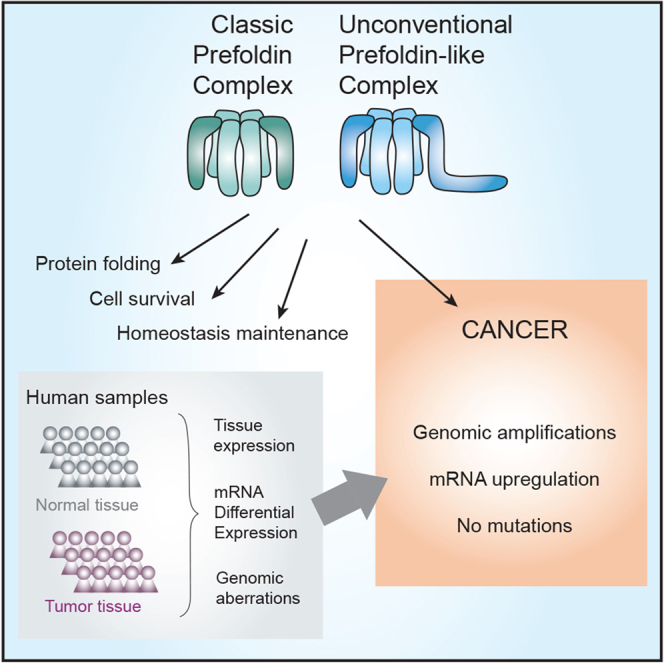
Summary
Prefoldins (PFDNs) are evolutionary conserved co-chaperones, initially discovered in archaea but universally present in eukaryotes. PFDNs are prevalently organized into hetero-hexameric complexes. Although they have been overlooked since their discovery and their functions remain elusive, several reports indicate they act as co-chaperones escorting misfolded or non-native proteins to group II chaperonins. Unlike the eukaryotic PFDNs which interact with cytoskeletal components, the archaeal PFDNs can bind and stabilize a wide range of substrates, possibly due to their great structural diversity. The discovery of the unconventional RPB5 interactor (URI) PFDN-like complex (UPC) suggests that PFDNs have versatile functions and are required for different cellular processes, including an important role in cancer. Here, we summarize their functions across different species. Moreover, a comprehensive analysis of PFDNs genomic alterations across cancer types by using large-scale cancer genomic data indicates that PFDNs are a new class of non-mutated proteins significantly overexpressed in some cancer types.
Subject areas: Biological sciences, Cell biology, Cancer
Graphical Abstract
Biological sciences; Cell biology; Cancer
Introduction
Proteins must fold into their 3D-structure to fulfill their biological functions (Balchin et al., 2016). Protein misfolding leads to the accumulation of unfolded proteins, causing the activation of stress response pathways, including the cytosolic and endoplasmic reticulum stress responses (Gregersen and Bross, 2010), which has toxic effects for the cell and has been associated with numerous degenerative diseases (Wang and Kaufman, 2016). Thus, it is not surprising that cellular organisms have developed a range of sophisticated strategies to assure a proper protein folding to maintain protein homeostasis or proteostasis (Kim et al., 2013). Molecular chaperones interact and aid in the folding of proteins or in the assembly of protein complexes, contributing to proteostasis (Hartl et al., 2011). The complex cellular network of chaperones is supported by co-chaperones: proteins that collaborate with chaperones by regulating and selecting the binding to their partners (Echtenkamp and Freeman, 2012). PFDNs, described more than two decades ago, are members of these co-chaperones, whose main functions were reported to be related to the assembly of cytoskeletal proteins in yeasts and archaebacteria (Geissler et al., 1998; Vainberg et al., 1998). Since then, PFDNs have been found in a wide range of organisms, from archaea to humans, and their spectrum of functions seems to go way beyond cytoskeletal protein folding, in line with the notion that proteins involved in folding have the ability to bind a wide spectrum of substrates (Chaves-Pérez et al., 2018). It is also not surprising that PFDNs are implicated in several pathologies such as cancer.
Discovery of the PFDN proteins
Vainberg et al. discovered in 1998 the mammalian PFDN complex by isolating by chromatography protein binding to nascent, non-folded actin from bovine testes. Purification of the mammalian actin-binding complex revealed the existence of six proteins of a molecular weight between 14 and 23 kDa, structurally related and named PFDN 1 to 6 (Vainberg et al., 1998). The binding of the mammalian PFDN complex to actin and tubulin was further confirmed in vitro, proposing that PFDNs bind to the nascent cytoskeletal unfolded proteins to transfer them to the cytosolic chaperonin containing TCP-1 (CCT) or T-complex-protein-1-ring (TRiC) (Hansen et al., 1999). Genes coding for five of these six PFDNs were previously found in yeast, although they were named as GIM1 to GIM5, forming the GimC complex (termed for genes involved in microtubule biogenesis complex), and were described to be also implicated in the folding of tubulin (Geissler et al., 1998).
Alignments of the yeast PFDN genes with other species’ genomes were the first evidences that PFDNs were also present in other organisms, ranging from archaea to mammals (Vainberg et al., 1998), but not in prokaryotes, indicating that eukaryotic PFDN likely evolved from archaea. These observations were confirmed by other studies corroborating the presence of PFDNs in plants (Hill and Hemmingsen, 2001). The first yeast PFDN homolog that was characterized was an archaeal PFDN (Leroux et al., 1999), which contains two different types of subunits, present in two and four copies, respectively, assembling into a hetero-hexametric complex like in eukaryotes. The PFDNs are thus a large family of proteins highly conserved through evolution which might have specific functions to maintain protein and cellular homeostasis (Cavalier-Smith, 2002).
Structure of the PFDNs
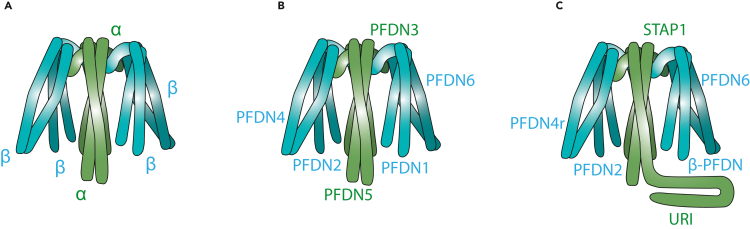
Regardless of the organism, PFDNs are divided into two classes: α and β (Leroux et al., 1999). The α class PFDNs are characterized by presenting two β hairpin loops connecting their two coiled-coil domains, whereas β class PFDNs have their two coiled-coil domains connected by only one β hairpin loop. Two α PFDNs assemble with four β PFDNs to form at least two different hetero-hexametric chaperone complexes in the cell: The classical or canonical PFDN complex (Siegert et al., 2000)—present in all archaea and eukaryotes—and the UPC (Gstaiger et al., 2003), found in higher organisms such as mammals.
The classic PFDN complex
Archaea
The molecular structure of the archaeal PFDN was elucidated two years after its discovery, based on crystallography evidences (Siegert et al., 2000) and mass spectrometry studies (Fandrich et al., 2000). Four PFDN genes have been found in archaea (two α and two β subunits), but different archaeal strains present different combinations of the PFDN genes (Table 1). In any case, the PFDNs always assemble into an α2β4 complex (Figure 1A), although the different combinations between the four genes may explain different regulations and roles of the resulting complexes (Iizuka et al., 2008). The α and β subunits are arranged by hydrophobic interactions with two β barrels at the center forming a central cavity from which protrude the coiled-coil α helices to form a jellyfish-like structure (Figure 1A). This structure provides a relatively high flexibility to the N-terminal region of the PFDN subunits, which conforms the tentacles of the structure, as demonstrated by a lower resolution of these regions obtained by x-ray crystallography analyses. This flexibility is maintained even when PFDN binds to the chaperonin complexes (Kurimoto et al., 2008). The tips of the tentacles are required for the formation of stable binary complexes of the PFDNs with target proteins such as actin and tubulin (Simons et al., 2004).
Table 1.
Mammal PFDN genes, names and orthologues from yeast and archaea
| Mammals |
Budding yeast | Archaea | ||||||
|---|---|---|---|---|---|---|---|---|
| Name in the text | Official protein name | Official gene name | Other names | Class | Classic complex | UPC | ||
| PFDN1 | PFDN1 | PDFN1 | β | x | PFDN1 | β | ||
| PFDN2 | PFDN2 | PFDN2 | β | x | x | Gim4 | ||
| PFDN4 | PFDN4 | PDFDN4 | Protein C-1 | β | x | x | Gim3 | |
| PFDN6 | PFDN6 | PFND6 | HKE2 | β | x | Gim1 | ||
| PFDN4r | PDRG1 | PDRG1 | C20orf126 | β | x | – | – | |
| ASDURF | – | ASDURF | ASNSD1, NS3TP1 | β | x | – | – | |
| PFDN3 | VBP1 | VBP1 | α | x | Gim2 | α | ||
| PFDN5 | PFDN5 | PFDN5 | MM1 | α | x | Gim5 | ||
| URI | URI1 | URI1 | RMP, C19orf2, NNX3 | α | x | Bud27 | – | |
| STAP1 | UXT | UXT | SKP2 | α | x | – | – | |
Figure 1.
Structure of PFDN
(A–C) Schematic representation of the structure of PFDN in Archaea (A), of the classic PFDN complex in eukaryotes (B) and of the eukaryotic UPC (C). Two α (in green) and four β-class (in blue) PFDNs assemble through hydrophobic interactions to form a jellyfish-like structure, as evidenced by x-ray crystallography analyses performed in Archaea. Similar structural model has been proposed for the classic eukaryotic PFDN complex. Yet, the structure of the UPC remains to be determined.
PFDNs have been proposed to have a general role in de novo protein folding and to assist in the biogenesis of nascent actin and tubulin monomers by escorting them specifically to the cytosolic chaperonin, a multisubunit toroidal chaperone complex which facilitates folding in an ATP-dependent manner. Chaperonins are organized in two groups. Class I chaperonins (Cpn60) are found in bacteria (named GroEL/GroES complex) and in organelles with endosymbiotic origin (mitochondria and chloroplasts) (Hemmingsen et al., 1988). Class II chaperonins are found in eukaryotes (CCT complex) and archaea (thermosome) but remain poorly characterized (Klunker et al., 2003). The exact mechanism by which chaperonins facilitate folding of substrate proteins remains unknown.
Thermococcus kodakaraensis present four PFDNs, named A to D, that specifically assemble to form two different complexes: PFDNA/PFDNB or α1β1, constitutively expressed and more related to the PFDN complex found in all archaea, and PFDNC/PFDND or α2β2, induced upon heat-stress conditions and phylogenetically more related to the eukaryotic PFDN (Danno et al., 2008). The α1β1 PFDN complex presents more affinity to the Cpn60 chaperonin subunit α, whereas the α2β2 bound to much higher affinity to the subunit β, whose expression is also induced by heat-shock exposure, because this interaction is stronger with higher temperatures (Sahlan et al., 2010b). Some β PFDN subunits are also able to form tetrameric complexes, such as the β1 PFDN from Thermococcus strain KS-1 (Kida et al., 2008). Other archaea, nevertheless, seem to only present a pair of α and β PFDN genes, like in Sulfolobus solfataricus (D'Amaro et al., 2008).
Substrate recognition of archaeal PFDN is driven mostly through hydrophobic forces, which explains the variety of PFDN substrates found in these organisms. It is important to note that for those archaea presenting two PFDN complexes, the β subunit of the α2β2 lacks the hydrophobic tip of the coiled coil, as demonstrated by X-ray crystallography, which further corroborates the hypothesis that this PFDN complex is closer to the eukaryotic one than the archaeal α1β1 (Sahlan et al., 2010a). Based on the structure and models, computational simulation studies corroborated the hypothesis that the hydrophobic forces in the tentacles of the PFDN complex are able to bind different unfolded proteins, β subunits being of highest relevance in this mechanism (Ohtaki et al., 2008).
Eukaryotes
The classic PFDN complex is formed by Gim1 to Gim6 proteins in the case of yeast, and by PFDN1 to PFDN6 proteins in the rest of the eukaryotes (Table 1). The structure of the eukaryotic PFDN complex has not been obtained yet, but its resolution by electron microscopy revealed that it shares similarities with its archaeal counterpart (Figure 1B), and it is also able to bind to nascent, unfolded actin which is transferred to CCT (Martin-Benito et al., 2002). Moreover, attempts to solve the structure of one of the β subunits of the eukaryotic yeast PFDN have been performed, although interpretation of the x-ray diffraction of the tetrameric crystals is still missing (Perez de Diego et al., 2008). Human PFDN complexes have been also expressed, purified, and crystallized, but without a 3D structure reconstruction (Aikawa et al., 2015). The six PFDNs forming the classical PFDN complex are able to self-assemble spontaneously (Simons et al., 2004). Even though the β subunits present a more crucial role in substrate binding, α PFDNs are required to achieve a proper hexamer structure, since β subunits alone aggregate in tetramers (Perez de Diego et al., 2008). In eukaryotes, the assembly has been proposed to occur clockwise in the following order: PFDN 3, 2, 1, 5, 6, and 4 (Miyazawa et al., 2011; Simons et al., 2004). In vitro experiments demonstrated that the formation of the PFDN complex prevents its degradation by the proteasome system. It has also been proposed that PFDN 2 and 6 present a longer half life than the rest of the PFDNs, which allows the assembly of the complex and prevents the degradation of each subunit (Miyazawa et al., 2011).
Since yeasts are eukaryotes, their genomic and proteomic landscape is closer to what is found in human cells, but yeasts are considerably simpler organisms, even though their chaperone system presents an already high level of complexity (Gong et al., 2009). This general statement also applies to the yeast PFDN family. Even though the complexity in yeast is increased compared to archaea, where the PFDN complex is formed by only two different proteins, the yeast PFDN complex is also arranged in a 2α4β manner, being Gim2 and Gim5 of the α-class and the rest of the PFDNs belonging to the β-class.
Unconventional PFDN-like complex
A few years after the discovery of the classic PFDN family, an additional α-class member was identified and termed URI, standing for unconventional PFDN RNA polymerase binding subunit 5 (RPB5) interactor (Gstaiger et al., 2003). URI was found as a PFDN protein via co-immunoprecipitation (co-IP) experiments combined with proteomic analysis in mammalian cells in an attempt to identify new cell cycle regulators bound to SKP2, the E3-ligase for p27. SKP2 co-IP first identified SKP2-associated α-PFDN (STAP1) also called the ubiquitously expressed transcript protein (UXT), which was found to bind to URI and other PFDNs, some of them identified later on: PFDN2, PDRG1 (or PFDN4r), and PFDN6 (Gstaiger et al., 2003; Sardiu et al., 2008) (Table 1). The model for the structure of the UPC would be similar to the classic PFDN complex: A heterohexameric structure composed of two α PFDN subunits (STAP1 and URI) and four β PFDNs (PFDN2, PFDN4R, PFDN6, and likely one of them duplicated) (Figure 1C). Very recently, a sixth β PFDN candidate has been identified by proximity-dependent biotinylation (BioID) forming a complex with other prefoldins and proposed as a member of this complex, termed ASDURF (Cloutier et al., 2020). Yet, it remains to be confirmed whether indeed ASDURF is part of the UPC or whether one additional β PFDN subunit is simply duplicated in this complex to form a heterohexameric structure. It is not excluded that the UPC can be an unconventional pentameric complex. Yeast also expresses a URI orthologue, termed Bud27 (Table 1), however the UPC is not found in yeast, since orthologues for STAP1 and PFDN4r are missing in these organisms (Martinez-Fernandez et al., 2018).
Gamma PFDN
In certain archaea organisms, such as Methanocaldococcus jannaschii, an additional type of PFDN was identified as an α class PFDN which results to be upregulated in response to heat shock (Boonyaratanakornkit et al., 2005). This α class PFDN was renamed afterward as γ PFDN and was described to form long filaments instead of being part of the PFDN complex (Whitehead et al., 2007). As for the classic PFDN complex, substrate recognition of the γ PFDN filament depends on the flexibility of the hydrophobic coiled-coil tentacles that emerge from the filament. This interaction model has been validated by docking molecular dynamic simulations and in vitro assays (Glover and Clark, 2015).
Functions of the PFDNs
Archaea
PFDNs are present in archaebacteria but not in conventional bacteria. Nevertheless, resolving the structure of the already known Skp protein from Escherichia coli by crystallography techniques showed its high structural similarity to the PFDN complex (Walton and Sousa, 2004), even though both complexes arise from very different topologies (Korndorfer et al., 2004). Skp functions as a jellyfish structure that captures unfolded proteins to prevent its aggregation and delivers them to the outer bacterial membrane to allow their folding and membrane insertion. That is not the only example of a protein complex resembling the jellyfish-like structure of PFDN. The mitochondrial chaperone TIM9-10 structure presents this kind of structure too (Webb et al., 2006), as does the Tim8-Tim13 complex (Beverly et al., 2008).
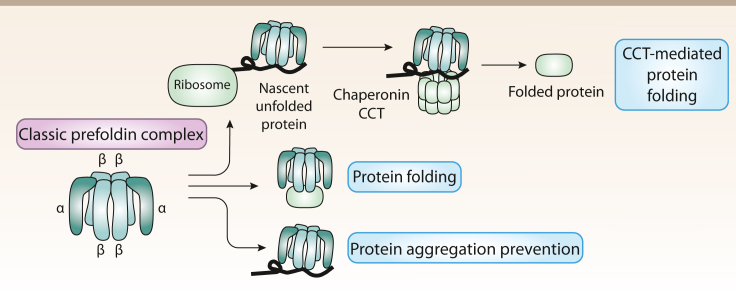
The most well studied role of the classic PFDN complex in archaea is related to its function as a co-chaperone for group II chaperonins (CCT), to which PFDN transfers different substrate proteins. Hence, PFDN molecules work as a transfer protein in conjunction with a molecule of chaperonin to form a chaperone complex acting to correctly fold other nascent proteins. The PFDN complex interacts with unfolded and nascent actin and tubulin proteins (Hansen et al., 1999) through the distal part of its tentacles (Lundin et al., 2004). The proposed function of this complex is to prevent protein aggregation in misfolded proteins and deliver them to the group II chaperonin CCT, so that folding of client protein can be completed (Figure 2).
Figure 2.
Functions of PFDNs in Archaea
Archaea present two different PFDNs, α and β, which assemble in a 2α4β ratio to form the classic PFDN complex. This complex binds to nascent unfolded proteins to deliver them to the CCT class II chaperonin to complete their folding. The classic PFDN complex also has intrinsic folding properties and can bind to a range of proteins to prevent their aggregation.
The PFDN hexamer directly binds to the chaperonin in a substrate-independent manner (Okochi et al., 2004). Binding of the PFDN-substrate complex to chaperonin facilitates the dissociation of PFDN to the protein substrates, facilitating the hand-off mechanism of substrate transfer to the chaperonin (Zako et al., 2005). This mechanism is ATP independent. The specific binding sites of the PFDN to the chaperonin have been described, highlighting the electrostatic interactions that make the formation of the complex possible (Zako et al., 2006), as well as the kinetics in the binding to the chaperonin Cpnβ of archaeas (Sahlan et al., 2009). It has been proposed that the β subunits of the PFDN interact with the helical protrusions found in the chaperonin ring complex (Sahlan et al., 2010b).
Further evidences of the importance of PFDN in the cytoskeletal proteins folding come from in vitro experiments, demonstrating that several mutations in the tubulin genes prevent the interaction between tubulin and PFDN, thus not achieving a proper folded structure (Tian et al., 2010). Nevertheless, the archaeal PFDN has been shown to promote the folding of other proteins besides the ones related to the cytoskeleton. Even though the PFDN complex does not have any ATPase activity, it has been proposed that PFDN may also contribute in a direct way to the folding of unfolded proteins when interacting with them at low affinity, as demonstrated in vitro for the PFDN from Pyrococcus horikoshii (Zako et al., 2010) and Pyrococcus furiosus (Hongo et al., 2012) (Figure 2). In Pyrococcus, PFDN prevents aggregation of citrate synthase upon thermal stress, and it also binds to GFP to promote a correct folding (Hongo et al., 2012; Okochi et al., 2002). Thus, it is interesting to note that the PFDN complex has chaperone activity by itself. Similar studies with other archaeal strains failed to find such a role of PFDN in promoting GFP folding, possibly because of thermal restrictions, but they described a higher folding efficiency of other proteins such as isopropylmalate dehydrogenase (Okochi et al., 2005). More evidences of the relationship between PFDNs and other proteins such as polymerases came from studies showing that addition of the PFDN complex together with HSP60 retained the DNA polymerase activity at 100° longer than in the controls (Laksanalamai et al., 2006).
Evidences from electron microscopy experiments revealed that the archaeal PFDN was able to bind proteins of different size and shape (Martin-Benito et al., 2007), unlike the eukaryotic PFDN which would have evolved towards a more specialized and selective machinery. Furthermore, some PFDN subunits are upregulated during heat-stress conditions. Tk-Phr was suggested as a possible transcription factor involved in this response, since its ablation in Thermococcus kodakaraensis leads to downregulation of archaeal PFDN in this strain, together with other heat-stress response genes (Kanai et al., 2010).
Other unexpected proteins that showed PFDN domains are found in the costa of protozoa of the Trichomonadidae family, a striated fiber that presents an undulating membrane (de Andrade Rosa et al., 2017). Several studies have shown that ectopic expression of archaeal PFDN leads to an enhancement of the folding efficiency of different proteins. This effect, together with the stability at high temperatures of the archaeal PFDN, brought some attention to the archaeal PFDN in the biotechnological field. It can be used to improve the production of proteins in recombinant bacteria. For example, expression of archaeal PFDN from P. furiousus in recombinant E. coli notably increased the soluble protein levels of alfa-amylase, having this protein a particular interest in industry (Peng et al., 2016). Ectopic archaea expression in bacteria also protects these organisms from several environmental insults. For instance, PFDN expression protects against organic solvents toxicity by sequestrating unfolded proteins and thus cooperating with the bacterial chaperone systems to reduce the protein stress caused by exposure to these types of solvents (Okochi et al., 2008). It also increases the tolerance to hyperthermia in these organisms, consistent with the role of archaeal PFDN in preventing protein aggregation and favoring a proper protein folding (Chen et al., 2010) (Figure 2). PFDN, unlike other archaeal chaperones, also rescues the cell growth inhibition caused by antibiotic (aminoglycoside) toxicity, probably through a decrease in the protein folding aggregation caused by this agent (Peng et al., 2017).
Furthermore, the γ PFDN present in some archaeal strains also has biotechnological applications, for example, as biotemplate in the generation of platinum nanowires (Dordick, 2013; Glover et al., 2013, 2016).
Yeast
Classic PFDN complex
The interaction between the PFDN complex and the CCT complex has been also assessed in eukaryotes. Human TRiC complex and yeast PFDN complex were expressed, purified, and mixed, allowing for determination of the macromolecular structure of the resulting macro-complex by Cryo-EM. A similar function as described for archaea was also proposed in eukaryotes: The two protein complexes bind through electrostatic interactions aligning the two substrate-binding cavities which allows the transfer of substrates such as actin to promote the folding (Figure 3). A role of PFDN as a processivity factor rather than a delivery partner of CCT was also discussed. One way or another, the interaction between the two complexes is essential to maintain proteostasis in vivo since disruption of this binding is strongly deleterious in yeast, accompanied by an important accumulation of protein aggregates (Gestaut et al., 2019).
Figure 3.
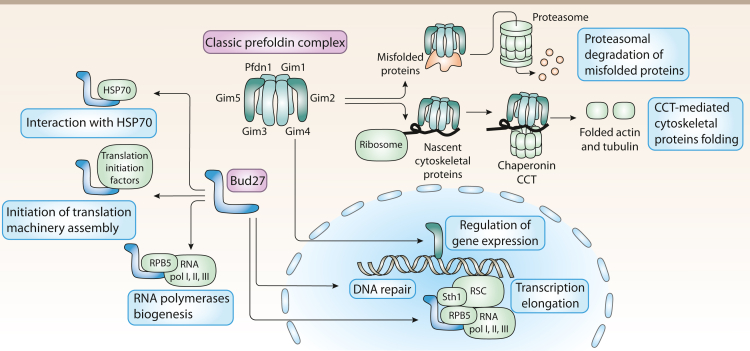
Functions of PFDNs in budding yeast
In budding yeast, the classic PFDN complex is formed by proteins Gim1 to Gim 5 and PFDN1. This complex binds to nascent cytoskeletal proteins, such as actin and tubulin, to deliver them to the chaperonin CCT where they complete their folding. It also binds to misfolded proteins to promote their degradation through the delivery to the proteasome machinery. Some components of the complex are also involved in nuclear regulation of gene expression. Yeast presents an atypical PFDN termed Bud27 which is not forming part of any PFDN complex in these organisms. Bud27 also contributes to protein folding by binding to HSP70. It also contributes to the biogenesis of the RNA polymerases I, II, and III thanks to its interaction with RPB5, a common component of the three of them; and to the assembly of the initiation of translation machinery. In the nucleus, Bud27 contributes to transcription elongation by interacting with the polymerases and with chromatin remodelling complexes such as RSC. Loss of URI also increases DNA damage.
One difference between the archaeal and eukaryotic PFDN complex is that the latter presents a higher charge density in the tips of its tentacles and also in the core cavity of the structure when compared to the archaeal homologue, meaning that the substrate binding might be different and pointing to a more specialized function of the eukaryotic complex (Simons et al., 2004). This is translated into a different mechanism of substrate binding compared to the one proposed in archaea: eukaryotic PFDN encapsulates the substrate inside its cavity (Martin-Benito et al., 2007). Although these differences could explain distinct substrate-binding capabilities of the archaeal and the eukaryotic PFDNs, a detailed study of the binding domains of the archaeal PFDN showing its flexibility and adaptation properties highlighted the possibility of the eukaryotic PFDNs to bind further substrates than the cytoskeletal proteins (Lundin et al., 2004). In agreement with this idea, eukaryotic PFDN from the thermophilic fungus Chaetomium thermophilum was successfully expressed and purified. Unlike archaeal PFDNs, it was not able to prevent citrate synthase aggregation, supporting the idea that eukaryotic PFDN has more specific functions than a regular chaperone as happens in archaea. Nevertheless, eukaryotic PFDN is also able to bind actin and tubulin (Morita et al., 2018), and genetic ablation of the prefoldin genes in yeast alters the microtubules phenotype (Chesnel et al., 2020; Geissler et al., 1998).
Other regulators of this system have been described, such as PhLP3 in yeasts, which acts as a negative regulator compared to the PFDN complex, although a balance of the two components seems to be required to achieve a functional cytoskeleton (Stirling et al., 2006). The role of PFDN in assisting the folding of cytoskeletal proteins increases the efficiency of the process at least five times (Siegers et al., 1999). Ablation of one PFDN gene can cause the disruption of the whole complex, arguing that PFDNs have no redundant functions (Warringer et al., 2003). In line with this non-redundancy concept, deletion of specific PFDN genes in Saccharomyces cerevisiae leads to different phenotypic defects upon exposure to cellular stressing agents that affect the cytoskeleton, further demonstrating additional roles of the PFDN proteins beyond actin and tubulin folding. It was also reported that Gim2, 3 and 6 are required for the induction of the expression of certain genes, corroborating the association of the PFDN genes in other complexes distinct to the classic PFDN complex, and the role of some of these complexes in gene transcription regulation (Amorim et al., 2017), even though the UPC is missing in yeast (Figure 3). Additional clients in these organisms have been also characterized. For example, in S. cerevisiae the PFDN complex functionally interacts with the phospholipase A2, although this relationship might be indirect (Mattiazzi et al., 2010).
The PFDN complex is required for proteasome-mediated quality control of misfolded proteins, as demonstrated in a yeast model consisting of a thermosensitive allele of the yeast Guk1 guanylate kinase (Comyn et al., 2016). PFDN binds and stabilizes misfolded proteins, enhancing their solubility and allowing them for proteasomal degradation (Figure 3). This also corroborates the additional substrates of yeast PFDN besides actin and tubulin. This notion was further evidenced by modeling protein complexes interactions from yeast protein complexes, showing that PFDN complex in S. cerevisiae is able to bind several other protein complexes, being an important number of these interactions lethal (Le Meur and Gentleman, 2008). Indeed, the hepatitis C virus F protein interacts with PFDN and prevents their assembly into a complex, resulting in an alteration of the tubulin cytoskeleton (Tsao et al., 2006).
It was proposed that arsenic, a pollutant and toxic agent present in the environment, inhibits the CCT/PFDN complex in yeasts and mammalian cells, affecting protein folding (Pan et al., 2010). Thus, it is not surprising that PFDN genes are among the genes upregulated upon arsenite exposure, probably as a survival mechanism (Guo et al., 2016).
Bud27, a yeast homolog of the mammalian URI
Yeasts also express Bud27, a homolog of the mammalian URI PFDN. Bud27 is required for the cytoplasmic assembly of the three RNA polymerases, a process that depends on RPB5 (a common subunit to the three RNA polymerases) and that occurs prior to their nuclear transportation (Miron-Garcia et al., 2013) (Figure 3). It has been demonstrated in yeast that Bud27 plays a role in transcription elongation in a different manner than the classic PFDN complex, although URI and the whole UPC would also interact with chromatin remodeling factors such as RSC, thereby modulating the elongation activity of the RNA polymerase (Miron-Garcia et al., 2014). Additionally, ablation of Bud27 prevents RNA polymerase III assembly and its interaction with the chromatin remodeling complex RSC, probably through disruption of the interaction with RPB5, affecting the polymerase transcription activity and gene expression (Vernekar and Bhargava, 2015) (Figure 3). Supporting this hypothesis, Bud27 is able to bind to Sth1, a member of RSC, a complex that contributes to gene transcription regulation due to its ability to modify the status and accessibility of the chromatin (Miron-Garcia et al., 2014).
Furthermore, Bud27 was found to form a complex with other proteins implicated in gene transcription and DNA repair with Bud27 being a central component required for the formation of such protein complex (Tronnersjo et al., 2007). Moreover, Bud27 has been linked to translation in yeast. It was proposed that Bud27 interacts with several translation initiation factors that promote the assembly of the translation initiation machinery and, thus, Bud27 increases the efficiency of this key cellular process (Deplazes et al., 2009) (Figure 3).
Plants
The PFDN family is also present in plants. Analysis of 14 species revealed that plant PFDNs can be divided in nine groups, resembling the nine PFDN genes found in mammals, and also that they probably originated by gene duplications, supporting the idea that these genes are highly conserved through evolution (Cao, 2016). The Arabidopsis genome encodes six PFDN genes that also assemble into a hexameric complex whose function is crucial for a proper stabilization of tubulin, among other proteins, since mutations in the PFDN6 gene causes impairments in microtubule dynamics as evidenced by reduced plant size and general disorganization in the plant (Gu et al., 2008). Similar results are obtained when generating null mutants for either PFDN3 or PFDN5, which result in decreased levels of actin and tubulin, altering microtubule homeostasis and some physiological processes, such as sensitivity to NaCl (Rodriguez-Milla and Salinas, 2009). Ablation of PFDN4 gene also leads to a similar phenotype involving abnormalities in microtubules (Perea-Resa et al., 2017). Thus, it seems that the classic PFDN complex presents similar functions in plants than in other eukaryotic organisms.
The subcellular localization of the PFDN complex in plants seems to be dependent on DELLA proteins, plant transcriptional regulators acting downstream of the phytohormones gibberellins (GA). In the presence of GA, DELLA proteins result degraded, which allows the translocation of the PFDN complex to the nucleus and thus prevents its role in tubulin folding (Locascio et al., 2013). This process is also regulated by the daily rhythms observed in plants. The translocation of the PFDNs to the nucleus has not only consequences in actin and tubulin folding, but also exerts specific roles in this subcellular compartment. For example, it interacts with HY5 to promote its ubiquitination-mediated degradation, downregulating its target genes and ensuring a proper response to cold exposure (Perea-Resa et al., 2017).
Conversely to what happens in bacteria, where ectopic expression of PFDN plant genes has a protective role to different environmental stresses, overexpression of PFDN2 did not enhance biomass production neither bud flush in Populus deltoides trees engineered for biofuel production (Macaya-Sanz et al., 2017). Nevertheless, these results could be due to the overexpression of only one PFDN gene, maybe disrupting the regulation of the whole complex and thus not presenting any beneficial effects. Later studies in the same Populus species showed that overexpressing PFDN2 causes alterations in certain metabolic pathways related to sugar production and release, thus having an impact on the cell wall properties. These effects could be beneficial for the biofuel conversion use of this species (Zhang et al., 2020).
Challenging siRNA-treated plants with parasites is a method to silence gene expression in the parasites, such as nematodes. Using this approach, PFDN2 was silenced in M. incognita through Nicotiana benthamiana, resulting in a detrimental effect on the parasites’ development, which proves the plausible use of this technique to silence PFDN expression in parasites to increase the resistance of plants to them (Ajjappala et al., 2015).
Caenorhabditis elegans
The role of PFDNs in this worm has not been deeply studied. Nevertheless, deletion of any of the PFDN genes (with the exception of PFDN4) causes lethality at embryonic stages due to defects in microtubules biogenesis (Lundin et al., 2008). Later studies showed that ablation of PFDN using siRNA disrupts actin cytoskeleton, whereas this same ablation is able to restore normal actin biogenesis when disrupted because of mutations in dynein, another protein involved in actin regulation (Gil-Krzewska et al., 2010).
Studies in C. elegans also allowed the determination of a relationship between URI and DNA damage. URI loss causes important DNA damage (comparable to the one caused by low-dose irradiation), which results in a cell-cycle arrest, preventing cell proliferation and thus causing sterility in adult worms (Parusel et al., 2006). UPC in worms was shown to mediate the crosstalk between HSF-1 and FOXO transcription factors. HSF-1 upregulates PFDN6, a member of the UPC, and this complex increases the transcriptional activity of FOXO (Son et al., 2018), promoting life span and corroborating previous results that showed a direct interaction between PFDN6 and this transcription factor (Riedel et al., 2013). This effect seems to have a special importance in intestine and hypodermis (Son et al., 2018).
Drosophila
Drosophila also expresses the PFDN family of proteins. The classic PFDN complex is crucial for a correct function of spindles and centrosomes in this organism, as highlighted by in vivo experiments depleting one of the PFDN subunits (Mgr, ortholog of PFDN3), that causes aberrant tubulin folding and stability. PFDN is not only able to bind to classic tubulin in Drosophila, but also to Mst (misato), a tubulin-like protein recently described (Palumbo et al., 2015). Nevertheless, this function seems to gain importance when higher tubulin levels are transcribed, such as in spermatocytes and neuroblasts (Delgehyr et al., 2012). Similar results were obtained when depleting the expression of PDFN2, which also leads to abnormalities in spindles and centrosomes (Zhang et al., 2016). This outcome was accompanied by neuroblasts overgrowth, suggesting a role of PFDNs as tumor suppressor proteins. Further evidences of the importance of PFDNs in the brain of Drosophila rely on the fact that at least some of them present a discrete and regulated expression pattern (Diao et al., 2007).
In Drosophila, URI is highly expressed in the cytoplasm of cells from embryos, pupae, and adult gonads, while its deletion is lethal in these animals, probably due to an imbalance in cell viability (Kirchner et al., 2008). Defects in DNA integrity following URI downregulation also occurs in Drosophila (Kirchner et al., 2008). Furthermore, URI was shown to localize in active chromatin regions, suggesting a role of this PFDN in transcription (Kirchner et al., 2008). URI in Drosophila was the first PP1α specific binding protein described (Kirchner et al., 2008).
Mammals
As genomic complexity increases along evolution, so does the diversity of the PFDN family of proteins. In mammals, ten PFDN genes have been found, apparently including the very recently identified ASDURF (Cloutier et al., 2020); that assembly to form both the classic PFDN complex and the UPC. Furthermore, the different PFDN genes are not redundant and have been linked to different functions apart from their contribution of the PFDN complexes.
Classic PFDN complex in mammals
The classic PFDN complex in mammals, formed by PFDNs 1 to 6, has been associated with protein folding. As in other organisms, it mediates actin and tubulin folding through their delivery to the CCT chaperonin complex. The importance of the classic PFDN complex in this process is highlighted by the aberrant microtubule assembly in human cell lines after siRNA-mediated downregulation of PFDNs (Chesnel et al., 2020). Furthermore, PFDN5 was recently found to bind β-actin, and its overexpression promotes the assembly of this monomer into filopodia, thus increasing filopodia density and length, which results in an increase in cell migration as evidenced by transwell experiments (Fan et al., 2020). This folding function is also shared with other proteins, such as for pVHL, since ablation of different prefoldin genes and especially PFDN3 causes aggregation of pVHL (Chesnel et al., 2020). Furthermore, in the nucleus, the classic PFDN complex has been proposed to deliver HDAC1 to the CCT complex for its proper folding, as evidenced by interaction analysis performed in human cells (Banks et al., 2018). In addition to CCT-mediated folding of proteins, the classic PFDN complex plays a role in protein binding to prevent their aggregation, as detailed later.
URI-R2TP complex or PAQosome
UPC was found to interact with different partners, arguing that UPC might have pleiotropic functions (Figure 4). A combination of proteomics analysis together with newly developed computational methods allowed to identify direct interaction partners between the human UPC and DNA polymerases-related genes (POL3A, RPAP3, RPB5, RPB5MP), components of the R2TP co-chaperone complex (RUVBL1, RUVBL2, RPAP3 and PIHD1), and associated proteins like WDR92, and proteins related to chromatin-remodeling complexes such as SMARCB1, among other interactors (Gstaiger et al., 2003; Sardiu et al., 2008). URI was also found to interact with other chaperones in yeast, such as HSP70 (Deplazes et al., 2009), that also interacts with the R2TP complex through RPAP3. The RUVBL1 and 2 proteins are ATPase proteins that might have helicase functions and are essential and present in all eukaryotes (Kanemaki et al., 1999), forming part of other protein complexes such as INO80, TIP60, SWR/SRCAP (von Morgen et al., 2015).
Figure 4.
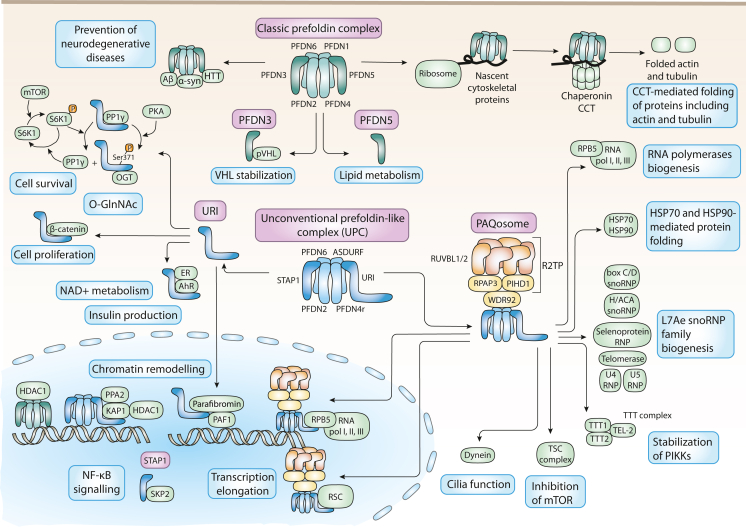
Role of PFDNs in mammals
In mammals, the classic PFDN complex is formed by PFDNs 1 to 6. As in yeasts, it binds to nascent actin and tubulin cytoskeletal proteins to deliver them to the chaperonin CCT to promote their folding. The chaperone activity of the complex has been also associated to proteins involved in neurodegenerative diseases, such as Aβ oligomers, α-synuclein, or huntingtin (HTT). Apart from these functions within the complex, PFDN 3 is also involved in the stabilization of pVHL; and PFDN 5 has been associated with lipid metabolism. Mammals also present the unconventional or URI PFDN-like complex (UPC). It is involved in chromatin remodelling functions, for example by binding KAP1 and PPA2 together to allow the regulation of the histone deacetylase HDAC1. UPC also binds to the R2TP module to form a complex termed PAQosome that has been involved in the assembly of other protein complexes, such as the RNA polymerases, L7Ae snoRNP family of ribonucleoproteins, or PIKKs through the interaction with the TTT complex. The PAQosome is also involved in protein folding since it interacts with the chaperones HSP70 and 90, and is related to other cellular processes such as inhibition of mTOR through interaction with the TSC complex or the cilia function though its interaction with dynein. In the nucleus, the PAQosome promotes transcription elongation by its interaction with the polymerases and with chromatin remodeling complexes such as RSC. The UPC component URI has also shown other functions besides its contribution to the UPC. URI affects cell survival thanks to its relationship with PP1γ, and also cell proliferation by binding to β-catenin and thus preventing its translocation to the nucleus to promote cell proliferation and by affecting O-Glucosyl N-Acetylation of proteins like c-Myc by its PKA-dependent interaction with OGT. Furthermore, loss of URI has been associated with an increase in DNA damage, at least in part by binding to AhR and ER receptors and thus affecting NAD+ metabolism. URI binding to these receptors also affects insulin production in β-cells in the pancreas. URI also affects chromatin remodeling in the nucleus by interacting with parafibromin and PAF1. The other alfa protein forming the UPC, STAP1, is also involved in other functions beyond the UPC, for example in NF-κB signaling.
The R2TP complex is known for its involvement in the assembly and stabilization of several multiprotein complexes such as L7Ae ribonucleoproteins, U5 small nuclear ribonucleoprotein, RNA polymerase II, phosphatidylinositol 3-kinase-related kinases (PIKKs), and the mTOR/tuberous sclerosis complex (TSC1-TSC2). Indeed, the R2TP/URI PFDN-like complex has been recently renamed as PAQosome, standing for particle for arrangement of quaternary structure (Houry et al., 2018) (Figure 4). Even though the list of interactors with the PAQosome is believed to be far from complete, the importance of the PAQosome in the complex stabilization is likely to reside in the optimization of the assembly process, especially under stress conditions.
Although the roles and functions of the two modules composing the PAQosome remain unclear in the protein complexes assembly, several studies show a contribution of the UPC in the assembly of RNA polymerases I to III, directly linked to the ability of URI to bind RPB5, a common subunit of the three RNA polymerases (Cloutier and Coulombe, 2010) (Figure 4). In this regard, URI reportedly shuttles between nuclear and cytoplasmic compartments of prostate cancer cells together with the RNA polymerase II complex in a CRM1 dependent manner (Mita et al., 2013). Moreover, UPC has been speculated to mediate the localization of the RNA polymerase II to the centrosomes (Hintermair et al., 2016), although further studies are still needed to clarify this function.
URI is not only crucial for RNA polymerases biogenesis, but also has important roles in their activity, as evidenced by studies performed in mammals and yeasts. For example, PFDN has been found in active chromatin sites in a transcription-dependent manner, and the classic prefoldin complex influences RNA polymerase elongation (Millan-Zambrano et al., 2013) (Figure 4). URI binds to the tumor suppressor parafibromin, which physically interacts with components of the PAF1 complex to regulate the transcription elongation and RNA processing pathway (Figure 4). Dysfunction of this pathway may be a general phenomenon in the majority of cases of hereditary parathyroid cancer (Yart et al., 2005). Loss of URI ameliorates the cellular response to AR agonists, which can be explained taking into account the role of the UPC in gene transcription by interacting with chromatin-remodeling complexes including RSC and SWR-C (Mita et al., 2011). A URI complex would be required for a proper chromatin remodeling state that ensures a correct translocation of AR to the AR response elements in the DNA (also known as AREs).
A link between URI, DNA damage and chromatin remodeling complexes has been also proposed (Parusel et al., 2006). Indeed, PFDNs 1 to 5 have been shown to interact in hepatocytes with HDAC1, an important histone deacetylase, supporting the notion that PFDNs have a role in chromatin remodeling (Farooq et al., 2013) (Figure 4). Overexpressing analysis coupled with mass spectrometry revealed that the transcription repressor HDAC complex could bind to PFDNs, preventing the formation of the functional chromatin remodeling complex. This leads to the hypothesis that PFDNs assist the folding of the HDAC complex subunits before they assemble together to form the fully active chromatin remodeling complex. Additional links between URI and chromatin remodeling complexes have been studied in prostate cancer cells. It has been proposed that UPC serves as a scaffold to bring together KAP1 and the phosphatase PP2A, which ensures the dephosphorylated state of KAP1 essential to bind to the HDAC1/2 repressor complex, thus inhibiting gene transcription (Mita et al., 2016) (Figure 4).
Even though the role of URI in the assembly, biogenesis, and activity of RNA polymerases is clearly proposed, we cannot exclude a function in conjunction with the R2TP complex, coordinated by the PAQosome macromolecular complex. Moreover, the mechanism of action by which the assembly and biogenesis of client complexes occur remains unclear. One possibility consists of the passive facilitation of the assembly by locating the different elements of a protein complex together in the space. Another model would involve an active role in the folding of client proteins for their proper assembly. In this regard, the heat shock protein 90 (HSP90), a chaperone common to both modules, URI and R2TP complexes, may assist in folding and stabilizing the client proteins properly (Figure 4). However, the module that has the most important function within the PAQosome system remains to be determined. Clearly, an important source of energy is needed, and it may depend either on the RUVBL1/2 proteins that hold some ATPase activity or on HSP90 that also binds with high affinity ATP and hydrolyses it.
Besides RNA polymerases, the PAQosome has also been associated with the biogenesis of the yeast small nucleolar ribonucleoprotein (snoRNP), a family of proteins important in RNA processing, with the snoRNPs containing RNA-binding proteins of the L7Ae-family in humans (Figure 4). The first evidence of the PAQosome as a mediator for snoRNPs assembly and maturation demonstrated that the R2TP complex was required for the accumulation of box C/D snoRNA (Zhao et al., 2008), one of the components of the box C/D snoRNPs. Indeed, depletion of different components of the R2TP complex reduces global C/D snoRNA levels (Gonzales et al., 2005; Kakihara et al., 2014; King et al., 2001; McKeegan et al., 2007). The PAQosome is also essential for H/ACA snoRNP complex assembly, as highlighted by the interaction between different proteins of the R2TP complex with H/ACA snoRNP components (Boulon et al., 2008; King et al., 2001; Machado-Pinilla et al., 2012). The R2TP module interacts with different components of the telomerase RNP complex, also containing the L7Ae protein, probably helping these subunits to bind together and thus helping its assembly (Boulon et al., 2008; Venteicher et al., 2008). Selenoprotein mRNPs include L7Ae protein as well, and SBP2 protein among other components. It has been described that the R2TP complex facilitates the SBP2 binding to selenoprotein mRNA to form the selenoprotein mRNP complex (Boulon et al., 2008). Additionally, a role of the R2TP complex has been proposed in the assembly of U4 snRNP, one of the factors in charge of cellular splicing, since it interacts with several components involved in its biogenesis, although its exact contribution requires further investigation (Bizarro et al., 2015). The PAQosome seems to play additional roles in the regulation of the splicing machinery by modulating the biogenesis of the splicing factor U5 snRNP and its assembly via the protein ZNHIT2, which has been shown to interact with several members of the PAQosome (Cloutier et al., 2017; Malinova et al., 2017).
Clearly, R2TP complex within the PAQosome plays a major role in RNA processing, although we cannot exclude that this action is mediated through the coordination with the URI PFDN module. Hence, together with the fact that URI regulates RNA polymerases assembly, biogenesis, and activity, these findings suggest a major role of URI machinery in RNP biology including tRNA processing, rRNA maturation, and RNA modification.
Additional assembly functions have been associated with this complex. PIKKs are kinases related to DNA damage, mTOR signalling, chromatin remodeling, and nonsense-mediated mRNA decay. This family of proteins requires the action of the TTT complex (TEL2, TTI1, and TTI2) to be stabilized. TTT complex was found binding to the PAQosome in pull-down and co-IP experiments in a TEL2-phosphorylated dependent manner (Horejsi et al., 2010). Importantly, inhibition of the PAQosome members resulted in a reduction of PIKK assembly and signaling (Izumi et al., 2012), highlighting the relevance of the PAQosome in the function of these proteins (Figure 4). Moreover, different components of the PAQosome have also been reported to interact with the tumor suppressor complex TSC and inhibitor of mTOR complex 1 (Cloutier et al., 2017; Malinova et al., 2017). Additionally, a possible role of the PAQosome as a scaffold to promote TSC-inhibition of HSP90 has been proposed (Woodford et al., 2017), suggesting an additional function of the PAQosome in the regulation of mTOR activity by mediating TSC assembly and stabilization (Figure 4).
Interestingly, a more direct relationship between URI and the nutrient sensing pathway has been reported. URI was found located on mitochondria, and in response to nutrient availability is directly phosphorylated by S6K1, a downstream kinase of the mTOR complex 1 (mTORC1) pathway (Djouder et al., 2007; Gstaiger et al., 2003). Unphosphorylated URI binds and inhibits PP1γ phosphatase activity in mammalian cells (Djouder et al., 2007). Nutrient surpluses activate S6K1 which phosphorylates URI at Ser 371 and which, in turn, causes disruption of URI/PP1γ complexes. Released PP1γ feedbacks to S6K1 to shut down its activity and control cell survival in response to nutrient surpluses (Figure 4). URI/PP1γ are central components of a mitochondria negative feedback program which restricts S6K1 survival signaling to set a threshold of apoptosis during periods of metabolic stress. Although URI regulates mTORC/S6K1 activity, this does not exclude an additional function of URI as part of the PAQosome to contribute to mTORC1 assembly. Clearly, the PAQosome plays a crucial role in the biogenesis, assembly, and activity of several macromolecular complexes, but further studies are required to better understand the contribution of both R2TP and URI modules in the assembly of these large complexes.
PFDN functions in the brain
PFDNs are highly expressed in the brain (Tebbenkamp and Borchelt, 2010) (Figure 5). Indeed, the gene encoding for the PFDN6 protein (CCDC30) was identified in human fetal brain (Zhang et al., 2006), suggesting a role in brain development. This is also supported by the fact that an unidentified PFDN shows a differential expression pattern according to the neuronal differentiation (Oh et al., 2006). Studies in mice have highlighted the importance of some of the PFDN genes present in the classic PFDN complex in neural tissue and cellular development, in line with the observations from Drosophila (Delgehyr et al., 2012). Ablation of PFDN1 in mice presents a phenotype that fits with the importance of PFDN in actin and tubulin folding, including neuronal loss (Cao et al., 2008), but also improper B and T cell development and mucus clearance defects. Deleterious mutations in PFDN5 by point mutations also cause an impaired function of the nervous system, along with hypogonadism, probably due to defects in actin and tubulin metabolism (Lee et al., 2011).
Figure 5.
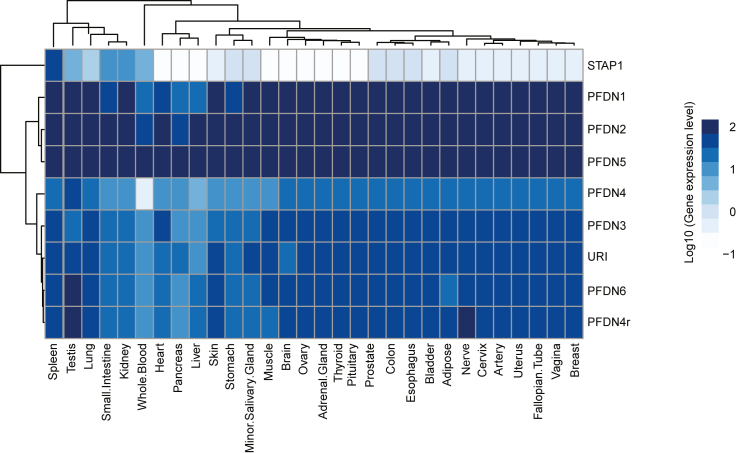
Gene expression level of PFDNs across tissues in humans
Heatmap showing the gene expression of the 9 PFDN genes in different human tissues. Gene expression levels are from 29 human tissues from GTEx Project data V8. Presented values are the mean of log10-transformed Transcript Per Million (TPM) of samples in each tissue.
Because of the role of PFDNs in protein folding, and the crucial part of this process in protein aggregation during different degenerative diseases, the effect of the complex in the formation of Alzheimer-related amyloid beta oligomers has been studied (Sakono et al., 2008). In vitro expression of archaeal PFDN leads to Aβ oligomers formation, which is hypothesized to contribute to Alzheimer’s disease (AD) (Sakono et al., 2008). This is consistent with the upregulation in PFDN2 found in AD brains (Loring et al., 2001); with the upregulation of PFDN complex in AD mouse models (Sorgjerd et al., 2013); with the association between this disease and both the SNPs found upstream of PFDN2, and the significant relationship with the HSP70 pathway and AD based on genomic studies (Broer et al., 2011). Other genomic studies also found AD-associated SNPs in PFDN1 (Jun et al., 2017). Nevertheless, in vitro studies using human PFDN showed that this complex inhibits the formation of αβ fibrils and instead traps the Aβ protein forming large soluble oligomers that are less toxic to neuronal cells (Sorgjerd et al., 2013) contradicting previous results found with the archaeal PFDN (Sakono et al., 2008), maybe because of the different binding properties between the PFDN complexes in the two species (Figure 4).
PFDN also prevents the accumulation of α-synuclein, a protein that forms aggregates in a familial form of AD (Figure 4). PFDN is co-localized with α-synuclein in the lysosomes and knockout of the complex results in pathogenic aggregation of this protein (Takano et al., 2014), although it remains unclear if this effect is direct or indirect through saturation of the quality control machinery of the cell. PFDN5 is downregulated in an age-dependent manner in the hippocampus of mouse models of AD. This protein is also linked to synaptic plasticity since its mRNA and protein levels are modulated alongside this process, proposing that PFDN5 is newly synthesized after induction of synaptic plasticity (Kadoyama et al., 2019).
Another pathology characterized by accumulation of protein aggregates is Huntington’s disease, in this case by huntingtin protein (HTT). Knockdown of PFDN2 or PFDN5 with the subsequent disruption of the PFDN complex leads to the accumulation of pathogenic aggregates of HTT, inducing cell death (Tashiro et al., 2013). This study suggested that PFDN complex would have a protective effect in Huntington’s disease, such as the one described previously for AD (Sorgjerd et al., 2013) (Figure 4). The importance of the folding pathways in neurodegenerative diseases is also highlighted in studies showing that antipsychotic treatment of patients upregulates the protein folding response, including the PFDN/CCT pathway (Mas et al., 2015).
In line with the preventive role of PFDN complex in protein aggregation, Abe et al. demonstrated that knockdown of the PFDN complex increased the amount of insoluble ubiquitinated proteins (Abe et al., 2013). Presumably, PFDN would recognize the polyubiquitinated proteins and bind to them, preventing their aggregation, and thus their toxic consequences for the cell, this mechanism being even more important under certain pathological conditions, such as in the context of neurodegenerative diseases. Furthermore, PFDN seems to upregulate under proteasomal inhibition conditions, which further links PFDN with the protein folding quality mechanism within the cell. They also demonstrated that PFDN complex also protects cells from different types of stress, such as ER stress. Indeed, PFDN2 is a target gene of some miRNAs in neuroblastoma cells, along with other proteins involved in neuronal processes (Patil et al., 2015). PFDN2 has also been proposed as an upstream modulator for the expression of gamma-synuclein gene, SNCG, in retinal ganglion cells, which loss contributes to glaucoma and other retinal neurodegenerative diseases (Chintalapudi et al., 2016).
PFDN functions in the testis
Drosophila studies highlighted the important function of PFDNs in gonads (Delgehyr et al., 2012; Parusel et al., 2006). Experiments in mice corroborated those observations. Impairing the function of PFDN5 with the missense mutation L110R in mice causes male infertility. This effect is due to a lack of differentiation and maturation of the germ cells (Yamane et al., 2015). The proposed mechanism goes through the downregulation of lipid metabolism genes upon PFDN5 mutation, causing a dysregulation in fatty acid metabolism, which is essential for the formation of functional sperm (Figure 4). The effects seem to be dependent on PFDN5 specifically, and not to the whole PFDN complex. Also related to lipid metabolism, PFDN5 is differentially expressed according to the adipose tissue type and animal of origin (Hishikawa et al., 2005). These results go in line with the fact that PFDN5 is highly expressed at the protein level in HUVEC cells, along with other proteins related to motility (Bruneel et al., 2003).
Recent studies propose a role of PAQosome in the regulation of the cellular microtubule network. Ablation of different members of the PAQosome results in disruption of cilia function, as demonstrated in planaria (Patel-King et al., 2019). Another link between the PAQosome and these cilia relies on the interaction between RUVBL1/2 and dynein, as demonstrated in zebrafish (Hartill et al., 2018; Li et al., 2017) (Figure 4). This function of regulation of the cilia would explain the importance of PFDNs in the gonads, since spermatozoids are among the cells presenting motile cilia.
PFDN functions in the pancreas
In porcine pancreas, PFDN presents a differential expression according to the stage of the development, being downregulated after birth, following similar expression patterns as other cytoskeletal regulatory proteins (Choi et al., 2009). Surprisingly, auto-antibodies against PFDN2 were found significantly enriched in type 2 diabetes patients, especially in those with an early onset of the disease (Chang et al., 2017). PFDN5 and anti-PFDN5 antibodies have been proposed as the first biomarkers for uveitis (inflammation of the medium layer in the eye) in ankylosing spondylitis patients (Kwon et al., 2019). Importantly, this could be a novel link between inflammation and the PFDNs.
Recent findings suggest that a class of enteroviruses, coxsackievirus B type 4 (CVB4), downregulates URI in human islet-engrafted mice and in rat insulinoma cells, affecting PDX1 expression and β cell function and identity. Likewise, genetic URI ablation in the mouse pancreas triggers the nuclear translocation of the DNA methyltransferase 1 (DNMT1) expression, which induces Pdx1 promoter hypermethylation and silencing in β cells causing diabetes. Hence, the loss URI provides a causal link between enterovirus infection and diabetes (Bernard et al., 2020).
PFDN functions in the intestine
The development of genetically engineered mouse models (GEMMs) as tools to modulate URI expression levels shed light into the in vivo role of URI in mammals, highlighting its pleiotropic function (Chaves-Pérez et al., 2018). In mice, deleterious mutants for classic PFDN proteins do not lead to specific phenotypes in the intestine. However, deletion of URI specifically in the intestine of adult mice destroys the organ architecture and leads to mouse lethality. A deep characterization of the epithelium revealed that only certain cell types express URI: the quiescent label-retaining (LR) or +4 stem cells, and the transient amplifying progenitor cells, both located in the crypt compartment. Importantly, heterozygous deletion of URI in the intestine makes the usually quiescent LR cells enter proliferation resulting in sensitization to ionizing radiation, thereby failing to regenerate the epithelium after radiation-induced damage (Chaves-Perez et al., 2019). However, complete URI ablation reportedly increases replicative stress-associated DNA damage in LR cells, causing the death of these cells, organ failure, and mouse death (Chaves-Perez et al., 2019). On the other hand, overexpression of URI protects against the deleterious effects of high dose irradiation by preserving the quiescent state of LR cells. A possible explanation of these results relies on the interaction between URI and β catenin: URI loss in LR cells promotes β catenin nuclear translocation and Myc transcription, activating a subset of genes involved in proliferation (Figure 4). Further data also reveal that URI is involved in double-strand break repair by non-homologous end-joining (Chaves-Perez et al., 2019). Thus, URI labels a population of cells that are considered to be the facultative stem cell pool required for intestinal regeneration following radiation-induced injury (Ayyaz et al., 2019; Takeda et al., 2011).
PFDN functions in the liver
While URI deletion in the intestine leads to DNA damage, specific URI overexpression in the liver induces a multistep hepatocarcinogenesis process leading to non-alcoholic steatohepatitis and hepatocellular carcinoma, the most common and one of the most aggressive liver cancers (Tummala et al., 2014). URI expression in hepatocytes increases DNA damage levels at early stages by reduction of NAD+ levels through cytoplasmic sequestration of AhR and the ER receptors implicated in transcription of enzymes involved in de novo NAD+ synthesis (Gomes et al., 2016; Tummala et al., 2014, 2017) (Figure 4). As shown in the intestine, URI expression in adult mice is essential since homozygous depletion leads to spontaneous death in one week (Tummala et al., 2014). Thus, homeostatic URI levels are required to maintain genome integrity and URI may thus have buffering activity functions.
Furthermore, under limited glucose conditions, PKA-mediated phosphorylation of URI at Ser-371 disassociates its interaction with PP1γ and allows the inhibition of OGT (O-linked N-acetylglucosamine transferase), altering the O-GlcNAc-mediated regulation in the cell and causing the turnover of c-Myc (Buren et al., 2016) (Figure 4). The turnover of c-Myc allows liver cancer cells to proliferate in low glucose conditions. These findings uncover URI as the first and unique OGT regulator maintaining glucose and liver homeostasis.
Other functions of PFDNs
The ubiquitously expressed PFDN, named STAP1 or UXT, was identified as an interaction partner of SKP2 (Gstaiger et al., 2003) and described as a nuclear chaperone involved in the formation of NF-κB signaling (Sun et al., 2007) (Figure 4). STAP1 is an α class PFDN that forms part of the UPC (Gstaiger et al., 2003). It has been shown that STAP1 interacts with many genes, including the androgen receptor (Mita et al., 2011), Als2 (Enunlu et al., 2011), NF-κB (Sun et al., 2007), and PIAS2 (Kong et al., 2015). In details, STAP1 interacts with Als2 in the cytoplasm, as demonstrated in vitro and in vivo in neuronal cells, and the expression of both proteins seems to be co-regulated (Enunlu et al., 2011).
Furthermore, STAP1 protein levels change during the cell cycle, being G0/G1 the state in which cells express STAP1 in a higher rate (Enunlu et al., 2011). The complete list of protein interaction partners of STAP1 is still unknown, though. Further studies regarding its interactions, both through the UPC and in a PFDN complex-independent manner, would help to characterize the role of this protein in the cell.
Another PFDN that has been especially studied is PFDN3, also known as VBP1, standing for von Hippel-Lindau (VHL) binding protein (Chesnel et al., 2020; Tsuchiya et al., 1996). PFDN3 is required for the stability of pVHL, as demonstrated in yeast (Le Goff et al., 2016) and in human cell lines (Chesnel et al., 2020) (Figure 4). The PFDN3-mediated stabilization of pVHL enhances the function of the latter on delivering client proteins to the ubiquitin ligase that will send them to proteasomal degradation. Through this mechanism, PFDN3 promotes the degradation of proteins such as HIF-1α (Kim et al., 2018), hMSH4, and p97 (Xu and Her, 2013) or HIV-1 integrase (Mousnier et al., 2007). This role of PFDN3 in the regulation of HIV-1 integrase degradation seems to be essential for the transition between the integration and transcription viral status.
Results
PFDN genomic aberrations in cancer
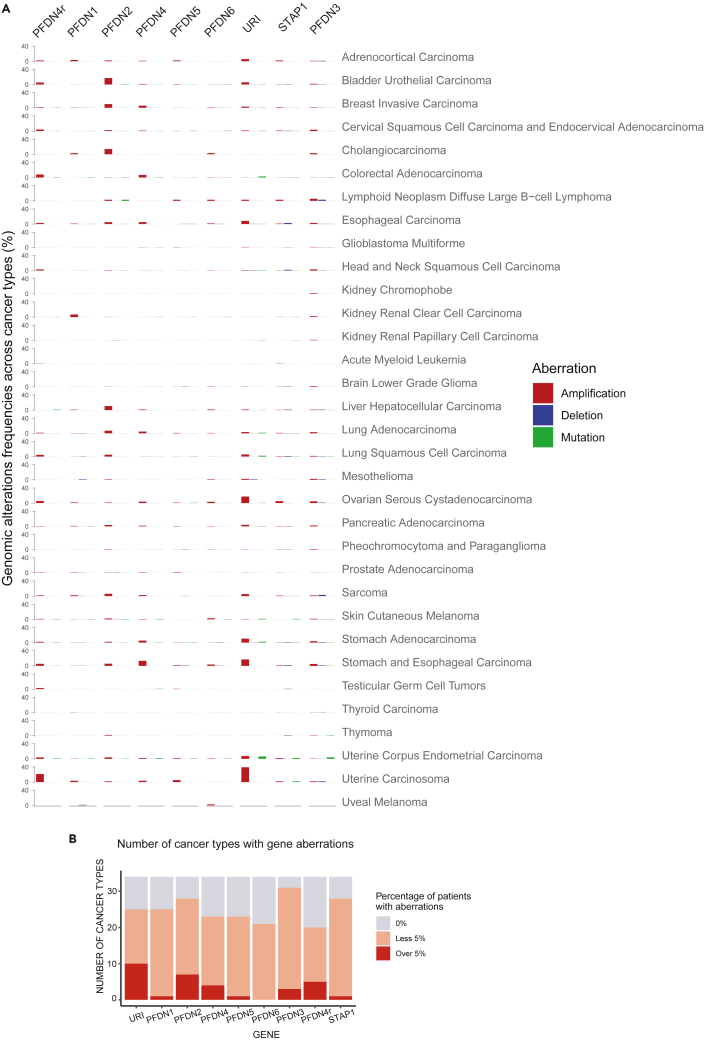
In order to elucidate what kinds of functional roles PFDN genes play in cancer, we first analyzed genomic aberrations (i.e., copy-number alterations either amplification or deletion, and somatic mutations) of PFDN genes using over 10,000 tumors from 33 cancer types sequenced as part of The Cancer Genome Atlas (TCGA) (Cerami et al., 2012; Gao et al., 2013) (Table 2). A significant percentage of patients showed high copy-number alteration (CNAs) amplifications in some cancer types across three genomic alterations (Figure 6A). More concisely, URI, PFDN2, and PDRG1 showed the highest CNAs amplification frequencies, showing alterations in more than 5% of the samples in between 5 and 10 cancer types (Figure 6B). These amplifications in the prefoldin genes are striking, since for example, only 6 cancer types of TCGA dataset show amplification in more than 5% of the patients in KRAS, a very well-known oncogene (data not shown). The cancer types with a higher frequency of amplifications compared to the other cancer types were uterine carcinosarcoma (UCS), bladder urothelial carcinoma (BLCA), stomach adenocarcinoma (STAD), esophageal carcinoma (ESCA), and ovarian serous cystadenocarcinoma (OV). Notably, a low frequency of point mutations in the PFDN genes was observed, demonstrating that they are not highly mutated genes in cancer.
Table 2.
The cancer genome atlas (TCGA) samples
| Cohort | Description | mRNA tumor | mRNA NT | DEA | N CNVs |
|---|---|---|---|---|---|
| ACC | Adrenocortical carcinoma | 79 | 0 | NP | 89 |
| BLCA | Bladder Urothelial Carcinoma | 408 | 19 | OK | 406 |
| BRCA | Breast invasive carcinoma | 1093 | 112 | OK | 996 |
| CESC | Cervical squamous cell carcinoma and endocervical adenocarcinoma | 304 | 3 | LN | 278 |
| CHOL | Cholangiocarcinoma | 36 | 9 | OK | 36 |
| COADREAD | Colorectal adenocarcinoma | 379 | 51 | OK | 526 |
| DLBC | Lymphoid Neoplasm Diffuse Large B-cell Lymphoma | 48 | 0 | NP | 37 |
| ESCA | Esophageal carcinoma | 184 | 11 | OK | 182 |
| GBM | Glioblastoma multiforme | 153 | 5 | LN | 378 |
| HNSC | Head and Neck squamous cell carcinoma | 520 | 44 | OK | 496 |
| KICH | Kidney Chromophobe | 66 | 25 | OK | 65 |
| KIRC | Kidney renal clear cell carcinoma | 533 | 72 | OK | 354 |
| KIRP | Kidney renal papillary cell carcinoma | 290 | 32 | OK | 274 |
| LAML | Acute Myeloid Leukemia | 0 | 0 | NP | 109 |
| LGG | Brain Lower Grade Glioma | 516 | 0 | NP | 507 |
| LIHC | Liver hepatocellular carcinoma | 371 | 50 | OK | 353 |
| LUAD | Lung adenocarcinoma | 515 | 59 | OK | 507 |
| LUSC | Lung squamous cell carcinoma | 501 | 51 | OK | 649 |
| MESO | Mesothelioma | 87 | 0 | NP | 82 |
| OV | Ovarian serous cystadenocarcinoma | 302 | 0 | NP | 398 |
| PAAD | Pancreatic adenocarcinoma | 178 | 4 | LN | 175 |
| PCPG | Pheochromocytoma and Paraganglioma | 179 | 3 | LN | 692 |
| PRAD | Prostate adenocarcinoma | 497 | 52 | OK | 488 |
| SARC | Sarcoma | 259 | 2 | NP | 232 |
| SKCM | Skin Cutaneous Melanoma | 103 | 1 | NP | 363 |
| STAD | Stomach adenocarcinoma | 415 | 35 | OK | 434 |
| STES | Stomach and Esophageal carcinoma | 599 | 46 | OK | 265 |
| TGCT | Testicular Germ Cell Tumors | 150 | 0 | NP | 144 |
| THCA | Thyroid carcinoma | 501 | 59 | OK | 482 |
| THYM | Thymoma | 120 | 2 | NP | 123 |
| UCEC | Uterine Corpus Endometrial Carcinoma | 176 | 24 | OK | 509 |
| UCS | Uterine Carcinosarcoma | 57 | 0 | NP | 56 |
| UVM | Uveal Melanoma | 80 | 0 | NP | 80 |
NT, Normal Tissue; DEA, Differential Expression Analysis; NP, Not Possible; LN, Low Number; N, Number of samples available; CNV, Copy Number Variation; OK, Possible.
Figure 6.
Genome aberrations of PFDN genes in cancer
(A) Barplot showing the percentage of patients with genomic aberrations (amplifications in red, deletions in blue and single mutations in green) in the PFDN genes in the different cancer types.
(B)URI, PFDN2 and PFDN4r are the PFDN genes found in a higher number of cancer types with an important percentage of patients with genomic aberrations (CNVs and mutations) in these genes. CNV, Copy Number Variation.
Amplifications of some PFDN genes have been already shown in many other cancer studies, such as PFDN2, in different cancer types including bladder cancer (in 21% of the patients) (Lopez et al., 2013), PFDN4 in breast cancer (Collins et al., 2001), URI in uterine carcinosarcoma (40%) (Wang et al., 2015c), ovarian (10%) (Theurillat et al., 2011), gastric (Leung et al., 2006) and esophageal cancers (23%) (Lin et al., 2000). Notably, PFDN2 is amplified in bladder cancer and this amplification correlated with increased protein levels and with poor prognosis (Lopez et al., 2013), suggesting PFDN2 is a potential target gene for bladder cancer. PFDN2 amplification was also detected in breast (10%), esophageal (5%), liver (10.5%), and lung (5-7%) carcinomas, and in cholangiocarcinoma (14%) and sarcoma (6%) (Figure 6A).
The amplicon containing PFDN4 (20q13.2) was identified and characterized in breast cancer and it was proposed to have oncogenic activities, although its mRNA levels were not increased in the primary tumor samples evaluated (Collins et al., 2001). Interestingly, our analysis revealed amplification of PFDN4 in stomach and esophageal carcinoma (13%) and colorectal cancer (7%) (Figure 6A) where it shows enhanced mRNA expression levels (Figure 7). PFDN4r also presents high CNA amplification frequencies in many cancer types, including uterine carcinosarcoma (21%), colorectal (8%), bladder (5%), lung (4.5%), and ovarian (5.5%) cancers (Figure 6A). Although we could not detect genomic aberrations in PFDN5(Figure 6A), deletion of PFDN5 was previously reported in canine mammary tumors (Beck et al., 2013; Hennecke et al., 2015).
Figure 7.
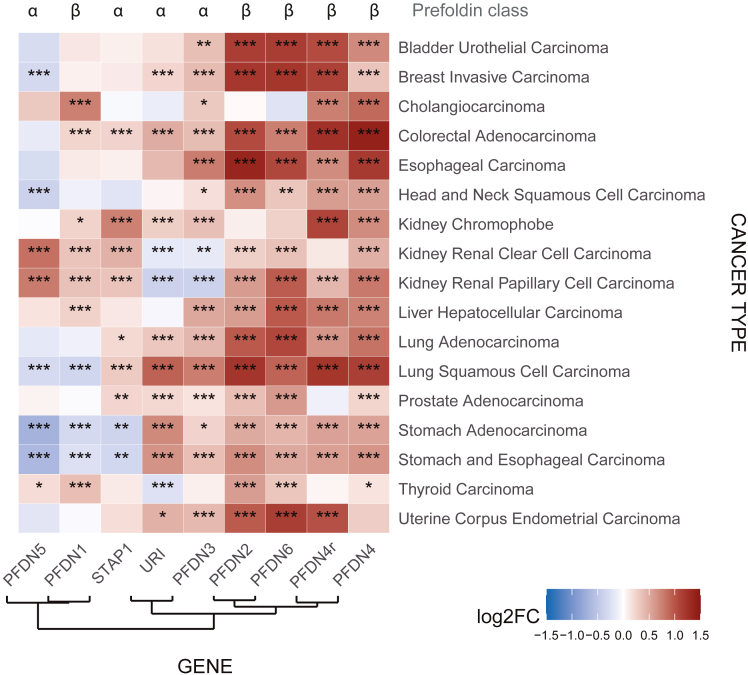
PFDNs are upregulated in cancer at mRNA level
Heatmap showing the log 2 of the fold change in mRNA levels of PFDNs in cancer tissues compared to adjacent normal tissue, as computed using moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 across different cancer types of TCGA. Stars represent the adjusted p value associated to the change assessed by the Wald test after normalization of RNA-seq counts (∗∗∗ = p < 0.005; ∗∗ = p < 0.01; ∗ = p < 0.05). PFDN rows are displayed according to clustering analysis of the log2FC. TCGA, The Cancer Genome Atlas; UPC, Unconventional PFDN-like Complex; FC, Fold Change
Notably, URI is one of the highest CNA amplification frequencies in various cancer types (Figure 6B). It is amplified in about 40% of patients with uterine carcinosarcoma (UCS) from TCGA and in 5.5% of uterine carcinomas on an independent dataset of 363 patients (Wang et al., 2015c). This amplification located in the 19q12 is associated with poor survival after primary treatment and with a worse response to treatments such as radio- or chemotherapy in this cancer type (Wang et al., 2015c). Consistent with the role of UPC in stabilization of PIKKs such as ATM, amplification of URI enhances ATM mRNA expression level, and thus cancer cells become more resistant to DNA damage stress (Wang et al., 2015c). A significantly high percentage of ovarian cancer showed amplifications in URI gene in TCGA, as described by Theurillat et al. (Theurillat et al., 2011), who reported 10% of CNA amplification in the samples from two different Swiss cohorts of 272 and 242 samples, respectively, of different ovarian cancer types. Amplifications correlated with increased URI protein levels and with poor disease-free survival (Theurillat et al., 2011). URI amplification in ovarian cancer was also shown to be required for tumor initiation and progression, likely through alterations in the signaling hub in which URI proteins inhibit PP1γ to maintain cancer cell survival-dependent S6K1 activity. Interestingly, in the URI amplicons also resides the cyclin E1 gene, which has been described as the tumorigenic driving force in many cancers, including gastric (Leung et al., 2006) and esophageal cancers (Lin et al., 2000). However, deep characterization of URI amplicon in various cancer cell lines and downregulation of several other genes located on this amplicon demonstrated that URI has an oncogenic effect and is required for tumorigenesis (Theurillat et al., 2011). This is corroborated by TCGA data showing that URI is also amplified in gastric and esophageal carcinomas (Figure 6A). Moreover, high level of amplification of URI gene was also detected in acute myeloid leukemia (5.5%), sarcoma (5.2%), bladder (6%), lung (5.5%), and pancreatic cancers (4%) (Figure 6A). URI was also reported to be targeted and downregulated by microRNA-598 in ovarian cancer cells, causing a reduction in cell proliferation, migration, and invasion (Xing et al., 2019).
PFDNmRNA expression in cancers
Tumoral tissues often present dysregulations in many different chaperones (Calderwood et al., 2006). Given the role of PFDNs in cytoskeletal protein folding (a process implicated in cell motility), assembly of the quaternary structure of protein complexes, and also in the regulation of gene transcription and chromatin remodeling, among others, it would not be surprising that PFDN gene expression level and functions were altered in cancer cells or cancer patients. The expression of the 9 human PFDN genes evaluated in over 10,000 cancer patient samples from The Cancer Genome Atlas (TCGA) across more than 30 cancer types showed a general upregulation of the PFDN genes in tumoral tissues compared to matched normal tissues (Hadizadeh Esfahani et al., 2018). Nevertheless, since PFDNs can assemble into two different protein complexes with distinct functions, together with the notion that some PFDNs are thought to exert part of their functions in a PFDN complex-independent manner, a more detailed analysis of the gene expression of the nine PFDN genes is required to understand their role in the different cancer types.
High mRNA or protein levels of PFDNs are already described in many different cancer types and are correlated with poor clinical outcomes, as described herein. Using publicly available mRNA expression data from TCGA, we analyzed the expression level of the nine PFDN genes across the 18 cancer types for which enough tumoral and normal adjacent tissue samples (sample size >8) were available to perform a differential expression analysis (total number of sample size more than 8,100 samples) (Table 2). Statistically significant mRNA expression level changes were found in most of the PFDN genes across cancer types (Figure 7; adjusted p value <0.005 after FDR correction of the p value from Wald test). Most of these changes represent an upregulation in the cancer tissue compared to the adjacent normal one, reaching up to a 3-fold increase. Clustering analysis of mRNA expression levels present two distinct clusters, where PDFN4, PDRG1 (PFDN4r), PFDN6, and PFDN2 share a very similar gene expression level in all the tested cancer types, suggesting a common gene expression regulation mechanism of these genes in cancer patients. Notably, these 4 PFDNs belong to the class β PFDNs. In the mRNA levels of the rest of the PFDNs, containing all the α-class prefoldins, this common upregulated pattern was not so clearly found.
As shown in Figure 7, not all the components of the classic PFDN complex followed the same regulation in the different cancer types. The same conclusions were obtained for the components of the UPC, suggesting that either the different components of the PFDN protein complexes are not regulated in the same fashion in cancer tissues or the expression of the mRNA of the different PFDNs is regulated by different mechanisms across cancer types. These results are in agreement with the fact that the each PFDN’s genes are located in different coordinates of the genome, even in different chromosomes, which also suggests that different regulatory mechanisms are very likely to be found for the different PFDN genes (Atkinson and Halfon, 2014).
Importantly, amplification in the PFDN genes results in a statistically significant increase of the mRNA levels as discussed previously. Taken together, these observations indicate that there is a cancer-type specific CNA amplification in certain PFDN genes, which causes the upregulation of the mRNA levels of those genes. Nevertheless, in most of the cancer types analyzed, an upregulation of the PFDN genes occurs, especially in the case of the β PFDNs, caused by mechanisms different from genomic amplifications.
mRNA expression levels of PFDNs from the classic complex
In a recent study, the higher expression level of the whole classic PFDN complex has shown bad prognostic value in gastric cancer (Yesseyeva et al., 2020), especially in the case of PFDN2. The classic PFDNs have been also studied in dysplastic nevus cells, which can develop into cutaneous melanomas (Goldstein and Tucker, 2013). Differential gene expression analysis revealed that dysplastic nevus tissues present a significant upregulation of the PFDN family when compared to normal adjacent tissues, probably as part of the protein folding response triggered by the increase in reactive oxygen species (Gao et al., 2012). However, our data were not conclusive since the number of control samples for skin cutaneous melanoma (SKCM) was too low (n = 1) to perform a differential expression analysis, and no control samples were available for uveal melanoma (UVM) (Table 2).
Protein level of PFDN1 is upregulated in colorectal cancers when compared to adjacent tissues and this upregulation positively correlated with tumor size and invasion, whereas negatively correlated with survival time (Wang et al., 2015a). Moreover, downregulation of PFDN1 in colorectal cancer cells inhibits proliferation due to cell cycle arrest, caused by defects in cytoskeletal remodeling, inhibits cell motility and decreases tumor burden in nude mice (Wang et al., 2015a). We were also able to detect a significant, although moderate, increase (fold change of 0.35 between the tumor tissue and the normal adjacent tissue; p value < 5.0E-3) of PFDN1 mRNA expression in TCGA colorectal cancer cohort (Figure 7). Enhanced PFDN1 expression was also observed in lung cancer cells undergoing EMT (Wang et al., 2017). In these cells, PFDN1 was shown to bind cyclin A promoter, suppressing its transcription, and cyclin A overexpression rescued the phenotype observed upon PFDN1 overexpression (Wang et al., 2017), consisting of an increase in cell proliferation, migration, and invasion, although we were not able detect any upregulation of PFDN1 in lung cancers as shown by our TCGA study (Figure 7).
PFDN3 was reported to be downregulated during differentiation in neuronal cells, and it has been proposed to act as an oncogene through its binding to the VHL tumor suppressor protein in neuroblastoma (Cimmino et al., 2007). We could not evaluate the differential expression of PFDNs in this cancer type due to the low number of control samples (Table 2).
Moreover, upregulation of PFDN4 was reportedly detected in B cell non-Hodgkin’s lymphoma, a cancer type for which TCGA samples are not available to evaluate the differential gene expression (Table 2). A common feature in this cancer type is the fusion of histone H4 and Bcl-6 as a result of chromosome rearrangements. Interestingly, overexpression of this fusion protein in kidney fibroblasts resulted in the upregulation of the PFDN4 gene, although the downstream effects of upregulation were not addressed (Kurata et al., 2002). Conversely, a higher expression of PFDN4 in colorectal samples correlated with a better patient survival but also associated with the development of metastasis (Miyoshi et al., 2010). Knockdown of PFDN4 resulted in increased cell proliferation and invasion in these samples (Miyoshi et al., 2010). Thus, the effects of the modulation of PFDN4 mRNA levels in cancer is still to be clarified.
PFDN5 has also been shown upregulated in pancreatic cancer cell lines when compared to primary cell lines (Alldinger et al., 2005). Although we could not corroborate this observation using our TCGA datasets because of the limited number of normal pancreatic tissue (n = 4) to make the comparison with the pancreatic tumors, a significant upregulation of PFDN5 in kidney renal papillary and clear cell carcinomas (KIRP and KIRC) was detected (fold change of 0.9; p value <0.005) that would deserve further characterization.
Additional studies in PFDN5 have been performed and its relationship with cancer has been evaluated. In parallel to its discovery as a prefoldin, PFDN5 was also identified as a Myc modulator by yeast two-hybrid screening, and termed MM-1 (Myc Modulator 1) (Mori et al., 1998). PFDN5 was proposed as a negative regulator of c-Myc by several mechanisms that include direct binding to c-Myc box II domain to repress its transcriptional activity (Kimura et al., 2007; Mori et al., 1998); recruitment of HDAC repression complex to c-Myc via interaction with KAP1/TIF1β (Satou et al., 2001, 2004); and c-Myc degradation through E3 ubiquitin ligase complexes involving Cullin2 and SKP2 (Kimura et al., 2007) and Rabring7 (Yoshida et al., 2008). Overexpression of PFDN5 repressed c-Myc transcriptional activity whereas its downregulation showed the opposite effects. In this regard, c-Myc repression abolishes cell proliferation and increases senescence (Chen et al., 2018). Furthermore, the mutated version of PFDN5 (mistakenly termed as A157R, which corresponds to the A144 in the wild type protein) found in cancer samples, especially in lymphoma, was shown to relieve its repressive effects on c-Myc (Fujioka et al., 2001). An additional link between PFDN5 and c-Myc was proposed by Yoshida et al., who demonstrated that PFDN5 downregulation increased the expression of wnt4, an activator of the Wnt signalling pathway known to activate c-Myc transcription (Yoshida et al., 2008).
Interestingly, different modulators of PFDN5 have been identified. Interaction of PFDN5 with Δp63α in human cells leads to proteasomal degradation of PFDN5 (Han et al., 2016), likely mediated by the E3 ubiquitin ligase HERC3 (Chen et al., 2018). Further evidences showed low expression of PFDN5 together with enhanced expression of p63, HERC3, and c-Myc in breast carcinoma samples (Chen et al., 2018).
Upregulation of PFDN6 protein level was detected in several cancers, including brain, colon, thyroid, breast, and ovarian (Ostrov et al., 2007), as consistent with our observations (Figure 7). In a different study using the DEXA-resistant cell line as a model for acute lymphoblastic leukemia (ALL) poor responding patients, mRNA PFDN6 levels were found downregulated, as it also happens in bone marrow samples of ALL patients (Dehghan-Nayeri et al., 2017). Hence, low levels of PFDN6 were proposed to serve as a targeting marker for poor prognosis in childhood ALL (Dehghan-Nayeri et al., 2017). However, we could not evaluate the mRNA expression of PFDNs in ALL due to a lack of the mRNA expression analysis in this cancer type from TCGA (Table 2). It is also important to consider that PFDN6 forms part of both the classic and the UPC, and therefore its differential regulation and expression might be linked either to classic PFDNs or to UPC.
Expression of PFDNs from the UPC
Several studies have focused on studying the role of URI in carcinogenesis, and increasing evidence exists supporting the idea of URI being an oncogene whose overexpression enhances cell proliferation, migration, and invasion, as demonstrated in multiple cancer types such as ovarian, cervical, liver, prostate, gastric, and colorectal, and in multiple myeloma (Fan et al., 2014; Gu et al., 2015; Hu et al., 2016; Lipinski et al., 2016; Luo et al., 2016; Mita et al., 2016; Theurillat et al., 2011; Tummala et al., 2014; Xing et al., 2019; Zhang et al., 2018).
In particular, in liver cancer URI mRNA and protein levels in hepatocellular carcinoma (HCC) samples are significantly increased when compared to peritumoral tissues and URI overexpression correlates with poor patient survival in HCC (Tummala et al., 2014). Moreover, this high URI expression level is associated with hepatitis B and C virus (HBV and HCV) infection, both well known risk factors for HCC (Tummala et al., 2014). Further evidences suggest that URI amplicon is not amplified in human HCC, but its promoter is regulated by HBV X protein (HBx), the oncoprotein of HBV, thereby leading to its expression and most likely tumorigenesis (Tummala et al., 2014). Indeed, overexpression of URI in mouse hepatocytes leads to tumor formation (Tummala et al., 2014). We also detect a significant URI increase in liver cancer (LIHC) in our analysis (Figure 7; p. value <5.0E-3). The first mechanism linking URI and oncogenesis was described by Tummala et al. in 2014 in HCC, showing that URI inhibits aryl hydrocarbon receptor (AhR) and estrogen receptor (ER) signaling, causing a decrease in transcription of NAD+ metabolism and thus increasing DNA damage. This increased DNA damage upon URI overexpression was proposed to trigger the transformation process in the cells. URI is also highly expressed in endometrioid adenocarcinoma and its expression is even increased in high grade tumors (Gu et al., 2013).
As described for URI in HCC, it has been proposed that in breast cancer cells, STAP1 binds to estrogen receptor (ER), preventing its translocation to the nucleus and thus its transcriptional activity. On the other hand, the tumor suppressor lysyl oxidase propeptide (LOX-PP) binds to STAP1 and promotes its degradation, allowing ER to activate transcription and protecting against the acquisition of oncogenic properties (Sanchez-Morgan et al., 2017).
Furthermore, URI is overexpressed in multiple myeloma (MM) patients and cell lines derived from MM (Fan et al., 2014), and leads to transcription of IL-6, which through autocrine mechanisms promote MM cell survival and resistance to chemotherapy (Fan et al., 2014). In vitro treatment of melanoma cell lines with a mutated-BRAF inhibitor used in the clinic caused the disruption of the interaction between the URI PFDN-like complex and the RNA polymerases that leads to their destabilization (Frischknecht et al., 2019).
Other studies revealed that certain colorectal cell lines, but not all, are URI-dependent since silencing of URI with shRNA leads to cell death (Lipinski et al., 2016). The URI-dependent colorectal cell lines are also PFDN3 dependent, indicating that the dependency of URI might be through its function in the UPC. Depletion of URI activates p53-mediated apoptosis and diminishes the tumor growth when URI-dependent colorectal cells were injected into nude mice, indicating that URI expression protects cells against apoptosis (Lipinski et al., 2016).
URI was also proposed to regulate transcriptionally LINE-1 retrotransposon in prostate cancer cells. URI binds to KAP1 to facilitate its dephosphorylation by PP2A and thus preventing transcriptional activity. Upon URI loss, transcription of LINE-1 retrotransposon is increased and can be integrated in various genomic locations, thereby causing mutation-driven cancer (Mita et al., 2016). In this case, it would be the loss of URI or its translocation to the cytoplasm that would have oncogenic properties.
The role of URI in gastric cancer has been also studied. Overexpressing URI in gastric cancer cells are protected against apoptosis induced by Adriamycin (Hu et al., 2016). Additional mechanisms linking URI and oncogenesis have been studied in gastric cancer cells. Potassium dichromate induces oxidative stress and DNA damage in cells, effects that were exacerbated upon URI knockout in vitro leading to apoptosis. Then, it is speculated that URI expression would protect cancer cells against DNA damage and apoptosis, contributing to chemoresistance (Luo et al., 2016). Furthermore, knockdown of URI in vitro induces autophagy, probably through the mTOR/p70S6K pathway to which URI has been previously linked (Zhang et al., 2018).
In ovarian cancer, amplification of URI was proved to be required for tumor initiation and progression, likely through alterations in the signaling hub in which URI participates together with PP1gamma and S6K1. Overexpression of URI causes the continuous activation of this signaling branch, conferring an enhanced survival signaling to the cancer cells (Theurillat et al., 2011). Additional mechanisms linking URI and ovarian cancer were discovered later on, thanks to in vitro experiments demonstrating that URI is a target for microRNA-598. Overexpression of this microRNA in the cells caused a reduction in proliferation, migration, and invasion; and microRNA-598 overexpression cells inoculated into mice to assess tumor growth validated those results in vivo (Xing et al., 2019). The role of URI as an oncogene has been also proved in cervical cancer cells, since overexpression of URI promotes cell proliferation, invasion, migration, EMT and cisplatin resistance, whereas its downregulation causes the opposite effect (Gu et al., 2015).
STAP1 is overexpressed in breast tumors and derived cell lines when compared to normal tissues and presents high protein levels in bladder, breast, ovary, and thyroid tumor tissues (Zhao et al., 2005), suggesting that STAP1 has oncogenic activities. However, this increase was not detected in our study (Figure 7) and recent data indicate that STAP1 expression was lost in high-grade prostatic intraepithelial neoplasia and prostate cancer samples when compared to normal tissues and in benign prostatic hyperplasia samples (Wang et al., 2019). In line with the above results describing regulation of LINE-1 by URI, knocking out STAP1 in murine prostate cells leads to intraepithelial neoplasias in the tissue, probably due to the promotion of retrotransposition of LINE-1 elements, likely mediated by the interaction of this protein with KAP1 (Wang et al., 2019).
Consistent with our observations (Figure 7), increased PFDN4r has been reported in a variety of tumors (reviewed in Pajares, 2017). Nevertheless, the function of PFDN4r has been only addressed in bladder cancer. The effect of PFDN4r in this cancer type was identified as it is a direct downstream target of the microRNA-214. Either microRNA-214 expression or PFDN4r knockdown attenuates cell proliferation and migration in vitro (Wang et al., 2015b).
Finally, treatment of prostate cancer cell lines with antitumoral drugs such as dictyoceratin-A and -C identified the UPC component RPAP3 as a target of these compounds. Treated cells showed an inhibition of RPAP3 and the whole R2TP/PFDN complex, preventing its function as a stabilizer for PIKKs such as mTOR and causing cell death under hypoxic conditions (Kawachi et al., 2019).
Discussion
The PFDN family of proteins is evolutionary conserved and contributes to maintain cellular homeostasis, not only by assuring a proper protein folding of cytoskeletal proteins and large macromolecular complexes within the cell but also by interacting with different proteins, which confers them a wider range of functions. Yet, the full list of PFDNs’ interactors, their functions in the cell and their regulation remains to be fully understood.
Most of the studies evaluating the role of the classic PFDN complex focused the investigations in single PFDN genes or proteins, making it difficult to distinguish if effects seen on tumorigenesis are caused by the contribution of the whole PFDN complex (or combination of members in PFDN complex) or by independent functions of PFDN either acting alone or with interactions between components. As a general trend, PFDNs are upregulated in various cancers and they might promote tumorigenesis, although organ-specific tumor suppressive or oncogenic functions cannot be excluded. The generation of genetically engineered mouse models for all PFDNs is critical to better decipher their functions during tumorigenesis.
URI is the PFDN protein that received more attention in recent years. Lessons from mice indicate that URI levels might be critical to perform either oncogenic or tumor suppressive functions, through its ability to directly bind and regulate the activity of diverse proteins including transcription factors (ERα and AhR), enzymes (PP1γ and OGT), tumor suppressors (parafibromin), and proteins with oncogenic roles (β catenin) (Figure 4).
The ability of URI to be considered as a co-chaperone and to modulate activity of various partner proteins suggests that URI uncovers buffering activities, surmising the hypothesis that PFDNs might be fundamental to neutralize the deleterious effects of environmental stress and maintain cellular and protein homeostasis (Figure 8). Perturbations in URI levels might affect the quality control mechanisms required to reduce or eliminate the impact of environmental stress-induced protein misfolding and avoid proteotoxicity, which could lead to cancer and many other diseases. To fully elucidate the functions of the classical PFDN complex or UPC, having more information on the organization and structure of both protein complexes will certainly help us to obtain a comprehensive mechanistic understanding of their functions and roles at cellular and tissue levels and in diseases such as cancer.
Figure 8.
PFDNs contribute to cellular adaptive responses to stress
Different pathology-related environmental factors disrupt cellular functions, thus inducing a cellular response in order to restore homeostasis. PFDNs contribute to this cellular buffering response, either by their functions as PFDN complexes (classic or UPC) or by their actions as single proteins, as demonstrated for URI. PFDNs ensure a proper protein folding, as well as a proper regulation of cell cycle and survival, thus contributing to restore cellular homeostasis. When PFDNs' buffering activities are not enough to compensate the stress or when PFDN protein levels are altered, cell functions result dysregulated, leading to different pathological conditions.
Limitations of the study
The analyses included in this manuscript were performed using the GTEx and TCGA datasets for RNA-Seq data. Although these datasets have already given us sufficient information, similar analyses using additional datasets could have been performed to corroborate our findings, in particular for checking gene aberrations, mutations and mRNA expression of PFDNs. Moreover, protein analysis and functional studies would have shed light on the biological relevance of our findings. Finally, information regarding the recently discovered protein ASDURF that is suggested to be a novel PFDN was not included in these analyses since it was not annotated as such in the used datasets, and further work is required to determine whether ASDURF is really part of the UPC.
Acknowledgments
The results published in this study are in part based on data generated by The Cancer Genome Atlas project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions that constitute the TCGA research network can be found at https://cancergenome.nih.gov/. We also acknowledge the Genotype-Tissue Expression (GTEx) program. This work was funded by the State Research Agency (AEI, https://doi.org/10.13039/501100011033) from the Spanish Ministry of Science and Innovation (projects SAF2016-76598-R, SAF2017-92733-EXP, RTI2018-094834-B-I00 and RED2018-102723-T), cofunded by European Regional Development Fund (ERDF). This work was developed at the CNIO funded by the Health Institute Carlos III (ISCIII) and the Spanish Ministry of Science and Innovation.
Author contributions
I.H.M. searched and analyzed most of the data for the article and drew all figures. S.P. assisted in the analysis of some data. I.H.M. and N.D. discussed the content and wrote the manuscript. N.D. supervised the study and secured funding.
Declaration of interests
The authors declare no competing interests.
References
- Abe A., Takahashi-Niki K., Takekoshi Y., Shimizu T., Kitaura H., Maita H., Iguchi-Ariga S.M., Ariga H. Prefoldin plays a role as a clearance factor in preventing proteasome inhibitor-induced protein aggregation. J. Biol. Chem. 2013;288:27764–27776. doi: 10.1074/jbc.M113.476358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikawa Y., Kida H., Nishitani Y., Miki K. Expression, purification, crystallization and X-ray diffraction studies of the molecular chaperone prefoldin from Homo sapiens. Acta Crystallogr. F Struct. Biol. Commun. 2015;71:1189–1193. doi: 10.1107/S2053230X15013990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajjappala H., Chung H.Y., Sim J.S., Choi I., Hahn B.S. Disruption of prefoldin-2 protein synthesis in root-knot nematodes via host-mediated gene silencing efficiently reduces nematode numbers and thus protects plants. Planta. 2015;241:773–787. doi: 10.1007/s00425-014-2211-2. [DOI] [PubMed] [Google Scholar]
- Alldinger I., Dittert D., Peiper M., Fusco A., Chiappetta G., Staub E., Lohr M., Jesnowski R., Baretton G., Ockert D. Gene expression analysis of pancreatic cell lines reveals genes overexpressed in pancreatic cancer. Pancreatology. 2005;5:370–379. doi: 10.1159/000086537. [DOI] [PubMed] [Google Scholar]
- Amorim A.F., Pinto D., Kuras L., Fernandes L. Absence of Gim proteins, but not GimC complex, alters stress-induced transcription. Biochim. Biophys. Acta Gene Regul. Mech. 2017;1860:773–781. doi: 10.1016/j.bbagrm.2017.04.005. [DOI] [PubMed] [Google Scholar]
- Atkinson T.J., Halfon M.S. Regulation of gene expression in the genomic context. Comput. Struct. Biotechnol. J. 2014;9:e201401001. doi: 10.5936/csbj.201401001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyaz A., Kumar S., Sangiorgi B., Ghoshal B., Gosio J., Ouladan S., Fink M., Barutcu S., Trcka D., Shen J. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature. 2019;569:121–125. doi: 10.1038/s41586-019-1154-y. [DOI] [PubMed] [Google Scholar]
- Balchin D., Hayer-Hartl M., Hartl F.U. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- Banks C.A.S., Miah S., Adams M.K., Eubanks C.G., Thornton J.L., Florens L., Washburn M.P. Differential HDAC1/2 network analysis reveals a role for prefoldin/CCT in HDAC1/2 complex assembly. Sci. Rep. 2018;8:13712. doi: 10.1038/s41598-018-32009-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J., Hennecke S., Bornemann-Kolatzki K., Urnovitz H.B., Neumann S., Strobel P., Kaup F.J., Brenig B., Schutz E. Genome aberrations in canine mammary carcinomas and their detection in cell-free plasma DNA. PLoS ONE. 2013;8:e75485. doi: 10.1371/journal.pone.0075485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard H., Teijeiro A., Chaves-Pérez A., Perna C., Satish B., Novials A., Wang J.P., Djouder N. Coxsackievirus B type 4 infection in β cells downregulates the chaperone prefoldin URI to induce a MODY4-like diabetes via Pdx1 silencing. Cell Rep Med. 2020;1:100125. doi: 10.1016/j.xcrm.2020.100125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverly K.N., Sawaya M.R., Schmid E., Koehler C.M. The Tim8-Tim13 complex has multiple substrate binding sites and binds cooperatively to Tim23. J. Mol. Biol. 2008;382:1144–1156. doi: 10.1016/j.jmb.2008.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizarro J., Dodre M., Huttin A., Charpentier B., Schlotter F., Branlant C., Verheggen C., Massenet S., Bertrand E. NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins. Nucl. Acids Res. 2015;43:8973–8989. doi: 10.1093/nar/gkv809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonyaratanakornkit B.B., Simpson A.J., Whitehead T.A., Fraser C.M., El-Sayed N.M., Clark D.S. Transcriptional profiling of the hyperthermophilic methanarchaeon Methanococcus jannaschii in response to lethal heat and non-lethal cold shock. Environ. Microbiol. 2005;7:789–797. doi: 10.1111/j.1462-2920.2005.00751.x. [DOI] [PubMed] [Google Scholar]
- Boulon S., Marmier-Gourrier N., Pradet-Balade B., Wurth L., Verheggen C., Jady B.E., Rothe B., Pescia C., Robert M.C., Kiss T. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J.Cell Biol. 2008;180:579–595. doi: 10.1083/jcb.200708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broer L., Ikram M.A., Schuur M., DeStefano A.L., Bis J.C., Liu F., Rivadeneira F., Uitterlinden A.G., Beiser A.S., Longstreth W.T. Association of HSP70 and its co-chaperones with Alzheimer's disease. J. Alzheimers Dis. 2011;25:93–102. doi: 10.3233/JAD-2011-101560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneel A., Labas V., Mailloux A., Sharma S., Vinh J., Vaubourdolle M., Baudin B. Proteomic study of human umbilical vein endothelial cells in culture. Proteomics. 2003;3:714–723. doi: 10.1002/pmic.200300409. [DOI] [PubMed] [Google Scholar]
- Buren S., Gomes A.L., Teijeiro A., Fawal M.A., Yilmaz M., Tummala K.S., Perez M., Rodriguez-Justo M., Campos-Olivas R., Megias D. Regulation of OGT by URI in response to glucose confers c-MYC-dependent survival mechanisms. Cancer Cell. 2016;30:290–307. doi: 10.1016/j.ccell.2016.06.023. [DOI] [PubMed] [Google Scholar]
- Calderwood S.K., Khaleque M.A., Sawyer D.B., Ciocca D.R. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem. Sci. 2006;31:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Cao J. Analysis of the prefoldin gene family in 14 plant species. Front Plant Sci. 2016;7:317. doi: 10.3389/fpls.2016.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Carlesso G., Osipovich A.B., Llanes J., Lin Q., Hoek K.L., Khan W.N., Ruley H.E. Subunit 1 of the prefoldin chaperone complex is required for lymphocyte development and function. J. Immunol. 2008;181:476–484. doi: 10.4049/jimmunol.181.1.476. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol. 2002;52:7–76. doi: 10.1099/00207713-52-1-7. [DOI] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.C., Piaggi P., Hanson R.L., Knowler W.C., Bogardus C., Krakoff J. Autoantibodies against PFDN2 are associated with an increased risk of type 2 diabetes: a case-control study. Diabetes Metab. Res. Rev. 2017;33:e2922. doi: 10.1002/dmrr.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves-Pérez A., Thompson S., Djouder N. Roles and functions of the unconventional prefoldin URI. Adv. Exp. Med. Biol. 2018;1106:95–108. doi: 10.1007/978-3-030-00737-9_7. [DOI] [PubMed] [Google Scholar]
- Chen H., Yang L., Zhang Y., Yang S. Over-expression and characterization of recombinant prefoldin from hyperthermophilic archaeum Pyrococcus furiosus in E. coli. Biotechnol. Lett. 2010;32:429–434. doi: 10.1007/s10529-009-0156-5. [DOI] [PubMed] [Google Scholar]
- Chaves-Perez A., Yilmaz M., Perna C., de la Rosa S., Djouder N. URI is required to maintain intestinal architecture during ionizing radiation. Science. 2019;364(6443) doi: 10.1126/science.aaq1165. [DOI] [PubMed] [Google Scholar]
- Chen Y., Li Y., Peng Y., Zheng X., Fan S., Yi Y., Zeng P., Chen H., Kang H., Zhang Y. ΔNp63α down-regulates c-Myc modulator MM1 via E3 ligase HERC3 in the regulation of cell senescence. Cell Death Differ. 2018;25:2118–2129. doi: 10.1038/s41418-018-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnel F., Couturier A., Alusse A., Gagné J.P., Poirier G.G., Jean D., Boisvert F.M., Hascoet P., Paillard L., Arlot-Bonnemains Y. The prefoldin complex stabilizes the von Hippel-Lindau protein against aggregation and degradation. Plos Genet. 2020;16:e1009183. doi: 10.1371/journal.pgen.1009183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintalapudi S.R., Morales-Tirado V.M., Williams R.W., Jablonski M.M. Multipronged approach to identify and validate a novel upstream regulator of Sncg in mouse retinal ganglion cells. FEBS J. 2016;283:678–693. doi: 10.1111/febs.13620. [DOI] [PubMed] [Google Scholar]
- Choi J.S., Cho Y.K., Yoon S.H., Kwon S.O., Koo D.B., Yu K. Proteomic analysis of porcine pancreas development. BMB Rep. 2009;42:661–666. doi: 10.5483/bmbrep.2009.42.10.661. [DOI] [PubMed] [Google Scholar]
- Cimmino F., Spano D., Capasso M., Zambrano N., Russo R., Zollo M., Iolascon A. Comparative proteomic expression profile in all-trans retinoic acid differentiated neuroblastoma cell line. J. Proteome Res. 2007;6:2550–2564. doi: 10.1021/pr060701g. [DOI] [PubMed] [Google Scholar]
- Cloutier P., Coulombe B. New insights into the biogenesis of nuclear RNA polymerases? Biochem.Cell Biol. 2010;88:211–221. doi: 10.1139/o09-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier P., Poitras C., Durand M., Hekmat O., Fiola-Masson E., Bouchard A., Faubert D., Chabot B., Coulombe B. R2TP/Prefoldin-like component RUVBL1/RUVBL2 directly interacts with ZNHIT2 to regulate assembly of U5 small nuclear ribonucleoprotein. Nat. Commun. 2017;8:15615. doi: 10.1038/ncomms15615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier P., Poitras C., Faubert D., Bouchard A., Blanchette M., Gauthier M.S., Coulombe B. Upstream ORF-encoded ASDURF is a novel prefoldin-like subunit of the PAQosome. J. Proteome Res. 2020;19:18–27. doi: 10.1021/acs.jproteome.9b00599. [DOI] [PubMed] [Google Scholar]
- Collins C., Volik S., Kowbel D., Ginzinger D., Ylstra B., Cloutier T., Hawkins T., Predki P., Martin C., Wernick M. Comprehensive genome sequence analysis of a breast cancer amplicon. Genome Res. 2001;11:1034–1042. doi: 10.1101/gr.174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comyn S.A., Young B.P., Loewen C.J., Mayor T. Prefoldin promotes proteasomal degradation of cytosolic proteins with missense mutations by maintaining substrate solubility. Plos Genet. 2016;12:e1006184. doi: 10.1371/journal.pgen.1006184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amaro A., Valenti A., Napoli A., Rossi M., Ciaramella M. The prefoldin of the crenarchaeon Sulfolobus solfataricus. Protein Pept. Lett. 2008;15:1055–1062. doi: 10.2174/092986608786071094. [DOI] [PubMed] [Google Scholar]
- Danno A., Fukuda W., Yoshida M., Aki R., Tanaka T., Kanai T., Imanaka T., Fujiwara S. Expression profiles and physiological roles of two types of prefoldins from the hyperthermophilic archaeon Thermococcus kodakaraensis. J. Mol. Biol. 2008;382:298–311. doi: 10.1016/j.jmb.2008.07.032. [DOI] [PubMed] [Google Scholar]
- de Andrade Rosa I., Caruso M.B., de Oliveira Santos E., Gonzaga L., Zingali R.B., de Vasconcelos A.T.R., de Souza W., Benchimol M. The costa of trichomonads: a complex macromolecular cytoskeleton structure made of uncommon proteins. Biol.Cell. 2017;109:238–253. doi: 10.1111/boc.201600050. [DOI] [PubMed] [Google Scholar]
- Dehghan-Nayeri N., Rezaei-Tavirani M., Omrani M.D., Gharehbaghian A., Goudarzi Pour K., Eshghi P. Identification of potential predictive markers of dexamethasone resistance in childhood acute lymphoblastic leukemia. J.Cell Commun Signal. 2017;11:137–145. doi: 10.1007/s12079-016-0357-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N., Wieland U., Rangone H., Pinson X., Mao G., Dzhindzhev N.S., McLean D., Riparbelli M.G., Llamazares S., Callaini G. Drosophila Mgr, a Prefoldin subunit cooperating with von Hippel Lindau to regulate tubulin stability. Proc. Natl. Acad. Sci. U S A. 2012;109:5729–5734. doi: 10.1073/pnas.1108537109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplazes A., Mockli N., Luke B., Auerbach D., Peter M. Yeast Uri1p promotes translation initiation and may provide a link to cotranslational quality control. EMBO J. 2009;28:1429–1441. doi: 10.1038/emboj.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao W.F., Chen W.Q., Hoger H., Shim K.S., Pollak A., Lubec G. The hippocampal protein machinery varies over the estrous cycle. Proteomics Clin. Appl. 2007;1:1462–1475. doi: 10.1002/prca.200700333. [DOI] [PubMed] [Google Scholar]
- Djouder N., Metzler S.C., Schmidt A., Wirbelauer C., Gstaiger M., Aebersold R., Hess D., Krek W. S6K1-mediated disassembly of mitochondrial URI/PP1gamma complexes activates a negative feedback program that counters S6K1 survival signaling. Mol.Cell. 2007;28:28–40. doi: 10.1016/j.molcel.2007.08.010. [DOI] [PubMed] [Google Scholar]
- Dordick J.S. Functional nanoscale biomolecular materials. Biotechnol. J. 2013;8:165–166. doi: 10.1002/biot.201200338. [DOI] [PubMed] [Google Scholar]
- Echtenkamp F.J., Freeman B.C. Expanding the cellular molecular chaperone network through the ubiquitous cochaperones. Biochim. Biophys. Acta. 2012;1823:668–673. doi: 10.1016/j.bbamcr.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Enunlu I., Ozansoy M., Basak A.N. Alfa-class prefoldin protein UXT is a novel interacting partner of Amyotrophic Lateral Sclerosis 2 (Als2) protein. Biochem. Biophys. Res. Commun. 2011;413:471–475. doi: 10.1016/j.bbrc.2011.08.121. [DOI] [PubMed] [Google Scholar]
- Fan J.L., Zhang J., Dong L.W., Fu W.J., Du J., Shi H.G., Jiang H., Ye F., Xi H., Zhang C.Y. URI regulates tumorigenicity and chemotherapeutic resistance of multiple myeloma by modulating IL-6 transcription. Cell Death Dis. 2014;5:e1126. doi: 10.1038/cddis.2014.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S., Chen Y., Jiang Y., Hu K., Li C. Prefoldin subunit MM1 promotes cell migration via facilitating filopodia formation. Biochem. Biophys. Res. Commun. 2020;533:613–619. doi: 10.1016/j.bbrc.2020.09.063. [DOI] [PubMed] [Google Scholar]
- Fandrich M., Tito M.A., Leroux M.R., Rostom A.A., Hartl F.U., Dobson C.M., Robinson C.V. Observation of the noncovalent assembly and disassembly pathways of the chaperone complex MtGimC by mass spectrometry. Proc. Natl. Acad. Sci.U S A. 2000;97:14151–14155. doi: 10.1073/pnas.240326597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M., Hozzein W.N., Elsayed E.A., Taha N.A., Wadaan M.A. Identification of histone deacetylase 1 protein complexes in liver cancer cells. Asian Pac. J. Cancer Prev. 2013;14:915–921. doi: 10.7314/apjcp.2013.14.2.915. [DOI] [PubMed] [Google Scholar]
- Frischknecht L., Britschgi C., Galliker P., Christinat Y., Vichalkovski A., Gstaiger M., Kovacs W.J., Krek W. BRAF inhibition sensitizes melanoma cells to alpha-amanitin via decreased RNA polymerase II assembly. Sci. Rep. 2019;9:7779. doi: 10.1038/s41598-019-44112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y., Taira T., Maeda Y., Tanaka S., Nishihara H., Iguchi-Ariga S.M., Nagashima K., Ariga H. MM-1, a c-Myc-binding protein, is a candidate for a tumor suppressor in leukemia/lymphoma and tongue cancer. J. Biol. Chem. 2001;276:45137–45144. doi: 10.1074/jbc.M106127200. [DOI] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci.Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., van Nieuwpoort F.A., Out-Luiting J.J., Hensbergen P.J., de Snoo F.A., Bergman W., van Doorn R., Gruis N.A. Genome-wide analysis of gene and protein expression of dysplastic naevus cells. J. Skin Cancer. 2012;2012:981308. doi: 10.1155/2012/981308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S., Siegers K., Schiebel E. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J. 1998;17:952–966. doi: 10.1093/emboj/17.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut D., Roh S.H., Ma B., Pintilie G., Joachimiak L.A., Leitner A., Walzthoeni T., Aebersold R., Chiu W., Frydman J. The chaperonin TRiC/CCT associates with prefoldin through a conserved electrostatic interface essential for cellular proteostasis. Cell. 2019;177:751–765 e715. doi: 10.1016/j.cell.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Krzewska A.J., Farber E., Buttner E.A., Hunter C.P. Regulators of the actin cytoskeleton mediate lethality in a Caenorhabditis elegans dhc-1 mutant. Mol. Biol.Cell. 2010;21:2707–2720. doi: 10.1091/mbc.E09-07-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover D.J., Clark D.S. Oligomeric assembly is required for chaperone activity of the filamentous gamma-prefoldin. FEBS J. 2015;282:2985–2997. doi: 10.1111/febs.13341. [DOI] [PubMed] [Google Scholar]
- Glover D.J., Giger L., Kim J.R., Clark D.S. Engineering protein filaments with enhanced thermostability for nanomaterials. Biotechnol. J. 2013;8:228–236. doi: 10.1002/biot.201200009. [DOI] [PubMed] [Google Scholar]
- Glover D.J., Giger L., Kim S.S., Naik R.R., Clark D.S. Geometrical assembly of ultrastable protein templates for nanomaterials. Nat. Commun. 2016;7:11771. doi: 10.1038/ncomms11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.M., Tucker M.A. Dysplastic nevi and melanoma. Cancer Epidemiol. Biomarkers Prev. 2013;22:528–532. doi: 10.1158/1055-9965.EPI-12-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes A.L., Teijeiro A., Buren S., Tummala K.S., Yilmaz M., Waisman A., Theurillat J.P., Perna C., Djouder N. Metabolic inflammation-associated IL-17A causes non-alcoholic steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2016;30:161–175. doi: 10.1016/j.ccell.2016.05.020. [DOI] [PubMed] [Google Scholar]
- Gong Y., Kakihara Y., Krogan N., Greenblatt J., Emili A., Zhang Z., Houry W.A. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol. Syst. Biol. 2009;5:275. doi: 10.1038/msb.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales F.A., Zanchin N.I., Luz J.S., Oliveira C.C. Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-interacting protein that is involved in Pre-rRNA processing. J. Mol. Biol. 2005;346:437–455. doi: 10.1016/j.jmb.2004.11.071. [DOI] [PubMed] [Google Scholar]
- Gregersen N., Bross P. Protein misfolding and cellular stress: an overview. Methods Mol. Biol. 2010;648:3–23. doi: 10.1007/978-1-60761-756-3_1. [DOI] [PubMed] [Google Scholar]
- Gstaiger M., Luke B., Hess D., Oakeley E.J., Wirbelauer C., Blondel M., Vigneron M., Peter M., Krek W. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science. 2003;302:1208–1212. doi: 10.1126/science.1088401. [DOI] [PubMed] [Google Scholar]
- Gu J., Liang Y., Qiao L., Li X., Li X., Lu Y., Zheng Q. Expression analysis of URI/RMP gene in endometrioid adenocarcinoma by tissue microarray immunohistochemistry. Int. J. Clin. Exp. Pathol. 2013;6:2396–2403. [PMC free article] [PubMed] [Google Scholar]
- Gu J., Liang Y., Qiao L., Lu Y., Hu X., Luo D., Li N., Zhang L., Chen Y., Du J. URI expression in cervical cancer cells is associated with higher invasion capacity and resistance to cisplatin. Am. J. Cancer Res. 2015;5:1353–1367. [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Deng Z., Paredez A.R., DeBolt S., Wang Z.Y., Somerville C. Prefoldin 6 is required for normal microtubule dynamics and organization in Arabidopsis. Proc. Natl. Acad. Sci.U S A. 2008;105:18064–18069. doi: 10.1073/pnas.0808652105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ganguly A., Sun L., Suo F., Du L.L., Russell P. Global fitness profiling identifies arsenic and cadmium tolerance mechanisms in fission yeast. G3 (Bethesda) 2016;6:3317–3333. doi: 10.1534/g3.116.033829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadizadeh Esfahani A., Sverchkova A., Saez-Rodriguez J., Schuppert A.A., Brehme M. A systematic atlas of chaperome deregulation topologies across the human cancer landscape. Plos Comput. Biol. 2018;14:e1005890. doi: 10.1371/journal.pcbi.1005890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A., Li J., Li Y., Wang Y., Bergholz J., Zhang Y., Li C., Xiao Zh X. p63α modulates c-Myc activity via direct interaction and regulation of MM1 protein stability. Oncotarget. 2016;7:44277–44287. doi: 10.18632/oncotarget.10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen W.J., Cowan N.J., Welch W.J. Prefoldin-nascent chain complexes in the folding of cytoskeletal proteins. J.CellBiol. 1999;145:265–277. doi: 10.1083/jcb.145.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartill V.L., van de Hoek G., Patel M.P., Little R., Watson C.M., Berry I.R., Shoemark A., Abdelmottaleb D., Parkes E., Bacchelli C. DNAAF1 links heart laterality with the AAA+ ATPase RUVBL1 and ciliary intraflagellar transport. Hum. Mol. Genet. 2018;27:529–545. doi: 10.1093/hmg/ddx422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.U., Bracher A., Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hemmingsen S.M., Woolford C., van der Vies S.M., Tilly K., Dennis D.T., Georgopoulos C.P., Hendrix R.W., Ellis R.J. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature. 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Hennecke S., Beck J., Bornemann-Kolatzki K., Neumann S., Murua Escobar H., Nolte I., Hammer S.C., Hewicker-Trautwein M., Junginger J., Kaup F.J. Prevalence of the prefoldin subunit 5 gene deletion in canine mammary tumors. PLoS ONE. 2015;10:e0131280. doi: 10.1371/journal.pone.0131280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.E., Hemmingsen S.M. Arabidopsis thaliana type I and II chaperonins. Cell Stress Chaperones. 2001;6:190–200. doi: 10.1379/1466-1268(2001)006<0190:attiai>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintermair C., Voss K., Forne I., Heidemann M., Flatley A., Kremmer E., Imhof A., Eick D. Specific threonine-4 phosphorylation and function of RNA polymerase II CTD during M phase progression. Sci. Rep. 2016;6:27401. doi: 10.1038/srep27401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishikawa D., Hong Y.H., Roh S.G., Miyahara H., Nishimura Y., Tomimatsu A., Tsuzuki H., Gotoh C., Kuno M., Choi K.C. Identification of genes expressed differentially in subcutaneous and visceral fat of cattle, pig, and mouse. Physiol. Genomics. 2005;21:343–350. doi: 10.1152/physiolgenomics.00184.2004. [DOI] [PubMed] [Google Scholar]
- Hongo K., Itai H., Mizobata T., Kawata Y. Varied effects of Pyrococcus furiosus prefoldin and P. furiosus chaperonin on the refolding reactions of substrate proteins. J. Biochem. 2012;151:383–390. doi: 10.1093/jb/mvr141. [DOI] [PubMed] [Google Scholar]
- Horejsi Z., Takai H., Adelman C.A., Collis S.J., Flynn H., Maslen S., Skehel J.M., de Lange T., Boulton S.J. CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol.Cell. 2010;39:839–850. doi: 10.1016/j.molcel.2010.08.037. [DOI] [PubMed] [Google Scholar]
- Houry W.A., Bertrand E., Coulombe B. The PAQosome, an R2TP-based chaperone for quaternary structure formation. Trends Biochem. Sci. 2018;43:4–9. doi: 10.1016/j.tibs.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Hu X., Zhang F., Luo D., Li N., Wang Q., Xu Z., Bian H., Liang Y., Lu Y., Zheng Q. URI promotes gastric cancer cell motility, survival, and resistance to adriamycin in vitro. Am. J. Cancer Res. 2016;6:1420–1430. [PMC free article] [PubMed] [Google Scholar]
- Iizuka R., Sugano Y., Ide N., Ohtaki A., Yoshida T., Fujiwara S., Imanaka T., Yohda M. Functional characterization of recombinant prefoldin complexes from a hyperthermophilic archaeon, Thermococcus sp. strain KS-1. J. Mol. Biol. 2008;377:972–983. doi: 10.1016/j.jmb.2008.01.070. [DOI] [PubMed] [Google Scholar]
- Izumi N., Yamashita A., Hirano H., Ohno S. Heat shock protein 90 regulates phosphatidylinositol 3-kinase-related protein kinase family proteins together with the RUVBL1/2 and Tel2-containing co-factor complex. Cancer Sci. 2012;103:50–57. doi: 10.1111/j.1349-7006.2011.02112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun G.R., Chung J., Mez J., Barber R., Beecham G.W., Bennett D.A., Buxbaum J.D., Byrd G.S., Carrasquillo M.M., Crane P.K. Transethnic genome-wide scan identifies novel Alzheimer's disease loci. Alzheimers Dement. 2017;13:727–738. doi: 10.1016/j.jalz.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoyama K., Matsuura K., Takano M., Maekura K., Inoue Y., Matsuyama S. Changes in the expression of prefoldin subunit 5 depending on synaptic plasticity in the mouse hippocampus. Neurosci. Lett. 2019;712:134484. doi: 10.1016/j.neulet.2019.134484. [DOI] [PubMed] [Google Scholar]
- Kakihara Y., Makhnevych T., Zhao L., Tang W., Houry W.A. Nutritional status modulates box C/D snoRNP biogenesis by regulated subcellular relocalization of the R2TP complex. Genome Biol. 2014;15:404. doi: 10.1186/s13059-014-0404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai T., Takedomi S., Fujiwara S., Atomi H., Imanaka T. Identification of the Phr-dependent heat shock regulon in the hyperthermophilic archaeon, Thermococcus kodakaraensis. J. Biochem. 2010;147:361–370. doi: 10.1093/jb/mvp177. [DOI] [PubMed] [Google Scholar]
- Kanemaki M., Kurokawa Y., Matsu-ura T., Makino Y., Masani A., Okazaki K., Morishita T., Tamura T.A. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem. 1999;274:22437–22444. doi: 10.1074/jbc.274.32.22437. [DOI] [PubMed] [Google Scholar]
- Kawachi T., Tanaka S., Fukuda A., Sumii Y., Setiawan A., Kotoku N., Kobayashi M., Arai M. Target identification of the marine natural products dictyoceratin-A and -C as selective growth inhibitors in cancer cells adapted to hypoxic environments. Mar. Drugs. 2019;17(163) doi: 10.3390/md17030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida H., Sugano Y., Iizuka R., Fujihashi M., Yohda M., Miki K. Structural and molecular characterization of the prefoldin beta subunit from Thermococcus strain KS-1. J. Mol. Biol. 2008;383:465–474. doi: 10.1016/j.jmb.2008.08.041. [DOI] [PubMed] [Google Scholar]
- Kim J.A., Choi D.K., Min J.S., Kang I., Kim J.C., Kim S., Ahn J.K. VBP1 represses cancer metastasis by enhancing HIF-1α degradation induced by pVHL. FEBS J. 2018;285:115–126. doi: 10.1111/febs.14322. [DOI] [PubMed] [Google Scholar]
- Kim Y.E., Hipp M.S., Bracher A., Hayer-Hartl M., Hartl F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- Kimura Y., Nagao A., Fujioka Y., Satou A., Taira T., Iguchi-Ariga S.M., Ariga H. MM-1 facilitates degradation of c-Myc by recruiting proteasome and a novel ubiquitin E3 ligase. Int. J. Oncol. 2007;31:829–836. [PubMed] [Google Scholar]
- King T.H., Decatur W.A., Bertrand E., Maxwell E.S., Fournier M.J. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol.CellBiol. 2001;21:7731–7746. doi: 10.1128/MCB.21.22.7731-7746.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner J., Vissi E., Gross S., Szoor B., Rudenko A., Alphey L., White-Cooper H. Drosophila Uri, a PP1alpha binding protein, is essential for viability, maintenance of DNA integrity and normal transcriptional activity. BMC Mol. Biol. 2008;9:36. doi: 10.1186/1471-2199-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunker D., Haas B., Hirtreiter A., Figueiredo L., Naylor D.J., Pfeifer G., Muller V., Deppenmeier U., Gottschalk G., Hartl F.U. Coexistence of group I and group II chaperonins in the archaeon Methanosarcina mazei. J. Biol. Chem. 2003;278:33256–33267. doi: 10.1074/jbc.M302018200. [DOI] [PubMed] [Google Scholar]
- Kong X., Ma S., Guo J., Ma Y., Hu Y., Wang J., Zheng Y. Ubiquitously expressed transcript is a novel interacting protein of protein inhibitor of activated signal transducer and activator of transcription 2. Mol. Med. Rep. 2015;11:2443–2448. doi: 10.3892/mmr.2014.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korndorfer I.P., Dommel M.K., Skerra A. Structure of the periplasmic chaperone Skp suggests functional similarity with cytosolic chaperones despite differing architecture. Nat. Struct. Mol. Biol. 2004;11:1015–1020. doi: 10.1038/nsmb828. [DOI] [PubMed] [Google Scholar]
- Kurata M., Maesako Y., Ueda C., Nishikori M., Akasaka T., Uchiyama T., Ohno H. Characterization of t(3;6)(q27;p21) breakpoints in B-cell non-Hodgkin's lymphoma and construction of the histone H4/BCL6 fusion gene, leading to altered expression of Bcl-6. Cancer Res. 2002;62:6224–6230. [PubMed] [Google Scholar]
- Kurimoto E., Nishi Y., Yamaguchi Y., Zako T., Iizuka R., Ide N., Yohda M., Kato K. Dynamics of group II chaperonin and prefoldin probed by 13C NMR spectroscopy. Proteins. 2008;70:1257–1263. doi: 10.1002/prot.21606. [DOI] [PubMed] [Google Scholar]
- Kwon O.C., Lee E.J., Lee J.Y., Youn J., Kim T.H., Hong S., Lee C.K., Yoo B., Robinson W.H., Kim Y.G. Prefoldin 5 and anti-prefoldin 5 antibodies as biomarkers for uveitis in ankylosing spondylitis. Front Immunol. 2019;10:384. doi: 10.3389/fimmu.2019.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laksanalamai P., Pavlov A.R., Slesarev A.I., Robb F.T. Stabilization of Taq DNA polymerase at high temperature by protein folding pathways from a hyperthermophilic archaeon, Pyrococcus furiosus. Biotechnol. Bioeng. 2006;93:1–5. doi: 10.1002/bit.20781. [DOI] [PubMed] [Google Scholar]
- Le Goff X., Chesnel F., Delalande O., Couturier A., Dreano S., Le Goff C., Vigneau C., Arlot-Bonnemains Y. Aggregation dynamics and identification of aggregation-prone mutants of the von Hippel-Lindau tumor suppressor protein. J.Cell Sci. 2016;129:2638–2650. doi: 10.1242/jcs.184846. [DOI] [PubMed] [Google Scholar]
- Le Meur N., Gentleman R. Modeling synthetic lethality. Genome Biol. 2008;9:R135. doi: 10.1186/gb-2008-9-9-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Smith R.S., Jordan W., King B.L., Won J., Valpuesta J.M., Naggert J.K., Nishina P.M. Prefoldin 5 is required for normal sensory and neuronal development in a murine model. J. Biol. Chem. 2011;286:726–736. doi: 10.1074/jbc.M110.177352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux M.R., Fandrich M., Klunker D., Siegers K., Lupas A.N., Brown J.R., Schiebel E., Dobson C.M., Hartl F.U. MtGimC, a novel archaeal chaperone related to the eukaryotic chaperonin cofactor GimC/prefoldin. EMBO J. 1999;18:6730–6743. doi: 10.1093/emboj/18.23.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung S.Y., Ho C., Tu I.P., Li R., So S., Chu K.M., Yuen S.T., Chen X. Comprehensive analysis of 19q12 amplicon in human gastric cancers. Mod. Pathol. 2006;19:854–863. doi: 10.1038/modpathol.3800593. [DOI] [PubMed] [Google Scholar]
- Li Y., Zhao L., Yuan S., Zhang J., Sun Z. Axonemal dynein assembly requires the R2TP complex component Pontin. Development. 2017;144:4684–4693. doi: 10.1242/dev.152314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Prescott M.S., Zhu Z., Singh P., Chun S.Y., Kuick R.D., Hanash S.M., Orringer M.B., Glover T.W., Beer D.G. Identification and characterization of a 19q12 amplicon in esophageal adenocarcinomas reveals cyclin E as the best candidate gene for this amplicon. Cancer Res. 2000;60:7021–7027. [PubMed] [Google Scholar]
- Lipinski K.A., Britschgi C., Schrader K., Christinat Y., Frischknecht L., Krek W. Colorectal cancer cells display chaperone dependency for the unconventional prefoldin URI1. Oncotarget. 2016;7:29635–29647. doi: 10.18632/oncotarget.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A., Blazquez M.A., Alabadi D. Dynamic regulation of cortical microtubule organization through prefoldin-DELLA interaction. Curr. Biol. 2013;23:804–809. doi: 10.1016/j.cub.2013.03.053. [DOI] [PubMed] [Google Scholar]
- Lopez V., Gonzalez-Peramato P., Suela J., Serrano A., Algaba F., Cigudosa J.C., Vidal A., Bellmunt J., Heredero O., Sanchez-Carbayo M. Identification of prefoldin amplification (1q23.3-q24.1) in bladder cancer using comparative genomic hybridization (CGH) arrays of urinary DNA. J. Transl Med. 2013;11:182. doi: 10.1186/1479-5876-11-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loring J.F., Wen X., Lee J.M., Seilhamer J., Somogyi R. A gene expression profile of Alzheimer's disease. DNA CellBiol. 2001;20:683–695. doi: 10.1089/10445490152717541. [DOI] [PubMed] [Google Scholar]
- Lundin V.F., Srayko M., Hyman A.A., Leroux M.R. Efficient chaperone-mediated tubulin biogenesis is essential for cell division and cell migration in C. elegans. Dev. Biol. 2008;313:320–334. doi: 10.1016/j.ydbio.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Lundin V.F., Stirling P.C., Gomez-Reino J., Mwenifumbo J.C., Obst J.M., Valpuesta J.M., Leroux M.R. Molecular clamp mechanism of substrate binding by hydrophobic coiled-coil residues of the archaeal chaperone prefoldin. Proc. Natl. Acad. Sci.U S A. 2004;101:4367–4372. doi: 10.1073/pnas.0306276101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Xu Z., Hu X., Zhang F., Bian H., Li N., Wang Q., Lu Y., Zheng Q., Gu J. URI prevents potassium dichromate-induced oxidative stress and cell death in gastric cancer cells. Am. J. Transl Res. 2016;8:5399–5409. [PMC free article] [PubMed] [Google Scholar]
- Macaya-Sanz D., Chen J.G., Kalluri U.C., Muchero W., Tschaplinski T.J., Gunter L.E., Simon S.J., Biswal A.K., Bryan A.C., Payyavula R. Agronomic performance of Populus deltoides trees engineered for biofuel production. Biotechnol. Biofuels. 2017;10:253. doi: 10.1186/s13068-017-0934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado-Pinilla R., Liger D., Leulliot N., Meier U.T. Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA. 2012;18:1833–1845. doi: 10.1261/rna.034942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinova A., Cvackova Z., Mateju D., Horejsi Z., Abeza C., Vandermoere F., Bertrand E., Stanek D., Verheggen C. Assembly of the U5 snRNP component PRPF8 is controlled by the HSP90/R2TP chaperones. J.CellBiol. 2017;216:1579–1596. doi: 10.1083/jcb.201701165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Benito J., Boskovic J., Gomez-Puertas P., Carrascosa J.L., Simons C.T., Lewis S.A., Bartolini F., Cowan N.J., Valpuesta J.M. Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J. 2002;21:6377–6386. doi: 10.1093/emboj/cdf640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Benito J., Gomez-Reino J., Stirling P.C., Lundin V.F., Gomez-Puertas P., Boskovic J., Chacon P., Fernandez J.J., Berenguer J., Leroux M.R. Divergent substrate-binding mechanisms reveal an evolutionary specialization of eukaryotic prefoldin compared to its archaeal counterpart. Structure. 2007;15:101–110. doi: 10.1016/j.str.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Martinez-Fernandez V., Garrido-Godino A.I., Cuevas-Bermudez A., Navarro F. The yeast prefoldin Bud27. Adv. Exp. Med. Biol. 2018;1106:109–118. doi: 10.1007/978-3-030-00737-9_8. [DOI] [PubMed] [Google Scholar]
- Mas S., Gasso P., Parellada E., Bernardo M., Lafuente A. Network analysis of gene expression in peripheral blood identifies mTOR and NF-kappaB pathways involved in antipsychotic-induced extrapyramidal symptoms. Pharmacogenomics J. 2015;15:452–460. doi: 10.1038/tpj.2014.84. [DOI] [PubMed] [Google Scholar]
- Mattiazzi M., Jambhekar A., Kaferle P., Derisi J.L., Krizaj I., Petrovic U. Genetic interactions between a phospholipase A2 and the Rim101 pathway components in S. cerevisiae reveal a role for this pathway in response to changes in membrane composition and shape. Mol. Genet. Genomics. 2010;283:519–530. doi: 10.1007/s00438-010-0533-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeegan K.S., Debieux C.M., Boulon S., Bertrand E., Watkins N.J. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol.CellBiol. 2007;27:6782–6793. doi: 10.1128/MCB.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan-Zambrano G., Rodriguez-Gil A., Penate X., de Miguel-Jimenez L., Morillo-Huesca M., Krogan N., Chavez S. The prefoldin complex regulates chromatin dynamics during transcription elongation. Plos Genet. 2013;9:e1003776. doi: 10.1371/journal.pgen.1003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron-Garcia M.C., Garrido-Godino A.I., Garcia-Molinero V., Hernandez-Torres F., Rodriguez-Navarro S., Navarro F. The prefoldin bud27 mediates the assembly of the eukaryotic RNA polymerases in an rpb5-dependent manner. Plos Genet. 2013;9:e1003297. doi: 10.1371/journal.pgen.1003297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron-Garcia M.C., Garrido-Godino A.I., Martinez-Fernandez V., Fernandez-Pevida A., Cuevas-Bermudez A., Martin-Exposito M., Chavez S., de la Cruz J., Navarro F. The yeast prefoldin-like URI-orthologue Bud27 associates with the RSC nucleosome remodeler and modulates transcription. Nucl. Acids Res. 2014;42:9666–9676. doi: 10.1093/nar/gku685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita P., Savas J.N., Briggs E.M., Ha S., Gnanakkan V., Yates J.R., 3rd, Robins D.M., David G., Boeke J.D., Garabedian M.J. URI regulates KAP1 phosphorylation and transcriptional repression via PP2A phosphatase in prostate cancer cells. J. Biol. Chem. 2016;291:25516–25528. doi: 10.1074/jbc.M116.741660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita P., Savas J.N., Djouder N., Yates J.R., 3rd, Ha S., Ruoff R., Schafler E.D., Nwachukwu J.C., Tanese N., Cowan N.J. Regulation of androgen receptor-mediated transcription by RPB5 binding protein URI/RMP. Mol.CellBiol. 2011;31:3639–3652. doi: 10.1128/MCB.05429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita P., Savas J.N., Ha S., Djouder N., Yates J.R., 3rd, Logan S.K. Analysis of URI nuclear interaction with RPB5 and components of the R2TP/prefoldin-like complex. PLoS ONE. 2013;8:e63879. doi: 10.1371/journal.pone.0063879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa M., Tashiro E., Kitaura H., Maita H., Suto H., Iguchi-Ariga S.M., Ariga H. Prefoldin subunits are protected from ubiquitin-proteasome system-mediated degradation by forming complex with other constituent subunits. J. Biol. Chem. 2011;286:19191–19203. doi: 10.1074/jbc.M110.216259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N., Ishii H., Mimori K., Nishida N., Tokuoka M., Akita H., Sekimoto M., Doki Y., Mori M. Abnormal expression of PFDN4 in colorectal cancer: a novel marker for prognosis. Ann. Surg. Oncol. 2010;17:3030–3036. doi: 10.1245/s10434-010-1138-5. [DOI] [PubMed] [Google Scholar]
- Mori K., Maeda Y., Kitaura H., Taira T., Iguchi-Ariga S.M., Ariga H. MM-1, a novel c-Myc-associating protein that represses transcriptional activity of c-Myc. J. Biol. Chem. 1998;273:29794–29800. doi: 10.1074/jbc.273.45.29794. [DOI] [PubMed] [Google Scholar]
- Morita K., Yamamoto Y.Y., Hori A., Obata T., Uno Y., Shinohara K., Noguchi K., Noi K., Ogura T., Ishii K. Expression, functional characterization, and preliminary crystallization of the cochaperone prefoldin from the thermophilic fungus Chaetomium thermophilum. Int. J. Mol. Sci. 2018;19(2452) doi: 10.3390/ijms19082452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousnier A., Kubat N., Massias-Simon A., Segeral E., Rain J.C., Benarous R., Emiliani S., Dargemont C. von Hippel Lindau binding protein 1-mediated degradation of integrase affects HIV-1 gene expression at a postintegration step. Proc. Natl. Acad. Sci.U S A. 2007;104:13615–13620. doi: 10.1073/pnas.0705162104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J.E., Karlmark K.R., Shin J., Hengstschlager M., Lubec G. Differentiation-dependent expression of hypothetical proteins in the neuroblastoma cell line N1E-115. Proteins. 2006;63:671–680. doi: 10.1002/prot.20807. [DOI] [PubMed] [Google Scholar]
- Ohtaki A., Kida H., Miyata Y., Ide N., Yonezawa A., Arakawa T., Iizuka R., Noguchi K., Kita A., Odaka M. Structure and molecular dynamics simulation of archaeal prefoldin: the molecular mechanism for binding and recognition of nonnative substrate proteins. J. Mol. Biol. 2008;376:1130–1141. doi: 10.1016/j.jmb.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Okochi M., Kanie K., Kurimoto M., Yohda M., Honda H. Overexpression of prefoldin from the hyperthermophilic archaeum Pyrococcus horikoshii OT3 endowed Escherichia coli with organic solvent tolerance. Appl. Microbiol. Biotechnol. 2008;79:443–449. doi: 10.1007/s00253-008-1450-1. [DOI] [PubMed] [Google Scholar]
- Okochi M., Matsuzaki H., Nomura T., Ishii N., Yohda M. Molecular characterization of the group II chaperonin from the hyperthermophilic archaeum Pyrococcus horikoshii OT3. Extremophiles. 2005;9:127–134. doi: 10.1007/s00792-004-0427-y. [DOI] [PubMed] [Google Scholar]
- Okochi M., Nomura T., Zako T., Arakawa T., Iizuka R., Ueda H., Funatsu T., Leroux M., Yohda M. Kinetics and binding sites for interaction of the prefoldin with a group II chaperonin: contiguous non-native substrate and chaperonin binding sites in the archaeal prefoldin. J. Biol. Chem. 2004;279:31788–31795. doi: 10.1074/jbc.M402889200. [DOI] [PubMed] [Google Scholar]
- Okochi M., Yoshida T., Maruyama T., Kawarabayasi Y., Kikuchi H., Yohda M. Pyrococcus prefoldin stabilizes protein-folding intermediates and transfers them to chaperonins for correct folding. Biochem. Biophys. Res. Commun. 2002;291:769–774. doi: 10.1006/bbrc.2002.6523. [DOI] [PubMed] [Google Scholar]
- Ostrov D.A., Barnes C.L., Smith L.E., Binns S., Brusko T.M., Brown A.C., Quint P.S., Litherland S.A., Roopenian D.C., Iczkowski K.A. Characterization of HKE2: an ancient antigen encoded in the major histocompatibility complex. Tissue Antigens. 2007;69:181–188. doi: 10.1111/j.1399-0039.2006.00730.x. [DOI] [PubMed] [Google Scholar]
- Pajares M.A. PDRG1 at the interface between intermediary metabolism and oncogenesis. World J. Biol. Chem. 2017;8:175–186. doi: 10.4331/wjbc.v8.i4.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo V., Pellacani C., Heesom K.J., Rogala K.B., Deane C.M., Mottier-Pavie V., Gatti M., Bonaccorsi S., Wakefield J.G. Misato controls mitotic microtubule generation by stabilizing the TCP-1 tubulin chaperone complex [corrected] Curr. Biol. 2015;25:1777–1783. doi: 10.1016/j.cub.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Reissman S., Douglas N.R., Huang Z., Yuan D.S., Wang X., McCaffery J.M., Frydman J., Boeke J.D. Trivalent arsenic inhibits the functions of chaperonin complex. Genetics. 2010;186:725–734. doi: 10.1534/genetics.110.117655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parusel C.T., Kritikou E.A., Hengartner M.O., Krek W., Gotta M. URI-1 is required for DNA stability in C. elegans. Development. 2006;133:621–629. doi: 10.1242/dev.02235. [DOI] [PubMed] [Google Scholar]
- Patel-King R.S., Sakato-Antoku M., Yankova M., King S.M. WDR92 is required for axonemal dynein heavy chain stability in cytoplasm. Mol. Biol.Cell. 2019;30:1834–1845. doi: 10.1091/mbc.E19-03-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil K.S., Basak I., Pal R., Ho H.P., Alves G., Chang E.J., Larsen J.P., Moller S.G. A proteomics approach to investigate miR-153-3p and miR-205-5p targets in neuroblastoma cells. PLoS One. 2015;10:e0143969. doi: 10.1371/journal.pone.0143969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Chu Z., Lu J., Li D., Wang Y., Yang S., Zhang Y. Co-expression of chaperones from P. furiosus enhanced the soluble expression of the recombinant hyperthermophilic alpha-amylase in E. coli. Cell Stress Chaperones. 2016;21:477–484. doi: 10.1007/s12192-016-0675-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Chu Z., Lu J., Li D., Wang Y., Yang S., Zhang Y. Heterologous expression of chaperones from hyperthermophilic archaea inhibits aminoglycoside-induced protein misfolding in Escherichia coli. Biochemistry (Mosc) 2017;82:1169–1175. doi: 10.1134/S0006297917100091. [DOI] [PubMed] [Google Scholar]
- Perea-Resa C., Rodriguez-Milla M.A., Iniesto E., Rubio V., Salinas J. Prefoldins negatively regulate cold Acclimation in Arabidopsis thaliana by promoting nuclear proteasome-mediated HY5 degradation. Mol. Plant. 2017;10:791–804. doi: 10.1016/j.molp.2017.03.012. [DOI] [PubMed] [Google Scholar]
- Perez de Diego R., Ortiz-Lombardia M., Bravo J. Crystallization and preliminary X-ray diffraction analysis of the beta subunit Yke2 of the Gim complex from Saccharomyces cerevisiae. Acta Crystallogr. Sect F Struct. Biol. Cryst. Commun. 2008;64:501–503. doi: 10.1107/S1744309108011846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C.G., Dowen R.H., Lourenco G.F., Kirienko N.V., Heimbucher T., West J.A., Bowman S.K., Kingston R.E., Dillin A., Asara J.M. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat.CellBiol. 2013;15:491–501. doi: 10.1038/ncb2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Milla M.A., Salinas J. Prefoldins 3 and 5 play an essential role in Arabidopsis tolerance to salt stress. Mol. Plant. 2009;2:526–534. doi: 10.1093/mp/ssp016. [DOI] [PubMed] [Google Scholar]
- Sahlan M., Kanzaki T., Yohda M. Construction and characterization of the hetero-oligomer of the group II chaperonin from the hyperthermophilic archaeon, Thermococcus sp. strain KS-1. Extremophiles. 2009;13:437–445. doi: 10.1007/s00792-009-0229-3. [DOI] [PubMed] [Google Scholar]
- Sahlan M., Kanzaki T., Zako T., Maeda M., Yohda M. Analysis of the interaction mode between hyperthermophilic archaeal group II chaperonin and prefoldin using a platform of chaperonin oligomers of various subunit arrangements. Biochim. Biophys. Acta. 2010;1804:1810–1816. doi: 10.1016/j.bbapap.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Sahlan M., Zako T., Tai P.T., Ohtaki A., Noguchi K., Maeda M., Miyatake H., Dohmae N., Yohda M. Thermodynamic characterization of the interaction between prefoldin and group II chaperonin. J. Mol. Biol. 2010;399:628–636. doi: 10.1016/j.jmb.2010.04.046. [DOI] [PubMed] [Google Scholar]
- Sakono M., Zako T., Ueda H., Yohda M., Maeda M. Formation of highly toxic soluble amyloid beta oligomers by the molecular chaperone prefoldin. FEBS J. 2008;275:5982–5993. doi: 10.1111/j.1742-4658.2008.06727.x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Morgan N., Kirsch K.H., Trackman P.C., Sonenshein G.E. UXT is a LOX-PP interacting protein that modulates estrogen receptor alpha activity in breast cancer cells. J.Cell Biochem. 2017;118:2347–2356. doi: 10.1002/jcb.25893. [DOI] [PubMed] [Google Scholar]
- Sardiu M.E., Cai Y., Jin J., Swanson S.K., Conaway R.C., Conaway J.W., Florens L., Washburn M.P. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc. Natl. Acad. Sci.U S A. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou A., Hagio Y., Taira T., Iguchi-Ariga S.M., Ariga H. Repression of the c-fms gene in fibroblast cells by c-Myc-MM-1-TIF1beta complex. FEBS Lett. 2004;572:211–215. doi: 10.1016/j.febslet.2004.07.034. [DOI] [PubMed] [Google Scholar]
- Satou A., Taira T., Iguchi-Ariga S.M., Ariga H. A novel transrepression pathway of c-Myc. Recruitment of a transcriptional corepressor complex to c-Myc by MM-1, a c-Myc-binding protein. J. Biol. Chem. 2001;276:46562–46567. doi: 10.1074/jbc.M104937200. [DOI] [PubMed] [Google Scholar]
- Siegers K., Waldmann T., Leroux M.R., Grein K., Shevchenko A., Schiebel E., Hartl F.U. Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO J. 1999;18:75–84. doi: 10.1093/emboj/18.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert R., Leroux M.R., Scheufler C., Hartl F.U., Moarefi I. Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell. 2000;103:621–632. doi: 10.1016/s0092-8674(00)00165-3. [DOI] [PubMed] [Google Scholar]
- Simons C.T., Staes A., Rommelaere H., Ampe C., Lewis S.A., Cowan N.J. Selective contribution of eukaryotic prefoldin subunits to actin and tubulin binding. J. Biol. Chem. 2004;279:4196–4203. doi: 10.1074/jbc.M306053200. [DOI] [PubMed] [Google Scholar]
- Son H.G., Seo K., Seo M., Park S., Ham S., An S.W.A., Choi E.S., Lee Y., Baek H., Kim E. Prefoldin 6 mediates longevity response from heat shock factor 1 to FOXO in C. elegans. Genes Dev. 2018;32:1562–1575. doi: 10.1101/gad.317362.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorgjerd K.M., Zako T., Sakono M., Stirling P.C., Leroux M.R., Saito T., Nilsson P., Sekimoto M., Saido T.C., Maeda M. Human prefoldin inhibits amyloid-beta (Abeta) fibrillation and contributes to formation of nontoxic Abeta aggregates. Biochemistry. 2013;52:3532–3542. doi: 10.1021/bi301705c. [DOI] [PubMed] [Google Scholar]
- Stirling P.C., Cuellar J., Alfaro G.A., El Khadali F., Beh C.T., Valpuesta J.M., Melki R., Leroux M.R. PhLP3 modulates CCT-mediated actin and tubulin folding via ternary complexes with substrates. J. Biol. Chem. 2006;281:7012–7021. doi: 10.1074/jbc.M513235200. [DOI] [PubMed] [Google Scholar]
- Sun S., Tang Y., Lou X., Zhu L., Yang K., Zhang B., Shi H., Wang C. UXT is a novel and essential cofactor in the NF-kappaB transcriptional enhanceosome. J.CellBiol. 2007;178:231–244. doi: 10.1083/jcb.200611081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano M., Tashiro E., Kitamura A., Maita H., Iguchi-Ariga S.M.M., Kinjo M., Ariga H. Prefoldin prevents aggregation of α-synuclein. Brain Res. 2014;1542:186–194. [PubMed] [Google Scholar]
- Takeda N., Jain R., LeBoeuf M.R., Wang Q., Lu M.M., Epstein J.A. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro E., Zako T., Muto H., Itoo Y., Sorgjerd K., Terada N., Abe A., Miyazawa M., Kitamura A., Kitaura H. Prefoldin protects neuronal cells from polyglutamine toxicity by preventing aggregation formation. J. Biol. Chem. 2013;288:19958–19972. doi: 10.1074/jbc.M113.477984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebbenkamp A.T., Borchelt D.R. Analysis of chaperone mRNA expression in the adult mouse brain by meta analysis of the Allen Brain Atlas. PLoS ONE. 2010;5:e13675. doi: 10.1371/journal.pone.0013675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurillat J.P., Metzler S.C., Henzi N., Djouder N., Helbling M., Zimmermann A.K., Jacob F., Soltermann A., Caduff R., Heinzelmann-Schwarz V. URI is an oncogene amplified in ovarian cancer cells and is required for their survival. Cancer Cell. 2011;19:317–332. doi: 10.1016/j.ccr.2011.01.019. [DOI] [PubMed] [Google Scholar]
- Tian G., Jaglin X.H., Keays D.A., Francis F., Chelly J., Cowan N.J. Disease-associated mutations in TUBA1A result in a spectrum of defects in the tubulin folding and heterodimer assembly pathway. Hum. Mol. Genet. 2010;19:3599–3613. doi: 10.1093/hmg/ddq276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronnersjo S., Hanefalk C., Balciunas D., Hu G.Z., Nordberg N., Muren E., Ronne H. The jmjN and jmjC domains of the yeast zinc finger protein Gis1 interact with 19 proteins involved in transcription, sumoylation and DNA repair. Mol. Genet. Genomics. 2007;277:57–70. doi: 10.1007/s00438-006-0171-3. [DOI] [PubMed] [Google Scholar]
- Tsao M.L., Chao C.H., Yeh C.T. Interaction of hepatitis C virus F protein with prefoldin 2 perturbs tubulin cytoskeleton organization. Biochem. Biophys. Res. Commun. 2006;348:271–277. doi: 10.1016/j.bbrc.2006.07.062. [DOI] [PubMed] [Google Scholar]
- Tsuchiya H., Iseda T., Hino O. Identification of a novel protein (VBP-1) binding to the von Hippel-Lindau (VHL) tumor suppressor gene product. Cancer Res. 1996;56:2881–2885. [PubMed] [Google Scholar]
- Tummala K.S., Brandt M., Teijeiro A., Grana O., Schwabe R.F., Perna C., Djouder N. Hepatocellular carcinomas originate predominantly from hepatocytes and benign lesions from hepatic progenitor cells. Cell Rep. 2017;19:584–600. doi: 10.1016/j.celrep.2017.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummala K.S., Gomes A.L., Yilmaz M., Grana O., Bakiri L., Ruppen I., Ximenez-Embun P., Sheshappanavar V., Rodriguez-Justo M., Pisano D.G. Inhibition of de novo NAD(+) synthesis by oncogenic URI causes liver tumorigenesis through DNA damage. Cancer Cell. 2014;26:826–839. doi: 10.1016/j.ccell.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Vainberg I.E., Lewis S.A., Rommelaere H., Ampe C., Vandekerckhove J., Klein H.L., Cowan N.J. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell. 1998;93:863–873. doi: 10.1016/s0092-8674(00)81446-4. [DOI] [PubMed] [Google Scholar]
- Venteicher A.S., Meng Z., Mason P.J., Veenstra T.D., Artandi S.E. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell. 2008;132:945–957. doi: 10.1016/j.cell.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernekar D.V., Bhargava P. Yeast Bud27 modulates the biogenesis of Rpc128 and Rpc160 subunits and the assembly of RNA polymerase III. Biochim. Biophys. Acta. 2015;1849:1340–1353. doi: 10.1016/j.bbagrm.2015.09.010. [DOI] [PubMed] [Google Scholar]
- von Morgen P., Horejsi Z., Macurek L. Substrate recognition and function of the R2TP complex in response to cellular stress. Front Genet. 2015;6:69. doi: 10.3389/fgene.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton T.A., Sousa M.C. Crystal structure of Skp, a prefoldin-like chaperone that protects soluble and membrane proteins from aggregation. Mol.Cell. 2004;15:367–374. doi: 10.1016/j.molcel.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Wang D., Shi W., Tang Y., Liu Y., He K., Hu Y., Li J., Yang Y., Song J. Prefoldin 1 promotes EMT and lung cancer progression by suppressing cyclin A expression. Oncogene. 2017;36:885–898. doi: 10.1038/onc.2016.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Zhang X., Wang L., Yang Y., Dong Z., Wang H., Du L., Wang C. MicroRNA-214 suppresses oncogenesis and exerts impact on prognosis by targeting PDRG1 in bladder cancer. PLoS ONE. 2015;10:e0118086. doi: 10.1371/journal.pone.0118086. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang M., Kaufman R.J. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:326–335. doi: 10.1038/nature17041. [DOI] [PubMed] [Google Scholar]
- Wang P., Zhao J., Yang X., Guan S., Feng H., Han D., Lu J., Ou B., Jin R., Sun J. PFDN1, an indicator for colorectal cancer prognosis, enhances tumor cell proliferation and motility through cytoskeletal reorganization. Med. Oncol. 2015;32:264. doi: 10.1007/s12032-015-0710-z. [DOI] [PubMed] [Google Scholar]
- Wang Y., Garabedian M.J., Logan S.K. URI1 amplification in uterine carcinosarcoma associates with chemo-resistance and poor prognosis. Am. J. Cancer Res. 2015;5:2320–2329. [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Schafler E.D., Thomas P.A., Ha S., David G., Adney E., Garabedian M.J., Lee P., Logan S.K. Prostate-specific loss of UXT promotes cancer progression. Oncotarget. 2019;10:707–716. doi: 10.18632/oncotarget.26573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warringer J., Ericson E., Fernandez L., Nerman O., Blomberg A. High-resolution yeast phenomics resolves different physiological features in the saline response. Proc. Natl. Acad. Sci.U S A. 2003;100:15724–15729. doi: 10.1073/pnas.2435976100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C.T., Gorman M.A., Lazarou M., Ryan M.T., Gulbis J.M. Crystal structure of the mitochondrial chaperone TIM9.10 reveals a six-bladed alpha-propeller. Mol.Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Whitehead T.A., Boonyaratanakornkit B.B., Hollrigl V., Clark D.S. A filamentous molecular chaperone of the prefoldin family from the deep-sea hyperthermophile Methanocaldococcus jannaschii. Protein Sci. 2007;16:626–634. doi: 10.1110/ps.062599907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodford M.R., Sager R.A., Marris E., Dunn D.M., Blanden A.R., Murphy R.L., Rensing N., Shapiro O., Panaretou B., Prodromou C. Tumor suppressor Tsc1 is a new Hsp90 co-chaperone that facilitates folding of kinase and non-kinase clients. EMBO J. 2017;36:3650–3665. doi: 10.15252/embj.201796700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing F., Wang S., Zhou J. The expression of MicroRNA-598 inhibits ovarian cancer cell proliferation and metastasis by targeting URI. Mol. Ther. Oncolytics. 2019;12:9–15. doi: 10.1016/j.omto.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Xu Y., Her C. VBP1 facilitates proteasome and autophagy-mediated degradation of MutS homologue hMSH4. FASEB J. 2013;27:4799–4810. doi: 10.1096/fj.13-235127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane T., Shimizu T., Takahashi-Niki K., Takekoshi Y., Iguchi-Ariga S.M.M., Ariga H. Deficiency of spermatogenesis and reduced expression of spermatogenesis-related genes in prefoldin 5-mutant mice. Biochem. Biophys. Rep. 2015;1:52–61. doi: 10.1016/j.bbrep.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yart A., Gstaiger M., Wirbelauer C., Pecnik M., Anastasiou D., Hess D., Krek W. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol.CellBiol. 2005;25:5052–5060. doi: 10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesseyeva G., Aikemu B., Hong H., Yu C., Dong F., Sun J., Zang L., Zheng M., Ma J. Prefoldin subunits (PFDN1-6) serve as poor prognostic markers in gastric cancer. Biosci. Rep. 2020;40(BSR20192712) doi: 10.1042/BSR20192712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Kitaura H., Hagio Y., Sato T., Iguchi-Ariga S.M., Ariga H. Negative regulation of the Wnt signal by MM-1 through inhibiting expression of the wnt4 gene. Exp.Cell Res. 2008;314:1217–1228. doi: 10.1016/j.yexcr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Zako T., Banba S., Sahlan M., Sakono M., Terada N., Yohda M., Maeda M. Hyperthermophilic archaeal prefoldin shows refolding activity at low temperature. Biochem. Biophys. Res. Commun. 2010;391:467–470. doi: 10.1016/j.bbrc.2009.11.081. [DOI] [PubMed] [Google Scholar]
- Zako T., Iizuka R., Okochi M., Nomura T., Ueno T., Tadakuma H., Yohda M., Funatsu T. Facilitated release of substrate protein from prefoldin by chaperonin. FEBS Lett. 2005;579:3718–3724. doi: 10.1016/j.febslet.2005.05.061. [DOI] [PubMed] [Google Scholar]
- Zako T., Murase Y., Iizuka R., Yoshida T., Kanzaki T., Ide N., Maeda M., Funatsu T., Yohda M. Localization of prefoldin interaction sites in the hyperthermophilic group II chaperonin and correlations between binding rate and protein transfer rate. J. Mol. Biol. 2006;364:110–120. doi: 10.1016/j.jmb.2006.08.088. [DOI] [PubMed] [Google Scholar]
- Zhang F., Hu X., Gu Y., Bian H., Xu Z., Wang Q., Chen J., Lu Y., Sun L., Zheng Q. URI knockdown induces autophagic flux in gastric cancer cells. Am. J. Cancer Res. 2018;8:2140–2149. [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liu L., Zhang X., Jin F., Chen J., Ji C., Gu S., Xie Y., Mao Y. Cloning and characterization of a novel human prefoldin and SPEC domain protein gene (PFD6L) from the fetal brain. Biochem. Genet. 2006;44:69–74. doi: 10.1007/s10528-006-9008-3. [DOI] [PubMed] [Google Scholar]
- Zhang J., Xie M., Li M., Ding J., Pu Y., Bryan A.C., Rottmann W., Winkeler K.A., Collins C.M., Singan V. Overexpression of a Prefoldin beta subunit gene reduces biomass recalcitrance in the bioenergy crop Populus. Plant Biotechnol. J. 2020;18:859–871. doi: 10.1111/pbi.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Rai M., Wang C., Gonzalez C., Wang H. Prefoldin and Pins synergistically regulate asymmetric division and suppress dedifferentiation. Sci. Rep. 2016;6:23735. doi: 10.1038/srep23735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Davey M., Hsu Y.C., Kaplanek P., Tong A., Parsons A.B., Krogan N., Cagney G., Mai D., Greenblatt J. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Zhao R., Kakihara Y., Gribun A., Huen J., Yang G., Khanna M., Costanzo M., Brost R.L., Boone C., Hughes T.R. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J.Cell Biol. 2008;180:563–578. doi: 10.1083/jcb.200709061. [DOI] [PMC free article] [PubMed] [Google Scholar]