Abstract
SUV39H1, a human homologue of the Drosophila position effect variegation modifier Su(var)3-9 and of the Schizosaccharomyces pombe silencing factor clr4, encodes a novel heterochromatic protein that transiently accumulates at centromeric positions during mitosis. Using a detailed structure-function analysis of SUV39H1 mutant proteins in transfected cells, we now show that deregulated SUV39H1 interferes at multiple levels with mammalian higher-order chromatin organization. First, forced expression of full-length SUV39H1 (412 amino acids) redistributes endogenous M31 (HP1β) and induces abundant associations with inter- and metaphase chromatin. These properties depend on the C-terminal SET domain, although the major portion of the SUV39H1 protein (amino acids 89 to 412) does not display affinity for nuclear chromatin. By contrast, the M31 interaction surface, which was mapped to the first 44 N-terminal amino acids, together with the immediately adjacent chromo domain, directs specific accumulation at heterochromatin. Second, cells overexpressing full-length SUV39H1 display severe defects in mitotic progression and chromosome segregation. Surprisingly, whereas localization of centromere proteins is unaltered, the focal, G2-specific distribution of phosphorylated histone H3 at serine 10 (phosH3) is dispersed in these cells. This phosH3 shift is not observed with C-terminally truncated mutant SUV39H1 proteins or with deregulated M31. Together, our data reveal a dominant role(s) for the SET domain of SUV39H1 in the distribution of prominent heterochromatic proteins and suggest a possible link between a chromosomal SU(VAR) protein and histone H3.
Higher-order chromatin is essential for epigenetic gene control and for the structural organization of chromosomes at centromeres and telomeres (21, 26, 35, 36). Despite these important functions, only very few components of higher-order chromatin, particularly for mammalian systems, have been identified. By contrast, genetic screens of Drosophila melanogaster (40) and Schizosaccharomyces pombe (4) characterized a subfamily of ≈30 to 40 loci which, collectively, can be referred to as Su(var) group genes. Since Su(var) genes suppress position effect variegation (PEV), their gene products are implicated in the establishment of repressive chromatin domains. Indeed, the majority of isolated family members encode either heterochromatic proteins or enzymes that can modify the basic unit (DNA and histones) of chromatin (49). For example, several histone deacetylases (11, 16) or protein phosphatase 1 (5) have been classified as Su(var) products. Moreover, Su(var)2–5 (which encodes heterochromatin protein 1 [HP1] [24, 12]), Su(var)3-7 (8, 39), and Su(var)3-9 (47) are all dose-dependent modifiers of PEV, suggesting that subtle differences in the concentration of chromosomal SU(VAR) proteins direct the extension of heterochromatin (21).
Su(var) gene function thus supports a simplified model in which modifications at the nucleosomal level, like changes in the acetylation (48) and phosphorylation (20, 50) of core histones, contribute to the definition of repressive chromatin domains. Heterochromatic SU(VAR) proteins would then stabilize such an altered structure and propagate higher-order chromatin. This model predicts that some SU(VAR) proteins will preferably interact with modified histones and/or be able to nucleate extended associations with nuclear chromatin. However, with the exception of the paradigm of SIR proteins (17, 18) in Saccharomyces cerevisiae, which can be regarded as analogues of SU(VAR) in budding yeast, a mechanistic link between histones and heterochromatic SU(VAR) proteins has not been described in multicellular organisms. Moreover, the interdependence of SU(VAR) proteins in regulating the association with heterochromatin is only poorly understood.
Although Su(var) genes affect epigenetic control of gene expression, their major function appears to reside in the coregulation of higher-order chromatin at centromeres and telomeres (26). For example, mutations in Su(var)2-5 have recently been shown to result in telomeric fusions (14). Similarly, mutations in swi6 (28) and clr4 (23), the respective Su(var)2-5 and Su(var)3-9 homologues in S. pombe, induce segregation defects and elevated rates of chromosome loss (13)—phenotypes that are accompanied by impaired proliferative potential. Interestingly, accumulation of SWI6p at fission yeast centromeres has been shown to depend on clr4 function (13).
Recently, we isolated human (SUV39H1) and murine (Suv39h1) homologues (1) of Su(var)3-9 and clr4. SUV39H1 and Suv39h1 encode novel heterochromatic proteins that accumulate at centromeric positions during mitosis. In addition, SUV39H1 associates with M31 (HP1β) (1), one member of the mammalian SU(VAR)2-5 protein (HP1) family. Su(var)3-9-related genes are of particular interest, since (i) Su(var)3-9 is dominant over most other PEV modifier mutations (47) and (ii) because their products combine two prominent domains of chromatin regulators, i.e., the chromo and SET domains. Interestingly, mutations in either the chromo or SET domain of clr4 have been shown to impair gene silencing, implicating both domains in the modulation of a repressive chromatin structure (23). The 60-amino-acid (aa) chromo domain (3, 27), initially identified in HP1 and POLYCOMB (34), represents a protein-specific interaction surface that resembles an ancient histone-like fold (6) and which directs eu- or heterochromatic associations (30, 37). By contrast, the molecular role of the 130-aa SET domain (47) remains enigmatic. Although SET domain motifs are present in over 140 gene sequences (44) and represent preferred sites for mutations (25), only a few SET domain interactions have been described using yeast two-hybrid and in vitro binding assays (7, 9, 41). However, the SET domain has recently been shown to be a target for dual-specificity phosphatases and their inhibitor Sbf1 (SET binding factor 1), suggesting involvement in phosphorylation-dependent signaling pathways (10). Since SUV39H1 is a phosphoprotein with mitosis-specific isoforms (2), the SET domain could provide a protein module to induce dynamic transitions in chromosomal associations and protein interactions.
We have been using a detailed structure-function analysis of mutant SUV39H1 proteins in transfected cells to uncover the functional roles of the chromo and SET domains. Whereas heterochromatin localization is mediated through the N terminus by an M31 interaction surface and the immediately adjacent chromo domain, an isolated C-terminal SET domain appears to be inactive in these assays. However, the SET domain modulates several properties of deregulated SUV39H1. For example, only SUV39H1 proteins with an intact SET domain disperse endogenous M31, abundantly associate with nuclear chromatin, induce growth and chromosome segregation defects, and interfere with the G2-specific distribution of phosphorylated histone H3 at serine 10 (phosH3). Our data indicate a modular nature for SUV39H1 protein function that is largely governed by the SET domain and suggest a possible link between a chromosomal SU(VAR) protein and histone H3.
MATERIALS AND METHODS
Epitope-tagged expression plasmids and transient transfections.
All of the mutant DNA encoding various SUV39H1 proteins (see Fig. 5) and the M31 cDNA were inserted as PCR amplicons into pKW2T (derivative of pRK7; Genentech), which directs overexpression under the control of the cytomegalovirus (CMV) enhancer-promoter. Modification of the 5′ end of the SUV39H1 cDNA by a NotI oligonucleotide encoding a (myc)3H6 epitope has been described previously (1). Point mutations and short deletions in the SUV39H1 cDNA were introduced by double PCR mutagenesis. Since the N44 and cysSET mutant proteins lack the putative nuclear localization signal (NLS) present at amino acid positions 105 to 109 in SUV39H1 (1), an oligonucleotide encoding the simian virus 40 (SV40) NLS (PKKKRKV) was additionally inserted between the triple myc tag and the start of the mutant cDNAs. To generate a flag-tagged version of SUV39H1, the (myc)3H6 epitope was replaced with a NotI oligonucleotide encoding a single flag (DYKDDDDK) peptide. For all expression plasmids, the cDNA inserts and correct reading frames were confirmed by sequencing. Cloning details are available upon request.
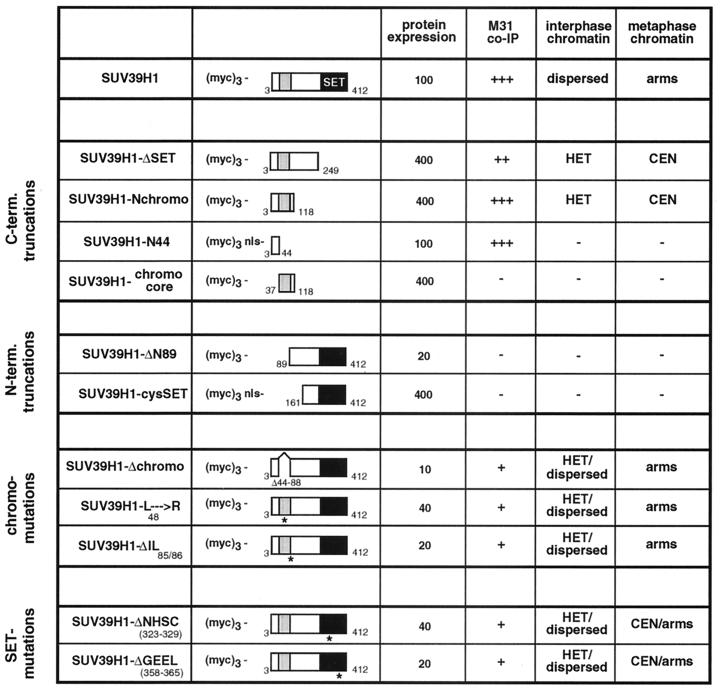
FIG. 5.
Summary of mutant SUV39H1 analysis. The names and schematic representations of mutant (myc)3-SUV39H1 are in the leftmost columns. The chromo domain is shown as a grey-shaded box, and the C-terminal SET domain is in black. Numbers refer to amino acid positions in the SUV39H1 protein (1). Mutant proteins are grouped as C-terminal (C-term.) and N-terminal (N-term.) truncations and chromo and SET domain mutations. The names of the two SET domain mutant proteins are abbreviated; they contain short deletions of seven (ΔNHSCDPN323-329) or eight (ΔGEELTFDY358-365) amino acids within the two most-conserved regions of the SET domain (25). The abbreviation nls denotes an SV40 NLS added to the N44 and cysSET mutant proteins. Average expression levels of ectopic proteins were estimated from Western blots of nuclear extracts after transient transfections of Cop8 and HeLa cells, setting the relative abundance of full-length (myc)3-SUV39H1 arbitrarily to 100%. The M31 coimmunoprecipitation (co-IP) data are from Fig. 4, and the M31 coprecipitation potentials are categorized as strong (+++), intermediate (++), weak (+), and negative (−). The data on association with nuclear chromatin are from Fig. 2 and 3. The different distributions in mouse interphase (after Triton X-100 extraction) are indicated as enriched at heterochromatic foci (HET) or broad staining throughout the entire nucleus (dispersed). Association with metaphase chromosomes is summarized as enriched at pericentric heterochromatin (CEN) or broad decoration along the arms (arms). Lack of chromatin association is indicated by a minus sign.
Expression plasmids (2 to 20 μg) were transiently transfected into murine Cop8 or human HeLa cells using either electroporation or LipofectAMINE (Gibco BRL). Cells were processed for immunofluorescence assay (IF) or immunoprecipitation 36 to 48 h after transfection.
Generation of stable cell lines.
HeLa cells were cotransfected with 10 μg of expression plasmid DNA and 1 μg of a vector conferring G418 resistance (pMex-neo; 29). For each expression plasmid, ≥100 G418r colonies were analyzed by IF and only colonies with significant expression in the majority of clonal cells were expanded in the presence of 200 μg of G418 (Gibco BRL) per ml. Whereas (myc)3-Nchromo and (myc)3-M31 clones could readily be obtained, we failed to generate HeLa cell lines that would overexpress full-length (myc)3-SUV39H1 with SET domain mutations, probably because of reduced protein stability (see coimmunoprecipitations below). By contrast, some full-length (myc)3-SUV39H1 clones with high expression could be identified in ≥300 G418r colonies screened, although they gradually lost their ectopic protein with increasing cell divisions. Therefore, aliquots of early-passage cells were stored in liquid nitrogen and only recultivated prior to experimentation.
Antibodies and immunocytochemistry.
The following antibodies were used at the indicated dilutions for IF: mouse monoclonal anti-myc (α-myc) (immunoglobulin G fraction of 9E10 hybridoma [1:100] or crude supernatant [1:10]), mouse monoclonal anti-flag (α-flag) M2 (Sigma; 1:15) rat monoclonal anti-M31 (α-M31) (51; crude MAC353 hybridoma supernatant; 1:5), rabbit polyclonal anti-phosH3 (α-phosH3) (20; 1:1,000), human anticentromeric antiserum (hACA; patient serum no. 2412; kindly provided by Günter Steiner, AKH, Vienna, Austria; 1:300), and mouse monoclonal, fluorescein isothiocyanate (FITC)-conjugated antitubulin (α-tubulin; Sigma; 1:100). Secondary antibodies were purchased from Jackson Immuno Research Laboratories.
IF of unfixed human chromosomal spreads from stable cell lines was done as described previously (1).
Immunolocalization of transfected proteins in mouse interphase and human metaphase chromatin.
At 24 h after electroporation, Cop8 cells were split onto coverslips and cultivated for another 12 h. Cells were washed twice in phosphate-buffered saline, fixed for 15 min at room temperature with 2% formaldehyde, and processed for IF (untreated). Alternatively, cells were washed twice in phosphate-buffered saline, extracted for 2 min in stabilization buffer containing 0.5% Triton X-100 (1), and then fixed for IF analysis. Ectopic proteins were visualized with α-myc or α-flag M2 antibodies, followed by incubation with FITC-conjugated secondary antibodies.
HeLa cells were transfected by electroporation, cultivated for ≈48 h, and incubated for 1 h with Colcemid (Gibco BRL; 0.1 μg/ml). Following trypsinization, cells were hypotonically swollen for 15 min in 10 mM Tris/HCl (pH 7.5)–10 mM NaCl–5 mM MgCl2 and spread onto microscope slides in a Cytospin 3 (Shandon). Spread chromosomes were fixed for 10 min at room temperature with 2% formaldehyde and processed for indirect IF with α-myc antibodies. DNA was counterstained with distamycin A (DA; Sigma) and 4′,6′-diamidino-2-phenylindole (DAPI; Sigma). Human chromosomes 1 were visually selected by their size and prominent pericentric block of DA-DAPI dense staining. Image analysis was performed using a Zeiss Axiophot microscope, a charge-coupled device camera (Photometrics), and Adobe Photoshop 4.0. Quantification of IF signals by chromosome scanning was done using IP-Lab Spectrum software.
Coimmunoprecipitations and protein blot analysis.
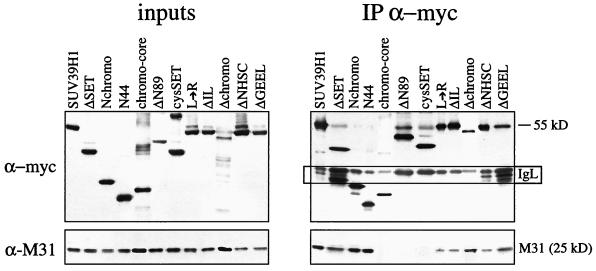
HeLa cells were transiently transfected with 20 μg of expression plasmids by electroporation. At 48 h after transfection, nuclear extracts were prepared. Since the expression levels and/or stability of the various mutant SUV39H1 proteins varied (particularly for the Δchromo mutant protein and for the chromo and SET domain mutations; see Fig. 5), the amounts of ectopic proteins were precalibrated by Western analysis and normalized by the addition of HeLa nuclear extract. Approximately 900 μg of adjusted nuclear extracts was then processed for coimmunoprecipitation with α-myc beads as described previously (1). Protein blots were probed with α-myc, α-M31, and anti-Suv39h1 (α-Suv39h1) (1) antibodies. Primary antibodies were visualized by peroxidase staining using Enhanced ChemiLuminescence (Amersham).
Growth curves and cell cycle analysis.
For growth curve analysis, HeLa cell clones were reseeded in triplicate at 105 cells per 10-cm-diameter dish. Cultures were incubated without a medium change for 1 week, and total cell numbers were determined every other day using a CASY1 cell counter. Synchronization of cells at the G1/S boundary by double thymidine block and analysis of cell cycle stages and DNA content by FACScan (Becton Dickinson) were done as previously described (2).
RESULTS
Deregulated SUV39H1 redistributes endogenous M31 (HP1β).
Suv39h1 has been shown to display significant colocalization with M31 (HP1β) at mouse interphase heterochromatin (1). To investigate the interdependence of M31 and the mammalian Suv39h1 or SUV39H1 protein, we analyzed the distribution of endogenous M31 in mouse Cop8 cells that had been transiently transfected with (myc)3-tagged SUV39H1. Following transfection, the cells were extracted with Triton X-100 to visualize chromatin-bound proteins. Surprisingly, in cells with a high ectopic protein content, endogenous M31 is displaced from heterochromatin and colocalizes with (myc)3-SUV39H1 in a rather uniform distribution throughout the entire nucleus (Fig. 1, top panel). By contrast, overexpressed (myc)3-M31 remains at heterochromatic foci in interphase (see Fig. 2, top panel).
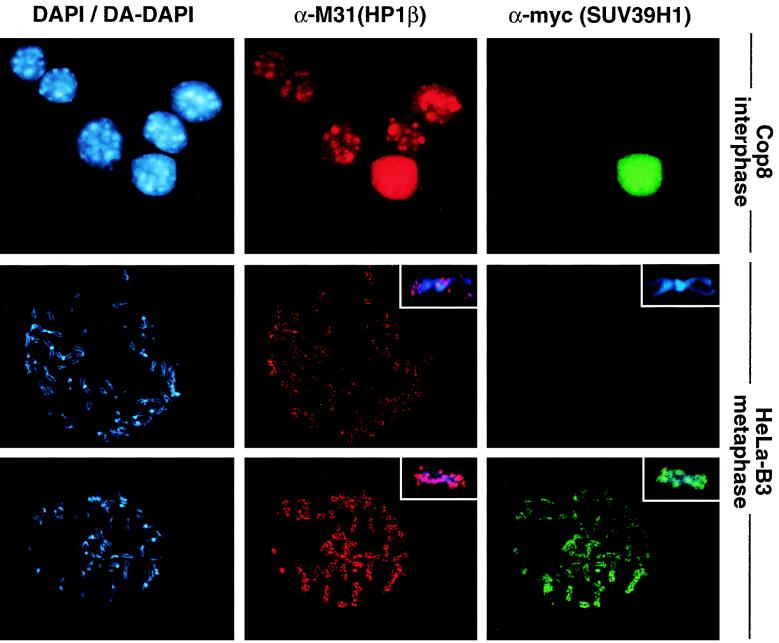
FIG. 1.
Ectopic SUV39H1 redistributes endogenous M31 (HP1β) in mouse interphase and human metaphase chromatin. (Top panel) Murine Cop8 cells were transiently transfected with a plasmid driving overexpression of (myc)3-tagged full-length SUV39H1 under the control of the CMV promoter-enhancer. Costaining for endogenous M31 and ectopic (myc)3-SUV39H1 was performed with Triton X-100-extracted cells by sequential incubation with (i) rat monoclonal α-M31 antibodies visualized with secondary CY3-conjugated antibodies (red) and (ii) mouse monoclonal α-myc antibodies that were detected with secondary FITC-conjugated antibodies (green). DNA was counterstained with DAPI, which highlights A/T-rich repeat sequences present in the prominent heterochromatic foci (bright blue patches). (Middle and bottom panels) Colocalization of M31 and (myc)3-SUV39H1 on unfixed metaphase-arrested spreads prepared from a “stably” transfected human cell line (HeLa-B3) that overexpresses (myc)3-SUV39H1 in most of the cells (1). Images in the middle panel were taken from a (myc)3-SUV39H1-negative spread, whereas the bottom panel shows the localization of M31 under conditions of (myc)3-SUV39H1 overexpression. Human DNA was counterstained with DA-DAPI. The inserts display the distribution of antigens at enlarged chromosomes.
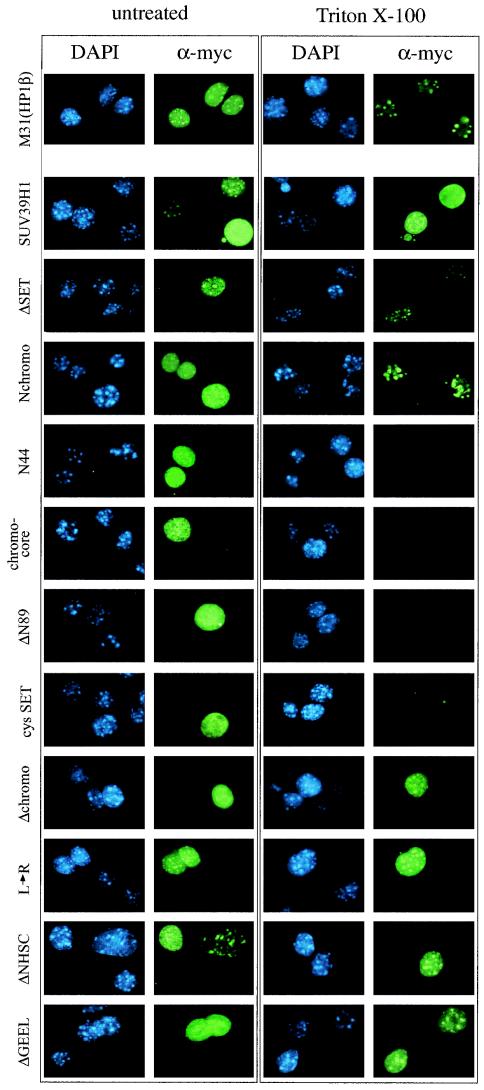
FIG. 2.
Distribution of ectopic M31, full-length SUV39H1, and SUV39H1 mutants in mouse interphase chromatin. Murine Cop8 cells were transiently transfected with overexpression plasmids encoding (myc)3-M31, full-length (myc)3-SUV39H1, and mutant (myc)3-SUV39H1 (indicated to the left; see Fig. 3 and 5 for mutant protein descriptions) and stained by indirect IF with α-myc antibodies (green). DNA was counterstained with DAPI (blue). Transfected cells were either directly processed for IF (left row) or extracted with Triton X-100 to visualize chromatin-associated proteins (right row). After Triton X-100 extraction, only full-length (myc)3-SUV39H1 displays a broad distribution that is not enriched for heterochromatic foci (second panel from the top).
We next examined whether SUV39H1 could also modulate the localization of M31 at mitotic chromosomes. We established several human HeLa cell lines that “stably” express (myc)3-SUV39H1 in the majority of cells within a given clonal population (see Materials and Methods). Following Colcemid arrest, unfixed chromosome spreads were analyzed by indirect IF with α-myc and α-M31 antibodies. In cells lacking (myc)3-SUV39H1, the mitotic fraction of endogenous M31 localizes to pericentric heterochromatin (43, 51) and to several discrete areas in the chromosomal arms (31) (Fig. 1, middle panel with insert). However, in the presence of ectopic SUV39H1 protein, staining for endogenous M31 is significantly enhanced and, together with (myc)3-SUV39H1, results in abundant signals along the entire chromosomes (Fig. 1, lower panel with inserts).
Abundant chromatin association of ectopic SUV39H1.
The extensive associations of deregulated SUV39H1 with inter- and metaphase chromatin suggested that the Suv39h1 or SUV39H1 protein may have much broader affinity for nuclear chromatin than previously anticipated. To identify domains in the SUV39H1 protein responsible for chromatin association, we generated a series of (myc)3-SUV39H1 mutant proteins (see Fig. 5) and analyzed their distribution in mouse interphase chromatin after transient transfection in Cop8 cells. In particular, we examined C-terminal truncations, N-terminal truncations, and chromo and SET domain mutations. Mutant proteins lacking the putative NLS at aa 105 to 109 (1) were modified by addition of the SV40 NLS, and nuclear accumulation of all mutant (myc)3-SUV39H1 proteins was verified by indirect IF (Fig. 2, left rows). To visualize chromatin-bound proteins, transfected Cop8 cells were extracted with Triton X-100 prior to fixation for indirect IF with α-myc antibodies (Fig. 2, right rows).
Since expression levels differed among mutant SUV39H1 proteins (see Fig. 5) and transfected cells vary in the abundance of ectopic protein, we evaluated subnuclear localization under conditions of both low and high overexpression. Indirect IF of low-expression Cop8 cells indicated accumulation of full-length (myc)3-SUV39H1 (412 aa with several nuclear patches that overlap the bright DAPI counterstaining (Fig. 2, two cells in the upper half of the second panel, left row). At higher levels of overexpression, however, (myc)3-SUV39H1 is no longer enriched at heterochromatic foci but associates with chromatin throughout the entire nucleus, even after after Triton X-100 extraction (Fig. 2, second panel, right row). Such broad affinity for nuclear chromatin was specific for full-length SUV39H1 and was not observed with truncated SUV39H1 proteins, whose subnuclear localizations were largely insensitive to protein expression levels (see below and data not shown). Similarly, ectopic (myc)3-M31 also did not display abundant affinity for nuclear chromatin but remained accumulated at heterochromatic foci (Fig. 2, top panel).
Identification of a heterochromatin targeting region in SUV39H1.
Analysis of mutant (myc)3-SUV39H1 proteins revealed a complex pattern of chromatin distribution. First, mutant proteins with truncations of the C-terminal SET domain (aa 3 to 249; ΔSET) or of the major portion of the protein (aa 3 to 118; Nchromo) resulted in heterochromatin-restricted associations. Second, the very N terminus (aa 3 to 44; N44), the chromo core (aa 37 to 118), or mutant proteins lacking the N terminus (aa 89 to 412 [ΔN89] and aa 161 to 412 [cysSET]) failed to remain chromatin bound. Third, deletion of the chromo domain (aa 44 to 88; Δchromo) induced extensive chromatin associations throughout the nucleus, whereas chromo domain mutations (L48→R and ΔIL85/86 [data not shown]) that are predicted to interfere with protein interactions mediated by the hydrophobic groove (6) still allowed some enriched distribution at heterochromatin. Fourth, short internal deletions (ΔNHSCDPN323-329 [ΔNHSC] and ΔGEELTFDY358-365 [ΔGEEL]) of the two most-conserved regions in the SET domain (25) significantly reduced the dispersed chromatin association of the full-length protein to more heterochromatin-enriched distributions. These data define the combination of the very N terminus and the chromo domain as a heterochromatin-targeting region (the first 118 aa of SUV39H1), whose specific accumulation at heterochromatin appears to be modulated by the presence of an intact SET domain.
The SET domain extends chromosomal associations in inter- and metaphase chromatin.
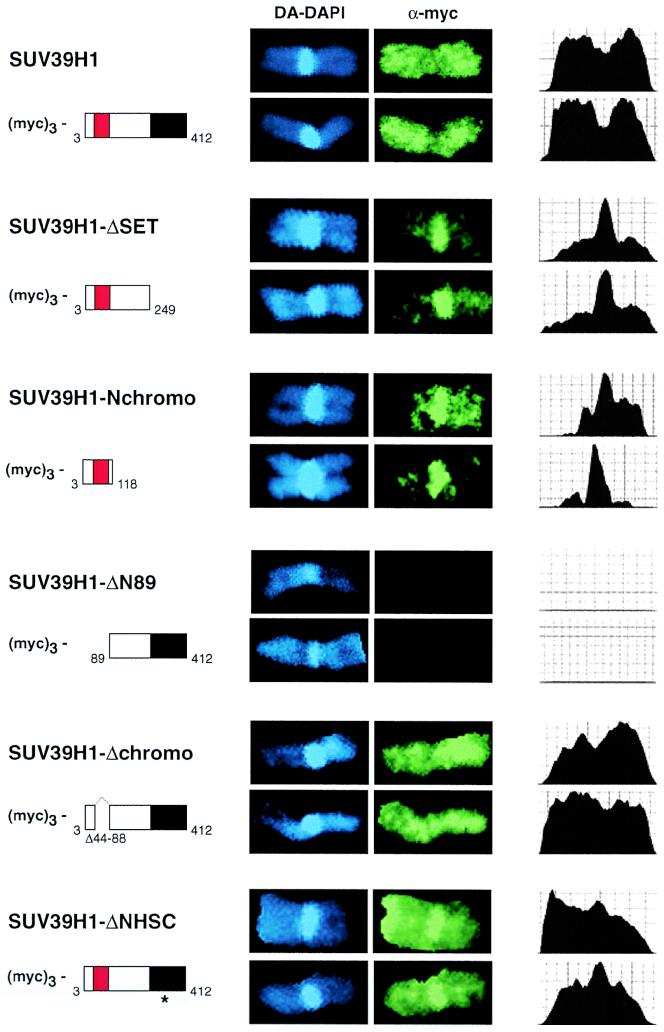
We next investigated the distribution of (myc)3-SUV39H1 mutant proteins in metaphase spreads of transiently transfected HeLa cells. To standardize this analysis for all of the transfectants, we selected human chromosome 1 because of its large size and prominent block of pericentric heterochromatin. To further quantify the resolution, stained chromosomes were also scanned along their entire length (see Materials and Methods). Similar to the dispersed distribution in interphase chromatin, full-length (myc)3-SUV39H1 abundantly decorates the chromosomal arms and is even slightly underrepresented at pericentric heterochromatin (Fig. 3, top panel; images and scans). By contrast, the ΔSET and Nchromo mutant proteins are specifically enriched at pericentric heterochromatin and show only residual staining in the arms. The chromosome 1 distribution of the Δchromo mutant protein or of the internal chromo mutations (data not shown) closely resembles that of the full-length protein. However, with mutations in the SET domain (ΔNHSC and ΔGEEL [data not shown]), a significant fraction of ectopic protein was present at pericentric heterochromatin. Thus, although an extended SET domain (e.g., ΔN89 and cysSET [data not shown]) lacks activity for interaction with nuclear chromatin, the SET domain appears to be a dominant protein module that extends and redirects the intrinsic heterochromatin affinity of SUV39H1.
FIG. 3.
Distribution of full-length and mutant SUV39H1 on human metaphase chromosome 1. Human HeLa cells were transiently transfected with overexpression plasmids encoding full-length and mutant (myc)3-SUV39H1 (indicated to the left) and stained by indirect IF with α-myc antibodies (green). DNA was counterstained with DA-DAPI (blue). Transfected cells were enriched for metaphase by Colcemid arrest, hypoton treated, spread by cytocentrifugation, and fixed prior to IF. Human chromosomes 1 were visually selected by their large size and prominent block of pericentric heterochromatin. To quantify the intensities of myc signals, chromosomes were also scanned along their entire length (right column).
M31 (HP1β) interaction is insufficient to direct chromatin association of SUV39H1.
The presence of the chromo domain in the heterochromatin-targeting region of SUV39H1 suggested that a possible interaction with M31 is involved in the regulation of chromosomal associations. Previous studies indicated that in vitro cotranslations or pull-down assays with recombinant SUV39H1 proteins are not suitable for detection of a direct interaction between SUV39H1 and M31 (1). We therefore performed coimmunoprecipitations with nuclear extracts of HeLa cells that had been transiently transfected with the (myc)3-SUV39H1 mutant constructs. Nuclear extracts were adjusted for ectopic protein (see Materials and Methods) (Fig. 4, left panel; inputs) and precipitated with α-myc beads, and bound proteins were probed with α-myc and α-M31 antibodies (Fig. 4, right panel). The results of these coimmunoprecipitations indicated that all of the C-terminal truncations efficiently precipitated endogenous M31, with N44 representing the smallest mutant SUV39H1 protein capable of associating with M31. By contrast, N-terminal truncations or, surprisingly, even the chromo core failed to interact. However, compared with the amount of M31 precipitated by full-length SUV39H1 or by the Nchromo and N44 mutant proteins, the Δchromo deletion and the two internal chromo domain mutations significantly weakened the M31 interaction. Similarly, the two SET domain mutations also reduced the amount of precipitated M31.
FIG. 4.
In vivo coimmunoprecipitation of mutant SUV39H1 with M31. Human HeLa cells were transiently transfected with overexpression plasmids encoding full-length and mutant (myc)3-SUV39H1 and processed for coimmunoprecipitation (IP) with α-myc antibody beads (right panel). Nuclear extracts were precalibrated and adjusted for comparable amounts of ectopic proteins prior to coimmunoprecipitation (left panel; inputs). Protein blots were probed with α-myc and α-M31 antibodies. The boxed area (marked IgL) indicates residual staining of immunoglobulin light chains that were retained in the precipitated material.
Figure 5 summarizes the results of our structure-function analysis of SUV39H1. Although chromatin association could not entirely be uncoupled from M31 interaction, the affinity of SUV39H1 for nuclear chromatin does not appear to be governed by M31. For example, the M31 interaction surface (aa 3 to 44) is not sufficient for localization to inter- or metaphase chromatin and SUV39H1 mutant proteins with a compromised M31 interaction still display broad chromosomal associations.
Forced expression of SUV39H1 interferes with growth control.
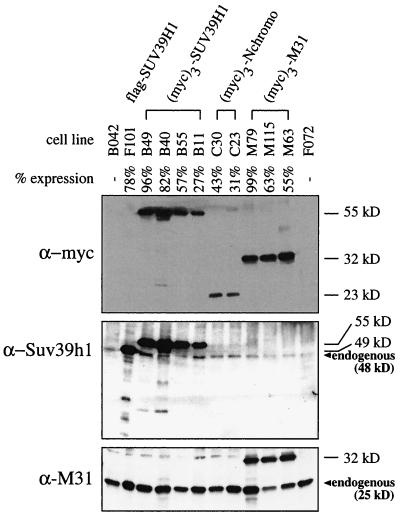
In the course of our studies, we realized that cells overexpressing full-length SUV39H1 display abnormal nuclear morphologies and compromised growth potential. To investigate SUV39H1-induced aberrations in more detail, we generated four HeLa cell lines that allow more long-term expression of full-length (myc)3-SUV39H1 driven by a CMV promoter-enhancer (see Materials and Methods). To exclude possible interference by the triple myc epitope, we also established one cell line overexpressing flag-SUV39H1. For comparison, we included two cell lines expressing only the heterochromatin-targeting domain of SUV39H1 [(myc)3-Nchromo] and three cell lines expressing (myc)3-M31. Following stable transfection, high-expression clones were selected by indirect IF and the abundance of epitope-tagged proteins was determined by Western analysis of nuclear extracts (Fig. 6). Since the percentage of full-length SUV39H1-expressing cells within a given clonal population declined with progressive cell divisions (data not shown), only early-passage cells with more than 50% expression were chosen for the following growth analyses.
FIG. 6.
Characterization of stable cell lines. Human HeLa cells were stably cotransfected with a vector conferring G418 resistance and overexpression plasmids encoding full-length flag-SUV39H1, full-length (myc)3-SUV39H1, or (myc)3-Nchromo and (myc)3-M31. Individual cell lines are grouped, with B042 and F072 representing control clones lacking transfected protein. The percentage of cells that stained positive for ectopic protein within a clonal population was determined weekly by indirect IF. The percentage below each cell line designation reflects the respective expression profile of early-passage cells used in the course of this study. Approximately 25 μg of nuclear extracts was processed for immunoblotting with α-myc, α-Suv39h1, and α-M31 antibodies. Positions of endogenous SUV39H1 (48 kDa) and M31 (25 kDa) proteins are indicated.
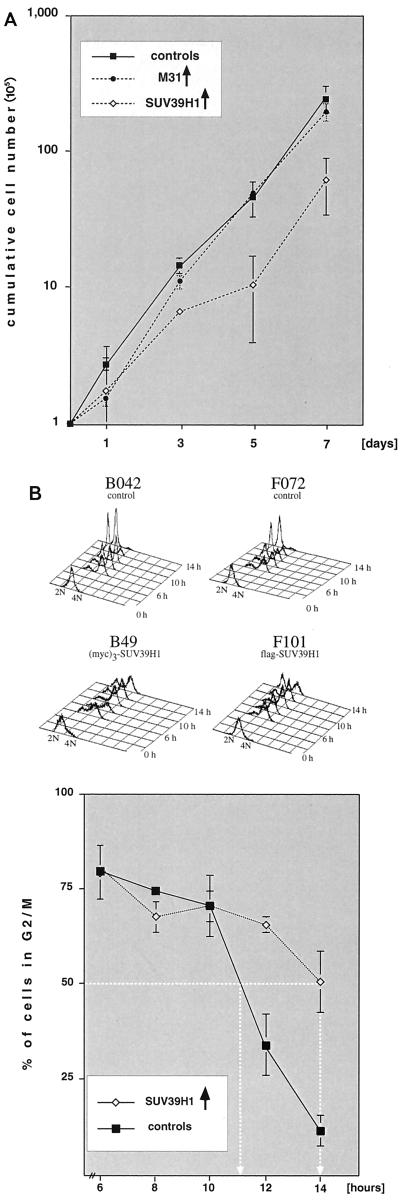
Comparative growth curves of four SUV39H1 [one flag- and three (myc)3-tagged clones], three (myc)3-M31, and three HeLa control cell lines indicated that overexpression of SUV39H1 results in severe growth retardation, with generation times (28.0 h) which are significantly longer than those of (myc)3-M31-expressing cells (23.7 h) or the HeLa controls (22.4 h) (Fig. 7A). Similarly, only the SUV39H1 clones, and not the (myc)3-M31 or the (myc)3-Nchromo clones, displayed a high degree of aberrant nuclear morphologies, like micro- and polynuclei (see below), suggesting that SUV39H1 overexpression is likely to induce defects during mitosis. Therefore, we synchronized one flag- and two (myc)3-SUV39H1 clones by double thymidine block at the G1/S boundary. Following release, cell cycle progression was monitored by fluorescence-activated cell sorter analysis and the relative percentage of cells in G2/M was determined. Whereas SUV39H1-overexpressing cells entered G2/M with kinetics similar to those of the HeLa controls, more than 50% of the cells in the three SUV39H1 clones were still in G2/M approximately 3 h after the HeLa control cells had reached the next G1 phase (Fig. 7B). These data demonstrate that forced expression of the full-length SUV39H1 protein interferes with growth control by inducing delayed G2/M progression.
FIG. 7.
Overexpression of SUV39H1 induces growth retardation and delayed mitotic progression. (A) Cumulative growth curves of three HeLa cell controls (untransfected HeLa, B042, and F072 cells) and three (myc)3-M31 (M79, M115, and M63), one flag-SUV39H1 (F101), and two (myc)3-SUV39H1 (B49 and B40) clones. For each cell line, the total cell numbers were determined in triplicate every other day after reseeding of 105 cells. The graph contains averaged growth curves, and deviations among the clones are indicated by bars. Average generation times for SUV39H1 clones (28.0 h) are approximately 4 or 5 h longer than those for (myc)3-M31-expressing cells (23.7 h) or for the HeLa controls (22.4 h). (B) The three SUV39H1-expressing and three HeLa control cell lines were synchronized at the G1/S boundary by a double thymidine block. Following release, cell cycle progression was monitored by fluorescence-activated cell sorter analysis (top panels; shown for only two cell lines each). The relative percentage of cells in G2/M (i.e., containing 4N DNA) was determined, and progression through G2/M is plotted (averaged graph at the bottom). SUV39H1 clones needed approximately 3 h longer than the HeLa controls to reach a cell population with 50% G2/M cells (indicated by white lines).
Aberrant chromosome segregation in SUV39H1-overexpressing cells.
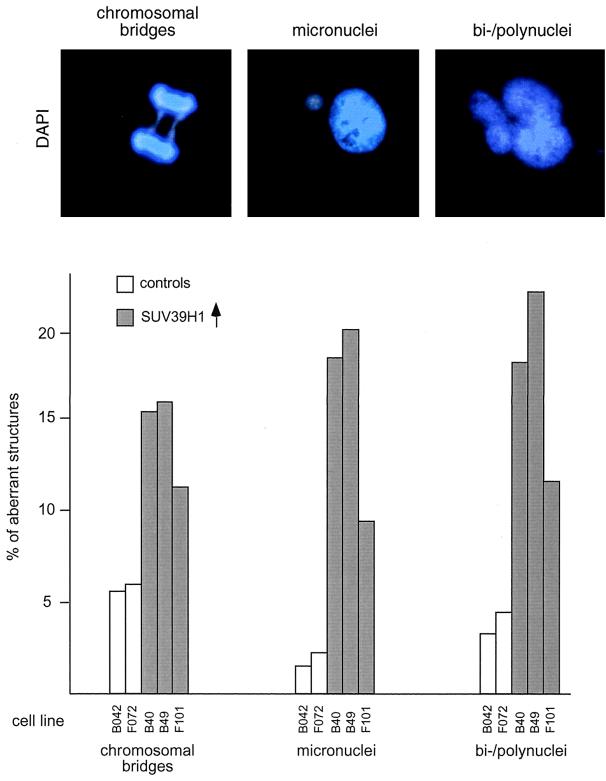
Statistical evaluation of the morphological and cytological abnormalities in SUV39H1-overexpressing cells further revealed severe chromosome segregation defects. For example, the numbers of cells with micronuclei and polynuclei (with and without multipolar spindles as detected by α-tubulin staining; data not shown) are 4- to 10-fold higher than those of HeLa control cells (Fig. 8). In addition, mitoses with chromosomal bridges and lagging chromosomes at ana- and telophase are present at two- to threefold elevated levels. This increased number of chromosome segregation defects including aberrant nuclear morphologies was not observed with the (myc)3-Nchromo and (myc)3-M31 clones (data not shown). Together with the above growth analysis, these results suggest that properties specific to full-length SUV39H1 are responsible for interfering with proliferation and chromosome segregation.
FIG. 8.
Aberrant chromosome segregation in SUV39H1-overexpressing cells. Logarithmically growing HeLa control cells (B042 and F072) and SUV39H1-expressing clones with a high percentage of overexpressing cells (B40, B49, and F101 cells; Fig. 6) were stained with α-tubulin antibodies (not shown) and DAPI and processed for IF (top panel). SUV39H1 clones display an increased number of cells with aberrant nuclear morphologies (micronuclei and bi- or polunuclei). Many polynuclei also contained multipolar spindles (not shown). At anaphase and telophase, the numbers of chromosomal bridges (including lagging DAPI-stained material between separating sets of chromosomes) are significantly enhanced compared to HeLa control clones. Plotted are the percentages of aberrant structures determined in two independent blind evaluations of ≈500 cells per clone (bottom panel).
Deregulated SUV39H1 disperses phosH3-positive G2 foci.
Although endogenous or ectopic SUV39H1 has failed to physically associate with centromere-specific proteins (CENPs) (1), altered localization of CENPs is one possible mechanism by which overexpression of SUV39H1 may induce growth and segregation defects. Therefore, we compared the nuclear distribution of epitopes detected by hACA in HeLa control cells and in SUV39H1 clones that display a high percentage (78 to 96%) of overexpressing cells. However, examination of the characteristically dotted hACA pattern did not reveal apparent differences between control and SUV39H1-overexpressing cells (data not shown).
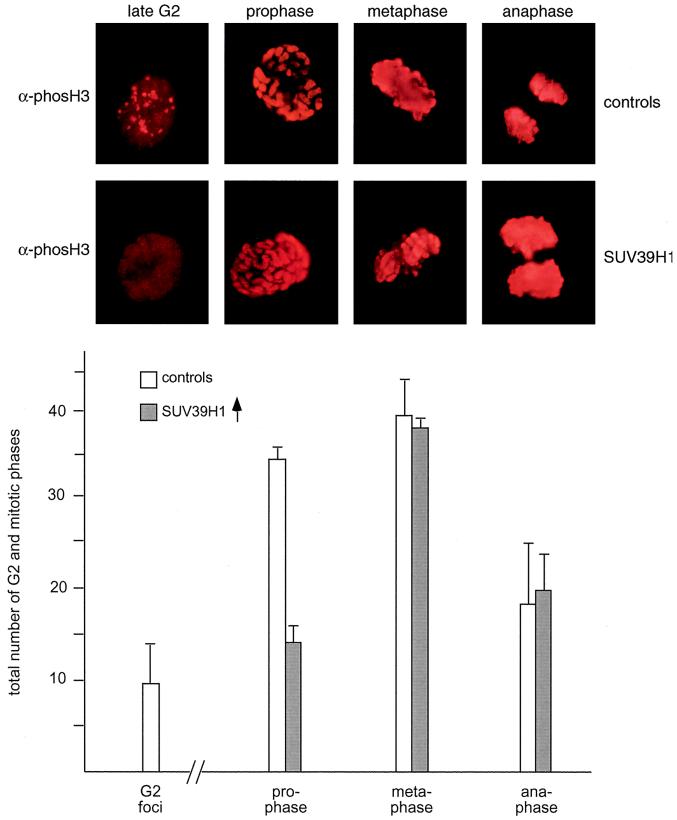
Recently, phosH3 has been shown to be required for the condensation and subsequent segregation of chromosomes (50). During the cell cycle, phosH3 suddenly accumulates at pericentric heterochromatin in late G2 and then is present along the entire chromosomes from prophase to anaphase (20). Since deregulated SUV39H1 also abundantly decorates mitotic chromosomes and induces segregation defects that can be correlated with delayed G2/M progression, we asked whether the distribution of phosH3-positive epitopes could possibly be changed in SUV39H1-overexpressing cells. Surprisingly, this analysis indicated that phosH3-positive G2 foci are nearly absent in the high-percentage (82 to 96% expressing cells) SUV39H1 clones (see Fig. 10), which, instead, display dispersed phosH3 staining in interphase (Fig. 9, top panel). By contrast, the broad phosH3 decoration of mitotic chromosomes from prophase to anaphase appears unaltered. Using DAPI staining and chromosomal morphology to define mitotic stages, statistical evaluation of mitotic phase indices further revealed that the number of prophases is significantly reduced, whereas the numbers of meta- and anaphases are very similar to those of the HeLa control cells (Fig. 9, bottom panel).
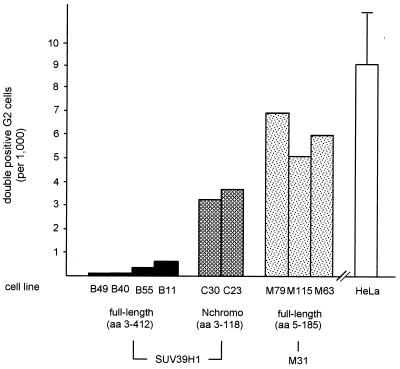
FIG. 10.
Dispersion of phosH3-positive G2 foci is specific for full-length SUV39H1. Logarithmically growing cells of the indicated clones were costained with rabbit polyclonal α-phosH3 and mouse monoclonal α-myc antibodies. Using two independent blind evaluations of ≥1,000 cells per clone, the total numbers of double-positive cells displaying the characteristic phosH3 foci in interphase (Fig. 9) were determined and are plotted. The HeLa bar represents the average number of phosH3-positive G2 foci detected in untransfected HeLa cells and in the two control clones (B042 and F072).
FIG. 9.
Deregulated SUV39H1 disperses phosH3-positive G2 foci. Logarithmically growing HeLa controls (B042 and F072 cells) and (myc)3-SUV3H1-expressing clones with a high percentage of overexpressing cells (B40 and B49 cells [Fig. 6]) were stained with α-phosH3 antibodies and CY3-conjugated secondary antibodies (red). DNA was counterstained with DAPI (not shown). The pericentric phosH3-positive foci in late G2 (20) and the prominent phosH3 decoration of mitotic chromosomes are shown in the top panel. Evaluation of ≥1,000 cells per (myc)3-SUV39H1-expressing clone only revealed dispersed phosH3 staining in interphase and failed to detect the characteristic phosH3-positive G2 foci. Also plotted are averaged phase indices of ≈200 mitotic cells per clone as determined by DAPI staining and chromosomal morphology (bottom panel).
phosH3 redistribution is specific for full-length SUV39H1 clones.
Finally, we investigated whether the surprising interference with the definition of phosH3-positive G2 foci could be linked to a particular domain in the SUV39H1 protein. We therefore determined the cell numbers displaying phosH3-positive G2 foci in the full-length (myc)3-SUV39H1, (myc)3-Nchromo, and (myc)3-M31 clones. Since the percentage of overexpressing cells varied between 27 and 96% [four (myc)3-SUV39H1 lines], between 31 and 43% [two (myc)3-Nchromo lines], and between 55 and 99% [three (myc)3-M31 lines] among the different clonal populations, we costained logarithmically growing cells with mouse α-myc and rabbit α-phosH3 antibodies. Using two independent blind evaluations, we then averaged the number of double-positive G2 cells which show myc signals and also display the characteristic phosH3 foci. Examination of the (myc)3-M31 clones indicated that five to seven double-positive G2 cells were present per ≈1,000 cells (Fig. 10), a number which is comparable to that of the HeLa controls. Similarly, clones expressing the heterochromatin-targeting region of SUV39H1 [(myc)3-Nchromo] contained three or four double-positive G2 cells and thus do not appear to significantly interfere with the distribution of phosH3. By contrast, overexpression of full-length (myc)3-SUV39H1 almost completely prevents the definition of phosH3-positive G2 foci, and only after evaluation of 3,000 cells in the low-percentage expression clones could one or two double-positive G2 cells be detected. From this analysis, we infer that properties responsible for the broad localization of full-length SUV39H1 are also involved in the dispersion of phosH3 from pericentric heterochromatin. Surprisingly then, since the SET domain induces extensive chromatin associations (Fig. 2 and 3), it is likely that the SET domain provides a function to modulate the phosphorylation pattern of histone H3.
DISCUSSION
Using a detailed analysis of mutant proteins in transfected cells, we found that deregulated SUV39H1 interferes at multiple levels with higher-order chromatin organization and induces growth and chromosome segregation defects. These functions largely depend on the C-terminal SET domain, which appears to modulate several activities of ectopic SUV39H1. Only SUV39H1 proteins with an intact SET domain abundantly associate with nuclear chromatin and redistribute endogenous M31 (HP1β). Surprisingly, deregulated SUV39H1 also disperses the G2-specific definition of phosH3, suggesting a possible link between a chromosomal SU(VAR) protein and histone H3.
Ectopic versus endogenous SUV39H1.
In previous studies, we showed that endogenous SUV39H1 is a heterochromatic protein during interphase that selectively accumulates at centromeric positions of metaphase chromosomes (1, 2). Similar to endogenous SUV39H1, low-abundance ectopic SUV39H1 shows the highest affinity for heterochromatic foci in interphase. However, at increased protein abundance, ectopic SUV39H1 localizes in a rather uniform distribution with inter- and metaphase chromatin (Fig. 1 to 3). These data suggest that chromosomal localizations of human SUV39H1 (and probably also of murine Suv39h1) are very sensitive to protein expression levels and imply a much broader affinity for nuclear chromatin than previously anticipated. Endogenous SUV39H1 and Suv39h1 are low-abundance proteins (1) which, indeed, display more extended but weak chromosomal staining after triple labeling (2). Because ectopic SUV39H1 disperses the distribution of phosH3, it is tempting to infer that some of the putative low-affinity binding sites could be provided by histone H3.
By contrast, overexpression may also induce a deregulated function(s) for ectopic SUV39H1 that is not reflected by the endogenous protein. For example, high levels of ectopic SUV39H1 could sequester essential factors that normally would not interact with endogenous SUV39H1, particularly if SUV39H1 were misexpressed during the cell cycle. Although this possibility cannot be excluded, analysis of clr4 mutants of S. pombe (13, 23) reveals phenotypes that are very similar to defects mediated by overexpressed SUV39H1 (see below).
Modular nature of SUV39H1 protein.
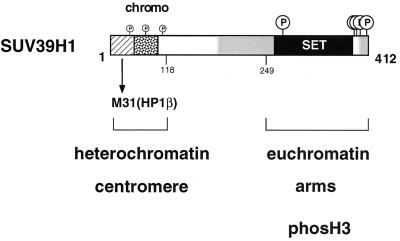
SU(VAR)3-9-related proteins combine the two most characteristic domains of chromatin regulators, i.e., the chromo and SET domains (see introduction). In addition, they contain a SET-associated cysteine-rich region (22), a highly conserved C-terminal tail, and an N terminus that is shared between Drosophila SU(VAR)3-9 (47) and mammalian SUV39H1 or Suv39h1 (1). Based on our structure-function analysis, we can assign a direct function only to the N terminus and the chromo domain. The first 44 aa represent an interaction surface for M31 (HP1β) (Fig. 4) which does not appear to contain a consensus peptide that has been described to confer preferred binding to the chromo shadow domain of mammalian HP1 proteins (32, 33, 42). Together with the immediately adjacent chromo domain, this N terminus is required to direct heterochromatic associations in interphase and accumulation at centromeric positions on metaphase chromosomes (Fig. 2 and 3).
By contrast, the major portion of the SUV39H1 protein (aa 89 to 412), including the cysteine-rich region and the C-terminal SET domain, does not display chromatin binding activity or interaction with M31, although ectopic full-length SUV39H1 broadly associates with inter- and metaphase chromatin. Surprisingly, SET domain mutations restrict these abundant chromosomal associations and also weaken the M31 interaction (Fig. 5). These data are consistent with a dominant role for the SET domain, which appears functional only in the context of other protein motifs. SUV39H1 therefore represents a modular protein (Fig. 11) with adjustable affinites for nuclear chromatin, suggesting multiple roles in higher-order chromatin organization.
FIG. 11.
Modular nature and functional domains of SUV39H1 protein. The 412-aa human SUV39H1 protein contains several conserved regions, including a chromo domain (stippled box), the C-terminal SET domain (black box), SET domain-associated cysteine-rich regions (grey shading), and an N terminus (hatched box) that is shared with Drosophila SU(VAR)3-9 (1). Based on our structure-function analysis, we can assign a direct function to the N-terminal 44 aa and the immediately adjacent chromo domain. The N terminus is an M31 interaction surface which, together with the chromo domain, defines the heterochromatin-targeting region (aa 3 to 118) of SUV39H1. By contrast, the SET domain appears to be functional only in the context of the full-length protein and represents a dominant module that regulates the chromatin association and M31 interaction potentials of SUV39H1. Because mutant SUV39H1 proteins lacking the SET domain do not significantly interfere with the distribution of phosH3-positive G2 foci (Fig. 10), the unexpected link between SUV39H1 and histone H3 is also proposed to be provided by functions of the SET domain. SUV39H1 has recently been shown to be a phosphoprotein with preferred serine phosphorylation of the C-terminal tail, the SET domain, and also, to a lesser extent, the chromo domain (2).
Interdependence of SUV39H1 and M31 (HP1β).
The chromatin association of SUV39H1 could not entirely be uncoupled from M31 interaction, although the broad distribution of overexpressed full-length SUV39H1 is clearly distinct from the heterochromatin-restricted staining pattern of endogenous or ectopic M31 (Fig. 2). Moreover, the strong M31 interaction surface (aa 3 to 44) present within the heterochromatin-targeting region (aa 3 to 118) of SUV39H1 does not bind nuclear chromatin, and localization of SUV39H1 is shifted to more heterochromatic positions in chromo and SET domain mutants, which are weakened in their M31 interaction potential (Fig. 5). These data indicate that M31 interaction does not govern chromosomal associations of SUV39H1, although the M31 interaction surface contributes to enrichment for heterochromatic localization of mutant SUV39H1 lacking an intact SET domain.
By contrast, ectopic full-length SUV39H1 redistributes endogenous M31 in inter- and metaphase chromatin (Fig. 1), demonstrating instead that SUV39H1 can modulate chromosomal associations of M31. This property depends on the C-terminal SET domain, because ΔSET, Nchromo, and N44 mutant proteins cannot disperse endogenous M31 (data not shown). Consistent with a dominant role of the SET domain in the control of chromatin distribution, a chimeric M31-SET construct (resembling the modular nature of SUV39H1) aberrantly localizes to chromatin (data not shown). Moreover, S. pombe clr4 mutants have been shown to disrupt heterochromatic associations of SWI6p (13). Together, these data indicate a potential for SUV39H1 in directing the localization of M31 and suggest that its deregulation can impart a dominant-negative function in modulating higher-order chromatin.
SUV39H1 interferes with growth control and chromosome segregation.
High levels of ectopic SUV39H1 protein, but not of overexpressed M31, induce severe growth retardation that is characterized by delayed progression through G2/M (Fig. 7). Moreover, SUV39H1-overexpressing cells display a high percentage of micro- and polynuclei and increased numbers of lagging chromosomes and chromosomal bridges at the meta- to anaphase transition (Fig. 8). Similar phenotypes have been described upon forced expression of clr4 in S. pombe (discussed in reference 23) or in clr4 null mutants (13). Evaluation of phase indices in full-length SUV39H1 clones indicates an ≃2-fold reduction in the total number of prophases, whereas the numbers of meta- and anaphases are similar to those of control clones (Fig. 9). These data suggest that deregulated SUV39H1 does not activate a known mitotic checkpoint but appears to interfere with chromosome segregation through a different mechanism.
Endogenous SUV39H1 has recently been shown to be a phosphoprotein with mitosis-specific isoforms, whose transient accumulation at centromeres is maximal from prometa- to late metaphase (2). Examination of the distribution of CENPs using hACA indicated no significant shift between SUV39H1-overexpressing and control cells (data not shown). Surprisingly, however, the pericentric definition of characteristic G2 foci that are visualized by antibodies specifically recognizing phosH3 (20) is dispersed in SUV39H1-overexpressing cells (Fig. 9). At later stages, the broad phosH3 decoration of mitotic chromosomes appears unchanged. Inability to phosphorylate serine 10 in histone H3 has recently been shown to cause chromosome condensation and segregation defects (50). Thus, our data support a model in which deregulated SUV39H1 may transiently perturb the nucleation and/or timing of phosH3-dependent chromosome condensation, resulting in delayed G2/M progression and aberrant chromosome alignment and/or segregation.
The SET domain is a dominant module which regulates SUV39H1 function.
In the context of full-length SUV39H1, an intact SET domain directs abundant chromosomal associations, redistributes endogenous M31, induces growth and chromosome segregation defects, and disperses the G2-specific definition of phosH3-positive foci. Although we have failed to generate cell lines stably overexpressing SUV39H1 proteins with SET domain mutations (see Materials and Methods), our comparative analysis of full-length SUV39H1 and the Nchromo and M31 clones (Fig. 10) is most consistent with a role for the SET domain in the modulation of the properties of ectopic SUV39H1.
Several mechanisms could explain how the SET domain may regulate SUV39H1 function, including altered protein folding or stability, induction of homomeric interactions, or covalent modification. Because SUV39H1 is a modular protein, it is unlikely that an intact SET domain is solely involved in protein folding. Further, although coexpression of flag-SUV39H1 and (myc)3-SUV39H1 mutant proteins indeed indicates a potential for intermolecular associations, it is the chromo core domain rather than the SET domain that appears to direct homomeric interactions (data not shown). Thus, we favor the view that covalent modifications of the SET domain are intrinsically responsible for modulation of the chromatin distribution and protein interaction potentials of SUV39H1. This interpretation is supported by the inability of recombinant SUV39H1 to complex with nuclear M31 (1). In addition, the presence of mitosis-specific phospho isoforms and the preferred phosphorylation of serine residues in the SUV39H1 protein, particularly in the SET domain and its extended C-terminal tail (2; Fig. 11), all point to a possible regulatory role(s) that is triggered by unknown phosphorylation pathways. Consistent with this model is the recent discovery that several SET domains are targets for dual-specificity phosphatases and their inhibitor Sbf1 (10). Interestingly, SUV39H1 also interacts with and seems to be modulated by Sbf1 (15). Using a variety of experimental approaches, we have, however, been unable to assign a direct function to the isolated SET domain of SUV39H1. In agreement with the modular nature of the SUV39H1 protein, the SET domain of SUV39H1 therefore appears to require targeting to nuclear chromatin via the N terminus in order to transduce its predicted regulatory signal(s).
SUV39H1 links a chromosomal SU(VAR) protein with histone H3.
The unexpected finding that deregulated SUV39H1 possibly interferes with phosH3-dependent chromatin condensation is surprising, particularly for a chromosomal SU(VAR) protein. Based on the above considerations, high concentrations of SUV39H1 could probably titrate components of phosphorylation pathways, some of which may be connected to putative kinases or phosphatases that act on histone H3 (46). Consistent with this interpretation is also the transient dispersion of phosH3, which is apparent in late G2 but not in the subsequent mitotic stages (Fig. 9). An altered phosphorylation pattern of the N terminus of histone H3 has been proposed to affect the binding of condensation factors or other non-histone proteins (46, 50).
Finally, our analysis reveals intriguing parallels between human SUV39H1 and the dominant S. cerevisiae SIR3 protein that may extend beyond a broader chromatin association of overexpressed SIR3 (19, 38), and its modification by phosphorylation (45). Regarding SIR3 as a chromosomal SU(VAR) analogue with histone binding activity (18) in budding yeast, SUV39H1 could have a more intrinsic link to histone H3 than that uncovered by interference with phosH3 distribution.
ACKNOWLEDGMENTS
M.M. and M.S. contributed equally to this work and should be considered joint first authors. We thank Dieter Schweizer (Institute of Botany, Vienna, Austria) for help and advice and critical reading of the manuscript, Frank Eisenhaber (IMP) for computer similarity searches of SET domain sequences, Prim Singh (The Roslin Institute, Roslin, United Kingdom) for the M31 cDNA and α-M31 antibodies, David Allis (University of Virginia, Charlottesville) for the α-phosH3 antibodies, and Günter Steiner (AKH, Vienna, Austria) for hACA. We are indebted to Stefan Schöftner and John Haskew for their help in generating stable SUV39H1 and M31 transfectants and to Lydia Sattler (Institute of Botany, Vienna, Austria) and Martina Stich for IF analyses. We also thank Ron Firestein and Michael Cleary (Stanford University) for sharing information on SUV39H1-Sbf1 interaction prior to submission.
Research in T.J.'s laboratory is funded by the IMP and by the Austrian Research Promotion Fund.
REFERENCES
- 1.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh P B, Reuter G, Jenuwein T. Functional mammalian homologues of the Drosophila PEV modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aagaard L, Schmid M, Warburton P, Jenuwein T. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J Cell Sci. 2000;113:817–829. doi: 10.1242/jcs.113.5.817. [DOI] [PubMed] [Google Scholar]
- 3.Aasland R, Stewart A F. The chromo shadow domain, a second chromodomain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 1995;23:3168–3173. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allshire R C, Nimmo E R, Ekwall K, Javerzat J P, Granston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 5.Baksa K, Morawietz H, Dombradi V, Axton M, Taubert H, Szabo G, Török I, Udvardy A, Gyuorkovics H, Szöör B, Glover D, Reuter G, Gausz J. Mutations in the protein phosphatase 1 gene at 87B can differentially affect suppression of position effect variegation and mitosis in Drosophila melanogaster. Genetics. 1993;135:117–125. doi: 10.1093/genetics/135.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball L J, Murzina N V, Broadhurst R W, Raine A R C, Archer S J, Stott F J, Murzin A G, Singh P B, Domaille P J, Laue E D. Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J. 1997;16:2473–2481. doi: 10.1093/emboj/16.9.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardoso C, Timsit S, Villard L, Khrestchatisky M, Fontes M, Colleaux L. Specific interaction between the XNP/ATR-X gene product and the SET domain of the human EZH2 protein. Hum Mol Genet. 1998;7:679–684. doi: 10.1093/hmg/7.4.679. [DOI] [PubMed] [Google Scholar]
- 8.Cléard F, Delattre M, Spierer P. SU(VAR)3-7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. EMBO J. 1997;16:5280–5288. doi: 10.1093/emboj/16.17.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corda Y, Schramke V, Longhese M P, Smokvina T, Paciotti V, Brevet V, Gilson E, Géli V. Interaction between Set1p and checkpoint protein Mec3p in DNA repair and telomere functions. Nat Genet. 1999;21:204–208. doi: 10.1038/5991. [DOI] [PubMed] [Google Scholar]
- 10.Cui X, De Vito I, Slany R, Miyamoto A, Firestein R, Cleary M L. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet. 1998;18:331–337. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- 11.De Rubertis F, Kadosh D, Henchoz S, Pauli S, Reuter G, Struhl G, Spierer P. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature. 1996;384:589–591. doi: 10.1038/384589a0. [DOI] [PubMed] [Google Scholar]
- 12.Eissenberg J C, Morris G D, Reuter G, Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992;131:345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekwall K, Nimmo E R, Javerzat J-P, Borgström B, Egel R, Cranston G, Allshire R. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci. 1996;109:2637–2648. doi: 10.1242/jcs.109.11.2637. [DOI] [PubMed] [Google Scholar]
- 14.Fanti L, Giovinazzo G, Berlocco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- 15.Firestein, R., X. Cui, P. Huie, and M. L. Cleary. SET domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3-9. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 16.Grewal S I, Bonaduce M J, Klar A J. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics. 1999;150:563–576. doi: 10.1093/genetics/150.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–328. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 18.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 19.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–95. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 20.Hendzel M J, Wei Y, Mancini M A, van Hooser A, Ranalli T, Brinkley B R, Bazett-Jones D P, Allis C D. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 21.Henikoff S. Position-effect-variegation in Drosophila: recent progress. In: Russo V E A, Martienssen R A, Riggs A D, editors. Epigenetic mechanisms of gene regulation. New York, N.Y: CSHL Press; 1996. pp. 319–334. [Google Scholar]
- 22.Huang N, vom Baur E, Garnier J-M, Lerouge T, Vonesch J-L, Lutz Y, Chambon P, Losson R. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J. 1998;17:3398–3412. doi: 10.1093/emboj/17.12.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanova A V, Bonaduce M J, Ivanov S V, Klar A J S. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat Genet. 1998;19:192–195. doi: 10.1038/566. [DOI] [PubMed] [Google Scholar]
- 24.James T C, Elgin S C R. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila and its gene. Mol Cell Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenuwein T, Laible G, Dorn R, Reuter G. SET-domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karpen G, Allshire R C. The case for epigenetic effects on centromere identity and function. Trends Genet. 1997;13:489–496. doi: 10.1016/s0168-9525(97)01298-5. [DOI] [PubMed] [Google Scholar]
- 27.Koonin E V, Zhou S, Lucchesi J C. The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 1995;23:4229–4232. doi: 10.1093/nar/23.21.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorentz A, Ostermann K, Fleck O, Schmidt H. Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene. 1994;143:139–143. doi: 10.1016/0378-1119(94)90619-x. [DOI] [PubMed] [Google Scholar]
- 29.Martin-Zanca D, Oskam R, Mitra G, Copeland T, Barbacid M. Molecular and biochemical characterisation of the human trk proto-oncogene. Mol Cell Biol. 1989;9:24–33. doi: 10.1128/mcb.9.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messmer S, Franke A, Paro R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992;6:1241–1254. doi: 10.1101/gad.6.7.1241. [DOI] [PubMed] [Google Scholar]
- 31.Minc E, Allory Y, Worman H J, Courvalin J-C, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 32.Murzina N, Verrault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen A L, Ortiz J A, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paro R, Hogness D S. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paro R, Harte P J. The role of Polycomb group and trithorax group complexes in the maintenance of determined cell states. In: Russo V E A, Martienssen R A, Riggs A D, editors. Epigenetic mechanisms of gene regulation. New York, N.Y: CSHL Press; 1996. pp. 507–528. [Google Scholar]
- 36.Pirrotta V. Stable chromatin states regulating homeotic genes in Drosophila. In: Russo V E A, Martienssen R A, Riggs A D, editors. Epigenetic mechanisms of gene regulation. New York, N.Y: CSHL Press; 1996. pp. 489–505. [Google Scholar]
- 37.Platero J S, Harnett T, Eissenberg J C. Functional analysis of the chromo domain of HP-1. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Renauld H, Aparicio O, Zierath P, Billington L, Chhablani S, Gottschling D. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 39.Reuter G, Giarre M, Farah J, Gausz J, Spierer A, Spierer P. Dependence of position-effect variegation in Drosophila on dose of a gene encoding an unusual zinc-finger protein. Nature. 1990;344:219–223. doi: 10.1038/344219a0. [DOI] [PubMed] [Google Scholar]
- 40.Reuter G, Spierer P. Position-effect variegation and chromatin proteins. Bioessays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- 41.Rosenblatt-Rosen O, Rozovskaia T, Burakov D, Sedkov Y, Tillib S, Blechman J, Nakamura T, Croce C M, Mazo A, Canaani E. The C-terminal SET domains of ALL-1 and TRITHORAX interact with the INI1 and SNR1 proteins, components of the SWI/SNF complex. Proc Natl Acad Sci USA. 1998;95:4152–4157. doi: 10.1073/pnas.95.8.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan R F, Schultz D C, Ayyanathan K, Singh P B, Friedman J R, Fredericks W J, Rauscher F J., III KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box–zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saunders W S, Chue C C, Goebl M, Craig C, Clark R F, Powers J A, Eissenberg J C, Elgin S C R, Rothfield N F, Earnshaw W C. Molecular cloning of a human homologue of Drosophila heterochromatin protein HP1 using anti-centromere autoantibodies with anti-chromo specificity. J Cell Sci. 1993;104:573–582. doi: 10.1242/jcs.104.2.573. [DOI] [PubMed] [Google Scholar]
- 44.Schultz J, Copley R R, Doerks T, Ponting C P, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stone E M, Pillus L. Activation of a MAP kinase cascade leads to Sir3p hyperphosphorylation and strengthens transcriptional silencing. J Cell Biol. 1996;135:571–583. doi: 10.1083/jcb.135.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 47.Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner B M. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci. 1998;54:21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallrath L L. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- 50.Wei Y, Lanlan Y, Bowen J, Gorovsky M A, Allis C D. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- 51.Wreggett K A, Hill F, James P S, Hutchings G W, Singh P B. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet Cell Genet. 1994;66:99–103. doi: 10.1159/000133676. [DOI] [PubMed] [Google Scholar]