Abstract
We have isolated one sorbitol-nonfermenting (SNF) Escherichia coli O157:H7 isolate and five sorbitol-fermenting (SF) E. coli O157:H− isolates that do not contain Shiga toxin (Stx) genes (stx). Isolates originated from patients with diarrhea (n = 4) and hemolytic-uremic syndrome (HUS) (n = 2). All isolates harbored a chromosomal eae gene encoding gamma-intimin as well as the plasmid genes E-hly and etp. The E. coli O157:H7 isolate was katP and espP positive. Respective sera obtained from the patient with HUS contained antibodies to the O157 lipopolysaccharide antigen. The stx-negative E. coli O157:H7 isolate is genetically related to stx-positive SNF E. coli O157:H7. All stx-negative SF E. coli O157:H− isolates belong to the same genetic cluster and are closely related to stx-positive SF E. coli O157:H− isolates. Our data indicate that stx-negative E. coli O157:H7/H− variants may occur at a low frequency and cannot be recognized by diagnostic methods that target Stx.
Shiga toxin (Stx)-producing Escherichia coli (STEC) of various serotypes has been linked to a spectrum of disorders, including watery diarrhea, bloody diarrhea (hemorrhagic colitis), and hemolytic-uremic syndrome (HUS) (37).
STEC has as its cardinal virulence trait the ability to produce one or more Stxs (Stx1, Stx2, or Stx2 variants). Stxs are toxic to cultured human colonic and ileal epithelial cells (25) and endothelial cells (27). Even in the absence of cytotoxicity, Stxs can stimulate the production of vasoactive factors by endothelial cells (5). Thus, the ability to produce an Stx is quite plausibly related to the intestinal and extraintestinal manifestations of human STEC infections.
E. coli strains that express the O157 antigen are the most commonly isolated STEC strains worldwide. Such organisms are easily detected by toxin-independent detection protocols, such as sorbitol-MacConkey (SMAC) agar screening (29), or the immunomagnetic separation (IMS) technique. Unlike approximately 80% of other E. coli strains, most O157 STEC isolates do not ferment d-sorbitol after overnight incubation. Therefore, SMAC agar was developed by substituting the carbohydrate sorbitol for lactose in MacConkey agar. SMAC agar has proved to be effective for the isolation of O157 STEC and is the most widely used medium for this purpose.
IMS can isolate sorbitol-fermenting (SF) E. coli O157:H− as well as sorbitol-nonfermenting (SNF) E. coli O157:H7 (19). However, SF non-O157:H7 STEC can also cause human disease (17, 38), and because of this, Stx detection systems have been used to identify such pathogens in human stools (10, 15, 21, 28).
We have isolated from humans nontoxigenic E. coli strains that express the O157 antigen and present data suggesting that Stx may not be obligatorily produced by E. coli O157 strains associated with human disease, including HUS.
MATERIALS AND METHODS
Bacterial strains.
The origins and characteristics of the E. coli O157 strains used as controls in this study are described in Table 1. The origins of all other strains are described in the text.
TABLE 1.
E. coli strains used as controls in this studya
| Strain | Serotype | DA | Relevant genetic markers | Fermentation of sorbitol | Reference |
|---|---|---|---|---|---|
| EDL933 | O157:H7 | HC | stx1, stx2, eae | − | 26 |
| 702/88 | O157:H− | HUS | stx2, eae | + | 1 |
| 1533/97 | O157:H− | HUS | stx2, eae | + | SC |
| 3817/96 | O157:H− | HUS | stx1, stx2c | − | 7 |
| E32511 | O157:H− | HUS | stx2, stx2c, eae | − | 35 |
| 693/91 | O157:H19 | WD | stx−, eae− | + | 3 |
| 241/88 | O157:H43 | WD | stx−, eae− | + | 1 |
| 1083/87 | O157:H45 | WD | stx−; eae,b EAF+ | + | 31 |
| 904/90 | O157:H45 | WD | stx−; eae,b EAF− | + | 1 |
Abbreviations: SC, strain collection (isolation during routine diagnostic work in 1997); HC, hemorrhagic colitis; DA, disease association; WD, watery diarrhea; stx− and eae−, negative for stx and eae, respectively; EAF+, positive for EAF plasmid; EAF−, negative for EAF plasmid.
Enteropathogenic E. coli eae.
Isolation of E. coli O157 from stool specimens.
A total of 2,785 stool specimens from patients with diarrhea or HUS were screened in 1996 and 1997 for the presence of Stx-producing E. coli in the Institute for Hygiene and Microbiology, University of Würzburg, Würzburg, Germany. The majority of the stool samples were obtained from hospitalized children throughout Germany. Detection of E. coli O157 was performed as described below.
A total of 10 ml of GN broth Hajna (Difco, Detroit, Mich.) was inoculated with 1 g of the stool sample, and the mixture was incubated for 6 h at 37°C. E. coli O157 was sought from 1 ml of this broth by the IMS technique as described previously (19). Fifty microliters of the bacterium-bead suspension was streaked onto SMAC agar and cefixime-tellurite SMAC (CT-SMAC) agar plates. Up to 1,500 colonies from both plates were scraped off with a sterile swab and were suspended in 1 ml of sterile 0.9% NaCl solution. The bacterial cell concentration was determined and was adjusted with the McFarland no. 3 turbidity standard. Fifteen microliters of a 1:25 dilution (106 bacterial cells) made from this suspension was subjected to PCR with primer pairs KS7 and KS8, LP43 and LP44, and SK1 and SK2 (34), which are specific for stx1, stx2, and eae, respectively. The remaining suspension was stored at 4°C until the PCR results were obtained.
PCR-positive samples were processed in the following manner: the bacterial suspension which was stored at 4°C was diluted and restreaked onto three SMAC agar plates to obtain 500 CFU/plate. Colony blot hybridization was then performed with stx1-, stx2-, and eae-specific probes (33) on these plates to detect and isolate STEC colonies.
Microbiological methods.
Detection of the enterohemolytic phenotype was performed on blood agar plates containing washed sheep erythrocytes (34). Fermentation of sorbitol was detected on SMAC agar plates after overnight incubation (29). Resistance to tellurite was determined on CT-SMAC agar containing 2.5 mg of tellurite per liter (40).
PCR techniques.
PCR was conducted with 106 bacteria. Amplification was performed in a total volume of 50 μl containing each deoxynucleoside triphosphate at 200 μM, 30 pmol of each primer, 5 μl of 10-fold-concentrated DNA polymerase synthesis buffer (Perkin-Elmer, Applied Biosystems, Weiterstadt, Germany), 3 μl of a 25 mM MgCl2 stock solution, and 2.0 U of AmpliTaq DNA polymerase (Perkin-Elmer, Applied Biosystems). For detection of stx genes in the isolated strains, primers LP30-LP31 (stxA1), KS7-KS8 (stxB1), and LP43-LP44 (stxA2) were used as described previously (34). JS1 (5′-CAT GAA GAA GAT GTT TAT GGC G-3′) and JS2 (5′-CTC AGT CAT TAT TAA ACT GCA C-3′) were used for the amplification of the entire stxB2 subunit gene by the protocol described previously for GK5 and GK6 (15). For detection of plasmid genes, PCRs for E-hly (34), etp (34), katP (34), and espP (9) were performed as described previously. The eae genes were amplified with primers SK1-SK2 (32) and with primers LP1-LP2 and LP1-LP3 (31). A 5-μl volume of each PCR sample was analyzed by gel electrophoresis on 1.5% agarose gels. The fliC gene was amplified with primers F-FLIC1 and R-FLIC2, and the PCR product was restricted with RsaI as described by Fields et al. (12). Restriction fragments were separated on a 2.5% (wt/vol) agarose gel.
Analysis of genomic DNA of E. coli O157 strains was performed by random amplification of polymorphic DNA (RAPD) PCR fingerprinting with a single primer, primer 1247 (5′-AAG AGC CCG T-3′) (16). Internal standards (PCR products with known sizes) were run in each lane for standardization of gels for analysis. Gels were stained with ethidium bromide and were digitized for computer-aided analysis. The GelCompar software package (Applied Maths, Kortrijk, Belgium) was used for analysis. Calculation of the similarity matrix was performed with the Jacquard algorithm after defining each single band. The hierarchic clustering was achieved by the unweighted pair group method with arithmetic averages clustering algorithm (36).
Standard DNA techniques.
Restriction endonuclease digestion (New England Biolabs) was performed according to the supplier's instructions. Restriction fragments were separated electrophoretically in 0.6 to 0.9% agarose gels in 0.5-fold TBE (Tris-borate-EDTA) buffer (30), stained in ethidium bromide, and analyzed. For Southern blot hybridization, DNA was prepared by the method of Heuvelink et al. (16): 100 ng of DNA was separated electrophoretically and was transferred from agarose gels to Zeta-probe nylon membranes (Bio-Rad, Munich, Germany) by standard methods (30). For hybridization assays, the nonradioactive DNA Labeling and Detection Kit (Boehringer GmbH, Mannheim, Germany) was used according to the manufacturer's instructions. The specific washing step was performed twice (5 min each time) in SSC buffer (sodium chloride, sodium citrate) and 0.1% (wt/vol) sodium dodecyl sulfate. The concentration of SSC and the temperature of the washings were calculated as described by Meinkoth and Wahl (24) to effect different stringencies. Probes were labeled with digoxigenin-11-dUTP by random hexamer labeling. The probes used included fragments of stx1 (amplified from EDL933 by PCR with primers KS7 and KS8), stx2 (amplified from EDL933 by PCR with primers GK3 and GK4), and eae (amplified from EDL933 with primers SK1 and SK2 [32]).
Vero cell assay.
The Vero cell toxicities of bacterial culture supernatants and fecal filtrates were determined as described previously (14, 20, 34). Ten, 100, and 1,000 50% cytotoxic doses of Stx2 per ml were added to filtrates to confirm that no inhibitors of cytotoxicity were present.
Serological and immunological techniques.
Isolation of lipopolysaccharide (LPS) and electroblotting were performed as described previously (4). For detection of immunoglobulin M (IgM) antibodies against O157 LPS by immunoblotting, the techniques of Bitzan et al. (4) were used. The E. coli O and H antigens were determined with the Wellcolex E. coli O157:H7 Rapid Latex Test Kit (Murex Diagnostica/Abbott Laboratories, Wiesbaden, Germany) according to the manufacturer's instructions. E. coli serotypes were confirmed by the method described by Bockemühl et al. (6).
An enzyme immunoassay (Premier EHEC EIA; Meridian Diagnostics Inc., Cincinnati, Ohio) was used to detect the expression of Stxs from isolated organisms according to the manufacturer's instructions, with minor modifications. Briefly, 4 ml of tryptic soy broth was supplemented with 0.05 μg of mitomycin C per ml and was inoculated with the test strain, and the mixture was incubated overnight at 37°C with shaking. A total of 50 μl of the overnight culture was mixed with 200 μl of sample dilution buffer, and 100 μl of this mixture was transferred to the well of a microtiter plate coated with polyclonal anti-Stx IgG antibodies. After 1 h of incubation at room temperature, the well was washed five times with washing buffer. Detection of bound antigen was performed as described in the manual of the Premier EHEC EIA. Stx-positive strain E. coli O157:H7 EDL933 and stx-negative strain E. coli O157:H19 693/91 were used as controls.
RESULTS
Identification of stx-negative E. coli O157 strains.
During routine diagnostic work in 1996 and 1997, tests for the detection of STEC were performed with 2,875 stool specimens. From 145 of these stool specimens, 145 STEC O157 (118 sorbitol-negative and 27 sorbitol-positive strains) could be isolated in our laboratory. Eighty-five strains were from patients with HUS and 60 were from patients with diarrhea without HUS. In addition, five stool specimens from five different patients (Table 2) did not show toxic activity for Vero cells, and PCR screening with stx1 and stx2-specific primers (stx PCRs) after enrichment by the IMS technique was negative. However, the PCR of the five stool specimens with eae-specific primers (eae PCR) was positive. Stool samples from diarrheal patients were taken 1 to 2 days after the onset of diarrhea and from both HUS patients during the acute phase of disease 7 days after the onset of diarrhea.
TABLE 2.
Characteristics of stx-negative E. coli O157 isolatesa
| Strain designation | Serotype | DA or origin | Age (yr)b | Presence of free fecal Verotoxin | Fermentation of sorbitol | Growth on CT-SMAC | Presence of the following:
|
Anti-O157 LPS immunoblotting result | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| E-hlyc | etp | katP | espP | ||||||||
| 19685 | O157:H7 | WD | 4 | − | − | + | + | + | + | + | ND |
| 2937 | O157:H− | WD | 3 | − | + | − | + | + | − | − | ND |
| 6790 | O157:H− | HUS | 6 | − | + | − | + | + | − | − | + |
| 431 | O157:H− | WD | 3 | − | + | − | + | + | − | − | ND |
| 659 | O157:H− | WD | 3 | − | + | − | + | + | − | − | ND |
| 2576 | O157:H− | HUSd | 1 | + | + | − | + | + | − | − | + |
Abbreviations: DA, disease association; WD, watery diarrhea; ND, not determined.
Age of patients.
All isolates were also enterohemolytic on blood agar plates.
STEC O103:H2 was found to be coisolate.
By colony blot hybridization, five E. coli O157 strains were isolated (Table 2). These isolates were also positive by the universal eae PCR but negative by the stx PCRs. Filtrates of stools from five of the six patients from which these bacteria were obtained were also negative by the Vero cell assay. To ensure that no inhibitors of Stx were present in the stools, the supernatant of an Stx2-positive culture was added at different concentrations to samples of all stools that were primarily negative by the Vero cell assay. All samples supplemented with Stx2 were cytotoxic by the Vero cell assay, indicating that no inhibitors were present in the stools.
A sixth, Stx-positive stool which contained an Stx-producing E. coli O103:H2 isolate was noticed. However, serum from the same patient reacted with E. coli O157 LPS. Therefore, the presence of E. coli O157 was likely. An enrichment culture of the stool sample was streaked onto blood agar plates, and the enterohemolytic phenotypes were investigated. Beside strongly hemolytic colonies, which were later shown to be E. coli O103:H2, colonies surrounded by the typical enterohemolytic phenotype were detected. Analysis with the Wellcolex E. coli O157:H7 Rapid Latex Test Kit revealed that these colonies were serotype O157:H−.
Sera from the two children with HUS were obtained during the acute phase of HUS 8 days after the onset of diarrhea and were tested by immunoblot analysis with O157 LPS. Both sera were positive by the IgM immunoblot assay.
Strain characteristics.
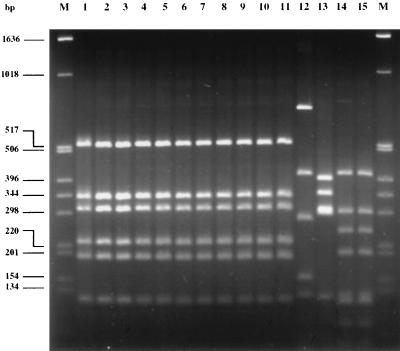
One of the Stx-negative isolates was of serotype O157:H7, and the other five were of serotype O157:H−. All isolates were from patients with primary cases of infection, and no temporal or geographic clustering could be observed. The origins of these isolates are described in Table 2. PCR was performed to characterize the fliC gene of the E. coli O157:H7 isolates and selected controls (see Fig. 1). Restriction fragment length polymorphism (RFLP) analysis of the PCR products after digestion with RsaI demonstrated a pattern identical to that obtained by the fliC PCR with strains of the H7 or H− type as described by Fields et al. (12). The results of fliC PCR and RFLP analysis of all tested strains are depicted in Fig. 1. E. coli O157:H7 strains EDL933 and 19685 and E. coli O157:H− control strains 702/88, 1533/97, 3817/96, and E32511 as well as stx-negative SF O157:H− isolates 2937, 6790, 431, 659, and 2576 demonstrated identical RFLP patterns after digestion with RsaI. This pattern is identical to that described by Fields et al. (12) and confirmed the presence of an H7/H− antigen in the respective isolates. E. coli control strains 693/91 (O157:H19), 241/88 (O157:H43), 1083/87 (O157:H45), and 904/90 (O157:H45) showed different patterns after digestion with RsaI, and these patterns were distinguishable from that of E. coli O157:H7/H−.
FIG. 1.
Agarose gel electrophoresis of fliC PCR products of E. coli O157 strains restricted with RsaI. Lanes M, molecular size marker (1-kb ladder; Gibco GmbH); lane 1, E. coli O157:H7 EDL933; lane 2, E. coli O157:H7 19685; lane 3, SF E. coli O157:H− 702/88; lane 4, SF E. coli O157:H− 1533/97; lane 5, SNF E. coli O157:H− 3817/96; lane 6, SNF E. coli O157:H− E32511; lane 7, SF E. coli O157:H− 2937; lane 8, SF E. coli O157:H− 6790; lane 9, SF E. coli O157:H− 431; lane 10, SF E. coli O157:H− 659; lane 11, SF E. coli O157:H− 2576; lane 12, E. coli O157:H19 693/91; lane 13, E. coli O157:H43 241/88; lane 14, E. coli O157:H45 1083/87; and lane 15, E. coli O157:H45 904/90.
Virulence determinants of the stx-negative E. coli O157 strains.
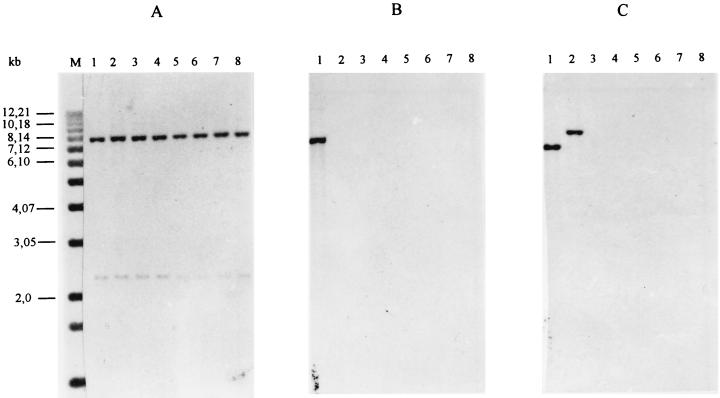
None of the E. coli O157 strains gave a PCR product with primers LP30-LP31 and LP43-LP44, which are complementary to the A-subunit genes of stx1 and stx2, respectively. To test the hypothesis that A-subunit genes different from stxA1 and stxA2 were present, stx PCR was repeated with primers KS7-KS8 and JS1-JS2, which are complementary to the B-subunit genes of stx1 and stx2, respectively, but these PCRs were also negative. Moreover, the chromosomal DNAs of all isolates were hybridized with probes complementary to stx1 and stx2 sequences under low-stringency conditions, but no signal could be demonstrated (Fig. 2B and C). Control hybridization with an eae-specific probe showed a signal in all cases (Fig. 2A). None of the strains reacted in the Premier EHEC EIA.
FIG. 2.
Southern blot hybridization of EcoRI-restricted chromosomal DNAs of control strains E. coli O157:H7 EDL933 (lanes 1) and E. coli O157:H− E32511 (lanes 2) and the O157 isolates 19685 (lanes 3), 2937 (lanes 4), 6790 (lanes 5), 431 (lanes 6), 659 (lanes 7), and 2576 (lanes 8) with probes specific for eae (A), stx1 (B), and stx2 (C) under low-stringency conditions. M, molecular size marker.
The E. coli O157:H7 strain was E-hly, etpP, katP, and espP positive by PCR. The five E. coli O157:H− isolates, however, contained only E-hly and etp and lacked espP and katP.
By PCR with primers LP1 and LP3 it could be shown that eae encoding gamma-intimin, which is typically found in Stx-producing E. coli O157, is present in all strains. The E. coli O157:H7 strain showed the typical enterohemolytic phenotype on blood agar plates after 18 to 24 h of incubation. However, of the E. coli O157:H− isolates, only 30% of the colonies were enterohemolytic when they were streaked for isolation onto blood agar plates.
Clonal analysis.
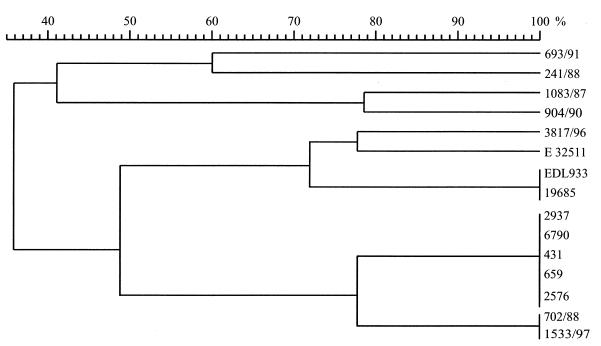
We compared the genetic relatedness of these strains by RAPD analysis. Figure 3 depicts a dendrogram constructed with GelCompar software. The dendrogram shows the genetic relatedness of the stx-negative E. coli O157 isolates and stx-positive E. coli O157:H7 and O157 isolates with other H antigens.
FIG. 3.
Dendrogram of E. coli O157 strains used in this study based on RAPD analysis-PCR pattern similarity. The percentages represent band pattern similarities as calculated with the Jacquard algorithm.
RAPD analysis demonstrated that the stx-negative isolate E. coli O157:H7 19685 is identical to the stx-positive reference strain E. coli O157:H7 EDL933. The SNF O157:H7 strains are more closely related to the SNF stx-positive E. coli O157:H− strains than to the SF O157:H− isolates. Moreover, as expected, the SF stx-negative E. coli O157:H− strains are clonal and are most closely related to the stx-positive E. coli O157:H− strains. E. coli O157 strains of the H− and H7 types are more closely related to each other than to E. coli strains of other H types.
DISCUSSION
Our finding of clinical E. coli O157:H7/H− isolates that lack stx genes was surprising. Such stx-negative E. coli O157 strains have rarely been identified, presumably because in strains of this serotype, unlike other toxigenic E. coli strains that produce Stx, the toxin genes are retained after multiple replications. It is probable that a progenitor of the organisms that we detected contained stx genes that were subsequently lost. The emergence of pathogenic E. coli O157:H7 proposed by Feng et al. (11) does not accommodate the existence of a nontoxigenic E. coli strain that expresses the O157 LPS antigen.
We are unable to state with certainty that Stx had no role in the diseases observed in the patients who we report on here. Specifically, we cannot prove that we did not inadvertently overlook the true toxigenic pathogens in the isolation process, and we cannot affirm that the organisms that were initially shed did not contain toxin genes that were lost during isolation or subculture, a phenomenon that has been seen among STEC strains belonging to serotypes O2:H5, O26:H11, O73:H34, and O100:H32 (18).
Several lines of evidence suggest that these explanations do not apply. First, had we neglected to recover toxin-producing E. coli O157:H7/H− strains that were actually present or if stx genes were lost on subculture, the probability of identification of toxin in the stool by Vero cell assay should have been high, especially as the stools from early in the course of the illness were analyzed. Second, our isolation protocol contains no obvious bias against the isolation of toxin-producing organisms. In addition, stx-negative E. coli O157:H7/H− presumably contains many of the virulence traits contained by nontoxigenic, enteropathogenic E. coli O55:H7 (11), an organism closely related to E. coli O157:H7/H−.
E. coli strains of this serotype have been recovered from children with nonbloody diarrhea in North America (8), and it is plausible that nontoxigenic E. coli O157:H7 is similarly pathogenic.
Our data have a variety of implications for the diagnosis of E. coli O157:H7/H− infections and for our understanding of the pathogenesis of diseases caused by this organism. First, screening on SMAC agar is inadequate for the detection of SF non-O157:H7 STEC strains, including stx2-positive SF E. coli O157:H−. To identify these organisms, it is necessary to detect the Stx phenotype by cytotoxicity assay or antigen detection or to detect stx genes. However, the presumptively pathogenic strains that we reported on above would be overlooked by protocols that rely on toxin detection. Thus, we strongly urge the performance of parallel and complementary tests that address a variety of nontoxin phenotypes and loci possessed by such organisms when performing thorough enteric microbiologic evaluations.
Second, our data support the role of Stx as a cause of bloody diarrhea in humans, in contrast to its questionable role as a cause of nonbloody diarrhea. None of the patients described in this report developed bloody diarrhea, suggesting that toxin production is necessary for hemorrhagic colitis in most patients.
Third, Stx does not appear to be necessary for all manifestations of the diseases associated with E. coli O157:H7. It is interesting that Shigella dysenteriae serotype 1, which does not have the ability to produce Stx, causes nonbloody diarrhea in monkeys (13) and that Stx production is not needed for diarrhea in gnotobiotic piglets challenged with STEC (39). Also, Li et al. (23) demonstrated that E. coli O157:H7 strains deficient in the ability to produce Stxs were able to induce abnormalities of colonic structure and ion transport in New Zealand White rabbits.
Fourth, HUS might result from non-Stx factors produced by E. coli O157:H7/H−. Other bacteria that do not produce Stx, such as Streptococcus pneumoniae and Neisseria meningitidis, have occasionally been associated with HUS (2, 22). Our data raise the possibility that other properties of E. coli O157:H7 might also be sufficient to produce HUS in susceptible hosts. However, we observed HUS caused by an Stx-negative E. coli O157 strain without evidence of the presence of other pathogenic E. coli strains in only a single patient, and we urge clinicians to use caution before attributing HUS to nontoxigenic E. coli. Certainly, additional reports of the isolation of such potential pathogens from children with HUS, without evidence of the presence of fecal Stx, would lend support to our speculation.
In summary, we have identified stx-deficient E. coli O157 strains from infected humans with diarrhea and HUS using stx-independent recovery techniques. Moreover, these patients had no evidence of fecal Stx. The infecting isolates had other characteristics of E. coli O157:H7/H− and were closely related to pathogenic toxigenic E. coli O157. Our findings suggest that Stx- and stx gene-based detection systems should be complemented by additional methods for the identification of stx-negative E. coli O157 in microbiologic evaluations and that the diseases in humans caused by E. coli O157:H7/H− do not uniformly require the production of Stx by these pathogens.
ACKNOWLEDGMENTS
We thank Phillip I. Tarr for helpful discussions and critical reading of the manuscript and Barbara Plaschke for excellent technical assistance.
This work was supported by grant Ka 717/3-1 (Ökologie bakterieller Krankheitserreger: Molekulare und evolutionäre Aspekte) from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Aleksic S, Karch H, Bockemühl J. A biotyping scheme for Shiga-like (Vero) toxin-producing Escherichia coli O157 and a list of serological cross-reactions between O157 and other gram-negative bacteria. Int J Med Microbiol Virol Parasitol Infect Dis. 1992;276:221–230. doi: 10.1016/s0934-8840(11)80009-5. [DOI] [PubMed] [Google Scholar]
- 2.Alon U, Adler S P, Chan J C. Hemolytic-uremic syndrome associated with Streptococcus pneumoniae. Report of a case and review of the literature. Am J Dis Child. 1984;138:496–499. doi: 10.1001/archpedi.1984.02140430072019. [DOI] [PubMed] [Google Scholar]
- 3.Bielaszewska M, Schmidt H, Karmali M A, Khakhria R, Janda J, Bláhová K, Karch H. Isolation and characterization of sorbitol-fermenting Shiga toxin (Verocytotoxin)-producing Escherichia coli O157:H− strains in the Czech Republic. J Clin Microbiol. 1998;36:2135–2137. doi: 10.1128/jcm.36.7.2135-2137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bitzan M, Moebius E, Ludwig K, Müller Wiefel D E, Heesemann J, Karch H. High incidence of serum antibodies to Escherichia coli O157 lipopolysaccharide in children with hemolytic-uremic syndrome. J Pediatr. 1991;119:380–385. doi: 10.1016/s0022-3476(05)82049-9. [DOI] [PubMed] [Google Scholar]
- 5.Bitzan M M, Wang Y, Lin J, Marsden P A. Verotoxin and ricin have novel effects on preproendothelin-1 expression but fail to modify nitric oxide synthase (ecNOS) expression and NO production in vascular endothelium. J Clin Invest. 1998;101:372–382. doi: 10.1172/JCI522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bockemühl J, Aleksic S, Karch H. Serological and biochemical properties of Shiga-like toxin (verocytotoxin)-producing strains of Escherichia coli, other than O-group 157, from patients in Germany. Int J Med Microbiol Virol Parasitol Infect Dis. 1992;276:189–195. doi: 10.1016/s0934-8840(11)80005-8. [DOI] [PubMed] [Google Scholar]
- 7.Bockemühl J, Karch H, Tschäpe H. Infektionen des Menschen durch enterohämorrhagische Escherichia coli (EHEC) in Deutschland, 1996. Bundesgesundhbl. 1997;6:194–197. doi: 10.1007/s00103-002-0458-4. [DOI] [PubMed] [Google Scholar]
- 8.Bokete T N, Whittam T S, Wilson R A, Clausen C R, O'Callahan C M, Moseley S L, Frotsche T R, Tarr P I. Genetic and phenotypic analysis of Escherichia coli with enteropathogenic characteristics isolated from Seattle children. J Infect Dis. 1997;175:1382–1389. doi: 10.1086/516470. [DOI] [PubMed] [Google Scholar]
- 9.Brunder W, Schmidt H, Frosch M, Karch H. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology. 1999;145:1005–1014. doi: 10.1099/13500872-145-5-1005. [DOI] [PubMed] [Google Scholar]
- 10.Cebula T A, Payne W L, Feng P. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol. 1995;33:248–250. doi: 10.1128/jcm.33.1.248-250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng P, Lampel K A, Karch H, Whittam T S. Genotypic and phenotypic changes in the emergence of Escherichia coli O157:H7. J Infect Dis. 1998;177:1750–1753. doi: 10.1086/517438. [DOI] [PubMed] [Google Scholar]
- 12.Fields P I, Blom K, Hughes H J, Helsel L O, Feng P, Swaminathan B. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J Clin Microbiol. 1997;35:1066–1070. doi: 10.1128/jcm.35.5.1066-1070.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontaine A, Arondel J, Sansonetti P J. Role of Shiga toxin in the pathogenesis of bacillary dysentery, studied by using a Tox− mutant of Shigella dysenteriae 1. Infect Immun. 1988;56:3099–3109. doi: 10.1128/iai.56.12.3099-3109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentry M K, Dalrymple J M. Quantitative microtiter cytotoxicity assay for Shigella toxin. J Clin Microbiol. 1980;12:361–366. doi: 10.1128/jcm.12.3.361-366.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunzer F, Böhm H, Rüssmann H, Bitzan M, Aleksic S, Karch H. Molecular detection of sorbitol-fermenting Escherichia coli O157 in patients with hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1807–1810. doi: 10.1128/jcm.30.7.1807-1810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heuvelink A E, van de Kar N C, Meis J F, Monnens L A, Melchers W J. Characterization of verocytotoxin-producing Escherichia coli O157 isolates from patients with haemolytic uraemic syndrome in Western Europe. Epidemiol Infect. 1995;15:1–14. doi: 10.1017/s0950268800058064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson R P, Clarke R C, Wilson J B, Read S C, Rahn K, Renwick S A, Sandhu K A, Alves D, Karmali M A, Lior H, McEwen S A, Spika J S, Gyles C L. Growing concerns and recent outbreaks involving non-O157:H7 serotypes of verotoxigenic Escherichia coli. J Food Prot. 1996;59:1112–1122. doi: 10.4315/0362-028X-59.10.1112. [DOI] [PubMed] [Google Scholar]
- 18.Karch H, Meyer T, Rüssmann H, Heesemann J. Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect Immun. 1992;60:3464–3467. doi: 10.1128/iai.60.8.3464-3467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karch H, Janetzki-Mittmann C, Aleksic S, Datz M. Isolation of enterohemorrhagic Escherichia coli O157 strains from patients with hemolytic-uremic syndrome by using immunomagnetic separation, DNA-based methods, and direct culture. J Clin Microbiol. 1996;34:516–519. doi: 10.1128/jcm.34.3.516-519.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karmali M A, Petric M, Lim C, Fleming P C, Arbus G S, Lior H. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis. 1985;151:775–782. doi: 10.1093/infdis/151.5.775. [DOI] [PubMed] [Google Scholar]
- 21.Kehl K S, Havens P, Behnke C E, Acheson D W. Evaluation of the premier EHEC assay for detection of Shiga toxin-producing Escherichia coli. J Clin Microbiol. 1997;35:2051–2054. doi: 10.1128/jcm.35.8.2051-2054.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khodasevich L S, Dobrodeev K G. Hemolytic-uremic syndrome in meningococcal infection. Pediatriia. 1991;1:86–88. [PubMed] [Google Scholar]
- 23.Li Z, Bell C, Buret A, Robins Browne R, Stiel D, O'Loughlin E. The effect of enterohemorrhagic Escherichia coli O157:H7 on intestinal structure and solute transport in rabbits. Gastroenterology. 1993;104:467–474. doi: 10.1016/0016-5085(93)90415-9. [DOI] [PubMed] [Google Scholar]
- 24.Meinkoth J, Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984;138:267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- 25.Moyer M P, Dixon P S, Rothman S W, Brown J E. Cytotoxicity of Shiga toxin for primary cultures of human colonic and ileal epithelial cells. Infect Immun. 1987;55:1533–1535. doi: 10.1128/iai.55.6.1533-1535.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Brien A D, Lively T A, Chen M E, Rothman S W, Formal S B. Escherichia coli O157:H7 strains associated with haemorrhagic colitis in the United States produce a Shigella dysenteriae 1 (SHIGA) like cytotoxin. Lancet. 1983;i:702. doi: 10.1016/s0140-6736(83)91987-6. [DOI] [PubMed] [Google Scholar]
- 27.Obrig T G. Interaction of Shiga toxins with endothelial cells. In: Kaper J B, O'Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C: American Society for Microbiology; 1998. pp. 303–311. [Google Scholar]
- 28.Ramotar K, Waldhart B, Church D, Szumski R, Louie T J. Direct detection of verotoxin-producing Escherichia coli in stool samples by PCR. J Clin Microbiol. 1995;33:519–524. doi: 10.1128/jcm.33.3.519-524.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ratnam S, March S B, Ahmed R, Bezanson G S, Kasatiya S. Characterization of Escherichia coli serotype O157:H7. J Clin Microbiol. 1988;26:2006–2012. doi: 10.1128/jcm.26.10.2006-2012.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Schmidt H, Rüssmann H, Karch H. Virulence determinants in nontoxinogenic Escherichia coli O157 strains that cause infantile diarrhea. Infect Immun. 1993;61:4894–4898. doi: 10.1128/iai.61.11.4894-4898.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt H, Plaschke B, Franke S, Rüssmann H, Schwarzkopf A, Karch H. Differentiation in virulence patterns of Escherichia coli possessing eae genes. Med Microbiol Immunol Berl. 1994;183:23–31. doi: 10.1007/BF00193628. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt H, Rüssmann H, Schwarzkopf A, Aleksic S, Heesemann J, Karch H. Prevalence of attaching and effacing Escherichia coli in stool samples from patients and controls. Int J Med Microbiol Virol Parasitol Infect Dis. 1994;281:201–213. doi: 10.1016/s0934-8840(11)80571-2. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt H, Geitz C, Tarr P I, Frosch M, Karch H. Non-O157:H7 pathogenic Shiga toxin-producing Escherichia coli: phenotypic and genetic profiling of virulence traits and evidence for clonality. J Infect Dis. 1999;179:115–123. doi: 10.1086/314537. [DOI] [PubMed] [Google Scholar]
- 35.Schmitt C K, McKee M L, O'Brien A D. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect Immun. 1991;59:1065–1073. doi: 10.1128/iai.59.3.1065-1073.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sneath P H, Sokal R R. Numerical taxonomy. The principles and practice of numerical classification. San Francisco, Calif: W. H. Freeman & Co.; 1973. [Google Scholar]
- 37.Tarr P I. Escherichia coli O157:H7: clinical, diagnostic, and epidemiological aspects of human infection. Clin Infect Dis. 1995;20:1–8. doi: 10.1093/clinids/20.1.1. [DOI] [PubMed] [Google Scholar]
- 38.Tarr P I, Neill M A. Perspective: the problem of non-O157:H7 Shiga toxin (Verocytotoxin)-producing Escherichia coli. J Infect Dis. 1996;174:1136–1139. doi: 10.1093/infdis/174.5.1136. [DOI] [PubMed] [Google Scholar]
- 39.Tzipori S, Karch H, Wachsmuth K I, Robins-Browne R M, O'Brien A D, Lior H, Cohen M L, Smithers J, Levine M M. Role of a 60-megadalton plasmid and Shiga-like toxins in the pathogenesis of infection caused by enterohemorrhagic Escherichia coli O157:H7 in gnotobiotic piglets. Infect Immun. 1987;55:3117–3125. doi: 10.1128/iai.55.12.3117-3125.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zadik P M, Chapman P A, Siddons C A. use of tellurite for the selection of verocytotoxigenic Escherichia coli O157. J Med Microbiol. 1993;39:155–158. doi: 10.1099/00222615-39-2-155. [DOI] [PubMed] [Google Scholar]