Abstract
Glioblastoma (GBM) is the most common, most aggressive and deadliest brain tumor. Recently, remarkable progress has been made towards understanding the cellular and molecular biology of gliomas. GBM tumor initiation, progression and relapse as well as resistance to treatments are associated with glioma stem cells (GSCs). GSCs exhibit a high proliferation rate and self-renewal capacity and the ability to differentiate into diverse cell types, generating a range of distinct cell types within the tumor, leading to cellular heterogeneity. GBM tumors may contain different subsets of GSCs, and some of them may adopt a quiescent state that protects them against chemotherapy and radiotherapy. GSCs enriched in recurrent gliomas acquire more aggressive and therapy-resistant properties, making them more malignant, able to rapidly spread. The impact of SOX transcription factors (TFs) on brain tumors has been extensively studied in the last decade. Almost all SOX genes are expressed in GBM, and their expression levels are associated with patient prognosis and survival. Numerous SOX TFs are involved in the maintenance of the stemness of GSCs or play a role in the initiation of GSC differentiation. The fine-tuning of SOX gene expression levels controls the balance between cell stemness and differentiation. Therefore, innovative therapies targeting SOX TFs are emerging as promising tools for combatting GBM. Combatting GBM has been a demanding and challenging goal for decades. The current therapeutic strategies have not yet provided a cure for GBM and have only resulted in a slight improvement in patient survival. Novel approaches will require the fine adjustment of multimodal therapeutic strategies that simultaneously target numerous hallmarks of cancer cells to win the battle against GBM.
Keywords: Glioblastoma, SOX transcription factors, Glioma stem cells, Stemness, Differentiation
Core Tip: Despite the remarkable progress that has been made in understanding the cellular and molecular biology of gliomas, current therapeutic strategies have not yet provided a significant benefit to patients or a cure. This review highlights the key functions of SOX transcriptional factors (TFs) in glioblastoma (GBM) and glioma stem cells (GSCs). SOX TFs influence stemness, self-renewal, proliferation, differentiation, viability, migration, invasion, apoptosis, therapy resistance, sphere-forming capacity and tumorigenicity. We emphasized that fine-tuning of the SOX expression level is required to control the balance between the stemness and differentiation of GSCs, making SOX TFs promising therapeutic targets for combatting GBM.
INTRODUCTION
Cancer is among the leading causes of death worldwide, representing a substantial public health burden affecting welfare and life expectancy globally, with enormous impacts on individuals, families and health systems. The global burden of cancer continues to grow with its increasing incidence in the 21st century, mainly due to the growth and ageing of populations, adoption of unhealthy behaviors and exposure to unhealthy environments.
Cancers comprise a large group of diseases that arise from cells that escape the normal checkpoints of cell division and are capable of uncontrolled growth and proliferation. Deregulation of the precise molecular networks operating at the molecular and cellular levels that control cell proliferation, differentiation and cell death leads to the transformation of normal cells into cancer. Despite decades of intensive research, the underlying mechanisms that transform normal cells into cancer cells and enable cancer cells to spread and metastasize other sites in the body, leading to a fatal outcome, are still not completely understood.
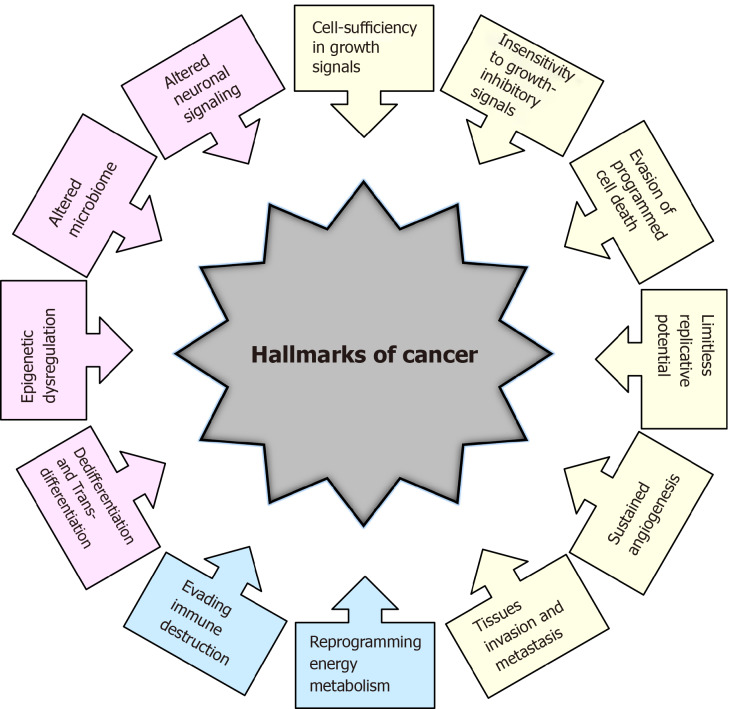
The transformation of normal cells into cancer cells is a complex multistep process, and in recent decades, tremendous efforts have been made to understand the underlying mechanisms. In a highly influential article published more than 20 years ago, Hanahan and Weinberg[1] elucidated six essential biological capabilities of cancer cell, widely accepted as the six hallmarks of cancer. Most, if not all, cancers acquire this set of capabilities during the multistep development of human tumors. The six hallmarks of cancer (Figure 1) include cell sufficiency in growth signals, insensitivity to growth-inhibitory signals, evasion of programmed cell death, limitless replicative potential, sustained angiogenesis, and tissue invasion and metastasis[1]. Together, these hallmarks constitute an organized framework for interpreting the remarkable diversity of neoplastic diseases[1]. It has been proposed that the genomic instability underlying these hallmarks generates genetic diversity, which contributes to multiple cancer hallmark functions[1].
Figure 1.
Historical overview of the hallmarks of cancer. Hallmarks of cancer proposed by Hanahan and Weinberg[1,2] in 2000 and 2011 are presented in yellow and blue colors, respectively. Additional hallmarks of cancer proposed by Senga and Grose[3] in 2021 are presented in pink.
In subsequent years, remarkable progress was made towards understanding the mechanisms underlying each of the six hallmarks. New technologies and significant advances in the understanding of the cellular and molecular biology of cancer cells provided better insight into the multiple events that allow cancer cells to acquire additional functional capabilities enabling them to survive, proliferate and disseminate. Thus, two additional enabling characteristics of cancer cells-genomic instability and inflammation via innate immune cells, have been defined[2]. Genomic instability in cancer cells generates random mutations and acquired genetic diversity, endowing cancer cells with genetic alterations that drive tumor progression[3]. Inflammation that fights infections and heals wounds under physiological conditions can assist tumor progression in premalignant and malignant lesions[4]. Research on the intersections between inflammation and cancer pathogenesis has provided overwhelming evidence of the essential effects that immune cells, largely of the innate immune system, have on tumor-promoting neoplastic progression[4-6].
In an updated article published by Hanahan and Weinberg[2] in 2011, two novel hallmarks were proposed and added to the list of core hallmarks of cancer: reprogramming energy metabolism and evading immune destruction. The first includes the major reprogramming of cellular energy metabolism that is required to support uncontrolled cell proliferation. Since the immune system serves as an important barrier to tumor formation, the second hallmark implicates the ability of cancer cells to evade attack and elimination by immune cells[2].
It has become necessary for the hallmarks of cancer to be revised and upgraded as a result of the progression of cancer research and the accumulation of knowledge over the last decade. Four novel hallmarks of cancer were recently proposed, justified and incorporated into the mainstream hallmark conceptualization. These hallmarks include dedifferentiation and transdifferentiation, epigenetic dysregulation, altered microbiome and altered neuronal signaling (Figure 1)[7].
STEM CELLS: DEDIFFERENTIATION AND TRANSDIFFERENTIATION
Stem cells are defined as cells that have self-renewing capacity and the ability to differentiate into multiple cell types[8]. Stem cells are present in mammalian embryos at the blastocyst stage as well as in the tissues and organs of adults. While embryonic stem cells (ESCs) have the ability to differentiate into any cell type present in the adult body, adult stem cells are capable of generating and replacing terminally differentiated cells in specific tissues and are involved in the continual maintenance and repair of tissues and organs throughout life[9,10].
An unidirectional developmental model suggesting that pluripotent stem cells progressively lose their pluripotency as they differentiate along developmental pathways until they reach a terminally differentiated state was widely accepted for several decades[11]. It was believed that adult stem cells have the ability to generate only the differentiated cell phenotypes of the tissue in which they reside. At the beginning of the 21st century, numerous studies challenged this strict hierarchy of stem cells and their unidirectional differentiation. The phenomenon of stem cell plasticity, or transdifferentiation, has emerged, suggesting that some adult stem cells have phenotypic potential that extends beyond the cell types of their resident tissue[12,13]. It was proposed that some stem cells under specific conditions might diverge from their predetermined pathway and generate cells of a different tissue by entering into a process of transdifferentiation, or that mature cells dedifferentiate into cells with a stem cell phenotype and eventually differentiate into cells of a different tissue[9].
In 1962, Gurdon[14] first proposed the hypothesis that the genome of every specialized cell of an adult organism has all the information required to develop into all different cell types. This hypothesis was proven in the seminal publication of Takahashi and Yamanaka[15], which demonstrated that adult differentiated cells could be reprogrammed into induced pluripotent stem cells that have the ability to differentiate into any of the endodermal, ectodermal and mesodermal cell lineages. This reprogramming is achieved by the overexpression of stem cell-associated genes, also known as stemness factors or Yamanaka factors, in the differentiated cells.
These findings validated the phenomenon of dedifferentiation and transdifferentiation, laying the groundwork for cancer stem cell (CSC) theory and the discovery of CSCs.
CSCs
The first indication of the existence of cancer cells with a stem cell phenotype came from the study of teratomas, which showed that undifferentiated cells preferably gave rise to nontumorigenic differentiated cells[16]. The CSC hypothesis was proposed in line with these data, suggesting that tumors comprise a mixture of malignant stem cells and their benign counterparts[17].
CSCs represent a small subpopulation of tumor cells with the capabilities of self-renewal, differentiation, and tumorigenicity when transplanted into an animal host[18]. CSCs and stem cells share similar properties, including self-renewal ability, unlimited growth potential, invasiveness and blockade of differentiation (reviewed in[19]), whereby indefinite self-renewal capability enables CSCs to initiate and maintain tumor growth.
CSCs were first identified in acute myeloid leukemia[20] and subsequently in a wide variety of tumor types, including melanoma, osteosarcoma, leukemia, breast, colorectal, brain, prostate, pancreatic, ovarian, liver and lung cancer[21-23].
Advances in whole-genome sequencing have revealed the remarkable genetic complexity of malignant tumors and the presence of subpopulations of cells with distinct genotypes and phenotypes[24]. The distinct genotypes of cancer cells within the tumor endow the cells with different biological features and phenotypes, providing the basis for intra- and inter-tumor heterogeneity[25]. The CSC model explains this phenotypic and functional heterogeneity among cancer cells[26-29].
It has been proposed that CSCs originate from either adult tissue-resident stem cells or from differentiated cells that have been reprogrammed to a pluripotent state by the process of dedifferentiation[30]. The CSC hypothesis proposes that many heterogenic cancers are organized in a hierarchical fashion based on the differentiation capacity of the cells comprising the tumor[31]. It has been suggested that this hierarchical order recapitulates the normal tissue hierarchy established by healthy stem cells. Thus, CSCs generate cellular heterogeneity by imposing a differentiation hierarchy by generating a range of distinct cell types present within the tumor[26]. However, the established hierarchy is not permanent, and under specific conditions, could be reversed as terminally differentiated cells become dedifferentiated and regain CSC properties[22,27].
CSCs have been shown to exhibit high plasticity, resulting in changes in their phenotypic and functional appearance in response to chemo- and radiotherapeutics that cause alterations in the tumor microenvironment (TME)[30]. The negative effects of senescence can directly promote cancer stemness by increasing CSC plasticity, activating stemness pathways in non-CSCs, and promoting senescence escape and subsequent activation of a stemness pathway[30].
CSCs share a number of unique features that distinguish them from other tumor cells. CSCs are believed to be responsible for cancer initiation, progression, metastasis, recurrence and drug resistance[32]. Epithelial CSCs express many genes/pathways typically associated with normal stem cells (reviewed in[31]). In many types of tumors, some CSCs acquire epithelial-to-mesenchymal transition profiles through the upregulation of the expression of specific genes driving metastasis. CSCs are suggested to be responsible for drug resistance and cancer relapse due to their ability to self-renew and differentiate into heterogeneous lineages of cancer cells[33]. Drug resistance has been linked to the ability of CSCs to become quiescent, upregulate the expression of enzymes such as aldehyde dehydrogenase, and upregulate the expression of anti-apoptotic proteins and multidrug resistance pumps that increase chemotherapeutic elimination from cells, resulting in low intracellular drug concentrations[31].
Accordingly, the majority of CSC features depend on the deregulation of signaling pathways that, in turn, rely on the altered activity of specific transcription factors (TFs).
SOX TFs AND CSCs
SOX (Sry-related HMG box) proteins constitute a large family of diverse and well-conserved TFs comprising at least 20 SOX family members in mammals[34]. They have been divided into eight distinct groups designated A-H based on their structure, expression profiles and homology (Table 1)[35]. The SOXB group is further subdivided into subgroup B1 comprising SOX1, SOX2 and SOX3 and subgroup B2 consisting of SOX14 and SOX21[36].
Table 1.
Classification of the human SOX genes[35]
| SOXA | SOXB | SOXC | SOXD | SOXE | SOXF | SOXG | SOXH | |
| SOXB1 | SOXB2 | |||||||
| SRY | SOX1 | SOX14 | SOX4 | SOX5 | SOX8 | SOX7 | SOX15 | SOX30 |
| SOX2 | SOX21 | SOX11 | SOX6 | SOX9 | SOX17 | |||
| SOX3 | SOX12 | SOX13 | SOX10 | SOX18 | ||||
SOX proteins display properties of both classical TFs and architectural components of chromatin (reviewed in[37]). SOX TFs possess a 79 amino acid HMG domain that enables their specific DNA binding and additional domains involved in transcriptional regulation (reviewed in[37]). SOX TFs exert regulatory functions to activate or repress gene transcription through specific interactions with their partner factor(s) and by establishing contacts with the basic transcription machinery[38].
Several essential roles have been attributed to SOX TFs since their discovery. SOX TFs are a component of a regulatory network and, together with other TFs, signaling pathways, epigenetic modifiers and microRNAs, govern diverse cellular processes during development, such as the maintenance of stem cell pluripotency, cell proliferation, cell fate decisions, germ layer formation and the terminal differentiation of cells into tissues and organs (reviewed in[39,40]). However, the roles of SOX TFs are not limited to development as they also influence cell survival, regeneration and death, and control homeostasis in adult tissues[41,42].
Numerous studies have reported the roles of SOX TFs in the preservation of stem cell characteristics, playing a part of regulatory network required to establish ESCs and to maintain their pluripotent and proliferative state. SOX2, OCT4 (octamer-binding TF 4) and NANOG (named after the mythological Celtic land of the ever-young, “Tir nan Og”)[43] comprise the core transcriptional circuit that orchestrates the maintenance of stem cell self-renewal and pluripotency[44].
Accumulating evidence has demonstrated that OCT4, SOX2 and NANOG are the core factors in a pluripotency gene network involved in the induction, maintenance and loss of pluripotency (reviewed in[45,46]). The state of pluripotency, displaying regulatory flexibility, is supported by a highly interconnected pluripotency gene regulatory network that functionally relies on a set of core pluripotency TFs. The state of pluripotency integrates external signals and exerts control over the decision between self-renewal and differentiation at the transcriptional, post-transcriptional and epigenetic levels[46]. Growing evidence shows that the overexpression of these core stemness-associated TFs occurs in various types of human cancers, and aberrant expression of these TFs is associated with tumor initiation, progression and therapy resistance (reviewed in[47]). Although core stemness-associated TFs play critical roles in maintaining pluripotency and self-renewal in both ESCs and CSCs, distinct mechanistic functions between them have been proposed[48].
Since the first reviews of the involvement of the SOX genes in cancer[49], the roles of particular members of this gene family in the development of multiple cancer types have been extensively studied. It has been shown that SOX genes may act as oncogenes, tumor suppressor genes, or both depending on the cellular context (reviewed in[50,51]). Oncogenic SOX TFs are overexpressed in multiple cancer types, exerting their oncogenic function via several mechanisms, including the promotion of proliferation, suppression of apoptosis, promotion of metastasis, and maintenance of CSCs[50,52-54].
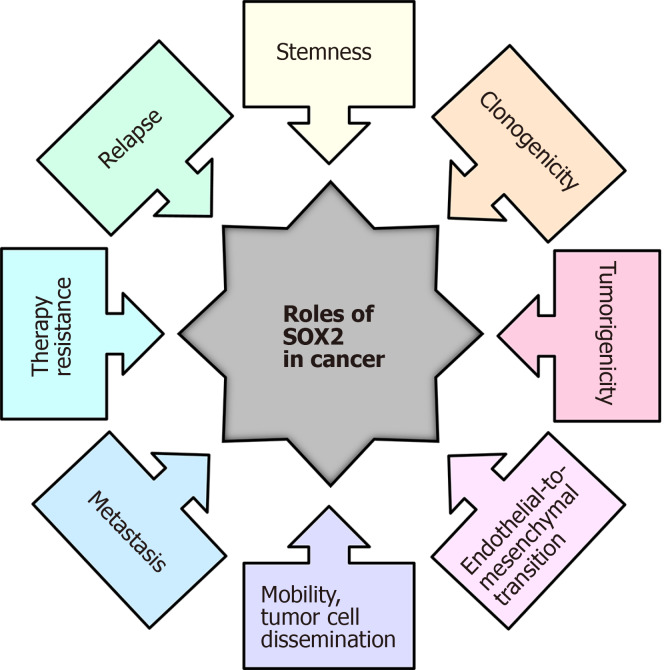
SOX2 TF is one of the most studied SOX proteins, and during the last decade, aberrant SOX2 expression has been associated with various types of cancer (reviewed in[55,56]). In various tumor types, SOX2 has been linked to cancer stemness, and elevated SOX2 expression has also been associated with chemotherapy resistance[57], endothelial-to mesenchymal transition[58], promotion of clonogenicity, and in vivo tumorigenicity[57,59]. In addition, elevated SOX2 expression in glioblastoma (GBM) is associated with increased cell motility and tumor spreading, and its expression is detected amongst circulating CSC islets[60-62]. A summary of the various roles that SOX2 plays in cancer is presented in Figure 2.
Figure 2.
The roles of the SOX2 transcription factor in cancer. Modified from[223].
The impact of SOX proteins on brain tumors has been extensively studied in the last few years, and significant contributions have been made to the understanding of the roles of SOX in the initiation, progression, dedifferentiation and spreading of glioma tumors.
GBM
GBM, classified as a grade IV glioma tumor, is the most common and malignant primary brain tumor[63]. GBM accounts for more than 60% of all brain tumors in adults and approximately 45% of all gliomas; the annual incidence of GBM is 3-4 cases per 100000 people[64-66]. The ratio of GBM occurrence is, to some extent, higher in men than in women (1.6:1)[63]. The median overall survival of patients with this type of tumor is approximately 12–15 months[67], with a median age at diagnosis of 65 years[68]. It has been demonstrated that the 5-year survival rate of GBM is 5.3%[69]. It is the most aggressive malignant brain tumor in adults, is among the most vascularized solid tumors (reviewed in[70,71]), consists of heterogeneous cell populations in different phases of differentiation[72], and recapitulates most of the hallmarks of cancer, including uncontrolled proliferation, resistance to apoptosis, dysregulation of the cell cycle and angiogenesis (reviewed in[73]).
GBMs are defined as primary, arising de novo, or secondary, developing from lower-grade glioma tumors (grade II or III astrocytoma) through malignant transformation[74]. Four clinically relevant subtypes of GBM have been identified based on molecular expression patterns: proneural, neural, classical and mesenchymal[75]. It has been demonstrated that various molecular subclasses can be detected within the same tumor[76]. It is reported in the literature that the proneural and neural GBM subtypes arise in or near the subventricular zone, while the mesenchymal and classical subtypes occur distal to the subventricular zone (reviewed in[77]). Furthermore, based on the gene expression profile, Park et al[78] identified three prognostic subtypes of GBM: mitotic (favorable), intermediate and invasive.
GBM persistently communicates with its TME, and the TME contributes to the tumorigenesis and progression of GBM (reviewed in[79,80]). The GBM TME is extremely heterogeneous, consisting of the extracellular matrix, tumor cells such as glioma stem cells (GSCs), and non-tumor cells, including endothelial cells, pericytes, microglia, immune cells, oligodendrocytes, neurons, astrocytes and myeloid-derived suppressor cells (reviewed in[81]). Extracellular vesicles containing soluble proteins, DNA, mRNA and noncoding RNAs enable communication between the GBM and the TME, and these vesicles contribute to angiogenesis, invasion, evasion of apoptosis and resistance to drugs (reviewed in[82,83]). Furthermore, it has been shown that GBM cells communicate by thin membrane channels (tunneling nanotubes), and the results obtained by Valdebenito et al[84] revealed that these nanotubes mediate the protection of GBM cells from temozolomide (TMZ) and ionizing radiation treatment.
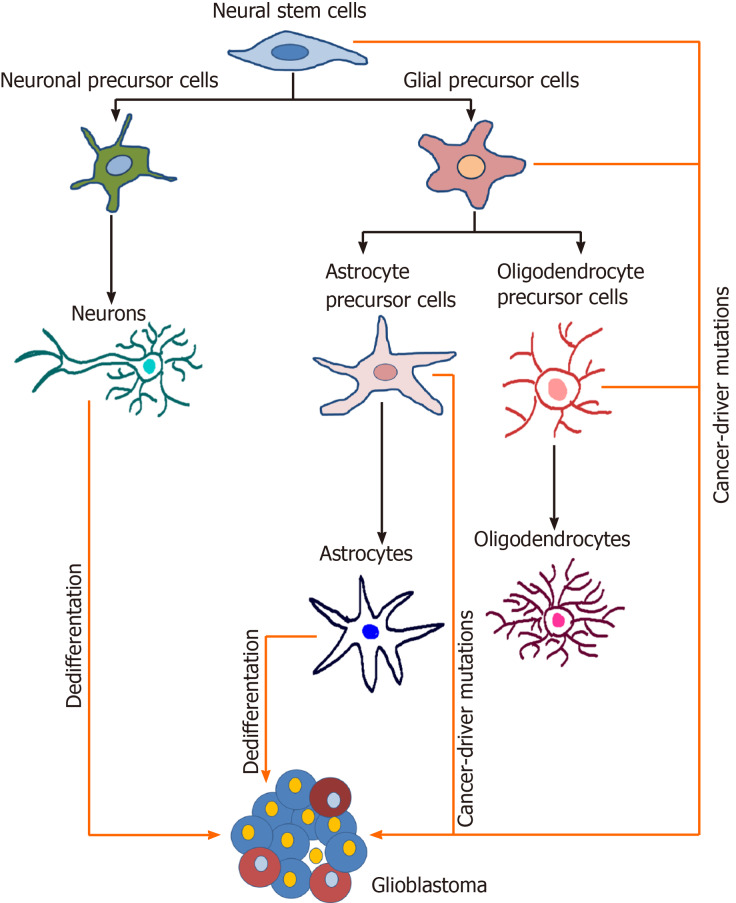
The precise cell of origin of GBM is still a controversial issue. It has been reported in the literature that neural stem cells (NSCs) of the subventricular zone and oligodendrocyte precursor cells represent two major candidates for the GBM cell of origin (Figure 3) (reviewed in[85]). Furthermore, several studies proposed that GBM might arise from the malignant transformation of glial and astrocyte precursor cells as well as from the dedifferentiation of astrocytes and neurons (reviewed in[85-87]).
Figure 3.
Potential cells of origin of glioblastoma. During the neural differentiation process (black arrows), neural stem cells differentiate into fate-restricted precursors that are capable of differentiating into neurons, astrocytes or oligodendrocytes. Glioblastoma cells can arise due to the accumulation of cancer driver mutations in different cell types or due to the dedifferentiation of mature cells (orange arrows). Modified from[224]. References are included in the main text.
The current standard therapeutic treatment of GBM includes tumor resection by surgery, followed by radiotherapy and concomitant chemotherapy with TMZ[67]. However, GBM still remains incurable and usually recurs due to a high level of intertumoral and intratumoral heterogeneity demonstrated at the histological, cellular and molecular levels (reviewed in[88-90]), the presence of GSCs, infiltration into the healthy brain parenchyma, the high rate of migration from the tumor core and the ability to generate secondary tumors (reviewed in[91]).
GBM tumor initiation, progression, relapse, and resistance to treatments are associated with GSCs (reviewed in[92,93]), and these cells represent one of the main therapeutic targets.
ROLE OF SOX GENES IN GBM AND CORRELATIONS WITH PROGNOSIS AND SURVIVAL OF PATIENTS
Almost all members of the SOX gene family contribute to the malignant phenotype of GBM by regulating cell proliferation, migration, invasion, apoptosis, stemness, tumorigenicity or angiogenesis. The correlation of SOX gene expression levels with patient clinical outcomes has been documented (Table 2).
Table 2.
SOX gene activity in glioblastoma and correlation with the clinical outcome of glioblastoma patients
|
Ref.
|
Gene
|
Activity
|
Clinical outcome
|
| [94] | SOX1 | Oncogenic | Poor |
| [98-101] | SOX2 | Oncogenic | Poor |
| [104,105] | SOX3 | Possible oncogenic | No correlation /poor |
| [129-131] | SOX4 | Oncogenic in glioma-initiating cells; tumor suppressor in GBM | Poor/good |
| [115,125] | SOX5 | Tumor suppressor | Poor |
| [116,125] | SOX6 | Tumor suppressor | Poor |
| [117,127] | SOX7 | Tumor suppressor; glioma angiogenesis | Poor |
| [106-108] | SOX9 | Oncogenic | Poor |
| [110,111] | SOX10 | Possible oncogenic | Poor |
| [119] | SOX11 | Tumor suppressor | Good |
Glioblastoma: GBM.
Various SOX genes exert oncogenic properties in GBM. SOX1 regulates the self-renewal and proliferation of patient-derived cells, tumor initiation and progression[94]. The expression of SOX1 in GBM is higher than that in the normal brain and low-grade glioma and correlates with shorter overall survival[94]. SOX2 is one of the most studied SOX genes and is overexpressed in tumor samples from patients with GBM and in GBM cell lines[95-97]. SOX2 gene exerts oncogenic activity in glioma cells[98]. High expression levels of SOX2, together with other stem cell markers, are observed in poorly differentiated GBM and are associated with its aggressiveness and poor prognosis[99-101]. It has been shown that recurrence of the tumor arises at the original site with a more invasive, aggressive phenotype and that is less sensitive to radiochemotherapy, and exhibits increased SOX2 expression levels compared to corresponding observations in the primary tumors[102]. In contrast, there are data showing that SOX2 expression is decreased in recurrent gliomas in comparison with its expression in primary gliomas and that patients with lower SOX2 expression have a poor prognosis after receiving chemoradiotherapy[103]. Furthermore, we have shown that SOX3 promotes the malignant behavior of GBM cells. SOX3 affects GBM cell viability, proliferation, migration, invasion, and autophagy, and its high expression does not correlate with the overall survival of patients with GBM[104]. However, another study revealed that higher SOX3 expression levels in glioma and glioma cell lines correlate with poor outcome[105]. SOX9 exhibits oncogenic functions; its silencing reduces proliferative capacity, arrests the cell cycle in the G2/M phase and increases glioma cell apoptosis[106]. The SOX9 gene is also involved in the survival, proliferation and senescence of glioma cells[107]. Compared to the survival time of patients with lower SOX9 expression, the survival of glioma patients with increased SOX9 gene expression is shorter[106,108]. In addition, SOX10 is expressed in gliomas[109] and, together with platelet-derived growth factor B, induces glioma development[110]. Moreover, high expression of SOX10 correlates with reduced overall survival[111].
Despite the well-established oncogenic activities of some SOX genes, others show tumor suppressive properties and are associated with good clinical outcomes. It was shown that SOX5 expression was high in glioma samples and glioma cell lines compared to the SOX5 expression in the normal adult brain and that the survival of patients with GBM with IgG antibodies against SOX5 in the sera was significantly prolonged compared to the survival of patients with no IgG antibodies[112]. Furthermore, SOX5 overexpression inhibits clone formation of human glioma cells[113]. Schlierf et al[114] showed that the expression level of SOX5 in gliomas was the same or lower than that in adult brain tissue. In contrast, Tian et al[115] revealed that high expression of SOX5 correlates with poor prognosis in patients with GBM. These contradictory data probably arose as a consequence of GBM heterogeneity. Furthermore, it was shown that low SOX6 expression in patients with GBM contributes to a better survival rate than that in patients who display high SOX6 expression[116]. Functional analyses have shown that SOX7 acts as a tumor suppressor. It has been demonstrated that SOX7 represses glioma cell proliferation and migration in vitro and suppresses tumor formation in vivo[117]. It has been reported that SOX11 is overexpressed in glioma samples[118]. Hide et al[119] showed that downregulation of SOX11 expression is associated with a decrease in the survival of patients with GBM. Another study showed that SOX11, together with neurogenin 2, can reprogram glioma cells into neuron-like cells, inhibiting tumor growth and improving survival[120]. Recent data confirmed the tumor suppressor activity of the SOX15 gene in glioma tumors. Specifically, SOX15 overexpression inhibits proliferation and invasion, while its upregulation delays tumor formation in vivo[121]. Furthermore, it has been demonstrated that SOX21 acts as a tumor suppressor[122] that inhibits SOX2 expression, induces apoptosis in human glioma cells[123] and leads to the differentiation of glioma cells, thus inhibiting glioma progression in vivo[124]. Moreover, it has been shown that simultaneous ectopic expression of the SOX5, SOX6 and SOX21 genes in human primary GBM cells induces senescence and apoptosis, thus reducing their malignant potential[125]. Their downregulation increases the capacity of stem cells from the subventricular zone of the mouse brain to form glioma tumors upon encountering oncogenic stimuli[125].
Angiogenesis represents one of the hallmarks of GBM[126]. Upregulation of SOX7 expression promotes tumor angiogenesis in a mouse model of high-grade glioma, and its high expression in GBM patients is associated with poor survival and early recurrence[127]. SOX13 silencing reduced the tube formation abilities of U87 glioma-exposed endothelial cells[128]. Downregulation of SOX17 expression inhibited tumor angiogenesis in a mouse model of high-grade glioma, while no correlation between its expression level and the clinical outcome of patients with GBM was documented[127].
Some SOX genes display both oncogenic and tumor suppressive functions depending on the cellular context. For instance, SOX4 exerts tumor suppressor activity in glioma cells by inducing GO/G1 cell cycle arrest and inhibiting growth[129]. In contrast, this gene is involved in maintaining the stemness of glioma-initiating cells[130]. There are also contradictory data regarding the correlation between SOX4 expression and the clinical outcome in patients with GBM. Galatro et al[131] reported that patients with GBM with combined high expression of ID4, SOX4 and OCT4 exhibited lower survival. On the other hand, Zhang et al[129] demonstrated that SOX4 expression is positively correlated with a good prognosis for patients with primary GBM.
Taken together, these reported findings demonstrate that almost all SOX genes are expressed in GBM and the expression levels of the majority of these genes are associated with the prognosis and survival of patients with GBM. SOXgenes that exert oncogenic roles are usually associated with a poor prognosis and shorter survival, making them potential candidates as prognostic biomarkers and future therapeutic targets.
GSCs
GSCs were among the first CSCs identified in solid tumors[132]. GSCs are one of the major contributors to GBM heterogeneity; these cells exhibit a high proliferation rate and self-renewal capacity (reviewed in[71]), an ability to differentiate into diverse cell types, the ability to recapitulate a whole tumor (reviewed in[133]), enhanced invasive capacity, increased angiogenic potential[134] and an ability to initiate tumor growth and progression shortly after the surgical removal of the primary GBM tumor (reviewed in[92]). GSCs usually express specific markers, such as CD133 (Prominin 1), CD44, NANOG, SOX2, OCT4, POU class 3 homeobox 2 (POU3F2), MYC proto-oncogene (c-Myc), Spalt Like TF 2 (SALL2) and KAT8 regulatory NSL complex subunit 2[135,136]. It has been reported that GBM cells are able to interconvert between GSC and non-GSC states[137]. Additionally, it has been found that a single GBM tumor contains different subsets of GSCs: proneural and mesenchymal[138].
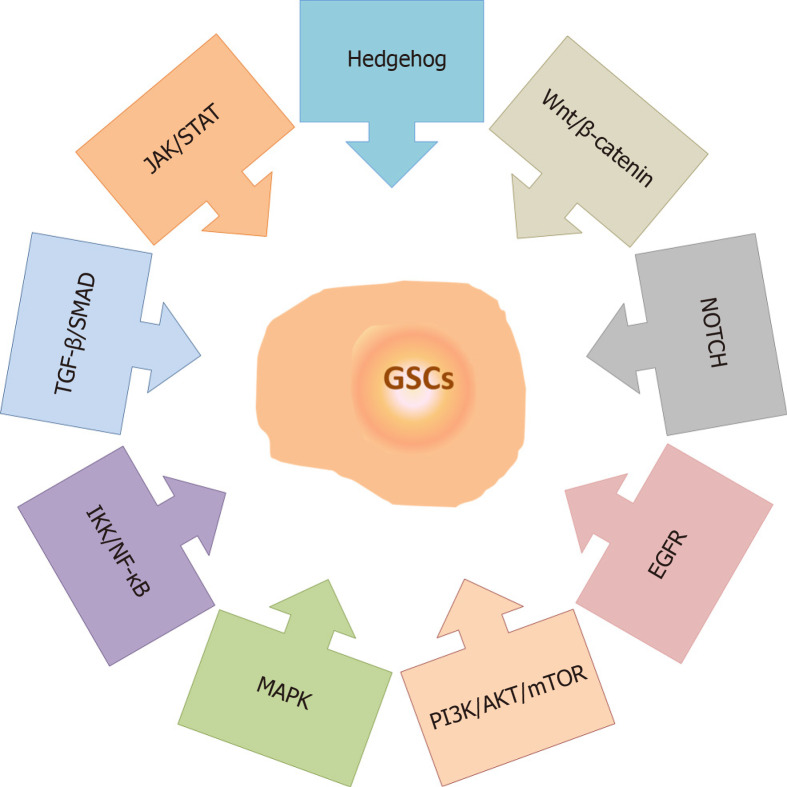
Numerous studies have demonstrated the importance of signaling pathways in the maintenance of GSC properties, including the Hedgehog (HH)[139], Wnt/β-catenin[140], NOTCH[141], Epidermal growth factor receptor[142], Phosphatidylinositol-3-phosphate kinase/AKT/Mammalian target of rapamycin[143], Mitogen-activated protein kinase (MAPK)[144], Inhibitory kappa B kinase/Nuclear factor-kappa B (NF-κB)[145], Transforming growth factor-beta (TGF-β)/Small mothers against decapentaplegic[130] and Janus kinase/Signal transducers and activators of transcription (STAT)[146] pathways (Figure 4). It has been proposed that the regulation of the GSC phenotype results from a complex interplay between multiple signaling cascades[143,147-149]. Accordingly, targeting several GSC-related signaling pathways at the same time seems to be necessary for the increased efficiency of anticancer treatments. A variety of small-molecule inhibitors of the HH, NOTCH and WNT/β-catenin signaling pathways have been identified/developed, along with inhibitors that target different pathways simultaneously, and the efficacy of some of these inhibitors have been demonstrated in preclinical and clinical studies[150,151].
Figure 4.
Signaling pathways involved in the maintenance of glioma stem cell properties. The main signaling pathways involved in the maintenance of glioma stem cell properties include Hedgehog, Wnt/β-catenin, NOTCH, Epidermal growth factor receptor, Phosphatidylinositol-3-phosphate kinase/AKT/Mammalian target of rapamycin, Mitogen-activated protein kinase, Inhibitory kappa B kinase/Nuclear factor-kappa B, Transforming growth factor-beta/Small mothers against decapentaplegic and Janus kinase/Signal transducers and activators of transcription. References are included in the main text. MAPK: Mitogen-activated protein kinase; JAK/STAT: Janus kinase/Signal transducers and activators of transcription; EGFR: Epidermal growth factor receptor; PI3K/AKT/mTOR: Phosphatidylinositol-3-phosphate kinase/AKT/Mammalian target of rapamycin; IKK/NF-κB: Inhibitory kappa B kinase/Nuclear factor-kappa B; TGF-β/SMAD: Transforming growth factor-beta/Small mothers against decapentaplegic; GSCs: Glioma stem cells.
GSCs are enriched in recurrent gliomas (reviewed in[152]), and they form more aggressive and diffuse tumors after intracranial transplantation in athymic nude mice than GSCs originating from the primary tumor of the same patient[153]. These factors make GSCs a primary therapeutic target. Cho et al[154] described five methods for targeting GSCs: development of GSC-specific chemotherapeutic agents, application of radiosensitizers, usage of immune cells capable of attacking GSCs, induction of GSC differentiation and gene therapy.
GSCs reside in the perivascular and perinecrotic GBM niches (reviewed in[79]). Perivascular niches contain normal and reactive astrocytes, pericytes, glioma-associated microglia/macrophages, myeloid cells, fibroblasts and normal NSCs in addition to GSCs and tumor cells (reviewed in[79]). GSCs residing in the perivascular niches divide slowly, are resistant to therapy and produce high levels of proangiogenic factors (reviewed in[155,156]). It has been reported that endothelial cells stimulate GSC self-renewal and tumorigenicity; on the other hand, GSCs may regulate and contribute to the tumor vasculature by producing cytokines and chemokines and by transdifferentiating into endothelial cells or pericytes (reviewed in[157]). Perinecrotic niches contain GSCs around necrotic foci induced by hypoxia (reviewed in[158]). Hypoxia induces signaling pathways that can impact GSC self-renewal, proliferation and invasion (reviewed in[158]). Cells expressing molecular markers of both hypoxia and GSCs (e.g., SOX2, NANOG, CD133) are largely found in perinecrotic niches (reviewed in[134]).
SOX GENES AND MAINTENANCE OF GSCs PROPERTIES
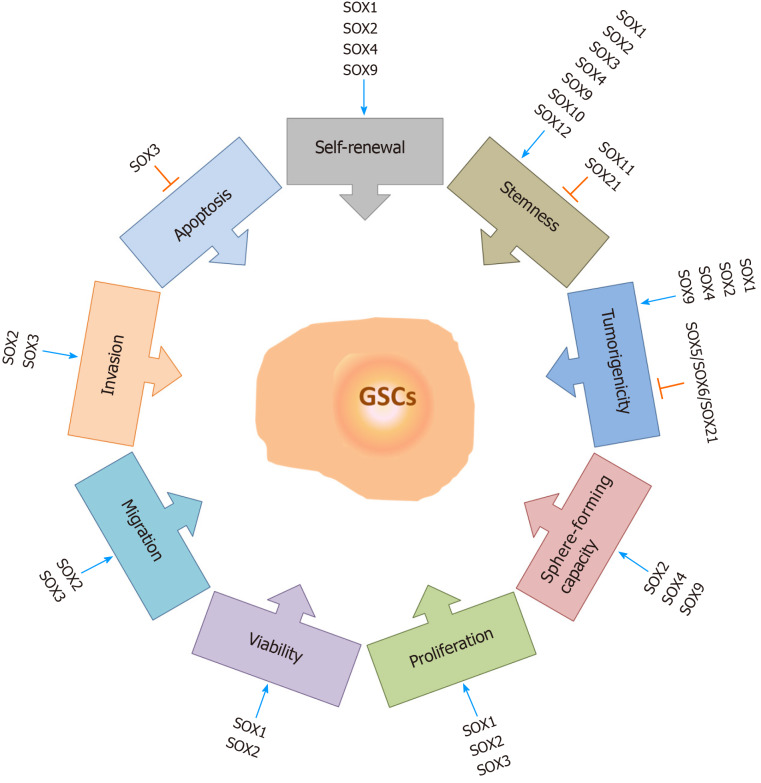
It has been reported that SOX genes play important roles in the maintenance of GSC properties (Figure 5). Knockdown of SOX1 expression in GSCs impairs their self-renewal, proliferation, viability and tumor-forming capacity, while SOX1 overexpression promotes their malignant phenotype[94]. Among the SOX TF family, SOX2 is considered to be the most important player in the maintenance of GSC properties, including their tumorigenicity. Gangemi et al[159] demonstrated that silencing the SOX2 gene in GSCs terminated cell proliferation, consequentially leading to the loss of tumorigenicity. The results obtained by Song et al[160] indicate that SOX2 plays a primary role in the regulation of CD133-positive GBM cell tumorigenicity. On the other hand, Cox et al[161] demonstrated that a 3-fold increase in SOX2 expression in GBM cells decreases their capacity to proliferate and generate spheres. These results suggest that the level of SOX2 expression must be precisely regulated by tumor cells during gliomagenesis. Furthermore, it has been found that co-expression of SOX2, oligodendrocyte TF 2 (OLIG2) and zinc finger E-box binding homeobox 2 is involved in the maintenance of GSCs[162]. Downregulation of SOX2 expression impairs sphere-forming capacity, self-renewal, viability, proliferation, migratory and invasive capabilities of brain tumor stem cells derived from GBM[98]. Furthermore, knockdown of SOX3 expression in GSCs decreases the proliferation, migration and invasion capabilities of these cells and enhances their apoptosis[163], while knockdown of SOX4 expression in glioma-initiating cells results in reduced sphere-forming and self-renewal capacities[130]. Silencing of SOX9 leads to inhibition of sphere-forming capacity[164], while SOX9 overexpression increases the sphere-forming capacity of GBM cells and induces the formation of larger tumors[165]. In glioma-initiating cells, inhibition of the midkine/anaplastic lymphoma kinase axis promotes SOX9 degradation, resulting in a decrease in the self-renewal and tumorigenic capacities of glioma-initiating cells[166]. Kurtsdotter et al[125] demonstrated that simultaneous forced expression of SOX5, SOX6 and SOX21 factors blocks the tumor-inducing capacity of human primary GBM cells.
Figure 5.
The roles of SOX transcription factors in glioma stem cells. SOX transcription factors exert numerous functions in glioma stem cells (GSCs). They influence GSC self-renewal, stemness, tumorigenicity, sphere-forming capacity, proliferation rate, viability, migration and invasion capacities, and apoptosis. Some of the members of the SOX gene family are involved in the resistance of GSCs to therapy. Blue arrows represent stimulation, and orange bars represent inhibition. References are included in the main text. GSCs: Glioma stem cells.
THE ROLES OF SOX GENES IN STEMNESS AND DIFFERENTIATION OF GSCs
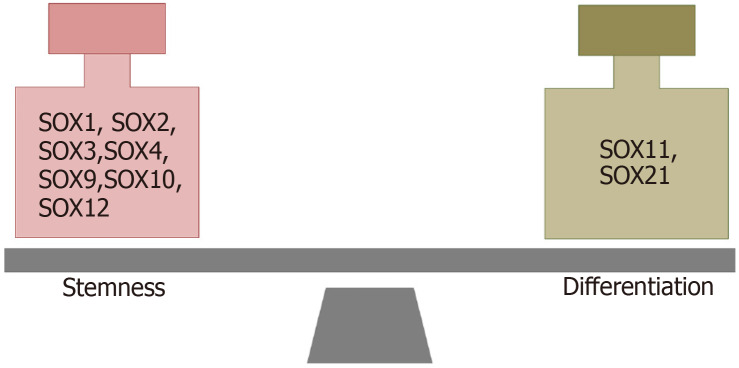
It has been reported that SOX TFs have important functions in both the maintenance of the stemness of GSCs and the dedifferentiation of glioma cells (Figure 6). The results obtained by Berezovsky et al[100] indicate that SOX2 may play a central role in the regulation of dedifferentiation and acquisition of GSC properties. They found that knockdown of SOX2 expression blocks the dedifferentiation of HF2303 GBM cells and reduces their tumorigenicity. Additionally, downregulation of SOX2 expression in brain tumor stem cells derived from GBM led to an increase in the expression of differentiation markers[98]. Furthermore, it was shown that the TGF-β/SOX4/SOX2 pathway plays a crucial role in the maintenance of stemness and tumorigenicity in glioma-initiating cells[130]. It has been shown that the combination of SOX2, POU3F2, SALL2 and OLIG2, detected in the proneural subtype of GBMs, is able to reprogram differentiated tumor cells into GSCs[136].
Figure 6.
SOX transcription factors involved in the maintenance of stemness and differentiation of glioma stem cells. References are included in the main text.
In addition to the overwhelming evidence of the importance of the SOX2 gene in the maintenance of the stem cell properties of GSCs, it was shown that the other two members of the SOXB1 subgroup also play important roles. Dedifferentiation of GBM cell lines is accompanied by an increase in SOX1 expression. Additionally, SOX1 expression was higher in patient-derived GSCs than in conventional glioma cell lines, and after the differentiation of these cells, the levels of SOX1 decreased dramatically[94]. Recently, we revealed that SOX3 expression was higher in patient-derived GSCs, as well as in oncospheres derived from GBM cell lines, compared to SOX3 expression in their differentiated counterparts, suggesting that SOX3 expression is associated with the undifferentiated state of GBM cells[104].
Additionally, the roles of other SOX genes in the regulation of stemness and differentiation of GSCs have been described. It was found that the number of differentiated cells is increased after SOX4 knockdown in glioma-initiating cells[130]. On the other hand, the expression of SOX9 is increased in oncospheres derived from GBM cell lines compared to its expression in their differentiated counterparts, and silencing of SOX9 downregulated the expression of the stem cell markers CD133, NESTIN and SOX2[164]. Overexpression of SOX10 in LN229 GBM cells increased the expression levels of the stemness markers CD133, NESTIN, OCT4, NANOG and CD44[111]. On the other hand, SOX11 overexpression in glioma-initiating cell-like cells and human glioma-initiating cells from malignant glioma blocked their tumorigenic ability by inducing neuronal differentiation[119]. The results obtained by Wu et al[167] indicate that SOX12 regulates the stemness of GBM cells and that SOX2, CD133, and OCT4 act as downstream effectors of SOX12 in these cells. Additionally, the results obtained by Caglayan et al[124] suggest that SOX21 overexpression decreases the stem-like cell properties of glioma cells and initiates their differentiation in vivo.
Together, these reported findings indicate that SOX TFs play important roles in GBM and GSCs. Since GSCs are the main drivers of GBM progression and therapy resistance, further studies are needed to comprehensively delineate the roles of SOX TFs in these cells.
SOX GENES AND CHEMO-AND RADIORESISTANCE OF GSCs
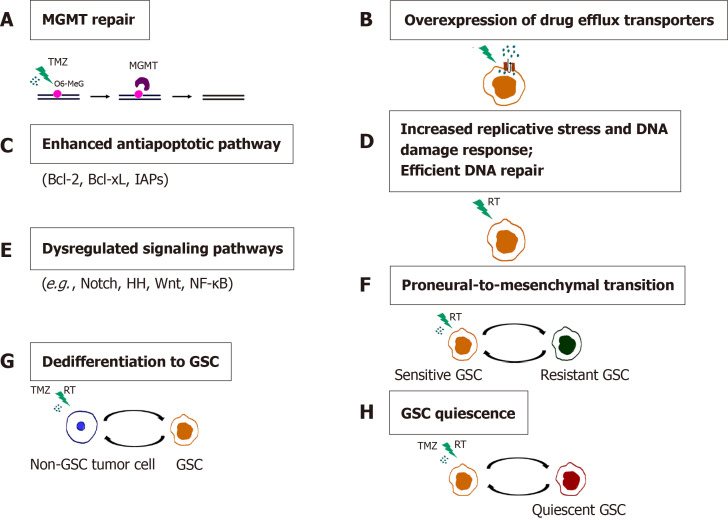
GBM inevitably recurs following the initial therapy as a result of intratumoral heterogeneity and resistance to chemo- and radiotherapy, which are considered to be promoted by GSCs (reviewed in[133,156]). Conventional chemo- and radiotherapies affect the proliferative cells within the tumor, while GSCs successfully evade their effects through multiple intrinsic GSC features and adaptive mechanisms (Figure 7). These mechanisms include enhanced expression of the DNA mismatch repair gene MGMT (O6-methylguanine-DNA methyltransferase)[168], high expression of drug efflux transporters[168], activation of the antiapoptotic pathway[168], increased replication stress leading to a constitutively active DNA damage response[169,170] and aberrant activation of multiple signaling pathways, such as Notch, HH, Wnt/β-catenin and NF-κB[171-174].
Figure 7.
Mechanisms of chemo- and radioresistance of glioma stem cells. A: O6-methylguanine-DNA methyltransferase (MGMT) repair; B: Overexpression of drug efflux transporters; C: Enhanced antiapoptotic pathway; D: Increased replicative stress and DNA damage response. Efficient DNA repair; E: Dysregulated signaling pathways; F: Proneural-to-mesenchymal transition; G: Dedifferentiation to GSC; H: Glioma stem cells (GSC) quiescence. Resistance of GSCs to temozolomide is achieved through increased expression of MGMT, drug efflux transporters and antiapoptotic proteins. Replicative stress, constitutively active DNA damage response and efficient DNA repair confer radioresistance to GSCs. Aberrantly activated signaling pathways, the ability of non-GSC tumor cells to dedifferentiate, proneural-to-mesenchymal transition and the capacity of GSCs to become quiescent contribute to the chemo- and radioresistance of glioblastoma. References are included in the main text. RT: radiation; O6-MeG: O-6-methylguanine; GSC: Glioma stem cell; TMZ: Temozolomide.
One of the essential features of cancer cells that confers resistance to therapy is their plasticity, i.e., the capability of going through phenotypic changes in response to different environmental signals without genomic alteration. It has been shown that extrinsic factors such as radiation[175] or TNF-α (tumor necrosis factor α) through activation of the NF-κB pathway[171] promote proneural to mesenchymal shift of GSCs leading to a radioresistant phenotype[171]. Segerman et al[176] demonstrated that GBMs contain a heterogeneous population of glioma-initiating cells that exhibit variable levels of resistance to radiation and drugs[176]. The observed variations in therapy responses are the result of the slow shifts between the proneural- and mesenchymal-like cell states regulated by the changes in DNA methylation of the cis-regulatory regions of mesenchymal master regulator genes.
Another important mechanism underlying GBM resistance to therapy is the ability of differentiated cancer cells to undergo dedifferentiation into cancer stem-like cells. Auffinger et al[177] showed that long-term exposure of glioma cells to clinically relevant doses of TMZ caused the phenotypic switch of non-GSCs towards GSCs that exhibited expression of pluripotency- and stemness-related markers CD133, SOX2, OCT4, and Nestin, a highly infiltrative phenotype and increased chemoresistance. Analyses of the long-term effects of sub-toxic doses of radiation on patient-derived differentiated GBM cells and the phenotypic and molecular changes in GBM towards stemness in vitro were demonstrated, including the expression of stem cell markers SOX2, Nestin and OLIG2, as well as increased tumorigenicity in vivo[178].
Quiescent, slow-cycling CSCs play a critical role in tumor recurrence, as they escape the effects of chemo- and radiotherapy that mostly eradicate proliferative cells in the tumor bulk. TMZ treatment in a transgenic mouse model of glioma demonstrated that recurrent tumors derived from dormant glioma cells exhibited stem cell properties[179]. Qazi et al[180] revealed that the combined chemoradiotherapy of primary human GBMs resulted in acquired resistance of brain tumor stem cells, which was accompanied by increased expression of stemness-associated factors SOX2 and BMI1 and an enhanced self-renewal capacity of primary GBM cells.
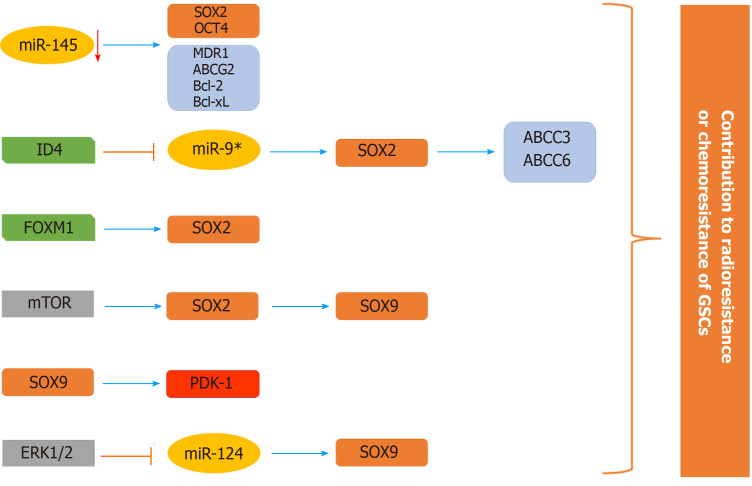
The roles of the SOX2 and SOX9 genes in the chemo- and radioresistance of GSCs have been well documented among the members of the SOX TF family (Figure 8). High expression of SOX2 in GBM patient-derived CD133-positive GSCs was shown to play a key role in promoting the self-renewal of GSCs, thereby contributing to their drug resistance[160]. Yang et al[181] showed that the miR-145/OCT4/SOX2 axis plays an important role in the chemoradioresistance of CD133-positive GSCs. Inhibition of stemness factors SOX2 and OCT4 by miR-145 was followed by an increased sensitivity of patient-derived CD133-positive GSCs to TMZ and radiation. The reduced chemoradioresistance of GSCs was also accompanied by a decrease in the expression of a few drug resistance (MDR1 - multidrug resistance protein 1 and ABCG2-ABC subfamily G member 2) and antiapoptotic genes (Bcl-2 - B-cell lymphoma 2 and Bcl-xL - B-cell lymphoma-extra large)[181]. Overexpression of ID-4 (inhibitor of differentiation 4) induced the dedifferentiation of human glioma cells into glioma stem-like cells and downregulated the expression of SOX2-repressing miR-9*, leading to enhanced SOX2 expression[182]. SOX2, as a direct transcriptional regulator, upregulates the expression of ABCC3 and ABCC6 transporters, conferring chemoresistance on ID4-induced glioma stem-like cells and patient-derived GSCs[182]. Furthermore, it was revealed that SOX2 is a direct transcriptional target of Forkhead box M1 (FOXM1) and that the FOXM1/SOX2 axis promotes the stemness and radioresistance of GBM[183].
Figure 8.
Schematic illustration of the involvement of SOX2 and SOX9 in the chemoresistance and radioresistance of glioma stem cells. The expression of stemness-related transcription factor SOX2 is elevated in glioma stem cells (GSCs) through various mechanisms (decreased expression of miR-145 and miR-9* that directly target SOX2, direct transcriptional activation by Forkhead box M1 and through mammalian target of rapamycin signaling) contributing to chemo- and radioresistance of GSCs. SOX2 directly activates the expression of ABCC3 and ABCC6 transporters. mTOR signaling affects the SOX2/SOX9 axis, contributing to chemoresistance. The ERK1/2/miR-124/SOX9 axis and direct targeting of PDK1 (pyruvate dehydrogenase kinase 1) by SOX9 have a role in resistance to radiation and temozolomide, respectively. References are included in the main text. GSCs: Glioma stem cells; FOXM1: Forkhead box M1; mTOR: Mammalian target of rapamycin; PDK1: Pyruvate dehydrogenase kinase 1.
A study by Garros-Regulez et al[165] demonstrated the correlation between high levels of SOX2 and SOX9 expression and resistance of GSCs to TMZ. The SOX2/SOX9 axis acts downstream of the mTOR signaling pathway and contributes to the observed chemoresistance. The authors proposed pharmacological inhibition of SOX2 by using specific inhibitors of the Sonic Hedgehog (SHH) and mTOR signaling pathways as a promising therapeutic approach to overcome TMZ resistance in a group of GBMs that display high expression levels of SOX2 and SOX9[165]. Wang et al[164] uncovered the PDK1 (pyruvate dehydrogenase kinase 1) gene as a direct target of SOX9 that was shown to play a role in the regulation of GSC self-renewal and resistance to TMZ. The silencing of SOX9 and inhibition of PDK1 both reduced the activity of the AKT pathway and sensitized GSCs to TMZ treatment in vivo, highlighting the SOX9/PDK1 axis as a new promising target for more efficient GBM therapy. Most recently, Sabelström et al[184] demonstrated the critical role of the ERK1/2/miR-124/SOX9 axis in triggering the differentiation of stem-like GBM cells towards a neuronal phenotype. Inhibition of ERK1/2 activation by using a MEK inhibitor caused the enhanced expression of miR-124 and a reduction in the expression of its target gene, SOX9, consequently leading to decreased tumorigenicity and radioresistance in patient-derived GBM xenografts. These results imply that the induction of neuronal differentiation of GSCs represents a promising therapeutic approach in anti-GBM therapy.
CONCLUSION
Combatting GBM has been a demanding and challenging goal for decades. Heterogeneity of cell types within the tumor, the contribution of patient-to-patient variability to the growth, the response to treatment driven by the genomics of each tumor and the transition between proliferative and non-proliferative phases make GBM extremely difficult to treat (reviewed in[185]). GBM is one of the deadliest cancers, and GBM studies face many challenges, some of which are unique to brain tumors (blood-brain barrier and immunosuppressive environment) and others that are shared with other tumors (tumor heterogeneity at the cellular and molecular levels, plasticity, and the presence of CSCs). One of the most essential hallmarks of GBM is tumor heterogeneity, which has been documented at the histological, cellular and molecular levels. Both intertumor heterogeneity (comprising distinct genetic alterations between different individual tumors) and intratumor heterogeneity (comprising diversity within individual tumors) have been identified in GBM (reviewed in[90]). The heterogeneity of GBM has been defined at multiple molecular levels using-omics technologies. Heterogeneity documented at the genomic and transcriptomic levels enables the molecular classification of histopathologically indistinguishable tumors into different subtypes. An advanced understanding of intratumor heterogeneity has been achieved by single-cell sequencing. Patel et al[186] performed the first single-cell RNA-seq and confirmed that GBM subtype classifiers are variably expressed across individual cells within a single tumor, while a recent study revealed that malignant cells in GBM exist in four main cellular states that are reminiscent of the canonical neurodevelopmental cell types[187]. Another level of heterogeneity is linked to treatment-induced plasticity and temporal heterogeneity. By sequencing exomes of initial low-grade gliomas and recurrent tumors from the same patients, Johnson et al[188] revealed that at least half of the mutations in the initial tumor were undetected at recurrence in 43% of cases. Recently, a database of initial tumor and tumor recurrence samples from patients assembled by the Glass consortium revealed that 35 out of 222 patients exhibited treatment-related hypermutation at recurrence, while 70% of the cohort had an increased mutational burden after recurrence compared with the mutational burden of their initial tumor[189]. Taken together, these findings indicate that GBM heterogeneity provides additional layers of complexity that must be taken into consideration in the development of more effective therapies aimed at improving survival in GBM patients.
Remarkable progress has been made towards understanding the cellular and molecular biology of gliomas in the last decade. Advanced technologies have provided novel insights into multiple events that allow GSCs to acquire functional capabilities, enabling them to survive, proliferate and disseminate.
CSCs are considered to be, at least in part, responsible for drug resistance and cancer relapse due to their ability to self-renew and differentiate into heterogeneous lineages of cancer cells. Accordingly, the eradication of CSCs has become one of the major goals in cancer therapies. However, CSCs are very often therapy-resistant, representing the main obstacle to their eradication. Therapy resistance of CSCs is mediated by many different mechanisms, including the acquisition of dormancy, decreased apoptosis, increased DNA repair and drug efflux capacity, interaction with their supporting microenvironment within the CSC niche and hypoxic stability (reviewed in[190]). GBM tumors may contain different subsets of GSCs[138]. GSCs may adopt a quiescent state that protects them from chemotherapy and radiotherapy (reviewed in[191]), and these treatments might increase the population of GSCs over time (reviewed in[192]). GSCs successfully evade the effects of conventional chemo- and radiotherapies through multiple intrinsic GSC features and adaptive mechanisms (Figure 7). GSCs enriched in recurrent gliomas acquire more aggressive and therapy-resistant properties than GSCs originating from primary tumors that become more malignant and rapidly spread, leading to therapeutic failures[152,153,185].
All these properties make GSCs a promising but very complex and challenging therapeutic target for GBM treatment. A number of approaches for targeting CSCs have been developed, including inhibition of drug efflux ABC transporters, targeting signaling pathways, the microenvironment and cell surface markers of CSCs, differentiation therapy, immunotherapy and application of radiosensitizers (reviewed in[133,193-196]).
Dedifferentiation of tumor cells into a stem cell-like state is considered an additional source of cellular heterogeneity, pointing to tumor cell plasticity as an important driver of GBM heterogeneity[90]. Gimple et al[137] proposed novel advanced therapies that are aimed at targeting tumor cell plasticity conferred by dedifferentiation (reviewed in[7,137]). One of the proposed approaches focuses on blocking dedifferentiation by applying a combination of therapies to prevent early resistance to therapeutics endowed by dedifferentiation and acquired lineage plasticity. Differentiation therapy is a novel approach that aims to target dedifferentiation by enhancing the conversion of dedifferentiated tumor cells towards permanently differentiated cells that are more sensitive to chemotherapy. The final proposed approach is focused on the differentiation of CSCs into harmless cell lineages that lack tumorigenic potency and metastatic potential by applying TFs or small molecules.
Significant conceptual and mechanistic similarities between cellular transformation in cancers and cellular reprogramming and dedifferentiation driven by various stem cell-associated pathways have been revealed[197]. The master regulators of stemness operate through transcriptional control of various stem cell-associated pathways during cellular reprogramming.
Since cell renewal is an ability that cancer cells must acquire to survive therapeutic treatment and reconstitute the tumor, master regulators of stemness have attracted significant attention in the treatment of GBM. SOX TFs are master regulators of stemness with known essential roles in GBM. Accordingly, SOX proteins and SOX2, in particular, have emerged as one of the major therapeutic targets for GBM. Overwhelming evidence of the importance of the SOX2 gene in the maintenance of the stem cell properties of GSCs has been revealed. Numerous studies have demonstrated that glioma cells rely on SOX2 to maintain their tumorigenic activity, with GSCs displaying high levels of SOX2 (reviewed in[198]). SOX2 overexpression has been found in approximately 90% of human biopsies in GBM patient samples with enrichment in undifferentiated GSC populations. Downregulation of SOX2 in GSCs impairs cell proliferation and the ability of the cells to form tumors in vivo.
It has also been shown that numerous SOX TFs are involved in the maintenance of the stemness of GSCs (SOX1, SOX2, SOX3, SOX4, SOX9, SOX10, SOX12) or play a role in the initiation of GSC differentiation (SOX11 and SOX21). Accordingly, the fine-tuning of SOX gene expression levels is associated with the control of the balance between stemness and differentiation.
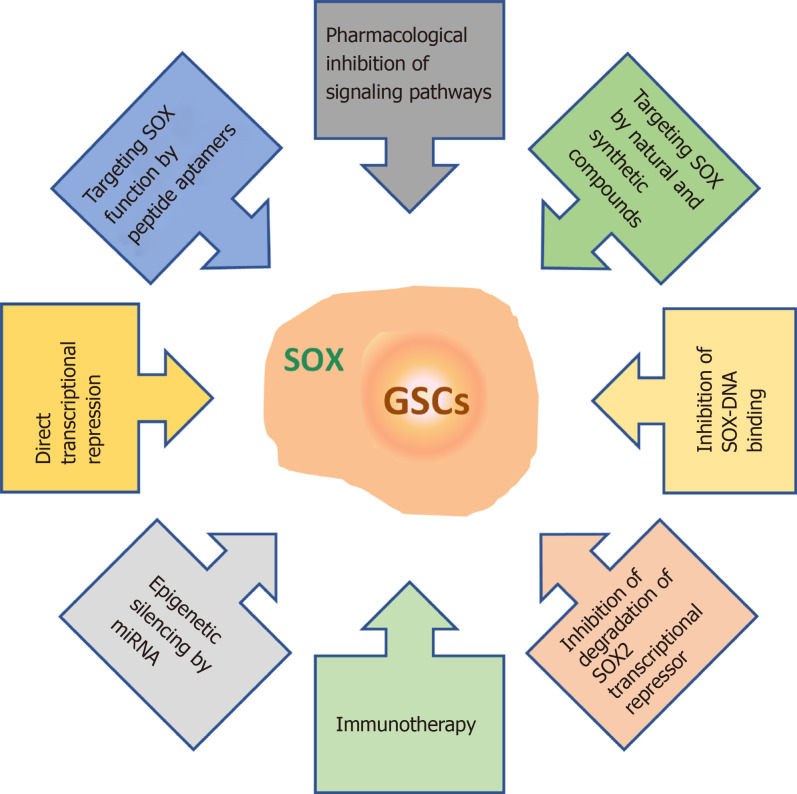
As outlined in this review, SOX TFs play multiple roles in GSCs. They influence GSC stemness, self-renewal, proliferation, differentiation, viability, migration, invasion, apoptosis, therapy resistance, sphere-forming capacity, and tumorigenicity (Figure 5). Therefore, innovative therapies aimed at targeting SOX TFs, and SOX2 in particular, are emerging as promising tools to combat GBM. These therapeutic approaches include the pharmacological inhibition of signaling pathways involving SOX genes[165,199,200], application of natural and synthetic compounds that control the expression of these genes[201-204], inhibition of SOX-DNA binding[205], inhibition of ubiquitylation and degradation of the SOX2 transcriptional repressor[206], immunotherapy[207,208], epigenetic silencing by miRNAs[181], blocking SOX function by peptide aptamers[209] and direct transcriptional repression of SOX gene expression[210] (Figure 9).
Figure 9.
Overview of the therapeutic strategies for targeting SOX in glioma stem cells. References are included in the main text. GSCs: Glioma stem cells.
Modulation of SOX gene expression in GSCs represents an attractive approach to combat GBM. SOX gene expression in GSCs might be targeted by applying different approaches. One of the approaches to modulate SOX gene expression is treatment with natural plant products. Studies on natural plant products have revealed that they can act as effective antitumor agents[211]. Forty-nine per cent of the small molecules approved for tumor treatment between 1940 and 2014 were natural plant products or their derivatives[211]. Some of these products have anti-GBM potential alone or in combination with other chemotherapeutics[211-214]. Our results revealed that extracts from Anthrisci cerefolii and Ononis spinosa L. plants have anti-GBM activity[215,216]. Furthermore, it has been reported that nuciferine obtained from the leaves of N. nucifera Gaertn. inhibited GBM progression via the SOX2/AKT/STAT3/Slug pathway[201]. On the other hand, it was found that curcumin, which inhibits the proliferation of glioma cells and induces apoptosis and autophagy, simultaneously induces glioma cells to become stem-like and enhances the expression of SOX2, SOX4 and OCT4[203]. In addition to natural plant products, antibiotics such as salinomycin and actinomycin D reduce SOX2 expression in GSCs, indicating that drug repurposing may provide additional options for GBM treatment[202,204].
The expression of SOX genes in GSCs could be targeted by pharmacological inhibitors of signaling pathways that regulate GSC properties. Rapamycin, an inhibitor of mTOR complex 1, and cyclopamine, an inhibitor of the SHH pathway, reduced the expression of SOX2 and SOX9 in GBM cell lines[165]. Additionally, SOX genes could be targeted by engineered artificial TFs that selectively modulate SOX gene expression. Stolzenburg et al[210] developed zinc finger-based TFs that reduced SOX2 expression, leading to reduced proliferation and colony formation in breast cancer cells.
Another approach is targeting SOX TFs in GSCs by the delivery of vectors that express miRNAs that were shown to specifically downregulate the expression of SOX proteins in GSCs. For example, the delivery of miRNA-145 to GBM patient-derived CSCs in vitro and in vivo resulted in the downregulation of the expression of stemness factors SOX2 and OCT4, suppression of tumorigenicity, increased sensitivity to TMZ and radiation and enhanced survival rate of xenotransplant mice when applied in combination with radiotherapy and TMZ[181].
GSCs could also be combated by vaccine approaches. It was demonstrated that vaccination of immunocompetent mice with Sox2 peptides significantly delayed tumor development[207]. Furthermore, Ueda et al[208] found that the SOX6 peptide induces SOX6 peptide-specific cytotoxic T lymphocytes, which are able to lyse GSCs derived from GBM.
In the last decade, immunotherapy that engages naturally occurring or genetically engineered oncolytic viruses has provided an additional strategy, especially for the treatment of cancers with poor prognosis, such as GBM. Although oncolytic viruses can infect both normal and cancer cells, they can only replicate in cancer cells[217]. Preclinical and clinical trials have pointed to the capability of oncolytic viruses to stimulate antitumor immune responses by recruiting T cells (reviewed in[218,219]). It has been reported that the Zika virus exerts oncolytic activity against GSCs. Compared to Zika virus infection of differentiated GBM cells, the Zika virus preferentially infects and selectively kills GSCs and stem-like cells in a SOX2-dependent manner[220]. It has also been reported that an immunotherapeutic approach in gliomas that uses a vesicular stomatitis virus expressing HIF-2a, SOX10 and c-Myc together with checkpoint inhibitors anti-PD1 and anti-CTLA-4 enhances the antitumor response[221].
The current therapeutic strategies for GBM have not yet provided a cure and have only resulted in a slight improvement in patient survival. Considering that GBM therapies result in poor outcomes and that GBM treatment is one of the most expensive cancer treatment per capita in the United States[222], there is a critical need for novel and more effective therapeutic options. Novel approaches to GBM treatment will require the fine-tuning of multimodal therapeutic strategies that simultaneously target numerous hallmarks of cancer cells to win the battle against GBM, one of the deadliest cancers.
Footnotes
Conflict-of-interest statement: The authors declare no conflict of interest for this article.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: Serbian Academy of Sciences and Arts, Full member; National Council for Scientific and Technological Development of the Republic of Serbia, Vice-president; Serbian Society for Molecular Biology, Steering Committee member; Serbian Neuroscience Society, Steering Committee member.
Peer-review started: March 26, 2021
First decision: July 4, 2021
Article in press: September 16, 2021
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: Serbia
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cenciarelli C S-Editor: Liu M L-Editor: A P-Editor: Yu HG
Contributor Information
Milena Stevanovic, Laboratory for Human Molecular Genetics, Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, Belgrade 11042, Serbia; Chair Biochemistry and Molecular Biology, Faculty of Biology, University of Belgrade, Belgrade 11158, Serbia; Department of Chemical and Biological Sciences, Serbian Academy of Sciences and Arts, Belgrade 11000, Serbia. milenastevanovic@imgge.bg.ac.rs.
Natasa Kovacevic-Grujicic, Laboratory for Human Molecular Genetics, Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, Belgrade 11042, Serbia.
Marija Mojsin, Laboratory for Human Molecular Genetics, Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, Belgrade 11042, Serbia.
Milena Milivojevic, Laboratory for Human Molecular Genetics, Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, Belgrade 11042, Serbia.
Danijela Drakulic, Laboratory for Human Molecular Genetics, Institute of Molecular Genetics and Genetic Engineering, University of Belgrade, Belgrade 11042, Serbia.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 4.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 5.DeNardo DG, Andreu P, Coussens LM. Interactions between lymphocytes and myeloid cells regulate pro- versus anti-tumor immunity. Cancer Metastasis Rev. 2010;29:309–316. doi: 10.1007/s10555-010-9223-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senga SS, Grose RP. Hallmarks of cancer-the new testament. Open Biol. 2021;11:200358. doi: 10.1098/rsob.200358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 9.Kørbling M, Estrov Z. Adult stem cells for tissue repair - a new therapeutic concept? N Engl J Med. 2003;349:570–582. doi: 10.1056/NEJMra022361. [DOI] [PubMed] [Google Scholar]
- 10.Young HE, Black AC Jr. Adult stem cells. Anat Rec A Discov Mol Cell Evol Biol. 2004;276:75–102. doi: 10.1002/ar.a.10134. [DOI] [PubMed] [Google Scholar]
- 11.Waddington CH. The Strategy of the Genes: A Discussion of Some Aspects of Theoretical Biology. London: George Allen and Unwin, 1957. [Google Scholar]
- 12.Krause DS. Plasticity of marrow-derived stem cells. Gene Ther. 2002;9:754–758. doi: 10.1038/sj.gt.3301760. [DOI] [PubMed] [Google Scholar]
- 13.Poulsom R, Alison MR, Forbes SJ, Wright NA. Adult stem cell plasticity. J Pathol. 2002;197:441–456. doi: 10.1002/path.1176. [DOI] [PubMed] [Google Scholar]
- 14.GURDON JB. Adult frogs derived from the nuclei of single somatic cells. Dev Biol. 1962;4:256–273. doi: 10.1016/0012-1606(62)90043-x. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Kleinsmith LJ, Pierce GB Jr. Multipotentiality of single embryonal carcinoma cells. Cancer Res. 1964;24:1544–1551. [PubMed] [Google Scholar]
- 17.Pierce GB, Speers WC. Tumors as caricatures of the process of tissue renewal: prospects for therapy by directing differentiation. Cancer Res. 1988;48:1996–2004. [PubMed] [Google Scholar]
- 18.Yu Z, Pestell TG, Lisanti MP, Pestell RG. Cancer stem cells. Int J Biochem Cell Biol. 2012;44:2144–2151. doi: 10.1016/j.biocel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvalho J. Cell Reversal From a Differentiated to a Stem-Like State at Cancer Initiation. Front Oncol. 2020;10:541. doi: 10.3389/fonc.2020.00541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 21.Chen W, Dong J, Haiech J, Kilhoffer MC, Zeniou M. Cancer Stem Cell Quiescence and Plasticity as Major Challenges in Cancer Therapy. Stem Cells Int. 2016;2016:1740936. doi: 10.1155/2016/1740936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medema JP. Cancer stem cells: the challenges ahead. Nat Cell Biol. 2013;15:338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- 23.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108:479–485. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prasetyanti PR, Medema JP. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer. 2017;16:41. doi: 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 27.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 29.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Walcher L, Kistenmacher AK, Suo H, Kitte R, Dluczek S, Strauß A, Blaudszun AR, Yevsa T, Fricke S, Kossatz-Boehlert U. Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front Immunol. 2020;11:1280. doi: 10.3389/fimmu.2020.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cole AJ, Fayomi AP, Anyaeche VI, Bai S, Buckanovich RJ. An evolving paradigm of cancer stem cell hierarchies: therapeutic implications. Theranostics. 2020;10:3083–3098. doi: 10.7150/thno.41647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 33.Phi LTH, Sari IN, Yang YG, Lee SH, Jun N, Kim KS, Lee YK, Kwon HY. Cancer Stem Cells (CSCs) in Drug Resistance and their Therapeutic Implications in Cancer Treatment. Stem Cells Int. 2018;2018:5416923. doi: 10.1155/2018/5416923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- 35.Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- 36.Uchikawa M, Kamachi Y, Kondoh H. Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech Dev. 1999;84:103–120. doi: 10.1016/s0925-4773(99)00083-0. [DOI] [PubMed] [Google Scholar]
- 37.Pevny LH, Lovell-Badge R. Sox genes find their feet. Curr Opin Genet Dev. 1997;7:338–344. doi: 10.1016/s0959-437x(97)80147-5. [DOI] [PubMed] [Google Scholar]
- 38.Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 39.Reiprich S, Wegner M. From CNS stem cells to neurons and glia: Sox for everyone. Cell Tissue Res. 2015;359:111–124. doi: 10.1007/s00441-014-1909-6. [DOI] [PubMed] [Google Scholar]
- 40.She ZY, Yang WX. SOX family transcription factors involved in diverse cellular events during development. Eur J Cell Biol. 2015;94:547–563. doi: 10.1016/j.ejcb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Mercurio S, Serra L, Nicolis SK. More than just Stem Cells: Functional Roles of the Transcription Factor Sox2 in Differentiated Glia and Neurons. Int J Mol Sci. 2019;20:4540. doi: 10.3390/ijms20184540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pevny L, Placzek M. SOX genes and neural progenitor identity. Curr Opin Neurobiol. 2005;15:7–13. doi: 10.1016/j.conb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 43.Cavaleri F, Schöler HR. Nanog: a new recruit to the embryonic stem cell orchestra. Cell. 2003;113:551–552. doi: 10.1016/s0092-8674(03)00394-5. [DOI] [PubMed] [Google Scholar]
- 44.Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 45.Chen CY, Cheng YY, Yen CY, Hsieh PC. Mechanisms of pluripotency maintenance in mouse embryonic stem cells. Cell Mol Life Sci. 2017;74:1805–1817. doi: 10.1007/s00018-016-2438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li M, Belmonte JC. Ground rules of the pluripotency gene regulatory network. Nat Rev Genet. 2017;18:180–191. doi: 10.1038/nrg.2016.156. [DOI] [PubMed] [Google Scholar]
- 47.Huang T, Song X, Xu D, Tiek D, Goenka A, Wu B, Sastry N, Hu B, Cheng SY. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics. 2020;10:8721–8743. doi: 10.7150/thno.41648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu A, Yu X, Liu S. Pluripotency transcription factors and cancer stem cells: small genes make a big difference. Chin J Cancer. 2013;32:483–487. doi: 10.5732/cjc.012.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong C, Wilhelm D, Koopman P. Sox genes and cancer. Cytogenet Genome Res. 2004;105:442–447. doi: 10.1159/000078217. [DOI] [PubMed] [Google Scholar]
- 50.Castillo SD, Sanchez-Cespedes M. The SOX family of genes in cancer development: biological relevance and opportunities for therapy. Expert Opin Ther Targets. 2012;16:903–919. doi: 10.1517/14728222.2012.709239. [DOI] [PubMed] [Google Scholar]
- 51.Thu KL, Becker-Santos DD, Radulovich N, Pikor LA, Lam WL, Tsao MS. SOX15 and other SOX family members are important mediators of tumorigenesis in multiple cancer types. Oncoscience. 2014;1:326–335. doi: 10.18632/oncoscience.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32:3397–3409. doi: 10.1038/onc.2012.506. [DOI] [PubMed] [Google Scholar]
- 54.Zhu Y, Li Y, Jun Wei JW, Liu X. The role of Sox genes in lung morphogenesis and cancer. Int J Mol Sci. 2012;13:15767–15783. doi: 10.3390/ijms131215767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grimm D, Bauer J, Wise P, Krüger M, Simonsen U, Wehland M, Infanger M, Corydon TJ. The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol. 2020;67:122–153. doi: 10.1016/j.semcancer.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 56.Wuebben EL, Rizzino A. The dark side of SOX2: cancer - a comprehensive overview. Oncotarget. 2017;8:44917–44943. doi: 10.18632/oncotarget.16570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schaefer T, Wang H, Mir P, Konantz M, Pereboom TC, Paczulla AM, Merz B, Fehm T, Perner S, Rothfuss OC, Kanz L, Schulze-Osthoff K, Lengerke C. Molecular and functional interactions between AKT and SOX2 in breast carcinoma. Oncotarget. 2015;6:43540–43556. doi: 10.18632/oncotarget.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, Zheng S, Hood L, Goodlett DR, Foltz G, Lin B. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics. 2011;12:11. doi: 10.1186/1471-2164-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bareiss PM, Paczulla A, Wang H, Schairer R, Wiehr S, Kohlhofer U, Rothfuss OC, Fischer A, Perner S, Staebler A, Wallwiener D, Fend F, Fehm T, Pichler B, Kanz L, Quintanilla-Martinez L, Schulze-Osthoff K, Essmann F, Lengerke C. SOX2 expression associates with stem cell state in human ovarian carcinoma. Cancer Res. 2013;73:5544–5555. doi: 10.1158/0008-5472.CAN-12-4177. [DOI] [PubMed] [Google Scholar]
- 60.Krol I, Castro-Giner F, Maurer M, Gkountela S, Szczerba BM, Scherrer R, Coleman N, Carreira S, Bachmann F, Anderson S, Engelhardt M, Lane H, Evans TRJ, Plummer R, Kristeleit R, Lopez J, Aceto N. Detection of circulating tumour cell clusters in human glioblastoma. Br J Cancer. 2018;119:487–491. doi: 10.1038/s41416-018-0186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mansouri S, Nejad R, Karabork M, Ekinci C, Solaroglu I, Aldape KD, Zadeh G. Sox2: regulation of expression and contribution to brain tumors. CNS Oncol. 2016;5:159–173. doi: 10.2217/cns-2016-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaefer T, Ramadoss A, Leu S, Tintignac L, Tostado C, Bink A, Schürch C, Müller J, Schärer J, Moffa G, Demougin P, Moes S, Stippich C, Falbo S, Neddersen H, Bucher H, Frank S, Jenö P, Lengerke C, Ritz MF, Mariani L, Boulay JL. Regulation of glioma cell invasion by 3q26 gene products PIK3CA, SOX2 and OPA1. Brain Pathol. 2019;29:336–350. doi: 10.1111/bpa.12670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thakkar JP, Dolecek TA, Horbinski C, Ostrom QT, Lightner DD, Barnholtz-Sloan JS, Villano JL. Epidemiologic and molecular prognostic review of glioblastoma. Cancer Epidemiol Biomarkers Prev. 2014;23:1985–1996. doi: 10.1158/1055-9965.EPI-14-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hottinger AF, Stupp R, Homicsko K. Standards of care and novel approaches in the management of glioblastoma multiforme. Chin J Cancer. 2014;33:32–39. doi: 10.5732/cjc.013.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh KM, Wrensch MR, Barnholtz-Sloan JS. The epidemiology of glioma in adults: a "state of the science" review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rock K, McArdle O, Forde P, Dunne M, Fitzpatrick D, O'Neill B, Faul C. A clinical review of treatment outcomes in glioblastoma multiforme--the validation in a non-trial population of the results of a randomised Phase III clinical trial: has a more radical approach improved survival? Br J Radiol. 2012;85:e729–e733. doi: 10.1259/bjr/83796755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 68.Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012-2016. Neuro Oncol. 2019;21:v1–v100. doi: 10.1093/neuonc/noz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O'Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L TCGA Research Network. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Conroy S, Wagemakers M, Walenkamp AM, Kruyt FA, den Dunnen WF. Novel insights into vascularization patterns and angiogenic factors in glioblastoma subclasses. J Neurooncol. 2017;131:11–20. doi: 10.1007/s11060-016-2269-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Escamilla-Ramírez A, Castillo-Rodríguez RA, Zavala-Vega S, Jimenez-Farfan D, Anaya-Rubio I, Briseño E, Palencia G, Guevara P, Cruz-Salgado A, Sotelo J, Trejo-Solís C. Autophagy as a Potential Therapy for Malignant Glioma. Pharmaceuticals (Basel) 2020;13:156. doi: 10.3390/ph13070156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ryskalin L, Gaglione A, Limanaqi F, Biagioni F, Familiari P, Frati A, Esposito V, Fornai F. The Autophagy Status of Cancer Stem Cells in Gliobastoma Multiforme: From Cancer Promotion to Therapeutic Strategies. Int J Mol Sci. 2019;20:3824. doi: 10.3390/ijms20153824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas L, Florio T, Perez-Castro C. Extracellular Vesicles Loaded miRNAs as Potential Modulators Shared Between Glioblastoma, and Parkinson's and Alzheimer's Diseases. Front Cell Neurosci. 2020;14:590034. doi: 10.3389/fncel.2020.590034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavaré S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jovčevska I. Genetic secrets of long-term glioblastoma survivors. Bosn J Basic Med Sci. 2019;19:116–124. doi: 10.17305/bjbms.2018.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park J, Shim JK, Yoon SJ, Kim SH, Chang JH, Kang SG. Transcriptome profiling-based identification of prognostic subtypes and multi-omics signatures of glioblastoma. Sci Rep. 2019;9:10555. doi: 10.1038/s41598-019-47066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schiffer D, Annovazzi L, Casalone C, Corona C, Mellai M. Glioblastoma: Microenvironment and Niche Concept. Cancers (Basel) 2018;11:5. doi: 10.3390/cancers11010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schiffer D, Annovazzi L, Mazzucco M, Mellai M. The Microenvironment in Gliomas: Phenotypic Expressions. Cancers (Basel) 2015;7:2352–2359. doi: 10.3390/cancers7040896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roberts LM, Munson J. Modulating microenvironments for treating glioblastoma. Curr Tissue Microenviron Rep. 2020;1:99–111. doi: 10.1007/s43152-020-00010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matarredona ER, Pastor AM. Extracellular Vesicle-Mediated Communication between the Glioblastoma and Its Microenvironment. Cells. 2019;9:96. doi: 10.3390/cells9010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simon T, Jackson E, Giamas G. Breaking through the glioblastoma micro-environment via extracellular vesicles. Oncogene. 2020;39:4477–4490. doi: 10.1038/s41388-020-1308-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Valdebenito S, Audia A, Bhat KPL, Okafo G, Eugenin EA. Tunneling Nanotubes Mediate Adaptation of Glioblastoma Cells to Temozolomide and Ionizing Radiation Treatment. iScience. 2020;23:101450. doi: 10.1016/j.isci.2020.101450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fan X, Xiong Y, Wang Y. A reignited debate over the cell(s) of origin for glioblastoma and its clinical implications. Front Med. 2019;13:531–539. doi: 10.1007/s11684-019-0700-1. [DOI] [PubMed] [Google Scholar]
- 86.Altmann C, Keller S, Schmidt MHH. The Role of SVZ Stem Cells in Glioblastoma. Cancers (Basel) 2019;11:448. doi: 10.3390/cancers11040448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Matarredona ER, Pastor AM. Neural Stem Cells of the Subventricular Zone as the Origin of Human Glioblastoma Stem Cells. Therapeutic Implications. Front Oncol. 2019;9:779. doi: 10.3389/fonc.2019.00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aum DJ, Kim DH, Beaumont TL, Leuthardt EC, Dunn GP, Kim AH. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg Focus. 2014;37:E11. doi: 10.3171/2014.9.FOCUS14521. [DOI] [PubMed] [Google Scholar]
- 89.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 2011;71:4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Friedmann-Morvinski D. Glioblastoma heterogeneity and cancer cell plasticity. Crit Rev Oncog. 2014;19:327–336. doi: 10.1615/critrevoncog.2014011777. [DOI] [PubMed] [Google Scholar]
- 91.Abadi B, Ahmadi-Zeidabadi M, Dini L, Vergallo C. Stem cell-based therapy treating glioblastoma multiforme. Hematol Oncol Stem Cell Ther. 2021;14:1–15. doi: 10.1016/j.hemonc.2020.08.001. [DOI] [PubMed] [Google Scholar]
- 92.Bryukhovetskiy I, Pak O, Khotimchenko Y, Bryukhovetskiy A, Sharma A, Sharma HS. Personalized therapy and stem cell transplantation for pro-inflammatory modulation of cancer stem cells microenvironment in glioblastoma: Review. Int Rev Neurobiol. 2020;151:67–98. doi: 10.1016/bs.irn.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 93.Vieira de Castro J, Gonçalves CS, Hormigo A, Costa BM. Exploiting the Complexities of Glioblastoma Stem Cells: Insights for Cancer Initiation and Therapeutic Targeting. Int J Mol Sci. 2020;21:5278. doi: 10.3390/ijms21155278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Garcia I, Aldaregia J, Marjanovic Vicentic J, Aldaz P, Moreno-Cugnon L, Torres-Bayona S, Carrasco-Garcia E, Garros-Regulez L, Egaña L, Rubio A, Pollard S, Stevanovic M, Sampron N, Matheu A. Oncogenic activity of SOX1 in glioblastoma. Sci Rep. 2017;7:46575. doi: 10.1038/srep46575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Annovazzi L, Mellai M, Caldera V, Valente G, Schiffer D. SOX2 expression and amplification in gliomas and glioma cell lines. Cancer Genomics Proteomics. 2011;8:139–147. [PubMed] [Google Scholar]
- 96.Hong X, Chedid K, Kalkanis SN. Glioblastoma cell line-derived spheres in serumcontaining medium versus serum-free medium: a comparison of cancer stem cell properties. Int J Oncol. 2012;41:1693–1700. doi: 10.3892/ijo.2012.1592. [DOI] [PubMed] [Google Scholar]
- 97.Schmitz M, Temme A, Senner V, Ebner R, Schwind S, Stevanovic S, Wehner R, Schackert G, Schackert HK, Fussel M, Bachmann M, Rieber EP, Weigle B. Identification of SOX2 as a novel glioma-associated antigen and potential target for T cell-based immunotherapy. Br J Cancer. 2007;96:1293–1301. doi: 10.1038/sj.bjc.6603696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alonso MM, Diez-Valle R, Manterola L, Rubio A, Liu D, Cortes-Santiago N, Urquiza L, Jauregi P, Lopez de Munain A, Sampron N, Aramburu A, Tejada-Solís S, Vicente C, Odero MD, Bandrés E, García-Foncillas J, Idoate MA, Lang FF, Fueyo J, Gomez-Manzano C. Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PLoS One. 2011;6:e26740. doi: 10.1371/journal.pone.0026740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Berezovsky AD, Poisson LM, Cherba D, Webb CP, Transou AD, Lemke NW, Hong X, Hasselbach LA, Irtenkauf SM, Mikkelsen T, deCarvalho AC. Sox2 promotes malignancy in glioblastoma by regulating plasticity and astrocytic differentiation. Neoplasia. 2014;16:193–206. doi: 10.1016/j.neo.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sathyan P, Zinn PO, Marisetty AL, Liu B, Kamal MM, Singh SK, Bady P, Lu L, Wani KM, Veo BL, Gumin J, Kassem DH, Robinson F, Weng C, Baladandayuthapani V, Suki D, Colman H, Bhat KP, Sulman EP, Aldape K, Colen RR, Verhaak RG, Lu Z, Fuller GN, Huang S, Lang FF, Sawaya R, Hegi M, Majumder S. Mir-21-Sox2 Axis Delineates Glioblastoma Subtypes with Prognostic Impact. J Neurosci. 2015;35:15097–15112. doi: 10.1523/JNEUROSCI.1265-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nandeesh BN, Naskar S, Shashtri AH, Arivazhagan A, Santosh V. Recurrent Glioblastomas Exhibit Higher Expression of Biomarkers with Stem-like Properties. J Neurosci Rural Pract. 2018;9:86–91. doi: 10.4103/jnrp.jnrp_417_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yu W, Ren X, Hu C, Tan Y, Shui Y, Chen Z, Zhang L, Peng J, Wei Q. Glioma SOX2 expression decreased after adjuvant therapy. BMC Cancer. 2019;19:1087. doi: 10.1186/s12885-019-6292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marjanovic Vicentic J, Drakulic D, Garcia I, Vukovic V, Aldaz P, Puskas N, Nikolic I, Tasic G, Raicevic S, Garros-Regulez L, Sampron N, Atkinson MJ, Anastasov N, Matheu A, Stevanovic M. SOX3 can promote the malignant behavior of glioblastoma cells. Cell Oncol (Dordr) 2019;42:41–54. doi: 10.1007/s13402-018-0405-5. [DOI] [PubMed] [Google Scholar]
- 105.Lu S, Yu Z, Zhang X, Sui L. MiR-483 Targeted SOX3 to Suppress Glioma Cell Migration, Invasion and Promote Cell Apoptosis. Onco Targets Ther. 2020;13:2153–2161. doi: 10.2147/OTT.S240619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang L, He S, Yuan J, Mao X, Cao Y, Zong J, Tu Y, Zhang Y. Oncogenic role of SOX9 expression in human malignant glioma. Med Oncol. 2012;29:3484–3490. doi: 10.1007/s12032-012-0267-z. [DOI] [PubMed] [Google Scholar]
- 107.Aldaz P, Otaegi-Ugartemendia M, Saenz-Antoñanzas A, Garcia-Puga M, Moreno-Valladares M, Flores JM, Gerovska D, Arauzo-Bravo MJ, Samprón N, Matheu A, Carrasco-Garcia E. SOX9 promotes tumor progression through the axis BMI1-p21CIP. Sci Rep. 2020;10:357. doi: 10.1038/s41598-019-57047-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gao J, Zhang JY, Li YH, Ren F. Decreased expression of SOX9 indicates a better prognosis and inhibits the growth of glioma cells by inducing cell cycle arrest. Int J Clin Exp Pathol. 2015;8:10130–10138. [PMC free article] [PubMed] [Google Scholar]
- 109.Bannykh SI, Stolt CC, Kim J, Perry A, Wegner M. Oligodendroglial-specific transcriptional factor SOX10 is ubiquitously expressed in human gliomas. J Neurooncol. 2006;76:115–127. doi: 10.1007/s11060-005-5533-x. [DOI] [PubMed] [Google Scholar]
- 110.Ferletta M, Uhrbom L, Olofsson T, Pontén F, Westermark B. Sox10 has a broad expression pattern in gliomas and enhances platelet-derived growth factor-B--induced gliomagenesis. Mol Cancer Res. 2007;5:891–897. doi: 10.1158/1541-7786.MCR-07-0113. [DOI] [PubMed] [Google Scholar]
- 111.Jin C, Zhao J, Zhang ZP, Wu M, Li J, Liu B, Bin Liao X, Liao YX, Liu JP. CircRNA EPHB4 modulates stem properties and proliferation of gliomas via sponging miR-637 and up-regulating SOX10. Mol Oncol. 2021;15:596–622. doi: 10.1002/1878-0261.12830. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 112.Ueda R, Yoshida K, Kawase T, Kawakami Y, Toda M. Preferential expression and frequent IgG responses of a tumor antigen, SOX5, in glioma patients. Int J Cancer. 2007;120:1704–1711. doi: 10.1002/ijc.22472. [DOI] [PubMed] [Google Scholar]
- 113.Tchougounova E, Jiang Y, Bråsäter D, Lindberg N, Kastemar M, Asplund A, Westermark B, Uhrbom L. Sox5 can suppress platelet-derived growth factor B-induced glioma development in Ink4a-deficient mice through induction of acute cellular senescence. Oncogene. 2009;28:1537–1548. doi: 10.1038/onc.2009.9. [DOI] [PubMed] [Google Scholar]
- 114.Schlierf B, Friedrich RP, Roerig P, Felsberg J, Reifenberger G, Wegner M. Expression of SoxE and SoxD genes in human gliomas. Neuropathol Appl Neurobiol. 2007;33:621–630. doi: 10.1111/j.1365-2990.2007.00881.x. [DOI] [PubMed] [Google Scholar]
- 115.Tian R, Wang J, Yan H, Wu J, Xu Q, Zhan X, Gui Z, Ding M, He J. Differential expression of miR16 in glioblastoma and glioblastoma stem cells: their correlation with proliferation, differentiation, metastasis and prognosis. Oncogene. 2017;36:5861–5873. doi: 10.1038/onc.2017.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jiang L, Yang H, Chen T, Zhu X, Ye J, Lv K. Identification of HMG-box family establishes the significance of SOX6 in the malignant progression of glioblastoma. Aging (Albany NY) 2020;12:8084–8106. doi: 10.18632/aging.103127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhao T, Yang H, Tian Y, Xie Q, Lu Y, Wang Y, Su N, Dong B, Liu X, Wang C, Jiang C. SOX7 is associated with the suppression of human glioma by HMG-box dependent regulation of Wnt/β-catenin signaling. Cancer Lett. 2016;375:100–107. doi: 10.1016/j.canlet.2016.02.044. [DOI] [PubMed] [Google Scholar]
- 118.Weigle B, Ebner R, Temme A, Schwind S, Schmitz M, Kiessling A, Rieger MA, Schackert G, Schackert HK, Rieber EP. Highly specific overexpression of the transcription factor SOX11 in human malignant gliomas. Oncol Rep. 2005;13:139–144. [PubMed] [Google Scholar]
- 119.Hide T, Takezaki T, Nakatani Y, Nakamura H, Kuratsu J, Kondo T. Sox11 prevents tumorigenesis of glioma-initiating cells by inducing neuronal differentiation. Cancer Res. 2009;69:7953–7959. doi: 10.1158/0008-5472.CAN-09-2006. [DOI] [PubMed] [Google Scholar]
- 120.Su Z, Zang T, Liu ML, Wang LL, Niu W, Zhang CL. Reprogramming the fate of human glioma cells to impede brain tumor development. Cell Death Dis. 2014;5:e1463. doi: 10.1038/cddis.2014.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang D, Guo S, Wang H, Hu Y. SOX15 exerts antitumor function in glioma by inhibiting cell proliferation and invasion via downregulation of Wnt/β-catenin signaling. Life Sci. 2020;255:117792. doi: 10.1016/j.lfs.2020.117792. [DOI] [PubMed] [Google Scholar]
- 122.de la Rocha AM, Sampron N, Alonso MM, Matheu A. Role of SOX family of transcription factors in central nervous system tumors. Am J Cancer Res. 2014;4:312–324. [PMC free article] [PubMed] [Google Scholar]
- 123.Ferletta M, Caglayan D, Mokvist L, Jiang Y, Kastemar M, Uhrbom L, Westermark B. Forced expression of Sox21 inhibits Sox2 and induces apoptosis in human glioma cells. Int J Cancer. 2011;129:45–60. doi: 10.1002/ijc.25647. [DOI] [PubMed] [Google Scholar]
- 124.Caglayan D, Lundin E, Kastemar M, Westermark B, Ferletta M. Sox21 inhibits glioma progression in vivo by forming complexes with Sox2 and stimulating aberrant differentiation. Int J Cancer. 2013;133:1345–1356. doi: 10.1002/ijc.28147. [DOI] [PubMed] [Google Scholar]
- 125.Kurtsdotter I, Topcic D, Karlén A, Singla B, Hagey DW, Bergsland M, Siesjö P, Nistér M, Carlson JW, Lefebvre V, Persson O, Holmberg J, Muhr J. SOX5/6/21 Prevent Oncogene-Driven Transformation of Brain Stem Cells. Cancer Res. 2017;77:4985–4997. doi: 10.1158/0008-5472.CAN-17-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xu Y, Yuan FE, Chen QX, Liu BH. Molecular mechanisms involved in angiogenesis and potential target of antiangiogenesis in human glioblastomas. GLIOMA. 2018;1:35–42. [Google Scholar]
- 127.Kim IK, Kim K, Lee E, Oh DS, Park CS, Park S, Yang JM, Kim JH, Kim HS, Shima DT, Hong SH, Cho YH, Kim YH, Park JB, Koh GY, Ju YS, Lee HK, Lee S, Kim I. Sox7 promotes high-grade glioma by increasing VEGFR2-mediated vascular abnormality. J Exp Med. 2018;215:963–983. doi: 10.1084/jem.20170123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.He Z, Ruan X, Liu X, Zheng J, Liu Y, Liu L, Ma J, Shao L, Wang D, Shen S, Yang C, Xue Y. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in Glioma. J Exp Clin Cancer Res. 2019;38:65. doi: 10.1186/s13046-019-1065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhang J, Jiang H, Shao J, Mao R, Liu J, Ma Y, Fang X, Zhao N, Zheng S, Lin B. SOX4 inhibits GBM cell growth and induces G0/G1 cell cycle arrest through Akt-p53 axis. BMC Neurol. 2014;14:207. doi: 10.1186/s12883-014-0207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009;5:504–514. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 131.Galatro TF, Uno M, Oba-Shinjo SM, Almeida AN, Teixeira MJ, Rosemberg S, Marie SK. Differential expression of ID4 and its association with TP53 mutation, SOX2, SOX4 and OCT-4 expression levels. PLoS One. 2013;8:e61605. doi: 10.1371/journal.pone.0061605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 133.Auffinger B, Spencer D, Pytel P, Ahmed AU, Lesniak MS. The role of glioma stem cells in chemotherapy resistance and glioblastoma multiforme recurrence. Expert Rev Neurother. 2015;15:741–752. doi: 10.1586/14737175.2015.1051968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Boyd NH, Tran AN, Bernstock JD, Etminan T, Jones AB, Gillespie GY, Friedman GK, Hjelmeland AB. Glioma stem cells and their roles within the hypoxic tumor microenvironment. Theranostics. 2021;11:665–683. doi: 10.7150/thno.41692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ferreyra Solari NE, Belforte FS, Canedo L, Videla-Richardson GA, Espinosa JM, Rossi M, Serna E, Riudavets MA, Martinetto H, Sevlever G, Perez-Castro C. The NSL Chromatin-Modifying Complex Subunit KANSL2 Regulates Cancer Stem-like Properties in Glioblastoma That Contribute to Tumorigenesis. Cancer Res. 2016;76:5383–5394. doi: 10.1158/0008-5472.CAN-15-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Suvà ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, Curry WT, Martuza RL, Rivera MN, Rossetti N, Kasif S, Beik S, Kadri S, Tirosh I, Wortman I, Shalek AK, Rozenblatt-Rosen O, Regev A, Louis DN, Bernstein BE. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gimple RC, Bhargava S, Dixit D, Rich JN. Glioblastoma stem cells: lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019;33:591–609. doi: 10.1101/gad.324301.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jin X, Kim LJY, Wu Q, Wallace LC, Prager BC, Sanvoranart T, Gimple RC, Wang X, Mack SC, Miller TE, Huang P, Valentim CL, Zhou QG, Barnholtz-Sloan JS, Bao S, Sloan AE, Rich JN. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat Med. 2017;23:1352–1361. doi: 10.1038/nm.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17:165–172. doi: 10.1016/j.cub.2006.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim KH, Seol HJ, Kim EH, Rheey J, Jin HJ, Lee Y, Joo KM, Lee J, Nam DH. Wnt/β-catenin signaling is a key downstream mediator of MET signaling in glioblastoma stem cells. Neuro Oncol. 2013;15:161–171. doi: 10.1093/neuonc/nos299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, Koh C, Zhang J, Li YM, Maciaczyk J, Nikkhah G, Dimeco F, Piccirillo S, Vescovi AL, Eberhart CG. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16. doi: 10.1002/stem.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Soeda A, Inagaki A, Oka N, Ikegame Y, Aoki H, Yoshimura S, Nakashima S, Kunisada T, Iwama T. Epidermal growth factor plays a crucial role in mitogenic regulation of human brain tumor stem cells. J Biol Chem. 2008;283:10958–10966. doi: 10.1074/jbc.M704205200. [DOI] [PubMed] [Google Scholar]
- 143.Nanta R, Shrivastava A, Sharma J, Shankar S, Srivastava RK. Inhibition of sonic hedgehog and PI3K/Akt/mTOR pathways cooperate in suppressing survival, self-renewal and tumorigenic potential of glioblastoma-initiating cells. Mol Cell Biochem. 2019;454:11–23. doi: 10.1007/s11010-018-3448-z. [DOI] [PubMed] [Google Scholar]
- 144.Riddick G, Kotliarova S, Rodriguez V, Kim HS, Linkous A, Storaska AJ, Ahn S, Walling J, Belova G, Fine HA. A Core Regulatory Circuit in Glioblastoma Stem Cells Links MAPK Activation to a Transcriptional Program of Neural Stem Cell Identity. Sci Rep. 2017;7:43605. doi: 10.1038/srep43605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rinkenbaugh AL, Cogswell PC, Calamini B, Dunn DE, Persson AI, Weiss WA, Lo DC, Baldwin AS. IKK/NF-κB signaling contributes to glioblastoma stem cell maintenance. Oncotarget. 2016;7:69173–69187. doi: 10.18632/oncotarget.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Almiron Bonnin DA, Havrda MC, Lee MC, Liu H, Zhang Z, Nguyen LN, Harrington LX, Hassanpour S, Cheng C, Israel MA. Secretion-mediated STAT3 activation promotes self-renewal of glioma stem-like cells during hypoxia. Oncogene. 2018;37:1107–1118. doi: 10.1038/onc.2017.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jin X, Yin J, Kim SH, Sohn YW, Beck S, Lim YC, Nam DH, Choi YJ, Kim H. EGFR-AKT-Smad signaling promotes formation of glioma stem-like cells and tumor angiogenesis by ID3-driven cytokine induction. Cancer Res. 2011;71:7125–7134. doi: 10.1158/0008-5472.CAN-11-1330. [DOI] [PubMed] [Google Scholar]
- 148.Peñuelas S, Anido J, Prieto-Sánchez RM, Folch G, Barba I, Cuartas I, García-Dorado D, Poca MA, Sahuquillo J, Baselga J, Seoane J. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–327. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 149.Sunayama J, Matsuda K, Sato A, Tachibana K, Suzuki K, Narita Y, Shibui S, Sakurada K, Kayama T, Tomiyama A, Kitanaka C. Crosstalk between the PI3K/mTOR and MEK/ERK pathways involved in the maintenance of self-renewal and tumorigenicity of glioblastoma stem-like cells. Stem Cells. 2010;28:1930–1939. doi: 10.1002/stem.521. [DOI] [PubMed] [Google Scholar]
- 150.Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, Zhang G, Wang X, Dong Z, Chen F, Cui H. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther. 2020;5:8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yang Y, Li X, Wang T, Guo Q, Xi T, Zheng L. Emerging agents that target signaling pathways in cancer stem cells. J Hematol Oncol. 2020;13:60. doi: 10.1186/s13045-020-00901-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Codrici E, Enciu AM, Popescu ID, Mihai S, Tanase C. Glioma Stem Cells and Their Microenvironments: Providers of Challenging Therapeutic Targets. Stem Cells Int. 2016;2016:5728438. doi: 10.1155/2016/5728438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Huang Q, Zhang QB, Dong J, Wu YY, Shen YT, Zhao YD, Zhu YD, Diao Y, Wang AD, Lan Q. Glioma stem cells are more aggressive in recurrent tumors with malignant progression than in the primary tumor, and both can be maintained long-term in vitro. BMC Cancer. 2008;8:304. doi: 10.1186/1471-2407-8-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cho DY, Lin SZ, Yang WK, Lee HC, Hsu DM, Lin HL, Chen CC, Liu CL, Lee WY, Ho LH. Targeting cancer stem cells for treatment of glioblastoma multiforme. Cell Transplant. 2013;22:731–739. doi: 10.3727/096368912X655136. [DOI] [PubMed] [Google Scholar]
- 155.Brooks MD, Sengupta R, Snyder SC, Rubin JB. Hitting Them Where They Live: Targeting the Glioblastoma Perivascular Stem Cell Niche. Curr Pathobiol Rep. 2013;1:101–110. doi: 10.1007/s40139-013-0012-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Prager BC, Bhargava S, Mahadev V, Hubert CG, Rich JN. Glioblastoma Stem Cells: Driving Resilience through Chaos. Trends Cancer. 2020;6:223–235. doi: 10.1016/j.trecan.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jhaveri N, Chen TC, Hofman FM. Tumor vasculature and glioma stem cells: Contributions to glioma progression. Cancer Lett. 2016;380:545–551. doi: 10.1016/j.canlet.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 158.Schiffer D, Mellai M, Bovio E, Bisogno I, Casalone C, Annovazzi L. Glioblastoma niches: from the concept to the phenotypical reality. Neurol Sci. 2018;39:1161–1168. doi: 10.1007/s10072-018-3408-0. [DOI] [PubMed] [Google Scholar]
- 159.Gangemi RM, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 160.Song WS, Yang YP, Huang CS, Lu KH, Liu WH, Wu WW, Lee YY, Lo WL, Lee SD, Chen YW, Huang PI, Chen MT. Sox2, a stemness gene, regulates tumor-initiating and drug-resistant properties in CD133-positive glioblastoma stem cells. J Chin Med Assoc. 2016;79:538–545. doi: 10.1016/j.jcma.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 161.Cox JL, Wilder PJ, Desler M, Rizzino A. Elevating SOX2 levels deleteriously affects the growth of medulloblastoma and glioblastoma cells. PLoS One. 2012;7:e44087. doi: 10.1371/journal.pone.0044087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Singh DK, Kollipara RK, Vemireddy V, Yang XL, Sun Y, Regmi N, Klingler S, Hatanpaa KJ, Raisanen J, Cho SK, Sirasanagandla S, Nannepaga S, Piccirillo S, Mashimo T, Wang S, Humphries CG, Mickey B, Maher EA, Zheng H, Kim RS, Kittler R, Bachoo RM. Oncogenes Activate an Autonomous Transcriptional Regulatory Circuit That Drives Glioblastoma. Cell Rep. 2017;18:961–976. doi: 10.1016/j.celrep.2016.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Su R, Cao S, Ma J, Liu Y, Liu X, Zheng J, Chen J, Liu L, Cai H, Li Z, Zhao L, He Q, Xue Y. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol Cancer. 2017;16:171. doi: 10.1186/s12943-017-0737-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang Z, Xu X, Liu N, Cheng Y, Jin W, Zhang P, Wang X, Yang H, Liu H, Tu Y. SOX9-PDK1 axis is essential for glioma stem cell self-renewal and temozolomide resistance. Oncotarget. 2018;9:192–204. doi: 10.18632/oncotarget.22773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Garros-Regulez L, Aldaz P, Arrizabalaga O, Moncho-Amor V, Carrasco-Garcia E, Manterola L, Moreno-Cugnon L, Barrena C, Villanua J, Ruiz I, Pollard S, Lovell-Badge R, Sampron N, Garcia I, Matheu A. mTOR inhibition decreases SOX2-SOX9 mediated glioma stem cell activity and temozolomide resistance. Expert Opin Ther Targets. 2016;20:393–405. doi: 10.1517/14728222.2016.1151002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.López-Valero I, Dávila D, González-Martínez J, Salvador-Tormo N, Lorente M, Saiz-Ladera C, Torres S, Gabicagogeascoa E, Hernández-Tiedra S, García-Taboada E, Mendiburu-Eliçabe M, Rodríguez-Fornés F, Sánchez-Domínguez R, Segovia JC, Sánchez-Gómez P, Matheu A, Sepúlveda JM, Velasco G. Midkine signaling maintains the self-renewal and tumorigenic capacity of glioma initiating cells. Theranostics. 2020;10:5120–5136. doi: 10.7150/thno.41450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wu B, Yang C, Fang Y, Ding W, Zhang Y. Long noncoding RNA DUXAP10 promotes the stemness of glioma cells by recruiting HuR to enhance Sox12 mRNA stability. Environ Toxicol. 2021;36:840–849. doi: 10.1002/tox.23087. [DOI] [PubMed] [Google Scholar]
- 168.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 170.Carruthers RD, Ahmed SU, Ramachandran S, Strathdee K, Kurian KM, Hedley A, Gomez-Roman N, Kalna G, Neilson M, Gilmour L, Stevenson KH, Hammond EM, Chalmers AJ. Replication Stress Drives Constitutive Activation of the DNA Damage Response and Radioresistance in Glioblastoma Stem-like Cells. Cancer Res. 2018;78:5060–5071. doi: 10.1158/0008-5472.CAN-18-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, Conroy S, Long L, Lelic N, Wang S, Gumin J, Raj D, Kodama Y, Raghunathan A, Olar A, Joshi K, Pelloski CE, Heimberger A, Kim SH, Cahill DP, Rao G, Den Dunnen WFA, Boddeke HWGM, Phillips HS, Nakano I, Lang FF, Colman H, Sulman EP, Aldape K. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hsieh A, Ellsworth R, Hsieh D. Hedgehog/GLI1 regulates IGF dependent malignant behaviors in glioma stem cells. J Cell Physiol. 2011;226:1118–1127. doi: 10.1002/jcp.22433. [DOI] [PubMed] [Google Scholar]
- 173.Kim Y, Kim KH, Lee J, Lee YA, Kim M, Lee SJ, Park K, Yang H, Jin J, Joo KM, Nam DH. Wnt activation is implicated in glioblastoma radioresistance. Lab Invest. 2012;92:466–473. doi: 10.1038/labinvest.2011.161. [DOI] [PubMed] [Google Scholar]
- 174.Wang J, Wakeman TP, Lathia JD, Hjelmeland AB, Wang XF, White RR, Rich JN, Sullenger BA. Notch promotes radioresistance of glioma stem cells. Stem Cells. 2010;28:17–28. doi: 10.1002/stem.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Halliday J, Helmy K, Pattwell SS, Pitter KL, LaPlant Q, Ozawa T, Holland EC. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc Natl Acad Sci U S A. 2014;111:5248–5253. doi: 10.1073/pnas.1321014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Segerman A, Niklasson M, Haglund C, Bergström T, Jarvius M, Xie Y, Westermark A, Sönmez D, Hermansson A, Kastemar M, Naimaie-Ali Z, Nyberg F, Berglund M, Sundström M, Hesselager G, Uhrbom L, Gustafsson M, Larsson R, Fryknäs M, Segerman B, Westermark B. Clonal Variation in Drug and Radiation Response among Glioma-Initiating Cells Is Linked to Proneural-Mesenchymal Transition. Cell Rep. 2016;17:2994–3009. doi: 10.1016/j.celrep.2016.11.056. [DOI] [PubMed] [Google Scholar]
- 177.Auffinger B, Tobias AL, Han Y, Lee G, Guo D, Dey M, Lesniak MS, Ahmed AU. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ. 2014;21:1119–1131. doi: 10.1038/cdd.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dahan P, Martinez Gala J, Delmas C, Monferran S, Malric L, Zentkowski D, Lubrano V, Toulas C, Cohen-Jonathan Moyal E, Lemarie A. Ionizing radiations sustain glioblastoma cell dedifferentiation to a stem-like phenotype through survivin: possible involvement in radioresistance. Cell Death Dis. 2014;5:e1543. doi: 10.1038/cddis.2014.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Qazi MA, Vora P, Venugopal C, McFarlane N, Subapanditha MK, Murty NK, Hassell JA, Hallett RM, Singh SK. A novel stem cell culture model of recurrent glioblastoma. J Neurooncol. 2016;126:57–67. doi: 10.1007/s11060-015-1951-6. [DOI] [PubMed] [Google Scholar]
- 181.Yang YP, Chien Y, Chiou GY, Cherng JY, Wang ML, Lo WL, Chang YL, Huang PI, Chen YW, Shih YH, Chen MT, Chiou SH. Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microRNA145 with cationic polyurethane-short branch PEI. Biomaterials. 2012;33:1462–1476. doi: 10.1016/j.biomaterials.2011.10.071. [DOI] [PubMed] [Google Scholar]
- 182.Jeon HM, Sohn YW, Oh SY, Kim SH, Beck S, Kim S, Kim H. ID4 imparts chemoresistance and cancer stemness to glioma cells by derepressing miR-9*-mediated suppression of SOX2. Cancer Res. 2011;71:3410–3421. doi: 10.1158/0008-5472.CAN-10-3340. [DOI] [PubMed] [Google Scholar]
- 183.Lee Y, Kim KH, Kim DG, Cho HJ, Kim Y, Rheey J, Shin K, Seo YJ, Choi YS, Lee JI, Lee J, Joo KM, Nam DH. FoxM1 Promotes Stemness and Radio-Resistance of Glioblastoma by Regulating the Master Stem Cell Regulator Sox2. PLoS One. 2015;10:e0137703. doi: 10.1371/journal.pone.0137703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sabelström H, Petri R, Shchors K, Jandial R, Schmidt C, Sacheva R, Masic S, Yuan E, Fenster T, Martinez M, Saxena S, Nicolaides TP, Ilkhanizadeh S, Berger MS, Snyder EY, Weiss WA, Jakobsson J, Persson AI. Driving Neuronal Differentiation through Reversal of an ERK1/2-miR-124-SOX9 Axis Abrogates Glioblastoma Aggressiveness. Cell Rep 2019; 28: 2064-2079, e11. [DOI] [PubMed] [Google Scholar]
- 185.Yool AJ, Ramesh S. Molecular Targets for Combined Therapeutic Strategies to Limit Glioblastoma Cell Migration and Invasion. Front Pharmacol. 2020;11:358. doi: 10.3389/fphar.2020.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suvà ML, Regev A, Bernstein BE. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, Richman AR, Silverbush D, Shaw ML, Hebert CM, Dewitt J, Gritsch S, Perez EM, Gonzalez Castro LN, Lan X, Druck N, Rodman C, Dionne D, Kaplan A, Bertalan MS, Small J, Pelton K, Becker S, Bonal D, Nguyen QD, Servis RL, Fung JM, Mylvaganam R, Mayr L, Gojo J, Haberler C, Geyeregger R, Czech T, Slavc I, Nahed BV, Curry WT, Carter BS, Wakimoto H, Brastianos PK, Batchelor TT, Stemmer-Rachamimov A, Martinez-Lage M, Frosch MP, Stamenkovic I, Riggi N, Rheinbay E, Monje M, Rozenblatt-Rosen O, Cahill DP, Patel AP, Hunter T, Verma IM, Ligon KL, Louis DN, Regev A, Bernstein BE, Tirosh I, Suvà ML. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019; 178: 835-849. :e21. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, Asthana S, Jalbert LE, Nelson SJ, Bollen AW, Gustafson WC, Charron E, Weiss WA, Smirnov IV, Song JS, Olshen AB, Cha S, Zhao Y, Moore RA, Mungall AJ, Jones SJM, Hirst M, Marra MA, Saito N, Aburatani H, Mukasa A, Berger MS, Chang SM, Taylor BS, Costello JF. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Barthel FP, Johnson KC, Varn FS, Moskalik AD, Tanner G, Kocakavuk E, Anderson KJ, Abiola O, Aldape K, Alfaro KD, Alpar D, Amin SB, Ashley DM, Bandopadhayay P, Barnholtz-Sloan JS, Beroukhim R, Bock C, Brastianos PK, Brat DJ, Brodbelt AR, Bruns AF, Bulsara KR, Chakrabarty A, Chakravarti A, Chuang JH, Claus EB, Cochran EJ, Connelly J, Costello JF, Finocchiaro G, Fletcher MN, French PJ, Gan HK, Gilbert MR, Gould PV, Grimmer MR, Iavarone A, Ismail A, Jenkinson MD, Khasraw M, Kim H, Kouwenhoven MCM, LaViolette PS, Li M, Lichter P, Ligon KL, Lowman AK, Malta TM, Mazor T, McDonald KL, Molinaro AM, Nam DH, Nayyar N, Ng HK, Ngan CY, Niclou SP, Niers JM, Noushmehr H, Noorbakhsh J, Ormond DR, Park CK, Poisson LM, Rabadan R, Radlwimmer B, Rao G, Reifenberger G, Sa JK, Schuster M, Shaw BL, Short SC, Smitt PAS, Sloan AE, Smits M, Suzuki H, Tabatabai G, Van Meir EG, Watts C, Weller M, Wesseling P, Westerman BA, Widhalm G, Woehrer A, Yung WKA, Zadeh G, Huse JT, De Groot JF, Stead LF, Verhaak RGW GLASS Consortium. Longitudinal molecular trajectories of diffuse glioma in adults. Nature. 2019;576:112–120. doi: 10.1038/s41586-019-1775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Steinbichler TB, Dudás J, Skvortsov S, Ganswindt U, Riechelmann H, Skvortsova II. Therapy resistance mediated by cancer stem cells. Semin Cancer Biol. 2018;53:156–167. doi: 10.1016/j.semcancer.2018.11.006. [DOI] [PubMed] [Google Scholar]
- 191.Fidoamore A, Cristiano L, Antonosante A, d'Angelo M, Di Giacomo E, Astarita C, Giordano A, Ippoliti R, Benedetti E, Cimini A. Glioblastoma Stem Cells Microenvironment: The Paracrine Roles of the Niche in Drug and Radioresistance. Stem Cells Int. 2016;2016:6809105. doi: 10.1155/2016/6809105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Garnier D, Renoult O, Alves-Guerra MC, Paris F, Pecqueur C. Glioblastoma Stem-Like Cells, Metabolic Strategy to Kill a Challenging Target. Front Oncol. 2019;9:118. doi: 10.3389/fonc.2019.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bindra RS, Chalmers AJ, Evans S, Dewhirst M. GBM radiosensitizers: dead in the water…or just the beginning? J Neurooncol. 2017;134:513–521. doi: 10.1007/s11060-017-2427-7. [DOI] [PubMed] [Google Scholar]
- 194.Cho Y, Kim YK. Cancer Stem Cells as a Potential Target to Overcome Multidrug Resistance. Front Oncol. 2020;10:764. doi: 10.3389/fonc.2020.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ghosh D, Nandi S, Bhattacharjee S. Combination therapy to checkmate Glioblastoma: clinical challenges and advances. Clin Transl Med. 2018;7:33. doi: 10.1186/s40169-018-0211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Weenink B, French PJ, Sillevis Smitt PAE, Debets R, Geurts M. Immunotherapy in Glioblastoma: Current Shortcomings and Future Perspectives. Cancers (Basel) 2020;12:751. doi: 10.3390/cancers12030751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Garros-Regulez L, Garcia I, Carrasco-Garcia E, Lantero A, Aldaz P, Moreno-Cugnon L, Arrizabalaga O, Undabeitia J, Torres-Bayona S, Villanua J, Ruiz I, Egaña L, Sampron N, Matheu A. Targeting SOX2 as a Therapeutic Strategy in Glioblastoma. Front Oncol. 2016;6:222. doi: 10.3389/fonc.2016.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hägerstrand D, He X, Bradic Lindh M, Hoefs S, Hesselager G, Ostman A, Nistér M. Identification of a SOX2-dependent subset of tumor- and sphere-forming glioblastoma cells with a distinct tyrosine kinase inhibitor sensitivity profile. Neuro Oncol. 2011;13:1178–1191. doi: 10.1093/neuonc/nor113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Singh S, Trevino J, Bora-Singhal N, Coppola D, Haura E, Altiok S, Chellappan SP. EGFR/Src/Akt signaling modulates Sox2 expression and self-renewal of stem-like side-population cells in non-small cell lung cancer. Mol Cancer. 2012;11:73. doi: 10.1186/1476-4598-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Li Z, Chen Y, An T, Liu P, Zhu J, Yang H, Zhang W, Dong T, Jiang J, Zhang Y, Jiang M, Yang X. Nuciferine inhibits the progression of glioblastoma by suppressing the SOX2-AKT/STAT3-Slug signaling pathway. J Exp Clin Cancer Res. 2019;38:139. doi: 10.1186/s13046-019-1134-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 202.Magrath JW, Raney WR, Kim Y. In vitro demonstration of salinomycin as a novel chemotherapeutic agent for the treatment of SOX2positive glioblastoma cancer stem cells. Oncol Rep. 2020;44:777–785. doi: 10.3892/or.2020.7642. [DOI] [PubMed] [Google Scholar]
- 203.Shi L, Wang Z, Sun G. Curcumin induces glioma stem-like cell formation. Neuroreport. 2015;26:167–172. doi: 10.1097/WNR.0000000000000320. [DOI] [PubMed] [Google Scholar]
- 204.Taylor JT, Ellison S, Pandele A, Wood S, Nathan E, Forte G, Parker H, Zindy E, Elvin M, Dickson A, Williams KJ, Karabatsou K, McCabe M, McBain C, Bigger BW. Actinomycin D downregulates Sox2 and improves survival in preclinical models of recurrent glioblastoma. Neuro Oncol. 2020;22:1289–1301. doi: 10.1093/neuonc/noaa051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Taniguchi J, Pandian GN, Hidaka T, Hashiya K, Bando T, Kim KK, Sugiyama H. A synthetic DNA-binding inhibitor of SOX2 guides human induced pluripotent stem cells to differentiate into mesoderm. Nucleic Acids Res. 2017;45:9219–9228. doi: 10.1093/nar/gkx693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yin Y, Xie CM, Li H, Tan M, Chen G, Schiff R, Xiong X, Sun Y. The FBXW2-MSX2-SOX2 axis regulates stem cell property and drug resistance of cancer cells. Proc Natl Acad Sci U S A. 2019;116:20528–20538. doi: 10.1073/pnas.1905973116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Favaro R, Appolloni I, Pellegatta S, Sanga AB, Pagella P, Gambini E, Pisati F, Ottolenghi S, Foti M, Finocchiaro G, Malatesta P, Nicolis SK. Sox2 is required to maintain cancer stem cells in a mouse model of high-grade oligodendroglioma. Cancer Res. 2014;74:1833–1844. doi: 10.1158/0008-5472.CAN-13-1942. [DOI] [PubMed] [Google Scholar]
- 208.Ueda R, Ohkusu-Tsukada K, Fusaki N, Soeda A, Kawase T, Kawakami Y, Toda M. Identification of HLA-A2- and A24-restricted T-cell epitopes derived from SOX6 expressed in glioma stem cells for immunotherapy. Int J Cancer. 2010;126:919–929. doi: 10.1002/ijc.24851. [DOI] [PubMed] [Google Scholar]
- 209.Liu K, Xie F, Zhao T, Zhang R, Gao A, Chen Y, Li H, Zhang S, Xiao Z, Li J, Hong X, Shang L, Huang W, Wang J, El-Rifai W, Zaika A, Chen X, Que J, Lan X. Targeting SOX2 Protein with Peptide Aptamers for Therapeutic Gains against Esophageal Squamous Cell Carcinoma. Mol Ther. 2020;28:901–913. doi: 10.1016/j.ymthe.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Stolzenburg S, Rots MG, Beltran AS, Rivenbark AG, Yuan X, Qian H, Strahl BD, Blancafort P. Targeted silencing of the oncogenic transcription factor SOX2 in breast cancer. Nucleic Acids Res. 2012;40:6725–6740. doi: 10.1093/nar/gks360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Abbas MN, Kausar S, Cui H. Therapeutic potential of natural products in glioblastoma treatment: targeting key glioblastoma signaling pathways and epigenetic alterations. Clin Transl Oncol. 2020;22:963–977. doi: 10.1007/s12094-019-02227-3. [DOI] [PubMed] [Google Scholar]
- 212.Kuete V, Sandjo LP, Ouete JL, Fouotsa H, Wiench B, Efferth T. Cytotoxicity and modes of action of three naturally occurring xanthones (8-hydroxycudraxanthone G, morusignin I and cudraxanthone I) against sensitive and multidrug-resistant cancer cell lines. Phytomedicine. 2014;21:315–322. doi: 10.1016/j.phymed.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 213.Ramachandran C, Portalatin G, Quirin KW, Escalon E, Khatib Z, Melnick SJ. Inhibition of AKT signaling by supercritical CO2 extract of mango ginger (Curcuma amada Roxb.) in human glioblastoma cells. J Complement Integr Med. 2015;12:307–315. doi: 10.1515/jcim-2015-0005. [DOI] [PubMed] [Google Scholar]
- 214.Suresh D, Srinivasan K. Tissue distribution & elimination of capsaicin, piperine & curcumin following oral intake in rats. Indian J Med Res. 2010;131:682–691. [PubMed] [Google Scholar]
- 215.Stojković D, Drakulić D, Gašić U, Zengin G, Stevanović M, Rajčević N, Soković M. Ononis spinosa L, an edible and medicinal plant: UHPLC-LTQ-Orbitrap/MS chemical profiling and biological activities of the herbal extract. Food Funct. 2020;11:7138–7151. doi: 10.1039/d0fo01595d. [DOI] [PubMed] [Google Scholar]
- 216.Stojković D, Drakulić D, Schwirtlich M, Rajčević N, Stevanović M, Soković MD, Gašić U. Extract of Herba Anthrisci cerefolii: Chemical Profiling and Insights into Its Anti-Glioblastoma and Antimicrobial Mechanism of Actions. Pharmaceuticals (Basel) 2021;14:55. doi: 10.3390/ph14010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sayed-Ahmed MZ MH, Elsherbini MM, Syed NK, Shoeib SM. Oncolytic Viruses: A gene Therapy for Treatment of Cancer in Companion Animals. Health Sci J. 2018;12:579. [Google Scholar]
- 218.Sostoa J, Dutoit V, Migliorini D. Oncolytic Viruses as a Platform for the Treatment of Malignant Brain Tumors. Int J Mol Sci. 2020;21:7449. doi: 10.3390/ijms21207449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Stavrakaki E, Dirven CMF, Lamfers MLM. Personalizing Oncolytic Virotherapy for Glioblastoma: In Search of Biomarkers for Response. Cancers (Basel) 2021;13:614. doi: 10.3390/cancers13040614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhu Z, Gorman MJ, McKenzie LD, Chai JN, Hubert CG, Prager BC, Fernandez E, Richner JM, Zhang R, Shan C, Tycksen E, Wang X, Shi PY, Diamond MS, Rich JN, Chheda MG. Zika virus has oncolytic activity against glioblastoma stem cells. J Exp Med. 2017;214:2843–2857. doi: 10.1084/jem.20171093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Cockle JV, Rajani K, Zaidi S, Kottke T, Thompson J, Diaz RM, Shim K, Peterson T, Parney IF, Short S, Selby P, Ilett E, Melcher A, Vile R. Combination viroimmunotherapy with checkpoint inhibition to treat glioma, based on location-specific tumor profiling. Neuro Oncol. 2016;18:518–527. doi: 10.1093/neuonc/nov173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Perry J, Okamoto M, Guiou M, Shirai K, Errett A, Chakravarti A. Novel therapies in glioblastoma. Neurol Res Int. 2012;2012:428565. doi: 10.1155/2012/428565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Schaefer T, Lengerke C. SOX2 protein biochemistry in stemness, reprogramming, and cancer: the PI3K/AKT/SOX2 axis and beyond. Oncogene. 2020;39:278–292. doi: 10.1038/s41388-019-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Agnihotri S, Burrell KE, Wolf A, Jalali S, Hawkins C, Rutka JT, Zadeh G. Glioblastoma, a brief review of history, molecular genetics, animal models and novel therapeutic strategies. Arch Immunol Ther Exp (Warsz) 2013;61:25–41. doi: 10.1007/s00005-012-0203-0. [DOI] [PubMed] [Google Scholar]