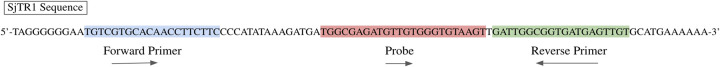Abstract
Background
Elimination and control of Schistosoma japonicum, the most virulent of the schistosomiasis-causing blood flukes, requires the development of sensitive and specific diagnostic tools capable of providing an accurate measurement of the infection prevalence in endemic areas. Typically, detection of S. japonicum has occurred using the Kato-Katz technique, but this methodology, which requires skilled microscopists, has been shown to radically underestimate levels of infection. With the ever-improving capabilities of next-generation sequencing and bioinformatic analysis tools, identification of satellite sequences and other highly repetitive genomic elements for use as real-time PCR diagnostic targets is becoming increasingly common. Assays developed using these targets have the ability to improve the sensitivity and specificity of results for epidemiological studies that can in turn be used to inform mass drug administration and programmatic decision making.
Methodology/Principal findings
Utilizing Tandem Repeat Analyzer (TAREAN) and RepeatExplorer2, a cluster-based analysis of the S. japonicum genome was performed and a tandemly arranged genomic repeat, which we named SjTR1 (Schistosoma japonicum Tandem Repeat 1), was selected as the target for a real-time PCR diagnostic assay. Based on these analyses, a primer/probe set was designed and the assay was optimized. The resulting real-time PCR test was shown to reliably detect as little as 200 ag of S. japonicum genomic DNA and as little as 1 egg per gram of human stool. Based on these results, the index assay reported in this manuscript is more sensitive than previously published real-time PCR assays for the detection of S. japonicum.
Conclusions/Significance
The extremely sensitive and specific diagnostic assay described in this manuscript will facilitate the accurate detection of S. japonicum, particularly in regions with low levels of endemicity. This assay will be useful in providing data to inform programmatic decision makers, aiding disease control and elimination efforts.
Author summary
Schistosomiasis is a Neglected Tropical Disease (NTD) estimated to infect more than 230 million people worldwide. Of the various species of schistosomes that cause disease in humans, Schistosoma japonicum is considered the most virulent. As such, this pathogen presents a crucial public health threat. Typically, diagnosis of S. japonicum has been performed via the Kato-Katz technique which has been shown to dramatically underestimate the burden of infection, resulting in a need for improved detection strategies. To address this need, we have employed bioinformatic tools in order to identify a tandemly arranged, highly repetitive, DNA sequence, SjTR1, in the S. japonicum genome. Utilizing this sequence as a real-time PCR assay target, we have developed a sensitive and specific assay for the detection of S. japonicum DNA. Employment of this assay in field settings will facilitate the accurate detection of S. japonicum and provide guidance capable of informing mass drug administration efforts targeting elimination.
Introduction
Schistosomiasis is a debilitating Neglected Tropical Disease (NTD) estimated to infect more than 230 million people worldwide. Of the blood flukes causing this disease, Schistosoma japonicum, which is endemic in China, Indonesia, Taiwan (zoophilic strain), and the Philippines, is the most virulent species [1–3]. S. japonicum is known to successfully infect 46 mammalian hosts [4] with each adult female schistosome producing approximately 1,000 eggs per day [5]. The deposition of S. japonicum eggs can result in the formation of tissue granulomas, in turn giving rise to a severe immune response that frequently leads to cognitive impairment, growth stunting, diarrhea, rectal bleeding and abdominal pain, with some people developing severe hepatosplenic disease [1,6].
While S. japonicum infection in both children and adults can be controlled through periodic administration of the preventive chemotherapeutic praziquantel [7], until now S. japonicum has only been successfully eliminated from Japan [8]. S. japonicum’s wide range of animal hosts make its elimination highly challenging [9]. This pathogen’s complicated life cycle, involving the excretion of eggs into freshwater environments followed by further growth in Oncomelania hupensis, a freshwater snail that serves as an intermediate host [9], results in the exposure of many reservoir hosts—such as bovines, rodents, goats, and dogs—to infection [10–11]. Given these challenges, elimination will require accurate, efficient, and sensitive diagnostic tools that can be applied to screening humans, other definitive hosts, and snail vectors.
Typically, detection of S. japonicum eggs from human-derived stool samples has occurred using the quantitative, coprological Kato-Katz technique [12–13]. However, this technique, which requires skilled microscopists, has been shown to have low reproducibility in determining egg counts [14] and to have poor sensitivity in low endemicity/infection intensity settings [15]. In contrast, molecular techniques such as real-time PCR can detect extremely limited concentrations of parasite-derived DNA, exhibiting greater sensitivity than microscopy-based approaches [16]. While a small number of S. japonicum-targeting real-time PCR diagnostic assays can be found in the literature, each assay has targeted comparatively low copy number sequences such as the mitochondrial NADH dehydrogenase I gene [17], a putative DNA photo-lyase gene [18], and the mitochondrial DNA 16S rRNA gene [19]. While these assays have exhibited greater sensitivity than microscopy-based techniques, targeting coding sequences inevitably raises concerns about target specificity as such genes frequently exhibit greater conservation among closely related species [20]. In contrast, non-coding tandemly repeated genomic regions make ideal real-time PCR targets, often providing high sensitivity and species-level specificity [20–21]. Here we report the development of a highly sensitive, specific real-time PCR assay for the detection of S. japonicum in human stool by targeting an abundant, tandemly-repeated genomic DNA sequence, which we have named SjTR1 (Schistosoma japonicum Tandem Repeat 1) (GenBank:MW631938).
Materials and methods
Ethics statement
Informed written consent was received from all human participants in the study and ethical approval was provided by the Ethics Committee of the Research Institute of Tropical Medicine (RITM), Manila, and the Queensland Institute of Medical Research (QIMR) Human Research Ethics Committee (Approval Number: H0309-058 (P524)).
Assay target identification
A library of raw, paired-end reads resulting from the next-generation sequencing of female S. japonicum-derived DNA (SRR6841388) was retrieved from the Sequence Read Archive at the National Center for Biotechnology Information (NCBI) website (www.ncbi.nlm.nih.gov). Following quality analysis using FASTQC, a random sampling of 500,000 sequences was analyzed using Tandem Repeat Analyzer (TAREAN) and RepeatExplorer2 software as implemented on the Galaxy-based web server [22–26]. The repetitive elements of the genome were characterized using a graph-based technique, which forms clusters of reads that demonstrate 90% or greater identity over 55% or greater of the longer sequence length. Clusters containing threshold-exceeding numbers of shared paired-end reads were then aligned to form superclusters, resulting in the identification of multiple, high-coverage consensus sequences [24–26]. From these genomic elements, the most repetitive sequence (SjTR1) was chosen as a putative real-time PCR assay target.
Real-time PCR assay design
A set of forward and reverse primers and a probe (Table 1) were designed using PrimerQuest Tool software (Integrated DNA Technologies, Coralville, IA) to target the SjTR1 sequence (Fig 1). The primers and probes were analyzed with the Primer-Blast tool, available from the NCBI website (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) to screen for possible off-target amplification sites. The probe was labeled with a 56-FAM fluorophore at the 5’ end and was double quenched with ZEN and 3IABkFQ.
Table 1. S. japonicum assay primers and probe.
| Forward Primer | 5’-TGT CGT GCA CAA CCT TCT TC-3’ |
| Reverse Primer | 5’-ACA ACT CAT CAC CGC CAA TC-3’ |
| Probe | 5’-/56-FAM/ TGG CGA GAT / ZEN/ GTT GTG GGT GTA AGT / 3IABkFQ/-3’ |
Fig 1. The amplified region of the SjTR1 genomic sequence.
Locations of the S. japonicum assay primer and probe binding sites are indicated.
Nucleic acid extraction and whole genome amplification of S. japonicum, S. mansoni, S. haematobium, and S. mekongi genomic DNA
DNA was extracted from samples of S. japonicum (Hubei, China), S. mansoni (Senegal River Basin), and S. haematobium (Zanzibar Island, Tanzania) single male worms graciously provided by the Natural History Museum, London, UK [27]. These extractions were performed using the Isolate II Genomic DNA kit from Meridian Bioscience (Memphis, TN) and extracted DNA was whole genome amplified using the REPLI-g UltraFast Mini Kit (Qiagen, Germantown, MD) in accordance with manufacturer protocols. DNA from S. mekongi worms (Maintained at Applied Malacology Laboratory, Faculty of Tropical Medicine, Mahidol University, Thailand) was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD).
Real-time PCR assay optimization and limit of detection
Real-time PCR assay optimization experiments were performed as described in previous studies [26] using the StepOnePlus Real-Time PCR System (Thermofisher Scientific, Waltham, MA). Briefly, the optimal annealing/extension temperature for the assay was determined by testing a range of annealing temperatures from 52°C to 62°C. We initially tested six temperatures at 2°C increments from 52°C to 62°C and then narrowed the range, testing six temperatures at 0.5°C increments from 56.5°C to 59°C. The annealing temperature yielding the lowest Cq value was chosen. Optimal primer concentrations were determined using 7 μL reactions containing 3.5 μL of TaqPath ProAmp Master Mix (Thermofisher Scientific), and employing doubling dilutions of forward and reverse primers at concentrations ranging from 62.5 nM to 1000 nM in all possible combinations. The limit of detection was determined under optimized assay conditions by testing ten-fold serial dilutions of a S. japonicum genomic DNA stock with template masses ranging from 100 pg to 1 ag. Negative controls for all experiments were prepared using the same master mix without any template DNA added.
Specificity panel for the S. japonicum assay
Specificity of the assay was assessed by testing against a variety of non-target DNA templates. All testing occurred in 7 μL reactions containing 3.5 μL of TaqPath ProAmp Master Mix and optimized primer/probe concentrations. All templates were added at a mass of 200 pg/reaction and included isolated DNA from S. mansoni, S. haematobium, S. mekongi, Trichuris trichiura, Strongyloides stercoralis, Ancylostoma duodenale, Ascaris lumbricoides, Baylisascaris procyonis, Ancylostoma caninum, Parascaris univalens, Anisakis typica, Necator americanus, Taenia solium, Taenia crassiceps, Giardia intestinalis, Mock Microbial Community B(HM-277D) (BEI resources, Manassas,VA), Candida albicans (strain L26), Escherechia coli, and human DNA. Origins of samples are provided in S1 Table.
Generation of plasmid control containing the assay target sequence
SjTR1 qPCR forward and reverse primers were used to amplify the target sequence from genomic S. japonicum DNA using conventional PCR. The target sequence was size selected using agarose gel electrophoresis, purified from the gel using the Monarch Genomic DNA Purification Kit (New England Biolabs, Ipswich, MA), and cloned using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen, Carlsbad, CA) as previously described [26]. The resulting plasmid was used to transform NEB Express Competent E. coli cells (New England Biolabs), which were plated on LB-kanamycin plates and grown overnight at 37°C. Following the selection of colonies, PCR and sequencing were performed to identify a plasmid containing a single copy of the assay’s target sequence which was subsequently used as a positive control and to calculate the assay’s efficiency.
Real-time PCR assay efficiency
To determine assay efficiency, the plasmid concentration was quantified using a Qubit 2.0 Fluorometer (Thermofisher Scientific), and serial dilutions were created resulting in stocks spanning 10-fold serial dilutions ranging from 100 pg/μL to 100 ag/μL. The number of plasmid copies was calculated for each dilution with 100 ag of S. japonicum DNA corresponding to 26 copies of the target sequence. The log of plasmid copies was plotted against the mean Cq resulting from the amplification of each concentration of plasmid (11 replicates per concentration) and the slope of the linear line was used to calculate the assay’s efficiency.
Testing naive human stool spiked with S. japonicum eggs
A series of Lysing Matrix E tubes (MP Biomedicals, Santa Ana, CA) were prepared according to the manufacturer’s recommendations. Three samples each of 1, 3 and 10 S. japonicum eggs were transferred to their respective Lysing Matrix E tubes. Using a sterile loop, 50 mg of naive stool (BioIVT, Westbury, NY) was then added to each tube, resulting in three samples containing 20, 60, and 200 eggs per gram (epg), respectively. Three samples were prepared for each concentration of eggs. Following preparation, all samples were homogenized for 40 seconds using the FastPrep-24 5G Instrument (MP Biomedicals) on a speed setting of 6, and DNA extraction was performed as described previously using the FastDNA Spin Kit for Soil (MP Biomedicals) [26]. To test the assay’s ability to detect as little as 1 epg of stool, additional replicates were prepared by spiking a single egg into each of ten 1 g stool samples. The weight of the stool was measured using an accurate balance and the full 1 g of stool was extracted. For preparation of these samples, a smaller volume of sodium phosphate buffer (528 μL) was used to reserve room in the tube for the additional stool. These samples were homogenized for 80 seconds using the FastPrep Instrument on a speed setting of 6 and were then subjected to DNA extraction as described above.
Comparison of index and previously published real-time PCR assay performance
To assess comparative performance, the sensitivity of our real-time PCR index assay was compared to that of previously published reference assays [17–19]. Testing was done using S. japonicum gDNA as template, as well as DNA extracted from human stool spiked with S. japonicum eggs. When conducting assays targeting SjTR1, the mitochondrial NADH dehydrogenase I [17] or putative DNA photo-lyase [18] genes, testing was performed in 7 μL reaction volumes containing 3.5 μL of TaqPath ProAmp Master Mix. When targeting the mitochondrial DNA 16S rRNA gene [19], reactions were performed in 5 μL volumes containing 2.5 μL of PrimeTime Gene Expression Master Mix (Integrated DNA Technologies). Testing using all published assays utilized primer and probe concentrations and annealing temperatures described in the literature. Each assay was tested against 2 ng and 200 pg of gDNA template with five [17–18] or three [19] replicates respectively run for each assay and each template mass. When testing against spiked stool samples, three reaction replicates were run for each assay.
Validating the SjTR1 assay using clinical samples collected from endemic area
A panel of 100 human stool samples collected from an S. japonicum endemic area in the Philippines, of which 38 were positive and 62 were negative based on Kato-Katz data, was used to validate the performance of our assay on clinical samples [28]. DNA from these samples was extracted using the FastDNA Spin Kit for Soil (MP Biomedicals). To evaluate the efficiency of each extraction, an internal control IAC plasmid (100 pg) was spiked into each extracted sample prior to the DNA binding step [29]. The recovery of the IAC plasmid was tested via real-time PCR in duplicate 7 uL reactions with 125 nM primer/probe concentrations and an annealing temperature of 59°C. All samples with mean Cq values that were >3 standard deviations from the mean were re-extracted and re-tested. To assess the performance of our index assay in comparison to a published real-time PCR reference assay, we tested all samples using the published mitochondrial NADH dehydrogenase I gene-targeting assay [17]. This assay was chosen for comparison due to its performance during the spiking experiments described above. For both assays, samples that showed inconsistent amplification (amplifying in one of two replicates) as well as samples that had a standard deviation >3 between tested replicates underwent re-testing, again in duplicate, and retest results were reported. Each experiment was run for 40 cycles and a sample was considered positive if it had a Cq of 40 or less.
Results
Target identification and real-time PCR assay design
RepeatExplorer2 and TAREAN analyses resulted in 5.95% of the analyzed S. japonicum sequence reads mapping to putative satellite repeats yielding a total of 40 clusters and superclusters. From among the 24 clusters classified as “high confidence” satellites, the cluster comprising the highest proportion of the analyzed sequences (1.7%) was chosen as our assay target. The consensus sequence resulting from this cluster, SjTR1, had a length of 101 base pairs and contained no protein-coding domains. The primers and probe designed for the assay had a GC content of 50% and produced an amplicon of 80 nucleotides (Table 1 and Fig 1).
Real-time PCR assay validation and optimization
Optimization reactions resulted in an ideal annealing/extension temperature of 59°C (S2 Table). Primer limiting reactions determined that primers performed optimally with the forward primer at a concentration of 62.5 nM and the reverse primer at a concentration of 500 nM (S3 Table). Dilutions of genomic DNA showed the assay to be reliably sensitive at template masses as low as 200 ag (Tables 2 and S4). The assay did not amplify the DNA of any of the common gut parasites, S. mansoni, S. haematobium, the Mock Microbial Community, C. albicans, E. coli, or human DNA in the specificity panel. The assay did, however, amplify S. mekongi DNA (S5 Table).
Table 2. Analytical limits of detection for the SjTR1 assay.
| Mass of S. japonicum DNA | Mean Cq +/- SD |
|---|---|
| 200 pg | 11.79 +/- 0.11 |
| 20 pg | 14.93 +/- 0.08 |
| 2 pg | 17.79 +/- 0.36 |
| 200 fg | 21.52 +/- 0.21 |
| 20 fg | 25.21+/- 0.62 |
| 2 fg | 28.71 +/- 0.18 |
| 200 ag | 32.14 +/-1.47 |
| 20 ag | N/A |
| 2 ag | N/A |
Real-time PCR assay efficiency
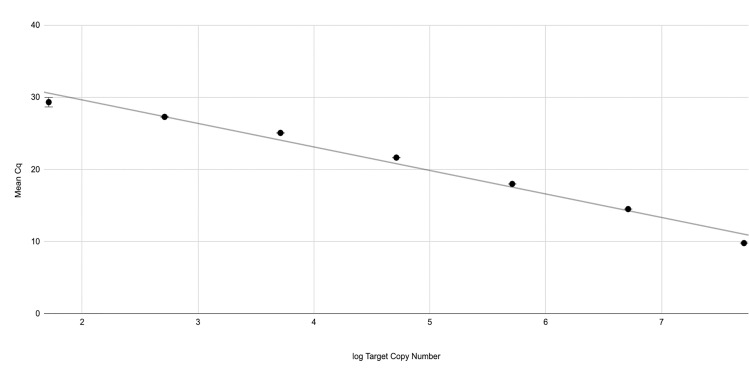
Using the size of the S. japonicum repeat-containing plasmid to estimate the number of target copies in successive 10-fold dilutions of the plasmid, a linear curve was constructed as described previously [26]. The slope of the line (-3.26) was used to calculate an assay efficiency of 102.7% with an amplification factor of 2. The R2 value of the linear curve was 0.983 (Fig 2).
Fig 2. Assay efficiency.
Ten-fold serial dilutions of the plasmid, ranging from 100 pg to 100 ag were prepared and the target copy number was estimated for each dilution (S6 Table). The log of the target copy number was plotted against the mean Cq of 11 replicates for each respective dilution. The slope of the line was used to determine the efficiency of the assay. Error bars are included but due to the small standard deviations resulting from each concentration of plasmid DNA tested, they are not distinguishable at most points.
Comparison of S. japonicum real-time PCR assay targets
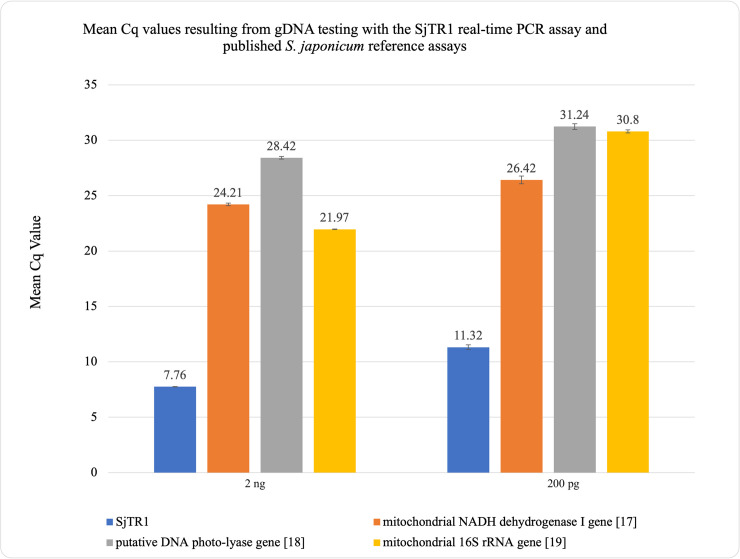
Real-time PCR comparing the detection of S. japonicum assay targets using S. japonicum gDNA showed that the mean Cq values produced by the SjTR1-targeting assay were 16.45, 20.66, and 14.21 cycles lower than those produced by previously published assay targets when testing 2 ng of template DNA and 15.10, 19.92, and 19.48 cycles lower when testing 200 pg of template DNA (Fig 3 and S7 Table) [17–19]. Although comparisons made using genomic DNA as template cannot be used to evaluate the comparative clinical sensitivities of the examined assays, these sizable differences in mean Cq values strongly suggest that the SjTR1 is considerably more abundant in copy number within the S. japonicum genome than previously published targets [17–19,21]. When testing stool spiked with S. japonicum eggs, the mean Cq values produced by the SjTR1-targeting assay were 12.70, 17.86, and 12.09 cycles lower than values produced by previously published assays on samples containing 20 epg of stool; 12.78, 17.77, and 12.27 cycles lower on samples containing 60 epg of stool; and 12.24, 17.74, and 11.75 cycles lower on samples containing 200 epg of stool [17–19] (Tables 3 and S8). Most notably, when testing 1 gram stool samples spiked with a single egg, the SjTR1-targeting assay detected S. japonicum DNA in all tested samples, while the mitochondrial NADH dehydrogenase I gene target assay [17] detected DNA in only eight of the ten samples, with sporadic detection in four of these replicates. All other assays failed to detect DNA from any of the “one egg” samples (Tables 4 and S9). Ten negative controls were analyzed in parallel, containing 1 gram of naive stool and no S. japonicum eggs. No negative control samples were amplified by any of the assays.
Fig 3. Comparison of mean Cq values.
Results from gDNA testing with the SjTR1 real-time PCR assay and previously published S. japonicum assays.
Table 3. Comparative detection of DNA extracted from naive human stool spiked with S. japonicum eggs.
| 1 egga = 20 EPG Mean Cq [Range] |
3 eggsa = 60 EPG Mean Cq [Range] |
10 eggsa = 200 EPG Mean Cq [Range] |
|
|---|---|---|---|
| SjTR1 | 16.68 [15.93–17.19] | 15.58 [14.89–15.99] | 14.06 [13.60–14.34] |
| mitochondrial NADH dehydrogenase I gene [17] | 29.38 [28.94–30.00] | 28.36 [26.73–29.46] | 26.30 [25.71–26.73] |
| putative DNA photo-lyase gene [18] | 34.54 [33.38–35.96] | 33.35 [32.36–33.88] | 31.80 [31.31–32.25]] |
| mitochondrial 16S rRNA gene [19] | 28.77 [28.44–29.17] | 27.85 [26.42–28.95] | 25.81[25.32–26.19] |
a Number of eggs spiked in a sample containing 0.05 g of stool.
Each concentration of eggs was tested using three biological replicates, and each replicate was analyzed by the SjTR1 assay in triplicate. The reported mean Cq value was calculated as a mean value of all component sample means. The reported range includes the smallest and greatest individual Cq values for each egg concentration. EPG = eggs per gram of stool.
Table 4. Comparative Detection of 1 gram stool samples spiked with an individual S. japonicum egg (positive samples are bolded; negative controls are in standard font).
| SjTR1* | mitochondrial NADH dehydrogenase I gene [17]* | putative DNA photo-lyase gene [18]* | mitochondrial 16S rRNA gene [19]* | |||||
|---|---|---|---|---|---|---|---|---|
| Mean Cq [Range]a | Total Detected | Mean Cq [Range]a | Total Detected | Mean Cq [Range]a | Total Detected | Mean Cq [Range]a | Total Detected | |
| Sample 1 | 38.45 [36.96–39.95] | 2/3 | 35.44 | 1/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 2 | 31.62 [31.42–31.95] | 3/3 | 36.58 | 1/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 4 | 30.05 [29.93–30.25] | 3/3 | 37.67 [35.02–39.31] | 3/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 5 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 6 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 7 | 30.90 [30.55–31.19] | 3/3 | 36.57 [36.27–36.86] | 2/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 8 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 9 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 10 | 33.45 [33.26–33.72] | 3/3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 11 | 31.93 [31.49–32.27] | 3/3 | 35.00 [33.42–36.20] | 3/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 12 | 30.34 [30.08–30.49] | 3/3 | 33.97 [33.58–34.40] | 3/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 13 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 14 | 33.08 [32.87–33.45] | 3/3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 15 | 31.06 [30.85–31.34] | 3/3 | 35.85 | 1/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 16 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 17 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 18 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 19 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 | N/A | 0/3 |
| Sample 20 | 33.15 [32.81–33.34] | 3/3 | 37.16 | 1/3 | N/A | 0/3 | N/A | 0/3 |
a. Each spiked sample was run in triplicate, and the mean Cq values are reported. The reported range includes the smallest and greatest individual Cq values for each individual sample. None of the negative control samples were amplified by any of the assays.
*The SjTR1 assay detected S. japonicum DNA in all 10 of the samples spiked with a single egg (amplification in three out of three replicates for 9 of the 10 samples and two out of three for one of the samples). The mitochondrial NADH dehydrogenase I real-time PCR assay [17] detected S. japonicum DNA in only eight of the ten samples tested, with sporadic detection (amplification of only one out of three replicates) in four of these samples. The real-time PCR assays targeting the putative DNA photo-lyase and mitochondrial 16S rRNA genes [18–19] did not detect S. japonicum DNA in any of the samples.
Validation of the assay on clinical samples
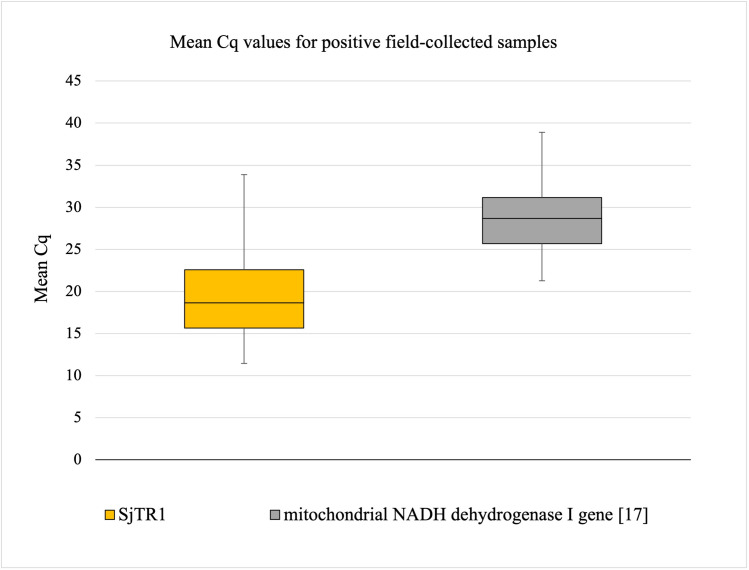
A panel of 100 field collected samples from endemic areas in the Philippines were blindly tested, of which 38 were positive and 62 were negative based on Kato-Katz data [28]. Testing of our SjTR1 real-time PCR assay resulted in 59 positive samples and 41 negative samples (S10 Table). On the other hand, testing using the NADH dehydrogenase I real-time PCR assay [17] resulted in 52 positive samples and 48 negative samples (S10 Table). Seven of the SjTR1 positive samples were not detected by the NADH dehydrogenase I assay while all 41 negatives by the SjTR1 assay were not detected by the NADH dehydrogenase I assay [17] (S10 Table). The same thirty-six of the thirty-eight Kato-Katz positive samples were amplified by both the SjTR1 and the NADH Dehydrogenase assays. The two unidentified Kato-Katz positive samples were not detected by any of the published real-time PCR assays discussed in this manuscript [17–19]. For the positive samples, the minimum, the median, the maximum, and the quartiles of mean Cq values are shown below for both assays (Fig 4). Among the positive samples, the mean Cq difference between the two assays was 10 cycles lower for the SjTR1 assay compared to the NADH dehydrogenase I assay (S10 Table).
Fig 4. Boxplots for mean Cq values of positive field collected samples.
Results for testing with the SjTR1 and mitochondrial NADH dehydrogenase I gene [17] assays.
Discussion
Endemic in 3 provinces in Indonesia, 28 provinces in the Philippines, in Taiwan (zoophilic strain), and in China, where its infection levels exceed 10% in high-risk populations, the diagnosis of S. japonicum has typically occurred using the Kato-Katz technique, a method which has been shown to underestimate infection levels by up to 70% [3,9,30–32]. Immunological tests, like antigen detection tests and enzyme-linked immunosorbent assays (ELISA), are another possible avenue for diagnosis of S. japonicum. These tests, however, often lack specificity and sensitivity [33–34]. Such detection methods may lead to insufficient intervention efforts, making the development of highly sensitive, specific molecular diagnostic tools imperative for the successful elimination of S. japonicum.
Within target areas, preventive chemotherapy efforts through praziquantel mass drug administration as well as eligibility of different age groups for treatment is determined based on prevalence of infection as assessed via positive parasitological diagnosis [35]. Thus, for the Western Pacific Region and South-East Asia, the WHO regional priorities for the 2012–2020 period included maintenance of high and/or regular coverage of preventive chemotherapy in the Philippines and Lao People’s Democratic Republic, intensification of preventive chemotherapy in Cambodia, China and Indonesia, and verification of status of elimination in Japan, Malaysia, Thailand, and India [35]. Of these areas, S. japonicum, whose elimination from its endemic areas is complicated by the presence of animal reservoir hosts, is endemic in the Philippines and China—the two countries with the highest proportion of people requiring treatment for schistosomiasis in the Western Pacific Region—and in Indonesia [9,35]. Furthermore, to determine the success of these treatment efforts, sensitive and specific diagnostic tools are needed [9,36–37].
The results reported here demonstrate that our repeat-targeting assay can reliably detect S. japonicum at concentrations as low as 1 egg per gram of human stool. Putting these results in context, S. japonicum infection intensity is defined by the World Health Organization as light (1–100 eggs per gram), moderate (101–400 eggs per gram), and heavy (more >400 eggs per gram) [38]. As such, this assay should greatly improve detection capabilities in areas of low infection intensity if used for determination of infection prevalence.
For the accurate estimation of parasite prevalence necessary to shape and guide mass treatment efforts, the specificity of a diagnostic method is also critical. Accordingly, one of our assay’s main limitations is its ability to detect the closely related Asian schistosome, S. mekongi. However, the range of S. mekongi is believed to be limited to select areas along the Mekong River Basin within the Lao People’s Democratic Republic and Cambodia [39–40]. In addition, S. japonicum and S. mekongi are only thought to overlap in a limited region in Myanmar [41]. Thus, in this area of possible co-endemicity, a positive test result from our highly sensitive assay would need to be confirmed using methods allowing for the molecular differentiation of S. mekongi and S. japonicum [42–43]. It is worth noting that our assay can reliably detect as little as 2 fg of S. mekongi gDNA, which provides evidence for the possible use of the SjTR1 assay for the detection of S. mekongi. The suitability of our assay for detection of S. mekongi in human stool as well as optimization of real-time PCR for differential detection of the two species will be explored in a future study. Further work will also test the SjTR1 assay against various S. japonicum and S. mekongi strains found in Asia. Although mass drug administration for all blood flukes causing schistosomiasis relies on the same drug, praziquantel, specificity of testing is nonetheless important for accurate surveillance of parasite endemicity and infection prevalence.
When compared to other published real-time PCR assays, testing of the SjTR1 real-time PCR assay described here demonstrated remarkable reductions in Cq values and therefore increased sensitivity. This suggests that the target of the assay, a putative tandem repeat, is more abundant in the S. japonicum genome than the target regions of the other qPCR assays tested. This provides additional support for improved analytical sensitivity via the use of satellite repeats instead of traditional targets such as ribosomal or mitochondrial DNA targets [20–21]. Evidence of the increased clinical sensitivity for our assay compared to previously published assays is supported by the results of the spiking study in which the SjTR1 assay was the only real-time PCR assay capable of reliably detecting 1 egg per gram in all samples (Table 4).
Providing further evidence of increased clinical sensitivity, our assay also showed superior performance when used to test field-collected clinical stool samples from an endemic area in the Philippines. Outperforming the most promising published real-time PCR assay through the detection of seven additional samples, our assay detected twenty-one samples more than the traditional Kato-Katz technique. We were not surprised to see that two Kato-Katz positive samples (containing 35 and 113.3 epg, respectively) were not detected by any of the four real-time PCR assays using various targets discussed in the study (S10 Table). Given the known specificity challenges associated with the Kato-Katz technique and the lack of amplification by four sensitive real-time PCR assays, we strongly suspect that those two samples represent Kato-Katz false positive results. Our assay was able to detect parasite DNA in clinical samples with as few as 3 epg, providing evidence that sensitivity of our assay seems like an unlikely cause for these results. Furthermore, the internal control results give us confidence that the quality of the extraction did not interfere with the results. Nonetheless, these two samples comprise an important avenue for further exploration and will be examined using next generation DNA sequencing in a future study. For the positive samples, the mean Cq difference between our assay and the NADH dehydrogenase assay [17] was 10 cycles, providing additional evidence for our assay’s superior sensitivity. Although these results provide strong evidence for the clinical utility of our assay, given S. japonicum’s wide range of hosts, further validation of the assay on stool samples from different endemic areas as well as multiple animal hosts will be important.
Given the acknowledged shortcomings of current coproscopic and immunological methods employed for schistosome detection, improved diagnostic methods for S. japonicum are urgently needed. Due to increased need for HIV, and more recently COVID-19, real-time PCR testing, the availability of updated PCR testing labs in endemic countries has greatly increased, facilitating the use of cost-effective, real-time PCR assays for detection of numerous infectious agents. In fact, it has been shown that the cost of PCR is comparable to that of Kato-Katz with both double-slide Kato-Katz and duplicate PCR testing of a sample costing around 2 US dollars [44–45]. In conclusion, this assay has the capacity to improve detection of S. japonicum, helping to guide programmatic decision-making efforts to control and eventually eliminate S. japonicum from endemic countries.
Supporting information
(DOCX)
(XLSX)
(XLSX)
The Cq values are provided for each replicate of forward and reverse primers diluted at concentrations ranging from 62.5 nM to 1000 nM in all possible combinations.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We would like to thank Ms. Kareen Seignon, Susan Haynes, Dr. Samantha D. Torquato, and Dr. Lori Saunders for their assistance. Computational resources were provided by the CERIT-SC Center (LM2015085) and ELIXIR-CZ project (LM2015047), part of the international ELIXIR infrastructure. Genomic DNA from the Microbial Mock Community B (Staggered, High Concentration), v5.2H, for Whole Genome Shotgun Sequencing, HM-277D was obtained through BEI Resources, NIAID, NIH as part of the Human Microbiome Project. S. japonicum eggs were provided by the NIAID Schistosomiasis Resource Center of the Biomedical Research Institute (Rockville, MD) through NIH-NIAID contract HHSN2722010000051. We’d also like to thank Dr. Aidan Emery, Dr. Fiona Allan, and Ms. Muriel Rabone at SCAN, Schistosomiasis Collection at the Natural History Museum, which is part-funded by the Wellcome Trust (grant 104958/Z/14/Z), for providing us with S. japonicum, S. mansoni, and S. haematobium samples. Material provided via SCAN was originally collected between 1993 and 2015 and we acknowledge the support and generosity of our partners from the countries concerned. We would like to thank Dr. Poom Adisakwattana at Mahidol University, Salaya, Thailand for graciously providing us with S. mekongi worms and Dr. Thomas Nutman at the National Institutes of Health (Bethesda, MD) for providing us with T. solium and T. crassiceps.
Data Availability
Sequence file is available from GenBank with accession number MW631938. All other relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the National Academy of Sciences, USA (through the Blakeslee Fund for Genetics Research awarded to Smith College). The Philippines field work study was supported by grants from UBS Optimus foundation (ID496600), the NHMRC program grant (ID1037304), and the NHMRC project grant (ID613671). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Colley DG, Bustinduy AL, Secor WE, King CH. Human schistosomiasis. The Lancet. 2014;383(9936):2253–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moendeg KJ, Angeles JMM, Nakao R, Leonardo LR, Fontanilla IKC, Goto Y, et al. Geographic strain differentiation of Schistosoma japonicum in the Philippines using microsatellite markers. PLoS Negl Trop Dis. 2017;11(7):e0005749. doi: 10.1371/journal.pntd.0005749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin M., Zheng HX., Su, J. et al. Co-dispersal of the blood fluke Schistosoma japonicum and Homo sapiens in the Neolithic Age. Sci Rep 5, 18058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He YX, Salafsky B, Ramaswamy K. Host–parasite relationships of Schistosoma japonicum in mammalian hosts. Trends Parasitol. 2001;17(7):320–324. doi: 10.1016/s1471-4922(01)01904-3 [DOI] [PubMed] [Google Scholar]
- 5.Cheever AW, Macedonia JG, Mosimann JE, Cheever EA. Kinetics of Egg Production and Egg Excretion by Schistosoma mansoni and S. japonicum in Mice Infected with a Single Pair of Worms. Am J Trop Med Hyg. 1994;50(3):281–295. doi: 10.4269/ajtmh.1994.50.281 [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Ross AG, Hou X, Lou Z, McManus DP. Oriental schistosomiasis with neurological complications: case report. Ann Clin Microbiol Antimicrob. 2011;10(1):5. doi: 10.1186/1476-0711-10-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schistosomiasis [Internet]. World Health Organization. World Health Organization; [cited 2021Jan27]. Available from: https://www.who.int/news-room/fact-sheets/detail/schistosomiasis
- 8.Rollinson D, Knopp S, Levitz S, Stothard JR, Tchuem Tchuenté LA, Garba A, et al. Time to set the agenda for schistosomiasis elimination. Acta Tropica. 2013;128(2):423–440. doi: 10.1016/j.actatropica.2012.04.013 [DOI] [PubMed] [Google Scholar]
- 9.Gordon CA, Kurscheid J, Williams GM, Clements ACA, Li Y, Zhou XN, et al. Asian Schistosomiasis: Current Status and Prospects for Control Leading to Elimination. J Travel Med. 2019;4(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray DJ, Williams GM, Li Y, Chen H, Forsyth SJ, Li RS, et al. A Cluster-Randomised Intervention Trial against Schistosoma japonicum in the Peoples’ Republic of China: Bovine and Human Transmission. PLoS ONE. 2009;4(6):e5900. doi: 10.1371/journal.pone.0005900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.VAN Dorssen CF, Gordon CA, Li Y, Williams GM, Wang Y, Luo Z, et al. Rodents, goats and dogs–their potential roles in the transmission of schistosomiasis in China. Parasitology. 2017;144(12):1633–1642. doi: 10.1017/S0031182017000907 [DOI] [PubMed] [Google Scholar]
- 12.Kato K, Miura M. Comparative examinations. Japanese Journal of Parasitology 3: 35, 1954. [Google Scholar]
- 13.Katz N, Chaves A, Pellegrino J. A simple device for quantitative stool thick-smear technique in Schistosomiasis mansoni. Rev Inst Med Trop Sao Paulo. 1972;14(6):397–400. [PubMed] [Google Scholar]
- 14.Kongs A, Marks G, Verle P, Van der Stuyft P. The unreliability of the Kato-Katz technique limits its usefulness for evaluating S. mansoni infections. Trop Med Int Health. 2001;6(3):163–169. doi: 10.1046/j.1365-3156.2001.00687.x [DOI] [PubMed] [Google Scholar]
- 15.Bärenbold O, Raso G, Coulibaly JT, N’Goran EK, Utzinger J, Vounatsou P. Estimating sensitivity of the Kato-Katz technique for the diagnosis of Schistosoma mansoni and hookworm in relation to infection intensity. PLoS Negl Trop Dis. 2017;11(10):e0005953. doi: 10.1371/journal.pntd.0005953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao GH, Li J, Blair D, Li XY, Elsheikha HM, Lin RQ, et al. Biotechnological advances in the diagnosis, species differentiation and phylogenetic analysis of Schistosoma spp. Biotechnol Adv. 2012;30(6):1381–1389. doi: 10.1016/j.biotechadv.2012.02.008 [DOI] [PubMed] [Google Scholar]
- 17.Lier T, Simonsen GS, Haaheim H, Hjelmevoll SO, Vennervald BJ, Johansen MV. Novel real-time PCr for detection of Schistosoma japonicum in stool. Southeast Asian J Trop Med Public Health. 2006;37(2):257–264. [PubMed] [Google Scholar]
- 18.Hung YW, Remais J. Quantitative detection of Schistosoma japonicum cercariae in water by real-time PCR. PLoS Negl Trop Dis. 2008;2(11):e337. doi: 10.1371/journal.pntd.0000337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alzaylaee H, Collins RA, Rinaldi G, Shechonge A, Ngatunga B, Morgan ER et al. Schistosoma species detection by environmental DNA assays in African freshwaters. PLoS Negl Trop Dis. 2020;14(3):e0008129. doi: 10.1371/journal.pntd.0008129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant JR, Pilotte N, Williams SA. A Case for Using Genomics and a Bioinformatics Pipeline to Develop Sensitive and Species-Specific PCR-Based Diagnostics for Soil-Transmitted Helminths. Front Genet. 2019;10:883. doi: 10.3389/fgene.2019.00883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zulch MF, Pilotte N, Grant JR, Minetti C, Reimer LJ, Williams SA. Selection and exploitation of prevalent, tandemly repeated genomic targets for improved real-time PCR-based detection of Wuchereria bancrofti and Plasmodium falciparum in mosquitoes. PLOS ONE. 2020;15(5):e0232325. doi: 10.1371/journal.pone.0232325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 23.Novák P, Ávila Robledillo LA, Koblížková A, Vrbová I, Neumann P, Macas J. TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017;45(12):e111. doi: 10.1093/nar/gkx257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novák P, Neumann P, Macas J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics. 2010;11:378. doi: 10.1186/1471-2105-11-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novák P, Neumann P, Pech J, Steinhaisl J, Macas J. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics. 2013;29(6):792–793. doi: 10.1093/bioinformatics/btt054 [DOI] [PubMed] [Google Scholar]
- 26.Pilotte N, Maasch JRMA, Easton AV, Dahlstrom E, Nutman TB, Williams SA. Targeting a highly repeated germline DNA sequence for improved real-time PCR-based detection of Ascaris infection in human stool. PLoS Negl Trop Dis. 2019;13(7):e0007593. doi: 10.1371/journal.pntd.0007593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emery AM, Allan FE, Rabone ME, Rollinson D. Schistosomiasis collection at NHM (SCAN). Parasit Vectors. 2012. Sep 3;5:185. doi: 10.1186/1756-3305-5-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon CA, Acosta LP, Gobert GN, Olveda RM, Ross AG, Williams GM, et al. Real-time PCR demonstrates high prevalence of Schistosoma japonicum in the Philippines: implications for surveillance and control. PLoS Negl Trop Dis. 2015;9(1):e0003483. doi: 10.1371/journal.pntd.0003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Papaiakovou M, Pilotte N, Baumer B, Grant J, Asbjornsdottir K, Schaer F, et al. A comparative analysis of preservation techniques for the optimal molecular detection of hookworm DNA in a human fecal specimen. PLoS Negl Trop Dis. 2017; 12(1): e0006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Budiono NG, Satrija F, Ridwan Y, Handharyani E, Murtini S. The contribution of domestic animals to the transmission of schistosomiasis japonica in the Lindu Subdistrict of the Central Sulawesi Province, Indonesia. Vet World. 2019;12(10):1591–1598. doi: 10.14202/vetworld.2019.1591-1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leonardo L, Chigusa Y, Kikuchi M, Kato-Hayashi N, Kawazu SI, Angeles JM, et al. Schistosomiasis in the Philippines: Challenges and some successes in control. Southeast Asian J Trop Med Public Health. 2016, 47(4): 651–666. [Google Scholar]
- 32.WHO. Expert consultation to accelerate elimination of Asian schistosomiasis. In Meeting Report WHO;WHO: Shanghai, China, 2017. [Google Scholar]
- 33.Wang W, Li Y, Li H, Xing Y, Qu G, Dai J, Liang Y. Immunodiagnostic efficacy of detection of Schistosoma japonicum human infections in China: a meta analysis. Asian Pac J Trop Med. 2012. Jan;5(1):15–23. doi: 10.1016/S1995-7645(11)60238-1 [DOI] [PubMed] [Google Scholar]
- 34.Polman K, Diakhate MM, Engels D, Nahimana S, Van Dam GJ, Falcão Ferreira ST, Deelder AM, Gryseels B. Specificity of circulating antigen detection for schistosomiasis mansoni in Senegal and Burundi. Trop Med Int Health. 2000. Aug;5(8):534–7. doi: 10.1046/j.1365-3156.2000.00600.x [DOI] [PubMed] [Google Scholar]
- 35.WHO. Schistosomiasis Progress Report (2001–2011) and Strategic Plan (2012–2020). Geneva: World Health Organization; 2013. [Google Scholar]
- 36.Gray D.J., Ross A.G., Li Y., McManus D.P. Diagnosis and management of schistosomiasis. Br. Med. J. 2011;342:d2651. doi: 10.1136/bmj.d2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gass K (2020) Time for a diagnostic sea-change: Rethinking neglected tropical disease diagnostics to achieve elimination. PLoS Negl Trop Dis 14(12): e0008933. doi: 10.1371/journal.pntd.0008933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balen J, Zhao ZY, Williams GM, McManus DP, Raso G, Utzinger J, et al. Prevalence, intensity and associated morbidity of Schistosoma japonicum infection in the Dongting Lake region, China. Bull World Health Organ. 2007;85(7):519–526. doi: 10.2471/blt.06.034033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khieu V, Sayasone S, Muth S, Kirinoki M, Laymanivong S, Ohmae H, et al. Elimination of Schistosomiasis Mekongi from Endemic Areas in Cambodia and the Lao People’s Democratic Republic: Current Status and Plans. Trop Med Infect Dis. 2019;4(1):30. doi: 10.3390/tropicalmed4010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohmae H, Sinuon M, Kirinoki M, Matsumoto J, Chigusa Y, Socheat D, et al. Schistosomiasis mekongi: from discovery to control. Parasitol Int. 2004;53(2):135–142. doi: 10.1016/j.parint.2004.01.004 [DOI] [PubMed] [Google Scholar]
- 41.Soe HZ, Oo CC, Myat TO, Maung NS. Detection of Schistosoma Antibodies and exploration of associated factors among local residents around Inlay Lake, Southern Shan State, Myanmar [published correction appears in Infect Dis Poverty. 2017 Jul 13;6(1):118]. Infect Dis Poverty. 2017;6(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thanchomnang T, Tantrawatpan C, Intapan PM, Sri-Aroon P, Limpanont Y, Lulitanond V, et al. Pyrosequencing for rapid molecular identification of Schistosoma japonicum and S. mekongi eggs and cercariae. Exp Parasitol. 2013. Sep;135(1):148–52. doi: 10.1016/j.exppara.2013.06.013 [DOI] [PubMed] [Google Scholar]
- 43.Kongklieng A, Kaewkong W, Intapan PM, Sanpool O, Janwan P, Thanchomnang T, et al. Molecular differentiation of Schistosoma japonicum and Schistosoma mekongi by real-time PCR with high resolution melting analysis. Korean J Parasitol. 2013. Dec;51(6):651–6. doi: 10.3347/kjp.2013.51.6.651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pilotte N, Papaiakovou M, Grant JR, Bierwert LA, Llewellyn S, McCarthy JS, et al. (2016) Improved PCR-Based Detection of Soil Transmitted Helminth Infections Using a Next-Generation Sequencing Approach to Assay Design. PLoS Negl Trop Dis 10(3): e0004578. doi: 10.1371/journal.pntd.0004578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Speich B, Knopp S, Mohammed KA, Khamis IS, Rinaldi L, Cringoli G, et al. Comparative cost assessment of the Kato-Katz and FLOTAC techniques for soil-transmitted helminth diagnosis in epidemiological surveys. Parasit Vectors. 2010. Aug 14;3:71. doi: 10.1186/1756-3305-3-71 [DOI] [PMC free article] [PubMed] [Google Scholar]