Acad. Sci. U.S.A 107, 17872–17879 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prinz M, Priller J, The role of peripheral immune cells in the CNS in steady state and disease. Nat. Neurosci 20, 136–144 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Cuyar LF, Sonnen JA, Montine KS, Keene CD, Montine TJ, Role of cerebrospinal fluid and plasma biomarkers in the diagnosis of neurodegenerative disorders and mild cognitive impairment. Curr. Neurol. Neurosci. Rep 11, 455–463 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janelidze S, Mattsson N, Stomrud E, Lindberg O, Palmqvist S, Zetterberg H, Blennow K, Hansson O, CSF biomarkers of neuroinflammation and cerebrovascular dysfunction in early Alzheimer disease. Neurology 91, e867–e877 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quintana FJ, Yeste A, Weiner HL, Covacu R, Lipids and lipid-reactive antibodies as biomarkers for multiple sclerosis. J. Neuroimmunol 248, 53–57 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuenz B, Lutterotti A, Ehling R, Gneiss C, Haemmerle M, Rainer C, Deisenhammer F, Schocke M, Berger T, Reindl M, Cerebrospinal fluid B cells correlate with early brain inflammation in multiple sclerosis. PLOS ONE 3, e2559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Link H, Huang Y-M, Oligoclonal bands in multiple sclerosis cerebrospinal fluid: An update on methodology and clinical usefulness. J. Neuroimmunol 180, 17–28 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Lovato L, Willis SN, Rodig SJ, Caron T, Almendinger SE, Howell OW, Reynolds R, O’Connor KC, Hafler DA, Related B cell clones populate the meninges and parenchyma of patients with multiple sclerosis. Brain 134, 534–541 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skulina C, Schmidt S, Dornmair K, Babbe H, Roers A, Rajewsky K, Wekerle H, Hohlfeld R, Goebels N, Multiple sclerosis: Brain-infiltrating CD8+ T cells persist as clonal expansions in the cerebrospinal fluid and blood. Proc. Natl. Acad. Sci. U.S.A 101, 2428–2433 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar BV, Connors TJ, Farber DL, Human T cell development, localization, and function throughout life. Immunity 48, 202–213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gate D, Saligrama N, Leventhal O, Yang AC, Unger MS, Middeldorp J, Chen K, Lehallier B, Channappa D, De Los Santos MB, McBride A, Pluvinage J, Elahi F, Tam GK-Y, Kim Y, Greicius M, Wagner AD, Aigner L, Galasko DR, Davis MM, Wyss-Coray T, Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577, 399–404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beltrán E, Gerdes LA, Hansen J, Flierl-Hecht A, Krebs S, Blum H, Ertl-Wagner B, Barkhof F, Kumpfel T, Hohlfeld R, Dornmair K, Early adaptive immune activation detected in monozygotic twins with prodromal multiple sclerosis. J. Clin. Invest 129, 4758–4768 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho EL, Ronquillo R, Altmeppen H, Spudich SS, Price RW, Sinclair E, Cellular composition of cerebrospinal fluid in HIV-1 infected and uninfected subjects. PLOS ONE 8, e66188 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hafler DA, Fox DA, Manning ME, Schlossman SF, Reinherz EL, Weiner HL, In vivo activated T lymphocytes in the peripheral blood and cerebrospinal fluid of patients with multiple sclerosis. N. Engl. J. Med 312, 1405–1411 (1985). [DOI] [PubMed] [Google Scholar]
- 20.Han S, Lin YC, Wu T, Salgado AD, Mexhitaj I, Wuest SC, Romm E, Ohayon J, Goldbach-Mansky R, Vanderver A, Marques A, Toro C, Williamson P, Cortese I, Bielekova B, Comprehensive immunophenotyping of cerebrospinal fluid cells in patients with neuroimmunological diseases. J. Immunol 192, 2551–2563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nemecek A, Zimmermann H, Rübenthaler J, Fleischer V, Paterka M, Luessi F, Müller-Forell W, Zipp F, Siffrin V, Flow cytometric analysis of T cell/monocyte ratio in clinically isolated syndrome identifies patients at risk of rapid disease progression. Mult. Scler. J 22, 483–493 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Farhadian SF, Mehta SS, Zografou C, Robertson K, Price RW, Pappalardo J, Chiarella J, Hafler DA, Spudich SS, Single-cell RNA sequencing reveals microglia-like cellsin cerebrospinal fluid during virologically suppressed HIV. JCI Insight 3, e121718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schafflick D, Xu CA, Hartlehnert M, Cole M, Schulte-Mecklenbeck A, Lautwein T, Wolbert J, Heming M, Meuth SG, Kuhlmann T, Gross CC, Wiendl H, Yosef N, Meyer Zu Horste G, Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nat. Commun 11, 247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szabo PA, Levitin HM, Miron M, Snyder ME, Senda T, Yuan J, Cheng YL, Bush EC, Dogra P, Thapa P, Farber DL, Sims PA, Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun 10, 4706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miragaia RJ, Gomes T, Chomka A, Jardine L, Riedel A, Hegazy AN, Whibley N, Tucci A, Chen X, Linderman I, Emerton G, Krausgruber T, Shields J, Haniffa M, Powrie F, Teichmann SA, Single-cell transcriptomics of regulatory T cells reveals trajectories of tissue adaptation. Immunity 50, 493–504.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon KR, van Dijk D, Wang Z, Gigante S, Burkhardt DB, Chen WS, Yim K, van der Elzen A, Hirn MJ, Coifman RR, Ivanova NB, Wolf G, Krishnaswamy S, Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol 37, 1482–1492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maggi L, Santarlasci V, Capone M, Peired A, Frosali F, Crome SQ, Querci V, Fambrini M, Liotta F, Levings MK, Maggi E, Cosmi L, Romagnani S, Annunziato F, CD161 is a marker of all human IL-17-producing T-cell subsets and is induced by RORC. Eur. J. Immunol 40, 2174–2181 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Zhu J, Yamane H, Cote-Sierra J, Guo L, Paul WE, GATA-3 promotes Th2 responses through three different mechanisms: Induction of Th2 cytokine production, selective growth of Th2 cells and inhibition of Th1 cell-specific factors. Cell Res. 16, 3–10 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Kunicki MA, Amaya Hernandez LC, Davis KL, Bacchetta R, Roncarolo M-G, Identity and diversity of human peripheral Th and T regulatory cells defined by single-cell mass cytometry. J. Immunol 200, 336–346 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Kivisäkk P, Mahad DJ, Callahan MK, Trebst C, Tucky B, Wei T, Wu L, Baekkevold ES, Lassmann H, Staugaitis SM, Campbell JJ, Ransohoff RM, Human cerebrospinal fluid central memory CD4+ T cells: Evidence for trafficking through choroid plexus and meninges via P-selectin. Proc. Natl. Acad. Sci. U.S.A 100, 8389–8394 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skon CN, Lee J-Y, Anderson KG, Masopust D, Hogquist KA, Jameson SC, Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat. Immunol 14, 1285–1293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groom JR, Luster AD, CXCR3 in T cell function. Exp. Cell Res 317, 620–631 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder ME, Finlayson MO, Connors TJ, Dogra P, Senda T, Bush E, Carpenter D, Marboe C, Benvenuto L, Shah L, Robbins H, Hook JL, Sykes M, D’Ovidio F, Bacchetta M, Sonett JR, Lederer DJ, Arcasoy S, Sims PA, Farber DL, Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci. Immunol 4, eaav5581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swarnalekha N, Schreiner D, Litzler LC, Iftikhar S, Kirchmeier D, Künzli M, King CG, Redefining CD4 T cell residency: Helper T cells orchestrate protective humoral immunity in the lung. bioRxiv 2020.02.28.963280 [Preprint]. 28 February 2020. 10.1101/2020.02.28.963280. [DOI] [Google Scholar]
- 35.Martin JC, Chang C, Boschetti G, Ungaro R, Giri M, Grout JA, Gettler K, Chuang L-S, Nayar S, Greenstein AJ, Dubinsky M, Walker L, Leader A, Fine JS, Whitehurst CE, Mbow ML, Kugathasan S, Denson LA, Hyams JS, Friedman JR, Desai PT, Ko HM, Laface I, Akturk G, Schadt EE, Salmon H, Gnjatic S, Rahman AH, Merad M, Cho JH, Kenigsberg E, Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178, 1493–1508.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, Senda T, Sun X, Ho S-H, Lerner H, Friedman AL, Shen Y, Farber DL, Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoidand mucosal sites. Cell Rep. 20, 2921–2934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hidalgo LG, Einecke G, Allanach K, Halloran PF, The transcriptome of human cytotoxic T cells: Similarities and disparities among allostimulated CD4+ CTL, CD8+ CTL and NK cells. Am. J. Transplant 8, 627–636 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Leipe J, Pirronello F, Klose A, Schulze-Koops H, Skapenko A, Increased plasticity of non-classic Th1 cells toward the Th17 phenotype. Mod. Rheumatol 30, 930–936 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Becattini S, Latorre D, Mele F, Foglierini M, De Gregorio C, Cassotta A, Fernandez B, Kelderman S, Schumacher TN, Corti D, Lanzavecchia A, Sallusto F, Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science 347, 400–406 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Restorick SM, Durant L, Kalra S, Hassan-Smith G, Rathbone E, Douglas MR, Curnow SJ, CCR6+ Th cells in the cerebrospinal fluid of persons with multiple sclerosis are dominated by pathogenic non-classic Th1 cells and GM-CSF-only-secreting Th cells. Brain Behav. Immun 64, 71–79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behr FM, Kragten NAM, Wesselink TH, Nota B, van Lier RAW, Amsen D, Stark R, Hombrink P, van Gisbergen K, Blimp-1 rather than hobit drives the formation of tissue-resident memory CD8+ T cells in the lungs. Front. Immunol 10, 400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackay LK, Minnich M, Kragten NAM, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, Braun A, Wynne-Jones E, Behr FM, Stark R, Pellici DG, Godfrey DI, Belz GT, Pellegrini M, Gebhardt T, Busslinger M, Shi W, Carbone FR, van Lier RAW, Kallies A, van Gisbergen KPJM, Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Wein AN, McMaster SR, Takamura S, Dunbar PR, Cartwright EK, Hayward SL, McManus DT, Shimaoka T, Ueha S, Tsukui T, Masumoto T, Kurachi M, Matsushima K, Kohlmeier JE, CXCR6 regulates localization of tissue-resident memory CD8 T cells to the airways. J. Exp. Med 216, 2748–2762 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milner JJ, Toma C, He Z, Kurd NS, Nguyen QP, McDonald B, Quezada L, Widjaja CE, Witherden DA, Crowl JT, Shaw LA, Yeo GW, Chang JT, Omilusik KD, Goldrath AW, Heterogenous populations of tissue-resident CD8+ T cells are generated in response to infection and malignancy. Immunity 52, 808–824.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Machado-Santos J, Saji E, Tröscher AR, Paunovic M, Liblau R, Gabriely G, Bien CG, Bauer J, Lassmann H, The compartmentalized inflammatory response in the multiple sclerosis brain is composed of tissue-resident CD8+ T lymphocytes and B cells. Brain 141, 2066–2082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smolders J, Heutinck KM, Fransen NL, Remmerswaal EBM, Hombrink P, ten Berge IJM, van Lier RAW, Huitinga I, Hamann J, Tissue-resident memory T cells populate the human brain. Nat. Commun 9, 4593 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dulken BW, Buckley MT, Navarro Negredo P, Saligrama N, Cayrol R, Leeman DS, George BM, Boutet SC, Hebestreit K, Pluvinage JV, Wyss-Coray T, Weissman IL, Vogel H, Davis MM, Brunet A, Single-cell analysis reveals T cell infiltration in old neurogenic niches. Nature 571, 205–210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burkhardt DB, Stanley III JS, Perdigoto AL, Gigante SA, Herold KC, Wolf G, Giraldez A, van Dijk D, Krishnaswamy S, Enhancing experimental signals in single-cell RNA-sequencing data using graph signal processing. bioRxiv 532846 [Preprint]. 28 January 2019. 10.1101/532846. [DOI] [Google Scholar]
- 49.Kivisäkk P, Trebst C, Liu Z, Tucky BH, Sørensen TL, Rudick RA, Mack M, Ransohoff RM, T-cells in the cerebrospinal fluid express a similar repertoire of inflammatory chemokine receptors in the absence or presence of CNS inflammation: Implications for CNS trafficking. Clin. Exp. Immunol 129, 510–518 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yaldizli Ö, Putzki N, Natalizumab in the treatment of multiple sclerosis. Ther. Adv. Neurol. Disord 2, 115–128 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kidani Y, Elsaesser H, Hock MB, Vergnes L, Williams KJ, Argus JP, Marbois BN, Komisopoulou E, Wilson EB, Osborne TF, Graeber TG, Reue K, Brooks DG, Bensinger SJ, Sterol regulatory element–binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat. Immunol 14, 489–499 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schreiner D, King CG, CD4+ memory T cells at home in the tissue: Mechanisms for health and disease. Front. Immunol 9, 2394 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Behr FM, Chuwonpad A, Stark R, van Gisbergen K, Armed and ready: Transcriptional regulation of tissue-resident memory CD8 T cells. Front. Immunol 9, 1770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goods BA, Hernandez AL, Lowther DE, Lucca LE, Lerner BA, Gunel M, Raddassi K, Coric V, Hafler DA, Love JC, Functional differences between PD-1+ and PD-1− CD4+ effector T cells in healthy donors and patients with glioblastoma multiforme. PLOS ONE 12, e0181538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tu AA, Gierahn TM, Monian B, Morgan DM, Mehta NK, Ruiter B, Shreffler WG, Shalek AK, Love JC, TCR sequencing paired with massively parallel 3′ RNA-seq reveals clonotypic T cell signatures. Nat. Immunol 20, 1692–1699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartolomé-Casado R, Landsverk OJB, Chauhan SK, Richter L, Phung D, Greiff V, Risnes LF, Yao Y, Neumann RS, Yaqub S, Øyen O, Horneland R, Aandahl EM, Paulsen V, Sollid LM, Qiao S-W, Baekkevold ES, Jahnsen FL, Resident memory CD8 T cells persist for years in human small intestine. J. Exp. Med 216, 2412–2426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J, Olshansky M, Carbone FR, Ma JZ, Transcriptional analysis of T cells resident in human skin. PLOS ONE 11, e0148351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, Yin W, Zhu L, Li J, Yao Y, Chen F, Sun M, Zhang J, Shen N, Song Y, Chang X, Iron drives T helper cell pathogenicity by promoting RNA-binding protein PCBP1-mediated proinflammatory cytokine production. Immunity 49, 80–92.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Cibrián D, Sánchez-Madrid F, CD69: From activation marker to metabolic gatekeeper. Eur. J. Immunol 47, 946–953 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shoda H, Fujio K, Yamaguchi Y, Okamoto A, Sawada T, Kochi Y, Yamamoto K, Interactions between IL-32 and tumor necrosis factor alpha contribute to the exacerbation of immune-inflammatory diseases. Arthritis Res. Ther 8, R166 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ota K, Matsui M, Milford EL, Mackin GA, Weiner HL, Hafler DA, T-cell recognition of an immunodominant myelin basic protein epitope in multiple sclerosis. Nature 346, 183–187 (1990). [DOI] [PubMed] [Google Scholar]
- 62.van Nierop GP, van Luijn MM, Michels SS, Melief M-J, Janssen M, Langerak AW, Ouwendijk WJD, Hintzen RQ, Verjans G, Phenotypic and functional characterization of T cells in white matter lesions of multiple sclerosis patients. Acta Neuropathol. 134, 383–401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu H, Leitenberg D, Li B, Flavell RA, Deficiency of small GTPase Rac2 affects T cell activation. J. Exp. Med 194, 915–926 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suga H, Sugaya M, Miyagaki T, Kawaguchi M, Fujita H, Asano Y, Tada Y, Kadono T, Sato S, The role of IL-32 in cutaneous T-cell lymphoma. J. Invest. Dermatol 134, 1428–1435 (2014). [DOI] [PubMed] [Google Scholar]
- 65.Louveau A, Plog BA, Antila S, Alitalo K, Nedergaard M, Kipnis J, Understanding the functions and relationships of the glymphatic system and meningeal lymphatics. J. Clin. Invest 127, 3210–3219 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sankowski R, Böttcher C, Masuda T, Geirsdottir L, Sagar, Sindram E, Seredenina T, Muhs A, Scheiwe C, Shah MJ, Heiland DH, Schnell O, Grün D, Priller J, Prinz M, Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat. Neurosci 22, 2098–2110 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Perlman K, Couturier CP, Yaqubi M, Tanti A, Cui Q-L, Pernin F, Stratton JA, Ragoussis J, Healy L, Petrecca K, Dudley R, Srour M, Hall JA, Kennedy TE, Mechawar N, Antel JP, Developmental trajectory of oligodendrocyte progenitor cells in the human brain revealed by single cell RNA sequencing. Glia 68, 1291–1303 (2020). [DOI] [PubMed] [Google Scholar]
- 68.Bohlen CJ, Bennett FC, Tucker AF, Collins HY, Mulinyawe SB, Barres BA, Diverse requirements for microglial survival, specification, and function revealed by defined-medium cultures. Neuron 94, 759–773.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Iijima N, Iwasaki A, Tissue instruction for migration and retention of TRM cells. Trends Immunol. 36, 556–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schenkel JM, Fraser KA, Casey KA, Beura LK, Pauken KE, Vezys V, Masopust D, IL-15–independent maintenance of tissue-resident and boosted effector memory CD8 T cells. J. Immunol 196, 3920–3926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salou M, Nicol B, Garcia A, Laplaud D-A, Involvement of CD8+ T cells in multiple sclerosis. Front. Immunol 6, 604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singhal T, O’Connor K, Dubey S, Pan H, Chu R, Hurwitz S, Cicero S, Tauhid S, Silbersweig D, Stern E, Kijewski M, DiCarli M, Weiner HL, Bakshi R, Gray matter microglial activation in relapsing vs progressive MS: A [F-18]PBR06-PET study. Neurol. Neuroimmunol. Neuroinflamm 6, e587 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baecher-Allan C, Kaskow BJ, Weiner HL, Multiple sclerosis: Mechanisms and immunotherapy. Neuron 97, 742–768 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Shwetank EL Frost TE Mockus HM Ren M Toprak MD Lauver CS Netherby-Winslow G Jin JM Cosby BD Evavold AE Lukacher, PD-1 dynamically regulates inflammation and development of brain-resident memory CD8 T cells during persistent viral encephalitis. Front. Immunol 10, 783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Filiano AJ, Xu Y, Tustison NJ, Marsh RL, Baker W, Smirnov I, Overall CC, Gadani SP, Turner SD, Weng Z, Peerzade SN, Chen H, Lee KS, Scott MM, Beenhakker MP, Litvak V, Kipnis J, Unexpected role of interferon-γ in regulating neuronal connectivity and social behaviour. Nature 535, 425–429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kipnis J, Cohen H, Cardon M, Ziv Y, Schwartz M, T cell deficiency leads to cognitive dysfunction: Implications for therapeutic vaccination for schizophrenia and other psychiatric conditions. Proc. Natl. Acad. Sci. U.S.A 101, 8180–8185 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goffin D, Allen M, Zhang L, Amorim M, Wang I-TJ, Reyes A-RS, Mercado-Berton A, Ong C, Cohen S, Hu L, Blendy JA, Carlson GC, Siegel SJ, Greenberg ME, Zhou Z, Rett syndrome mutation MeCP2 T158A disrupts DNA binding, protein stability and ERP responses. Nat. Neurosci 15, 274–283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M, Waldman J, Sud M, Andrews E, Velonias G, Haber AL, Jagadeesh K, Vickovic S, Yao J, Stevens C, Dionne D, Nguyen LT, Villani A-C, Hofree M, Creasey EA, Huang H, Rozenblatt-Rosen O, Garber JJ, Khalili H, Desch AN, Daly MJ, Ananthakrishnan AN, Shalek AK, Xavier RJ, Regev A, Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730.e22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr AJ, Burdziak C, Moon KR, Chaffer CL, Pattabiraman D, Bierie B, Mazutis L, Wolf G, Krishnaswamy S, Pe’er D, Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e27 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levine JH, Simonds EF, Bendall SC, Davis KL, Amirel AD, Tadmor MD, Litvin O, Fienberg HG, Jager A, Zunder ER, Finck R, Gedman AL, Radtke I, Downing JR, Pe’er D, Nolan GP, Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, Gottardo R, MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Collier SP, Henderson MA, Tossberg JT, Aune TM, Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J. Immunol 193, 3959–3965 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Serroukh Y, Gu-Trantien C, Hooshiar Kashani B, Defrance M, Manh TPV, Azouz A, Detavernier A, Hoyois A, Das J, Bizet M, Pollet E, Tabbuso T, Calonne E, van Gisbergen K, Dalod M, Fuks F, Goriely S, Marchant A, The transcription factors Runx3 and ThPOK cross-regulate acquisition of cytotoxic function by human Th1 lymphocytes. eLife 7, e30496 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bishu S, El Zaatari M, Hayashi A, Hou G, Bowers N, Kinnucan J, Manoogian B, Muza-Moons M, Zhang M, Grasberger H, Bourque C, Zou W, Higgins PDR, Spence JR, Stidham RW, Kamada N, Kao JY, CD4+ tissue-resident memory T cells expand and are a major source of mucosal tumour necrosis factor α in active Crohn’s disease. J. Crohns Colitis 13, 905–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chihara N, Madi A, Kondo T, Zhang H, Acharya N, Singer M, Nyman J, Marjanovic ND, Kowalczyk MS, Wang C, Kurtulus S, Law T, Etminan Y, Nevin J, Buckley CD, Burkett PR, Buenrostro JD, Rozenblatt-Rosen O, Anderson AC, Regev A, Kuchroo VK, Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 558, 454–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krishnaswamy S, Spitzer MH, Mingueneau M, Bendall SC, Litvin O, Stone E, Pe’er D, Nolan GP, Conditional density-based analysis of T cell signaling in single-cell data. Science 346, 1250689 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park J-E, Stephenson E, Polański K, Goncalves A, Gardner L, Holmqvist S, Henriksson J, Zou A, Sharkey AM, Millar B, Innes B, Wood L, Wilbrey-Clark A, Payne RP, Ivarsson MA, Lisgo S, Filby A, Rowitch DH, Bulmer JN, Wright GJ, Stubbington MJT, Haniffa M, Moffett A, Teichmann SA, Single-cell reconstruction of the early maternal–fetal interface in humans. Nature 563, 347–353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Cell quality and lineage identification of healthy and MS PBMCs and CSF.
Fig. S2. PHATE representation of T cells.
Fig. S3. Chemokine receptor flow cytometry of healthy PBMCs and CSF.
Fig. S4. Gating strategy for total T cell selection for surface and intracellular staining.
Fig. S5. Analysis of healthy TCRs.
Fig. S6. Analysis of MS TCRs.
Fig. S7. Sample sort strategy for in vitro experiments.
Fig. S8. Sample sort strategy for non-neuronal nuclei enrichment.
Fig. S9. Cell quality and lineage identification in the brain parenchyma in snRNA-seq and scRNA-seq datasets.
Table S1. Donor information.
Table S2. Healthy T cell clusters.
Table S3. Gene clusters and canonical pathways.
Table S4. Healthy clonal group differentially expressed genes.
Table S5. T cell cluster distribution statistics.
Table S6. MS versus healthy T cell clusters differentially expressed genes.
Table S7. MS versus healthy expanded cells differentially expressed genes.
Table S8. T cells in brain parenchyma differentially expressed genes.
Table S9. Cell-cell interactions using CellPhoneDB.
Table S10. Raw data.


