Abstract
We have identified a new murine transforming growth factor β superfamily member, growth-differentiation factor 15 (Gdf15), that is expressed at highest levels in adult liver. As determined by Northern analysis, the expression of Gdf15 in liver was rapidly and dramatically up-regulated following various surgical and chemical treatments that cause acute liver injury and regeneration. In situ hybridization analysis revealed distinct patterns of Gdf15 mRNA localization that appeared to reflect the known patterns of hepatocyte injury in each experimental treatment. In addition, treatment of two hepatocyte-like cell lines with either carbon tetrachloride or heat shock induced Gdf15 mRNA expression, indicating that direct cellular injury can induce Gdf15 expression in the absence of other cell types, such as inflammatory cells. In order to investigate the potential functions of Gdf15, we created Gdf15 null mice by gene targeting. Homozygous null mice were viable and fertile. Despite the dramatic regulation of Gdf15 expression observed in the partial-hepatectomy and carbon tetrachloride injury models, we found no differences in the injury responses between homozygous null mutants and wild-type mice. Our findings suggest either that Gdf15 does not have a regulatory role in liver injury and regeneration or that Gdf15 function within the liver is redundant with that of other signaling molecules.
The transforming growth factor β (TGF-β) superfamily consists of a diverse group of structurally related proteins involved in the growth, differentiation, and repair of many tissues (reviewed by McPherron and Lee [26]). For mammals, more than 30 different members of this superfamily have been reported, including the TGF-βs, bone morphogenetic proteins, inhibins and activins, Müllerian inhibiting substance, nodal, leftys, TGF-β-related neurotrophic factors (GDNF, neurturin, persephin, and artemin), and a heterogeneous group of proteins referred to as growth-differentiation factors.
Each member of the TGF-β superfamily is synthesized as a large precursor protein that undergoes two proteolytic processing steps. The first involves removal of the N-terminal hydrophobic signal sequence. The second cleavage event occurs at a conserved RXXR site approximately 120 amino acids from the C terminus, generating an N-terminal proregion and a biologically active C-terminal region. The C-terminal regions of all superfamily members are structurally related, and the various TGF-β superfamily members can be classified into distinct subgroups based on sequence homology in this region. C-terminal amino acid identities within a particular subgroup generally range from 70 to 90%, although homologies between subgroups are considerably lower. For most of the family members, the active species appears to be a disulfide-linked homodimer of C-terminal fragments. Heterodimers of some family members, such as the TGF-βs and inhibins and activins, have also been shown to be biologically active, although in some cases with biological properties distinct from those of either homodimeric form.
We have been using degenerate PCR and low-stringency screening methods to identify new TGF-β family members that may play important regulatory roles during embryonic development or adult tissue homeostasis. Using a human sequence previously reported as human TGF-βPL (hTGF-βPL) (42), hMIC-1 (2), hPDF (32), hPLAB (16), and hPTGFB (21) (for simplicity, we refer to this gene as hTGF-βPL) as a probe, we identified a novel sequence which we designated Gdf15. While our studies on this clone were being completed, the murine and rat Gdf15 sequences and their expression patterns were published (3, 4). Here we describe the regulation of Gdf15 expression in multiple models of acute liver and bile duct injury and the characterization of Gdf15 null mice.
MATERIALS AND METHODS
Library screening, hybridization, and in situ analysis.
A murine 129/SvJ genomic library (25) and a liver cDNA library (Stratagene, La Jolla, Calif.) were screened as described previously (22), using the region corresponding to the entire C terminus of hTGF-βPL (2, 16, 21, 32, 42) as a probe. Low-stringency genomic Southern analysis was carried out as described before (8, 35). The blots were hybridized at 33°C below the melting temperature of the probes. Northern analysis of RNA samples prepared using the one-step RNAzol B method (Tel-Test, Inc., Friendswood, Tex.) was carried out as described previously (35). In situ hybridization using digoxigenin-labeled cRNA probes corresponding to the 5′ untranslated region and propeptide-coding region of Gdf15 was carried out essentially as described before (20, 36), except that slides were washed overnight prior to detection and that BM-purple (Boehringer Mannheim, Indianapolis, Ind.) was used as the colored precipitant. Color development reactions were carried out for 16 to 24 h.
In vivo injury models.
All animal studies were approved by the Animal Care and Use Committee at The Johns Hopkins University School of Medicine. Partial hepatectomy was performed on 5- to 7-week-old male CD-1 mice (Charles River, Wilmington, Mass.) as previously described (15), using methoxyflurane (Mallinckrodt Veterinary, Inc., Mundelein, Ill.) as an inhaled anesthetic. Each sham-operated animal was anesthetized, followed by incision of the peritoneal cavity, gentle manipulation of the liver, and closure of the abdomen with sutures. Carbon tetrachloride (CCl4), d-galactosamine (GalN), and methylenedianiline (DAPM) (all from Sigma Chemical Corporation, St. Louis, Mo.) were administered to 5- to 6-week-old male C57BL/6J mice (Jackson Laboratories, Bar Harbor, Maine) by intraperitoneal injection in a volume of 0.1 ml. Doses for these chemicals were 20 μl of CCl4 in soy oil (Wesson vegetable oil; Hunt-Wesson, Inc., Fullerton, Calif.) per mouse, 0.7 g of GalN in saline per kg of body weight, and 50 mg of DAPM in 50% ethanol in saline per kg of body weight. Ethanol was delivered by intraperitoneal injection of 4 g of absolute ethanol per kg of body weight, diluted with water to a final volume of 0.3 ml. Control samples for each chemical injury were isolated from animals that received injection of the carrier alone. Serum samples were collected by cardiac puncture of anesthetized animals just prior to euthanasia and analyzed by Antech Diagnostics (Farmingdale, N.Y.).
Dividing cells were labeled for 2 h with 5-bromo-2′-deoxyuridine (BrdU) by injecting mice with 200 mg (per kg body weight) of a 10:1 molar ratio of BrdU (Sigma) and 5-fluoro-2′-deoxyuridine (Sigma) resuspended in phosphate-buffered saline (11). BrdU-labeled nuclei were detected on frozen sections using the FLUOS in situ cell proliferation kit (Boehringer Mannheim) according to the manufacturer's instructions, counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (Molecular Probes, Eugene, Oreg.), and mounted in ProLong antifade mounting medium (Molecular Probes). Terminal deoxynucleotidyltransferase-mediated dUTP-X nick end labeling (TUNEL)-positive cells were detected on frozen sections using the fluorescein in situ cell death detection kit (Boehringer Mannheim) and counterstained and mounted as described above.
Cell isolation and culture.
Peritoneal macrophages were isolated from 5- to 6-week-old C57BL/6J female mice as described before (6). Briefly, reactive macrophages were isolated from mice injected with thioglycolate (Sigma) 6 days before harvest and allowed to adhere to plastic tissue culture dishes for 3 h at 37°C in 5% CO2. The culture medium was removed and replaced with either fresh medium or fresh medium supplemented with 160 nM phorbol-12-myristate-13-acetate (PMA) (Sigma) for an additional 3 h.
Separation of liver cells into parenchymal and nonparenchymal fractions was carried out as described previously (13), using Liver Perfusion and Liver Digest media from Life Technologies (Gibco BRL, Rockville, Md.). NMuLi cells (ATCC CRL-1638) and AML12 cells (ATCC CRL-2254) from the American Type Culture Collection (Manassas, Va.) were grown to near confluence in plastic flasks. CCl4 was added directly to the culture medium at a final concentration of 20 or 40 mM. The flasks were sealed and incubated on an orbital platform at 37°C and 30 rpm. For the heat shock experiments, near-confluent cells were incubated in 5% CO2 at 44°C for 30 min and then allowed to recover at 37°C in 5% CO2.
Gdf15 null mice.
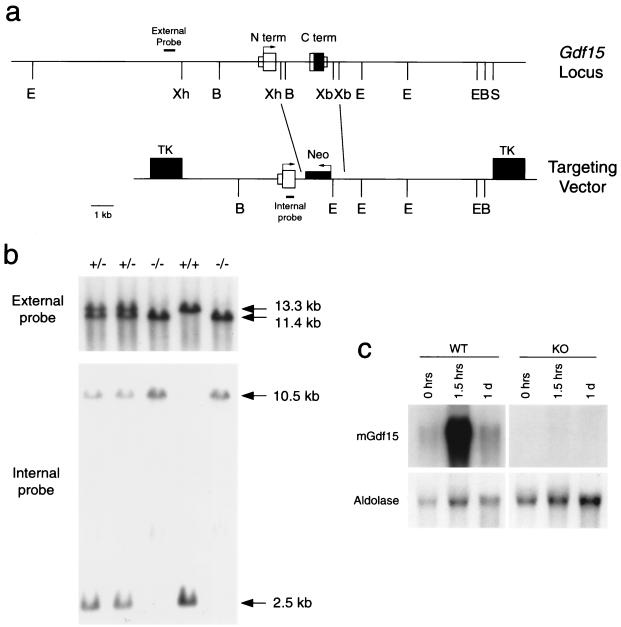
The genomic structure of the murine Gdf15 gene was determined using restriction mapping and sequence analysis of lambda phage clones isolated from a 129/SvJ genomic library. A targeting construct was designed and transfected into R1 embryonic stem cells (30) and selected in 2 μM ganciclovir (Roche Discovery Welwyn, Hertfordshire, England) and 200 μg of G418 (Gibco BRL) per ml following established protocols (34). Injection of targeted clones into C57BL/6 blastocysts and embryo transfer into pseudopregnant females were carried out by the Johns Hopkins Transgenic Core Facility (Baltimore, Md.). The Gdf15 null mice and their wild-type littermates were maintained on a hybrid C57BL/6/129/SvJ background. Mice were screened using probes both external and internal to the targeted vector (see Fig. 6).
FIG. 6.
Targeting construct and genomic identification of Gdf15 null mice. (a) Restriction map of the Gdf15 genomic locus (top line) and of the targeting vector (bottom line). S, SalI; B, BamHI; E, EcoRI; Xh, XhoI; Xb, XbaI; TK, thymidine kinase cassette; Neo, neomycin resistance cassette. The external and internal probes used for Southern blot analysis are indicated. (b) Representative Southern blots of DNA isolated from wild-type, heterozygous, and homozygous Gdf15 null mice. The upper blot shows genomic DNA digested with EcoRI and probed with the external probe. The endogenous Gdf15 allele is contained within the 13.3-kb fragment, and the null allele is contained within the 11.4-kb fragment. The lower blot shows genomic DNA digested with BamHI and probed with the internal probe. The endogenous Gdf15 allele gives a 2.5-kb fragment, while the null allele produces a 10.5-kb fragment. (c) Northern analysis of 20 μg of total RNA isolated from livers of wild-type (WT) and null (KO) mice treated with carbon tetrachloride and probed with a fragment corresponding to the C-terminal region of Gdf15.
Nucleotide sequence accession number.
The nucleotide sequence of Gdf15 is available under GenBank accession number AF159571.
RESULTS
Identification of Gdf15.
We identified Gdf15 in a screen for novel murine TGF-β superfamily members while probing a murine genomic library with the hTGF-βPL sequence. While the experiments described here were being completed, the cloning and expression pattern of Gdf15 in the mouse and rat were published (3, 4).
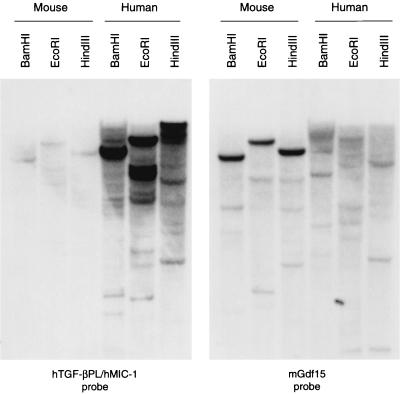
Pairwise comparisons between the C-terminal regions of Gdf15 and the other TGF-β superfamily members revealed that Gdf15 is most closely related to hTGF-βPL (67% amino acid identity and 72% nucleotide identity [data not shown]). Because most other TGF-β family members are more highly conserved across species, we attempted to identify the human ortholog of Gdf15 and the murine ortholog of the hTGF-βPL gene by screening both human and murine genomic DNA libraries with these probes. Despite extensive screening of these libraries, we were unable to identify a murine sequence more closely related to the hTGF-βPL gene than Gdf15 or a human sequence more closely related to Gdf15 than the hTGF-βPL gene. To further investigate the possibility that Gdf15 and hTGF-βPL are orthologs, we performed genomic Southern analysis of murine and human DNA using probes corresponding to the C-terminal regions of the two proteins. As shown in Fig. 1, the most intensely hybridizing bands identified by the hTGF-βPL probe on murine genomic DNA corresponded to the Gdf15 gene. The results of the converse experiment (hybridization of the Gdf15 probe to human genomic DNA) were inconclusive.
FIG. 1.
Genomic Southern analysis of human and mouse DNA probed with fragments corresponding to the C-terminal regions of Gdf15 and hTGF-βPL.
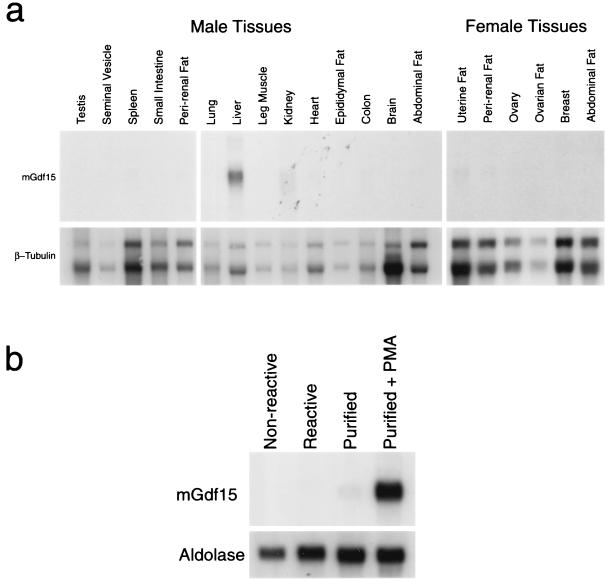
We further examined the relationship between Gdf15 and hTGF-βPL by comparing their expression patterns. hTGF-βPL was previously shown to be expressed widely in human tissues, with the highest levels being detected in the prostate (32) and placenta (16, 21, 42). By Northern analysis, we detected Gdf15 in a variety of different adult mouse tissues (Fig. 2a), with highest levels in the adult liver. In contrast to hTGF-βPL, Gdf15 mRNA was barely detectable in mouse prostate and placenta (data not shown). We also determined whether Gdf15 expression is up-regulated in activated macrophages, as previously reported for hTGF-βPL (2). Reactive peritoneal macrophages were isolated from thioglycolate-injected C57BL/6J mice and purified by adhesion to plastic tissue culture surfaces. As shown in Fig. 2b, Gdf15 mRNA levels increased significantly in macrophages activated with PMA relative to untreated control macrophages. Hence, while Gdf15 and hTGF-βPL showed distinct expression patterns in adult tissues, their expression patterns were similar following macrophage activation.
FIG. 2.
(a) Northern analysis (20 μg of total RNA) of adult mouse tissues. (b) Northern analysis (10 μg of total RNA) of peritoneal macrophages isolated from untreated (nonreactive) or thioglycolate-injected (reactive) mice. Macrophages were purified by adhesion to dishes and incubated in the absence or presence of PMA.
Induction of Gdf15 expression during liver injury and regeneration.
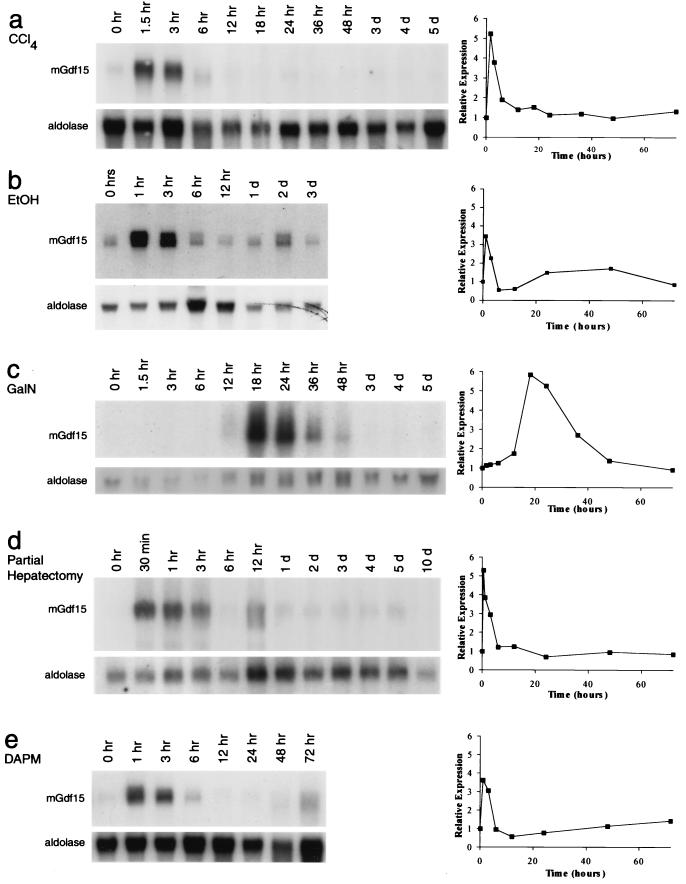
In order to determine the potential roles of Gdf15 in the liver, we examined the regulation of Gdf15 mRNA expression in several models of acute liver injury and regeneration. We first examined Gdf15 expression in the liver following administration of chemicals known to injure hepatocytes. Administration of CCl4 to animals is known to cause acute centrilobular necrosis of hepatocytes, followed by a regenerative response in the surviving hepatocytes (7). As shown in Fig. 3a, Gdf15 mRNA levels in the liver increased rapidly and dramatically following CCl4 administration. Gdf15 mRNA levels were highest within 1.5 h following CCl4 treatment and then returned to baseline levels by 6 to 12 h. We also observed a rapid and dramatic induction of Gdf15 following intraperitoneal administration of a single dose of ethanol (Fig. 3b). As in the case of CCl4 injury, Gdf15 RNA levels were increased 1 to 3 h after ethanol administration and returned to baseline levels by 12 h. We also examined the expression of Gdf15 following treatment with GalN, which causes hepatocyte injury by inhibiting RNA synthesis (9). Unlike for CCl4 treatment, liver regeneration following GalN injury has been shown to involve recruitment of liver stem cells, or oval cells (1, 7, 23). As shown in Fig. 3c, Gdf15 expression was also increased following treatment with GalN, although the time course of induction was considerably delayed compared to that of CCl4 injury.
FIG. 3.
Expression of Gdf15 following liver and bile duct injury. Northern analysis was carried out using 20 μg of total RNA, except for panel c, in which 5 μg of twice-poly(A)-selected RNA was used. Graphs to the right of the Northern blots show Gdf15 expression normalized to aldolase expression and plotted relative to uninduced levels.
In order to determine whether the induction of Gdf15 mRNA was restricted to chemical injury of hepatocytes, we examined the expression of Gdf15 following two-thirds hepatectomy. Partial hepatectomy initiates a regenerative response in all cell types in the liver remnant (5, 38), including a large hepatocyte proliferative response similar to that seen after CCl4 injury. As shown in Fig. 3d, Gdf15 expression was induced following partial hepatectomy, and the time course of induction was similar to that observed following CCl4 treatment. Gdf15 mRNA levels were increased dramatically at the earliest time point examined (30 min following surgery) and remained elevated for up to 12 h, after which the Gdf15 RNA levels decreased to baseline. In 5 of 13 sham-operated animals, Gdf15 mRNA levels showed a transient increase at 1 h, but the magnitude of induction was significantly lower than that following partial hepatectomy (data not shown).
We next examined the expression of Gdf15 following chemical bile duct injury to determine whether Gdf15 expression could be induced by injury to other cell types within the liver. Following treatment with DAPM, which selectively injures bile duct epithelial cells (17), Gdf15 mRNA levels were increased at 1 h and returned to baseline levels by 6 to 12 h (Fig. 3e). While levels of Gdf15 mRNA were also mildly elevated at 1 h in animals receiving the ethanol carrier only, the magnitude of Gdf15 induction in these control animals was lower than that seen with DAPM, and Gdf15 mRNA levels returned to baseline by 3 h (data not shown). Hence, Gdf15 expression appears to be induced rapidly not only following direct injury to hepatocytes but also following injury to bile duct epithelial cells.
Patterns of Gdf15 mRNA localization following liver injury.
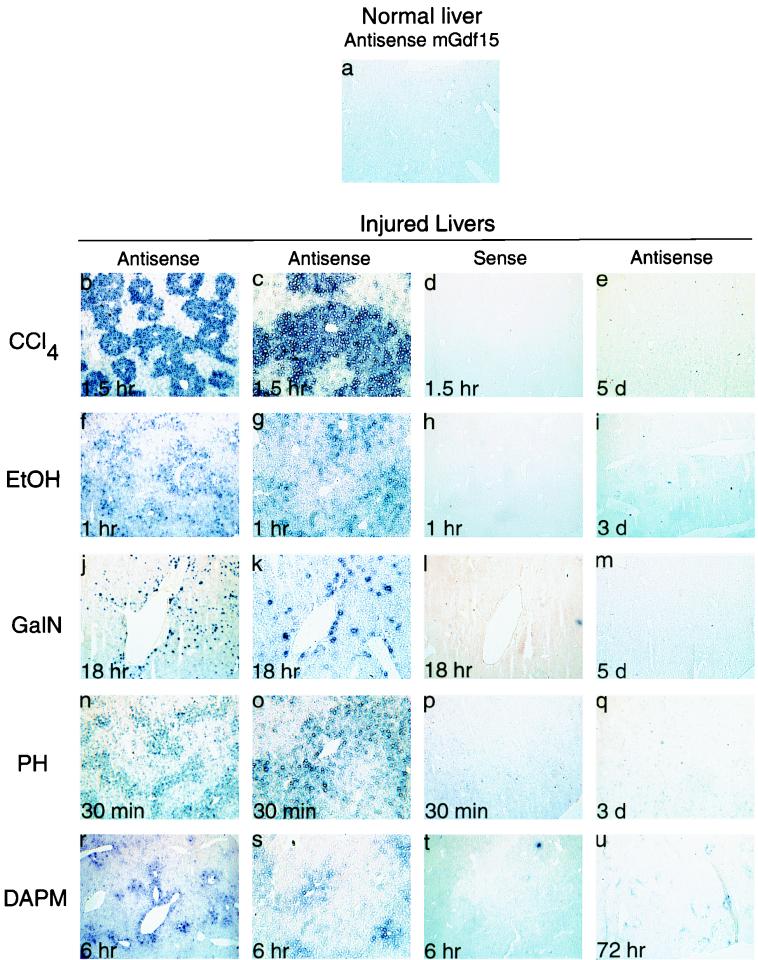
As shown above, Gdf15 expression in the liver was rapidly induced following various chemical and surgical treatments that cause liver injury and regeneration. We used in situ hybridization to determine the pattern of Gdf15 expression in these various injury models. The most dramatic results were obtained in livers taken from animals that had been treated with CCl4. As shown in Fig. 4b to e, Gdf15 mRNA was selectively localized to centrilobular regions, which coincides with the known pattern of hepatocyte injury induced by CCl4 (28, 33). Following ethanol administration, Gdf15 was again expressed by centrilobular hepatocytes but in a more diffuse pattern, with various levels of expression between individual hepatocytes (Fig. 4f to i). The centrilobular expression of Gdf15 correlates with the centrilobular fatty change, necrosis, and fibrosis previously described for rodent models of acute and chronic liver injury by ethanol (14, 18). In contrast, GalN treatment caused Gdf15 mRNA to be expressed in a punctate pattern distributed throughout the parenchyma, with a large number of binucleate hepatocytes expressing Gdf15 (Fig. 4j to m). A similar punctate pattern has been described for the parenchymal distribution of necrotic cells following GalN administration (27).
FIG. 4.
In situ localization of Gdf15 mRNA in liver. All panels were photographed at ×40 magnification, except for panels c, g, k, o, and s, which are ×100 magnifications of the sections shown in panels b, f, j, n, and r, respectively.
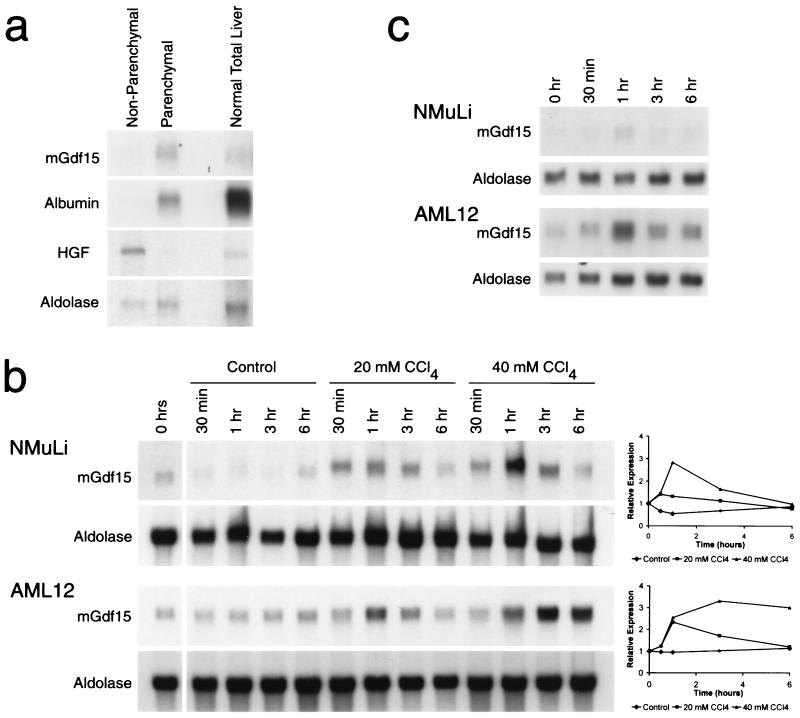
In order to confirm that the Gdf15-expressing cells were hepatocytes, we separated liver cells from GalN-treated mice into parenchymal and nonparenchymal populations by low-speed centrifugation (13) and then isolated RNA from each group. As controls for the cell separation procedure, we carried out Northern analysis on these RNA samples using probes for murine hepatocyte growth factor (19) and albumin as markers for nonparenchymal and parenchymal cells, respectively. Northern analysis showed that Gdf15 mRNA was restricted to the parenchymal cell fraction (Fig. 5a), consistent with the in situ hybridization results. Similar results were also obtained using separated cell populations obtained from untreated livers (data not shown).
FIG. 5.
(a) Expression of Gdf15 in parenchymal cells. Liver cells isolated from GalN-treated mice were separated into nonparenchymal and parenchymal fractions. HGF, hepatocyte growth factor. (b) Expression of Gdf15 in NMuLi and AML12 cells treated with CCl4 in culture. Control flasks containing cells were sealed and incubated at 37°C in the absence of CCl4. Graphs to the right of the Northern blots show Gdf15 expression normalized to aldolase expression and plotted relative to uninduced levels. (c) Expression of Gdf15 following heat shock treatment of NMuLi and AML12 cells in culture. Cells were incubated at 44°C for 30 min starting at time zero and then allowed to recover at 37°C.
Following partial hepatectomy, Gdf15 mRNA was induced in cells throughout the liver parenchyma, with more intense staining around central veins (Fig. 4n to q). Unlike the CCl4 and GalN injuries, the centrilobular pattern we observed following partial hepatectomy did not correspond to the previously reported periportal distribution of injured hepatocytes in the early period following surgery (5, 38). The pattern of Gdf15 expression at 6 h following DAPM treatment was also centrilobular (Fig. 4r to u). Notably, no Gdf15 mRNA was detected in the periportal or bile duct regions, which presumably represent the primary sites of DAPM-mediated injury.
Induction of Gdf15 mRNA in cultured liver cells.
As shown above, a variety of acute liver injuries in vivo can result in up-regulation of Gdf15 expression in hepatocytes. Because many complex cellular interactions can occur in vivo in the liver, we asked whether direct injury to a uniform population of cells in tissue culture could also induce Gdf15 expression. We examined the response of NMuLi cells (31), which are believed to represent early differentiated liver epithelial cells, and AML12 cells (40), which are derived from mature hepatocytes, to CCl4 treatment in culture. As shown in Fig. 5b, Gdf15 mRNA levels were rapidly elevated in a CCl4 dose-dependent manner in both cell types.
We also exposed AML12 and NMuLi cells to a transient 44°C heat shock to determine whether Gdf15 could be induced by a nonchemical stressor. As seen in Fig. 5c, levels of Gdf15 mRNA were mildly elevated in both cell lines at 1 h following heat shock. These in vitro data indicate that Gdf15 expression can be induced in hepatocytes in response to direct stress and injury. In addition, the induction of Gdf15 expression can occur in the absence of other cell types, such as inflammatory cells.
Generation of Gdf15 null mice.
In order to address the potential function of Gdf15 in vivo, we used gene targeting to generate Gdf15 null mice. As shown in Fig. 6a, the Gdf15 gene is composed of two exons spanning approximately 4 kb of DNA. The region corresponding to the entire mature C terminus, which is carried within exon 2, was replaced by the neomycin resistance cassette in our targeting construct. The targeted construct was electroporated into R1 embryonic stem cells, and clones doubly resistant to ganciclovir and G418 were isolated. Using Southern analysis with a probe from outside the targeting construct (Fig. 6a), we identified eight targeted clones among 143 screened colonies. All subsequent analysis was carried out on offspring of a single male chimera derived from blastocyst injection of one of the clones.
Gdf15 homozygous null mice were obtained by crosses of F1 heterozygotes. The mice were genotyped using a probe external to the targeted region and by a second probe internal to the targeted site (Fig. 6b). Absence of Gdf15 expression in the homozygous null mice was confirmed by Northern analysis of liver RNA isolated from untreated mice and mice treated with CCl4 for 1.5 h or 1 day. No expression of Gdf15 was detected in the livers of null mice (Fig. 6c). Analysis of 775 adult offspring from heterozygous matings showed a ratio of wild-type, heterozygous, and homozygous mice that approximated 1:2:1 (196:392:184). Homozygous null mice were viable and fertile. Analysis of serum chemistries and examination of various tissues by gross pathology and microscopic techniques revealed no obvious abnormalities in the Gdf15 null mice (data not shown).
Characterization of Gdf15 null mice after liver injury.
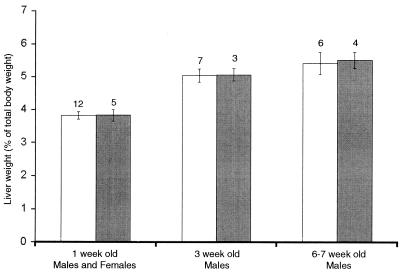
Based on the high level of Gdf15 expression in the liver and the dramatic regulation of Gdf15 expression following liver injury, we focused most of our detailed analysis on liver function. Liver weights of homozygous null mice at 1, 3, and 6 to 7 weeks were indistinguishable from those of wild-type littermates (Fig. 7). Therefore, we tested the Gdf15 null mice in the CCl4 and partial-hepatectomy injury models.
FIG. 7.
Mean liver weight as a percent of total body weight of wild-type (open bars) and Gdf15 null (solid bars) mice. Numbers above each category are the numbers of mice per point. Error bars represent 1 standard error of the mean.
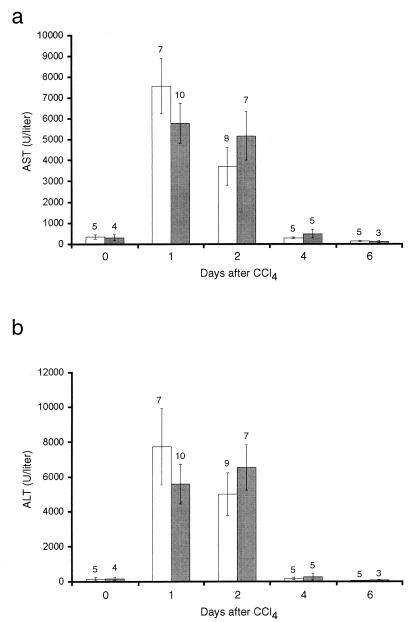
As described above, we injected the Gdf15 homozygous null mice and their wild-type counterparts with CCl4 and assessed hepatocyte injury by measuring serum transaminase levels as described previously (10). As shown in Fig. 8, wild-type mice showed peak levels of serum AST and ALT levels at 1 day after administration of CCl4, which normalized by 4 days after injury. Gdf15 null mice showed elevations in AST and ALT levels comparable to those of wild-type mice. In addition, comparison of liver sections from wild-type and null mice by microscopic histology showed no significant differences in the size of the necrotic zones around the central veins (data not shown). Examination of the time course of DNA synthesis following CCl4 administration by BrdU labeling also showed no significant differences between wild-type and null mice (data not shown). Finally, TUNEL analysis of the same livers showed no significant differences at 1 day after CCl4 injury, which coincided with the peak of TUNEL labeling seen in our experiments (data not shown).
FIG. 8.
Mean serum aspartate transferase (AST) and alanine transferase (ALT) levels in wild-type (open bars) and Gdf15 null (solid bars) mice following CCl4 treatment. Numbers above each category are the numbers of mice per point. Error bars represent 1 standard error of the mean.
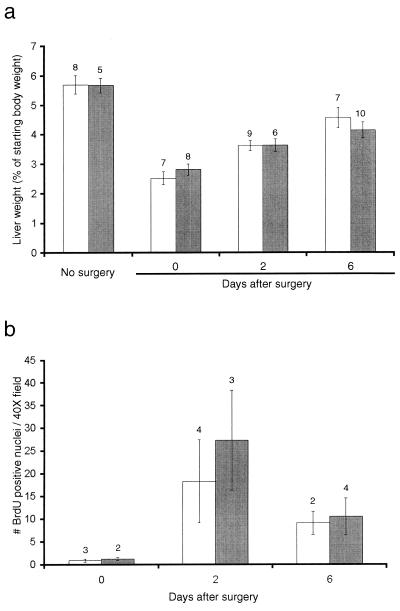
In order to examine if Gdf15 is involved in liver regeneration, we performed partial hepatectomies on male Gdf15 null and wild-type mice. As shown in Fig. 9a, the liver masses of the wild-type mice recovered to approximately 50 and 90% of the starting liver mass after 2 and 6 days, respectively, as previously described (41). The Gdf15 null mice regenerated their liver mass at the same rate as the wild-type mice. Comparison of cell proliferation by BrdU incorporation at these time points (Fig. 9b) also showed no significant difference between wild-type and knockout mice.
FIG. 9.
Mean liver weights following partial hepatectomy in wild-type (open bars) and Gdf15 null (solid bars) mice. (a) Liver weights as a percent of starting body weights in untreated mice, mice immediately following partial hepatectomy and at 2 and 6 days after partial hepatectomy. (b) Number of BrdU-labeled nuclei in liver sections following partial hepatectomy. The number above each category is the number of mice per point. Error bars represent 1 standard error of the mean.
DISCUSSION
In this report, we describe our initial characterization of a new murine TGF-β family member, Gdf15. Among the known family members, Gdf15 is most closely related to hTGF-βPL (42) (also known as hMIC-1, hPDF, hPLAB, and hPTGFB), which was identified independently by several different groups searching for novel secreted proteins (42), for genes expressed by activated macrophages (2), for TGF-β-like molecules in expressed sequence tag databases (16, 32), and for novel genes expressed in placenta (21). Although Gdf15 and hTGF-βPL are significantly more divergent than is usually observed for other murine and human TGF-β ortholog pairs, the results of our comparative genomic Southern analysis and the fact that Gdf15 and the hTGF-βPL gene are both expressed by activated macrophages raise the possibility that these genes may be orthologs. Further functional studies will be required to determine whether these two genes play analogous roles in their respective species.
In the adult mouse, Gdf15 mRNA is expressed at highest levels in the liver, with lower levels seen in other tissues. This expression pattern is similar to that previously found using reverse transcription-PCR detection methods (3), although we did not observe strong Gdf15 signals in the brain or prostate. We have also shown that Gdf15 expression in the liver is rapidly and dramatically up-regulated in various models of liver injury and regeneration. Although a number of different experimental treatments can induce Gdf15 expression, each type of injury induces a distinct pattern of Gdf15 expression in the liver. Perhaps the most striking pattern is produced by CCl4, which results in expression of Gdf15 exclusively in centrilobular hepatocytes. Because CCl4 causes centrilobular liver necrosis (28, 33) and because the expression of Gdf15 mRNA extended to cells closest to the central veins, at least some, if not all, of the hepatocytes that express Gdf15 in response to CCl4 must be ones that will eventually undergo necrosis.
Similarly, it seems likely that the hepatocytes expressing Gdf15 in the other chemical and surgical models have also been injured. GalN causes a reversible inhibition of RNA synthesis (9), leading to diffuse liver damage with individual necrotic cells distributed within the hepatic lobules (27). The regenerative response following GalN injury is believed to be mediated by the proliferation of oval cells located in the periportal regions (7, 23). Indeed, the expression pattern of Gdf15 following GalN administration is diffuse and does not show any obvious localization to centrilobular or periportal regions. The centrilobular expression of Gdf15 after a single dose of ethanol correlates with the necrosis, fibrosis, and fatty change that is seen in centrilobular hepatocytes after chronic ethanol injury (14, 18). Acute doses of ethanol are known to cause changes in lipid metabolism within the injured liver, presumably contributing to the morphologic changes that are observed in chronic injury (29). Given the dramatic expression of Gdf15 after ethanol administration, we would hypothesize that Gdf15 is expressed by the cells in the centrilobular area that are being injured by the ethanol.
In the case of DAPM treatment, we would speculate that the centrilobular hepatocytes expressing Gdf15 are ones that have been injured secondarily by cholestasis caused by the loss of bile duct epithelial cells (24, 37). Similarly, given that the pattern of Gdf15 expression following partial hepatectomy does not coincide with the periportal distribution of proliferating hepatocytes during the initial wave of regeneration (12), it seems possible that Gdf15 expression is a response to the acute stresses of hepatectomy on the hepatic remnant. One of our striking findings is that Gdf15 expression can be induced in the absence of inflammatory cells. As we have shown with CCl4 and heat shock treatments of hepatocyte-like cells in culture, direct cellular injury or stress appears to be sufficient to induce Gdf15 expression.
Given the dramatic up-regulation of Gdf15 following multiple types of liver injury, we hypothesized that Gdf15 may be involved in the regulation of the acute injury response in hepatocytes. Two recent reports have suggested that Gdf15 and hTGFβ-PL could be regulated by multiple transcription factors that are known to regulate cell growth and injury responses (3), including p53 (39). To determine the in vivo function of Gdf15, we generated mice in which the Gdf15 gene had been deleted. Homozygous null mice were viable and fertile and had no gross abnormalities in any of the major organs. Our studies also showed that the Gdf15 null mice and wild-type mice had comparable degrees of liver injury and recovery after CCl4 administration. Finally, the Gdf15 null mice were able to regenerate their liver mass at the same rate as wild-type mice after partial hepatectomy.
Our results suggest that Gdf15 is not essential for proper regeneration of the liver following surgical or chemical injury. It is possible that the Gdf15 mutant mice have subtle defects that were not revealed in our analysis. It is also possible that the loss of Gdf15 function is compensated by other secreted growth factors or by other regulatory pathways. A full elucidation of the roles of Gdf15 during the injury response awaits additional analysis of the null mice and the analysis of the biological activities of the Gdf15 protein both in vitro and in vivo.
ACKNOWLEDGMENTS
We thank Paul Dunlap for his assistance in the maintenance of mice and Tracie E. Bunton, John K. Boitnott, and Daniel Nathans for their helpful discussions.
T.Z.-K. and L.G.K. were supported by NIH training grant 5 T32 CA09139 to the Department of Molecular Biology and Genetics, The Johns Hopkins University School of Medicine. E.C.H. is a trainee of the Medical Training Scientist Program (Public Health Service grant 5 T32 GM07309). This work was supported by grants from MetaMorphix, Inc., and American Home Products, Inc. (to S.-J.L.). Under an agreement among Johns Hopkins University, MetaMorphix, Inc., and American Home Products, Inc., all of the authors are entitled to a share of royalties received by the university from sales of Gdf15. S.-J.L., T.V.H., and the university also own MetaMorphix stock, which is subject to certain restrictions under university policy. The terms of these arrangements are being managed by the university in accordance with its conflict of interest policies. S.-J.L. is a consultant to MetaMorphix.
ADDENDUM IN PROOF
Radiation hybrid mapping (Research Genetics, Inc.) linked Gdf15 to the mouse framework marker D8Mit233. This region of mouse chromosome 8 is syntenic to a portion of human chromosome 19p13.1 containing TGF-βPL (Bottner et al., Gene 237:105–111, 1999; see also mapping data from the Human Genome Center at Lawrence Livermore Laboratory), consistent with mGdf15 and hTGF-βPL being orthologs.
REFERENCES
- 1.Alison M R, Golding M, Sarraf C E. Liver stem cells: when the going gets tough they get going. Int J Exp Pathol. 1997;78:365–381. doi: 10.1046/j.1365-2613.1997.500375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bootcov M R, Bauskin A R, Valenzuela S M, Moore A G, Bansal M, He X Y, Zhang H P, Donnellan M, Mahler S, Pryor K, Walsh B J, Nicholson R C, Fairlie W D, Por S B, Robbins J M, Breit S N. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc Natl Acad Sci USA. 1997;94:11514–11519. doi: 10.1073/pnas.94.21.11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böttner M, Laaff M, Schechinger B, Rappold G, Unsicker K, Suter-Crazzolara C. Characterization of the rat, mouse, and human genes of growth/differentiation factor-15/macrophage inhibiting cytokine-1 (GDF-15/MIC-1) Gene. 1999;237:105–111. doi: 10.1016/s0378-1119(99)00309-1. [DOI] [PubMed] [Google Scholar]
- 4.Böttner M, Suter-Crazzolara C, Schober A, Unsicker K. Expression of a novel member of the TGF-beta superfamily, growth/differentiation factor-15/macrophage-inhibiting cytokine-1 (GDF-15/MIC-1), in adult rat tissues. Cell Tissue Res. 1999;297:103–110. doi: 10.1007/s004410051337. [DOI] [PubMed] [Google Scholar]
- 5.Bucher N L R. Regeneration of mammalian liver. Int Rev Cytol. 1963;15:245–300. doi: 10.1016/s0074-7696(08)61119-5. [DOI] [PubMed] [Google Scholar]
- 6.Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W. Current protocols in immunology. New York, N.Y: John Wiley and Sons, Inc.; 1994. [Google Scholar]
- 7.Dabeva M D, Alpini G, Hurston E, Shafritz D A. Models for hepatic progenitor cell activation. Proc Soc Exp Biol Med. 1993;204:242–252. doi: 10.3181/00379727-204-43660. [DOI] [PubMed] [Google Scholar]
- 8.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 9.Decker K, Keppler D. Galactosamine hepatitis: key role of the nucleotide deficiency period in the pathogenesis of cell injury and cell death. Rev Physiol Biochem Pharmacol. 1974;71:77–106. doi: 10.1007/BFb0027661. [DOI] [PubMed] [Google Scholar]
- 10.Doolittle D J, Muller G, Scribner H E. Relationship between hepatotoxicity and induction of replicative DNA synthesis following single or multiple doses of carbon tetrachloride. J Toxicol Environ Health. 1987;22:63–78. doi: 10.1080/15287398709531051. [DOI] [PubMed] [Google Scholar]
- 11.Ellwart J, Dormer P. Effect of 5-fluoro-2′-deoxyuridine (FdUrd) on 5-bromo-2′-deoxyuridine (BrdUrd) incorporation into DNA measured with a monoclonal BrdUrd antibody and by the BrdUrd/Hoechst quenching effect. Cytometry. 1985;6:513–520. doi: 10.1002/cyto.990060605. [DOI] [PubMed] [Google Scholar]
- 12.Fabrikant J I. The kinetics of cellular proliferation in regenerating liver. J Cell Biol. 1968;36:551–565. doi: 10.1083/jcb.36.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujio K, Evarts R P, Hu Z, Marsden E R, Thorgeirsson S S. Expression of stem cell factor and its receptor, c-kit, during liver regeneration from putative stem cells in adult rat. Lab Investig. 1994;70:511–516. [PubMed] [Google Scholar]
- 14.Goldin R D, Wickramasinghe S N. Hepatotoxicity of ethanol in mice. Br J Exp Pathol. 1987;68:815–824. [PMC free article] [PubMed] [Google Scholar]
- 15.Higgins G M, Anderson R M. Experimental pathology of the liver. I. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186–202. [Google Scholar]
- 16.Hromas R, Hufford M, Sutton J, Xu D, Li Y, Lu L. PLAB, a novel placental bone morphogenetic protein. Biochim Biophys Acta. 1997;1354:40–44. doi: 10.1016/s0167-4781(97)00122-x. [DOI] [PubMed] [Google Scholar]
- 17.Kanz M F, Gunasena G H, Kaphalia L, Hammond D K, Syed Y A. A minimally toxic dose of methylene dianiline injures biliary epithelial cells in rats. Toxicol Appl Pharmacol. 1998;150:414–426. doi: 10.1006/taap.1998.8382. [DOI] [PubMed] [Google Scholar]
- 18.Keegan A, Martini R, Batey R. Ethanol-related liver injury in the rat: a model of steatosis, inflammation and pericentral fibrosis. J Hepatol. 1995;23:591–600. doi: 10.1016/0168-8278(95)80067-0. [DOI] [PubMed] [Google Scholar]
- 19.Kinoshita T, Tashiro K, Nakamura T. Marked increase of HGF mRNA in non-parenchymal liver cells of rats treated with hepatotoxins. Biochem Biophys Res Commun. 1989;165:1229–1234. doi: 10.1016/0006-291x(89)92733-2. [DOI] [PubMed] [Google Scholar]
- 20.Komminoth P. Detection of mRNA in tissue sections using DIG-labeled RNA and oligonucleotide probes. In: Grunewald-Janho S, Keesey J, Leous M, van Miltenburg R, Schroeder C, editors. Nonradioactive in situ hybridization application manual. 2nd ed. Mannheim, Germany: Boehringer Mannheim GmbH, Biochemica; 1996. pp. 126–135. [Google Scholar]
- 21.Lawton L N, Bonaldo M F, Jelenc P C, Qiu L, Baumes S A, Marcelino R A, de Jesus G M, Wellington S, Knowles J A, Warburton D, Brown S, Soares M B. Identification of a novel member of the TGF-beta superfamily highly expressed in human placenta. Gene. 1997;203:17–26. doi: 10.1016/s0378-1119(97)00485-x. [DOI] [PubMed] [Google Scholar]
- 22.Lee S-J. Identification of a novel member (GDF-1) of the transforming growth factor-beta superfamily. Mol Endocrinol. 1990;4:1034–1040. doi: 10.1210/mend-4-7-1034. [DOI] [PubMed] [Google Scholar]
- 23.Lemire J M, Shiojiri N, Fausto N. Oval cell proliferation and the origin of small hepatocytes in liver injury induced by d-galactosamine. Am J Pathol. 1991;139:535–552. [PMC free article] [PubMed] [Google Scholar]
- 24.Marucci L, Baroni G S, Mancini R, Benedetti A, Jezequel A M, Orlandi F. Cell proliferation following extrahepatic biliary obstruction. Evaluation by immunohistochemical methods. J Hepatol. 1993;17:163–169. doi: 10.1016/s0168-8278(05)80032-7. [DOI] [PubMed] [Google Scholar]
- 25.McPherron A C, Lawler A M, Lee S-J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 26.McPherron A C, Lee S-J. The transforming growth factor-beta superfamily. In: Leroith D, Bondy C, editors. Growth factors and cytokines in health and disease. 1B. Stamford, Conn: JAI Press, Inc.; 1996. pp. 357–393. [Google Scholar]
- 27.Medline A, Schaffner F, Popper H. Ultrastructural features in galactosamine-induced hepatitis. Exp Mol Pathol. 1970;12:201–211. doi: 10.1016/0014-4800(70)90050-x. [DOI] [PubMed] [Google Scholar]
- 28.Mourelle M, Rubalcava B. Regeneration of the liver after carbon tetrachloride. Differences in adenylate cyclase and pancreatic hormone receptors. J Biol Chem. 1981;256:1656–1660. [PubMed] [Google Scholar]
- 29.Muramatsu M, Kuriyama K, Yuki T, Ohkuma S. Hepatic lipogenesis and mobilization of peripheral fats in the formation of alcoholic fatty liver. Jpn J Pharmacol. 1981;31:931–940. doi: 10.1254/jjp.31.931. [DOI] [PubMed] [Google Scholar]
- 30.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Owens R B, Smith H S, Hackett A J. Epithelial cell cultures from normal glandular tissue of mice. J Natl Cancer Inst. 1974;53:261–269. doi: 10.1093/jnci/53.1.261. [DOI] [PubMed] [Google Scholar]
- 32.Paralkar V M, Vail A L, Grasser W A, Brown T A, Xu H, Vukicevic S, Ke H Z, Qi H, Owen T A, Thompson D D. Cloning and characterization of a novel member of the transforming growth factor-beta/bone morphogenetic protein family. J Biol Chem. 1998;273:13760–13767. doi: 10.1074/jbc.273.22.13760. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds E S. Liver parenchymal cell injury. I. Initial alterations of the cell following poisoning with carbon tetrachloride. J Cell Biol. 1963;19:139–157. doi: 10.1083/jcb.19.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson E J, editor. Teratocarcinomas and embryonic stem cells. Oxford, England: IRL Press, Ltd.; 1987. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 37.Schaffner F, Popper H. Morphologic studies of cholestasis. Gastroenterology. 1968;54(Suppl.):750–751. [PubMed] [Google Scholar]
- 38.Steer C J. Liver regeneration. FASEB J. 1995;9:1396–1400. doi: 10.1096/fasebj.9.14.7589980. [DOI] [PubMed] [Google Scholar]
- 39.Tan M, Wang Y, Guan K, Sun Y. PTGF-beta, a type beta transforming growth factor (TGF-beta) superfamily member, is a p53 target gene that inhibits tumor cell growth via TGF-beta signaling pathway. Proc Natl Acad Sci USA. 2000;97:109–114. doi: 10.1073/pnas.97.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J C, Merlino G, Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc Natl Acad Sci USA. 1994;91:674–678. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokoyama H O, Wilson M E, Tsuboi K K, Stowell R E. Regeneration of mouse liver after partial hepatectomy. Cancer Res. 1953;13:80–85. [PubMed] [Google Scholar]
- 42.Yokoyama-Kobayashi M, Saeki M, Sekine S, Kato S. Human cDNA encoding a novel TGF-beta superfamily protein highly expressed in placenta. J Biochem. 1997;122:622–626. doi: 10.1093/oxfordjournals.jbchem.a021798. [DOI] [PubMed] [Google Scholar]