Abstract
Comprehensive characterization of post-translationally modified histone proteoforms is challenging due to their high isobaric and isomeric content. Trapped ion mobility spectrometry (TIMS), implemented on a quadrupole/time-of-flight (Q-ToF) mass spectrometer, has shown great promise in discriminating isomeric complete histone tails. The absence of electron activated dissociation (ExD) in the current platform prevents from a comprehensive characterization of unknown histone proteoforms. In the present work, we report for the first time the use of an electromagnetostatic (EMS) cell devised for non-ergodic dissociation based on electron capture dissociation (ECD), implemented within a nESI-TIMS-Q-ToF mass spectrometer for the characterization of acetylated (AcK18 and AcK27) and trimethylated (TriMetK4, TriMetK9 and TriMetK27) complete histone tails. The integration of the EMS cell in a TIMS-q-TOF MS permitted fast mobility-selected top-down ECD fragmentation with near 10% efficiency overall. The potential of this coupling was illustrated using isobaric (AcK18/TriMetK4) and isomeric (AcK18/AcK27 and TriMetK4/TriMetK9) binary H3 histone tail mixtures, and the H3.1 TriMetK27 histone tail structural diversity (e.g., three IMS bands at z = 7+). The binary isobaric and isomeric mixtures can be separated in the mobility domain with RIMS>100 and the non-ergodic ECD fragmentation permitted the PTM localization (sequence coverage of ~86%). Differences in the ECD patterns per mobility band of the z=7+ H3 TriMetK27 molecular ions suggested that the charge location is responsible for the structural differences observed in the mobility domain. This coupling further enhances the structural toolbox with fast, high resolution mobility separations in tandem with non-ergodic fragmentation for effective proteoform differentiation.
Graphical Abstract

Post-translational modifications (PTMs) of histones (H2A, H2B, H3, and H4), which are nucleosome core particles, play a major role in regulating chromatin dynamics and influence processes such as transcription and DNA replication.1–6 While histones consist of ~100–150 residues, most of the PTMs are located in the N-terminal part of the proteins, so-called histone tails that protrude from the nucleosome.4 These histone tails can be heavily modified with acetylation, methylation, phosphorylation, trimethylation and others on arginine, lysine, serine and/or threonine residues at different and/or multiple sites.5–12 These modifications in the histone code substantially alter the nature of the chromatin domains and then have an impact on the function of the whole genome.12–14 The nature and position of each PTM is crucial to this histone code and must be elucidated to decipher how such a code is translated into biological response.2 The analytical challenge consist in discriminating between isobaric and/or isomeric proteoforms that differ in the type and/or position site of PTM(s).
Analysis of histone proteoforms by conventional proteomic approaches, involving tandem mass spectrometry (MS/MS), have become a fundamental tool for the characterization of isobaric and isomeric PTMs.15–17 Traditional bottom-up proteomic strategies showed potential in identifying and quantifying individual PTMs of short peptides, derived from enzymatic digestion, but usually failed to elucidate the proteoform specific connectivity which is the key to the histone functions.18, 19 Instead, middle-down and top-down mass spectrometry methods are gaining momentum and enable detailed characterization of histones that are decorated with multiple co-occurring PTMs.20, 21 Such co-existing PTMs exhibit inter- and intra-molecular interplay and crosstalk over longer distances and as such, combinatorial histone marks are of great biological interest.22 Liquid chromatography (LC) can achieve separation in the bottom-up mass range (peptides <2.5 kDa) but struggle with the middle-down mass range (histone tails, 5–6 kDa) or intact histones, especially for isomeric PTMs.23–28 Advances in non-ergodic electron-based fragmentation (ExD) and ultraviolet photodissociation (UVPD) methods have significantly advanced top-down and middle-down proteomics by providing much more detailed and accurate proteoform characterization.19, 29–36
Ion mobility spectrometry (IMS) coupled to MS has emerged as an alternative to LC-MS for the characterization of biomolecules, particularly for the case of the separation of isomeric species.37–39 In particular, linear IMS (e.g., trapped IMS, TIMS)40, 41 and non-linear IMS (e.g., field asymmetric waveform IMS, FAIMS)42 exhibited great potential in separating histone tail proteoforms.43, 44 While FAIMS has been coupled to ExD for investigating histone tail PTMs,44, 45 TIMS in tandem with ECD has only been demonstrated for the case of effective mobility separation and identification of isomeric glycans in an FT-ICR MS platform.46 The recent introduction of the electromagnetostatic (EMS)47, 48 cell capable of performing ECD without the need for long reaction times or ultrahigh vacuum has opened new avenues for top-down analysis using MS/MS (e.g., triple quadrupole,49–51 q-TOF52–54 MS and Orbitrap55–58), and more recently IMS-ToF-MS/MS platforms.54
In the present work, we describe the first implementation of the EMS cell into a TIMS-q-ToF MS platform. The potential of fast, high resolution mobility separation in tandem with non-ergodic top-down dissociation is illustrated for the separation and identification of proteoforms (H3.1 histone tails) with known PTM locations from binary isomeric and isobaric mixtures. The potential of this novel platform is also shown for structural analysis of conformers/protomers.
EXPERIMENTAL SECTION
Materials and Reagents.
A set of five histone H3.1 tail peptides (ARTKQTARKSTGGKAPRKQLATKAARK-SAPATGGVKKPHRYRPGTVALRE) was probed incorporating acetylation (AcK18 and AcK27) and trimethylation (TriMetK4, TriMetK9 and TriMetK27) PTMs in biologically relevant positions (MW 5381 Da).44 These histone tails are proteotypic for the endoproteinase Glu-C derived peptides from histone H3. The synthetic histone H3.1 tails were purchased from GenScript (Piscataway, NJ). H3.1 tail solutions were analyzed at a concentration of 5 μM in 50:50 water/methanol (H2O/MeOH) solvent conditions. A low concentration tuning mix standard (G1969–85000, Agilent Technologies, Santa Clara, CA) was used for external mobility and mass calibration.
TIMS-MS Instrumentation.
Ion mobility experiments were carried out on a custom built nanoESI-TIMS-q-ToF MS (Bruker Daltonics Inc., Billerica, MA).40, 41 The nanoESI emitters were pulled in-house from quartz capillaries (O.D. = 1.0 mm and I.D. = 0.70 mm) using a Sutter Instrument Co. P2000 laser puller. A capillary voltage of ~800 V relative to the interface entrance were applied to sample solutions. The TIMS unit is controlled by an in-house software in LabView (National Instruments) and synchronized with the MS platform controls.40
The general fundamentals of TIMS as well as the calibration procedure have been described in the literature.59–64 Briefly, an rf is applied to the electrodes of the TIMS analyzer to generate a radially confining pseudopotential, while an axial electric field gradient is produced across the electrodes to counteract the drag force exerted by the gas flow, effectively leading to the trapping of the ions. Ions are then eluted from the TIMS analyzer by decreasing the axial electric field.
TIMS-MS experiments were performed using nitrogen (N2) at ambient temperature (T) with a gas velocity (vg) defined by the funnel entrance (P1 = 2.6 mbar) and exit (P2 = 0.8 mbar) pressure differences. An rf voltage of 250 Vpp at 880 kHz was applied to all electrodes. A voltage ramp (Vramp) of −100 to −40 V, deflector voltage (Vdef) of 80 V and base voltage (Vout) of 60 V were used for the mobility separations. Collision-induced dissociation (CID) experiments were performed in the collision cell located after the TIMS analyzer. The mass-selected [M+7H]7+ ions were fragmented using nitrogen as collision gas at a collision voltage of ~42 V. The scan rate (Sr = ΔVramp/tramp) was selected to trap all charge states in a single experiment and optimized for mobility separation. All resolving power (R) and resolution (r) values reported herein were determined as R = Ω/w and r = 1.18*(Ω2-Ω1)/(w1+w2), where Ω and w are collision cross sections (CCS) and the full peak width at half maximum (FWHM) of the IMS profile.
TIMS-q-ECD-ToF MS Platform.
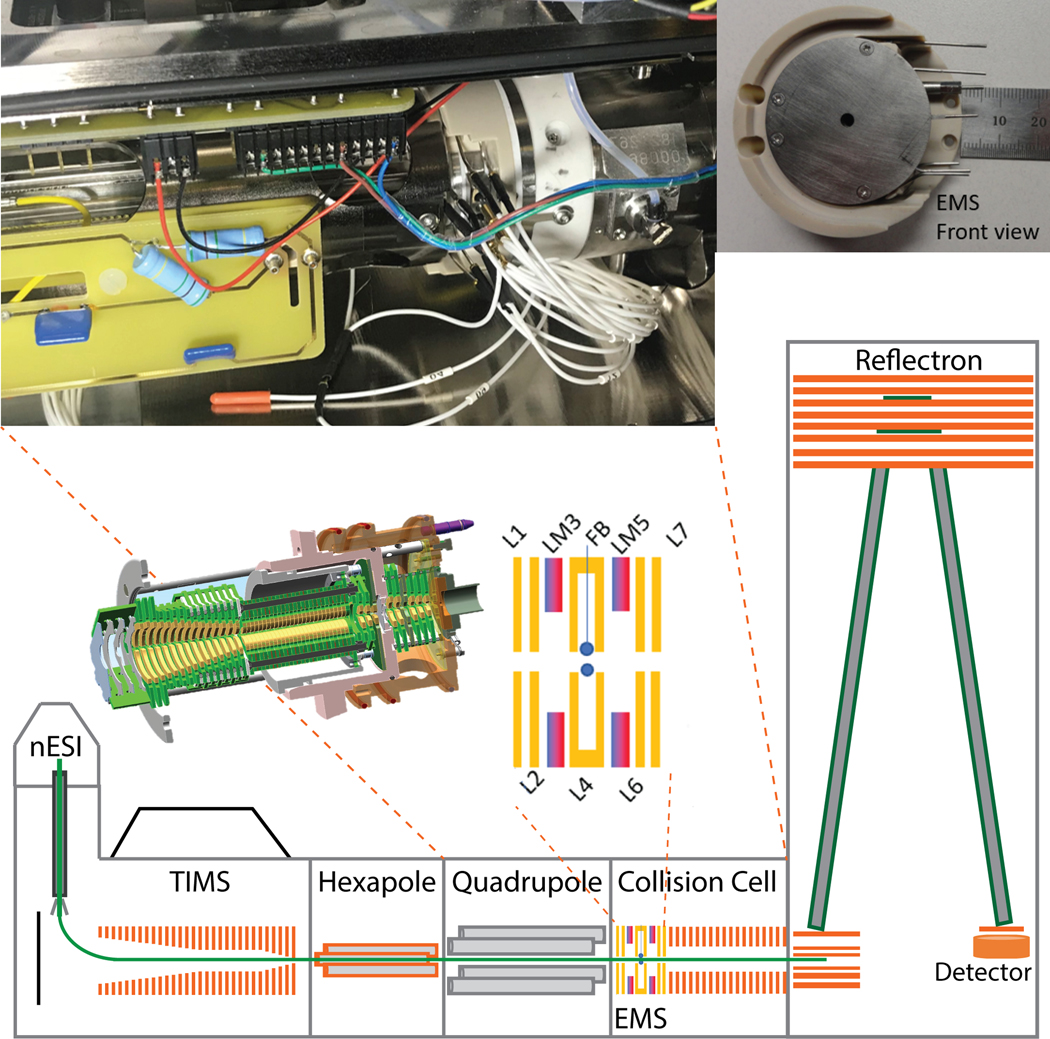
A custom built EMS (e-MSion Inc., Corvallis, OR) was attached to a custom built collision cell and mounted between the quadrupole exit and the pulsing plates of the ToF MS (Figure 1). The shortened collision cell was operated at 1500 Vpp at 2.3 MHz. The 19 mm long EMS cell is composed of seven cylindrical electrostatic lenses (L1-L7), two ring magnets and a heated rhenium filament (Scientific Instrument Services, Ringoes, NJ) housed in L4, where electrons are generated at the center of the cell (Figure 1). Electrons are confined along the ion longitudinal axis. The filament was operated at a current of 2.5 A. The electrostatic lenses applied to the EMS cell were tuned to get maximum ion intensity in the transmission mode (non-ECD mode) and optimal ECD fragmentation events in ECD mode (Table S1). The filament was turned off (0 A) during transmission mode and then manually turned on using the ExDControl software (e-MSion) when acquiring ECD data. The collision cell was operated using high purity argon (oxygen free) to enhance the cooling of the ions. No changes to the TIMS operation were required due to the fast speed of the ECD events (~ 10 μs). 2D-IMS-MS spectra were summed every 12.5s (100 acquisitions of 125 ms). Due to the low ECD efficiency (typical fragment signal <1% intensity relative to the parent ion), a minimum of 15 frames (~3 min) for abundant fragment ion signal was used with 75 frames (~15 min) as a standard protocol. ECD events are aligned with the mobility scan steps which allows for precursor-fragment mobility alignment.
Figure 1.
Schematic of the TIMS-q-EMS-CC-oToF MS instrument. In details, pictures of the q-EMS-CC interface and the EMS front view. The EMS lenses (L1-L7), magnet (LM3 and LM5) and filament (FB) arrangement is detailed.
The MS fragment ion annotations were performed using a custom excel table with all theoretical combinations of fragments based on the peptide sequence. The fragment ions were assigned with a mass error of ~15 ppm average with S/N of >3–4 in the ECD spectra. Note that Tuning Mix calibration 2D-IMS-MS were found similar when comparing before and after the implementation of the EMS cell (Figure S1). Complementary CID, higher-energy collisional dissociation (HCD) and electron transfer dissociation (ETD) tandem mass spectra acquired using an Orbitrap Eclipse Tribrid mass spectrometer (Thermo Fisher Scientific) are shown in the supporting information. Data was deconvoluted with the Xtract algorithm (Thermo) and annotated using the Mascot search engine (v2.7, Matrix Science, UK). The ionization source as well as MS/MS parameters are detailed in Supporting Information.
RESULTS AND DISCUSSION
TIMS-MS Analysis of Histone Tail Proteoforms.
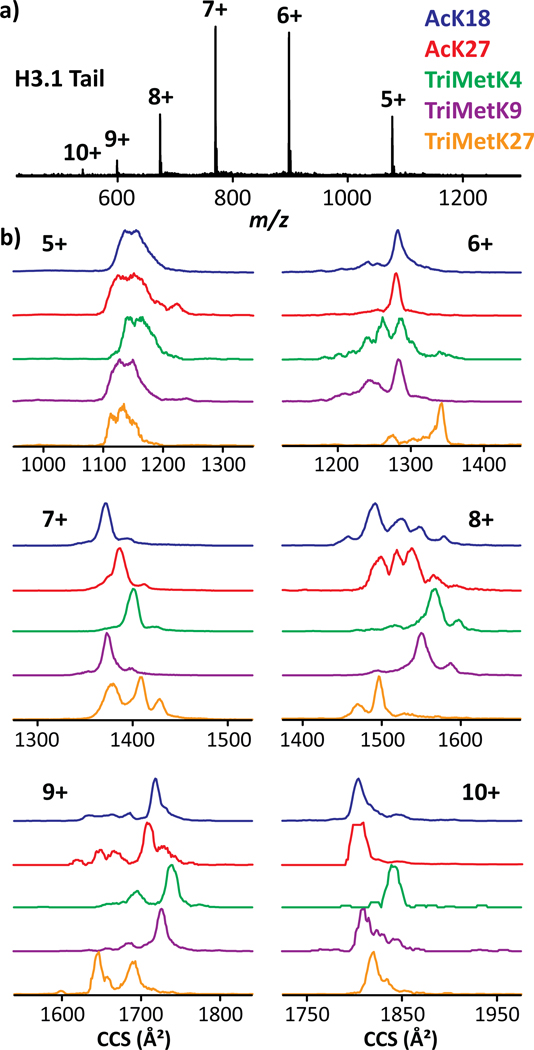
The mass spectrometric analysis of the investigated H3.1 tail proteoforms in denaturing solution conditions (i.e., 50:50 H2O/MeOH) exhibited a charge state distribution ranging from [M + 5H]5+ to [M + 10H]10+ molecular ion species centered at 7+ (Figure 2a). Inspection of the TIMS profiles at Sr = 0.48 V/ms exhibited a large structural heterogeneity (Figure 2b). Most histone proteoforms displayed characteristic IMS features (position and/or relative abundance of the IMS bands), for which the PTM location influences the intramolecular network leading to significant conformational changes. The CCS values were also found increasing with the charge state, which can be associated to more extended structures induced by the increase of the coulombic interactions with the net charge.
Figure 2.
TIMS-MS analysis showing (a) MS and (b) TIMS profiles for the multiply protonated species of H3.1 AcK18 (blue), AcK27 (red), TriMetK4 (green), TriMetK9 (purple) and TriMetK27 (orange) tails.
All the investigated H3.1 tail proteoforms, including AcK18 (blue), AcK27 (red), TriMetK4 (green), TriMetK9 (purple) and TriMetK27 (orange) PTMs, were separated in the mobility domain at a given charge state except for z = 5+ (Figure 2b). Although the isomeric H3.1 AcK18 and AcK27 tails exhibited similar CCS profiles for the z= 6+, 9+ and 10+, they can be separated in the 7+ and 8+ (based on the most intense IMS bands) charge states; the conformational multiplicity of TriMetK27 in 7+ will obstruct separations in mixture with other PTMs. The TriMetK4, TriMetK9 and TriMetK27 can be discriminated from the two isobaric AcK18 and AcK27 PTMs in 7+/8+/9+/10+, 8+/9+ and 6+/9+/10+ charge states, respectively (Figure 2b). Overall, the investigated H3.1 tail PTMs can be mobility separated in at least one charge state.
TIMS-q-ECD-ToF MS of Isobaric/Isomeric H3.1 PTM Tails.
The potential of TIMS-q-ECD-ToF MS is illustrated for the [M + 7H]7+ molecular species using binary mixtures of isobaric and isomeric histone tail proteoforms (Figures 3). The charge state 7+ was selected based on the relative simplicity of the mobility profiles (mainly one IMS band), differences in CCS between the two components, and highest abundance (Figure 2). Note that the 10+ could also have been a good choice for certain histone tails, but appears at lower intensity. Overall, a sequence coverage of ~86% and ECD fragmentation efficiency of ~0.05–0.4% per fragment ion (~10% efficiency overall, Table S2) was observed for all investigated histone tails. Additional CID, HCD and ETD spectra obtained from an Orbitrap instrument as well as TIMS mobility-selected CID spectra on the selected [M + 7H]7+ molecular species of the investigated isobaric and isomeric histone tail mixtures can be found in Figures S2–S3 and S4–S7, respectively, for direct comparison with the ECD spectra (Table S3).
Figure 3.
TIMS-ECD analysis showing the (a, c) TIMS profiles of H3.1 AcK18 (blue), TriMetK4 (green) and AcK27 (red) with the binary mixtures (black) and (b, d) mobility-selected ECD spectra of the selected [M +7H]7+ species of AcK18, TriMetK4 and AcK27 tails (m/z 769.7). The fragments comprising the PTM are highlighted in blue, green and red for AcK18, TriMetK4 and AcK27, respectively.
Figure 3a displays the mobility profiles for AcK18 (blue), TriMetK4 (green) and their binary mixture (black). A scan rate of Sr = 0.24 V/ms was sufficient for minimal mobility separation of the two components, with an apparent mobility R ~ 115 and r ~ 1.05. Note that slower scan rate experiments (Sr = 0.13 V/ms) allowed to baseline separate the two components, with an apparent mobility R ~ 175 and r ~ 1.6 (Figure S4). Results from the mobility-selected ECD experiments are shown in (Figure 3b). The charge-reduced [M + 7H]•6+ ions (m/z 898.9) were detected as the major species for the two histone PTMs. In addition, similar ECD fragmentation patterns were observed between AcK18 and TriMetK4 for [c18′ to c49′] and [z4• to z32• / z47• to z49•] product ions corresponding to either fragments not containing the PTM or fragments comprising the PTM (Figure 3b). However, specific product ions were observed for the two histone tails, where a shift of 42 Da were obtained for [c4′ to c17′] and [z33• to z46•] fragments, confirming the PTM localization at position 18 for the IMS band 1 (ECD 1) and at position 4 for IMS band 2 (ECD 2, Figure 3b). Although ultrahigh resolution mass analyzer (FT-ICR or Orbitrap) are needed to discriminate between acetylation and trimethylation PTMs, we previously demonstrated that histone tails can be discriminated based on their mobility profiles.43
Figures 3c and S8a display the mobility profiles for the isomeric AcK18 (blue)/AcK27 (red) and TriMetK4 (green)/TriMetK9 (purple) with their binary mixtures (black), respectively. A scan rate of Sr = 0.24 V/ms was sufficient for minimal separation of the two components, with an apparent mobility R ~ 160 (r ~ 0.9) and R ~ 130 (r ~ 0.8), respectively. Slower scan rate experiments (Sr = 0.07/0.13 V/ms) allowed to quasi-baseline resolve the two components, with an apparent mobility R ~ 200 (r ~ 1.0) and R ~ 220 (r ~ 1.4) for the AcK18/AcK27 and TriMetK4/TriMetK9 binary mixtures, respectively (Figures S5–S6). Mobility-selected ECD experiments yielded the charge-reduced [M + 7H]•6+ ions (m/z 898.9) as the more abundant species for the two histone PTM pairs (Figures 3d and S2b). Common ci′/zj• series were observed between AcK18/AcK27 and TriMetK4/TriMetK9, including [c4′ to c17′ / c27′ to c49′]/[z4• to z23• / z33• to z49•] and [c9′ to c49′]/[z4• to z41• / z47• to z49•] product ions, respectively. However, specific ci′/zj• fragments were observed for the two histone PTM pairs, where a shift of 42 Da were obtained for [c18′ to c26′]/[z24• to z32•] and [c4′ to c8′]/[z42• to z46•] product ions, respectively (Figures 3d and S2b). These ECD data were consistent with PTM localization at position 18 for the IMS band 1 (ECD 1) and at position 27 for IMS band 2 (ECD 2) of AcK18/AcK27 (Figure 3d) as well as at position 4 for IMS band 2 (ECD 2) and at position 9 for IMS band 1 (ECD 1) of TriMetK4/TriMetK9 (Figure S2b).
The mobility-selected ECD spectra allowed clear mobility separation and localization of the PTM sites for the isobaric and isomeric histone proteoforms. Closer inspection showed that the fragment ions correspond to ECD-like processes (e.g., ci′/zj• series); CID events were not observed under the optimized instrumental settings. Although similar bi′/yj• series using CID/HCD, corresponding to either fragments not containing the PTM or fragments comprising the PTM, were observed for the investigated histone tail mixtures, specific product ions were observed, where a shift of 42 Da were obtained corresponding to either an acetylation or trimethylation at this specific location (Figures S2–S7, Table S3). This implies that the acetylation and trimethylation PTMs are not labile enough and can be detected using CID/HCD techniques. However three major concerns using CID/HCD were observed as compared to ECD: i) the sequence coverage provided by the CID was found lower (~ 68–74%) than with ECD (~ 86%), ii) the amount together with the intensity of fragments containing the PTM, which were lower for CID than ECD and iii) the localization of the fragments containing the PTM, which were not across the entire sequence as in the case of ECD that could induce ambiguities because of the large number of lysine residues present in the histone tails. All of these features make ECD more suitable for investigating histone tail mixtures including acetylation and trimethylation PTMs.
TIMS-q-ECD-ToF MS of Heterogeneous Mobility Profiles.
The mobility analysis of H3.1 TriMetK27 tail resulted in the observation of three IMS bands for the [M + 7H]7+ species with an apparent mobility resolution of R ~ 120 using a Sr = 0.48 V/ms (Figure 4a and b). Slower scan rate experiments (Sr = 0.29 V/ms) allowed to quasi-baseline separate the three IMS bands, with an apparent mobility R ~ 175 (Figure S7). Mobility-selected ECD product ion spectra are shown in Figure 4c. The charge-reduced [M + 7H]•6+ ions (m/z 898.9) were detected as major species for all three IMS bands. Classic ci′/zj• series with a shift of 42 Da, including [c27′ to c49′] and [z24• to z49•] product ions, were obtained for the three IMS bands and were consistent with PTM localization at position 27 (Figure 4c). A sequence coverage of ~86% was observed together with an ECD fragmentation efficiency of ~0.05–0.4% per fragment ion (~10% efficiency overall, Table S2).
Figure 4.
TIMS-ECD analysis showing the (a) TIMS profiles of H3.1 TriMetK27, (b) isolation window of the mass-selected [M + 7H]7+ ions (m/z 769.7).and (c) mobility-selected ECD spectra of the three observed IMS bands The fragments comprising the PTM as well as potential charged residues are highlighted in orange.
Closer inspection showed differences in the mobility selected ECD fragmentation pattern. A charge location analysis per IMS band was investigated based on the ECD profiles. In the IMS band 1 (ECD 1, Figure 4c), singly charged species were only observed for [c4′ to c6′] fragments, suggesting that Arg2 was protonated. An additional charge was only observed for [c8′2+ to c16′2+] fragments, suggesting the protonation of the Arg8 residue. Triply charged species were become more abundant for [c17′3+ to c25′3+] fragments, suggesting the protonation of Arg17 residue. An additional charge was observed from [c26′4+ to c38′4+], suggesting the protonation of Arg26 residue. From [c39′5+ to c46′5+], mainly 5+ fragments ions were observed, for which the His39 residue probably carry the charge. An additional charge for [c47′6+ to c48′6+] product ions suggested the protonation of Arg42 residue. Finally, the presence of c49′7+ species suggested the protonation of Arg49 residue. These observations were also consistent with the zj• series. The IMS band 2 (ECD 2, Figure 4c) analysis resulted similar to the one observed for the IMS band 1, suggesting that these structures are probably conformers with similar protonation scheme. (Figure 4c).
Comparison of the IMS band 3 (ECD 3, Figure 4c) and that of IMS band 1/2 (ECD 1/2) showed differences in the ECD fragmentation in the [c39′ to c49′] / [z4• to z11•] regions. The presence of [c44′7+ to c49′7+] together with the absence of [z4• to z6•] fragments suggest that the Arg49 residue is not protonated (Figure 4c). In addition, z9• showed as single charged ions for IMS band 3 while IMS bands 1/2 showed z9•2+ fragments doubly charged, suggesting that the Arg42 residue is probably protonated. IMS band 3 showed fragments with 5+ charges starting at c40′5+ as compared to c39′5+ for IMS bands 1/2 suggesting that an additional charge is probably located on the Arg40 residue. Moreover, the observation of [z16•3+ to z23•3+] fragments implied that one of the two Lys36 and Lys37 residues probably carry a charge. These differences in the ECD fragmentation pattern suggest the presence of different proton distributions between IMS bands 1/2 and IMS band 3. The distribution of the charge residues along the sequence is in good agreement with the mobility distribution of the bands. For example, the closer proximity of the charged residues in the case of the IMS band 3 relative to 1 and 2 leads to a more extended structure (larger CCS) due to the stronger coulombic repulsion. In addition, inspection of the mobility-selected CID fragmentation pattern appeared similar across the IMS bands (Figure S7). This indicates that beside ECD being more convenient over CID by yielding higher sequence coverage, amount and localization of fragments containing the PTM, mobility-selected ECD experiments also provide fingerprints associated with the gas-phase conformational isomers of the same species.
CONCLUSION
An EMS cell has been successfully integrated in a TIMS-q-ToF MS instrument for the characterization of histone tail proteoforms. The high mobility resolving power of the TIMS analyzer permitted the separation of the H3.1 isobaric/isomeric tails with varying PTM locations in at least one charge state. The observed fragment ions correspond to ECD-like processes (e.g., ci′/zj• series), for which CID events were not observed under the optimized instrumental settings. These experiments also allowed the identification of structural differences evidenced by different mobility bands caused by different charge distributions along the H3.1 tails (resulting in different ECD fragment ions). All histone PTM locations were successfully detected and high sequence coverage (~86%) was observed. Typical ECD fragment ion abundances were in the order of 0.1–0.4% per ion, corresponding to ~10% efficiency overall. The potential of this coupling was effectively illustrated using isobaric (AcK18/TriMetK4) and isomeric (AcK18/AcK27 and TriMetK4/TriMetK9) binary H3.1 histone tail mixtures, and the H3.1 TriMetK27 histone tail structural diversity (e.g., three IMS bands at z = 7+). Mobility-selected ECD approaches exhibited advantages over mobility-selected CID fragmentation, for which higher sequence coverage, amount and localization of fragments containing the PTM as well as fingerprints associated with the gas-phase conformational isomers were provided. In addition to the analytical power for the separation and identification of histone positional isomers, we anticipate that this platform will find wide application in structural biology. The capability to perform mobility measurements and non-ergodic fragmentations can further enable molecular modeling for the characterization of the conformational space of biomolecules. This platform shows promises for the analysis of intact histones with varying PTMs as well as other biological systems. Potential challenges for mobility-based top-down ExD analysis are 1) need for pre-concentration due to the relatively low ECD fragmentation efficiency, and 2) need for high resolution mobility to separate higher MW isomeric species.
Supplementary Material
ACKNOWLEDGEMENTS
The authors at FIU acknowledge the financial support from the National Science Foundation Division of Chemistry, under CAREER award CHE-1654274, with co-funding from the Division of Molecular and Cellular Biosciences to FFL and funding from National Institutes of General Medicine (R01GM134247). We thank Dr. Mark E. Ridgeway and Dr. Melvin A Park for helpful discussions during the TIMS-q-EMS-ToF MS development.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at http://pubs.acs.org. Table of the optimized electrostatic potentials, filament current and collision energy settings in the transmission and ECD modes for the EMS cell. 2D-TIMS-MS spectra for Tuning Mix obtained before and after implementation of the EMS cell. TIMS-ECD/CID analysis showing the TIMS profiles and mobility-selected ECD/CID spectra of the selected [M +7H]7+ species of H3.1 TriMetK9 and TriMetK4 tails. Table gathering the list and relative intensities of the observed ECD/CID fragments for AcK18, AcK27, TriMetK4, TriMetK9 and TriMetK27.
The authors declare no competing financial interest.
REFERENCES
- 1.Britton LM; Gonzales-Cope M; Zee BM; Garcia BA, Breaking the histone code with quantitative mass spectrometry. Expert Rev. Proteomics 2011, 8 (5), 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenuwein T; Allis CD, Translating the histone code. Science 2001, 293 (5532), 1074–1080. [DOI] [PubMed] [Google Scholar]
- 3.Zhang T; Cooper S; Brockdorff N, The interplay of histone modifications - writers that read. EMBO Rep. 2015, 16 (11), 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister AJ; Kouzarides T, Regulation of chromatin by histone modifications. Cell Res. 2011, 21 (3), 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sidoli S; Garcia BA, Middle-down proteomics: a still unexploited resource for chromatin biology. Expert Rev. Proteomics 2017, 14 (7), 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jung HR; Pasini D; Helin K; Jensen ON, Quantitative mass spectrometry of histones H3.2 and H3.3 in Suz12-deficient mouse embryonic stem cells reveals distinct, dynamic post-translational modifications at Lys-27 and Lys-36. Mol. Cell. Proteomics 2010, 9 (5), 838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wisniewski JR; Zougman A; Mann M, Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res. 2008, 36 (2), 570–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y; Wysocka J; Sayegh J; Lee YH; Perlin JR; Leonelli L; Sonbuchner LS; McDonald CH; Cook RG; Dou Y; Roeder RG; Clarke S; Stallcup MR; Allis CD; Coonrod SA, Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 2004, 306 (5694), 279–283. [DOI] [PubMed] [Google Scholar]
- 9.Turner BM, Defining an epigenetic code. Nat. Cell. Biol. 2007, 9 (1), 2–6. [DOI] [PubMed] [Google Scholar]
- 10.Mosammaparast N; Shi Y, Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 2010, 79, 155–179. [DOI] [PubMed] [Google Scholar]
- 11.Tvardovskiy A; Schwammle V; Kempf SJ; Rogowska-Wrzesinska A; Jensen ON, Accumulation of histone variant H3.3 with age is associated with profound changes in the histone methylation landscape. Nucleic Acids Res. 2017, 45 (16), 9272–9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouzarides T, Chromatin modifications and their function. Cell 2007, 128 (4), 693–705. [DOI] [PubMed] [Google Scholar]
- 13.Taverna SD; Li H; Ruthenburg AJ; Allis CD; Patel DJ, How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007, 14 (11), 1025–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strahl BD; Allis CD, The language of covalent histone modifications. Nature 2000, 403 (6765), 41–45. [DOI] [PubMed] [Google Scholar]
- 15.McLachlin DT; Chait BT, Analysis of phosphorylated proteins and peptides by mass spectrometry. Curr. Opin. Chem. Biol. 2001, 5 (5), 591–602. [DOI] [PubMed] [Google Scholar]
- 16.Benevento M; Tonge PD; Puri MC; Nagy A; Heck AJ; Munoz J, Fluctuations in histone H4 isoforms during cellular reprogramming monitored by middle-down proteomics. Proteomics 2015, 15 (18), 3219–3231. [DOI] [PubMed] [Google Scholar]
- 17.Trelle MB; Jensen ON, Functional proteomics in histone research and epigenetics. Expert Rev. Proteomics 2007, 4 (4), 491–503. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y; Fonslow BR; Shan B; Baek MC; Yates JR 3rd, Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 2013, 113 (4), 2343–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung HR; Sidoli S; Haldbo S; Sprenger RR; Schwammle V; Pasini D; Helin K; Jensen ON, Precision mapping of coexisting modifications in histone H3 tails from embryonic stem cells by ETD-MS/MS. Anal. Chem. 2013, 85 (17), 8232–8239. [DOI] [PubMed] [Google Scholar]
- 20.Sidoli S; Cheng L; Jensen ON, Proteomics in chromatin biology and epigenetics: Elucidation of post-translational modifications of histone proteins by mass spectrometry. J. Proteomics 2012, 75 (12), 3419–3433. [DOI] [PubMed] [Google Scholar]
- 21.Lu C; Coradin M; Porter EG; Garcia BA, Accelerating the Field of Epigenetic Histone Modification Through Mass Spectrometry-Based Approaches. Mol. Cell. Proteomics 2020, 20, 100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwammle V; Aspalter CM; Sidoli S; Jensen ON, Large scale analysis of co-existing post-translational modifications in histone tails reveals global fine structure of cross-talk. Mol. Cell. Proteomics 2014, 13 (7), 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan ZF; Lin S; Molden RC; Cao XJ; Bhanu NV; Wang X; Sidoli S; Liu S; Garcia BA, EpiProfile Quantifies Histone Peptides With Modifications by Extracting Retention Time and Intensity in High-resolution Mass Spectra. Mol. Cell. Proteomics 2015, 14 (6), 1696–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sidoli S; Schwammle V; Ruminowicz C; Hansen TA; Wu X; Helin K; Jensen ON, Middle-down hybrid chromatography/tandem mass spectrometry workflow for characterization of combinatorial post-translational modifications in histones. Proteomics 2014, 14 (19), 2200–2211. [DOI] [PubMed] [Google Scholar]
- 25.Lindner H; Sarg B; Meraner C; Helliger W, Separation of acetylated core histones by hydrophilic-interaction liquid chromatography. J. Chromatogr. A 1996, 743 (1), 137–144. [DOI] [PubMed] [Google Scholar]
- 26.Pesavento JJ; Bullock CR; LeDuc RD; Mizzen CA; Kelleher NL, Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J. Biol. Chem. 2008, 283 (22), 14927–14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalli A; Sweredoski MJ; Hess S, Data-dependent middle-down nano-liquid chromatography-electron capture dissociation-tandem mass spectrometry: an application for the analysis of unfractionated histones. Anal. Chem. 2013, 85 (7), 3501–3507. [DOI] [PubMed] [Google Scholar]
- 28.Young NL; DiMaggio PA; Plazas-Mayorca MD; Baliban RC; Floudas CA; Garcia BA, High throughput characterization of combinatorial histone codes. Mol. Cell. Proteomics 2009, 8 (10), 2266–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greer SM; Sidoli S; Coradin M; Schack Jespersen M; Schwammle V; Jensen ON; Garcia BA; Brodbelt JS, Extensive Characterization of Heavily Modified Histone Tails by 193 nm Ultraviolet Photodissociation Mass Spectrometry via a Middle-Down Strategy. Anal. Chem. 2018, 90 (17), 10425–10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chi A; Huttenhower C; Geer LY; Coon JJ; Syka JE; Bai DL; Shabanowitz J; Burke DJ; Troyanskaya OG; Hunt DF, Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. USA 2007, 104 (7), 2193–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molina H; Horn DM; Tang N; Mathivanan S; Pandey A, Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 2007, 104 (7), 2199–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riley NM; Hebert AS; Durnberger G; Stanek F; Mechtler K; Westphall MS; Coon JJ, Phosphoproteomics with Activated Ion Electron Transfer Dissociation. Anal. Chem. 2017, 89 (12), 6367–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siuti N; Roth MJ; Mizzen CA; Kelleher NL; Pesavento JJ, Gene-specific characterization of human histone H2B by electron capture dissociation. J. Proteome Res. 2006, 5 (2), 233–239. [DOI] [PubMed] [Google Scholar]
- 34.Thomas CE; Kelleher NL; Mizzen CA, Mass spectrometric characterization of human histone H3: a bird’s eye view. J. Proteome Res. 2006, 5 (2), 240–247. [DOI] [PubMed] [Google Scholar]
- 35.Boyne MT 2nd; Pesavento JJ; Mizzen CA; Kelleher NL, Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J. Proteome Res. 2006, 5 (2), 248–253. [DOI] [PubMed] [Google Scholar]
- 36.Coon JJ; Ueberheide B; Syka JE; Dryhurst DD; Ausio J; Shabanowitz J; Hunt DF, Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 2005, 102 (27), 9463–9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burnum-Johnson KE; Zheng X; Dodds JN; Ash J; Fourches D; Nicora CD; Wendler JP; Metz TO; Waters KM; Jansson JK; Smith RD; Baker ES, Ion Mobility Spectrometry and the Omics: Distinguishing Isomers, Molecular Classes and Contaminant Ions in Complex Samples. Trends Anal. Chem. 2019, 116, 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeanne Dit Fouque K; Fernandez-Lima F, Recent advances in biological separations using trapped ion mobility spectrometry – mass spectrometry. Trends Anal. Chem. 2019, 116, 308–315. [Google Scholar]
- 39.Wu Q; Wang J-Y; Han D-Q; Yao Z-P, Recent advances in differentiation of isomers by ion mobility mass spectrometry. Trends Anal. Chem. 2020, 124, 115801. [Google Scholar]
- 40.Fernandez-Lima FA; Kaplan DA; Park MA, Note: Integration of trapped ion mobility spectrometry with mass spectrometry. Rev. Sci. Instrum. 2011, 82 (12), 126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandez-Lima FA; Kaplan DA; Suetering J; Park MA, Gas-phase separation using a Trapped Ion Mobility Spectrometer. Int. J. Ion Mobil. Spectrom. 2011, 14 (2–3), 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shvartsburg A, Differential ion mobility spectrometry: nonlinear ion transport and fundamentals of FAIMS. CRC Press: Boca Raton, 2009. [Google Scholar]
- 43.Garabedian A; Baird MA; Porter J; Jeanne Dit Fouque K; Shliaha PV; Jensen ON; Williams TD; Fernandez-Lima F; Shvartsburg AA, Linear and Differential Ion Mobility Separations of Middle-Down Proteoforms. Anal. Chem. 2018, 90 (4), 2918–2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shliaha PV; Baird MA; Nielsen MM; Gorshkov V; Bowman AP; Kaszycki JL; Jensen ON; Shvartsburg AA, Characterization of Complete Histone Tail Proteoforms Using Differential Ion Mobility Spectrometry. Anal. Chem. 2017, 89 (10), 5461–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baird MA; Shliaha PV; Anderson GA; Moskovets E; Laiko V; Makarov AA; Jensen ON; Shvartsburg AA, High-Resolution Differential Ion Mobility Separations/Orbitrap Mass Spectrometry without Buffer Gas Limitations. Anal. Chem. 2019, 91 (10), 6918–6925. [DOI] [PubMed] [Google Scholar]
- 46.Pu Y; Ridgeway ME; Glaskin RS; Park MA; Costello CE; Lin C, Separation and Identification of Isomeric Glycans by Selected Accumulation-Trapped Ion Mobility Spectrometry-Electron Activated Dissociation Tandem Mass Spectrometry. Anal. Chem. 2016, 88 (7), 3440–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voinov VG; Deinzer ML; Barofsky DF, Electron capture dissociation in a linear radiofrequency-free magnetic cell. Rapid Commun. Mass Spectrom. 2008, 22 (19), 3087–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voinov VG; Beckman JS; Deinzer ML; Barofsky DF, Electron-capture dissociation (ECD), collision-induced dissociation (CID) and ECD/CID in a linear radio-frequency-free magnetic cell. Rapid Commun. Mass Spectrom. 2009, 23 (18), 3028–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voinov VG; Deinzer ML; Barofsky DF, Radio-frequency-free cell for electron capture dissociation in tandem mass spectrometry. Anal. Chem. 2009, 81 (3), 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voinov VG; Bennett SE; Beckman JS; Barofsky DF, ECD of tyrosine phosphorylation in a triple quadrupole mass spectrometer with a radio-frequency-free electromagnetostatic cell. J. Am. Soc. Mass Spectrom. 2014, 25 (10), 1730–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voinov VG; Bennett SE; Barofsky DF, Electron-induced dissociation of peptides in a triple quadrupole mass spectrometer retrofitted with an electromagnetostatic cell. J. Am. Soc. Mass Spectrom. 2015, 26 (5), 752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voinov VG; Deinzer ML; Beckman JS; Barofsky DF, Electron capture, collision-induced, and electron capture-collision induced dissociation in Q-TOF. J. Am. Soc. Mass Spectrom. 2011, 22 (4), 607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voinov VG; Hoffman PD; Bennett SE; Beckman JS; Barofsky DF, Electron Capture Dissociation of Sodium-Adducted Peptides on a Modified Quadrupole/Time-of-Flight Mass Spectrometer. J. Am. Soc. Mass Spectrom. 2015, 26 (12), 2096–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams JP; Morrison LJ; Brown JM; Beckman JS; Voinov VG; Lermyte F, Top-Down Characterization of Denatured Proteins and Native Protein Complexes Using Electron Capture Dissociation Implemented within a Modified Ion Mobility-Mass Spectrometer. Anal. Chem. 2020, 92 (5), 3674–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fort KL; Cramer CN; Voinov VG; Vasil’ev YV; Lopez NI; Beckman JS; Heck AJR, Exploring ECD on a Benchtop Q Exactive Orbitrap Mass Spectrometer. J. Proteome Res. 2018, 17 (2), 926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaw JB; Malhan N; Vasil’ev YV; Lopez NI; Makarov A; Beckman JS; Voinov VG, Sequencing Grade Tandem Mass Spectrometry for Top-Down Proteomics Using Hybrid Electron Capture Dissociation Methods in a Benchtop Orbitrap Mass Spectrometer. Anal. Chem. 2018, 90 (18), 10819–10827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw JB; Liu W; Vasil Ev YV; Bracken CC; Malhan N; Guthals A; Beckman JS; Voinov VG, Direct Determination of Antibody Chain Pairing by Top-down and Middle-down Mass Spectrometry Using Electron Capture Dissociation and Ultraviolet Photodissociation. Anal. Chem. 2020, 92 (1), 766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou M; Liu W; Shaw JB, Charge Movement and Structural Changes in the Gas-Phase Unfolding of Multimeric Protein Complexes Captured by Native Top-Down Mass Spectrometry. Anal. Chem. 2020, 92 (2), 1788–1795. [DOI] [PubMed] [Google Scholar]
- 59.Hernandez DR; Debord JD; Ridgeway ME; Kaplan DA; Park MA; Fernandez-Lima F, Ion dynamics in a trapped ion mobility spectrometer. Analyst 2014, 139 (8), 1913–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ridgeway ME; Lubeck M; Jordens J; Mann M; Park MA, Trapped ion mobility spectrometry: A short review. Int. J. Mass Spectrom. 2018, 425, 22–35. [Google Scholar]
- 61.Michelmann K; Silveira JA; Ridgeway ME; Park MA, Fundamentals of trapped ion mobility spectrometry. J. Am. Soc. Mass Spectrom. 2015, 26 (1), 14–24. [DOI] [PubMed] [Google Scholar]
- 62.Silveira JA; Michelmann K; Ridgeway ME; Park MA, Fundamentals of Trapped Ion Mobility Spectrometry Part II: Fluid Dynamics. J. Am. Soc. Mass Spectrom. 2016, 27 (4), 585–595. [DOI] [PubMed] [Google Scholar]
- 63.Chai M; Young MN; Liu FC; Bleiholder C, A Transferable, Sample-Independent Calibration Procedure for Trapped Ion Mobility Spectrometry (TIMS). Anal. Chem. 2018, 90 (15), 9040–9047. [DOI] [PubMed] [Google Scholar]
- 64.Silveira JA; Ridgeway ME; Park MA, High resolution trapped ion mobility spectrometery of peptides. Anal. Chem. 2014, 86 (12), 5624–5627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.