SUMMARY
The Ebola virus matrix protein VP40 forms distinct structures linked to distinct functions in the virus life cycle. Dimeric VP40 is a structural protein associated with virus assembly, while octameric, ring-shaped VP40 is associated with transcriptional control. In this study, we show that suitable nucleic acid is sufficient to trigger a dynamic transformation of VP40 dimer into the octameric ring. Deep sequencing reveals a binding preference of the VP40 ring for the 3′ untranslated region of cellular mRNA and a guanine- and adenine-rich binding motif. Complementary analyses of the nucleic-acid-induced VP40 ring by native mass spectrometry, electron microscopy, and X-ray crystal structures at 1.8 and 1.4 Å resolution reveal the stoichiometry of RNA binding, as well as an interface involving a key guanine nucleotide. The host factor-induced structural transformation of protein structure in response to specific RNA triggers in the Ebola virus life cycle presents unique opportunities for therapeutic inhibition.
Graphical abstract
In brief
Landeras-Bueno et al. find that the Ebola virus matrix protein VP40 transforms to its essential octameric ring form when triggered by binding sequences in the 3′ UTR of host cell mRNA. Small RNAs/DNAs are sufficient to induce this transformation, suggesting a potential for therapeutic disruption of the virus life cycle.
INTRODUCTION
The World Health Organization has declared six global public health emergencies due to RNA virus epidemics from 2009 through 2020, two of which have been epidemics of the emerging pathogen Ebola virus (Rabi et al., 2020). Ebola virus encodes just seven genes but has more than seven functions in the virus life cycle (Sanchez et al., 1993; Kuhn, 2008), suggesting a need for multi-functional viral proteins. Protein structure can be dynamic, with molecules changing their conformation to activate one or more functions. This structural plasticity has been proposed for the Ebola virus matrix protein VP40, which adopts distinct structures to carry out multiple essential functions (Radzimanowski et al., 2014; Wasserman and Saphire, 2016).
The hypothesis of structural transformation reconciles distinct forms of VP40 discovered in cellular lysates and solved by X-ray crystallography, including a butterfly-shaped dimer and an octameric ring (Dessen et al., 2000; Gomis-Ruth et al., 2003; Bornholdt et al., 2013). Ebola virus VP40 comprises an N-terminal domain (NTD; residues 1–194) and a C-terminal domain (CTD; residues 195–326). The NTD and the CTD adjoin in monomeric or dimeric VP40; this is also true for CTDs that dimerize with each other at structurally important flexible interfaces in the virion matrix (Dessen et al., 2000; Bornholdt et al., 2013; Pavadai et al., 2019). The NTD and the CTD are, however, not joined in the VP40 ring. The ring is instead formed by eight NTDs that assemble by interfaces not found in the dimer. The CTDs occupy unknown positions in the ring, apparently at the end of flexible linkers (Scianimanico et al., 2000; Gomis-Rüth et al., 2003). The VP40 dimer is trafficked through the host cell to the plasma membrane, where VP40 induces budding and becomes a structural component of the virion (Jasenosky et al., 2001; Noda et al., 2002; Bornholdt et al., 2013; Gc et al., 2016). The VP40 octameric ring, in contrast, does not localize to the plasma membrane or incorporate into the virion (Ruigrok et al., 2000; Panchal et al., 2003), but instead carries out a regulatory function essential to the virus life cycle (Hoenen et al., 2005, 2010b, 2010a; Bornholdt et al., 2013).
The VP40 octameric ring has been observed bound to RNA from the E. coli expression system (Gomis-Rüth et al., 2003). The VP40 monomer and dimer, in contrast, do not bind RNA. Comparison of dimer and ring structures suggests that RNA may disrupt both the dimer interface and the NTD-CTD interface within each VP40 protomer, hypothetically triggering transformation of dimer into ring (Gomis-Rüth et al., 2003; Bornholdt et al., 2013). Previous studies have shown that the VP40 NTD expressed alone (in the absence of the CTD) can be induced to form the ring both with and without RNA (Timmins et al., 2003; Bornholdt et al., 2013), but whether complete VP40 can form the ring without RNA is unknown. Hence, questions have remained of whether a transformation from the dimer to the ring can occur in the cell, and whether RNA is the natural trigger for this transformation.
In this study, we characterize the RNA bound to the VP40 ring expressed in a human cell line. We discover a binding preference of VP40 for purine-rich sequences located in the 3′ untranslated region (UTR) of cellular mRNA. Moreover, we demonstrate a transformation of VP40 monomer/dimer to the ring in response to specific nucleic acid triggers. We analyze the resulting ring by native mass spectrometry, X-ray crystallography, and electron microscopy. We show that the VP40 ring that we have generated in vitro by triggering the RNA-free dimer using nucleic acid oligonucleotides (oligos) of known sequence corresponds closely in stoichiometry and structure to the VP40 ring that is purified from cells and is already in complex with cellular RNA. These data clarify the role of molecular transformation in the virus life cycle and provide templates for antiviral development.
RESULTS AND DISCUSSION
The VP40 ring binds the 3′ UTR of cellular mRNA
To clarify the role of the VP40 ring and its bound RNA in the Ebola virus life cycle, we set out to determine whether there are specific cellular or viral RNAs that the ring preferentially binds. We transfected the HEK293T human cell line with plasmid encoding VP40, then pulled down the VP40 and deep sequenced the RNA found in each protein sample (Figure 1A). Specifically, we transfected Strep-tagged VP40 in wild-type (WT), a ring-free mutant form (VP40-R134A), and two highly ring-forming mutant forms (VP40-I307R, VP40ΔCTD) (Ruigrok et al., 2000; Gomis-Rüth et al., 2003; Hoenen et al., 2005; Bornholdt et al., 2013). We also transfected Strep-tag alone as a control. The VP40-R134A and VP40-I307R mutants were each produced in three biological replicates. With the VP40 we co-transfected an Ebola minigenome replicon system in order to more closely model an Ebola virus infection (Jasenosky et al., 2010). We omitted this co-transfection from one sample (biological replicate I307R-3) in order to observe how significantly the presence of the Ebola minigenome replicon system affected our experimental results. We pulled down the VP40 from the cellular lysates using Strep-Tactin resin, then verified the pull-down eluates using a blue native protein gel (Schägger and von Jagow, 1991) (Figure 1B). The VP40-WT appears in the gel as monomer, dimer, and a small fraction of ring, the VP40-R134A appears as monomer and dimer only, and the VP40-I307R and VP40ΔCTD appear primarily as ring.
Figure 1. The VP40 ring binds the 3′ untranslated region (UTR) of cellular mRNA.
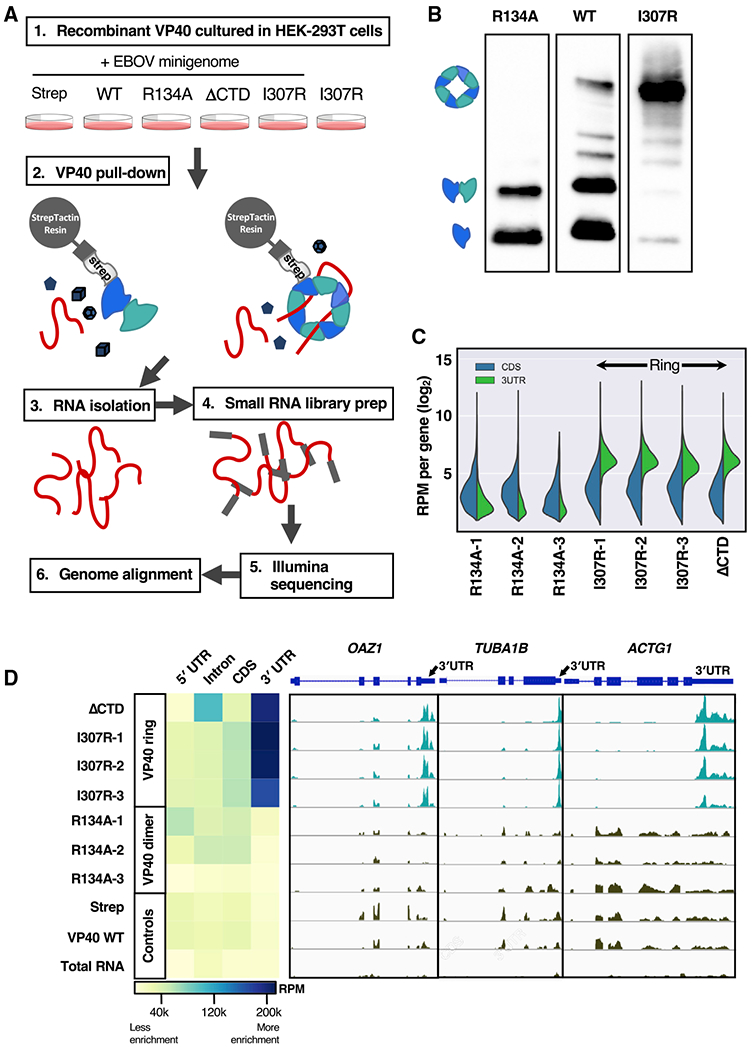
(A) HEK293T cells were transfected with Strep-tagged VP40-WT, three VP40-R134A biological replicates, VP40ΔCTD, three VP40-I307R biological replicates, and Strep-tag alone as a control. An Ebola minigenome replicon system was co-transfected in all cases, with the exception of one I307R biological replicate (I307R-3) as a control. The VP40 was pulled down from the cellular lysate by Strep-Tactin resin, after which the RNA was extracted from each VP40 sample and deep sequenced.
(B) Three of our Strep-Tactin purified VP40 samples run in a blue native protein gel and blotted for Strep-tag. The display as separate images is solely to remove irrelevant lanes. We identify the dominant protein bands seen here as the VP40 monomer, dimer, and octameric ring.
(C) For seven of our RNA samples, the number of reads that align with the coding sequence (CDS) or with the 3′ UTR, respectively, of each of the 1,986 cellular mRNAs that are highest expressed in our data. We observe 3′ UTR enrichment in the ring-forming VP40 mutant samples (VP40-I307R biological replicates, VP40ΔCTD). Read counts were normalized to reads per million (RPM).
(D) For each of our samples, the heatmap displays the RNA RPM that align with each mRNA component (5′ UTR, intron, CDS, or 3′ UTR). For the genes OAZ1, TUBA1B, and ACTG1, we also display the per-gene read peaks. We observed 3′ UTR enrichment in the ring-forming VP40 mutant samples (VP40-I307R biological replicates, VP40ΔCTD). An Ebola minigenome replicon system was co-transfected in all cases except I307R-3 and total RNA (RNA extracted from the HEK293T cellular lysate).
We isolated the RNA from each VP40 eluate and used the NEXTflex small RNA sequencing (RNA-seq) kit (PerkinElmer) to prepare libraries. We targeted ~4 million sequencing reads per sample on a MiSeq sequencer (Illumina). After aligning to the human genome (hg19) and filtering to remove the duplicate reads and the low-quality alignments, this yielded at least 0.7 million reads for each of the samples, with the average being 1.4 million reads per sample (Table S1).
We normalized to reads per million (RPM) and screened for enrichment of reads across the cellular RNA features defined by UCSC Known Genes annotation (Hsu et al., 2006). This annotation includes several types of non-protein-coding RNA, as well as mRNA components as follows: the 5′ UTR, the coding sequence (CDS), the introns, and the 3′ UTR. Our ring-forming VP40 mutant samples (VP40-I307R, VP40ΔCTD) exhibit a significant enrichment of reads that align to the 3′ UTR of cellular mRNA (Mann-Whitney p < 0.01) (Figures 1C, 1D, 2A, and 2B). This enrichment is observed for all of our ring-forming VP40 mutant samples relative to all the other samples (Figure S1). A visual genome alignment confirms the presence of read peaks within the 3′ UTR for our ring-forming VP40 mutant samples (Figure 1D).
Figure 2. The VP40 ring’s RNA binding preference is specific to the mRNA 3′ UTR.
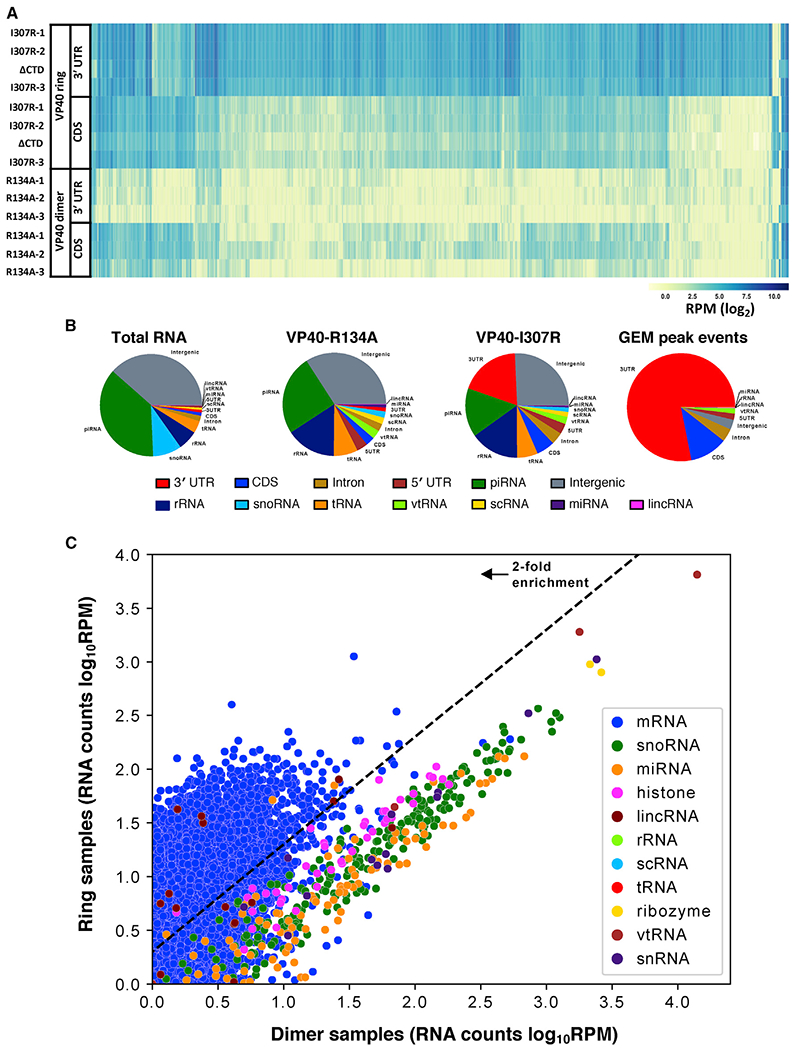
(A) The number of reads that align with the 3′ UTR and with the CDS, respectively, of each of the 1,986 cellular mRNAs that are highest expressed in our data. We observed that the ring-formingVP40 mutant samples (VP40-I307R biological replicates, VP40ΔCTD) have more 3′ UTR reads than CDS reads, while for the dimer-forming VP40 mutant samples (VP40-R134A biological replicates) the opposite is true. Read counts were normalized to RPM.
(B) Reads categorized by RNA feature (the components of mRNA including the 3′ UTR, the CDS, the introns, and the 5′ UTR; also, several forms of non-coding RNA) for a ring-forming VP40 mutant sample (biological replicate VP40-I307R-1) and two non-ring-forming samples (biological replicate VP40-R134A-1, total RNA control). Also included is a similar categorization of the VP40-RNA binding event loci called by the GEM peak-finding algorithm (Figure 3). See also Figure S1.
(C) The cellular RNA transcript abundances in our VP40 samples, comparing our ring-forming VP40 mutant samples versus our dimer-forming VP40 mutant samples. Reads were aligned with the human genome (hg19), filtered to remove duplicates and low-quality alignments, and normalized to RPM. We then averaged over our three VP40-I307R biological replicates and one VP40ΔCTD sample to determine the ring samples value, and over our three VP40-R134A biological replicates to determine the dimer samples value. 7,072 genes are plotted, representing all UCSC Known Genes for which both averages are at least 1 RPM. See also Figure S2.
To investigate whether the 3′ UTR is consistently enriched across individual genes, we quantified the number of reads aligning to the 3′ UTR and to the CDS, respectively, for each mRNA in the UCSC Known Genes. For the 1,986 cellular mRNAs that are highest expressed in our data, there is an average of 84.9 reads per 3′ UTR for the ring-forming VP40 mutant samples (VP40-I307R, VP40ΔCTD), compared to 5.2 reads per 3′ UTR for the dimer-forming VP40 mutant samples (VP40-R134A). Read counts were normalized to RPM. This 16.3-fold enrichment in 3′ UTR reads is statistically significant (p < 0.0001) and is consistently seen when plotting the per-gene read counts (Figures 1C and 2A). We also observed the statistically significant distinction (p < 0.0001) that our ring-forming VP40 mutant samples have more 3′ UTR reads than CDS reads, while for our dimer-forming VP40 mutant samples the opposite is true.
To determine whether any type of RNA other than mRNA is enriched, we quantified reads across the 23,710 unique coding and non-coding transcripts of the UCSC Known Genes annotation. This analysis confirms the preferential enrichment of mRNA in our ring-forming VP40 mutant samples (Figures 2C and S2). Among the 15,480 UCSC Known Genes with nonzero reads in our data, 7,673 are enriched at least 2-fold in our ring-forming VP40 mutant samples, and of these, 99% (7,590) are mRNAs.
Intriguingly, results are different for histone mRNAs, which are unusual in their structure, metabolism, and protein associations (Marzluff and Koreski, 2017). Of the 42 histone mRNAs that are non-trivially expressed in our data (Figure 2C), only 1 (HIST1H2AE) is enriched at least 2-fold in our ring-forming VP40 mutant samples. We also observe a comparative lack of reads aligning to introns (Figures 1D, 2B, and S1), which confirms that VP40 encounters mature, spliced mRNA in the cytoplasm. We conclude that the VP40 ring binds the 3′ UTR of mature mRNA, and that this preference extends across most cellular mRNAs.
Our VP40-WT sample yields sequencing results similar to those for our R134A VP40 mutant samples that remain dimeric and do not transition to ring (Figures 1D, S1, and S2). We hypothesize that our transfection-based experimental model did not provide all of the conditions necessary for VP40-WT to form substantial amounts of the VP40 ring in the host cell. This calls for the experiment to be repeated in the context of an Ebola virus infection, where VP40 would be present at all stages of the Ebola virus life cycle and would have the chance to interact with all possible viral protein partners.
Analysis reveals a guanine- and adenine-rich 3′ UTR binding motif
We have found that the VP40 ring binds the 3′ UTR, which is a known mRNA regulatory domain having distinctive base frequencies and binding motifs (Kuersten and Goodwin, 2003). This led us to speculate that the ring may identify RNA via preferred binding sequences. It has previously been observed that VP40ΔCTD transfected in E. coli forms the VP40 ring bound to E. coli RNA fragments with the sequence 5′-UGA-3′ (Gomis-Rüth et al., 2003). We sought fuller information from our deep-sequencing data.
We used the genome-wide event finding and motif discovery (GEM) algorithm to identify nucleotide-binding motifs (Guo et al., 2012; Thomas et al., 2017). This algorithm works in two phases. First, the genome positioning system (GPS) phase uses read peaks to identify binding events and binding motifs. Second, the GEM phase iteratively shifts and filters the binding event loci to optimize the binding motifs. We used our ring-forming VP40 mutant samples (VP40-I307R, VP40ΔCTD) to call read peaks, and our dimer-forming VP40 mutant samples (VP40-R134A) as controls. GEM identified 29,059 binding events across the human genome, 25,175 of which could be aligned within a specific RNA feature; of these, 78% (19,663) occur within a 3′ UTR (Figure 2B). This confirms our finding of a 3′ UTR binding preference for our ring-forming VP40 mutant samples. To minimize the effect of false positives, for the final GEM motif-optimization phase we limited analysis to binding events within 3′ UTRs. GEM then identified as its highest-ranked binding motif a6-ntG/A-rich motif (Figure 3A). A similar G/A preference is found if we instead analyze all of the GEM binding events, or the binding events initially identified by GPS.
Figure 3. Analysis reveals a guanine- and adenine-rich 3′ UTR binding motif.
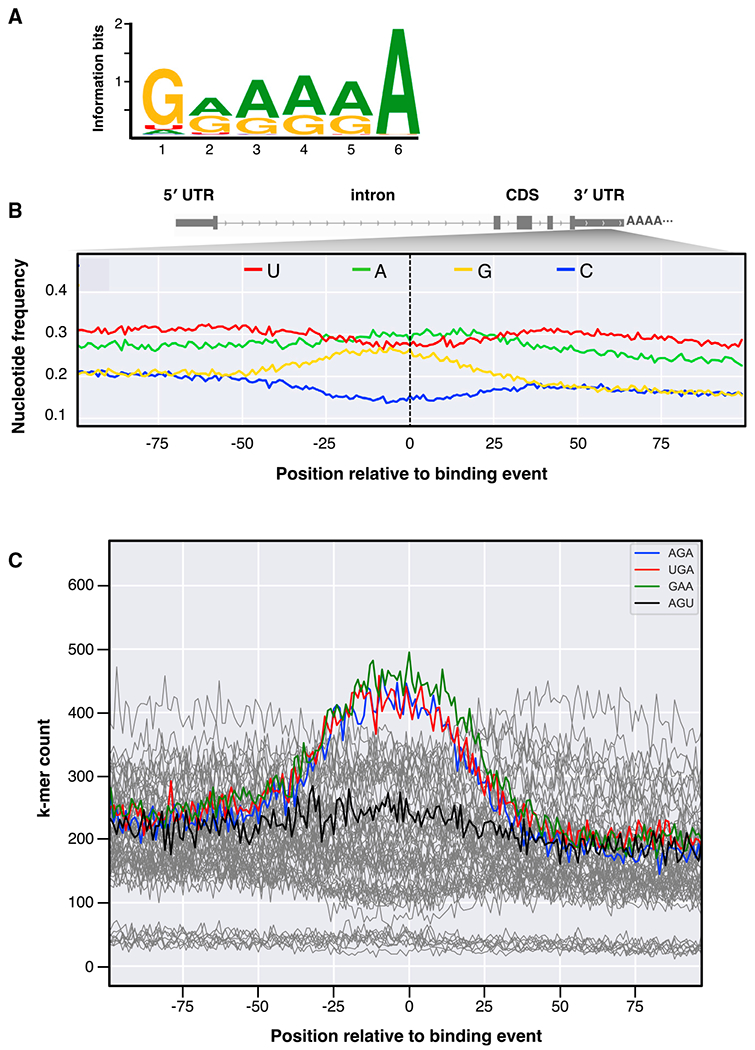
(A) The highest ranked binding motif called by the genome-wide event finding and motif discovery (GEM) analysis of the VP40 binding events in mRNA 3′ UTRs. Letter heights indicate the nucleotide base frequencies (guanine, yellow; adenine, green; uracil, red; cytosine, blue).
(B) The average nucleotide base frequencies within 100 nt of the genome positioning system (GPS)-identified binding events in 3′ UTRs. Human 3′ UTRs are on average U/A-rich, G/C-poor (left), but G/A enrichment extends ~25–50 nt to either side of the binding events (center).
(C) For each possible 3-nt k-mer (with the data for the k-mers AGA, UGA, GAA, and AGU highlighted), the number of times the k-mer occurs in the human genome at the specified offset relative to any GPS-identified 3′ UTR binding event.
We also quantified the nucleotide frequencies within 100 nt of all GPS-identified binding events in 3′ UTRs. This analysis reveals an increase of guanine and adenine frequencies within ~25–50 bases of binding events (Figure 3B). Human 3′ UTRs are on average U/A-rich, G/C-poor (Figure 3B, left); hence, the G/A preference discovered for VP40 ring binding is distinct from the overall characteristics of the 3′ UTR. We conclude that distinctive G/A-rich expanses of the 3′ UTR are favored for VP40 ring binding.
We similarly quantified the frequencies of all 3-nt k-mers (that is, all 64 possible sequences of three adjacent RNA bases) within 100 nt of the GPS-identified binding events in 3′ UTRs. We found the k-mers AGA, UGA, and GAA to be most enriched (Figure 3C). These k-mers are all G/A-rich, and AGA and GAA are high-probability subsequences of the binding motif specified above. The enrichment of UGA may corroborate the previous finding of UGA RNA fragments bound to the VP40 ring (Gomis-Rüth et al., 2003). The fact that simple variants such as the reversed k-mer AGU are not enriched supports the significance of this finding.
Our 6-nt G/A-rich binding motif (Figure 3A) is not in itself sufficient to explain the VP40 ring’s binding preference for the 3′ UTR. Indeed, this brief motif is commonly matched throughout the human genome, and 3′ UTRs are not G/A-rich overall (Figure 3B, left). We speculate that factors such as localization in the cell and interactions with RNA-binding proteins may help pair VP40 with 3′ UTRs.
To determine whether the VP40 ring also binds viral RNA, we aligned our reads to the Zaire ebolavirus genome. We did not observe region-specific enrichment or well-defined read peaks (Figure S3). When we applied the GEM algorithm to the Zaire ebolavirus alignment data, no binding events were identified. This suggests that VP40 interacts primarily with cellular rather than viral RNA. However, this result is based on our use of an Ebola minigenome replicon system that does not include all of the Ebola virus genes and in which the natural Ebola UTRs have been replaced by mammalian-inspired UTRs (Jasenosky et al., 2010). This again calls for the experiment to be repeated in the context of an Ebola virus infection, where the full set of authentic Ebola virus RNAs would be present.
3′ UTR-inspired nucleic acids are sufficient to transform VP40 into the ring
Having determined that the VP40 octameric ring binds the 3′ UTRs of cellular mRNA, we next investigated whether this favored RNA may serve as a sufficient trigger for a VP40 transformation from monomer/dimer to the ring. This hypothesis motivated experiments in which we incubated the purified VP40 monomer/dimer with nucleic acids, including synthetic nucleic acids whose sequences were inspired by favored 3′ UTRs.
For these experiments we used VP40-WT (Δ43, HIS-tagged), which can be expressed in E. coli and purified by a two-step protocol, using HIS-tag affinity followed by size-exclusion chromatography (SEC). This protocol yields two SEC peaks, which have been identified as the VP40 octameric ring and VP40 dimer (Bornholdt et al., 2013). We used blue native protein gels to verify the oligomeric state of these peak samples (Figure 4A, lanes 2–3). The high-weight SEC peak forms a ring-size gel band, while the low-weight peak appears to be a mixture of monomer and dimer. A260/280 ratios confirm the co-purification of RNA with the VP40 ring: ~1.4 for the ring, ~0.65 for the monomer/dimer.
Figure 4. 3′ UTR-inspired nucleic acids are sufficient to transform VP40 into the ring.
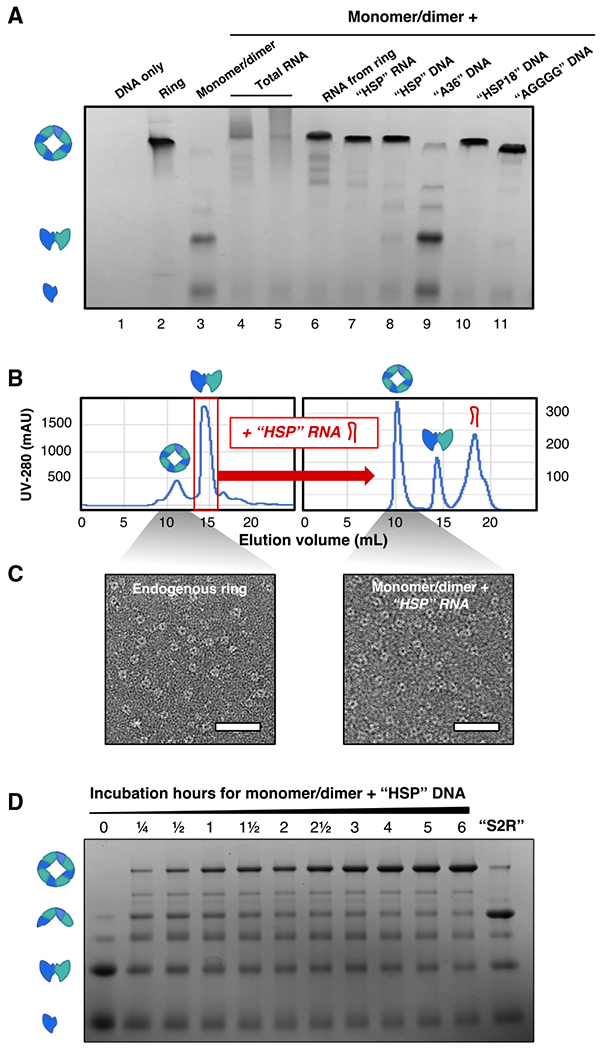
(A) VP40-WT(Δ43, HIS-tagged) in a blue native protein gel. The first three lanes are a no-protein control (1 μg of “HSP” DNA) (lane 1) and VP40 samples from the ring (lane 2) and monomer/dimer (lane 3) SEC fractions. For the remaining lanes, the monomer/dimer was incubated with nucleic acids as follows: the total RNA extracted from E. coli (lanes 4–5); the RNA extracted from the VP40 ring SEC fraction (lane 6); and 3′ UTR-inspired RNA (lane 7) or DNA (lanes 8–11) oligos as specified in Table S2. The protein bands are identified as the VP40 monomer, dimer, and ring by analogy to our native mass spectrometry (nMS) results (Figure 6).
(B) Left: the SEC purification of ~20 mg of E. coli-expressed VP40-WT (Δ43, HIS-tagged), yielding endogenous ring and monomer/dimer peaks. Right: we have incubated the purified monomer/dimer with the HSP RNA oligo (Table S2), then carried out a second SEC purification, revealing peaks of generated VP40 ring, residual monomer/dimer, and unbound RNA.
(C) VP40 samples stained and visualized by electron microscopy. Left: the endogenous ring fraction from the left panel of (B). Right: the RNA-generated ring fraction from the right panel of (B). Oligomers visibly similar to endogenous ring are generated if and only if the monomer/dimer is incubated with nucleic acid. Scale bars, 50 nm.
(D) Lanes 1–11: a gradient of incubation times for the VP40 monomer/dimer incubated with the 3′ UTR-inspired HSP DNA oligo (Table S2). Lane 12: the VP40 monomer/dimer incubated 6 h with the “S2R” DNA oligo (Table S2). The protein bands are identified as the VP40 monomer, dimer, tetramer, and ring by analogy to our nMS results (Figure 6).
We next incubated the VP40 monomer/dimer with several forms of purified nucleic acid. First, we isolated the RNA from the ring peak sample using a QIAGEN miRNeasy micro kit. This RNA that the VP40 ring itself has selected from the cellular milieu should be well-suited for VP40 interaction. Indeed, incubating the VP40 monomer/dimer with this ring-extracted RNA transformed most of the VP40 into ring (Figure 4A, lane 6). Significantly, this shows that selective binding to RNA and a consequent transformation to the ring occurs for VP40-WT, not just for the mutant forms such as VP40-I307R.
To determine which nucleic acids are sufficient to generate this transformation, we next experimented with uniform oligos of single-stranded RNA or DNA (Table S2). Our main test-case RNA is a 35-nt oligo that we named “HSP” because it is based on a sequence within the 3′ UTR of the HSPA1B mRNA, which is one of the most highly enriched genes in our deep-sequencing data. Incubation with this HSP RNA transformed the VP40 monomer/dimer into the ring (Figure 4A, lane 7). Surprisingly, the equivalent HSP sequence in DNA worked equally well (lane 8). This lack of RNA specificity suggests that the nucleic acid-protein interface may be mediated by the nucleotide base or the backbone phosphate rather than the ribose. This is confirmed by previous crystal structures of the VP40 ring (Gomis-Rüth et al., 2003). Above we reported evidence that VP40 in fact interacts with mature, spliced mRNA in the cytoplasm. DNA is however useful for experimental purposes due to its higher stability.
We found multiple DNA oligos sufficient to generate the VP40 ring in this in vitro experimental model: in particular, an 18-nt subsequence of HSP (termed HSP18, Table S2), as well as the very short oligo with sequence AGGGG, whose testing was motivated by the determination above of a G/A-rich binding preference. Oligos that did not efficiently generate the ring include polyadenylate DNA, double-stranded DNA, and several oligos of length shorter than ~12 nt (with AGGGG being an exception). The negative result for polyadenylate suggests that VP40 binding is specific to the 3′ UTR, not to the adjacent mRNA polyadenylate tail.
Our synthetic oligos such as HSP RNA or DNA produced a sharp VP40 ring band, suggesting a well-defined stoichiometry (Figure 4A, lanes 7–8 and 10–11). The ring originally discovered in E. coli also produced a clear band, as did the ring generated using RNA extracted from this original ring (lanes 2 and 6). In contrast, attempts to generate the ring using total E. coli RNA produce extremely heterogeneous bands (lanes 4–5). This suggests that unknown factors in the cell may assist in RNA selection to form homogeneous VP40-RNA complexes.
For further mechanistic studies we chose HSP, our nucleic acid oligo based on a subsequence of the human HSPA1B 3′ UTR that is favored for VP40 ring binding (Table S2). We confirmed that the incubation of the VP40 monomer/dimer with the HSP RNA generates the VP40 ring, using techniques including SEC (Figure 4B) and negative-stain electron micrographs (Figure 4C). These results suggest that the nucleic acid-generated ring resembles the endogenous ring that has previously been studied.
VP40 oligomers of a size intermediate between the dimer and the ring may represent intermediate forms in the transformation to the ring. Incubation of the VP40 monomer/dimer with a G/C-poor DNA oligo that we have named “S2R” (Table S2) generates an intermediate-weight VP40 oligomer (Figure 4D, last lane). We have determined this oligomer to be a VP40 tetramer using native mass spectrometry (nMS) (Figure 6G). When the VP40 monomer/dimer was incubated with the HSP DNA, bands of the VP40 tetramer and other intermediate-weight oligomers first increased and then decreased as the ring band developed (Figure 4D), suggesting that these oligomers may be transformation intermediates.
Figure 6. The VP40-nucleic acid complexes have well-defined stoichiometry.
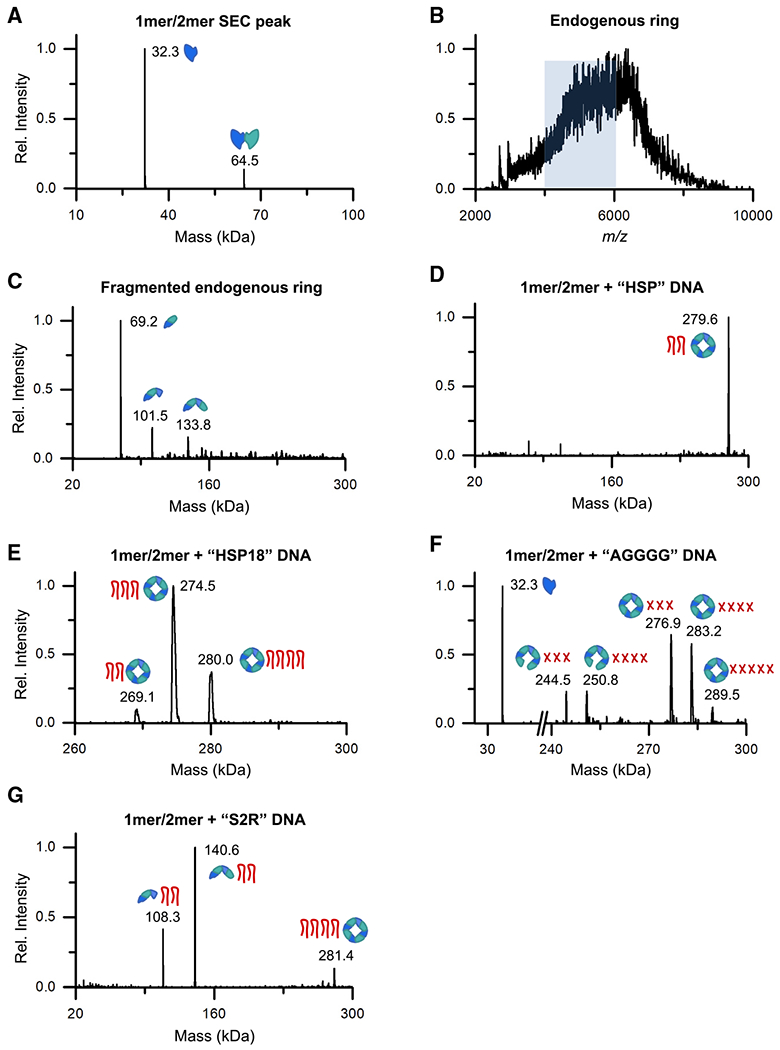
(A) The deconvoluted mass spectrum of the VP40 monomer/dimer SEC fraction as measured by size-exclusion chromatography-nMS (SEC-nMS).
(B) The mass spectrum of the VP40 endogenous ring SEC fraction as measured by nMS, yielding a broad m/z signal that could not be resolved. The m/z range highlighted in blue was mass selected for dissociation in (C).
(C) The deconvoluted mass spectrum of the heterogeneous ring signal highlighted in (B) after mass-selection and fragmentation by surface-induced dissociation (SID).
(D-G) The deconvoluted mass spectra of the VP40 monomer/dimer after incubation with 3′ UTR-inspired DNA oligos (Table S2), as measured by SEC-nMS. For each detected complex, cartoons indicate the VP40 oligomeric state and the number of bound DNA oligos. In (F), each red X denotes a putative tetramolecular G-quadruplex of AGGGG oligos.
A low salt concentration is required for the nucleic acid-induced transformation of the VP40 monomer/dimer into the ring. An NaCl concentration of ~30–60 mM appears to be optimal, while at 300 mM the transformation is inhibited (Figure S4). We theorize that a concentration of anions higher than that typically found in the cytosol outcompetes the nucleic acid for binding to the positively charged surface residues of the VP40.
The VP40 ring generated using 3′ UTR-inspired nucleic acids aligns with known ring structures
In order to visualize the nucleic acid-generated VP40 ring at the atomic level, and to compare this ring with the endogenous ring that has previously been studied, we next determined structures of the nucleic acid-generated ring by X-ray crystallography. We used here our HSP RNA and HSP DNA oligos (Table S2), which are based on a subsequence of the human HSPA1B 3′ UTR that is favored for VP40 ring binding.
The VP40 CTDs are flexibly positioned in the ring structure (Scianimanico et al., 2000) and inhibit crystallization. Hence, previous VP40 ring structures (Gomis-Rüth et al., 2003; Bornholdt et al., 2013) have used VP40ΔCTD (i.e., the VP40 NTD alone). This approach presents difficulties here, because VP40ΔCTD expresses as ring and immediately binds cellular RNA (Ruigrok et al., 2000; Bornholdt et al., 2013). Hence, we modified our plasmid for VP40-WT (Δ43, HIS-tagged) by inserting a tobacco etch virus (TEV) protease recognition site between the NTD and the CTD. We expressed this modified VP40, purified its RNA-free monomer/dimer fraction, incubated it with the HSP RNA or DNA to generate ring, and then applied TEV protease to delete the CTDs (Figures 5A and 5B).
Figure 5. The VP40 ring generated using 3′ UTR-inspired nucleic acids aligns with known ring structures.
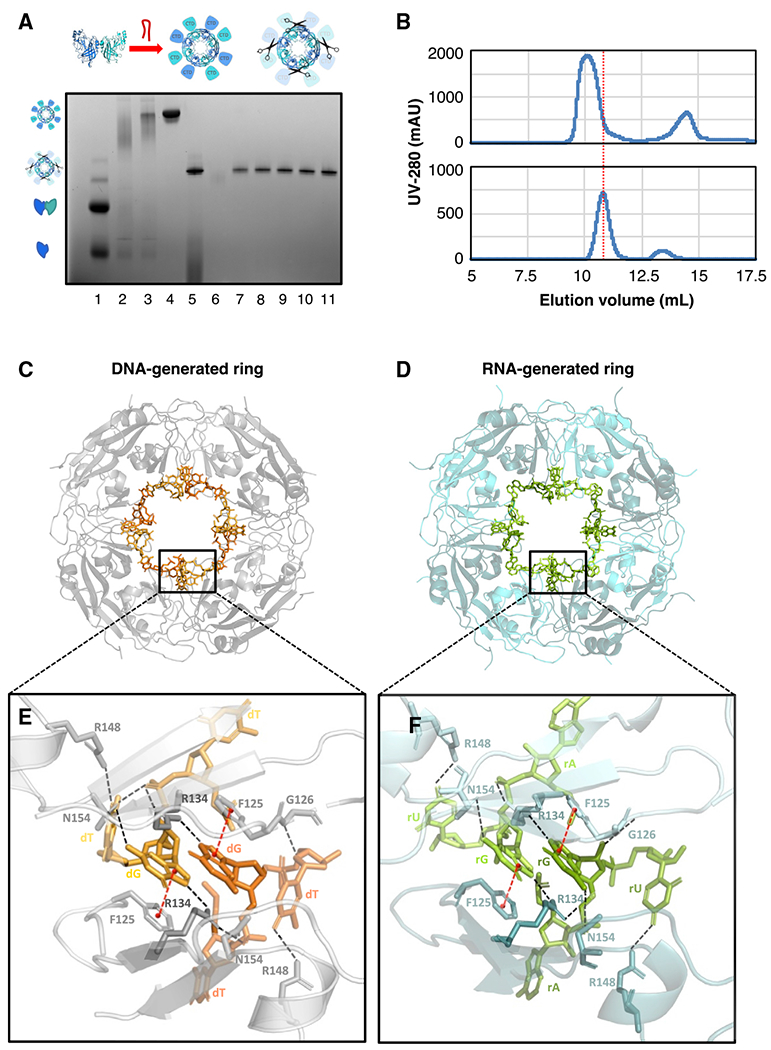
(A) VP40 including a TEV protease recognition site was expressed, after which the purified VP40 monomer/dimer was incubated with RNA or DNA to generate the ring, and finally TEV protease was applied to delete the C-terminal domains (CTDs). A blue native protein gel verifies the creation of the CTD-deleted ring using the 3′ UTR-inspired HSP DNA oligo (Table S2). The VP40 monomer/dimer (lane 1), the VP40 endogenous ring (lane 2), the ring generated from the monomer/dimer by the DNA incubation (lane 3), the DNA-generated ring after SEC purification (lane 4), the CTD-deleted ring after the TEV protease digestion (lane 5), and the fractions after a final SEC purification (lanes 6–11) are shown.
(B) Two SEC traces from the above protocol. Top: after the incubation of the VP40 monomer/dimer with the HSP DNA, the full-length VP40 ring elutes at 10.1 mL. Bottom: after the subsequent TEV protease digestion, the CTD-deleted VP40 ring elutes at 10.8 mL.
(C) Our crystal structure at 1.78-Å resolution of the VP40 octameric ring generated using the HSP DNA oligo (PDB: 7K5D). Eight symmetry-related DNA fragments are seen bound to the ring.
(D) Our crystal structure at 1.38-Å resolution of the VP40 octameric ring generated using the HSP RNA oligo (PDB: 7K5L). Eight symmetry-related RNA fragments are seen bound to the ring. The structures in (C) and (D) align closely, with protein backbone RMSD of 0.10 Å.
(E) A detail from (C) of two DNA fragments bound to the ring. The two DNA fragments are symmetrical to each other and share a binding cleft between two VP40 molecules that are also symmetrical to each other. Each DNA fragment has been built as 5′-TGT-3′. Hydrogen bonds are indicated with dashed black lines, pi stacking with red lines. The central guanine nucleotide is key to the DNA-protein interface, interacting with both of the adjacent VP40 molecules: its phosphate interfaces with R134 and N154, while its base interfaces with F125 and with R134 and N154 again. The third nucleotide (thymine) interfaces with R148, G126, and N154 again. See also Figure S5A.
(F) A detail from (D) of two RNA fragments bound to the ring. Each RNA fragment has been built as 5′-AGU-3′. The nucleic acid-protein interface closely resembles that seen in (E), particularly with regard to the central guanine nucleotide. See also Figure S5B.
This CTD-deleted VP40 ring yielded crystals, allowing us to solve structures at 1.38-Å resolution for the HSP RNA-generated ring and 1.78-Å resolution for the HSP DNA-generated ring (Table S3). These structures align closely, with protein backbone root-mean-square deviation (RMSD) of 0.10 Å (Figures 5C and 5D). Both structures also align with the previous structure PDB: 1H2C of the endogenous VP40 ring bound to E. coli RNA (Gomis-Rüth et al., 2003), with protein backbone RMSD of at most 0.39 Å. This proves the near-identity of the endogenous and nucleic acid-generated ring structures.
Each ring structure consists of eight VP40 NTDs, and of its eight NTD-NTD interfaces, every second interface forms a cleft that binds nucleic acid. At each such cleft, two symmetry-related 3-nt fragments of nucleic acid are visible (Figures 5E and 5F). In both structures there is a strong electron density for a central guanine base that interfaces with the VP40 R134 residue (Figure S5). This correlates with our findings of a guanine- and adenine-rich binding sequence (Figure 3A) and efficient ring generation by the AGGGG oligo (Figure 4A). Two contiguous nucleotides are observed 5′ and 3′ of this central guanine, but these nucleotides have incomplete or indeterminate electron density (Figure S5). The 5′ nucleotide has been built as thymine in our DNA-generated ring structure, while the 5′ nucleotide density in our RNA-generated ring structure appears to best match adenine. The 3′ nucleotides in both structures have only partial base densities, yet they appear to significantly interface with the protein (Figures 5E and 5F). We have built these 3′ nucleotides as thymine/uracil by reference to the HSP nucleic acid sequence. In many of these details, including the central guanine-R134 interface and the partial densities for the 5′ and 3′ nucleotides, our structures closely resemble the previous structures of the endogenous VP40 ring (Gomis-Ruth et al., 2003). This suggests that the previous structures, which originated from VP40ΔCTD bound to E. coli RNA, also reflect the structure adopted by WT VP40 when it is triggered by 3′ UTR sequences to form the ring in the human cell.
The VP40-nucleic acid complexes have well-defined stoichiometry
In order to understand the biologically possible VP40-nucleic acid complexes, we set out to determine the VP40 oligomeric states corresponding to protein bands visible in our blue native protein gels, as well as the stoichiometry by which nucleic acid oligos can combine with VP40. This led us to examine our VP40 samples using native mass spectrometry (nMS) (Katta and Chait, 1991; Heck, 2008; Liko et al., 2016; Sarni et al., 2020).
We first examined the monomer/dimer and ring fractions of the SEC-purified VP40-WT (Δ43, HIS-tagged). The low-weight SEC fraction was confirmed to be an RNA-free mixture of VP40 monomer and dimer (Figure 6A). The detected VP40 mass matched the theoretical mass minus the N-terminal methionine. The ring fraction of VP40-WT yielded a broad, unresolvable m/z spectrum (Figure 6B); this is as expected if the endogenous VP40 ring binds a variety of cellular RNA. To further investigate this ring sample, we partially disrupted it by tandem MS using surface-induced dissociation (SID) (VanAernum etal., 2019). This yielded dimer, trimer, and tetramer fragments (Figure 6C), suggesting that the original sample included larger VP40 oligomers (presumably the ring). The subcomplexes produced in this study contained an additional mass of 4.7 kDa, suggesting that a ligand of unknown nature may be present.
We next measured samples in which the VP40 monomer/dimer had been incubated with DNA oligos. The incubation of the monomer/dimer with our 3′ UTR-inspired 35-nt DNA oligo HSP (Table S2) yielded a VP40 octameric ring with two bound HSP oligos (Figure 6D). The shorter HSP18 DNA, 18 nt long and representing a central portion of the HSP sequence, yielded a stoichiometry of two, three, or four DNA oligos per ring (Figure 6E). Given that there are four nucleic acid binding clefts per ring (Gomis-Rüth et al., 2003), this may indicate that each 35-nt HSP oligo binds two binding clefts, while each 18-nucleotide HSP18 oligo can bind either one or two binding clefts. For the very short AGGGG oligo, the number of oligos per ring is 12, 16, or 20. Short nucleic acid strands containing several guanine bases in a row are known to form tetramolecular structures called G-quadruplexes (Sen and Gilbert, 1988), and we hypothesize that what we are seeing here are 3, 4, or 5 G-quadruplexes bound per ring (Figure 6F). VP40 heptamer and complementary monomer are also present in the AGGGG sample, suggesting that unintentional ion activation during ionization and transmission led to some dissociation of the gas-phase labile ring.
Incubating the VP40 monomer/dimer with the S2R DNA oligo is known to generate an intermediate-weight VP40 oligomer (Figure 4D). nMS suggests that this oligomer is VP40 tetramer bound to two S2R oligos (Figure 6G). VP40 trimer bound to two S2R oligos and VP40 octamer bound to four S2R oligos are also observed. This octamer is presumably the ring, and we hypothesize that the VP40 tetramer represents an intermediate stage in the transformation from the monomer/dimer to the ring.
The gel bands visible in Figure 4D can now be identified as follows: VP40 monomer, dimer, trimer, tetramer, pentamer (a faint band), hexamer, and octamer (ring). The study of the full set of VP40 oligomers may clarify the dynamic process by which the VP40 monomer/dimer transforms into the ring.
Our nMS data provide intriguing details of the nucleic acid oligos bound to the VP40 ring. Eight nucleic acid binding sites are available, grouped into a pair of sites at each of four binding clefts (Gomis-Ruth et al., 2003). However, when substantial-length oligos such as the 35-nt HSP DNA are used, as few as two oligos bind per ring, suggesting that each oligo loops through multiple binding sites and sterically blocks the binding of additional strands. The 3′ UTRs of human mRNA are on average ~1,000 nt long (Zhao et al., 2011), and we speculate that, in the cell, a single 3′ UTR is sufficient to trigger the transformation to the ring and to satisfy all eight of the ring’s RNA binding sites (Figure 7).
Figure 7. The hypothesized interaction of an mRNA 3′ UTR with VP40 dimers to generate the VP40 ring.
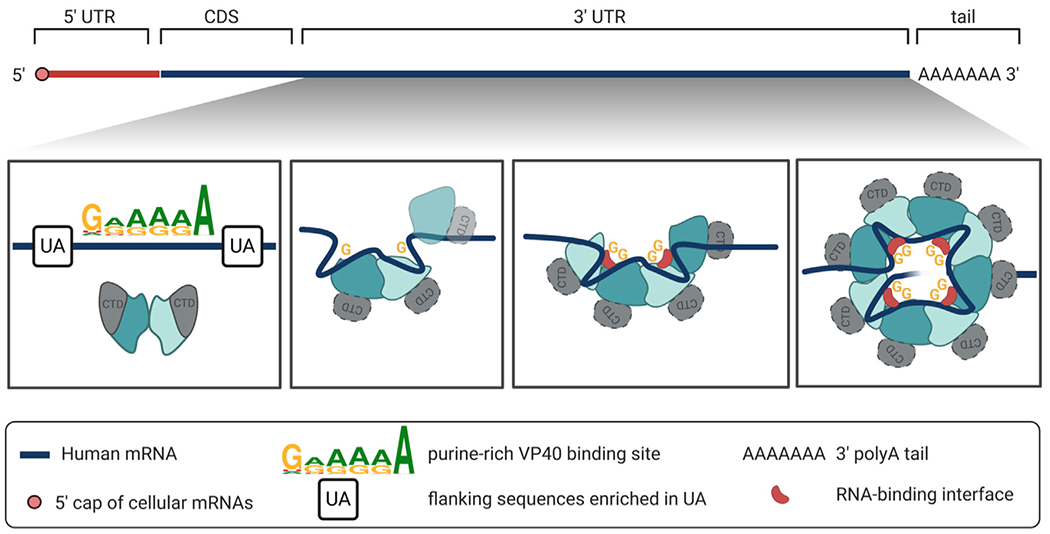
Top: a human mRNA comprises a 5′ UTR, a CDS, a 3 UTR, and a polyadenylate tail. First panel: as long as the VP40 remains in its butterfly-shaped dimer configuration, it does not bind RNA. Second panel: however, when a VP40 dimer encounters suitable RNA, namely a G/A-rich binding sequence within the generally U/A-rich mRNA 3′ UTR, the dimer interface is disrupted, as well as the interface between the NTD and the CTD of each VP40 protomer. The protein-protein interfaces are rearranged, while the RNA remains bound to each NTD via a central guanine base. Third panel: the 3′ UTR strand may now assist in bringing rearranged VP40 dimers together. A half-ring may form as a transformation intermediate. Fourth panel: the octameric VP40 ring is assembled. A single 3′ UTR strand may loop through the ring to bind at the eight known binding sites, two at each of the ring’s four RNA binding clefts.
We have shown suitable nucleic acids such as those based on the HSPA1B mRNA to be a sufficient trigger of the VP40 transformation from monomer/dimer to the ring. Guanine nucleotides that can interface with the R134 residue appear to be of primary importance here: this is evidenced by our finding of a guanine-and adenine-rich binding motif (Figure 3) and efficient ring generation for the AGGGG oligo (Figure 4A), as well as by our crystal structures (Figure 5) and closely aligned previous structures (Gomis-Rüth et al., 2003). The behavior in the cell is more specific, exhibiting a binding preference for the 3′ UTR of cellular mRNA (Figures 1 and 2). This is likely due to cellular factors that we have not modeled, such as VP40 localizing proximate to mRNA.
We have shown that VP40 can transform from one structure to another, and that host RNA may be both necessary and sufficient for the transformation of VP40 from its structural role to its essential non-structural role in the virus life cycle. This host-triggered structural transformation allows the Ebola virus gene products to carry out a greater number of functions in the virus life cycle than would otherwise be possible. The requirement to support structural transformation may limit mutational change in viral proteins such as VP40, reducing the danger of escape mutations and thus representing an Achilles’ heel of the virus. Thus, as we increasingly understand VP40 and its structures supporting molecular transformation, this transformer protein may emerge as an important target for antiviral drug design.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Erica Ollmann Saphire (erica@lji.org).
Materials availability
All reagents generated in this study are available from the Lead Contact upon request.
Data and code availability
The atomic coordinates generated during this study are deposited to the RCSB Protein Data Bank. EBOV VP40 RNA-generated ring structure, PDB: 7K5L. EBOV VP40 DNA-generated ring structure, PDB: 7K5D.
Deep sequencing data can be found in the Sequence Read Archive database under project PRJNA665696.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial strains
E. coli strain Rosetta DE3 (Novagen) was grown in lysogeny broth. The genotype is: F-ompT hsdSB(rB- mB-) gal dcm (DE3) pRARE (CamR). Selection markers were used at the indicated concentrations: Ampicillin (100 μg/mL); Chloramphenicol (28.3 μg/mL).
Human cell lines
HEK293T cells (ATCC) were cultured in DMEM medium (GIBCO 31966021) supplemented with 10% Fetal Bovine Serum and incubated at 37°C and 5% CO2.
METHOD DETAILS
VP40 expression in HEK293T cells
VP40 was expressed in Human Embryonic Kidney (HEK293T) cells using the pTriEx-5 vector (Novagen). An N-terminal Strep-tag prefix (sequence MASWSHPQFEKGADDDDK) is followed by the full-length VP40 of Zaire Ebola virus (Ebola virus/H.sapiens-tc/COD/1976/Yambuku-Mayinga, GenBank KR063671), employed as wild-type or as the mutants R134A, I307R, and ΔCTD (truncated after residue 194). A control plasmid expressed the Strep-tag prefix only. Except as stated, we co-transfected an Ebola minigenome replicon system (Jasenosky et al., 2010). Each 100 mm dish was transfected using TransIT-LT1 Transfection Reagent (Mirus), the VP40 vector (2 μg), and the vectors of the minigenome replicon system: pCEZ-NP-2A-VP35 (2.07 μg), pCEZ-VP30 (1.24 μg), pCEZ-L (12.4 μg), and pHH21–3E5E-Luc (1.04 μg). Cells were lysed after 19.5 hours using CytoBuster Protein Extraction Reagent (Millipore 71009) mixed with RNase Inhibitor (New England BioLabs M0307S). The lysate was combined with BioLock (IBA 2–0205-250), incubated 60 minutes with ~100 μL QIAGEN Strep-Tactin Superflow Plus beads using the batch method, washed twice with 50 mM sodium phosphate monobasic pH 8, 100 mM NaCl, and eluted twice for ~2 minutes using 10 mM biotin.
RNA/DNA
We extracted RNA from the protein samples using a QIAGEN miRNeasy Micro Kit and suspended the RNA in DEPC-treated water. The RNA/DNA oligonucleotides specified in Table S2 were ordered from Integrated DNA Technologies (IDT).
Deep sequencing
In order to assay RNA features of all sizes, we adapted the NEXTFLEX Small RNA-Seq Kit protocol (PerkinElmer). Specifically, RNA was sheared naturally during the cell lysis and Strep-Tactin bead incubation stages described above (Figure S6A); and in order to include RNA fragments up to ~300 nucleotides, we omitted the upper-bound bead purification from protocol step F. Multiplexed libraries were sequenced using a version 3 paired-end 150-cycle MiSeq flow cell (Illumina).
VP40 expression in E. coli
We used the pET-46 Ek/LIC Vector (Novagen) to express VP40-WT (Δ43, HIS-tagged): an N-terminal prefix (sequence MAHHHHHHVDDDDKM) followed by residues 44 to the end of the Yambuku-Mayinga VP40 sequence specified above. Rosetta E. coli cells (Novagen) were transformed, grown overnight at 37°C in 6 mL lysogeny broth with 600 mg Ampicillin, 170 μg Chloramphenicol, then grown to ODA600~0.4 in 1 L lysogeny broth with 100 mg Ampicillin. Induction with 0.5 mM IPTG was overnight at 25°C. Pellets were resuspended in 50 mM sodium phosphate monobasic pH 8, 300 mM NaCl, 10 mM imidazole, and lysed using a Microfluidics M-110P microfluidizer. The clarified lysate was applied to 2.5 mL QIAGEN Ni-NTA Agarose beads using the batch method, incubated 60 minutes, washed twice using 30 mM imidazole, and eluted 30 minutes using 250 mM imidazole. The eluate was dialyzed overnight into 10 mM Tris pH 8,300 mM NaCl. Size exclusion chromatography was performed using a Superdex 200 Increase 10/300 GL column, yielding ring and monomer/dimer fractions.
VP40-nucleic acid incubation
The incubation conditions were as follows: the VP40-WT (Δ43, HIS-tagged) purified monomer/dimer was combined with 8%−25% as much nucleic acid by weight, 10 mM Tris pH 8,30 mM NaCl, and incubated overnight at 4°C. We note that a low salt concentration is needed (Figure S4). 8% nucleic acid was used for Figures 4B, 4C, and 5; 25% nucleic acid was used for Figures 4A, 4D, and S4.
Blue native protein gels
Sample preparation:
The samples requiring nucleic acid incubation used incubation conditions as described above, with 4 μg VP40, 1 μg DNA or RNA, in 7 μL total volume. For Figure 4A, lane 2 used 8 μg VP40 and lane 5 used 10 μg RNA.
Gel electrophoresis:
Blue native electrophoresis (Schägger and von Jagow, 1991) employed an Invitrogen NativePAGE 4%−16% Bis-Tris Gel and an Invitrogen Novex Mini-Cell electrophoresis tank. Each sample was mixed with an equal quantity of 100 mM Bis-Tris pH 7,100 mM NaCl, 20%v/v glycerol, 0.08%w/v Coomassie Brilliant Blue G-250, after which 10 μL was put in each well. Cathode buffer was 15 mM Bis-Tris pH 7, 50 mM Tricine, 0.02%w/v Coomassie Brilliant Blue G-250. Anode buffer was 50 mM Bis-Tris pH 7. Electrophoresis was 3 hours at 150 Volts.
Post-electrophoresis:
For Figure 1B the gel was transferred to an Immobilon-P PVDF transfer membrane (Millipore) and Western probed with an anti-Strep-tag antibody (GeneTex GT517). The other gels (Figures 4A, 4D, 5A, and S4) were destained as follows: the gel was placed in 10% acetic acid, 40% methanol, brought to 100°C, incubated 15 minutes, transferred to 8% acetic acid, brought to 100°C, incubated ~30 minutes, and imaged with a Bio-Rad ChemiDoc MP Imaging System.
Negative stain microscopy
6 μL 0.05 mg/mL protein was applied to a carbon film 400 mesh grid for 1 minute, after which the grid was three times washed with drops of water, then twice stained with 20 μL drops of 1% uranyl formate hydrate, the first time briefly, the second time for 1 minute. The grid was blotted with filter paper after each application of liquid. Imaging was with a Titan Halo transmission electron microscope equipped with a Falcon 3EC detector at 75,000x magnification.
Crystallography
We modified the VP40-WT (Δ43, HIS-tagged) plasmid described above by inserting a TEV protease recognition sequence (ENLYFQG) after VP40 residue 194. We expressed VP40 in E. coli and purified it as described above, except using 500 mM NaCl buffers. We combined the VP40 from the monomer/dimer SEC peak with 8% as much “HSP” DNA or “HSP” RNA by weight (Table S2). We incubated ~6 hours at 4°C, then continued incubation another ~10 hours while dialyzing into 10 mM Tris pH 8,30 mM NaCl. We used SEC again to purify the generated VP40 ring fraction, then combined the ring with 1.67% as much TEV protease by weight and incubated 36–48 hours at 4°C. We used SEC a third time to purify the CTD-deleted ring fraction.
The “HSP” DNA-generated ring was crystallized at 20°C for 1.5 months in a hanging drop containing 1 μL 3.5 mg/mL protein, 1 μL 75 mM sodium citrate tribasic pH 6.6,150 mM ammonium acetate. The cryoprotectant used 15% ethylene glycol. The structure was solved to 1.78 Å resolution at the 12–2 beamline of the Stanford Synchrotron Radiation Lightsource. The “HSP” RNA-generated ring was crystallized similarly, except at pH 8.4. The cryoprotectant used 25% ethylene glycol and 12.5% sucrose. The structure was solved to 1.38 Å resolution at the Eli Lilly LRL-CAT beamline of the Advanced Photon Source at Argonne National Laboratory.
Native mass spectrometry (nMS)
Sample preparation:
VP40-WT (Δ43, HIS-tagged) was expressed in E. coli and purified as described above, with the SEC peaks yielding ring and monomer/dimer fractions in 10 mM Tris pH 8, 300 mM NaCl. For samples requiring nucleic acid incubation, monomer/dimer was diluted to attain 30 μM VP40,10 mM Tris pH 8,30 mM NaCl and mixed with DNA oligonucleotide (Table S2).The molar ratio of VP40 to oligonucleotide was optimized to 1:1 for “HSP,” “HSP18,” and “S2R,” and to 1:4 for “AGGGG.” Incubation was ~24 hours at 4°C. Samples were buffer exchanged into 20 mM ammonium acetate (Sigma) adjusted to pH 8 with ammonium hydroxide (Fisher) using P6 Micro Bio-Spin columns (Bio-Rad) immediately before nMS analysis.
Liquid chromatography and mass spectrometry:
The VP40 monomer/dimer and ring fractions were first screened by online buffer exchange coupled to mass spectrometry (VanAernum et al., 2020). Each sample was then measured over a range of protein concentrations (0.020–2.5 mg/mL) by size exclusion chromatography coupled to native mass spectrometry (SEC-nMS). A Vanquish LC system equipped with a variable wavelength UV detector was coupled to a Q Exactive UHMR mass spectrometer. A MAbPac SEC-1 column (5 μm, 2.1 mm × 300 mm, Thermo Scientific) was used for separation. The sample was injected (2 μL for Figures 6A and 6B and 5 μL for Figures 6D–6G) and eluted with an isocratic flow of 20 mM ammonium acetate pH 8 at 50 μL/min. The mass spectrometer was tuned as follows: ESI voltage: 4.0 kV, source temperature: 350°C, S-lens RF amplitude: 200 V, high m/z ion transfer mode, low m/z detector mode. The in-source trapping desolvation voltage on the Q Exactive UHMR was optimized per-sample to remove non-specific adducts while maintaining protein-protein and protein-nucleic acid interactions.
For Figure 6C, the ring fraction was measured by direct infusion into a Q Exactive UHMR instrument modified with a surface-induced dissociation (SID) device (VanAernum et al., 2019). Sample ionization was accomplished using a Nanospray Flex ion source and in-house pulled nanospray emitters. Electrical contact between the ion source and the nanospray emitter was made using a platinum wire. SID spectra of the endogenous ring were recorded by selecting a wide section of the heterogenous ring signal using the selection quadrupole and performing SID at 85 V on the mass-selected ions.
QUANTIFICATION AND STATISTICAL ANALYSIS
Deep sequencing data analysis
The NEXTFLEX adaptor sequences were trimmed using Cutadapt (Martin, 2011). Alignment to the human (hg19) and Zaire ebolavirus (AF086833.2) genomes used BWA (Li and Durbin, 2009). We used the 4-nucleotide random sequences present on the NEXTFLEX adapters and the coordinates of paired read start sites to identify unique molecules and remove duplicates. Low-quality hg19 alignments (< Q40) were removed using Cutadapt. The Integrative Genomics Viewer was used to display read depths (Thorvaldsdóttir et al., 2013). Regions in the human genome were defined using the UCSC Table Browser, and Bedtools intersect was used to remove overlapping and ambiguous regions (Karolchik et al., 2004; Hsu et al., 2006; Quinlan and Hall, 2010). Custom scripts employing Bedtools intersect and Samtools (Li et al., 2009) were used to count the reads aligning to each region. We used HTSeq to count the reads aligning to known human genes (Anders et al., 2015). To compensate for the differing numbers of reads per sample, the read count data was normalized to reads per million (RPM). The GEM software suite was used to call binding events and binding motifs (Guo et al., 2012). To minimize false positives, for the GEM motif-optimization phase we limited the analysis to binding events within 3′ UTRs. Figure S6B summarizes our analysis workflow.
Crystallography data analysis
We used XDS (Kabsch, 2010), AIMLESS (Evans and Murshudov, 2013), CCP4 (Winn et al., 2011), autoPROC (Vonrhein et al., 2011), and PHENIX(Liebschner et al., 2019). Molecular replacement using Phaser (McCoy et al., 2007) was by reference to PDB:4LDM, after which protein structure was corrected and nucleic acid built using Coot (Emsley et al., 2010), with refinement by phenix.refine (Adams et al., 2010). Noncovalent interactions between protein and nucleic acid were identified using the Protein-Ligand Interaction Profiler (PLIP) (Salentin et al., 2015). Structures were aligned and figures were created using the PyMOL Molecular Graphics System, Version 2.3 Schrödinger, LLC. Figures were created using UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311 (Pettersen et al., 2004).
Native mass spectrometry (nMS) data analysis
The mass spectrometry data were collected using Xcalibur version 4.2 (Thermo Scientific) and deconvolved using Intact Mass version 2.7 (Protein Metrics) (Bern et al., 2018). The theoretical masses for all possible VP40-DNA complexes were entered as target masses in the Intact Mass software. The following parameters were used for mass spectral deconvolution and identification: minimum difference between peaks: 500 Da, charge vector spacing: 0.2, baseline radius: 15 m/z, smoothing sigma: 0.02 m/z, spacing: 0.04 m/z, mass smoothing sigma: 3, mass spacing: 0.5, charge range: 5–100, mass matching tolerance: 200 Da. The raw and deconvolved mass spectra were replotted using Origin.
Figure 7 and the graphical abstract were created with BioRender.com.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Strep epitope Tag antibody (GT517) | GeneTex | Cat#GTX628899; RRID: AB_2538748 |
| Bacterial and virus strains | ||
| E. coli Rosetta (DE3) | Novagen | Cat#70954-3 |
| Chemicals, peptides, and recombinant proteins | ||
| Ampicillin Sodium Salt | BioPioneer | Cat#C0029 |
| Chloramphenicol | Fisher | Cat#50-213-255 |
| DMEM | GIBCO | Cat#31966021 |
| IPTG | BioPioneer | Cat#C0012 |
| Biotin | Sigma | Cat#B4501 |
| DEPC-Treated Water | Invitrogen | Cat#750023 |
| Coomassie Brilliant Blue G-250 | Bio-Rad | Cat#1610406 |
| Tricine | Sigma | Cat#T0377 |
| Sodium citrate tribasic | Sigma | Cat#C8532 |
| Ethylene glycol | Hampton Research | Cat#HR2-621 |
| Ammonium acetate | Sigma | Cat#372331 |
| Ammonium hydroxide | Fisher | Cat#A470-250 |
| Uranyl formate hydrate | EMS | Cat#22450 |
| Critical commercial assays | ||
| TransIT-LT1 Transfection Reagent | Mirus Bio | Cat#MIR2305 |
| BioLock | IBA | Cat#2-0205-250 |
| miRNeasy Micro Kit | QIAGEN | Cat#217084 |
| P6 Micro Bio-Spin columns | Bio-Rad | Cat#7326221 |
| NativePAGE 4-16% Bis-Tris Gel | Invitrogen | Cat#BN1002BOX |
| Immobilon-P PVDF Membrane | Millipore | Cat#IPVH07850 |
| CytoBuster Protein Extraction Reagent | Millipore | Cat#71009 |
| RNase Inhibitor | New England Biolabs | Cat#M0307S |
| Strep-Tactin Superflow Plus beads | QIAGEN | Cat#30004 |
| Ni-NTA Agarose beads | QIAGEN | Cat#30230 |
| Superdex 200 Increase 10/300 GL | Sigma | Cat#GE28-9909-44 |
| NEXTFLEX Small RNA-Seq Kit v3 | PerkinElmer | Cat#NOVA-5132-05 |
| MiSeq Reagent Kit v3 (150-cycle) | Illumina | Cat#MS-102-3001 |
| MAbPac SEC-1 column | Thermo Scientific | Cat#088789 |
| Carbon film 400 mesh grid | EMS | Cat#CF400-Cu |
| Deposited data | ||
| EBOV VP40 RNA-generated ring structure | This paper | PDB: 7K5L |
| EBOV VP40 DNA-generated ring structure | This paper | PDB: 7K5D |
| RNA deep sequencing data | This paper | Sequence Read Archive PRJNA665696 |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | Cat#CRL-3216 |
| Oligonucleotides | ||
| ssDNA oligos (see Table S2 for sequences) | IDT | N/A |
| ssRNA oligos (see Table S2 for sequences) | IDT | N/A |
| Recombinant DNA | ||
| pTriex5-strep | This paper | N/A |
| pTriex5-strep-VP40-WT | This paper | N/A |
| pTriex5-strep-VP40-R134A | This paper | N/A |
| pTriex5-strep-VP40-I307R | This paper | N/A |
| pET-46 Ek/LIC Vector | Novagen | Cat#71335 |
| pET-46 VP40-WT (Δ43, HIS-tagged) | Bornholdt et al., 2013 | N/A |
| pCEZ-NP-2A-VP35 | Jasenosky et al., 2010 | N/A |
| pCEZ-VP30 | Jasenosky et al., 2010 | N/A |
| pCEZ-L | Jasenosky et al., 2010 | N/A |
| pHH21-3E5E-Luc | Jasenosky et al., 2010 | N/A |
| Software and algorithms | ||
| Cutadapt | Martin, 2011 | RRID: SCR_011841;https://cutadapt.readthedocs.io/en/stable/ |
| BWA | Li and Durbin, 2009 | RRID: SCR_010910;http://bio-bwa.sourceforge.net |
| Integrative Genomics Viewer | Thorvaldsdóttir et al., 2013 | RRID: SCR_011793;https://www.broadinstitute.org/igv/ |
| UCSC Table Browser | Karolchik et al., 2004 | https://genome.ucsc.edu/cgi-bin/hgTables |
| Bedtools | Quinlan and Hall, 2010 | RRID: SCR_006646; https://github.com/arq5x/bedtools2 |
| Samtools | Li et al., 2009 | RRID: SCR_002105; https://htslib.org/ |
| HTSeq | Anders et al., 2015 | RRID: SCR_005514; https://htseq.readthedocs.io/en/master/ |
| GEM | Guo et al., 2012 | RRID: SCR_005339;https://cgs.csail.mit.edu/gem/ |
| XDS | Kabsch, 2010 | RRID: SCR_015652;https://xds.mr.mpg.de/ |
| AIMLESS | Evans and Murshudov, 2013 | RRID: SCR_015747; https://www.mrc-lmb.cam.ac.uk/harry/pre/aimless.html |
| CCP4 | Winn et al., 2011 | RRID: SCR_007255; https://www.ccp4.ac.uk/ |
| autoPROC | Vonrhein et al., 2011 | RRID: SCR_015748; https://www.globalphasing.com/autoproc/ |
| Phaser | McCoy et al., 2007 | RRID: SCR_014219; https://www.phenix-online.org/documentation/reference/phaser.html |
| Coot | Emsley et al., 2010 | RRID: SCR_014222; https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PHENIX | Liebschner et al., 2019 | RRID: SCR_014224;https://www.phenix-online.org |
| Phenix.refine | Adams et al., 2010 | RRID: SCR_016736 |
| Protein-Ligand Interaction Profiler (PLIP) | Salentin et al., 2015 | https://plip-tool.biotec.tu-dresden.de/plip-web/plip/index |
| PyMOL version 2.3 | Schrödinger, LLC | RRID: SCR_000305;https://www.schrodinger.com/pymol |
| UCSF Chimera | Pettersen et al., 2004 | RRID: SCR_004097;https://www.cgl.ucsf.edu/chimera/ |
| Xcalibur version 4.2 | Thermo Scientific | RRID: SCR_014593 |
| Intact Mass version 2.7 | Bern et al., 2018 | https://proteinmetrics.com/resources/intact-mass-calculations-feature-finder-report/ |
| Origin | OriginLab | RRID: SCR_014212;https://www.originlab.com/index.aspx?go=PRODUCTS/Origin |
| BioRender | BioRender | RRID: SCR_018361;https://BioRender.com |
Highlights.
Ebola virus matrix protein VP40 can form a dimer or an RNA-binding octameric ring
3′ UTRs of host mRNA are favored for ring binding, with G/A-rich binding motifs
Small RNAs/DNAs with related motifs are sufficient to transform VP40 dimer to ring
This transformed ring has well-defined stoichiometry aligning with endogenous ring
ACKNOWLEDGMENTS
We thank Emilia Arturo for preparing protein crystals for structure determination. We thank the beamline scientists at the Eli Lilly LRL-CAT beamline of the Advanced Photon Source at Argonne National Laboratory and at the 12–2 beamline of the Stanford Synchrotron Radiation Lightsource. Native MS development was supported by NIH grant P41 GM128577 to V.H.W. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02–06CH11357. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02–76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41 GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
DECLARATION OF INTERESTS
Z.L.V. is now affiliated with Analytical Research and Development Mass Spectrometry, Merck & Co., and F.B. is now affiliated with Bruker Daltonik GmbH. The remaining authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108986.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bern M, Caval T, Kil YJ, Tang W, Becker C, Carlson E, Kletter D, Sen KI, Galy N, Hagemans D, et al. (2018). Parsimonious charge deconvolution for native mass spectrometry. J. Proteome Res. 17, 1216–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornholdt ZA, Noda T, Abelson DM, Halfmann P, Wood MR, Kawaoka Y, and Saphire EO (2013). Structural rearrangement of Ebola virus VP40 begets multiple functions in the virus life cycle. Cell 154, 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessen A, Volchkov V, Dolnik O, Klenk HD, and Weissenhorn W (2000). Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 19, 4228–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR, and Murshudov GN (2013). How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gc JB, Gerstman BS, Stahelin RV, and Chapagain PP (2016). The Ebola virus protein VP40 hexamer enhances the clustering of PI(4,5)P2 lipids in the plasma membrane. Phys. Chem. Chem. Phys. 18, 28409–28417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Rüth FX, Dessen A, Timmins J, Bracher A, Kolesnikowa L, Becker S, Klenk HD, and Weissenhorn W (2003). The matrix protein VP40 from Ebola virus octamerizes into pore-like structures with specific RNA binding properties. Structure 11, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Mahony S, and Gifford DK (2012). High resolution genome wide binding event finding and motif discovery reveals transcription factor spatial binding constraints. PLoS Comput. Biol. 8, e1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck AJR (2008). Native mass spectrometry: A bridge between interactomics and structural biology. Nat. Methods 5, 927–933. [DOI] [PubMed] [Google Scholar]
- Hoenen T, Volchkov V, Kolesnikova L, Mittler E, Timmins J, Ottmann M, Reynard O, Becker S, and Weissenhorn W (2005). VP40 octamers are essential for Ebola virus replication. J. Virol. 79, 1898–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Biedenkopf N, Zielecki F, Jung S, Groseth A, Feldmann H, and Becker S (2010a). Oligomerization of Ebola virus VP40 is essential for particle morphogenesis and regulation of viral transcription. J. Virol. 84, 7053–7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Jung S, Herwig A, Groseth A, and Becker S (2010b). Both matrix proteins of Ebolavirus contribute to the regulation of viral genome replication and transcription. Virology 403, 56–66. [DOI] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, and Haussler D (2006). The UCSC Known Genes. Bioinformatics 22, 1036–1046. [DOI] [PubMed] [Google Scholar]
- Jasenosky LD, Neumann G, Lukashevich I, and Kawaoka Y (2001). Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 75, 5205–5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasenosky LD, Neumann G, and Kawaoka Y (2010). Minigenome-based reporter system suitable for high-throughput screening of compounds able to inhibit Ebolavirus replication and/or transcription. Antimicrob. Agents Chemother. 54,3007–3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W (2010). XDS. A 66, 125–132, cta Crystallogr. D Biol. Crystallogr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, and Kent WJ (2004). The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katta V, and Chait BT (1991). Observation of the heme-globin complex in native myoglobin by electrospray-ionization mass spectrometry. J. Am. Chem. Soc. 113, 8534–8535. [Google Scholar]
- Kuersten S, and Goodwin EB (2003). The power of the 3′ UTR: Translational control and development. Nat. Rev. Genet. 4, 626–637. [DOI] [PubMed] [Google Scholar]
- Kuhn JH (2008). Filoviruses. A compendium of 40 years of epidemiological, clinical, and laboratory studies. Arch. Virol. Suppl. 20, 13–360. [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Bur-rows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Sub-group (2009). The Sequence Alignment/Map format and SAM tools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liko I, Allison TM, Hopper JT, and Robinson CV (2016). Mass spectrometry guided structural biology. Curr. Opin. Struct. Biol. 40, 136–144. [DOI] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. [Google Scholar]
- Marzluff WF, and Koreski KP (2017). Birth and death of histone mRNAs. Trends Genet. 33, 745–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Sagara H, Suzuki E, Takada A, Kida H, and Kawaoka Y (2002). Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 76, 4855–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal RG, Ruthel G, Kenny TA, Kallstrom GH, Lane D, Badie SS, Li L, Bavari S, and Aman MJ (2003). In vivo oligomerization and raft localization of Ebola virus protein VP40 during vesicular budding. Proc. Natl. Acad. Sci. USA 100, 15936–15941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavadai E, Bhattarai N, Baral P, Stahelin RV, Chapagain PP, and Gerstman BS (2019). Conformational flexibility of the protein-protein interfaces of the ebola virus VP40 structural matrix filament. J. Phys. Chem. B 123, 9045–9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabi FA, Al Zoubi MS, Kasasbeh GA, Salameh DM, and Al-Nasser AD (2020). Sars-cov-2 and coronavirus disease 2019: What we know so far. Pathogens 9, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzimanowski J, Effantin G, and Weissenhorn W (2014). Conformational plasticity of the Ebola virus matrix protein. Protein Sci. 23, 1519–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruigrok RW, Schoehn G, Dessen A, Forest E, Volchkov V, Dolnik O, Klenk HD, and Weissenhorn W (2000). Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J. Mol. Biol. 300, 103–112. [DOI] [PubMed] [Google Scholar]
- Salentin S, Schreiber S, Haupt VJ, Adasme MF, and Schroeder M (2015). PLIP: Full yautomated protein-ligand interaction profiler. Nucleic Acids Res. 43 (W1), W443–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A, Kiley MP, Holloway BP, and Auperin DD (1993). Sequence analysis of the Ebola virus genome: Organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 29, 215–240. [DOI] [PubMed] [Google Scholar]
- Sarni S, Biswas B, Liu S, Olson ED, Kitzrow JP, Rein A, Wysocki VH, and Musier-Forsyth K (2020). HIV-1 Gag protein with or without p6 specifically dimerizes on the viral RNA packaging signal. J. Biol. Chem. 295, 14391–14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, and von Jagow G (1991). Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Bio-chem. 199, 223–231. [DOI] [PubMed] [Google Scholar]
- Scianimanico S, Schoehn G, Timmins J, Ruigrok RH, Klenk HD, and Weissenhorn W (2000). Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J. 19, 6732–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D, and Gilbert W (1988). Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334, 364–366. [DOI] [PubMed] [Google Scholar]
- Thomas R, Thomas S, Holloway AK, and Pollard KS (2017). Features that define the best ChIP-seq peak calling algorithms. Brief. Bioinform. 18, 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer(IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmins J, Schoehn G, Kohlhaas C, Klenk HD, Ruigrok RW, and Weissenhorn W (2003). Oligomerization and polymerization of the filovirus matrix protein VP40. Virology 312, 359–368. [DOI] [PubMed] [Google Scholar]
- VanAernum ZL, Gilbert JD, Belov ME, Makarov AA, Horning SR, and Wysocki VH (2019). Surface-induced dissociation of noncovalent protein complexes in an extended mass range Orbitrap mass spectrometer. Anal. Chem. 91, 3611–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanAernum ZL, Busch F, Jones BJ, Jia M, Chen Z, Boyken SE, Sa-hasrabuddhe A, Baker D, and Wysocki VH (2020). Rapid online buffer exchange for screening of proteins, protein complexes and cell lysates by native mass spectrometry. Nat. Protoc. 15, 1132–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, and Bricogne G (2011). Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman H, and Saphire EO (2016). More than meets the eye: Hidden structures in the proteome. Annu. Rev. Virol. 3, 373–386. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Blagev D, Pollack JL, and Erle DJ (2011). Toward a systematic understanding of mRNA 3′ untranslated regions. Proc. Am. Thorac. Soc. 8, 163–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates generated during this study are deposited to the RCSB Protein Data Bank. EBOV VP40 RNA-generated ring structure, PDB: 7K5L. EBOV VP40 DNA-generated ring structure, PDB: 7K5D.
Deep sequencing data can be found in the Sequence Read Archive database under project PRJNA665696.


