Abstract
Wastewater surveillance of SARS-CoV-2 has become a promising tool to estimate population-level changes in community infections and the prevalence of COVID-19 disease. Although many studies have reported the detection and quantification of SARS-CoV-2 in wastewater, remarkable variation remains in the methodology. In this study, we validated a molecular testing method by concentrating viruses from wastewater using ultrafiltration and detecting SARS-CoV-2 using one-step RT-qPCR assay. The following parameters were optimized including sample storage condition, wastewater pH, RNA extraction and RT-qPCR assay by quantification of SARS-CoV-2 or spiked human coronavirus strain 229E (hCoV-229E). Wastewater samples stored at 4 °C after collection showed significantly enhanced detection of SARS-CoV-2 with approximately 2–3 PCR-cycle threshold (Ct) values less when compared to samples stored at −20 °C. Pre-adjustment of the wastewater pH to 9.6 to aid virus desorption followed by pH readjustment to neutral after solid removal significantly increased the recovery of spiked hCoV-229E. Of the five commercially available RNA isolation kits evaluated, the MagMAX-96 viral RNA isolation kit showed the best recovery of hCoV-229E (50.1 ± 20.1%). Compared with two-step RT-qPCR, one-step RT-qPCR improved sensitivity for SARS-CoV-2 detection. Salmon DNA was included for monitoring PCR inhibition and pepper mild mottle virus (PMMoV), a fecal indicator indigenous to wastewater, was used to normalize SARS-CoV-2 levels in wastewater. Our method for molecular detection of SARS-CoV-2 in wastewater provides a useful tool for public health surveillance of COVID-19.
Keywords: SARS-CoV-2, Wastewater, RT-qPCR, hCoV-229E, PMMoV
Graphical abstract
1. Introduction
The finding of fecal shedding of SARS-CoV-2 by infected individuals (Chen et al., 2020; Tang et al., 2020; Wang et al., 2020; Wolfel et al., 2020; Wu et al., 2020b; Xiao et al., 2020) and the worldwide detection of SARS-CoV-2 RNA in wastewater (Ahmed et al., 2020a; Kumar et al., 2020; Medema et al., 2020; Wu et al., 2020a) have made the wastewater surveillance (WWS) of SARS-CoV-2 a promising approach to monitor the dynamics of COVID-19 prevalence in the community. Unlike clinical diagnostic tests, which are mainly performed on individuals with symptoms, wastewater monitoring of SARS-CoV-2 assesses the burden of all COVID-19 cases, including asymptomatic, pre/post-symptomatic and symptomatic individuals, who have and have not been tested in the targeted population (Daughton, 2020). Some evidences also showed that WWS provided an early indication of appearance or increased COVID-19 infection in a community (Ahmed et al., 2020a; Gonzalez et al., 2020; Weidhaas et al., 2021). Therefore, WWS has the potential to become a complementary approach to clinical testing by providing evidence-based support for public health policy and actions for infection prevention and control.
Since the first report of detecting SARS-CoV-2 in wastewater in the Netherlands (Medema et al., 2020), the international water community has made great efforts towards developing and standardizing methods for measuring SARS-CoV-2 in wastewater. Although numerous studies have reported the detection and quantification of SARS-CoV-2 RNA in wastewater in different countries, there are still some variations in specific methodologies used (Cervantes-Aviles et al., 2021; Jafferali et al., 2021; Kantor et al., 2021). Challenges, such as the virus concentration and recovery, assay sensitivity and PCR inhibition need to be resolved (Kitajima et al., 2020; Michael-Kordatou et al., 2020). This prompted us to perform this study in order to develop a reliable, specific and sensitive method for detection of SARS-CoV-2 in wastewater. Different interpretations of results derived from various methods have introduced uncertainties in how to use the data, which hinders the application of WWS as a reliable laboratory measuring and predicting tool for SARS-CoV-2 (McClary-Gutierrez et al., 2021). In the present study, the parameters that potentially affect the results of RT-qPCR-based detection of SARS-CoV-2 signal in wastewater were assessed and optimized, including sample storage conditions, pH of wastewater matrix during virus concentration, various RNA extraction kits and different gene targets for RT-qPCR assays. The aim of this study is to optimize and standardize the procedures for molecular detection of SARS-CoV-2 in wastewater through validating these parameters. The human coronavirus strain 229E (hCoV-229E) was spiked into the wastewater sample as a surrogate to monitor virus recovery. A set of criteria for interpretation of results was also defined regarding the PCR inhibition, recovery efficiency and normalization to fecal load in wastewater.
2. Materials and methods
2.1. Wastewater sample collection
Five hundred millilitres of post-grit raw influent wastewater samples were subsampled from the daily 24-hour composite samples from two wastewater treatment plants (WWTPs) located in Edmonton and Calgary, Canada for a period of two weeks in May 2020 on a daily base. Samples were frozen at −20 °C upon collection and shipped to the lab on a weekly base. Once received by the laboratory, samples were stored at −20 °C if not processed within 72 h. Otherwise, samples were stored at 4 °C until processing. All the wastewater samples were processed directly without heat inactivation. The processing of wastewater samples was conducted in the biosafety cabinet to reduce aerosol production.
2.2. Various parameters for robust SARS-CoV-2 detection in wastewater
2.2.1. Virus concentration by adjusting pH of wastewater matrix
It has been suggested that high pH (9.6–10) may aid the desorption of virus from sludge/solid in wastewater samples (Grohmann et al., 1993; Hurst et al., 1991). To determine the effect of pH adjustment on virus recovery from wastewater, 200 μl cultured hCoV-229E (5.7E + 06 copies/ml, ATCC® VR-740™) virus was spiked into eight 200 ml wastewater samples, each of which was aliquoted into two 100 ml spiked samples for parallel assessment of pH adjustment. The pH of one aliquot was adjusted to 9.6–10 using 5 N NaOH and the sample was centrifuged at 4500 ×g for 10 min to remove the solids. Considering that high pH may cause the degradation of viral RNA, the supernatant pH was further adjusted back to neutral (7–7.5) using 1.2 N HCl. Another aliquot of the wastewater was processed without pH adjustment in parallel. The supernatant was added into Centricon Plus-70 centrifugal ultrafilter cup (30-kDa MWCO, Millipore) and centrifuged at 3000 ×g for 10 min using a refrigerated centrifuge (Allegra X-15R, Beckman Coulter) as previously described (Qiu et al., 2016). The filtrate was discarded, and the same procedure was repeated until all of the 100 ml supernatant was filtered. Filtrate collection cup was then removed and the concentration cup was placed on top of the sample filter cup. The whole device was inverted carefully and centrifuged at 800 ×g for 2 min. The concentrated sample was collected from the concentration cup and made up to a final volume of 1 ml with PBS. The concentrates of the sample were stored at −70 °C until later use.
2.2.2. Different commercial kits for viral RNA extraction
Ten wastewater samples (100 ml/each) were spiked with the same amount of hCoV-229E and concentrated followed by RNA extraction using five different commercial nucleic acid isolation kits, including kit A: RNeasy PowerMicrobiome kit (Qiagen, ON, CA); kit B: MagMAX-96 viral RNA isolation kit (ThermoFisher, ON, CA); kit C: MagMAX Viral/Pathogen nucleic acid isolation kit (ThermoFisher, ON, CA); kit D: QIAamp viral RNA mini kit (Qiagen, ON, CA) and kit E: ReliaPrep™ RNA Miniprep System (Promega, WI, USA). For each extraction method, RNA was isolated from 200 μl of wastewater concentrate and eluted at a final volume of 50 μl according to the manufacturers' instructions. MagMAX-96 viral RNA isolation and MagMAX Viral/Pathogen nucleic acid isolation were processed using the automated KingFisher™ Flex and KingFisher™ mL Purification System (Thermofisher, ON, CA), respectively. The other three RNA isolations were performed manually.
2.2.3. One-step versus two-step RT-qPCR assays for detecting SARS-CoV-2 RNA
To assess whether the separated step of reverse transcription (RT) could improve the sensitivity of qPCR assay, one-step RT-qPCR assay was compared with two-step RT-qPCR assay using the RNA isolated from a clinical respiratory specimen tested positive for SARS-CoV-2. The clinical RNA sample was tested with both RT-qPCR assays in a 10-fold serial dilution from neat to 10−4 in duplicate. Both qPCR assays were carried out using the ABI 7500Fast PCR instrument.
The one-step RT-qPCR reaction contains 5 μl of RNA template, 2.5 μl of 4× Taqman Fast Virus One-Step RT-PCR Master Mix (Thermofisher, ON, CA), 800 nM each of forward and reverse primer along with 200 nM probe in a total volume of 10 μl. The one-step RT-qPCR program includes RT reaction at 50 °C for 5 min, enzyme activation at 95 °C for 20 s and 45 cycles of PCR amplification of 95 °C for 3 s and 60 °C for 30 s. A threshold of 0.05 was set for data analysis. For each one-step RT-qPCR reaction, an equivalent of 2 ml of the original wastewater sample was assayed based on the calculation as below:
where VRNA in each PCR reaction is the volume of RNA assayed in a PCR reaction, Vextracted RNA is the total volume of extracted RNA, Vwastewater concentrate for RNA extraction is the volume of wastewater concentrate used for RNA extraction, V wastewater concentrate is the sample volume after concentration, Vsample is the volume of original wastewater sample processed.
The two-step RT-qPCR assay includes a separate RT reaction as previously described (Pang et al., 2012). Briefly, 5 μl of RNA was preheated at 95 °C for 5 min and quickly chilled on ice, followed by adding 15 μl of the mixture containing 5 μl of transcript buffer, 5 mM DTT, 20 units of RNaseOut™ recombinant ribonuclease inhibitor, 100 units of SuperScript™ II reverse transcriptase, 2.5 mM each of dATP, dCTP, dGTP, and dTTP, 300 ng random primer. The RT reaction was performed at 42 °C for 1 h and 72 °C for 15 min. The qPCR reaction was performed in a total volume of 10 μl containing 2 × TaqMan Fast Universal MasterMix (Thermofisher, ON, CA), 900 nM of each primer, 250 nM of specific probe, and 2.5 μl cDNA. Amplification consists of initial incubation at 95 °C for 20 s followed by 45 cycles of 3 s at 95 °C, 30 s at 60 °C. Sequences of primers and probes for RNA-dependent RNA polymerase (RdRp) gene, E gene, N1 and N2 gene are summarized in Table 1 (Corman et al., 2020; Pabbaraju et al., 2020; Vogels et al., 2020).
Table 1.
Primer and probe sequences of target genes for SARS-CoV-2, hCoV-229E and PMMoV.
| Target | Sequence (5′–3′) | |
|---|---|---|
| RdRp gene (Pabbaraju et al., 2021) | Forward primer | TTTTAACATTTGTCAAGCTGTCACG |
| Reverse primer | GTTGTAAATTGCGGACATACTTATCG | |
| Probe | VIC-CACTTTTATCTACTGATGGTAAC-MGB | |
| E gene (Pabbaraju et al., 2021) | Forward primer | GAGACAGGTACGTTAATAGTTAATAGCG |
| Reverse primer | CAATATTGCAGCAGTACGCACAC | |
| Probe | NED-CTAGCCATCCTTACTGCG-MGB | |
| N1 gene (2019-nCoV CDC) | Forward primer | GACCCCAAAATCAGCGAAAT |
| Reverse primer | TCTGGTTACTGCCAGTTGAATCTG | |
| Probe | FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1 | |
| N2 gene (2019-nCoV CDC) | Forward primer | TTACAAACATTGGCCGCAAA |
| Reverse primer | GCGCGACATTCCGAAGAA | |
| Probe | FAM-ACAATTTGCCCCCAGCGCTTCAG-BHQ1 | |
| hCoV-229E (Vijgen et al., 2005) | Forward primer | TTCCGACGTGCTCGAACTTT |
| Reverse primer | CCAACACGGTTGTGACAGTGA | |
| Probe | FAM-TCCTGAGGTCAATGCA-MGB | |
| PMMoV (Lee et al., 2018) | Forward primer | GAGTGGTTTGACCTTAACGTTGA |
| Reverse primer | TTGTCGGTTGCAATGCAAGT | |
| Probe | FAM-CCTACCGAAGCAAATG-MGB | |
2.2.4. Storage temperature for wastewater samples
To evaluate the effect of sample storage temperature on detection of SARS-CoV-2, duplicate wastewater samples were collected from the Edmonton WWTP on 7 different dates between December 20, 2020 and January 31, 2021, with one bottle stored at 4 °C and one frozen at −20 °C upon collection. The samples were shipped to the lab in cooler with ice pack. Both non-frozen and frozen samples were observed in their original conditions when received in the laboratory and processed in parallel upon arrival. Another five paired frozen and non-frozen wastewater samples collected between Oct 15 and Oct 18, 2021 from five different WWTPs across Alberta were also tested. Samples were concentrated after pH adjustment using Centricon Plus-70 centrifugal ultrafilter. RNA was extracted using MagMAX-96 viral RNA isolation kit along with automated KingFisher™ Flex Purification System. One-step RT-qPCR targeting N1, N2 and E gene of SARS-Cov-2 were performed.
2.3. One-step RT-qPCR for hCoV-229E and PMMoV
Primers and probes targeting to PMMoV and the membrane protein gene of hCoV-229E were described previously (Table 1) (Lee et al., 2018; Vijgen et al., 2005). RT-qPCR reactions for hCoV-229E and PMMoV are the same as one-step RT-qPCR for SARS-CoV-2, except using 400 nM and 500 nM of the primers for hCoV-229E and PMMoV, respectively. The program of one-step RT-qPCR for SARS-Cov-2 was also used for hCoV-229E and PMMoV as described above.
2.4. Standard curve for quantification of SARS-CoV-2, hCoV-229E and PMMoV
Long oligonucleotide sequences (gblocks) including the regions flanking T7 and SP6 RNA polymerase promoter binding sites were designed for the E-gene target of SARS-CoV-2 (IDT, Iowa, USA). RNA transcription was performed using the RiboMAX™ SP6 RNA Production System (Promega, WI, USA). The transcribed RNA was spectrophotometrically quantified using NanoDrop™ for the calculation of copy numbers (Pabbaraju et al., 2021). A series of 10-fold dilutions (1.66 to 1.66E + 06 copies) were analyzed by qPCR to identify the dynamic ranges and establish the standard curve for quantification of the E gene of SARS-CoV-2 (Supplementary Fig. S1). As the efficiency of RT-qPCR for SARS-CoV-2 E, N1, N2, hCoV-229E and PMMoV were similar, this standard curve for E gene has been used for quantification of all five targets. The virus concentration was expressed as genome equivalent (GE) copies/100 ml of wastewater and calculated as below:
where Vwastewater analyzed in a PCR reaction is the volume of wastewater sample tested in each one-step RT-qPCR reaction (2 ml in this study), Vsample is the volume of original wastewater sample processed (100 ml in this study).
2.5. Salmon DNA as an internal control for monitoring PCR inhibition
To assess the presence of PCR inhibition in the wastewater concentrated samples, qPCR assay for salmon DNA was performed as previously described (Qiu et al., 2013). An appropriate amount of salmon DNA identified at Ct = 30 using qPCR was used for monitoring inhibition of RT-qPCR. Briefly, 5 μl salmon DNA (Ct = 30) was added into 200 μl concentrated sample followed by RNA extraction and qPCR was performed for detection of salmon DNA. Inhibition was defined as a delay of Ct by 3 cycles as compared to a distilled water control spiked with the same amount of salmon DNA followed by RNA extraction.
2.6. Evaluation of pre-analytic sensitivity, specificity, and precision of one-step RT-qPCR for SARS-CoV-2
The specificity of one-step RT-qPCR for SARS-CoV-2 was evaluated by clinical samples confirmed with common coronavirus strains, including 229E, OC43, NL63 and HKU1. The sensitivity of our assay was assessed by 10-fold serial dilutions (1.66 to 1.66 × 106 copies) of the RNA fragment that was used as standard. The precision of RT-qPCR was analyzed using Ct values generated from replicates of standard curve performed on different days.
2.7. Determination of hCoV-229E recovery
HCoV-229E strain purchased from ATCC (VR-740) was propagated in human fibroblast cell line MRC-5. The concentration of cultured hCoV-229E virus stock was measured by TCID50 method to determine the infectious unit (IU) per ml of the stock. Each wastewater sample (100 ml) was spiked with 100 μl of cultured hCoV-229E (4.8E + 05 IU/ml) and concentrated as described above to monitor the virus recovery. The same aliquot of the virus was added to 900 μl of PBS as baseline control and tested in parallel with the concentrated spiked water samples using RT-qPCR. The recovery (%) was calculated as follows:
2.8. Statistical analysis
The recovery of hCoV-229E in experiments was compared using Wilcoxon signed-rank test for paired samples. The Ct value between non-frozen and frozen paired samples was also compared using Wilcoxon signed-rank test. The precision of the RT-qPCR was expressed as coefficient of variation (CV) and 95% confidence interval. The p value less than 0.05 was considered as statistically significant.
3. Results
3.1. Effect of wastewater pH adjustment on viral recovery
Using the original wastewater sample without pH adjustment, the recovery of hCoV-229E ranged from 0.2 to 2.4% with the median of 0.8%, while the recovery increased to a range between 1.4 and 5.8% with the median of 2.1% after pH adjustment to 9.6–10 before solid removal (Table 2 ). Due to unknown inhibitory substances present in the samples (assessed by inhibition of spiked salmon DNA), 10-fold dilution of the RNA extracted from wastewater samples with and without pH adjustment was used for the RT-qPCR assays. The results showed that hCoV-229E was not detected in 7 out of the 8 samples. However, it was detected in 7/8 samples with mean recovery of 3.5 ± 2% after pH adjustment (Table 2), which was significantly improved compared to the group without pH adjustment (p < 0.05). Furthermore, three wastewater samples were tested in parallel with and without pH readjusted back to 7.0 after solid removal. The mean hCoV-229E recovery increased from 5 ± 0.95% to 17.2 ± 4.8% in the original samples without dilution and from 5.7 ± 2.8% to 73 ± 23% in 10-fold diluted samples with pH readjustment to 7.0 (Table 3 ). These results provide evidence that pH adjustment of the wastewater samples before and after solids removal during the wastewater concentration step can facilitate the desorption of virus from solids and prevent viral RNA from degradation in high pH.
Table 2.
Comparison of hCoV-229E recovery in wastewater samples with or without high pH adjustment.
| Sample no | hCoV-229E recovery (%) |
|||
|---|---|---|---|---|
| Without pH adjustment |
High pH adjustment before solid removal |
|||
| No dilution | 1:10 dilution | No dilution | 1:10 dilution | |
| 1 | 0.97 | ND | 1.90 | 3.06 |
| 2 | 0.90 | 2.55 | 1.72 | 2.50 |
| 3 | 1.68 | ND | 5.83 | 7.27 |
| 4 | 2.38 | ND | 2.21 | 1.37 |
| 5 | 0.70 | ND | ND | ND |
| 6 | 0.16 | ND | ND | 2.33 |
| 7 | 0.18 | ND | 1.42 | 2.67 |
| 8 | 0.31 | ND | 5.43 | 5.29 |
ND: not detectable.
Table 3.
Comparison of hCoV-229E recovery in wastewater samples with or without pH readjusted to neutral. Wastewater sample pH was adjusted to 9.6 followed by solid removal and the supernatant pH was readjusted to neutral or remain 9.6.
| Sample no | hCoV-229E recovery (%) |
|||
|---|---|---|---|---|
| pH = 9.6–10 |
pH readjusted to 7.0 |
|||
| No dilution | 1:10 dilution | No dilution | 1:10 dilution | |
| 1 | 4.1 | 8.2 | 14.9 | 60.8 |
| 2 | 6.0 | 2.7 | 22.7 | 100 |
| 3 | 4.9 | 6.1 | 13.9 | 59.2 |
3.2. Nucleic acid isolation kits for RNA extraction
The recoveries of hCoV-229E using five commercial nucleic acid extraction kits are shown in Fig. 2. Overall, the median recovery of the five kits ranged from 24.3–47.8%. Kit B provided the highest hCoV-229E recovery with a range of 29.4–91.7%, followed by kit E (18.8–73.4%), kit D (11.6–59.8%) and kit A (9.8–40.9%). Kit C showed the lowest recovery with a range of 7.5–41.2%. Although there is no significant difference for the hCoV-229E recovery between kit B and E, kit B (MagMAX-96 viral RNA isolation kit) compatible with Kingfisher Flex automatic purification system was used as the viral RNA extraction method in our protocol considering the time effectiveness.
Fig. 2.
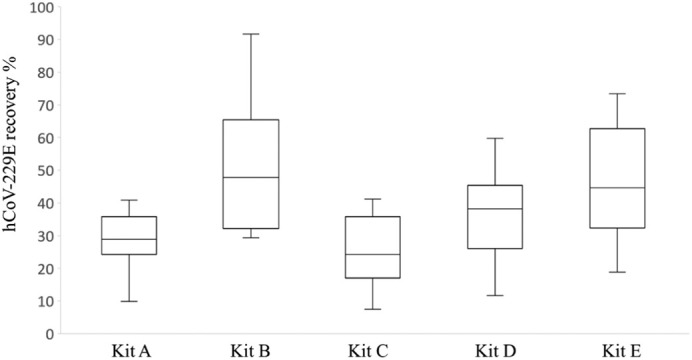
hCoV-229E recovery in wastewater samples using five commercial RNA extraction kits. Kit A: RNeasy PowerMicrobiome kit; Kit B: MagMAX-96 viral RNA isolation kit; Kit C: MagMAX Viral/Pathogen nucleic acid isolation kit; Kit D: QIAamp viral RNA mini kit; Kit E: ReliaPrep™ RNA Miniprep System.
3.3. Comparison of one-step and two-step RT-qPCR for SARS-CoV-2 detection
One-step RT-qPCR detected both the RdRp and E gene until sample dilution up to 10−4, while two-step RT-qPCR assay could only detect RdRp gene at sample dilution to 10−3 and E gene at sample dilution to 10−2, respectively (Table 4 ). The Ct value of two-step RT-qPCR assay increased by 2.93 ± 0.26 for RdRp gene and 5.88 ± 0.54 for E gene compared to one-step, indicating that separation of RT reaction from qPCR including the preheating did not improve the sensitivity of qPCR assay for SARS-CoV-2. Thus, one-step RT-qPCR was applied in our standard method for detection of SARS-CoV-2 in wastewater. In addition, E gene showed enhanced sensitivity with the Ct value of 2.64 ± 0.35 lower than that of RdRp gene when one-step RT-qPCR assay was performed using the four dilutions of the samples. Therefore, E gene was included in our protocol as one of the target genes for SARS-CoV-2 detection in wastewater.
Table 4.
The mean Ct value of SARS-CoV-2 RdRP and E gene (mean ± standard deviation) using one-step versus two-step RT-qPCR.
| Sample dilutions | Ct, RdRP gene |
Ct, E gene |
||
|---|---|---|---|---|
| one-step | two-step | one-step | two-step | |
| 10−1 | 26.22 ± 0.28 | 29.44 ± 0.11 | 23.38 ± 0.11 | 28.88 ± 0.04 |
| 10−2 | 29.88 ± 0.01 | 32.71 ± 0.17 | 27.28 ± 0.51 | 33.54 ± 0.42 |
| 10−3 | 32.88 ± 0.29 | 35.61 ± 0.16 | 30.71 ± 0.21 | ND |
| 10−4 | 36.32 ± 0.26 | ND | 33.35 ± 0.66 | ND |
ND: not detectable.
3.4. SARS-CoV-2 detection in non-frozen and frozen wastewater samples
All three target genes of SARS-CoV-2 including N1, N2 and E were detected in all twelve non-frozen samples, with the mean Ct of 29.7, 30.3 and 32.7, respectively. However, for the frozen samples, N1 and N2 genes were only detected in ten samples and E gene was detected in nine samples. The mean Ct of the three target genes for frozen samples were significantly higher than non-frozen samples (2.4–3.6 cycles) (p < 0.05), indicating that freeze-thaw cycle may cause degradation of SARS-CoV-2 virus (Table 5 ). Therefore, a samples storage temperature of 4 °C upon collection was selected for our protocol.
Table 5.
Comparison of SARS-CoV-2 gene detection in frozen and non-frozen wastewater samples.
| Non-frozen samples (4 °C) |
Frozen samples (−20 °C) |
|||||
|---|---|---|---|---|---|---|
| N1 gene | N2 gene | E gene | N1 gene | N2 gene | E gene | |
| Positive samples detected | 12/12 | 12/12 | 12/12 | 10/12 | 10/12 | 9/12 |
| Ct (mean) | 29.7 | 30.3 | 32.7 | 33.3 | 33.5 | 35.0 |
| SD | 1.34 | 1.74 | 1.75 | 2.74 | 2.70 | 2.51 |
SD: standard deviation.
3.5. The sensitivity, specificity and precision of one-step RT-qPCR for SARS-CoV-2 detection in wastewater
Good sensitivity of the one-step RT-qPCR assay for quantitation of SARS-CoV-2 was revealed in the linear log range from 1.66 to 1.66E + 06 copies/reaction with E gene primer/probes. The limit of detection (LOD) of the one-step RT-qPCR assay for SARS-CoV-2 is 1.6 copies/PCR, which is equivalent to 80 copies/100 ml of water. The efficiency was observed as 99% from the standard curve. There was no cross-reaction of SARS-CoV-2 RT-qPCR with other common hCoV strains, including 229E, OC43, NL63 and HKU-1. The coefficient of variation for the Ct generated from 20 replicates of RT-qPCR was determined to be 1.48% for 1.66E + 03 copies/reaction and 1.76% for 1.66E + 05 copies/reaction, respectively.
Since the N1 and N2 primer/probe sets developed by the United States Centers for Disease Control and Prevention (CDC) have demonstrated good sensitivity in previous studies (Vogels et al., 2020), both targets are included in our protocol for SARS-CoV-2 detection. To maximize the detection of SARS-CoV-2 RNA signals in wastewater, each sample was tested for E, N1 and N2 genes in duplicate. A SARS-CoV-2 positive sample was defined when two or more positive results out of the six PCR runs for three SARS-CoV-2 genes were observed. Also, hCoV-229E was integrated as a surrogate for monitoring the process of sample concentration, RNA extraction and RT-qPCR procedures. Virus recovery was calculated based on hCoV-229E performance for quality control and the recovery ranged from 1 to 10% for the majority of samples tested. Salmon DNA was spiked to the sample concentrates before RNA extraction to monitor the extraction process and PCR inhibition which was observed in approximately 7% of the wastewater samples. PMMoV was used as a fecal indicator to normalize the SARS-CoV-2 concentration in wastewater, which showed consistent Ct value between 18 and 20. Positive and negative controls for RNA extraction and RT-qPCR reaction were also included for quality control purposes.
4. Discussion
Virus concentration process is a key step of SARS-CoV-2 detection because of the low level of target viral RNA presence in the environmental water matrices. Various concentration methods have been used for SARS-CoV-2 detection in wastewater including polyethylene glycol (PEG) precipitation, ultrafiltration, ultracentrifugation, and adsorption-elution with electronegative membranes (Jafferali et al., 2021; Medema et al., 2020; Prado et al., 2021; Wu et al., 2020a). These methods have demonstrated effective concentration of SARS-CoV-2 from wastewater although the recovery of the virus yield occurred at different levels (Ahmed et al., 2020b; Kantor et al., 2021). In our previous study, the ultrafiltration method for enteric virus concentration and detection in wastewater using Centricon plus-70 ultrafilter has been validated, which showed comparable and consistent results to that obtained using the virus adsorption-elution method (Qiu et al., 2016). Considering the ultrafiltration method provides fast processing and is less labor intensive, which is critical for rapid response in the pandemic management regardless its relatively higher cost, it was chosen for SARS-CoV-2 concentration from wastewater in our protocol. Several studies have successfully detected SARS-CoV-2 in wastewater using Centricon ultrafilter for viral concentration (Fores et al., 2021; Gerrity et al., 2021, Medema et al., 2020), supporting the feasibility and effectiveness of Centricon ultrafilter for SARS-CoV-2 concentration from wastewater. The recovery of hCoV-229E could reach over 90% when it was spiked into pure water and concentrated by Centricon plus-70 ultrafilter (data not shown). Over time we noticed that there were variations in the recovery of hCoV-229E, which might be due to several reasons such as the composition of wastewater matrix, sample processing, as well as the lot change of filters.
Concentration of large volumes of water sample might increase the opportunity of detecting viruses with low abundance in water matrix (Philo et al., 2021). However, increasing the water sample volume may also introduce more inhibitors in the sample concentrates, which can interfere with the RT-qPCR reaction. Various amounts of wastewater samples with volumes between 0.25 ml and 500 ml have been reported by different laboratories for SARS-CoV-2 detection (Nemudryi et al., 2020; Pecson et al., 2021). Most of laboratories concentrated less than 200 ml of wastewater (Ahmed et al., 2020a; Chik et al., 2021; Kumar et al., 2020; Medema et al., 2020; Wu et al., 2020a). We tested three different volumes of wastewater samples (50, 100 and 200 ml) with spiked hCoV-229E, and no significant difference was found for hCoV-229E recovery among all three volumes (Supplementary Table S1). Thus, to balance between detection sensitivity and the amount of PCR inhibitors, the volume of 100 ml wastewater was chosen in our protocol. Ahmed et al. reported that addition of MgCl2 to the wastewater sample prior to filtration step yielded the best recovery of murine hepatitis virus (Ahmed et al., 2020b), which may be caused by the increased virus adsorption to the filter under high MgCl2 concentration (Ikner et al., 2012; Villar et al., 2006). To evaluate the effect of MgCl2, we added MgCl2 to the wastewater sample without pH adjustment to obtain a final concentration of 25 mM after solid removal. The results showed that pre-treatment with MgC12 did not increase the recovery of hCoV-229E (data not shown). Thus, MgC12 treatment was not included in our protocol.
Desorption of solid-associated viruses from sewage sludge using high pH was first experimented in 1970s for enterovirus (Hurst et al., 1978). The rationale of high pH to disperse virions from particulate matter in environmental samples is based on the fact that most viruses have an isoelectric point (pI) ranging from 3.5 to 7 (Michen and Graule, 2010). Although currently there is no experimental data available for the pI of SARS-CoV-2, we hypothesized that high pH may facilitate the dissociation of SARS-CoV-2 from solids in wastewater as no viruses have been reported with pI value in the strongly basic range (Michen and Graule, 2010). For the majority of the wastewater samples we have tested, the pH ranged from 7.5 to 8.5. After pH adjustment to 9.6, the recovery of hCoV-229E increased, especially in 10 fold-diluted samples, indicating that high pH can improve virus desorption from solids in wastewater. Because RNA is not stable in high alkaline conditions (Lemire et al., 2016), the pH was re-adjusted back to neutral after solid removal. The recovery of hCoV-229E significantly increased after pH adjusted to neutral, with a range between 60 and 100% in the 1:10 diluted samples. Furthermore, we measured the amount of hCoV-229E in solids after pH adjustment and found that the level of hCoV-229E was much lower in solids compared to the supernatant (Hasing et al., 2021). We also observed lower recovery of hCoV-229E and lower detection rate for SARS-CoV-2 in solids with pH adjustment compared to without pH adjustment (Hasing et al., 2021). This observation was also supported by the study that reported low levels of bovine CoV in the solids portion using spiked samples (Gerrity et al., 2021). These results indicated that changing the wastewater pH during the viral concentration process facilitated detection of SARS-CoV-2 in wastewater.
Based on our knowledge of enteric virus detection in wastewater (Qiu et al., 2015, Qiu et al., 2018), two-step RT-qPCR assay showed better detection for norovirus in wastewater compared to one-step RT-qPCR assay, which may be due to the disruption of the secondary structure of viral RNA prior to cDNA synthesis by preheating for better interaction between random primer and targets. Since SARS-CoV-2 is also an RNA virus, two-step RT-qPCR was evaluated for the sensitivity of SARS-CoV-2 detection. The results showed that two-step RT-qPCR had a 3–6 Ct delay in detection of RdRp and E gene of SARS-CoV-2 compared to the one-step method. This is consistent with previous observations that one-step RT-qPCR was more sensitive in detection of low abundance genes but showed little difference in detection of genes expressed at relatively high levels compared to two-step (Wacker and Godard, 2005). Considering the low level of SARS-CoV-2 in wastewater, one-step RT-qPCR was applied in the protocol for SARS-CoV-2 detection because of its better performance. By using one-step RT-qPCR, SARS-CoV-2 RNA was detected at levels as low as 1.6 virus copies/PCR in wastewater samples. Excellent reproducibility was also observed with one-step RT-qPCR for all the targets including SARS-CoV-2 genes, hCoV-229E and PMMoV.
Virus nucleic acid extraction is another critical step that impacts the effectiveness of virus detection, which often uses three main techniques: organic extraction, silica-membrane based spin column and paramagnetic particles (Michael-Kordatou et al., 2020). Various extraction systems have been utilized for the SARS-CoV-2 detection in wastewater (Chik et al., 2021; Pecson et al., 2021). However, no study to-date has evaluated the SARS-CoV-2 RNA yield from wastewater using different RNA extraction methods. In our study, we compared five commercially available kits for the SARS-CoV-2 RNA extraction from wastewater using either spin columns or magnetic beads. Overall, the five kits all showed good recovery for hCoV-229E in wastewater, with recovery ranging from 25 to 50%. MagMAX-96 viral RNA isolation kit (kit B) and ReliaPrep™ RNA Miniprep System (kit E) showed comparably good performance with the average recovery of 50%, suggesting that magnetic beads and silica spin column-based extraction methods are preferred for SARS-CoV-2 RNA isolation from wastewater. With the automated KingFisher™ Flex purification system in our laboratory, we chose MagMAX-96 viral RNA isolation kit as the standard RNA extraction method in our protocol.
Wastewater storage condition could be another important factor influencing SARS-CoV-2 detection. In our study, SARS-CoV-2 was detected in more samples with higher copy numbers when the samples were stored at 4 °C compared to −20 °C storage. This was consistent with other findings that increased viral degradation occurred at −20 °C rather than at 4 °C and −80 °C, presumably due to the formation of large ice crystals that damage virions (Michael-Kordatou et al., 2020; Olson et al., 2004). Thus, starting from January 2021, we requested all the wastewater samples collected for SARS-CoV-2 test were stored at 4 °C upon collection until further analysis.
The levels of SARS-CoV-2 in wastewater are affected by many factors, including the daily fecal discharge, wastewater flow rate, total suspended solids, the types of sample and weather conditions (Ahmed et al., 2021). Considering the variability caused by these factors, an important aspect of wastewater surveillance of SARS-CoV-2 is to normalize the detection results, which has been reported in several studies using different approaches such as biomarkers and daily mass flux (D'Aoust et al., 2021; Gerrity et al., 2021; Wu et al., 2020a). PMMoV is an abundant RNA virus in human feces and is very stable in the wastewater exhibiting little variation (Kitajima et al., 2018; Rosario et al., 2009). PMMoV has been used by many laboratories as a fecal indicator to normalize the SARS-CoV-2 level in wastewater (D'Aoust et al., 2021; Haramoto et al., 2020; Jafferali et al., 2021; Wu et al., 2020a). Normalization of SARS-CoV-2 signal using PMMoV has demonstrated a strong correlation between SARS-CoV-2 level in wastewater and COVID-19 clinical cases (D'Aoust et al., 2021). Therefore, we also incorporated the PMMoV test in our protocol as a reliable reference for data normalization and analysis. In addition, we used salmon DNA in our protocol to monitor PCR inhibition. Although USCDC recommends to spike viral RNA for inhibition assessment, salmon DNA has been broadly used for inhibition monitoring in many water-related studies (Ahmed et al., 2020b; Qiu et al., 2013; Sylvestre et al., 2021).
5. Conclusions
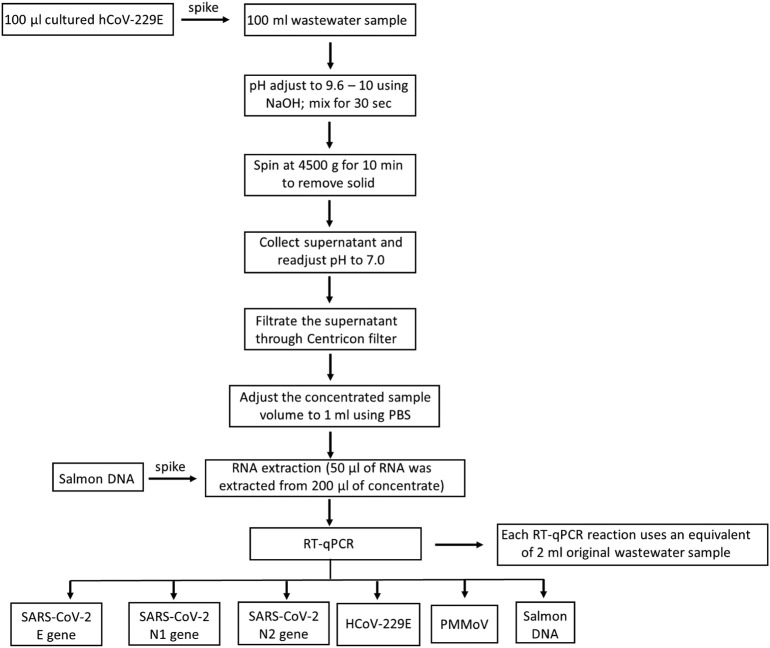
Taken together, we validated a standard protocol for analytic procedures of one-step RT-qPCR based detection of SARS-CoV-2 in wastewater. The final standardized protocol is outlined in Fig. 1 . Specifically, multiple key steps, including sample storage, virus concentration, RNA extraction and RT-qPCR assays, were optimized and good sensitivity and reliability were achieved by our method in detecting SARS-CoV-2 in wastewater. This method provides a useful tool for public health surveillance of COVID-19, as well as support for public health policy and actions for infection prevention and control.
Fig. 1.
The flow chart of standardized procedure for detection and quantification of SARS-CoV-2 in wastewater.
The following are the supplementary data related to this article.
The standard curve of E gene RNA fragment used for quantification in one-step RT-qPCR for SARS-CoV-2, PMMoV and hCoV-229E.
hCoV-229E recovery in wastewater samples using different volume of wastewater (50 ml, 100 ml and 200 ml).
CRediT authorship contribution statement
Yuanyuan Qiu: Methodology, Data analysis, Writing – original draft, Writing – review & editing. Jiaao Yu: Methodology, Writing – review & editing. Kanti Pabbaraju: Resources, Writing – review & editing. Bonita E. Lee: Conceptualization, Investigation, Writing – review & editing. Tiejun Gao: Project coordination, Writing – review & editing. Nicholas J. Ashbolt: Conceptualization, Writing – review & editing. Steve E. Hrudey: Conceptualization, Writing – review & editing. Mathew Diggle: Writing – review & editing. Graham Tipples: Writing – review & editing. Rasha Maal-Bared: Writing – review & editing. Xiaoli Pang: Conceptualization, Supervision, Project administration, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Emma Zwaigenbaum and Melissa Wilson for their technical support. We would like to thank Norma Ruecker and staff from City of Calgary, Stephen Craik and Jeff Charrois from EPCOR Water Canada for providing the wastewater samples. We also want to thank the Alberta Precision Laboratory - Public Health Laboratory for their assistants with samples transportation. This study was supported by research grants from the Canadian Institutes of Health Research, Alberta Innovates and Alberta Health [RES0051047].
The Alberta COVID-19 Wastewater surveillance team (ACWST) members are: Xiaoli Pang (Public Health Laboratory, Alberta Precision Laboratories and Department of Laboratory Medicine and Pathology, University of Alberta, Edmonton, Alberta, Canada), Nicholas Ashbolt (Faculty of Science and Engineering, Southern Cross University, Lismore, New South Wales, Australia), Deena Hinshaw (Chief Medical Officer of Health, Alberta Health, Edmonton, Alberta, Canada), James Talbot, Bonita Lee (Department of Pediatrics, University of Alberta, Edmonton, Alberta, Canada), Norman Neumann (School Public Health, University of Alberta, Edmonton, Alberta, Canada), Kimberly Simmonds (Alberta Health, Alberta, Canada), Graham Tipples (Public Health Laboratory, Alberta Precision Laboratories, Edmonton, Alberta, Canada), Mathew Diggle (Public Health Laboratory, Alberta Precision Laboratories, Edmonton, Alberta, Canada), Stephen Craik (EPCOR, Edmonton, Alberta, Canada), Norma Ruecker (City of Calgary, Alberta, Canada), Lyndon Gyurek (Drinking Water and Wastewater, Alberta Environment and Parks, Edmonton, Alberta, Canada), Qiaozhi Li (School of Public Health, University of Alberta, Edmonton, Alberta, Canada).
Editor: Kevin V. Thomas
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Gyawali P., Sherchan S.P., Simpson S.L., Thomas K.V., Verhagen R., Kitajima M., Mueller J.F., Korajkic A. Intraday variability of indicator and pathogenic viruses in 1-h and 24-h composite wastewater samples: implications for wastewater-based epidemiology. Environ. Res. 2021;193 doi: 10.1016/j.envres.2020.110531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Aviles P., Moreno-Andrade I., Carrillo-Reyes J. Approaches applied to detect SARS-CoV-2 in wastewater and perspectives post-COVID-19. J. Water Process Eng. 2021:40. doi: 10.1016/j.jwpe.2021.101947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C., Yang J., Ye G., Cheng Z. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92(7):833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Chik A.H.S., Glier M.B., Servos M., Mangat C.S., Pang X., Qiu Y., D’Aoust P.M., Burnet J.B., Delatolla R., Dorner S., Geng Q., Giesy J.P., Jr., McKay R.M., Jr., Mulvey M.R., Jr., Prystajecky N., Jr., Srikanthan N., Jr., Xie Y., Jr., Conant B., Jr., Hrudey S.E., Jr., Consortium C.S.-C.-I.-L. Comparison of approaches to quantify SARS-CoV-2 in wastewater using RT-qPCR: Results and implications from a collaborative inter-laboratory study in Canada. J. Environ. Sci. 2021;107:218–229. doi: 10.1016/j.jes.2021.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Mercier E., Montpetit D., Jia J.J., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Langlois M.A., Servos M.R., MacKenzie M., Figeys D., MacKenzie A.E., Graber T.E., Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fores E., Bofill-Mas S., Itarte M., Martinez-Puchol S., Hundesa A., Calvo M., Borrego C.M., Corominas L.L., Girones R., Rusinol M. Evaluation of two rapid ultrafiltration-based methods for SARS-CoV-2 concentration from wastewater. Sci. Total Environ. 2021;768:144786. doi: 10.1016/j.scitotenv.2020.144786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res. X. 2021;10 doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann G.S., Ashbolt N.J., Genova M.S., Logan G., Cox P., Kueh C.S.W. Detection of viruses in coastal and river water-systems in Sydney, Australia. Water Sci. Technol. 1993;27(3–4):457–461. [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasing M., Yu J.A., Qiu Y.Y., Maal-Bared R., Bhavanam S., Lee B., Hrudey S., Pang X.L. Comparison of detecting and quantitating SARS-CoV-2 in wastewater using moderate-speed centrifuged solids versus an ultrafiltration method. Water. 2021;13(16) [Google Scholar]
- Hurst C.J., Farrah S.R., Gerba C.P., Melnick J.L. Development of quantitative methods for the detection of enteroviruses in sewage sludges during activation and following land disposal. Appl. Environ. Microbiol. 1978;36(1):81–89. doi: 10.1128/aem.36.1.81-89.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst C.J., Schaub S.A., Sobsey M.D., Farrah S.R., Gerba C.P., Rose J.B., Goyal S.M., Larkin E.P., Sullivan R., Tierney J.T., et al. Multilaboratory evaluation of methods for detecting enteric viruses in soils. Appl. Environ. Microbiol. 1991;57(2):395–401. doi: 10.1128/aem.57.2.395-401.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikner L.A., Gerba C.P., Bright K.R. Concentration and recovery of viruses from water: a comprehensive review. Food Environ. Virol. 2012;4(2):41–67. doi: 10.1007/s12560-012-9080-2. [DOI] [PubMed] [Google Scholar]
- Jafferali M.H., Khatami K., Atasoy M., Birgersson M., Williams C., Cetecioglu Z. Benchmarking virus concentration methods for quantification of SARS-CoV-2 in raw wastewater. Sci. Total Environ. 2021;755(Pt 1) doi: 10.1016/j.scitotenv.2020.142939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor R.S., Nelson K.L., Greenwald H.D., Kennedy L.C. Challenges in measuring the recovery of SARS-CoV-2 from wastewater. Environ. Sci. Technol. 2021;55(6):3514–3519. doi: 10.1021/acs.est.0c08210. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Sassi H.P., Torrey J.R. Pepper mild mottle virus as a water quality indicator. NPJ Clean Water. 2018:1. [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.W., Lee H.M., Yoon S.R., Kim S.H., Ha J.H. Pretreatment with propidium monoazide/sodium lauroyl sarcosinate improves discrimination of infectious waterborne virus by RT-qPCR combined with magnetic separation. Environ. Pollut. 2018;233:306–314. doi: 10.1016/j.envpol.2017.10.081. [DOI] [PubMed] [Google Scholar]
- Lemire K.A., Rodriguez Y.Y., McIntosh M.T. Alkaline hydrolysis to remove potentially infectious viral RNA contaminants from DNA. Virol. J. 2016;13:88. doi: 10.1186/s12985-016-0552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClary-Gutierrez J., Mattioli M., Marcenac P., Silverman A., Boehm A., Bibby K., Balliet M., de los Reyes F., III, Gerrity D., III, Griffith J., III, Holden P., III, Katehis D., III, Kester G., III, LaCross N., III, Lipp E., III, Meiman J., III, Noble R., III, Brossard D., III, McLellan S., III . Preprints; 2021. Sars-Cov-2 Wastewater Surveillance for Public Health Action: Connecting Perspectives From Wastewater Researchers and Public Health Officials During a Global Pandemic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Michael-Kordatou I., Karaolia P., Fatta-Kassinos D. Sewage analysis as a tool for the COVID-19 pandemic response and management: the urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020;8(5) doi: 10.1016/j.jece.2020.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michen B., Graule T. Isoelectric points of viruses. J. Appl. Microbiol. 2010;109(2):388–397. doi: 10.1111/j.1365-2672.2010.04663.x. [DOI] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1(6) doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.R., Axler R.P., Hicks R.E. Effects of freezing and storage temperature on MS2 viability. J. Virol. Methods. 2004;122(2):147–152. doi: 10.1016/j.jviromet.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Pabbaraju K., Wong A.A., Douesnard M., Ma R., Gill K., Dieu P., Fonseca K., Zelyas N., Tipples G.A. A public health laboratory response to the pandemic. J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.01110-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabbaraju K., Wong A.A., Douesnard M., Ma R., Gill K., Dieu P., Fonseca K., Zelyas N., Tipples G.A. Development and validation of RT-PCR assays for testing for SARS-CoV-2. Off. J. Assoc. Med. Microbiol. Infect. Dis. Can. 2021;6(1):16–22. doi: 10.3138/jammi-2020-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang X.L., Lee B.E., Pabbaraju K., Gabos S., Craik S., Payment P., Neumann N. Pre-analytical and analytical procedures for the detection of enteric viruses and enterovirus in water samples. J. Virol. Methods. 2012;184(1-2):77–83. doi: 10.1016/j.jviromet.2012.05.014. [DOI] [PubMed] [Google Scholar]
- Pecson B.M., Darby E., Haas C.N., Amha Y., Bartolo M., Danielson R., Dearborn Y., Giovanni G.D., Ferguson C., Fevig S., Gaddis E., Gray D., Lukasik G., Mull B., Olivas L., Olivieri A., Qu Y., Consortium S.-C.-I. Reproducibility and sensitivity of 36 methods to quantify the SARS-CoV-2 genetic signal in raw wastewater: findings from an interlaboratory methods evaluation in the U.S. Environ. Sci.: Water Res. Technol. 2021;7:504–520. doi: 10.1039/d0ew00946f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo S.E., Keim E.K., Swanstrom R., Ong A.Q.W., Burnor E.A., Kossik A.L., Harrison J.C., Demeke B.A., Zhou N.A., Beck N.K., Shirai J.H., Meschke J.S. Sci. Total Environ. 760. 2021. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Resende P.C., Motta F.C., Eppinghaus A.L.F., Chagas do Vale V.H., Braz R.M.S., de Andrade J., Maranhao A.G., Miagostovich M.P. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 2021;191:116810. doi: 10.1016/j.watres.2021.116810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Yuan T., Kon T., Zurawell R., Huang Y., Graham M., Gabos S., Pang X. Rapid detection and quantitation of microcystin-producing microcystis using real-time PCR. Mol. Biomark. Diagn. 2013;S5(006) [Google Scholar]
- Qiu Y., Lee B.E., Neumann N., Ashbolt N., Craik S., Maal-Bared R., Pang X.L. Assessment of human virus removal during municipal wastewater treatment in Edmonton, Canada. J. Appl. Microbiol. 2015;119(6):1729–1739. doi: 10.1111/jam.12971. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Lee B.E., Ruecker N.J., Neumann N., Ashbolt N., Pang X. A one-step centrifugal ultrafiltration method to concentrate enteric viruses from wastewater. J. Virol. Methods. 2016;237:150–153. doi: 10.1016/j.jviromet.2016.09.010. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Li Q., Lee B.E., Ruecker N.J., Neumann N.F., Ashbolt N.J., Pang X. UV inactivation of human infectious viruses at two full-scale wastewater treatment plants in Canada. Water Res. 2018;147:73–81. doi: 10.1016/j.watres.2018.09.057. [DOI] [PubMed] [Google Scholar]
- Rosario K., Symonds E.M., Sinigalliano C., Stewart J., Breitbart M. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol. 2009;75(22):7261–7267. doi: 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre E., Prevost M., Burnet J.B., Pang X., Qiu Y., Smeets P., Medema G., Hachad M., Dorner S. Demonstrating the reduction of enteric viruses by drinking water treatment during snowmelt episodes in urban areas. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2021.100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Tong Z.D., Wang H.L., Dai Y.X., Li K.F., Liu J.N., Wu W.J., Yuan C., Yu M.L., Li P., Yan J.B. Detection of novel coronavirus by RT-PCR in stool specimen from asymptomatic child, China. Emerg. Infect. Dis. 2020;26(6):1337–1339. doi: 10.3201/eid2606.200301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L., Keyaerts E., Moes E., Maes P., Duson G., Van Ranst M. Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. J. Clin. Microbiol. 2005;43(11):5452–5456. doi: 10.1128/JCM.43.11.5452-5456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar L.M., de Paula V.S., Diniz-Mendes L., Lampe E., Gaspar A.M. Evaluation of methods used to concentrate and detect hepatitis a virus in water samples. J. Virol. Methods. 2006;137(2):169–176. doi: 10.1016/j.jviromet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J., Klein J., Lu P., Lu-Culligan A., Jiang X., Kim D.J., Kudo E., Mao T., Moriyama M., Oh J.E., Park A., Silva J., Song E., Takahashi T., Taura M., Tokuyama M., Venkataraman A., Weizman O.E., Wong P., Yang Y., Cheemarla N.R., White E.B., Lapidus S., Earnest R., Geng B., Vijayakumar P., Odio C., Fournier J., Bermejo S., Farhadian S., Dela Cruz C.S., Iwasaki A., Ko A.I., Landry M.L., Foxman E.F., Grubaugh N.D. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat. Microbiol. 2020;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker M.J., Godard M.P. Analysis of one-step and two-step real-time RT-PCR using SuperScript III. J. Biomol. Tech. 2005;16(3):266–271. [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., Torgersen K., Laan J.V., LaCross N. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775 doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brunink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., Bonneau R., Brown M.A., Bushman M., Chai P.R., Duvallet C., Erickson T.B., Foppe K., Ghaeli N., Gu X., Hanage W.P., Huang K.H., Lee W.L., Matus M., McElroy K.A., Nagler J., Rhode S.F., Santillana M., Tucker J.A., Wuertz S., Zhao S., Thompson J., Alm E.J. medRxiv; 2020. SARS-CoV-2 Titers in Wastewater Foreshadow Dynamics and Clinical Presentation of New COVID-19 Cases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., Yin H., Xiao Q., Tang Y., Qu X., Kuang L., Fang X., Mishra N., Lu J., Shan H., Jiang G., Huang X. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020;5(5):434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao J., Huang J., Zhao J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26(8):1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The standard curve of E gene RNA fragment used for quantification in one-step RT-qPCR for SARS-CoV-2, PMMoV and hCoV-229E.
hCoV-229E recovery in wastewater samples using different volume of wastewater (50 ml, 100 ml and 200 ml).