Abstract
Objective
The potential correlation between the ε2/ε3/ε4 variants of the ApoE (Apolipoprotein E) gene and the odds of mesial temporal lobe epilepsy was investigated.
Methods
The database searching for eligible studies was performed in October 2020. A series of pooling analyses were conducted.
Results
We enrolled a total of twelve case-control studies for pooling. Within the pooling analysis of ε4, there was an increased risk of mesial temporal lobe epilepsy in cases under the models of carrier ε4 vs. ε3, ε3ε4 vs. ε3ε3, and ε3ε4+ε4ε4 vs. ε3ε3 [P < 0.05, odds ratio (OR) > 1], compared with controls. Moreover, we observed similar positive results in the subgroup analyses of “China” and “Population-based control” under the genetic models of ε4 (P < 0.05, OR > 1). Nevertheless, we did not detect the significant difference between the mesial temporal lobe epilepsy cases and controls in the pooling analyses of ε2 (all P > 0.05).
Conclusion
The ε3ε4 genotype of ApoE seems to be linked to the risk of mesial temporal lobe epilepsy for patients in China. More sample sizes are required to confirm the potential role of ApoE isoforms in the susceptibility to diverse types of epilepsy from different origins.
Keywords: Epilepsy, ApoE, isoforms, susceptibility
Introduction
Epilepsy is a disease of the nervous system with disabling neurologic conditions, characterized by at least two unprovoked seizures more than twenty-four hours apart1–4. As the most common form of partial epilepsy with focal seizures, TLE (temporal lobe epilepsy) is characterized by recurrent, unprovoked focal seizures in the temporal lobe of the brain5–7. The MTLE (mesial temporal lobe epilepsy) is a highly prevalent indication for the surgical treatment5, 8. The pathophysiological mechanism of TLE or MTLE remains elusive. A growing number of genes and the relevant genetic variants are reportedly associated with the odds of clinical epilepsy disease, which contribute to the therapeutic advice during the personalized medicine1, 9.
Human ApoE (Apolipoprotein E) protein, encoded by the ApoE gene on chromosome 19, contains three protein isoforms (E2, E3, and E4) and is related to the transformation and metabolism of lipoproteins10–12. There are three common allelic forms of the human ApoE gene (ε2, ε4, and ε3), and six genotypes, namely ε3ε3, ε3ε2, ε2ε2, ε3ε4, ε4ε4, and ε2ε4, are generated by the combination of two different polymorphisms rs429358 and rs741212–15. Several meta-analyses reported the statistical genetic relationship between the ApoE ε4 carrier and the risk of PD (Parkinson disease)13 or FTLD (frontotemporal lobar degeneration)14. Herein, we are interested in investigating whether ε2/ε3/ε4 isoforms of the ApoE gene is associated with the odds of TLE/MTLE, based on the available evidence16–27.
In the present study, we pooled the data of twelve eligible case-control studies to analyze the genetic correlation between ApoE ε2/ε3/ε4 isoforms and the susceptibility to the mesial temporal lobe epilepsy.
Materials and methods
Study identification
We tried to retrieve four databases, including PubMed, Embase (Excerpta medica database), Wanfang, CNKI (china national knowledge infrastructure), for the identification of relevant case-control studies, until October 2020. The searching terms were shown in Table S1.
Screening criteria
Then, we excluded the records using the following criteria: (1) duplicate studies; (2) case report, meta-analysis, or review article; (3) meeting abstract or animal data; (4), not ApoE isoforms, or not TLE/MTLE data; (5) without full genotype of genotypic or allelic frequency data. We tried to send emails to the authors for the missing data. The included studies should contain the distribution data of ε2, ε3, ε4 allele, or the genotype frequencies of “ε2/ε2”, “ε2/ε3”, “ε2/ε4”, “ε3/ε3”, “ε3/ε4”, “ε4/ε4” in both TLE/MTLE cases and negative controls. Besides, after the assessment of the NOS (Newcastle-Ottawa Scale) system, only the studies with high quality (NOS score >=5) were included.
Pooling analysis
We extracted the basic information of the first author, publication year, country, ethnicity, genotype frequency, control source, genotyping assay, and sample size in each study. Then, we performed a series of pooling analyses under the genetic models of allelic ε4 vs. total (ε3+ε2+ε4), allelic ε4 vs. ε3, allelic ε2 vs. total (ε3+ε2+ε4), allelic ε2 vs. ε3, carrier ε4 vs. total, carrier ε4 vs. ε3, carrier ε2 vs. total, carrier ε2 vs. ε3, ε4ε4 vs. ε3ε3 (homozygote), ε3ε4 vs. ε3ε3 (heterozygote), ε2ε2 vs. ε3ε3 (homozygote), ε3ε2 vs. ε3ε3 (heterozygote), ε3ε4+ε4ε4 vs. ε3ε3 (dominant), ε4ε4 vs. ε3ε3+ε3ε4 (recessive), ε3ε2+ε2ε2 vs. ε3ε3 (dominant), and ε2ε2 vs. ε3ε3+ε3ε2 (recessive). After pooling analysis of at least three case-control studies, we obtained the PA (P-value of the association test) and the value of the OR (95% CI) [odds ratio (95 % confidence interval)].
For the heterogeneity test, we obtained the PH (P-value of Cochran's Q statistic) and I2 value. When PH < 0.05 or I2 > 50%, the heterogeneity between studies was considered, and a random-effect model was applied for the DerSimonian and Laird statistics. If not, a fixed-effect model was for the Mantel-Haenszel statistics. Additionally, the subgroup analyses stratified by control source and country were performed.
Sensitivity and publication bias
To assess the statistical stability of our pooling results, we performed a group of sensitivity analyses, in which each study was excluded sequentially. Besides, we employed both the Begg's test and Egger's test to evaluate publication bias. The presence of potential publication bias was considered when the P-value of Begg's /Egger's test (PB / PE) was larger than 0.05. Stata software (Stata Corporation, College Station, USA) was applied for the above analysis.
Results
Study inclusion
As indicated in Figure 1, we obtained 91 records from PubMed, 235 records from the Embase, 26 records from the Wanfang, 15 records from the CNKI database. Then, based on our exclusion criteria, we excluded the 96 duplicates and other 235 unsuitable records. In total, 36 full-text articles were evaluated for eligibility. We then removed 24 articles because of “without full genotypic or allelic frequency data”. Finally, twelve eligible case-control studies16–27 with high-quality (NOS score >=5) were included. Of them, NOS scores of nine studies were larger than seven. We listed the basic information in Table 1. It should be noted that only the data of allelic frequency of ε3/ε2/ε4 was extracted from one study16, which was only used for the pooling analysis under the allelic model.
Figure 1.

Flow chart for study identification.
Table 1.
Basic information data
| First author, Year |
Country | Ethnicity | ε2ε2/ε2ε3/ε2ε4/ ε3ε3/ε3ε4/ε4ε4 |
Disease | ε2ε2/ε2ε3/ε2ε4/ ε3ε3/ε3ε4/ε4ε4 |
Control Source |
genotyping assay |
NOS |
| Cavalleri, 2005 | UK | Caucasian | 230/20/36* | TLE | 469/57/108* | PB | gene sequencing |
6 |
| Fu, 2010 | China | Asian | 6/91/9/358/88/8 | TLE | 8/106/6/344/91/3 | PB | PCR-RFLP | 8 |
| Gambardella, 2005 |
Italy | Caucasian | 0/13/2/101/21/1 | TLE | 1/38/3/227/27/1 | PB | one-stage PCR |
8 |
| Gambardella, 1999 |
Italy | Caucasian | 0/8/0/50/5/0 | TLE | 1/31/2/166/19/1 | PB | PCR-RFLP | 8 |
| Huang, 2015 | China | Asian | 3/2/0/27/13/1 | MTLE | 0/3/0/13/3/0 | HB | PCR-RFLP | 5 |
| Kumar, 2006 | India | Asian | 0/1/0/46/9/2 | TLE | 0/3/0/46/7/1 | PB | PCR-RFLP | 8 |
| Leal, 2017 | Portugal | Caucasian | 0/15/3/133/37/0 | MTLE | 0/40/3/248/50/1 | PB | PCR-RFLP | 7 |
| Li, 2007 | China | Asian | 1/12/0/64/17/0 | MTLE | 0/11/1/78/12/0 | PB | gene sequencing | 7 |
| Li, 2016 | China | Asian | 3/39/2/209/55/0 | MTLE | 1/33/2/230/36/0 | PB | gene sequencing | 7 |
| Salzmann, 2008 |
France | Caucasian | 0/9/1/72/27/0 | MTLE | 0/25/5/151/43/3 | PB | PCR-RFLP | 7 |
| Song, 2016 | China | Asian | 0/8/51/0/10/0 | TLE | 0/15/12/0/18/3 | PB | gene sequencing | 8 |
| Yeni, 2005 | Turkey | Asian | 5/4/1/30/6/1 | MTLE | 10/13/0/30/4/5 | HB | PCR-RFLP | 6 |
TLE, temporal lobe epilepsy; MTLE, mesial temporal lobe epilepsy; PB, population-based; HB, hospital-based; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; NOS: Newcastle-Ottawa Scale
the allelic frequency of ε3 /ε2 /ε4.
Meta-analysis data of ε4
As shown in Table S2, there were a total of twelve studies (1,823 cases and 2,551 controls) in the pooling analysis of TLE under the models of allelic ε4 vs. total and allelic ε4 vs. ε3. No significant statistical difference between the TLE patients and negative controls was detected (Table S2, PA >0.05). For the meta-analysis under the carrier ε4 vs. total and carrier ε4 vs. ε3 models (Table 2), eleven studies with 1,680 cases and 2,234 controls were enrolled. We observed an increased risk of TLE in cases, compared with controls, under the genetic models of carrier ε4 vs. total (Table 2, PA = 0.009, OR=1.24), carrier ε4 vs. ε3 (Table 2, PA = 0.001, OR=1.32), ε3ε4 vs. ε3ε3 (Table 3, PA = 0.011, OR=1.27), ε3ε4+ε4ε4 vs. ε3ε3 (Table S3, PA = 0.008, OR=1.28). These suggested that the ε3ε4 genotype of the ApoE gene was likely to be linked to the odds of TLE. Two factors of control source (population-based, PB), country (China) were then applied in our subgroup analyses. As shown in Table 2, Table 3, Table S2, and Table S3, we observed similar significant statistical differences between TLE cases and controls in the subgroups of “TLE/PB” under the models of carrier ε4 vs. total, carrier ε4 vs. ε3, ε3ε4 vs. ε3ε3, ε3ε4+ε4ε4 vs. ε3ε3 (PA < 0.05, OR > 1). In the subgroup analysis of “TLE/China”, there is an increased risk of TLE in cases under the models of carrier ε4 vs. ε3 (Table 2, PA = 0.007, OR=1.35), allelic ε4 vs. total (Table S2, PA = 0.045, OR=1.23), and allelic ε4 vs. ε3 (Table S2, PA = 0.018, OR=1.58), compared with controls. The forest plots for the subgroup analyses of TLE by country were shown in Figure 2.
Table 2.
Pooling data under the carrier model
| Comparison | Group | study | Association test | case | control | ||
|
| |||||||
| OR (95% CI) | P A | z | |||||
| carrier ε4 vs. total | TLE | 11 | 1.24 (1.06, 1.47) | 0.009 | 2.64 | 1,680 | 2,234 |
| TLE/PB | 9 | 1.24(1.05, 1.46) | 0.013 | 2.48 | 1,587 | 2,153 | |
| TLE/China | 5 | 1.22 (0.98, 1.51) | 0.071 | 1.81 | 1,077 | 1,029 | |
| MTLE | 6 | 1.35 (1.06, 1.72) | 0.015 | 2.44 | 792 | 1,054 | |
| MTLE/PB | 4 | 1.34(1.04, 1.73) | 0.022 | 2.28 | 699 | 973 | |
| MTLE/China | 3 | 1.49 (1.03, 2.15) | 0.033 | 2.13 | 448 | 423 | |
|
| |||||||
| carrier ε4 vs. ε3 | TLE | 11 | 1.32(1.11, 1.56) | 0.001 | 3.23 | 1,680 | 2,234 |
| TLE/PB | 9 | 1.31 (1.11, 1.56) | 0.002 | 3.13 | 1,587 | 2,153 | |
| TLE/China | 5 | 1.35(1.08, 1.68) | 0.007 | 2.68 | 1,077 | 1,029 | |
| MTLE | 6 | 1.34(1.05, 1.71) | 0.017 | 2.39 | 792 | 1,054 | |
| MTLE/PB | 4 | 1.34(1.04, 1.72) | 0.024 | 2.26 | 699 | 973 | |
| MTLE/China | 3 | 1.51(1.05, 2.18) | 0.028 | 2.20 | 448 | 423 | |
|
| |||||||
| carrier ε2 vs. total | TLE | 11 | 0.91(0.76, 1.09) | 0.296 | 1.05 | 1,680 | 2,234 |
| TLE/PB | 9 | 0.94(0.78, 1.12) | 0.467 | 0.73 | 1,587 | 2,153 | |
| TLE/China | 5 | 1.03(0.83, 1.27) | 0.803 | 0.25 | 1,077 | 1,029 | |
| MTLE | 6 | 0.88(0.68, 1.18) | 0.425 | 0.80 | 792 | 1,054 | |
| MTLE/PB | 4 | 0.96(0.71, 1.29) | 0.779 | 0.28 | 699 | 973 | |
| MTLE/China | 3 | 1.15(0.78, 1.71) | 0.483 | 0.70 | 448 | 423 | |
|
| |||||||
| carrier ε2 vs. ε3 | TLE | 11 | 0.96 (0.71, 1.30) | 0.776 | 0.28 | 1,680 | 2,234 |
| TLE/PB | 9 | 1.02(0.74, 1.41) | 0.906 | 0.12 | 1,587 | 2,153 | |
| TLE/China | 5 | 1.34(0.79, 2.26) | 0.280 | 1.08 | 1,077 | 1,029 | |
| MTLE | 6 | 0.89 (0.67, 1.17) | 0.392 | 0.86 | 792 | 1,054 | |
| MTLE/PB | 4 | 0.96(0.71, 1.29) | 0.775 | 0.29 | 699 | 973 | |
| MTLE/China | 3 | 1.16(0.78, 1.73) | 0.452 | 0.75 | 448 | 423 | |
TLE, temporal lobe epilepsy; PB, population-based control; MTLE, mesial temporal lobe epilepsy; OR, odds ratio; CI, confidence interval; PA, P-value in association test.
Table 3.
Pooling data under the homozygotic and heterozygotic models
| Comparison | Group | study | Association test | case | control | ||
|
| |||||||
| OR (95% CI) | P A | z | |||||
| ε4ε4 vs. ε3ε3 | TLE | 7 | 1.53(0.67, 3.47) | 0.312 | 1.01 | 999 | 1,445 |
| TLE/PB | 6 | 1.53(0.66, 3.58) | 0.324 | 0.99 | 958 | 1,429 | |
| MTLE | 3 | 0.56(0.11, 3.06) | 0.518 | 0.65 | 310 | 512 | |
|
| |||||||
| ε3ε4 vs. ε3ε3 | TLE | 9 | 1.27(1.06, 1.54) | 0.011 | 2.54 | 1,344 | 1,801 |
| TLE/PB | 8 | 1.26(1.04, 1.53) | 0.016 | 2.41 | 1,303 | 1,785 | |
| TLE/China | 4 | 1.21(0.94, 1.55) | 0.135 | 1.50 | 840 | 810 | |
| MTLE | 5 | 1.52(1.17, 1.97) | 0.002 | 3.12 | 655 | 868 | |
| MTLE/PB | 4 | 1.50(1.15, 1.96) | 0.003 | 1.02 | 614 | 852 | |
| MTLE/China | 3 | 1.73(1.18, 2.54) | 0.006 | 2.76 | 386 | 372 | |
|
| |||||||
| ε2ε2 vs. ε3ε3 | TLE | 6 | 1.21(0.56, 2.63) | 0.630 | 0.48 | 987 | 1,291 |
| TLE/PB | 5 | 1.09(0.48, 2.46) | 0.842 | 0.20 | 955 | 1,275 | |
| TLE/China | 4 | 1.26(0.55, 2.89) | 0.585 | 0.55 | 815 | 827 | |
| MTLE | 3 | 3.42(0.70, 16.73) | 0.129 | 1.52 | 360 | 369 | |
| MTLE/China | 3 | 3.42(0.70, 16.73) | 0.129 | 1.52 | 360 | 369 | |
|
| |||||||
| ε3ε2 vs. ε3ε3 | TLE | 9 | 0.87(0.71, 1.07) | 0.188 | 1.32 | 1,263 | 1,804 |
| TLE/PB | 8 | 0.88 (0.72, 1.08) | 0.231 | 1.20 | 1,231 | 1,788 | |
| TLE/China | 4 | 0.95(0.74, 1.22) | 0.685 | 0.41 | 815 | 827 | |
| MTLE | 5 | 0.97(0.71, 1.33) | 0.853 | 0.19 | 589 | 833 | |
| MTLE/PB | 4 | 1.00(0.73, 1.38) | 0.243 | 0.01 | 557 | 817 | |
| MTLE/China | 3 | 1.22(0.80, 1.86) | 0.362 | 0.91 | 360 | 369 | |
TLE, temporal lobe epilepsy; PB, population-based control; MTLE, mesial temporal lobe epilepsy; OR, odds ratio; CI, confidence interval; PA, P-value in association test.
Figure 2.
Subgroup analysis of TLE by country under the models of ε4. (a) allelic ε4 vs. ε3; (b) carrier ε4 vs. ε3; (c) ε3ε4 vs. ε3ε3; (d) ε3ε4+ε4ε4 vs. ε3ε3. The data of the “China” subgroup was marked with a rectangle.
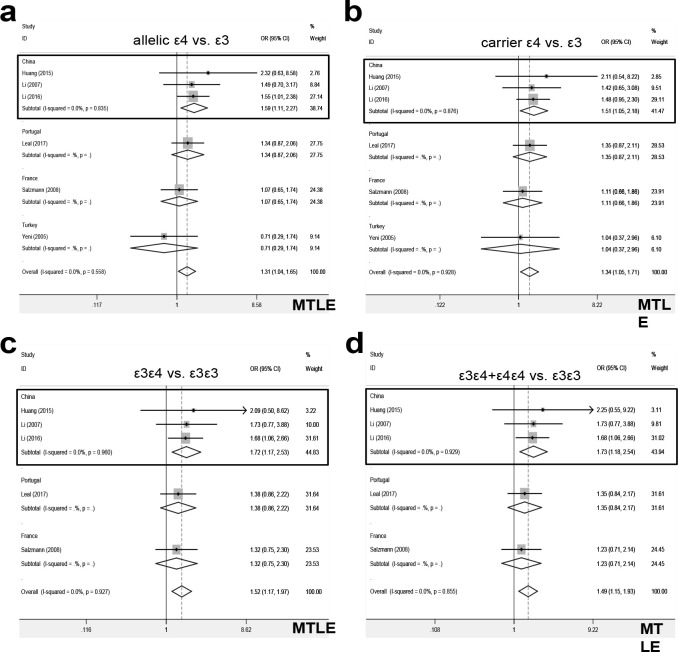
Further, we performed a series of pooling analyses of ε4, only including the data of MTLE cases. Compared with controls, there was an increased risk of MTLE in cases under the models of carrier ε4 vs. total (Table 2, PA = 0.015, OR =1.35), carrier ε4 vs. ε3 (Table 2, PA = 0.017, OR =1.34), ε3ε4 vs. ε3ε3 (Table 3, PA = 0.002, OR =1.52), allelic ε4 vs. total (Table S2, PA = 0.026, OR =1.29), allelic ε4 vs. ε3 (Table S2, PA = 0.020, OR =1.31), ε3ε4+ε4ε4 vs. ε3ε3 (Table S3, PA = 0.003, OR =1.49). Also, we observed similar positive conclusions in the subgroup analysis of “MTLE/PB” and “MTLE/China” (Table 2-3, Table S2-S3, all PA <0.05, OR >1). The forest plots for the subgroup analyses of MTLE by country were shown in Figure 3. Thus, ε3ε4 genotype is more likely to be associated with the susceptibility of Chinese patients to the mesial temporal lobe epilepsy.
Figure 3.
Subgroup analysis of MTLE by country under the models of ε4. (a) allelic ε4 vs. ε3; (b) carrier ε4 vs. ε3; (c) ε3ε4 vs. ε3ε3; (d) ε3ε4+ε4ε4 vs. ε3ε3. The data of the “China” subgroup was marked with a rectangle.
Meta-analysis data of ε2
For the pooling analysis of ε2, we did not detect a significant difference between the TLE/MTLE cases and negative controls under the models of carrier ε2 vs. total, carrier ε2 vs. ε3, ε2ε2 vs. ε3ε3, ε3ε2 vs. ε3ε3, allelic ε2 vs. total, allelic ε2 vs. ε3, ε3ε2+ε2ε2 vs. ε3ε3, ε2ε2 vs. ε3ε3+ε3ε2 (Table 2–3, Table S2-S3, all PA > 0.05). Also, no positive conclusions were observed in the subgroup analyses by the control source or country under any genetic model of ε2 (Table 2–3, Table S2-S3, all PA > 0.05). The forest plots for the subgroup analyses by country were shown in Figure S1-S2. These suggested that ε2 allele, or ε3ε2, ε2ε2 genotype may not be strongly linked to the odds of TLE or MTLE.
Heterogeneity analysis
As shown in Table S4, we utilized a random-effect model (DerSimonian and Laird statistics) for the association test under the genetic models of carrier ε2 vs. ε3 (PH < 0.021, I2 = 52.6%), allelic ε4 vs. ε3 (PH = 0.023, I2 = 50.3%), and allelic ε2 vs. ε3 (PH = 0.009, I2 = 56.4%), respectively. And a fixed-effect model (Mantel-Haenszel statistics) was applied for others, due to the lack of between-study heterogeneity (Table 4, PH > 0.05 and I2 < 50.0 %).
Sensitivity and publication bias
Our results of sensitivity analysis indicated the statistical stability of the above conclusions. We showed the data of the carrier models (carrier ε4 vs. total; carrier ε4 vs. ε3; carrier ε2 vs. total; carrier ε2 vs. ε3.) as examples in Figure S3. As shown in Table S4, we did not observe significant publication bias in all comparisons (PB>0.05, PE>0.05). Figure S4 presents the publication bias plots in Egger's test under the carrier models (carrier ε4 vs. total; carrier ε4 vs. ε3; carrier ε2 vs. total; carrier ε2 vs. ε3) as examples.
Discussion
No statistical differences in ApoE ε 4 allelic frequencies between MTLE-HS (mesial temporal lobe epilepsy with hippocampal sclerosis) cases and patients and healthy controls were detected; ApoEε 4 carriers may be related to earlier MTLE-HS onset in Portugal22. ApoE ε 4 allele was reportedly associated with the odds of Chinese NLMTLE (nonlesional mesial temporal lobe epilepsy)23, and TLE with prior trauma17. Nevertheless, the ApoEε 4 allele was reportedly unrelated to the onset age of epilepsy, duration, or the silent period in the refractory TLE group17. Also, no genetic correlation between ApoEε 4 isoform and the onset age or outcome after surgery of MTLE-HS was observed in Turkey27. The lack of the genetic role of ApoE isoform in the occurrence of nonlesional TLE cases in Italy was reported19. Thus, this issue merits the preformation of a meta-analysis.
There were eight studies included in a relevant meta-analysis of Kauffman, M. A. et al. in 2010, which evaluated the effect of ApoEε 4 isoform on the age at onset of temporal lobe epilepsy28. In 2019, another meta-analysis containing nine studies reported that ApoE ε 4 isoform is associated with a high susceptibility to Asian epilepsy cases29. In the present study, we enrolled the available eligible studies and used the different analysis strategies to explore the genetic role of the allelic and genotypic frequencies of ApoE ε2/ε3/ε4 isoforms in the risk of TLE or MTLE. After the database searching, we enrolled a total of twelve eligible case-control studies for the pooling analysis under a series of genetic models, namely allelic ε4 vs. total, allelic ε4 vs. ε3, allelic ε2 vs. total, allelic ε2 vs. ε3, carrier ε4 vs. total, carrier ε4 vs. ε3, carrier ε2 vs. total, carrier ε2 vs. ε3, ε4ε4vs. ε3ε3, ε3ε4 vs. ε3ε3, ε2ε2 vs. ε3ε3, ε3ε2 vs. ε3ε3, ε3ε4+ε4ε4 vs. ε3ε3, ε4ε4 vs. ε3ε3+ε3ε4, ε3ε2+ε2ε2 vs. ε3ε3, and ε2ε2 vs. ε3ε3+ε3ε2. Our findings revealed that the ε3ε4 genotype of the ApoE gene is more likely to be linked to the odds of mesial temporal lobe epilepsy cases in China, which was considered statistically credible by the preformation of sensitivity analyses.
Despite this, we should consider the findings of our pooling analyses with precaution. There are insufficient cases and controls in some comparisons. For instance, even though we observed a statistical association between the ε3ε4 genotype of ApoE and an increased MTLE susceptibility for Chinese cases, only three case-control studies20, 23, 24 were included for the pooling analysis. Although the lack of more considerable publication bias in all comparisons, less than ten case-control studies were included for the pooling analysis under the models of ε4ε4 vs. ε3ε3, ε3ε4 vs. ε3ε3, ε2ε2 vs. ε3ε3, ε3ε2 vs. ε3ε3, ε3ε4+ε4ε4 vs. ε3ε3, ε4ε4 vs. ε3ε3+ε3ε4, ε3ε2+ε2ε2 vs. ε3ε3, ε2ε2 vs. ε3ε3+ε3ε2. We observed a high level of between-study heterogeneity under the genetic models of carrier ε2 vs. ε3, allelic ε4 vs. ε3, and allelic ε2 vs. ε3.
Besides, the potential effect of non-ε2/ε3/ε4 ApoE isoforms or the combined impact of ApoE isoforms with other variants, [e.g., ABCA7 (ATP Binding Cassette Subfamily A Member 7) rs4147929 or CD33 rs3865444, etc.], on the odds of TLE/MTLE should be considered when the more sample sizes were available. In addition, temporal lobe epilepsy is often accompanied by some other neurological pathologies, such as hippocampal sclerosis27, 30. The factors of clinical features should be fully considered for the adjusted estimation in the future as well.
Conclusion
Taken together, our data suggested that the ε3ε4 genotype of the ApoE gene may be related to enhanced susceptibility to mesial temporal lobe epilepsy for patients in China. Large-scale publications are required to verify the role of more ApoE variants in the risk of cases with different types of epilepsy in other regions.
Conflict of interest
We declare that we have no conflict of interest.
References
- 1.Weber YG, Biskup S, Helbig KL, Von Spiczak S, Lerche H. The role of genetic testing in epilepsy diagnosis and management. Expert Rev Mol Diagn. 2017;17:739–750. doi: 10.1080/14737159.2017.1335598. [DOI] [PubMed] [Google Scholar]
- 2.Singh A, Trevick S. The Epidemiology of Global Epilepsy. Neurol Clin. 2016;34:837–847. doi: 10.1016/j.ncl.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 3.Stafstrom CE, Carmant L. Seizures and epilepsy: an overview for neuroscientists. Cold Spring Harb Perspect Med. 2015;5:a022426. doi: 10.1101/cshperspect.a022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerreiro CA. Epilepsy: Is there hope? Indian J Med Res. 2016;144:657–660. doi: 10.4103/ijmr.IJMR_1051_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muzumdar D, Patil M, Goel A, Ravat S, Sawant N, Shah U. Mesial temporal lobe epilepsy - An overview of surgical techniques. Int J Surg. 2016;36:411–419. doi: 10.1016/j.ijsu.2016.10.027. [DOI] [PubMed] [Google Scholar]
- 6.Tramoni-Negre E, Lambert I, Bartolomei F, Felician O. Long-term memory deficits in temporal lobe epilepsy. Rev Neurol (Paris) 2017;173:490–497. doi: 10.1016/j.neurol.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Alonso Vanegas MA, Lew SM, Morino M, Sarmento SA. Microsurgical techniques in temporal lobe epilepsy. Epilepsia. 2017;58(Suppl 1):10–18. doi: 10.1111/epi.13684. [DOI] [PubMed] [Google Scholar]
- 8.Chong S, Phi JH, Lee JY, Kim SK. Surgical Treatment of Lesional Mesial Temporal Lobe Epilepsy. J Epilepsy Res. 2018;8:6–11. doi: 10.14581/jer.18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orsini A, Zara F, Striano P. Recent advances in epilepsy genetics. Neurosci Lett. 2018;667:4–9. doi: 10.1016/j.neulet.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Moreno-Grau S, Hernandez I, Heilmann-Heimbach S, Ruiz S, Rosende-Roca M, Mauleon A, Vargas L, Rodriguez-Gomez O, Alegret M, Espinosa A, Ortega G, Aguilera N, Abdelnour C, Neuroimaging Initiative AD. Gil S, Maier W, Sotolongo-Grau O, Tarraga L, Ramirez A, Lopez-Arrrieta J, Antunez C, Serrano-Rios M, Boada M, Ruiz A. Genome-wide significant risk factors on chromosome 19 and the APOE locus. Oncotarget. 2018;9:24590–24600. doi: 10.18632/oncotarget.25083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kockx M, Traini M, Kritharides L. Cell-specific production, secretion, and function of apolipoprotein. E. J Mol Med (Berl) 2018;96:361–371. doi: 10.1007/s00109-018-1632-y. [DOI] [PubMed] [Google Scholar]
- 12.Seripa D, D'Onofrio G, Panza F, Cascavilla L, Masullo C, Pilotto A. The genetics of the human APOE polymorphism. Rejuvenation Res. 2011;14:491–500. doi: 10.1089/rej.2011.1169. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Luo J, Liu L, Fu H, Tang L. The genetic association between apolipoprotein E gene polymorphism and Parkinson disease: A meta-Analysis of 47 studies. Medicine (Baltimore) 2018;97:e12884. doi: 10.1097/MD.0000000000012884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su WH, Shi ZH, Liu SL, Wang XD, Liu S, Ji Y. Updated meta-analysis of the role of APOE epsilon2/epsilon3/epsilon4 alleles in frontotemporal lobar degeneration. Oncotarget. 2017;8:43721–43732. doi: 10.18632/oncotarget.17341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braga LW, Borigato EV, Speck-Martins CE, Imamura EU, Gorges AM, Izumi AP, Dantas RC, Nunes LG. Apolipoprotein E genotype and cerebral palsy. Dev Med Child Neurol. 2010;52:666–671. doi: 10.1111/j.1469-8749.2009.03465.x. [DOI] [PubMed] [Google Scholar]
- 16.Cavalleri GL, Lynch JM, Depondt C, Burley MW, Wood NW, Sisodiya SM, Goldstein DB. Failure to replicate previously reported genetic associations with sporadic temporal lobe epilepsy: where to from here? Brain. 2005;128:1832–1840. doi: 10.1093/brain/awh524. [DOI] [PubMed] [Google Scholar]
- 17.Fu YH, Lv RJ, Jin LR, Lu Q, Shao XQ, He JS, Wu LW, Zhang LS, Hu HG. Association of apolipoprotein E polymorphisms with temporal lobe epilepsy in a Chinese Han population. Epilepsy Res. 2010;91:253–259. doi: 10.1016/j.eplepsyres.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 18.Gambardella A, Aguglia U, Chifari R, Labate A, Manna I, Serra P, Romeo N, Sibilia G, Lepiane E, Russa AL, Ventura P, Cittadella R, Sasanelli F, Colosimo E, Leggio U, Zappia M, Quattrone A. ApoE epsilon4 allele and disease duration affect verbal learning in mild temporal lobe epilepsy. Epilepsia. 2005;46:110–117. doi: 10.1111/j.0013-9580.2005.15804.x. [DOI] [PubMed] [Google Scholar]
- 19.Gambardella A, Aguglia U, Cittadella R, Romeo N, Sibilia G, LePiane E, Messina D, Manna I, Oliveri RL, Zappia M, Quattrone A. Apolipoprotein E polymorphisms and the risk of nonlesional temporal lobe epilepsy. Epilepsia. 1999;40:1804–1807. doi: 10.1111/j.1528-1157.1999.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 20.Huang C, Yan B, Lei D, Si Y, Li H, Chen MW, Li L, Chen F, Zhou Q, Zhou D, Li JM. Apolipoprotein 4 may increase viral load and seizure frequency in mesial temporal lobe epilepsy patients with positive human herpes virus 6B. Neurosci Lett. 2015;593:29–34. doi: 10.1016/j.neulet.2014.12.063. [DOI] [PubMed] [Google Scholar]
- 21.Kumar A, Tripathi M, Pandey RM, Ramakrishnan L, Srinivas M, Luthra K. Apolipoprotein E in temporal lobe epilepsy: a case-control study. Dis Markers. 2006;22:335–342. doi: 10.1155/2006/951632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leal B, Chaves J, Carvalho C, Bettencourt A, Freitas J, Lopes J, Ramalheira J, Costa PP, Mendonça D, Silva AM, Silva BM. Age of onset of mesial temporal lobe epilepsy with hippocampal sclerosis: the effect of apolipoprotein E and febrile seizures. International Journal of Neuroscience. 2017;127:800–804. doi: 10.1080/00207454.2016.1264396. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Ding C, Gong X, Wang X, Cui T. Apolipoprotein E epsilon4 Allele was Associated With Nonlesional Mesial Temporal Lobe Epilepsy in Han Chinese Population. Medicine (Baltimore) 2016;95:e2894. doi: 10.1097/MD.0000000000002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Wang W. Study on clinical characteristics, polymorphisms of ApoE genotype, quality of life and cognitive functions in patients with temporal lobe epilepsy. Hebei Medical University. 2007:33–39. [Google Scholar]
- 25.Salzmann A, Perroud N, Crespel A, Lambercy C, Malafosse A. Candidate genes for temporal lobe epilepsy: a replication study. Neurol Sci. 2008;29:397–403. doi: 10.1007/s10072-008-1060-9. [DOI] [PubMed] [Google Scholar]
- 26.Song X. The relevant research in ApoE gene polymorphism, temporal lobe epilepsy and cognitive dysfunction in patients with epilepsy. Jilin University. 2016:13–18. [Google Scholar]
- 27.Yeni SN, Ozkara C, Buyru N, Baykara O, Hanoglu L, Karaagac N, Ozyurt E, Uzan M. Association between APOE polymorphisms and mesial temporal lobe epilepsy with hippocampal sclerosis. Eur J Neurol. 2005;12:103–107. doi: 10.1111/j.1468-1331.2004.00956.x. [DOI] [PubMed] [Google Scholar]
- 28.Kauffman MA, Consalvo D, Moron DG, Lereis VP, Kochen S. ApoE epsilon4 genotype and the age at onset of temporal lobe epilepsy: a case-control study and meta-analysis. Epilepsy Res. 2010;90:234–239. doi: 10.1016/j.eplepsyres.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Liang Y, Zhou Z, Wang H, Cheng X, Zhong S, Zhao C. Association of apolipoprotein E genotypes with epilepsy risk: A systematic review and meta-analysis. Epilepsy Behav. 2019;98:27–35. doi: 10.1016/j.yebeh.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 30.Kauffman MA, Consalvo D, Gonzalez MD, Kochen S. Transcriptionally less active prodynorphin promoter alleles are associated with temporal lobe epilepsy: a case-control study and meta-analysis. Dis Markers. 2008;24:135–140. doi: 10.1155/2008/723723. [DOI] [PMC free article] [PubMed] [Google Scholar]