Abstract
The ongoing pandemic of COVID-19 caused by the SARS-COV2 virus has triggered millions of deaths around the globe. Emerging several variants of the virus with increased transmissibility, the severity of disease, and the ability of the virus to escape from the immune system has a cause for concerns. Here, we compared the spike protein sequence of 91 human SARS CoV2 strains of Iraq to the first reported sequence of SARS-CoV2 isolate from Wuhan Hu-1/China. The strains were isolated between June 2020 and March 2021. Twenty-two distinct mutations were identified within the spike protein regions which were: L5F, L18F, T19R, S151T, G181A, A222V, A348S, L452 (Q or M), T478K, N501Y, A520S, A522V, A570D, S605A, D614G, Q675H, N679K, P681H, T716I, S982A, A1020S, D1118H. The most frequently mutations occurred at the D614G (87/91), followed by S982A (50/91), and A570D (48/91), respectively. In addition, a distinct shift was observed in the type of SARS-COV2 variants present in 2020 compared to 2021 isolates. In 2020, B.1.428.1 lineage was appeared to be a dominant variant (85%). However, the diversity of the variants increased in 2021, and the majority (73%) of the isolated were appeared to belong to B.1.1.7 lineage (VOC/alpha variants). To our knowledge, this is the first major genome analysis of SARS-CoV2 in Iraq. The data from this research could provide insights into SARS-CoV2 evolution, and can be potentially used to recognize the effective vaccine against the disease.
Abbreviations: SARS-CoV2, Severe acute respiratory syndrome coronavirus 2; ACE2, Angiotensin-Converting Enzyme 2; RBD, Receptor-Binding Domain; VOC, Virus of Concern; GISAID, Global Initiative on Sharing All Influenza Data; PDB, Protein Data Bank; SP, Signal Peptide; NTD, N-terminal domain; FP, Fusion peptide; HR1, Heptad Repeat 1; HR2, Heptad Repeat 2; TM, Transmembrane Domain; CP, Cytoplasmic Peptide
Keywords: SARS-COV2, Covid-19, Spike protein, Receptor binding domain, Mutation, Iraq
1. Introduction
Highly contagious coronavirus disease 2019 (COVID-19) is a human respiratory system infection which is caused by SARS-coronavirus-2 (SARS-CoV2). The disease was firstly reported in the city of Wuhan, China in December 2019 (Wu et al., 2020) and it was declared to be pandemic by WHO on March 11, 2020 (World Health Organization, 2020a). Inhalation and/or direct contact with infected droplets is the main route of disease transmission (Mahmud et al., 2020). In Iraq, the first confirmed case of COVID-19 was reported on the 24th of February 2020 in Al-Najaf city in the south of Baghdad (Alsayed et al., 2020). As of 30 June 2021, more than 1.6 million people have contracted the disease in Iraq with more than 18,000 deaths (World Health Organization, 2020b).
The etiology of COVID-19 is SARS-Cov-2; positive single-stranded RNA viruses belong to β-genus of the Coronaviridae family (Wu et al., 2020; Sabir et al., 2020). The size of the genome is about 29 kbp, having 14 open reading frames (ORFs) and 27 proteins, including four structural proteins which are spike (S), envelop protein (E), membrane protein (M), and nucleocapsid (N) (Sabir et al., 2020; Chan et al., 2020). Spike protein is a key target for diagnosis and developing vaccines (Wrapp et al., 2020) since it plays important role in the pathogenicity of the virus by interacting with cellular receptors such as the angiotensin-converting enzyme 2 (ACE2) to enter the human body (Wu et al., 2020). It is made up of 1273 amino acid residues and the structure of the protein was first determined using cryo-electron microscopy (PDB: 6VSB) (Wrapp et al., 2020). The protein is composed of two S1 and S2 sub-domains. Residues from 1 to 685 were located in the S1 subdomain, whereas the rest were located in the S2 subdomain that is used to fuse and enter into the target cells (Tegally et al., 2021; Prevention ECfD, Control, 2021). The receptor-binding domain (RBD) (residue 319 to 541) is an important region of the S1 subdomain which facilitate the virus to interact with ACE2 of the human cells (Thomson et al., 2021; Wise, 2020).
Several variants of SARS-COV2 have been reported around the world resulted possessing one or more mutations in the virus genome that can affect the transmissibility of the virus and also the immunological response of the host cells (Lauring and Hodcroft, 2021). A variant that carries D614G mutation in the spike protein of the virus, also known as G clade or B.1, was diagnosed during the early days of the pandemic in the north of America, and then reported in many European countries (Korber et al., 2021). Another variant possesses N439K mutation in the receptor-binding domain (RBD) of spike protein has also been reported independently in many countries in Europe and the USA. The N439K variant has shown to have increased pathogenicity and can escape from neutralizing monoclonal antibodies and reducing the activities of some polyclonal responses (Thomson et al., 2021). B.1.1.7 lineage (Alpha variant, also known as VOC (Virus of Concern)) is a variant of the SARS-COV2 which was first identified in the January of 2020 in the UK (Wise, 2020). The variant is characterized by having a deletion in the amino acid position of 69/70 (nucleotide number: 21,765–21,770), and 17 mutations in the total viral genome including N501Y in the receptor-binding domain of spike protein (Wise, 2020). Beta variant (B.1.351) lineage is another variant of concern (VOC), which was first reported in South Africa in October 2020, and characterized by eight mutations in the spike protein region, including K417N, E484K, and N501Y (Tegally et al., 2021). Unlike B.1.1.7 lineage, B.1.351 variant does not have a deletion at the position of 69/70 (Tegally et al., 2021). Delta variant (B.1.617.2) lineage, a current dominant variant of the virus in many countries around the globe, was first identified in India in December 2020 (Prevention ECfD, Control, 2021; Planas et al., 2021). This variant is characterized by having several mutations in the spike proteins including L452R, T478K and P681R (Planas et al., 2021). A study has shown that this variant of SARS-COV2 is 60% more transmissible than the Alpha variant (B.1.1.7) (Planas et al., 2021). There are other variants, such as P.1 (Gamma) lineage (20J/501Y.V3) which is mainly spread in Brazil, carrying 17 mutations; three of them are located in the spike protein regions (K417T, E484K, and N501Y) (Faria et al., 2021).
In this study, we describe the different variants of SARS-COV2 spread from June 2020 to March 2021 in Iraq based on a comprehensive analysis of 91 spike protein sequences of the virus. This study should give an insight into the evolution and the transmissibility of the virus in the region.
2. Materials and methods
Spike protein sequences of Iraqis' SARS CoV2 strains were obtained from the publicly accessible Global Initiative on Sharing All Influenza Data (GISAID) databank on 18 June 2021. Multiple sequence alignments were created using online program Clustal-Omega at the European Bioinformatics Institute database (https://www.ebi.ac.uk/). The alignments were carefully inspected to recognize the mutations within the 91 Iraqi isolates relative to the Wuhan-Hu-1 (accession number: NC_045512.2) (Wu et al., 2020). The location of the amino acids within the structure of the spike protein was mapped based on the structural information of the S protein available in the Protein Data Bank (PDB: 6VXX). Visualization of the protein structure was carried out using UCSF Chimera software.
3. Results and discussion
3.1. Variants of SARS-COV2 of Iraq
At the time of writing, a total of 91 SARS-COV2 genome sequences of Iraq were available in the GISAID database. The strains GISAID accession numbers; collection dates, age and gender of the patients are shown in Table 1 .
Table 1.
The accession numbers of the viral genome, and collection date, gender, and age of the patients who samples were taken.
| No. | Accession numbers | Collection date | Gender | Age |
|---|---|---|---|---|
| 1 | EPI_ISL_812258 | 06 June 2020 | Female | 33 |
| 2 | EPI_ISL_812281 | 07 June 2020 | Male | 47 |
| 3 | EPI_ISL_812280 | 07 June 2020 | Male | 32 |
| 4 | EPI_ISL_812276 | 07 June 2020 | Male | 43 |
| 5 | EPI_ISL_812275 | 07 June 2020 | Male | 48 |
| 6 | EPI_ISL_812274 | 07 June 2020 | Male | 33 |
| 7 | EPI_ISL_812272 | 07 June 2020 | Male | 37 |
| 8 | EPI_ISL_812270 | 07 June 2020 | Male | 33 |
| 9 | EPI_ISL_812267 | 07 June 2020 | Male | 40 |
| 10 | EPI_ISL_812262 | 07 June 2020 | Male | 40 |
| 11 | EPI_ISL_812260 | 11 June 2020 | Male | 20 |
| 12 | EPI_ISL_812282 | 16 June 2020 | Male | 24 |
| 13 | EPI_ISL_812279 | 16 June 2020 | Female | 36 |
| 14 | EPI_ISL_812278 | 16 June 2020 | Male | 34 |
| 15 | EPI_ISL_812277 | 16 June 2020 | Male | 41 |
| 16 | EPI_ISL_812273 | 16 June 2020 | Male | 30 |
| 17 | EPI_ISL_812271 | 16 June 2020 | Male | 26 |
| 18 | EPI_ISL_812269 | 16 June 2020 | Male | 29 |
| 19 | EPI_ISL_812268 | 16 June 2020 | Female | 24 |
| 20 | EPI_ISL_812266 | 16 June 2020 | Male | 36 |
| 21 | EPI_ISL_812265 | 16 June 2020 | Male | 41 |
| 22 | EPI_ISL_812264 | 16 June 2020 | Male | 38 |
| 23 | EPI_ISL_812263 | 16 June 2020 | Male | 30 |
| 24 | EPI_ISL_812261 | 16 June 2020 | Male | 37 |
| 25 | EPI_ISL_812259 | 16 June 2020 | Male | 31 |
| 26 | EPI_ISL_812285 | 17 June 2020 | Male | 33 |
| 27 | EPI_ISL_812284 | 17 June 2020 | Male | 27 |
| 28 | EPI_ISL_582030 | 30 June 2020 | Female | 26 |
| 29 | EPI_ISL_907075 | 01 October 2020 | Unknown | Unknown |
| 30 | EPI_ISL_956332 | 01 October 2020 | Unknown | Unknown |
| 31 | EPI_ISL_2383317 | 14 December 2020 | Male | 37 |
| 32 | EPI_ISL_1524332 | 27 December 2020 | Male | 38 |
| 33 | EPI_ISL_1524344 | 29 December 2020 | Male | 38 |
| 34 | EPI_ISL_1524330 | 20 January 2021 | Female | 72 |
| 35 | EPI_ISL_885143 | 20 January 2021 | Female | Unknown |
| 36 | EPI_ISL_1524347 | 23 January 2021 | Female | 65 |
| 37 | EPI_ISL_2383206 | 07 February 2021 | Female | 52 |
| 38 | EPI_ISL_2467919 | 08 February 2021 | Female | 65 |
| 39 | EPI_ISL_1524338 | 10 February 2021 | Male | 33 |
| 40 | EPI_ISL_1524359 | 10 February 2021 | Male | 50 |
| 41 | EPI_ISL_1524349 | 10 February 2021 | Male | 54 |
| 42 | EPI_ISL_1524345 | 10 February 2021 | Male | 36 |
| 43 | EPI_ISL_2467924 | 11 February 2021 | Female | 23 |
| 44 | EPI_ISL_1524369 | 11 February 2021 | Female | 24 |
| 90 | EPI_ISL_2467929 | 11 February 2021 | Male | 30 |
| 91 | EPI_ISL_2467909 | 11 February 2021 | Male | 23 |
| 45 | EPI_ISL_1524351 | 15 February 2021 | Female | 49 |
| 46 | EPI_ISL_1524346 | 15 February 2021 | Male | 45 |
| 47 | EPI_ISL_1524374 | 16 February 2021 | Male | 30 |
| 48 | EPI_ISL_1524372 | 16 February 2021 | Female | 30 |
| 49 | EPI_ISL_1524333 | 16 February 2021 | Male | 35 |
| 50 | EPI_ISL_1524364 | 16 February 2021 | Male | 42 |
| 51 | EPI_ISL_1524362 | 16 February 2021 | Male | 31 |
| 52 | EPI_ISL_2467911 | 17 February 2021 | Male | 86 |
| 53 | EPI_ISL_1524361 | 18 February 2021 | Male | 29 |
| 54 | EPI_ISL_1524356 | 19 February 2021 | Female | 25 |
| 55 | EPI_ISL_1524353 | 19 February 2021 | Male | 45 |
| 56 | EPI_ISL_1524343 | 19 February 2021 | Female | 70 |
| 57 | EPI_ISL_1524340 | 19 February 2021 | Male | 42 |
| 58 | EPI_ISL_1524337 | 20 February 2021 | Male | 50 |
| 59 | EPI_ISL_1524336 | 20 February 2021 | Male | 42 |
| 60 | EPI_ISL_2467915 | 20 February 2021 | Male | 25 |
| 61 | EPI_ISL_2467914 | 20 February 2021 | Female | 26 |
| 62 | EPI_ISL_1524335 | 20 February 2021 | Male | 40 |
| 63 | EPI_ISL_1524334 | 20 February 2021 | Male | 85 |
| 64 | EPI_ISL_1524378 | 20 February 2021 | Male | 60 |
| 65 | EPI_ISL_1524377 | 20 February 2021 | Male | 33 |
| 66 | EPI_ISL_1524375 | 20 February 2021 | Male | 31 |
| 67 | EPI_ISL_1524371 | 20 February 2021 | Female | 39 |
| 68 | EPI_ISL_1524370 | 20 February 2021 | Female | 56 |
| 69 | EPI_ISL_1524368 | 20 February 2021 | Female | 30 |
| 70 | EPI_ISL_1524367 | 20 February 2021 | Male | 43 |
| 71 | EPI_ISL_1524366 | 20 February 2021 | Female | 65 |
| 72 | EPI_ISL_1524365 | 20 February 2021 | Male | 15 |
| 73 | EPI_ISL_1524363 | 20 February 2021 | Male | 48 |
| 74 | EPI_ISL_1524358 | 20 February 2021 | Male | 63 |
| 75 | EPI_ISL_1524357 | 20 February 2021 | Female | 35 |
| 76 | EPI_ISL_1524354 | 20 February 2021 | Male | 52 |
| 77 | EPI_ISL_1524352 | 20 February 2021 | Male | 13 |
| 78 | EPI_ISL_1524350 | 20 February 2021 | Male | 57 |
| 79 | EPI_ISL_1524331 | 20 February 2021 | Male | 31 |
| 80 | EPI_ISL_1524342 | 20 February 2021 | Male | 48 |
| 81 | EPI_ISL_1524341 | 20 February 2021 | Female | 22 |
| 82 | EPI_ISL_1524339 | 20 February 2021 | Female | 42 |
| 83 | EPI_ISL_2467916 | 21 February 2021 | Female | 53 |
| 84 | EPI_ISL_1524380 | 22 February 2021 | Female | 42 |
| 85 | EPI_ISL_1524379 | 22 February 2021 | Female | 51 |
| 86 | EPI_ISL_1524355 | 22 February 2021 | Female | 42 |
| 87 | EPI_ISL_1524360 | 25 February 2021 | male | 40 |
| 88 | EPI_ISL_1524348 | 25 February 2021 | male | 27 |
| 89 | EPI_ISL_2379893 | 26 February 2021 | male | 72 |
Overall, an increase in the genome diversity and different viral lineages was observed in the SARS-COV2 isolates in 2021 compared to 2020. Such an increase in viral genome diversity is derived from the mutations which occur as the result of viral replication (Wise, 2020). Despite coronaviruses have a genetic proofreading mechanism to maintain their genome (Smith et al., 2013) and SARS-COV2 has low genetic diversity (Fauver et al., 2020), still, several variants of the virus have been recorded around the world since the first identification of the virus in December 2019 (Morais et al., 2020; Li et al., 2020). Knowing the viral variant that circulating in the region is important to develop effective use of the vaccines and therapeutic agents.
Among the 33 genome sequences from 2020, B.1.428.1 appeared to be a dominant lineage (85%, 28 strains). This is followed by 6% (2 strains) of B.1.1.7 lineages (Alpha/VOC), and 3% (1 strain) of each B.1 (clade GH), and B.4 (O clade), and B.1.177.80 lineages (Fig. 1 ).
Fig. 1.
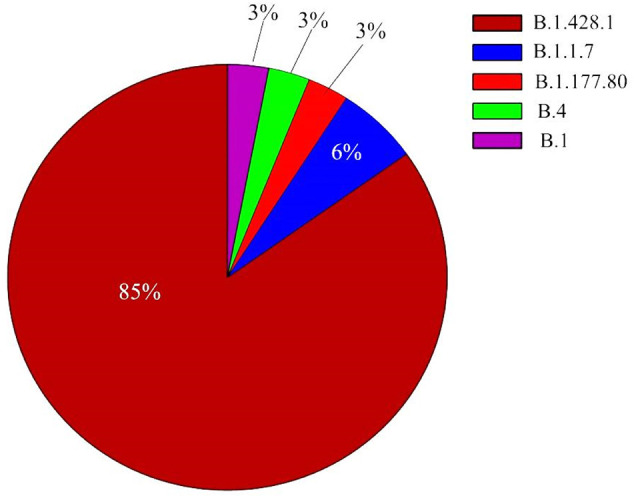
Percentage of the different lineages of the SARS-COV2 that appears in Iraq during late 2020.
B.1.428.1 lineage, which is designated as Iraqi lineage, is a subtype of B.1.428 (known as Danish lineage according to Pango lineages database:https://cov-lineages.org/) (Rambaut et al., 2020). The sub-lineage of B.1.428.1 was first reported on 27 April of 2020 in Qatar (Benslimane et al., 2021). This lineage is suggested to be highly transmissible and highly contributed towards the first pandemic waves in Qatar (Benslimane et al., 2021). Also, B.1.428 is categorized as an Epsilon (Variants of Interest or VOI) by WHO (2021). In terms of genome diversity, B.1.428.1 lineage is characterized by possessing several mutations in the genome including two mutations in the spike protein region which are A522V, D614G (https://www.gisaid.org/). These mutations can increase transmissibility and infectivity of the virus compared to the wild type (Korber et al., 2021; Zhang et al., 2020) (this variant is 10 times more infectious than the original SARS-COV2 (Li et al., 2020)). In addition, these mutations in the spike protein might affect the correct diagnosis of the virus, and also the severity of the disease (Control CfD, 2021). However, it does not increase the affinity of the protein to the ACE2, nor affects the neutralization action of the antibodies (Zhang et al., 2020). Thus, a vaccine produced based on the wild type spike protein should equally be effective against this variant (Zhang et al., 2020).
Only one of the 2020's isolates was classified as a B.4 lineage. The strain was isolated in October 2020 and has an accession number of EPI_ISL_907075. Finding B.4 lineage among Iraqi strains is not surprising since this lineage has shown to be originated from Iran (Organization WH, 2020; Eden et al., 2020), and is mainly distributed in Asian countries between March to mid-May of 2020 (https://cov-lineages.org/). Additionally, people returning from Iran was thought to be the main factor to initiate spreading the SARS-COV2 virus in Iraq (Alsayed et al., 2020). Moreover, since the mutations of this lineage are located in the non-structural proteins (NPs), which are either enzymes or functional proteins involves in viral replication and methylation, it is less likely the variants can affect the pathogenicity of the virus or the host immune responses (Khateeb et al., 2021). Feghali et al. (2021) (Feghali et al., 2021) was also reported that the predominant lineage of SARS-COV2 strains isolated between 2 February 2020 and 15 March 2020 in Lebanon was B.1, and then B.4. Moreover, another one of the 2020 isolates was found to belong to B.1 lineage (or 20 clade). This lineage is characterized by having a mutation in NPS and D614G mutation in the spike protein region (Hodcroft et al., 2020). It has increased transmissibility and it first appeared in early 2020 in Europe then it became a predominant variant in many countries around the world (Hodcroft et al., 2020).
There were three genome sequences of SARS-CoV2 isolated in December of 2020. These strains showed an increase in the genome diversity compared to other strains isolated earlier in the same year. The two strains which were belonging to B.1.1.7 variants had accession numbers of EPI_ISL_1524332 and EPI_ISL_1524344, and the third strain was belonging to B.1.177.80 variant with an accession number of EPI_ISL_2383317.
B.1.1.7 variant (Alpha variant/VOC (Wise, 2020; Frampton et al., 2021)) was first identified in the UK. The variant was also reported among Karnataka/India isolates in December 2020 (Pattabiraman et al., 2021). This variant is characterized by higher transmissibility compared to most other variants (Frampton et al., 2021). However, there was no indication that the B.1.1.7 variation was linked to the severity of the illness or mortality (Frampton et al., 2021). On the other hand, B.1.177.80 lineage, which was identified among 2020's strain, is also called (Scandinavian lineage) has been reported in several countries around the world mainly in Sweden, Norway, Denmark, but also in Iraq and Switzerland (https://www.isitzen.com/). The variant possesses several mutations including A222V, D614G, and L18F mutations in the spike proteins. To our knowledge, there is no information about transmissibility and severity of the disease caused by this variant; however, it is expected to spread faster than the wild type since it is having D614G mutation.
In early 2021, 58 SARS-COV2 genome sequences from Iraq belonging to 9 different lineages were submitted to the GISAID databank. The lineage of B.1.1.7 (Alpha variant or VOC) was appeared to be dominant with 73%, followed by B.1.1.337 (7%), B.1.438 (5%), and B.1.36.1 (5%). One variant (2%) of each B.1.1, B.1.1.374, B.1.177.7, B.1.36, and B.1.441 were also recorded among 2021's of Iraqi's SARS-COV2 variants (Fig. 2 ).
Fig. 2.
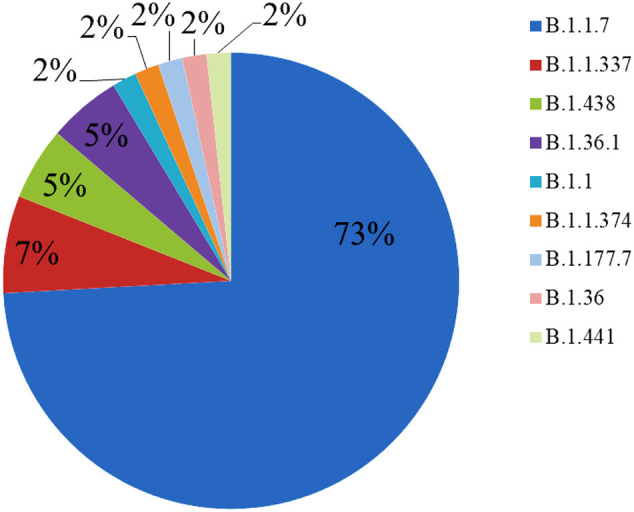
Percentage of the different lineages of the SARS-COV2 appears in Iraq during the early of 2021.
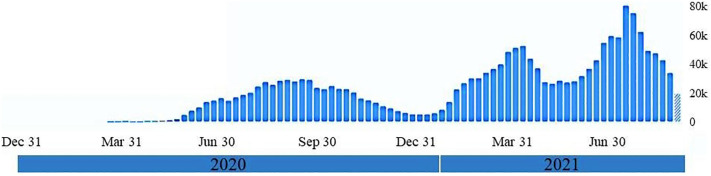
An increase in the B.1.1.7 (Alpha, VOC) lineage can be observed among the early 2021's strains (73%) compared to the late 2020's strains (6%). However, the lineage B.1.428.1 which appeared to be dominant in 2020, was not detected among the sequenced samples of 2021. Considering that B.1.1.7 has an increased transmissibility (Frampton et al., 2021), perhaps this could explain the reason for increasing COVID-19 cases in early of 2021 in Iraq compared to 2020 (Fig. 3 ). Other factors like stopping lockdown and back people to routine social life practice such as shaking hands, gathering during funeral and wedding ceremonies (Alsayed et al., 2020) might also play a role in increasing COVD-19 cases in Iraq.
Fig. 3.
Weekly confirmed cases of COVID-19 in Iraq from the beginning of the pandemic. An increase in COVID-19 cases can be observed from the data of early 2021 compared to the second half of 2020. Data extracted from World Health Organization (WHO).
Another variant was B.1.36 and its sublineages B.1.36.1 appeared among 2021 strains. The B.1.36 lineage was first reported in Saudi Arabia in February 2020 (Pattabiraman et al., 2021) and it was characterized by possessing D614G and L452M mutations, whereas, sublineage B.1.36.1 has an additional mutation of N679K in the spike protein region. Overall, several new lineages can be detected among 2021 strains of SARS-COV2 from Iraq, with higher genome diversity and might be pathogenicity.
3.2. Variations of the spike protein
SARS-COV2 is an RNA virus and mutations can naturally occur during viral replications, which can lead to variation (Wise, 2020). These mutations, particularly if occurs in the spike protein region, can change the transmissibility, pathogenicity, and also the effect of the vaccine and therapeutic antibodies against the virus (Frampton et al., 2021; Plante et al., 2021).
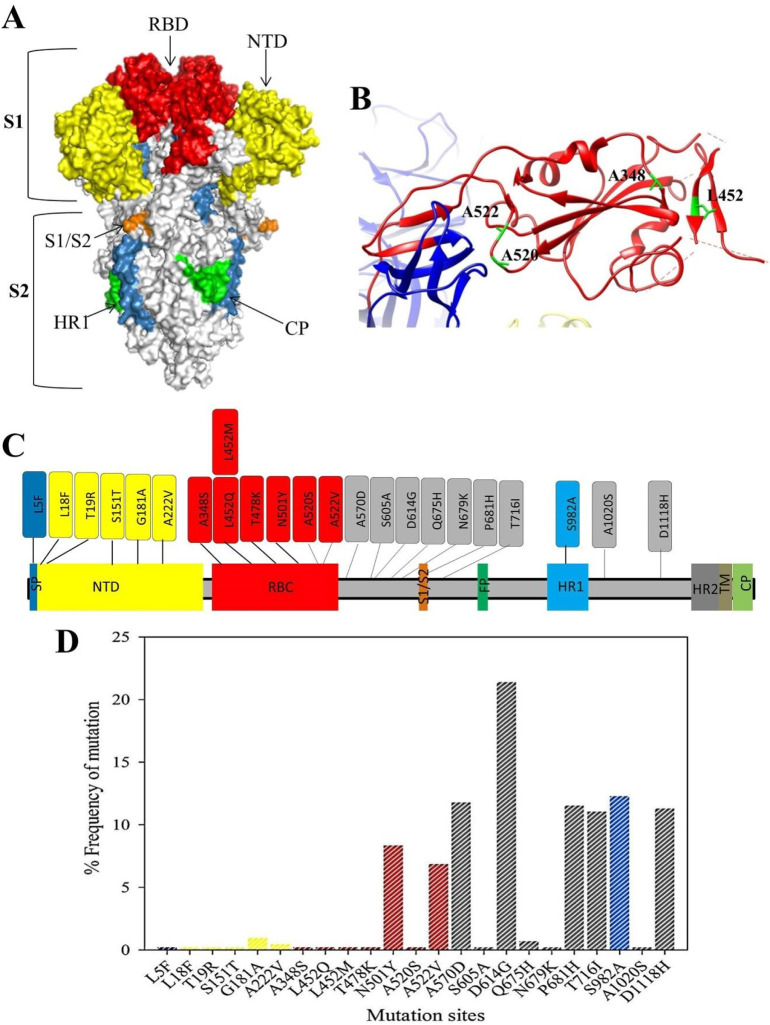
Non-synonymous mutations were analysed among the genome of 91 SARS-COV2 isolates in the spike protein region. At least one or more mutations were found in all strains, except in the genome sequence of the EPI_ISL_907075 strain which did not have a mutation in the spike protein region. However, mutations V198I in NSP2, L37F in SNP6, and T113I in NSP14 were recorded in the genome sequence of the strain. In total, 23 non-synonymous mutations were distributed in 22 distinct sites of the spike protein. D614G was found to be the most predominant mutation as 96% (87/91 strains) of the sequenced strains were carrying the mutation (Fig. 4 ).
Fig. 4.
Structure and the frequency of the mutations in the spike protein region among SARS-COV2 strains from Iraq. (A) Trimer structure of SARS-COV2 protein (PDB: 6VXX), the RBD (residue 319–541) is coloured red. (B): The ribbon structure of RBD of the monomeric structure of the protein and the locations of 5 mutations in the region of the protein. (C) Schematic representation of the different regions of SARS-COV2 and approximate mutations in each of the regions. SP: signal peptide; NTD: N-terminal domain; RBD: Ribosomal binding domain; S1/ S2 protease cleavage site; FP: fusion peptide; HR1: heptad repeat 1; HR2: heptad repeat 2; TM: transmembrane domain; CP: cytoplasmic peptide. (D) Percentage of the mutations of each of the residues located in the Spike proteins among 91 Iraqi isolates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
D614G mutation was also reported to be the dominant (84.20%) among 2634 genome sequence data of SARS-COV2 from Qatar (Benslimane et al., 2021). SARS-COV2 variant carrying D614G mutation has been shown to alter the virus fitness and increased transmissibility of the virus compared to the wild type (Korber et al., 2021; Plante et al., 2021; Yurkovetskiy et al., 2020; Volz et al., 2021). However, such mutation is not affecting the severity of the disease caused by the variant (Korber et al., 2021; Plante et al., 2021; Yurkovetskiy et al., 2020; Volz et al., 2021). Other predominant mutations which appeared frequently among Iraq isolates were S982A (55%), A570D (53%), P681H (52%), D1118H (51%). Limited researches have been carried out to study the effects of each mutations separatly. However, P681H, which can be found in Alpha (20I, V1), Kappa (21A), and Delta (21B) variants, and the residue is located near to the furin cleavage; mutation of P618 to H can increase the transmissibility of the virus (Khateeb et al., 2021). Similarly, D1118H, which is found in Alpha (20I, V1, B.1.1.7) lineage, can increase the transmissibility of the virus (Khateeb et al., 2021).
Within the spike protein region, six of the mutations were located in the ribosomal binding domain (RBD; 319–541). These mutations were L452 (Q, M), T478K, N501Y, A520S, A522V. Residue 452 in the RBD of the spike protein had two variants (L452Q in the sequence accession number of EPI_ISL_1524379) and (L452M in the sequence accession number of EPI_ISL_1524347). Mutations in the RBD region can alter the sensitivity of the variants towards neutralizing monoclonal or polyclonal antibodies. Previously, L452R variant was shown to become resistant to monoclonal antibodies (mAbs) X593 and P2B-2F6 (Li et al., 2020; Khateeb et al., 2021), however, mutating leucine (L) to glutamine (Q) or methionine (M) among the Iraqi strains could affect differently on the sensitivity of the strain towards the host and/or vaccine-induced immunity. Particularly, in the case of mutating leucine to glutamine since these two amino acids have different biochemical properties (leucine has a hydrophobic side chain, whereas glutamine has a polar uncharged side chain). Mutation of T478K in the RBD can be found in both Delta and Kappa (B.1.617.2/1) variants of the virus, but not in Alpha (B.1.1.7), beta (B.1.351), or Gamma (P.1) (Khateeb et al., 2021). This mutation has been shown to increase the transmissibility and affinity to ACE2. In addition, it helps the virus to escape from the host immune response (Khateeb et al., 2021). Both N501Yand A522V were the most frequent mutations that occurred at the RBS of the Iraqi strains with 37% and 53%, respectively (Fig. 4). N501Y mutation can be found in Alpha, Beta, or Gamma, but not in Delta and Kappa variants. Such mutation, same as T478K, increases the transmissibility of the virus (Khateeb et al., 2021). However, N501T reduces the binding affinity of spike protein to the human ACE2 (Shang et al., 2020).
In summary, our data emphasise the increased genome diversities of the SARS-COV2 in Iraq. B.1.428.1 variant was appeared to be prevalent linage in the second half of 2020, whereas B.1.1.7 was appeared to be the dominant lineage in early 2021. Twenty-three mutations were detected in 22 sides of the spike protein of the Iraqi strains.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
None.
References
- Alsayed R., Ali A., Makia R., Kadhom M., Raheem R., Al-Obaidi O., et al. Challenges facing Iraq to tackle the spread of COVID-19: an overview. J. Uni. Anbar Pure Sci. 2020;14(2):22–27. [Google Scholar]
- Benslimane F.M., AlKhatib H.A., Al-Jamal O., Albatesh D., Boughattas S., Ahmed A.A., et al. One year of SARS-CoV-2: genomic characterization of COVID-19 outbreak in Qatar. medRxiv. 2021 doi: 10.1101/2021.05.19.21257433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Control CfD, Prevention . Vol. 16. 2021. SARS-CoV-2 Variant Classifications and Definitions; p. 2020. Retrieved September. [Google Scholar]
- Eden J.-S., Rockett R., Carter I., Rahman H., De Ligt J., Hadfield J., et al. An emergent clade of SARS-CoV-2 linked to returned travellers from Iran. Virus Evol. 2020;6(1) doi: 10.1093/ve/veaa027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., DdS Candido, Mishra S., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372(6544):815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauver J.R., Petrone M.E., Hodcroft E.B., Shioda K., Ehrlich H.Y., Watts A.G., et al. Coast-to-coast spread of SARS-CoV-2 during the early epidemic in the United States. Cell. 2020;181(5):990–996.e5. doi: 10.1016/j.cell.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feghali R., Merhi G., Kwasiborski A., Hourdel V., Ghosn N., Tokajian S. Genomic characterization and phylogenetic analysis of the first SARS-CoV-2 variants introduced in Lebanon. PeerJ. 2021;9 doi: 10.7717/peerj.11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton D., Rampling T., Cross A., Bailey H., Heaney J., Byott M., et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B. 1.1. 7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021;21:1246–1255. doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B., Zuber M., Nadeau S., Crawford K.H., Bloom J.D., Veesler D., et al. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. MedRxiv. 2020 [Google Scholar]
- Khateeb J., Li Y., Zhang H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit. Care. 2021;25(1):1–8. doi: 10.1186/s13054-021-03662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B., Zuber M., Nadeau S., Vaughan T.G., Crawford K.H.D., Althaus C.L., Reichmuth M.L., Bowen J.E., Walls A.C., Corti D., Bloom J.D., Veesler D., Mateo D., Hernando A., Comas I., González-Candelas F., SeqCOVID-SPAIN consortium, Stadler T., Neher R.A. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature. 2021;595(7869):707–712. doi: 10.1038/s41586-021-03677-y. [DOI] [PubMed] [Google Scholar]
- Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2—what do they mean? JAMA. 2021;325(6):529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmud P.K., Ali S.M., Sabir D.K. Impacts of novel pandemic coronavirus (COVID-19) outbreak on dental practice: a review of the current literature. Edorium J. Dent. 2020;7 [Google Scholar]
- Morais I.J., Polveiro R.C., Souza G.M., Bortolin D.I., Sassaki F.T., Lima A.T.M. The global population of SARS-CoV-2 is composed of six major subtypes. Sci. Rep. 2020;10(1):1–9. doi: 10.1038/s41598-020-74050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization WH . 2020. COVID-19 WHO African Region: External Situation Report 19. [Google Scholar]
- Organization WH . 2021. COVID-19 Weekly Epidemiological Update, 08 June 2021. [Google Scholar]
- Pattabiraman C., Prasad P., George A.K., Sreenivas D., Rasheed R., Reddy N.V.K., et al. Importation, circulation, and emergence of variants of SARS-CoV-2 in the South Indian State of Karnataka. Wellcome Open Res. 2021;6(110):110. doi: 10.12688/wellcomeopenres.16768.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., et al. Reduced sensitivity of SARS-CoV-2 variant delta to antibody neutralization. Nature. 2021:1–7. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592(7852):116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevention ECfD, Control . 2021. Threat Assessment Brief: Emergence of SARS-CoV-2 B. 1.617 Variants in India and Situation in the EU/EEA. [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5(11):1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabir D.K., Khwarahm N.R., Ali S.M., Abdoul H.J., Mahmood K.I., Kodzius R. Children protection against COVID-19 at the pandemic outbreak. J. Immunol. Sci. 2020;4(2) [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Blanc H., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Path. 2013;9(8) doi: 10.1371/journal.ppat.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592(7854):438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184(5):1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O’Toole Á., et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184(1):64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise J. Covid-19: new coronavirus variant is identified in UK. BMJ [Internet] 2020;371:m4857. doi: 10.1136/bmj.m4857. Dec 16 [cited 2020 Dec 23] [DOI] [PubMed] [Google Scholar]
- World Health Organization Coronavirus disease 2019 (COVID-19) situation report–103. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200311-sitrep-51-covid-19.pdf?sfvrsn=1ba62e57_10
- World Health Organization . World Health Organization; Geneva, Switzerland: 2020. Coronavirus Disease 2019 (COVID-19) Situation Report – 59.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200319-sitrep-59-covid-19.pdf?sfvrsn=c3dcdef9_2 [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183(3):739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020;11(1):1–9. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]