Abstract
Structural biologists provide direct insights into the molecular bases of human health and disease. The open-access Protein Data Bank (PDB) stores and delivers three-dimensional (3D) biostructure data that facilitate discovery and development of therapeutic agents and diagnostic tools. We are in the midst of a revolution in vaccinology. Non-infectious mRNA vaccines have been proven during the coronavirus disease 2019 (COVID-19) pandemic. This new technology underpins nimble discovery and clinical development platforms that use knowledge of 3D viral protein structures for societal benefit. The RCSB PDB supports vaccine designers through expert biocuration and rigorous validation of 3D structures; open-access dissemination of structure information; and search, visualization, and analysis tools for structure-guided design efforts. This resource article examines the structural biology underpinning the success of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) mRNA vaccines and enumerates some of the many protein structures in the PDB archive that could guide design of new countermeasures against existing and emerging viral pathogens.
Keywords: mRNA vaccine, structural biology, virus structure, structure-facilitated design, surface glycoprotein, carbohydrate, SARS-CoV-2, COVID-19
Graphical abstract

Goodsell and Burley examine the structural biology underpinning the success of SARS-CoV-2 mRNA vaccines and present freely available resources at the RCSB Protein Data Bank that could guide the structure-facilitated design of new countermeasures against existing and emerging viral pathogens.
Introduction
Structural biology represents an essential tool in our quest to understand fundamental biology, biomedicine, bioenergy, and biotechnology/bioengineering in 3D at atomic resolution (Burley et al., 2018). The Protein Data Bank (PDB) was established in 1971 with just seven X-ray crystal structures of proteins as the first open-access digital data resource in biology (Protein Data Bank, 1971). Now in its 50th year of continuous operations, the PDB is the single global archive of >180,000 experimentally determined 3D structures of proteins, DNA, and RNA. Since 2003, the global PDB archive has been managed jointly by the Worldwide PDB partnership (wwPDB; Berman et al., 2003; wwPDB consortium, 2019). Current wwPDB member organizations include the US-funded RCSB PDB (RCSB PDB; Berman et al., 2000; Burley et al., 2021), the PDB in Europe (PDBe; Armstrong et al., 2020), PDB Japan (PDBj; Kinjo et al., 2018), the Electron Microscopy Data Bank (EMDB; Abbott et al., 2018), and the Biological Magnetic Resonance Bank (BMRB; Romero et al., 2020). In addition to the PDB archive, wwPDB members are also jointly responsible for global management of the EMDB and BMRB archives. The wwPDB supports tens of thousands of structural biologists from all inhabited continents, who freely contribute their data to the archive and many millions of PDB data consumers (e.g., researchers, educators, students, policy makers, science funders, and the curious public) living and working in every sovereign nation and territory around the world (Burley et al., 2018).
Structural biologists and the PDB are playing critical roles in efforts to improve global health and fight disease in humans, animals, and agricultural crops (Burley et al., 2018; Goodsell et al., 2020). Approximately 90% of United States Food and Drug Administration (US FDA) new drug approvals between 2010 and 2016 were facilitated by open access to PDB data, much of it contributed by researchers in universities, government laboratories, and not-for-profit research institutes largely funded by taxpayer monies (Galkina Cleary et al., 2018; Westbrook and Burley, 2019). Structural biologists and the PDB have had particularly significant impacts on discovery and development of antineoplastic agents (Westbrook et al., 2020). More than 70% of new small-molecule anticancer drugs approved by US FDA in 2010–2018 were products of structure-guided drug discovery in biopharmaceutical companies (reviewed in Burley, 2021). In the vast majority of cases, these for-profit drug discovery efforts were enabled by open access to PDB structures of the drug target contributed by publicly funded researchers. Every major drug company and many smaller biotechnology companies maintain copies of the PDB archive inside their firewalls for interoperation with proprietary information. The charter governing wwPDB operations (https://www.wwpdb.org/about/agreement) expressly forbids charging PDB, EMDB, and BMRB data depositors and data consumers, and provides access to all archival information under the most permissive Creative Commons License CC0 1.0 Universal (https://creativecommons.org/publicdomain/zero/1.0/). Every structure housed in the PDB archive is identified with a unique code (currently four alphanumeric character codes; e.g., PDB: 6lu7, the first deposited structure of the severe acute respiratory syndrome coronavirus-2 [SARS-CoV-2] main protease). Minimal information regarding each PDB structure can be accessed using its dedicated wwPDB landing page at https://doi.org/10.2210/pdb6lu7/pdb. These DOIs may be used to provide citations to individual structures (strongly recommended for citing PDB structures lacking primary literature references describing structure determinations). Links on each landing page in turn provide access to wwPDB partner structure summary pages (e.g., https://www.rcsb.org/structure/6LU7, hosted by RCSB PDB).
With the growing realization that emerging viral pathogens pose an increasing threat to global health, structural biologists have aggressively explored the basic principles of virus biology, methods for using structure-guided drug discovery to develop antiviral agents, and new ways to apply knowledge of viral structure to create safe and effective vaccines. Since late January 2020, more than 1,500 structures of SARS-CoV-2 proteins have been deposited into the PDB (https://rcsb.org/covid19). Of central importance to this resource article, as of October 2021 there are ∼600 PDB structures of the viral surface glycoprotein. These data are informing our understanding of SARS-CoV-2 variants and enabling 3D characterization of neutralizing antibodies generated in response to infection or vaccination, or engineered for passive immunization of infected individuals.
Vaccines represent one of the great successes of medical science, providing long-term protection against multiple life-threatening infections and saving hundreds of millions of lives (Pollard and Bijker, 2021; Rappuoli et al., 2021). The tried and true approach in the fight against viral diseases has been protein based, administering viral antigens to stimulate an immune response. Given the phenomenal success of early vaccines, many variations on this approach have been developed and deployed, including inactivated viruses (e.g., poliovirus); live cell-culture adapted viruses (e.g., measles, mumps, rubella); empty viral capsids (e.g., human papillomavirus); recombinant viral proteins (e.g., hepatitis B virus surface antigen); and, more recently, engineered nanoparticles that display viral proteins (e.g., Novavax). The protein-based approach to vaccine development, however, is a long and expensive process that must be customized to each virus. A typical vaccine may require >10 years for discovery, development, and testing before regulatory approval (Kowalzik et al., 2021). New approaches using nucleic acids are currently being developed to shorten this timeline. Rather than challenging the immune system directly with viral antigens, these vaccines deliver genetic material that encodes immunogens. Once the delivered gene is transcribed and/or translated, viral proteins are displayed on host cell surfaces and presented to the cellular immune surveillance system. Current gene-based approaches include DNA-based vaccines using engineered adenoviruses and messenger RNA (mRNA) vaccines (Pardi et al., 2018).
Recent successes of the non-infectious Pfizer-BioNTech and Moderna mRNA vaccines discovered and developed for the coronavirus disease 2019 (COVID-19) pandemic have demonstrated the promise of this new, more nimble approach to vaccine development (Figure 1 ) (Park et al., 2021). mRNA vaccines have many advantages: they elicit both humoral (i.e., antibody) and cell-mediated immune responses, while being well tolerated by healthy individuals with few side effects and minimal risk of anaphylaxis. They are also far less expensive and time consuming to develop. Next-generation mRNA vaccines may be rapidly deployed by simply changing the sequence of the mRNA (e.g., for protection against SARS-CoV-2 variants). For existing or newly emerging diseases, recent experience suggests that vaccine discovery and development timelines can be substantially reduced going forward.
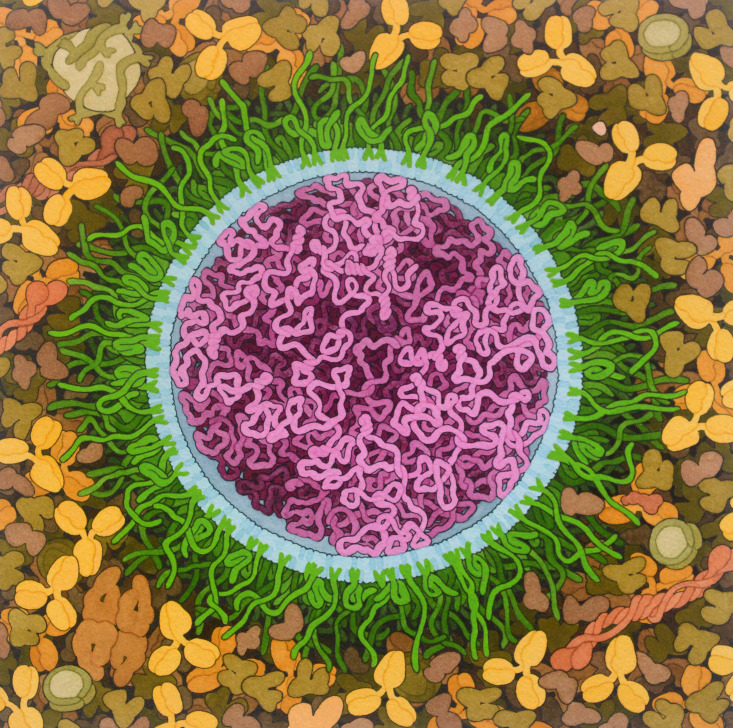
Figure 1.
Idealized artistic conception of a SARS-CoV-2 mRNA vaccine
mRNA (magenta) is surrounded by a specialized lipid membrane, which typically includes PEGylated lipids (green), which protect the surface, and ionizable lipids (blue), which are neutral at physiological pH but become charged upon acidification of the endosome, thereby facilitating mRNA delivery into the cytoplasm. Surrounding the vaccine particle, IgG antibodies and various human plasma proteins are depicted. Original painting
(https://doi.org/10.2210/rcsb_pdb/goodsell-gallery-027) was created with traditional watermedia based on shapes and sizes of molecular structures taken from the PDB archive.
This resource article is the product of the US-funded RCSB PDB. The RCSB PDB delivers PDB data using two web portals: research-focused (https://rcsb.org) and education/outreach-focused (https://pdb101.rcsb.org). Herein, we briefly introduce a short history of modern mRNA vaccines, and then present resources that are available from the RCSB.org web portal to facilitate structure-guided approaches to mRNA vaccine design against existing and emerging viral pathogens. Several case studies are presented that exemplify use of structural data in vaccine design. We also briefly describe how the structure-facilitated design of mRNA vaccines is currently informing advances in the adjacent medical field of cancer therapy.
History and initial deployment of mRNA vaccines
Prior to the COVID-19 pandemic, a number of viral pathogens had become the focus of mRNA vaccine design and development efforts (e.g., respiratory syncytial virus [RSV], rabies virus, Zika virus, and human cytomegalovirus [CMV]) (Pardi et al., 2018). Perfecting this promising new vaccine design technology became imperative in the face of the COVID-19 global public health emergency.
A little more than 12 months after individuals infected with SARS-CoV-2 were first identified in Wuhan in the People’s Republic of China, the Pfizer-BioNTech and Moderna mRNA vaccines against SARS-CoV-2 received Emergency Use Authorization in the US and other developed countries. The initial design of the Moderna mRNA-1273 vaccine was finalized 42 days after the sequence of the SARS-CoV-2 mRNA genome was made publicly available (Hodgson, 2020). During the design phase, researchers at both Pfizer-BioNTech and Moderna had open access to a total of 18 3D structures of the extracellular portion of the SARS-CoV-1 spike protein (first PDB: 5x5b, publicly released 5/3/2017; Yuan et al., 2017). At approximately 78% amino acid sequence identity to its SARS-CoV-2 counterpart, they would have known that the 3D structures of the two spike proteins were highly similar (Sander and Schneider, 1991). For reference, the first PDB structure of a SARS-CoV-2 spike protein (PDB: 6vsb) was publicly released on 2/26/2020 (Wrapp et al., 2020) revealing root-mean-square deviation of about 1.5Å (PDB: 5x5b) (Yuan et al., 2017) for 917 equivalent α-carbon pairs. Since PDB: 6vsb became publicly available, ∼600 structures of SARS-CoV-2 spike proteins have been deposited into the PDB, including those of the spike protein bound to its cellular receptor, angiotensin-converting enzyme 2 (ACE2), and various neutralizing antibodies.
By mid-2021, mass vaccination programs in much of the developed world (utilizing both mRNA vaccines described above and two adenovirus-based DNA vaccines: Oxford-AstraZeneca ChAdOx1 nCoV-19 or AZD1222; Johnson & Johnson JNJ-78436735 or Ad26.COV2.S) began to turn the tide of the pandemic. Israel, for example, had administered more than 10 million doses of the Pfizer vaccine (sufficient to fully vaccinate approximately 59% of the country’s estimated population of more than nine million). Also, by mid-2021, new infections in Israel had declined from a peak of more than 10,000 per day in mid-January 2021 to less than 100, and daily fatalities in Israel had declined from a peak of more than 60 per day in late January 2021 to zero (7-day averages). In Israel and around the globe, the race is now on to vaccinate as many medically eligible individuals as possible before existing and emerging hyper-transmissible variants of SARS-CoV-2 cause entirely preventable deaths in geographies with low vaccination rates and put those around the world who cannot be vaccinated for medical or religious reasons at needless risk of serious illness requiring hospitalization or death.
Visualizing viral surface proteins and structure/function relationships in 3D
Structural biology is playing a central role in the design of new vaccines, by revealing the structure/function relationships of the viral targets of vaccines and by providing ways to optimize the effectiveness of the target antigens used in vaccines. mRNA viral vaccine candidates are typically designed to elicit both humoral and cellular immune responses against viral surface proteins that are readily susceptible to antibody neutralization (Figure 2 ). Macromolecular crystallography (MX) and single-particle cryoelectron microscopy tools (3D electron microscopy or 3DEM) are being used routinely to visualize viral proteins in 3D at the atomic level. Not surprisingly, these studies have revealed structural features of the viral surface proteins that present challenges for both structural biologists and vaccine designers.
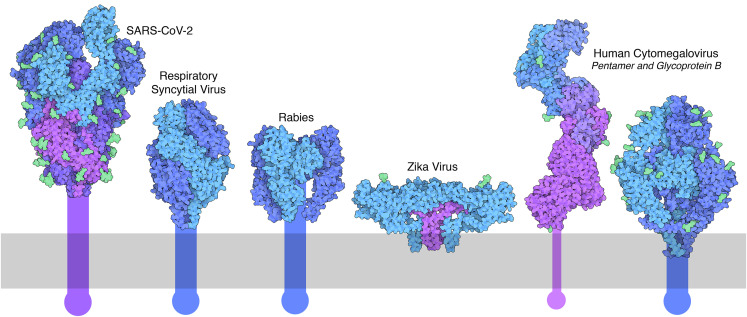
Figure 2.
Selected viral surface glycoproteins currently being targeted with mRNA vaccines
Structures are available in the PDB archive for ectodomains, with proteins shown in shades of blue and purple and surface glycans (when included in the atomic coordinates) in green. The extent of the lipid bilayer is indicated in gray, and transmembrane portions are shown schematically. SARS-CoV-2 spike (PDB: 6vyb; Walls et al., 2020); RSV fusion glycoprotein (PDB: 4jhw; McLellan et al., 2013b); rabies virus glycoprotein (PDB: 6lgx; Yang et al., 2020) with trimeric assembly (based on PDB: 5i2s; Roche et al., 2007); Zika virus E (blue) and M (purple) proteins (PDB: 5ire; Sirohi et al., 2016); cytomegalovirus (CMV) pentamer with subunits depicted in shades of blue and purple (PDB: 5vob; Chandramouli et al., 2017); and CMV glycoprotein B (PDB: 7kdp; Liu et al., 2021b). Figure created with Illustrate software (ccsb.scripps.edu/illustrate).
First, many of these proteins undergo significant structural transitions during the course of viral infection. Viruses typically use their surface antigens to recognize and bind to one or more cellular receptors under physiologic conditions (i.e., pH 7.4). Following endocytosis, spike protein structures can change dramatically (triggered by acidification of the local environment that is mediated by proton pumps pre-positioned in the endosomal membrane) into a fusion-competent conformation (Sollner, 2004). These phenomena make for interesting extra work by structural biologists, requiring determination of multiple structures of different conformational states and computational modeling of putative conformations that cannot be captured and studied with current experimental methods. In addition, both conformationally dynamic loops and membrane-spanning regions frequently pose experimental challenges for structure determination, particularly with MX, wherein well-ordered crystals are required. Troublesome segments of the full-length polypeptide chain are often removed or replaced with more stable, engineered sequences before they can be visualized in 3D.
Second, many viral surface proteins are glycoproteins (i.e., they are decorated with carbohydrate moieties resulting from enzymatic post-translational modification). Glycosylation plays an important functional role in shielding portions of these proteins from immune surveillance (Julien et al., 2012). Glycosylation, however, can pose difficulties in structure determination because of the static or dynamic disorder and chemical heterogeneity of glycan chains. Frequently for MX studies, wherein carbohydrates are notorious for interfering with crystallization, sites of glycosylation are mutated to eliminate post-translational modification. Single-particle 3DEM does not require crystallization, permitting structural studies of glycoproteins in their native states. For vaccine designers, however, the entire glycan may not be visible in the structure-determination experiment using either MX or 3DEM, because of dynamic disorder. As a result, atomic-level 3D structures of glycoproteins in the PDB do not always provide information regarding the full complement of covalently bound sugars.
The RCSB PDB provides a number of resources to help PDB data consumers navigate these challenges. The research-focused RCSB.org web portal maintains strong and accessible connections to over 50 biodata resources, such as UniProt (UniProt, 2021), NCBI/RefSeq (Li et al., 2021), GlyTouCan (Tiemeyer et al., 2017), GlyCosmos (Yamada et al., 2020), and GlyGen (York et al., 2020), allowing ready access to authoritative sequence and functional information. External sequence data integration can be useful, for example, in identifying regions of the polypeptide chain that are not represented in the atomic coordinates and learning more about their functional roles. wwPDB partners recently performed a remediation of carbohydrate-containing structures across the entire PDB archive (Shao et al., 2021). Nearly 15,000 PDB structures (∼10% of archival holdings at the time) were remediated, including many viral glycoproteins. These remediated structures (and those of every glycoprotein that will be deposited to the PDB in future) use standardized atom and residue nomenclature for all carbohydrates based on the 1996 International Union of Pure and Applied Chemistry (IUPAC) recommendations (McNaught, 1996). All glycoproteins in the PDB are now properly annotated for glycosylation sites and all glycans are uniformly represented as branched oligosaccharides, utilizing the Symbol Nomenclature for Glycans (SNFG) representation standard (Varki et al., 2015). Improved representation and annotation now support glycosylation-specific searching and analyses using the RCSB.org web portal.
PDB archival holdings for viral surface glycoproteins
Given the importance of a structural understanding of the mechanisms of viral entry and immune neutralization, the structural biology community has launched a comprehensive effort to characterize many of the viruses currently posing risks to global health. Table 1 and Figure 2 include a representative selection of the many viral glycoprotein structures currently housed in the PDB archive. Well-studied exemplars include hundreds of individual PDB structures, providing a detailed portrait of the structure and function of each glycoprotein. The RCSB.org web portal includes powerful tools for streamlining exploration of the current holdings for a particular protein. The Structure Summary Page (SSP) of a representative PDB entry may be easily found through a simple text search using the main search bar. Once a relevant text search hit is identified and selected, the user is taken to the SSP. Therein, several tools allow enumeration of related entries. Because of the diversity of viruses (many of which have variants with proteins differing slightly in amino acid sequence and structure), use of several search options may be necessary to get a comprehensive view of current archival holdings for a particular spike protein. The simplest search option supports finding all structures corresponding to a given UniProt ID. For viruses that encode multiple proteins within one or more polyproteins, however, searching on UniProt ID may return PDB structure hits other than the desired spike protein. A more consistently reliable approach allows the PDB data consumer to search for all structures with a desired sequence identity to the representative entry (available sequence identity options: 100%, 95%, 90%, 80%, 70%, 60%, 40%, 30%). One-click sequence searching at 100% identity will not return structures of variants. When searching for variants of a particular glycoprotein, we recommend performing sequence searching at 95% identity, which should eliminate false-positives corresponding to related viruses (e.g., SARS-CoV-2 and SARS-CoV spike proteins are ∼78% identical). Occasionally, PDB data depositors have studied chimeric structures composed of sequence segments from related viruses. Sequence searching at lower sequence identity can help reveal such cases. (Note that care must be taken to exclude false-positive structure search results that encompass only a small fraction of the protein sequence, such as affinity purification tags.) When studying the structures of surface glycoproteins from related viruses (e.g., from SARS-CoV-2 and other coronaviruses), one-click structure similarity searching from an RCSB.org SSP using our Zernike polynomial-based system (Guzenko et al., 2020) can be very effective.
Table 1.
Selected viral pathogen glycoprotein structures in the PDB
| Virus name | Protein name | PDB ID | UniProt hitsa | 80% hitsb | References |
|---|---|---|---|---|---|
| Coronaviruses | |||||
| SARS-CoV | spike glycoprotein | 6crz | 49 | 43 | Kirchdoerfer et al. (2018) |
| SARS-CoV-2 | spike glycoprotein | 6vsb | 444 | 438 | Wrapp et al. (2020) |
| MERS-CoV | spike glycoprotein | 5w9n | 38 | 32 | Pallesen et al. (2017) |
| Paramyxoviruses | |||||
| RSV A | fusion glycoprotein F0 | 4jhw | 63 | 82 | McLellan et al. (2013b) |
| Measles virus | hemagglutinin | 2zb5 | 6 | 8 | Hashiguchi et al. (2007) |
| Mumps virus | hemagglutinin-neuraminidase | 5b2d | 4 | 4 | Kubota et al. (2016) |
| Rhabdoviruses | |||||
| Rabies virus | rabies glycoprotein | 6lgx | 2 | 3 | Yang et al. (2020) |
| Vesicular stomatitis virus | glycoprotein G | 5i2s | 3 | 5 | Roche et al. (2007) |
| Orthomyxoviruses | |||||
| Influenza virus | hemagglutinin | 1ruz | 17 | 81,432c | Gamblin et al. (2004) |
| neuraminidase | 1nn2 | 11 | 35,218c | Varghese and Colman (1991) | |
| Herpesviruses | |||||
| Herpes simplex virus | envelope glycoprotein B | 3nw8 | 12 | 12 | Stampfer et al. (2010) |
| Human cytomegalovirus | envelope glycoprotein B | 7kdp | 6 | 6 | Liu et al. (2021b) |
| Flaviviruses | |||||
| Zika virus | envelope protein E | 5ire | 48 | 42 | Sirohi et al. (2016) |
| Dengue virus | envelope protein E | 1tg8 | 2 | 45 | Zhang et al. (2004) |
| West Nile virus | envelope protein E | 2i69 | 1 | 13 | Kanai et al. (2006) |
| Tick-borne encephalitis virus | envelope protein E | 1svb | 5 | 14 | Rey et al. (1995) |
| Hepatitis C virus | envelope glycoprotein E2 | 4mwf | ndd | 23d | Kong et al. (2013) |
| Retroviruses | |||||
| HIV-1 | Envelope glycoprotein | 4nco | 93 | 121 | Julien et al. (2013) |
Number of PDB structures with identical UniProt IDs versus the representative PDB ID, evaluated July 15, 2021.
Number of PDB structures obtained using an 80% sequence identity search versus the representative PDB ID, then filtered using the "Scientific Name of Source Organism" refinement on the SSP. (the non-intuitive cases where 80% sequence identity number is smaller than the UniProt ID search number is due in large part to the presence of PDB structures with small peptide fragments of the proteins, which are not recognized by the sequence similarity search, or inclusion of other domains in cases where the UniProt ID corresponds to a polyprotein.)
Because influenza virus proteins are so diverse, a fuller representation of PDB holdings was evaluated using the “Structure” similarity search versus the representative PDB ID.
A 50% sequence identity search versus the representative PDB ID was used to capture multiple subtypes of hepatitis C virus glycoprotein E2; the UniProt entry includes the entire genome polyprotein, so results for the UniProt search are not included here.
RCSB PDB and open-access data
To make structure-enabled mRNA vaccine design possible, and indeed all structure-enabled science, standardized structure archiving, rigorous validation, expert biocuration, and facile data delivery are essential. Ample evidence of the central role played by the PDB archive has been published in peer-reviewed scientific journals (Burley et al., 2018; Feng et al., 2020; Goodsell et al., 2020; Markosian et al., 2018), going well beyond the fields of structure-guided drug discovery (Burley, 2021; Westbrook and Burley, 2019; Westbrook et al., 2020) and protein structure prediction (Burley and Berman, 2021). The RCSB PDB and its wwPDB partners are dedicated to timely archiving of new results, continuing the 50-year PDB tradition of supporting scientific discovery and technical innovation based on experimental data freely contributed by structural biologists. Making good on this commitment involves complementary activities, including timely validation/biocuration and archiving of newly deposited information, open access to 3D structure data with no limitations on usage, provision of effective tools for searching and downloading archival data, and enabling web-based visualization and analysis of PDB structures.
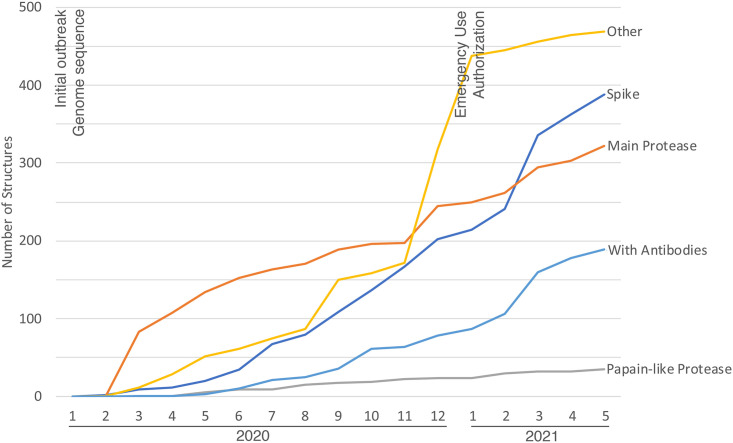
The RCSB PDB response to the COVID-19 global pandemic highlights the PDB’s long-standing adherence to the FAIR principles of findability, accessibility, interoperability, and reusability (Wilkinson et al., 2016). It is no exaggeration to state that the PDB was “walking the walk” decades before people began “talking the talk” about concepts such as FAIR and fairness, accuracy, confidentiality, and transparency (FACT) (van der Aalst et al., 2017). As illustrated in Figure 3 , the first SARS-CoV-2 protein structure was deposited to the PDB within months of the initial outbreak. The shared commitment of the scientific community, including structural biologists, the PDB, and most scientific publishers, was to make pandemic-related research results immediately accessible. This unprecedented level of cooperation, and our ability to build on abundant and freely available structure data from previous coronavirus outbreaks, is supporting rapid discovery and development of multiple vaccines, neutralizing antibodies, and small-molecule drugs targeting SARS-COV-2.
Figure 3.
PDB archival holdings related to SARS-CoV-2 proteins accumulated during the COVID-19 global pandemic
In particular, we are enjoying the fruits of a “resolution revolution” in 3DEM (Kuhlbrandt, 2014), which is providing structural results for challenging biological systems at a pace far exceeding the capabilities of more established structure-determination techniques (e.g., MX). Improved sample preparation and cryo-preservation techniques, cryogenically cooled electron microscopes, direct electron detectors, and advances in software for data processing and structure determination are together providing new opportunities and posing new challenges to the scientific community, the wwPDB, and RCSB PDB. Of immediate concern is the need for new methods for assessing and validating 3DEM structural results and methods for interpretable display and exploration of large macromolecular assemblies. The wealth of structural information now available for the SARS-CoV-2 spike protein and its interactions with cell-surface receptors and antibodies, described in more detail below, is testament to the power of 3DEM.
Recent admission of EMDB to the wwPDB partnership formalized a long-standing arrangement, wherein the OneDep global system for PDB structure deposition, validation, and biocuration served the needs of the 3DEM community. OneDep is the “one-stop shop” for deposition of atomic coordinates and supporting experimental data for structures determined using MX, 3DEM, and nuclear magnetic resonance (NMR) spectroscopy. Atomic coordinates for structures determined by MX, 3DEM, or NMR are stored in the PDB archive. Supporting experimental data are stored in the core archives jointly managed by the wwPDB; MX data are stored in the PDB archive, 3DEM maps are stored in the EMDB archive, and NMR data are stored in the BMRB archive. All three wwPDB core archives interoperate with one another.
The PDB is one of the most highly curated biological data archives. The PDB and individual structures therein are trusted by data depositors and data consumers alike. For this reason, and others discussed below, PDB usage is among the highest of any data repository in biology (Read et al., 2015).
Free availability of rigorously validated and expertly biocurated 3D structures from the PDB archive has enabled progress in the fields of structural biology and structural bioinformatics (reviewed in Burley and Berman, 2021) in myriad ways, including development of new structure-determination methods, structure-guided drug discovery, predicting the impact of point mutations in proteins, comparative or homology protein structure modeling, protein-ligand pose prediction and scoring, prediction of protein-protein interactions, molecular dynamics simulations, and de novo protein structure prediction.
Open access to 3D biostructures supports established research efforts broadly in fundamental biology, biomedicine, bioenergy, and biotechnology/bioengineering, and studies of newly emerging topics. Because the vast majority of PDB data consumers are not structural biologists (and are indeed unlikely to ever contribute a structure to the archive), our RCSB.org web portal has been engineered (1) to enable searching for relevant structure(s), (2) to provide integrated complementary information from trusted external data sources, and (3) to support facile 3D visualization of structures:
-
(1)
Multi-dimensional approaches to structure searching allow RCSB.org web portal users to pinpoint the data they need for their particular research or teaching needs. Simple text searching available at the top of every RCSB.org web page often suffices. A powerful, intuitive interface is also available for performing specialized searches across hundreds of structural features and descriptors with the option of using Boolean logic to further narrow the results set. Once an initial set of structures is returned by the search system, the user can further narrow the results. A variety of options are available to manage examination of a results set, such as easy "Refinement" checkboxes, which are particularly useful in cases with large numbers of structures, such as viral structures or structures related to immunoglobulin structure and binding. Browsing and detailed examination of individual structures is supported by dedicated SSPs, as described above for viral surface glycoproteins.
-
(2)
On every dedicated SSP, extensive interoperation with trusted external data sources facilitates exploration of multiple sources of information for the protein of interest. Currently more than 40 external data resources are integrated with PDB data. A robust pipeline connects individual PDB structures to corresponding UniProt (UniProt, 2021) and NCBI/RefSeq (Li et al., 2021) accession codes with mapping at the amino acid level between the 3D structure and the protein sequence in both cases. Sequence and function information in UniProt is available graphically in the RCSB Protein Explorer, and links embedded in the SSP allow direct access to UniProt pages and other PDB entries with the same UniProt ID. Reciprocally, UniProt provides a simple browser and viewing capability for PDB holdings, and direct links to wwPDB member web sites. Links are also provided to CATH (Sillitoe et al., 2021), SCOP (Andreeva et al., 2019), DrugBank (Wishart et al., 2018), PubChem (Kim et al., 2021), BindingDB (Gilson et al., 2016), and Pharos (Nguyen et al., 2017), all of which are relevant to the problem of discovery and development of viral pathogen countermeasures.
-
(3)
Turnkey molecular visualization is accessible on the RCSB.org web portal as a stand-alone feature and from within every dedicated SSP, using the web-native 3D molecular graphics display system Mol∗ (Sehnal et al., 2021). The principal advantage of Mol∗ versus other currently available web-native molecular graphic tools stems from deep integration of protein sequence information with the 3D atomic coordinates made possible by its reliance on the PDBx/mmCIF data standard that underpins the PDB archive. This unique feature of Mol∗ enables navigation of PDB structures and communication with one-dimensional (1D) protein sequence features integrated from external data resources. The Mol∗ software library provides a technology stack for state-of-the-art data delivery, web-native molecular graphics, and analysis tools for interrogating 3D macromolecular structure data. Mol∗ is the collaboratively developed successor of the RCSB PDB NGL Viewer (Rose et al., 2018) and PDB in Europe LiteMol (Sehnal et al., 2017). It works entirely within the user’s web browser, avoiding the need to license, download, install, or maintain external software.
Mol∗ supports routine display of atoms and interatomic bonds, metal ions, and bound ligands in a variety of commonly used biomolecular representational styles and rendering of molecular surfaces for depiction of protein-protein interfaces and small-molecule binding sites. It also supports graphical display and analyses (structural interrogation) of intra- and inter-molecular contacts, and more generally inter-molecular contacts between any number of polymer chains and ligands in macromolecular assemblies of any size (e.g., larger than an entire ribosome). Mol∗ provides a user-friendly and rapid way to visualize structures “on-the-fly,” streamlining exploration of many structures during the initial search phase of a study and providing advanced options for detailed study and analysis of the most salient structures.
Vaccine design case studies: prefusion state stabilization of viral glycoproteins
The power of structure-facilitated vaccine design is being demonstrated in an ongoing effort to discover and develop a vaccine against RSV, which is the most common cause of bronchiolitis and pneumonia in children under 1 year of age in the US. Infants, young children, and older adults with chronic medical conditions are at risk of severe disease from RSV infection. Annually in the US, RSV is responsible on average for approximately 58,000 hospitalizations, with 100–500 deaths among children younger than 5 years old and 177,000 hospitalizations with 14,000 deaths among adults aged 65 years or older. RSV, a member of the Orthopneumovirus family, is an enveloped virus carrying a negative-sense single-stranded RNA genome that encodes 11 proteins (∼15,000 nucleotides in length). Although there is currently no vaccine against RSV, passive immunization with monoclonal antibodies (MAbs) is available to prevent RSV infection and hospitalization in infants at highest risk. F and G glycoproteins are the two major surface proteins that control viral attachment and the initial stages of infection. They are the primary targets for neutralizing antibodies during natural infection (Battles and McLellan, 2019). The G protein is produced as either a membrane-bound form that mediates viral attachment or a secreted form involved in immune evasion. It is largely disordered, but structures of a central region have revealed details of its interactions with antibodies The most successful anti-RSV MAb palivizumab (Astra Zeneca, brand name Synagis) is directed against the A antigenic site of the surface fusion (F) glycoprotein of RSV. The fusion glycoprotein is also the target in a structure-facilitated design effort for vaccine development.
Analysis of antibody binding revealed that the most effective antigenic sites are present on the metastable prefusion state of the F glycoprotein, suggesting that stabilization of this conformation would lead to a more effective vaccine (Gilman et al., 2016; Magro et al., 2012). Based on structures of the glycoprotein ectodomain in pre- and post-fusion conformations, a series of stabilizing amino acid changes were predicted and tested (Figure 4 ). These substitutions included addition of a disulfide bridge between a pair of amino acids that are 4.4 Å apart in the prefusion form but 124.2 Å apart in the post-fusion conformation, plus several changes intended to fill small cavities and a T4-phage fibritin trimerization domain (foldon) to stabilize the C terminus (McLellan et al., 2013a). The resulting prefusion-stabilized form of the protein showed much improved RSV-neutralizing activity. This landmark study has led to subsequent work demonstrating a proof of concept for effectiveness of this approach in a subunit-based vaccine in humans (Crank et al., 2019) and use in an mRNA vaccine tested in rodent models (Espeseth et al., 2020).
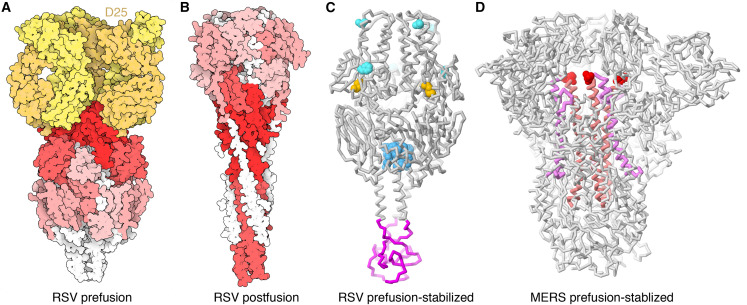
Figure 4.
Stabilization of prefusion conformations of viral surface glycoproteins
(A) Space-filling representation of the prefusion conformation of the RSV glycoprotein F ectodomain (PDB: 4jhw; McLellan et al., 2013b), with antibody epitopes colored by neutralization sensitivity (dark red, highest; light red, intermediate; pink, lowest; white, antibody inaccessible) and a neutralizing antibody D25 Fab shown in yellow.
(B) Space-filling representation of the post-fusion conformation of the RSV glycoprotein F ectodomain (PDB: 3rrr; McLellan et al., 2011), showing that the most neutralization-sensitive epitopes are inaccessible to antibodies in the altered conformation.
(C) Polypeptide chain backbone representation of the RSV F glycoprotein stabilized in the prefusion conformation (PDB: 4mmv; McLellan et al., 2013a) with an engineered disulfide bridge (yellow), several sites of mutation to fill pockets (turquoise and blue), and a foldon to stabilize the homotrimer (magenta).
(D) Polypeptide chain backbone representation of the MERS-CoV virus spike protein homotrimer stabilized in a prefusion conformation (PDB: 5w9j; Pallesen et al., 2017) with two prolines (red spheres) linking the heptad repeat (HR1, magenta) and the central helix (pink). (A) and (B) created with Illustrate software; (C) and (D) created with Mol∗ (Sehnal et al., 2021) at the RCSB PDB website.
Building on this work, the idea of stabilizing prefusion conformational states of surface glycoproteins has been applied to coronaviruses. Analysis of RSV and HIV surface glycoproteins identified a structurally critical region in their interiors, linking the heptad repeat (HR1) to the central α helices. Adding two prolines to this loop was found to stabilize the prefusion form of Middle East respiratory syndrome coronavirus (MERS-CoV) glycoprotein and other coronaviruses (Pallesen et al., 2017). Similar stabilizing prolines are included in both the Moderna and BioNTech/Pfizer mRNA vaccines against SARS-CoV-2, and combined with inactivation of the furin cleavage site in the virus-vectored vaccine from Janssen/Johnson&Johnson and a subunit vaccine from Novavax (reviewed in Dai and Gao, 2021). We expect that a similar approach will be applied to all manner of viral targets as the body of structural information grows. For example, the recent structure of hepatitis C glycoprotein E2 in complex with its cellular receptor (PDB: 7mwx; Kumar et al., 2021), compared with a decade of previous structural studies of the glycoprotein alone and with neutralizing antibodies, reveals conformational changes upon acidification in preparation for membrane fusion. These structural insights will potentially allow targeted design of prefusion-stabilized glycoprotein in future vaccine development programs.
Looking ahead: the challenge of SARS-CoV-2 variants
One of the great promises of mRNA vaccines is that they will provide a timely and effective way of responding to newly emerging variants of SARS-CoV-2. Every time the virus replicates in a human (or animal) host, there is a non-zero likelihood that one or more of the viral protein sequences (and consequently 3D structures) will change. Coronaviruses have the longest RNA virus genomes of all known single-stranded RNA viruses (∼30,000 nucleotides). SARS-CoV-2 RNA-dependent RNA polymerase (multi-subunit enzymes composed of non-structural proteins [nsps] 7, 8, and 12) acts in concert with an RNA helicase (nsp13) and a proofreading exonuclease (nsp14) to carry out efficient and relatively faithful copying of the lengthy genome (Denison et al., 2011). Proofreading notwithstanding, coronavirus genome replication is not perfect, and coronaviruses evolve as they passage serially from one host to the next (Harvey et al., 2021). A recently published study of SARS-CoV-2 protein evolution in 3D during the first 6 months of the pandemic examined amino acid changes in >48,000 viral isolates and documented how each one of the 29 viral proteins underwent amino acid changes (Lubin et al., 2020).
Of greatest concern at the time of writing is the Delta variant of SARS-CoV-2 (B.1.617.2; https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/) with spike glycoprotein substitutions T19R, V70F, T95I, G142D, E156-, F157-, R158G, A222V, W258L, K417N, L452R, T478K, D614G, P681R, D950N (https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html). These substitutions include a critical change in the receptor-binding domain (position 452) that is thought to confer a stronger binding to the ACE2. An additional change at position 681 affects the rate of cleavage of the spike protein precursor (Cherian et al., 2021). Figure 5 illustrates the locations of some amino acid changes in the Delta variant spike protein. Initial reports suggest that both the Pfizer-BioNTech and Moderna mRNA vaccines offer high levels of protection against serious illness requiring hospitalization and death for doubly vaccinated individuals (but not for those who are singly vaccinated or previously infected with another SARS-CoV-2 variant; Callaway, 2021). Experts around the world concur that continued infections in geographies with low rates of complete vaccination represent potential breeding grounds for new variants that could threaten a second global pandemic. Both Pfizer and Moderna have reported progress in developing booster mRNA vaccines, which could be re-designed to include Delta and/or possibly newer post-Delta variants. Structural biologists are providing critical information archived in the PDB that contribute to our understanding of new SARS-CoV-2 spike protein variants and how substitutions at various positions within the homotrimer impact function, infectivity, and virus neutralization by antibodies of either natural or human-made origin.
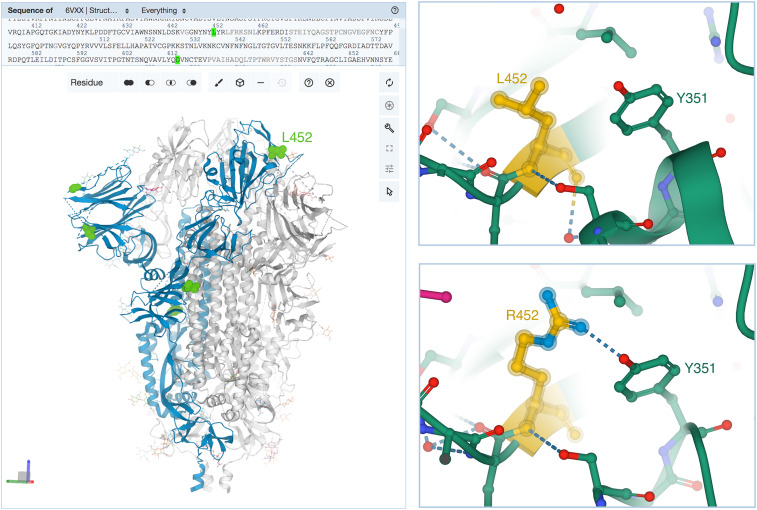
Figure 5.
Sites of mutation in the SARS-CoV-2 spike protein Delta variant
(Left) Ribbon representation of the SARS-CoV-2 spike protein homotrimer (one protomer in blue) with attached carbohydrates (pink atomic stick figures) created using the RCSB.org web portal with Mol∗. Selected sites of amino acid substitutions in the Delta variant (versus the original viral isolate) are highlighted in green from the upper sequence selection panel (PDB: 6vxx; Walls et al., 2020). (Upper right) Close-up view of L452 occurring in the receptor-binding domain was generated in Mol∗ by clicking on the position, triggering automatic Mol∗ display of the location showing interactions with neighboring amino acids (PDB: 7ora; Liu et al., 2021a). (Lower right) Consequences of the L452R substitution showing a new hydrogen bond (dashed line) with Y351 (PDB: 7orb; Liu et al., 2021a). Atom color coding: C, green or yellow; N, blue; O, red. Figure created with Mol∗ (Sehnal et al., 2021) at the RCSB PDB website.
The RCSB.org web portal 3D Protein Feature View (PFV) is equipped with tools that assist users in relating changes in 3D structure due to sequence/structure variation with changes in function. Based on data integrated from UniProt, the PFV includes mappings of variant locations with assorted protein sequence features, including domains, proteolytic processing sites, and glycosylation sites. The 1D graphical view is also linked to a Mol∗ structure viewer, allowing protein sequence features to be visualized in the context of atomic-level 3D structure.
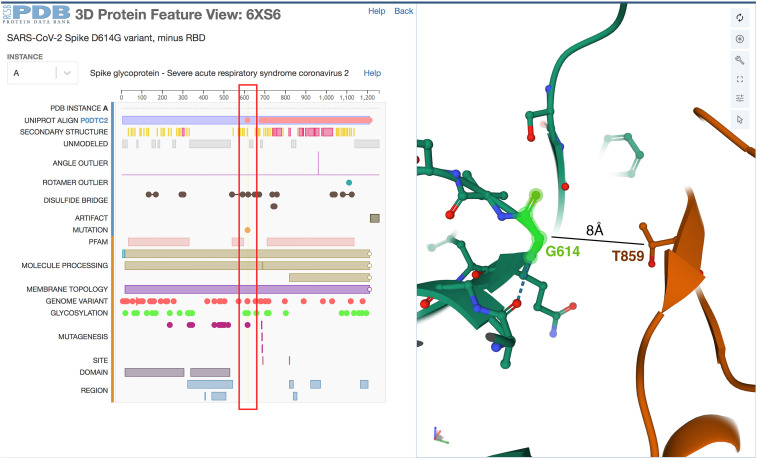
Figure 6 exemplifies uses of the 3D PFV, illustrating the structure of the SARS-CoV-2 spike protein D614G variant implicated in increased infectivity. Analysis of >33,000 viral genomes sequenced before late June 2020 revealed that ∼74% of the viral isolates possessed the D614G amino acid substitution in their spike proteins (Lubin et al., 2020). Structural characterization of an engineered D614G spike protein by 3DEM revealed a significantly increased population of conformations in which the ACE2 receptor-binding domains occur in the open state, presumed to be due to loss of an interaction with T859 in a neighboring subunit (PDB: 6xs6; Yurkovetskiy et al., 2020). Alteration of the equilibrium between closed and open states is thought to explain increased infectivity. Figure 6 (right) shows that G614 is ∼8Å from the sidechain of T859 of another copy of the protein within the trimer. In the structure of the spike protein from the original viral isolate, D614 makes a hydrogen bond with T859, stabilizing the structure in a closed state that is unable to interact with ACE2 (data not shown).
Figure 6.
Screenshot of the RCSB PDB 3D PFV for the D614G substituted form of the SARS-CoV-2 spike protein
(Left) The location of the substitution is selected using the sequence-based display in the PFV (red box). (Right) The amino acid G614 is highlighted with a green halo using Mol∗ structure viewer, revealing loss of a stabilizing interaction with T859 in a neighboring protomer within the homotrimer. Structure is shown from PDB: 6xs6 (Yurkovetskiy et al., 2020).
Design of future antiviral mRNA vaccines and potential anticancer therapies
The wealth of 3D structure information available for the SARS-CoV-2 spike protein and its potential to influence design of second-generation COVID-19 mRNA vaccines suggests that there is much to be optimistic about for design of additional mRNA vaccines against other viral pathogens. mRNA vaccines are non-infectious, do not become integrated into the host genome, stimulate both humoral and cell-based immunity, are well tolerated by healthy individuals with few side effects, can be rapidly designed, and are less expensive to develop and manufacture. They have enormous potential as vehicles for addressing viral diseases more broadly. A number of candidate mRNA vaccines are currently being evaluated in clinical trials for Zika virus, metapneumovirus, parainfluenzavirus, cytomegalovirus, rabies, and others (Wadhwa et al., 2020), while contributing to a multi-pronged effort to combat the COVID-19 pandemic (Park et al., 2021).
mRNA vaccines are also making inroads as experimental agents for treating human cancers. The past decade has witnessed a revolution in cancer immunotherapy with approval of monoclonal antibodies that recognize cell-surface antigens (first CTLA-4, followed by PD-1, and then PD-L1) that are responsible for downregulation of T cell responses to cancers expressing neoantigens specific to the tumor. In a subset of antibody-treated individuals (with the exact proportion depending on the type of cancer), interdiction of the T cell immune checkpoint prevents the malignant cell from “persuading” the T cell that it should not be killed. President Jimmy Carter was on the verge of death due to late-stage melanoma when he received pembrolizumab (Merck, brand name Keytruda). At the time of writing, Carter was in long-term remission and may well have been cured. Current challenges with immune checkpoint therapies include their relatively low response rates and ascertaining why some individuals respond to antibody treatment while others do not. mRNA vaccines represent a potentially important means of increasing the likelihood of long-term remissions for some cancers.
Both Moderna and BioNTech have been active in this arena with their respective lipid nanoparticle formulations for mRNA delivery. Other cancer vaccine approaches include naked synthetic mRNA, an individual’s own dendritic cells that have been manipulated and expanded ex vivo, protamine formulations, and self-amplifying mRNA (SAM) vaccines (reviewed in Miao et al. (2021). A SAM vaccine vehicle carries viral replication machinery capable of self-amplifying over 1–2 months and inducing more potent and persistent immune responses. SAM platforms are expected to support significant antigen production following vaccination with very low doses (versus those used for non-replicating mRNA vaccines). As for design of antiviral mRNA vaccines, the challenge will be to deliver a genetic payload that encodes the right antigen. Indeed, selection of tumor-associated or tumor-specific antigens (TAAs or TSAs) preferentially expressed in malignant cells appears to be the problem. BioNTech’s BNT111 vaccine encoding four TAAs identified in melanoma cells has yielded T cell responses in early-stage clinical trials. Alphavax has also reported encouraging results for their AVX701 SAM, which encodes carcinoembryonic antigen (CEA). While certainly promising, mRNAs encoding one or more TAAs may not prove to be broadly effective. Individuals with highly variable TAAs present in their polyclonal tumors due to errors in DNA replication may not respond to vaccines that deliver mRNAs encoding wild-type proteins. There is also the problem of autoimmune attack of normal tissues that may encode TAAs.
An alternative approach to antigen selection focuses on neoantigens, which derive from random somatic mutations in malignant cells. The success of the immune checkpoint antibodies is thought to result from T cell recognition of neoantigens as non-self-proteins. The challenge of this approach is identification of oligopeptide fragments of mutant proteins that have high immunogenicity. Moderna and Merck, a leader in antibody immunotherapy, are collaborating on development of mRNA-5671, which encodes four well-characterized somatic mutations of KRAS affecting amino acid glycine 12 (see PDB: 4l8g for the 3D structure of KRAS G12C; Ostrem et al., 2013), for use as a monotherapy and in combination with pembrolizumab in participants with KRAS mutant advanced or metastatic non-small cell lung cancer, colorectal cancer, or pancreatic cancer. BioNTech is currently collaborating with Genentech (a member of the Roche group) on individualized neoantigen-specific therapy using its mRNA delivery platform, which aims to customize vaccines according to the repertoire of immunogenic neoantigens detected in a particular tumor.
In all of these exciting developments, the PDB archive has played an essential role by providing open access to the global corpus of structural knowledge, and the RCSB PDB has provided the resources to find and utilize this information. This has streamlined the structural understanding of the basic mechanisms of carcinogenesis, facilitated the structure-based design of antineoplastic drugs, and provided detailed structural understanding of antibodies and their neutralization of cancer antigens. Indeed, a recent analysis revealed that open access to 3D structural information facilitated the discovery and development of the majority of recent antineoplastic agents, including 25 biologics (Westbrook et al., 2020).
While no one expects that every new effort involving an mRNA vaccine will succeed, it appears likely that at least some of them will work and improve the range of options available to intervene medically and improve the human condition. Strategic investments by Pfizer/BioNTech and in parallel by Moderna (with considerable financial assistance from the US government and philanthropic contributors) in the face of the global pandemic transformed the landscape for mRNA vaccine design and development. The public now knows much more about the science and technology of mRNA vaccines and how they can generate live-saving results in a short time. Absent COVID-19, it would have taken much longer for the technology to mature and become broadly available. There is much more to come and the RCSB PDB team looks forward to ensuring that the PDB contributes to the good of humankind for another 50 years in this arena and more broadly across the biological and biomedical sciences.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| SARS-CoV-1 spike protein | Yuan et al. (2017) | 5x5b |
| SARS-CoV-1 spike protein | Kirchdoerfer et al. (2018) | 6crz |
| SARS-CoV-2 spike protein | Walls et al. (2020) | 6vyb |
| SARS-CoV-2 spike protein | Wrapp et al. (2020) | 6vsb |
| SARS-CoV-2 spike protein D614G | Yurkovetskiy et al. (2020) | 6xs6 |
| SARS-CoV-2 spike protein | Walls et al. (2020) | 6vxx |
| SARS-CoV-2 spike protein | Liu et al. (2021a) | 7ora |
| SARS-CoV-2 spike protein | Liu et al. (2021a) | 7orb |
| MERS-CoV spike protein | Pallesen et al. (2017) | 5w9j |
| MERS-CoV spike protein | Pallesen et al. (2017) | 5w9n |
| RSV glycoprotein F | McLellan et al. (2013b) | 4jhw |
| RSV glycoprotein F | McLellan et al. (2011) | 3rrr |
| RSV glycoprotein F | McLellan et al. (2013a) | 4mmv |
| VSV glycoprotein G | Roche et al. (2007) | 5i2s |
| Rabies virus glycoprotein | Yang et al. (2020) | 6lgx |
| Zika virus | Sirohi et al. (2016) | 5ire |
| CMV pentamer | Chandramouli et al. (2017) | 5vob |
| CMV glycoprotein B | Liu et al. (2021b) | 7kdp |
| Measles hemagglutinin | Hashiguchi et al. (2007) | 2zb5 |
| Mumps hemagglutinin-neuraminidase | Kubota et al. (2016) | 5b2d |
| Influenza hemagglutinin | Gamblin et al. (2004) | 1ruz |
| Influenza neuraminidase | Varghese and Colman (1991) | 1nn2 |
| Herpes simplex envelope glycoprotein B | Stampfer et al. (2010) | 3nw8 |
| Dengue envelope protein E | Zhang et al. (2004) | 1tg8 |
| West nile envelope protein E | Kanai et al. (2006) | 2i69 |
| Tick-borne encephalitis envelope protein E | Rey et al. (1995) | 1svb |
| Hepatitis C glycoprotein E2 | Kong et al. (2013) | 4mwf |
| Hepatitis C glycoprotein E2 | Kumar et al. (2021) | 7mwx |
| HIV-1 envelope glycoprotein | Julien et al. (2013) | 4nco |
| KRAS G12C | Ostrem et al. (2013) | 4l8g |
| Software and algorithms | ||
| Mol∗ | RCSB PDB | rcsb.org/3d-view |
| Illustrate | Scripps Research, Center for Computational Structural Biology | ccsb.scripps.edu/illustrate |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Stephen K. Burley (stephen.burley@rcsb.org).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
All data are generated from the datasets provided in the key resources table.
Method details
This manuscript describes resources that are available at the RCSB Protein DataBank. Atomic structures used for Figure 2 were downloaded from RCSB.org and visualized with the stand-alone version of Illustrate (ccsb.scripps.edu/illustrate). All other figures were generated with Mol∗ at the RCSB.org site (rcsb.org/3d-view). Hit values for Table 1 were evaluated based on the PDB archive in August 2021 using the search tools available in the “Macromolecules” section of the RCSB Structure Summary Page (for example, www.rcsb.org/structure/6CRZ), with several exceptions described in the Table footnotes. PDB accession codes for all structures used in the figures and Table 1 are given in the captions and table, and summarized in the key resources table.
Quantification and statistical analysis
This study does not utilize quantification or statistical analysis methods.
Additional resources
Not applicable.
Acknowledgments
First and foremost, the authors thank the tens of thousands of structural biologists who deposited structures to the PDB since 1971 and the many millions around the world who consume PDB data. We also thank Christine Zardecki for assistance with manuscript preparation. The authors gratefully acknowledge contributions to the success of the PDB archive made by all members of RCSB PDB (past and present) and our PDBe, PDBj, EMDB, and BMRB wwPDB partners. RCSB PDB is jointly funded by the National Science Foundation (DBI-1832184; PI. S.K. Burley), the US Department of Energy (DE-SC0019749; PI, S.K. Burley), and the National Cancer Institute, National Institute of Allergy and Infectious Diseases, and National Institute of General Medical Sciences of the National Institutes of Health under grant R01GM133198 (PI, S.K. Burley). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
D.S.G. and S.K.B. contributed to the conceptualization, investigation, visualization, and writing of this article. S.K.B. is responsible for supervision, project administration, and securing funding for the RCSB PDB.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community.
Published: November 4, 2021
Data and code availability
-
•
This paper analyzes existing, publicly available data. The accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abbott S., Iudin A., Korir P.K., Somasundharam S., Patwardhan A. EMDB web resources. Curr. Protoc. Bioinformatics. 2018;61:5.10.11–15.10.12. doi: 10.1002/cpbi.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A., Kulesha E., Gough J., Murzin A.G. The SCOP database in 2020: expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 2019;48:D376–D382. doi: 10.1093/nar/gkz1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong D.R., Berrisford J.M., Conroy M.J., Gutmanas A., Anyango S., Choudhary P., Clark A.R., Dana J.M., Deshpande M., Dunlop R., et al. PDBe: improved findability of macromolecular structure data in the PDB. Nucleic Acids Res. 2020;48:D335–D343. doi: 10.1093/nar/gkz990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battles M.B., McLellan J.S. Respiratory syncytial virus entry and how to block it. Nat. Rev. Microbiol. 2019;17:233–245. doi: 10.1038/s41579-019-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Henrick K., Nakamura H. Announcing the Worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S.K. Impact of structural biologists and the Protein Data Bank on small-molecule drug discovery and development. J. Biol. Chem. 2021;296:100559. doi: 10.1016/j.jbc.2021.100559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S.K., Berman H.M. Open-access data: a cornerstone for artificial intelligence approaches to protein structure prediction. Structure. 2021;29:515–520. doi: 10.1016/j.str.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S.K., Berman H.M., Christie C., Duarte J.M., Feng Z., Westbrook J., Young J., Zardecki C. RCSB Protein Data Bank: sustaining a living digital data resource that enables breakthroughs in scientific research and biomedical education. Protein Sci. 2018;27:316–330. doi: 10.1002/pro.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chen L., Crichlow G., Christie C.H., Dalenberg K., Costanzo L.D., Duarte J.M., et al. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering, and energy sciences. Nucleic Acid Res. 2021;49:D437–D451. doi: 10.1093/nar/gkaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. Delta coronavirus variant: scientists brace for impact. Nature. 2021;595:17–18. doi: 10.1038/d41586-021-01696-3. [DOI] [PubMed] [Google Scholar]
- Chandramouli S., Malito E., Nguyen T., Luisi K., Donnarumma D., Xing Y., Norais N., Yu D., Carfi A. Structural basis for potent antibody-mediated neutralization of human cytomegalovirus. Sci. Immunol. 2017;2:eaan1457. doi: 10.1126/sciimmunol.aan1457. [DOI] [PubMed] [Google Scholar]
- Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., Rakshit P., Singh S., Abraham P., Panda S., team N. Convergent evolution of SARS-CoV-2 spike mutations, L452R, E484Q and P681R. bioRxiv. 2021 doi: 10.1101/2021.04.22.440932. the second wave of COVID-19 in Maharashtra, India. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crank M.C., Ruckwardt T.J., Chen M., Morabito K.M., Phung E., Costner P.J., Holman L.A., Hickman S.P., Berkowitz N.M., Gordon I.J., et al. A proof of concept for structure-based vaccine design targeting RSV in humans. Science. 2019;365:505–509. doi: 10.1126/science.aav9033. [DOI] [PubMed] [Google Scholar]
- Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeseth A.S., Cejas P.J., Citron M.P., Wang D., DiStefano D.J., Callahan C., Donnell G.O., Galli J.D., Swoyer R., Touch S., et al. Modified mRNA/lipid nanoparticle-based vaccines expressing respiratory syncytial virus F protein variants are immunogenic and protective in rodent models of RSV infection. NPJ Vaccin. 2020;5:16. doi: 10.1038/s41541-020-0163-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Verdiguel N., Di Costanzo L., Goodsell D.S., Westbrook J.D., Burley S.K., Zardecki C. Impact of the Protein Data Bank across scientific disciplines. Data Sci. J. 2020;19:1–14. doi: 10.5334/dsj-2020-025. [DOI] [Google Scholar]
- Galkina Cleary E., Beierlein J.M., Khanuja N.S., McNamee L.M., Ledley F.D. Contribution of NIH funding to new drug approvals 2010–2016. Proc. Natl. Acad. Sci. U. S. A. 2018;115:2329–2334. doi: 10.1073/pnas.1715368115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamblin S.J., Haire L.F., Russell R.J., Stevens D.J., Xiao B., Ha Y., Vasisht N., Steinhauer D.A., Daniels R.S., Elliot A., et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- Gilman M.S., Castellanos C.A., Chen M., Ngwuta J.O., Goodwin E., Moin S.M., Mas V., Melero J.A., Wright P.F., Graham B.S., et al. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci. Immunol. 2016;1:eaaj1879. doi: 10.1126/sciimmunol.aaj1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson M.K., Liu T., Baitaluk M., Nicola G., Hwang L., Chong J. BindingDB in 2015: a public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016;44:D1045–D1053. doi: 10.1093/nar/gkv1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell D.S., Zardecki C., Di Costanzo L., Duarte J.M., Hudson B.P., Persikova I., Segura J., Shao C., Voigt M., Westbrook J.D., et al. RCSB Protein Data Bank: enabling biomedical research and drug discovery. Protein Sci. 2020;29:52–65. doi: 10.1002/pro.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzenko D., Burley S.K., Duarte J.M. Real time structural search of the Protein Data Bank. Plos Comput. Biol. 2020;16:e1007970. doi: 10.1371/journal.pcbi.1007970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Consortium C.-G.U., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiguchi T., Kajikawa M., Maita N., Takeda M., Kuroki K., Sasaki K., Kohda D., Yanagi Y., Maenaka K. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson J. The pandemic pipeline. Nat. Biotechnol. 2020;38:523–532. doi: 10.1038/d41587-020-00005-z. [DOI] [PubMed] [Google Scholar]
- Julien J.P., Cupo A., Sok D., Stanfield R.L., Lyumkis D., Deller M.C., Klasse P.J., Burton D.R., Sanders R.W., Moore J.P., et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J.P., Lee P.S., Wilson I.A. Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol. Rev. 2012;250:180–198. doi: 10.1111/imr.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai R., Kar K., Anthony K., Gould L.H., Ledizet M., Fikrig E., Marasco W.A., Koski R.A., Modis Y. Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 2006;80:11000–11008. doi: 10.1128/JVI.01735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B., et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49:D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo A.R., Bekker G.J., Wako H., Endo S., Tsuchiya Y., Sato H., Nishi H., Kinoshita K., Suzuki H., Kawabata T., et al. New tools and functions in data-out activities at Protein Data Bank Japan (PDBj) Protein Sci. 2018;27:95–102. doi: 10.1002/pro.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Wang N., Pallesen J., Wrapp D., Turner H.L., Cottrell C.A., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018;8:15701. doi: 10.1038/s41598-018-34171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L., Giang E., Nieusma T., Kadam R.U., Cogburn K.E., Hua Y., Dai X., Stanfield R.L., Burton D.R., Ward A.B., et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalzik F., Schreiner D., Jensen C., Teschner D., Gehring S., Zepp F. mRNA-based vaccines. Vaccines. 2021;9:390. doi: 10.3390/vaccines9040390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota M., Takeuchi K., Watanabe S., Ohno S., Matsuoka R., Kohda D., Nakakita S.I., Hiramatsu H., Suzuki Y., Nakayama T., et al. Trisaccharide containing alpha2,3-linked sialic acid is a receptor for mumps virus. Proc. Natl. Acad. Sci. U. S. A. 2016;113:11579–11584. doi: 10.1073/pnas.1608383113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- Kumar A., Hossain R.A., Yost S.A., Bu W., Wang Y., Dearborn A.D., Grakoui A., Cohen J.I., Marcotrigiano J. Structural insights into hepatitis C virus receptor binding and entry. Nature. 2021 doi: 10.1038/s41586-021-03913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., O'Neill K.R., Haft D.H., DiCuccio M., Chetvernin V., Badretdin A., Coulouris G., Chitsaz F., Derbyshire M.K., Durkin A.S., et al. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021;49:D1020–D1028. doi: 10.1093/nar/gkaa1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236.e13. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Heim K.P., Che Y., Chi X., Qiu X., Han S., Dormitzer P.R., Yang X. Prefusion structure of human cytomegalovirus glycoprotein B and structural basis for membrane fusion. Sci. Adv. 2021;7:eabf3178. doi: 10.1126/sciadv.abf3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin J.H., Zardecki C., Dolan E.M., Lu C., Shen Z., Dutta S., Westbrook J.D., Hudson B.P., Goodsell D.S., Williams J.K., et al. Evolution of the SARS-CoV-2 proteome in three dimensions (3D) during the first six months of the COVID-19 pandemic. bioRxiv. 2020 doi: 10.1101/2020.12.01.406637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro M., Mas V., Chappell K., Vazquez M., Cano O., Luque D., Terron M.C., Melero J.A., Palomo C. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc. Natl. Acad. Sci. U. S. A. 2012;109:3089–3094. doi: 10.1073/pnas.1115941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markosian C., Di Costanzo L., Sekharan M., Shao C., Burley S.K., Zardecki C. Analysis of impact metrics for the Protein Data Bank. Sci. Data. 2018;5:180212. doi: 10.1038/sdata.2018.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Chen M., Joyce M.G., Sastry M., Stewart-Jones G.B., Yang Y., Zhang B., Chen L., Srivatsan S., Zheng A., et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Chen M., Leung S., Graepel K.W., Du X., Yang Y., Zhou T., Baxa U., Yasuda E., Beaumont T., et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan J.S., Yang Y., Graham B.S., Kwong P.D. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J. Virol. 2011;85:7788–7796. doi: 10.1128/JVI.00555-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNaught A.D. International union of pure and applied chemistry and International Union of Biochemistry and Molecular Biology - Joint Commission on Biochemical nomenclature - nomenclature of carbohydrates - recommendations 1996. Pure Appl. Chem. 1996;68:1919–2008. doi: 10.1351/pac199668101919. [DOI] [PubMed] [Google Scholar]
- Miao L., Zhang Y., Huang L. mRNA vaccine for cancer immunotherapy. Mol. Cancer. 2021;20:41. doi: 10.1186/s12943-021-01335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.T., Mathias S., Bologa C., Brunak S., Fernandez N., Gaulton A., Hersey A., Holmes J., Jensen L.J., Karlsson A., et al. Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Res. 2017;45:D995–D1002. doi: 10.1093/nar/gkw1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem J.M., Peters U., Sos M.L., Wells J.A., Shokat K.M. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.W., Lagniton P.N.P., Liu Y., Xu R.H. mRNA vaccines for COVID-19: what, why and how. Int. J. Biol. Sci. 2021;17:1446–1460. doi: 10.7150/ijbs.59233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard A.J., Bijker E.M. A guide to vaccinology: from basic principles to new developments. Nat. Rev. Immunol. 2021;21:83–100. doi: 10.1038/s41577-020-00479-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protein Data Bank Crystallography: Protein Data Bank. Nat. (London) New Biol. 1971;233:223. doi: 10.1038/newbio233223b0. [DOI] [Google Scholar]
- Rappuoli R., De Gregorio E., Del Giudice G., Phogat S., Pecetta S., Pizza M., Hanon E. Vaccinology in the post-COVID-19 era. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2020368118. e2020368118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read K.B., Sheehan J.R., Huerta M.F., Knecht L.S., Mork J.G., Humphreys B.L., N.I.H. Big Data Annotator Group Sizing the problem of improving discovery and access to NIH-funded data: a preliminary study. PLoS One. 2015;10:e0132735. doi: 10.1371/journal.pone.0132735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey F.A., Heinz F.X., Mandl C., Kunz C., Harrison S.C. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Roche S., Rey F.A., Gaudin Y., Bressanelli S. Structure of the prefusion form of the vesicular stomatitis virus glycoprotein G. Science. 2007;315:843–848. doi: 10.1126/science.1135710. [DOI] [PubMed] [Google Scholar]
- Romero P.R., Kobayashi N., Wedell J.R., Baskaran K., Iwata T., Yokochi M., Maziuk D., Yao H., Fujiwara T., Kurusu G., et al. BioMagResBank (BMRB) as a resource for structural biology. Methods Mol. Biol. 2020;2112:187–218. doi: 10.1007/978-1-0716-0270-6_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A.S., Bradley A.R., Valasatava Y., Duarte J.M., Prlić A., Rose P.W. NGL viewer: web-based molecular graphics for large complexes. Bioinformatics. 2018;34:3755–3758. doi: 10.1093/bioinformatics/bty419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander C., Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins Struct. Funct. Genet. 1991;9:56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- Sehnal D., Bittrich S., Deshpande M., Svobodova R., Berka K., Bazgier V., Velankar S., Burley S.K., Koca J., Rose A.S. Mol∗ Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021;49:W431–W437. doi: 10.1093/nar/gkab314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnal D., Deshpande M., Varekova R.S., Mir S., Berka K., Midlik A., Pravda L., Velankar S., Koca J. LiteMol suite: interactive web-based visualization of large-scale macromolecular structure data. Nat. Methods. 2017;14:1121–1122. doi: 10.1038/nmeth.4499. [DOI] [PubMed] [Google Scholar]
- Shao C., Feng Z., Westbrook J.D., Peisach E., Berrisford J., Ikegawa Y., Kurisu G., Velankar S., Burley S.K., Young J.Y. Modernized uniform representation of carbohydrate molecules in the Protein Data Bank. Glycobiology. 2021;31:1204–1218. doi: 10.1093/glycob/cwab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillitoe I., Bordin N., Dawson N., Waman V.P., Ashford P., Scholes H.M., Pang C.S.M., Woodridge L., Rauer C., Sen N., et al. CATH: increased structural coverage of functional space. Nucleic Acids Res. 2021;49:D266–D273. doi: 10.1093/nar/gkaa1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi D., Chen Z., Sun L., Klose T., Pierson T.C., Rossmann M.G., Kuhn R.J. The 3.8 A resolution cryo-EM structure of Zika virus. Science. 2016;352:467–470. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner T.H. Intracellular and viral membrane fusion: a uniting mechanism. Curr. Opin. Cell Biol. 2004;16:429–435. doi: 10.1016/j.ceb.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Stampfer S.D., Lou H., Cohen G.H., Eisenberg R.J., Heldwein E.E. Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1. J. Virol. 2010;84:12924–12933. doi: 10.1128/JVI.01750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiemeyer M., Aoki K., Paulson J., Cummings R.D., York W.S., Karlsson N.G., Lisacek F., Packer N.H., Campbell M.P., Aoki N.P., et al. GlyTouCan: an accessible glycan structure repository. Glycobiology. 2017;27:915–919. doi: 10.1093/glycob/cwx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Aalst W.M.P., Bichler M., Heinzl A. Responsible data science. Bus. Inf. Syst. Eng. 2017;59:311–313. doi: 10.1007/s12599-017-0487-z. [DOI] [Google Scholar]
- Varghese J.N., Colman P.M. Three-dimensional structure of the neuraminidase of influenza virus A/Tokyo/3/67 at 2.2 A resolution. J. Mol. Biol. 1991;221:473–486. doi: 10.1016/0022-2836(91)80068-6. [DOI] [PubMed] [Google Scholar]
- Varki A., Cummings R.D., Aebi M., Packer N.H., Seeberger P.H., Esko J.D., Stanley P., Hart G., Darvill A., Kinoshita T., et al. Symbol nomenclature for graphical representations of glycans. Glycobiology. 2015;25:1323–1324. doi: 10.1093/glycob/cwv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadhwa A., Aljabbari A., Lokras A., Foged C., Thakur A. Opportunities and challenges in the delivery of mRNA-based vaccines. Pharmaceutics. 2020;12:102. doi: 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook J.D., Burley S.K. How structural biologists and the Protein Data Bank contributed to recent FDA new drug approvals. Structure. 2019;27:211–217. doi: 10.1016/j.str.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook J.D., Soskind R., Hudson B.P., Burley S.K. Impact of Protein Data Bank on anti-neoplastic approvals. Drug Discov. Today. 2020;25:837–850. doi: 10.1016/j.drudis.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., da Silva Santos L.B., Bourne P.E., et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data. 2016;3:1–9. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- wwPDB consortium Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res. 2019;47:D520–D528. doi: 10.1093/nar/gky949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada I., Shiota M., Shinmachi D., Ono T., Tsuchiya S., Hosoda M., Fujita A., Aoki N.P., Watanabe Y., Fujita N., et al. The GlyCosmos portal: a unified and comprehensive web resource for the glycosciences. Nat. Methods. 2020;17:649–650. doi: 10.1038/s41592-020-0879-8. [DOI] [PubMed] [Google Scholar]
- Yang F., Lin S., Ye F., Yang J., Qi J., Chen Z., Lin X., Wang J., Yue D., Cheng Y., et al. Structural analysis of rabies virus glycoprotein reveals pH-dependent conformational changes and interactions with a neutralizing antibody. Cell Host Microbe. 2020;27:441–453 e447. doi: 10.1016/j.chom.2019.12.012. [DOI] [PubMed] [Google Scholar]
- York W.S., Mazumder R., Ranzinger R., Edwards N., Kahsay R., Aoki-Kinoshita K.F., Campbell M.P., Cummings R.D., Feizi T., Martin M., et al. GlyGen: computational and informatics resources for glycoscience. Glycobiology. 2020;30:72–73. doi: 10.1093/glycob/cwz080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751.e738. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang W., Ogata S., Clements D., Strauss J.H., Baker T.S., Kuhn R.J., Rossmann M.G. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
-
•
This paper analyzes existing, publicly available data. The accession numbers for the datasets are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.