Abstract
Objective
Autoimmune diseases affect women disproportionately more than men. Estrogen is implicated in immune cell dysfunction, yet its precise molecular roles are not fully known. We recently identified new roles for serine/arginine‐rich splicing factor 1 (SRSF1) in T cell function and autoimmunity. SRSF1 levels are decreased in T cells from patients with systemic lupus erythematosus (SLE) and are associated with active disease and comorbidity. However, the molecular mechanisms that control SRSF1 expression are unknown. Srsf1 messenger RNA (mRNA) has a long 3′‐untranslated region (3′‐UTR), suggesting posttranscriptional control. This study was undertaken to investigate the role of estrogen and posttranscriptional mechanisms of SRSF1 regulation in T cells and SLE.
Methods
In silico bioinformatics analysis of Srsf1–3′‐UTR revealed multiple microRNA (miRNA; miR)–binding sites. Additional screening and literature searches narrowed down hsa‐miR‐10b‐5p for further study. Peripheral blood T cells from healthy individuals and SLE patients were evaluated for mRNA and miRNA expression by quantitative reverse transcription–polymerase chain reaction, and SRSF1 protein levels were assessed by immunoblotting. T cells were cultured with β‐estradiol, and transient transfections were used to overexpress miRNAs. Luciferase assays were used to measure 3′‐UTR activity.
Results
We demonstrated that estrogen increased hsa‐miR‐10b‐5p expression in human T cells, and hsa‐miR‐10b‐5p down‐regulated SRSF1 protein expression. Mechanistically, hsa‐mir‐10b‐5p regulated SRSF1 posttranscriptionally via control of its 3′‐UTR activity. Importantly, hsa‐miR‐10b‐5p expression levels were elevated in T cells from healthy women compared to healthy men and also elevated in T cells from SLE patients.
Conclusion
We identified a previously unrecognized molecular link between estrogen and gene regulation in immune cells, with potential relevance to systemic autoimmune disease.
INTRODUCTION
Systemic lupus erythematosus (SLE) is a debilitating autoimmune disease that disproportionately affects women (1), and is the fifth leading cause of death in young women (2). Loss of immune tolerance to self antigens leads to a hyperactive immune response and destruction of target organs. SLE typically develops in women during the reproductive years, and flares frequently occur during pregnancy; therefore, sex hormones are implicated in SLE pathogenesis. Estrogens contribute to the immune dysregulation underlying autoimmunity and disease manifestations (3), yet the precise molecular events controlled by estrogen are unknown.
Besides transcriptional control through classic estrogen receptor–linked pathways and nonclassic pathways, estrogen also controls target genes through posttranscriptional mechanisms (4, 5). Posttranscriptional regulation is a powerful mechanism of gene control and involves modulation of messenger RNA (mRNA) stability and/or translation through regulatory elements frequently found within the 3′‐untranslated region (3′‐UTR) and conserved through species. Genes with a long 3′‐UTR are therefore considered to be significantly regulated at the posttranscriptional level. Besides cis elements, trans‐factors include proteins and noncoding RNA, which bind to and control mRNA expression. MicroRNAs (miRNAs; miRs) are short noncoding RNAs (~23 nucleotides in length) that regulate genes by binding complementary sites within the 3′‐UTR of target genes and consequently destabilize mRNA and/or hinder translation. In T cells, miRNAs are crucial for regulating T cell development, differentiation, and function. Therefore, defects in miRNA levels lead to immune dysfunction (6). Aberrant expression of miRNAs is implicated in autoimmune diseases, including SLE. While studies have demonstrated sex‐based factors in miRNA expression and have implicated miRNAs in SLE, the role of estrogen in regulating specific miRNA and downstream genes involved in T cell dysfunction is not fully known.
We have recently identified novel roles of the prototypical splicing regulator, serine/arginine‐rich splicing factor 1 (SRSF1), in T cells and systemic autoimmunity. Using discovery approaches, we identified SRSF1 in a pull‐down assay of proteins binding the mRNA of the T cell receptor–associated CD3 ζ‐chain (7). We demonstrated that SRSF1 controls genes involved in T cell signaling and cytokine production (7, 8), and mice with T cell–restricted deficiency of SRSF1 display T cell hyperactivity and develop systemic autoimmunity (9). SRSF1 levels are decreased in SLE patients and are associated with active disease and comorbidity (8, 10). These studies have demonstrated the relevance of low SRSF1 levels in SLE. Yet, how SRSF1 is regulated in T cells, and whether hormones contribute to its expression, is unknown. Besides transcriptional mechanisms, SRSF1 is controlled via posttranscriptional mechanisms, and this is exemplified by its long 3′‐UTR.
In the present study, we used in silico bioinformatics analysis and literature search approaches to screen for miRNAs targeting SRSF1, and we investigated the influence of estrogen on the selected hsa‐miR‐10b‐5p. We demonstrated that hsa‐miR‐10b‐5p overexpression leads to down‐regulation of SRSF1 through control of its 3′‐UTR activity. Estrogen exposure leads to increased hsa‐miR‐10b‐5p expression in human T cells. Importantly, T cells from healthy women exhibit significantly increased levels of hsa‐miR‐10b‐5p compared to healthy men. Furthermore, hsa‐miR‐10b‐5p levels are elevated in SLE patients regardless of disease activity. Thus, our study provides evidence of hormone‐ and miRNA‐linked posttranscriptional regulation of a key regulator (SRSF1) and insight into its reduced expression in systemic autoimmune disease. These findings reveal molecular links between hormones and gene regulation in the immune system.
MATERIALS AND METHODS
Human subjects
Subjects were recruited from the rheumatology clinic at Beth Israel Deaconess Medical Center. All SLE patients fulfilled 4 of the European Alliance of Associations for Rheumatology/American College of Rheumatology criteria for SLE (11). Written informed consent was obtained from all participants. The study was approved by the local institutional review board.
Cell lines, antibodies, and reagents
HEK 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (complete DMEM), at 37°C with 5% CO2. SRSF1 antibody (clone 96) was obtained from Life Technologies, horseradish peroxidase (HRP)–conjugated secondary antibodies from Santa Cruz Biotechnology, anti‐CD3 (clone OKT3) from Bio X Cell, anti‐CD28 from BioLegend, goat anti‐mouse crosslinkers from EMD Millipore, β‐estradiol powder from Sigma‐Aldrich, and mirVana mimics from ThermoFisher.
Estrogen experiments and T cell activation
Peripheral blood was collected from premenopausal healthy women during the follicular phase of the menstrual cycle. Total T cells were purified by negative selection using a RosetteSep Human T cell kit (StemCell Technologies) and cultured in RPMI 1640 without phenol red, supplemented with charcoal‐dextran–stripped FBS (ThermoFisher), and cultured with reconstituted β‐estradiol at 37°C. To activate T cells, cells were cultured in complete RPMI and stimulated with soluble anti‐CD3 (5 μg/ml), anti‐CD28 (2.5 μg/ml), and crosslinker (2.5 μg/ml).
MicroRNA transfections
HEK 293T cells were seeded in complete DMEM overnight. During transfection, complete DMEM was replaced with DMEM without FBS. MicroRNA mimics were transfected using Lipofectamine 2000 (Invitrogen). FBS was added again to cells 2 hours prior to collection.
Luciferase assays
Srsf1 3′‐UTR was cloned into Topo‐TA cloning vector and subcloned into pmirGLO luciferase vector (Promega). HEK 293T cells were cotransfected with 1 μg of pmirGLO‐SRSF1‐3′‐UTR luciferase construct and 10 nM of miRNA mimic. As an internal control, pRL‐TK Renilla luciferase construct (25 ng) was included. Cells were lysed, and luciferase activity was measured by dual‐luciferase assay (Promega).
Quantitative reverse transcription–polymerase chain reaction (qRT‐PCR)
Total RNA was isolated using a miRNeasy kit (Qiagen) and reverse‐transcribed with RNA‐to‐complementary DNA EcoDry premix (Clontech). Real‐time qRT‐PCR was performed with LightCycler 480 SYBR Green I Master Mix (Roche) under the following conditions: initial denaturation at 95°C for 5 minutes, 40 cycles of amplification; denaturation at 95°C for 15 seconds, annealing at 60°C for 15 seconds, extension at 72°C for 30 seconds; 1 cycle of melting curves at 95°C for 5 seconds, 65°C for 2 minutes, and 97°C (continuously); and final cooling. Threshold cycle (Ct) values were used to calculate relative expression by the ΔCt quantification method. The following primer sequences were used: SRSF1 forward 5ʹ‐TCTCTGGACTGCCTCCAAGT‐3ʹ, reverse 5ʹ‐GGCTTCTGCTACGACTACGG‐3ʹ; hsa‐miR‐10b‐5p forward 5ʹ‐CGCCTGCTTGGTAACCCTGACC‐3ʹ, reverse 5ʹ‐GGGTCCCACCCAGAGTGAGGT‐3ʹ; U6 forward 5ʹ‐CTCGCTTCGGCAGCACA‐3ʹ, reverse 5ʹ‐AACGCTTCACGAATTTGCGT‐3ʹ.
Western blotting
Cells were lysed in radioimmunoprecipitation assay buffer (Boston BioProducts) with protease inhibitors (Roche). Lysates were resolved in 4–12% Bis‐Tris sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and transferred to PVDF membranes. Membranes were blocked with 5% nonfat milk in Tris buffered saline–Tween 20 (0.05%) (TBST) for 1 hour, incubated with a primary antibody (1:1,000 or 1:4,000 β‐actin) overnight at 4°C or for 1 hour at room temperature (β‐actin), washed 3 times with TBST, incubated with an HRP‐conjugated secondary antibody (1:2,000) for 1 hour at room temperature, washed 3 times with TBST, developed using enhanced chemiluminescence reagents, and analyzed with a Bio‐Rad ChemiDoc‐XRS imaging system. Densitometry analysis was performed using Quantity One software (Bio‐Rad).
Statistical analysis
Data analysis was performed using GraphPad Prism software. Student’s 2‐tailed t‐test and linear regression were used for statistical analysis. P values less than 0.05 were considered significant.
RESULTS
In silico analysis and screening for miRNAs targeting Srsf1
The Srsf1 mRNA has a long (~4,000 bp) 3′‐UTR suggesting that its expression is controlled at the posttranscriptional level. MicroRNAs silence genes posttranscriptionally either by destabilizing mRNA or by hindering protein translation through binding complementary sites within target mRNA. Therefore, we investigated the role of miRNA on Srsf1 gene regulation. We performed a bioinformatics analysis of the Srsf1 3′‐UTR to identify miRNA‐binding sites using online prediction tools (TargetScan and the miRSystem), combined with a literature search using NCBI/PubMed (Figures 1A and D). First, we used TargetScan to screen the Srsf1 3′‐UTR, which yielded a set of putative miRNAs targeting SRSF1. The Context++ Score percentile provided by TargetScan combines information, including site type, 3ʹ supplementary pairing, distance from the end of the 3′‐UTR, and AU content, to predict the strength of binding between the miRNA and target mRNA. Of the miRNA–mRNA seed sequence match sites, the 8mer is the strongest site type. To supplement this investigation, we performed a literature search using specific search criteria limited to curate miRNAs that have previously been implicated in SLE, that are up‐regulated by estrogen or its metabolites, that are up‐regulated in T cells, or that are shown to control SRSF1. To further narrow down miRNAs of interest, we used the miRSystem, which evaluates the miRNA–mRNA relationship by integrative analyses of 7 different miRNA prediction databases. The “hits” column in Figure 1D represents the number of databases, according to the miRSystem, in which a relationship between Srsf1 and the miRNA is mentioned.
Figure 1.
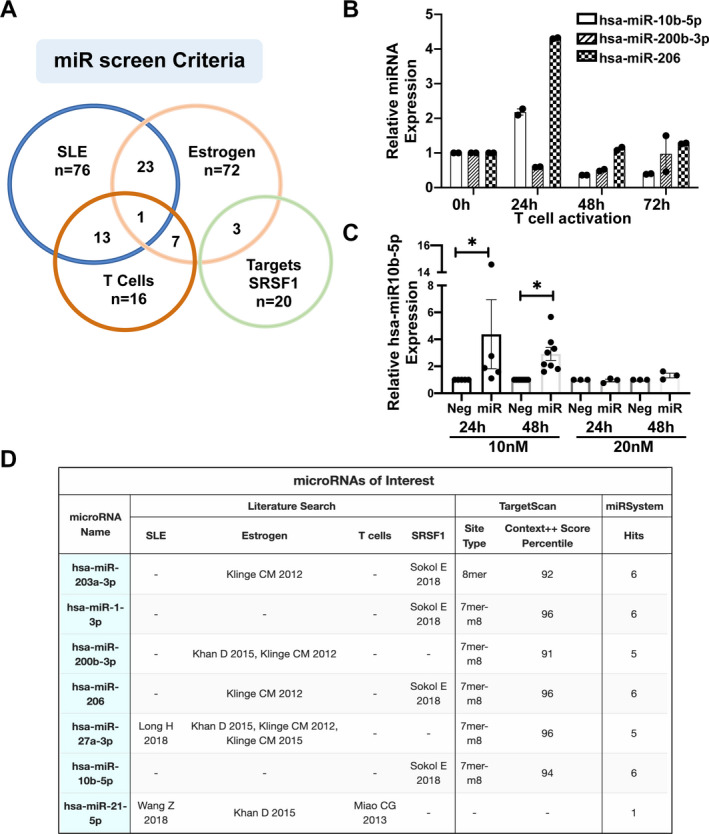
In silico screening and selection of microRNA (miRNA; miR) targeting Srsf1. A, Web‐based bioinformatics prediction tools (TargetScan and miRSystem) were used to screen for microRNAs targeting Srsf1, combined with a literature search to curate miRNAs that are up‐regulated in systemic lupus erythematosus (SLE), up‐regulated by estrogen, up‐regulated in T cells, and/or shown to target Srsf1. B, Quantitative reverse transcription–polymerase chain reaction was performed for 3 selected miRNAs in resting T cells (0 hours) and in activated T cells (24 hours, 48 hours, and 72 hours). Each symbol represents an individual subject; bars show the mean ± SEM relative miRNA expression from duplicates. C, Negative control (Neg) or hsa‐miR‐10b‐5p mimics were transiently transfected into HEK 293T cells in 10 nM or 20 nM concentrations, and cells were collected at 24 hours or 48 hours posttransfection. Expression levels of miRNA were measured by reverse transcription–polymerase chain reaction. Each symbol represents an individual subject; bars show the mean ± SEM relative miRNA expression from at least 3 independent experiments. * = P < 0.05. D, The chart shows the miRNAs narrowed for further study based on the above‐mentioned criteria. SRSF = serine/arginine‐rich splicing factor 1.
Among the putative miRNAs identified, we selected 7 for further study based on a combination of occurrence in multiple categories of the literature search (4, 12, 13, 14, 15, 16), Context++ Score >90th percentile, and the number of database hits from the miRSystem (Figure 1D). We tested the expression of these miRNAs by real‐time qRT‐PCR in T cells. Of these, hsa‐miR‐10b‐5p, hsa‐miR‐200b‐3p, and hsa‐miR‐206 displayed quantifiable levels (Figure 1B). We further tested these 3 miRNAs for overexpression by transfection of miRNA mimics into HEK 293T cells. Of these, hsa‐miR‐10b‐5p showed reliable and consistently robust overexpression under the conditions tested (Figure 1C). Therefore, we selected hsa‐miR‐10b‐5p for further study.
Overexpression of hsa‐miR‐10b‐5p leads to decreased SRSF1 levels
To investigate the effect of hsa‐miR‐10b‐5p on SRSF1 expression, we used miRVana mimics in transient transfection experiments. We overexpressed hsa‐miR‐10b‐5p by transient transfection of the miRNA mimic into HEK 293T cells. As controls, cells were transfected with a negative control miRNA mimic. Cells were collected 48 hours posttransfection, and hsa‐miR‐10b‐5p mRNA levels were quantified by real‐time qRT‐PCR, and SRSF1 protein levels were quantified by Western blotting. We confirmed the increased levels of hsa‐miR‐10b‐5p and found that its overexpression led to decreased SRSF1 protein levels compared to controls (Figures 2A and B). These results show that hsa‐miR‐10b‐5p induces the down‐regulation of SRSF1 expression.
Figure 2.
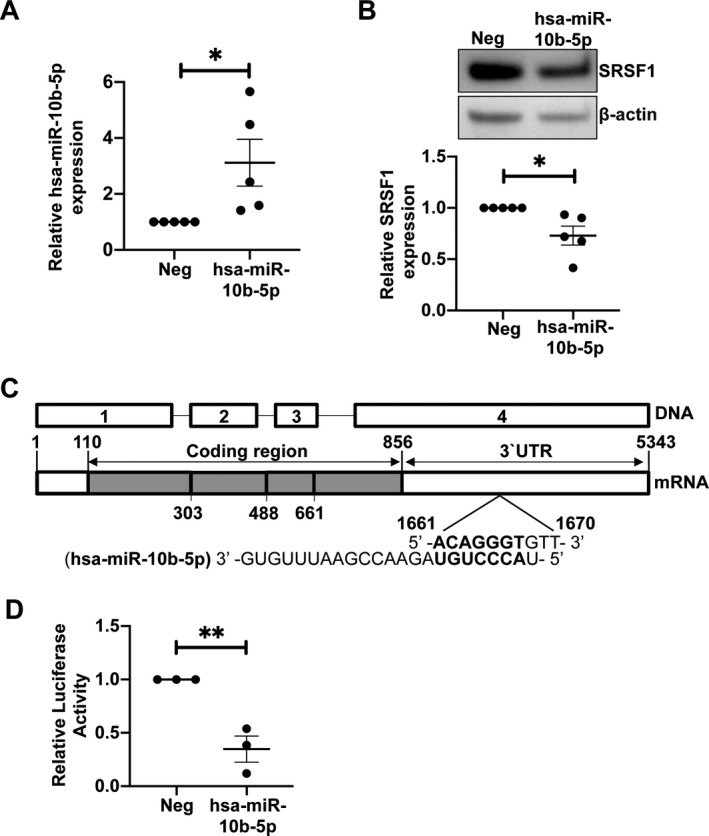
Overexpression of hsa‐miR‐10b‐5p leads to down‐regulation of SRSF1 via its 3′‐untranslated region (3′‐UTR). A, Negative control miR mimics or hsa‐miR‐10b‐5p mimics were transiently transfected into HEK 293T cells. Quantitative reverse transcription–polymerase chain reaction was performed to assess the levels of hsa‐miR‐10b‐5p. Each symbol represents an individual subject; bars show the mean ± SEM from 4 experiments. * = P < 0.05. B, Levels of SRSF1 total protein in transfected cells were assessed by Western immunoblotting. Representative blots are shown (top). Densitometric results from Western blotting were quantified as the expression of SRSF1 protein normalized to β‐actin (bottom). Each symbol represents an individual subject; bars show the mean ± SEM from 4 experiments. * = P < 0.05. C, Schematic shows DNA and mRNA sequence of Srsf1. The hsa‐miR‐10b‐5p sequence and its complementary binding site within the 3′‐UTR are shown. D, HEK 293T cells were cotransfected with the Srsf1 3ʹ‐UTR–luciferase plasmid and 10 nM of hsa‐miR‐10b‐5p mimic or negative control mimic. Cells were collected and 3ʹ‐UTR activity was measured by luciferase assay. Each symbol represents an individual subject; bars show the mean ± SEM from 3 experiments. ** = P < 0.01. See Figure 1 for other definitions.
MicroRNA hsa‐miR‐10b‐5p controls the 3′‐UTR activity of SRSF1
MicroRNAs mediate their effect at the posttranscriptional level by binding cognate sites within the 3′‐UTR and targeting the mRNA for degradation and/or inhibition of translation. Because the overexpression of hsa‐miR‐10b‐5p led to decreased SRSF1 expression (Figure 2B) and Srsf1 mRNA has an hsa‐miR‐10b‐5p binding site within its 3′‐UTR (Figure 2C), we questioned whether hsa‐miR‐10b‐5p regulates the activity of the Srsf1 3′‐UTR. To evaluate this, we used an Srsf1 3′‐UTR–luciferase reporter plasmid construct and cotransfected it with either a negative control mimic or an hsa‐miR‐10b‐5p mimic. We collected cells 48 hours posttransfection and measured the 3′‐UTR activity by luciferase reporter assays. We found that hsa‐miR‐10b‐5p overexpression led to significantly decreased 3′‐UTR luciferase activity compared to the negative control (Figure 2D), suggesting that hsa‐miR‐10b‐5p binds to and controls the activity of the Srsf1 3′‐UTR, and thus controls SRSF1 expression.
Estrogen up‐regulates hsa‐miR‐10b‐5p in human T cells
Given that estrogen is known to control miRNAs in immune cells and our findings showed that hsa‐miR‐10b‐5p controls SRSF1 expression (Figure 2), we wished to examine the relationship between estrogen, miRNA, and SRSF1. To evaluate the influence of estrogen on hsa‐miR‐10b‐5p, we obtained peripheral blood T cells from healthy women during the follicular phase of the menstrual cycle. Circulating estrogen levels are lowest during this phase, and ensuring this criterion enabled comparison between individuals. We treated T cells in vitro with increasing concentrations of β‐estradiol and measured the expression of hsa‐miR‐10b‐5p by qRT‐PCR. Our results demonstrate that exposure to β‐estradiol increases hsa‐miR‐10b‐5p expression levels in a dose‐dependent manner (Figure 3A). These results indicate that hsa‐miR‐10b‐5p is responsive to estrogen and may be involved in the downstream gene regulation mediated by estrogen in T cells.
Figure 3.
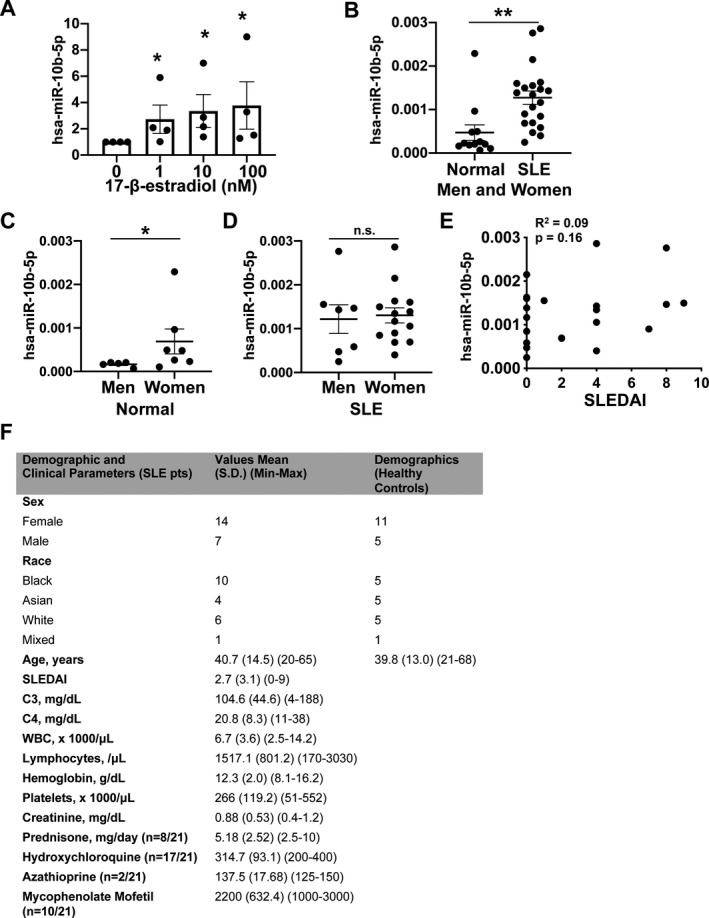
Induction of hsa‐miR‐10b‐5p by estrogen and elevated levels of hsa‐miR‐10b‐5p in T cells from healthy women and systemic lupus erythematosus (SLE) patients. A, T cells from healthy women were treated with increasing concentrations of β‐estradiol, and hsa‐miR‐10b‐5p levels were assessed by real‐time quantitative reverse transcription–polymerase chain reaction. Results are from 4 experiments. * = P < 0.05 versus no β‐estradiol. B, Levels of hsa‐miR‐10b‐5p were measured by quantitative reverse transcription–polymerase chain reaction in T cells from SLE patients (n = 21) and age‐, race‐, and sex‐matched healthy controls (n = 12). ** = P < 0.01. C, Levels of hsa‐miR‐10b‐5p in healthy men and healthy women are shown. * = P < 0.05. D, Levels of hsa‐miR‐10b‐5p in men and women with SLE are shown. In A‐D, each symbol represents an individual subject; bars show the mean ± SEM. E, Correlations between Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) scores and hsa‐miR‐10b‐5p levels in patients with SLE are shown. F, Demographic and clinical features of the SLE patients and healthy controls are shown. NS = not significant; WBC = white blood cells.
Elevation of hsa‐miR‐10b‐5p in T cells from healthy women and SLE patients
We have previously shown that T cells from SLE patients display lower levels of SRSF1 compared to healthy individuals, which is associated with higher SLE Disease Activity Index (SLEDAI) scores (17) and comorbidity in patients (8, 10). To date, hsa‐miR‐10b‐5p has not been implicated in SLE. Thus, we investigated hsa‐miR‐10b‐5p expression in T cells from SLE patients compared to age‐, race‐, and sex‐matched healthy individuals. We found significantly increased levels of hsa‐miR‐10b‐5p in SLE patients compared to healthy controls (Figure 3B). To evaluate sex‐based differences, we compared hsa‐miR‐10b‐5p levels between men and women. Levels of hsa‐miR‐10b‐5p were significantly increased in healthy women compared to healthy men (Figure 3C). However, levels were similar in men and women with SLE (Figure 3D). Furthermore, hsa‐miR‐10b‐5p levels did not correlate with SLEDAI scores in patients (Figure 3E). These results indicate that hsa‐miR‐10b‐5p is up‐regulated in healthy women and in patients with SLE, and may contribute to the observed decrease in SRSF1 levels in patients with SLE. Overall, our results indicate that estrogen contributes to the induction of hsa‐miR‐10b‐5p, which down‐regulates SRSF1. Elevated hsa‐miR‐10b‐5p levels in SLE patients may therefore contribute to decreased SRSF1 expression.
DISCUSSION
In this study, we obtained a number of interesting findings. We demonstrated hsa‐mir‐10b‐5p–mediated posttranscriptional regulation of the prototypical splicing regulator SRSF1 (Figures 1 and 2). Mechanistically, we showed that hsa‐miR‐10b‐5p controls the 3′‐UTR activity of SRSF1 (Figure 2C). Furthermore, estrogen increases the expression of hsa‐miR‐10b‐5p in human T cells (Figure 3A). Finally, we showed that hsa‐miR‐10b‐5p levels are elevated in T cells from SLE patients and in T cells from healthy women compared to healthy men (Figures 3B and C).
These findings are significant because SRSF1 controls genes involved in T cell function, and its expression is decreased in SLE patients, which correlates with higher SLEDAI scores (8) and comorbidity (10). Therefore, understanding how SRSF1 is regulated would guide strategies to correct its expression and potentially improve disease. SRSF1 is a relatively understudied molecule in the immune system. The present study elucidates mechanisms of its regulation in immune cells with implications for autoimmune disease. We showed, for the first time to our knowledge, that T cells from SLE patients display higher expression of hsa‐miR‐10b‐5p. These results imply that hsa‐miR‐10b‐5p may contribute to low SRSF1 levels in SLE. Furthermore, we showed that hsa‐miR‐10b‐5p is estrogen‐responsive in human T cells, thus highlighting its role in the immune system. We also demonstrated that hsa‐miR‐10b‐5p levels are elevated in healthy women compared to healthy men. Elevated hsa‐miR‐10b‐5p levels in SLE implicate its contribution to autoimmune disease. Given its regulation by estrogen, our data showing that hsa‐miR‐10b‐5p is elevated in healthy women compared to healthy men identify a sex‐based difference in the physiologic regulation of this miRNA. After disease onset in SLE patients, multiple factors may be involved, and therefore the sex‐based difference may be obviated. Furthermore, hsa‐miR‐10b‐5p expression in SLE does not correlate with disease activity, suggesting that this may be inherent to the disease process rather than due to nonspecific immune activation.
Besides posttranscriptional regulation of miRNA, posttranslational mechanisms may be involved in regulating SRSF1 expression. We have previously demonstrated ubiquitin‐proteasome–mediated degradation of SRSF1 and ubiquitinated SRSF1 forms in SLE T cells. Lysosome‐mediated proteolysis is another mechanism that may be involved in SRSF1 regulation. Recently, the X‐linked gene Cxorf21, which is involved in lysosomal pH regulation and proteolysis, was found to have single‐nucleotide polymorphisms associated with SLE (18). SRSF1 is also known to autoregulate its expression via binding to its 3′‐UTR which leads to nonproductive splicing or non–sense‐mediated mRNA decay. In addition, SRSF1 controls miRNA expression, and these miRNAs may contribute to a feedback regulatory loop. It would be interesting to investigate whether these proteolytic mechanisms or autoregulatory feedback loops occur in T cells and in systemic autoimmunity.
One limitation of the present study is that we focused on hsa‐miR‐10b‐5p, and other miRNAs may be involved in SRSF1 regulation and controlled by estrogen. Indeed, our miRNA screen revealed potential hits that may be relevant for future studies. Differences in hsa‐miR‐10b‐5p levels in total SLE T cells may result from differences in proportions of CD4/CD8 and other cell lineages, and therefore examination of subset‐specific expression is important. Evaluating hsa‐miR‐10b‐5p in other autoimmune diseases would elucidate whether this is specific to SLE or shared by multiple autoimmune diseases.
In conclusion, we have identified sex hormone–linked control of hsa‐miR‐10b‐5p and SRSF1 in immune cells. This finding reveals molecular links between hormones and the immune system with relevance to autoimmunity.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Ms Ramanujan had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design
Ramanujan, Moulton.
Acquisition of data
Ramanujan, Cravens, Krishfield, Kyttaris, Moulton.
Analysis and interpretation of data
Ramanujan, Cravens, Krishfield, Kyttaris, Moulton.
ACKNOWLEDGMENTS
We would like to thank Dr. George C. Tsokos for critical reading of the manuscript and access to patient samples, and Dr. Nobuya Yoshida for helpful technical advice.
Supported by the NIH (National Institute of Arthritis and Musculoskeletal and Skin Diseases grant R01‐AR‐068974 and National Institute of Allergy and Infectious Diseases grant R01‐AI‐049954).
Dr. Kyttaris has received research support from Exagen Diagnostics, AbbVie, and Takeda. No other disclosures relevant to this article were reported.
References
- 1. Moulton VR. Sex hormones in acquired immunity and autoimmune disease. Front Immunol 2018;9:2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yen EY, Singh RR. Lupus—an unrecognized leading cause of death in young females: a population‐based study using nationwide death certificates, 2000–2015. Arthritis Rheumatol 2018;70:1251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Corradetti C, Jog NR, Cesaroni M, Madaio M, Caricchio R. Estrogen receptor α signaling exacerbates immune‐mediated nephropathies through alteration of metabolic activity. J Immunol 2018;200:512–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klinge CM. miRNAs regulated by estrogens, tamoxifen, and endocrine disruptors and their downstream gene targets [review]. Mol Cell Endocrinol 2015;418:273–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rider V, Abdou NI, Kimler BF, Lu N, Brown S, Fridley BL. Gender bias in human systemic lupus erythematosus: a problem of steroid receptor action? Front Immunol 2018;9:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jeker LT, Bluestone JA. microRNA regulation of T‐cell differentiation and function [review]. Immunol Rev 2013;253:65–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moulton VR, Tsokos GC. Alternative splicing factor/splicing factor 2 regulates the expression of the ζ subunit of the human T cell receptor‐associated CD3 complex. J Biol Chem 2010;285:12490–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moulton VR, Grammatikos AP, Fitzgerald LM, Tsokos GC. Splicing factor SF2/ASF rescues IL‐2 production in T cells from systemic lupus erythematosus patients by activating IL‐2 transcription. Proc Natl Acad Sci U S A 2013;110:1845–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Katsuyama T, Li H, Comte D, Tsokos GC, Moulton VR. Splicing factor SRSF1 controls T cell hyperactivity and systemic autoimmunity. J Clin Invest 2019;129:5411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Katsuyama T, Martin‐Delgado IJ, Krishfield SM, Kyttaris VC, Moulton VR. Splicing factor SRSF1 controls T cell homeostasis and its decreased levels are linked to lymphopenia in systemic lupus erythematosus. Rheumatology (Oxford) 2020;59:2146–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aringer M, Costenbader K, Daikh D, Brinks R, Mosca M, Ramsey‐Goldman R. 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Arthritis Rheum 2019;71:1400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Long H, Wang X, Chen Y, Wang L, Zhao M, Lu Q. Dysregulation of microRNAs in autoimmune diseases: pathogenesis, biomarkers and potential therapeutic targets [review]. Cancer Lett 2018;428:90–103. [DOI] [PubMed] [Google Scholar]
- 13. Wang Z, Heid B, Dai R, Ahmed SA. Similar dysregulation of lupus‐associated miRNAs in peripheral blood mononuclear cells and splenic lymphocytes in MRL/lpr mice. Lupus Sci Med 2018;5:e000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khan D, Dai R, Ahmed SA. Sex differences and estrogen regulation of miRNAs in lupus, a prototypical autoimmune disease. Cell Immunol 2015;294:70–9. [DOI] [PubMed] [Google Scholar]
- 15. Miao C, Yang Y, He X, Huang C, Huang Y, Zhang L, et al. The emerging role of microRNAs in the pathogenesis of systemic lupus erythematosus. Cell Signal 2013;25:1828–36. [DOI] [PubMed] [Google Scholar]
- 16. Sokół E, Kędzierska H, Czubaty A, Rybicka B, Rodzik K, Tański Z, et al. microRNA‐mediated regulation of splicing factors SRSF1, SRSF2 and hnRNP A1 in context of their alternatively spliced 3′UTRs. Exp Cell Res 2018;363:208–17. [DOI] [PubMed] [Google Scholar]
- 17. Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH, and the Committee on Prognosis Studies in SLE . Derivation of the SLEDAI: a disease activity index for lupus patients. Arthritis Rheum 1992;35:630–40. [DOI] [PubMed] [Google Scholar]
- 18. Harris VM, Harley ITW, Kurien BT, Koelsch KA, Scofield RH. Lysosomal pH is regulated in a sex dependent manner in immune cells expressing CXorf21 . Front Immunol 2019;10:578. [DOI] [PMC free article] [PubMed] [Google Scholar]


