Abstract
Background & Aim
Coronary artery disease (CAD) is the primary cause of mortality in patients with end stage renal disease (ESRD). MicroRNA profiling is proven as a powerful tool in the diagnosis of any disease at the molecular level. Hence, the present study aimed to profile the microRNA expression for CAD especially coronary artery calcification in CKD patients.
Materials and Methods
Two hundread patients with CKD stages 3 to 5 without dialysis and healthy controls were included in this study. All two hundred patients underwent 1024 multi sliceardiac computed tomography (CT) scan for calcium scoring. The calcium scoring more than 100 have been included in the study. We performed miRNA microarray analysis from serum samples of seven high calcium scored with CKD patients and one control patients.
Results
Seven patients have observed circulating miRNAs has significantly upregulated and downregulated when compared with control patients. mir21, mir 67, mir 390, mir 56, mir 250, mir 65 and mir 13 were up regulated and mir235, mir256, mir226, mir207, mir255, mir193 were downregulated. There was no significant difference in left ventricle function.
Conclusion
13 microRNAs play a potential role in coronary artery calcification in CKD patients.
Keywords: CKD, CAD, microRNA, coronary artery calcification
Introduction
Chronic kidney disease is a major global health problem1 and it is an independent risk factor for the development of coronary artery disease (CAD).2 Coronary artery disease is the leading cause of morbidity and mortality in patients with CKD.3 Data from prospective studies support that cardiovascular diseases (CVD) remain the most common cause of morbidity and mortality in patients with CKD and end-stage renal disease (ESRD).4 The spectrum of CVD not only involves obstructive coronary artery disease (CAD), but also involves other disease states such as chronic heart failure, sudden death, and arrhythmias. The main factors for the sensitive risk in this population, beside advanced age and a high proportion of diabetes and hypertension, are malnutrition, chronic inflammation, accelerated atherosclerosis, highly prevalent endothelial dysfunction (ED), coronary artery calcification (CAC), left-ventricular structural and functional abnormalities and bone mineral disorders (BMD).5–7 Coronary artery calcium (CAC) score is an independent predictor of cardiac events in both the general population and CKD patients.1 The prevalence of CAC in different stages of CKD varies from 13.9% in stages I and II, up to 83% in stages III-V. The prevalence and extent of CAC are increased in patients with ESRD even in young adults.8 The Dallas Heart Study showed that CAC scores >400 were 8-fold more prevalent in stages III-V CKD compared with patients without CKD. This association is substantially stronger in diabetics.9 The calcification could be reduced by using magnesium chloride and sodium thiosulfate. However, there are no pharmaceutical drugs available to reduce calcium deposits. Experiments using calcium channel blockers or statins have not been convincing.10,11 Some current medical treatments, however, may be associated with an increased risk of calcification. For example, treatment of patients for recurrent thrombosis with coumarins results in accelerated calcification.12,13 Similarly, in end stage renal disease, treatment of hyperphosphatemia with phosphate binders that contain calcium have been associated with more CAC compared to treatments with non-calcium-based phosphate binders.14 In patient groups with a strongly elevated risk for arterial calcification, patient-specific measures could possibly prevent arterial calcifications. CAD is hard to diagnose without the help of the well-established invasive coronary angiogram (CAG) technique. Although ECG and ETT have been widely used, there is no specific plasma biomarker for the clinical diagnosis of CAD especially calcium deposition on arteries, particularly for the early diagnosis of CAD. Therefore, there is a clinical demand for specific and reliable non-invasive, innovative, molecular biomarkers for the early diagnosis of CAD specifically calcification on arteries as well as therapeutic targets.
Circulating microRNAs (miRNAs) have attracted major interest as novel biomarkers for the early diagnosis of CAD.15 miRNAs are a class of small (B22 nucleotides long), highly specific, endogenous, single-stranded, non-coding RNAs that regulate the expression of target genes by binding to the 3′ untranslated region and degrading or inhibiting the translation of mRNAs.16 It is well established that miRNAs play critical roles in physiological and pathological processes in the cardiovascular system, such as endothelial dysfunction, inflammation, apoptosis, angiogenesis, atherosclerosis, and neointimal hyperplasia or restenosis.17–20 However, there are no reports regarding circulating miRNAs as non-invasive biomarkers for the diagnosis of coronary artery calcification in CKD patients.
The purpose of the present study was to use human CKD serum samples to profile the circulatory microRNA expression which have a putative involvement in calcification and provide new platform for these microRNAs may serve as targets for subsequent diagnostic and recognised therapeutic studies in CKD patients.
Materials and methods
Study population and design
Two hundred patients with CKD stages 3 to 5 age ranged from 48 years to 65 years and healthy control were included in this case control observational study from Dr.Giri's out patient clinic (OPD), Tiruchirappalli, India during January 2018 to December 2018. The study participants who were age >30 years, calcium score >100, willing to provide written and informed consent were included and age >65 years, history or clinical features of cardiac failure, previous heart surgeries, active malignancies and non willing participants were excluded from the study.
Calcium scoring
All two hundred patients underwent multi slice cardiac computed tomography (CT) scan for calcium scoring and it was done by the same radiologist. Administration of 20 mg metoprolol if patients had a heart rate more than 70 beats/min prior to the scan and administration of sublingual nitroglycerin 0.8 mg for all patients. Performed a scan without contrast dye to calculate total calcium score (Iwasaki K et al., 2011) and expressed as Agatston score (Agatston AS et al.,1990). The participants whose coronary calcium scoring ≥100 has been at very high cardio-vascular risk (Valensi P et al., 2018).
10 ml of blood samples were drawn from patients those who have calcium score more than 100 and centrifuged for serum according to a standardized protocol. This study was approved by the hospital ethics committee and written informed consent has been obtained from all the study population prior to the study. Also, the study was performed according to the principles laid out in the Declaration of Helsinki. Similarly, patients samples were analysed the routine parameters like haemoglobin, blood sugar, serum urea, creatinine, eGFR, Total protein, albumin, globulin, calcium, phosphorous, Ca*PO4, Alakline phosphatase, uric acid, electrolytes and 2D echo cardiography has been done all the study participants by a same cardiologist for assessment of ejection fraction.
miRNA Microarray profiling Using Agilent Platform
Microarray: Labelling and hybridization
The miRNA labelling was performed using miRNA Complete Labelling and Hyb Kit (Agilent Technologies, Part Number: 5190-0456). The total RNA sample was diluted to 100ng/ul in nuclease free water. About 200ng of total RNA was dephosphorylated using Calf Intestinal Alkaline Phosphatase (CIP) master mix (Agilent Technologies, Part Number: 5190-0456) by incubating at 37°C for 30 minutes. The dephosphorylated miRNA sample was denatured by adding Dimethyl Sulfoxide and heating at 100°C for 10 minutes and transferred to ice-water bath. The Ligation master mix (Agilent Technologies, Part Number: 5190-0456) containing Cyanine 3-pCp was added to the denatured miRNA sample and incubated at 16°C for 2 hours. The Cyanine 3-pCp labelled miRNA sample was dried completely in the vacuum concentrator (Eppendorf, Concentrator Plus, Catlog Number 5305000) at 45°C for 2 hour. The dried sample was suspended in nuclease free water and mixed with Hybridization Mix containing blocking solution (Agilent Technologies, Part Number: 5190-0456) and Hi-RPM Hybridization Buffer (Agilent Technologies, Part Number: 5190-0456) and incubated at 100°C for 5 minutes followed by snap chill on ice for 5 minutes. The samples were hybridized on the Human miRNA 8x60K Arrays. The hybridization was carried out at 55°C for 20 hours. After hybridization, the slides were washed using Gene Expression Wash Buffer1 (Agilent Technologies, Part Number 5188-5325) at room temperature for 5 minutes and Gene Expression Wash Buffer 2 (Agilent Technologies, Part Number 5188-5326) at 37oC for 5 minutes. The microarray slide was scanned on a G2600D scanner (Agilent Technologies)
Microarray Data Analysis
Data extraction from Images was done using Feature Extraction software v 11.5.1.1 of Agilent. Feature extracted raw data was analyzed using Gene Spring GX Version 12.0 software from Agilent. Normalization of the data was done in Gene Spring GX using the 90th percentile shift (Percentile shift normalization is a global normalization, where the locations of all the spot intensities in an array are adjusted. This normalization takes each column in an experiment independently and computes the ηth percentile of the expression values for this array, across all spots (where n has a range from 0–100 and n=90 is the median). It subtracts this value from the expression value of each entity) and normalized to Specific control Samples. Significant miRNA up and down regulated in test samples with respect to control sample were identified. Statistical T-test p-value was calculated based on volcano Plot. The gene targets for the differentially regulated miRNA were identified using Target Scan database http://www.targetscan.org/ which is integrated in Gene Spring GX software. The differentially expressed miRNA were clustered using hierarchical clustering based on Pearson coefficient correlation algorithm to identify significant miRNA expression patterns across the different conditions of the experiment.
Results
There were no significant correlation with ejection fraction and other parameters. The baseline characteristics of the study participants were shown in the table 1.
Table 1.
Patient's baseline characteristics
| Parameters | Patient Group (N:07) |
| Age years | 56.57±6.02 |
| Calcium score | 843 ±190.23 |
| EF % | 54.66 ± 21.93 |
| HB gms% | 7.35 ± 1.58 |
| Sugar mg/dl | 202.2 ±104.85 |
| Urea mg/dl | 160.33 ±112.75 |
| Creatinine mg/dl | 4.97 ±2.87 |
| eGFR ml/min | 26.71 ± 14.64 |
| Total Protein gms% | 5.53 ±0.70 |
| Albumin gms% | 2.43 ±0.76 |
| Globulin gms% | 3.1 ± 0.55 |
| Calcium mg/dl | 7.8 ±1.32 |
| Phosphorous mg/dl | 5.77 ± 1.24 |
| Ca*PO4 | 44.62 ±10.12 |
| Alkaline Phosphatase U/L | 167.25 ±96.93 |
| Uric Acid mg/dl | 6.67 ± 3.50 |
| Na mEq/L | 136.5 ±8.96 |
| K mEq/L | 4.55 ± 1.04 |
| HCO3 mmol/L | 11.66 ± 2.88 |
| Cl mmol/L | 99 ±14.52 |
The microarray cohort of subjects included 8 individuals and their Samples were labelled using the Agilent's Quick-Amp labelling Kit. Quality control was performed using Nanodrop (Table 2). The results were submitted in NCBI and obtained miRNA gene expression omnibus number (GSE89699) successfully.
Table 2.
Nanodrop Analysis of labelled cRNA
| Sample ID | Dye | RNA Concentration ng/µl |
Absorbance value 260/280 |
Absorbance value 260/230 |
Total yield Ng |
| 1A | pCp-Cy3 | 32.00 | 1.39 | 0.33 | 480 |
| 2A | pCp-Cy3 | 18.30 | 1.63 | 0.06 | 274.5 |
| 3A | pCp-Cy3 | 46.80 | 1.50 | 0.57 | 702 |
| 4A | pCp-Cy3 | 14.60 | 1.26 | 0.34 | 365 |
| 5A | pCp-Cy3 | 89.10 | 1.44 | 0.52 | 1336.5 |
| 6A | pCp-Cy3 | 9.90 | 1.32 | 0.45 | 247.5 |
| 7A | pCp-Cy3 | 68.30 | 1.75 | 0.71 | 1024.5 |
| 9A Dnase treated | pCp-Cy3 | 10.1 | 1.70 | 0.2 | 252.5 |
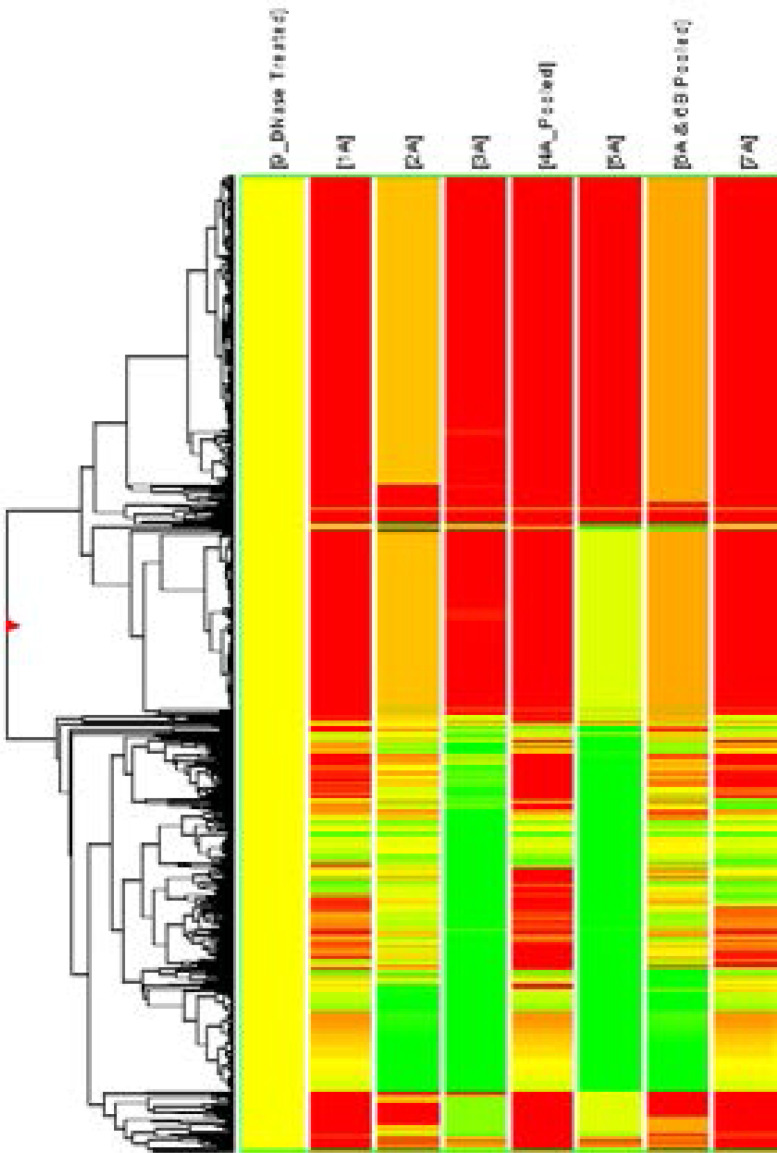
Expression profiles of microRNAs in the plasma of individuals with CAC
To evaluate the differential miRNA levels in individuals with CAC, we comparatively profiled plasma miRNA expression of 7 and 1 individuals with and without CAC, respectively. The levels of circulating miRNAs significantly upregulated and downregulated among CAC patients, as illustrated in the heat map shown in Fig. 1. Levels of 7 miRNAs were upregulated and those of 6 miRNAs were down regulated in the CAC patients (Table 3). After evaluating the differential miRNA expression pattern, it showed significant positive correlation with CACS.
Fig. 1.
Heat map of microRNA (miRNA) microarray expression data from blood samples of Individuals with (n = 7) and without (n = 1) coronary artery calcification. Hierarchical clustering of miRNA is based on similar expression profiles in test vs. Control. Clustering analysis was performed using GeneSpringGX Software using Average Linkage rule with Pearson centered Distance Metric. Red color in the cluster indicates up regulation in test and green color in the cluster indicate down regulation in test compare to control. The Fold expression values represented in the cluster are in terms of log base 2.
Table 3.
Differentially regulated miRNA
| Samples | Up Regulated | Down Regulated |
| 1A | 21 | 255 |
| 2A | 67 | 236 |
| 3A | 390 | 226 |
| 4A | 56 | 207 |
| 5A | 250 | 255 |
| 6A | 65 | 193 |
| 7A | 13 | 256 |
Discussion
MiRNAs have been demonstrated to play crucial roles in many physiological and pathophysiological processes.21 miRNAs contribute to different forms of CVD, and the changes in circulating miRNAs can be detected because of pathological changes.22,23 Circulating miRNAs have been identified as biomarkers for various physiological and pathological conditions.22,24 The microarray chip for miRNA provides a powerful approach for global circulating miRNA characterization, and it is simple to universally perform quantitative validation using real time-PCR.23 It has been suggested that the discovery-validation procedure for circulating miRNA biomarkers will be more efficient than that for traditional proteomic biomarker identification. To best of our knowledge, the function of miR-67, miR-390, miR-56, miR-250, miR-65, miR-13, miR-226, miR-236, miR-207, miR-256 remains unknown. Functional analysis in target Scan 6.0 showed that miR-21 and miR-255 could target different genes involved in increased the Wnt pathway in hepatocellular carcinoma. miR-193 involved in cause of primary segmental glomerulosclerosis.
The study provided significant clinical significance, to our knowledge, this is the first study to evaluate the circulating miRNA profile of CAC in patients with CKD. Calcification of the coronary arteries is highly correlated with atherosclerosis.25,26 Early detection of CAC is important for identifying subclinical atherosclerosis and predict the risk of CAD.25,27 By identifying specific circulating miRNAs for CAC, we are providing a novel way of identifying the severity of CAC which can also function as potential biomarkers for the presence of obstructive CAD. Moreover, the results of the current study highlight that further insight into the function of circulating miRNAs in the process and progression of CAC.
Conclusion
All these 13 micro RNAs are play a key role in coronary artery calcification in patients with CKD. However, larger population studies are required to confirm our results.
Compliance with Ethical Standards
Funding
Nil.
Conflict of Interest
Nil.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
References
- 1.Qiangjun Cai, Venkata K, Mukku Masood, Ahmad Coronary Artery Disease in Patients with Chronic Kidney Disease: A Clinical Update. Current Cardiology Reviews. 2013;9(4):331–339. doi: 10.2174/1573403X10666140214122234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarnak MJ, Levey AS, Schoolwerth AC. Kidney disease as a risk factor for development of cardiovascular disease: a statement from the American Heart Association Councils on Kidney in Cardiovascular Disese, High Blood Pressure Research, Clinical Cardiology, and Epidemiology and Prevention. Circulation. 2003;108:2154–2169. doi: 10.1161/01.CIR.0000095676.90936.80. [DOI] [PubMed] [Google Scholar]
- 3.Collins AJ, Foley RN, Chavers B. United States Renal Data System 2011 Annual Data Report: Atlas of chronic kidney disease & end-stage renal disease in the United States. Am J Kidney Dis. 2012;59:1–420. doi: 10.1053/j.ajkd.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 4.United States Renal Data System, author. “Atlas of end-stage renal disease in United States,” USRDS 2006 Annual Data Report. Bethesda, Md, USA: National Institute of Diabetes and digestive and Kidney Diseases; 2006. [Google Scholar]
- 5.Parfrey PS, Foley RN, Harnett JD, Kent GM, Murray DC, Barre PE. Outcome and risk factors of ischemic heart disease in chroic uremia. Kidney International. 1996;49(5):1428–1434. doi: 10.1038/ki.1996.201. [DOI] [PubMed] [Google Scholar]
- 6.Collins AJ. Cardiovascular mortality in endstage renal disease. American Journal of the Medical Sciences. 2003;325(4):163–167. doi: 10.1097/00000441-200304000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Parfrey PS, Foley RN, Harnett JD, Kent GM, Murray DC, Barre PE. Outcome and risk factors for left ventricular disorders in chronic uraemia. Nephrology Dialysis Transplantation. 1996;11(7):1277–1285. [PubMed] [Google Scholar]
- 8.Goodman WG, Goldin J, Kuizon BD. Coronary-artery calcification in young adults with end-stage renal disease who are undergoing dialysis. N Engl J Med. 2000;342:1478–1483. doi: 10.1056/NEJM200005183422003. [DOI] [PubMed] [Google Scholar]
- 9.Kramer H, Toto R, Peshock R. Association between chronic kidney disease and coronary artery calcification: the Dallas Heart Study. J Am Soc Nephrol. 2005;16:507–513. doi: 10.1681/ASN.2004070610. [DOI] [PubMed] [Google Scholar]
- 10.Motro M, Shemesh J. Calcium channel blocker nifedipine slows down progression of coronary calcification in hypertensive patients compared with diuretics. Hypertension. 2001;37:1410–1413. doi: 10.1161/01.hyp.37.6.1410. [DOI] [PubMed] [Google Scholar]
- 11.Wong ND, Kawakubo M, LaBree L. Relation of coronary calcium progression and control of lipids according to National Cholesterol Education Program guidelines. Am J Cardiol. 2004;94:431–436. doi: 10.1016/j.amjcard.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Koos R, Mahnken AH, Muhlenbruch G. Relation of oral anticoagulation to cardiac valvular and coronary calcium assessed by multislice spiral computed tomography. Am J Cardiol. 2005;96:747–749. doi: 10.1016/j.amjcard.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Schurgers LJH, Vermeer C. Oral anticoagulant treatment: friend or foe in cardiovascular disease. Blood. 2004;104:3231–3232. doi: 10.1182/blood-2004-04-1277. [DOI] [PubMed] [Google Scholar]
- 14.Chertow GM, Burke SK, Raggi P. Sevelamer attenuates the progression of coronary and aortic calcification in hemodialysis patients. Kidney Int. 2002;62:245–252. doi: 10.1046/j.1523-1755.2002.00434.x. [DOI] [PubMed] [Google Scholar]
- 15.Gupta SK, Bang C, Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ Cardiovasc Genet. 2010;3(5):484–488. doi: 10.1161/CIRCGENETICS.110.958363. [DOI] [PubMed] [Google Scholar]
- 16.Diehl P, Fricke A, Sander L, Stamm J, Bassler N, Htun N. Microparticles: major transport vehicles for distinct microRNAs in circulation. Cardiovasc Res. 2012;93(4):633–644. doi: 10.1093/cvr/cvs007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng Y, Song L, Zhao M, Harmelink C, Debenedittis P, Cui X. Critical roles of miRNA-mediated regulation of TGFb signalling during mouse cardiogenesis. Cardiovasc Res. 2014;103(2):258–267. doi: 10.1093/cvr/cvu126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469(7330):336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thum T, Catalucci D, Bauersachs J. MicroRNAs: novel regulators in cardiac development and disease. Cardiovasc Res. 2008;79(4):562–570. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 20.Dangwal S, Bang C, Thum T. Novel techniques and targets in cardiovascular microRNA research. Cardiovasc Res. 2012;93(4):545–554. doi: 10.1093/cvr/cvr297. [DOI] [PubMed] [Google Scholar]
- 21.Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, Li D, Hou Z, Lv K, Kan G, Cao H, Wu H, Song J, Pan X, Sun Q, Ling S, Li Y, Zhu M, Zhang P, Peng S, Xie X, Tang T, Hong A, Bian Z, Bai Y, Lu A, Li Y, He F, Zhang G, Li Y. miR-214 targets ATF4 to inhibit bone formation. Nat Med. 2013;19(1):93–100. doi: 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- 22.McManus DD, Ambros V. Circulating MicroRNAs in cardiovascular disease. Circulation. 2011;124:1908–1910. doi: 10.1161/CIRCULATIONAHA.111.062117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, Ma X, Lau WB, Rong R, Yu X, Wang B, Li Y, Xiao C, Zhang M, Wang S, Yu L, Chen AF, Yang X, Cai J. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. 2011;124(2):175–184. doi: 10.1161/CIRCULATIONAHA.110.012237. [DOI] [PubMed] [Google Scholar]
- 24.Era L, Pogosova-Agadjanyan Amelia Peterson, Noteboom Jennifer, Kathy C, O'Briant April Allen, Lin Daniel W, Urban Nicole, Drescher Charles W, Knudsen Beatrice S, Stirewalt Derek L, Gentleman Robert, Vessella Robert L, Nelson Peter S, Martin Daniel B, Tewari Muneesh. Circulating microRNAs as stable bloodbased markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McEvoy JW, Blaha MJ, Defilippis AP, Budoff MJ, Nasir K, Blumenthal RS, Jones SR. Coronary artery calcium progression: an important clinical measurement? A review of published reports. J Am Coll Cardiol. 2010;56(20):1613–1622. doi: 10.1016/j.jacc.2010.06.038. [DOI] [PubMed] [Google Scholar]
- 26.Erbel R, Lehmann N, Churzidse S, Rauwolf M, Mahabadi AA, Möhlenkamp S, Moebus S, Bauer M, Kälsch H, Budde T, Montag M, Schmermund A, Stang A, Führer-Sakel D, Weimar C, Roggenbuck U, Dragano N, Jöckel KH. Progression of coronary artery calcification seems to be inevitable, but predictable-results of the Heinz Nixdorf Recall (HNR) study dagger. Eur Heart J. 2014;35:2960–2971. doi: 10.1093/eurheartj/ehu288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bourantas CV, Zhang YJ, Garg S, Iqbal J, Valgimigli M, Windecker S, Mohr FW, Silber S, Vries TD, Onuma Y, Garcia-Garcia HM, Morel MA, Serruys PW. Prognostic implications of coronary calcification in patients with obstructive coronary artery disease treated by percutaneous coronary intervention: a patient-level pooled analysis of 7 contemporary stent trials. Heart. 2014;100(15):1158–1164. doi: 10.1136/heartjnl-2013-305180. [DOI] [PubMed] [Google Scholar]