Abstract
Subacute ruminal acidosis (SARA) is a common metabolic disease in the dairy farming industry which is usually caused by an excessive amount of high concentrate diet. SARA not only threatens animal welfare but also leads to economic losses in the farming industry. The liver plays an important role in the distribution of nutritional substances and metabolism; however, a high concentrate diet can cause hepatic metabolic disorders and liver injury. Recently, noncoding RNA has been considered as a critical regulator of hepatic disease, however, its role in the bovine liver is limited. In this study, 12 mid-lactating dairy cows were randomly assigned to a control (CON) group (40% concentrate of dry matter, n = 6) and a SARA group (60% concentrate of dry matter, n = 6). After 21 d of treatment, all cows were sacrificed, and liver tissue samples were collected. Three dairy cows were randomly selected from the CON and SARA groups respectively to perform whole transcriptome analysis. More than 20,000 messenger RNA (mRNA), 10,000 long noncoding RNA (lncRNA), 3,500 circular RNA (circRNA) and 1,000 micro RNA (miRNA) were identified. Furthermore, 43 mRNA, 121 lncRNA and 3 miRNA were differentially expressed, whereas no obvious differentially expressed circRNA were detected between the 2 groups. Gene Ontology (GO) annotation revealed that the differentially expressed genes were mainly enriched in oxidoreductase activity, stress, metabolism, the immune response, cell apoptosis, and cell proliferation. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis showed that the deferentially expressed genes were highly enriched in the phosphatidylinositol 3 kinase (PI3K)-serine/threonine kinase (AKT) signaling pathway (P < 0.05). According to KEGG pathway analysis, the differentially expressed lncRNA (DElncRNA) target genes were mainly related to proteasomes, peroxisomes, and the hypoxia-inducible factor-1 signaling pathway (P < 0.005). Further bioinformatics and integrative analyses revealed that the lncRNA were strongly correlated with mRNA; therefore, it is reasonable to speculate that lncRNA potentially play important roles in the liver dysfunction induced by SARA. Our study provides a valuable resource for future investigations on the mechanisms of SARA to facilitate an understanding of the importance of lncRNA, and offer functional RNA information.
Keywords: Whole transcriptome sequencing, Liver, High concentrate diet, Dairy cow
1. Introduction
The dairy industry is gradually developing towards a large-scale, intensive farming model in China with the increasing demand for dairy products. Although the production performance of dairy cows has improved gradually, it has become a great challenge for researchers and breeders to more effectively meet the nutritional needs of cows (Sundrum, 2015). In order to meet the nutritional requirement of cows, farmers have increased the usage of high concentrate diets (Patel, 2012). A high concentrate diet improves the production performance of dairy cows, however, it can also induce a variety of metabolic diseases (Kleen et al., 2003). Subacute ruminal acidosis (SARA) is one of the most representative metabolic diseases. SARA decreases rumen pH, which leads to microbiota disorders and chronic metabolic diseases (Ametaj et al., 2010). More importantly, SARA can cause a series of secondary diseases, increasing the elimination rate of cows (McCann et al., 2016). The incidence of SARA is increasing annually. Therefore, SARA not only poses a threat to the health of high-yielding cows, but also results in great economic losses to the cattle industry (Kitkas et al., 2013).
RNA-seq-based transcriptome data provide a helpful platform for investigating the molecular aspects of metabolic diseases in animals. Therefore, we concentrated on the transcriptomic data to analyze the molecular aspects of SARA in the present study. Previous studies have suggested that eukaryotic genomes encode a large number of functional noncoding RNA (ncRNA) transcripts, including housekeeping and regulatory RNA (Huttenhofer et al., 2005; Brosnan and Voinnet, 2009). miRNA are the most widely known small/short ncRNA, which can regulate the expression of targeted genes (Gindin et al., 2015; Mogilyansky et al., 2016). circRNA is a new type of ncRNA that eliminates miRNA (Hansen et al., 2013; Memczak et al., 2013). Research in lncRNA has rapidly emerged and an increasing number of long noncoding RNA (lncRNA) have been reported in cows (Sun et al., 2016). Moreover, previous studies have revealed that the regulatory function of lncRNA at the transcriptional and post-transcriptional levels is related to disease (Gutschner and Diederichs, 2012; He et al., 2015). However, limited research has been conducted on ncRNA in dairy cows. Studies have indicated that the hepatic metabolic disorders caused by a high concentrate diet occur at the transcriptional level (Dong et al., 2013; Xu et al., 2015). Therefore, we speculated that ncRNA are responsible for hepatic metabolic abnormalities in dairy cows. Understanding the pathogenic mechanisms of a high concentrate diet by studying ncRNA would be helpful.
The liver is an important metabolic organ responsible for glucose and lipid distribution (Knegsel et al., 2005; Xu et al., 2015). In ruminants, the liver is the major site for gluconeogenesis which is the main way to maintain an adequate glucose supply in mammary tissue; and it is also the major site for fat synthesis which is important for the physical properties and quality of milk (Knegsel et al., 2005; Zhao and Keating, 2007; Dong et al., 2013). Therefore, it is of great significance to study the molecular mechanism of SARA by exploring the hepatic transcriptome in dairy cows. Our study used RNA-seq technology for the first time to conduct a joint analysis of mRNA and ncRNA to provide useful information for further research in liver metabolic disorders of dairy cows with SARA.
2. Materials and methods
2.1. Ethics statement
In this study, all animal experiments were approved by the Institutional Animal Care and Use Committee of Nanjing Agricultural University. All procedures of the Guidelines on Ethical Treatment of Experimental Animals (2006) No. 398 set by the Ministry of Science and Technology, China and the Regulation regarding the Management and Treatment of Experimental Animals'’ (2008) No. 45 set by the Jiangsu Provincial People's Government, was strictly followed during the slaughter and sampling procedures.
2.2. Animals, diets and experimental design
Twelve healthy Chinese Holstein cows (mean body weight, 651 ± 54 kg) during mid-lactation (mean milk yeild, 17.43 ± 4.04 kg/d) were raised in dairy farm (Taizhou, Jiangsu Province, China) for the study. All the cows were in their second parity or third parity when the liver tissues were collected. These cows were kept in freestall housing in the same cubicle partition of the barn during the experimental period. After a week of prefeeding, they were randomly assigned to the CON or SARA group. The CON group received a low concentrate diet (40% of dry matter) (n = 6), and the SARA group received a high concentrate diet (60% of dry matter) (n = 6) for 3 weeks. The experimental diets were formulated according to the lactation nutritional requests of a 30 kg milk/d producing cow. The ingredients in the diets and the nutritional composition are shown in Appendix Table 1. Crude protein, starch, ash and other extracts were measured according to the AOAC methods (Helrich and Helrich, 1990). Acid detergent fiber (ADF) and neutral detergent fiber (NDF) were measured by following the methods of Van Soest et al. (Van Soest et al., 1991). The amounts of NEL were calculated using NRC (2001). Ca and P were assayed by inductively coupled plasma spectrometry. The NFC content of the grain was determined by calculation. Cows were given free access to both food and water throughout the experimental time. Dry matter intake (DMI) (kg/d) was recorded daily (CON = 23.72, SARA = 20.82, SEM = 0.68, P = 0.639).
2.3. Sample collection and illumina deep sequencing
After fasting, all cows were sacrificed using electric stunning slaughtering. Samples of about 40 mL of the ruminal fluid were immediately collected through spontaneous efflux after aspirating with a modified 60-mL syringe, and strained through 4 layers of cheesecloth. The ruminal fluid pH was measured directly using a calibrated pH meter (Starter 300; Ohaus Instruments Co. Ltd., Nanjing, China). The liver samples were collected from the same sites and stored under −80 °C for further RNA extraction.
Three dairy cows were randomly selected from the CON and SARA groups respectively. Total RNA was extracted from 50 mg of liver tissue using total RNA extraction reagent following the manufacturer's protocol (Sangon, Shanghai, China). RNA degradation and contamination was monitored on 1.5% agarose gels. RNA concentration and purity was measured using advanced molecular biology equipment to ensure the high quality of samples for transcriptome sequencing.
1.5 μg RNA per sample was used for rRNA removal using the Ribo-Zero rRNA removal kit (Epicentre, USA). Sequencing libraries were generated by an ultra directional RNA library preparation kit for Illumina RNA library preparation (NEB, USA), following manufacturer's recommendations and index codes were added to attribute sequences to each sample. Divalent cations were used to carry out fragmentation under elevated temperature in first strand synthesis reaction buffer (NEB, USA). Random hexamer primer and reverse transcriptase were used to synthesize first strand cDNA. Then second strand cDNA was synthesized by DNA polymerase I and RNase H. Through exonuclease/polymerase activities, the remaining overhangs were converted into blunt ends. After adenylation of 3′ ends of DNA fragments, NEBNext adaptors with hairpin loop structures were ligated to prepare for hybridization. For preferentially selecting insert fragments of 150 to 200 bp in length, AMPure XP beads (Beckman, USA) were used to purify the library fragments. Then 3 μL USER enzyme (NEB, USA) was used with size-selected, adaptor-ligated cDNA at 37 °C for 15 min before PCR. Next, high-fidelity DNA polymerase, universal PCR primers and index primer were used to perform PCR (NEB, USA). Lastly, PCR products were purified and library quality was assessed on the Agilent 2100 bioanalyzer (Agilent, USA).
The clustering of the index-coded samples was performed on a cBot cluster generation system (Illumia, USA) according to the manufacturer's instructions. Then the library preparations were sequenced on an Illumina Hiseq platform and paired-end reads were generated.
2.4. Bioinformatics analysis
2.4.1. Quality control
First of all, raw data of fastq format were cleaned by self-developed Perl scripts. Through removing ploy-N-containing, low-quality reads, and adapter-containing reads from the raw data, the clean data were acquired. The Q20, Q30 and GC contents of the clean data were calculated. All the subsequent analyses were based on the high-quality clean data.
2.4.2. ncRNA identity
For lncRNA identity, the transcriptome was assembled using the StringTie based on the reads mapped to the reference genome. The assembled transcripts were annotated using the gffcompare program. CPC2/CNCI/Pfam/CPAT were combined to sort nonprotein coding RNA candidates. Putative protein-coding RNA were filtered out using a minimum length and exon number threshold. Transcripts with lengths more than 200 nt and 2 exons were selected as lncRNA candidates. Then, the protein-coding genes could be distinguished from the noncoding genes by CPC2/CNCI/Pfam/CPAT. Furthermore, cuffcompare was used to select the different types of lncRNA including lincRNA, intronic lncRNA, anti-sense lncRNA, and sense lncRNA; For miRNA identity, we compared the known reads to the miRBase (v22) database, allowing at most one mismatch, so that the identified reads were considered to be the known miRNA. For the sequences not identified to known miRNA, we used miRDeep2 software to predict novel miRNA; for circRNA identity, we used circRNA identifier (CIRI) tools. During the first scanning of SAM alignment, CIRI detected junction reads with PCC signals that reflect a circRNA candidate. Preliminary filtering was implemented using paired-end mapping (PEM) and GT-AG splicing signals for the junctions. After clustering junction reads and recording each circRNA candidate, CIRI scaned the SAM alignment again to detect additional junction reads. At the same time, CIRI performed further filtering to eliminate false positive candidates produced from incorrectly mapped reads of homologous genes or repetitive sequences.
2.4.3. Quantification of gene expression levels
FPKM of both mRNA and ncRNA were calculated using StringTie (1.3.1) in each sample. FPKM were calculated through summing the FPKM of transcripts in each group. FPKM means fragments per kilo-base of exon per million fragments mapped, calculated based on the length of the fragments and reads count mapped to this fragment.
2.4.4. Differential expression analysis
Analysis of 2 deferentially expressed conditions/groups was implemented using the DESeq R package (1.10.1). The resulting P-values were adjusted using the Benjamini and Hochberg's approach for controlling the false discovery rate (FDR). RNA absolute value of FDR < 0.05 and |(fold change)| ≥ 2 found by DESeq were assigned as deferentially expressed.
2.5. Function annotations of RNA in bovine liver
Gene function was annotated based on the following databases: Nr (NCBI non-redundant protein sequences); Pfam (Protein family); KOG/COG (Clusters of Orthologous Groups of proteins); Swiss-Prot (A manually annotated and reviewed protein sequence database); KEGG (Kyoto Encyclopedia of Genes and Genomes); GO (Gene Ontology). GO enrichment analysis of the differentially expressed genes (DEG) was implemented by the topGO R packages. We used KOBAS (Mao et al., 2005) software to test the statistical enrichment of differential expression genes in KEGG pathways. The sequences of the DEG was blast (blastx) to the genome of a related species (the protein interaction of which exists in the STRING database:http://stringdb.org/) to get the predicted protein–protein interaction (PPI) of these DEG. Then the PPI of these DEG were visualized in Cytoscape (Shannon et al., 2003).
2.6. Validation of RNA-seq results analysed by quantitative PCR (qPCR)
In order to validate the gene expression data obtained from RNA sequencing, 11 DEG and 12 differentially expressed lncRNA were radomly selected to perform qPCR. Two micrograms of total RNA was treated with RNase-free DNase (M6101; Promega, Madison, WI, USA) and reverse transcribed according to manufacturer's instructions. 2 μL of diluted cDNA (1:40, vol/vol) was used for qPCR, which was performed in an Mx3000 P (Stratagene, USA). SYBR green qPCR master mix was used for qPCR analysis (Vazyme, Nanjing, China). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which is not affected by the experimental factors, was chosen as the reference gene. All the primers, listed in Appendix Table 2, were synthesized by Tsingke Company (Nanjing, China). The relative gene expression level was calculated using the 2 −ΔΔCt method. The data of gene expression are shown as the mean ± standard error. Statistical significance of the results was tested using the paired t-test.
2.7. Plasma triglyceride and glucose detection
During slaughter, the jugular vein was cut and blood samples were collected using 5 mL Vacutainer tubes (Jiangsu Kangjian Medical Apparatus Co., Ltd., Nanjing, China) with sodium heparin as an anticoagulant. Plasma was isolated from the blood samples by centrifugation at 3,000 × g at 4 °C for 15 min and stored at − 20 °C. Plasma glucose and triglyceride were measured using an automatic biochemical analyzer (7020, HITACHI, Tokyo, Japan).
2.8. Biochemical analysis of oxidative and antioxidative biomarkers in the liver
Hepatic catalase (CAT, cat. no. A009), malonaldehyde (MDA, cat. no. A003-1), superoxide dismutase (SOD, cat. no. A001-3) and total antioxidant capacity (TAOC, cat. no. A015) were determined using commercial kits (Jiancheng Co. Ltd, Nanjing, China). Absorbance of supernatants was detected by microplate reader (Biotek, Synergy H1, USA) at 405, 532, 550, and 593 nm, respectively.
3. Results
3.1. Overview of identified ncRNA and mRNA in Holstein cow livers
Changes in the expression of ncRNA and mRNA were examined based on the transcriptomes of Holstein cow liver samples. The Q30 proportion of lncRNA, miRNA, and circRNA exceeded 95.28%, 94.48%, and 98.41%, respectively. Then we mapped clean reads to the UMD 3.1 reference genome (http://www.ncbi.nlm.nih.gov/genome/guide/cow/index.html), alignment efficiency of lncRNA, miRNA and circRNA exceeded 96.80%, 66.55%, and 99.80%, respectively (Appendix Table 3).
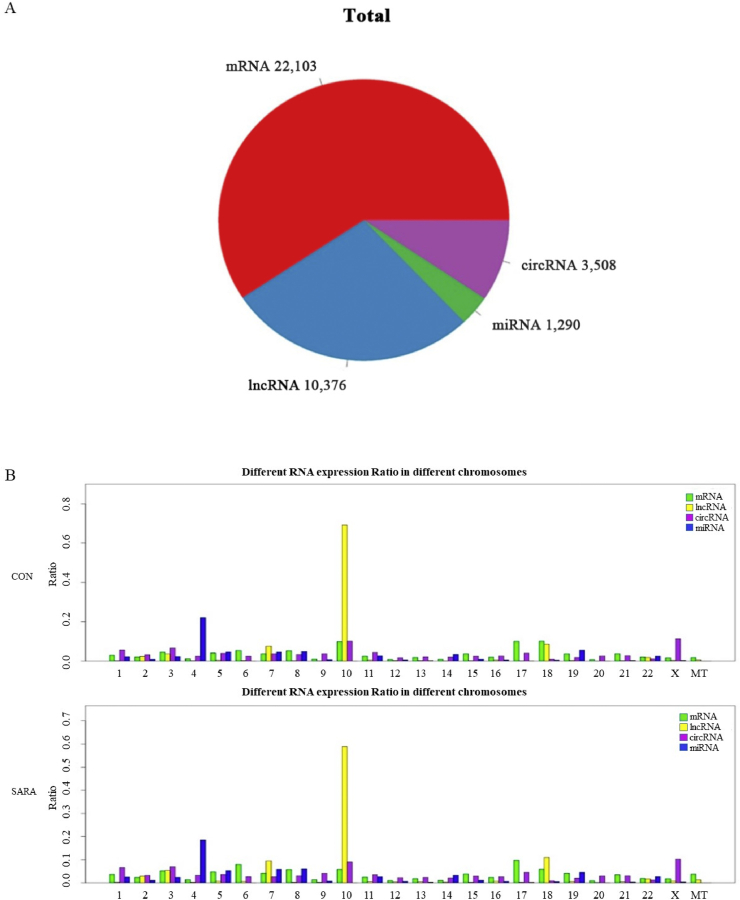
In this study, a total of 22,103 mRNA were identified. To identify ncRNA in bovine livers, transcripts were filtered following a rigorous set of criteria. Finally 10,376 lncRNA, 1,290 miRNA and 3,508 circRNA were obtained (Fig. 1A). To assess the distribution of the putative RNA, we analyzed the RNA expression ratio across the chromosomes. Description of Fig. 1B, the putative RNA were not equally distributed across the bovine chromosomes. Most mRNA and lncRNs were found on chromosome 10, whereas most miRNA were found on chromosome 4. In addition, most circRNA were found on chromosome 10 and X.
Fig. 1.
Global hepatic gene expression analysis using whole transcriptome sequencing. (A) The number of different types of RNA identified in this study. (B) Different RNA expression ratio in different chromosomes. Different colors represent different RNA types. X-axis, different chromosomes; Y-axis, proportion of RNA expressed on the chromosome. Gene expression is represented by logarithm. lncRNA = long noncoding RNA; circRNA = circular RNA; miRNA = micro RNA; CON = control; SARA = subacute ruminal acidosis.
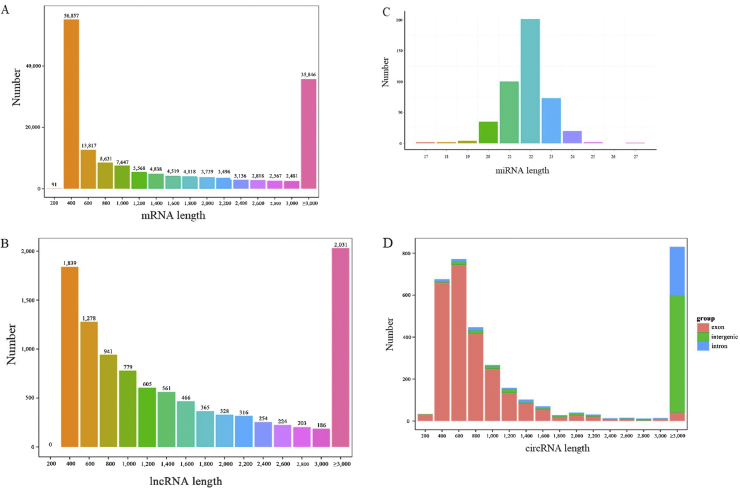
The length distribution of mRNA and ncRNA showed that mRNA and lncRNA have similar length distribution within 3,000 nt. As described in Fig. 2A and B, the length of most mRNA and lncRNA ranged from 200 to 400 nt. With the increase of the length, the number of mRNA and lncRNA gradually reduced. Different from mRNA and lncRNA, the length of most circRNA ranged from 400 to 600 nt, followed by 200 to 400 nt. As the length exceeded 600 nt, the number of circRNA decreased gradually (Fig. 2D). The final generated mature miRNA length mainly ranged from 20 to 24 nt, especially those enriched in 22 nt (Fig. 2C).
Fig. 2.
The length distribution of different RNA on chromosomes. (A) mRNA length distribution on chromosomes. X-axis, the length of mRNA. Y-axis, the number of mRNA. (B) lncRNA length distribution on chromosomes. X-axis, the length of lncRNA. Y-axis, the number of lncRNA. (C) miRNA length distribution on chromosomes. X-axis, the length of miRNA. Y-axis, the number of miRNA. (D) circRNA length distribution on chromosomes. X-axis, the length of circRNA. Y-axis, the number of circRNA. lncRNA = long noncoding RNA; miRNA = micro RNA; circRNA = circular RNA.
3.2. Analysis of differentially expressed RNA
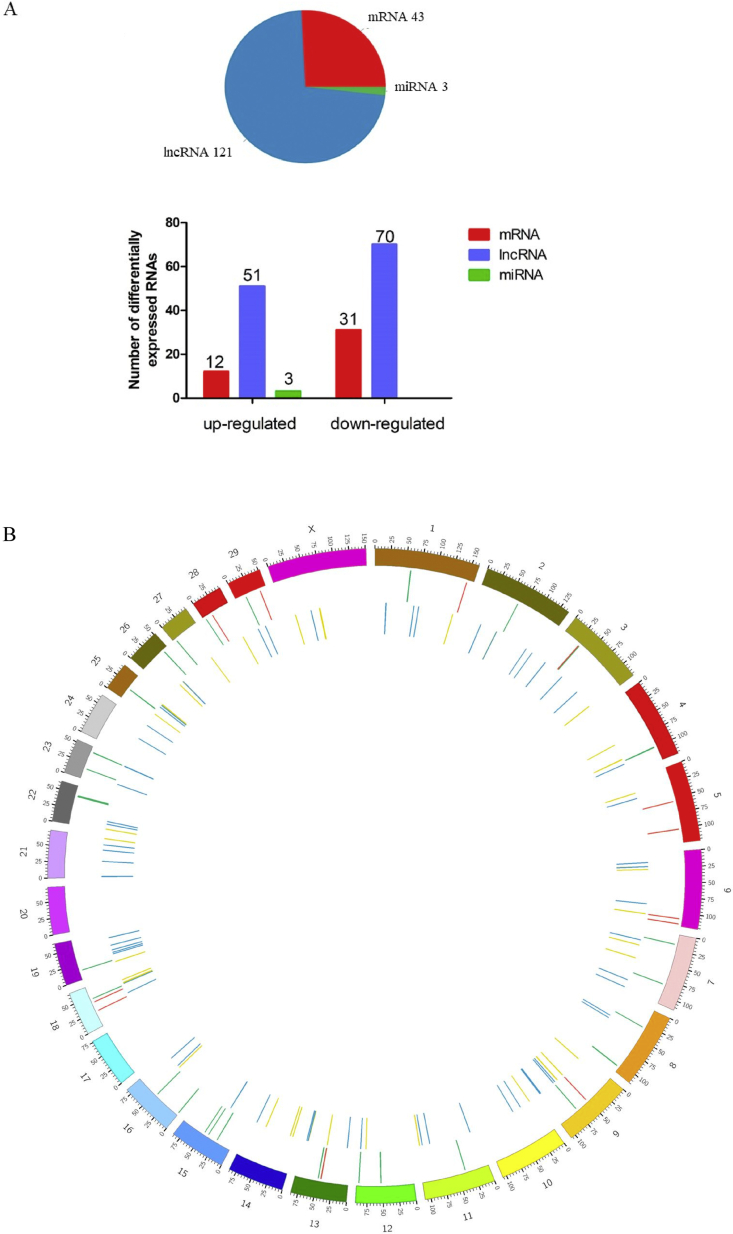
As shown in Fig. 3A, under the requirement of FDR < 0.05 and |(fold change)| ≥ 2, we acquired 167 deferentially expressed RNA, including 43 mRNA, 121 lncRNA, and 3 miRNA. However, no deferentially expressed circRNA was found. The 121 DElncRNA consisted of 70 down-regulated and 51 up-regulated lncRNA transcripts. Among the 43 DEG, 12 mRNA up-regulated whereas 31 mRNA down-regulated. Moreover, 3 DEmiRNA were up-regulated compared to the CON group. The distribution of DEG and DElncRNA across the chromosomes showed that most DEG were located on chromosome 15 and 18, and most DElncRNA were located on chromosome 6 and 19 (Fig. 3B).
Fig. 3.
Statistics on the numbers and distribution of differentially expressed RNA. (A) Schematic representation of the differentially expressed RNA between CON group and SARA group. Red, blue, and green represent mRNA, lncRNA, and miRNA respectively. Transcripts that were identified as significantly differentially expressed at most 0.05 false discovery rate and at least 2-fold changes. (B) Distribution of differentially expressed mRNA and lncRNA on chromosomes. The outermost circle indicates chromosome information; the middle circle indicates differentially expressed genes; the innermost circle indicates DElncRNA. miRNA = micro RNA; lncRNA = long noncoding RNA.
3.3. Analysis of ncRNA target genes
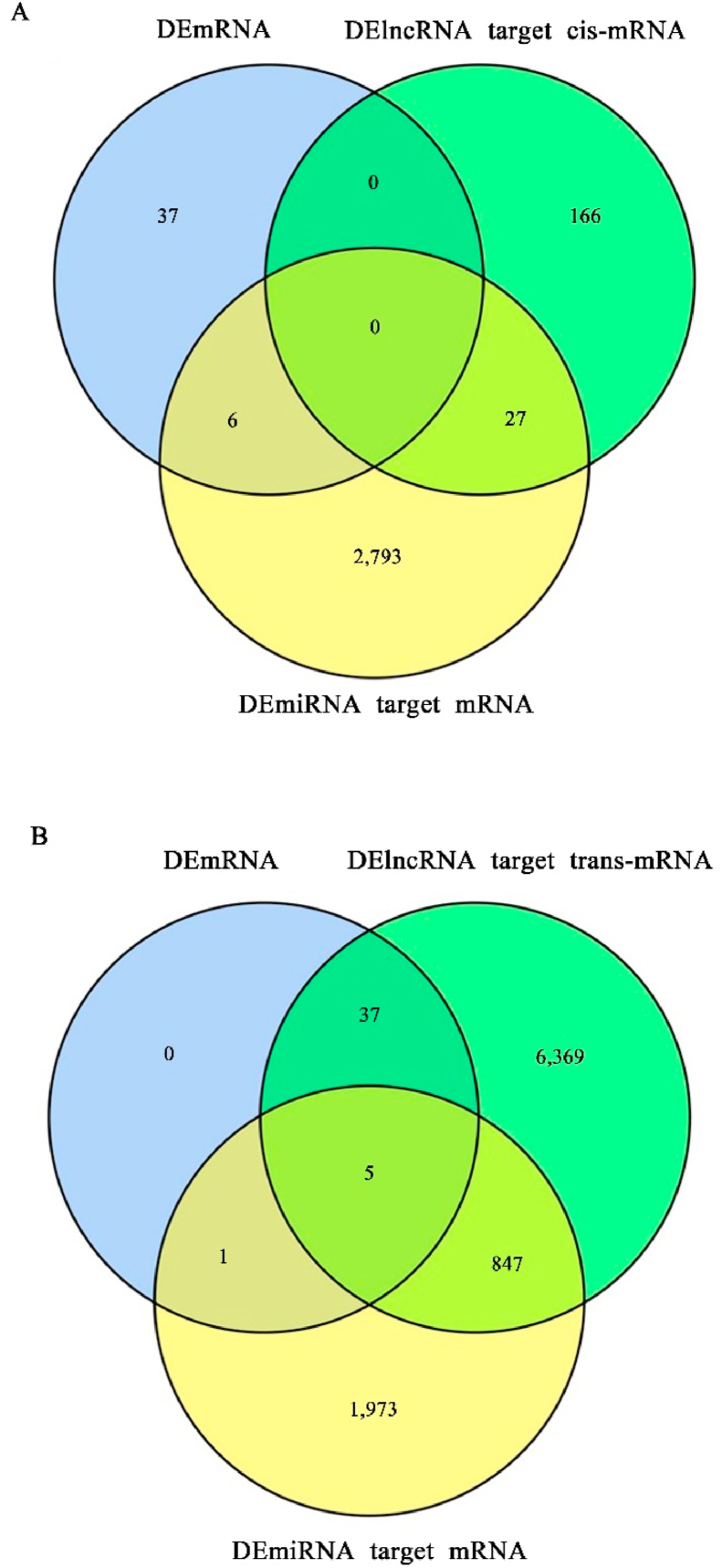
In order to find the relationship between DEG and ncRNA, 2 methods, position (cis-acting) and correlation between lncRNA and mRNA (trans-acting), were used to predict the lncRNA target genes. The results are shown in Fig. 4A and B, respectivelly. A total of 7,451 lncRNA and 2,826 miRNA target genes were found. All 42 DEG were the lncRNA target genes and they were trans-acted by lncRNA. Six DEG were the miRNA target genes and 5 DEG were both lncRNA and miRNA target genes. These results indicated that DEG tend to be directly trans-acted by lncRNA.
Fig. 4.
The intersection of DEG, DElncRNA target genes and DEmiRNA target genes shown in Venn diagram. (A) The intersection of DEG (blue), DElncRNA target genes (cis-acting, green) and DEmiRNA target genes (yellow). (B) The intersection of DEG (blue), DElncRNA target genes (trans-acting, green) and DEmiRNA target genes (yellow). DEG = differentially expressed genes; DElncRNA = differentially expressed long noncoding RNA; DEmiRNA = differentially expressed micro RNA.
3.4. Functional annotation and classification of DEG
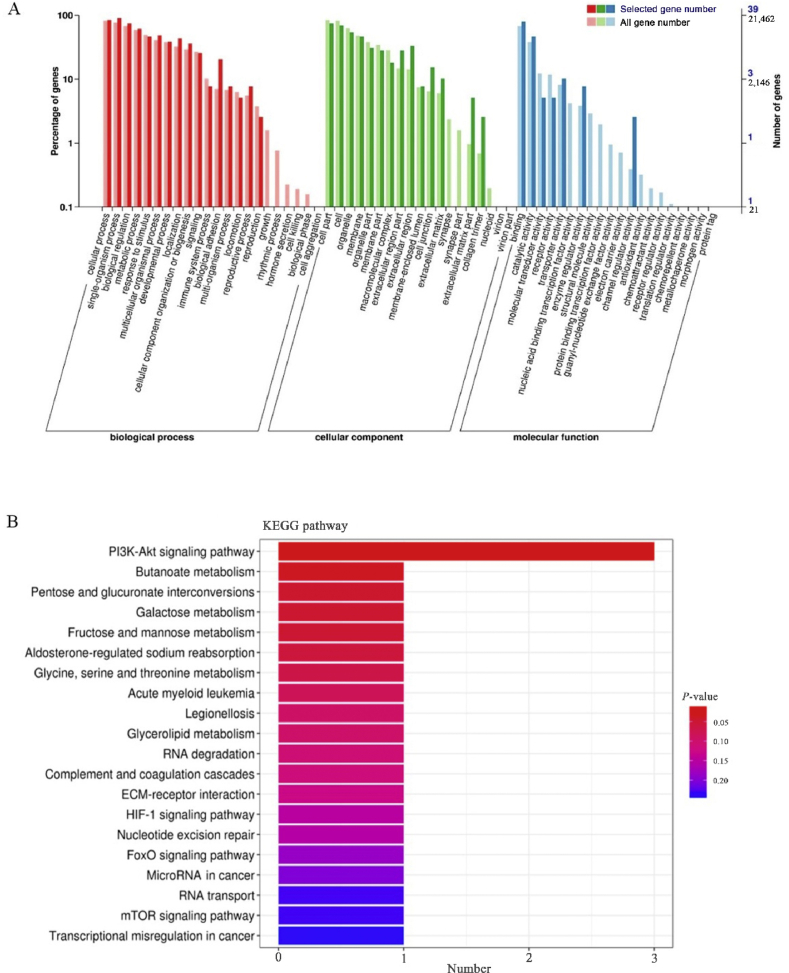
A total of 39 DEG were annotated on the GO database. In the biological process ontology, cellular process, single-organism, and biological regulation were the most abundant terms. In the cellular component ontology, cell part, cell, and organelle were the most abundant terms. In the molecular function ontology, binding, catalytic activity, and transporter activity were the most abundant terms. Furthermore, the biological adhesion, extracellular matrix part, collagen trimer and channel regulator activity were significantly different in proportion between the DEG and all genes (Fig. 5A). A total of 24 DEG were annotated to KEGG pathway database. We found that most annotated genes were involved in phosphatidylinositol 3 kinase (PI3K)-serine/threonine kinase (AKT) signaling pathway. Moreover, KEGG enrichment analysis showed that DEG were highly enriched in PI3K-AKT signaling pathway (P < 0.05) (Fig. 5B).
Fig. 5.
GO and KEGG analysis of DEG. (A) GO classification of the Holstein cow genes. Genes were assigned to GO categories, and the terms were summarized into 3 main GO categories and 61 sub-categories. Light color indicates all genes; deep color indicates DEG. Left Y-axis, percentage of genes; right Y-axis, number of genes. (B) KEGG pathway enrichment analysis of DEG. X-axis represents the number of DEG annotated in the pathway; Y-axis indicates the pathway name. Color of columns represent P-value, based on hypergeometric tests. GO = Gene Ontology; KEGG = Kyoto Encyclopedia of Genes and Genomes; DEG = differentially expressed genes; PI3K = phosphatidylinositol 3 kinase; Akt = RAC-alpha serine/threonine-protein kinase; ECM = extracellular matrix protein; HIF-1 = hypoxia-inducible factor-1; FoxO = forkhead box O; mTOR = mammalian target of rapamycin.
3.5. Functional annotation and classification of DElncRNA target genes
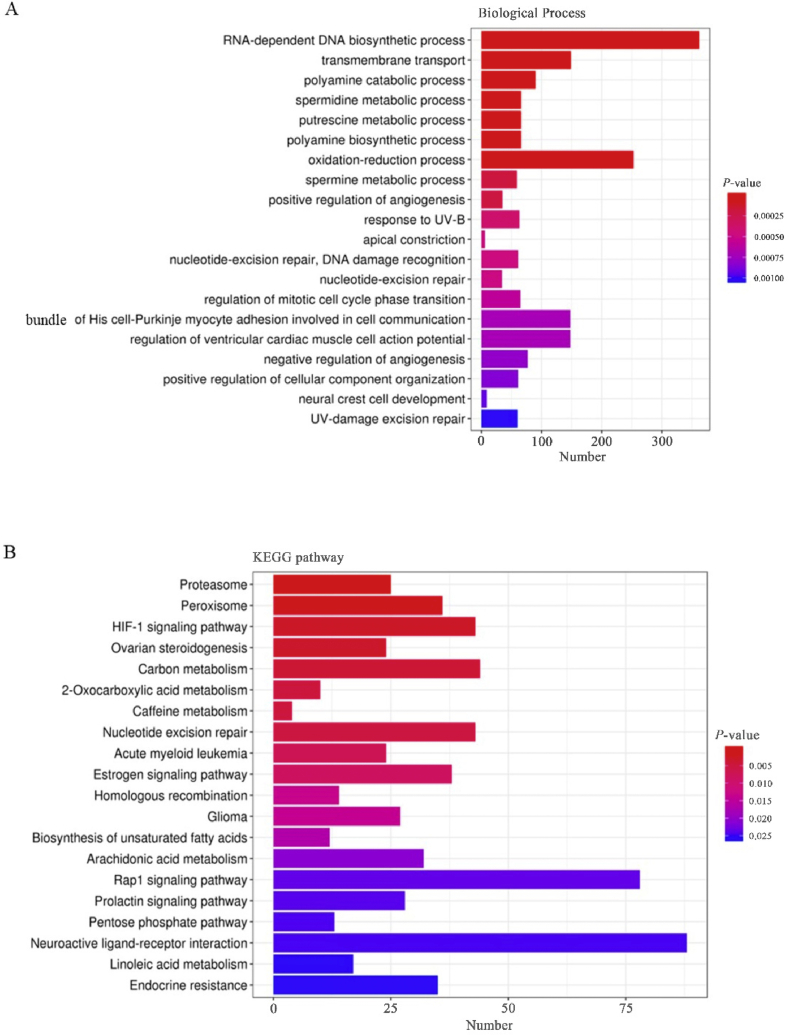
To infer the biological function of the identified DElncRNA, genes trans-acted by DElncRNA were annotated to the GO and KEGG databases. GO enrichment analysis showed that RNA-dependent DNA biosynthetic process, transmembrane transport and polyamine catabolic process were the most enriched terms. RNA-dependent DNA biosynthetic process and oxidation-reduction process were the most abundant terms (P < 0.001) (Fig. 6A). KEGG enrichment analysis showed that proteasome, peroxisome, and hypoxia inducible factor-1 (HIF-1) signaling pathway were the most enriched pathways (P < 0.005) (Fig. 6B).
Fig. 6.
Biological process and KEGG analyses of DElncRNA target genes (trans-acing). (A) X-axis indicates the number of genes; Y-axis indicates term name. Color of columns represent P-value, based on hypergeometric tests. (B) KEGG pathway enrichment analysis of DEG. X-axis represents the number of genes annotated in the pathway; Y-axis indicates the pathway name. Color of columns represent P-value, based on hypergeometric tests. KEGG = Kyoto Encyclopedia of Genes and Genomes; DEG = differentially expressed genes; HIF-1 = hypoxia-inducible factor-1.
3.6. Detailed information of selected mRNA and lncRNA
In order to find out potential mRNA and lncRNA involved in SARA, we screened 20 DEG and 60 DElncRNA with the minimum FDR for further investigation. Except for unannotated new genes, 20 DEG sorted by FDR are shown in Table 1. Among these DEG, only 4 DEG up-regulated and the others were down-regulated. GO annotation showed that AKR1B10 and DDH3 were related to the oxidation-reduction process; 6 DEG (AKR1B10, ACSM3, SGK1, CHI3L2, KRTAP5-AS1 and CCDC80) were related to substance metabolism including lipid, carbohydrate and protein metabolism; 5 DEG (C1orf198, SGK1, C21orf5, NEDD9 and PLEKHF1) were related to response to stimulus; 4 DEG (AKR1B10, SGK1, ZBTB16 and PLEKHF1) were related to cell proliferation or apoptotic process; 3 DEG (FERMT1, SGK1 and CCDC80) were related to cell adhesion; and 5 DEG (A2M, SGK1, ICAM3, CHI3L2 and HAMP) were related to inflammation or immune defense. Furthermore, the DElncRNA which have at least 3 target DEG were sorted by FDR, and 20 DElncRNA with the minimum FDR are shown in Table 2. From Table 2, we found LYPD1, DDH3, and ACSM3 were the most annotated genes. In addition, we analyzed the distribution of these DElncRNA on chromosomes. There were 3 DElncRNA on chromosome 5, 9, and 19, 2 DElncRNA on chromosome 1 and 15.
Table 1.
Detailed information of the 20 DEG with the lowest FDR.
| Gene symbol | GO annotation | FDR | log2FC |
|---|---|---|---|
| AKR1B10 | NADP+1-oxidoreductase activity, fatty acid metabolic process, positive regulation of cell death, lipid catabolic process, oxidation-reduction process, regulation of lipid biosynthetic process | 3.43E-31 | −3.33444 |
| C1orf198 | Response to stimulus | 2.63E-19 | −2.39293 |
| DDH3 | NADP+1-oxidoreductase activity, positive regulation of cell proliferation, oxidoreductase activity, cellular response to reactive oxygen species, positive regulation of reactive oxygen species metabolic process | 1.02E-08 | −1.89136 |
| ACSM3 | Fatty acid biosynthetic process, phospholipid biosynthetic process, fatty acid ligase activity | 7.37E-08 | −1.02436 |
| FERMT1 | Cell adhesion, regulation of cell–cell adhesion mediated by integrin, establishment of epithelial cell polarity | 9.98E-06 | −1.01197 |
| A2M | Negative regulation of complement activation, lectin pathway, negative regulation of endopeptidase activity, stem cell differentiation | 1.90E-05 | −1.55168 |
| SGK1 | Apoptotic process, defense response, cellular response to DNA damage stimulus, regulation of cell adhesion, lipopolysaccharide-mediated signaling pathway, regulation of cellular protein metabolic process, regulation of cell cycle | 5.81E-05 | −1.19952 |
| ICAM3 | Immune system process, integrin binding, single organismal cell–cell adhesion | 6.48E-05 | −1.0813 |
| LYPD1 | Unknown | 0.000104 | −1.31005 |
| PLLP | Ion transport, integral component of membrane | 0.000212 | 1.428531 |
| C21orf5 | Macromolecule metabolic process, cellular response to stimulus | 0.000235 | 1.287287 |
| CHI3L2 | Carbohydrate metabolic process, inflammatory response, positive regulation of angiogenesis | 0.000276 | −1.40479 |
| NEDD9 | Animal organ morphogenesis, regulation of cellular process, cellular response to stimulus | 0.000349 | −1.34656 |
| CLRN2 | Integral component of membrane | 0.000432 | 1.16737 |
| ZBTB16 | Negative regulation of cell proliferation, positive regulation of apoptotic process | 0.000793 | −1.15451 |
| KRTAP5-AS1 | Regulation of metabolic process | 0.00088 | −1.28645 |
| CCDC80 | Fibronectin binding, protein binding, positive regulation of cell-substrate adhesion, cellular metabolic process | 0.00295 | −1.2423 |
| LOC514257 | Transporter activity, transmembrane transport | 0.00295 | −1.09758 |
| PLEKHF1 | Apoptotic process, positive regulation of autophagy, cellular response to stimulus | 0.00295 | 1.144043 |
| HAMP | Hormone activity, cellular iron ion homeostasis, defense response to bacterium | 0.003941 | −1.14331 |
DEG = differentially expressed genes; FDR = false discovery rate; GO = Gene Ontology; FC = fold change.
Table 2.
Detailed information about 20 candidate lncRNA involved in SARA.
| lncRNA ID | Target DEG symbol | Chromosome | FDR | log2FC |
|---|---|---|---|---|
| MSTRG.87868.4 | DDH3, LYPD1, FERMT1 | 5 | 1.07E-26 | −7.84486 |
| MSTRG.92851.2 | ICAM3, HAMP, SGK1, SCGB1D2 | 19 | 3.74E-26 | −8.32203 |
| MSTRG.111771.1 | DDH3, LYPD1, CHI3l2 | X | 8.18E-18 | 3.900421 |
| MSTRG.497.2 | DDH3, ACSM3, FERMT1, LYPD1, CLRN2, SCGB1D2 | 1 | 2.78E-15 | −6.42199 |
| MSTRG.87074.3 | DDH3, LYPD1, PLEKHF1, SLC26A10 | 5 | 1.26E-14 | 6.474252 |
| MSTRG.7842.1 | ICAM3, PLLP, ZBTB16, LURAP1L, ANGPT2 | 10 | 4.98E-14 | −5.29469 |
| MSTRG.87074.11 | DDH3, LYPD1, PLEKHF1, ACSM3, SLC26A10 | 5 | 7.61E-14 | 6.234416 |
| MSTRG.74107.3 | DDH3, ACSM3, LYPD1, CLRN2, SCGB1D2 | 29 | 4.42E-11 | −5.73536 |
| MSTRG.18357.3 | ACSM3, SGK1, LYPD1, NEDD9, SCGB1D2, CPXM2 | 12 | 2.90E-10 | −5.61773 |
| MSTRG.41223.2 | DDH3, ACSM3, LYPD1, PLEKHF1, SLC26A10, SCGB1D2 | 19 | 9.13E-10 | 5.468157 |
| MSTRG.27428.8 | C1orf198, FERMT1, ICAM3, ANGPT2 | 15 | 4.66E-09 | −4.65541 |
| MSTRG.27428.1 | AKR1B10, A2M, CHI3l2, NEDD9, CCDC80, CPXM2, BTG2, MICAL2 | 15 | 1.13E-08 | −4.75513 |
| MSTRG.42738.6 | DDH3, FERMT1, LYPD1, CLRN2, SCGB1D2 | 19 | 1.05E-07 | −4.68639 |
| MSTRG.110548.10 | A2M, CHI3l2, NEDD9, CCDC80, CPXM2, BTG2, MICAL2 | 9 | 1.38E-07 | −5.20861 |
| MSTRG.110548.7 | DDH3, ACSM3, LYPD1, CLRN2, SCGB1D2 | 9 | 3.66E-07 | 4.076852 |
| MSTRG.18737.7 | DDH3, FERMT1, LYPD1, CLRN2 | 13 | 4.32E-07 | −4.84714 |
| MSTRG.61972.4 | A2M, CHI3l2, NEDD9, CCDC80, CPXM2, BTG2, MICAL2 | 23 | 4.42E-07 | −5.14377 |
| MSTRG.2436.11 | DDH3, LYPD1, KRTAP5-AS1 | 1 | 5.97E-07 | −4.87173 |
| MSTRG.108870.21 | C1of198, FERMT1, ICAM3, ANGPT2 | 9 | 1.00E-06 | −3.06608 |
| MSTRG.66136.2 | DDH3, ACSM3, LYPD1, CLRN2 | 25 | 1.31E-06 | −4.73371 |
lncRNA = long noncoding RNA; DEG = differentially expressed genes; FDR = false discovery rate; FC = fold change.
3.7. Validation of RNA-seq results by qPCR
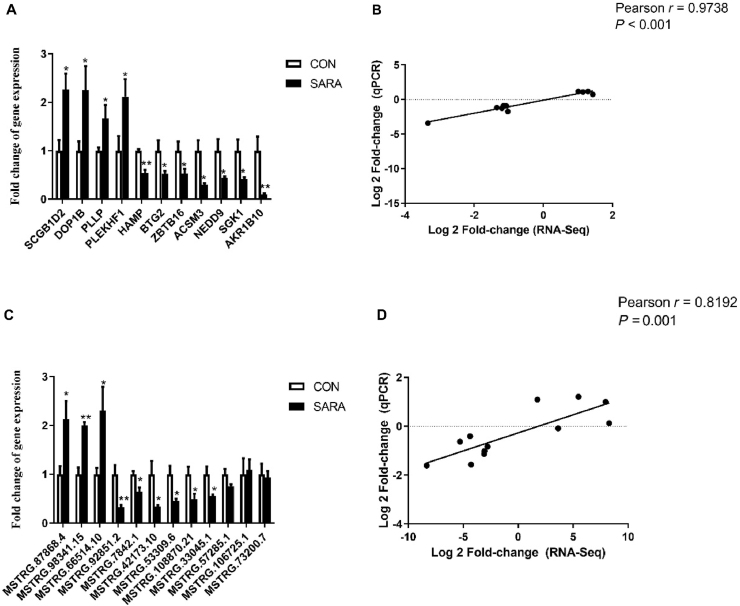
Eleven DEG and 12 DElncRNA were subjected to qPCR analysis to validate the RNA-seq result. As is shown in Fig. 7A and C, the expression profiling of these DEG and lncRNA using qPCR had similar trends with the RNA-seq samples. The Pearson's correlation coefficients of the differential expression ratios between RNA-seq and qPCR were 0.9738 (P < 0.001) and 0.8192 (P = 0.001), respectivelly (Fig. 7B and D). It indicated that the expression differences of mRNA and lncRNA observed in RNA-seq between the CON and SARA groups were highly reliable.
Fig. 7.
qPCR validation of RNA-seq results. (A) Expression of mRNA in the liver. (B) Pearson correlation analysis for mRNA between RNA-seq and qPCR. Fold change indicates the ratio of average expression of SARA-affected mRNA relative to controls. (C) Expression of lncRNA in the liver. (D) Pearson correlation analysis for lncRNA between RNA-seq and qPCR. Fold change indicates the ratio of average expression of SARA-affected lncRNA relative to controls. Fold difference for each gene was calculated via the FPKM values of CON and SARA. GAPDH was used as the reference gene for genes expression. The data were analyzed by an independent-samples t-test using the Compare Means of SPASS 11.0 for Windows (StaSoft Inc, Tulsa, OK, USA). Values are mean ± SEM. ∗P < 0.05, ∗∗P < 0.01 (CON vs SARA). qPCR = quantitative PCR; FPKM = fragments per kilo-base of exon per million fragments; CON = control; SARA = subacute ruminal acidosis; GAPDH = glyceraldehyde-3-phosphate dehydrogenase.
3.8. Detection of plasma glucose, triglyceride and ruminal pH
Plasma glucose showed no significant difference between the CON and SARA groups (P > 0.05). However, the SARA group had a lower plasma triglyceride concentration than the CON group (P < 0.05). Compared with the CON group, the ruminal pH was lower in the SARA group (P < 0.01) (Table 3).
Table 3.
Plasma biochemical parameters and ruminal pH1.
| Item | Group |
P-value | |
|---|---|---|---|
| CON | SARA | ||
| Plasma triglyceride, mmol/L | 0.102 ± 0.002 | 0.0875 ± 0.006 | 0.046 |
| Plasma glucose, mmol/L | 3.35 ± 0.16 | 3.30 ± 0.20 | 0.842 |
| Ruminal pH | 6.31 ± 0.87 | 5.79 ± 0.92 | 0.002 |
CON = control; SARA = subacute ruminal acidosis.
The data were analyzed by an independent-samples t-test using the Compare Means of SPASS 11.0 for Windows (StaSoft Inc, Tulsa, OK, USA). Values are mean ± SEM.
3.9. Oxidative stress increased in SARA group
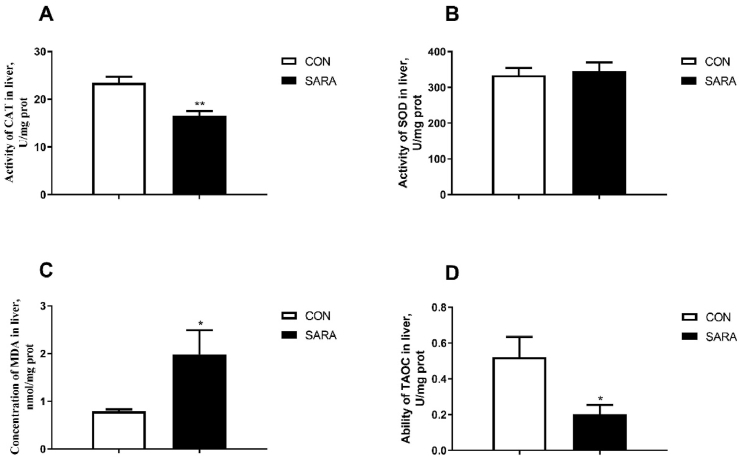
The antioxidant and lipid peroxide profiles in the livers of the dairy cows are presented in Fig. 8. The SOD levels were not changed (P > 0.05) by a high concentrate diet (Fig. 8B). However, CAT and TAOC dramatically decreased in the SARA group (P < 0.01) (Fig. 8A and D). In addition, there was an increase in MDA content in the SARA group (P < 0.05) (Fig. 8C).
Fig. 8.
Effect of high and low concentrate diet on oxidative stress in the liver. (A) Catalase (CAT) activity; (B) superoxide dismutase (SOD) activity; (C) malonaldehyde (MDA) concentration; (D) ability of total antioxidant capacity (TAOC). All enzyme activity and biomarkers were evaluated spectrophotometrically, and the data were analyzed by an independent-samples t-test using the Compare Means of SPASS 11.0 for Windows (StaSoft Inc, Tulsa, OK, USA). Values are mean ± SEM. ∗P<0.05, ∗∗P<0.01. CON = control; SARA = subacute ruminal acidosis.
4. Discussion
A high concentrate diet improves the performance of dairy cows over the short-term. However, long-term usage of a high concentrate diet could result in a series of nutritional and metabolic diseases (Jouany, 2006). SARA is one of these representative diseases, which leads to reduced dry matter intake and milk production, and increased elimination rate of cows (McCann et al., 2016). SARA has received increasing attention in recent years because of its damaging effects on animal welfare and profits. Therefore, investigating the occurrence and mechanisms of SARA is of great importance. Most studies attribute SARA to abnormal fermentation, volatile fatty acid accumulation, and reduced chewing frequency induced by a high concentrate diet (Yun and Han, 1989; Shaani et al., 2017). The liver plays important roles in nutrition metabolism and distribution and is key for maintaining milk fat, lactose, and lactoprotein levels. Although a high concentrate diet can induce liver dysfunction, the mechanism remains unclear. Thus, we investigated whole-transcriptome changes in the liver via high-throughput RNA sequencing.
The quality control and alignment efficiency results indicated that a successful library was constructed with high sequencing quality. In this study, 22,103 mRNA, 10,376 lncRNA, 1,290 miRNA, and 3,508 circRNA were obtained from the clean data. Furthermore, we screened 43 DEG, 121 lncRNA, and 3 miRNA. These results indicate that lncRNA play an important role in SARA at the transcriptional level. Analysis of ncRNA target genes also confirmed that 42 of 43 DEG were DElncRNA target genes. In contrast, only 6 DEG were DEmiRNA target genes. We performed a qPCR analysis to validate the RNA-seq results. The qPCR results were highly consistent with the RNA-seq results, suggesting that the RNA-seq data were reliable. In addition, we counted the number of exons and the length of the open reading frames (ORF) of the mRNA and lncRNA. An overwhelming majority of the mRNA had only one exon, whereas most of the lncRNA had 2 exons (Appendix Fig. 1A and B). The ORF length ranged from 100 to 400 nt and was dominated by mRNA. The ORF length of the lncRNA was shorter and most were <150 nt (Appendix Fig. 1C and D). In general, the lncRNA in the liver were expressed at lower levels, with fewer exons and shorter ORF compared to the mRNA. This result was similar to previous studies on bovine rumen (Zheng et al., 2018).
We conducted further research on the DEG and DElncRNA. The KEGG analysis revealed that most of the DEG were annotated in the PI3K-AKT signaling pathway. AKT is an important regulatory factor in cell signal transduction pathways. It is involved in biological signal transduction, such as nutrient metabolism, cell apoptosis, and inflammation. Previous studies have reported that the PI3K-AKT pathway is essential for the synthesis of milk components, such as lipids and lactose (Oliver and Watson, 2013). Furthermore, a high concentrate diet can increase lipopolysaccharide (LPS) from the gastrointestinal tract into the peripheral circulation (Emmanuel et al., 2008). Studies on dairy cow mammary epithelial cells have confirmed that LPS significantly disturbs cell proliferation and lipid metabolism via the PI3K-AKT signaling pathway (Liu et al., 2015). Little information is available about the roles of lncRNA in the liver during SARA. Hence, we annotated the DElncRNA target genes to the GO and KEGG databases. The GO enrichment analysis showed that oxidation–reduction processes were highly enriched and contained the second most targeted mRNA. Similar to the DEG results, the KEGG classification for the DElncRNA target genes indicated that most were classified to the PI3K-AKT pathway. The KEGG enrichment analysis also showed that the DElncRNA target genes were highly enriched in the proteasome, peroxisome, and the HIF-1 signaling pathways. These pathways are strongly correlated with inflammation and energy metabolism; therefore, they could be potentially involved in the regulation of SARA. For example, Xu et al. (2015) reported that LPS derived from the rumen upregulates mRNA expression of peroxisome proliferator-activated receptor-α, carnitine palmitoyl transferase 1, and acyl-CoA oxidase 1 in the liver, possibly due to the increased energy demand to resist inflammation during SARA. Kim et al. (2007) showed that activated HIF-1 induced by LPS binds directly to the tumor necrosis factor-α (TNF-α) promoter to increase TNF-α expression in hepatocytes. However, the specific roles of such pathways during SARA require further study.
Subsequently, we selected 20 candidate DEG and DElncRNA with regulatory potential for further study. Table 1 shows that these DEG were mainly involved in oxidoreductase activity, stress, metabolism, the immune response, cell adhesion, apoptosis, and proliferation. This result was highly consistent with our previous studies. Our previous studies on dairy goats reported that a high concentrate diet increases plasma acute phase protein and cortisol concentrations, and induces activation of the hypothalamic–pituitary–adrenal axis. This result suggests that goats with SARA experience stress (Dong et al., 2013; Jia et al., 2014). Furthermore, many studies have found that a high concentrate diet causes liver metabolic abnormalities, including glucose metabolism, lipid metabolism, and protein metabolism (Dong et al., 2013; Xie et al., 2015). In addition, a high concentrate diet induces oxidative stress in the liver. The enzyme activities of SOD, glutathione peroxidase, and catalase change during SARA. TAOC significantly decreases, whereas malondialdehyde notably increases (Abaker et al., 2017). Our comparative proteomic results suggested that a high concentrate diet activates the inflammatory response and decreases antioxidant capacity (Duanmu et al., 2016). Some of these DEG are worthy of note, as they were strongly correlated with the disease process. For example, AKR1B10, which had the greatest fold-change, not only participates in cell proliferation but also participates in lipid metabolism (Cao et al., 2014). AKR1B10 promotes cell growth and proliferation but it regulates lipid synthesis via stabilizing acetyl-coenzyme A carboxylase α (Ma et al., 2008). However, its role in SARA has not been reported.
Most of the target genes of the 20 candidate DElncRNA involved in SARA overlapped with the 20 DEG in Table 1, suggesting that such DEG may be regulated by lncRNA. It would be important to explore and validate the functions of the lncRNA to alleviate the negative effects of SARA. For example, DDH3, which is a DElncRNA target gene, has a strong ability to resist reactive oxygen species production in vitro, and inhibiting its expression can lead to cell death (Chen et al., 2008). Therefore, increased DDH3 expression in the liver via lncRNA may be an effective strategy to reduce oxidative stress that has been induced by a high concentrate diet. Although lncRNA research in ruminants is limited compared to humans and rodents, the findings of the present study suggest it is of value for further studies on SARA.
The result of whole transcriptome indicated that SARA induced metabolic disorders and oxidative stress. Therefore, we detected plasma triglyceride and glucose concentrations, as well as hepatic antioxidant and lipid peroxide profiles. Although plasma glucose did not show an obvious difference, there was a significant decrease in plasma triglyceride concentration. In accordance with the result of whole transcriptome, decreased TAOC, CAT and increased MDA suggested that the SARA group suffered from hepatic oxidative stress. In addition, there was no significant difference in the dry matter intake, daily yields of milk, milk lactose and protein. However, milk fat dramatically reduced in the SARA group (data are not shown). Such results were highly consistent with our transcriptome data. For example, AKR1B10, DDH3 and ACSM3, which are related to fatty acid synthesis or antioxidation significantly decreased in the SARA group.
In this study, transcriptome analysis was used to reveal the DEG in the liver of SARA dairy cows compared to normal counterparts. The DElncRNA target genes were mainly related to some cellular process or signaling pathways by KEGG pathway analysis. However, some limitations of this study should be mentioned. Considering the animal welfare and the 3Rs principles, the number of dairy cows used in this study was limited. Moreover, the underlying mechanism of altered lncRNA expression in the liver of SARA animals is still unclear. The target molecules as well as the regulatory functions of these DElncRNA still need further investigation.
5. Conclusion
This study identified 10,376 lncRNA, 1,290 miRNA, 3,508 circRNA, and 22,103 mRNA in Holstein cow livers. Among these RNA, 121 lncRNA, 3 miRNA, and 43 mRNA were differentially expressed. Our results indicate that mRNA and lncRNA rather than miRNA or circRNA play major roles in SARA at the transcriptional level. Moreover, a bioinformatics analysis showed that the mRNA were strongly correlated with the lncRNA. The GO and KEGG analysis revealed that the DEG were mainly related to oxidoreductase activity, stress, metabolism, the immune response, cell apoptosis and proliferation. Similarly, DEG and DElncRNA target genes were enriched in pathways, such as the PI3K-AKT, proteasome, and HIF-1 signaling pathways. Such pathways are related to inflammation, metabolism, and cell apoptosis and proliferation. Taken together, our results suggest that DEG, mainly regulated by lncRNA at the transcriptional level, and lncRNA play an important regulatory role in SARA. Above all, the findings from the current study provide valuable resources for further studies on SARA.
Author contributions
Qu Chen: Investigation, Writing-Reviewing and Editing, Visualization, Software, Formal analysis, Data Curation; Chen Wu: Investigation; Zhihao Yao: Investigation, Liuping Cai: Investigation; Yingdong Ni: Conceptualization, Supervision, Writing- Reviewing and Editing, Project administration, Funding acquisition, Methodology, Resources; Shengyong Mao: Conceptualization, Project administration, Funding acquisition Methodology, Resources, Software; Ruqian Zhao: Supervision, Resources.
Conflict of interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
The authors acknowledge the financial contributions of the Fundamental Research Funds for the Central Universities (JCQY201905) and National Key Research and Development Project (2016YFD0501203).
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2021.10.002.
Appendix.
The following is the Supplementary data to this article:
References
- Abaker J.A., Xu T.L., Jin D., Chang G.J., Zhang K., Shen X.Z. Lipopolysaccharide derived from the digestive tract provokes oxidative stress in the liver of dairy cows fed a high-grain diet. J Dairy Sci. 2017;100(1):666–678. doi: 10.3168/jds.2016-10871. [DOI] [PubMed] [Google Scholar]
- Ametaj B.N., Zebeli Q., Saleem F., Psychogios N., Lewis M.J., Dunn S.M., Xia J., Wishart D.S. Metabolomics reveals unhealthy alterations in rumen metabolism with increased proportion of cereal grain in the diet of dairy cows. Metabolomics. 2010;6(4):583–594. [Google Scholar]
- Brosnan C.A., Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009;21(3):416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Cao Z., Zhou B., Chen X., Huang D., Zhang X., Wang Z., Huang H., Wang Y., Cao D. Statil suppresses cancer cell growth and proliferation by the inhibition of tumor marker AKR1B10. Anti Cancer Drugs. 2014;25(8):930–937. doi: 10.1097/CAD.0000000000000121. [DOI] [PubMed] [Google Scholar]
- Chen J., Adikari M., Pallai R., Parekh H.K., Simpkins H. Dihydrodiol dehydrogenases regulate the generation of reactive oxygen species and the development of cisplatin resistance in human ovarian carcinoma cells. Cancer Chemother Pharmacol. 2008;61(6):979–987. doi: 10.1007/s00280-007-0554-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Wang S., Jia Y., Ni Y., Zhang Y., Zhuang S., Shen X., Zhao R. Long-term effects of subacute ruminal acidosis (SARA) on milk quality and hepatic gene expression in lactating goats fed a high-concentrate diet. PLoS One. 2013;8(12):e82850. doi: 10.1371/journal.pone.0082850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu Y., Cong R., Tao S., Tian J., Dong H., Zhang Y., Ni Y., Zhao R. Comparative proteomic analysis of the effects of high-concentrate diet on the hepatic metabolism and inflammatory response in lactating dairy goats. J Anim Sci Biotechnol. 2016;7:5. doi: 10.1186/s40104-016-0065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmanuel D.G., Dunn S.M., Ametaj B.N. Feeding high proportions of barley grain stimulates an inflammatory response in dairy cows. J Dairy Sci. 2008;91(2):606–614. doi: 10.3168/jds.2007-0256. [DOI] [PubMed] [Google Scholar]
- Gindin Y., Jiang Y., Francis P., Walker R.L., Abaan O.D., Zhu Y.J., Meltzer P.S. miR-23a impairs bone differentiation in osteosarcoma via down-regulation of GJA1. Front Genet. 2015;6:233. doi: 10.3389/fgene.2015.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- He Y., Ding Y., Zhan F., Zhang H., Han B., Hu G., Zhao K., Yang N., Yu Y., Mao L., Song J. The conservation and signatures of lincRNAs in Marek's disease of chicken. Sci Rep. 2015;5:15184. doi: 10.1038/srep15184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helrich K., Helrich K. 1990. Official methods of analysis of the AOAC. [Google Scholar]
- Huttenhofer A., Schattner P., Polacek N. Non-coding RNAs: hope or hype? Trends Genet : TIG (Trends Genet) 2005;21(5):289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Jia Y.Y., Wang S.Q., Ni Y.D., Zhang Y.S., Zhuang S., Shen X.Z. High concentrate-induced subacute ruminal acidosis (SARA) increases plasma acute phase proteins (APPs) and cortisol in goats. Animal. 2014;8(9):1433–1438. doi: 10.1017/S1751731114001128. [DOI] [PubMed] [Google Scholar]
- Jouany J.P. Optimizing rumen functions in the close-up transition period and early lactation to drive dry matter intake and energy balance in cows. Anim Reprod Sci. 2006;96(3–4):250–264. doi: 10.1016/j.anireprosci.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Kim H.Y., Kim Y.H., Nam B.H., Kong H.J., Kim H.H., Kim Y.J., An W.G., Cheong J. HIF-1alpha expression in response to lipopolysaccaride mediates induction of hepatic inflammatory cytokine TNFalpha. Exp Cell Res. 2007;313(9):1866–1876. doi: 10.1016/j.yexcr.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Kitkas G.C., Valergakis G.E., Karatzias H., Panousis N. vol. 14. 2013. pp. 183–189. (Subacute ruminal acidosis: prevalence and risk factors in Greek dairy herds). 3. [Google Scholar]
- Kleen J.L., Hooijer G.A., Rehage J., Noordhuizen J.P. Subacute ruminal acidosis (SARA): a review. J Veterinary Med A Physiol Pathol Clin Med. 2003;50(8):406. doi: 10.1046/j.1439-0442.2003.00569.x. [DOI] [PubMed] [Google Scholar]
- Knegsel A.T. M.v., Brand H.V.D., Dijkstra J., Tamminga S., Kemp B. Effect of dietary energy source on energy balance, production, metabolic disorders and reproduction in lactating dairy cattle. Reprod Nutr Dev. 2005;45(6):665–688. doi: 10.1051/rnd:2005059. [DOI] [PubMed] [Google Scholar]
- Liu L., Lin Y., Liu L., Bian Y., Zhang L., Gao X., Li Q. 14-3-3gamma regulates lipopolysaccharide-induced inflammatory responses and lactation in dairy cow mammary epithelial cells by inhibiting NF-kappaB and MAPKs and up-regulating mTOR signaling. Int J Mol Sci. 2015;16(7):16622–16641. doi: 10.3390/ijms160716622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Yan R., Zu X., Cheng J.M., Rao K., Liao D.F., Cao D. Aldo-keto reductase family 1 B10 affects fatty acid synthesis by regulating the stability of acetyl-CoA carboxylase-alpha in breast cancer cells. J Biol Chem. 2008;283(6):3418–3423. doi: 10.1074/jbc.M707650200. [DOI] [PubMed] [Google Scholar]
- Mao X., Cai T., Olyarchuk J.G., Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21(19):3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- McCann J.C., Luan S., Cardoso F.C., Derakhshani H., Khafipour E., Loor J.J. Induction of subacute ruminal acidosis affects the ruminal microbiome and epithelium. Front Microbiol. 2016;7:701. doi: 10.3389/fmicb.2016.00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., le Noble F., Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Mogilyansky E., Clark P., Quann K., Zhou H., Londin E., Jing Y., Rigoutsos I. Post-transcriptional regulation of BRCA2 through interactions with miR-19a and miR-19b. Front Genet. 2016;7:143. doi: 10.3389/fgene.2016.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver C.H., Watson C.J. Making milk: a new link between STAT5 and Akt1. JAK-STAT. 2013;2(2):e23228. doi: 10.4161/jkst.23228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M. Effects of increasing the proportion of high-quality grass silage in the diet of dairy cows. [Doctor Degree Thesis Dissertation] Swedish University of Agricultural Science; 2012. [Google Scholar]
- Shaani Y., Nikbachat M., Yosef E., Ben-Meir Y., Friedman N., Miron J., Mizrahi I. Effect of wheat hay particle size and replacement of wheat hay with wheat silage on rumen pH, rumination and digestibility in ruminally cannulated non-lactating cows. Animal. 2017;11(3):426–435. doi: 10.1017/S1751731116001865. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Li M., Sun Y., Cai H., Lan X., Huang Y., Bai Y., Qi X., Chen H. The developmental transcriptome sequencing of bovine skeletal muscle reveals a long noncoding RNA, lncMD, promotes muscle differentiation by sponging miR-125b. Biochim Biophys Acta. 2016;1863(11):2835–2845. doi: 10.1016/j.bbamcr.2016.08.014. [DOI] [PubMed] [Google Scholar]
- Sundrum A. Metabolic disorders in the transition period indicate that the dairy cows' ability to adapt is overstressed. Animals : an open access journal from MDPI. 2015;5(4):978–1020. doi: 10.3390/ani5040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest P.J., Robertson J.B., Lewis B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. 1991;74(10):3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- Xie Z.L., Ye P.S., Zhang Y.S., Shen X.Z. Effect of high-concentrate diet on amino acid transporter expression and milk quality in Holstein dairy cows. Genet Mol Res : GMR. 2015;14(2):5246–5257. doi: 10.4238/2015.May.18.16. [DOI] [PubMed] [Google Scholar]
- Xu T., Tao H., Chang G., Zhang K., Xu L., Shen X. Lipopolysaccharide derived from the rumen down-regulates stearoyl-CoA desaturase 1 expression and alters fatty acid composition in the liver of dairy cows fed a high-concentrate diet. BMC Vet Res. 2015;11:52. doi: 10.1186/s12917-015-0360-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S.K., Han J.D. Effect of feeding frequence of concentrate to milking cow in early lactation on pH and VFA-concentration in rumen fluid and on milk composition and milk yield. Asian-Australas J Anim Sci. 1989;2(3):418–420. [Google Scholar]
- Zhao F.Q., Keating A.F. Expression and regulation of glucose transporters in the bovine mammary gland. J Dairy Sci. 2007;90(Suppl 1):E76–E86. doi: 10.3168/jds.2006-470. [DOI] [PubMed] [Google Scholar]
- Zheng X., Ning C., Zhao P., Feng W., Jin Y., Zhou L., Yu Y., Liu J. Integrated analysis of long noncoding RNA and mRNA expression profiles reveals the potential role of long noncoding RNA in different bovine lactation stages. J Dairy Sci. 2018;101(12):11061–11073. doi: 10.3168/jds.2018-14900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.