SUMMARY
The human genome encodes tens of thousands circular RNAs (circRNAs) with mostly unknown functions. Circular RNAs requires internal ribosome entry sites (IRES) if they are to undergo translation without 5’ cap. Here, we develop a high-throughput screen to systematically discover RNA sequences that can direct circRNA translation in human cells. We identify over 17,000 endogenous and synthetic sequences as candidate circRNA IRES. 18S rRNA complementarity and a structured RNA element positioned on the IRES are important for driving circRNA translation. Ribosome profiling and peptidomic analyses show extensive IRES-ribosome association, hundreds of circRNA-encoded proteins with tissue -specific distribution, and antigen presentation. We find that circFGFR1p, a protein encoded by circFGFR1 that is down regulated in cancer, functions as a negative regulator of FGFR1 oncoprotein to suppress cell growth during stress. Systematic identification of circRNA IRES elements may provide important links among circRNA regulation, biological function, and disease.
eTOC Blurb
Chen et al. identified thousands RNA elements that initiate circRNA translation and discovered key sequence and structural features for their activity. They identify hundreds of circRNA-encoded proteins, and characterized circFGFR1 as a dominant negative FGF receptor that suppresses proliferation during cell stress.
INTRODUCTION
Over the past decade, deep sequencing and computational analysis suggested circular RNAs (circRNAs) are a large class of RNAs in mammalian cells that play important roles in various biological processes (Hansen et al., 2013; Jeck and Sharpless, 2014; Jeck et al., 2013; Memczak et al., 2013; Salzman et al., 2013; Salzman et al., 2012). Disruption of circRNA expression was found to be associated with human diseases, such as Alzheimer’s disease, diabetes, and cancers (Bachmayr-Heyda et al., 2015; Dou et al., 2016; Lukiw, 2013; Panda et al., 2016; Shang et al., 2016; Wang et al., 2016; Xu et al., 2015; Zhong et al., 2016). While most of the studies demonstrated circRNAs functioning in a non-coding manner, such as sponging miRNAs (Ashwal-Fluss et al., 2014; Hansen et al., 2013; Kulcheski et al., 2016; Memczak et al., 2013; Xu et al., 2015; Zheng et al., 2016), regulating mRNA splicing machinery (Liu et al., 2019), sequestering RNA-binding proteins (RBPs), regulating RBP interaction (Ashwal-Fluss et al., 2014; Memczak et al., 2013), and activating immune response (Chen et al., 2019; Chen et al., 2017; Li et al., 2017b; Liu et al., 2019), emerging evidence suggests that circRNAs can function through their encoded proteins (Legnini et al., 2017; Liang et al., 2019; Pamudurti et al., 2017; Yang et al., 2018; Zhang et al., 2018a; Zhang et al., 2018b; Zheng et al., 2019). Proteins translated from circRNAs regulate cell proliferation, differentiation, migration, and myogenesis (Legnini et al., 2017; Liang et al., 2019; Pamudurti et al., 2017; Yang et al., 2018; Zhang et al., 2018a; Zhang et al., 2018b; Zheng et al., 2019). Dysregulation of circRNA-encoded proteins was found to cause tumorigenesis of certain cancers, suggesting circRNA-encoded proteins as important links between circRNA and diseases (Liang et al., 2019; Yang et al., 2018; Zhang et al., 2018a; Zhang et al., 2018b; Zheng et al., 2019). Understanding the mechanism of circRNA translation may provide some valuable clinical insights. However, the molecular mechanism behind circRNA translation remains largely unknown. Given the important role circRNA translation plays in various cellular functions, it is imperative that we determine the key elements regulating circRNA translation.
Because circRNA is generated by spliceosome-mediated head-to-tail joining of pre-mRNAs (Chen et al., 2015; Chen and Shan, 2015; Vicens and Westhof, 2014), it does not contain the 5’ cap for cap-dependent translation (Both et al., 1975; Shafritz et al., 1976). Thus, circRNA translation requires an internal ribosome entry site (IRES) sequence on the RNA to initiate cap-independent translation (Chen et al., 2016; Granados-Riveron and Aquino-Jarquin, 2016; Jang et al., 1988; Legnini et al., 2017; Li et al., 2017a; Pamudurti et al., 2017; Pelletier and Sonenberg, 1988; Schneider et al., 2016; Wang and Wang, 2015). Most of the endogenous circRNAs are exported to the cytoplasm (Huang et al., 2018; Jeck et al., 2013; Salzman et al., 2012) and introducing IRES on synthetic circRNAs is sufficient to produce proteins (Chen and Sarnow, 1995), suggesting that endogenous circRNAs containing the IRES may have translation potential. Systematic identification of the RNA sequence and structural elements for facilitating circRNA translation on the IRES can further help us determine the potential endogenous protein-coding circRNAs. Here, we developed a high-throughput reporter assay to systematically screen and quantify the IRES activity of the RNA sequence that can facilitate circRNA translation. With this assay, we identified the regulatory elements in the primary and secondary structure on circRNA IRES that are important for facilitating circRNA translation. By mapping the circRNA IRES sequences to the human endogenous circRNA, we determined the putative protein-coding circRNAs and the peptide sequences encoded by these circRNAs. Moreover, mass spectrometry (MS) peptidomic analysis further validates that the identified circRNA IRES can indeed initiation translation of endogenous circRNAs and lead to detectable protein production. Finally, we characterized a circRNA-encoded protein, circFGFR1p, that functions as a negative regulator of FGFR1 through dominant negative mechanism to suppress cell growth under stress conditions. Together, our findings provide a resource to recognize and manipulate circRNA translation, which may provide valuable insights into both circRNA and proteomic research.
RESULTS
Systematic identification of RNA sequences that facilitate cap-independent circRNA translation
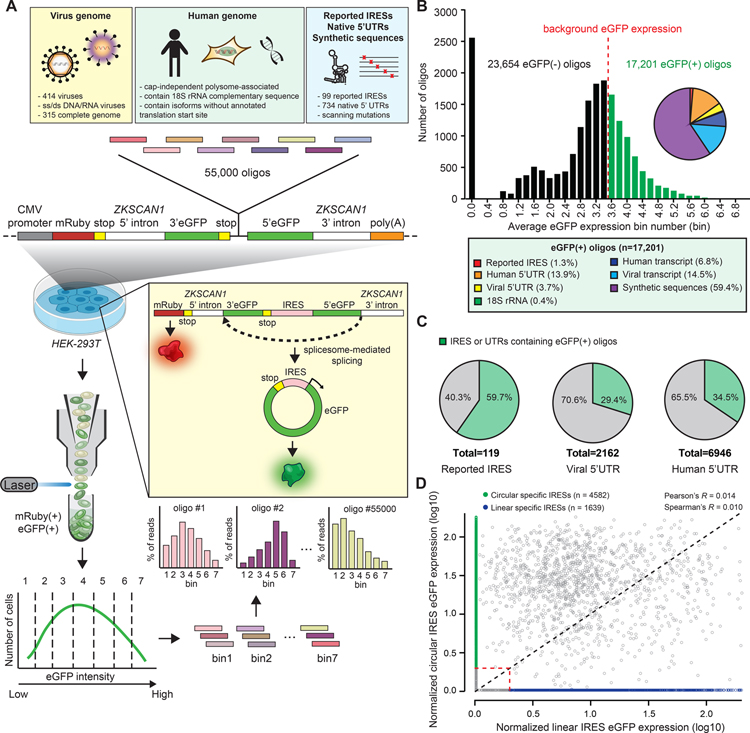
To systematically identify the RNA sequence that can facilitate cap-independent translation on circRNAs, we developed an oligo-split-eGFP-circRNA reporter construct to screen and quantify the cap-independent translation activity of synthetic oligonucleotide inserts (“oligos” hereafter) on circRNA in a high-throughput manner (Figure 1A, Figure S1). We then cloned a synthetic oligo library into the construct to drive the expression of eGFP reporter. The library contained 55,000 oligos of native and synthetic sequences (Figure 1A) (library design is detailed in Weingarten-Gabbay et al., 2016). We captured 40,855 out of 55,000 oligos from the library (~74.3%). To quantify the eGFP expression level of each oligo, we calculated the mean weighted rank distribution of the reads across the bins. (Figure 1B). Our screening assay revealed three groups of oligos - a group (~2,500) of oligos which showed no eGFP expression (eGFP expression (bin) = 0.0), and two groups of oligos showing a bimodal distribution of eGFP expression (eGFP expression (bin) = 0.8–2.2 and 2.4–7.0, respectively). To determine the oligos with cap-independent translation activity (eGFP(+) oligos), we calculated the weighted rank distribution of the eGFP intensity of the cells transfected with no-IRES-inserted reporter plasmid as the background eGFP expression (Figure S2D). With this approach, we identified 17,201 eGFP(+) oligos from our screening assay (Figure 1B, Table S1).
Figure 1. High-throughput identification of RNA sequences that can facilitate circRNA cap-independent translation.
(A) A schematic overview of the high-throughput split-eGFP circRNA reporter screening assay for identifying circRNA IRES. (B) The eGFP expression distribution of captured synthetic oligos. Pie chart: the composition of different categories among eGFP(+) oligos. (C) Quantification of the percentage of captured eGFP(+) oligo sequences from different orgin. (D) Normalized eGFP expression of each captured oligo in the screening assay performed on the circular RNA or the linear RNA system. Red dashed lines: normalized eGFP expression threshold.
To validate our identified eGFP(+) oligos, all reported IRES sequences from the IRESite database were included in the synthetic oligo library (Mokrejš et al., 2009; Weingarten-Gabbay et al., 2016). Our assay successfully identified 71 of 119 reported IRESs as having circRNA translation activity (~59.7%), as well as identifying the functional region within some long IRESs such as the encephalomyocarditis virus (EMCV) IRES. However, our library, which is limited to ~200 nt length sequences, cannot detect some long complex IRESs such as the ectropis obliqua picorna-like virus (EoPV) IRES (Figure 1C, Figure S3A–S3C, Table S2). Because a previous study has utilized the same synthetic oligo library on a linear bicistronic eGFP reporter screening assay to identify oligos with cap-independent translation activity on linear RNA (Weingarten-Gabbay et al., 2016), we were able to compare the cap-independent translation activity of each oligo sequence on linear RNA and circular RNA respectively. We found that a large number of the oligos showing cap-independent translation activity in both linear and circular RNA screening system (n = 7,424) (Figure S4A). However, there was little correlation between the overall IRES activity of circular vs. linear RNA (Figure 1D). Interestingly, we also captured some oligos showing IRES activity specifically in either linear or circular screening system (linear IRES (n = 1,639) and circular IRES (n = 4,582), respectively) (Figure 1D, Figure S4A, Table S3). Together, these results demonstrate that our high-throughput screening assay utilizing the circRNA reporter construct is able to systematically identify the RNA sequences harboring IRES activity that can facilitate cap-independent translation on circRNA.
Synthetic circRNAs containing the identified eGFP(+) oligo sequences are actively translated
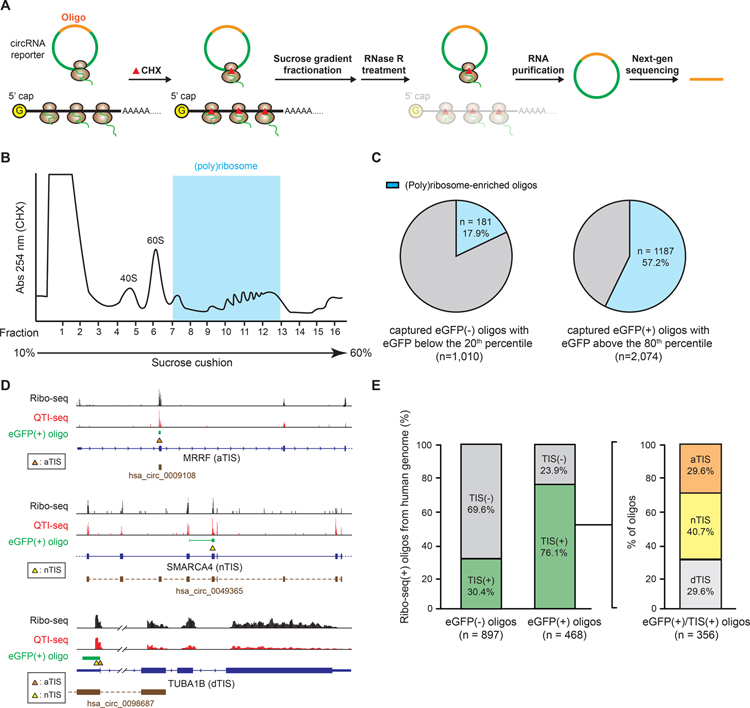
We used polysome profiling (Chasse et al., 2016; Chen et al., 2011; Del Prete et al., 2007; Kang and Pomerening, 2012; Kronja et al., 2014; Kuersten et al., 2013; Larsson et al., 2013) to examine whether circRNAs containing the identified eGFP(+) oligo sequence are actively translated and engaged on the ribosome (Figure 2A and 2B, Figure S5A). To avoid the result being confounded by the weakly translated circRNAs (Methods S1), we compared the ratio of (poly)ribosome-enriched oligos among the eGFP(+) oligos with eGFP expression above the 80th percentile and the eGFP(−) oligos with eGFP expression below the 20th percentile. The eGFP(+) oligos are more enriched in the (poly)ribosome fractions (57.2%) than the eGFP(−) oligos (17.9%) (Figure 2C). This result suggests that the circRNAs containing the eGFP(+) oligo sequences are more actively translated. We further examined published QTI-seq data (Gao et al., 2015), and discovered that among the oligos derived from the human genome with Ribo-seq coverage, the majority of the eGFP(+) oligos (~76%) overlaps with the identified translation initiation sites (TIS) on human transcripts (TIS(+) oligos), while only 30% of the eGFP(−) oligos are TIS(+) (Figure 2E, Figure S6A), suggesting that the eGFP(+) oligos are more likely to initiate translation at those TIS than the eGFP(−) oligos. Interestingly, we identified three types of eGFP(+)/TIS(+) oligos - annotated TIS (aTIS), non-annotated TIS (nTIS), and dual TIS (dTIS) (Figure 2E). These different types of TIS(+) oligos may suggest that the oligos utilized different mechanisms for initiating translation (Methods S2). Interestingly, we found 15 eGFP(+)/TIS(+) oligos are located within the genomic regions that encode annotated circRNAs (Figure 2D), which suggests that these circRNA may utilize the TIS on the oligo to initiate translation. Together, the results above provide strong evidence supporting that our screening assay can identify the oligo sequences that facilitate cap-independent translation activity on circRNAs.
Figure 2. CircRNAs containing eGFP(+) oligo have higher cap-independent translation activity.
(A) A schematic of the circRNA polysome profiling for translating circRNAs. (B) (Poly)ribosome fractionations of cells transfected with oligo library split-eGFP circRNA reporter followed by CHX treatment. (C) Quantification of the percentage of (poly)ribosome-enriched oligos of captured eGFP(−) or eGFP(+) oligos. (D) Sequencing reads from Ribo-seq and QTI-seq plotted on the genes showing eGFP(+) oligos harboring aTIS, nTIS, and dTIS with overlapping annotated circRNAs (brown segments). (E) Quantification of the percentage of eGFP(−) or eGFP(+) oligos harboring no TIS (TIS(−)) (left) or more than one TIS (TIS(+)) (right), and the percentage of aTIS, nTIS, or dTIS oligos among eGFP(+)/TIS(+) oligos.
Identification of 18S rRNA complementary sequence that facilitates circRNA translation
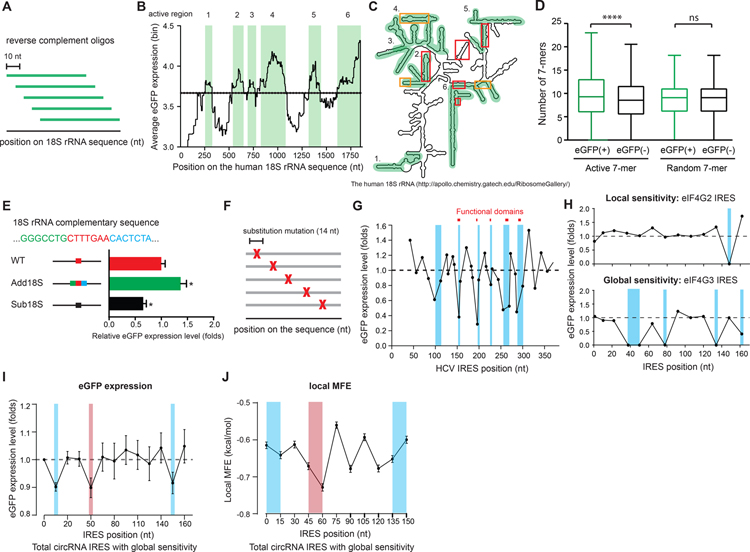
Watson-Crick base-pairing between IRES and 18S ribosomal RNA (18S rRNA) has been demonstrated to facilitate cap-independent translation of linear mRNAs (Malygin et al., 2013; Meng et al., 2010; Nicholson et al., 1991; Owens et al., 2001; Quade et al., 2015; Zeenko and Gallie, 2005). With the sliding window method, we identified six “active regions” on the 18S rRNA (Figure 3A–3C, Table S4, Method S3). Since the 7-mers derived from active region 4 have been shown to be enriched in reported IRES for linear RNAs (Weingarten-Gabbay et al., 2016), we extracted all 7-mers from the sequence complementary to the active regions of 18S rRNA (active 7-mers), and compared the number of active 7-mers harbored by the eGFP(+) and eGFP(−) oligos respectively. Indeed, we found that eGFP(+) oligos have higher enrichment of active 7-mers than eGFP(−) oligos (Figure 3D). In contrast, when we compared the random 7-mers which do not overlap with the active 7-mers between eGFP(+) and eGFP(−) oligos, we did not find any significant difference (Figure 3D), suggesting that the higher enrichment of active 7-mers in eGFP(+) oligos we observed here is specific to the 18S rRNA complementary sequence. To further validate the results, we perturbed the 18S rRNA complementarity of the IRES by either substituting the 18S rRNA complementary sequence with a random 7-mer or adding the flanking 18S rRNA complementary sequence to the IRES and measured their circRNA translation activity. We indeed observed diminished IRES activity with lower 18S rRNA complementarity on the IRES, and conversely programmed stronger IRES activity with higher 18S rRNA complementarity added to the IRES (Figure 3E). These results suggest that the circRNA IRES containing RNA sequence complementary to the active regions on 18S rRNA is one of the regulatory elements that can facilitate circRNA translation.
Figure 3. 18S rRNA complementary sequence on the IRES facilitates circRNA translation.
(A) A schematic of the sliding-window approach for mapping the active regions on the human 18S rRNA. (B) Quantification of the mean eGFP expression of the oligos overlapping with the corresponding position across the human 18S rRNA. Dashed line: background eGFP expression. Green shaded: active regions on the 18S rRNA. (C) An illustration of the secondary structure of human 18S rRNA showing the active regions (green) and reported mRNA (red) or IRES RNA (orange) contact regions. (D) Quantification of the number of the 18S rRNA active 7-mers or the random 7-mers harbored by eGFP(+) or eGFP(−) oligos plotted on a Tukey box-plot. (E) Quantification of the IRES activity for the oligo with higher or lower 18S rRNA complementarity. Error bar: SEM. (F) A schematic of the synthetic oligos for systematic scanning mutagenesis. (G) The eGFP expression of each oligo containing the random substitution mutation at the corresponding position on HCV IRES. The eGFP expression for each oligo was normalized to the mean eGFP expression of all the oligos on the HCV IRES. (H) Examples of circRNA IRES with local and global sensitivity identified by scanning mutagenesis. Blue shaded: the identified essential elements on the IRES. (I) The mean eGFP expression of all the circRNA IRES oligos with global sensitivity at each mutation position across the IRES (blue: 5–15 nt and 135–165 nt; red: 40–60 nt). (J) Quantification of the local MFE in a 15 nt sliding window on the IRES (blue: 5–15 nt and 135–165 nt; red: 40–60 nt).
Identification of essential elements on circRNA IRES with scanning mutations
We next employed scanning mutagenesis to define the essential elements on circRNA IRES (Figure 3F). We used hepatitis C virus (HCV) IRES to validate scanning mutagenesis assay and we indeed observed that the known functional domains on HCV IRES co-localized with the mutation positions that have dramatically reduction in IRES activity (Bhat et al., 2015; Lukavsky, 2009) (Figure 3G). By examining the identified circRNA IRES with scanning mutagenesis, we captured two classes of circRNA IRES – circRNA IRES with local sensitivity, which shows reduction in circRNA IRES activity only when a specific position in mutated (Figure 3H; top), and circRNA IRES with global sensitivity, which mutation in most positions can cause reduction in IRES activity (Figure 3H; bottom). By examing all the captured circRNA IRESs with global sensitivity, we identified three regions (5–15 nt, 40–60 nt, and 135–165 nt) on the IRES, where when the mutation hit these regions, the IRES activity decreased significantly (Figure 3I), suggesting that these regions may harbor the key elements for circRNA IRES activity. We calculated the local MFE along the circRNA IRESs with global sensitivity in a 15 nt non-overlapping window and discovered that local MFE of the 40–60 nt region shows significantly lower local MFE (Figure 3J), suggesting that it may harbor the local structural element that facilitates circRNA translation. The results show that our scanning mutagenesis assay can determine circRNA IRES with local or global sensitivity, and systematically identify essential elements that are required for circRNA IRES activity.
Identification of structured RNA element that facilitates circRNA translation
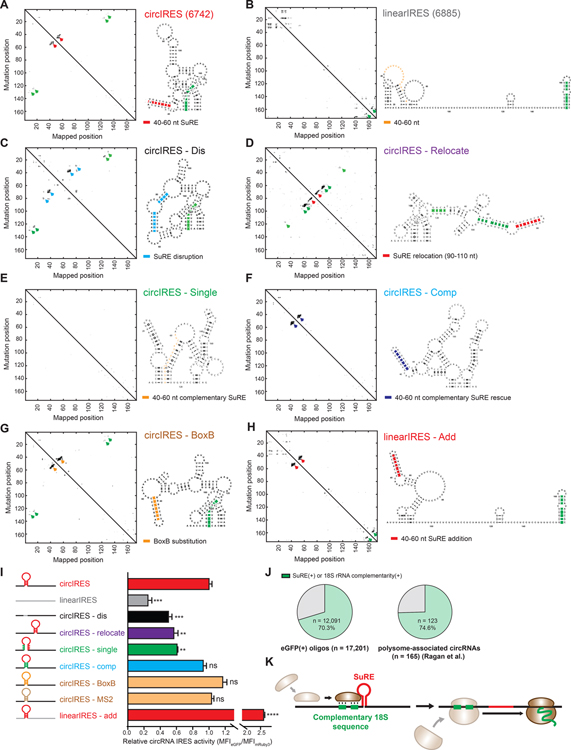
By comparing the screening results of our circRNA reporter with the previous linear RNA reporter using the same synthetic oligo library (Weingarten-Gabbay et al., 2016), we identified two distinct groups of oligos that harbor IRES activity on either linear or circular RNA specifically (linear IRES and circular IRES, respectively). We first examined their primary sequences, and discovered that circular IRES contain higher GC-content and lower MFE than linear IRES (Figure S7A), while the number of AUG, the Kozak consensus sequence, and the m6A motif showed no difference (Figure S7B–S7D), suggesting that structural element may play a role in facilitating circular IRES translation activity (Gruber et al., 2008; Hofacker et al., 1994; Jaeger et al., 1989). We then characterized the IRES secondary structure with M2-seq of four circular IRES and linear IRES (Cheng et al., 2017) (Figure S7E). The M2-seq revealed that the circular IRES are in general more structured than linear IRES. (Figure 4A and 4B, Figure S7F and S7G). Among all four circular IRES we examined, we observed that they all contain a stem-loop structured RNA element (SuRE) on the IRES at a distinct position (40–60 nt position from the first nucleotide (+1) of the IRES), while all the linear IRES we examined did not contain such a structure at this position. In line with our previous systematic scanning mutation profiling, which also suggests that this distinct position on IRES contains structural element for facilitating circRNA translation (Figure 3I and 3J), we proposed that the 40–60 nt SuRE on the IRES can facilitate cap-independent translation activity of circular IRES.
Figure 4. 40–60 nt SuRE on the IRES can facilitate circular IRES activity.
(A)-(H) The secondary structure of the mutated IRESs determined by M2-seq. Red arrowheads: 40–60 nt SuRE. CircIRES-dis: circular IRES with the SuRE disrupted by sequence substitution. CircIRES-relocate: circular IRES with the SuRE relocated to 90–110 nt region. CircIRES-single and circIRES-comp: circular IRES with single complementary mutations and compensatory double complementary mutations, respectively. circIRES-BoxB: circular IRES with the SuRE substituted by BoxB stem-loop. linearIRES-add: linear IRES with 40–60 nt region substituted by the 40–60 nt SuRE on the circular IRES. (I) Quantification of the IRES activity for each mutated IRES normalized to the linear IRES. An unpaired two-sample t-test relative to the linear IRES was performed. Error bar: SEM. (J) Quantification of the percentage of the eGFP(+) oligos (left) and endogenous translated circRNAs (right) harboring 18S rRNA complementarity or the SuRE element. (K) An illustration of two key regulatory elements facilitating circRNA translation.
To test our hypothesis, we disrupted the 40–60 nt SuRE on the circular IRES by substituting it with the sequence extracted from the same position on the linear IRES and found that it indeed leads to a reduced IRES activity (Figure 4C nd 4I). Moreover, we relocated the SuRE to 90–110 nt position by swapping the sequences of these two regions on the IRES, and we also observed a decreased translation activity of the IRES, suggesting the element is position-dependent (Figure 4D and 4I). To further validate that the SuRE is structural dependent rather than sequence-dependent, we performed compensatory mutagenesis of the SuRE. Specifically, we mutated each of the seven base pairs on the stem region of the SuRE to disrupt its duplex structure, and observed lower translation activity of the IRES (Figure 4E and 4I); The translation activity of the IRES can be rescued by compensatory double complementary mutations to restore each of the seven base pairs on the stem region (Figure 4F and 4I). Moreover, when we substituted the SuRE with MS2 or BoxB, which has a similar RNA structure, we observed similar IRES activity (Figure 4G and 4I), suggesting that the SuRE is indeed structure-dependent rather than sequence-dependent. Finally, we were able to convert the linear IRES into a circular IRES by transplanting the 40–60 nt SuRE from the circular IRES to the linear IRES (Figure 4H and 4I).
Together, the results above along with the 18S rRNA profiling suggest that two key regulatory elements on circRNA IRES, the 18S rRNA complementarity and 40–60 nt SuRE on the IRES, can facilitate cap-independent translation on circRNA. In line with our model, among the eGFP(+) oligos we identified and the polysome-associated circRNAs (translating circRNAs) captured in HEK-293 cells (Ragan et al., 2019), a large proportion of them are 18S rRNA complementarity(+) or 40–60 nt SuRE(+) (Figure 4J). It suggests that these two regulatory elements are common features among the exogenous and endogenous translating circRNAs. Nevertheless, we did not observe a preferential localization of the 18S complementary sequence to the 5’ or 3’ of the SuRE (Figure S6C); hence we speculate that the SuRE may cause a pause for RNA unwinding, increasing the chance for the 18S complementary sequence on the IRES to interact with the 18S rRNA on the ribosome to facilitate circRNA translation (Figure 4K).
IRES elements facilitate endogenous circRNA translation initiation
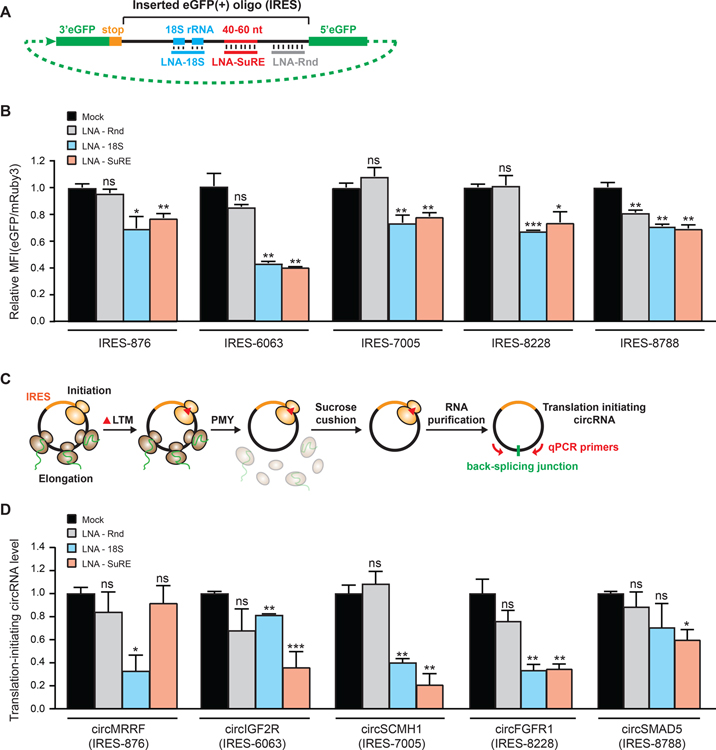
To validate if the 18S rRNA complementarity and the 40–60 nt SuRE can facilitate circRNA translation, we utilized locked nucleic acid (LNA) to disrupt these key elements on the IRES (Tuplin et al., 2015). We designed anti-sense LNAs targeting the 18S rRNA complementary sequence (LNA-18S), 40–60 nt SuRE (LNA-SuRE), or random position (LNA-Rnd) of 5 identified IRESs (Figure 5A). We discovered that co-transfecting LNA-18S or LNA-SuRE can indeed disrupt the cap-independent translation activity of all 5 IRESs, while most of the LNA-Rnd co-transfection did not affect the translation activity of the IRES (Figure 5B and Figure S8A). The result suggests that disrupting the key elements on the IRES with LNA can affect the cap-independent translation activity of the exogenous reporter circRNA. To further examine if the key elements can also facilitate endogenous circRNA translation, we transfected the cells with the corresponding anti-sense LNA and quantified translating circRNAs by QTI method (Gao et al., 2015; Ingolia et al., 2012) (Figure 5C). We found that disrupting the key regulatory elements by LNA-18S or LNA-SuRE on the IRES of the endogenous circRNA can in general cause decreased translation activity of the endogenous circRNAs, while all the LNA-Rnd has no effect on endogenous circRNA tranlsation (Figure 5D). Since QTI method specifically captured the RNAs that are at the initiation stage of translation, it suggested that the decrease of endogenous circRNA translation we observed upon LNA transfection came from the decrease in translation initiation. We further validated the results by quantifying the protein level produced from the endogenous circRNAs (Figure S8C, Methods S5). The fact that a few of the LNAs did not affect the translation activity of the endogenous circRNAs (LNA-SuRE of IRES-876; LNA-18S of IRES-8788) indicates that there may be other regulatory elements on the IRES that can independently facilitate endogenous circRNA translation. Nevertheless, the results above suggest that the key elements such as the 18S rRNA complementarity and the 40–60 nt SuRE are important for facilitating translation initiation of endogenous circRNAs.
Figure 5. IRES elements facilitate translation initiation of endogenous circRNAs.
(A) A schematic of disrupting the key regulatory elements on the IRES by anti-sense LNAs targeting specific regions. (B) Quantification of the normalized eGFP fluorescence intensity of the cells co-transfected with the corresponding LNA and the reporter plasmid carrying the corresponding IRES. The number represents the index number of the oligo. (C) A schematic of QTI-qRT-PCR quantification of the level of translation-initiating endogenous circRNAs. (D) Quantification of the translation-initiating RNA level of the human endogenous circRNAs containing the corresponding IRES upon corresponding LNA disruption. The circRNA level was normalized to the GAPDH mRNA. (B) and (D): An unpaired two-sample t-test relative to mock transfection was performed. Error bar: SEM.
Identification of potential endogenous protein-coding circRNAs
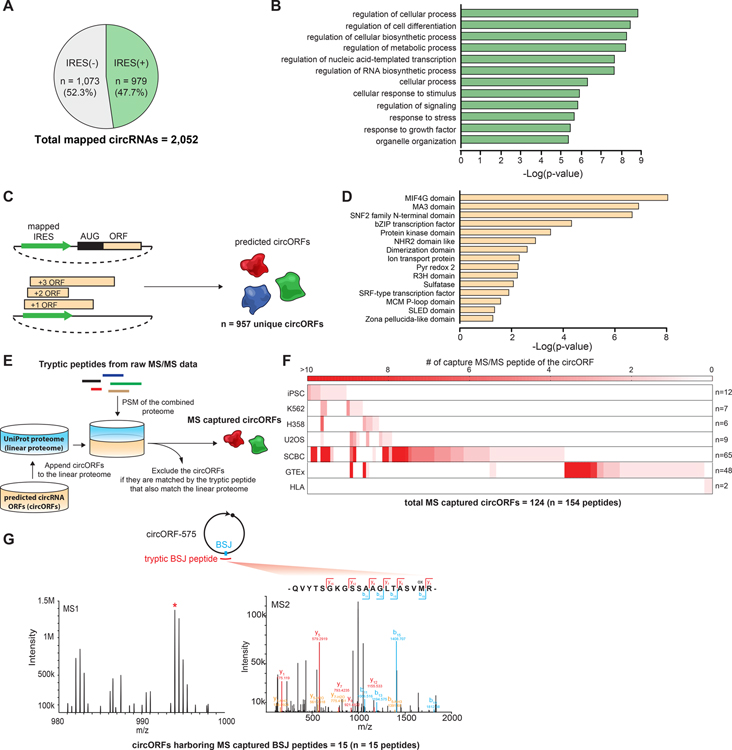
Introducing synthetic IRES on circRNAs is sufficient to initiate cap-independent translation (Chen and Sarnow, 1995), suggesting that endogenous circRNAs harboring the active circular IRES may have protein-coding potential. Thus, to determine the potential protein-coding circRNAs, we mapped the eGFP(+) oligo sequences captured in our screens to the human circRNA database (circBase) (Glazar et al., 2014) to identify the endogenous circRNAs which harbor the active IRES. Out of 2,052 endogenous circRNAs containing the sequences from the oligo library, 979 circRNAs (~48%) contain one or more eGFP(+) oligo sequences (IRES(+) circRNAs) (Figure 6A, Figure S9A, Table S5). These circRNAs were generated from various parent genes which showed a fairly homogenous distribution across the genome (Figure S9B). We examined the cancer-specific circRNA database (CSCD) (Xia et al., 2018) and discovered that 294 of 979 IRES(+) circRNAs (~30%) are expressed specifically in either non-transformed cell lines (n = 141 cell lines) or cancer cell lines (n = 87 cell lines across 19 cancer types), respectively (Figure S9C), indicating their potential association with cancer progression. In line with our observation, many oncogenes possess transcript isoforms that encode circRNAs, and dysregulation of several circRNA-encoded proteins was found to enhance tumorigenesis and metastasis in several cancer types (Liang et al., 2019; Yang et al., 2018; Zhang et al., 2018a; Zhang et al., 2018b; Zheng et al., 2019).
Figure 6. Identification of putative endogenous circRNA-encoded proteins.
(A) Quantification of the percentage of IRES-mapped human endogenous circRNAs. (B) Top 12 represented biological processes from GO term analysis that are enriched in the parent genes of IRES(+) circRNAs. (C) A schematic of generating the putative endogenous circORF list. (D) Top 15 represented conserved motifs from Pfam analysis that are enriched in the predicted circORFs. (E) A schematic of peptidomic validation of the putative circORFs. (F) The heat map showing the number of the unique MS/MS peptides detected in different peptidomic datasets for each of the peptidomic detected circORF. The number on the right indicates the total number of different circORFs detected in the corresponding dataset. (G) The MS1 and MS2 spectra of a representative tryptic BSJ peptide captured from circORF_575.
We discovered that most IRES(+) circRNAs contain only one IRES and most eGFP(+) oligos map to only one circRNA (Figure S9D and S9E), suggesting a specific one-to-one relationship between these IRES(+) circRNAs and the proteins encoded by them. Thus, circRNA IRES would have been difficult, to discover by comparative sequence analysis across circRNAs. The result also suggests that circRNA IRES activity may require long RNA sequences which are more likely to show up once per transcript, rather than very short or repetitive sequences. Furthermore, we discovered that the position of mapped eGFP(+) oligos on circRNAs is most frequently near the back-splicing junction of the circRNA,. This result suggests that the cap-independent translation activity of the IRES on the circRNA is back-splicing dependent – the IRES element or its downstream open reading frame (ORF) is only assembled upon back-splicing (Figure S9F), where such phenomenon has been reported in the IRES of some protein-coding circRNAs (Legnini et al., 2017). Finally, gene ontology (GO) analysis of the parent genes of these circRNAs suggested that they are enriched in stress-response and translation regulation (Figure 6B), where these two functions have also been discovered in many circRNA-encoded proteins (Legnini et al., 2017; Liang et al., 2019; Pamudurti et al., 2017; Yang et al., 2018; Zhang et al., 2018a; Zhang et al., 2018b; Zheng et al., 2019). Above all, these results demonstrated that with our identified eGFP(+) oligo sequences, we are able to determine potential endogenous protein-coding circRNAs.
Identification of potential endogenous circRNA-encoded peptides
Our data can map the position of eGFP(+) oligo sequences on the circRNA, which allows us to determine the regions on circRNAs where the translation start sites may be located. With IRES mapping and ORF analysis, we generated a list of 957 predicted peptide sequences encoded by human endogenous circRNAs (circORFs) (Figure 6C, Table S6). We discovered that some circRNAs contain the IRES sequence overlapping the translated region of the ORF (n = 457; ~48%) (Figure S9G). The IRES-overlapping ORF has been observed in some endogenous circRNA-encoded proteins, which suggests some regulatory mechanisms may exist between the initiation and the elongation of circRNA translation (Legnini et al., 2017; Zhang et al., 2018a). Interestingly, some circRNAs contain in-frame ORFs without stop codons (n = 82; ~18%), forming recursive ORFs which may be a mechanism for amplifying the expression level of the circORFs (Figure S9H and S9I).
We then characterized the general functions of the potential circORFs by searching for conserved motifs on the predicted peptide sequences with Pfam analysis. The top motifs are DNA binding motifs, translation elongation factors binding motifs, protein kinase domains, and protein dimerization domains (Figure 6D), suggesting that circORFs may play roles in regulating various biological functions including signal transduction, transcription, and translation. The size of most of these potential circORFs was small (<100 amino acids) (Figure S9J), which suggests that the majority of them may be the truncated forms of the proteins generated from their parent linear transcripts. We further examined the short open reading frame (sORFs) database (Olexiouk et al., 2017), which contains the peptide sequences (<100 amino acids) from the identified sORFs aggregated by multiple ribosome profiling studies, to validate the potential circORFs. In comparison to traditional ORF analysis, which takes all possible translation initiation locations our IRES-mapped can identify endogenous circORFs more efficiently where ~33% of the potential circORFs can be captured by the sORF studies (Figure S9K).
We further performed peptidomic analyses on the tandem mass spectrometry (MS/MS) datasets to validate the endogenous expression of the circORFs (Figure 6E). We captured 124 circORFs harboring MS-matched unique tryptic peptides, including 15 circORFs harboring MS-matched tryptic peptides spanning across the circRNA back-splicing junction (BSJ) (Figure 6F and 6G, Table S6). Because circRNAs are expressed in a tissue-specific and cell line-specific manner (Rybak-Wolf et al., 2015), to accurately estimate the circORF discovery rate (the number of circORF detected in each dataset), we consider the IRES-containing circRNAs expressed in the corresponding cell line as the denominator. By examing the IRES-containing circRNAs expressed in human iPSC (Lei et al., 2018) and K562 (Okholm et al., 2020), we observe circORF discovery rates much higher than the 1% false-discovery rate (FDR) we set for the PSM (iPSC: 12/115, 10.4%; K562: 7/180, 3.9%). To further validate the circORF are expressed in K563 and U2OS, we perform parallel reaction monitoring-MS (PRM-MS) and confirmed the detection of 4 of 7 unique circORF peptides found in MS/MS peptidomics (Figure S10, Table S6). These peptidomic data provide evidence supporting that the circORFs are indeed expressed endogenously.We further analyzed the human leukocyte antigen I (HLA1)-associated peptidomics (Bassani-Sternberg et al., 2015) and identify two HLA1-associated circORFs (Figure 6F). In silico HLA1 binding predictor NetMHC4.1 analysis (Reynisson et al., 2020) suggests that these two circORFs are indeed strong HLA1 binder to the HLA1 variants expressed in the cell line (Table S6). The result indicates a new functional role of circORFs that some of the circORFs may enter the HLA-I presentation pathway and contribute to the antigen repertoire.
Given the limitations of circORF peptidomics (Methods S6), we interpret identification of circORFs as positive validation; the absence of detection in MS proteomic data does not rule out the translational potential for circRNA candidates. In line with these consideration, when we applied the same limitations to proteins encoded by lowly expressed mRNAs, matching for expression level and down-sampled reference space, we found that current peptidomic data can only recover ~5% peptides of the protein encoded by the mRNA with the same RPKM as the mean circRNA RPKM (Figure S9L and S9M). The fact that we are able to validate ~13% (124 out of 957) circORFs with peptidomics supports the contention that circRNAs widely encode peptides similar to lowly expressed mRNAs. Together, these results suggest that we are able to build a putative circORF list using our circRNA IRES screening assay which can be validated by genomic and peptidomic analyses.
circFGFR1p suppresses cell proliferation under stress conditions through a dominant-negative regulation
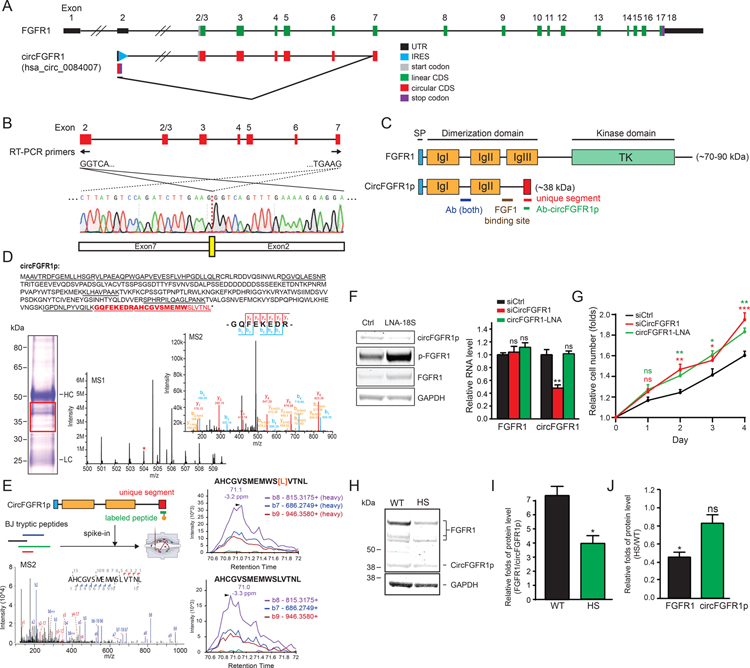
To evaluate the functions of the circORFs, we chose an example of a potential protein-coding circRNA (hsa_circ_0084007) and examine its encoded protein functions. The circRNA is generated from exon 7 to exon 2 back-splicing of Fibroblast Growth Factor Receptor 1 (FGFR1) transcript (Glazar et al., 2014; Salzman et al., 2013); hence we suggest the name circFGFR1 and circFGFR1p for this circRNA and its encoded protein, respectively. CircFGFR1 harbors an IRES with strong translation activity in our screening assay (top 2%), located in the 5’ UTR region of FGFR1 (Figure 7A). ORF analysis using the immediate downstream AUG revealed that the back-splicing generates a frameshift in exon 2 followed by a de novo stop codon within the IRES of circFGFR1, resulting in an ORF that partially overlaps with the IRES (circORF_949) (Figure 7A). To characterize circFGFR1 functions, we utilized a non-transformed human cell line, BJ fibroblasts, which has a diploid genome for better phenotypic analysis and high FGFR1 expression, for subsequent analyses.
Figure 7. CircRNA-encoded circFGFR1p suppresses cell proliferation under stress conditions.
(A) A schematic of FGFR1 and circFGFR1 transcript. (B) Sanger sequencing results detecting the back-splicing junction (yellow box) of circFGFR1. (C) A schematic of the conserved motifs on FGFR1 and circFGFR1p. Ab (both): the antibody which can detect both FGFR1 and circFGFR1p. Ab-circFGFR1p: custom circFGFR1p antibody. (D) A schematic of the peptides captured by IP-MS (underline) that matched the circFGFR1p unique region (red) and the region overlaping with FGFR1 (black). The antigen peptide of the circFGFR1p custom antibody was labeled in bold. Red box: the extracted region for IP-LC-MS/MS. (E) The MS2 and the top 3 rank PRM-MS transition ions spectra of the spike-in heavy isotope labeled peptide (top) and the BJ tryptic peptide (bottom) of circFGFR1p. [L]: heavy isotope labeled leucine. (F) Western blots showing circFGFR1p and FGFR1 protein level (Ab-both), and the quantification of FGFR1 and circFGFR1 RNA level by qRT-PCR of cells transfected with siRNA or LNA. An unpaired two-sample t-test relative to siCtrl was performed. (G) Quantification of cell proliferation in cells with the knockdown of siCircFGFR1 or circFGFR1-LNA with FGF1 addition. An unpaired two-sample t-test relative to siCtrl was performed. (H) Western blots showing FGFR1 protein and circFGFR1p level with or without the heat-shock. (I) Quantification of the Western blot of circFGFR1p protein level relative to FGFR1 (all isoforms) under normal (WT) and the heat-shock (HS) condition. An unpaired two-sample t-test relative to WT was performed. (J) Quantification of the Western blot showing the change of the protein level of FGFR1 and circFGFR1p under the heat-shock condition. Protein level is normalized to the GAPDH. A one-sample t-test relative to 1 was performed. The error bars in (F), (G), (I), and (J) represent SEM.
We first confirmed circFGFR1 expression in BJ cell by reverse transcription PCR (RT-PCR) and Sanger sequencing using the divergent primers flanking the back-splicing junction of the exon 2 and the exon 7 on circFGFR1 (Figure 7B). Analysis of the predicted circFGFR1p sequence indicates that it is a truncated form of FGFR1, which has an intact extracellular fibroblast growth factor 1 (FGF1) ligand-binding site, part of the dimerization domain (IgI, IgII, and partial N’ terminus of IgIII) but lacks the intracellular FGFR1 tyrosine kinase domain (Plotnikov et al., 2000) (Figure 7C). CircFGFR1p also harbor a unique region due to circFGFR1 back-splicing, where the peptide sequence of this region does not exist in the linear proteome database (UniProt) (Figure 7C). Western blotting using an antibody against the common region of circFGFR1p and FGFR1 (Ab-both) showed signals at the corresponding size of circFGFR1p (~38 kDa) and FGFR1 (70–90 kDa) (Figure 7H). ENCODE data demonstrated an absence of chromatin signature of promoters (H3K4me3) near circFGFR1p IRES (Figure S11A), suggesting the protein was not generated from the truncated linear transcript due to hidden promoters located in the exon 2 of FGFR1. In line with the observation above, the circFGFR1 IRES we identified (oligo index: 8228) does not display promoter activity from the linear RNA IRES reporter screens (score = 0) (Weingarten-Gabbay et al., 2016).
To validate endogenous circFGFR1p expression, we first generated a custom antibody against the unique region of circFGFR1p and performed immunoprecipitation liquid chromatography with tandem mass spectrometry (IP-LC-MS/MS). While no circFGFR1p peptides were detected in the IgG control sample, we were able to detect the tryptic peptides of the unique region of circFGFR1p as well as the tryptic peptides overlapping with the linear FGFR1 in the IP-LC-MS/MS sample, suggestubg that circFGFR1p is indeed expressed and can be captured by our circFGFR1p antibody (Figure 7D). To further confirm circFGFR1p expression in high resolution, we performed PRM-MS using a synthetic heavy isotope-labeled reference peptide of the circFGFR1p unique region. We can indeed identify the corresponding transition ions of the labeled reference peptide and the sampling tryptic peptide from BJ cells (Figure 7E). Collectively, the IP-LC-MS/MS and the PRM-MS provide the strong evidence demonstrating endogenous circFGFR1p expression.
Upon binding to FGF, full-length FGFR1 dimerizes and autophosphorylates the kinase domain, which triggers the downstream signaling pathway and facilitates cell proliferation (Huret, 2009). By co-expressing HA-tagged FGFR1 and FLAG-tagged circFGFR1p in HEK-293T cells, we confirmed that circFGFR1p is localized at the cell membrane and endosomes similarly to FGFR1 (Figure S11B and S11C). CircFGFR1p contains the FGFR1 dimerization/ligand-binding domain but lacks the kinase domain, suggesting circFGFR1p may function as a dominant-negative regulator of FGFR1 that suppresses cell proliferation. To test this hypothesis, we knocked down circFGFR1 with siRNAs targeting the back-splicing junction and discovered that knockdown of circFGFR1 can indeed facilitate cell proliferation upon FGF addition (Figure 7G), suggesting that circFGFR1 negatively regulates FGFR1 function. Interestingly, we also observe the same phenotype upon knock down of circFGFR1p using anti-sense LNA (LNA-18S of IRES-8228) to disrupt circFGFR1p IRES without altering the level of circFGFR1 or FGFR1 RNA, suggesting that the higher cell proliferation rate is caused by knocking down circFGFR1p but not circFGFR1 RNA. Also, circFGFR1p knockdown leads higher level of total and active FGFR1 (phosphorylated FGFR1; p-FGFR1) (Figure 7F and 7G), suggesting that circFGFR1p functions not solely as a dominant-negative of FGFR1 signal transduction, but circFGF1p also inhibits FGFR1 accumulation, perhaps by increasing FGFR1 turnover or degradation (Saffell et al., 1997). Conversely, we observed that circFGFR1p overexpression (circFGFR1pOE) suppressse cell proliferation, and co-overexpressing FGFR1 and circFGFR1p (FGFR1OE + circFGFR1pOE) can partially rescue the phenotype of cell proliferation suspension (Figure S11F), suggesting an antagonistic function of circFGFR1p for FGFR1. These results show that circRNA-encoded circFGFR1p can suppress cell growth by interacting with FGFR1 through a dominant-negative mechanism.
In comparison to FGFR1, the expression level of circFGFR1p is relatively low (Figure 7H), indicating that circFGFR1p may not be a strong regulator under normal conditions. Nevertheless, many IRESs have been reported to have steady cap-independent translation activity under stress conditions (Bushell et al., 2006; Holcik et al., 2000; Johannes and Sarnow, 1998; Nevins et al., 2003; Schepens et al., 2005; Stoneley et al., 2000; Warnakulasuriyarachchi et al., 2004), including the IRES of some endogenous protein-coding circRNAs, such as circZNF-609 (Legnini et al., 2017). Thus, we further examined circFGFR1 IRES under stress conditions, such as under heat-shock, and found out that the cap-independent translation activity of FGFR1 IRES remains steady during the heat-shock (Figure S11G). We then examined the FGFR1 and circFGFR1p protein levels under the heat-shock condition. We discovered that FGFR1 protein level was downregulated after the heat-shock (Figure 7H and 7I), which is likely due to the global reduction in cap-dependent translation during the heat-shock (Cuesta et al., 2000; Rhoads and Lamphear, 1995). On the other hand, the circFGFR1p level, which is regulated by cap-independent translation, remained steady after the heat-shock (Figure 7H and 7J, Figure S11H and S11I). The result suggests that during heat shock although the global FGFR1 cap-dependent translation reduction is not directly caused by the circFGFR1p level, the reduced FGFR1 level and stable circFGFR1p level enhances the circFGFR1p dominant negative effect and further lower the cell growth rate. Moreover, FGFR1 has been shown to form homomultimers when induced by cell adhesion molecules (Zamai et al., 2019). The nature of FGFR1 oligomerization may further enhance the dominant-negative effect of circFGFR1p. These phenomena may explain how lowly expressed circFGFR1p can effectively regulate highly expressed FGFR1 and suppress cell proliferation under heat shock (Figure S11J and S11K). In sum, our findings above demonstrated that with our methods, we not only identify a novel circRNA-encoded protein, circFGFR1p, that negatively regulates FGFR1 and suppresses cell proliferation through a dominant-negative mechanism under stress conditions, but also reveal an important regulatory mechanism of circRNAs and their encoded proteins.
DISCUSSION
IRES elements suggests pervasive circRNA translation
Here we describe a high-throughput reporter assay that identified a family of 17,201 RNA elements capable of driving circRNA translation. Although our IRES screening was performed on an ectopic circRNA reporter, which in general shows higher translation activity than endogenous circRNA (Fan et al., 2019; Wang and Wang, 2015), the number of identified human 5’ UTR IRES contribute to ~34% of all the human 5’ UTR oligos in our screening is close to the percentage that has been reported in previous IRES screening studies using a linear bicistronic reporter (Hershey et al., 2012; Weingarten-Gabbay et al., 2016). Moreover, Pamudurti et al. (2017b) reported 122 endogenous protein-coding circRNAs based on ribosome profiling of Drosophila heads. Considering the genome-wide nature of our screen which is applicable to circRNAs from multiple human cell and tissue types and the increased complexity of the human transcriptome compared to the fly transcriptomes, the ten-fold increase in detection of protein-coding circRNAs in our assay is within a reasonable scale. Thus, along with the validation by many orthogonal approaches, our study suggests pervasive circRNA translation driven by circRNA IRES.
A bipartite model of circRNA IRES organization and function
We discover that 18S rRNA complementarity and a distinct secondary structure (SuRE) on the IRES can facilitate circRNA translation. These new elements thus potentially mediate a significant but previously unexplained output of the human gene expression program. While m6A modification and A-U rich sequences have been shown to facilitate circRNA translation (Fan et al., 2019; Yang et al., 2017), we did not find enrichment of these two elements in the identified circRNA IRES (Figure S6D). Since m6A on endogenous circRNAs can prevent immune recognition and activation of the PKR translation inhibition pathway (Chen et al., 2019), it is possible that the role of m6A and A-U rich sequences on circRNA translation were confounded by circRNA immunity. We performed our IRES screen in HEK-293T cells which lack RIG-I activity (Chen et al., 2017) so that the result was not affected by the effects of m6A on circRNA immunity. Nevertheless, we do not rule out other regulatory mechanisms that can collaboratively facilitate cap-independent translation on the circRNA.
Although the mechanism for IRES to distinguish circular and linear RNA remains unclear, we suggest that the mechanism may be associated with different biogenesis process between circular and linear RNA. One possible mechanism is that the recruitment of IRES trans-acting factors (ITAFs) on circular IRES is back-splicing dependent. Our study revealed that many IRESs on the endogenous circRNAs are located near the back-splicing junction further supports this mechanism. Another possible mechanism is that the recruitment of ITAFs on the circular IRES depends on the circRNA-specific nuclear export pathway. Other mechanisms such as the RNA methylation patterns on the circRNA near the back-splicing junction may also play roles in regulating circRNA-specific IRES activity (Chen et al., 2019). Nevertheless, further investigation is needed to identify the detailed mechanism regulating specific translation activity of circular IRES and linear IRES.
Multiple outcomes of circRNA translation
The act of circRNA translation may also lead to a number of possible outcomes beyond producing functional proteins, such as generating rapidly degraded peptides that regulate immune surveillance (Goldberg, 2007; Rock et al., 1994). Moreover, circRNA translation may engage additional mechanisms such as nonsense-mediated RNA decay analogous to lncRNA translation (D’Lima et al., 2017; Hartford and Lal, 2020; Kurosaki and Maquat, 2016; Smith and Baker, 2015; Wery et al., 2016), RNA quality control via ribosome movement (D’Orazio and Green, 2021), and inhibiting the translation of other RNAs by competing with standby ribosomes (Darfeuille et al., 2007). In these scenarios, circRNA maybe actively translated and regulating important cellular functions, neither of which requires the accumulation of stable proteins to a detectable level. These alternative outcomes may also explain the limitation of utilizing MS datasets for validating circRNA translation. Our discovery of circRNA IRES elements can facilitate the discovery and characterization of the multiple outcomes of endogenous circRNA translation which opens the door to study circRNA’s engagement in these diverse mechanisms.
CircRNA-encoded proteins as functional regulators of signaling circuitry
Our findings highlight an important regulatory mechanism of how cells utilize different translation machinery to respond to stress conditions. While cells predominantly utilize cap-dependent linear mRNA translation to produce proteins, they can shift the RNA source of translation toward circRNA by downregulating the cap-dependent translation activity and maintain the same or higher cap-independent translation activity of circRNA IRES under stress conditions (Methods S7). The depletion of circFGFR1 in cancer samples may occur to downregulate circFGFR1p and increase the proliferative signaling through FGF signaling cancers (Nair et al., 2016; Vo et al., 2019; Xia et al., 2018) (Methods S8). More generally, we speculate circRNA-encoded proteins may be a useful mechanism to elaborate modules from within multi-domain proteins that endow cells with the ability to independently control their translation. Although further investigation is needed to understand how cells coordinate the linear and circular RNA translation machinery, our findings identify a model of how circRNA translation is regulated by a different mechanism from linear mRNA translation and how cells utilize circRNA-encoded proteins to respond to a dynamic environment.
Limitations of the study
Several limitations set the stage for future studies. First, our oligo library did not tile all human circRNA sequences. The ~17,000 candidate circRNA IRES were identified from an artificial oligo library that represented only a subset of endogenous circRNA sequences. The landscape of endogenous circRNAs continue to be refined and expanded, and there are circRNAs for which we have no functional data on IRES activity. Second, although the BSJ tryptic peptide provides strong evidence that these circORFs are expressed in cells, unique BSJ peptide was not available in many cases. Further genomic and proteomic validation to exclude possible alternative origins of circORF-matching peptide is recommended. Third, our screen was conducted in one cell type. Whether and how circRNA translation is differentially regulated in different cell types remain to be explored. Future studies aimed at comprehensive circRNA coverage and endogenous circRNA translation will shed light on the biological significance, if any, when circular RNAs meet ribosomes.
STAR★METHODS
Lead Contact
Further information and questions for methods and resources should be directed to the Lead Contact, Howard Y. Chang (howchang@stanford.edu).
Materials Availability
Plasmids and reagents generated in this study are available upon request to the Lead Contact.
Data and Code Availability
The IRES screening, polysome profiling, and M2-seq sequencing data generated in this paper were deposited in Gene Expression Omnibus. The MS/MS and the PRM-MS data generated in this paper are deposited in PRIDE. Accession numbers are listed in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-GFP antibody - ChIP Grade (1:300 for WB) | Abcam | Cat# ab290, RRID:AB_303395 |
| CD4 [EPR6855] antibody (1:500 for WB) | Abcam | Cat# ab133616, RRID:AB_2750883 |
| Cre recombinase antibody [7.23] (1:200 for WB) | Abcam | Cat# ab24607, RRID:AB_448179 |
| anti-SMAD5 antibody[3H9] (1:200 for WB) | Novus Biologicals | Cat# NBP2–37648 |
| FGFR1 Antibody (1:200 for WB) | Novus | Cat# NB100–2079, RRID:AB_10144921 |
| FGFR1 (phospho Y654) antibody (1:200 for WB) | Abcam | Cat# ab59194, RRID:AB_941585 |
| Monoclonal ANTI-FLAG® M2 antibody (1:400 for WB; 1:150 for IF) | Sigma-Aldrich | Cat# F1804, RRID:AB_262044 |
| HA tag antibody - ChIP Grade (1:400 for WB; 1:150 for IF) | Abcam | Cat# ab9110, RRID:AB_307019 |
| beta Tubulin antibody - Loading Control (1:500 for WB) | Abcam | Cat# ab6046, RRID:AB_2210370 |
| Anti-GAPDH antibody [EPR16891] (1:500 for WB) | Abcam | Cat# ab181602, RRID:AB_2630358 |
| IRDye 680RD Goat anti-Mouse IgG antibody (1:5,000 for WB) | LI-COR Biosciences | Cat# 926–68070, RRID:AB_10956588 |
| IRDye® 800CW Goat anti-Rabbit IgG (H + L) (1:5000 for WB) | LI-COR Biosciences | Cat# 926–32611 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:1,500 for IF) | Thermo Fisher Scientific | Cat# A-11029, RRID:AB_2534088 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (1:150 for IF) | Thermo Fisher Scientific | Cat# A-11012, RRID:AB_2534079 |
| Customize rabbit anti-circFGFR1p | Thermo Fisher Scientific | Project# VJ2580 |
|
| ||
| Bacterial and Virus Strains | ||
|
| ||
| NEB® 10-beta Competent E. coli | New England Biolabs | C3019H |
|
| ||
| Chemicals, Peptides, and Recombinant Proteins | ||
|
| ||
| SYBR™ Gold Nucleic Acid Gel Stain | Thermo Fisher Scientific | Cat# S11494 |
| BrightStar™-Plus Positively Charged Nylon Membrane | Thermo Fisher Scientific | Cat# AM10100 |
| Millennium™ RNA Markers | Thermo Fisher Scientific | Cat# AM7150 |
| Cycloheximide,≥93.0% (HPLC) | Sigma-Aldrich | Cat# 01810 |
| Puromycin Dihydrochloride | Thermo Fisher Scientific | Cat# A11138–03 |
| Pierce™ IP Lysis Buffer | Thermo Fisher Scientific | Cat# 87788 |
| Pierce™ Protein A/G Magnetic Beads | Thermo Fisher Scientific | Cat# 88803 |
| NuPAGE™ 4 to 12%, Bis-Tris, 1.5 mm, Mini Protein Gel, 10-well | Thermo Fisher Scientific | Cat# NP0335BOX |
| Corning® BioCoat™ Poly-D-Lysine 12 mm #1 German Glass Coverslip | Corning | Cat# 354086 |
| ON-TARGET plus non-targeting pool | Dharmacon | Cat# D-001810–10-05 |
| SMARTpool: ON-TARGETplus FGFR1 siRNA | Dharmacon | Cat# L-003131–00-0005 |
| Gibco FGF1 Recombinant Human Protein | Thermo Fisher Scientific | Cat# 13–241-013 |
| PrestoBlue™ Cell Viability Reagent | Thermo Fisher Scientific | Cat# A13261 |
| E-Gel™ EX Agarose Gels, 2% | Thermo Fisher Scientific | Cat# G402022 |
| NEB® 10-beta/Stable Outgrowth Medium | New England Biolabs | Cat# B9035S |
| Opti-MEM™ I Reduced Serum Medium | Thermo Fisher Scientific | Cat# 31985070 |
| Q5® Hot Start High-Fidelity 2X Master Mix | New England Biolabs | Cat# M0409L |
| TrypLE™ Express Enzyme (1X), no phenol red | Thermo Fisher Scientific | Cat# 12604013 |
| NEBNext® Ultra™ II Q5® Master Mix | New England Biolabs | Cat# M0544L |
| Invitrogen Ambion SUPERase·In RNase Inhibitor (20U/μL) | Thermo Fisher Scientific | Cat# AM2694 |
| SuperBlock Blocking Buffers | Thermo Fisher Scientific | Cat# 37517 |
| TURBO DNase (2U/μL) | Thermo Fisher Scientific | Cat# AM2238 |
| SuperScript IV Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18090050 |
| Normal Rabbit IgG | Sigma-Aldrich | Cat# 12–370 |
| RIPA Lysis and Extraction Buffer | Thermo Fisher Scientific | Cat# 89900 |
| Halt Protease and Phosphatase Inhibitor Cocktails | Thermo Fisher Scientific | Cat# 78441 |
| RNaseR | MCLAB Products | Cat# RNASR-200 |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| Lipofectamine™ 3000 Transfection Reagent | Thermo Fisher Scientific | Cat# L3000015 |
| Quick-DNA Microprep Plus Kit | Zymo Research | Cat# D4074 |
| NorthernMax™-Gly Kit | Thermo Fisher Scientific | Cat# AM1946 |
| RNeasy Mini Kit | Qiagen | Cat# 74004 |
| SuperScript™ IV One-Step RT-PCR System | Thermo Fisher Scientific | Cat# 12594100 |
| HiScribe™ T7 High Yield RNA Synthesis Kit | New England Biolabs | Cat# E2040S |
| GeneMorph II Random Mutagenesis Kit | Agilent | Cat# 200550 |
| DNA Clean & Concentrator-25 | Zymo Research | Cat# D4033 |
| Zymoclean Gel DNA Recovery Kit (uncapped columns) | Zymo Research | Cat# D4002 |
| Direct-zol RNA Microprep | Zymo Research | Cat# R2061 |
| RNA Clean & Concentrator-25 | Zymo Research | Cat# R1017 |
| RNA Clean & Concentrator-5 | Zymo Research | Cat# R1013 |
| Zymoclean Gel RNA Recovery Kit | Zymo Research | Cat# R1011 |
| NEBuilder® HiFi DNA Assembly Master Mix | New England Biolabs | Cat# E2621L |
| ZymoPURE II Plasmid Maxiprep Kit | Zymo Research | Cat# D4203 |
| Complete kit (optimized for Roche® LightCycler 480) | Kapa Biosystems | Cat# KK4854 |
| NEBuilder® HiFi DNA Assembly Master Mix | New England Biolabs | Cat# E2621L |
|
| ||
| Deposited Data | ||
|
| ||
| IRES screening, polysome profiling, and M2-seq sequencing | This paper | GEO: GSE178718 |
| Ribosome footprinting | Chen et al., 2020 | GEO: GSM3791725, GSM3791726, GSM3791727, GSM3791728, GSM3791729, and GSM3791730 |
| iPSC peptidomics | Chen et al., 2020 | PRIDE: PXD014031 |
| SCBC peptidomics | Orre et al., 2019 | ProteomeXchange: PXD006895 |
| GTEx peptidomics | Jiang et al., 2020 | ProteomeXchange: PXD016999 |
| K562/U2OS/H358 peptidomics | This paper | ProteomeXchange: PXD025233 |
| HLA class I peptidomes | Bassani-Sternberg et al., 2015 | PRIDE: PXD014031 |
| circFGFR1p IP-MS | This paper | ProteomeXchange: PXD024993 |
| K562/U2OS PRM-MS | This paper | ProteomeXchange: PXD025235 |
| circFGFR1p PRM-MS | This paper | ProteomeXchange: PXD025203 |
| QTI-seq | Gao et al., 2015 | SRA: 160745 |
|
| ||
| Experimental Models: Cell Lines | ||
|
| ||
| HEK293T | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| HeLa | ATCC | Cat# CCL-2; RRID: CVCL_0030 |
| BJ | ATCC | Cat# CRL-2522; RRID:CVCL_3653 |
| U2OS | ATCC | Cat# HTB-96, RRID:CVCL_0042 |
|
| ||
| Oligonucleotides | ||
|
| ||
| qRT-PCR primers | This paper | Table S7 |
| IRES screening oligo library | Weingarten-Gabbay et al., 2016 | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| Plasmid: pcDNA3.1(+) ZKSCAN1 Sense | Addgene | Cat# 60631 |
| Plasmid: pHAGE-FGFR1 | Addgene | Cat# 116740 |
| Plasmid: pUC19 Vector | New England Biolabs | Cat# N3041L |
| Plasmid: pLV-mCherry | Addgene | Cat# 36084 |
| Plasmid: mRuby-ZKSCAN-split-eGFP reporter | This paper | N/A |
| Plasmid: mRuby-IRES-eGFP reporter | This paper | N/A |
| Plasmid: autocatalytic-splicing circEGFP | This paper | N/A |
|
| ||
| Software and Algorithms | ||
|
| ||
| ZEN (blue edition) | Carl Zeiss Microscopy | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| FlowJo_V10 | FlowJo, LLC | https://flowjo.com/solutions/flowjo/downloads |
| Bowtie2 | Langmead and Salzberg, 2012 | https://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.4.4/ |
| TopHat2 | Kim et al., 2013 | https://ccb.jhu.edu/software/tophat/downloads/tophat-2.1.1.tar.gz |
| M2-seq and M2-net | Cheng et al., 2017 | https://ribokit.github.io/M2seq/ |
| ViennaRNA | Lorenz et al., 2011 | https://www.tbi.univie.ac.at/RNA/ |
| MaxQuant | Cox and Mann, 2008 | https://www.maxquant.org/download_asset/maxquant/latest |
| Trans Proteomic Pipeline | Collaborative open-source | http://tools.proteomecenter.org/wiki/index.php?title=Software:TPP |
| Byonic | Protein Metrics | https://proteinmetrics.com/byos/ |
| Skyline 19.1.0.193 | MacLean et al., 2010 | https://skyline.ms/wiki/home/software/Skyline/page.view?name=install-administator-64 |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental Model and Subject Details
Cell lines and maintenance
Human HeLa (cervical adenocarcinoma, ATCC CCL-2), human HEK293T (embryonic kidney, ATCC CRL-3216) cells, human BJ (fibroblast, ATCC CRL-2522), and U2OS (osteosarcoma, ATCC HTB-96) were grown in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, 11995–073) supplemented with 100 units/ml penicillin-streptomycin (GIBCO, 15140–163) and 10% (v/v) fetal bovine serum (Thermo Fisher Scientific, SH30071.03). Cell growth was maintained at 37°C in a 5% CO2 atmosphere, passaging at a 1:10 dilution every 2–3 days.
Cell culture and transient transfection
Cells were plated 24 hours prior to transfection. Cells were at 70 to 80% confluence and 500 ng of RNA was transfected into one well of a 24-well plate using Lipofectamine 3000 (Thermo Fisher Scientific, L3000008). The nucleic acids with P3000 and Lipofectamine 3000 were diluted in Opti-MEM (Thermo Fischer Scientific, 31985088) per manufacturer’s instructions, and incubated for 3 minutes at room temperature. The nucleic acids and Lipofectamine 3000 were the mixed together, incubated for 20 minutes at room temperature and then the nucleic acids-Lipofectamine 3000 complexes were applied dropwise to the monolayer cultures. For NEON transfection, cells were electroporated with NEON Transfection System (Thermo Fisher Scientific MPK5000S) per manufacturer’s instructions. In most cases, cells were resuspended in buffer R at 2×107/mL and 5 μg of DNA plasmid was electroporated with a 100 μL NEON tip.
Method Details
Generating mRuby-ZKSCAN1-split-eGFP reporter construct
The mRuby-ZKSCAN1-split-eGFP reporter construct contains a bicistronic mRuby reporter followed by a permuted split-eGFP reporter flanked by human ZKSCAN1 introns, where during transcription, the pre-mRNA of the construct will undergo spliceosome-mediated back-splicing and reconstitute full-length eGFP on the circRNA (Liang and Wilusz, 2014). Because full-length eGFP is only reconstituted upon back-splicing, the eGFP fluorescence signal can only come from the circRNA through cap-independent translation. Two well-known concerns for bicistronic IRES screens are cryptic promoters or splice sites that activate transcription or readthrough of the downstream open reading frame (ORF), respectively (Baranick et al., 2008). Our design obviates both concerns because ectopic transcription of only the 5’ fragment of the split-eGFP cannot produce fluorescence signal. Northern blots, quantitative reverse transcription polymerase chain reaction (qRT-PCR), RNase R treatment, and reporter gene experiments confirmed that the eGFP signal we detected did not come from trans-splicing or the nicking of the eGFP circRNA (Figure S1). Following transfection into human embryonic kidney (HEK) 293T cells, we sorted the transfected cells by the ratio of eGFP to mRuby fluorescence into seven bins, and deconvoluted the frequencies of oligo sequences in each pool by deep sequencing (Sharon et al., 2012; Weingarten-Gabbay et al., 2016).
Specifically, the mRuby-ZKSCAN-split-eGFP reporter plasmid was generated using pcDNA3.1(+) ZKSCAN1 Sense as the backbone vector (addgene: 60631) (Liang and Wilusz, 2014). Specifically, the eGFP reporter was split into two parts and cloned into the backbone vector flanked by the ZKSCAN1 intron 2 and 3, where the full-length eGFP can only be reconstituted when the RNA is circularized. An EcoRV cloning site was inserted right before the start codon of the split eGFP reporter for oligo library insertion. The mRuby construct was then inserted between the CMV promoter and the upstream of the ZKSCAN1 intron to generate mRuby-ZKSCAN1-split-eGFP reporter plasmid.
Oligo library amplification
The oligo library was obtained as a gift from Dr. Eran Segal, Weizmann Institute of Science. The detail desgin of the oligo library were described in Weingarten-Gabbay et al. (Weingarten-Gabbay et al., 2016). Specifically, the synthetic oligo library contains 55,000 different single-stranded 210 nt oligos. Each oligo contains a 174 nt unique sequence flanked by common priming sequences (5’-CTAGGGCGCGCCAGTCCT-(N)174-CGACTCGGACCGATGGTG-3’). For the oligo library amplification, we set up 16 individual PCR reactions using Q5® Hot Start High-Fidelity 2X Master Mix (NEB: M4049). Each 50 μL PCR reaction contained 0.3 ng library DNA, 2.5 μL 20 μM forward primer, and 2.5 μL 20 μM reverse primer (Fw: ACGAGCTGTACAAGTAAGATGGCGCGCCAGTCCT; Rv: TCGCCCTTGCTCACCATGATCATCGGTCCGAGTCG; underlined sequence represents the sequence complementary to the priming sequence of oligos in the library). The parameters for PCR were 95°C for 1 min, 20 cycles of 95°C for 20 s, 63°C for 20 s, and 72°C for 1 min, each, and finally one cycle of 72°C for 5 min. The PCR products from all 16 reactions were pooled and purified using DNA Clean & Concentrator-25 (Zymo Research: D4033).
Cloning oligo library into reporter construct
The mRuby-ZKSCAN1-split-eGFP reporter plasmid was first digested with EcoRV-HF in CutSmart Buffer (NEB) at 37°C for 2 h. The digested plasmid was then separated on a 2% E-Gel™ EX Agarose Gel (Invitrogen: G402022) and purified using Zymoclean Gel DNA Recovery Kit (Zymo Research: D4002). To clone the oligo library into the reporter plasmid, we set up 4 assembly reactions using NEBuilder® HiFi DNA Assembly (NEB: E2621). Each 10 μL assembly reaction contained 100 ng digested reporter plasmid and 6 ng amplified oligo library. The assembly reactions were incubated at 50°C for 1 h. We then set up 8 transformation reactions. For each transformation, 5 μL of assembly product was transformed into 25 μL NEB® 10-beta Competent E. coli (NEB: C3019H) by heat shock. The transformed competent cells were recovered in 1 mL 10-beta/Stable Outgrowth Medium (NEB: B9035) at 37°C for 1 h. We then pooled all the recovered competent cells and plated them on LB AMP-100 Agar plates (Teknova) (400 μL each). The plates were incubated at 37°C for 16 h. We then scraped the plates into LB medium and purified the pooled oligo library reporter plasmids (mRuby-ZKSCAN1-oligo-split-eGFP) using ZymoPURE II Plasmid Maxiprep (Zymo Research: D4203).
Transfection of mRuby-ZKSCAN1-oligo-split-eGFP reporter construct
We first used the percentage of mRuby(+) cells to determine the multiplicity of infection (MOI). Specifically, we performed serial dilutions that the pooled mRuby-ZKSCAN1-oligo-split-eGFP reporter plasmids were diluted in dummy plasmids (pUC19 Vector; NEB: N3041) of 1:5, 1:10, 1:50, 1:100, 1:500, 1:1000, 1:5000, and 1:50000 in molar ratio. We then transfected HEK-293T cells on a 15-cm plate with each dilution respectively using Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher Scientific) according to manufacturer’s protocol. After five days, we collected the cells from each plate and determine the MOI of each dilution by analyzing the percentage of mRuby(+) cells using flow cytometry. We used the dilution of 1:500 for the subsequent transfection as it showed the MOI close to 0.1. Six 15-cm plates of HEK-293T cells were then transfected with diluted mRuby-ZKSCAN1-oligo-split-eGFP reporter plasmids as described above. Each 15-cm plate transfection contained 45 μg total DNA, 90 μL P3000, and 90 μL Lipofectamine Reagent in 4.5 mL Opti-MEM™ Reduced Serum Medium (Thermo Fisher Scientific: 31985070). We chose to conduct our circRNA IRES screen in HEK-293 cells which lack RIG-I activity so that the IRES screening result will not be confounded by effects of m6A on circRNA immunity.
Sorting cells transfected with mRuby-ZKSCAN1-oligo-split-eGFP reporter construct by FACS
Transfected cells were grown for five days. Cells were then trypsinized with TrypLE Express (Thermo Fisher Scientific: 12604013), centrifuged, and resuspended in FACS sorting buffer (2 mM EDTA and 2% Fetal Bovine Serum in PBS). Sorting was performed with BD FACS Aria II Sorter. The mRuby gene allowed us to normalize for transduction efficiency by translation of a regular linear mRNA. mRuby(+)/eGFP(+) cells were sorted into seven expression bins according to their mRuby normalized eGFP signal intensity. We collected ~2 million mRuby(+)/eGFP(+) in total. In addition, we collected ~1 million mRuby(+) cells to determine the representation of the oligo library.
Generating library for next-generation sequencing
Total DNA from each expression bin was extracted using Quick-DNA Microprep Plus Kit (Zymo Research: D4074). Three rounds of PCR using NEBNext Ultra II Q5 Master Mix (NEB: M0544) were performed to generate the library for next-generation sequencing. For the first PCR, oligo library sequence was amplified using the following primer set – Fw: GGGATCACTCTCGGCATGGA; Rv: GCTCCTCGCCCTTGCTCAC. Each 50 μL PCR reaction contained 200 ng total DNA, 500 nM forward primer, and 500 nM reverse primer. The parameters for the PCR were 98°C for 1 min, 24 cycles of 98°C for 30 s, 65°C for 30 s, 72°C for 40 s, each, and finally one cycle of 72°C for 5 min. For the second PCR, adapter priming sequences were added to the oligo library using the following primer set – Fw: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGGGATCACTCTCGGCATGGA; Rv: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGCTCCTCGCCCTTGCTCAC (underlined sequence represents the sequence for illumina adapters priming). Each 50 μL PCR reaction contained 1.25 μL of the first PCR product, 500 nM forward primer, and 500 nM reverse primer. The parameters for the PCR were 98°C for 1 min, 18 cycles of 98°C for 30 s, 70°C for 30 s, 72°C for 40 s, each, and finally one cycle of 72°C for 5 min. For the third PCR, custom barcodes adapters were used for the reaction (Buenrostro et al., 2015a). Each 50 μL PCR reaction contained 1 μL of 1:20 diluted second PCR product, 1 μM Ad1 adapter, and 1 μM Ad2 adapter. The parameters for the PCR were 98°C for 1 min, 15 cycles of 98°C for 30 s, 72°C for 30 s, 72°C for 40 s, each, and finally one cycle of 72°C for 5 min. The PCR product was then separated on a 2% E-Gel™ EX Agarose Gel (Invitrogen: G402022) and the DNA fragments with expected size were purified using Zymoclean Gel DNA Recovery Kit (Zymo Research). The concentration of library was determined using KAPA Library Quantification Kit (Kapa Biosystems: KK4854). The library of total mRuby(+) cells was prepared and generated using the same method described above. The library of each expression bin and the total mRuby(+) library were sequenced on MiSeq (Illumina) respectively. We collected ~1.2 million reads for each expression bin and ~2 million reads for the total mRuby(+) library.
Quantification of mean eGFP expression bin for each designed oligo
To determine the number of reads for each designed oligo in each expression bin, we first generated an artificial genome by concatenating the first 37 nucleotide sequence of all 55,000 designed oligos with spacers of 50 N’s. For each single-end MiSeq reads, the common priming sequences were trimmed, and the first 37 nucleotide sequence of the trimmed reads were extracted and mapped to the artificial genome. The number of reads of each designed oligo was then counted in each expression bin respectively. The multi-mappers were excluded from the analysis. To quantify the eGFP expression level of each oligo, we calculated the mean weighted rank distribution of the reads acorss the bins. The weight of each bin is the fraction of the number of reads in this bin of its total reads in all seven bins. The rank is the bin number from the bin with the lowest eGFP (bin #1) to the bin with the highest eGFP signal (bin #7). We performed two independent biological replicates and we used the average eGFP expression of the two replicates as the mean eGFP expression bin for each designed oligo. The oligos were determined as not presented in the library if the oligos had no reads detected in two adjacent bins or had less than five reads in the total mRuby(+) library or any of the two replicates. We assigned the value of −1 to the oligos that were not presented in the library. To determine the oligos with cap-independent translation activity (eGFP(+) oligos), we calculated the weighted rank distribution of cells transfected with no-IRES-inserted reporter plasmid as the background eGFP expression. The background eGFP expression was calculated based on the distribution of the reads across the bins rather than a simple cut off value, which is a more conservative approach to avoid possible false positive events because the empty circRNA eGFP reporter could have weak translation activity (Abe et al., 2015) (Figure S2D). The oligos were defined as eGFP(+) oligo as the oligo with eGFP expression higher than the background eGFP expression (eGFP expression (bin) = 3.466387). ). We further verify that the eGFP(+) oligos we identified were able to initiate circRNA translation of different coding sequences (CDSs) (Figure S2E). We also observed substantially weaker cap-independent translation activity of circRNA compared to linear RNA translation driven by the cap-dependent translation (Figure S2F). In addition to the 5’ UTRs of the human genes that have been recently discovered by the linear RNA bicistronic IRES screening assay, such as EIF4 and BID (Weingarten-Gabbay et al., 2016), our assay further captured ~900 sequences from the 5’ UTR of the human genes that can initiate cap-independent translation on the circRNA reporter but do not show cap-independent translation activity in the previous linear RNA bicistronic IRES screening assay (Weingarten-Gabbay et al., 2016). The quantification of translation activity was highly reproducible between two independent biological replicates (Figure S2A), and we confirmed that the results were not confounded by the change of circRNA back-splicing efficiency due to different oligo inserts (Figure S2B and S2C).
Northern blotting
One 15-cm plate of HEK-293T cells were transfected with diluted mRuby-ZKSCAN1-oligo-split-eGFP reporter plasmids using Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher Scientific) as described in the IRES screening. Transfected cells were grown for five days, trypsinized, and sorted by eGFP signal intensity. mRuby(+)/eGFP(+) cells were collected and the total RNA of the cells was purified using Quick-RNA Midiprep Kit (Zymo Research). 30 μg of total RNA was then treated with 30 U RNase R (MACLAB: RNASR-200) at 37°C for 30 min in 1X RNase R Buffer (0.2 M Tris-HCl (pH8.0), 1 mM MgCl2 and 1 M LiCl). Another 30 μg of total RNA was incubated in 1X RNase R Buffer without RNase R addition at 37°C for 30 min as the RNase R(−) sample. Total RNA with or without RNase R treatment was purified using RNA Clean & Concentrator-25 (Zymo Research: R1017), respectively. Northern blotting was then performed using NorthernMax®-Gly Kit (Thermo Fisher Scientific: AM1946) according to manufacturer’s protocol. Specifically, purified RNA was incubated with equal volume of Glyoxal Load Dye (Thermo Fisher Scientific: AM8551) at 50°C for 30 min, loaded into 1% agarose gels made in 1X Gel Prep/ Gel Running buffer (Thermo Fisher Scientific: AM8678), and ran at 75V for 40 min. The gels were then stained in 1X SYBR™ Gold Nucleic Acid Gel Stain (Thermo Fisher Scientific: S11494) diluted in 1X Gel Prep/ Gel Running buffer at room temperature for 5 min, and transferred to BrightStar™-Plus Positively Charged Nylon Membranes (Thermo Fisher Scientific: AM10100) using iBlot™ 2 Gel Transfer Device (Thermo Fisher Scientific: IB21001) with the following parameters: 20V for 2 min, 23V for 2 min, and 25V for 3 min. We then crosslinked the blots with Stratalinker 1800 (Stratagene) using autocrosslink Mode. RNA ladders on the blots (Millennium™ RNA Markers, Invitrogen: AM7150) were then visualized and marked on a E-Gel™ Imager (Thermo Fisher Scientific). The blots were then blocked in ULTRAhyb Buffer (Thermo Fisher Scientific: AM8670; pre-warmed warmed to 68°C) with 20 U SUPERase•In™ RNase Inhibitor (Invitrogen: AM2694) at 65°C for 30 min. 0.1 nM biotinylated ssDNA probes synthesized by IDT against the corresponding region on the reporter RNA (mRuby: /5BiosG/AGAAATCAGGGATGTCGGCCGGGTACTTGATAAAAGTACGGCTGCCATACATGAACGACGTGGCAAGAATGTCAAAGGCAAATGGCAGGG; 3’eGFP: /5BiosG/ACAGCTCGTCCATGCCGAGAGTGATCCCGGCGGCGGTCACGAACTCCAGCAGGACCATGTGATCGCGCTTCTCGTTGGGGTCTTTGCTCA; eGFP-junction: /5Biosg/GTAGTGGTCGGCGAGCTGCACGCTGCCGTCCTCGATGTTGTGGCGGATCTTGAAGTTCAC) were added to the blots, respectively, in ULTRAhyb Buffer and incubated at 65°C overnight. The blots were then washed with Northern Blot Wash Solutions (Thermo Fisher Scientific: AM8673), followed by blocking in SuperBlock™ (PBS) Blocking Buffer (Thermo Fisher Scientific: 37517) at room temperature for 40 min. After the blocking, the blots were incubated with IRDye® 800CW Streptavidin (LI-COR Biosciences; 1:2000) in SuperBlock™ (PBS) Blocking Buffer at room temperature for 40 min, washed with Northern Blot Wash Solutions and PBS. The blots were imaged with Odyssey® CLx Imager (LI-COR Biosciences). Our reporter produces a ~3000 nucleotide (nt) primary linear transcript and a ~900 nt eGFP circRNA.
Normalization of eGFP expression level in linear and circular RNA reporter system
For each oligo, we calculated the normalized eGFP expression from the circRNA vs. from the linear RNA template. The eGFP expression level for each synthetic oligo in a linear bicistronic reporter screening assay was obtained from Weingarten-Gabbay et al. study (Weingarten-Gabbay et al., 2016). Their study utilized a similar eGFP reporter system and the same synthetic oligo library for screening IRES activity on the linear RNA. We normalized the data by first calculating the fold differences of eGFP level for each oligo relative to the background eGFP level (no-IRES reporter plasmid) in each study respectively ((eGFPoligo - eGFPbackground)/eGFPbackground). Since the maximum eGFP level obtained from Weingarten-Gabbay et al. study was higher than ours, we further normalized our data by multiplying the ratio of their maximum eGFP level over our maximum eGFP level (eGFPmax(Weingarten-Gabbay et al)/eGFPmax(our data)). Only the oligos that are captured in both studies were included in the analysis. To define linear and circular IRES, we took a more conservative approach, where linear IRES represents the oligo showing cap-independent translation activity in the linear RNA screening system only (Weingarten-Gabbay et al., 2016); and circular IRES represents the oligo showing cap-independent translation activity in the circular RNA screening system only (our study). When we examined the distribution of the human and viral IRES among circular IRES, linear IRES, and the IRES showing translation activity in both linear and circular RNA, respectively, we did not find any significant difference among the IRES (Figure S4B–S4D).
Cell lysis for polysome fractionation
Two 15-cm plates of HEK-293T cells were transfected with mRuby-ZKSCAN1-oligo-split-eGFP for 48 h. Each 15-cm plate transfection contained 45 μg total DNA, 90 μL P3000, and 90 μL Lipofectamine Reagent in 4.5 mL Opti-MEM™ I Reduced Serum Medium (Thermo Fisher Scientific). We then added final concentration of 0.1 mg/mL of cycloheximide to one of the 15-cm plate and incubate it at 37°C for 3 min. The plate was placed on ice immediately and wash with 10 mL ice-cold PBS for three times. We then added 600 μL of lysis buffer (20 mM Tris-HCl (pH7.5), 150 mM NaCl, 5 mM MgCl2, 1mM DTT, and 0.5% Triton X-100) containing 0.1 mg/mL of cycloheximide to the plate and collected the cell lysate. For the other plate, we added 600 μL of lysis buffer containing 3.3 mM of puromycin and collected the cell lysate. Both samples were then incubated on ice for 10 min, and centrifuged at 14,000 rpm at 4°C for 10 min. The supernatant from the samples were collected for sucrose gradient fractionation.
Sucrose gradient fractionation and circRNA purification
700 μg of cycloheximide or puromycin treated cell lysates were carefully layered over linear sucrose gradient (10–60% for cycloheximide sample and 10–50% for puromycin sample), and centrifuged at 35,000 rpm at 4°C in a Beckman SW41 rotor for 2 h and 45 min. The gradients were then fractionated using an ISCO gradient fractionation system equipped with a UA-6 detector. The light RNP, 40S, 60S, 80S, and polysome fractions were monitored by the absorption of A254, and collected by a fixed time interval. 16 tubes of fractions were collected in total (#1 to #16). The total RNAs of each fraction were purified respectively using RNeasy Mini Kit with DNase treatment (Qiagen: 74004). We then performed the RNase R treatment in conditions that allows RNase R to digest potential G-quadruplex containing linear RNA efficiently (Xiao and Wilusz, 2019). We also optimized the RNase R digestion duration to obtain >100 fold circRNA enrichment over linear RNA (Figure S5B). The RNAs were treated with RNase R (MCLAB; 20 U/20 μg RNA) in 1X RNase R Buffer (0.2 M Tris-HCl (pH8.0), 1 mM MgCl2 and 1 M LiCl) at 37°C for 30 min to obtain circRNAs, and the circRNAs were purified using RNA Clean & Concentrator-25 (Zymo Research: R1013). In comparison to the CHX treatment, treating transfected cells with puromycin (PMY) caused translated circRNAs shifting from the poly(ribosome)-associated fractions to the 40S fraction (Figure S5C), suggesting that CHX treatment is able to capture translated circRNAs. We confirmed that the higher enrichment of (poly)ribosome-associated eGFP(+) oligos was not caused by the capturing efficiency or the expression level of the oligos (Figure S5D and S5E).
Quantification of (poly)ribosome-enriched circRNAs
The oligo sequences from each sample were amplified by RT-PCR with SuperScript™ IV One-Step RT-PCR System (Thermo Fisher Scientific: 12594100) using primers flanking the oligo sequence (Fw: GGGATCACTCTCGGCATGGA; Rv: GCTCCTCGCCCTTGCTCAC). cDNA libraries were generated and sequenced with HiSeq 4000 using the library generation method described above. We pooled the cDNA libraries from two independent replicates of polysome fractionation for the subsequent analysis. The reads of each designed oligo were mapped and calculated as eGFP expression quantification. For each captured oligo, we calculated the total RPKM in the (poly)ribosome fractions (#7 to #12) as the (poly)ribosome RPKM. We then defined an oligo as (poly)ribosome enriched if the proportion of (poly)ribosome RPKM ((poly)ribosome RPKM divided by the total RPKM (RPKM(poly)ribosome/RPKMtotal)) is greater than the proportion of number of (poly)ribosome fractions (the number of (poly)ribosome fractions divided by the total number of fractions (6/16 = 0.375)) of the oligo.
Analyzing translation initiation sites from QTI-seq
The Ribo-seq and QTI-seq data of HEK293 cell was obtained from Gao et al. study (SRA160745) (Gao et al., 2015). The raw reads were filtered, trimmed, and rRNA depleted as described in the study. The processed reads of Ribo-seq and QTI-seq were then mapped to the human genome (hg19) using Tophat2 and visualized on IGV. The positions of the TIS on the captured oligos were determined by mapping the TIS positions identified in the Gao et al. study to the positions of the oligos on the human genome. We further identified three types of eGFP(+)/TIS(+) oligos: (1) oligos containing the translated initiation site that have been annotated on linear transcripts (annotated TIS; aTIS), (2) oligos containing the translated initiation site that were not annotated on linear transcripts (non-annotated TIS; nTIS) which can be located at the 5’ UTR, CDS or 3’ UTR region of the transcripts, and (3) oligos containing both aTIS and nTIS signal (dual TIS; dTIS). The aTIS was defined as the translated initiation site that has been annotated on the linear transcript within 5 nucleotides downstream of the position of the mapped oligo, and the nTIS was defined as the translated initiation site that was not annotated on the linear transcript. Only the oligos with Ribo-seq reads and can be mapped to the human genome were included in the analysis. When we mapped the position of translation initiation sites on each oligo, we did not observe any translation initiation hot spots on the oligo (Figure S6B), suggesting that the translation initiation is not influenced by the position on the oligo.
18S rRNA Active Regions Identification
The synthetic oligo library was designed to contain 171 oligos with sequences complementary to human 18S rRNA, with a 10-nt sliding window between each consecutive oligo that reconstitutes the entire 1869-nt full-length 18S rRNA sequence. For each position on the 18S rRNA, we calculated the average eGFP expression of all the oligos containing the corresponding complementary sequence. We defined active regions as the complementary sequence within these active regions shows average eGFP expression higher than the background eGFP expression. We did not observe any hot spot positions of the active 7-mers located on the circRNA IRES (Figure S6C).
Scanning Mutagenesis Analysis
We included oligos designed for scanning mutagenesis of 99 reported IRESs and 734 native 5’ UTRs in our synthetic oligo library (Weingarten-Gabbay et al., 2016). The oligos were designed as non-overlapping sliding windows of 14-nt random substitution mutation tiling across the entire IRES or 5’ UTR. We defined the essential elements (Figure 3G; shaded in blue) as the region from the start position of the mutation (Figure 3G; black dots) where there was a sharp decrease of the IRES activity to the next start position of the mutation where the IRES activity was resumed or above the mean eGFP expression level. We further defined the local and global sensitive IRES as whether the IRES activity was affected by a single mutation or multiple mutations, respectively. The global sensitive IRESs have more structured sequences (i.e., significantly lower minimum free energy (MFE) value) compared to local sensitive IRESs (Weingarten-Gabbay et al., 2016); hence we suggest that the overall secondary structure of global sensitive IRESs are crucial for their IRES activity, because the more structured sequences are more likely to be affected by the mutation regardless of the position of the mutation. On the other hand, local sensitive IRESs have less structured sequences and are more resistant to the mutation; hence we suggest that the IRES activity of local sensitive IRESs may be regulated by a short sequence as the essential element.
Quantification of IRES activity of designed oligos
All designed oligos were ordered from IDT gBlocks Gene Fragments. Designed oligos were first cloned into the mRuby-ZKSCAN1-split-eGFP plasmid using NEBuilder® HiFi DNA Assembly (NEB BioLabs) to generate mRuby-ZKSCAN1-oligo-split-eGFP reporter plasmid. The reporter plasmid was then transfected into cells using Lipofectamine 3000 (Thermo Fisher Scientific) for HEK-293T cells (100 ng DNA) or Neon™ Transfection System 10 μL Kit (Thermo Fisher Scientific: MPK5000) for BJ cells (1 μg DNA) according to manufacturer’s protocol. Transfected cells were grown for five days. Cells were then trypsinized, centrifuged, and resuspended in FACS sorting buffer (2 mM EDTA and 2% Fetal Bovine Serum in PBS). The measurement of mRuby and eGFP fluorescence signal intensity was performed on LSRII Analyzer. We analyzed 10,000–20,000 cells for each sample. The IRES activity was determined by the ratio of the median fluorescence intensity (MFI) of eGFP and mRub3 in mRuby(+)/eGFP(+) cells (MFIeGFP/MFImRuby). The analysis was performed using FlowJo V10. We used mock transfection cells as the mRuby(−)/eGFP(−) double negative control for gating mRuby(+)/eGFP(+) cells. N = 4–6 independent replicates.
Generating circRNA containing oligo of interest for M2-seq
All designed oligos used in M2-seq were ordered from IDT gBlocks Gene Fragments. The M2-seq protocol was adapted from Cheng et al. (Cheng et al., 2017). Specifically, the sequence of oligo of interest was flanked with strong hairpins to insulate the oligo sequence (5’- GAACGACTCGAGTAGAGTCGAAAAGGCGCGCCAGTCCT -(N)174- GTTGTTGTTGTTGTTTCTTTTGTTGGAGTCTACTCGACTCCTTTC −3’). We then performed error-prone PCR with GeneMorph II Random Mutagenesis Kit (Agilent Technologies: 200550) according to manufacturer’s protocol using the primer set containing the overhang sequences for subsequent cloning (Fw: CATATGCCAGTGTTATGGATGAACGACTCGAGTAGAGTCGAAAAGGCGCGCCAGTCCT; Rv: AAAACATCTACTGAGGTTGTTGTTGTTGTTTCTTTTGTTGGAGTCTACTCGACTCCTTTC; underlined sequence represents the overhang sequence). The error-prone PCR oligos were cloned into a circRNA producing plasmid described in Chen et al. (Chen et al., 2017). Specifically, the autocatalytic-splicing circEGFP vector was pre-digested with ScaI-HF (NEB: R3122) and EcoRV-HF (NEB: R3195) and the error-prone PCR product was cloned into the vector using NEBuilder® HiFi DNA Assembly (NEB: E2621). It generated vectors containing the error-prone PCR oligos flanked by the permuted Td intron from T4 bacteria phage. The circRNAs of error-prone PCR oligos was produced using PCR with 500 nM T7 promoter overhang primers (Fw: TAATACGACTCACTATAGGGgggaattctagagaaaatttcgt; Rv: ctgcaggtcgactctagagaaag; underlined: T7 promoter overhang) followed by in vitro transcription using HiScribe™ T7 High Yield RNA Synthesis (NEB: E2040). During in vitro transcription, the permuted Td intron underwent autocatalytic back-splicing, producing circRNA of error-prone PCR oligos (Chen et al., 2017). The circRNAs were then purified with DNase digestion (TURBO DNase; Thermo Fisher Scientific: AM2238) at 37°C for 15 min and RNase R digestion (MACLAB; 20 U RNase R per 20 μg of RNA) in 1X RNase R Buffer (0.2 M Tris-HCl (pH8.0) at 37°C for 30 min. The samples were then separated on a 2% E-Gel™ EX Agarose Gel (Invitrogen) and the circRNA fragments with expected size were purified using Zymoclean Gel RNA Recovery Kit (Zymo Research: R1011).
Generating library for M2-seq and M2-net analysis
We first performed DMS mutagenesis on the circRNA generated above and purified the circRNA as described in Cheng et al. (Cheng et al., 2017). Specifically, the circRNAs were denatured at 95°C for 2 min and placed on ice for 1 min. We then allowed the circRNAs to refold in 5 μL of folding buffer (1.5 M sodium cacodylate (pH7.0) and 50 mM MgCl2) and 14.5 μL of water at 37°C for 30 min. The circRNAs were then treated with 2.5 μL DMS (15% DMS in ethanol) at 37°C for 6 min followed by quenching with 25 μL β-mercaptoethanol and 50 μL of water and circRNA purification by ethanol precipitation. The complimentary single-stranded DNA of the circRNA was generated by reverse transcription using SuperScript™ IV (Thermo Fisher Scientific: 18090050) with oligo sequence specific primer (TCTTTTGTTGGAGTCTACTCGACTCCTTT) and purified with DNA Clean & Concentrator-5 (Zymo Research: D4013). Three rounds of PCR using NEBNext Ultra II Q5 Master Mix (NEB) were performed to generate the cDNA library for next-generation sequencing. For the first PCR, oligo library sequence was amplified using the following primer set – Fw: GAACGACTCGAGTAGAGTCGAAAA; Rv: TCTTTTGTTGGAGTCTACTCGACTCCTTT. Each 50 μL PCR reaction contained 200 ng total DNA, 500 nM forward primer, and 500 nM reverse primer. The parameters for the PCR were 98°C for 1 min, 15 cycles of 98°C for 30 s, 67°C for 30 s, 72°C for 40 s, each, and finally one cycle of 72°C for 5 min. For the second PCR, sequences for adapters priming were added to the oligo library using the following primer set – Fw: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGAACGACTCGAGTAGAGTCGAAAA; Rv: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTCTTTTGTTGGAGTCTACTCGACTCCTTT (underline represents the sequence for illumina adapters priming). Each 50 μL PCR reaction contained 1.25 μL of the first PCR product, 500 nM forward primer, and 500 nM reverse primer. The parameters for the PCR were 98°C for 1 min, 12 cycles of 98°C for 30 s, 67°C for 30 s, 72°C for 40 s, each, and finally one cycle of 72°C for 5 min. For the third PCR, custom barcodes adapters were used for the reaction (Buenrostro et al., 2015b). Each 50 μL PCR reaction contained 1 μL of 1:20 diluted second PCR product, 1 μM Ad1 adapter, and 1 μM Ad2 adapter. The parameters for the PCR were 98°C for 1 min, 15 cycles of 98°C for 30 s, 72°C for 30 s, 72°C for 40 s, each, and finally one cycle of 72°C for 5 min. The PCR product was then separated on a 2% E-Gel™ EX Agarose Gel (Invitrogen) and the DNA fragments with expected size were purified using Zymoclean Gel DNA Recovery Kit (Zymo Research). The concentration of library was determined using KAPA Library Quantification Kit (Kapa Biosystems). The library was sequenced on MiSeq (illumina). We collected 1–1.5 million reads for each library. For each single-end MiSeq reads, the hairpins sequences and the common priming sequences were trimmed to extract the 174 nt oligo sequence. We then performed M2-net analysis, mutate-and-map visualization, RNA structural analysis, and RNA structure visualization as described in Cheng et al. (Cheng et al., 2017). Specifically, we utilize M2-seq software package to analyze the sequencing data of circRNA mutational profiling experiments and generate a 2D datasets. We then performed automated helix recognition of the 2D datasets using the M2-net algorithm included in the M2-seq software package. The secondary RNA structure was predicted based on the M2-net detected helics and visualized using the MATLAB-based wrapper rna_structure.m which is also included in the M2-seq software package.
18S rRNA complementarity and 40–60 nt SuRE analysis
We defined the IRES oligo as 18S rRNA complementarity(+) if it contains the number of 18S rRNA complementary 7-mers higher than the lower 95% confidence interval (CI) of the mean number of 18S rRNA complementary 7-mers of the eGFP(+) oligos (10.66). For SuRE analysis, we used the local MFE to quantify the SuRE element. We defined the IRES oligo as SuRE(+) if its local 40–60 nt MFE is lower than the lower 95% CI of the mean local 40–60 nt MFE of the eGFP(+) oligos (−0.748 kcal/mol). For endogenous translated circRNAs, we first determined the translated circRNA sequences from Ragan et al. (2019) by extracting the annotated circRNA sequences overlapped with the circRNA genomic coordinates. We then calculated the total number of 18S rRNA complementary 7-mers in the endogenous translated circRNAs and normalized it to 174 nt in length. We defined the circRNA as 18S rRNA complementarity(+) if the normalized number of 18S rRNA complementary 7-mers is higher than 10.66. To quantify the 40–60 SuRE of the circRNA, we split the circRNA into non-overlapping 174 nt windows and calculated the local MFE of 40–60 nt region for each of the window on the circRNA. We then defined the circRNA as SuRE(+) if the mean MFE of all the windows of the circRNA is lower than −0.748 kcal/mol.
Disrupt IRES with locked nucleic acid (LNA)
Anti-sense LNA oligos (20 nt) against specific regions on the IRES were synthesized from IDT Affinity Plus DNA & RNA Oligonucleotides. LNA-18S: the LNA targeting the 18S rRNA complementary sequence on the IRES; LNA-SuRE: the LNA targeting the SuRE at the 40–60 nt position on the IRES; LNA-Rnd: the LNA targeting the random position downstream of LNA-18S or LNA-SuRE on the IRES. The corresponding targeted regions on each IRES were IRES-876: 41–60 nt (SuRE), 97–116 nt (Rnd), and 152–171 nt (18S); IRES-6063: 5–24 nt (18S), 41–60 nt (SuRE), and 80–99 nt (Rnd); IRES-7005: 41–60 nt (SuRE), 93–112 nt (Rnd), and 143–162 nt (18S); IRES-8228: 41–60 nt (SuRE), 88–107 nt (Rnd), and 154–173 nt (18S); IRES-8788: 41–60 nt (SuRE), 95–114 nt (18S), and 135–154 nt (Rnd). HEK-293T cells seeded in a 24-well plate were co-transfected with 300 nmol anti-sense LNA and 100 ng mRuby-ZKSCAN-IRES-split-eGFP reporter plasmid carrying the corresponding IRES using Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher Scientific) according to manufacturer’s protocol. Transfected cells were grown for five days. Cells were then trypsinized with TrypLE Express (Thermo Fisher Scientific), centrifuged, and resuspended in FACS sorting buffer (2 mM EDTA and 2% Fetal Bovine Serum in PBS). The measurement of mRuby and eGFP fluorescence signal intensity was performed on a LSRII Analyzer. We analyzed 10,000–20,000 cells for each sample. The IRES activity was determined by the ratio of the median fluorescence intensity (MFI) of eGFP and mRuby in mRuby(+)/eGFP(+) cells (MFIeGFP/MFImRuby). The analysis was performed using FlowJo V10. We used mock transfection cells as a mRuby(−)/eGFP(−) double negative control for gating mRuby(+)/eGFP(+) cells. We also confirmed that the result was not confounded by the change of circRNA expression level because co-transfecting the LNAs generally did not change the circRNA expression level (Figure S8A and S8B).
Quantification of LNA-Quantitative Translation Initiation (LNA-QTI)
The LNA-QTI method was adapted from ribosome profiling and QTI-seq (Gao et al., 2015; Ingolia et al., 2012). For LNA transfection, BJ cells (for IRES #8228) or HeLa cells (for IRES #876, #6063, #7005, and #8788) seeded on a 6-well plate were transfected with 1.5 μmol anti-sense LNAs using the Neon transfection system (Thermo Fisher Scientific; for BJ cells) or Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher Scientific; for HeLa cells). Transfected cells were grown for 2–3 days. For QTI, to isolate translating RNAs, we treated LNA-transfected cells with lactimidomycin (LTM) followed by puromycin (PMY) treatment, sedimented ribosome-associated RNAs with sucrose cushion, and purified the translating RNAs (Figure 5C). We then quantified the level of translating endogenous circRNA which contains the LNA-targeted IRES by qRT-PCR using the divergent primers spanning across the back-splicing junction of the circRNA. Specifically, 50 μM final concentration of LTM was added to the media and incubated at 37°C for 30 min. Cells were wash in 1 mL cold PBS twice and lysed in 500 μL of lysis buffer (20 mM Tris-HCl (pH7.5), 150 mM NaCl, 5 mM MgCl2, 1mM DTT, and 0.5% Triton X-100) containing 3.3 mM of PMY. The samples were then incubated on ice for 10 min, and centrifuged at 14,000 rpm at 4°C for 10 min. The supernatant from the samples were carefully layered over 1 M sucrose cushion with 10 μL SUPERase·In RNase inhibitor (Thermo Fisher Scientific) in 10.4 mL polycarbonate ultracentrifuge tubes, and centrifuged at 50,000 rpm at 4°C in a Beckman 70.1 Ti rotor for 3 h to pellet ribosomes. Supernatant was gently removed from the tube and the ribosomal pellet was resuspended in 400 μL nuclease-free water. Ribosome-associated RNA was purified using RNeasy Mini Kit with DNase treatment (Qiagen: 74004) according to manufacturer’s protocol. The level of the corresponding circRNA of each sample was quantified by qRT-PCR using divergent primers spanning across the back-splicing junction of the circRNA.
Determine peptide sequences of circRNA-encoded proteins (circORFs)
We first determined potential endogenous protein-coding circRNAs by mapping the eGFP(+) oligo sequences (IRES) to all of the human endogenous circRNAs on circBase (Glazar et al., 2014). We gated against false positive by only considering circRNAs that have been annotated by two different circRNA prediction algorithms, and only including circRNAs with high mapping score in our analysis (Hansen, 2018). The circRNAs harboring perfect-matched IRES sequence(s) (IRES(+) circRNAs) were determined as potential protein-coding circRNAs. The circRNAs were separated as different circRNAs if they contain more than one IRES. To further determine the sequences of the peptide encoded by these circRNAs, we utilize the position of the mapped IRES on the circRNA to determine the region on the circRNA where the translation start site was located. We then generated the list of predicted circORF sequences by ORF analysis from the immediate downstream translation initiation codon (AUG) of the mapped IRES region for each circRNA. In the event of translation initiating from non-canonical start codons, we also performed ORF analysis from the top three reading-frames (+1, +2, and +3) with non-canonical start codons from the mapped IRES position and generated the predicted circORF sequences accordingly. The non-canonical start codons were collected from the translation initiation codons identified from the ribosome profiling (Ingolia et al., 2011) and the QTI-seq (Gao et al., 2015). In addition, in the event of translation initiating within the IRESs, we also included all possible circORFs that may start translation within the IRESs for all top three reading-frames. For the circORFs with infinite translation due to in-frame IRES, we capped the circORFs at 5 repeating peptide sequences. We combined circRNAs together if the same circORF sequence was encoded by multiple circRNAs. We merged different circRNA isoforms into one single circORF (the circORF generated from the longest circRNA isoform). To be conservative, we also examined micropeptides encoded by linear RNAs (Chen et al., 2020) and excluded any overlapped circORFs in our final list (n = 5 overlapped circORFs). Traditional ORF analysis was done by performing ORF analysis with both canonical and non-canonical start codons from any position on the IRES(+) circRNA. Peptides with less than eight amino acids were excluded from the list.
GO term and Pfam analysis
GO term analysis was performed with PANTHER (v14.1) Overrepresentation Test (Fisher’s exact test with FDR correction) using the annotation of the parent genes of IRES(+) circRNAs. The reference list was the parent genes of total synthetic oligos mapped circRNAs. The organism was set as Homo sapiens, and the annotation dataset was set as biological process. Pfam analysis was performed using HMMSCAN program from EMBL-EBI with the peptide sequences of all possible circRNA-coding proteins determined as described above. The biological functions of conserved domains were determined using the annotations from NCBI Conserved Domains database.
sORF peptides analysis
The sORF sequences were obtained from sorfs.org database (Olexiouk et al., 2017). We first extracted the sORFs with good FLOSS-score determined by the FLOSS algorithm on the database. The sORF sequences were then matched against the peptide sequences in the current human proteome database (UniProt). The matched sORFs were excluded from the subsequent analysis. The remaining sORFs were then matched against the peptide sequences in our circORF list. The circORF was determined as sORF(+) if it overlaps with the sORF sequences.
Mass spectrometry (MS)-based proteomic analysis
The MS-based proteomic analysis was performed on a previously published dataset (iPSC: PXD014031 (PRIDE) (Chen et al., 2020); SCBC: PXD006895 (ProteomeXchange) (Orre et al., 2019); GTEx: PXD016999 (ProteomeXchange) (Jiang et al., 2020)) or LC-MS/MS data of the protein/cell extract: PXD025233 (ProteomeXchange) (K562: digested protein extract (Promega); U2OS: ATCC HTB-96; H358: ATCC CRL-5807). The MS/MS datasets include a wide range of cell lines, including K562, H358, U2OS, 5 subcellular compartments SubCellBarCode (SCBC) database, and 32 normal human tissues from the GTEx collection. In addition to the transformed cell lines, we are able to capture circORFs in the peptidomics of normal human tissues (Jiang et al., 2020), suggesting that these circORFs are expressed in normal human cells. We first appended our potential circORF sequences to the human proteome database (UniProt: UP000005640_9606.fa) to generated a combined proteome database (circORF + linear proteome). According to the nature of the MS dataset, we performed peptide-spectrum match (PSM) with MaxQuant (iPSC and SCBC), Byonic (Protein Metrics) (K562, U2OS and h358) or Trans Proteomic Pipeline (TPP) (GTEx) on the combined proteome database. The PSM threshold was set as 1% peptide false discovery rate (FDR) for MaxQuant, score > 300 for Byonic, and protein group probability > 0.95 for TPP. For general PSM parameters, peptides with a minimum length of seven amino acids were considered for the search including N-terminal acetylation and methionine oxidation as variable modifications and cysteine carbamidomethylation as fixed modification. Enzyme specificity was set to trypsin cleaving C-terminal to arginine and lysine. A maximum of two missed cleavages were allowed. Maximum precursor and fragment ion mass tolerance was set to 4.5 and 20 ppm. The contaminant database containing 245 frequently observed contaminants such as human keratins, bovine serum proteins, and protease was included for the PSM. Further PSM configureation for each software is decribed in Table S6 respectively. To distinguish circORF from the linear proteome, we excluded the circORF that are matched by the tryptic peptide that also match the linear proteome. For the K562, U2OS and H358 datasets, we repeated the above procedure with same parameters using the circORF library only and added resulting peptide identifications without corresponding matches in the previous analysis using the combined library. It is noteworthy that we observed some circORF peptides overlapping with the linear proteome peptides after the PSM. We carefully examined these peptides and found out that all of these peptides can only be generated from the circORFs because the linear proteome peptides do not contain the trypsin cleavage site required to generate the captured tryptic peptides for PSM. On the other hand, the matching circORFs contain the novel trypsin cleavable sites that can generate these peptides due to different reading-frames from the back-splicing; hence we believe that these peptides were specifically captured from circORFs. To calculate the circORF discovery rate, we overlapped the circRNAs identified in iPSC (Lei et al., 2018) and K562 (Okholm et al., 2020) with the IRES-containing circRNAs we identified (Table S7), and calculated the percentage of the number of circORFs detected by peptidomics over the number of overlapping IRES-containing circRNAs expressed in the corresponding cell line. The discovery rate of lowly expressed mRNA-encoded proteins was estimated by calculating the proportion of the MS captured tryptic peptides (Chen et al., 2020) among total possible tryptic peptides of the mRNAs with the RPKM close to the estimated mean circRNA RPKM (0.065) (Dang et al., 2016).
The human leukocyte antigen (HLA) peptidome analysis
The HLA peptidome analysis was performed on a previously published HLA class I dataset, describing the HLA class I peptidomes of six allotype-resolved cell lines (PRIDE: PXD014031) (Bassani-Sternberg et al., 2015). The HLA peptidomics was performed with MaxQuant using the parameters as described in proteomic analysis (1% peptide FDR). Protease specificity was set to unspecific, possible peptide identifications were restricted from 8 to 15 amino acids, maximum peptide mass was set to 1500 Da, and modification was set to without fixed modifications.
Ribosome footprinting (RFP) analysis
The RFP data was obtained from Chen et al., (2020) (GSM3791725, GSM3791726, GSM3791727, GSM3791728, GSM3791729, and GSM3791730). The reads were pooled, rRNA depleted, and mapped to the hg19 genome as previously described (Chen et al., 2020). Specifically, the pooled reads were aligned to the human rRNA sequences, discard the rRNA alignments, and collect unaligned reads using Bowtie2. The rRNA depleted reads were further aligned to the hg19 genome, discard the transcriptome alignments, and collect unaligned reads using TopHat2. The unaligned reads were then mapped with Bowtie2 aligner to an artificial genome containing the +/− 40-nt (the maximum read length of RFP) from the back-splicing junction of the circRNAs producing circROFs that can be captured by peptidomics with spacers of 50 N’s. The reads that do not span across the back-splicing junction were excluded from the analysis.
IP-LC-MS/MS analysis
A 15-cm plate of BJ cells were lysed in 1 mL Pierce™ IP Lysis Buffer (ThermoFisher Scientific: 87788) on ice for 10 min. The lysate was then sonicated with Bioruptor UCD-200 at high intensity for 15 min (30 seconds on, 30 seconds off) at 4°C and pre-cleared in 50 μL Pierce™ Protein A/G Magnetic Beads (ThermoFisher Scientific: 88803) at room temperature for 30 min. 7 uL of custom circFGFR1p antibodies (ThermoFisher Scientific; antigen peptide: GQFEKEDRAHCGVSMEMW) or normal rabbit IgG (Sigma-Aldrich: 12–370) was added and incubated at 4°C overnight. We then added 70 uL Pierce™ Protein A/G Magnetic Beads and incubate at 4°C for 2–3 h. The samples were washed four times in Pierce™ IP Lysis Buffer and once in PBS. Samples were eluted in sample buffer and boiled at 95°C for 5 min. Proteins were separated on a NuPAGE 4–12% Bis-Tris Protein Gel (ThermoFisher Scientific: NP0335BOX). Bands with 30–45 kDa were extracted according to the protein marker to separate circFGFR1p from FGFR1 and were sent to Vincent Coates Foundation Mass Spectrometry Laboratory, Stanford University Mass Spectrometry for LC/MS analysis with their LC-MS/MS pipeline on Thermo QE-HFX (ThermoFisher Scientific). The PSM was performed using Byonic (Protein Metrics) as described above for the proteomic analysis (score > 300) on the combined proteome database (circFGFR1p + UniProt proteome). The tryptic peptides that match the circFGFR1p (both the unique region and the region overlapped with FGFR1) were considered to be captured circFGFR1p peptides (ProteomeXchange: PXD024993).
Parallel reaction monitoring-mass spectrometry (PRM-MS)
K562, U2OS and BJ cell lysates were prepared as described in IP-LC-MS/MS. 40 μg total proteins were separated on a NuPAGE 4–12% Bis-Tris Protein Gel (ThermoFisher Scientific). Regions of interest (35–45 kDa for BJ and whole gel for K562 and U2OS) were extracted according to the protein marker. The excised gel pieces were then reduced with 5 mM DTT in 50 mM ammonium bicarbonate at 55°C for 30 min. The gel pieces were rinsed 2 times with 50% acetonitrile, 50 mM ammonium bicarbonate and placed in a speed vacuum for 5 min. Digestion was performed with Trypsin/LysC (Promega) in the presence of 0.02% protease max (Promega) in a standard overnight digestion at 37°C. Samples were then centrifuged and the solvent containing the peptides was collected. Further peptide extraction was performed by the addition of 60% acetonitrile, 39.9% water, 0.1% formic acid and incubation for 10–15 min. The peptide pools were dried in a speed vacuum. Samples were then reconstituted in 20 μL reconstitution buffer (2% acetonitrile with 0.1% formic acid) spiked with 5 pM of the heavy isotope labeled reference peptide and 2 μL of it was injected on the instrument. Mass spectrometry was performed on a Q Exactive HF-X Hybrid Quadrupole - Orbitrap mass spectrometer (ThermoFisher Scientific) with liquid chromatography performed using a Nanoacquity UPLC (Waters Corporation). For PRM-MS, peptides were directly injected onto a 50 cm μPAC analytical column (Pharmafluidics) using a gradient (3–65% B, followed by a high-B wash) of 80 min. The mass spectrometer was set using a PRM method. HCD fragmentation was used for MS/MS spectra generation. MS/MS resolution was 120,000 (at m/z 200) with an AGC target value of 1 × 106 ions, a maximum fill time of 250 ms and an isolation window of 4.0 m/z. For PRM-MS analysis, the .RAW data files were processed using Byonic (Protein Metrics) for peptide identification. Proteolysis with Trypsin/LysC was assumed to be semi-specific allowing for N-ragged cleavage with up to two missed cleavage sites. Precursor and fragment mass accuracies were held within 12 ppm. Proteins were held to a false discovery rate of 1%. The .RAW data files were further imported into Skyline 19.1.0.193 (MacCoss Lab) to generate XIC and perform peak integration (ProteomeXchange: PXD025235 and PXD025203).
Immunofluorescence
Cells were fixed on poly-d lysine coated coverslips (Corning: 354086) with 4% formaldehyde at room temperature for 20 min, and blocked with 10% normal goat serum (Thermo Fisher Scientific: 50062Z) at room temperature for 20 min. Cells were then incubated with primary antibodies in blocking buffer at 4°C overnight, followed by incubating with secondary antibodies at room temperature for 1 h. Primary antibodies were diluted in blocking buffer as follows: anti-FLAG® M2 (Sigma-Aldrich; F1804) (1:150), and anti-HA (Abcam; ab9110) (1:150). Secondary antibodies were diluted in blocking buffer as following: Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (ThermoFisher Scientific: A-11029) (1:1500), and Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (ThermoFisher Scientific: A-11012) (1:1500). Cells were imaged using Inverted Zeiss LSM 880 Laser Scanning Confocal Microscope with the PLAN APO for AiryScan 63x/1.40 oil objective.
siRNA knock down and protein expression
For siRNA knockdown experiments, 500,000 BJ cells were transfected with 200 nM siRNAs using the Neon transfection system according to manufacturer’s protocol with suggested settings. Cells were processed and analyzed after 48–72 h. The siRNAs used were ON-TARGET plus non-targeting pool (Dharmacon: D-001810–10-05) and ON-TARGET plus custom circFGFR1 specific siRNA (Sense sequence: GAAGGGUCAGUUUGAAAAGGUU) (Dharmacon: L-003131–00-0005). For protein expression, FGFR1 (cloned from pHAGE-FGFR1 plasmid (Addgene: 116740)) or FLAG tagged circFGFR1p (synthesized from IDT gBlocks Gene Fragments) were cloned into a CMV expressing plasmid (backbone vector: Addgene: 36084). 200–500 ng of plasmids were transfected into the cells using the Neon transfection system. Cells were then processed and analyzed after 48–72 h.
Western blotting
BJ cells were lysed in RIPA Lysis Buffer (Thermo Fisher Scientific) containing 25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS and Halt Protease and Phosphatase Inhibitor Cocktails (Thermo Fisher Scientific), and sonicated with Bioruptor UCD-200 at high intensity for 10 min (30 seconds on, 30 seconds off) at 4°C. 15–30 μg of total protein was separated by SDS-PAGE and transferred to nitrocellulose membranes using iBlot™ 2 Gel Transfer Device (P0) followed by blocking with 4% non-fat milk in 0.05% Tween-20 diluted in PBS. For heat shock experiments, cells were cultured at 45°C for three hours and the total protein was collected as described above. Primary antibodies were diluted in blocking buffer as follows: anti-GFP antibody (Abcam: ab290) (1:300), anti-CD4 antibody (Abcam: ab133616) (1:500), anti-Cre recombinase antibody (Abcam: ab24607) (1:200), anti-SMAD5 antibody[3H9] (Novus Biologicals: NBP2–37648) (1:200), anti-FGFR1 (M17A3) (Novus: NB100:2079; Ab-both) (1:200), anti-FGFR1 (phospho Y654) antibody (Abcam: ab59194) (1:200), anti-FLAG® M2 (Sigma-Aldrich; F1804) (1:400), anti-HA (Abcam: ab9110) (1:400), anti-beta Tubulin antibody (Abcam: ab6046) (1:500), and anti-GAPDH [EPR16891] (Abcam: ab181602) (1:500). Secondary antibodies were diluted in blocking buffer as following: IRDye® 680RD Goat anti-Mouse IgG (H + L) (LI-COR; 926–68070) (1:5,000) and IRDye® 800CW Goat anti-Rabbit IgG (H + L) (LI-COR; 926–32611) (1:5,000). Blots were imaged with Odyssey® CLx Imager (LI-COR Biosciences). N = 3 independent replicates for the Western blot quantification.
Cell proliferation assay
Transfected cells were seeded on 96-well black cell culture plates (Stellar Scientific) at a density of 2,000 cells/well in DMEM (Thermo Fisher Scientific: 11995–073) medium supplemented with 10% FBS (Thermo Fisher Scientific: SH30071.03) After 4–6 hours of incubation, the medium was replaced with DMEM containing 0.1% FBS overnight. FGF-1 at a final concentration of 50 ng/mL was then added to the medium and the cells were further incubated for 1–4 days. The number of viable cells was quantified using PrestoBlue™ Cell Viability Reagent (Thermo Fisher Scientific: A13261) according to manufacturer’s protocol by measuring the fluorescence intensity of the well (Ex/Em: 560/590) on SpectraMax® M5 Microplate Reader (Molecular Devices LLC). N = 3–5 independent replicates.
Quantification and Statistical Analysis
All statistical analysis was done with the software Graphpad Prism (GraphPad Software, La Jolla, CA). Outliers were not shown on a Tukey box-plot. Information about the statistical analysis and sample size are described in in the figure legends or in the corresponding Method details section. Data in graphs are represented as mean ± SEM. Ns: not significant, *: p < 0.05, **: p < 0.01, ***: p < 0.005, ****: p < 0.0001.
Supplementary Material
Table S2. The IRES activity of oligos among different categories, related to Figure 1.
Table S3. The full-length oligo sequences of the circular IRESs and the linear IRESs, related to Figure 1.
Table S4. IRES activity of the oligos harboring the human 18S rRNA complementary sequence, related to Figure 3.
Table S1. The sequences and their corresponding IRES activity of the oligo in the synthetic oligo library, related to STAR Methods
Table S5. The human endogenous IRES-containing circRNAs, related to Figure 6.
Table S7. The RT-qPCR primer sequences, related to STAR Methods.
Table S6. The putative circORF sequences and the peptidomic analysis, related to Figure 6.
HIGHLIGHTS.
A family of IRES elements drives extensive circRNA translation
Focal RNA structure and 18S complementarity facilitate endogenous circRNA translation
IRES elements nominate candidate circRNA-encoded proteins
circFGFR1p is a dominant negative FGF receptor and suppresses proliferation during stress
ACKNOWLEDGMENTS
We thank Dr. Peter Sarnow and Dr. Kuo-Feng Weng for assisting the polysome profiling experiments; Dr. Rhiju Das and Ved Topkar for helping the M2-seq and M2-net analyses; Dr. Matthias Mann and Dr. Andreas-David Brunner for the MS analytical pipeline. Cell sorting/flow cytometry analysis was done in the Stanford Shared FACS Facility. Imaging was done in the Stanford Cell Sciences Imaging Facility. Mass spectrometry was done in the Stanford University Mass Spectrometry. J.C. is funded by the NIH K99/R00 Pathway to Independence Award (K99-GM134154). J.S.W. is an Investigator of the Howard Hughes Medical Institute. C.-K.C. is supported by the Helen Hay Whitney Foundation Fellowship. This work was funded by NIH (R01-HG004361, RM1-HG007735, R35-CA209919 to H.Y.C. and R01-CA250534 to P.K.J.). H.Y.C. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
DECLARATION OF INTERESTS
Stanford University has filed patent applications based on this work, and H.Y.C. and C.-K.C. are named as co-inventors. H.Y.C. is a co-founder and advisor of Accent Therapeutics, Boundless Bio, Cartography Biosciences and Circ Bio. H.Y.C. is an advisor of 10X Genomics, Arsenal Biosciences, and Spring Discovery. H.Y.C. is a member of the Molecular Cell advisory board.
REFERENCES
- Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, and Ito Y (2015). Rolling circle translation of circular RNA in living human cells. Scientific reports 5, 16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken CE, and Lorsch JR (2012). A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 19, 568–576. [DOI] [PubMed] [Google Scholar]
- Alkemar G, and Nygård O (2004). Secondary structure of two regions in expansion segments ES3 and ES6 with the potential of forming a tertiary interaction in eukaryotic 40S ribosomal subunits. RNA 10, 403–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. (2010). Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc Natl Acad Sci U S A 107, 19748–19753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, and Kadener S (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56, 55–66. [DOI] [PubMed] [Google Scholar]
- Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, and Pils D (2015). Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Scientific reports 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranick BT, Lemp NA, Nagashima J, Hiraoka K, Kasahara N, and Logg CR (2008). Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proceedings of the National Academy of Sciences 105, 4733–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, and Mann M (2015). Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Molecular & Cellular Proteomics 14, 658–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat P, Shwetha S, Sharma DK, Joseph AP, Srinivasan N, and Das S (2015). The beta hairpin structure within ribosomal protein S5 mediates interplay between domains II and IV and regulates HCV IRES function. Nucleic Acids Res 43, 2888–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Both GW, Furuichi Y, Muthukrishnan S, and Shatkin A (1975). Ribosome binding to reovirus mRNA in protein synthesis requires 5’ terminal 7-methylguanosine. Cell 6, 185–195. [DOI] [PubMed] [Google Scholar]
- Bourougaa K, Naski N, Boularan C, Mlynarczyk C, Candeias MM, Marullo S, and Fåhraeus R (2010). Endoplasmic reticulum stress induces G2 cell-cycle arrest via mRNA translation of the p53 isoform p53/47. Molecular cell 38, 78–88. [DOI] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, and Weissman JS (2012). High-resolution view of the yeast meiotic program revealed by ribosome profiling. science 335, 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, and Greenleaf WJ (2015a). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, and Greenleaf WJ (2015b). Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, and Willis AE (2006). Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Molecular cell 23, 401–412. [DOI] [PubMed] [Google Scholar]
- Chandramouli P, Topf M, Ménétret J-F, Eswar N, Cannone JJ, Gutell RR, Sali A, and Akey CW (2008). Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure 16, 535–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasse H, Mulner-Lorillon O, Boulben S, Glippa V, Morales J, and Cormier P (2016). Cyclin B Translation Depends on mTOR Activity after Fertilization in Sea Urchin Embryos. PLoS One 11, e0150318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, and Sarnow P (1995). Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268, 415–417. [DOI] [PubMed] [Google Scholar]
- Chen I, Chen CY, and Chuang TJ (2015). Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA 6, 563–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Brunner A-D, Cogan JZ, Nuñez JK, Fields AP, Adamson B, Itzhak DN, Li JY, Mann M, and Leonetti MD (2020). Pervasive functional translation of noncanonical human open reading frames. Science 367, 1140–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, and Conti M (2011). Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes & development 25, 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, and Shan G (2015). Circular RNAs remain peculiarly unclear in biogenesis and function. Sci China Life Sci 58, 616–618. [DOI] [PubMed] [Google Scholar]
- Chen X, Han P, Zhou T, Guo X, Song X, and Li Y (2016). circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep 6, 34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, Broughton JP, Kim J, Cadena C, and Pulendran B (2019). N6-methyladenosine modification controls circular RNA immunity. Molecular cell 76, 96–109. e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, and Chang HY (2017). Sensing Self and Foreign Circular RNAs by Intron Identity. Molecular Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Kladwang W, Yesselman JD, and Das R (2017). RNA structure inference through chemical mapping after accidental or intentional mutations. Proceedings of the National Academy of Sciences 114, 9876–9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, and Beyaert R (2000). Identification and characterization of a novel cell cycle–regulated internal ribosome entry site. Molecular cell 5, 597–605. [DOI] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nature biotechnology 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cuesta R, Laroia G, and Schneider RJ (2000). Chaperone hsp27 inhibits translation during heat shock by binding eIF4G and facilitating dissociation of cap-initiation complexes. Genes & development 14, 1460–1470. [PMC free article] [PubMed] [Google Scholar]
- D’Lima NG, Ma J, Winkler L, Chu Q, Loh KH, Corpuz EO, Budnik BA, Lykke-Andersen J, Saghatelian A, and Slavoff SA (2017). A human microprotein that interacts with the mRNA decapping complex. Nature chemical biology 13, 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orazio KN, and Green R (2021). Ribosome states signal RNA quality control. Molecular cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Yan L, Hu B, Fan X, Ren Y, Li R, Lian Y, Yan J, Li Q, and Zhang Y (2016). Tracing the expression of circular RNAs in human pre-implantation embryos. Genome biology 17, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille F, Unoson C, Vogel J, and Wagner EGH (2007). An antisense RNA inhibits translation by competing with standby ribosomes. Molecular cell 26, 381–392. [DOI] [PubMed] [Google Scholar]
- Del Prete MJ, Vernal R, Dolznig H, Müllner EW, and Garcia-Sanz JA (2007). Isolation of polysome-bound mRNA from solid tissues amenable for RT-PCR and profiling experiments. Rna 13, 414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, and Patton JG (2016). Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Scientific reports 6, 37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Yang Y, and Wang Z (2019). Pervasive translation of circular RNAs driven by short IRES-like elements. BioRxiv, 473207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Wan J, Liu B, Ma M, Shen B, and Qian SB (2015). Quantitative profiling of initiating ribosomes in vivo. Nat Methods 12, 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazar P, Papavasileiou P, and Rajewsky N (2014). circBase: a database for circular RNAs. RNA 20, 1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg A (2007). Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochemical Society Transactions 35, 12–17. [DOI] [PubMed] [Google Scholar]
- Granados-Riveron JT, and Aquino-Jarquin G (2016). The complexity of the translation ability of circRNAs. Biochim Biophys Acta 1859, 1245–1251. [DOI] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, and Hofacker IL (2008). The Vienna RNA websuite. Nucleic Acids Res 36, W70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, and Lander ES (2013). Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154, 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB (2018). Improved circRNA identification by combining prediction algorithms. Frontiers in cell and developmental biology 6, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, and Kjems J (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384. [DOI] [PubMed] [Google Scholar]
- Hartford CCR, and Lal A (2020). When long noncoding becomes protein coding. Molecular and cellular biology 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbreteau CH, Weill L, Décimo D, Prévôt D, Darlix J-L, Sargueil B, and Ohlmann T (2005). HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nature structural & molecular biology 12, 1001. [DOI] [PubMed] [Google Scholar]
- Hershey JW, Sonenberg N, and Mathews MB (2012). Principles of translational control: an overview. Cold Spring Harb Perspect Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG (2006). eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 31, 553–562. [DOI] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, and Schuster P (1994). Fast folding and comparison of RNA secondary structures. Monatshefte für Chemie/Chemical Monthly 125, 167–188. [Google Scholar]
- Holcik M, Yeh C, Korneluk RG, and Chow T (2000). Translational upregulation of X-linked inhibitor of apoptosis (XIAP) increases resistance to radiation induced cell death. Oncogene 19, 4174–4177. [DOI] [PubMed] [Google Scholar]
- Huang C, Liang D, Tatomer DC, and Wilusz JE (2018). A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev 32, 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huret J-L (2009). FGFR1 (fibroblast growth factor receptor 1). Atlas of Genetics and Cytogenetics in Oncology and Haematology. [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, and Weissman JS (2014). Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell reports 8, 1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, and Weissman JS (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, and Weissman JS (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger JA, Turner DH, and Zuker M (1989). Improved predictions of secondary structures for RNA. Proc Natl Acad Sci U S A 86, 7706–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, and Wimmer E (1988). A segment of the 5’ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62, 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, and Sharpless NE (2014). Detecting and characterizing circular RNAs. Nat Biotechnol 32, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, and Sharpless NE (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Wang M, Lin S, Jian R, Li X, Chan J, Dong G, Fang H, Robinson AE, and Aguet F (2020). A quantitative proteome map of the human body. Cell 183, 269–283. e219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G, and Sarnow P (1998). Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. Rna 4, 1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Q, and Pomerening JR (2012). Punctuated cyclin synthesis drives early embryonic cell cycle oscillations. Mol Biol Cell 23, 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology 14, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I, Yuan B, Eichhorn SW, Dzeyk K, Krijgsveld J, Bartel DP, and Orr-Weaver TL (2014). Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell reports 7, 1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S, Radek A, Vogel C, and Penalva LO (2013). Translation regulation gets its ‘omics’ moment. Wiley Interdisciplinary Reviews: RNA 4, 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulcheski FR, Christoff AP, and Margis R (2016). Circular RNAs are miRNA sponges and can be used as a new class of biomarker. Journal of biotechnology 238, 42–51. [DOI] [PubMed] [Google Scholar]
- Kurosaki T, and Maquat LE (2016). Nonsense-mediated mRNA decay in humans at a glance. Journal of cell science 129, 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O, Tian B, and Sonenberg N (2013). Toward a genome-wide landscape of translational control. Cold Spring Harb Perspect Biol 5, a012302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, and Wade M (2017). Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Molecular cell 66, 22–37. e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei W, Feng T, Fang X, Yu Y, Yang J, Zhao Z-A, Liu J, Shen Z, Deng W, and Hu S (2018). Signature of circular RNAs in human induced pluripotent stem cells and derived cardiomyocytes. Stem cell research & therapy 9, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LJ, Leng RX, Fan YG, Pan HF, and Ye DQ (2017a). Translation of noncoding RNAs: Focus on lncRNAs, pri-miRNAs, and circRNAs. Exp Cell Res 361, 1–8. [DOI] [PubMed] [Google Scholar]
- Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L, and Chen LL (2017b). Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol Cell 67, 214–227 e217. [DOI] [PubMed] [Google Scholar]
- Liang D, and Wilusz JE (2014). Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28, 2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W-C, Wong C-W, Liang P-P, Shi M, Cao Y, Rao S-T, Tsui SK-W, Waye MM-Y, Zhang Q, and Fu W-M (2019). Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome biology 20, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, Xue W, Cui Y, Dong K, Ding H, et al. (2019). Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 177, 865–880 e821. [DOI] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Zu Siederdissen CH, Tafer H, Flamm C, Stadler PF, and Hofacker IL (2011). ViennaRNA Package 2.0. Algorithms for molecular biology 6, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ (2009). Structure and function of HCV IRES domains. Virus Res 139, 166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ (2013). Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet 4, 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, and MacCoss MJ (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malygin AA, Kossinova OA, Shatsky IN, and Karpova GG (2013). HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res 41, 8706–8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O, and Kimchi A (2008). DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Molecular cell 30, 447–459. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, and Munschauer M (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333. [DOI] [PubMed] [Google Scholar]
- Meng Z, Jackson NL, Shcherbakov OD, Choi H, and Blume SW (2010). The human IGF1R IRES likely operates through a Shine-Dalgarno-like interaction with the G961 loop (E-site) of the 18S rRNA and is kinetically modulated by a naturally polymorphic polyU loop. J Cell Biochem 110, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian S-B, and Jaffrey SR (2015). 5′ UTR m6A promotes cap-independent translation. Cell 163, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokrejš M, Mašek T, Vopálenský V, Hlubuček P, Delbos P, and Pospíšek M (2009). IRESite--a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic acids research 38, D131–D136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair AA, Niu N, Tang X, Thompson KJ, Wang L, Kocher J-P, Subramanian S, and Kalari KR (2016). Circular RNAs and their associations with breast cancer subtypes. Oncotarget 7, 80967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefs JM, Van de Peer Y, De Rijk P, Goris A, and De Wachter R (1991). Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res 19 Suppl, 1987–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins TA, Harder ZM, Korneluk RG, and Holčík M (2003). Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. Journal of Biological Chemistry 278, 3572–3579. [DOI] [PubMed] [Google Scholar]
- Nicholson R, Pelletier J, Le S, and Sonenberg N (1991). Structural and functional analysis of the ribosome landing pad of poliovirus type 2: in vivo translation studies. Journal of Virology 65, 5886–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okholm TLH, Sathe S, Park SS, Kamstrup AB, Rasmussen AM, Shankar A, Chua ZM, Fristrup N, Nielsen MM, and Vang S (2020). Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome medicine 12, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olexiouk V, Van Criekinge W, and Menschaert G (2017). An update on sORFs. org: a repository of small ORFs identified by ribosome profiling. Nucleic acids research 46, D497–D502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orre LM, Vesterlund M, Pan Y, Arslan T, Zhu Y, Woodbridge AF, Frings O, Fredlund E, and Lehtiö J (2019). SubCellBarCode: proteome-wide mapping of protein localization and relocalization. Molecular cell 73, 166–182. e167. [DOI] [PubMed] [Google Scholar]
- Owens GC, Chappell SA, Mauro VP, and Edelman GM (2001). Identification of two short internal ribosome entry sites selected from libraries of random oligonucleotides. Proc Natl Acad Sci U S A 98, 1471–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, and Ramberger E (2017). Translation of circRNAs. Molecular Cell 66, 9–21. e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda AC, Grammatikakis I, Kim KM, De S, Martindale JL, Munk R, Yang X, Abdelmohsen K, and Gorospe M (2016). Identification of senescence-associated circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic acids research 45, 4021–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, and Sonenberg N (1988). Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325. [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, and Pestova TV (2008). Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J 27, 1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikov AN, Hubbard SR, Schlessinger J, and Mohammadi M (2000). Crystal structures of two FGF-FGFR complexes reveal the determinants of ligand-receptor specificity. Cell 101, 413–424. [DOI] [PubMed] [Google Scholar]
- Qin X, and Sarnow P (2004). Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. Journal of Biological Chemistry 279, 13721–13728. [DOI] [PubMed] [Google Scholar]
- Quade N, Boehringer D, Leibundgut M, Van Den Heuvel J, and Ban N (2015). Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-Å resolution. Nature communications 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan C, Goodall GJ, Shirokikh NE, and Preiss T (2019). Insights into the biogenesis and potential functions of exonic circular RNA. Scientific reports 9, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisson B, Alvarez B, Paul S, Peters B, and Nielsen M (2020). NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads R, and Lamphear B (1995). Cap-independent translation of heat shock messenger RNAs. In Cap-Independent Translation (Springer; ), pp. 131–153. [DOI] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, and Goldberg AL (1994). Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771. [DOI] [PubMed] [Google Scholar]
- Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, and Ashwal-Fluss R (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Molecular cell 58, 870–885. [DOI] [PubMed] [Google Scholar]
- Saffell JL, Williams EJ, Mason IJ, Walsh FS, and Doherty P (1997). Expression of a dominant negative FGF receptor inhibits axonal growth and FGF receptor phosphorylation stimulated by CAMs. Neuron 18, 231–242. [DOI] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, and Brown PO (2013). Cell-type specific features of circular RNA expression. PLoS Genet 9, e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, and Brown PO (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7, e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepens B, Tinton SA, Bruynooghe Y, Beyaert R, and Cornelis S (2005). The polypyrimidine tract-binding protein stimulates HIF-1α IRES-mediated translation during hypoxia. Nucleic acids research 33, 6884–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Hung LH, Schreiner S, Starke S, Eckhof H, Rossbach O, Reich S, Medenbach J, and Bindereif A (2016). CircRNA-protein complexes: IMP3 protein component defines subfamily of circRNPs. Sci Rep 6, 31313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafritz DA, Weinstein JA, Safer B, Merrick WC, Weber LA, Hickey ED, and Baglioni C (1976). Evidence for role of m7G5’-phosphate group in recognition of eukaryotic mRNA by initiation factor IF-M3. Nature 261, 291. [DOI] [PubMed] [Google Scholar]
- Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, and Wang C (2016). Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E, Kalma Y, Sharp A, Raveh-Sadka T, Levo M, Zeevi D, Keren L, Yakhini Z, Weinberger A, and Segal E (2012). Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nature biotechnology 30, 521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JE, and Baker KE (2015). Nonsense‐mediated RNA decay–a switch and dial for regulating gene expression. Bioessays 37, 612–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, and Frank J (2001). Structure of the 80S ribosome from Saccharomyces cerevisiae--tRNA-ribosome and subunit-subunit interactions. Cell 107, 373–386. [DOI] [PubMed] [Google Scholar]
- Stoneley M, Chappell SA, Jopling CL, Dickens M, MacFarlane M, and Willis AE (2000). c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Molecular and cellular biology 20, 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuplin A, Struthers M, Cook J, Bentley K, and Evans DJ (2015). Inhibition of HCV translation by disrupting the structure and interactions of the viral CRE and 3′ X-tail. Nucleic acids research 43, 2914–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q, and Westhof E (2014). Biogenesis of Circular RNAs. Cell 159, 13–14. [DOI] [PubMed] [Google Scholar]
- Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al. (2019). The Landscape of Circular RNA in Cancer. Cell 176, 869–881 e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Nazarali AJ, and Ji S (2016). Circular RNAs as potential biomarkers for cancer diagnosis and therapy. American journal of cancer research 6, 1167. [PMC free article] [PubMed] [Google Scholar]
- Wang Y, and Wang Z (2015). Efficient backsplicing produces translatable circular mRNAs. RNA 21, 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnakulasuriyarachchi D, Cerquozzi S, Cheung HH, and Holcik M (2004). Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J Biol Chem 279, 17148–17157. [DOI] [PubMed] [Google Scholar]
- Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, and Segal E (2016). Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351, aad4939. [DOI] [PubMed] [Google Scholar]
- Wery M, Descrimes M, Vogt N, Dallongeville A-S, Gautheret D, and Morillon A (2016). Nonsense-mediated decay restricts LncRNA levels in yeast unless blocked by double-stranded RNA structure. Molecular cell 61, 379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong W, Bai XC, Brown A, Fernandez IS, Hanssen E, Condron M, Tan YH, Baum J, and Scheres SH (2014). Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J, Van de Peer Y, and De Wachter R (2001). Distribution of substitution rates and location of insertion sites in the tertiary structure of ribosomal RNA. Nucleic acids research 29, 5017–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, Jin Y, Gao Y, Xia L, Chang H, et al. (2018). CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res 46, D925–D929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M-S, and Wilusz JE (2019). An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3′ ends. Nucleic acids research 47, 8755–8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Lin J, Qin Y, Zhou J, and Bu W (2011). Structural diversity of eukaryotic 18S rRNA and its impact on alignment and phylogenetic reconstruction. Protein Cell 2, 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Guo S, Li W, and Yu P (2015). The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Scientific reports 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. (2017). Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 27, 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, et al. (2018). Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamai M, Trullo A, Giordano M, Corti V, Cuesta EA, Francavilla C, Cavallaro U, and Caiolfa VR (2019). Number and brightness analysis reveals that NCAM and FGF2 elicit different assembly and dynamics of FGFR1 in live cells. J Cell Sci 132, jcs220624. [DOI] [PubMed] [Google Scholar]
- Zeenko V, and Gallie DR (2005). Cap-independent translation of tobacco etch virus is conferred by an RNA pseudoknot in the 5’-leader. Journal of Biological Chemistry 280, 26813–26824. [DOI] [PubMed] [Google Scholar]
- Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, Chen W, Gao X, Zhao K, and Zhou H (2018a). A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene, 1. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, Liu H, Xu J, Xiao F, Zhou H, et al. (2018b). A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 9, 4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, and Shi G (2016). Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature communications 7, 11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, Wu C, Zhou Q, Hu W, Wu C, et al. (2019). A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer 18, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Lv M, and Chen J (2016). Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Scientific reports 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. The IRES activity of oligos among different categories, related to Figure 1.
Table S3. The full-length oligo sequences of the circular IRESs and the linear IRESs, related to Figure 1.
Table S4. IRES activity of the oligos harboring the human 18S rRNA complementary sequence, related to Figure 3.
Table S1. The sequences and their corresponding IRES activity of the oligo in the synthetic oligo library, related to STAR Methods
Table S5. The human endogenous IRES-containing circRNAs, related to Figure 6.
Table S7. The RT-qPCR primer sequences, related to STAR Methods.
Table S6. The putative circORF sequences and the peptidomic analysis, related to Figure 6.
Data Availability Statement
The IRES screening, polysome profiling, and M2-seq sequencing data generated in this paper were deposited in Gene Expression Omnibus. The MS/MS and the PRM-MS data generated in this paper are deposited in PRIDE. Accession numbers are listed in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-GFP antibody - ChIP Grade (1:300 for WB) | Abcam | Cat# ab290, RRID:AB_303395 |
| CD4 [EPR6855] antibody (1:500 for WB) | Abcam | Cat# ab133616, RRID:AB_2750883 |
| Cre recombinase antibody [7.23] (1:200 for WB) | Abcam | Cat# ab24607, RRID:AB_448179 |
| anti-SMAD5 antibody[3H9] (1:200 for WB) | Novus Biologicals | Cat# NBP2–37648 |
| FGFR1 Antibody (1:200 for WB) | Novus | Cat# NB100–2079, RRID:AB_10144921 |
| FGFR1 (phospho Y654) antibody (1:200 for WB) | Abcam | Cat# ab59194, RRID:AB_941585 |
| Monoclonal ANTI-FLAG® M2 antibody (1:400 for WB; 1:150 for IF) | Sigma-Aldrich | Cat# F1804, RRID:AB_262044 |
| HA tag antibody - ChIP Grade (1:400 for WB; 1:150 for IF) | Abcam | Cat# ab9110, RRID:AB_307019 |
| beta Tubulin antibody - Loading Control (1:500 for WB) | Abcam | Cat# ab6046, RRID:AB_2210370 |
| Anti-GAPDH antibody [EPR16891] (1:500 for WB) | Abcam | Cat# ab181602, RRID:AB_2630358 |
| IRDye 680RD Goat anti-Mouse IgG antibody (1:5,000 for WB) | LI-COR Biosciences | Cat# 926–68070, RRID:AB_10956588 |
| IRDye® 800CW Goat anti-Rabbit IgG (H + L) (1:5000 for WB) | LI-COR Biosciences | Cat# 926–32611 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (1:1,500 for IF) | Thermo Fisher Scientific | Cat# A-11029, RRID:AB_2534088 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 (1:150 for IF) | Thermo Fisher Scientific | Cat# A-11012, RRID:AB_2534079 |
| Customize rabbit anti-circFGFR1p | Thermo Fisher Scientific | Project# VJ2580 |
|
| ||
| Bacterial and Virus Strains | ||
|
| ||
| NEB® 10-beta Competent E. coli | New England Biolabs | C3019H |
|
| ||
| Chemicals, Peptides, and Recombinant Proteins | ||
|
| ||
| SYBR™ Gold Nucleic Acid Gel Stain | Thermo Fisher Scientific | Cat# S11494 |
| BrightStar™-Plus Positively Charged Nylon Membrane | Thermo Fisher Scientific | Cat# AM10100 |
| Millennium™ RNA Markers | Thermo Fisher Scientific | Cat# AM7150 |
| Cycloheximide,≥93.0% (HPLC) | Sigma-Aldrich | Cat# 01810 |
| Puromycin Dihydrochloride | Thermo Fisher Scientific | Cat# A11138–03 |
| Pierce™ IP Lysis Buffer | Thermo Fisher Scientific | Cat# 87788 |
| Pierce™ Protein A/G Magnetic Beads | Thermo Fisher Scientific | Cat# 88803 |
| NuPAGE™ 4 to 12%, Bis-Tris, 1.5 mm, Mini Protein Gel, 10-well | Thermo Fisher Scientific | Cat# NP0335BOX |
| Corning® BioCoat™ Poly-D-Lysine 12 mm #1 German Glass Coverslip | Corning | Cat# 354086 |
| ON-TARGET plus non-targeting pool | Dharmacon | Cat# D-001810–10-05 |
| SMARTpool: ON-TARGETplus FGFR1 siRNA | Dharmacon | Cat# L-003131–00-0005 |
| Gibco FGF1 Recombinant Human Protein | Thermo Fisher Scientific | Cat# 13–241-013 |
| PrestoBlue™ Cell Viability Reagent | Thermo Fisher Scientific | Cat# A13261 |
| E-Gel™ EX Agarose Gels, 2% | Thermo Fisher Scientific | Cat# G402022 |
| NEB® 10-beta/Stable Outgrowth Medium | New England Biolabs | Cat# B9035S |
| Opti-MEM™ I Reduced Serum Medium | Thermo Fisher Scientific | Cat# 31985070 |
| Q5® Hot Start High-Fidelity 2X Master Mix | New England Biolabs | Cat# M0409L |
| TrypLE™ Express Enzyme (1X), no phenol red | Thermo Fisher Scientific | Cat# 12604013 |
| NEBNext® Ultra™ II Q5® Master Mix | New England Biolabs | Cat# M0544L |
| Invitrogen Ambion SUPERase·In RNase Inhibitor (20U/μL) | Thermo Fisher Scientific | Cat# AM2694 |
| SuperBlock Blocking Buffers | Thermo Fisher Scientific | Cat# 37517 |
| TURBO DNase (2U/μL) | Thermo Fisher Scientific | Cat# AM2238 |
| SuperScript IV Reverse Transcriptase | Thermo Fisher Scientific | Cat# 18090050 |
| Normal Rabbit IgG | Sigma-Aldrich | Cat# 12–370 |
| RIPA Lysis and Extraction Buffer | Thermo Fisher Scientific | Cat# 89900 |
| Halt Protease and Phosphatase Inhibitor Cocktails | Thermo Fisher Scientific | Cat# 78441 |
| RNaseR | MCLAB Products | Cat# RNASR-200 |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| Lipofectamine™ 3000 Transfection Reagent | Thermo Fisher Scientific | Cat# L3000015 |
| Quick-DNA Microprep Plus Kit | Zymo Research | Cat# D4074 |
| NorthernMax™-Gly Kit | Thermo Fisher Scientific | Cat# AM1946 |
| RNeasy Mini Kit | Qiagen | Cat# 74004 |
| SuperScript™ IV One-Step RT-PCR System | Thermo Fisher Scientific | Cat# 12594100 |
| HiScribe™ T7 High Yield RNA Synthesis Kit | New England Biolabs | Cat# E2040S |
| GeneMorph II Random Mutagenesis Kit | Agilent | Cat# 200550 |
| DNA Clean & Concentrator-25 | Zymo Research | Cat# D4033 |
| Zymoclean Gel DNA Recovery Kit (uncapped columns) | Zymo Research | Cat# D4002 |
| Direct-zol RNA Microprep | Zymo Research | Cat# R2061 |
| RNA Clean & Concentrator-25 | Zymo Research | Cat# R1017 |
| RNA Clean & Concentrator-5 | Zymo Research | Cat# R1013 |
| Zymoclean Gel RNA Recovery Kit | Zymo Research | Cat# R1011 |
| NEBuilder® HiFi DNA Assembly Master Mix | New England Biolabs | Cat# E2621L |
| ZymoPURE II Plasmid Maxiprep Kit | Zymo Research | Cat# D4203 |
| Complete kit (optimized for Roche® LightCycler 480) | Kapa Biosystems | Cat# KK4854 |
| NEBuilder® HiFi DNA Assembly Master Mix | New England Biolabs | Cat# E2621L |
|
| ||
| Deposited Data | ||
|
| ||
| IRES screening, polysome profiling, and M2-seq sequencing | This paper | GEO: GSE178718 |
| Ribosome footprinting | Chen et al., 2020 | GEO: GSM3791725, GSM3791726, GSM3791727, GSM3791728, GSM3791729, and GSM3791730 |
| iPSC peptidomics | Chen et al., 2020 | PRIDE: PXD014031 |
| SCBC peptidomics | Orre et al., 2019 | ProteomeXchange: PXD006895 |
| GTEx peptidomics | Jiang et al., 2020 | ProteomeXchange: PXD016999 |
| K562/U2OS/H358 peptidomics | This paper | ProteomeXchange: PXD025233 |
| HLA class I peptidomes | Bassani-Sternberg et al., 2015 | PRIDE: PXD014031 |
| circFGFR1p IP-MS | This paper | ProteomeXchange: PXD024993 |
| K562/U2OS PRM-MS | This paper | ProteomeXchange: PXD025235 |
| circFGFR1p PRM-MS | This paper | ProteomeXchange: PXD025203 |
| QTI-seq | Gao et al., 2015 | SRA: 160745 |
|
| ||
| Experimental Models: Cell Lines | ||
|
| ||
| HEK293T | ATCC | Cat# CRL-3216; RRID: CVCL_0063 |
| HeLa | ATCC | Cat# CCL-2; RRID: CVCL_0030 |
| BJ | ATCC | Cat# CRL-2522; RRID:CVCL_3653 |
| U2OS | ATCC | Cat# HTB-96, RRID:CVCL_0042 |
|
| ||
| Oligonucleotides | ||
|
| ||
| qRT-PCR primers | This paper | Table S7 |
| IRES screening oligo library | Weingarten-Gabbay et al., 2016 | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| Plasmid: pcDNA3.1(+) ZKSCAN1 Sense | Addgene | Cat# 60631 |
| Plasmid: pHAGE-FGFR1 | Addgene | Cat# 116740 |
| Plasmid: pUC19 Vector | New England Biolabs | Cat# N3041L |
| Plasmid: pLV-mCherry | Addgene | Cat# 36084 |
| Plasmid: mRuby-ZKSCAN-split-eGFP reporter | This paper | N/A |
| Plasmid: mRuby-IRES-eGFP reporter | This paper | N/A |
| Plasmid: autocatalytic-splicing circEGFP | This paper | N/A |
|
| ||
| Software and Algorithms | ||
|
| ||
| ZEN (blue edition) | Carl Zeiss Microscopy | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| FlowJo_V10 | FlowJo, LLC | https://flowjo.com/solutions/flowjo/downloads |
| Bowtie2 | Langmead and Salzberg, 2012 | https://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.4.4/ |
| TopHat2 | Kim et al., 2013 | https://ccb.jhu.edu/software/tophat/downloads/tophat-2.1.1.tar.gz |
| M2-seq and M2-net | Cheng et al., 2017 | https://ribokit.github.io/M2seq/ |
| ViennaRNA | Lorenz et al., 2011 | https://www.tbi.univie.ac.at/RNA/ |
| MaxQuant | Cox and Mann, 2008 | https://www.maxquant.org/download_asset/maxquant/latest |
| Trans Proteomic Pipeline | Collaborative open-source | http://tools.proteomecenter.org/wiki/index.php?title=Software:TPP |
| Byonic | Protein Metrics | https://proteinmetrics.com/byos/ |
| Skyline 19.1.0.193 | MacLean et al., 2010 | https://skyline.ms/wiki/home/software/Skyline/page.view?name=install-administator-64 |
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.