Abstract
Herpes simplex virus type 1 (HSV-1)-related disease ranges from a localized, self-limiting illness to fatal disease in immunocompromised individuals. The corneal disease herpetic keratitis may develop after reactivation of a latent virus or reinfection with an exogenous herpesvirus. Molecular analysis of the virus involved may allow distinction between these two options. The HSV-1 genome contains several hypervariable regions that vary in numbers of reiterating regions (reiterations I to VIII [ReI to ReVIII]) between individual strains. Twenty-four HSV-1 clones, derived by subcloning of HSV-1 (strain F) twice in limiting dilutions, were tested in a PCR-based assay to analyze the stabilities of ReI, ReIII, ReIV, and ReVII. ReI and ReIII proved to vary in size upon subcloning, whereas ReIV and ReVII were stable. Subsequently, 37 unrelated isolates and 10 sequential isolates from five patients, all with HSV-1-induced keratitis, were genotyped for ReIV and ReVII. Of the 37 unrelated samples, 34 (92%) could be discriminated, while the genotypes of the viruses in sequential samples were identical for each individual. Conclusively, the data show that the approach presented allows the rapid and accurate discrimination of HSV-1 strains in studies that address the transmission and pathogenesis of HSV-1 infections.
Herpes simplex virus (HSV) type 1 (HSV-1) infections are widespread in the human population and may cause a variety of disease symptoms, including localized recurrent ocular lesions like uveitis and keratitis (16). Clinical manifestations associated with herpetic corneal infections are herpetic epithelial keratitis and the development of the potentially cornea-blinding disease herpetic stromal keratitis.
It may be of clinical importance to know whether recurrent corneal HSV-1 infections are caused by reactivation of a latent virus or reinfection with an exogenous virus. Genetically different HSV-1 strains can induce different types of ocular lesions (8, 33). Intratypic differences between HSV strains have been demonstrated by plaque morphology, serology, and DNA restriction analysis (5, 15, 22). The method generally used to discriminate HSV-1 strains is restriction fragment length polymorphism (RFLP) analysis (7, 15, 17–19, 24, 29–31). Since this technique depends on virus culture to obtain sufficient quantities of viral DNA, it is unsuitable for rapid diagnosis or when no virus can be isolated. Vogel et al. (31) reported on an alternative method for clinical HSV strain differentiation that uses PCR amplification and subsequent RFLP analysis.
We have chosen to develop a different strategy, based on the variability of reiterated sequences within the HSV-1 genome. The genome of HSV-1 consists of a unique long (UL) and a unique short (US) sequence, each of which is flanked by inverted repeat sequences (14, 32). Several hypervariable regions, designated reiterations I to VIII (ReI to ReVIII), have been identified within the HSV-1 genome (Fig. 1). These regions contain multiple repeating sequences, which vary in numbers between unrelated HSV-1 strains (3, 9, 10, 13, 24, 25, 28, 34, 35). The stability of these regions varies. ReI, ReIII, ReIV, and ReVII have been demonstrated to be relatively stable during a short period of viral replication, and it has been suggested that several of these hypervariable regions could be used as markers to discriminate HSV-1 strains (27, 28). ReI and ReIII are located within the “a” sequence of the repeat regions that flank the unique short sequence. ReIV is present twice within the HSV-1 genome and is located within introns of both the genes US1 and US12, whereas ReVII is located within the protein-coding region of US10 and US11 (3, 11, 13).
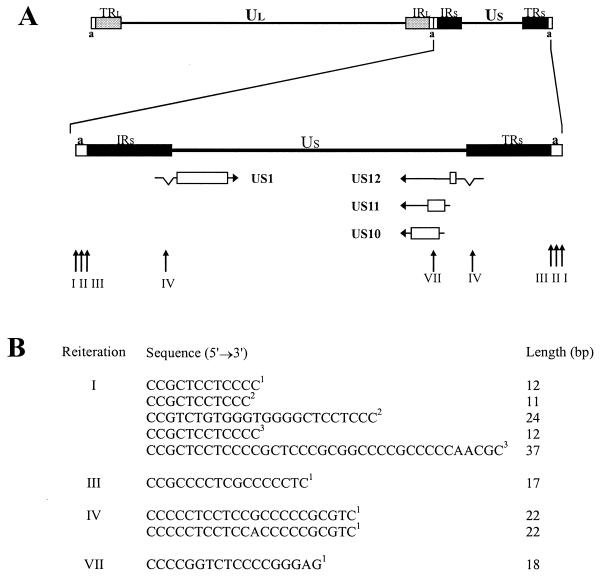
FIG. 1.
Map of HSV-1 genome and location and sequences of the reiterations tested. (A) The HSV-1 genome contains two covalently linked components (L and S), each of which consists of unique sequences (UL and US) flanked by inverted repeat sequences (IR and TR). The short “a” sequence is located at both termini of the genome and in the inverse orientation at the L-S junction (14). The enlargement of the S component shows the 5′→3′ orientations of mRNA species as horizontal arrows, with introns indicated as V-shaped indents. Protein-coding regions are shown as open boxes. Vertical arrows indicate locations of reiterations, and Roman numerals indicate locations as defined by Rixon et al. (13). (B) Reiteration-specific sequences, as indicated by the superscript numbers, were derived from the indicated HSV-1 strains: 1, MP17; 2, USA-8; 3, F.
This report describes the development of a PCR method that is used to discriminate HSV-1 strains and that is based on the variability of reiterated sequences within the HSV-1 genome. This approach was successfully used to discriminate 37 unrelated corneal HSV-1 isolates obtained from patients with herpetic corneal disease. Additionally, sequential HSV-1 isolates from five herpetic keratitis patients were compared.
MATERIALS AND METHODS
Clinical samples and viruses.
Corneal swab specimens were obtained from 37 patients with herpetic keratitis at the Rotterdam Eye Hospital (Rotterdam, The Netherlands) for diagnostic purposes. Sequential samples (n = 2) were obtained (mean time interval, 19 months; range, 9 to 38 months) from 5 patients: from the same eye for four patients and from different eyes for one patient. Virus was grown on human embryonic lung fibroblasts and was harvested when approximately 75% of the monolayer displayed a cytopathic effect. All culture samples were confirmed to be HSV-1 positive by PCR (data not shown). To determine the stability of the hypervariable regions, 24 subclones were generated from HSV-1 F (ATCC VR-733) by subcloning twice in limiting dilution as described before (26).
Nucleic acid extraction.
DNA was extracted from 100 μl of virus culture samples by a guanidinium thiocyanate-Celite binding method, as described before (1). Briefly, a sample was added to a tube containing 1 ml of lysis buffer and 40 μl of Celite suspension (Fischer Scientific, Den Bosch, The Netherlands), mixed, and incubated for 10 min at room temperature. The Celite-bound DNA was washed twice with wash buffer, twice with 70% (vol/vol) ethanol, and once with acetone and was subsequently dried. DNA was extracted by resuspending the pellet in 150 μl of water at 56°C for 10 min. A volume of 5 μl of the resulting DNA suspension was used per PCR mixture.
PCR amplification.
Primers were designed to amplify distinct regions in the HSV-1 genome that contained ReI, ReIII, ReIV, or ReVII. PCR amplification was performed with several combinations of primers (Table 1). The PCRs were performed in 50-μl volumes. The reaction mixture contained 1.25 U of cloned Pfu DNA polymerase (Stratagene Europe, Amsterdam, The Netherlands), corresponding buffer supplemented with 5% (vol/vol) dimethyl sulfoxide (DMSO), each of the primers at a concentration of 1 μM, and each deoxynucleoside triphosphate, including equimolar amounts of dGTP and 7-deaza-2′-dGTP (Boehringer Mannheim, Mannheim, Germany), at a concentration of 200 μM. A 5-μl sample of the DNA suspension was added, and the reaction mixtures were overlaid with 50 μl of mineral oil. PCR amplification was carried out as follows: an initial denaturation step of 95°C for 5 min, followed by 45 cycles of alternating denaturation (1 min, 95°C), primer annealing (1 min at the appropriate temperature; Table 1), and primer extension (1 min, 72°C). A final extension step of 7 min at 72°C was included. For negative control samples, the DNA suspension was replaced by water. All PCRs were performed in a Perkin-Elmer 480 thermocycler (PE Biosystems, Nieuwerkerk a/d IJssel, The Netherlands).
TABLE 1.
Primers used for amplification and detection of HSV-1 reiterations
| Genome region | Primera | Optimal annealing temp (°C) | Sequence (5′→3′) | Position in genomeb |
|---|---|---|---|---|
| “a” sequence | ReIF | 72 | GCCGCCACCGCTTTAAAGGGCCGC | 125976–125999 and 152234–152257 |
| ReIR | GTGCTCTGTTGGTTTCACCTGTGGCAGC | 126368–126395 and 151838–151865 | ||
| “a” sequence | ReIIIF | 72 | TCTCTACCTCAGTGCCGCCAATCTCAGGTC | 126742–126771 and 151462–151491 |
| ReIIIR | CGAAGACGCAATAAACGGCAACAACCTG | 127171–127198 and 151035–151062 | ||
| US1 | ReIVUS1F | 64 | TCCGACGACAGAAACCCACC | 132333–132352 |
| ReIVUS1R | GTCCCGGAGGACCACAGTGG | 132615–132634 | ||
| US12 | ReIVUS12F | 58 | TTTTTGCACGGGTAAGCAC | 145853–145871 |
| ReIVUS12R | TGGTGTCCAGGAAGGTGTCC | 145535–145554 | ||
| US10-US11 | ReVIIUS1011F | 56 | AGCGTATGCTCCATGTTGTG | 144697–144716 |
| ReVIIUS1011R | CGAGAACCTAGGGAACCCA | 144928–144946 | ||
| ReI | ReI probe | 37 | CCGCTCCTCCCC | |
| ReIII | ReIII probe | 37 | CCGCCCCTCGCCCCCTC | |
| ReIV | ReIV probe | 37 | CCCCCTCCTCCACCCCCGCGTC | |
| ReVII | ReVII probe | 37 | CCCCGGTCTCCCCGGGAG |
F, forward; R, reverse.
Positions correspond to the genomic HSV-1 sequence HE1CG (accession no. X14112).
Detection of amplified products.
Amplicons were size fractionated in 2% agarose gels and were visualized by ethidium bromide staining. The specificities of the amplicons were confirmed by Southern blotting (20). Briefly, the electrophoresed samples were transferred onto Hybond N+ membranes (Amersham, Pharmacia Biotech). Hybridization was performed overnight at 37°C with [γ-32P]ATP-labeled Re-specific oligonucleotides (Table 1). Posthybridization washes were performed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate at 37°C for 10 min. The filters were exposed with intensifying screens at −80°C. In case of small differences in length between amplicons from individual samples, the DNA fragments were electrophoresed on denaturing (8 M urea) 6% acrylamide gels (20). The lengths of the amplicons were estimated by comparison to a 100-bp DNA ladder (Gibco BRL). To confirm differences in amplicon length, all samples tested were finally electrophoresed in order of increasing length.
RESULTS
Amplification of hypervariable genomic HSV-1 regions containing ReI, ReIII, ReIV, and ReVII.
On the basis of documented variability and stability (27, 28), hypervariable regions containing ReI, ReIII, ReIV, and ReVII were selected as candidate templates for PCR-mediated discrimination of unrelated HSV-1 strains.
Amplification of these regions was not possible or was insufficient under standard PCR conditions (data not shown). Alternative conditions, selected to decrease the formation of secondary structures due to the high G+C contents of these sequences, improved amplification of the target sequences and allowed direct visualization of the amplicons with ethidium bromide. The specificities of the amplicons were confirmed by hybridization with a γ-32P-labeled Re-specific probe following Southern blotting (Fig. 2). Consistent results were obtained in all cases in subsequent experiments.
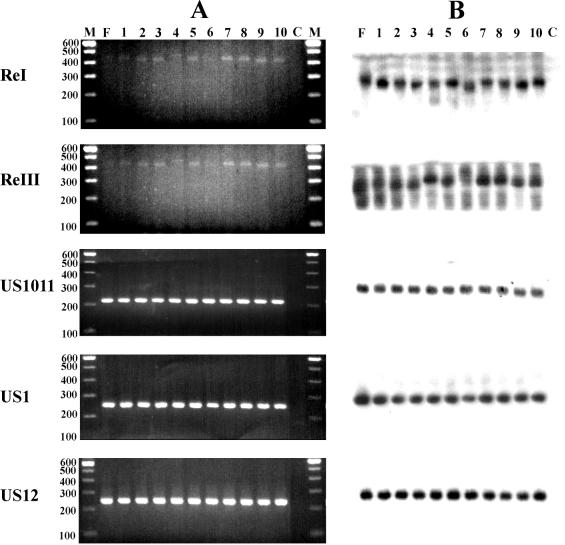
FIG. 2.
Amplification of hypervariable regions within the HSV-1 genome. (A) PCR amplification of regions containing ReI, ReIII, ReIV, and ReVII was performed with DNA from various HSV-1 (strain F) subclones. Amplicons were electrophoresed on a 2% agarose gel and were visualized by ethidium bromide staining. Ten representative samples from 24 subclones analyzed are shown. Lane F, parental strain HSV-1 F; lanes 1 to 10, HSV-1 F subclones; lane C, water control; lanes M, 100-bp molecular size marker. Numbers on the left are in base pairs. (B) Autoradiogram of DNA in gel from panel A after Southern blot transfer and hybridization with a Re-specific probe.
To test the stabilities of ReI, ReIII, ReIV, and ReVII, PCR amplification of these regions was performed with 24 separate subclones of HSV-1 F, and the sequences of these regions were compared with those of the amplicons of the parental strains (Fig. 2). For the regions containing ReIV and ReVII, amplicons from all 24 subclones were identical in size to those of their parental strains, indicating the stability of ReIV and ReVII during the two limiting dilution rounds. For the regions containing ReI and ReIII, not all separate subclones showed the same amplicon length as their parental strains, differences being greatest for ReIII (Fig. 2). Consequently, ReIV and ReVII were further used to discriminate 37 unrelated HSV-1 isolates obtained from keratitis patients. The results of the analyses performed with all 37 clinical corneal HSV-1 isolates are summarized in Table 2. As an example, differences in amplicon lengths between unrelated clinical isolates from 10 patients are shown in Fig. 3A. The variability in the US10-US11 region (ReVII) was low, showing only three different alleles. Regions US1 and US12 (ReIV) showed a wider variety of alleles, with 14 and 15 different alleles detected among the 37 samples analyzed, respectively (Table 2). Combination of the results for the three amplified regions showed that 34 of the 37 isolates (92%) displayed unique combinations of amplicons. For some clinical samples, no PCR product could be detected by ethidium bromide staining or multiple fragments appeared. This was probably due to the poor quality of the template DNA. Hybridization with the labeled probe, however, readily enabled the detection of the Re-specific amplicon in these samples (data not shown).
TABLE 2.
Length of reiteration-specific amplicons of corneal HSV-1 isolates
| Isolate or sample and patient no.a | Estimated amplicon length (bp)
|
||
|---|---|---|---|
| Region US10-US11 (ReVII) | Region US1 (ReIV) | Region US12 (ReIV) | |
| Unrelated HSV-1 isolates | |||
| 1 | 215 | 270 | 370 |
| 2 | 215 | 280 | 220 |
| 3 | 215 | 280 | 270 |
| 4 | 215 | 290 | 280 |
| 5 | 215 | 295 | 230 |
| 6 | 215 | 305 | 300 |
| 7 | 220 | 210 | 310 |
| 8 | 220 | 220 | 220 |
| 9 | 220 | 220 | 220 |
| 10 | 220 | 260 | 260 |
| 11 | 220 | 260 | 290 |
| 12 | 220 | 260 | 290 |
| 13 | 220 | 280 | 260 |
| 14 | 220 | 280 | 260 |
| 15 | 220 | 290 | 260 |
| 16 | 220 | 290 | 280 |
| 17 | 220 | 305 | 230 |
| 18 | 220 | 305 | 310 |
| 19 | 220 | 370 | 370 |
| 20 | 220 | 380 | 300 |
| 21 | 220 | 380 | 390 |
| 22 | 220 | 410 | 220 |
| 23 | 220 | 410 | 460 |
| 24 | 220 | 420 | 300 |
| 25 | 220 | 420 | 420 |
| 26 | 225 | 260 | 270 |
| 27 | 225 | 260 | 280 |
| 28 | 225 | 290 | 260 |
| 29 | 225 | 290 | 270 |
| 30 | 225 | 295 | 220 |
| 31 | 225 | 295 | 290 |
| 32 | 225 | 320 | 220 |
| 33 | 225 | 320 | 320 |
| 34 | 225 | 320 | 380 |
| 35 | 225 | 340 | 220 |
| 36 | 225 | 340 | 340 |
| 37 | 225 | 370 | 370 |
| Sequential samples | |||
| 1a | 215 | 280 | 270 |
| 1b | 215 | 280 | 270 |
| 2a | 215 | 280 | 220 |
| 2b | 215 | 280 | 220 |
| 3a | 215 | 295 | 230 |
| 3b | 215 | 295 | 230 |
| 4a | 215 | 305 | 300 |
| 4b | 215 | 305 | 300 |
| 5a | 225 | 220 | 220 |
| 5b | 225 | 220 | 220 |
Three pairs of patients (patients 8 and 9, patients 11 and 12, and patients 13 and 14) were infected with unrelated clinical isolates with identical DNA patterns.
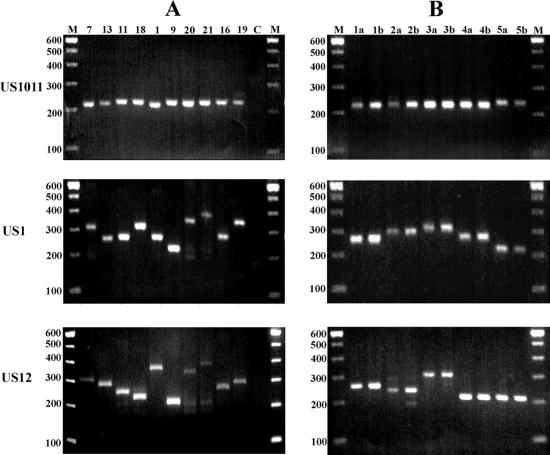
FIG. 3.
Variability of Re-containing regions US1, US12, and US10-US11 between unrelated and sequential corneal HSV-1 isolates. PCR amplification was performed with DNA from corneal HSV-1 isolates. Amplicons were analyzed as described in the legend to Fig. 2. (A) Results for 10 representative samples among the 37 samples analyzed. (B) Amplicons from sequential samples from five individuals. Lane C, water control; lanes M, 100-bp molecular size marker. Numbers on the left are in base pairs.
Analysis of sequential corneal HSV-1 isolates.
Sequential corneal HSV-1 isolates obtained from five patients with recurrent herpetic corneal infections were analyzed (Fig. 3; Table 2). The ReIV- and ReVII-specific amplicons showed interindividual variations in length. However, the amplicons from the sequential samples from each individual were identical.
DISCUSSION
In the present paper, we present a PCR-based approach that allows the rapid and accurate discrimination of unrelated HSV-1 strains. The method generally used to discriminate HSV-1 strains is RFLP analysis (7, 15, 17–19, 24, 29–31). This method requires virus culture, is time-consuming, and is highly labor-intensive. Furthermore, culture requires viable virus, which is not always obtainable from certain types of clinical samples (e.g., cerebrospinal and intraocular fluids). More recently, a system that uses PCR amplification and subsequent RFLP analysis has been developed to facilitate discrimination of HSV-1 strains, eliminating the necessity of virus culture. This method, however, is not significantly less time-consuming or labor-intensive than conventional strain differentiation (31).
Conventional RFLP analysis with restriction endonucleases that recognize 6 bp (6-bp REs) is insufficient for differentiation of HSV-1 strains of a predominant genotype. The use of 4-bp REs and RFLP analyses of reiterated sequences greatly improved the differentiation rate (28). As in our study, the RFLP analysis of reiterated sequences was based on various numbers of repeats. Use of both techniques generated similar results, verifying the applicability of either method in molecular epidemiological studies (27, 28). Similar hypervariable regions have been used successfully to discriminate strains of other herpesviruses like Epstein-Barr virus and human cytomegalovirus (23, 36).
To be applicable in a PCR-based assay for discrimination of different HSV-1 strains, these regions should show a considerable degree of variability and should remain stable during a relatively short time of replication. We tested the suitability of several HSV-1 hypervariable regions for discrimination of unrelated HSV-1 strains.
Due to their G+C-rich sequences, standard PCR protocols failed to reproducibly amplify the regions tested. The high G+C content increases the formation of secondary structures, preventing consistent amplification of the repeats. We tested a number of PCR conditions in order to obtain consistent DNA amplification. Addition of DMSO as a cosolvent to the reaction mixture has previously been shown to facilitate DNA amplification of G+C-rich sequences (12). Introduction of the exonuclease activity of the Pfu DNA polymerase enzyme in the PCR mixture prevents “skipping” of the repeats, which could result in the formation of products smaller than the actual size of the template repeat (2). Another modification was the introduction of 7-deaza-2′-dGTP. This analogue of dGTP is equally well incorporated into DNA but exerts a lesser binding strength to dCTP than normal dGTP (6, 21). The use of Pfu polymerase, 50% 7-deaza-2′-dGTP as a replacement for 100% dGTP, and 5% DMSO resulted in the most consistent amplification of the large alleles. The specificities of the amplicons were confirmed by hybridization with Re-specific probes after Southern blotting.
Analysis of subclones of HSV-1 F showed that the stability of the ReI and ReIII sequences was too low to be useful for discrimination of HSV-1 strains. In contrast, ReIV and ReVII were shown to be stable during this procedure. Thus, regions US1 (ReIV), US12 (ReIV), and US10-US11 (ReVII) were chosen for use in the discrimination of unrelated corneal HSV-1 isolates.
In agreement with previous studies, the variability in the US10-US11 region was found to be relatively low (27–29). We detected only three different alleles among 37 unrelated clinical HSV-1 isolates, which is not surprising since ReVII is located within a protein-coding region, making it a target for selective pressure. More drastic changes in the length of US10-US11 could influence the translation or function of the proteins encoded by genes US10 and US11. In contrast, the ReIV-containing sequences are located in the introns of genes US1 and US12. We found 14 and 15 different alleles for regions US1 and US12, respectively, in the 37 corneal HSV-1 isolates analyzed. Comparison of the alleles from the three regions for all 37 corneal HSV-1 isolates revealed 34 unique combinations. The isolates with identical combinations were obtained at different time points, indicating that this was most likely not due to contamination during virus isolation or culture procedures.
Sequential corneal isolates from five individuals with recurrent herpetic corneal infections were analyzed. For each individual, sequential samples showed identical DNA patterns, while the patterns for samples from different patients were different. These results indicate that the recurrent infections were most likely caused by the same virus. A comparative sequence database search revealed several point mutations between different HSV-1 strains, in addition to various numbers of repeats. More detailed analysis, like sequencing of the amplicons, might provide more conclusive evidence for this assumption. This also demonstrates that these hypervariable regions remain stable during reactivation and replication of latent HSV-1 in the corneas of these individuals.
Additionally, we have also analyzed clinical samples in which no viable virus can usually be detected (4). Re sequence-specific PCR analyses were performed with DNA isolated from affected corneal buttons and rims obtained from patients with herpetic stromal keratitis during therapeutic keratoplasty. The PCR approach proved to be sensitive enough for amplification of the low levels of viral DNA present in these samples (unpublished data). The major advantage of the approach presented is that it provides the opportunity to discriminate HSV-1 strains without virus culture or RFLP analysis, making it convenient for rapid diagnostic testing. Although not suitable for classification of HSV-1 strains, it provides a powerful tool that can be used to address questions regarding reactivation and the modes of transmission of HSV-1. For example, it could be used to assess the risk of HSV-1 transmission through cornea transplantation and other manifestations of recurrent HSV-1 infections.
ACKNOWLEDGMENTS
This work was supported by grants “Fischer Stichting” (to J.M.) and “Stichting Wetenschappelijk Onderzoek Oogziekenhuis” (to G.M.G.M.V. and L.R.).
REFERENCES
- 1.Boom R, Sol C J, Salimans M M, Jansen C L, Wertheim-van Dillen P M, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chong S S, Eichler E E, Nelson D L, Hughes M R. Robust amplification and ethidium-visible detection of the fragile X syndrome CGG repeat using Pfu polymerase. Am J Med Genet. 1994;51:522–526. doi: 10.1002/ajmg.1320510447. [DOI] [PubMed] [Google Scholar]
- 3.Davison A J, Wilkie N M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981;55:315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- 4.Easty D L, Shimeld C, Claoue C M, Menage M. Herpes simplex virus isolation in chronic stromal keratitis: human and laboratory studies. Curr Eye Res. 1987;6:69–74. doi: 10.3109/02713688709020071. [DOI] [PubMed] [Google Scholar]
- 5.Ejercito P M, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on the social behavior of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 6.Fernandez-Rachubinski F, Eng B, Murray W W, Blajchman M A, Rachubinski R A. Incorporation of 7-deaza dGTP during the amplification step in the polymerase chain reaction procedure improves subsequent DNA sequencing. DNA Seq. 1990;1:137–140. doi: 10.3109/10425179009016041. [DOI] [PubMed] [Google Scholar]
- 7.Haugen T H, Alden B, Matthey S, Nicholson D. Restriction enzyme fragment length polymorphisms of amplified herpes simplex virus type-1 DNA provide epidemiologic information. Virology. 1993;17:129–133. doi: 10.1016/0732-8893(93)90023-z. [DOI] [PubMed] [Google Scholar]
- 8.Kintner R L, Allan R W, Brandt C R. Recombinants are isolated at high frequency following in vivo mixed ocular infection with two avirulent herpes simplex virus type 1 strains. Arch Virol. 1995;140:231–244. doi: 10.1007/BF01309859. [DOI] [PubMed] [Google Scholar]
- 9.McGeoch D J, Dolan A, Donald S, Brauer D H K. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 1986;14:1727–1745. doi: 10.1093/nar/14.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mocarski E S, Post L E, Roizman B. Molecular engineering of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci USA. 1980;78:7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murchie M-J, McGeoch D J. DNA sequence analysis of an immediate-early gene region of the herpes simplex virus type 1 genome (map coordinates 0.950 to 0.0978) J Gen Virol. 1982;62:1–15. doi: 10.1099/0022-1317-62-1-1. [DOI] [PubMed] [Google Scholar]
- 12.Paragas J, Blaho J A. Cosolvents facilitate DNA synthesis in the herpes simplex virus 1 unique short (Us) inverted repeat. J Virol Methods. 1998;73:53–58. doi: 10.1016/s0166-0934(98)00036-6. [DOI] [PubMed] [Google Scholar]
- 13.Rixon F J, Campbell M E, Clements J B. A tandemly reiterated DNA sequence in the long repeat region of herpes simplex virus type 1 found in close proximity to immediate-early mRNA 1. J Virol. 1984;52:715–718. doi: 10.1128/jvi.52.2.715-718.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roizman B. The structure and isomerization of herpes simplex virus genomes. Cell. 1979;16:481–494. doi: 10.1016/0092-8674(79)90023-0. [DOI] [PubMed] [Google Scholar]
- 15.Roizman B, Tognon M. Restriction endonuclease patterns of herpes simplex virus DNA: application to diagnosis and molecular epidemiology. Curr Top Microbiol Immunol. 1983;104:273–286. doi: 10.1007/978-3-642-68949-9_17. [DOI] [PubMed] [Google Scholar]
- 16.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Vol. 1. New York, N.Y: Raven Press; 1996. pp. 2231–2295. [Google Scholar]
- 17.Sakaoka H, Aomori T, Saito H, Sato S, Kawana R, Hazlett D T, Fujinaga K. A comparative analysis by restriction endonucleases of herpes simplex virus type 1 isolated in Japan and Kenya. J Infect Dis. 1986;153:612–616. [PubMed] [Google Scholar]
- 18.Sakaoka H, Saito H, Sekine K, Aomori T, Grillner L, Wadell G, Fujinaga K. Genomic comparison of herpes simplex virus type 1 isolates from Japan, Sweden, and Kenya. J Gen Virol. 1987;68:749–764. doi: 10.1099/0022-1317-68-3-749. [DOI] [PubMed] [Google Scholar]
- 19.Sakaoka H, Kurita K, Iida Y, Takada S, Umene K, Kim Y T, Ren C S, Nahmias A J. Quantitative analysis of genomic polymorphism of herpes simplex virus type 1 strains from six countries: studies of molecular evolution and molecular epidemiology of the virus. J Gen Virol. 1994;75:513–527. doi: 10.1099/0022-1317-75-3-513. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Seela F, Rolling A. 7-Deazapurine containing DNA: efficiency of c7GdTP, c7AdTP and c7IdTP incorporation during PCR amplification and protection from endo deoxyribonuclease hydrolysis. Nucleic Acids Res. 1992;20:55–61. doi: 10.1093/nar/20.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seth P, Rawls W E, Duff R, Rapp F, Adam E, Melnick J L. Antigenic differences between isolates of herpesvirus type 2. Intervirology. 1974;3:1–14. doi: 10.1159/000149738. [DOI] [PubMed] [Google Scholar]
- 23.Triantos D, Boulter A W, Leao J C, Alberti L D, Porter S R, Scully C M, Birnbaum W, Johnson N W, Teo C G. Diversity of naturally occurring Epstein-Barr virus revealed by nucleotide sequence polymorphism in hypervariable domains in the BamHI K and N subgenomic regions. J Gen Virol. 1998;79:2809–2817. doi: 10.1099/0022-1317-79-11-2809. [DOI] [PubMed] [Google Scholar]
- 24.Umene K, Eto T, Mori R, Takagi Y, Enquist L W. Herpes simplex virus type 1 restriction fragment polymorphism determined using Southern hybridization. Arch Virol. 1984;80:275–290. doi: 10.1007/BF01311219. [DOI] [PubMed] [Google Scholar]
- 25.Umene K, Watson R J, Enquist L W. Tandem repeat DNA in an intergenic region of herpes simplex virus type 1 (Patton) Gene. 1984;30:33–39. doi: 10.1016/0378-1119(84)90102-1. [DOI] [PubMed] [Google Scholar]
- 26.Umene K, Enquist L W. Isolation of novel herpes simplex virus type 1 derivatives with tandem duplications of DNA sequences encoding immediate-early mRNA-5 and an origin of replication. J Virol. 1985;53:607–615. doi: 10.1128/jvi.53.2.607-615.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umene K, Yoshida M. Reiterated sequences of herpes simplex virus type 1 (HSV-1) genome can serve as physical markers for the differentiation of HSV-1 strains. Arch Virol. 1989;106:281–299. doi: 10.1007/BF01313958. [DOI] [PubMed] [Google Scholar]
- 28.Umene K, Sakaoka H. Homogeneity and diversity of genome polymorphism in a set of herpes simplex type 1 strains classified as the same genotypic group. Arch Virol. 1991;119:53–65. doi: 10.1007/BF01314323. [DOI] [PubMed] [Google Scholar]
- 29.Umene K, Yoshida M. Genomic characterization of two predominant genotypes of herpes simplex virus type 1. Arch Virol. 1993;131:29–46. doi: 10.1007/BF01379078. [DOI] [PubMed] [Google Scholar]
- 30.Umene K, Yoshida M. Preparation of herpes simplex virus type 1 genomic markers to differentiate strains of predominant genotypes. Arch Virol. 1994;138:55–69. doi: 10.1007/BF01310038. [DOI] [PubMed] [Google Scholar]
- 31.Vogel J-U, Weber B, Doerr H W. Typing and strain differentiation of clinical herpes simplex virus type 1 and 2 isolates by polymerase chain reaction and subsequent fragment length polymorphism analysis. Int J Med Microbiol Virol Parasitol Infect Dis. 1994;281:502–512. doi: 10.1016/s0934-8840(11)80338-5. [DOI] [PubMed] [Google Scholar]
- 32.Wadsworth S, Jacob R J, Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975;15:1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wander A H, Centifanto Y M, Kaufman H E. Strain specificity of clinical isolates of herpes simplex virus. Arch Ophthalmol. 1980;98:1458–1461. doi: 10.1001/archopht.1980.01020040310020. [DOI] [PubMed] [Google Scholar]
- 34.Watson R J, Umene K, Enquist L W. Reiterated sequences within the intron of an immediate-early gene of herpes simplex virus type 1. Nucleic Acids Res. 1981;9:4189–4199. doi: 10.1093/nar/9.16.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watson R J, Van de Woude G F. DNA sequence of an immediate-early gene (IE mRNA-5) of herpes simplex virus type 1. Nucleic Acids Res. 1982;10:979–991. doi: 10.1093/nar/10.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaia J A, Gallez-Hawkings G, Churchill M A, Morton-Blackshere A, Pande H, Adler S P, Schmidt G M, Forman S J. Comparative analysis of human cytomegalovirus a-sequence in multiple clinical isolates by using polymerase chain reaction and restriction fragment length polymorphism assays. J Clin Microbiol. 1990;28:2602–2607. doi: 10.1128/jcm.28.12.2602-2607.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]