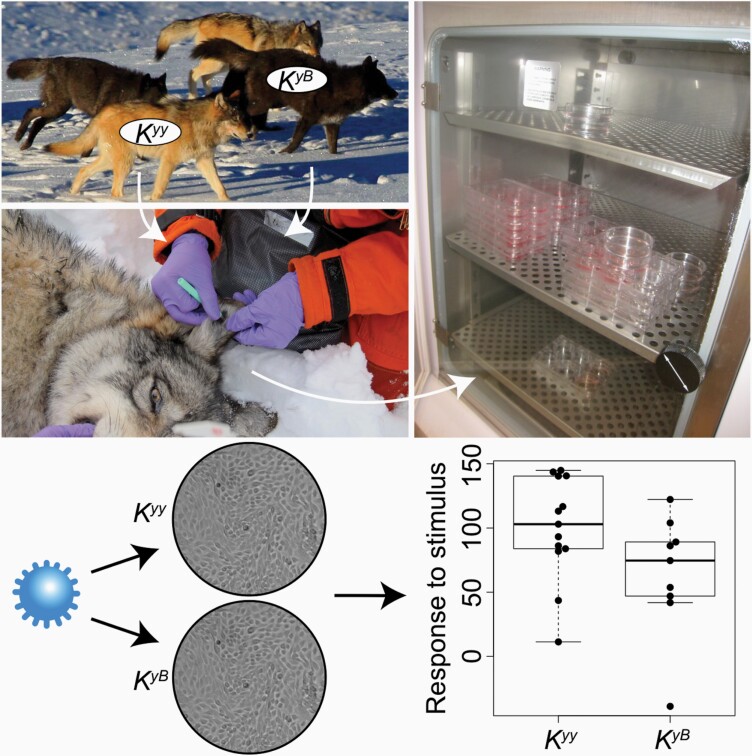
Abstract
In North American gray wolves, black coat color is dominantly inherited via a 3 base pair coding deletion in the canine beta defensin 3 (CBD103) gene. This 3 base pair deletion, called the KB allele, was introduced through hybridization with dogs and subsequently underwent a selective sweep that increased its frequency in wild wolves. Despite apparent positive selection, KBB wolves have lower fitness than wolves with the KyB genotype, even though the 2 genotypes show no observable differences in black coat color. Thus, the KB allele is thought to have pleiotropic effects on as-yet unknown phenotypes. Given the role of skin-expressed CBD103 in innate immunity, we hypothesized that the KB allele influences the keratinocyte gene expression response to TLR3 pathway stimulation and/or infection by canine distemper virus (CDV). To test this hypothesis, we developed a panel of primary epidermal keratinocyte cell cultures from 24 wild North American gray wolves of both Kyy and KyB genotypes. In addition, we generated an immortalized Kyy line and used CRISPR/Cas9 editing to produce a KyB line on the same genetic background. We assessed the transcriptome-wide responses of wolf keratinocytes to the TLR3 agonist polyinosinic:polycytidylic acid (polyI:C), and to live CDV. K locus genotype did not predict the transcriptional response to either challenge, suggesting that variation in the gene expression response does not explain pleiotropic effects of the KB allele on fitness. This study supports the feasibility of using cell culture methods to investigate the phenotypic effects of naturally occurring genetic variation in wild mammals.
Keywords: Coat color, pigmentation, pleiotropy, immunity, viral challenge, keratinocyte
Graphical Abstract
In mammals, much of the variation in pigmentation traits, especially fur and hair color, is controlled by the agouti-melanocortin receptor 1 (MC1R) pathway (Nordlund et al. 2006). North American gray wolves exemplify this pattern: coat color can be gray or black, where black coat color is controlled by a dominant 3 base pair deletion in the coding sequence of the gene canine beta defensin 3 (CBD103) (Figure 1A) (Candille et al. 2007). This coding deletion, known as the KB allele, increases the binding affinity of the CBD103 protein to MC1R. CBD103-MC1R binding in turn competitively excludes the Agouti protein from binding to MC1R, shifting the balance of pigment production to the dark pigment eumelanin. The same set of interactions explains, in part, coat color polymorphism in domestic dogs (Candille et al. 2007).
Figure 1.
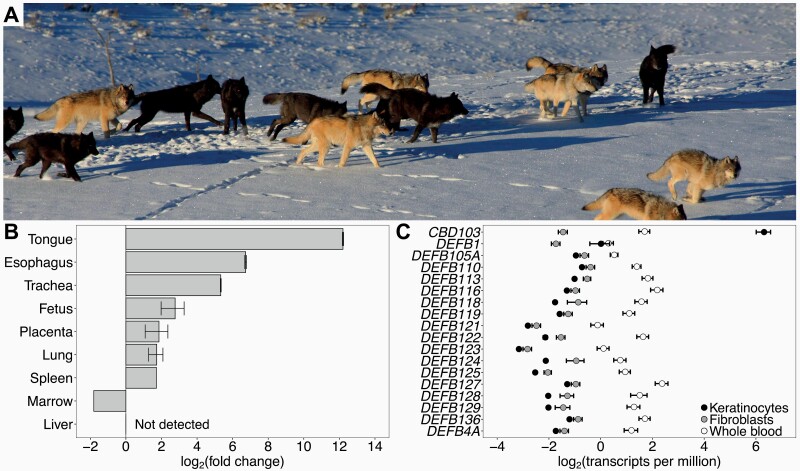
CBD103 gene expression in North American gray wolves. (A) Coat color polymorphism in North American gray wolves in Yellowstone National Park, in which coat color can be gray or black (photo credit Dan Stahler/National Park Service photo). Black coat color is dominantly inherited, conferred by a 3 base pair coding deletion in CBD103. (B) CBD103 expression, relative to expression in dog testis, across tissues of a recently deceased pregnant female Kyy wolf. Error bars represent standard errors across RT-qPCR replicates (2–3 replicates per tissue). A single tissue sample was collected for each tissue except fetus and placenta, which each represent 2 tissue samples (i.e., from 2 fetuses). (C) Absolute expression of annotated beta defensins in epidermal keratinocytes (N = 23), fibroblasts (N = 6), and whole blood (N = 25; (Charruau et al. 2016)) from North American gray wolves. Only CBD103 is highly expressed in keratinocytes.
In wolves, the CBD103 polymorphism is notable not only for being one of the few well-understood genotype-phenotype relationships in wild mammals, but also for its adaptive importance. The deletion allele was introduced to North American gray wolves from domestic dogs through hybridization circa 1000–10 000 years ago, likely with dogs brought with Native Americans. After it was introduced, it subsequently underwent a selective sweep, either due to effects on coat color or pleiotropic effects on other phenotypes (Anderson et al. 2009; Coulson et al. 2011; Schweizer et al. 2018). However, despite evidence of positive selection for the KB allele (Anderson et al. 2009; Schweizer et al. 2018), individuals that are homozygous for the KB allele (KBB) experience lower annual recruitment and survival rates and much lower mean lifetime reproductive success than KB allele heterozygotes (KyB, where the wild-type allele is conventionally abbreviated Ky for “yellow”). Moreover, in the well-sampled Yellowstone National Park population, fewer KBB individuals have been detected than expected based on observed allele frequencies (Stahler, unpublished data). The reduced fitness of KBB individuals relative to KyB individuals, as well as extensive population genetic simulations, thus suggest that both KB and Ky alleles are currently being maintained in wild wolves as a consequence of balancing selection (Coulson et al. 2011; Schweizer et al. 2018).
The phenotypic costs of KB homozygosity remain unclear. Although the coat color phenotype is the most readily apparent consequence of the KB allele, both KBB and KyB wolves exhibit the same black coat color (Coulson et al. 2011). The low recruitment of KBB animals, and resultant balancing selection, may be explained in part by disassortative mating, in which wolf pairs that successfully produce offspring tend to include one parent with black, and one parent with gray, coat color (Hedrick et al. 2016). However, the apparent survival and lifespan differences between KyB and KBB animals, and rarity of KBB individuals even from pairs of KyB parents, suggest that the KB allele also confers as-yet unknown pleiotropic effects in North American gray wolves that account for markedly lower fitness in KB homozygotes (Coulson et al. 2011; Schweizer et al. 2018). Indeed, pleiotropy of KB is indirectly supported by the reported effects of coat color on aggression (Cassidy et al. 2017) and on pup survival (Stahler et al. 2013).
The most obvious class of candidate phenotypes is immune defense. Defensin genes are named for their role in pathogen defense, immune signaling, and generation of antimicrobial peptides (García et al. 2001; Harder et al. 2001; Ganz 2003; Wilson et al. 2013). The human homolog of CBD103, DEFB103, is expressed in epithelial cells (especially tonsil, skin, trachea, and tongue) as a first line of defense against pathogens (García et al. 2001; Harder et al. 2001; Wilson et al. 2013). For example, its protein product, HBD3, is known to inhibit herpes simplex virus infection by deterring binding and entry of the virus into cells (Hazrati et al. 2006), and inhibits human immunodeficiency virus (HIV) replication via direct binding with virions (Quiñones-Mateu et al. 2003). Similarly, in domestic dogs, CBD103 is expressed in skin and tongue (particularly in keratinocytes), and the CBD103 protein shows potent antibacterial activity (Candille et al. 2007; Leonard et al. 2012). Based on antimicrobial assays against 5 gram-negative and gram-positive bacterial taxa, antibacterial activity of CBD103 shows no clear differentiation between the Ky and KB forms (Leonard et al. 2012). However, the impacts of CBD103 KB/Ky genotype on other aspects of immunity, such as antiviral activity, are not yet known.
In the present study, we tested the hypothesis that the gray wolf KB allele has pleiotropic effects on immunity via influencing the transcriptome-wide gene expression response to viral infection. To do so, we first identified the tissue and cell types that show the highest expression of CBD103 in North American gray wolves. We then established a panel of primary epidermal keratinocyte cultures from a genetically diverse set of wild gray wolves (14 Kyy and 10 KyB wolves; note that KBB homozygotes are rare due to their large fitness disadvantage). Based on these cultures, we challenged both primary and immortalized wolf keratinocytes with polyinosinic:polycytidylic acid (polyI:C), which is a synthetic analog of dsRNA that stimulates the TLR3-mediated antiviral response (Lebre et al. 2007). We also challenged a subset of the wolf keratinocyte cultures with a live, ecologically relevant pathogen, canine distemper virus (CDV), which is thought to be an important pathogen in explaining temporal spikes in wild wolf mortality (Almberg et al. 2009). If the CBD103 KB allele alters the transcriptional response to CDV, then such an effect could contribute to the complex, unexplained relationship between CBD103 genotype and host fitness. Finally, we demonstrated the utility of establishing new cell line resources from wild mammals by immortalizing Kyy wild-type keratinocyte cells and using CRISPR/Cas9 gene editing to introduce a targeted Ky to KB mutation. For the primary, immortalized, and edited cell lines, we then evaluated whether CBD103 genotype predicts the gene expression response to polyI:C or CDV.
Materials and Methods
Cross-Tissue Evaluation of CBD103 Expression
To identify the wolf tissue with the highest CBD103 expression, we leveraged opportunistically sampled tissue biopsies from a recently deceased Kyy female wolf (animal ID: 759F; tissues collected <48 h from time of death). These samples were collected by D.R.S. and National Park Service members working in Yellowstone National Park. A small piece of each tissue (<1 cm2 each of bone marrow, esophagus, fetus, liver, lung, placenta, skin, spleen, tongue, and trachea) was transferred with sterile forceps into a PAXGene Blood RNA tube and shipped to the University of California-Los Angeles (UCLA) on dry ice.
Total RNA was extracted from tissues using the Trizol Plus RNA Purification Kit with DNAse treatment (RNAse-Free DNAse Set, Ambion) and column cleanup (PureLink RNA Mini Kit, Invitrogen). As a reference RNA sample, we used commercially available total RNA from dog testis (Zyagen), because RNA from wolves was extremely limited prior to our establishment of immortalized cell cultures. For quantitative RT-qPCR, complementary DNA (cDNA) was synthesized using SuperScript II Reverse Transcriptase (ThermoFisher Scientific) on 200 ng RNA of each sample following the manufacturer’s instructions. The cDNA products were then purified using the QIAquick PCR Purification Kit (Qiagen). RT-qPCR reactions were performed on 10 ng cDNA using LightCycler 480 SYBR Green Master Mix (Roche) with 10 μl reaction volumes, using previously published CBD103 and rps5 primers and cycling conditions (Leonard et al. 2012) (Supplementary Table S1). The following program was used on a LightCycler 480 Instrument: 95 °C for 5 min; 45 cycles of 95 °C (10 s) and 58 °C (45 s); subsequent melting curve that ramps the temperature from 72 °C to 95 °C in increments of 0.1 °C/s. RT-qPCR values were standardized to the expression level of rps5.
Collection of Skin Biopsies for Primary Cell Culture
Our cross-tissue survey indicated that CBD103 was highly expressed in epithelial tissues (see Results), which are most readily sampled in wolves via skin biopsies from the ear. To collect skin biopsies from the field in a manner that allowed us to establish primary cell cultures, we pre-prepared culture medium nutrients in single sample aliquots (“MEFcubes”), which were stored frozen until use. Each MEFcube was comprised of 1 ml bovine calf serum (BCS; Hyclone, Cytiva Life Sciences), 0.1 ml L-glutamine (ThermoFisher Scientific), 20 μl Primocin antibiotic (Invivogen), and 252 μl 1 M HEPES buffer (ThermoFisher Scientific), and stored frozen up to 1 year. We also pre-prepared and refrigerated aliquots of 9 ml Dulbecco’s Modified Eagle’s Medium (DMEM; ThermoFisher Scientific) in 15 ml conical tubes (up to 1 year refrigeration). 0–3 days prior to tissue collection, collection medium was prepared for each sample by thawing a MEFcube and transferring it to a 15 ml tube containing 9 ml DMEM, and refrigerating the freshly prepared medium until the day of tissue collection. When storage space was expected to be limited, the 10 ml freshly prepared medium was aliquoted into smaller, 1 ml aliquots in 1.7 ml tubes.
Skin biopsies were opportunistically collected from wolves of both black and gray coat color phenotypes during Yellowstone National Park’s and Idaho Department of Fish and Game’s annual wolf capture, handling, and radio-collaring operations in 2015–2016. Each study subject was anesthetized prior to collection of a single 6 mm skin biopsy by the National Park Service following protocols approved by the National Park Service Institutional Animal Care and Use Committee (Permit IMR_YELL_Smith_wolves_2012) and the Idaho Department of Fish and Game following United States national guidelines for handling animals. Upon collection, each 6 mm skin biopsy was immediately transferred to either a 15 ml tube containing 10 ml collection medium or a 1.7 ml tube containing 1 ml collection medium, and kept in a chilled cooler (Tovar et al. 2008) (Supplementary Table S2). Biopsies that were collected into 1.7 ml tubes were transferred to 15 mL tubes containing 10 ml collection medium at the end of the field day. All samples were transferred to a refrigerator until overnight shipment to UCLA on wet ice. In total, we collected 24 biopsies, representing 14 Kyy and 10 KyB wolves (1 biopsy sample was taken per individual wolf).
Primary Culture of Wolf Keratinocytes and Fibroblasts
Working in a Biosafety Class II cabinet, each skin biopsy was transferred to a p100 Petri dish containing 50 μl collection medium. The biopsy was then minced into approximately 0.5 mm pieces using curved iris scissors and evenly dispersed across the dish. A small drop of medium (ca., 20 μl) was placed around each skin piece, and 2–3 ml medium was added around the bottom of the dish’s rim. Samples were then incubated at 37 °C under 5% CO2 and atmospheric oxygen levels. The medium was changed every 2–3 days. We evaluated 4 culture medium formulations for their promotion of wolf keratinocyte cell growth at initial plating (Supplementary Materials and Methods; Supplementary Figures S1–S3). From those results, we used the following optimized medium (Wu et al. 1982) (hereafter referred to as “FAD medium”) to initiate primary keratinocyte cultures from the biopsies: 1:1 DMEM:F12 base media (ThermoFisher Scientific) + 5% BCS + 0.4 μg/ml hydrocortisone + 10 ng/ml epidermal growth factor (EGF) + 1% Penicillin-Streptomycin (ThermoFisher Scientific). After 2 days incubation, 10 μM ROCK inhibitor Y-27632 (hereafter referred to as RI) (Cayman Chemical) was added (Liu et al. 2012). All images of cells were taken using an AxioCamMRm camera (Zeiss).
To separate fibroblasts from keratinocytes after initial plating (ca., 1 week post-plating), dishes were first washed once with 1× phosphate buffer solution (PBS; ThermoFisher Scientific). 1 ml 0.25% trypsin-EDTA (ThermoFisher Scientific) was then added to each well and quickly pipetted up and down 3–5 times, which detached fibroblasts but not keratinocytes. This initial cell suspension, containing fibroblasts, was transferred to a 15 ml tube containing 1 ml DMEM + 10% BCS. The dish was then quickly rinsed 2 times with PBS to remove any residual fibroblasts and then was incubated with 1 ml 0.25% trypsin-EDTA at 37 °C for 5–10 min to detach keratinocytes for cell passage. After this initial separation, selective growth of each cell type (keratinocytes and fibroblasts) was performed using cell type-specific media. Upon passage of cells, we compared 12 growth treatments for culturing primary wolf keratinocytes (Supplementary Materials and Methods). We found that primary wolf keratinocytes grow best in FAD medium (formula above) + RI, with 3T3 feeder cells (Todaro and Green 1963), which are known to promote growth of human epidermal keratinocytes (Rheinwald and Green 1975) (Supplementary Materials and Methods). We also cultured the separated fibroblasts, growing them in M199/M106 medium, a formulation optimal for human fibroblasts (Dickson et al. 2000: 1:1 [vol/vol] medium M199 and medium M106 [ThermoFisher Scientific] + 15% BCS + 10 ng/ml EGF + 0.4 μg/ml hydrocortisone + 1% Penicillin-Streptomycin), to permit quantification of CBD103 expression levels in this cell type.
Cells were cryopreserved in DMEM/F12 + 10% BCS + 10% dimethyl sulfoxide (DMSO; Sigma) by transferring cryotubes to a room temperature Mr. Frosty Freezing Container (ThermoFisher Scientific) which was placed in a −80 °C freezer overnight. Cryotubes were then transferred to a liquid nitrogen freezer for storage.
K Locus Genotyping
Genotyping of the K locus was performed on extracted DNA using a high resolution melt (HRM) assay as previously performed (Coulson et al. 2011; Schweizer et al. 2018). 10 μL HRM reactions contained 2.25 μL H2O, 1.75 μL MgCl2, 0.5 μL 4 µM primer mix (primers Klocus_F_HRM: 5′-GACCGCTCCTTATTCTGCAAT-3′ and Klocus_R_HRM 5′-CTTTCCGGCACGTTCTGTTT-3′, Supplementary Table S1), 5 μL Roche HRM master mix, and 0.5 μL DNA. Melting analysis was performed on a Roche LightCycler 480 following published PCR conditions (Wittwer et al. 2003), with genotypes showing melting curves at the following temperatures: Kyy 78.7 °C, KyB 78.0 °C, and KBB 77.7 °C. Genotyping was performed with 2 positive control samples for each genotype (i.e., samples of known genotype). All wolves with gray and black coat colors had the genotypes Kyy and KyB, respectively.
Immortalization of Wolf Keratinocytes and Generation of a KyB Cell Line Using CRISPR/Cas9
To investigate the effects of CBD103 genotype on a common genetic background, we used CRISPR/Cas9 editing to introduce the 3 base pair KB allele deletion into Kyy cells. Although reciprocal introduction of the Ky allele into KBB cells would have been ideal, we were unable to obtain KBB keratinocytes, given the rarity of the genotype in our wolf population. Wildtype Kyy cells were immortalized and edited to generate a KyB line by Applied StemCell, Inc (Milpitas, CA). Generation of a KBB line was also attempted, but was not successful. Keratinocytes from a single CBD103 wildtype Kyy individual (animal ID 15071) were first immortalized by infecting 1 × 105 cells with freshly packaged lentivirus encoding simian virus 40 (SV40) T antigen following Applied StemCell’s standard protocol. The cells were selected with puromycin and passaged for 5–8 passages. SV40 transgene expression of puromycin-selected cells was determined by PCR amplification of a 112 base pair amplicon of SV40 using the following primers: SV40-F: 5′-GGGAGGTGTGGGAGGTTTTT-3′, SV40-R: 5′-TCAAGGCTCATTTCAGGCCC-3′ (Supplementary Table S1). The resultant wolf immortalized keratinocyte (WIK) cell line was subsequently cultured in DMEM containing 10% fetal bovine serum, 1× non-essential amino acids, 1× L-glutamine, 1× sodium pyruvate, and 1% Penicillin-Streptomycin. Immortalized cells were plated at 2 × 104 cells per well of a 6-well plate. Medium was changed every day, and cells were passaged every 4–5 days or when confluent.
To generate the KB allele, which is the deletion of the 3 base pairs TCC/AGG at chr16:58965448-58965450 (canfam3.1), the following guide RNA (gRNA) sequence was selected based on proximity to the target 3 base pair deletion: CCTGCAGAGGTATTATTGCAGA. To prevent the gRNA/Cas9 complex from recognizing and cutting sequence after the single-stranded donor oligonucleotide (ssODN) was used as a repair template, a silent single nucleotide polymorphism (SNP), corresponding to the first base pair of the gRNA, was incorporated into the ssODN (C to T). Additionally, an intronic SNP (C to G) was introduced by the ssODN at chr16:58965507, as the dog genome was used to design the ssODN and is fixed for G at that genomic coordinate.
To screen clones for the 3 base pair deletion, the following primers were developed to amplify a 298 base pair region containing the 3 base pair CBD103 deletion and 2 TspEI digestion sites: CBD103-screenF: 5′-GTGAGGTGTACAATGAGGATTATAACTGAACTCC-3′, CBD103-screenR: 5′-GGAAGAACAGCGGCCTATCTGC-3′ (Supplementary Table S1). TspEI digestion of the PCR product of Ky yielded 3 fragments of lengths 112 base pairs, 103 base pairs, and 83 base pairs, whereas TspEI digestion of the PCR product of KB yielded 2 fragments, of lengths 112 base pairs and 183 base pairs (Supplementary Figure S4). Clones thought to carry the intended 3 base pair deletion were sequenced with Sanger sequencing. Sanger sequencing suggested that one clone (clone ID: P2H9) was heterozygous for the intended 3 base pair deletion.
To confirm K locus genotype and characterize the immortalized Kyy cell line and the KyB edited cell line, we performed whole-genome sequencing. We extracted DNA from each cell line using the DNeasy Blood and Tissue Kit (Qiagen) and prepared DNA-Seq libraries from 100 ng DNA using the NEBNext Ultra II FS DNA Library Prep Kit for Illumina (New England Biolabs; 12 min enzymatic shearing, 4 PCR cycles). 100 base pair, paired end sequencing was performed on a NovaSeq SP at the Duke Sequencing and Genomic Technologies Shared Resource core facility. Reads were trimmed with Trim Galore (Krueger 2015) to remove adapter sequence and base pairs with Phred score < 20 from the ends of reads, keeping only reads ≥ 20 basepairs long after trimming. We mapped reads to the dog genome (canfam3.1) using bwa with default settings and removed reads with MAPQ less than 10 (Li and Durbin 2009). We performed indel realignment and base recalibration using the Genome Analysis Toolkit (GATK) (McKenna et al. 2010). Base recalibration was based on genotypes with GQ ≥4 from initial genotyping of the data with the UnifiedGenotyper function. Final genotyping was performed with HaplotypeCaller. We removed variants that did not pass the following thresholds: QUAL < 100, QD < 2, MQ < 35, FS > 30, HaplotypeScore > 13, MQRankSum < −12.5, ReadPosRankSum < −8. Variants were then filtered to only retain biallelic variants with a genotype quality of at least 20, with ≥30× coverage in both cell lines. From the filtered dataset, we defined potential off-target edits as variants that had been called in the KyB edited cell line that had no reads supporting that variant in the parent immortalized Kyy cell line.
We performed quantitative RT-qPCR to measure CBD103 expression in the immortalized Kyy and edited KyB cell lines. We collected cells into Buffer RLT (Qiagen) and performed RNA extractions using the Qiagen RNeasy Mini Kit. cDNA was synthesized using SuperScript II Reverse Transcriptase (ThermoFisher Scientific) on 1.5 μg RNA of each sample following the manufacturer’s instructions. The cDNA products were purified using the QIAquick PCR Purification Kit (Qiagen). RT-qPCR reactions were performed on 20 ng cDNA using PerfeCTa SYBR Green FastMix ROX with 20 μL reaction volumes, with 1 μL 1 μM primer mix (CBD103_F: 5′-GCCAAAGGAGGAGCAGATAG-3′ and CBD103_R: 5′-ACTTTACAACGCTCCCATCC-3′; Supplementary Table S1). The following program was used on a StepOnePlus Real-Time PCR System: 95 °C for 2 min; 45 cycles of 95 °C (10 s), 58 °C (30 s), and 72 °C (30 s). RT-qPCR values were standardized to the expression level of ACTB using 1 μL 1 μM primer mix (ACTB_F: 5′-ATGCAGAAGGAGATCACTGC-3′ and ACTB_R: 5′-CTGCGCAAGTTAGGTTTTGT-3′; Supplementary Table S1). Reactions were performed in triplicate.
PolyI:C and Live CDV Challenge Experiments
Each of the 24 keratinocyte cultures (representing 14 Kyy and 10 KyB wolves) was plated on 2 wells of a 12-well plate at 1.5 × 104 cells per well, with 3T3 feeder cells in FAD + 10 μM RI (N = 48 wells in total; 2 wells per individual/cell culture). When cells were at approximately 30% confluence, they were switched to a low-serum medium (FAD + 0.5% BCS) without RI and incubated for 24 h. For each pair of wells (representing 1 individual/cell culture), 1 well was treated with 1 μg/ml polyI:C (Sigma-Aldrich) and 1 well was treated with vehicle control (sterile, endotoxin-free water) for 24 h. Wells were quickly washed once with 0.25% trypsin (by pipetting up and down 3 times) and twice with PBS to remove 3T3 cells before collecting keratinocytes into 1 ml Trizol (Invitrogen). Each well is considered a “sample,” such that the dataset is composed of paired samples for each individual wolf (i.e., N = 48 samples originating from 24 individuals/cell cultures). Details of the samples used for polyI:C challenge across the 24 wolf keratinocyte cultures are provided in Supplementary Table S2.
CDV challenges were performed on a subset of 6 primary keratinocyte cultures (from 3 Kyy and 3 KyB animals; N = 12 samples, 2 samples per individual/cell culture) and on the Kyy and KyB immortalized cell lines using CDV 5804PEH-eGFP, a recombinant wild-type CDV that expresses green fluorescent protein (GFP) (Von Messling et al. 2004). CDV was propagated in VerodogSLAMtag cells (Von Messling et al. 2004), and TCID50/cell was calculated following the Spearman-Kärber method (Finney 1952). Keratinocytes were plated in duplicate on 24-well plates at 8 × 104 cells per well in FAD + 5% BCS + RI, without feeder cells. After a 24 h incubation, the medium was changed to FAD + 0.5% BCS. After an additional 24 h, cells were infected at a multiplicity of infection (MOI) of 20 TCID50/ml or 100 TCID50/ml. At varying time points (5 days post-infection for primary cells, and 0, 1, 2, 3, 4, and 5 days post-infection for the immortalized cell lines), cells were collected in 1 ml Trizol for RNA preservation. Total RNA was extracted from cells using the Trizol Plus RNA Purification Kit with DNAse treatment (RNAse-Free DNAse Set, Ambion) and column cleanup (PureLink RNA Mini Kit, Invitrogen). For the primary and immortalized keratinocytes, images of CDV-infected cells (which express GFP) were captured using an AxioCamMRm camera (Zeiss).
RNA-Seq Data Generation and Analysis
We used RNA-seq to quantify CBD103 gene expression in primary keratinocytes and fibroblasts, and to profile the immune response to polyI:C and CDV in the primary keratinocyte cultures and the Kyy and KyB immortalized cell lines. RNA-seq libraries were generated on high-quality RNA extracts (RNA Integrity number [RIN] was measured for the 48 polyI:C and control samples; mean = 9.83 ± 0.52 s.d., Supplementary Table S2) using half-reactions from the TruSeq RNA Sample Preparation Kit (Illumina), following the manufacturer’s instructions. RNA-seq libraries were indexed and pooled at 12 samples per lane and sequenced as 100 base pair single-end reads on the Illumina HiSeq 4000 at the Vincent J. Coates Genomics Sequencing Laboratory at University of California-Berkeley.
The resulting reads (mean 29.62 million per sample ± 4.65 million s.d.; Supplementary Table S2) were trimmed with Trim Galore (Krueger 2015) to remove adapter sequence and base pairs with Phred score <20 from the ends of reads, keeping only reads ≥20 basepairs long after trimming. Reads were mapped to the dog genome (canfam3.1) using 2-pass mapping with STAR (Dobin et al. 2013). Only uniquely mapped reads were retained. We quantified gene expression using HT-Seq (Anders et al. 2014) with the “union” mode using the Canis_familiaris.CanFam3.1.94.gtf file from Ensembl. We transformed the raw counts to transcripts per million (TPM) (Wagner et al. 2012) and removed genes that had an average TPM < 2 in either condition within each RNA-Seq data set (i.e., null or polyI:C; null or CDV). For each data set (the polyI:C data set and the CDV data set), we also removed the 5% of genes with the lowest variance within each condition. We performed voom normalization (Law et al. 2014) within each data set using the voomWithQualityWeights function, with the trimmed mean of M-values (TMM) method (Robinson and Oshlack 2010) in DESeq (Anders et al. 2014) to estimate normalization factors. For the data set of primary keratinocytes challenged with polyI:C, we controlled for the technical effects of RNA-Seq library yield, sample provider (Yellowstone National Park Service or Idaho Department of Fish and Game), and PC2 (which improved power to detect cis-eQTL) by regressing them out from the expression level data for each gene using limma (Smyth 2005). Two samples (polyI:C treated 1005F, and null control 14 387) did not cluster in principal component space according to condition (null versus polyI:C-treated) and were removed from downstream analyses.
We modeled variation in the expression levels of each gene using mixed-effects models fit in emmreml (Akdemir and Okeke 2015). For each gene in each data set, we first ran a model in which normalized, batch-corrected expression levels were modeled as a function of the fixed effect of condition (null or stimulated) and a random effect to account for samples from the same individual and genetic relatedness across individuals (see Supplementary Materials and Methods for estimation of the relatedness matrix K). We then ran 2 additional models: 1) gene expression as a function of the fixed effects of condition and CBD103 genotype nested within condition, and a random effect to control for repeated samples and genetic relatedness; and 2) gene expression as a function of the fixed effects of condition, CBD103 genotype, and the interaction of condition and CBD103 genotype, and a random effect to control for repeated samples and genetic relatedness. False discovery rate (FDR) was calculated following the q-value method of (Storey and Tibshirani 2003). For the polyI:C data set, the observed P-value distributions were compared to an empirical null P-value distribution derived from re-running the same models but permuting the variable of interest (either condition or CBD103 genotype) across 100 permutations of the data. For the CDV data set, the sample size was too small to calculate FDR based on the permutation-derived empirical null. We therefore estimated FDR by comparison to a uniform null, using the R package qvalue (Storey et al. 2017) with the pfdr argument set to true. Because the assumption of a uniform null is likely anti-conservative for this analysis, we used more stringent criteria for significance by setting the FDR threshold to 1% and requiring that the absolute gene expression fold change be greater than 2.
To assess possible pathway or gene set enrichment among differentially expressed genes, we performed Gene Ontology enrichment analysis using g:ProfileR with the Ensembl 93 database (Raudvere et al. 2019). We required a minimum of 5 and maximum of 350 genes in each tested biological process, and a minimum of 5 genes overlapping the biological process category and each of our gene sets of interest. We applied the “Best per parent group” hierarchical filtering option and used a Benjamini-Hochberg false discovery rate significance threshold of 1%.
To quantify CDV transcription, we remapped the trimmed RNA-Seq reads of the CDV data set to a reference fasta file containing both the dog genome (canfam3.1) and the CDV strain 5804 genome (GenBank accession AY386315.1), using the mapping parameters described above. As above, only uniquely mapped reads were retained and gene expression was quantified with HT-Seq. Gene counts of CDV genes were summed to quantify total CDV transcription. Counts were then normalized to counts per million counts mapped (CPM) to either genome to control for library sequencing depth. The Integrated Genomics Viewer (IGV) (Robinson et al. 2011) version 2.9.2 was used to visualize read coverage across the CDV strain 5804 genome. To test for the effects of treatment (CDV or vehicle control) or CBD103 genotype on CDV transcript levels, we performed a mixed model with normalized CDV expression level as the dependent variable, treatment and CBD103 genotype (nested within treatment) as fixed effects, and animal ID as a random effect.
Results
Expression of CBD103 Across Tissues and Cell Types
To identify tissues where CBD103 is expressed, and is therefore most likely to be involved in immune defense, we first assessed CBD103 expression across a set of 9 gray wolf tissues. We predicted that CBD103 would be expressed in the same tissues in wolves as dogs, and therefore focused on tissues for which CBD103 expression levels are known for dogs (Leonard et al. 2012). CBD103 was most highly expressed in epithelial tissues, including tongue and esophagus, consistent with expression patterns described for both dogs and humans (Figure 1B) (Harder et al. 2001; Leonard et al. 2012). Based on published RNA-Seq data from whole blood (Charruau et al. 2016) and newly generated RNA-Seq data for the 2 major skin cell types, keratinocytes and fibroblasts, CBD103 has a unique expression pattern relative to other wolf beta defensins. Specifically, while all beta defensins we tested are very lowly expressed in fibroblasts and lowly expressed in whole blood, CBD103 is uniquely highly expressed in keratinocytes (Figure 1C). These results led us to focus on challenge experiments in keratinocytes for the remainder of our study.
Establishment of a Panel of Primary Wolf Keratinocytes
To assess the effects of CBD103 genotype on wolf keratinocytes, we established a panel of primary wolf keratinocyte cultures. We first optimized conditions for growth of wolf keratinocytes (Supplementary Materials and Methods). In brief, relative to 3 alternative conditions (FAD only, M199/M106 medium, or M199/M106 + RI), keratinocyte survival and proliferation rates were greatest when skin biopsies were plated in FAD medium and switched to FAD + RI 2 days post-plating, and supplemented with 3T3 feeder cells during serial passaging (Supplementary Figures S1–S3). One culture (from individual 15 071) was used to estimate keratinocyte lifespan under these conditions and produced 28.2 cell population doublings over the course of 80 days (Supplementary Figure S5). We therefore used this protocol to culture keratinocytes from all subsequent samples (N = 24). The 24 keratinocyte cultures represented 14 and 10 wolves with the Kyy and KyB genotypes, respectively (Supplementary Table S2).
Establishment of a CBD103-Edited Wolf Keratinocyte Line
To isolate possible effects of the CBD103 KB allele on the keratinocyte response to virus, we immortalized a single Kyy wildtype keratinocyte culture from individual 15 071 (see Materials and Methods). We then used CRISPR/Cas9 editing to generate a line that was designed to be identical to the wildtype line, except for deletion of the 3 base pair codon that distinguishes Ky from KB, as well as a silent site substitution (G to A) incorporated as part of the gene-editing process, and a single nucleotide intron substitution (C to G) that differentiates gray wolves from domestic dogs (because the donor oligonucleotide was designed based on the dog genome). Whole-genome sequencing confirmed this sequence in and around the CBD103 gene, as well as 323 potential off-target changes genome-wide (see Materials and Methods). Unexpectedly, however, RNA-Seq analyses revealed that CBD103 expression was fully abolished in the edited KyB line relative to the original immortalized line (Supplementary Figure S6). RT-qPCR of CBD103 in the immortalized Kyy line and the edited KyB line confirmed this pattern (KyB relative to Kyy log2(fold change) = −11.194 ± 0.249 standard error). Although we attempted to also quantify CBD103 protein levels using an ELISA targeted to human beta defensin 3 [Peprotech], we observed low inter-replicate repeatability of CBD103 quantification with this method, suggesting insufficient conservation of the epitope (84% between human and wolf) to generate accurate estimates.
Response of Wolf Keratinocytes to PolyI:C
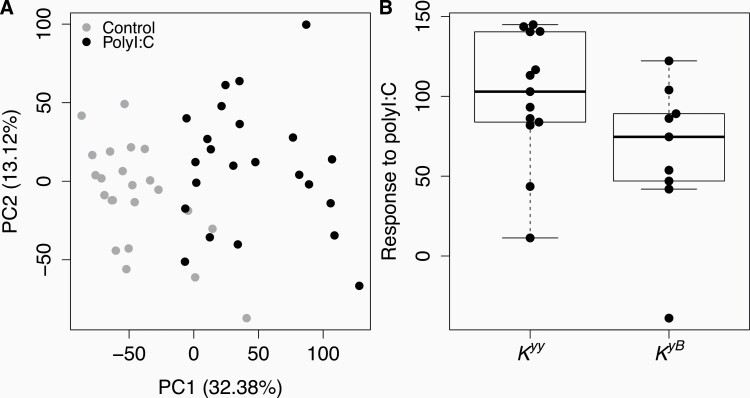
Given the role of CBD103 in innate immunity, including in viral defense, we evaluated whether the CBD103 Ky/KB genotype affects the gene expression response to polyI:C. To do so, we used RNA-seq to measure genome-wide gene expression in our 24 primary keratinocyte cultures, in 2 samples for each culture: 1) cells cultured in medium only (null); and 2) cells exposed to 1 μg/mL polyI:C. PolyI:C treatment elicited a strong gene expression response in wolf keratinocytes, such that the first principal component of overall gene expression levels separated null samples from polyI:C challenged samples (Figure 2A; R2 = 0.596, Pearson correlation P = 3.272 × 10−10). A total of 3505 genes were up-regulated and 4143 genes were down-regulated upon polyI:C stimulation (1% FDR; N = 10 665 genes tested; Supplementary Table S3). Genes up-regulated in response to polyI:C were enriched for 73 diverse biological processes, with the 3 most significantly enriched processes being “response to other organism,” “MAPK cascade,” and “inflammatory response” (all FDR < 1%; Supplementary Table S4).
Figure 2.
PolyI:C elicits a strong immune response in primary wolf keratinocytes that is not predicted by CBD103 K locus genotype. (A) Each dot represents a primary culture of wolf keratinocytes stimulated with polyI:C (black) or vehicle control (gray) for 24 h. Values in parentheses in the x and y axis labels represent the percent of overall gene expression variation explained by principal components 1 and 2, respectively. PolyI:C treatment is strongly associated with PC1 of the overall gene expression data (R2 = 0.596, Pearson correlation P = 3.272 × 10−10). (B) The global gene expression response to polyI:C does not differ between Kyy and KyB wolf keratinocytes (R2 = 0.154, Pearson correlation P = 0.070). Each dot represents the difference in PC1 between a matched pair of control and polyI:C stimulated samples. Each box represents the interquartile range, with a horizontal line depicting the median value. Whiskers indicate the most extreme values within 1.5× of the interquartile range.
In contrast, CBD103 genotype was not associated with the overall response to polyI:C (as represented by the distance separating null and polyI:C samples for the same individual, on PC1 of the full gene expression data set: R2 = 0.154, Pearson correlation P = 0.070; Figure 2B). CBD103 also did not predict gene expression for individual genes in either the null condition or in the polyI:C condition (all FDR > 10%; Supplementary Table S5). Finally, CBD103 genotype did not predict individual gene responses to poly I:C (modeled as an interaction effect between CBD103 genotype and polyI:C condition; 0 genes FDR < 1%, 7 genes FDR < 10%; Supplementary Table S6). This observation remained consistent even among genes that were most responsive to polyI:C stimulation (N = 7648 genes up-regulated by polyI:C at FDR < 0.1%; 0 genes FDR < 1%, 2 genes FDR < 10%; Supplementary Table S7).
Response of Wolf Keratinocytes to Live Canine Distemper Virus
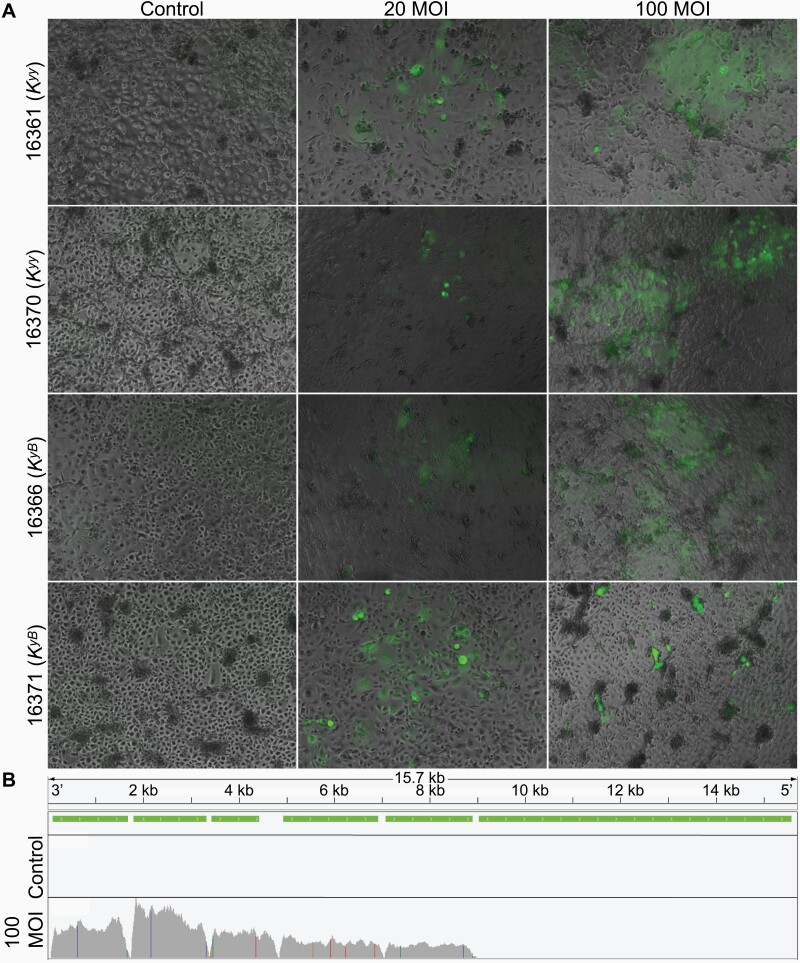
Given that the human homolog of CBD103 has direct antiviral activity (Wilson et al. 2013), we also tested whether CBD103 genotype affects the keratinocyte gene expression response to infection by live CDV (using the GFP expressing strain 5804PEH-eGFP; Von Messling et al. 2004), an important pathogen in North American gray wolves (Almberg et al. 2009). We observed that both primary keratinocytes and the immortalized Kyy and KyB lines are susceptible to infection, as evident by GFP and CDV expression within infected cells; increased rates of irregular cell shape, cell death and detachment; and up-regulation of key immune defense cytokines post-infection (Figure 3 and Supplementary Figures S7–S9).
Figure 3.
Primary wolf keratinocytes are vulnerable to canine distemper virus infection. (A) Kyy and KyB primary wolf keratinocytes infected with live canine distemper virus for 5 days. Keratinocytes were infected at an MOI of 20 and 100 TCID50/cell. Fluorescence images were captured to visualize CDV (expressing GFP) and overlaid on phase-contrast images. Animal IDs and corresponding CBD103 genotypes are indicated on the y axis. (B) Visualization of RNA-Seq read pile-ups (gray) across the full CDV genome from a representative wolf cell culture (animal ID 16361) treated for 5 days with CDV at 100 MOI or vehicle control. CDV transcription is evident from RNA extracted from infected cells, but not from control cells. Green bars indicate the genomic locations of CDV genes. Colored vertical lines indicate polymorphism relative to the reference genome.
We challenged a subset of primary keratinocyte cultures (N = 6; 3 Kyy and 3 KyB primary cultures) with live CDV (MOI of 100 TCID50/cell) for 5 days. CDV transcription was detected in RNA extracted from the infected keratinocytes, as evidenced by expected patterns of higher RNA-Seq read coverage in CDV genes compared to CDV intergenic regions, and higher expression levels in genes nearer to the 3’ end of the CDV genome (Figure 3B) (Wignall-Fleming et al. 2019). Treatment condition (null vs. CDV-challenged) was correlated with PC2 of overall gene expression (R2 = 0.397, Pearson correlation P = 0.028). A total of 348 genes were differentially expressed in response to infection by CDV (FDR < 1% and absolute fold change ≥ 2; Supplementary Table S8). The 156 genes up-regulated in response to CDV were enriched for “response to external biotic stimulus” (including key antiviral- and interferon-signaling genes such as ISG15, ISG20, OAS1, and OAS3) and “negative regulation of viral process” (FDR < 1%; Supplementary Table S9). As expected, the multi-day response to CDV, a live single-stranded RNA virus, provides distinct information from the 24 h response to polyI:C, a synthetic analog for dsRNA that binds to TLR3. Specifically, wolf genes that were responsive to CDV do not significantly overlap with those that were responsive to polyI:C (Fisher’s exact test OR (95% CI) = 1.007 (0.758, 1.351), P = 1).
CBD103 genotype did not predict keratinocyte gene expression levels after CDV treatment (modeled as CBD103 genotype nested in CDV; all FDR > 1%; Supplementary Table S10) or in response to CDV (modeled as an interaction term between CBD103 genotype and CDV; all FDR > 1%; Supplementary Table S11). Overall CDV transcript levels, however, were slightly lower in challenged KyB cells than in Kyy cells (β = −8591.9313, P = 0.039, Supplementary Figure S9).
Discussion
Previous research indicates that the CBD103 Ky/KB polymorphism in North American gray wolves not only affects fur color, but also has pleiotropic effects on other fitness-related traits (Coulson et al. 2011; Stahler et al. 2013; Cassidy et al. 2017; Schweizer et al. 2018). Most notably, KBB and KyB wolves have indistinguishable black fur color, but KBB wolves have much lower fitness than KyB wolves (Coulson et al. 2011). Given that CBD103 is a member of a gene family that is associated with innate immune function, we hypothesized that the KB allele might also alter immune defense, and specifically, the transcriptional response to CDV, in a manner that contributes to its intriguing evolutionary dynamics. Indeed, there is some evidence that immune function can be affected by pigmentation-related pathways. For example, the MC1R agonist alpha-melanocyte-stimulating hormone (α-MSH) also has microbicidal activity (Singh and Mukhopadhyay 2014), and experimental manipulation of the amino acid sequence of α-MSH affects this activity (Grieco et al. 2003). However, pleiotropic effects on immune function have yet to be explored for pigmentation variants that are naturally occurring.
Our data provide no strong support for the hypothesis that CBD103 genotype alters gene expression levels of keratinocytes in response to polyI:C or CDV. Further, CBD103 genotype was not clearly associated with variation in baseline gene expression levels. Additionally, expression levels of CBD103 itself did not differ depending on the K locus genotype. We note that, because our small sample size limits statistical power, particularly for the CDV data set, our results are limited to excluding the possibility that CBD103 genotype effects are moderate to large. It therefore remains possible that CBD103 has small effects on gene expression and/or the response to CDV infection that would require larger sample sizes to detect.
Our findings suggest that, if the fitness consequences of CBD103 genotype in wolves are indeed due to differences in immune function, they do not arise from simple differences in the immediate gene regulatory response to TLR3 pathway activation or CDV infection in keratinocytes. However, CBD103 genotype could influence immunity through mechanisms not evaluated here. First, given evidence that CBD103 can directly interact with pathogens (García et al. 2001; Harder et al. 2001; Wilson et al. 2013), the codon deletion in the KB allele could alter the strength of direct protein-pathogen interactions. In the case of viruses like CDV, such a mechanism could affect viral entry rates through non-regulatory routes that would not be apparent through measuring gene expression. In preliminary support of this possibility, we observed moderately lower levels of CDV transcript levels in KyB cells relative to Kyy cells. However, we interpret this result with caution given our small sample size, and note that even if robust, this result does not differentiate between CDV entry, versus post-entry events (e.g., replication). Quantification of intracellular viral titers and/or copy number over time using plaque assays and RT-qPCR, in a larger sample size, will be important to further investigate the observed pattern. Second, cellular interactions with other types of viruses, pathogens, or parasites known to affect North American gray wolf populations may be relevant (e.g., wolves are vulnerable to canine parvovirus, canine adenovirus, and Sarcoptes scabiei-induced sarcoptic mange: Almberg et al. 2009). Finally, in humans, beta defensin 3 appears to have more pronounced antiviral activity in low salt concentrations similar to the environment of the oral cavity (Quiñones-Mateu et al. 2003). Such concentrations would be lethal to the keratinocytes we studied here (derived from skin from ear punches), but could potentially be tested using epithelial cells established from the oral cavity or by incubating the virus with beta defensin 3 prior to cell infection (as in Quiñones-Mateu et al. 2003). Importantly, the methods we applied here provide a basis for testing several of these possibilities in vitro.
Although our results do not resolve the mechanisms explaining the fitness costs of the KB allele in wolves, they do exemplify new approaches for studying genotype-phenotype relationships in non-model organisms. Specifically, we show that methods that were originally developed to culture primary human keratinocytes can also be applied to samples collected from a wild mammal population (Rheinwald and Green 1975; Wu et al. 1982). This approach makes it feasible to generate population samples of keratinocyte lines for experimental studies that cannot be conducted in vivo. The ability to produce a >225-fold expansion of cells from small skin biopsies demonstrates the potential to cryopreserve many vials of early passage cells (on the order of >109 total cells), permitting hundreds of experiments. Such methods could be useful for studying cellular responses to cutaneous pathogens, such as the white-nose syndrome epidemic that has devastated wild bat populations (Cryan et al. 2010). With respect to pigmentation phenotypes, the cellular approaches we describe here could also be adapted for melanocyte culture (Costin et al. 2004) or melanocyte-keratinocyte coculture (Lei et al. 2002) to study the effects of genetic variation on the molecular basis of pigmentation phenotypes.
More broadly, cell culture methods provide a much-needed approach for testing the explosion of hypotheses now being generated in evolutionary genomics. The reduced costs of high-throughput sequencing make genome-wide association studies, selection scans, transcriptomic studies, and comparative genomic analyses feasible in a broad spectrum of species. For example, analyses of selective sweeps in wolves have revealed many potential target genes, including those related to hypoxia and cold adaptation (Schweizer et al. 2016a, 2016b). However, genetic manipulations to functionally dissect these signatures are not ethical or feasible for most mammals, especially those that are declining or already rare or endangered. Yet many of these species can be sampled, non-lethally, for culturable cells and tissues such as skin, and live cells can even be collected from recently deceased individuals. Although fibroblast culture has long been used as a method of preserving genetic material (e.g., the “Frozen Zoo”; [Benirschke 1984]), primary cell culture has not yet been widely applied to study inter-individual variation within or among populations. Such an approach is likely to become more attractive as immortalization, gene editing, and induced pluripotent stem cell (iPSC) generation becomes more feasible (e.g., Ben-Nun et al. 2011; Marchetto et al. 2013; Romero et al. 2015). These approaches may be particularly valuable for isolating the causal effects of specific variants of interest, especially if those variants are rare (as in the case of KBB, estimated to be at 2% frequency in the Yellowstone gray wolf population) (Coulson et al. 2011).
Supplementary Material
Acknowledgments
We thank the wolf team at Yellowstone National Park, Jennifer Struthers, Jason Husseman, and their teams at the Idaho Department of Fish and Game for their hard work to collect samples for this project. We thank Jessica Cinkornpumin for guidance on lab work, and Steven Nguyen, Kashif Iqbal, Nadia Riabkova, and Joey Curtiss for their help with much of the molecular and cell work. We also thank Dr. Veronika von Messling, who kindly provided us the CDV 5804PEH-eGFP strain.
Funding
National Science Foundation (DEB-1257716 to R.K.W. and B.M.V., DEB-1245373 to D.R.S.); the Howard Hughes Medical Institute (Gilliam Fellowship to R.A.J.); a UCLA Dissertation Year Fellowship (to R.A.J.); the National Institutes of Health (F32HD095616 to R.A.J.); and Yellowstone Forever.
Data Availability
RNA sequencing data generated for this study are available in the NCBI Gene Expression Omnibus (GEO series accession GSE163163). RNA sequencing data from blood were previously published and available under GEO series accession GSE80440. DNA sequencing data of the immortalized cell lines are available in the NCBI Sequence Read Archive (BioProject accession number PRJNA688221).
References
- Akdemir D, Okeke U. 2015. EMMREML: Fitting mixed models with known covariance structures. GPL-2. https://cran.r-project.org/web/packages/EMMREML/index.html.
- Almberg ES, Mech LD, Smith DW, Sheldon JW, Crabtree RL. 2009. A serological survey of infectious disease in Yellowstone National Park’s canid community. PLoS One. 4:e7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2014. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TM, vonHoldt BM, Candille SI, Musiani M, Greco C, Stahler DR, Smith DW, Padhukasahasram B, Randi E, Leonard JA, et al. 2009. Molecular and evolutionary history of melanism in North American gray wolves. Science. 323:1339–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benirschke K. 1984. The frozen zoo concept. Zoo Biol. 3:325–328. [Google Scholar]
- Ben-Nun IF, Montague SC, Houck ML, Tran HT, Garitaonandia I, Leonardo TR, Wang YC, Charter SJ, Laurent LC, Ryder OA, et al. 2011. Induced pluripotent stem cells from highly endangered species. Nat Methods. 8:829–831. [DOI] [PubMed] [Google Scholar]
- Candille SI, Kaelin CB, Cattanach BM, Yu B, Thompson DA, Nix MA, Kerns JA, Schmutz SM, Millhauser GL, Barsh GS. 2007. A β-defensin mutation causes black coat color in domestic dogs. Science. 318:1418–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy KA, Mech LD, MacNulty DR, Stahler DR, Smith DW. 2017. Sexually dimorphic aggression indicates male gray wolves specialize in pack defense against conspecific groups. Behav Processes. 136:64–72. [DOI] [PubMed] [Google Scholar]
- Charruau P, Johnston RA, Stahler DR, Lea A, Snyder-Mackler N, Smith DW, vonHoldt BM, Cole SW, Tung J, Wayne RK. 2016. Pervasive effects of aging on gene expression in wild wolves. Mol Biol Evol. 33:1967–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costin GE, Vieira WD, Valencia JC, Rouzaud F, Lamoreux ML, Hearing VJ. 2004. Immortalization of mouse melanocytes carrying mutations in various pigmentation genes. Anal Biochem. 335:171–174. [DOI] [PubMed] [Google Scholar]
- Coulson T, MacNulty DR, Stahler DR, vonHoldt B, Wayne RK, Smith DW. 2011. Modeling effects of environmental change on wolf population dynamics, trait evolution, and life history. Science. 334:1275–1278. [DOI] [PubMed] [Google Scholar]
- Cryan PM, Meteyer CU, Boyles JG, Blehert DS. 2010. Wing pathology of white-nose syndrome in bats suggests life-threatening disruption of physiology. BMC Biol. 8:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, Rheinwald JG. 2000. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 20:1436–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney DJ. 1952. Statistical method in biological assay. London: Charles Griffin & Co., Ltd. [Google Scholar]
- Ganz T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 3:710–720. [DOI] [PubMed] [Google Scholar]
- García JR, Jaumann F, Schulz S, Krause A, Rodríguez-Jiménez J, Forssmann U, Adermann K, Klüver E, Vogelmeier C, Becker D, et al. 2001. Identification of a novel, multifunctional beta-defensin (human beta-defensin 3) with specific antimicrobial activity. Its interaction with plasma membranes of Xenopus oocytes and the induction of macrophage chemoattraction. Cell Tissue Res. 306:257–264. [DOI] [PubMed] [Google Scholar]
- Grieco P, Rossi C, Colombo G, Gatti S, Novellino E, Lipton JM, Catania A. 2003. Novel alpha-melanocyte stimulating hormone peptide analogues with high candidacidal activity. J Med Chem. 46:850–855. [DOI] [PubMed] [Google Scholar]
- Harder J, Bartels J, Christophers E, Schröder JM. 2001. Isolation and characterization of human β-defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 276:5707–5713. [DOI] [PubMed] [Google Scholar]
- Hazrati E, Galen B, Lu W, Wang W, Ouyang Y, Keller MJ, Lehrer RI, Herold BC. 2006. Human α-and β-defensins block multiple steps in herpes simplex virus infection. J Immunol. 177:8658–8666. [DOI] [PubMed] [Google Scholar]
- Hedrick PW, Smith DW, Stahler DR. 2016. Negative-assortative mating for color in wolves. Evolution. 70:757–766. [DOI] [PubMed] [Google Scholar]
- Krueger F. 2015. Trim Galore. Available from: http://wwwbioinformaticsbabrahamacuk/projects/trim_galore/.
- Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebre MC, van der Aar AM, van Baarsen L, van Capel TM, Schuitemaker JH, Kapsenberg ML, de Jong EC. 2007. Human keratinocytes express functional Toll-like receptor 3, 4, 5, and 9. J Invest Dermatol. 127:331–341. [DOI] [PubMed] [Google Scholar]
- Lei TC, Virador VM, Vieira WD, Hearing VJ. 2002. A melanocyte-keratinocyte coculture model to assess regulators of pigmentation in vitro. Anal Biochem. 305:260–268. [DOI] [PubMed] [Google Scholar]
- Leonard BC, Marks SL, Outerbridge CA, Affolter VK, Kananurak A, Young A, Moore PF, Bannasch DL, Bevins CL. 2012. Activity, expression and genetic variation of canine β-defensin 103: a multifunctional antimicrobial peptide in the skin of domestic dogs. J Innate Immun. 4:248–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, et al. 2012. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 180:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MCN, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola ACM, Desai KN, Herai RH, Weitzman MD, et al. 2013. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 503:525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund JJ, Boissy RE, Hearing VJ, King RA, Oetting WS, Ortonne JP. 2006. The pigmentary system: physiology and pathophysiology. Blackwell Publishing. [Google Scholar]
- Quiñones-Mateu ME, Lederman MM, Feng Z, Chakraborty B, Weber J, Rangel HR, Marotta ML, Mirza M, Jiang B, Kiser P, et al. 2003. Human epithelial beta-defensins 2 and 3 inhibit HIV-1 replication. AIDS. 17:F39–F48. [DOI] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. 2019. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47:W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald JG, Green H. 1975. Serial cultivation of strains of human epidermal keratinocytes: the formation of keratinizing colonies from single cells. Cell. 6:331–343. [DOI] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol. 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero IG, Pavlovic BJ, Hernando-Herraez I, Zhou X, Ward MC, Banovich NE, Kagan CL, Burnett JE, Huang CH, Mitrano A, et al. 2015. A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics. Elife. 4:e07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer RM, Durvasula A, Smith J, Vohr SH, Stahler DR, Galaverni M, Thalmann O, Smith DW, Randi E, Ostrander EA, et al. 2018. Natural selection and origin of a melanistic allele in North American gray wolves. Mol Biol Evol. 35:1190–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer RM, Robinson J, Harrigan R, Silva P, Galverni M, Musiani M, Green RE, Novembre J, Wayne RK. 2016a. Targeted capture and resequencing of 1040 genes reveal environmentally driven functional variation in grey wolves. Mol Ecol. 25:357–379. [DOI] [PubMed] [Google Scholar]
- Schweizer RM, vonHoldt BM, Harrigan R, Knowles JC, Musiani M, Coltman D, Novembre J, Wayne RK. 2016b. Genetic subdivision and candidate genes under selection in North American grey wolves. Mol Ecol. 25:380–402. [DOI] [PubMed] [Google Scholar]
- Singh M, , MukhopadhyayK. 2014. Alpha-melanocyte stimulating hormone: an emerging anti-inflammatory antimicrobial peptide. Biomed Res Int. 2014:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. 2005. Limma: linear models for microarray data. In: Gentleman R, Carey VJ, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer. p. 397–420. doi: 10.1007/0-387-29362-0 [DOI] [Google Scholar]
- Stahler DR, MacNulty DR, Wayne RK, VonHoldt B, Smith DW. 2013. The adaptive value of morphological, behavioural and life-history traits in reproductive female wolves. J Anim Ecol. 82:222–234. [DOI] [PubMed] [Google Scholar]
- Storey JD, Bass AJ, Dabney A, Robinson D. 2017. qvalue: Q-value estimation for false discovery rate control. Available from: http://githubcom/StoreyLab/qvalue.
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 100:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todaro GJ, Green H. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 17:299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar H, Navarrete F, Rodríguez L, Skewes O, Castro FO. 2008. Cold storage of biopsies from wild endangered native Chilean species in field conditions and subsequent isolation of primary culture cell lines. In Vitro Cell Dev Biol Anim. 44:309–320. [DOI] [PubMed] [Google Scholar]
- von Messling V, Milosevic D, Cattaneo R. 2004. Tropism illuminated: lymphocyte-based pathways blazed by lethal morbillivirus through the host immune system. Proc Natl Acad Sci U S A. 101:14216–14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ. 2012. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131:281–285. [DOI] [PubMed] [Google Scholar]
- Wignall-Fleming EB, Hughes DJ, Vattipally S, Modha S, Goodbourn S, Davison AJ, Randall RE. 2019. Analysis of paramyxovirus transcription and replication by high-throughput sequencing. J Virol. 93:e00571–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SS, Wiens ME, Smith JG. 2013. Antiviral mechanisms of human defensins. J Mol Biol. 425:4965–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. 2003. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 49:853–860. [DOI] [PubMed] [Google Scholar]
- Wu YJ, Parker LM, Binder NE, Beckett MA, Sinard JH, Griffiths CT, Rheinwald JG. 1982. The mesothelial keratins: a new family of cytoskeletal proteins identified in cultured mesothelial cells and nonkeratinizing epithelia. Cell. 31:693–703. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data generated for this study are available in the NCBI Gene Expression Omnibus (GEO series accession GSE163163). RNA sequencing data from blood were previously published and available under GEO series accession GSE80440. DNA sequencing data of the immortalized cell lines are available in the NCBI Sequence Read Archive (BioProject accession number PRJNA688221).