Abstract
Among putative periodontal pathogens, Actinobacillus actinomycetemcomitans, Bacteroides forsythus, and Porphyromonas gingivalis are most convincingly implicated as etiological agents in periodontitis. Therefore, techniques for detection of those three species would be of value. We previously published a description of a multiplex PCR that detects A. actinomycetemcomitans and P. gingivalis. The present paper presents an improvement on that technique, which now allows more sensitive detection of all three periodontal pathogens. Sensitivity was determined by testing serial dilutions of A. actinomycetemcomitans, B. forsythus, and P. gingivalis cells. Primer specificity was tested against (i) all gene sequences from the GenBank-EMBL database, (ii) six A. actinomycetemcomitans, one B. forsythus, and four P. gingivalis strains, (iii) eight different species of oral bacteria, and (iv) supra- and subgingival plaque samples from 20 healthy subjects and subgingival plaque samples from 10 patients with periodontitis. The multiplex PCR had a detection limit of 10 A. actinomycetemcomitans, 10 P. gingivalis, and 100 B. forsythus cells. Specificity was confirmed by the fact that (i) none of our forward primers were homologous to the 16S rRNA genes of other oral species, (ii) amplicons of predicted size were detected for all A. actinomycetemcomitans, B. forsythus, and P. gingivalis strains tested, and (iii) no amplicons were detected for the eight other bacterial species. A. actinomycetemcomitans, B. forsythus, and P. gingivalis were detected in 6 of 20, 1 of 20, and 11 of 20 of supragingival plaque samples, respectively, and 4 of 20, 7 of 20, and 13 of 20 of subgingival plaque samples, respectively, from periodontally healthy subjects. Among patients with periodontitis, the organisms were detected in 7 of 10, 10 of 10, and 7 of 10 samples, respectively. The simultaneous detection of three periodontal pathogens is an advantage of this technique over conventional PCR assays.
Periodontitis describes an inflammation of the supporting tissues of the teeth (2). It exhibits a destructive change that leads to the loss of bone and connective tissue attachment. It is generally accepted that periodontal diseases are infectious diseases (30). Approximately a dozen oral bacterial species are associated with periodontitis. However, to date, the most convincing data implicate three microorganisms as etiologic agents in periodontitis (30). Those are Actinobacillus actinomycetemcomitans, Bacteroides forsythus, and Porphyromonas gingivalis. Techniques such as immunoassays, enzyme assays, and nucleic acid probe assays have been developed for the identification of A. actinomycetemcomitans, B. forsythus, and P. gingivalis (31). However, the techniques mentioned above require approximately between 103 to 105 targets per sample specimen.
PCR can lower the limit of bacterial detection. In recent years, there has been great interest in PCR-based tests which use the bacterial small-subunit 16S rRNA gene (16S rDNA) to detect bacterial pathogens. Nucleotide sequences of some portions of 16S rDNA have been highly conserved. However, other regions of this gene are hypervariable. Most tests have emphasized the detection of only a single species. However, sets of 16S rDNA-based primers can be combined to detect more than one species in a single patient sample. The general approach of combining multiple primers in a single reaction mixture is called multiplex PCR (5).
Multiplex PCR-based assays for the detection of periodontal pathogens have been reported (11, 27, 28). However, none of those assays can simultaneously detect A. actinomycetemcomitans, B. forsythus, and P. gingivalis. We previously published a description of a multiplex PCR that could detect A. actinomycetemcomitans and P. gingivalis (27). This paper presents an improvement on that technique, which now allows the more sensitive detection of all three periodontal pathogens by using one specific forward primer per species in combination with a single conserved reverse primer (i.e., a total of four primers) (Fig. 1).
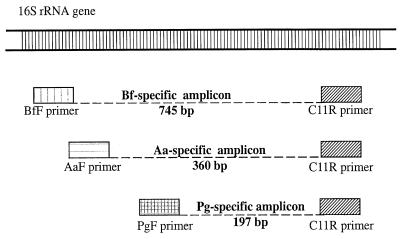
FIG. 1.
Multiplex PCR with conserved and species-specific 16S rDNA primers for simultaneous detection of A. actinomycetemcomitans (Aa), B. forsythus (Bf), and P. gingivalis (Pg). The drawing is a schematic of the location where the primers anneal to the bacterial 16S rDNA. The approximate sizes of the species-specific amplicons generated are also depicted. The 16S rDNA forward primer specific for A. actinomycetemcomitans is labeled AaF. BfF is the 16S rDNA forward primer specific for B. forsythus. PgF is the 16S rDNA forward primer specific for P. gingivalis. C11R is the 16S rDNA conserved (universal) reverse primer.
MATERIALS AND METHODS
Primer design and selection.
A literature search found a total of four published A. actinomycetemcomitans 16S rRNA-specific oligonucleotide probes, four B. forsythus 16S rRNA-specific oligonucleotide probes, and eight P. gingivalis 16S rRNA-specific oligonucleotide probes (1, 3, 6, 7, 9, 10, 17, 26). These probes were selected as possible species-specific forward primers. For the selection of the reverse primer, a total of seven prospective conserved (“universal”) 16S rDNA primers were identified (25). These reverse primers can hybridize to any bacterial 16S rDNA and can be combined with each species-specific forward primer to produce amplicons of different sizes that can be subsequently resolved on an agarose gel.
Suitable primers and PCR products were defined by using the program PRIME (Genetics Computer Group, Madison, Wis.). All 16S rDNA sequences of A. actinomycetemcomitans, B. forsythus, and P. gingivalis strains stored in the GenBank-EMBL database were used as DNA templates in PRIME. The strains used (GenBank accession numbers) were (i) A. actinomycetemcomitans ATCC 29522 (M75036), 29523 (M75038), 29524 (M75037), 33384T (M75039), and FDC Y4 (M75035), (ii) B. forsythus FDC 338 (L16495 and X73962), and (iii) P. gingivalis ATCC 33277 (L16492 and X73964). The specificities of the prospective forward primers were tested with the program FastA (Genetics Computer Group) against all existing DNA sequence information stored in two databases: GenBank-EMBL and the Ribosomal Database Project (16). No sequences completely homologous to prospective A. actinomycetemcomitans-, B. forsythus-, and P. gingivalis-specific forward primer sequences were found in 16S rDNA for over 100 other oral bacterial species in these databases. The conserved reverse primer C11R (see DNA sequence below) was selected because it had minimal differences in melting temperature compared with those of the three forward primers.
We identified a set of forward primers with minimal differences in their annealing temperatures. The calculated optimal annealing temperatures were 58.4, 58.9, and 60.7°C for the A. actinomycetemcomitans-, B. forsythus-, and P. gingivalis-specific primers, respectively. The expected product lengths were 360 bp for A. actinomycetemcomitans, 745 bp for B. forsythus, and 197 bp for P. gingivalis (Fig. 1). The nucleotide sequences of the four selected and modified 16S rDNA primers were as follows: A. actinomycetemcomitans-specific forward primer (AaF), 5′-ATT GGG GTT TAG CCC TGG TG-3′ (Escherichia coli positions 889 to 911); B. forsythus-specific forward primer (BfF), 5′-TAC AGG GGA ATA AAA TGA GAT ACG-3′ (E. coli positions 494 to 520); P. gingivalis-specific forward primer (PgF), 5′-TGT AGA TGA CTG ATG GTG AAA ACC-3′ (E. coli positions 1054 to 1078); and conserved reverse primer (C11R), 5′-ACG TCA TCC CCA CCT TCC TC-3′ (E. coli positions 1227 to 1246). The nucleotide positions given were obtained by aligning the sequences of A. actinomycetemcomitans ATCC 29522 (accession no. M75036), B. forsythus FDC 338 (accession no. L16495), and P. gingivalis ATCC 33277 (accession no. L16492) with that of E. coli (accession no. J01695) by using the subalign command from the Ribosomal Database Project (16). The selected oligonucleotide primers were synthesized by a commercial vendor (Life Technologies, Grand Island, N.Y.).
Preparation of bacterial DNA templates for multiplex PCR.
Bacteria and growth conditions were as described previously (27) (for the list of bacterial strains used, see the section Validation of Primer Specificity). DNA from bacterial cultures or plaque samples was extracted by use of the QIAamp Tissue Kit according to the manufacturer's instructions (Protocols for Bacteria; Qiagen Inc., Valencia, Calif.). Each plaque sample was eluted with 70 μl of Qiagen AE Buffer and was stored at −80°C.
Multiplex PCR with conserved and species-specific primers.
The approaches previously described by Chamberlain and Chamberlain (4) and Henegariu et al. (14) were used to optimize our modified multiplex PCR for the detection of A. actinomycetemcomitans, B. forsythus, and P. gingivalis. Optimization of the primer concentration was performed by mixing primers AaF, BfF, PgF, and C11R together at several concentrations between 0.25 and 1.2 μM with 106 cells each of A. actinomycetemcomitans, B. forsythus, and P. gingivalis. The primer concentrations given below yielded easily detectable amplicons for all three bacterial species. The AmpliTaq Gold (PE Biosystems, Foster City, Calif.) DNA polymerase concentration was optimized by testing concentrations between 2.5 and 12 U. Amplification bands other than the expected 197-, 360-, and 745-bp products did not occur with pure cultures. Therefore, we decided to keep the calculated optimal annealing temperature of 61°C used in our previous two-species multiplex PCR (27). The protocol described below was then used for all further experiments.
The volume of each multiplex PCR mixture was 53.6 μl (33.6 μl for the master mixture and 20 μl of extracted DNA stored in Qiagen AE buffer). A 50-μl mineral oil overlay was added. A hot-start step was included in our protocol by the use of AmpliTaq Gold. The amplification reaction mixture (master mixture) contained 10.3 mM Tris-HCl, 51.3 mM KCl (10× PCR Buffer II; PE Biosystems), 2.9 mM MgCl2, 0.15 μM primer AaF, 0.74 μM primer BfF, 0.49 μM primer PgF, 0.47 μM primer C11R, and 10 U of AmpliTaq Gold. The deoxynucleoside triphosphates included dATP, dCTP, and dGTP at each 200 μM and 600 μM dUTP (all deoxynucleoside triphosphates were from Boehringer Mannheim, Indianapolis, Ind.). The cycling parameters (cycling was performed with the model 480 Perkin-Elmer DNA thermal cycler) consisted of 40 cycles at the fastest ramp time available: 95°C for 1 min (except 10 min for the first cycle), 61°C for 1 min, and 72°C for 5 min (except 10 min for the last cycle). The samples were held at 4°C until analysis by agarose gel electrophoresis.
Each experiment included triplicate negative controls without template DNA and triplicate positive controls containing cells from pure cultures. For initial experiments, 100 cells of each species were used. For plaque samples, 50 A. actinomycetemcomitans and P. gingivalis cells and 500 B. forsythus cells were used. An additional positive control for the presence of bacteria was added in experiments with plaque samples (see below).
Post-multiplex PCR gel electrophoresis.
A 20-μl aliquot of amplified samples from each PCR tube was electrophoresed through a 2.3% agarose gel (NuSieve 3:1 Agarose; FMC Bioproducts, Rockland, Maine) in TBE (Tris-borate-EDTA) buffer for 1.8 h at 110 V. The amplification products were visualized and photographed under a UV light transilluminator (Fotodyne, Hartlands, Wis.) after 30 min of ethidium bromide (1 μg/ml) staining. The molecular sizes of the amplicons were determined by comparison to a commercial DNA molecular size marker (number XIV; Boehringer Mannheim). The photograph (Polapan 55 PN; Polaroid) of the gel was scored for the presence or absence of A. actinomycetemcomitans, B. forsythus, and P. gingivalis amplicons.
Determination of limit of detection.
The lower limit of detection was defined as the smallest number of bacteria in a sample that could be detected by our modified multiplex PCR. This was determined by 10-fold serial dilutions of mixed pure cultures of A. actinomycetemcomitans ATCC 29522, B. forsythus ATCC 43037, and P. gingivalis ATCC 33277 cells in phosphate-buffered saline (PBS). Dilutions were based on triplicate viable and microscopic counts. They ranged from 105 to 0 bacterial cells for each species.
Validation of primer specificity.
Primer specificity was defined as the ability of the primers to anneal specifically only to A. actinomycetemcomitans, B. forsythus, and P. gingivalis 16S rDNAs. The specificities of the primers were tested against the following organisms: (i) six A. actinomycetemcomitans strains (ATCC 29522, ATCC 29524, ATCC 33384, ATC 43717, ATCC 43718, and ATCC 43719), one B. forsythus strain (ATCC 43037), and four P. gingivalis strains (ATCC 33277 and clinical isolates W-50, 381, and 17-5) and (ii) eight different species of oral bacteria (Actinomyces naeslundii ATCC 12104, Campylobacter rectus ATCC 33238, Eikenella corrodens ATCC 43278, Fusobacterium nucleatum ATCC 10953, Prevotella intermedia ATCC 25611, Streptococcus crista ATCC 51110, Streptococcus sanguis ATCC 10556, and Treponema denticola ATCC 33520). The organisms were tested in suspensions containing greater than 106 cells of each species.
Plaque samples.
Supra- and subgingival plaque samples were collected from 20 dentists by procedures approved by the University of Minnesota Institutional Review Board. For each subject, one supragingival and one subgingival plaque sample were collected from sites of two anterior teeth that met all of the following criteria for good periodontal health: gingival index, 0 or 1; probing depth, ≤3 mm; and attachment level, ≤2 mm. Plaque samples were taken with sterile curettes, placed in 500 μl of PBS, and dispersed by sonication and vortexing. DNA was then extracted with the QIAamp Tissue Kit (Qiagen) as described above. Subgingival plaque samples from 10 subjects with early adult periodontitis were also analyzed. Those samples had been collected 10 years previously and had been stored at −20°C.
Each plaque sample was run in five PCRs as follows: (i) PCR with plaque sample alone (in triplicate tubes), (ii) PCRs with plaque sample spiked with 50 A. actinomycetemcomitans and P. gingivalis cells and 500 B. forsythus cells, and (iii) a nonmultiplex control PCR containing a pair of primers that target the Actinomyces naeslundii fimbrial gene (S1 primer, 5′-GTC CAC GTC TAC CCC AAG AAC-3′; AS1 primer, 5′-CAG GAA GAT GAC GCC GTT GGC A-3′; expected product length, 993 bp) (8). Because A. naeslundii is highly prevalent in supra- and subgingival plaque (22), its detection in a plaque sample would rule out the possibility that a sample was negative for A. actinomycetemcomitans, B. forsythus, and P. gingivalis because no bacteria were present.
RESULTS
Limit of detection.
In pure cultures, the multiplex PCR simultaneously detected as few as 10 A. actinomycetemcomitans and P. gingivalis cells and 100 B. forsythus cells (Fig. 2). Based on eight replicates, the reproducibility was as follows: at the 1-cell level, A. actinomycetemcomitans was detected five of eight times, B. forsythus was detected zero of eight times, and P. gingivalis was detected one of eight times; at the 10-cell level, A. actinomycetemcomitans was detected eight of eight times, B. forsythus was detected three of eight times, and P. gingivalis was detected eight of eight times; and at the 100-cell level, B. forsythus was detected eight of eight times.
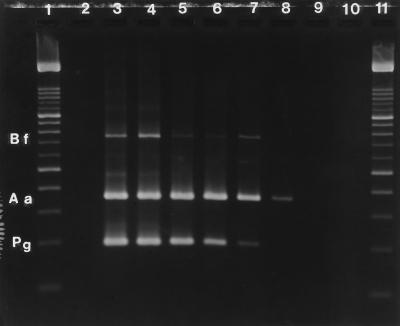
FIG. 2.
Limit of detection of multiplex PCR in pure cultures. Lane 2, negative control (PBS); lane 3, 105 cells each of A. actinomycetemcomitans, B. forsythus, and P. gingivalis diluted in PBS; lane 4, 104 cells of each species; lane 5, 1,000 cells of each species; lane 6, 100 cells of each species; lane 7, 10 cells of each species; lane 8, 1 cell of each species; lane 9, 0.1 cell of each species. The observed amplicons are at 360 bp for A. actinomycetemcomitans (Aa), 745 bp for B. forsythus (Bf), and 197 bp for P. gingivalis (Pg). In this gel, 1 A. actinomycetemcomitans cell, 10 B. forsythus cells, and 10 P. gingivalis cells are detected by the multiplex PCR. Lane 10, negative control (H2O); lanes 1 and 11, DNA molecular size marker XIV (Boehringer Mannheim) with bands at 100, 200, 300, 400, 500, 600, 700, 800, 900, 1,000, 1,100, 1,200, 1,300, 1,400, 1,500, and 2,642 bp. The 500- and 1,000-bp bands are two to three times brighter, as depicted on the gel.
We found an approximately fivefold decrease in sensitivity when B. forsythus was diluted into clinical plaque samples. For that reason we used 50 A. actinomycetemcomitans and P. gingivalis cells and 500 B. forsythus cells as a positive control when working with plaque samples (see below).
Specificities of primers.
Species-specific amplicons were observed for all A. actinomycetemcomitans, B. forsythus, and P. gingivalis strains tested. No A. actinomycetemcomitans-, B. forsythus-, or P. gingivalis-specific amplicons were seen for any of the other eight bacterial species. Furthermore, when 100 cells each of A. actinomycetemcomitans, B. forsythus, and P. gingivalis were added to mixtures of these eight species, A. actinomycetemcomitans-, B. forsythus-, and P. gingivalis-specific amplicons were detected (Fig. 3).
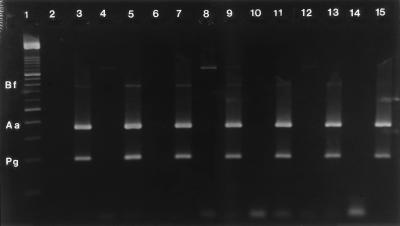
FIG. 3.
Specificity of multiplex PCR against other American Type Culture Collection strains of oral species. Lane 2, negative control (H2O); lane 3, positive control with 100 cells each of A. actinomycetemcomitans (amplicon observed at 360 bp), B. forsythus (745 bp), and P. gingivalis (197 bp); lanes 4, 6, 8, 10, 12, and 14, other species of oral bacteria at concentrations greater than 106 cells; lane 4, A. naeslundii ATCC 12104; a nonspecific (not expected) amplicon is observed at 1,200 bp; however, this does not affect the interpretation of the multiplex PCR; lane 6, F. nucleatum ATCC 10953; lane 8, P. intermedia ATCC 25611; lane 10, S. crista ATCC 51110; lane 12, S. sanguis ATCC 10556; lane 14, T. denticola ATCC 33520; lanes 8, 9, and 12, nonspecific amplicon observed at 1,200 bp; lanes 5, 7, 9, 11, 13, and 15, other species of oral bacteria, as mentioned above, spiked with 100 cells each of A. actinomycetemcomitans, B. forsythus, and P. gingivalis; for example, lane 5 contains 106 cells of A. naeslundii spiked with 100 cells each of A. actinomycetemcomitans, B. forsythus, and P. gingivalis; lane 1, DNA molecular size marker with bands at 100, 200, 300, 400, 500, 600, 700, 800, 900, 1,000, 1,100, 1,200, 1,300, 1,400, 1,500, and 2,642 bp.
Plaque samples.
A. actinomycetemcomitans, B. forsythus, and P. gingivalis were detected in 6 of 20, 1 of 20, and 11 of 20 supragingival plaque samples from periodontally healthy sites, respectively (Fig. 4). For the subgingival plaque samples, the results were 4 of 20, 7 of 20, and 13 of 20 samples, respectively. A. actinomycetemcomitans, B. forsythus, and P. gingivalis were detected in 7 of 10, 10 of 10, and 7 of 10 of the subgingival plaques obtained from early adult periodontitis subjects, respectively. By Fisher's exact test, there were statistically significant differences between subgingival sites for healthy subjects and subjects with periodontitis for the prevalence of A. actinomycetemcomitans (P = 0.01) and B. forsythus (P = 0.0006) but not P. gingivalis (P = 0.56). To confirm primer specificity, all plaque samples were spiked with 50 A. actinomycetemcomitans and P. gingivalis cells and 500 B. forsythus cells. Amplicons specific for A. actinomycetemcomitans, B. forsythus, and P. gingivalis were detected in 95% of these artificially infected supra- and subgingival plaque samples.
FIG. 4.
Multiplex PCR of supra- and subgingival plaque samples. Each plaque sample was run in three different ways (as described in the Materials and Methods section). Lane 2, negative control (H2O); lane 3, positive control with 50 cells of A. actinomycetemcomitans (Aa amplicon observed at 360 bp) and P. gingivalis (Pg amplicon observed at 197 bp) and 500 cells B. forsythus (Bf amplicon observed at 745 bp); lane 4, a supragingival plaque sample showing no A. actinomycetemcomitans, B. forsythus, or P. gingivalis amplicon; lane 5, the same supragingival plaque sample used for lane 4 run with A. naeslundii-specific primers, this shows that A. naeslundii (An) is present in that plaque sample at 993 bp; a faint nonspecific amplicon appears at about 600 bp; lane 6, the same supragingival plaque sample artificially infected (spiked) with 50 cells each of A. actinomycetemcomitans and P. gingivalis and 500 cells of B. forsythus; all three bacterial species spiked are detected; lane 7, a subgingival plaque sample showing the presence of A. actinomycetemcomitans and B. forsythus amplicons; lane 8, the same plaque sample run with A. naeslundii-specific primers; lane 9, the same plaque sample spiked with 50 cells each of A. actinomycetemcomitans and P. gingivalis and 500 cells of B. forsythus; lane 10, a subgingival plaque showing the presence of B. forsythus and P. gingivalis amplicons; lane 11, the same plaque sample run with A. naeslundii primers; lane 12, the same plaque sample spiked with 50 cells each of A. actinomycetemcomitans and P. gingivalis and 500 cells of B. forsythus; lane 13, a subgingival plaque sample showing the presence of A. actinomycetemcomitans and P. gingivalis amplicons; lane 14, the same plaque sample used for lane 13 run with A. naeslundii-specific primers; lane 15, negative control (H2O) with A. naeslundii-specific primers; lane 1, DNA molecular size marker with bands at 100, 200, 300, 400, 500, 600, 700, 800, 900, 1,000, 1,100, 1,200, 1,300, 1,400, 1,500, and 2,642 bp.
DISCUSSION
Time, effort, and cost can be saved by amplifying multiple sequences in a single reaction tube. Wahlfors and coworkers (28) reported a multiplex PCR that can detect 5 to 50 A. actinomycetemcomitans and P. gingivalis cells per sample. In a subsequent publication (21), they tried to add a B. forsythus-specific primer pair to their two-species multiplex PCR mixture. However, they were unable to consistently detect that species. The approach that they were pursuing would have required a total of six primers for the detection of A. actinomycetemcomitans, B. forsythus, and P. gingivalis. We introduced the technique of using one specific forward primer per species, in combination with a single conserved (universal) reverse primer (i.e., a total of four primers) (27). This strategy has been embraced successfully by other multiplex PCR-based assays (11, 13).
To date, in the field of periodontology, only Garcia and coworkers (11) have succeeded in simultaneously detecting three periodontal pathogens (A. actinomycetemcomitans, P. gingivalis, and Prevotella intermedia). However, their assay does not detect B. forsythus. This periodontal pathogen has been reported to be an important prognostic factor in longitudinal studies (20). Subjects who have periodontitis and who harbor B. forsythus at the baseline were found to be at seven times greater risk for increased pocket depth. Our modified multiplex PCR assay could detect as few as 10 A. actinomycetemcomitans and P. gingivalis cells and 100 B. forsythus cells in pure cultures and could detect 50 A. actinomycetemcomitans and P. gingivalis cells and 500 B. forsythus cells in plaque samples. Our approach is simple, cost-effective, highly sensitive, and specific and conserves samples in limited supply, while it allows the simultaneous detection of A. actinomycetemcomitans, B. forsythus, and P. gingivalis. Our modifications have also made this three-species multiplex PCR more sensitive than our original two-species assay. Previously, we could detect only A. actinomycetemcomitans and P. gingivalis in 2 of the 20 dentists' subgingival plaque samples also used here (27). This increased sensitivity might be due to the use of a greater amount of AmpliTaq Gold in a larger volume of plaque with a more sensitive DNA extraction kit (QIAamp Tissue Kit) (15, 19, 23).
Our present multiplex PCR assay does not have the capability for precise quantitative measurements of cell numbers in plaque samples. As an example of variation between runs, the bands that represent 50 cells of A. actinomycetemcomitans and P. gingivalis seen in Fig. 4 (lane 3) were brighter than the bands that represent 100 cells of A. actinomycetemcomitans and P. gingivalis seen in Fig. 3 (lane 3). In the same lanes, a fivefold difference in the number of B. forsythus cells was detectable as a difference in band intensities. Although crude estimates of cell numbers in plaque samples can be made by comparing band intensities to those observed for cell dilution series, a truly quantitative multiplex PCR would require internal standards designed to be added to every sample (18, 24).
Our main reason for analyzing both supra- and subgingival plaque samples from healthy subjects and subgingival plaque samples from patients with periodontitis was to test the multiplex assay under conditions that might be encountered clinically. Although the numbers of samples were small, the prevalence of two putative pathogens was significantly higher in periodontitis sites. That finding generally agrees with clinical studies that have used other techniques (12, 29). The fact that healthy and periodontitis sites were distinguished for the presence or absence of A. actinomycetemcomitans and B. forsythus suggests that the multiplex PCR will give useful information in future clinical studies.
It is still unclear if A. actinomycetemcomitans, B. forsythus, and P. gingivalis are exogenous pathogens or endogenous members of the microflora that could become pathogens in the presence of other host factors such as genetic susceptibility and smoking (30). In order to investigate this question, it is important to know whether small numbers of A. actinomycetemcomitans, B. forsythus, and P. gingivalis are typically found in healthy periodontal sites. Because these three bacterial species are anaerobes, they could become nonviable after sampling, during transport and analysis; thus, a technique such as culturing would underestimate the presence (numbers) of those pathogens in periodontal pockets. PCR-based assays have the advantage of detecting both viable and nonviable bacteria, whereas culturing techniques can detect only viable organisms. The high degree of sensitivity of PCR-based techniques allow us to investigate the natural distribution and localize reservoirs of A. actinomycetemcomitans, B. forsythus, and P. gingivalis in clinically healthy subjects. The information obtained can be used to select appropriate and effective preventive and interventional therapies to control the progression and the transmission of periodontal diseases. For example, if A. actinomycetemcomitans, B. forsythus, and P. gingivalis are “exogenous” pathogens, tests with a low limit of detection (such as PCR) would be valuable. The detection of these pathogens in the plaques of healthy patients could identify those subjects who are at a higher risk for periodontitis. This reservoir of putative pathogens can constitute a risk for transmission to other periodontally healthy partners of the infected patient.
Uses for our modified multiplex PCR-based technique thus include identification of subjects (or teeth) at risk for the onset or progression of periodontitis and evaluation of the success of periodontal therapy. Also, it may be possible for detection of other periodontal pathogens (e.g., Campylobacter rectus, Fusobacterium nucleatum, P. intermedia, Peptostreptococcus micros, and Treponema denticola) by adding more species-specific primers to this multiplex PCR mixture. In conclusion, our findings show that the multiplex PCR with conserved and specific 16S rDNA primers is highly sensitive and specific, while it allows the simultaneous detection of A. actinomycetemcomitans, B. forsythus, and P. gingivalis. The simultaneous detection of those three periodontal pathogens is an advantage of this technique over conventional PCR-based assays.
ACKNOWLEDGMENTS
This work was supported by NIH/NIDCR grants 2R01 DE07233 and 2T32 DE07014.
Special thanks go to Christopher Larson for technical assistance with the early stages of this project.
REFERENCES
- 1.Albandar J M, Lyngstadaas S P. PCR primers for the detection of Actinobacillus actinomycetemcomitans. J Dent Res. 1995;74(Special issue, abstracts of papers):174. . (Abstr. 1299.) [Google Scholar]
- 2.American Academy of Periodontology. Glossary of periodontal terms. 3rd ed. Chicago, Ill: American Academy of Periodontology; 1992. [Google Scholar]
- 3.Ashimoto A, Chen C, Bakker I, Slots J. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol Immunol. 1996;11:266–273. doi: 10.1111/j.1399-302x.1996.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 4.Chamberlain J S, Chamberlain J R. Optimization of multiplex PCRs. In: Mullis K B, Ferre F, Gibbs R A, editors. The polymerase chain reaction. Boston, Mass: Birkhauser; 1994. pp. 38–46. [Google Scholar]
- 5.Chamberlain J S, Gibbs R A, Ranier J E, Caskey C T. Multiplex PCR for the diagnosis of Duchenne muscular dystrophy. In: Innis M, Gelfand D, Sninski J, White T, editors. PCR protocols: a guide to methods and applications. Orlando, Fla: Academic Press, Inc.; 1990. pp. 272–281. [Google Scholar]
- 6.Choi B K, Paster B J, Dewhirst F E, Gobel U B. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect Immun. 1994;62:1889–1895. doi: 10.1128/iai.62.5.1889-1895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuba P J, Pelz K, Krekeler G, De Isele T S, Gobel U. Synthetic oligodeoxynucleotide probes for the rapid detection of bacteria associated with human periodontitis. J Gen Microbiol. 1988;134:1931–1938. doi: 10.1099/00221287-134-7-1931. [DOI] [PubMed] [Google Scholar]
- 8.Dale P, Johnson J, Schachtele C. Detection of fimbrial genes in Actinomyces with polymerase chain reaction. J Dent Res. 1996;75(Special Issue, abstracts of papers):94. . (Abstr. 609.) [Google Scholar]
- 9.Dewhirst F E, Paster B J. DNA probe analyses for the detection of periodontopathic bacteria in clinical samples. In: Hamada S, Holt S C, McGhee J R, editors. Periodontal disease: pathogens and host immune responses. Tokyo, Japan: Quintessence; 1991. pp. 367–377. [Google Scholar]
- 10.Dix K, Watanabe S M, McArdle S, Lee D I, Randolph C, Moncla B, Schwartz D E. Species-specific oligodeoxynucleotide probes for the identification of periodontal bacteria. J Clin Microbiol. 1990;28:319–323. doi: 10.1128/jcm.28.2.319-323.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia L, Tercero J C, Legido B, Ramos J A, Alemany J, Sanz M. Rapid detection of Actinobacillus actinomycetemcomitans, Prevotella intermedia, and Porphyromona gingivalis by multiplex PCR. J Periodont Res. 1998;33:59–64. doi: 10.1111/j.1600-0765.1998.tb02292.x. [DOI] [PubMed] [Google Scholar]
- 12.Haffajee A D, Cugini M A, Tanner A, Pollack R P, Smith C, Kent R L, Jr, Socransky S S. Subgingival microbiota in healthy, well-maintained elder and periodontitis subjects. J Clin Periodontol. 1998;25:346–353. doi: 10.1111/j.1600-051x.1998.tb02454.x. [DOI] [PubMed] [Google Scholar]
- 13.Hendolin P H, Markkanen A, Ylikoski J, Wahlfors J J. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J Clin Microbiol. 1997;35:2854–2858. doi: 10.1128/jcm.35.11.2854-2858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henegariu O, Heerema N A, Dlouhy S R, Vance G H, Vogt P H. Multiplex PCR: critical parameters and step-by-step protocol. BioTechniques. 1997;23:504–511. doi: 10.2144/97233rr01. [DOI] [PubMed] [Google Scholar]
- 15.Kebelmann-Betzing C, Seeger K, Dragon S, Schmitt G, Moricke A, Schild T A, Henze G, Beyermann B. Advantages of a new Taq DNA polymerase in multiplex PCR and time-release PCR. BioTechniques. 1998;24:154–158. doi: 10.2144/98241pf01. [DOI] [PubMed] [Google Scholar]
- 16.Larsen N, Olsen G J, Maidak B L, McCaughey M J, Overbeek R, Macke T J, Marsh T L, Woese C R. The ribosomal database project. Nucleic Acids Res. 1993;21(Suppl.):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leys E J, Griffen A L, Strong S J, Fuerst P A. Detection and strain identification of Actinobacillus actinomycetemcomitans by nested PCR. J Clin Microbiol. 1994;32:1288–1294. doi: 10.1128/jcm.32.5.1288-1294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lie Y S, Petropoulos C J. Advances in quantitative PCR technology: 5′ nuclease assays. Curr Opin Biotechnol. 1998;9:43–48. doi: 10.1016/s0958-1669(98)80082-7. [DOI] [PubMed] [Google Scholar]
- 19.Loffler J, Hebart H, Schumacher U, Reitze H, Einsele H. Comparison of different methods for extraction of DNA of fungal pathogens from cultures and blood. J Clin Microbiol. 1997;35:3311–3312. doi: 10.1128/jcm.35.12.3311-3312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Machtei E E, Dunford R, Hausmann E, Grossi S G, Powell J, Cummins D, Zambon J J, Genco R J. Longitudinal study of prognostic factors in established periodontitis patients. J Clin Periodontol. 1997;24:102–109. doi: 10.1111/j.1600-051x.1997.tb00474.x. [DOI] [PubMed] [Google Scholar]
- 21.Meurman J H, Wahlfors J, Korhonen A, Alakuijala P, Vaisanen P, Torkko H, Janne J. Identification of Bacteroides forsythus in subgingival dental plaque with the aid of a rapid PCR method. J Dent Res. 1997;76:1376–1380. doi: 10.1177/00220345970760070701. [DOI] [PubMed] [Google Scholar]
- 22.Moore W E C, Moore L V H. The bacteria of periodontal diseases. Periodontology 2000. 1994;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 23.Moretti T, Koons B, Budowle B. Enhancement of PCR amplification yield and specificity using AmpliTaqTM DNA polymerase. BioTechniques. 1998;25:716–722. [PubMed] [Google Scholar]
- 24.Orlando C, Pinzani P, Pazzagli M. Developments in quantitative PCR. Clin Chem Lab Med. 1998;36:255–269. doi: 10.1515/CCLM.1998.045. [DOI] [PubMed] [Google Scholar]
- 25.Relman D A. Universal bacterial 16S rDNA amplification and sequencing. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 489–495. [Google Scholar]
- 26.Socransky S S, Smith C, Martin L, Paster B J, Dewhirst F E, Levin A E. “Checkerboard” DNA-DNA hybridization. BioTechniques. 1994;17:788–792. [PubMed] [Google Scholar]
- 27.Tran S D, Rudney J D. Multiplex PCR using conserved and species-specific 16S rRNA gene primers for simultaneous detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. J Clin Microbiol. 1996;34:2674–2678. doi: 10.1128/jcm.34.11.2674-2678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wahlfors J, Meurman J H, Vaisanen P, Alakuijala P, Korhonen A, Torkko H, Janne J. Simultaneous detection of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis by a rapid PCR method. J Dent Res. 1995;74:1796–1801. doi: 10.1177/00220345950740111301. [DOI] [PubMed] [Google Scholar]
- 29.Wolff L F, Aeppli D M, Pihlstrom B L, Anderson L, Stoltenberg J L, Osborn J B, Hardie N A, Shelburne C E, Fisher G E. Natural distribution of 5 bacteria associated with periodontal disease. J Clin Periodontol. 1993;20:699–706. doi: 10.1111/j.1600-051x.1993.tb00694.x. [DOI] [PubMed] [Google Scholar]
- 30.Zambon J J. Periodontal diseases: microbial factors. Ann Periodontol. 1996;1:879–925. doi: 10.1902/annals.1996.1.1.879. [DOI] [PubMed] [Google Scholar]
- 31.Zambon J J, Haraszthy V I. The laboratory diagnosis of periodontal infections. Periodontology 2000. 1995;7:69–82. doi: 10.1111/j.1600-0757.1995.tb00037.x. [DOI] [PubMed] [Google Scholar]