Highlights
-
•
High intensity ultrasound technology was used to modify soy protein isolate-pectin.
-
•
The fluorescence intensity increased by high intensity ultrasound treatment.
-
•
The particle size of the complex reduced by high intensity ultrasound treatment.
-
•
The structural properties and solubility of protein were improved at 450 W for 15 min.
Keywords: High intensity ultrasound, Soy protein isolate, Pectin, Structural properties, Solubility
Abstract
In this study, a soy protein isolate (SPI)-pectin (PC) complex was prepared, and the effects of different high intensity ultrasound (HIU) powers on the structure and solubility of the complex were studied. Fourier transform infrared (FTIR) spectroscopy analysis exhibited that with increasing HIU power, the α-helix content of the SPI in the complex was significantly reduced, and the random coil content increased; however, an opposite trend appeared after higher power treatments. Fluorescence spectra showed that HIU treatment increased the fluorescence intensity of the complex, and the surface hydrophobicity was increased. The trend of the protein structure studied by Raman spectroscopy was similar to that of FTIR and fluorescence spectroscopy. When the HIU treatment was performed for 15 min and at 450 W power, the particle size of the complex was 451.85 ± 2.17 nm, and the solubility was 89.04 ± 0.19 %, indicating that the HIU treatment caused the spatial conformation of the protein to loosen and improved the functional properties of the complex. Confocal laser scanning microscopy (CLSM) revealed that the complex after HIU treatment exhibited improved dispersibility in water and smaller particle size. Gel electrophoresis results indicated that HIU treatment did not affect the protein subunits of the complex. Therefore, the selection of a suitable HIU treatment power can effectively improve the structural properties and solubility of SPI in the complex, and promote the application of the SPI-PC complex in food processing and industries.
1. Introduction
Soybean is one of the world’s major crops. It is rich in nutrients and has a high protein content of approximately 40 %, making it a high-quality plant protein resource [1]. Soy protein isolate (SPI) is the main component of soy protein, and its protein content is generally higher than 90 %. Studies have shown that SPI has several functional properties, such as gelation, emulsification, and foaming [2], [3], and is widely used in the field of food processing [4], [5]. However, SPI is easily affected by harsh processing conditions during processing and storage, resulting in quality deterioration. Especially at the pH of the isoelectric point, SPI with a net charge of zero is prone to aggregation and precipitation [6], which limits the performance of functional properties and its application in food production.
Pectin (PC) is the polysaccharide with the most complex structure and function in plant cell walls [7]. Its primary structure is mainly α-1,4-linked D-Galacturonic acid and its methyl ester. Due to the wide range of PC sources and low prices, PC is often used as an emulsifier, gelling, and stabilizing agent in food industries [8], [9]. However, the performance of PC alone is poor after being dissolved in water [10], [11]; therefore, PC is often combined with protein and other biological macromolecular substances to prepare complexes to improve their functional properties [12], [13].
Recently, the complex formed by the interaction of proteins and polysaccharides has been explored to have better functional properties and stability than the two alone. The interaction is generally divided into two types: one is covalent binding, mainly through Maillard reaction [14], [15]; the other is non-covalent binding through electrostatic attraction, hydrophobic interaction, and hydrogen bonding between proteins and polysaccharides [16], [17]. Mao et al. [18] used whey protein isolate and PC to produce mixed layer emulsions. Their findings indicated that mixed-layer emulsions could be applied to modulate the release of volatiles by changing the environmental conditions. Ma et al. [19] studied the electrostatic interaction between SPI and citrus pectin (CP) under the action of a high intensity ultrasonic field. They found that ultrasound enhanced the electrostatic interaction between SPI and CP, which contributed to the improvement of the more desirable emulsifying properties of the complex.
Although the complex can improve the emulsification properties of a single substance, the particle size of the complex will increase and the bond is unstable. Therefore, it is necessary to improve the functional properties and stability of the complex by a suitable modification method. However, some methods have limitations. For example, high-pressure homogenization is not suitable for high-viscosity materials, and the equipment is easy to damage when homogenizing high-hardness particles, and maintenance is difficult. High-shear emulsification equipment has a large volume, which is not suitable for small-scale preparation and production. Moreover, the design pressure of microjet homogenization equipment is high, the flow rate is low, and the cost is relatively high. Compared with the previous modification methods, high intensity ultrasound (HIU) technology has high efficiency and wide material applicability. HIU technology is characterized by a frequency between 20 and 100 kHz and an intensity greater than 1 W/cm2, which has been applied in the food industry [20], [21], [22]. The cavitation effect caused by ultrasound will generate strong pressure and shear force, and is the main power factor of ultrasound [23], [24]. Currently, many scholars have used ultrasonic technology to modify proteins and polysaccharides to improve the physical, chemical, and functional properties of foods [25], [26]. Cui et al. [27] explored the characteristics of SPI and SPI-glucose (SPI-G) complexes by ultrasonic treatment. The molecular flexibility of SPI and complexes after treatment was significantly related to protein emulsification activity and emulsion stability. Li et al. [28] prepared a peanut protein isolate-glucomannan complex by ultrasound technology. Compared with the complex prepared by classical heating, the solubility and emulsification properties of the complex obtained by ultrasonic treatment were improved.
Herein, HIU technology was used to prepare the SPI-PC complex, and the effect of HIU treatment power on the structure and solubility of the complex was investigated. Fourier transform infrared (FTIR) spectroscopy, fluorescence spectroscopy, Raman spectroscopy, changes in functional group content, and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) were used to explore the effects of HIU treatment on the spatial structure and chemical structure of the protein in the complex. In addition, confocal laser scanning microscopy (CLSM) was used to observe the dispersion state and solubility of the protein in the complex in water to improve the solubility of SPI and broaden the application range of the SPI-PC complex in food processing.
2. Materials and methods
2.1. Materials
SPI was prepared in the laboratory (protein content: 90.62 ± 0.84 %). High methoxymethyl ester pectin (derived from citrus, degree of esterification (DE) greater than 50%) was purchased from Yuanye Biotechnology Co., Ltd. (Shanghai, China). First-grade soybean oil was obtained from Jiusan Huikang Food Co., Ltd. (Harbin, China). All other chemical reagents were of analytical grade.
2.2. Preparation of SPI
SPI extraction was based on the method of Sui et al. [29] and improved. Defatted soybean meal was mixed with deionized water (DI), and then pH was adjusted to 8.0 with NaOH solution. The suspension was magnetically stirred at 400 rpm for 2 h, and centrifugated at 10000 × g for 30 min. The supernatant was adjusted to pH 4.5 with HCl solution and centrifuged at 10000 × g for 30 min. Then, the precipitate was taken and washed with deionized water three times. The suspension after each wash was centrifuged at 8000 × g for 15 min, and then the pH was adjusted to 7.0 with NaOH solution. The neutral protein solution was vacuum freeze-dried to obtain SPI. The final protein content measured by the Kjeldahl method was 90.62 ± 0.84 %.
2.3. Preparation of SPI-PC complex
First, the self-made SPI and commercial PC were dispersed in 0.01 M phosphate buffer solution (pH 7.0). After magnetic stirring at 400 rpm for 2 h, the 1% SPI and 0.3% PC solutions were mixed. Then, the pH value of the mixed solution was adjusted to 3.5 with citric acid. The mixed solution was magnetically stirred for 2 h, and 0.01% sodium azide was added. The mixed solution was then stored in a refrigerator at 4 °C overnight to prepare the SPI-PC complex.
2.4. HIU treatment of SPI-PC complex
The complex solution was placed in a beaker with ice to maintain the temperature below 20 °C. The ultrasonic treatments were performed using a Scientz-II D ultrasound generator (Scientz Biotechnology Co., Ltd., Ningbo, China). The ultrasonic frequency was 20 kHz, the pulse working time was 4 s, the intermittent time was 2 s, and the ultrasonic titanium probe with a diameter of 6 mm was immersed in the SPI-PC complex solution at a depth of 1 cm from the bottom, followed by treatments with different ultrasonic powers (150, 300, 450, and 600 W) for 15 min.
2.5. Characterization of SPI-PC complex treated by HIU
2.5.1. Fourier transform infrared (FTIR) spectroscopy analysis
The FTIR spectroscopy measurements of the SPI-PC complex before and after HIU treatment were performed by the KBr tablet method. First, 1 mg of the sample was extracted and 100 mg of KBr was added to fully grind, then the tablets were pressed. The sample was evaluated using an 8400S Fourier infrared spectrometer, and Peakfit 4.12 was used to perform Gaussian area fitting to calculate the change in secondary structure content [30].
2.5.2. Fluorescence spectra analysis
The samples of the SPI-PC complexes before and after HIU treatment were diluted with deionized water, and then the fluorescence spectra of the samples were measured with the F-4500 fluorescence spectrophotometer. The voltage was 700 mV, the excitation wavelength was 290 nm, the emission wavelength was 300–400 nm, the slit width was 5 nm, and the scan rate was 200 nm/min.
2.5.3. Surface hydrophobicity (H0) analysis
The SPI-PC complex solutions before and after HIU treatment were centrifuged at 10000 × g for 30 min to remove the precipitate. The supernatant was taken and diluted with phosphate buffer (0.01 M, pH 7.0) to the concentration range of 0.05–0.4 mg/mL. Moreover, ANS solution with a concentration of 8 mmol/L was added; afterward, the sample was mixed by vortexing and the fluorescence intensity was measured. The fluorescence intensity and the protein concentration in the complex were linearly fitted, and the slope was the value of H0 [31].
2.5.4. Raman spectroscopy analysis
Raman spectroscopy of the SPI-PC complex before and after HIU treatment was determined by the Raman Station 400 Raman Spectrometer. The excitation wavelength was 785 nm and the scan range was 400–2000 cm−1 with a spectral resolution of 3 cm−1. Peakfit 4.12 software was used to process the experimental data according to the peak intensity of the phenylalanine band at 1003 ± 1 cm−1, and analyze the changes in the protein tertiary structure in the complex sample.
2.5.5. Particle size and ζ-potential analysis
A Malvern Zetasizer Nano ZS potential and particle size distribution meter was used to measure the particle size distribution and ζ-potential of the sample. The sample solution was diluted with 0.01 M phosphate buffer (pH 7.0) as a dispersant. The particle size distribution and ζ-potential value of the sample were measured simultaneously.
2.5.6. CLSM analysis
A Leica TCS SP8® microscope was used to observe the microstructure of the samples, and the CLSM analysis was performed following the method of Zhong et al. [32] with slight modifications. First, Nile blue was dissolved in isopropanol to prepare 1% staining solutions, and then mixed uniformly and filtered. Next, 25 μL Nile blue solution was added to 0.5 mL of the sample solution treated with different ultrasound powers and allowed to label for 30 min. Consequently, 5 μL of the labeled complex solution was placed on the cover glass and sealed. Complex solution observation was carried out at excitation wavelengths of 488 nm.
2.5.7. Solubility analysis
The SPI-PC complex solution before and after HIU treatment was centrifuged at 3000 × g for 15 min to obtain the supernatant. The protein content in the supernatant was determined by the Lowry method [33]. A standard curve was drawn using bovine serum albumin as the standard substance, and the SPI solubility in the complex before and after HIU treatment was expressed as the percentage of the protein concentration in the supernatant to the total protein concentration.
2.5.8. SDS-PAGE analysis
The SDS-PAGE analysis was performed following the method of Jiang et al. [34] with slight modifications. The SPI-PC complex samples before and after HIU treatment were dissolved in deionized water to prepare a 3 mg/mL solution. Afterward, the complex solution was mixed with loading buffer with and without β-mercaptoethanol, heated in a boiling water bath for 5 min, and centrifuged at 3000 × g for 5 min. Subsequently, the supernatant was taken and the sample was loaded. After electrophoresis, the sample was labeled with Coomassie Brilliant Blue R-250 for 30 min, and decolorized with a decolorizing solution. Afterward, a gel imaging system was used to scan the image to analyze and evaluate the protein subunit composition in the complex sample.
2.6. Statistical analysis
All measurements were conducted at least in triplicate and mean values and standard deviations were analyzed. Origin 9.0 and Peakfit 4.12 were used to analyze the data. Statistics17 was used to analyze variance, and Duncan’s test (p < 0.05) was used to evaluate the significance of differences between data.
3. Results and discussion
3.1. FTIR spectroscopy analysis of SPI-PC complex under different HIU powers
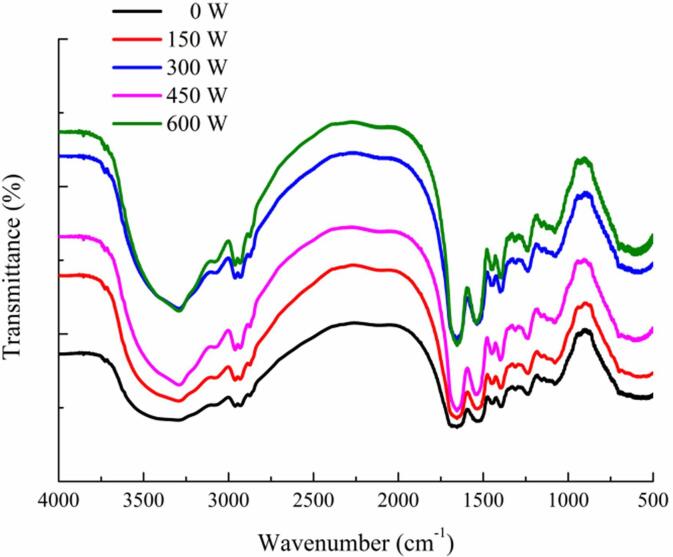
FTIR spectroscopy is an optical detection method to analyze the secondary structure of proteins. The amide I band (1600–1700 cm−1) is most commonly used to reflect changes in protein secondary structure [35]. The FTIR spectra of the SPI-PC complex under different HIU powers are shown in Fig. 1. The Gaussian integration method was used to fit and calculate the secondary structure content of the SPI in the complex, and the result is shown in Table 1. Compared with the untreated SPI-PC complex, the secondary structure of SPI in the complex after HIU treatment significantly changed. When the treatment power was between 150 and 450 W, the content of α-helix and β-sheet decreased, and the content of β-turn and random coil structures increased. The hydrogen bond that stabilized the protein structure was broken, the content of ordered structures in the protein molecule decreased, and the SPI unfolded, causing the molecular structure to loosen. When the treatment power was 450 W, the content of α-helix in SPI was the lowest, and the content of β-turn and random coil was the highest. The cavitation effect of HIU changed the protein secondary structure by breaking the intermolecular hydrogen bond and increasing protein flexibility [36]. When the treatment power exceeded 450 W, the α-helix and β-sheet structures of the SPI-PC complex increased and the β-turn and random coil structures decreased because the higher power treatment caused some protein aggregation.
Fig. 1.
FTIR spectra of HIU-treated SPI-PC complex under different powers.
Table 1.
Changes in the secondary structure content of soy protein isolate (SPI) under different high-intensity ultrasound (HIU) powers.
| HIU power (W) | α -helix (%) | β-sheet (%) | β-turn (%) | random coil (%) |
|---|---|---|---|---|
| 0 | 33.20 ± 0.07a | 19.37 ± 0.26e | 19.02 ± 0.29c | 28.41 ± 0.24e |
| 150 | 27.85 ± 0.13b | 28.74 ± 0.05a | 13.85 ± 0.41e | 29.56 ± 0.15c |
| 300 | 25.30 ± 0.06c | 26.74 ± 0.18b | 14.52 ± 0.08d | 33.44 ± 0.23b |
| 450 | 19.53 ± 0.12e | 24.61 ± 0.21d | 21.08 ± 0.11a | 34.78 ± 0.17a |
| 600 | 24.94 ± 0.11d | 25.39 ± 0.09c | 20.45 ± 0.09b | 29.22 ± 0.08d |
Note: The different superscript letters in the same column indicate significant differences among the data (p < 0.05)
3.2. Fluorescence spectrum analysis of SPI-PC complex under different HIU powers
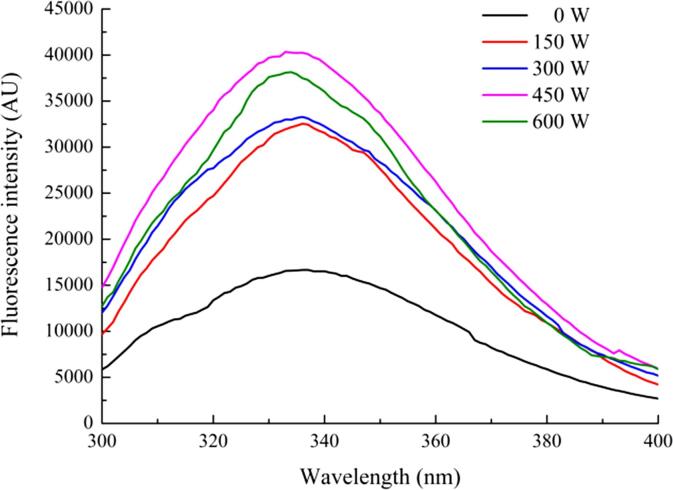
Endogenous fluorescence spectroscopy is an effective method to detect changes in the tertiary structure of proteins. As shown in Fig. 2, compared with the untreated SPI-PC complex, the maximum wavelength (λmax) of the HIU-treated complex did not have a significant blue-shift or red-shift, both were approximately 334 nm. The increase in fluorescence intensity indicates that the protein structure is unfolded, the originally buried aromatic amino acid residues are exposed to the surface of the protein, and the polarity of the microenvironment is increased [37]. As the treatment power increased, the fluorescence intensity of the SPI-PC complex increased first and then decreased. The fluorescence intensity of the complex treated with 450 W was the highest, while that of the complex treated with 600 W decreased significantly. The decrease in fluorescence intensity may be because the protein formed aggregates. Because of the cavitation effect caused by the formation and collapse of bubbles in the HIU treatment, the shear force and temperature increased, the protein structure was fully unfolded, and the hydrophobic interaction between molecules was promoted, leading to the formation of aggregates and the exposed aromatic amino acids being reburied, resulting in a decrease in fluorescence intensity.
Fig. 2.
Fluorescence spectra of HIU-treated SPI-PC complex under different powers.
3.3. H0 analysis of SPI-PC complex under different HIU powers
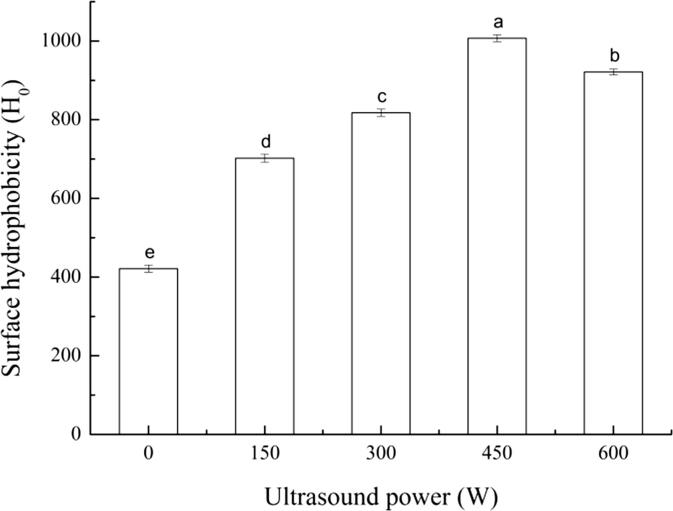
It can be seen from Fig. 3 that the H0 of the untreated SPI-PC complex was the lowest (421.36 ± 9.12), and the H0 of the samples after HIU treatment significantly increased (p < 0.05). As the treatment power increased, H0 gradually increased. This may be because after natural SPI interacted with PC to form a complex, the hydrophobic group was buried inside the molecule, reducing the contact between the hydrophobic group and the fluorescent probe during the measurement, so H0 was low. After HIU treatment, the structure of the protein unfolded, and the hydrophobic group inside the molecule was initially exposed to the polar environment, thereby increasing the H0 of the complex [38]. In addition, as power increased, H0 decreased, similar to the change in fluorescence intensity. The possible reason was that the HIU treatment under high-power conditions made the protein unfold excessively, and many hydrophobic groups migrated to the surface of the molecule, which promoted the hydrophobic interaction between the molecules, and the aggregation of proteins led to a decrease in H0. Therefore, the increase in H0 can increase the adsorption rate of the complex to the interface, so an appropriate HIU treatment helps to improve the emulsification properties of the SPI-PC complex. However, when the treatment power is too high, the emulsification properties of the complex will be reduced.
Fig. 3.
Surface hydrophobicity of HIU-treated SPI-PC complex under different powers Note: The superscript letters in the same group indicate significant differences among the data (p < 0.05).
3.4. Raman spectroscopy analysis of SPI-PC complex under different HIU powers
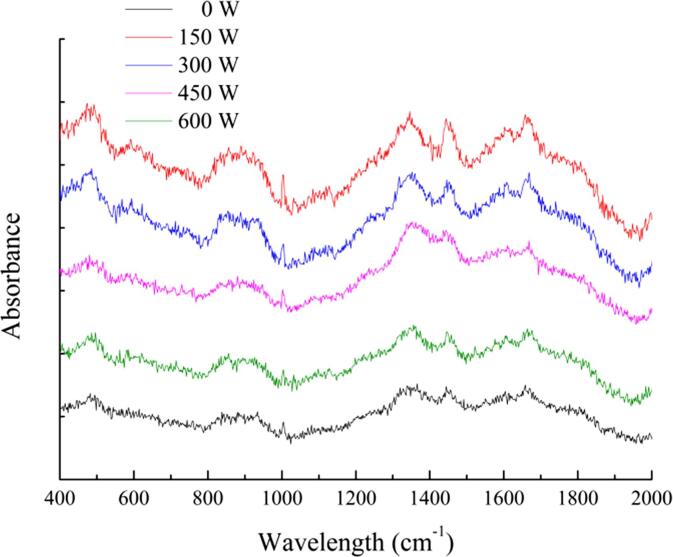
Fig. 4 shows the Raman spectra of the SPI-PC complex after HIU treatments under different powers. The conformational changes of the protein in the complex can be inferred by the changes in the scattering intensity and frequency of the Raman spectrum. The characteristic peaks of the amide I band (1630–1700 cm−1) in the Raman spectrum can be used to quantitatively analyze the secondary structure content of proteins. Peakfit 4.12 software is used to fit and analyze the amide I band of the Raman spectrum. The secondary structure content obtained was slightly different from that of FTIR spectroscopy, however, the trend of each secondary structure content was similar. This demonstrated that HIU treatment could effectively destroy the hydrogen bonds that maintained the stable structure of the α-helix. However, the high-power HIU treatment had an adverse effect on the change of SPI structure.
Fig. 4.
Raman spectra of HIU-treated SPI-PC complex under different powers.
In addition, some characteristic spectral frequencies in Raman spectroscopy can be applied to analyze the strength of protein hydrophobic interactions. The ratio of the bimodal intensity at 850 cm−1 and 830 cm−1 in the Raman spectrum can be used to monitor the hydrogen bonding force and polarity of the microenvironment around tyrosine (Tyr) residues, and to characterize the buried and exposed state of Tyr residues in protein molecules. When the ratio of I850 cm–1 to I830 cm–1 is between 0.900 and 1.450, it indicates that the Tyr residues in the protein structure are exposed to water or other polar environments; when the ratio of I850 cm–1 to I830 cm–1 is between 0.700 and 1.000, it indicates that the Tyr residues are buried in a hydrophobic environment [39]. Table 2 shows the Tyr bimodal ratio and C–H vibration band changes in the SPI-PC complex under HIU treatment. The I850 cm–1/I830 cm–1 ratio was between 0.824 and 1.033, indicating that the SPI Tyr residues changed from buried to unfolded during HIU treatment, and the hydrogen bond that maintained the SPI tertiary structure was destroyed to some extent. The Raman spectrum at 1450 cm−1 is the bending vibration of CH2 and CH3 with aliphatic amino acids. The intensity at this wavelength changed significantly with the HIU treatment power (p < 0.05), indicating that the environment of the SPI hydrophobic group changed from polarity to micropolarity. The change in the intensity of the C–H bond may be related to the state of the hydrophobic group and the unfolding of the protein structure.
Table 2.
Tyr band intensity and C–H band intensity of soy protein isolate (SPI) under different high-intensity ultrasound (HIU) powers.
| HIU power (W) | I850 cm-1/830 cm-1 | I1450 cm-1/1003 cm-1 |
|---|---|---|
| 0 | 0.905 ± 0.012bc | 1.156 ± 0.014d |
| 150 | 0.892 ± 0.008c | 1.183 ± 0.007c |
| 300 | 0.824 ± 0.011d | 1.199 ± 0.008bc |
| 450 | 0.915 ± 0.007b | 1.223 ± 0.005a |
| 600 | 1.033 ± 0.005a | 1.212 ± 0.016ab |
Note: The different superscript letters in the same column indicate significant differences among the data (p < 0.05)
3.5. Particle size and PDI analysis of SPI-PC complex under different HIU powers
Particle size is an important indicator reflecting the functional properties of the SPI-PC complex, and PDI is used to evaluate the particle size distribution of the complex particles in the solution. The effects of different treatment powers on the average particle size (D43) and PDI of the SPI-PC complex particles are shown in Table 3. The average particle size of the untreated complex was the largest, and was significantly reduced after HIU treatment (p < 0.05), and PDI showed that the particle size distribution range was more uniform. This is because physical forces such as turbulence and shear caused by cavitation during HIU treatment can destroy the non-covalent interaction of the SPI-PC complex; under vigorous agitation, the protein aggregates dissociate, thereby reducing the particle size of the complex [40]. However, the higher power treatment increased the average particle size of the complex. The formation and collapse of bubbles caused by HIU treatment led to cavitation effects, which increased physical forces such as turbulence and shear force. Moreover, the HIU treatment modified the arrangement of hydrogen bonds in the protein molecules, thereby changing the secondary structure of the proteins. With increasing treatment power, a considerable amount of heat was generated, thereby promoting SPI aggregation. Therefore, the selection of appropriate HIU treatment conditions can effectively improve the particle size of the SPI-PC complex [41].
Table 3.
Changes of average particle size (D43) and polymer dispersity index (PDI) of soy protein isolate (SPI)-pectin (PC) complex under different high-intensity ultrasound (HIU) powers.
| HIU power (W) | D43 (nm) | PDI |
|---|---|---|
| 0 | 1951.52 ± 7.52a | 0.576 ± 0.059a |
| 150 | 792.80 ± 6.31b | 0.394 ± 0.045d |
| 300 | 462.24 ± 4.28d | 0.467 ± 0.022b |
| 450 | 451.85 ± 2.17e | 0.427 ± 0.018c |
| 600 | 598.29 ± 1.98c | 0.366 ± 0.056d |
Note: The different superscript letters in the same column indicate significant differences among the data (p < 0.05)
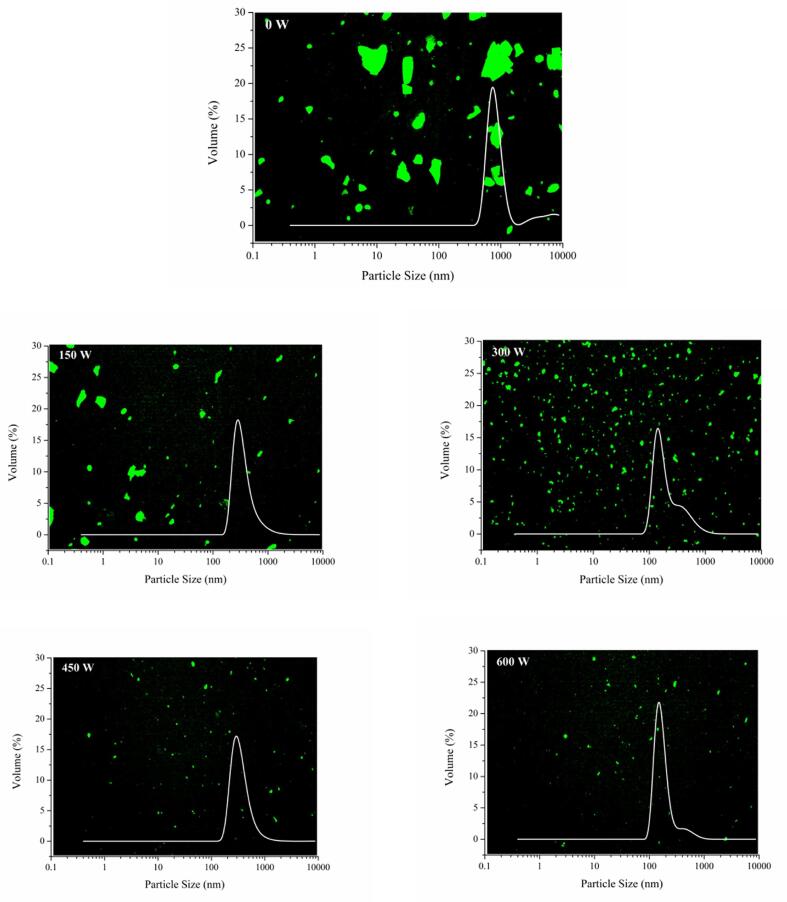
3.6. CLSM and particle size distribution analysis of SPI-PC complex under different HIU powers
CLSM was used to observe the distribution of protein in an aqueous solution in the HIU-treated SPI-PC complex under different powers. As the PC content was small, the dispersion state of the complex particles can be observed only by labeling the protein. SPI was labeled with Nile Blue and then excited green fluorescence; the result is shown in Fig. 5. The samples of the untreated SPI-PC complex in the aqueous solution were distributed in larger fragments with larger particle sizes. This is because SPI extracted by the freeze-drying method usually has a fragmented structure. HIU treatment can effectively reduce the particle size of SPI in the complex. As the treatment power increased, large pieces of SPI were gradually decomposed into smaller particles, which were evenly dispersed in the aqueous solution, and the particle size distribution peaks moved to the left. The turbulence effect of HIU treatment accelerated the collision of protein molecules and formed smaller particles under the action of cavitation. The CLSM image showed that the overall distribution of proteins in the complex was gradually more uniform, and the SPI particles were significantly smaller. This is consistent with the average particle size results. Gülseren et al. [42] reported that HIU treatment could effectively reduce the particle size of bovine serum albumin. The shear force caused by the ultrasound destroyed the non-covalent interactions in protein molecules, resulting in the collisions between proteins, and the particle size decreased. Therefore, HIU treatment, as an effective physical modification method, can destroy the protein structure, resulting in a significant reduction in protein particle size in aqueous solution.
Fig. 5.
CLSM and particle size distribution of HIU-treated SPI-PC complex under different powers.
3.7. ζ-potential and solubility analysis of SPI-PC complex under different HIU powers
ζ-potential is an important indicator to measure the net charge on the surface of complex particles, and is usually used to characterize the stability of the solution system. The greater the absolute value of the ζ-potential in the solution system, the stronger the repulsion between molecules and the less prone to aggregation. Therefore, the solution system is considered to be more stable. Table 4 shows the changes in ζ-potential and protein solubility of the SPI-PC complex treated with HIU under different powers. The untreated SPI-PC complex had the lowest absolute potential value (7.53 ± 0.58 mV). In contrast, all HIU-treated samples showed significantly higher absolute potential values (p < 0.05). The increase of the absolute potential value provided good electrostatic repulsion, which was beneficial to improve the stability of the SPI-PC complex solution. Mechanical forces, such as cavitation generated by HIU treatment, acted on the SPI-PC complex, which changed the structure of the complex. The polar groups originally located inside were exposed to the surface of the protein particles, so that the SPI and PC in the solution had more net charges, and the electrostatic interaction made the complexes more tightly bound and improved the dispersion and stability of the complex. The absolute potential value of the SPI-PC complex treated with HIU for 15 min at 450 W was the highest, indicating that the electrostatic repulsion between the particles in solution was the strongest at this time, and the dispersibility of the protein in water was better (Fig. 5). However, the absolute potential value decreased when the HIU was performed with higher power (600 W). Hu et al. [43] believed that this phenomenon could be due to the formation of protein aggregates, which masked the polar sites on the protein surface. This is consistent with the results of this study.
Table 4.
Changes of ζ-potential and solubility of soy protein isolate (SPI)-pectin (PC) complex under different high-intensity ultrasound (HIU) powers.
| HIU power (W) | ζ-potential (mV) | Solubility (%) |
|---|---|---|
| 0 | −7.53 ± 0.58e | 46.42 ± 0.48e |
| 150 | −14.65 ± 0.51d | 73.01 ± 0.31d |
| 300 | −18.33 ± 0.32b | 76.72 ± 0.22c |
| 450 | −19.87 ± 0.61a | 89.04 ± 0.29b |
| 600 | −16.96 ± 1.48c | 89.88 ± 0.27a |
Note: The different superscript letters in the same column indicate significant differences among the data (p < 0.05).
Protein solubility can to some extent reflect changes in protein structure. Generally, proteins with good solubility have better functional properties, which is conducive to the processing and utilization of the protein. As listed in Table 4, the solubility of SPI in the untreated complex was lower (46.42 ± 0.48 %). Protein solubility in the complexes after HIU treatment increased significantly (p < 0.05). The increase in power can enhance the solubility of SPI in the complexes. Among them, the solubility of the SPI sample treated at 600 W for 15 min was the highest. It was speculated that the strong physical force generated by cavitation destroyed the non-covalent bonds that maintained the protein spatial structure, which promoted the unfolding of the SPI structure and reduced the particle size. Concurrently, the hydrophobic and polar groups inside the SPI molecules were exposed to the surface, increasing the interaction between the protein particles and water, thereby improving the solubility of SPI [44].
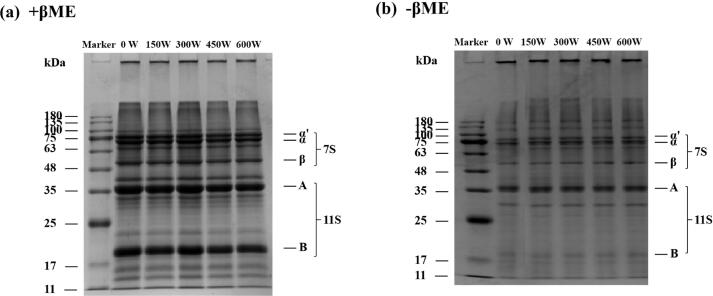
3.8. SDS-PAGE analysis of SPI-PC complex under different HIU powers
To further explore the effect of HIU treatment on the structure of SPI-PC complexes, reduced (with β-mercaptoethanol) and non-reduced (without β-mercaptoethanol) SDS-PAGE electrophoresis analyses were carried out. The results are shown in Fig. 6. All SPI-PC complex samples showed typical SPI electrophoresis bands with and without β-mercaptoethanol; where α', α, and β were subunits of β-conglobulin (7S), A and B were the acidic and basic subunits of glycinin (11S), respectively. This is similar to the results of Ma et al. [45]. They performed gel electrophoresis on SPI and SPI-PC complexes, and found that the electrophoretic band distribution of the complex was similar to that of natural SPI. β-mercaptoethanol could open and dissociate the disulfide bonds of the protein molecule into subunits. Under non-reducing conditions, all SPI-PC complex samples had aggregates at the relative molecular mass of 100–180 kDa. Under reducing conditions, the bands of these aggregates disappeared, indicating that disulfide bonds were the main force for the formation of these aggregates. In addition, compared with the untreated complex sample, the reduced and non-reduced profiles of the complex after HIU treatment did not change significantly, demonstrating that the HIU treatment under this condition did not destroy the covalent bond of the SPI-PC complex, nor did it form a new covalent bond, further proving that the HIU treatment did not change the subunit composition of the complex. This is similar to the results of Hu et al. [46]. They found that the electrophoresis of SPI samples treated with ultrasound for 15 and 30 min under different powers did not change significantly.
Fig. 6.
SDS-PAGE of HIU-treated SPI-PC complex under different powers (a) with β-mercaptoethanol, (b) without β-mercaptoethanol.
4. Conclusions
The SPI-PC complex was modified by HIU treatment to improve its functional properties. The results showed that with increasing treatment power, the α-helix and β-sheet content of SPI in the complex decreased, the content of β-turn and random coil increased, and the fluorescence intensity increased. The hydrophobic groups originally buried in the molecule were exposed, and H0 increased. The solubility and dispersibility of protein in water increased. However, when the HIU treatment power was too high, the content of α-helix and β-sheet increased, the content of β-turn and random coil decreased, H0 decreased, and the particle size of the complex tended to increase again. In addition, the SDS-PAGE electrophoresis analysis results found that HIU treatment under this condition could not change the subunit composition of the complex. In summary, suitable HIU treatment can unfold the protein structure in the SPI-PC complex and significantly reduce the particle size of the complex, thereby improving its solubility and dispersion in aqueous solution, and improving the stability of the SPI-PC complex.
CRediT authorship contribution statement
Ning Wang: Methodology, Investigation, Writing – original draft. Xiaonan Zhou: Investigation, Validation, Data curation. Weining Wang: Visualization. Liqi Wang: Supervision, Funding acquisition. Lianzhou Jiang: Supervision, Conceptualization. Tianyi Liu: Supervision, Conceptualization. Dianyu Yu: Conceptualization, Methodology, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (NSFC): Construction of a three-dimensional enzyme electrode and its correlation with phospholipid content in oil (No: 32072259).
Contributor Information
Tianyi Liu, Email: Ltyone80@neau.edu.cn.
Dianyu Yu, Email: dyyu2000@126.com.
References:
- 1.Ma L., Li B., Han F., Yan S., Wang L., Sun J. Evaluation of the chemical quality traits of soybean seeds, as related to sensory attributes of soymilk. Food Chem. 2015;173:694–704. doi: 10.1016/j.foodchem.2014.10.096. [DOI] [PubMed] [Google Scholar]
- 2.Nishinari K., Fang Y., Guo S., Phillips G.O. Soy proteins: A review on composition, aggregation and emulsification. Food Hydrocolloid. 2014;39:301–318. doi: 10.1016/j.foodhyd.2014.01.013. [DOI] [Google Scholar]
- 3.Li R., Cui Q., Wang G., Liu J., Chen S., Wang X., Wang X., Jiang L. Relationship between Surface Functional Properties and Flexibility of Soy Protein Isolate-Glucose Conjugates. Food Hydrocolloid. 2019;95:349–357. doi: 10.1016/j.foodhyd.2019.04.030. [DOI] [Google Scholar]
- 4.Keerati-u-rai M., Wang Z., Corredig M. Adsorption of Soy Protein Isolate in Oil-in-Water Emulsions: Difference Between Native and Spray Dried Isolate. J. Am. Oil Chem. Soc. 2011;88(10):1593–1602. doi: 10.1007/s11746-011-1818-8. [DOI] [Google Scholar]
- 5.Li Y., Sukmanov V., Kang Z.L., Ma H. Effect of soy protein isolate on the techno-functional properties and protein conformation of low: odium pork meat batters treated by high pressure. J Food Process. Eng. 2019;43 doi: 10.1111/jfpe.13343. [DOI] [Google Scholar]
- 6.Freitas M.L.F., Albano K.M., Telis V.R.N. Characterization of biopolymers and soy protein isolate-high-methoxyl pectin complex. Polímeros. 2017;27(1):62–67. doi: 10.1590/0104-1428.2404. [DOI] [Google Scholar]
- 7.MOHNEN D. Pectin structure and biosynthesis. Curr. Opin. Plant. Biol. 2008;11(3):266–277. doi: 10.1016/j.pbi.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Kyomugasho C., Christiaens S., Davy V., Loey A.V., Dewettinck K., Hendrickx M.E. Evaluation of cation-facilitated pectin-gel properties: Cryo-SEM visualisation and rheological properties. Food Hydrocolloid. 2016;61:172–182. doi: 10.1016/j.foodhyd.2016.05.018. [DOI] [Google Scholar]
- 9.Lutz R., Aserin A., Wicker L., Garti N. Structure and physical properties of pectins with block-wise distribution of carboxylic acid groups. Food Hydrocolloid. 2009;23(3):786–794. doi: 10.1016/j.foodhyd.2008.04.009. [DOI] [Google Scholar]
- 10.Vityazev F.V., Khramova D.S., Saveliev N.Y., Ipatova E.A., Burkov A.A., Beloserov V.S., Belyi V.A., Kononov L.O., Martinson E.A., Litvinets S.G., Markov P.A., Popov S.V. Pectin-glycerol gel beads: preparation, characterization and swelling behaviour. Carbohydr. Polym. 2020;238:116166. doi: 10.1016/j.carbpol.2020.116166. [DOI] [PubMed] [Google Scholar]
- 11.Jiang W.-X., Qi J.-R., Huang Y.-X., Zhang Y., Yang X.-Q. Emulsifying properties of high methoxyl pectins in binary systems of water-ethanol. Carbohydr. Polym. 2020;229:115420. doi: 10.1016/j.carbpol.2019.115420. [DOI] [PubMed] [Google Scholar]
- 12.Guo Q., Shu X., Su J., Li Q., Tong Z., Yuan F., Mao L., Gao Y. Interfacial properties and antioxidant capacity of pickering emulsions stabilized by high methoxyl pectin-surfactant-pea protein isolate-curcumin complexes: Impact of different types of surfactants. LWT-Food Sci. Technol. 2022;153:112453. doi: 10.1016/j.lwt.2021.112453. [DOI] [Google Scholar]
- 13.W.F. Xiong, Y. Li, C. Ren, J. Li, B. Li, F. Geng, Thermodynamic parameters of gelatin-pectin complex coacervation, Food Hydrocolloid. 120 (2021) 106958. https://doi.org/ 10.1016/j.foodhyd.2021.106958.
- 14.Chen W., Lv R., Wang W., Ma X., Muhammad A.I., Guo M., Ye X., Liu D. Time effect on structural and functional properties of whey protein isolate-gum acacia conjugates prepared via Maillard reaction. J. Sci. Food Agr. 2019;99(10):4801–4807. doi: 10.1002/jsfa.2019.99.issue-1010.1002/jsfa.9735. [DOI] [PubMed] [Google Scholar]
- 15.You Y., Liu F., Du K.J., Wen G.B., Lin Y.W. Structural and functional alterations of myoglobin by glucose-protein interactions. J. Mol. Model. 2014;3:2358. doi: 10.1007/s00894-014-2358-6. [DOI] [PubMed] [Google Scholar]
- 16.Wang T., Chen K., Zhang X., Yu Y., Yu D., Jiang L., Wang L. Effect of ultrasound on the preparation of soy protein isolate-maltodextrin embedded hemp seed oil microcapsules and the establishment of oxidation kinetics models. Ultrason. Sonochem. 2021;77:105700. doi: 10.1016/j.ultsonch.2021.105700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albano K.M., Cavallieri Â.L.F., Nicoletti V.R. Electrostatic interaction between proteins and polysaccharides: Physicochemical aspects and applications in emulsion stabilization. Food Rev. Int. 2019;35(1):54–89. doi: 10.1080/87559129.2018.1467442. [DOI] [Google Scholar]
- 18.Mao L., Boiteux L., Roos Y.H., Miao S. Evaluation of volatile characteristics in whey protein isolate-pectin mixed layer emulsions under different environmental conditions. Food Hydrocolloid. 2014;41:79–85. doi: 10.1016/j.foodhyd.2014.03.025. [DOI] [Google Scholar]
- 19.Ma X., Yan T., Hou F., Chen W., Miao S., Liu D. Formation of soy protein isolate (SPI)-citrus pectin (CP) electrostatic complexes under a high-intensity ultrasonic field: Linking the enhanced emulsifying properties to physicochemical and structural properties. Ultrason. Sonochem. 2019;59:104748. doi: 10.1016/j.ultsonch.2019.104748. [DOI] [PubMed] [Google Scholar]
- 20.Costa M.G.M., Fonteles T.V., de Jesus A.L.T., Almeida F.D.L., de Miranda M.R.A., Fernandes F.A.N., Rodrigues S. High-Intensity Ultrasound Processing of Pineapple Juice. Food Bioprocess. 2013;6(4):997–1006. doi: 10.1007/s11947-011-0746-9. [DOI] [Google Scholar]
- 21.Li Z.-X., Lin H., Cao L.-M., Jameel K. Effect of high intensity ultrasound on the allergenicity of shrimp. J. ZheJiang Univ-Sc. 2006;7(4):251–256. doi: 10.1631/jzus.2006.B0251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu H., Keh M.Z.M., Seow Y.X., Ong P.K.C., Zhou W. Kinetic Study of High-Intensity Ultrasound-Assisted Maillard Reaction in a Model System of D-Glucose and L-Methionine. Food Bioprocess. Tech. 2017;10:1984–1996. doi: 10.1016/j.foodchem.2018.07.053. [DOI] [Google Scholar]
- 23.Khadhraoui B., Ummat V., Tiwari B.K., Fabiano-Tixier A.S., Chemat F. Review of ultrasound combinations with hybrid and innovative techniques for extraction and processing of food and natural products. Ultrason. Sonochem. 2021;76:105625. doi: 10.1016/j.ultsonch.2021.105625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalla-Bertholdt A.-M., Nguyen P.-V., Baier A.K., Rauh C. Influence of dietary fiber on in-vitro lipid digestion of emulsions prepared with high-intensity ultrasound. Innov. Food Sci. Emerg. 2021;73:102799. doi: 10.1016/j.ifset.2021.102799. [DOI] [Google Scholar]
- 25.Huang L., Ding X., Li Y., Ma H. The aggregation, structures and emulsifying properties of soybean protein isolate induced by ultrasound and acid. Food Chem. 2019;279:114–119. doi: 10.1016/j.foodchem.2018.11.147. [DOI] [PubMed] [Google Scholar]
- 26.Gu J., Li Q., Liu J., Ye Z., Feng T., Wang G.e., Wang W., Zhang Y. Ultrasonic-assisted extraction of polysaccharides from Auricularia auricula and effects of its acid hydrolysate on the biological function of Caenorhabditis elegans. Int. J. Biol. Macromol. 2021;167:423–433. doi: 10.1016/j.ijbiomac.2020.11.160. [DOI] [PubMed] [Google Scholar]
- 27.Cui Q., Zhang A., Li R., Wang X., Sun L., Jiang L. Ultrasonic treatment affects emulsifying properties and molecular flexibility of soybean protein isolate-glucose conjugates. Food Biosci. 2020;38:100747. doi: 10.1016/j.fbio.2020.100747. [DOI] [Google Scholar]
- 28.Li C., Huang X., Peng Q., Shan Y., Xue F. Physicochemical properties of peanut protein isolate-glucomannan conjugates prepared by ultrasonic treatment. Ultrason. Sonochem. 2014;21(5):1722–1727. doi: 10.1016/j.ultsonch.2014.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Sui X., Bi S., Qi B., Wang Z., Zhang M., Li Y., Jiang L. Impact of ultrasonic treatment on an emulsion system stabilized with soybean protein isolate and lecithin: Its emulsifying property and emulsion stability. Food Hydrocolloid. 2017;63:727–734. doi: 10.1016/j.foodhyd.2016.10.024. [DOI] [Google Scholar]
- 30.Yu D., Zhao Y., Li T., Li D., Chen S., Wu N., Jiang L., Wang L. Effect of electrochemical modification on the structural characteristics and emulsion storage stability of soy protein isolate. Process Biochem. 2018;75:166–172. doi: 10.1016/j.procbio.2018.10.001. [DOI] [Google Scholar]
- 31.Li D., Zhao Y., Wang X.u., Tang H., Wu N., Wu F., Yu D., Elfalleh W. Effects of (+)-catechin on a rice bran protein oil-in-water emulsion: Droplet size, zeta-potential, emulsifying properties, and rheological behavior. Food Hydrocolloid. 2020;98:105306. doi: 10.1016/j.foodhyd.2019.105306. [DOI] [Google Scholar]
- 32.Zhong M., Xie F., Zhang S., Sun Y., Qi B., Li Y. Preparation and digestive characteristics of a novel soybean lipophilic protein-hydroxypropyl methylcellulose-calcium chloride thermosensitive emulsion gel. Food Hydrocolloid. 2020;106:105891. doi: 10.1016/j.foodhyd.2020.105891. [DOI] [Google Scholar]
- 33.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951;193:265–275. doi: 10.1515/bchm2.1951.286.1-6.270. [DOI] [PubMed] [Google Scholar]
- 34.Jiang L., Wang J., Li Y., Wang Z., Liang J., Wang R., Chen Y., Ma W., Qi B., Zhang M. Effects of ultrasound on the structure and physical properties of black bean protein isolates. Food Res. Int. 2014;62:595–601. doi: 10.1016/j.foodres.2014.04.022. [DOI] [Google Scholar]
- 35.Zhao X., Chen F., Xue W., Lee L. FTIR spectra studies on the secondary structures of 7S and 11S globulins from soybean proteins using AOT reverse micellar extraction. Food Hydrocolloid. 2008;22(4):568–575. doi: 10.1016/j.foodhyd.2007.01.019. [DOI] [Google Scholar]
- 36.Zhu Z., Zhu W., Yi J., Liu N., Cao Y., Lu J., Decker E.A., Mcclements D.J. Effects of sonication on the physicochemical and functional properties of walnut protein isolate. Food Res. Int. 2018;106:853–861. doi: 10.1016/j.foodres.2018.01.060. [DOI] [PubMed] [Google Scholar]
- 37.Li S., Yang X., Zhang Y., Ma H., Liang Q., Qu W., He R., Zhou C., Mahunu G.K. Effects of ultrasound and ultrasound assisted alkaline pretreatments on the enzymolysis and structural characteristics of rice protein. Ultrason. Sonochem. 2016;31:20–28. doi: 10.1016/j.ultsonch.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 38.Jiang S., Ding J., Andrade J., Rababah T.M., Almajwal A., Abulmeaty M.M., Feng H. Modifying the physicochemical properties of pea protein by pH-shifting and ultrasound combined treatments. Ultrason. Sonochem. 2017;38:835–842. doi: 10.1016/j.ultsonch.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 39.Zheng T., Li X., Taha A., Wei Y., Hu T., Fatamorgana P.B., Zhang Z., Liu F., Xu X., Pan S., Hu H. Effect of high intensity ultrasound on the structure and physicochemical properties of soy protein isolates produced by different denaturation methods. Food Hydrocolloid. 2019;97:105216. doi: 10.1016/j.foodhyd.2019.105216. [DOI] [Google Scholar]
- 40.Wen C., Zhang J., Yao H., Zhou J., Duan Y., Zhang H., Ma H. Advances in renewable plant-derived protein source: The structure, physicochemical properties affected by ultrasonication. Ultrason. Sonochem. 2018;53:83–98. doi: 10.1016/j.ultsonch.2018.12.036. [DOI] [PubMed] [Google Scholar]
- 41.Li Y., Cheng Y.u., Zhang Z., Wang Y., Mintah B.K., Dabbour M., Jiang H., He R., Ma H. Modification of Rapeseed Protein by Ultrasound-assisted pH Shift Treatment: Ultrasonic Mode and Frequency Screening, Changes in Protein Solubility and Structural Characteristics. Ultrason. Sonochem. 2020;69:105240. doi: 10.1016/j.ultsonch.2020.105240. [DOI] [PubMed] [Google Scholar]
- 42.Gülseren İ., Güzey D., Bruce B.D., Weiss J. Structural and functional changes in ultrasonicated bovine serum albumin solutions. Ultrason. Sonochem. 2007;14(2):173–183. doi: 10.1016/j.ultsonch.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 43.Hu S., Wu J., Zhu B., Du M., Wu C., Yu C., Song L., Xu X. Low oil emulsion gel stabilized by defatted Antarctic krill (Euphausia superba) protein using high-intensity ultrasound. Ultrason. Sonochem. 2021;70:105294. doi: 10.1016/j.ultsonch.2020.105294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higuera-Barraza O.A., Toro-Sanchez C.D., Ruiz-Cruz S., Márquez-Ríos E. Effects of high-energy ultrasound on the functional properties of proteins. Ultrason. Sonochem. 2016;31:558–562. doi: 10.1016/j.ultsonch.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Ma X., Chen W., Yan T., Wang D., Hou F., Miao S., Liu D. Comparison of citrus pectin and apple pectin in conjugation with soy protein isolate (SPI) under controlled dry-heating conditions. Food Chem. 2020;309:125501. doi: 10.1016/j.foodchem.2019.125501. [DOI] [PubMed] [Google Scholar]
- 46.Hu H., Wu J., Eunice C.Y., Chan L., Zhu L., Zhang F., Xu X., Fan G., Wang L., Huang X., Pan S. Effects of ultrasound on structural and physical properties of soy protein isolate (SPI) dispersions. Food Hydrocolloid. 2013;30:647–655. doi: 10.1016/j.foodhyd.2012.08.001. [DOI] [Google Scholar]