SMARCB1 is retained in PBAF and SS18–SSX-containing canonical BAF complexes and directs tumors toward synovial sarcomagenesis, while SS18–SSX promotes CBAF destruction and BAF subfamily (CBAF, PBAF, GBAF) rebalancing.
Abstract
Reduced protein levels of SMARCB1 (also known as BAF47, INI1, SNF5) have long been observed in synovial sarcoma. Here, we show that combined Smarcb1 genetic loss with SS18–SSX expression in mice synergized to produce aggressive tumors with histomorphology, transcriptomes, and genome-wide BAF-family complex distributions distinct from SS18–SSX alone, indicating a defining role for SMARCB1 in synovial sarcoma. Smarcb1 silencing alone in mesenchyme modeled epithelioid sarcomagenesis. In mouse and human synovial sarcoma cells, SMARCB1 was identified within PBAF and canonical BAF (CBAF) complexes, coincorporated with SS18–SSX in the latter. Recombinant expression of CBAF components in human cells reconstituted CBAF subcomplexes that contained equal levels of SMARCB1 regardless of SS18 or SS18–SSX inclusion. In vivo, SS18–SSX expression led to whole-complex CBAF degradation, rendering increases in the relative prevalence of other BAF-family subtypes, PBAF and GBAF complexes, over time. Thus, SS18–SSX alters BAF subtypes levels/balance and genome distribution, driving synovial sarcomagenesis.
Significance:
The protein level of BAF component SMARCB1 is reduced in synovial sarcoma but plays a defining role, incorporating into PBAF and SS18–SSX-containing canonical BAF complexes. Reduced levels of SMARCB1 derive from whole-complex degradation of canonical BAF driven by SS18–SSX, with relative increases in the abundance of other BAF-family subtypes.
See related commentary by Maxwell and Hargreaves, p. 2375
This article is highlighted in the In This Issue feature, p. 2355
Introduction
Malignant transformation represents the arrival at a particular cellular state with the specific potential to adapt, proliferate, and develop into a tissue with deadly consequences for its host organism. Mechanisms by which normal cells regulate differentiation and stemness (the capacity for subsequent differentiation) are often repurposed or dysregulated in oncogenesis. Chromatin remodeling, particularly the functions performed by the SWI/SNF family of complexes (also termed—and herein referred to generally as—BAF-family complexes in mammals, for BRG1 and Associated Factors), plays a major role in differentiation, stemness, and malignant transformation in many cellular contexts (1). Mutations in components of BAF-family complexes have been identified in a significant portion of cancers of many tissue origins (2). A few malignancies appear to be driven at the genetic level primarily (if not solely) by BAF-family derangements (3). One of these is malignant rhabdoid tumor (MRT), a pediatric cancer that consistently involves homozygous loss of function in the gene encoding the BAF component SMARCB1 (also called hSNF5, INI1, or BAF47). There are very few, if any, secondary alterations in the genomes of MRTs, suggesting that loss of SMARCB1 is an independent driver of the MRT oncogenic program (4). Smarcb1 loss in the Mx1 lineage in the mouse has also proven sufficient to induce tumors mimicking MRT (5).
Another cancer, epithelioid sarcoma (EpS), also shares the homozygous loss of SMARCB1 function at either the gene or protein level (6). Unlike MRT, EpS rarely afflicts children and typically bears a more complex genome, with additional genetic aberrations from normal cells (7). EpS shares with MRT particular cellular features, such as rhabdoid bodies (paranuclear inclusions) in the cytoplasm and epithelioid morphology (8).
A third malignancy, synovial sarcoma, involves a t(X;18) chromosomal translocation that creates a fusion to an SSX gene from SS18, a gene whose product is a known BAF-family component (9–13). All three malignancies, MRT, EpS, and synovial sarcoma, share the unusual combination of mesenchymal origins (MRT in only a subset of cases) and the ultimate expression of some epithelial marker proteins (14–19). However, only synovial sarcoma takes this mesenchymal-to-epithelial transition a step further by generating histomorphologically distinct gland-like structures in some tumors.
Synovial sarcoma also has a unique relationship with SMARCB1, with recognition in pathology comparative series that synovial sarcoma tissues have reduced levels of nuclear IHC staining for the protein, contrasted with the absence of staining in MRT and EpS and the full presence of staining in the nuclei of most other tissues and tumor types (20–23). One proposed explanation of the reduced levels of SMARCB1 in synovial sarcoma is that incorporation of SS18–SSX into the canonical BAF complex drives ejection/exclusion of the SMARCB1 protein, causing SMARCB1 instability/degradation (24). Since that model was described, a new subtype of BAF-family complexes was identified, which brought attention to a potential role in sarcomagenesis for all three major subtypes: GBAF (also known as ncBAF), canonical BAF (herein termed CBAF), and polybromo-BAF (PBAF, which provides a second host complex for SMARCB1; refs. 25, 26). As SS18–SSX-mediated synovial sarcomagenesis is robustly recapitulated in the mouse (27, 28), we determined to test this model through the impact of genetic manipulation of Smarcb1 on the characteristic synovial sarcoma tissue phenotypes, and to follow-up our results with genomic, molecular, and biochemical assays.
Results
Genetic Silencing of Smarcb1 in Mesenchyme Induces Tumors
To test the tumorigenic potential of Smarcb1 loss alone in mesenchyme, we injected TATCre protein into the paws of mice homozygous for a conditional disruption allele of Smarcb1 (5). Injected Smarcb1fl/fl mice developed a limited incidence of paw tumors over latencies consistently longer than 12 months (Fig. 1A; Supplementary Fig. S1A).
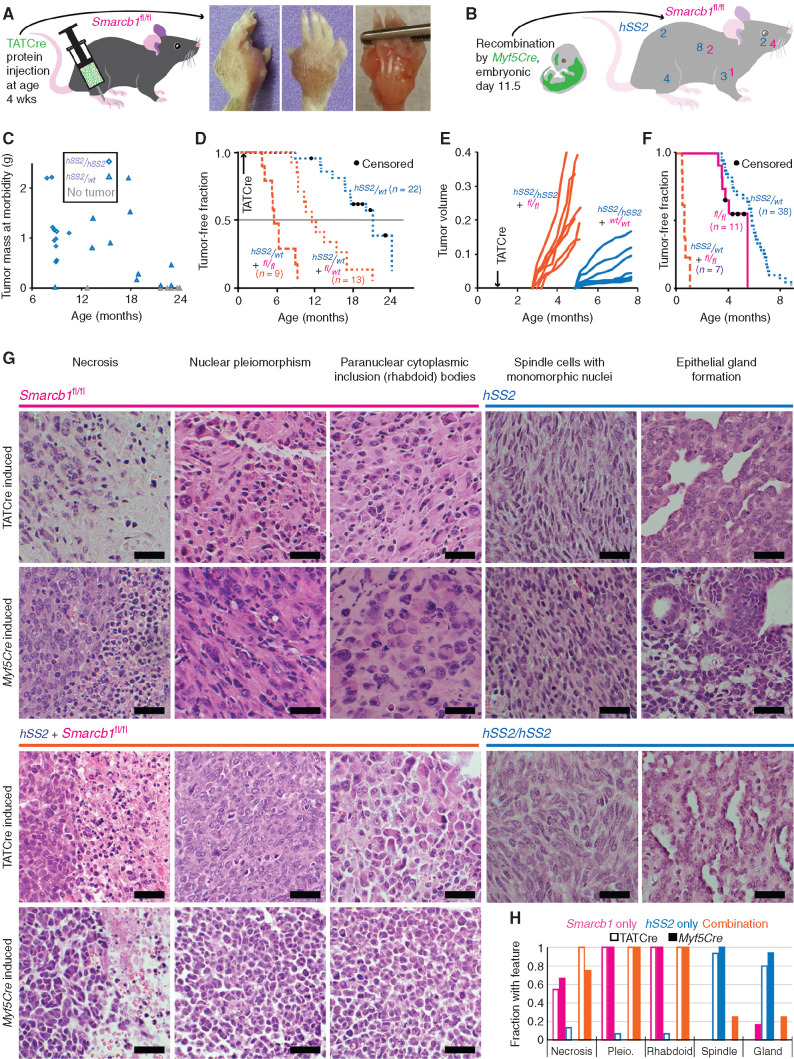
Figure 1.
SS18–SSX and Smarcb1 compete in the balance between sarcomagenesis and tumor suppression, driving different phenotypes. A, Schematic for TATCre protein injection into the paw and gross images of a tumor that developed over a year later in a Smarcb1fl/fl mouse. B, Schematic of the expression and recombination pattern from Myf5Cre and a mouse with anatomic distribution of Myf5Cre; Smarcb1fl/fl and Myf5Cre;hSS2 tumors from 16 consecutive mice in each group (two-tailed Fisher exact test for incidence of tumors in each cohort, P = 0.0002; χ2 = 17.03 with four degrees of freedom and P = 0.0019 for comparing distributions of tumors in each cohort). C, Distribution of tumor size versus time at morbidity among cohorts of hSS2/wt and hSS2/hSS2 mice in which sarcomagenesis was initiated at age 4 weeks by hind-limb injection of TATCre (two-tailed t test, P = 6.6 × 10−5 for time to tumorigenesis). D, Kaplan–Meier (KM) plot of the tumor-free fraction among cohorts of hSS2-heterozygous mice bearing each possible Smarcb1-floxed genotype (in magenta) after TATCre injection into the hind limb at age 4 weeks. (The cohort of hSS2-het–alone is data from the cohort depicted in Fig. 2A for reference; the other two cohorts are littermate-controlled cohorts: Smarcb1fl/wt compared with Smarcb1fl/fl, log-rank test z = 4.43, P < 0.001.) E, Tumor growth by caliper measurements in littermate cohorts of mice homozygous for hSS2 and either wild-type or Smarcb1fl/fl genotype following TATCre injection at age 4 weeks (two-tailed t test, P = 1.3 × 10−8 for time to detectable tumor). F, KM plot of Myf5Cre-induced combination genotype tumors compared with either hSS2 or Smarcb1fl/fl alone (log-rank tests comparing combination genotype to Smarcb1fl/fl alone, z = 4.62, P < 0.0001; to hSS2 alone, z = 8.15, P < 0.0001). G, Representative hematoxylin and eosin photomicrographs of tumors from Smarcb1fl/fl, hSS2 heterozygous, combination hSS2 and Smarcb1fl/fl, and hSS2 homozygous mice activated by TATCre limb injection or Myf5Cre (scale bars = 20 μm). H, Graph of the fraction of tumors by each induction method demonstrating each of the histologic features (TATCre + Smarcb1fl/fl, n = 11; Myf5Cre;Smarcb1fl/fl, n = 8; TATCre + hSS2, n = 15; Myf5Cre;hSS2, n = 17; TATCre + hSS2;Smarcb1fl/fl, n = 13; Myf5Cre;hSS2;Smarcb1fl/fl, n = 8). Pleio., pleiomorphism.
To test the hypothesis that SS18–SSX-mediated ejection/loss of SMARCB1 from BAF complexes was a stand-alone mechanism of synovial sarcomagenesis, we disrupted Smarcb1 in a tissue lineage with strong originating potential for SS18–SSX-mediated synovial sarcomagenesis (Supplementary Fig. S1A; refs. 27, 28). Penetrance of tumorigenesis in Myf5Cre;Smarcb1fl/fl mice was approximately 40%. Expression of SS18–SSX2 (conditionally expressed from the hSS2 allele; Supplementary Fig. S1A) in the same lineage demonstrated 100% penetrance of sarcomagenesis. The distribution of tumors in these two groups differed significantly (Fig. 1B).
Increasing Expression of SS18–SSX Enhances Synovial Sarcomagenesis
Copy-number gain in hSS2 is the only common secondary genetic change present in tumors initiated in Myf5Cre;hSS2 mice (28). As the hSS2 allele in our mouse model resides at the Rosa26 locus, both Ss18 loci are retained in their native state—unperturbed by chromosomal translocation—likely contributing more SS18 to compete with the fusion for occupancy in BAF-family complexes. To directly assess the impact of hSS2 copy number, we bred mice to be either heterozygous or homozygous for hSS2 at the Rosa26 locus, then injected TATCre protein in the hind limb at age 4 weeks. Homozygous hSS2 mice demonstrated higher (complete) penetrance of tumorigenesis. The resulting tumors grew to similar size at morbidity-determined euthanasia but in a shorter growth period (Fig. 1C).
Genetic Silencing of Smarcb1 Enhances Sarcomagenesis When Accompanying SS18–SSX Expression
To test specifically for potential competition between SS18–SSX and SMARCB1, we combined variably increased hSS2 copy number and reduced Smarcb1 copy number in several distinct models of synovial sarcomagenesis. First, an assay of tumorigenesis initiated by TATCre limb injections at age 4 weeks compared three genotypes of mice that were all heterozygous for hSS2, and wild-type, heterozygous, or homozygous for Smarcb1 floxed. Notably, heterozygosity for Smarcb1 genetic inactivation enhanced tumorigenesis in the presence of conditional fusion expression (Fig. 1D). This was tested again in a more tightly controlled littermates cohort of mice all homozygous for Rosa26hSS2/hSS2 and either heterozygous floxed or homozygous wild-type for Smarcb1. Injection of TATCre at day 8 led to a significantly shorter latency to tumorigenesis in the heterozygous Smarcb1 mice (Supplementary Fig. S1B). These TATCreinitiated Rosa26hSS2/hSS2; Smarcb1fl/wt tumors did not generally lose heterozygosity at Smarcb1 (Supplementary Fig. S1C), suggesting that Smarcb1 haploinsufficiency or reduced SMARCB1 protein stoichiometry alone may contribute to tumorigenesis. Tumorigenesis and growth rates were also compared after injection at 4 weeks of life into littermates homozygous for hSS2 and either wild-type or homozygous for conditional disruption of Smarcb1, demonstrating significantly more rapid development and growth of the latter cohort (Fig. 1E).
Expression of the fusion with homozygous conditional disruption of Smarcb1 in the Myf5Cre lineage led to aggressive tumorigenesis at short latency compared with either genetic manipulation alone. Remarkably, no Myf5Cre;Rosa26hSS2/wt; Smarcb1fl/fl mouse survived beyond the age of 1 month, and most developed morbid tumors by age 2 weeks (Fig. 1F; Supplementary Fig. S1D–S1F). Metastasis was histologically confirmed in the lungs of five of the seven Myf5Cre-initiated combination genotype mice and none of the other two genotypes.
Homozygous Silencing of Smarcb1 with or without SS18–SSX Expression Generates EpS-Related Histologic Features
Histologic evaluation of tumors from each of the Smarcb1-loss initiation methods identified EpS features, including necrosis, nuclear pleiomorphism, and paranuclear cytoplasmic inclusion bodies, distinct from the gland forming epithelial cells and spindle cells with monomorphic nuclei that characterize the synovial sarcomas from SS18–SSX–expressing hSS2 mice (both heterozygous and homozygous at Rosa26; Fig. 1G and H; Supplementary Fig. S1G). We observed no histologic difference between hSS2-induced tumors with wild-type Smarcb1 or heterozygous inactivation of Smarcb1 (Supplementary Fig. S1H), but homozygous ablation of Smarcb1 accompanying fusion expression initiated by TATCre generated distinct histology that included the same EpS features (Fig. 1G and H).
The histology of tumors with combined homozygous silencing of Smarcb1 and expression of SS18–SSX initiated by Myf5Cre also demonstrated EpS features, although with more of a poorly differentiated, MRT-like appearance overall (Fig. 1G and H).
Expression of SS18–SSX Impacts Tumor Transcriptomes and Combined Loss of Smarcb1 Confers Additional Changes
The transcriptomes of MRT and synovial sarcoma differ significantly (29), whether examining data sets from human tumors or derived cell lines. To explore the transcriptional impact of Smarcb1 genetic silencing, SS18–SSX expression, or the combination of the two in mice, RNA sequencing (RNA-seq) was performed for mouse tumors with either presence or absence of heterozygosity for hSS2 and wild-type or homozygous floxed alleles of Smarcb1. Sample tumors included those derived from Myf5Cre or TATCre initiation. Tumors of each genotype clustered most closely with themselves in pairwise comparisons of whole transcriptomes and in principal component analysis (Fig. 2A and B). For hSS2 activation alone, TATCre- and Myf5Cre-initiated tumors intermingled. For each of the genotypes that included Smarcb1 homozygous loss, TATCre and Myf5Cre tumors each clustered distinctly but in adjacent groups. Overall, there was slightly stronger correlation between all transcriptomes that shared hSS2 expression (with or without Smarcb1 genetic silencing) than with those that shared Smarcb1 loss (with or without hSS2 expression.)
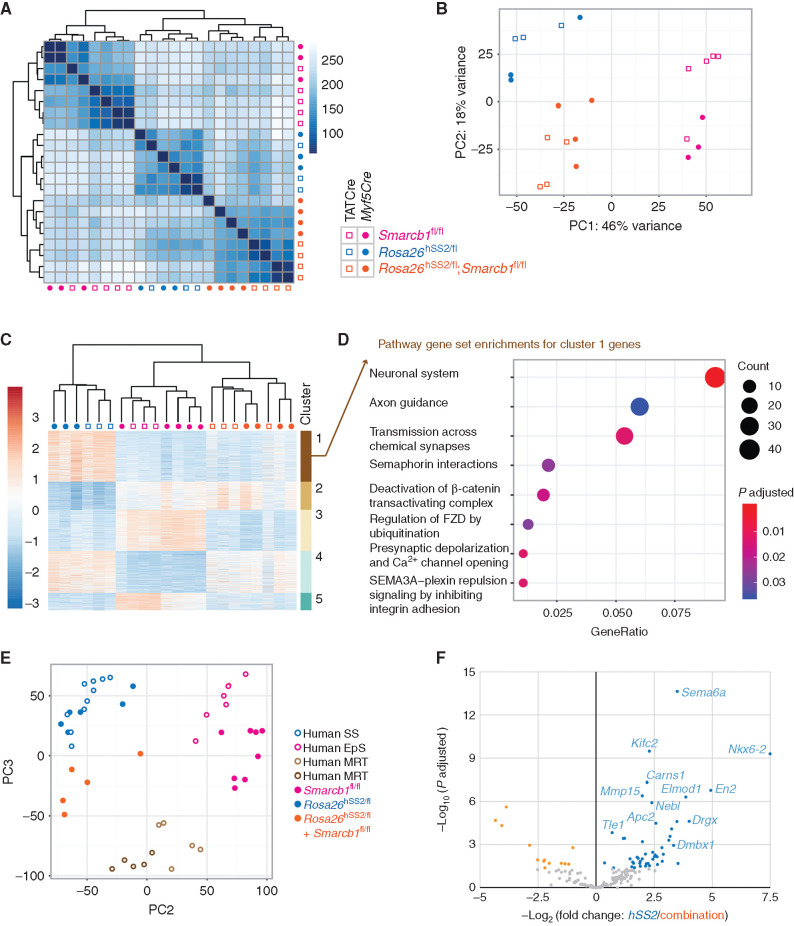
Figure 2.
Genetic silencing of Smarcb1 added to SS18–SSX expression results in distinct tumor transcriptomes. A, Euclidean distance of samples heat map of nonhierarchical clustering of transcriptomes from tumors in mice bearing hSS2, homozygosity for Smarcb1-floxed, or combination genotypes following Myf5Cre or TATCre induction. B, Two-dimensional principal component analysis (PCA) for the transcriptomes of these mouse tumors using the top 500 most differentially expressed genes. C, Heat map hierarchical clustering of the most differentially expressed genes between the six groups of tumors, with k-means clustering. D, Reactome pathway analysis of cluster 1 genes, specifically expressed at higher levels in fusion-only expressing tumors, compared with those that have silencing of Smarcb1 alone or in combination with the fusion. E, Two-dimensional PCA of transcriptomes of mouse and human synovial sarcoma, MRT, and EpS tumors, after separating principal component 1 (see Supplementary Fig. S2B), which represented the species-specific differences between these groups. F, Differential expression between hSS2-only and combination genotype tumors from mice for homologs of a human gene set defined as direct targets of the fusion in a human synovial sarcoma cell line.
Unsupervised hierarchical (k-means) clustering identified clusters of genes that discriminated between the groups (Fig. 2C). Additional interrogation of the Reactome pathways represented in the differential expression of the first cluster, genes relatively highly expressed in fusion-expressing tumors with wild-type Smarcb1 compared with either genotype with silencing of Smarcb1, indicated involvement of many pathways considered to be characteristic of target genes of the SS18–SSX fusion (Fig. 2D; refs. 29–31). These included axon guidance, β-catenin, and Frizzled pathways. The other clusters demonstrated some expected variations between synovial sarcoma and MRT/EpS (Supplementary Fig. S2A). Comparative genomics, using principal component analysis with published human tumor transcriptomes, clustered fusion-only mouse tumors with human synovial sarcomas and Smarcb1-loss–only mouse tumors with human EpSs, with combination genotype tumors clustering closer to human MRTs (Fig. 2E; Supplementary Fig. S2B). Evaluation of a gene set defined as direct fusion targets by chromatin immunoprecipitation sequencing (ChIP-seq) and reduced expression upon knockdown of SS18–SSX in a human synovial sarcoma cell line, as well as increased expression in synovial sarcomas over other sarcoma types in The Cancer Genome Atlas (30), showed differential expression in fusion-only tumors over combination genotype tumors (Fig. 2F), suggesting that even targets of the oncogenic BAF complex may be affected by genetic loss of Smarcb1, challenging the prior model that held that SMARCB1 is excluded from that complex by inclusion of SS18–SSX.
BAF-Family Genomic Locations Demonstrate Variable Impacts of Fusion Expression and Smarcb1 Silencing
In human MRT cancer cell lines, SMARCB1 loss diminishes BAF complex occupancy of many enhancers alongside relative retention of BAF at superenhancers (32). In human synovial sarcoma cell lines, BAF-family complexes reside at actively transcribed chromatin and can extend across gene bodies in broader or longer peaks than otherwise typical for BAF localization (29–31).
To test how BAF-family complexes distribute across the mouse genome in these different tumor genotypes, we performed ChIP-seq in tumor tissues, using antibodies raised against “core” BAF-family subunits shared by all BAF-family subtypes—SMARCC1 (also known as BAF155) and SMARCA4 (also known as BRG1)—and conducted comparisons (Supplementary Fig. S3A and S3B).
The ChIP-seq enrichment of SMARCC1 and SMARCA4 across regions defined by intersection of the two in each tumor genotype group was stronger in the fusion-only tumors compared with either tumor genotype that included genetically silenced Smarcb1 (Fig. 3A). Although the position of BAF-family peaks with respect to genes was similar between fusion-only and combination genotype tumors, there was an increased prevalence of intronic and decreased prevalence of promoter BAF-family binding sites in Smarcb1-loss–alone tumors (Fig. 3B; Supplementary Fig. S3C). The higher prevalence of combined SMARCA4 and SMARCC1 peaks near transcription start sites (TSS) in the presence of the fusion may have derived from the extended breadth of BAF peaks in fusion-expressing tumors regardless of Smarcb1 status, fitting with the previous descriptions from human synovial sarcoma cell lines (Fig. 3C and D; refs. 29–31). Although there were relative losses of BAF-family enrichment in promoters by the addition of Smarcb1 genetic silencing to fusion expression, the fusion-only tumor-defined distal intergenic SMARCA4 peaks were even more profoundly reduced in the other groups (Fig. 3E).
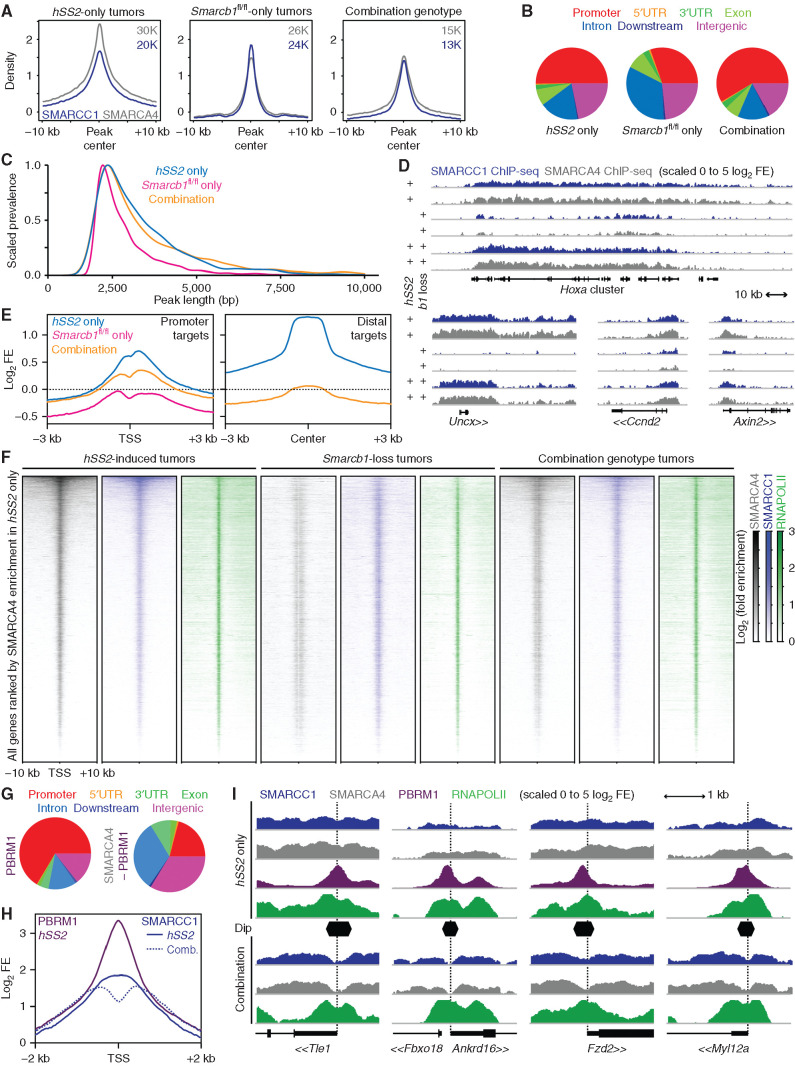
Figure 3.
Smarcb1 loss and SS18–SSX expression drive aberrant BAF-family complex distributions across chromatin genome-wide. A, ChIP-seq enrichment plots for SMARCA4 (gray) and SMARCC1 (blue) and centered on intersection peaks for combined SMARCA4 and SMARCC1 enrichment in tumors of fusion-only, Smarcb1-loss–only, or combination genotypes. B, Distribution of SMARCA4 ChIP-seq peaks genome-wide with respect to gene features in each group. UTR, untranslated region. C, Plot of the normalized prevalence of BAF-family peaks of different length in each of the groups after filtering out peaks less than 2 kb in length. (Tukey range test adjusted P value for comparing fusion-only to combination genotype tumor BAF-family peak lengths was 0.179, not significant. The comparison of either combination genotype or fusion-only tumor to Smarcb1-loss–alone tumor BAF-family peak lengths had P values below the detectable value.) D, Example ChIP-seq enrichment tracks of lengthened BAF-family peaks in tumors expressing the fusion contrasted to Smarcb1 loss alone (b1 indicates Smarcb1). E, Enrichment plots for log-transformed fold enrichments (FE) of SMARCA4 ChIP-seq for all three groups at promoter sites enriched in hSS2-only tumors or distal peaks in the same. F, Heat maps of ChIP-seq distributions around TSSs across the whole genome, ordered by enrichment for SMARCA4 in hSS2-only tumors. G, Distribution of SMARCA4 peaks that coincide with or are independent of PBRM1 peaks in fusion-only tumors with respect to gene annotations across the genome. H, Enrichment plot of SMARCC1 for fusion-only and combination (Comb.) genotype tumors at TSSs defined as having a steep dip in the combination tumors, with overlaid enrichment plot for PBRM1 in hSS2-only tumors. I, Example tracks of BAF component enrichments at TSSs (dotted vertical lines) among target genes contrasting hSS2-only and combination hSS2;Smarcb1fl/fl tumors.
We next performed ChIP-seq for RNA Polymerase II (RNAPOLII) to determine the BAF-family relationship with transcription in each tumor genotype. First, BAF ChIP-seq enrichments demonstrated a very similar overall pattern in combination and fusion-only genotype tumors at TSSs across the genome, albeit with reduced enrichment generally in the combination group (Fig. 3F). When we tested for differential expression of genes annotated by differential enrichment for SMARCC1 and SMARCA4, we found that reduced BAF-family enrichment in a group of tumor genotypes associated with reduced expression of the annotated genes (Supplementary Fig. S3D and S3E).
Focally at TSSs, we observed a striking reduction of BAF enrichment in both groups that bore genetic silencing of Smarcb1—a feature not present in the fusion-only group—visualized as a “stripe” of diminished BAF-family complexes at the TSS (Fig. 3F). Given that PBAF is the BAF-family complex subtype that is described to be specifically abundant at promoters (33), we performed another ChIP-seq for PBRM1 (also known as BAF180), a PBAF-specific component, in fusion-only tumors—but not in combination tumors—as PBAF was not significantly present on chromatin after genetic silencing of Smarcb1 (Supplementary Fig. S3F and S3G). PBAF peaks were mostly in promoters, whereas BAF-family peaks with PBAF omitted localize to distal sites (Fig. 3G). TSSs with strong focal loss of BAF-family ChIP enrichment in combination genotype tumors were also found to have strong promoter focal enrichment for PBAF in fusion-only tumors (Fig. 3H and I).
SMARCB1 Remains Present at Reduced Levels in Synovial Sarcoma Tumors, Integrated in BAF-Family Complexes
To test if in synovial sarcoma, SMARCB1 might incorporate into PBAF complexes, a subset of BAF complexes that are thought to exclude both native SS18 and the fusion SS18–SSX but include SMARCB1 (13), we performed immunoprecipitation (IP) of a human synovial sarcoma cell line, HSSYII, and HEK293T cells as a control, using an antibody against PBRM1 and SMARCB1, and then performed Western blots (WB) for each. These experiments demonstrated that in synovial sarcoma cells, SMARCB1 protein is present and co-IPs with a PBAF subunit (Fig. 4A). We next tested for SMARCB1 participation in CBAF, using co-IP for DPF2, a CBAF-specific component, and found that both proteins were able to co-IP the other from synovial sarcoma cells (Fig. 4B).
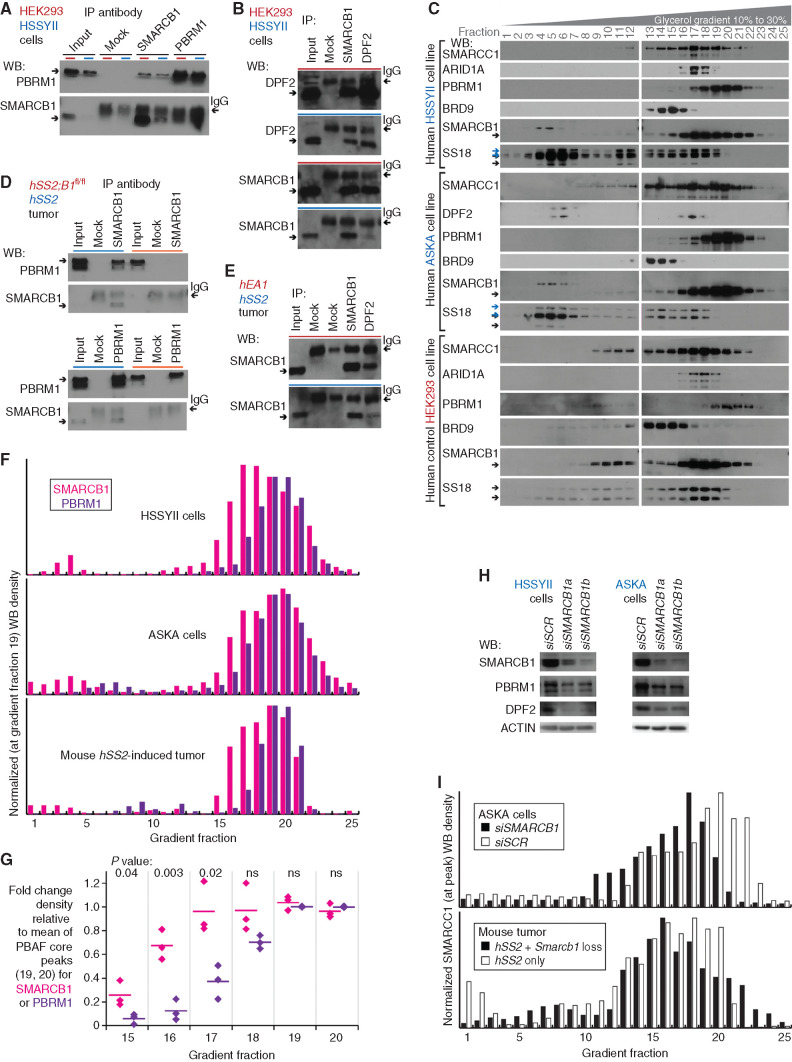
Figure 4.
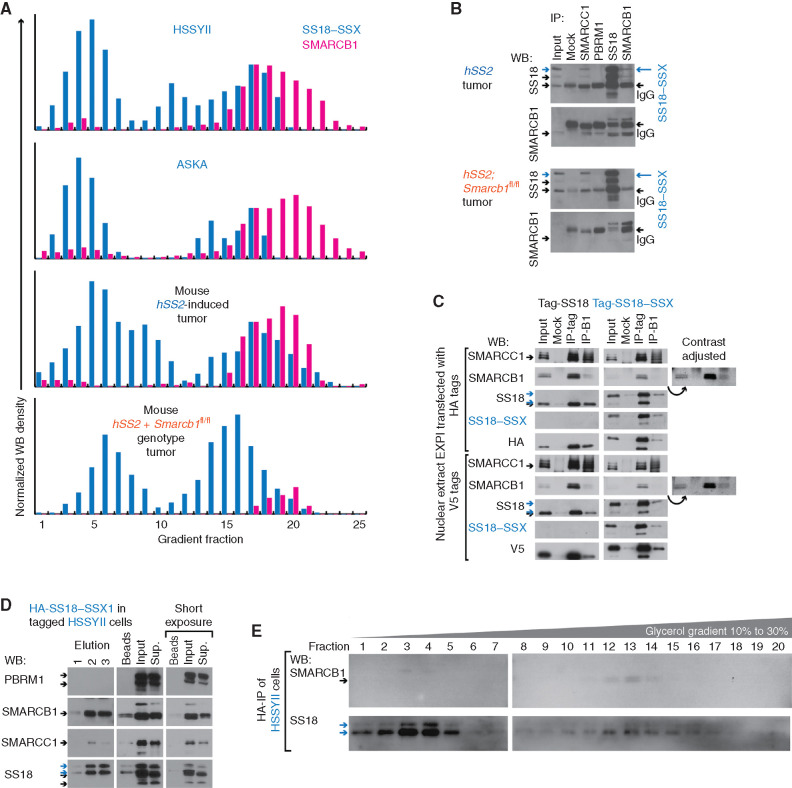
SMARCB1 at reduced protein levels in synovial sarcoma cells resides in BAF complexes. A, WBs for reciprocal IP in human synovial sarcoma cell line HSSYII and control HEK293T cells, for PBAF components SMARCB1, and PBRM1. B, WBs for reciprocal IP in human cells for CBAF components DPF2 and SMARCB1. C, WBs for BAF-family components in glycerol gradients for human synovial sarcoma cell lines HSSYII and ASKA, as well as control HEK293T cells. D, WBs for reciprocal IP in fusion-only and combination genotype mouse tumors for PBAF components SMARCB1 and PBRM1. E, WBs for CBAF component IP in fusion-only and control (EA1 = EWSR1–ATF1-induced mouse tumor) tumors. F, Optical densitometry–quantified gradients of SMARCB1 and PBRM1 depict overlap among the glycerol gradient fractions in two human synovial sarcoma cell lines and a mouse synovial sarcoma tumor. G, Quantified fraction densities of each protein compared with the mean density of PBAF core fractions 19 and 20 of itself (two-tailed t test P values listed at top; the three sample sources are HSSYII, ASKA, and mouse synovial sarcoma tumor gradients). ns, not significant. H, WBs for BAF-family components after application of scrambled versus SMARCB1-targeting siRNA (siSCR and siSMARCB1). I, Optical densitometry–quantified gradients of SMARCC1 demonstrate shifts in the relative abundance of PBAF-sized complexes with added disruption of SMARCB1 in the ASKA cell line or mouse tumors.
Glycerol gradients of nuclear extracts were then utilized to identify the size of SMARCB1-containing complexes, using two human synovial sarcoma cell lines, HSSYII and ASKA, with HEK293T cells as a control. SMARCB1 from synovial sarcoma nuclei cofractionated with PBRM1, sized appropriately for PBAF, and CBAF-specific components, DPF2 or ARID1A. Another smaller amount of protein, revealed by anti-SMARCB1 WB, appeared in gradient fractions consistent with BAF-family subunit monomers, running at a higher size than the predicted 47 kDa (Fig. 4C).
The same approaches of co-IP and glycerol gradients of nuclear extracts were tested in mouse tumors, using combination genotype tumors with genetically silenced Smarcb1 as a negative control. Similarly, synovial sarcoma tumors demonstrated strong coprecipitation of SMARCB1 with PBRM1 and DPF2 and the reciprocal coprecipitations, albeit with low levels of SMARCB1 present in the input (Fig. 4D and E; Supplementary Fig. S4A and S4B). There was a similar distribution of SMARCB1 across both PBAF and CBAF fractions in the mouse gradients. To quantify this distribution, appreciating that gradient WBs with different antibodies cannot be compared with one another but only to the relative distribution of any single protein across the different sized fractions, we compared the relative abundance of SMARCB1 in the CBAF-sized fractions compared with PBRM1 in the same fractions, each normalized against PBAF-peak-sized fractions. In both human synovial sarcoma cell lines and the mouse hSS2-only tumors, SMARCB1 was significantly more abundant than PBRM1 in gradient fractions 15 through 17, each relative to fractions 19 and 20 (Fig. 4F and G).
To test the interaction between SMARCB1 and BAF complexes in synovial sarcoma cells further, we applied short interfering RNA (siRNA) against SMARCB1 to reduce its presence in HSSYII and ASKA cells. Depletion of SMARCB1 by siRNA reduced PBRM1 levels (Fig. 4H), indicating that PBRM1 stability in synovial sarcoma is dependent on SMARCB1 and providing an orthogonal line of evidence that SMARCB1 incorporates into PBAF complexes in synovial sarcoma. SMARCB1 knockdown in synovial sarcoma cells also reduced the stability of the CBAF-specific subunit, DPF2, which only assembles into SMARCB1-containing CBAF complexes (26). This similarly suggested that SMARCB1 in synovial sarcoma cells without added siSMARCB1 was also incorporating into CBAF complexes.
Glycerol gradients of nuclear extracts were also performed for the ASKA cell line exposed to siSMARCB1 or control scrambled siRNA. These confirmed a drastic reduction of PBRM1 in PBAF-sized complexes (Supplementary Fig. S4C and S4D). Also, SMARCC1 gradient WBs were quantitated as a representative measure of the relative distribution of GBAF, CBAF, and PBAF, because SMARCC1 incorporates into each of these BAF subtypes. Compared with controls, cells or a tumor with forced reduction in SMARCB1 by RNA interference or genetic silencing reduced the relative presence of SMARCC1 in PBAF-sized fractions (Fig. 4I).
SMARCB1 Associates with CBAF Complexes That Contain the SS18–SSX Fusion
Distributions of SS18–SSX and SMARCB1 in human synovial sarcoma cell lines and mouse hSS2-induced tumors overlapped in CBAF-sized gradient fractions (Fig. 5A). To more directly test their potential interaction, we performed IP and WB for SMARCB1 and SS18, as well as SMARCC1 and PBRM1. In hSS2-induced tumors, but not those that also had undergone genetic ablation of Smarcb1, anti-SMARCB1 coprecipitated a fusion-sized band on SS18 WB (Fig. 5B). We therefore tested the ability of SMARCB1 and a V5- or HA-tagged SS18–SSX fusion transfected into EXPI293 cells (a HEK293T variant that grows in suspension culture) to reciprocally co-IP with each other. Here, we found clear coprecipitation, although we also observed reduced levels of both SMARCB1 and SMARCC1 proteins in the nuclear extracts (input sample) following expression of the fusion compared with controls that expressed tagged SS18 (Fig. 5C). Next, we attempted co-IP of SMARCB1 with a CRISPR-HA–tagged endogenous SS18–SSX1 in the human cell line HSSYII (30). Indeed, IP of HA-SS18–SSX1 coprecipitated SMARCB1, leading to SMARCB1 depletion from the supernatant, strongly suggesting that the two interact, either directly or within a CBAF complex (Fig. 5D). To test whether these two interact in BAF-family–sized complexes, we performed glycerol gradients to size the eluted complexes from the HA-IP. Although the yield of intact complexes was very low (even for SS18–SSX itself), most of the detectable SMARCB1 was found in the same fractions as SS18–SSX and ran in the fractions predicted for CBAF-sized complexes (Fig. 5E). Again, we noted a faint SMARCB1-staining band, running at larger size than 47 kDa in the monomer-sized fractions, possibly indicating SMARCB1 ubiquitylation during incorporation with SS18–SSX in CBAF complexes.
Figure 5.
SMARCB1 associates with SS18–SSX in CBAF complexes. A, Optical densitometry–quantified glycerol gradients of SS18–SSX and SMARCB1 in two human synovial sarcoma cell lines, as well as tumors from fusion-only and combination genotype mice. B, WBs for reciprocal IP of SS18 and SMARCB1, as well as other BAF components in fusion-only and combination genotype mouse tumors. C, HEK293 cells transfected with HA-SS18, HA-SS18–SSX, V5-SS18, or V5-SS18–SSX, followed by tag-IP or SMARCB1-IP and WB for each. (Additional contrast-adjusted images demonstrate the clearer presence of auto-IPed SMARCB1.) D, WB for BAF components after elution following IP-HA for SS18–SSX in HA-tagged HSSYII. Sup., supernatant. E, WBs for SS18 and SMARCB1 in glycerol gradient fractions (only 20 total for glycerol 10% to 30%) for HA-purified complexes.
Synovial Sarcomas Display Reduced Abundance of CBAF
Because SMARCB1 protein levels are reduced in synovial sarcoma, but SMARCB1 is not excluded from CBAF complexes that contain SS18–SSX, we hypothesized that changes in SMARCB1 abundance could be alternatively attributed to changes in the relative abundance of each BAF-family complex subtype (Fig. 6A). SS18–SSX overexpression led to a reduction in SMARCC1 protein levels in EXPI293 nuclear extracts (Fig. 5C), suggesting that the presence of the fusion leads to an overall reduction in BAF-family complexes in those cells. Further, we developed an analysis algorithm to use SMARCC1 distribution as a measure of the relative abundances of each complex subtype (described in detail in the Supplementary Detailed Methods). For this, we performed glycerol gradients of nuclear extracts, collected in 25 fractions, blotting the higher molecular weight fractions (12–24) for BRD9 to define the distribution of GBAF, DPF2 to define the distribution of CBAF, and PBRM1 to define the distribution of PBAF. We calculated the relative abundance of each complex type to explain the overall SMARCC1 distribution across those fractions. On mouse tumors, this algorithm revealed the almost complete obliteration of PBAF in combination genotype tumors but low abundance of CBAF overall in both hSS2-only and combination tumors (Fig. 6B, compared with the typical predominance of CBAF). To scale up the quantitative use of this algorithm, we performed gradients in triplicate for each of five human synovial sarcoma cell lines and three control cell lines. In synovial sarcoma cells, CBAF relative abundance was significantly reduced (Fig. 6C; Supplementary Fig. S5). Two possible explanations for lower CBAF abundance in synovial sarcoma are that (i) the fusion prevents CBAF assembly, or (ii) the fusion promotes CBAF degradation.
Figure 6.
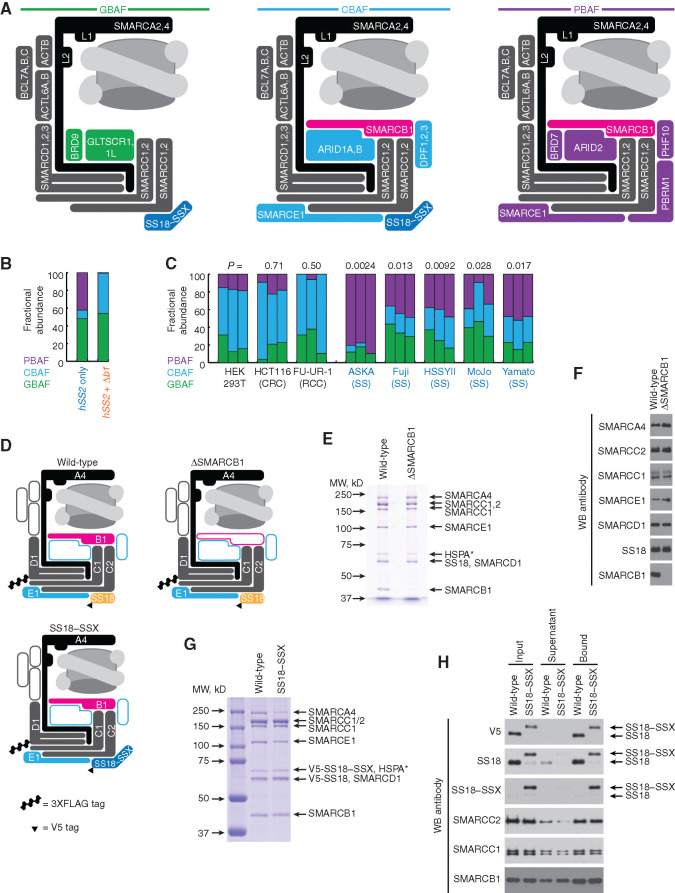
Purified mammalian recombinant CBAFs coincorporate SMARCB1 with SS18–SSX. A, Schematic of BAF-family complex subtypes and their varied componentry in synovial sarcoma. (L1 and L2 refer to the lobes by which SMARCA2/4 translocates DNA around the nucleosome.) B, The relative abundance of BAF subtypes determined by SMARCC1 distribution in gradients as a product of GBAF, CBAF, and PBAF contributions to each fraction. C, The fractional abundances of BAF subtypes calculated by optical densitometry–quantified gradients of SMARCC1 overlaid onto the normal distributions of PBAF, CBAF, and GBAF defined by PBRM1, DPF2, and BRD9, respectively, in synovial sarcoma and control cell lines. CRC, colorectal carcinoma; RCC, renal cell carcinoma. (P values at top are the comparison of each cell line to HEK293T for CBAF fractional abundance, by two-tailed heteroscedastic t test.) D, Schematic of canonical BAF components in the variant complexes, demonstrating which components were overexpressed in a mammalian cell-based recombinant system. E, Coomassie stain of a denaturing polyacrylamide gel electrophoresis of FLAG-purified recombinant CBAF complexes generated with or without the coexpression of SMARCB1. (HSPA* represents the substoichiometric presence of a heat shock protein chaperone.) MW, molecular weight. F, WBs of purified complexes with antibodies against each overexpressed component in either SMARCB1-including or excluding variants. G, Coomassie stain of a denaturing gel of FLAG-purified recombinant CBAF complexes generated from expression of SS18 or SS18–SSX. H, WBs of BAF components in input (FLAG-purified complexes), supernatant, and anti-V5 IP of complexes from coexpression of V5-tagged SS18 or SS18–SSX.
Recombinant CBAF Complexes Assemble Avidly, Coincorporating SS18–SSX and SMARCB1
To directly test assembly of CBAFs containing the fusion SS18–SSX, as well as coassembly with SMARCB1 in a context where degradation would not reduce CBAF levels, we developed a system to coexpress multiple CBAF components (including most conserved core subunits) in human EXPI293 cells. These components included SMARCA4, SMARCC1, SMARCC2, 3XFLAG-tagged SMARCD1, SMARCE1, and SS18—either with or without SMARCB1 (Fig. 6D). Anti-FLAG purification of SMARCD1 and associated CBAF subcomplexes from cells coexpressing SMARCB1 identified the position of the SMARCB1 band in SDS-PAGE gels, and revealed its loss when omitted from the expression system (Fig. 6E and F). To test whether SMARCB1 assembly was compatible with the SS18–SSX fusion, we expressed CBAF components (including SMARCB1) and either V5-tagged SS18–SSX or SS18 itself, followed by anti-FLAG enrichment and elution of complexes; this revealed the clear presence of SMARCB1 in both purified complexes (Fig. 6G). A subsequent IP with anti-V5, using the FLAG-purified complexes from each type as the input, demonstrated an equivalent amount of SMARCB1 protein in the purified recombinant CBAF complexes containing SS18–SSX as those with SS18 itself (Fig. 6H). This demonstrates that assembly of SMARCB1 into CBAF was not hampered by the presence of the fusion in this system.
SS18–SSX Incorporation in CBAF Complexes Leads to Their Degradation
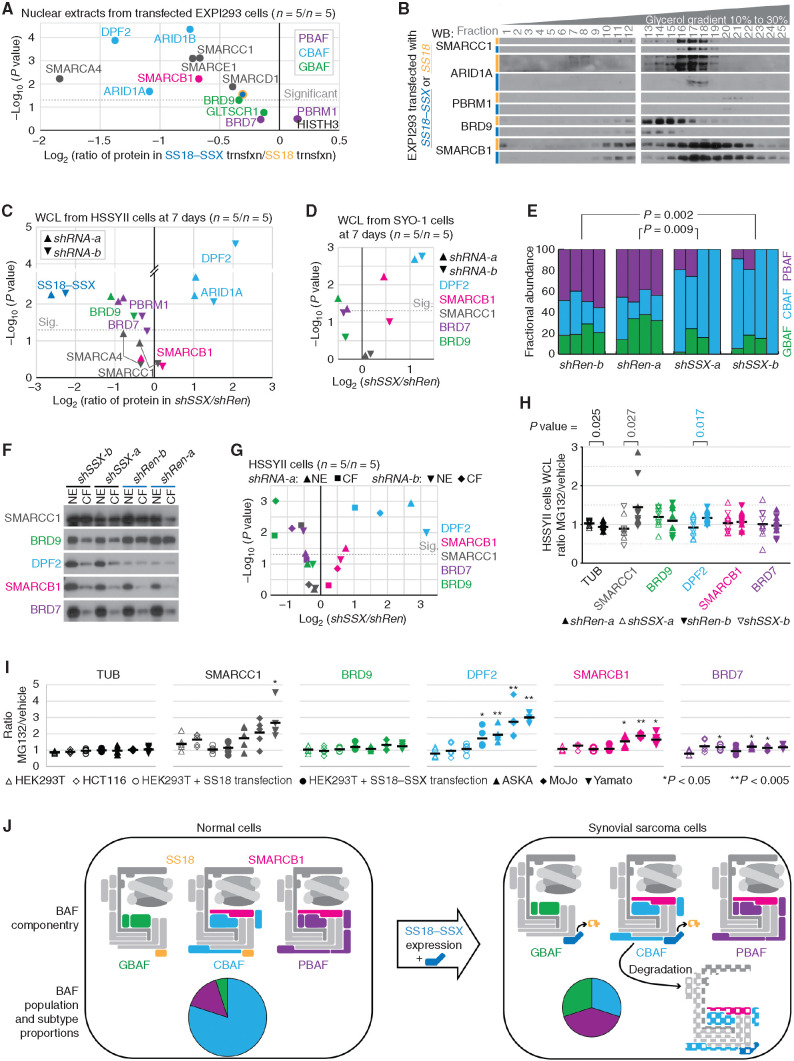
To investigate the alternate hypothesis, that degradation of CBAF drives its lower abundance in cells, LICOR quantitative WBs were performed for nuclear extracts from EXPI293 cells transfected to express either SS18–SSX or SS18 (n = 5, each), comparing the ratios in abundance of BAF-family components. The core components participate in all three BAF-family subtypes; SMARCB1 and SMARCE1 participate in both CBAF and PBAF, and SS18 or SS18–SSX can participate in both GBAF and CBAF (see Fig. 6A). Strikingly, every component that participates in CBAF was significantly reduced in abundance in fusion-expressing cells (Fig. 7A; Supplementary Fig. S6A). CBAF-exclusive components were among the most significantly reduced in cells expressing the fusion. WBs of the fractions from glycerol gradients of these two groups demonstrated a drastic reduction of components in all complex-sized fractions, but no significant redistribution of SMARCC1 to PBAF or GBAF fractions (Fig. 7B). As this suggested that the expression of the fusion drives a degradation of CBAF, it was next necessary to test if manipulation of the presence of the fusion in synovial sarcoma cells would also alter the relative abundances of different complex subtypes.
Figure 7.
Expression of SS18–SSX leads to CBAF complex reductions and relative overabundance of PBAFs and GBAFs. A, Quantitative LICOR WBs of BAF components in nuclear extracts of EXPI293 cells transfected with either SS18–SSX or SS18 (n = 5 each) presenting the log-transformed two-tailed Student t test P value of the difference between and the ratio of protein in the fusion-transfected versus SS18-transfected cells. trnsfxn, transfection. B, WBs of glycerol gradient fractions of EXPI293T cells transfected with SS18–SSX or SS18. C, LICOR quantitative WBs from whole-cell lysates (WCL) collected from HSSYII human synovial sarcoma cells subjected to 7 days of shRNAs (two sequences each; n = 5 for each sequence of each shRNA) directed against control (Renilla, shRen) or the fusion (SS18–SSX, shSSX), with BAF subunits color coded by BAF, presented as log-transformed paired two-tailed t test P values and ratios of fusion knockdown over control knockdown. Sig., significant. D, LICOR quantitative WBs of WCLs from SYO-1 human synovial sarcoma cells after knockdown of the fusion or control for 7 days. E, Fractional abundances of BAF subtypes defined by optical densitometry–quantified gradients of SMARCC1 (as in Fig. 6C) for HSSYII cells subjected to shRNAs against the fusion or control. (P values from two-tailed paired t tests; n = 4 for each shRNA.) F, WBs of nuclear extract (NE) with the paired chromatin fraction (CF; protein that stays with the insoluble chromatin pellet after NE) of proteins after 7 days of fusion or control knockdown. G, LICOR quantitative WB abundances presented as paired t test P values and ratios of fusion over control knockdown in each of the NE and CF components of HSSYII cells after 7 days. H, LICOR quantitative WB–defined proteins in WCLs presented as the ratios of MG132-treated over DMSO vehicle–treated cells after weeklong shRNA depletion of SS18–SSX or control Renilla (n = 5 for each condition, n = 10 for each group; two-tailed heteroscedastic t test comparing the ratios for each protein by knockdown group). I, LICOR quantitative WB–defined proteins in WCLs presented as of the ratios of MG132-treated over DMSO vehicle–treated cells from synovial sarcoma (or SS18–SSX-transfected) and control (or SS18-transfected) cell lines (n = 5 each; P values are from two-tailed Student t tests comparing each to HEK293T untransfected control cells). J, Model schematic of the impact of SS18–SSX expression on BAF componentry and relative abundance of BAF subtypes.
The relative abundances of BAF-family components were compared by quantitative WBs after shRNA depletion for 7 days of the fusion or control Renilla in two synovial sarcoma cell lines. Concerned that redistribution due to increased affinity for chromatin could also contribute to a depletion of CBAF in nuclear extracts, we first collected whole-cell lysates, harvested with high stringency, instead. These demonstrated significantly increased presence of CBAF-specific components after depletion of the fusion but also a less marked decrease of GBAF-specific and PBAF-specific components (Fig. 7C and D; Supplementary Fig. S6B and S6C).
Density gradient WBs from nuclear extracts for quantitation of complex subtype abundances in the fusion-depleted or control cells also demonstrated a significant increase in the relative abundance of CBAF complexes and decrease in the relative abundance of GBAF and PBAF in weeklong fusion depletion in HSSYII cells (Fig. 7E; Supplementary Fig. S7A–S7C).
From similarly manipulated cells, the protein remaining bound to the insoluble chromatin fraction following the nuclear extraction protocol demonstrated very little PBAF or CBAF, but nearly as much or more GBAF as that harvested from the nuclear extract (Fig. 7F). This prompted another round of quantitative WBs to compare these components in each of these nuclear “compartments” between fusion-depleting and control shRNAs. Despite a relative increase in the chromatin-bound fraction of CBAF in the presence of the fusion, the vast majority of CBAF still partitioned to the nuclear extract. Further, even the absolute level of CBAF in that insoluble chromatin-bound fraction was reduced in control knockdown cells compared with fusion knockdown, suggesting that changes in CBAF localization cannot explain the reduction in CBAF components (Fig. 7G; Supplementary Fig. S7D). Notably, the insoluble chromatin-bound fraction of GBAF was vastly depleted upon knockdown of the fusion, even while the nuclear extract fraction changed less significantly, suggesting that stronger affinity for chromatin in the presence of the fusion profoundly affects nuclear extract levels of GBAF.
Because no assembly defect was observed, and greater affinity for insoluble chromatin could not account for the depletion of CBAF and increased abundance of GBAF and PBAF in the presence of the fusion, we next tested the hypothesis that if CBAF depletion involved some fusion-promoted degradation mechanism, then this depletion would be blunted at least partly by proteasome inhibition. In the scrambled control knockdown HSSYII cells, in HEK293T cells expressing the fusion, and in a few additional synovial sarcoma cell lines, exposure to MG132, a proteasome inhibitor, decreased the depletion of CBAF-specific DPF2, contrasted with fusion-depleted HSSYII cells, HEK293T cells expressing SS18, and non–synovial sarcoma cell lines, respectively (Fig. 7H and I).
Discussion
The observation that SS18–SSX fusion oncoproteins interact with BAF-family complexes provided early mechanistic insight into synovial sarcoma (10–13, 24) and has here been corroborated (Fig. 4D and E; Supplementary Fig. S4A) in mice that recapitulate overall fusion-driven sarcomagenesis (27, 28). Advancing understanding of the BAF family of complexes related to synovial sarcoma, MRT, EpS, and other cancers has increasingly ascribed oncogenic and tumor-suppressive functions to particular components and BAF-family subtypes. Here, loss of Smarcb1 in mesenchyme, including the Myf5Cre cell lineage with strong origination potential for synovial sarcoma (27, 28), drove tumorigenesis that instead resembled EpS by histologic features and transcriptome (Figs. 1 and 2). This corroborates literature that retracted the briefly held concept in the synovial sarcoma field that SS18–SSX's only oncogenic function was ejection of SMARCB1 from BAF-family complexes (24, 29–31).
Adding Smarcb1 silencing to SS18–SSX expression synergized in tumorigenesis that only partly recapitulated synovial sarcoma features (Figs. 1–3). This finding was incompatible with the prevailing model in the field that expression of SS18–SSX renders BAF-family complexes to a SMARCB1-lacking state (24). SMARCB1 loss, by itself or added to fusion expression, affected BAF-family complexes in two ways: it reduced general BAF-family affinity for and distribution across chromatin and functionally obliterated PBAF assembly, altering BAF-family distribution to promoters and TSSs specifically (Figs. 3F–I and 4H and I).
PBAF is shown here to be prominent and active in synovial sarcoma but likely disrupted in MRT and EpS. The reportedly recovered distribution of BAF-family complexes to particular TSSs following SMARCB1 reexpression in MRT cell lines may derive from recovery of PBAF (34). In synovial sarcoma, at TSSs, PBAF exerts a tumor-suppressive function, likely enabling the expression of differentiation genes that affords synovial sarcoma its unique features of both mesenchymal and epithelial differentiation, which is lost upon Smarcb1 silencing/loss. PBAF stability has previously been shown to be compromised by loss of other components (35). A role for SMARCB1 and PBAF in TSS-specific binding may indicate their affinity for the H2A.Z-modified histones that reside at the TSS and +1 nucleosome positions (36). RSC (the most abundant BAF-related complex in yeast) binds to and ejects H2A.Z-containing nucleosomes preferentially (37).
We find that SMARCB1 incorporates into CBAF in synovial sarcoma (Fig. 4), including CBAF containing SS18–SSX (Figs. 5 and 6). As CBAF is the dominant BAF-family complex type in most cells, alterations in its abundance affect the detectable levels of each component, also impacting overall BAF-family distribution across chromatin. SS18–SSX expression reduces SMARCB1 levels; however, SMARCB1 levels are reduced to a less pronounced degree than CBAF-specific components, as the overall abundance of SMARCB1 is rescued partly by an increased presence of PBAF in synovial sarcoma (Fig. 7J).
The near disappearance of CBAF from nuclear extracts upon expression of the fusion SS18–SSX could derive from one of three mechanisms. First, SS18–SSX could drive a CBAF assembly defect, only subtly different from the SMARCB1 ejection model. Equivalent recombinant complex assembly observed here with SS18 or SS18–SSX (Fig. 6G and H) argues against fusion-mediated CBAF assembly failure, as does the observation in prior literature (24, 29–31) and glycerol gradient WBs presented here (Fig. 4C; Fig. 5A; Supplementary Figs. S4A; S7A and S7B) that SS18–SSX-containing CBAFs far outnumber those containing native SS18, though both proteins are similarly expressed.
Second, SS18–SSX could sequester CBAF in other cellular compartments, reducing its detection in nuclear extracts. Indeed, we observed a slightly increased portion of CBAF remaining on insoluble chromatin after nuclear extraction in the presence of the fusion (Fig. 7F), fitting the increased affinity for chromatin conferred by the fusion's SSX tail (38). However, the absolute abundance of CBAF associated with chromatin was also depleted (Fig. 7G) due to the fusion eliciting far lower CBAF total levels. In contrast, redistribution to chromatin from SS18–SSX incorporation profoundly affected GBAF abundance in nuclear extracts. Approximately half of GBAF remains bound to insoluble chromatin following nuclear extraction from synovial sarcoma cells without fusion depletion (Fig. 7F). This argues that BAF-family subtype abundances (based on nuclear extracts to isolate intact complexes) vastly underestimate relative GBAF abundances in synovial sarcoma specifically.
In the third model, degradation targets SS18–SSX-containing CBAF. This model is supported by the observation that proteasome inhibition blunts CBAF-specific component reductions in the presence of the fusion (Fig. 7H and I). Critically, fully assembled complexes appear to be the target of degradation, because all components involved in CBAF demonstrate some reduction in abundance (Fig. 7A), suggesting that CBAF components are not merely disassembling to be recycled for reincorporation into alternative CBAFs with SS18 (full model in Fig. 7J). The precise mechanism of fusion-mediated CBAF degradation deserves additional investigation. Others have shown that the fusion's SSXRD domain interacts with a ubiquitin E3 ligase with otherwise established BAF-family component targeting capacity (39). Here, we further speculate that the basic tail on SMARCB1 (40), coincorporated in a complex with the SSXRD domain on SS18–SSX, may offer tempting substrates for other E3 ligases as well.
Implications of oncoprotein-mediated CBAF complex degradation are broad. First, it is important to note that this particular effect of SS18–SSX on BAF-family complexes is not likely to be oncogenic directly. Instead, this better explains why expression of SS18–SSX in most cell types is quickly lethal (27, 28). If cells are rendered CBAF-deficient, most will not be capable of achieving a selectable epigenetic state that rescues general BAF-family function with GBAF and PBAF upregulation. Second, CBAF degradation explains the BRD9 dependency in synovial sarcoma that has been highlighted recently (31, 41). BRD9 is the only GBAF component without a paralog (see Fig. 6A). Notably, only drugs degrading BRD9, not functional inhibitors, have affected synovial sarcoma, making this dependence relationship more about GBAF function than about BRD9 specifically. Here, we show that GBAF is by far the more abundant SS18–SSX-containing BAF-family complex on synovial sarcoma chromatin, fitting the hypothesis of Brien and colleagues (31), who suggested that GBAF is the fusion-bearing complex, rather than that of Michel and colleagues (41), who suggested that GBAF is a synovial sarcoma dependency but not the host complex for the fusion. Importantly, although reduced in abundance, CBAF is not absent from synovial sarcoma chromatin, where it includes the fusion and SMARCB1. Because the mechanism for CBAF depletion relates to the extent of proteasomal degradation, predicted variations in penetrance may provide opportunities for selection of epigenetic states that promote cancer cell survival. In keeping, synovial sarcoma cell lines vary in levels of retained CBAF (Fig. 6C). Our work predicts that therapeutic strategies that target GBAF stability will lead to resistance mechanisms that downregulate CBAF degradation as cells strive to promote CBAF maintenance.
In summary, these experiments have demonstrated an essential, defining role for SMARCB1 in synovial sarcomagenesis, revealed its incorporation into both PBAF and SS18–SSX-containing CBAF complexes, and identified CBAF degradation coupled with GBAF and PBAF upregulation as a major effect of SS18–SSX expression. This work enhances our understanding of how SS18–SSX disrupts BAF-family complexes and provides additional conceptual support (as well as potential resistance mechanisms tumors will use) for the targeting of GBAF as a therapeutic strategy in synovial sarcoma. Future efforts will continue to pursue the relative contributions of changes in the abundance, localization, and activity of each BAF-family complex subtype in synovial sarcomagenesis.
Methods
Please see the Supplementary Detailed Methods for more detailed protocols and reagents used (Key Resources Table).
Cell Lines
HSSYII, ASKA, Yamato, SYO1, and Fuji were provided by T. Nielsen (42–45). HSSYII-HA–tagged was provided by A. Banito (30). MoJo was generated in the laboratory (46). HEK293T was purchased from ATCC and EXPI293 from Thermo. Cells were authenticated by Mycoplasma testing and STR DNA profiling every half-year of culture.
Animal Studies
Mouse experiments were approved by the University of Utah animal care committee in accordance with international legal and ethical standards. Smarcb1fl mice were obtained from Dana-Farber Cancer Institute (5). Rosa26hSS2 and Myf5Cre mice were previously described (27). Mouse strains were maintained on a mixed C57BL/6 and SvJ background with littermate controls and roughly equivalent distribution of sexes.
Recombinant BAF Expression in Mammalian Bigbac
The pFastBac1-CMV vector was a gift from Dr. Erhu Cao (University of Utah, Salt Lake City, UT). cDNA's of each of the BAF subunits were cloned into pFastBac1-CMV and then into pBig1 and pBig2 vectors with custom oligonucleotides (47). All BAF subunits were assembled into a single pBig2 vector prior to transfection into EXPI293 cells.
RNAi
SS18–SSX-specific shRNA expression vectors were provided by A. Banito and delivered by lentiviral transfection (shREN-a = shRen-713, shRen-b = shRen-660, shSSX-a = shSSX-1045, and shSSX-b = shSSX-1274) as previously described (30). Human SMARCB1-specific siRNAs (Invitrogen) were delivered with Lipofectamine RNAiMAX Transfection Reagent (Thermo).
Sequencing
The accession numbers for the RNA-seq and ChIP-seq data are GSE153856 and GSE153857, respectively.
Rigor, Reproducibility, and Statistical Analysis
For every assessment of data that invoked judgment of any kind, samples were randomized in order of assessment and raters were blinded as to the group identity of each sample.
Statistical comparisons between two groups used two-tailed Student t test (unless a paired or heteroscedastic t test was needed and noted in the figure legend) and were performed in GraphPad Prism software 7.0; statistical significance was set at P < 0.05 or 0.01 as indicated in the figure legends. Data are presented as mean ± standard deviation unless otherwise noted. The sample size was determined by power analysis using the results of preliminary experiments to estimate variance, and the number (n) indicated in the figures or figure legends represents biological replicates. All WB experiments were repeated to confirm the relationships presented and discussed.
Authors' Disclosures
J.J. Barrott reports personal fees from Specicare, Inc. and non-financial support from H2 Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Figures and Legends, Methods, and Key Resources Table
Acknowledgments
The authors thank Brian Dalley and the High-Throughput Genomics Core for their assistance, as well as the Research Histology Core at Huntsman Cancer Institute. Charlie Roberts kindly provided the mouse strain with floxed Smarcb1. This work was supported by R01CA201396 (to K.B. Jones and B.R. Cairns), U54CA231652 (to K.B. Jones, B.R. Cairns, and A. Banito), and 2P30CA042014-31 from the NCI/NIH; the Paul Nabil Bustany Fund for Synovial Sarcoma Research (to K.B. Jones); and the Sarcoma Foundation of America (to J.J. Barrott).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors' Contributions
J. Li: Investigation, writing–review and editing. T.S. Mulvihill: Investigation, writing–original draft, writing–review and editing.L. Li: Data curation, formal analysis, visualization, writing–review and editing. J.J. Barrott: Investigation, writing–review and editing. M.L. Nelson: Investigation, writing–review and editing. L. Wagner: Investigation. I.C. Lock: Investigation. A. Pozner: Investigation. S.L. Lambert: Investigation. B.B. Ozenberger: Data curation, formal analysis. M.B. Ward: Data curation, formal analysis.A.H. Grossmann: Data curation, formal analysis. T. Liu: Data curation, formal analysis, investigation, methodology. A. Banito: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration, writing–review and editing. B.R. Cairns: Conceptualization, resources, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing. K.B. Jones: Conceptualization, resources, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 2011;21:396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish Jet al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013;45:592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 2011;11:481–92. [DOI] [PubMed] [Google Scholar]
- 4. Lee RS, Stewart C, Carter SL, Ambrogio L, Cibulskis K, Sougnez Cet al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest 2012;122:2983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell 2002;2:415–25. [DOI] [PubMed] [Google Scholar]
- 6. Modena P, Lualdi E, Facchinetti F, Galli L, Teixeira MR, Pilotti Set al. SMARCB1/INI1 tumor suppressor gene is frequently inactivated in epithelioid sarcomas. Cancer Res 2005;65:4012–9. [DOI] [PubMed] [Google Scholar]
- 7. Jamshidi F, Bashashati A, Shumansky K, Dickson B, Gokgoz N, Wunder JSet al. The genomic landscape of epithelioid sarcoma cell lines and tumours. J Pathol 2016;238:63–73. [DOI] [PubMed] [Google Scholar]
- 8. Chbani L, Guillou L, Terrier P, Decouvelaere AV, Gregoire F, Terrier-Lacombe MJet al. Epithelioid sarcoma: a clinicopathologic and immunohistochemical analysis of 106 cases from the French sarcoma group. Am J Clin Pathol 2009;131:222–7. [DOI] [PubMed] [Google Scholar]
- 9. Crew AJ, Clark J, Fisher C, Gill S, Grimer R, Chand Aet al. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J 1995;14:2333–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. dos Santos NR, de Bruijn DR, Balemans M, Janssen B, Gartner F, Lopes JMet al. Nuclear localization of SYT, SSX and the synovial sarcoma-associated SYT-SSX fusion proteins. Hum Mol Genet 1997;6:1549–58. [DOI] [PubMed] [Google Scholar]
- 11. Thaete C, Brett D, Monaghan P, Whitehouse S, Rennie G, Rayner Eet al. Functional domains of the SYT and SYT-SSX synovial sarcoma translocation proteins and co-localization with the SNF protein BRM in the nucleus. Hum Mol Genet 1999;8:585–91. [DOI] [PubMed] [Google Scholar]
- 12. Kato H, Tjernberg A, Zhang W, Krutchinsky AN, An W, Takeuchi Tet al. SYT associates with human SNF/SWI complexes and the C-terminal region of its fusion partner SSX1 targets histones. J Biol Chem 2002;277:5498–505. [DOI] [PubMed] [Google Scholar]
- 13. Middeljans E, Wan X, Jansen PW, Sharma V, Stunnenberg HG, Logie C. SS18 together with animal-specific factors defines human BAF-type SWI/SNF complexes. PLoS One 2012;7:e33834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pinkus GS, Kurtin PJ. Epithelial membrane antigen–a diagnostic discriminant in surgical pathology: immunohistochemical profile in epithelial, mesenchymal, and hematopoietic neoplasms using paraffin sections and monoclonal antibodies. Hum Pathol 1985;16:929–40. [DOI] [PubMed] [Google Scholar]
- 15. Daimaru Y, Hashimoto H, Tsuneyoshi M, Enjoji M. Epithelial profile of epithelioid sarcoma: an immunohistochemical analysis of eight cases. Cancer 1987;59:134–41. [DOI] [PubMed] [Google Scholar]
- 16. Kodet R, Newton WA Jr, Sachs N, Hamoudi AB, Raney RB, Asmar Let al. Rhabdoid tumors of soft tissues: a clinicopathologic study of 26 cases enrolled on the Intergroup Rhabdomyosarcoma Study. Hum Pathol 1991;22:674–84. [DOI] [PubMed] [Google Scholar]
- 17. Parham DM, Weeks DA, Beckwith JB. The clinicopathologic spectrum of putative extrarenal rhabdoid tumors. An analysis of 42 cases studied with immunohistochemistry or electron microscopy. Am J Surg Pathol 1994;18:1010–29. [DOI] [PubMed] [Google Scholar]
- 18. Miettinen M, Lehto VP, Virtanen I. Monophasic synovial sarcoma of spindle-cell type: epithelial differentiation as revealed by ultrastructural features, content of prekeratin and binding of peanut agglutinin. Virchows Arch B Cell Pathol Incl Mol Pathol 1983;44:187–99. [DOI] [PubMed] [Google Scholar]
- 19. Mukai M, Torikata C, Iri H, Hanaoka H, Kawai T, Yakumaru Ket al. Cellular differentiation of epithelioid sarcoma: an electron-microscopic, enzyme-histochemical, and immunohistochemical study. Am J Pathol 1985;119:44–56. [PMC free article] [PubMed] [Google Scholar]
- 20. Arnold MA, Arnold CA, Li G, Chae U, El-Etriby R, Lee CCet al. A unique pattern of INI1 immunohistochemistry distinguishes synovial sarcoma from its histologic mimics. Hum Pathol 2013;44:881–7. [DOI] [PubMed] [Google Scholar]
- 21. Hoot AC, Russo P, Judkins AR, Perlman EJ, Biegel JA. Immunohistochemical analysis of hSNF5/INI1 distinguishes renal and extra-renal malignant rhabdoid tumors from other pediatric soft tissue tumors. Am J Surg Pathol 2004;28:1485–91. [DOI] [PubMed] [Google Scholar]
- 22. Kohashi K, Oda Y, Yamamoto H, Tamiya S, Matono H, Iwamoto Yet al. Reduced expression of SMARCB1/INI1 protein in synovial sarcoma. Mod Pathol 2010;23:981–90. [DOI] [PubMed] [Google Scholar]
- 23. Mularz K, Harazin-Lechowska A, Ambicka A, Kruczak A, Rozmus-Pieton M, Marchinska-Osika Uet al. Specificity and sensitivity of INI-1 labeling in epithelioid sarcoma. Loss of INI1 expression as a frequent immunohistochemical event in synovial sarcoma. Pol J Pathol 2012;63:179–83. [DOI] [PubMed] [Google Scholar]
- 24. Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell 2013;153:71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alpsoy A, Dykhuizen EC. Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J Biol Chem 2018;293:3892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mashtalir N, D'Avino AR, Michel BC, Luo J, Pan J, Otto JEet al. Modular organization and assembly of SWI/SNF family chromatin remodeling complexes. Cell 2018;175:1272–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell 2007;11:375–88. [DOI] [PubMed] [Google Scholar]
- 28. Jones KB, Barrott JJ, Xie M, Haldar M, Jin H, Zhu JFet al. The impact of chromosomal translocation locus and fusion oncogene coding sequence in synovial sarcomagenesis. Oncogene 2016;35:5021–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McBride MJ, Pulice JL, Beird HC, Ingram DR, D'Avino AR, Shern JFet al. The SS18-SSX fusion oncoprotein hijacks BAF complex targeting and function to drive synovial sarcoma. Cancer Cell 2018;33:1128–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Banito A, Li X, Laporte AN, Roe JS, Sanchez-Vega F, Huang CHet al. The SS18-SSX oncoprotein hijacks KDM2B-PRC1.1 to drive synovial sarcoma. Cancer Cell 2018;33:527–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brien GL, Remillard D, Shi J, Hemming ML, Chabon J, Wynne Ket al. Targeted degradation of BRD9 reverses oncogenic gene expression in synovial sarcoma. Elife 2018;7:e41305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang X, Lee RS, Alver BH, Haswell JR, Wang S, Mieczkowski Jet al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat Genet 2017;49:289–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pan J, McKenzie ZM, D'Avino AR, Mashtalir N, Lareau CA, St Pierre Ret al. The ATPase module of mammalian SWI/SNF family complexes mediates subcomplex identity and catalytic activity-independent genomic targeting. Nat Genet 2019;51:618–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuwahara Y, Wei D, Durand J, Weissman BE. SNF5 reexpression in malignant rhabdoid tumors regulates transcription of target genes by recruitment of SWI/SNF complexes and RNAPII to the transcription start site of their promoters. Mol Cancer Res 2013;11:251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AMet al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 2018;359:770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bagchi DN, Battenhouse AM, Park D, Iyer VR. The histone variant H2A.Z in yeast is almost exclusively incorporated into the +1 nucleosome in the direction of transcription. Nucleic Acids Res 2020;48:157–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cakiroglu A, Clapier CR, Ehrensberger AH, Darbo E, Cairns BR, Luscombe NMet al. Genome-wide reconstitution of chromatin transactions reveals that RSC preferentially disrupts H2AZ-containing nucleosomes. Genome Res 2019;29:988–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McBride MJ, Mashtalir N, Winter EB, Dao HT, Filipovski M, D'Avino ARet al. The nucleosome acidic patch and H2A ubiquitination underlie mSWI/SNF recruitment in synovial sarcoma. Nat Struct Mol Biol 2020;27:836–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Patel N, Wang J, Shiozawa K, Jones KB, Zhang Y, Prokop JWet al. HDAC2 regulates site-specific acetylation of MDM2 and its ubiquitination signaling in tumor suppression. iScience 2019;13:43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Valencia AM, Collings CK, Dao HT, St Pierre R, Cheng YC, Huang Jet al. Recurrent SMARCB1 mutations reveal a nucleosome acidic patch interaction site that potentiates mSWI/SNF complex chromatin remodeling. Cell 2019;179:1342–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Michel BC, D'Avino AR, Cassel SH, Mashtalir N, McKenzie ZM, McBride MJet al. A non-canonical SWI/SNF complex is a synthetic lethal target in cancers driven by BAF complex perturbation. Nat Cell Biol 2018;20:1410–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sonobe H, Manabe Y, Furihata M, Iwata J, Oka T, Ohtsuki Yet al. Establishment and characterization of a new human synovial sarcoma cell line, HS-SY-II. Lab Invest 1992;67:498–505. [PubMed] [Google Scholar]
- 43. Naka N, Takenaka S, Araki N, Miwa T, Hashimoto N, Yoshioka Ket al. Synovial sarcoma is a stem cell malignancy. Stem Cells 2010;28:1119–31. [DOI] [PubMed] [Google Scholar]
- 44. Kawai A, Naito N, Yoshida A, Morimoto Y, Ouchida M, Shimizu Ket al. Establishment and characterization of a biphasic synovial sarcoma cell line, SYO-1. Cancer Lett 2004;204:105–13. [DOI] [PubMed] [Google Scholar]
- 45. Nojima T, Wang YS, Abe S, Matsuno T, Yamawaki S, Nagashima K. Morphological and cytogenetic studies of a human synovial sarcoma xenotransplanted into nude mice. Acta Pathol Jpn 1990;40:486–93. [DOI] [PubMed] [Google Scholar]
- 46. Jin H, Barrott JJ, Cable MG, Monument MJ, Lerman DM, Smith-Fry Ket al. The impact of microenvironment on the synovial sarcoma transcriptome. Cancer Microenviron 2017;10:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weissmann F, Petzold G, VanderLinden R, Huis In't Veld PJ, Brown NG, Lampert Fet al. biGBac enables rapid gene assembly for the expression of large multisubunit protein complexes. Proc Natl Acad Sci U S A 2016;113:E2564–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Legends, Methods, and Key Resources Table