Abstract
Integration of human papillomavirus (HPV) DNA occurs early in cancer development and is an important event in malignant transformation of cervical cancer. Integration of HPVs preferentially disrupts or deletes the E2 open reading frame, which results in the loss of its expression. The preferential disruption of the E2 gene causes the absence of the E2 gene sequences in the PCR product following integration. Twenty-two carcinomas positive for HPV type 16 (HPV-16) DNA were first tested for the disruption of the E2 gene by PCR. A specific fragment of the E2 gene was not amplified in 10 cases, suggesting integration of HPV DNA into the host genome. Next, multiplex PCR for the HPV E2 and E6 genes was carried out in the remaining 12 cases. Copy numbers of both genes should be equivalent in episomal forms, while the E2 gene copy number will be smaller than that for E6 following the preferential disruption of the E2 gene in concominant forms. Although relative ratios of HPV E2 to E6 PCR products (E2/E6 ratios) ranged from 1.40 to 2.34 in 10 of 12 cases, multiplex PCR products from 2 cases displayed extremely low ratios of 0.69 and 0.61. Southern blot hybridization with an HPV-16 probe revealed that only in these two cases was both episomal and integrated HPV DNA being carried simultaneously. Thus, multiplex PCR for the E2 and E6 genes of HPV-16 DNA following PCR for the E2 gene can distinguish the pure episomal form from a mixed form of episomal and integrated HPV DNA. Clinical application of this technique will help researchers to understand the implication of the integration of HPV DNA for cervical carcinogenesis and cervical cancer progression.
A strong association between specific human papillomavirus (HPV) types and anogenital cancer has been well established. Certain types of HPV, including types 16, 18, 31, 33, 35, 45, 52, 56, and 58, play a pivotal role in the carcinogenesis of cervical cancer (33). In fact, the HPV viral DNA is identified in at least 90% of cervical carcinomas by PCR (2, 16, 30). The viral DNA is integrated into the cellular genome in cell lines derived from cervical carcinomas (3, 13, 27, 32) and in the majority of malignant tumors (6, 28). In contrast, integration of HPV DNA, regardless of type, occurs infrequently in preneoplastic-lesion, cervical intraepithelial neoplasia (CIN). Thus, integration has been proposed as an activation mechanism for progression from preinvasive lesions to cervical cancers (2, 6, 28).
It is known that integration usually disrupts or deletes either the E1 or E2 open reading frame (ORF), which results in the loss of expression of the corresponding gene products. Disruption of the E1 and E2 genes also leads to overexpression of the E6 and E7 oncoproteins (11, 15), since the E2 gene product can repress activities from the HPV promoters that direct the expression of the E6 and E7 genes (1, 21, 26). The preferential disruption of the E2 gene will cause the absence of the E2 gene sequences in the PCR product following integration. Thus, accurate detection of the integration of HPV DNA was achieved by using a simple PCR technique to amplify the E2 gene (8, 19). This rapid method, however, has limitations for distinguishing pure episomal forms of HPV DNA from mixed forms of episomal and integrated HPV DNA. Here, we report an accurate method of multiplex PCR for E2 and E6 to differentiate all physical types of HPV type 16 (HPV-16) DNA.
MATERIALS AND METHODS
Tissue specimens and DNA extraction.
Primary lesions were screened for the presence of HPV DNA as described previously (17). Twenty-two invasive carcinoma specimens positive for HPV-16 DNA were included in the present study. Specimens were obtained at the time of admission for surgery at the Department of Obstetrics and Gynecology, Okayama University Medical School Hospital, Okayama, Japan. DNA was extracted from tissue specimens and CaSki cell lines (20) by a routine procedure of proteinase K digestion and phenol extraction.
PCRs for E2 and a mixture of E2 and E6.
For detection of integration, the E2 ORF of the HPV-16 genome between nucleotides 2810 and 3836 was amplified according to the PCR conditions described by Park et al. (19). Next, multiplex PCR for the HPV E2 and E6 genes, both of which were in the same reaction tube, was performed. The primers for each sequence were 5′-CTTGGGCACCGAAGAAACAC-3′ (nucleotides 3438 to 3457) and 5′-TTGGTCACGTTGCCATTCAC-3′ (nucleotides 3770 to 3789) for the E2 gene and 5′-AAGGGCGTAACCGAAATCGGT-3′ (nucleotides 26 to 46) and 5′-CATATACCTCACGTCGCAG-3′ (nucleotides 215 to 233) for the E6 gene. These primers yielded 352- and 208-bp fragments for the E2 and E6 sequences, respectively. The conditions for multiplex PCR were the same as those previously described for the c-erbB-2 gene and the mdm-2 genes (23, 24). All oligodeoxynucleotides were synthesized with a model 394 DNA synthesizer (Applied Biosystems, Foster City, Calif.). PCR products were electrophoresed on a 2% agarose gel and stained with ethidium bromide. The UV-illuminated gels were photographed with Polaroid negatives (type 665), quantitated with an image scanner (GT8000; Epson, Suwa, Japan), and analyzed with Intelligent Quantifier software (Bio Image, Ann Arbor, Mich.). The relative ratio of HPV E2 to E6 PCR products (E2/E6 ratio) was calculated. In order to verify the constancy of the E2/E6 ratio, the amount of template DNA (0.5 to 20 pg) or the number of amplification cycles (20 to 35) was altered by using HPV-16 plasmid DNA as the template DNA. E2/E6 ratios in this preliminary experiment always exceeded 1.28 (data not shown).
Southern blot hybridization.
Ten micrograms of genomic DNA was digested with BamHI or PstI (New England Biolabs, Inc., Beverly, Mass.), electrophoresed in 1% agarose gels, and transferred onto nylon membranes (Hybond N; Amersham, Little Chalfont, Buckinghamshire, United Kingdom) by Southern blot procedures (25). BamHI has one cleavage site for HPV-16 DNA, while PstI is a multicut enzyme yielding the characteristic cleavage pattern. The membranes were sequentially hybridized with a 32P-labelled HPV-16 probe (11).
RESULTS
E2 PCR and multiplex PCR for E2 and E6.
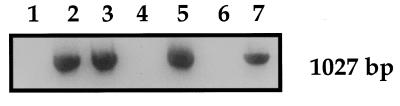
All invasive cervical cancer specimens were screened for the presence of HPV-16 DNA by nested PCR for the E6 gene (data not shown). Twenty-two carcinomas positive for HPV-16 DNA were then tested for the disruption of the HPV-16 E2 gene by PCR for E2. The specific fragment of the E2 gene (described above) was not amplified in 10 cases, suggesting integration of HPV DNA into the host genome in these cases. In contrast, the expected fragment of 1,027 bp was abundantly amplified in the remaining 12 cases (Fig. 1). In these cases, therefore, it was postulated that HPV DNA was present in episomal form without any disruption of the E2 gene. It was possible, however, that HPV DNA in these cancers exists in pure episomal form or in mixed episomal and integration forms.
FIG. 1.
PCR for the E2 ORF of HPV-16 DNA. Cervical carcinomas positive for HPV-16 DNA were examined for the disruption of the HPV-16 E2 gene by PCR. No amplification of a specific fragment of the E2 gene (1,027 bp) suggested the integration of HPV DNA into the host genome (lanes 1, 4, and 6). When the E2 gene was abundantly amplified, it was postulated that HPV DNA is present in episomal form without any disruption of the E2 gene (lanes 2, 3, 5, and 7).
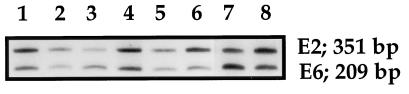
Multiplex PCR for the HPV E2 and E6 genes was carried out in these 12 cases. Specific fragments of the E2 and E6 genes were successfully coamplified in all cases (Fig. 2). Although E2/E6 ratios ranged from 1.40 to 2.34 in 10 of 12 cases, multiplex PCR products from 2 cases displayed extremely low ratios of 0.69 and 0.61.
FIG. 2.
Multiplex PCR for the E2 and E6 genes of HPV-16 DNA. Cervical carcinomas with intact E2 genes were subjected to multiplex PCR for the E2 and E6 genes. Specific fragments of the E2 (352 bp) and E6 genes (208 bp) were successfully coamplified in all cases. The E2/E6 ratio was obtained by densitometry. E2/E6 ratios for two cases were extremely low (0.69 and 0.61) compared to others (lanes 3 and 7).
Southern blot hybridization to analyze the physical status of HPV-16 DNA.
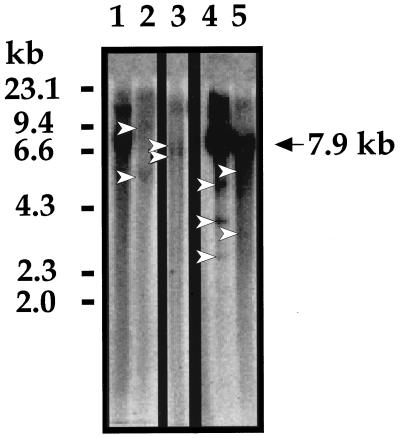
Southern blot hybridization was carried out with an HPV-16 DNA probe for confident detection of the physical status of HPV-16 DNA in these cervical carcinomas. Ten micrograms of tumorous DNA was first digested with single-cut enzyme BamHI and hybridized with an HPV-16 DNA probe. A single 7.9-kb band is supposed to appear when HPV DNA is episomal, and an off-sized fragment will appear when HPV DNA is integrated into the host genome. Figure 3 is a representative autoradiograph for five tumors. Lane 1 shows the single 7.9-kb band that was totally converted from the episomal DNA to a linear form upon BamHI digestion. Lanes 2 and 3 exhibit the presence of a couple of off-sized fragments and the absence of any 7.9-kb band, indicating that these are purely integrated HPV DNAs. Lanes 4 and 5 show the single 7.9-kb band as well as off-sized fragments smaller than 7.9 kb, suggesting that HPV DNA exists in mixed episomal and integration forms in these cancers. These lanes represent two cancers from which multiplex PCR products displayed E2/E6 ratios much lower than those for the PCR products in the other lanes.
FIG. 3.
Southern blot hybridization to analyze physical status of HPV-16 DNA. Ten micrograms of tumorous DNA was digested with single-cut enzyme BamHI and hybridized with an HPV-16 DNA probe. A single 7.9-kb band is shown when HPV DNA is episomal (lane 1). Off-sized fragments (arrowheads) appear when HPV DNA is integrated into the host genome (lanes 2 and 3). Lanes 4 and 5 show the single 7.9-kb band as well as off-sized fragments smaller than 7.9 kb, suggesting that HPV DNA exists as mixed episomal and integration forms in these cancers. Molecular size standards, in kilobases, are shown to the left.
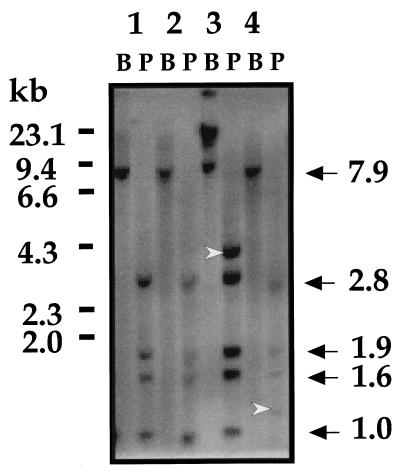
For confirmation of the simultaneous existence of episomal and integrated HPV DNA, genomic DNAs from these two cases were next digested with PstI, a multicut enzyme, to yield the authentic cleavage pattern. Genomic DNAs that exhibited the purely episomal HPV DNA were also digested with PstI and served as controls (Fig. 4). These controls show characteristic BamHI and PstI cleavage patterns (lanes 1 and 2). In contrast, the two cancers with extremely low E2/E6 ratios showed additional off-sized fragments along with authentic PstI fragments (lanes 3 and 4). This confirmed that HPV DNA exists in mixed episomal and integration forms in these cancers.
FIG. 4.
Southern blot hybridization following digestion with single-cut enzyme BamHI (B) and multicut enzyme PstI (P). PstI digestion is supposed to yield the authentic cleavage pattern, i.e., 2.8, 1.9, 1.6, and 1.0 kb (indicated at the right) and 0.5 kb (invisible in this panel). DNAs representing purely episomal HPV DNA were loaded in lanes 1 and 2. The two specimens with extremely low E2/E6 ratios showed additional off-sized fragments (arrowheads) along with authentic PstI fragments (lanes 3 and 4).
DISCUSSION
The viral genomes are exclusively maintained as episomes in benign lesions induced by HPVs such as HPV-6 and -11 (10, 17). Only episomal HPV DNA is detected in CIN type I (CIN-I), and integrated sequences are rarely found in CIN-II and -III (6, 8). In contrast, the viral DNA is usually integrated into the cellular genome in cell lines derived from cervical carcinomas (3, 13, 27, 32) and in the majority of malignant tumors (6, 28). Thus, integration occurs early in cancer development and is an important event in malignant transformation (2, 6, 28). Several studies have also demonstrated the stable persistence of integrated HPV DNA in the invasive tumor cells or the presence of HPV DNA sequences in cancer cells that exist within metastatic lymph nodes (5, 7, 12, 18, 31). This phenomenon made it possible to predict the unexpected recurrence of cervical cancer in patients with histologically negative nodes by a sensitive nested PCR for HPV DNA (17).
Thus, accurate detection of the physical status of HPV DNA is very important for a better understanding of the mechanisms of cervical carcinogenesis and of cervical cancer progression and metastasis. To analyze the physical status of HPV, a number of Southern blots and, sometimes, two-dimensional gel electrophoresis are essential. These procedures require large quantities of high-molecular-weight DNA, which is an obstacle when working with tiny intraepithelial lesions.
It is widely known that the transforming properties of the E6 and E7 oncoproteins are due, in part, to their capacity to bind to the p53 and the retinoblastoma tumor suppressor proteins and to inactivate the functions of their putative checkpoint controls (29). Integration of the viral genome into the human chromosome in the cancer cells usually disrupts or deletes the E2 ORF, which results in the loss of expression of the E2 gene, but high levels of E6 and E7 expression are maintained (11, 15). The E2 gene encodes a site-specific DNA-binding protein that is involved in the regulation of the HPV promoter that directs E6 and E7 expression (1, 21, 26). The reintroduction of the E2 gene into cervical carcinoma cell lines thereby leads to the inhibition of cell proliferation (10, 14).
The preferential disruption of the E2 genes will cause an absence of the E2 gene sequences in the PCR product following integration. Accurate detection of the integration of HPV DNA was achieved by a PCR technique to amplify the E2 gene (8, 19). First, we explored the presence of the E2 gene by amplifying the E2 ORF. A specific fragment of the E2 gene was not amplified in 10 of 22 cases (45.5%) with HPV-16 DNA. These cases involved the integrated form of HPV DNA. The frequency of viral integration that we found is slightly lower than those from preceding studies using the same method (8, 19). Since PCR for E2 and conventional Southern blot analysis in these previous studies were in complete concordance concerning the detection of the integration of HPV DNA, we presume such a small difference is due to epidemiological differences among the population studied. However, to rule out the possibility of integration outside the E2 ORF, including the E1 ORF, additional sets of primers from these regions may have to be used for further confirmation.
In the remaining 12 cases with an intact E2 gene, there are two possibilities: a pure episomal form or the concomitant presence of episomal and integration forms. We then tried multiplex PCR for the E2 and E6 genes to differentiate between these possibilities. The copy numbers of both genes should be equivalent in episomal forms, while the E2 gene copy number will be smaller than that for E6 following the preferential disruption of the E2 gene in concomitant forms. As expected, in a preliminary experiment with HPV-16 plasmid DNA, E2/E6 ratios were independent of the amount of template DNA and the number of amplification cycles. Next, we applied this technique to a survey of the differences in the copy numbers of both genes in 12 cases with an intact E2 gene. Multiplex PCR products from two cases displayed extremely low ratios of 0.69 and 0.61 compared to those for the other cases (1.40 to 2.34) (Fig. 2). These two cases might involve mixed episomal and integration forms.
In order to verify the simultaneous existence of episomal and integration forms of HPV DNA, conventional Southern hybridization was carried out with an HPV-16 DNA probe. Digestion of genomic DNA with BamHI or PstI is generally used for the accurate detection of the physical status of HPV-16 DNA (22). BamHI has one cleavage site for HPV-16 DNA, while PstI is a multicut enzyme yielding the authentic cleavage pattern. Control DNAs representing the purely episomal HPV DNA showed the linearized single 7.9-kb band upon BamHI digestion (Fig. 3, lane 1, and Fig. 4, lanes 1 and 2). In contrast, integrated HPV DNA exhibited the presence of a couple of off-sized fragments and the absence of any 7.9-kb band (Fig. 3, lanes 2 and 3). When DNAs from two cases with extremely low E2/E6 ratios were digested with BamHI, the single 7.9-kb band and off-sized fragments smaller than 7.9 kb were observed in the Southern blots, suggesting the simultaneous existence of episomes and integration forms of HPV DNA in these cancers (Fig. 3, lanes 4 and 5). To further confirm these results, DNAs were digested with PstI. Additional off-sized fragments along with PstI-specific restriction fragments were clearly observed in the Southern blots (Fig. 4, lanes 3 and 4). These two cancers are undoubtedly carrying both episomal and integrated HPV forms.
PCR amplifying the E2 ORF is a useful complement to prove the integrated form of HPV DNA (8). Reverse transcription-PCR for the E2 gene seems to be more sensitive than PCR for DNA (19). These methods, nevertheless, have limitations for distinguishing the pure episomal form of HPV DNA from mixed forms of episomal and integrated HPV DNA, because the E2 sequence is retained on the episomes in both cases. Our multiplex PCR for E2 and E6 is expected to solve this problem. It is possible that this tool cannot determine the physical status of HPV DNA accurately when concomitant episomal forms with relatively small amounts of HPV DNA exist due to limitations in the sensitivity for quantitation of the PCR products. HPV multimers carrying some deletions or duplications have been also reported (4, 9). Although a range of E2/E6 ratios for determination of the purely episomal forms will be required, we could not determine a cutoff value from the present study. It is widely believed that integrated sequences are rarely found in CIN-II and -III, but it is likely that the simultaneous presence of episomal and integrated forms in CINs and invasive carcinomas is more frequent. These lesions are generally small, and a sufficient amount of DNAs is not always available. A PCR-based analysis of the physical status of HPV DNA is applicable to small amounts of DNA purified from tiny lesions, formalin-fixed paraffin-embedded tissues, or cervical cytological specimens. Clinical application of this technique will help researchers to understand the implications of the integration of HPV for cervical carcinogenesis and the progression of cervical cancer.
Similar investigations for other types of HPV DNA will be easy to establish. In fact, we have found PCR primers that can coamplify the E2 and E6 genes of HPV-18 with satisfactory sensitivity and efficacy. Unfortunately, all carcinomas revealed exclusively integrated HPV except for one, which presented both episomal and integration forms (data not shown).
In conclusion, multiplex PCR for the E2 and E6 genes of HPV-16 DNA following PCR for the E2 gene can help researchers to analyze the physical status of HPV DNA.
ACKNOWLEDGMENTS
This work was supported in part by grants-in-aid 09671684 and 09771277 from the Ministry of Education, Sport, Science and Culture, Japan.
We thank A. Dusso, Renal Division, Washington University School of Medicine, for assistance in the preparation of the manuscript.
REFERENCES
- 1.Bernard B A, Bailly C, Lenoir M-C, Darmon M, Thierry F, Yaniv M. The human papillomavirus type 18 (HPV18) E2 gene product is a repressor of the HPV 18 regulatory region in human keratinocytes. J Virol. 1989;63:4317–4324. doi: 10.1128/jvi.63.10.4317-4324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch F X, Manos M M, Munoz N, Sherman M, Jansen A M, Peto J, Schiffman M H, Moreno V, Kurman R, Shah K V. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 3.Boshart M, Gissmann L, Ikenberg H, Kleinheinz A, Scheurlen W, zur Hausen H. A new type of papillomavirus DNA, its presence in genital cancer biopsies and in cell lines derived from cervical cancer. EMBO J. 1984;3:1151–1157. doi: 10.1002/j.1460-2075.1984.tb01944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choo K, Cheung W, Liew L, Lee H, Han S. Presence of catenated human papillomavirus type 16 episomes in a cervical carcinoma cell line. J Virol. 1989;63:782–789. doi: 10.1128/jvi.63.2.782-789.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Claas E C J, Melchers W J G, van der Linden H C, Lindeman J, Quint W G V. Human papillomavirus detection in paraffin-embedded cervical carcinomas and metastases of the carcinomas by the polymerase chain reaction. Am J Pathol. 1989;135:703–709. [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen A P, Reid R, Campion M, Lorincz A T. Analysis of the physical state of different human papillomavirus DNAs in intraepithelial and invasive cervical neoplasms. J Virol. 1991;65:606–612. doi: 10.1128/jvi.65.2.606-612.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czeglédy J, Póka R, Veress G, Gergely L. Amplification of human papillomavirus type 16 transforming genes from cervical cancer biopsies and lymph nodes of Hungarian patients. J Clin Microbiol. 1992;30:233–236. doi: 10.1128/jcm.30.1.233-236.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das B C, Sharma J K, Gopalakrishna V, Luthra U K. Analysis by polymerase chain reaction of the physical state of human papillomavirus type 16 DNA in cervical preneoplastic and neoplastic lesions. J Gen Virol. 1992;73:2327–2336. doi: 10.1099/0022-1317-73-9-2327. [DOI] [PubMed] [Google Scholar]
- 9.Di Luca D, Monini P, Rotola A, Saviolli A, Cassai E. Episomal HPV 16 DNA isolated from a cervical carcinoma presents a partial duplication of the early region. Virus Res. 1989;14:49–56. doi: 10.1016/0168-1702(89)90068-3. [DOI] [PubMed] [Google Scholar]
- 10.Dowhanick J J, McBride A A, Howley P M. Suppression of cellular proliferation by the papillomavirus E2 protein. J Virol. 1995;69:7791–7799. doi: 10.1128/jvi.69.12.7791-7799.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dürst M, Kleinheinz A, Hotz M, Gissman L. The physical state of human papillomavirus type 16 DNA in benign and malignant genital tumors. J Gen Virol. 1985;66:1515–1522. doi: 10.1099/0022-1317-66-7-1515. [DOI] [PubMed] [Google Scholar]
- 12.Fuchs P G, Girardi F, Pfister H. Human papillomavirus 16 DNA in cervical cancers and in lymph nodes of cervical cancer patients: a diagnostic marker for early metastases? Int J Cancer. 1989;43:41–44. doi: 10.1002/ijc.2910430110. [DOI] [PubMed] [Google Scholar]
- 13.Howley P M. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am J Pathol. 1985;119:361–366. [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang E S, Riese II D J, Settleman J, Nilson L A, Honig J, Flynn S, DiMaio D. Inhibition of cervical carcinoma cell line proliferation by the introduction of a bovine papillomavirus regulatory gene. J Virol. 1993;67:3720–3729. doi: 10.1128/jvi.67.7.3720-3729.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeon S, Lambert P F. Integration of HPV-16 DNA into the human genome leads to increased stability of E6/E7 mRNAs: implications for cervical carcinogenesis. Proc Natl Acad Sci USA. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsen F, Kalantari M, Jenkins A, Pettersen E, Kristensen G, Holm R, Johansson B, Hagmar B. Use of multiple PCR primer sets for optimal detection of human papillomavirus. J Clin Microbiol. 1996;34:2095–2100. doi: 10.1128/jcm.34.9.2095-2100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi Y, Yoshinouchi M, Tianqi G, Nakamura K, Hongo A, Kamimura S, Mizutani Y, Kodama J, Miyagi Y, Kudo T. Presence of human papilloma virus DNA in pelvic lymph nodes can predict unexpected recurrence of cervical cancer in patients with histologically negative lymph nodes. Clin Cancer Res. 1998;4:979–983. [PubMed] [Google Scholar]
- 18.Lewandowski G, Delgado G, Holloway R W, Farrell M, Jenson B, Lancaster W D. The use of in situ hybridization to show human papillomavirus deoxyribonucleic acid in metastatic cancer cells within lymph nodes. Am J Obstet Gynecol. 1990;163:1333–1337. doi: 10.1016/0002-9378(90)90715-j. [DOI] [PubMed] [Google Scholar]
- 19.Park J S, Hwang E S, Park S N, Ahn H K, Um S J, Kim C J, Kim S J. Physical status and expression of HPV genes in cervical cancers. Gynecol Oncol. 1997;65:121–129. doi: 10.1006/gyno.1996.4596. [DOI] [PubMed] [Google Scholar]
- 20.Pattilo R A, Hussa R O, Story M T, Ruckert A C F, Shalaby M R, Mattingly R F. Tumor antigen and human chorionic gonadotropin in CaSki cells: a new epidermoid cervical cancer cell line. Science. 1977;196:1456–1458. doi: 10.1126/science.867042. [DOI] [PubMed] [Google Scholar]
- 21.Romanczuk H, Thierry F, Howley P M. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J Virol. 1990;64:2849–2859. doi: 10.1128/jvi.64.6.2849-2859.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seedorf K, Krämmer G, Dürst M, Suhai S, Röwekamp W G. Human papillomavirus-type-16 DNA sequence. Virology. 1985;145:181–185. doi: 10.1016/0042-6822(85)90214-4. [DOI] [PubMed] [Google Scholar]
- 23.Seki A, Kodama J, Miyagi Y, Kamimura S, Yoshinouchi M, Kudo T. Amplification of the mdm-2 gene and P53 abnormalities in uterine sarcomas. Int J Cancer. 1997;73:33–37. doi: 10.1002/(sici)1097-0215(19970926)73:1<33::aid-ijc6>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Seki A, Nakamura K, Kodama J, Miyagi Y, Yoshinouchi M, Kudo T. A close correlation between c-erbB-2 gene amplification and local progression in endometrial adenocarcinoma. Eur J Gynaecol Oncol. 1998;19:90–92. [PubMed] [Google Scholar]
- 25.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 26.Thierry F, Howley P M. Functional analysis of E2 mediated repression of the HPV 18 P105 promotor. New Biol. 1991;3:90–100. [PubMed] [Google Scholar]
- 27.Tsunokawa Y, Takebe N, Nozawa S, Kasamatsu T, Gissmann L, zur Hausen H, Terada M, Sugimura T. Presence of human papillomavirus type-16 and type-18 DNA sequences and their expression in cervical cancers and cell lines from Japanese patients. Int J Cancer. 1986;37:499–503. doi: 10.1002/ijc.2910370405. [DOI] [PubMed] [Google Scholar]
- 28.Vernon S D, Unger E R, Miller D L, Lee D R, Reeves W C. Association of human papillomavirus type 16 integration in the E2 gene with poor disease-free survival from cervical cancer. Int J Cancer. 1997;74:50–56. doi: 10.1002/(sici)1097-0215(19970220)74:1<50::aid-ijc9>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 29.Vousden K. Interactions of human papillomavirus transforming proteins with the products of tumor suppressor genes. FASEB J. 1993;7:872–879. doi: 10.1096/fasebj.7.10.8393818. [DOI] [PubMed] [Google Scholar]
- 30.Walboomers J M, Meijer C J. Do HPV-negative cervical carcinomas exist? J Pathol. 1997;181:253–254. doi: 10.1002/(SICI)1096-9896(199703)181:3<253::AID-PATH755>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 31.Walboomers J M M, Fokke H E, Polak M, Volkers H, Houthoff H J, Barents J, van der Noordaa J, ter Schegget J. In situ localization of human papilloma virus type 16 DNA in a metastasis of an endocervical adenocarcinoma. Intervirology. 1987;27:81–85. doi: 10.1159/000149723. [DOI] [PubMed] [Google Scholar]
- 32.Yee C, Krishnan-Hewlett I, Baker C C, Schlegel R, Howley P M. Presence and expression of human papillomavirus sequences in human cervical carcinoma cell lines. Am J Pathol. 1985;119:361–366. [PMC free article] [PubMed] [Google Scholar]
- 33.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:55–78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]