Abstract
Antibiotic resistance is a global health challenge, involving the transfer of bacteria and genes between humans, animals and the environment. Although multiple barriers restrict the flow of both bacteria and genes, pathogens recurrently acquire new resistance factors from other species, thereby reducing our ability to prevent and treat bacterial infections. Evolutionary events that lead to the emergence of new resistance factors in pathogens are rare and challenging to predict, but may be associated with vast ramifications. Transmission events of already widespread resistant strains are, on the other hand, common, quantifiable and more predictable, but the consequences of each event are limited. Quantifying the pathways and identifying the drivers of and bottlenecks for environmental evolution and transmission of antibiotic resistance are key components to understand and manage the resistance crisis as a whole. In this Review, we present our current understanding of the roles of the environment, including antibiotic pollution, in resistance evolution, in transmission and as a mere reflection of the regional antibiotic resistance situation in the clinic. We provide a perspective on current evidence, describe risk scenarios, discuss methods for surveillance and the assessment of potential drivers, and finally identify some actions to mitigate risks.
Subject terms: Antimicrobial resistance, Microbial ecology, Bacterial infection
In this Review, Larsson and Flach discuss the drivers of and bottlenecks for environmental evolution and transmission of antibiotic resistance, and they explore environmental surveillance strategies that could complement clinical surveillance systems.
Introduction
Many bacterial species evolved the ability to tolerate antibiotics long before humans started to mass-produce them to prevent and treat infectious diseases1,2. Isolated caves2, permafrost cores1, and other environments and specimens that have been preserved from anthropogenic bacterial contamination3,4 can provide insights into the resistance mechanisms that prevailed during the pre-antibiotic era. An important driver of the ancient and still ongoing evolution of resistance mechanisms is likely to be the never-ending competition for resources among microorganisms, including the natural production of secondary metabolites that are similar to many of the antibiotics used today as pharmaceuticals5–7. The relatively recent introduction of antibiotics as clinical agents radically changed the preconditions for the evolution and spread of resistance by providing unprecedented selection pressures, especially on members of the microbiota of humans and domestic animals, but also in environments heavily polluted with antibiotics. This selection pressure has promoted the mobilization and horizontal transfer of a large range of antibiotic resistance genes (ARGs)8 to many bacterial species, particularly to those causing disease. The ultimate, well-known consequences of such accumulating evolutionary events are gradually increasing difficulties to prevent and treat bacterial infections. As bacteria and genes often cross environments and species boundaries, it is critical to understand and acknowledge the connections between the human, animal and environmental microbiota (the One Health Concept9,10) to manage this global health challenge11–23. In this Review, we describe our current understanding of the role of the environment in the evolution of resistance and as a route for transmission of resistant bacteria that already circulate in humans. We elaborate on how studies of resistance in the environment could provide a reflection of the regional clinical resistance situation, thereby complementing traditional surveillance. Furthermore, we provide a critical account of the methods used to study antibiotic resistance in the environment, particularly with regard to the assessment of selection pressures. Finally, we identify some principles that could guide strategies to reduce risks, with particular focus on challenges in low- and middle-income countries (Box 1) and emissions from antibiotic manufacturing.
Box 1 The antibiotic resistance crisis in low- and middle-income countries.
Many low- and middle-income countries (LMICs) are particularly vulnerable to the antibiotic resistance crisis. This is because of, for example, limited surveillance and diagnostic opportunities, less-controlled use of antibiotics in both humans and animals, overcrowding in hospitals, insufficient hygiene control, often rapidly growing meat and fish production, an overall greater infection burden, and limited access to expensive, second-line or third-line antibiotics167. The environmental dimensions can also be more important in these regions, for example, as a consequence of inferior infrastructure for managing human and animal waste streams, leading to greater environmental emissions of both resistant faecal bacteria and residual antibiotics168,169. Owing to low production costs, China and India have become the world’s largest producers of antibiotics. Insufficient waste management and excessive emissions of antibiotic residues from manufacturing have been reported in those countries, as well as in other regions of the world58,61. Management is often more challenging in LMICs than in high-income countries because of more limited resources, other pressing basal needs that need to be addressed, and a weaker governance of and trust in the public sector. Resolving the resistance crisis in LMICs is needed, not only for the LMICs themselves but also because resistant bacteria do not recognize borders. Resource-efficient management should therefore include intensified actions in LMICs. Such initiatives may often overlap with strategies to improve water quality, sanitation and hygiene21. Sewage surveillance to assess the clinical resistance situation probably also has its greatest future potential in LMICs being less resource-demanding than traditional clinical surveillance systems.
Resistance evolution in the environment
Antibiotic resistance can arise both from mutations in the pre-existing genome of a bacterium and from the uptake of foreign DNA. Mutations readily occur and become fixed in the patient or animal treated with the antibiotic. Such a strong selection pressure on pathogens is rarer elsewhere. The process is also independent of the genetic reservoir in other species. Hence, external environments are generally less likely to provide a major contribution to mutation-based evolution of resistance for most pathogens. With regard to uptake of novel resistance factors, water, soil and other environments with highly variable ecological niches provide an unmatched gene pool with a diversity that greatly exceeds that of the human and domestic animal microbiota24,25. Indeed, the most striking feature of the environmental microbiome is its immense diversity, providing numerous genes that potentially could be acquired and used by pathogens to counteract the effect of antibiotics26–30. All approved antibiotic classes so far, whether they be natural, semi-synthetic or synthetic compounds, have been met by resistance in at least some of the pathogens they target. This suggests that external environments already harbour resistance factors for all antibiotics that will ever be developed, unless we start thinking radically differently about how antibiotics are designed.
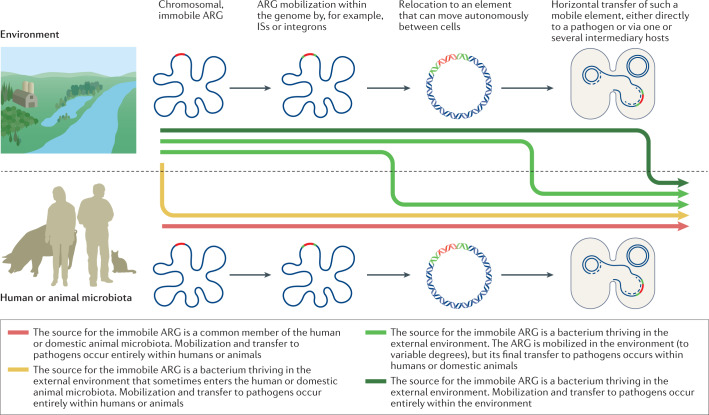
Over the eons, most ARGs have probably evolved gradually from genes with other functions31,32. The more recent evolutionary events responsible for their widespread occurrence in pathogens are mainly a result of transfer events from ancestral species in which the overall functionality of the genes was shaped. Starting from a chromosomal, immobile ARG, there is typically a stepwise evolution that leads to acquired resistance in a pathogen (Fig. 1). The first step is often the ability of an ARG to move within the genome, attained, for example, by association with insertion sequences33,34, or the formation of gene cassettes and incorporation into integrons35,36. The second step involves the relocation of the gene to an element that can move autonomously between cells, such as a plasmid or an integrative conjugative element. Some environments are probably more likely than others to provide the various genetic elements typically involved in mobilization and transfer of ARGs, either through the presence of faecal bacteria known to often carry such elements or possibly because the conditions (including reoccurring stress) favour frequent gene exchanges37,38. The third step is the horizontal transfer of a mobilized resistance gene, either directly to a pathogen or via one or several intermediary bacterial hosts. The fourth step, which may occur at any time in the process, is the physical transfer of the bacterium carrying the ARG to the human or domestic animal microbiota, an ability described by the term ‘ecological connectivity’39. High metabolic activity and extensive cell-to-cell contact (such as in biofilms) are probably increasing the rate of most steps. All of these steps, including mobilization by, for example, insertion sequences40 or integrons41, increases in donor cell abundance and thus transfer opportunities, and the rate of horizontal gene transfer (HGT)42,43, may be promoted by antibiotics. Importantly, though, most if not all steps also occur in the absence of antibiotics, but at different rates44,45. Hence, it is crucial to understand where the bottlenecks are in the evolution towards resistance in pathogens. A critical bottleneck is likely to be the selection of the rare genotypes with acquired resistance that result from mobilization and/or HGT, genotypes that otherwise would disappear46. At all stages, compensatory mutations somewhere in the genome of the bacterium carrying the ARG may occur, lowering potential fitness costs, either through reducing niche overlap or by increasing competitive ability47. The emergence of new ARGs in the clinic occurs only when all events align in time and space48.
Fig. 1. The role of the environment in the emergence of new resistance genes in pathogens.
Conceptual illustration of how evolution leading to the emergence of a new antibiotic resistance gene (ARG; red) in pathogens can involve the environment and/or the human/domestic animal microbiota to different extents. The evolution typically occurs in steps, as indicated by the grey arrows. The first can be the association of a chromosomal ARG (red) with, for example, insertions sequences (ISs; green), which provide intracellular mobility. Intracellular relocation to, for example, a plasmid allows the ARG to move horizontally across strains and species. The mobilized ARG can then be transferred to a pathogen in one or several steps. In the most extreme cases, all genetic steps occur in either the environment (top) or in the human or domestic animal microbiota (bottom). However, at any stage bacteria carrying the ARG may move physically from the environment to the human or domestic animal microbiota, as illustrated by the differently coloured, thick arrows. The genetic reservoir is considerably larger in the environment, suggesting that the source for new ARGs is often environmental bacteria. By contrast, reoccurring, strong antibiotic selection pressures and close contact with pathogens are more common in humans and domestic animals, although some external environments also share those drivers. Environmental release of faecal bacteria may also boost the evolutionary process by providing genetic elements that are adapted to capture and transfer ARGs. How common the different depicted scenarios are is still largely unknown. A better understanding of how often the different evolutionary steps occur in the environment versus the human or domestic animal microbiota and what drivers are most important would enable more efficient resource allocation to limit or delay the emergence of new ARGs in pathogens.
In principle, all, some or none of the evolutionary steps could occur in the external environment. Of 22 ARGs with strong evidence for their recent origin down to species level, 21 came from species that at least occasionally are associated with infections in humans and/or domestic animals49. This strong over-representation is coherent with the hypothesis that human and/or domestic animals provide the most important environments for resistance evolution under a selection pressure from antibiotics. That said, for the overwhelming majority of ARGs, their recent origin is not known, quite possibly because they originate from environmental species that have not been sequenced yet. This alternative hypothesis speaks in favour of a much greater role of the external environment.
While the introduction of new ARGs to pathogens is worrying, changes in the genetic context around ARGs that affect the level of resistance, co-selection opportunities, or virulence or transmission potential can also add to the resistance challenge. The consequences of evolutionary events leading to the emergence of pathogens with new, successful resistance genotypes through any of these routes differ profoundly from those of transmission events of already widely circulating genotypes (as described later). Even single events can lead to the irreversible50 global spread of a new genotype that is it more challenging to treat. Compared with transmission, critical evolutionary events are rare and to some extent unique in nature, which is why they are more difficult to predict. Nonetheless, the benefits from being able to delay or prevent their emergence can be substantial.
Pollution as a driver
Although the natural production of antibiotic molecules most likely contributed to the (more ancient) evolution of ARGs5, it is not responsible for the rapid evolutionary expansion and spread of resistance factors across strains, species and environments that we have observed since the introduction of antibiotics as therapeutic agents. Antibiotics produced by environmental microorganisms are widespread, but act largely on a microscale, as concentrations characteristically would be expected to drop rapidly around the producing organisms, hence limiting exposure. Man-made antibiotics, on the other hand, act on a macroscale and are typically associated with selection pressures across entire microbial communities.
Antibiotics reach the environment via excretions (urine and faeces) from humans and domestic animals51–53, through improper disposal and/or handling of unused drugs54, through direct environmental contamination in aquaculture55,56 or plant production57, and via waste streams from the production of antibiotics58–62 (Fig. 2). Undoubtedly, the most widespread emissions, and quite plausibly the largest proportion of released antibiotics, are the result of use and excretion. At the same time, exposure levels via this route are always limited by, for example, the proportion of the population that is using the antibiotic at a given time, the doses used and metabolism in the human or domestic animal.
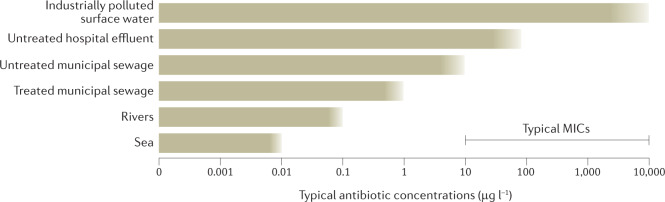
Fig. 2. Antibiotic concentrations in selected aquatic environments.
Different types of sources of antibiotic pollution typically give rise to different levels of exposure to aquatic bacterial communities. This, in turn, provides a reflection of the probability of environmental selection. Although very much a simplification, the ranges of typical antibiotic concentrations in aquatic environments exposed to excreted antibiotics from human use are depicted for the sea, rivers, treated and untreated municipal sewage effluents and untreated hospital effluents. Sea and river environments refer to those contaminated with treated municipal sewage. In addition, surface waters polluted directly by wastewater from drug manufacturing are included. As a comparison, typical minimal inhibitory concentrations (MICs) for many antibiotic–pathogen combinations often fall within the 10–10,000 µg l−1 range. As both depicted environmental concentrations and typical MICs are simplified illustrations representing many different antibiotics, an overlap between the two is not necessarily evidence of selection, unless there is overlap also for individual antibiotics. Note also that selection may occur at concentrations below the MIC.
With some exceptions, environmental concentrations of antibiotics are low, much lower than the minimal inhibitory concentrations (MICs) and most often lower than concentrations predicted (or shown) to select for resistant strains in the laboratory63,64. Although the concentration of antibiotics and the abundance of ARGs often correlate in environmental samples, in many cases this can be explained simply by different levels of pollution with human excreta, which is a source for both, rather than on-site selection of resistant bacteria in the environment by the antibiotic residues65. Still, the concentrations suspected to select for resistance (see later) are exceeded in many places, such as in sewage treatment plants14,63 suggesting, but not proving, that antibiotic pollution plays a role in the evolution of resistance. Although some studies report increases in the relative abundance of certain ARGs in such environments, it is difficult to distinguish whether this is a result merely of taxonomic changes, unrelated to antibiotic selection pressures, or from direct selection of resistant strains within species66–68. Although plausible, definite evidence for such direct selection in sewage treatment plants is still lacking, and some evidence points to the opposite69. A recent study on sterile-filtered wastewaters indicated no selective effect of the investigated treated municipal effluent and a small selective effect by untreated influent. By contrast, untreated hospital wastewater strongly selected for multiresistant Escherichia coli in different controlled exposure experiments with individual isolates and communities70. The exact selective agents responsible therein could not be identified, but the relatively high levels of antibiotics in hospital wastewater make them plausible drivers of resistance selection.
The collective evidence for resistance selection in environments with very high levels of antibiotics (pollution from antibiotic manufacturing) is considerably stronger than that for excreted antibiotics. This is based on the exceedance of selective concentrations by orders of magnitude at industrially polluted sites58,60–62, increased relative abundance of resistant bacteria61,71, and considerable increases in the number of ARGs60,72, including previously unknown ARGs73, which are not accompanied by increases in faecal contamination65.
In solid or semi-solid media, such as sediments, soils, and sewage sludge, reported concentrations can often be much higher than in aqueous media51,55,74,75. Still, in most cases, only a minor fraction is bioavailable. Despite, for example, ciprofloxacin being found in sewage sludge at milligram per kilogram concentrations, ciprofloxacin-sensitive strains are very common in sludge76,77, which suggests that the antibiotic is largely biologically unavailable here. Bioavailability in solid or semi-solid media can be inconsistent and depends strongly on physicochemical characteristics, including, for example, organic content and structure as well as the nature of the antibiotic78. Estimating the bioavailable fraction in such samples is challenging79, but genetically engineered reporter strains may provide a partial solution80.
Numerous studies have shown that the abundance of resistant bacteria and/or ARGs increase after manure from antibiotic-treated animals is added as a fertilizer to farmland. However, in many cases it is not possible to assign such increases to selective effects of antibiotic residues in the soil, as the added manure also carries resistant bacteria. In a recent study81, collected manure was spiked with antibiotics after collection, and the researchers observed selection of a fluorescently labelled Acinetobacter baylyi strain carrying a resistance plasmid compared with a similarly labelled non-resistant strain in the amended soil. Owing to the experimental design, it was possible to demonstrate within species selection by the antibiotics in the soil, although the study authors note that added antibiotic concentrations were higher than under normal fertilization regimens.
Metals and antibacterial biocides can, in many cases, co-select for antibiotic-resistant strains via cross-resistance (that is, via the same mechanism) or co-resistance (that is, via genetically linked mechanisms)82,83. However, evidence suggests that it is the historical exposure to antibiotics, rather than metal or biocide exposure, that has led to the current co-occurrence of metal and biocide resistance genes and ARGs on plasmids, as abundant co-occurrence is largely restricted to communities that have been shaped by strong antibiotic selection pressures — the human and domestic animal microbiota84. This does not exclude the possibility that metals and biocides could have an important role in maintaining strains that have already developed co-resistance, regardless of their prior evolutionary history. The concentrations needed for selection or co-selection are even less studied for metals and biocides, and need further attention. Some biocides can accelerate the rate of HGT42,85 as is the case for certain antibiotics43,86 and other pharmaceuticals87. Still, many naturally occurring stressors also accelerate HGT44. As stress-induced HGT is not a new phenomenon, it is unclear whether the induction of HGT by environmental pollutants has a discernible role in the rapid development of resistance in pathogens observed during the antibiotic era. Direct selection and co-selection are likely to be more critical.
Environmental contamination with faecal bacteria provides physical contact, and thus increased opportunities for gene exchange between resident environmental bacteria and bacteria adapted to the intestinal tract of human or domestic animals. Many intestinal bacteria are also known carriers of genetic elements (plasmids, integrative conjugative elements, insertion sequences, transposons or integrons) that can facilitate the acquisition of genes and their transfer to pathogens88,89. Experiments with fluorescently labelled E. coli cells added to soils demonstrate their ability to rapidly acquire resistance determinants from the soil microbiota90.
In addition, it is also a possibility that ARGs present in faecal bacteria that are introduced into the environment could contribute to clinically relevant evolution of resistance by being transferred horizontally in one or several steps to pathogens, which might ultimately infect humans. However, the probability of these events lining up is likely to be much higher within the human or domestic animal microbiota, as selection pressures, commensals and pathogens are more commonly encountered together, and there are no environmental transmission barriers that need to be overcome91. We therefore argue that the clinical significance of resistance evolution resulting from the environmental release of ARGs that are already commonly encountered in the human microbiota is probably limited91.
Environmental transmission
The environment can provide a route for some resistant bacteria to colonize or infect hosts92–94. In this Review, we refer to this as a ‘transmission event’, whereas changes in their DNA sequence, including genetic transmission across bacterial species, are categorized as ‘evolution events’ (see earlier). For a resistant pathogen that is already widely circulating among humans, the consequence of a single transmission event to another individual is much more limited than for an evolutionary event leading to the emergence of a new, successful resistance genotype in pathogens, with potentially global consequences (Fig. 3). By contrast, wherever such transmission events become common, as is probably the case in many low- and middle income countries with inferior infrastructure for handling faecal waste (Box 1), environmental transmission may have profound effects on the overall resistance situation95.
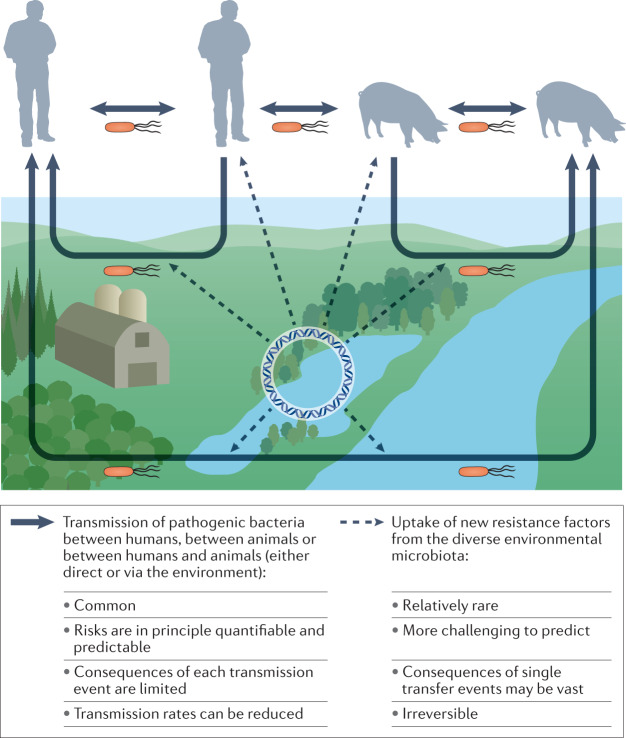
Fig. 3. Pathways for transmission of bacterial pathogens and recruitment of resistance genes from the environmental microbiota.
The dominating routes for transmission of (resistant) pathogens (solid arrows) are between humans, between domestic animals and sometimes between animals and humans. These transmission routes can be direct or indirect via the external environment (lower part of the figure), often through faecal contamination. The consequences of each transmission event are limited, and the risks are in principle quantifiable. There are also rarer and less predictable evolutionary events where new resistance factors are recruited to pathogens by horizontal gene transfer from the diverse, environmental microbiota (dashed arrows). Such transfer events may occur either in the environment or within the human or domestic animal microbiota. The consequences of single gene transfer events may be vast and are irreversible.
There is a vast literature on how certain resistant bacteria can spread via food and through contamination on surfaces, not the least in hospitals96. Exposure to surface waters heavily contaminated by faecal residues can also lead to various infections97. This has led, for example, to the bathing water directive in the European Union, which uses the levels of faecal indicator bacteria as surrogate exposure thresholds. It is plausible that resistant strains would have similar opportunities for transmission via contaminated water as sensitive strains of the same species. Accordingly, correlative analyses have suggested that the overall sanitation and waste infrastructure is a better predictor of national burdens of resistance than is the reported use of antibiotics95. Still, there are only a few dedicated studies indicating that environmental exposure could be important for colonization by and/or infection with specific resistant bacteria. A survey of faecal swabs from surfers from Britain, who are more likely to ingest seawater than non-surfers, found they were more prone to carry cephalosporin-resistant E. coli92. A different study found that recreational swimming might be a risk factor for urinary tract infections with extended-spectrum β-lactamase-producing E. coli or Klebsiella pneumoniae98. More studies of this type, in both high-income and low-income settings, are needed to estimate the role of contaminated water in the transmission of resistant bacteria.
Food, including raw vegetables, is another possible exposure route99. Infections caused by, for example, Salmonella spp., enterohaemorrhagic E. coli and Campylobacter jejuni as a result of consumption of contaminated fresh produce occur frequently100, but we are not aware of studies showing that consumers of contaminated produce have an increased probability of being colonized by resistant strains. However, it has been shown that fresh produce often carries various resistant bacteria with diverse mobile elements100–102. Hence, the safe use of human and animal faecal matter (manure, sludge and effluents from wastewater treatment plants) on farmland is warranted both to control transmission and to control resistance evolution103. Urban air can contain a high diversity of ARGs104, but there is very limited support for long-range aerial transmission of bacterial pathogens105.
The risks for transmission of resistant bacteria are, in principle, quantifiable and possible to model with data on environmental emissions, fate and exposure. Still, accurate models describing the environmental route are scarce106,107. As the bacterial host and the genetic context of mobile ARGs are critical for transmission risks, models based on specific resistant bacteria are more likely to become predictive than are ARG-based models. A major knowledge gap is that we still do not know what doses are required for colonization by many bacteria.
The role of selective agents in environmental transmission, if any, is unclear. Although some pathogens (for example, Legionella spp. or Vibrio spp.) thrive in the environment, for most it is a more hostile environment than a human or domestic animal host. For those pathogens, growth in the environment is often limited. It is thus conceivable that small growth differences between resistant and non-resistant strains, caused by exposure to sub-MICs of antibiotics, are a minor determinant for the possibility that environmental exposure becomes sufficiently high for colonization or infection of a human or animal host. Other biotic and abiotic factors, such as temperature, oxygen pressure, nutrients, predation, and competition with other species, all unrelated to the antibiotic resistance profile of the bacteria, are likely to be much more important for environmental transmission opportunities for both resistant and non-resistant strains.
The environment reflecting the clinic
Although the environment contributes to the problem of antibiotic resistance, both during the evolution of resistance and as a transmission route, it can also provide means to manage it. Environmental microorganisms, including both fungi and bacteria, have been a source for many novel candidate antibiotic molecules, thereby advancing drug development108. In addition, there is continuous and widespread environmental emission of human- and animal-associated bacteria through different waste streams. Analysing the abundance and pattern of resistance in the environmental microbiota could therefore provide an opportunity to predict the regional resistance situation109–114 and indirectly also provide indications of historical antibiotic use115. This overlaps with the main objectives of classic, clinical resistance surveillance, which is critical for guiding empirical treatment, for evaluating interventions, and for identifying regional and temporal trends of resistance.
Although analyses of different environmental matrices may be informative of the regional resistance situation in humans or domestic animals, samples taken as close to the emission sources as possible are advantageous as they are more representative116. Monitoring of raw sewage is particularly promising for large-scale surveillance, at it contains pooled faecal bacteria from large populations. The possibility to screen bacteria from up to millions of people in the same sample makes the approach considerably less resource demanding than traditional surveillance of clinical isolates, a feature which would have a particularly high value in low-income regions116. The broad coverage also open opportunities to provide an ‘early warning’ for the emergence or early spread of rare resistance factors117,118, which would otherwise require extensive surveillance of individuals.
Several recent studies highlight the possibility of using sewage monitoring as a complement to clinical surveillance of resistance, either via phenotypic analyses of isolates110,111,113 or via analyses of ARGs112,114,119 via quantitative PCR or shotgun metagenomics. The latter is the method of choice of the Global Sewage Surveillance Project115, a large research initiative that covers more than 100 countries. The two approaches have distinct advantages and disadvantages (Table 1). For example, isolate-based sewage surveillance has a greater potential to inform empirical treatment than metagenomics, as only the former can provide species-specific phenotypic data with certainty, but the simplicity of metagenomics could make it attractive for monitoring regional and temporal trends in resistance112,119. Both approaches have advantages over traditional clinical surveillance as they cannot be linked to individuals, and hence the risk of ethical dilemmas is minimized112. Also, both approaches are easy to standardize, facilitating comparisons across time and space. Indeed, regional differences in strategies for when patient samples are collected for resistance determination may introduce substantial bias when comparisons are made with and between traditional surveillance datasets120.
Table 1.
Comparison of sewage-based resistance surveillance with traditional clinical resistance surveillance
| Attribute | Sewage-based resistance surveillance (gene-based) | Sewage-based resistance surveillance (isolate-based) | Clinical resistance surveillance (isolate-based) |
|---|---|---|---|
| Potential bias comparing trends over time and space | Standardization of sampling easy, enables comparisons with limited bias | Standardization of sampling easy, enables comparisons with limited bias | Differences in sampling strategies often bias comparisons |
| Risk that the end points studied are influenced by a non-human bacterial population | High risk | Low to high risk depending on species | No risk |
| Reflects intestinal carriage or infections | Reflects carriage, but may correlate well with infection | Reflects carriage, but may correlate well with infection | Reflects infection or carriage depending on sample type |
| Reflects resistance in sick or healthy part of population | Reflects both, but to steer the focus, surveillance may target municipal or hospital sewage | Reflects both, but to steer the focus, surveillance may target municipal or hospital sewage | Reflects the resistance in people who are infected and seek care |
| Interpretation of numbers | Represents the average abundance of a selected gene or genes across the faecal microbiota | Represents the percentage of carriers times the average proportion of resistant strains within a species in the faecal microbiota of the carriers | Represents the percentage of infected individuals or the percentage of carriers depending on the sample type |
| Identification of resistance phenotypes | Predicts resistance phenotypes broadly from individual, acquired genes | Identifies resistance phenotypes | Identifies resistance phenotypes |
| Ability to link resistance to species | Difficult to link genes and thus predicted resistances to specific species | Links resistance to specific pathogen species | Links resistance to specific pathogen species |
| Ability to identify multiresistance | Does not enable the identification of multiresistance patterns | Identifies multiresistance patterns | Identifies multiresistance patterns |
| Ability to identify rare types of resistance | Possible via targeted analyses (PCR) | Possible via selective culturing | Challenging |
| Provides patient-specific information | No | No | Yes |
| Ability to inform empirical treatment | Unlikely | Possibly, after evaluation | Informs empirical treatment |
| Prospect for acceptance in clinical community | Very different from current surveillance, major challenges | Different from current surveillance, but also bears similarities, challenging | The accepted standard among the clinical community |
| Ethical issues | No ethical issues with sampling | No ethical issues with sampling | Ethical issues may arise when carriers are identified |
| Cost | Inexpensive | Rather inexpensive | Expensive |
| Simplicity of sample collection and processing | Very simple sampling | Simple, but more elaborate sampling compared with gene-based sewage surveillance | Resource-demanding to process samples from many individual patients |
| Need for many samples | A single sample can (to some extent) reflect the resistance situation in an entire community | A single sample can (to some extent) reflect the resistance situation in an entire community | A large number of samples are needed to reflect the resistance situation |
| Need for calibration against clinical resistance prevalence | More calibration against clinical resistance needed | More calibration against clinical resistance needed | Considered ‘gold standard’ but suffers from, for example, sampling bias |
| Need for development of sampling protocol | One sampling protocol covers all enteric species (but without separation) | Efficient, specific sampling method evaluated for Escherichia coli, not yet for other species | Sampling method exists for almost all bacterial pathogens |
| Need for local health care infrastructure | No local health care infrastructure needed | No local health care infrastructure needed | Local health care infrastructure needed |
| Need for local sewage collection system | Sewage collection system needed | Sewage collection system needed | No sewage collection system needed |
| Need for analytical infrastructure | Advanced infrastructure (DNA sequencing, bioinformatic competence) is needed, but can be performed elsewhere | Low-level to medium-level technological analyses required (culturing), can be done independently in any standard microbiology laboratory | Low-level to medium-level technological analyses required (culturing), can be done independently in any standard microbiology laboratory |
Sewage analyses with the objective to predict the regional, clinical resistance situation, based on either culture-independent analyses of genes (metagenomics or quantitative PCR arrays) or phenotypic resistance patterns of isolates, may provide an approach complementary to traditional, clinical, isolate-based resistance surveillance (compared from a conceptual point of view).
Still, although resistance surveillance in sewage provides several benefits over traditional surveillance, more research and extensive benchmarking are required to understand the potential and limitations, and to bring this surveillance strategy into practical use. Lessons may be learned from sewage surveillance of poliovirus, which has been in place for decades121. Similarly, over the past year, numerous research groups across the world have monitored the dynamics of severe acute respiratory syndrome coronavirus 2 in sewage122,123, increasing the awareness of the approach.
Studying resistance in the environment
Quantitative analyses of ARGs, resistant bacteria and selective agents in environmental samples differ in terms of how informative data are for assessing risks for evolution or transmission, or as a reflection of the regional clinical resistance situation. These distinct objectives are also best informed by analyses of different environmental matrices, as outlined recently116. Having such limitations in mind is critical for both designing and interpreting environmental surveillance studies. Findings of, for example, increased levels of ARGs can be a result of an on-site selection pressure, thereby indicating an increased risk of resistance evolution. When one is assessing evidence for on-site selection, any ARG, regardless of its clinical relevance, can be informative, as an antibiotic selection pressure would be expected to favour resistant strains of many species and ARGs in parallel. As an increased relative abundance of a species that tends to carry a given ARG can be unrelated to an antibiotic selection pressure, increased abundance of resistant strains over non-resistant strains within the same species adds to the evidence (see later). Increased levels of several ARGs providing resistance to the same antibiotic class, but less pronounced changes in the levels of ARGs providing resistance to other antibiotic classes, also supports a specific selection pressure as the driver behind their increase124. Alternatively, increased ARG abundances could be merely the result of faecal pollution104, which primarily would be informative about risks for transmission. Concurrent finding of selective agents at concentrations known to select for resistant bacteria in complex communities would support the former explanation and risk scenario, whereas high abundances of crAssphage, a bacteriophage that indicates human faecal pollution, would support the latter65. ARGs that occur predominantly in pathogens are also more informative about risks for transmission than those that tend to reside to a greater extent in non-pathogenic bacteria. Both aquatic and terrestrial environments polluted with residual faecal matter often harbour increased levels of ARGs65,125. Indeed, the spread of faecal material around the globe (for example, via sewage effluents, animal waste, and birds transporting bacteria from urban sites) has contributed to ARG contamination of almost the entire planet, including freshwater systems14,65,126, estuaries127, farmland soils128–130, arctic areas131,132, and air104,133, as a few examples.
Most bacterial species do not cause disease and are not associated with clinical breakpoint concentrations. Accordingly, environmental microbiologists most often define ‘resistance’ as a decreased susceptibility to an antibiotic compared with other strains of the same species. As most species of environmental bacteria are difficult to culture with standard methods134, environmental microbiologists, more often so than clinical microbiologists, also tend to study ARGs rather than resistant bacteria. It is important to appreciate the differences, particularly because the genetic context and host of the detected ARGs in most instances remain unknown. Linking ARGs to their hosts and/or mobile genetic element is often critical, both for assessing risks for evolution and transmission, and for predicting the resistance situation in the clinic from sewage analyses.
When feasible, cultivation-based approaches are still superior in terms of providing insights into both the phenotype and the context, particularly if combined with whole-genome sequencing. Long-read sequencing of DNA from bacterial communities might overcome some challenges in placing ARGs into context135, as short-read assemblies of ARG-containing contigs from complex metagenomes are notoriously uncertain. However, for plasmid-borne ARGs, other approaches are needed. EpicPCR (emulsion, paired isolation and concatenation PCR) could be applied to link plasmid-borne ARGs to hosts, but still sensitivity and specificity are limiting factors136. Another possible technology involves genomic crosslinking137. It should also be noted that detected ARGs in environmental samples may represent extracellular DNA138. Free DNA could potentially be taken up and incorporated in genomes, but compared with ARGs already present in living cells, the opportunities to propagate are still very small.
Shotgun metagenomics can be used to detect and quantify ARGs, with the main advantage over PCR being that any ARG present in available databases can be identified, also in retrospect. The chief disadvantage, even with short reads, is the limitation in sensitivity139. Quantitative PCR arrays can be a good compromise between coverage and sensitivity140. Analyses of other genetic elements or genes, such as the integrase of class 1 integrons, can often provide a good surrogate for the overall presence of anthropogenic pollution, including resistant bacteria in polluted environments141,142.
Characterizing the environmental resistome, particularly the already mobilized and thus more easily transferrable fraction, is important to understand the role of the environment as a source for new resistance factors. This includes understanding the host range and ecology of the vectors involved. Identifying those ARGs that are at risk of emerging or have just emerged in pathogens enables early detection in the clinic. Identifying such ARGs may also inform interventions to limit their spread, and enables gene-based diagnostics. Possibly, such knowledge could also guide future drug development by providing information on emerging resistance mechanisms143. Strategies to identify emerging resistance threats involve computational methods, including hidden Markov models27,29, as well as functional metagenomics screens26,144, which can be adapted to focus on already mobilized genetic elements36,117.
Environmental analyses of antibiotics advanced greatly in the past few decades; however, it can be challenging to accurately identify and quantify antibiotics that often occur at nanogram per litre levels in complex matrices53,145. The concentrations of antibiotics and other pharmaceuticals in sewage are usually reasonably stable over time, but can show diurnal patterns146. Emissions from production are often much more erratic and considerably more difficult to predict. This irregular discharge pattern was used in a recent study to attribute a large portion of various drugs found in a Swiss river to industrial emissions147. As antibiotics and resistant bacteria often have the same source (excreta from humans or domestic animals), correlations between the two in environmental samples provide, without additional data, very weak evidence for on-site selection by antibiotics. Study design and careful interpretation are therefore always key concerns.
Studying selection in the laboratory
The basis for environmental risk assessments is to compare exposure levels (predicted or measured in the environment) with effect levels; that is, concentrations that are known or predicted to cause a certain response, most commonly derived from simplified laboratory experiments. The ultimate concern with antibiotic pollution is that it will contribute to the evolution of new, successful, resistant genotypes in pathogens, causing difficult-to-treat infections and eventually higher morbidity and mortality. This chain of events is very difficult to trace back, and also to study in controlled experiments. Therefore, analyses of selection pressures, being the most well recognized driver of resistance development, are commonly used as surrogates for risk. However, it is possible that even evident selection pressures have no or little contribution to the end points of ultimate concern. Note that it is not yet exactly known for any ARG in what place, in what environment or under what conditions the ARG was mobilized or transferred to a pathogen for the first time. Hence, any actions to reduce selection pressures to mitigate the risk of emergence of new forms of resistance, whether in humans, animals or the environment, are based on the precautionary principle.
How to best assess the selective potential of antibiotics and co-selective agents in the environment is still an open question. Although the most critical form of selection for the evolution of resistance is between strains within species (largest niche overlap), transmission risks for a given resistant pathogen can increase as a consequence of both within-species and between-species selection. This should ideally be taken into account when one is designing assays for environmental selection and defining environmental selective concentrations. Assays based on analyses of ARGs may be sensitive but can rarely distinguish within-species selection from between-species selection with certainty, and should therefore be interpreted with some caution. Similar concerns apply to culture-based analyses where bacteria are not identified to the species level. Although single-species competition experiments are simple and easier to reproduce, they may not reflect the situation in more complex communities148. Culture-based assays often suffer from only one or a few species within a community being studied at a time. Still, culture-based assays, where the proportion of resistant bacteria within species in complex communities is studied, are probably the most relevant, but not necessarily the most sensitive, approach149.
Minimal selective concentrations (MSC) is an extrapolation of generated competition data, reflecting the concentration at which cost and benefit are predicted to be balanced64. If costs are very low, the estimation of cost, the estimation of balancing benefits and thus estimates of the MSC become more sensitive to noise. The confidence interval associated with such estimates could be high150 unless there is good replication around the MSC. The generation of lowest observed effect concentrations (LOECs) and the corresponding no observed effect concentrations (NOECs) for resistance selection are in that sense more robust measures.
A simplified approach to generate predicted no effect concentrations (PNECs) for resistance selection from available MICs assumes that selection must occur in at least some communities at the lowest reported or predicted MIC for a given antibiotic. The proposed approach63 has rapidly been applied widely, also in regulatory151 and industrial152 initiatives to curb risks for selection in the environment. There is potential to refine MSC predictions from MICs by taking into account also the shape of the dose–response curves153. Recently, it was also proposed that LOECs and NOECs for resistance selection could be based on the lowest concentration that affects growth of entire communities154. From a theoretical point of view, the lowest concentration affecting growth of any species in a community represents the lower boundary for possible resistance selection. But as overall community growth was assessed rather than the growth of individual strains, reduced growth of some strains could easily be compensated by other strains growing more rapidly. Nor would lowered growth of any but the most abundant strains be easy to detect, leading to limited sensitivity of such assays.
For most widely circulating ARGs, the costs are indeed low in the contexts in which they have become adapted, otherwise the resistant strains would disappear very quickly as soon as antibiotic exposure ceases155. As costs are strongly dependent on the genetic context, the presence of other strains and species in a community, and abiotic factors, it is difficult to set up tests that accurately reflect costs for a broader set of contexts and exposure scenarios. A recent study149 accordingly proposed basing concentration thresholds preventing environmental selection solely on the lowest concentration that provides a benefit to resistant strains, ignoring costs in the given test system. This may be achieved by comparing ratios of resistant strains versus non-resistant strains after exposure to different concentrations of antibiotics (given that costs are independent of the exposure concentration), rather than comparing ratios before exposure with those after exposure. Another study156 came to a similar conclusion on how to derive concentration thresholds, but not from the standpoint that costs are context dependent; the study authors argued that even if costs are not fully compensated, the persistence of resistant strains, and hence risks for transmission, would increase. Although we agree with the principle, we believe that other abiotic and biotic factors that determine the survival of most pathogens in environmental media, and hence exposure opportunities, are likely to be of much greater importance for transmission risks (see the section entitled “Environmental transmission”).
Outlook
The environment has a role both in evolution and transmission of resistance, possibly more so than has generally been recognized11–16. A remaining and pressing knowledge gap is our limited understanding of where and under what circumstances the critical steps occur that lead to the emergence of new forms of resistance in clinically important bacteria. Exploring the recent history of resistance factors that already have become clinical problems is one possible strategy to reveal patterns and enable generalizations49. Future genome sequencing of many more environmental species is likely to provide a much better foundation for such investigations. Understanding the role of pollution with selective agents in the emergence and evolution of resistance is particularly important, as neglecting an important driver could have major health consequences. A major knowledge gap is still what role the low or moderately high levels of excreted antibiotics have, and what methods are most suitable to reflect risks for environmental selection. This is in contrast to high-level industrial antibiotic pollution, for which many stakeholders already consider the risks unacceptable, and the core challenge now is rather how to accomplish change157. The use of antibiotics on crops is only rarely studied158, but could potentially be a source of very high concentrations, particularly in low- and middle-income countries57. With regard to risks for transmission, we know considerably more about the flow of resistant bacteria to the environment than to what extent resistant bacteria from environmental sources lead to colonization and disease.
On the basis of current evidence, many policymakers advocate the precautionary principle and call for actions to reduce exposures17–21. At the same time, actions to reduce pollution on a broad scale are expected to be both difficult and expensive, making prioritization necessary. Shaping mitigation strategies places additional demands on recognizing what is feasible from, for example, political, economic and geographical standpoints. It is also important to identify which actors can drive appropriate mitigations, including what their specific incentives and counterincentives for actions are157. While not neglecting the technological needs involved, creating socio-economic and legal drivers for change is often even more challenging. To reduce discharges from antibiotic manufacturing, a number of such actions either were initiated recently or have been proposed by governments151,159–161 or multinational organizations, such as the European Union20, the G7 (ref.162), the United Nations Environment Programme17, and the Word Health Organization, the World Organisation for Animal Health and the Food and Agriculture Organization of the United Nations21. These include pollution control as an award criterion in procurement processes20,160,161 and decisions on which products to subsidize159, demanding increased transparency on production sites and emission levels to increase accountability20,163, amending pollution control in the framework for good manufacturing practice162, and applying legally binding limits for discharges151. Many pharmaceutical companies also acknowledge the need for change and have collectively endorsed voluntary emission targets63,152. Although this must be seen as a positive initiative, the near-complete lack of disclosure of both production sites for active ingredients and emission levels18 makes it difficult to judge progress, reinforcing that industrial pollution is rarely resolved without active interference from public institutions.
On the technical side, installing basic treatment of waste streams, whether industrial, municipal or from animal sources, should have high priority as it addresses many types of risks, not the least substantial transmission risks for several pathogens (including resistant bacteria) and deterioration of valuable water resources164. Additional treatment of wastewaters with more advanced methods (such as ozonation or activated carbon) is a second step that would remove not only many selective agents but also a large range of additional contaminants165. Hence, an important motivation for more advanced treatment, particularly of municipal wastewater, is the ‘collateral benefit’ such treatment can provide by reducing the risks of many pollutants, known and unknown ones.
Measures to limit the risk for transmission of antibiotic-resistant bacteria could involve both reducing emissions to the environment and/or reducing exposure166. Such actions often coincide with measures to reduce infections in general. Given what is at stake, there could be good reasons also to take specific actions to reduce the risk for evolution of resistance, despite large uncertainties. To mitigate such risks, we ought to prioritize actions where risks are high and where changes can feasibly be achieved in a limited time frame. Prioritization should ideally be done on a global level, as the consequences of inaction will affect everyone in the long run, regardless of where a resistance factor emerges. Radically reducing emissions of exceptionally high concentrations from drug manufactures is one such apparent starting point17,19,59.
To assign appropriate measures to reduce the risk of resistance evolution associated with human waste streams, it is critical to understand where selection primarily occurs (hospital sewers, community sewers, wastewater treatment plants, recipients and so on). In contrast to managing risks with most other pollutants, the relevant protection target related to antibiotic resistance development is bacteria, rather than (aquatic) wildlife or humans. Sewers and wastewater treatment plant environments harbour dense, complex bacterial communities that often include pathogens. Furthermore, as antibiotic levels are often higher than in receiving waters, they may be more likely a spawning ground for resistance evolution than the recipient waterways. Measures with the intention to control resistance selection should therefore also take into account risks for selection that occur before discharge of the wastewater.
Sewage epidemiology is still in its infancy, at least with regard to how well the regional clinical resistance situation can be predicted. It is critical to benchmark environmental resistance data, whether based on isolates or metagenomes, against high-quality clinical data. Still, it may take more than evidence of good correlations to bring environmental analyses of resistance into clinical policy.
Acknowledgements
Work in the authors’ laboratories was funded by the Swedish research councils VR (2018-02835 and 2018-05771) and FORMAS (2108-00787) and the Region Västra Götaland under the ALF agreement (grant number ALFGBG-717901) to D.G.J.L. as well as FORMAS (2018-00833) to C.-F.F.
Author contributions
D.G.J.L. and C.-F.F. researched data for the article, contributed to the discussion of the content, and reviewed and edited the manuscript before submission. D.G.J.L. wrote the article.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Microbiology thanks J. Martínez, K. Smalla and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.D’Costa VM, et al. Antibiotic resistance is ancient. Nature. 2011;477:457–461. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 2.Bhullar K, et al. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE. 2012;7:e34953. doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lugli GA, et al. Ancient bacteria of the Ötzi’s microbiome: a genomic tale from the Copper Age. Microbiome. 2017;5:5. doi: 10.1186/s40168-016-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry J, Waglechner N, Wright G. The prehistory of antibiotic resistance. Cold Spring Harb. Perspect. Med. 2016;6:a025197. doi: 10.1101/cshperspect.a025197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 2010;74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen HK, et al. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 7.Martinez JL. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. R. Soc. B Biol. Sci. 2009;276:2521–2530. doi: 10.1098/rspb.2009.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alcock BP, et al. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackenzie JS, Jeggo M. The one health approach — why is it so important? Trop. Med. Infect. Dis. 2019;4:88. doi: 10.3390/tropicalmed4020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buschhardt T, et al. A one health glossary to support communication and information exchange between the human health, animal health and food safety sectors. One Health. 2021;13:100263. doi: 10.1016/j.onehlt.2021.100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berendonk TU, et al. Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 2015;13:310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 12.Wellington EM, et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect. Dis. 2013;13:155–165. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 13.Bengtsson-Palme J, Kristiansson E, Larsson DGJ. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 2017 doi: 10.1093/femsre/fux053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow LKM, Ghaly TM, Gillings MR. A survey of sub-inhibitory concentrations of antibiotics in the environment. J. Environ. Sci. 2021;99:21–27. doi: 10.1016/j.jes.2020.05.030. [DOI] [PubMed] [Google Scholar]
- 15.Andersson DI, et al. Antibiotic resistance: turning evolutionary principles into clinical reality. FEMS Microbiol. Rev. 2020;44:171–188. doi: 10.1093/femsre/fuaa001. [DOI] [PubMed] [Google Scholar]
- 16.Singer AC, Shaw H, Rhodes V, Hart A. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 2016 doi: 10.3389/fmicb.2016.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.United Nations Environment Programme. Frontiers 2017: emerging issues of environmental concern, https://www.unenvironment.org/resources/frontiers-2017-emerging-issues-environmental-concern (2017).
- 18.Access to Medicines Foundation. 2020 antimicrobial resistance benchmark, https://accesstomedicinefoundation.org/publications/2020-antimicrobial-resistance-benchmark (2020).
- 19.Review on Antimicrobial Resistance. Antimicrobials in agriculture and the environment: reducing unnecessary waste, https://amr-review.org/Publications.html (2015).
- 20.European Parliament. Strategic approach to pharmaceuticals in the environment, https://www.europarl.europa.eu/doceo/document/TA-9-2020-0226_EN.pdf (2020).
- 21.WHO. Technical brief on water, sanitation, hygiene (WASH) and wastewater management to prevent infections and reduce the spread of antimicrobial resistance (AMR)., https://www.who.int/water_sanitation_health/publications/wash-wastewater-management-to-prevent-infections-and-reduce-amr/en/ (2020).
- 22.Graham DW, et al. Complexities in understanding antimicrobial resistance across domesticated animal, human, and environmental systems. Ann. N. Y. Acad. Sci. 2019;1441:17–30. doi: 10.1111/nyas.14036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smalla K, Cook K, Djordjevic SP, Klümper U, Gillings M. Environmental dimensions of antibiotic resistance: assessment of basic science gaps. FEMS Microbiol. Ecol. 2018 doi: 10.1093/femsec/fiy195. [DOI] [PubMed] [Google Scholar]
- 24.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 25.Schulz F, et al. Towards a balanced view of the bacterial tree of life. Microbiome. 2017 doi: 10.1186/s40168-017-0360-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forsberg KJ, et al. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berglund F, et al. Identification of 76 novel B1 metallo-beta-lactamases through large-scale screening of genomic and metagenomic data. Microbiome. 2017;5:134. doi: 10.1186/s40168-017-0353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dantas G, Sommer MOA, Oluwasegun RD, Church GM. Bacteria subsisting on antibiotics. Science. 2008;320:100–103. doi: 10.1126/science.1155157. [DOI] [PubMed] [Google Scholar]
- 29.Berglund F, et al. Comprehensive screening of genomic and metagenomic data reveals a large diversity of tetracycline resistance genes. Microb. Genomics. 2020 doi: 10.1099/mgen.0.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pawlowski AC, et al. A diverse intrinsic antibiotic resistome from a cave bacterium. Nat. Commun. 2016;7:13803. doi: 10.1038/ncomms13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morar M, Wright GD. The genomic enzymology of antibiotic resistance. Annu. Rev. Genet. 2010;44:25–51. doi: 10.1146/annurev-genet-102209-163517. [DOI] [PubMed] [Google Scholar]
- 32.Andersson DI, Jerlström-Hultqvist J, Näsvall J. Evolution of new functions de novo and from preexisting genes. Cold Spring Harb. Perspect. Biol. 2015;7:a017996. doi: 10.1101/cshperspect.a017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Razavi M, Kristiansson E, Flach C-F, Larsson DGJ. The association between insertion sequences and antibiotic resistance genes. mSphere. 2020 doi: 10.1128/msphere.00418-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018 doi: 10.1128/cmr.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gillings M, et al. The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 2008;190:5095–5100. doi: 10.1128/JB.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Razavi M, et al. Discovery of the fourth mobile sulfonamide resistance gene. Microbiome. 2017 doi: 10.1186/s40168-017-0379-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flach C-F, et al. Does antifouling paint select for antibiotic resistance? Sci. Total Environ. 2017;590–591:461–468. doi: 10.1016/j.scitotenv.2017.01.213. [DOI] [PubMed] [Google Scholar]
- 38.Shintani M, et al. Plant species-dependent increased abundance and diversity of IncP-1 plasmids in the rhizosphere: new insights into their role and ecology. Front. Microbiol. 2020;11:590776. doi: 10.3389/fmicb.2020.590776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baquero F, Coque TM, Martínez J-L, Aracil-Gisbert S, Lanza VF. Gene transmission in the one health microbiosphere and the channels of antimicrobial resistance. Front. Microbiol. 2019 doi: 10.3389/fmicb.2019.02892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vandecraen J, Chandler M, Aertsen A, Van Houdt R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017;43:709–730. doi: 10.1080/1040841X.2017.1303661. [DOI] [PubMed] [Google Scholar]
- 41.Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P. Modes and modulations of antibiotic resistance gene expression. Clin. Microbiol. Rev. 2007;20:79–114. doi: 10.1128/CMR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jutkina J, Marathe NP, Flach CF, Larsson DGJ. Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci. Total Environ. 2018;616-617:172–178. doi: 10.1016/j.scitotenv.2017.10.312. [DOI] [PubMed] [Google Scholar]
- 43.Scornec H, Bellanger X, Guilloteau H, Groshenry G, Merlin C. Inducibility of Tn916 conjugative transfer in Enterococcus faecalis by subinhibitory concentrations of ribosome-targeting antibiotics. J. Antimicrob. Chemother. 2017;72:2722–2728. doi: 10.1093/jac/dkx202. [DOI] [PubMed] [Google Scholar]
- 44.Aminov RI. Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2011 doi: 10.3389/fmicb.2011.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knöppel A, Näsvall J, Andersson DI. Evolution of antibiotic resistance without antibiotic exposure. Antimicrob. Agents Chemother. 2017 doi: 10.1128/aac.01495-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimura M, Ohta T. The average number of generations until fixation of a mutant gene in a finite population. Genetics. 1969;61:763–771. doi: 10.1093/genetics/61.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Letten AD, Hall AR, Levine JM. Using ecological coexistence theory to understand antibiotic resistance and microbial competition. Nat. Ecol. Evol. 2021;5:431–441. doi: 10.1038/s41559-020-01385-w. [DOI] [PubMed] [Google Scholar]
- 48.Waglechner N, Wright GD. Antibiotic resistance: it’s bad, but why isn’t it worse? BMC Biol. 2017 doi: 10.1186/s12915-017-0423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ebmeyer S, Erik K, Larsson DGJ. A framework for identifying the recent origins of mobile antibiotic resistance genes. Commun. Biol. 2021 doi: 10.1038/s42003-020-01545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersson DI, Hughes D. Persistence of antibiotic resistance in bacterial populations. FEMS Microbiol. Rev. 2011;35:901–911. doi: 10.1111/j.1574-6976.2011.00289.x. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Chu L, Wojnárovits L, Takács E. Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: an overview. Sci. Total. Environ. 2020;744:140997. doi: 10.1016/j.scitotenv.2020.140997. [DOI] [PubMed] [Google Scholar]
- 52.Tran NH, Reinhard M, Gin KY-H. Occurrence and fate of emerging contaminants in municipal wastewater treatment plants from different geographical regions-a review. Water Res. 2018;133:182–207. doi: 10.1016/j.watres.2017.12.029. [DOI] [PubMed] [Google Scholar]
- 53.Szymańska U, et al. Presence of antibiotics in the aquatic environment in Europe and their analytical monitoring: recent trends and perspectives. Microchem. J. 2019;147:729–740. doi: 10.1016/j.microc.2019.04.003. [DOI] [Google Scholar]
- 54.Anwar M, Iqbal Q, Saleem F. Improper disposal of unused antibiotics: an often overlooked driver of antimicrobial resistance. Expert Rev. Antiinfect Ther. 2020 doi: 10.1080/14787210.2020.1754797. [DOI] [PubMed] [Google Scholar]
- 55.Cabello FC, et al. Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ. Microbiol. 2013;15:1917–1942. doi: 10.1111/1462-2920.12134. [DOI] [PubMed] [Google Scholar]
- 56.Cabello FC, Godfrey HP, Buschmann AH, Dölz HJ. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 2016;16:e127–e133. doi: 10.1016/S1473-3099(16)00100-6. [DOI] [PubMed] [Google Scholar]
- 57.Taylor P, Reeder R. Antibiotic use on crops in low and middle-income countries based on recommendations made by agricultural advisors. CABI Agric. Biosci. 2020 doi: 10.1186/s43170-020-00001-y. [DOI] [Google Scholar]
- 58.Larsson DGJ. Pollution from drug manufacturing: review and perspectives. Philos. Trans. R. Soc. B Biol. Sci. 2014;369:20130571. doi: 10.1098/rstb.2013.0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larsson DGJ, De Pedro C, Paxeus N. Effluent from drug manufactures contains extremely high levels of pharmaceuticals. J. Hazard. Mater. 2007;148:751–755. doi: 10.1016/j.jhazmat.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 60.Milaković M, et al. Pollution from azithromycin-manufacturing promotes macrolide-resistance gene propagation and induces spatial and seasonal bacterial community shifts in receiving river sediments. Environ. Int. 2019;123:501–511. doi: 10.1016/j.envint.2018.12.050. [DOI] [PubMed] [Google Scholar]
- 61.Bielen A, et al. Negative environmental impacts of antibiotic-contaminated effluents from pharmaceutical industries. Water Res. 2017;126:79–87. doi: 10.1016/j.watres.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 62.Fick J, et al. Contamination of surface, ground, and drinking water from pharmaceutical production. Environ. Toxicol. Chem. 2009;28:2522–2527. doi: 10.1897/09-073.1. [DOI] [PubMed] [Google Scholar]
- 63.Bengtsson-Palme J, Larsson DGJ. Concentrations of antibiotics predicted to select for resistant bacteria: proposed limits for environmental regulation. Environ. Int. 2016;86:140–149. doi: 10.1016/j.envint.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 64.Gullberg E, et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011;7:e1002158. doi: 10.1371/journal.ppat.1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karkman A, Pärnänen K, Larsson DGJ. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019 doi: 10.1038/s41467-018-07992-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Y, Li B, Zou S, Fang HHP, Zhang T. Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Res. 2014;62:97–106. doi: 10.1016/j.watres.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 67.Bengtsson-Palme J, et al. Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Sci. Total Environ. 2016;572:697–712. doi: 10.1016/j.scitotenv.2016.06.228. [DOI] [PubMed] [Google Scholar]
- 68.Manaia CM, et al. Antibiotic resistance in wastewater treatment plants: tackling the black box. Environ. Int. 2018;115:312–324. doi: 10.1016/j.envint.2018.03.044. [DOI] [PubMed] [Google Scholar]
- 69.Flach CF, Genheden M, Fick J, Joakim Larsson DG. A comprehensive screening of Escherichia coli isolates from Scandinavia’s largest sewage treatment plant indicates no selection for antibiotic resistance. Environ. Sci. Technol. 2018;52:11419–11428. doi: 10.1021/acs.est.8b03354. [DOI] [PubMed] [Google Scholar]
- 70.Kraupner N, et al. Evidence for selection of multi-resistant E. coli by hospital effluent. Environ. Int. 2021;150:106436. doi: 10.1016/j.envint.2021.106436. [DOI] [PubMed] [Google Scholar]
- 71.Flach CF, et al. Isolation of novel IncA/C and IncN fluoroquinolone resistance plasmids from an antibiotic-polluted lake. J. Antimicrob. Chemother. 2015;70:2709–2717. doi: 10.1093/jac/dkv167. [DOI] [PubMed] [Google Scholar]
- 72.Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ. Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Front. Microbiol. 2014 doi: 10.3389/fmicb.2014.00648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marathe NP, et al. Functional metagenomics reveals a novel carbapenem-hydrolyzing mobile beta-lactamase from Indian river sediments contaminated with antibiotic production waste. Environ. Int. 2018;112:279–286. doi: 10.1016/j.envint.2017.12.036. [DOI] [PubMed] [Google Scholar]
- 74.Thiele-Bruhn S. Pharmaceutical antibiotic compounds in soils–a review. J. Plant Nutr. Soil Sci. 2003;166:145–167. doi: 10.1002/jpln.200390023. [DOI] [Google Scholar]
- 75.Li W, Shi Y, Gao L, Liu J, Cai Y. Occurrence, distribution and potential affecting factors of antibiotics in sewage sludge of wastewater treatment plants in China. Sci. Total. Environ. 2013;445–446:306–313. doi: 10.1016/j.scitotenv.2012.12.050. [DOI] [PubMed] [Google Scholar]
- 76.Reinthaler FF, et al. Resistance patterns of Escherichia coli isolated from sewage sludge in comparison with those isolated from human patients in 2000 and 2009. J. Water Health. 2013;11:13–20. doi: 10.2166/wh.2012.207. [DOI] [PubMed] [Google Scholar]
- 77.Rutgersson C, et al. Long-term application of Swedish sewage sludge on farmland does not cause clear changes in the soil bacterial resistome. Environ. Int. 2020;137:105339. doi: 10.1016/j.envint.2019.105339. [DOI] [PubMed] [Google Scholar]
- 78.Jechalke S, Heuer H, Siemens J, Amelung W, Smalla K. Fate and effects of veterinary antibiotics in soil. Trends Microbiol. 2014;22:536–545. doi: 10.1016/j.tim.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 79.Boxall AB, et al. Pharmaceuticals and personal care products in the environment: what are the big questions? Environ. Health Perspect. 2012;120:1221–1229. doi: 10.1289/ehp.1104477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Song J, Rensing C, Holm PE, Virta M, Brandt KK. Comparison of metals and tetracycline as selective agents for development of tetracycline resistant bacterial communities in agricultural soil. Environ. Sci. Technol. 2017;51:3040–3047. doi: 10.1021/acs.est.6b05342. [DOI] [PubMed] [Google Scholar]
- 81.Jechalke S, et al. Plasmid-mediated fitness advantage of Acinetobacter baylyi in sulfadiazine-polluted soil. FEMS Microbiol. Lett. 2013;348:127–132. doi: 10.1111/1574-6968.12284. [DOI] [PubMed] [Google Scholar]
- 82.Pal C, et al. Metal resistance and its association with antibiotic resistance. Adv. Microb. Physiol. 2017;70:261–313. doi: 10.1016/bs.ampbs.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 83.Wales A, Davies R. Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics. 2015;4:567–604. doi: 10.3390/antibiotics4040567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics. 2015 doi: 10.1186/s12864-015-2153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klümper U, et al. Metal stressors consistently modulate bacterial conjugal plasmid uptake potential in a phylogenetically conserved manner. ISME J. 2017;11:152–165. doi: 10.1038/ismej.2016.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jutkina J, Rutgersson C, Flach CF, Joakim Larsson DG. An assay for determining minimal concentrations of antibiotics that drive horizontal transfer of resistance. Sci. Total. Environ. 2016;548–549:131–138. doi: 10.1016/j.scitotenv.2016.01.044. [DOI] [PubMed] [Google Scholar]
- 87.Wang Y, et al. Non-antibiotic pharmaceuticals enhance the transmission of exogenous antibiotic resistance genes through bacterial transformation. ISME J. 2020;14:2179–2196. doi: 10.1038/s41396-020-0679-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klumper U, et al. Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 2015;9:934–945. doi: 10.1038/ismej.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gillings MR, Paulsen IT, Tetu SG. Genomics and the evolution of antibiotic resistance. Ann. N. Y. Acad. Sci. 2017;1388:92–107. doi: 10.1111/nyas.13268. [DOI] [PubMed] [Google Scholar]
- 90.Heuer H, Smalla K. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 2012;36:1083–1104. doi: 10.1111/j.1574-6976.2012.00337.x. [DOI] [PubMed] [Google Scholar]
- 91.Bengtsson-Palme J, Larsson DG. Antibiotic resistance genes in the environment: prioritizing risks. Nat. Rev. Microbiol. 2015;13:396. doi: 10.1038/nrmicro3399-c1. [DOI] [PubMed] [Google Scholar]
- 92.Leonard AFC, et al. Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: environmental surveillance, exposure assessment, and epidemiological study (Beach Bum Survey) Environ. Int. 2018;114:326–333. doi: 10.1016/j.envint.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 93.Manaia CM. Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends Microbiol. 2017;25:173–181. doi: 10.1016/j.tim.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 94.Schijven JF, Blaak H, Schets FM, De Roda Husman AM. Fate of extended-spectrum β-lactamase-producing Escherichia coli from faecal sources in surface water and probability of human exposure through swimming. Environ. Sci. Technol. 2015;49:11825–11833. doi: 10.1021/acs.est.5b01888. [DOI] [PubMed] [Google Scholar]
- 95.Collignon P, Beggs JJ, Walsh TR, Gandra S, Laxminarayan R. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planet. Health. 2018;2:e398–e405. doi: 10.1016/S2542-5196(18)30186-4. [DOI] [PubMed] [Google Scholar]
- 96.Dancer SJ. Controlling hospital-acquired infection: focus on the role of the environment and new technologies for decontamination. Clin. Microbiol. Rev. 2014;27:665–690. doi: 10.1128/CMR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weber DJ, Anderson D, Rutala WA. The role of the surface environment in healthcare-associated infections. Curr. Opin. Infect. Dis. 2013;26:338–344. doi: 10.1097/QCO.0b013e3283630f04. [DOI] [PubMed] [Google Scholar]
- 98.Søraas A, Sundsfjord A, Sandven I, Brunborg C, Jenum PA. Risk factors for community-acquired urinary tract infections caused by ESBL-producing Enterobacteriaceae –a case–control study in a low prevalence country. PLoS ONE. 2013;8:e69581. doi: 10.1371/journal.pone.0069581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhou S-Y-D, et al. Prevalence of antibiotic resistome in ready-to-eat salad. Front. Public Health. 2020 doi: 10.3389/fpubh.2020.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Uyttendaele M, et al. Microbial hazards in irrigation water: standards, norms, and testing to manage use of water in fresh produce primary production. Compr. Rev. Food Sci. Food Saf. 2015;14:336–356. doi: 10.1111/1541-4337.12133. [DOI] [Google Scholar]
- 101.Reid CJ, Blau K, Jechalke S, Smalla K, Djordjevic SP. Whole genome sequencing of Escherichia coli from store-bought produce. Front. Microbiol. 2020;10:3050. doi: 10.3389/fmicb.2019.03050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Blau K, et al. The transferable resistome of produce. mBio. 2018;9:e01300-18. doi: 10.1128/mBio.01300-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhu Y-G, et al. Soil biota, antimicrobial resistance and planetary health. Environ. Int. 2019;131:105059. doi: 10.1016/j.envint.2019.105059. [DOI] [PubMed] [Google Scholar]
- 104.Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ. The structure and diversity of human, animal and environmental resistomes. Microbiome. 2016;4:54. doi: 10.1186/s40168-016-0199-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kozajda A, Jeżak K, Kapsa A. Airborne Staphylococcus aureus in different environments — a review. Environ. Sci. Pollut. Res. 2019;26:34741–34753. doi: 10.1007/s11356-019-06557-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ashbolt NJ, et al. Human health risk assessment (HHRA) for environmental development and transfer of antibiotic resistance. Environ. Health Perspect. 2013;121:993–1001. doi: 10.1289/ehp.1206316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Franz E, Schijven J, De Roda Husman AM, Blaak H. Meta-regression analysis of commensal and pathogenic Escherichia coli survival in soil and water. Environ. Sci. Technol. 2014;48:6763–6771. doi: 10.1021/es501677c. [DOI] [PubMed] [Google Scholar]
- 108.Lewis K. Platforms for antibiotic discovery. Nat. Rev. Drug. Discov. 2013;12:371–387. doi: 10.1038/nrd3975. [DOI] [PubMed] [Google Scholar]
- 109.Linton KB, Richmond MH, Bevan R, Gillespie WA. Antibiotic resistance and R factors in coliform bacilli isolated from hospital and domestic sewage. J. Med. Microbiol. 1974;7:91–103. doi: 10.1099/00222615-7-1-91. [DOI] [PubMed] [Google Scholar]
- 110.Huijbers P, Joakim Larsson DG, Flach CF. Surveillance of antibiotic resistant Escherichia coli in human populations through urban wastewater in ten European countries. Environ. Pollut. 2020;261:114200. doi: 10.1016/j.envpol.2020.114200. [DOI] [PubMed] [Google Scholar]
- 111.Hutinel M, et al. Population-level surveillance of antibiotic resistance in Escherichia coli through sewage analysis. Euro Surveill. 2019 doi: 10.2807/1560-7917.es.2019.24.37.1800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Aarestrup FM, Woolhouse MEJ. Using sewage for surveillance of antimicrobial resistance. Science. 2020;367:630–632. doi: 10.1126/science.aba3432. [DOI] [PubMed] [Google Scholar]
- 113.Kwak YK, et al. Surveillance of antimicrobial resistance among Escherichia coli in wastewater in Stockholm during 1 year: does it reflect the resistance trends in the society? Int. J. Antimicrob. Agents. 2015;45:25–32. doi: 10.1016/j.ijantimicag.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 114.Parnanen KMM, et al. Antibiotic resistance in European wastewater treatment plants mirrors the pattern of clinical antibiotic resistance prevalence. Sci. Adv. 2019;5:eaau9124. doi: 10.1126/sciadv.aau9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hendriksen RS, et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. 2019;10:1124. doi: 10.1038/s41467-019-08853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huijbers PMC, Flach CF, Larsson DGJ. A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 2019;130:104880. doi: 10.1016/j.envint.2019.05.074. [DOI] [PubMed] [Google Scholar]
- 117.Böhm M-E, Razavi M, Marathe NP, Flach C-F, Larsson DGJ. Discovery of a novel integron-borne aminoglycoside resistance gene present in clinical pathogens by screening environmental bacterial communities. Microbiome. 2020 doi: 10.1186/s40168-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Flach C-F, Hutinel M, Razavi M, Åhrén C, Larsson DGJ. Monitoring of hospital sewage shows both promise and limitations as an early-warning system for carbapenemase-producing Enterobacterales in a low-prevalence setting. Water Res. 2021;200:117261. doi: 10.1016/j.watres.2021.117261. [DOI] [PubMed] [Google Scholar]
- 119.Karkman A, Berglund F, Flach C-F, Kristiansson E, Larsson DGJ. Predicting clinical resistance prevalence using sewage metagenomic data. Commun. Biol. 2020 doi: 10.1038/s42003-020-01439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.European Centre for Disease Prevention and Control. Surveillance of antimicrobial resistance in Europe 2017 (Stockholm, Sweden, 2018).
- 121.Hovi T, et al. Role of environmental poliovirus surveillance in global polio eradication and beyond. Epidemiol. Infect. 2012;140:1–13. doi: 10.1017/S095026881000316X. [DOI] [PubMed] [Google Scholar]
- 122.Agrawal S, Orschler L, Lackner S. Long-term monitoring of SARS-CoV-2 RNA in wastewater of the Frankfurt metropolitan area in southern Germany. Sci. Rep. 2021 doi: 10.1038/s41598-021-84914-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Medema G, Heijnen L, Elsinga G, Italiaander R, Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 124.Lundstrom SV, et al. Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Sci. Total Environ. 2016;553:587–595. doi: 10.1016/j.scitotenv.2016.02.103. [DOI] [PubMed] [Google Scholar]
- 125.McCann CM, et al. Understanding drivers of antibiotic resistance genes in High Arctic soil ecosystems. Environ. Int. 2019;125:497–504. doi: 10.1016/j.envint.2019.01.034. [DOI] [PubMed] [Google Scholar]
- 126.Pruden A, Arabi M, Storteboom HN. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ. Sci. Technol. 2012;46:11541–11549. doi: 10.1021/es302657r. [DOI] [PubMed] [Google Scholar]
- 127.Zhu Y-G, et al. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2017;2:16270. doi: 10.1038/nmicrobiol.2016.270. [DOI] [PubMed] [Google Scholar]
- 128.Zhu Y-G, et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl Acad. Sci. USA. 2013;110:3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Knapp CW, Dolfing J, Ehlert PAI, Graham DW. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ. Sci. Technol. 2010;44:580–587. doi: 10.1021/es901221x. [DOI] [PubMed] [Google Scholar]
- 130.Nesme J, Simonet P. The soil resistome: a critical review on antibiotic resistance origins, ecology and dissemination potential in telluric bacteria. Environ. Microbiol. 2015;17:913–930. doi: 10.1111/1462-2920.12631. [DOI] [PubMed] [Google Scholar]
- 131.Finley RL, et al. The scourge of antibiotic resistance: the important role of the environment. Clin. Infect. Dis. 2013;57:704–710. doi: 10.1093/cid/cit355. [DOI] [PubMed] [Google Scholar]
- 132.Sjölund M, et al. Dissemination of multidrug-resistant bacteria into the Arctic. Emerg. Infect. Dis. 2008;14:70–72. doi: 10.3201/eid1401.070704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhu G, et al. Air pollution could drive global dissemination of antibiotic resistance genes. ISME J. 2020 doi: 10.1038/s41396-020-00780-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nichols D, et al. Use of Ichip for high-throughput in situ cultivation of “Uncultivable” microbial species. Appl. Environ. Microbiol. 2010;76:2445–2450. doi: 10.1128/AEM.01754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ashton PM, et al. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol. 2015;33:296–300. doi: 10.1038/nbt.3103. [DOI] [PubMed] [Google Scholar]
- 136.Spencer SJ, et al. Massively parallel sequencing of single cells by epicPCR links functional genes with phylogenetic markers. ISME J. 2016;10:427–436. doi: 10.1038/ismej.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rice EW, Wang P, Smith AL, Stadler LB. Determining hosts of antibiotic resistance genes: a review of methodological advances. Environ. Sci. Technol. Lett. 2020;7:282–291. doi: 10.1021/acs.estlett.0c00202. [DOI] [Google Scholar]
- 138.Sivalingam P, Poté J, Prabakar K. Extracellular DNA (eDNA): neglected and potential sources of antibiotic resistant genes (ARGs) in the aquatic environments. Pathogens. 2020;9:874. doi: 10.3390/pathogens9110874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bengtsson-Palme J, Larsson DGJ, Kristiansson E. Using metagenomics to investigate human and environmental resistomes. J. Antimicrob. Chemother. 2017;72:2690–2703. doi: 10.1093/jac/dkx199. [DOI] [PubMed] [Google Scholar]
- 140.Karkman, A. et al. High-throughput quantification of antibiotic resistance genes from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 92, 10.1093/femsec/fiw014 (2016). [DOI] [PubMed]
- 141.Gillings MR, et al. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015;9:1269–1279. doi: 10.1038/ismej.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gaze WH, Abdouslam N, Hawkey PM, Wellington EMH. Incidence of Class 1 integrons in a quaternary ammonium compound-polluted environment. Antimicrob. Agents Chemother. 2005;49:1802–1807. doi: 10.1128/AAC.49.5.1802-1807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sommer MOA, Munck C, Toft-Kehler RV, Andersson DI. Prediction of antibiotic resistance: time for a new preclinical paradigm? Nat. Rev. Microbiol. 2017;15:689–696. doi: 10.1038/nrmicro.2017.75. [DOI] [PubMed] [Google Scholar]
- 144.Pehrsson EC, Forsberg KJ, Gibson MK, Ahmadi S, Dantas G. Novel resistance functions uncovered using functional metagenomic investigations of resistance reservoirs. Front. Microbiol. 2013 doi: 10.3389/fmicb.2013.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kim C, Ryu H-D, Chung EG, Kim Y, Lee J-K. A review of analytical procedures for the simultaneous determination of medically important veterinary antibiotics in environmental water: sample preparation, liquid chromatography, and mass spectrometry. J. Environ. Manag. 2018;217:629–645. doi: 10.1016/j.jenvman.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 146.Fahrenfeld N, Bisceglia KJ. Emerging investigators series: sewer surveillance for monitoring antibiotic use and prevalence of antibiotic resistance: urban sewer epidemiology. Environ. Sci. Water Res. Technol. 2016;2:788–799. doi: 10.1039/C6EW00158K. [DOI] [Google Scholar]
- 147.Anliker S, et al. Assessing emissions from pharmaceutical manufacturing based on temporal high-resolution mass spectrometry data. Environ. Sci. Technol. 2020;54:4110–4120. doi: 10.1021/acs.est.9b07085. [DOI] [PubMed] [Google Scholar]
- 148.Klümper U, et al. Selection for antimicrobial resistance is reduced when embedded in a natural microbial community. ISME J. 2019;13:2927–2937. doi: 10.1038/s41396-019-0483-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kraupner N, et al. Selective concentrations for trimethoprim resistance in aquatic environments. Environ. Int. 2020;144:106083. doi: 10.1016/j.envint.2020.106083. [DOI] [PubMed] [Google Scholar]
- 150.Murray AK, et al. Novel insights into selection for antibiotic resistance in complex microbial communities. mBio. 2018 doi: 10.1128/mbio.00969-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Government of India. Environment (Protection) Amendment Rules, 2020 – Inviting comments/suggestions on Environmental Standards for Bulk Drug and Formulation (Pharmaceutical) Industry, http://moef.gov.in/g-s-r-44-e-date-23-01-2020-environment-protection-amendment-rules-2020-inviting-commentssuggestions-on-environmental-standards-for-bulk-drug-and-formulation-pharmaceutical-indu/ (2020).
- 152.Tell J, et al. Science-based targets for antibiotics in receiving waters from pharmaceutical manufacturing operations. Integr. Environ. Assess. Manag. 2019;15:312–319. doi: 10.1002/ieam.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Greenfield BK, et al. Modeling the emergence of antibiotic resistance in the environment: an analytical solution for the minimum selection concentration. Antimicrob. Agents Chemother. 2018 doi: 10.1128/aac.01686-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Murray AK, et al. The ‘Selection end points in Communities of bacTeria’ (SELECT) method: a novel experimental assay to facilitate risk assessment of selection for antimicrobial resistance in the environment. Environ. Health Perspect. 2020;128:107007. doi: 10.1289/EHP6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 156.Stanton IC, Murray AK, Zhang L, Snape J, Gaze WH. Evolution of antibiotic resistance at low antibiotic concentrations including selection below the minimal selective concentration. Commun. Biol. 2020 doi: 10.1038/s42003-020-01176-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nijsingh N, Munthe C, Larsson DGJ. Managing pollution from antibiotics manufacturing: charting actors, incentives and disincentives. Environ. Health. 2019;18:95. doi: 10.1186/s12940-019-0531-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sundin GW, Wang N. Antibiotic resistance in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2018;56:161–180. doi: 10.1146/annurev-phyto-080417-045946. [DOI] [PubMed] [Google Scholar]
- 159.Government of Sweden. Uppdrag angående försöksverksamhet för en miljöpremie i läkemedelsförmånssystemet, https://www.regeringen.se/499677/contentassets/36dcec65be904fd58e5e6b01c2f99709/uppdrag-angaende-forsoksverksamhet-for-en-miljopremie-i-lakemedelsformanssystemet-tlv.pdf (2021).
- 160.Norwegian Hospital Procurement Trust. New environmental criteria for the procurement of pharmaceuticals, https://sykehusinnkjop.no/nyheter/new-environmental-criteria-for-the-procurement-of-pharmaceuticals (2019).
- 161.Swedish Procurement Agency. Pharmaceuticals, https://www.upphandlingsmyndigheten.se/kriterier/sjukvard-och-omsorg/lakemedel/ (2021).
- 162.G7. G7 Health Ministers’ Declaration, Oxford, 4 June 2021, https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/992268/G7-health_ministers-communique-oxford-4-june-2021_5.pdf (2021).
- 163.Årdal C, et al. Supply chain transparency and the availability of essential medicines. Bull. World Health Organ. 2021;99:319–320. doi: 10.2471/BLT.20.267724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Graham D, Giesen M, Bunce J. Strategic approach for prioritising local and regional sanitation interventions for reducing global antibiotic resistance. Water. 2018;11:27. doi: 10.3390/w11010027. [DOI] [Google Scholar]
- 165.Margot J, et al. Treatment of micropollutants in municipal wastewater: ozone or powdered activated carbon? Sci. Total. Environ. 2013;461–462:480–498. doi: 10.1016/j.scitotenv.2013.05.034. [DOI] [PubMed] [Google Scholar]
- 166.Larsson DGJ, et al. Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environ. Int. 2018;117:132–138. doi: 10.1016/j.envint.2018.04.041. [DOI] [PubMed] [Google Scholar]
- 167.Laxminarayan R, et al. The Lancet Infectious Diseases Commission on antimicrobial resistance: 6 years later. Lancet Infect. Dis. 2020;20:e51–e60. doi: 10.1016/S1473-3099(20)30003-7. [DOI] [PubMed] [Google Scholar]
- 168.Ahammad ZS, Sreekrishnan TR, Hands CL, Knapp CW, Graham DW. Increased waterborne blaNDM-1 resistance gene abundances associated with seasonal human pilgrimages to the upper Ganges River. Environ. Sci. Technol. 2014;48:3014–3020. doi: 10.1021/es405348h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kookana RS, et al. Potential ecological footprints of active pharmaceutical ingredients: an examination of risk factors in low-, middle- and high-income countries. Philos. Trans. R. Soc. B Biol. Sci. 2014;369:20130586. doi: 10.1098/rstb.2013.0586. [DOI] [PMC free article] [PubMed] [Google Scholar]