Abstract
Background
The role of methylation in pancreatic cancer (PC) risk remains unclear. We integrated genome and methylome data to identify CpG sites (CpGs) with the genetically predicted methylation to be associated with PC risk. We also studied gene expression to understand the identified associations.
Methods
Using genetic data and white blood cell methylation data from 1,595 subjects of European descent, we built genetic models to predict DNA methylation levels. After internal and external validation, we applied prediction models with satisfactory performance to the genetic data of 8,280 PC cases and 6,728 controls of European ancestry to investigate the associations of predicted methylation with PC risk. For associated CpGs, we compared their measured levels in pancreatic tumor vs benign tissue.
Results
We identified 45 CpGs at nine loci showing an association with PC risk, including 15 CpGs showing an association independent from identified risk variants. We observed significant correlations between predicted methylation of 16 of the 45 CpGs and predicted expression of eight adjacent genes, of which six genes showed associations with PC risk. Of the 45 CpGs, we were able to compare measured methylation of 16 in pancreatic tumor versus benign pancreatic tissue. Of them, six showed differentiated methylation.
Conclusions
We identified methylation biomarker candidates associated with PC using genetic instruments and added additional insights into the role of methylation in regulating gene expression in PC development.
Impact:
A comprehensive study using genetic instruments identifies 45 CpG sites at nine genomic loci for PC risk.
Keywords: DNA methylation, genetic instrument, pancreatic cancer
Introduction
As the most fatal malignancy of all major cancers, pancreatic cancer is the third leading cause of cancer death in the United States (US) with an overall 5-year survival rate of only 9% [1]. Furthermore, distinct from other common cancers, the mortality from pancreatic cancer is expected to continue to increase and may develop into the second leading cause of cancer death before 2030 [2]. One of the major reasons for the lethality of this disease is that most pancreatic cancer patients are diagnosed late due to nonspecific symptoms in earlier stages. Unfortunately, up till now, there are no effective screening tests available for pancreatic cancer. Serum CA 19–9 is the only validated biomarker that is clinically used for pancreatic cancer diagnosis in symptomatic patients or for prognostic surveillance in predicting tumor stage or overall survival. However, this biomarker alone cannot serve as an effective screening tool given its unsatisfactory sensitivity (75.5%) and specificity (77.6%), as well as the inferior positive predictive value (0.5%−0.9%) [3]. There are urgent needs to identify additional biomarkers for improved risk assessment of pancreatic cancer.
DNA methylation, an important epigenetic modification that regulates gene expression, has been shown to be potentially related to pancreatic cancer. A number of studies evaluating DNA methylation levels in blood or pancreas tissue have identified multiple candidate DNA methylation markers for pancreatic cancer, including methylation at VHL, MYF3, TMS, GPC3, SRBC, HYAL2, ADAMTS1, BNC1, SERPINB5, and B3GALT5 [4–8]. However, many of these earlier studies involved a small sample size and only investigated a few CpG sites (CpGs), resulting in insufficient statistical power and limited scope for identifying discriminant DNA methylation markers. More importantly, previous studies using a conventional study design would be difficult to establish causality.
It has been increasingly recognized that one potential strategy for reducing several of these limitations is to evaluate the associations of interest using genetic instruments. The genetically determined proportion of DNA methylation levels should be less susceptible to these biases, given the random assortment of alleles from parents to offspring during the production of gametes. Studies have suggested there is high heritability for a large portion of CpGs, and multiple associations have been identified between genetic variants and DNA methylation levels of CpGs [9–12]. In a large study with sufficient power, many of the DNA methylation associated genetic variants are likely to serve as strong instrument variables for assessing the association between DNA methylation and pancreatic cancer risk. In the current study, we employed such a novel strategy to identify DNA methylation biomarker candidates associated with pancreatic cancer risk.
Besides identifying promising biomarkers, the findings of such a study may also help better understand the etiology of pancreatic cancer. So far, genome-wide association studies (GWAS) have identified 20 independent common susceptibility loci for pancreatic cancer in individuals of European ancestry, however, together these variants can only explain a small proportion of the total risk [13–18]. Recent work estimated the heritability of pancreatic cancer to be 21.2% [19]. A large proportion of the pancreatic cancer heritability remains unexplained [20]. Recently, two large transcriptome-wide association studies (TWAS) of pancreatic cancer were conducted. In these studies 31 candidate susceptibility genes, of which the genetically-predicted expression was associated with pancreatic cancer risk, were identified [21]. The current study represents another endeavor focusing on studying DNA methylation, the findings of which may contribute to additional understanding of pancreatic cancer genetics. These CpGs may influence pancreatic cancer risk either through regulating expression of pancreatic cancer susceptibility genes or through other mechanisms. In the current work we also studied gene expression aiming to characterize whether some of the identified associated CpGs may influence pancreatic cancer risk through regulating expression of their target genes.
As far as we know, this study is the first large study to evaluate the association between genetically-predicted DNA methylation and pancreatic cancer risk, using data of 8,280 cases and 6,728 controls of European descendants from Pancreatic Cancer Cohort Consortium (PanScan) and Pancreatic Cancer Case-Control Consortium (PanC4). For the identified associated DNA methylation biomarker candidates, we further compared their directly measured levels in pancreatic tumor tissue specimens (n=18) versus benign pancreatic tissue specimens (n=18).
Methods
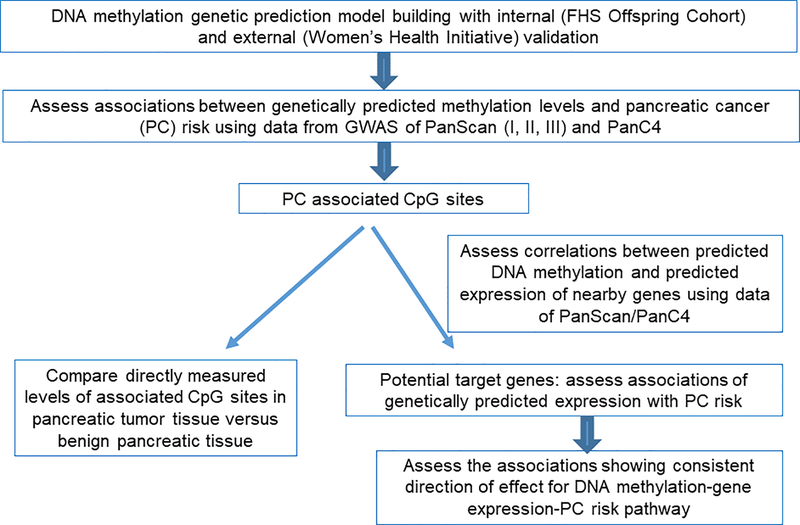
The overall study design is shown in Figure 1. Firstly, we developed genetic prediction models for DNA methylation levels by leveraging data of the Framingham Heart Study (FHS). After external validation, we selected DNA methylation models with satisfactory prediction performance for assessing associations of genetically predicted methylation levels with pancreatic cancer risk, by using data of the PanScan/PanC4 consortia which involves 8,280 cases and 6,728 controls. For CpGs showing an association with pancreatic cancer risk, we assessed correlations between their predicted methylation and predicted expression of adjacent genes (PanScan/PanC4), to identify potential target genes of these CpGs. For the identified candidate target genes, we further evaluated associations of their genetically predicted expression with pancreatic cancer risk. For the associated CpGs, we also compared their directly measured levels in pancreatic tumor tissue versus benign pancreatic tissue. Additional description of relevant studies was included in the Supplementary Material.
Figure 1.
Study design flow chart. PanScan, Pancreatic Cancer Cohort Consortium (PanScan); PanC4, Pancreatic Cancer Case-Control Consortium.
DNA methylation prediction models
Genetic data and white blood cell DNA methylation data of a total of 1,595 unrelated subjects from the FHS Offspring Cohort were used for methylation genetic prediction model building. The detailed information for the datasets and data quality control (QC), has been described elsewhere [22–24]. The genetic data were imputed to the Haplotype Reference Consortium reference panel. Single nucleotide polymorphisms (SNPs) with high imputation quality (R2 ≥ 0.8), minor allele frequency (MAF) ≥ 5%, and those included in the HapMap Phase 2 version and not strand ambiguous were retained. The R package “minfi” was used for the quality control (QC) and normalization of the DNA methylation data [25]. For the methylation level at each CpG site, a prediction model was built following the elastic net method (α = 0.50) using in-cis SNPs (flanking a 2 Mb window) with adjustment for age, sex, six cell type composition variables, and top ten genetic principal components (PCs). Ten-fold cross-validation was used to choose the penalty parameter lambda and validate the models internally [26]. Performance of established prediction models were also examined externally by using data from Women’s Health Initiative (WHI) (N=883), which were downloaded from dbGaP (accession numbers phs001335, phs000675 and phs000315). Identical methods were used for the imputation and QC as it was described for FHS data. DNA methylation data was processed following a similar procedure as for FHS data. We calculated the predicted DNA methylation for each CpG site using the models that were established using FHS data, and then compared the predicted methylation with the measured levels using Spearman’s correlation. DNA methylation prediction models with both internal and external performance R2 ≥ 0.01 (correlation between predicted and measured DNA methylation level > 0.1) were used for downstream association analyses. This is one of the standard criteria used in TWAS for gene expression [27–29], heritability of which is in similar range to that of DNA methylation in blood [30, 31]. Importantly, in our work we aimed to capture the genetically regulated component of DNA methylation levels, and thus it is expected that the model performance R2 will not necessarily always be high for different CpGs. Indeed, the upper limit for such R2 should be the heritability of each CpG. We further excluded CpGs with SNPs within their probes in the Illumina 450K Beadchip because of potential bias for the measurement of DNA methylation levels of such CpGs [32].
Evaluation of the association between genetically predicted DNA methylation levels and pancreatic cancer risk
For evaluating associations of predicted DNA methylation levels with pancreatic cancer risk, we used data of GWAS conducted in PanScan and PanC4. Detailed information on these consortia has been described elsewhere [13–18]. For the current analyses, the genetic and covariate data were accessed from dbGaP (dbGaP Study Accession: phs000206.v5.p3 and phs000648.v1.p1). We performed subject and SNP level QC based on guidelines recommended by the consortia [33]. Briefly, in PanC4 dataset, we excluded subjects who were related to each other, with missing call rate ≥ 2%, or with missing information on covariates age and sex; we excluded SNPs with missing call rate ≥ 2%, positional duplicates, more than two discordant calls in study duplicates, more than one mendelian error in HapMap control trios, Hardy-Weinberg equilibrium (HWE) P < 1 × 10−4, sex difference in allele frequency > 0.2 for autosomes/XY in subjects of European ancestry, and/or sex difference in heterozygosity > 0.3 for autosomes/XY for European ancestry subjects, or with MAF < 0.005. In PanScan datasets, we excluded subjects with sex discordance, related with each other, or with a call rate < 94%; we further excluded SNPs with a call rate < 94% or HWE P < 1 × 10−7. In our analyses we only retained subjects with genetic ancestry of Europeans evaluated using principal component analysis. The genotype data from all sources were imputed together to the Haplotype Reference Consortium reference panel (r1.1 2016) [34] using Minimac3 for imputation and SHAPEIT for prephasing [35, 36], by using the Michigan Imputation Server (https://imputationserver.sph.umich.edu). Only imputed data with an imputation quality of at least 0.3 were retained in the association analyses. The final dataset included 8,280 cases and 6,728 controls.
The S-PrediXcan method [37] was used to evaluate the associations between genetically predicted DNA methylation levels and pancreatic cancer risk, using summary statistics of SNP-pancreatic cancer associations generated with adjustments of age, sex, and top PCs. The Z-score for the association between predicted DNA methylation levels at each CpG and pancreatic cancer risk was estimated based on the formula of:
Here wsm represents the weight of SNP s on the methylation levels of the CpG m. and refer to the GWAS-estimated effect size and standard error of SNP s on pancreatic cancer risk, respectively. and are the estimated variances of SNP s and the predicted methylation level at CpGs m, respectively. For the present study, the correlations between predicting SNPs were estimated based on the data of European descendants from 1000 Genomes Project Phase 3. Considering that a large number of CpGs may have correlated DNA methylation and predicted methylation levels, a false discovery rate (FDR)-adjusted P value < 0.05 was used to determine significant associations. For identified associated CpGs, GCTA-COJO analyses were conducted to examine whether the observed associations were independent of previously identified risk variants of pancreatic cancer [38]. Briefly, for each SNP that was included in the prediction models of the identified CpGs, we used GCTA-COJO to estimate the modified and conditioning on nearby GWAS-identified pancreatic cancer risk SNPs. Then we re-performed the S-PrediXcan analysis using the modified values of and to assess the associations between genetically predicted DNA methylation levels and pancreatic cancer risk after adjusting for previously reported GWAS risk SNPs. Only associated CpGs with a large proportion of predicting SNPs (>50%) in the corresponding models used in association analyses were reported here, to decrease possibility of false positive findings. We further performed analyses using individual level genetic data for these CpGs, and conducted analyses to examine whether the identified significant associations were consistent cross study phases (PanScan I, II, III; PanScan I, II; PanC4 and PanScan I, II; and PanC4), especially for PanScan III which included only cases.
Potential target genes of associated CpGs
The identified CpGs associated with pancreatic cancer risk were annotated with ANNOVAR (29). To determine potential target genes of these CpGs, we assessed whether genetically predicted DNA methylation levels of these CpGs were significantly correlated with genetically predicted expression of their adjacent genes in 8,280 cases and 6,728 controls of European ancestry included in PanScan I-III and PanC4. We estimated genetically predicted gene expression using prediction models built with data from the Genotype-Tissue Expression (GTEx) project focusing on blood tissue (N=338). Only gene expression prediction models with R2 ≥ 0.01 were used for the analyses. For genes showing a correlation (P < 0.05), we further assessed whether their genetically predicted expression was significantly associated with pancreatic cancer risk. Finally, we assessed the consistency of the direction of identified associations in the DNA methylation-gene expression-pancreatic cancer risk pathway.
Directly measured levels of associated CpGs in pancreatic tumor tissue specimens versus benign pancreatic tissue specimens
RRBS was performed on DNA extracted from 18 pancreatic tumor tissue specimens and 18 benign pancreatic tissue specimens, as described previously [39]. Sequencing was performed using the Illumina HiSeq 2000 in the Mayo Clinic Medical Genome Facility. SAAP-RRBS was used for sequence alignment and methylation extraction [40]. We compared the DNA methylation levels of identified associated CpGs in pancreatic tumor tissue specimens versus benign pancreatic tissue specimens. For this exploratory analysis, P<0.05 was used to determine significant differences.
Results
DNA methylation prediction models
Using data from the FHS, we were able to establish DNA methylation prediction models for a total of 223,959 CpGs, of which 70,269 showed a prediction performance (R2) ≥ 0.01 in both internal and external validation. Among them, 62,994 CpGs have no SNPs within their probes. The prediction models for these 62,994 CpGs showed similar performance in external and internal validation (Supplementary Figure 1). The correlation coefficient between R2 in FHS and WHI was 0.95.
Associations between genetically predicted DNA methylation and pancreatic cancer risk
Of the 62,994 CpGs examined, 45 at nine genomic loci showed significant associations with pancreatic cancer risk for their genetically predicted methylation levels after FDR adjustment (Supplementary Figure 2). Fifteen of the 45 CpGs were located > 500 kb away from any risk variant reported in previous GWAS of pancreatic cancer. Positive associations between predicted DNA methylation level and pancreatic cancer risk were observed for cg02871659, cg18279742, cg01554064, cg04520704, cg19586165, cg16557858, cg02944084, and cg20930114; in contrast, inverse associations were identified for cg24483576, cg24520381, cg22833065, cg17288560, cg19439043, cg03013999, and cg15445000. After conditioning on previously identified pancreatic cancer risk variants, associations for all of these 15 CpGs at five novel loci remained largely unchanged (Table 1), suggesting that the identified associations represent novel associations independent of previously identified risk SNPs. On the other hand, for the other 30 identified CpGs located at four known pancreatic cancer risk loci, their associations with pancreatic cancer risk were all significantly attenuated after conditioning on adjacent risk SNPs (Table 2), suggesting that the identified associations may be influenced by the risk SNPs. Based on subgroup analyses, the associations of the identified 45 CpGs tended to be robust across different subsets ((PanScan I, II, and III; PanScan I and II; PanC4 and PanScan I, II; and PanC4) (Supplementary Table 1).
Table 1.
CpG sites with genetically predicted DNA methylation to be independently associated with pancreatic cancer risk after adjustment for previously identified risk SNPs
| CpG site | Chr | Position (build37) | Number of SNPs used for prediction | Classification | R2a | OR (95% CI)b | P valuec | P value after FDR | Risk SNP adjusted for | P value after adjusting for risk SNP |
|---|---|---|---|---|---|---|---|---|---|---|
| cg20930114 | 2 | 110372285 | 15 | exonic | 0.02 | 1.94 (1.44–2.61) | 1.28 × 10−5 | 0.019 | rs1486134 | 1.28 × 10−5 |
| cg01554064 | 9 | 106855171 | 27 | upstream | 0.20 | 1.22 (1.12–1.32) | 1.75 × 10−6 | 0.003 | rs505922 | 1.77 × 10−6 |
| cg02871659 | 16 | 2014063 | 7 | intronic | 0.32 | 1.18 (1.09–1.28) | 3.34 × 10−5 | 0.045 | rs7190458 | 3.41 × 10−5 |
| cg18279742 | 16 | 2015703 | 46 | upstream/downstream | 0.21 | 1.20 (1.10–1.30) | 2.89 × 10−5 | 0.040 | rs7190458 | 2.94 × 10−5 |
| cg15445000 | 17 | 37608096 | 50 | upstream | 0.28 | 0.85 (0.80–0.91) | 2.42 × 10−6 | 0.005 | rs4795218 | 1.16 × 10−6 |
| cg03013999 | 17 | 37608204 | 21 | upstream | 0.18 | 0.81 (0.74–0.89) | 4.02 × 10−6 | 0.007 | rs4795218 | 1.63 × 10−6 |
| cg19439043 | 17 | 37719913 | 27 | intergenic | 0.04 | 0.64 (0.53–0.76) | 6.76 × 10−7 | 0.002 | rs4795218 | 2.51 × 10−7 |
| cg17288560 | 17 | 37720009 | 18 | intergenic | 0.05 | 0.62 (0.52–0.75) | 3.41 × 10−7 | 0.001 | rs4795218 | 1.35 × 10−7 |
| cg24520381 | 17 | 37784694 | 20 | intronic | 0.02 | 0.54 (0.43–0.69) | 3.71 × 10−7 | 0.001 | rs4795218 | 1.10 × 10−7 |
| cg24483576 | 17 | 37792770 | 13 | UTR3 | 0.03 | 0.51 (0.38–0.68) | 7.31 × 10−6 | 0.012 | rs4795218 | 4.23 × 10−6 |
| cg19586165 | 17 | 37814072 | 10 | exonic | 0.08 | 1.38 (1.19–1.59) | 1.26 × 10−5 | 0.019 | rs4795218 | 2.86 × 10−6 |
| cg02944084 | 17 | 37827057 | 22 | downstream | 0.03 | 1.81 (1.44–2.29) | 5.82 × 10−7 | 0.001 | rs4795218 | 1.47 × 10−7 |
| cg16557858 | 17 | 37879740 | 23 | intronic | 0.06 | 1.47 (1.25–1.74) | 4.98 × 10−6 | 0.009 | rs4795218 | 1.23 × 10−6 |
| cg22833065 | 17 | 38095691 | 14 | intergenic | 0.03 | 0.59 (0.46–0.76) | 3.14 × 10−5 | 0.043 | rs4795218 | 1.86 × 10−5 |
| cg04520704 | 22 | 18325160 | 18 | intronic | 0.08 | 1.36 (1.18–1.57) | 2.63 × 10−5 | 0.038 | rs16986825 | 2.65 × 10−5 |
R2: model prediction performance (R2) derived using FHS data
OR (odds ratio) and CI (confidence interval) per one standard deviation increase in genetically predicted DNA methylation
P value: derived from association analyses of 8,282 cases and 6,728 controls; FDR-adjust p≤0.05 considered statistically significant
Table 2.
CpG sites with genetically predicted DNA methylation to be associated with pancreatic cancer risk that are potentially influenced by previously identified risk SNPs
| CpG site | Chr | Position (build37) | Number of SNPs used for prediction | Classification | R2b | OR (95% CI)c | P valued | P value after FDR | Risk SNP adjusted for | P value after adjusting for risk SNP |
|---|---|---|---|---|---|---|---|---|---|---|
| cg10015974 | 1 | 199827580 | 87 | intergenic | 0.13 | 0.80 (0.73–0.87) | 1.28 × 10−7 | 3.84 × 10−4 | rs16986825; rs3790844 |
0.02 |
| cg10098523 | 1 | 200002343 | 40 | intronic | 0.22 | 0.83 (0.78–0.90) | 1.29 × 10−6 | 2.73 × 10−3 | rs16986825; rs3790844 |
0.52 |
| cg07926895 | 1 | 200005833 | 24 | intronic | 0.03 | 0.61 (0.49–0.77) | 1.89 × 10−5 | 2.77 × 10−2 | rs16986825; rs3790844 |
0.32 |
| cg17804356 | 1 | 200009927 | 3 | intronic | 0.01 | 3.38 (2.12–5.39) | 2.81 × 10−7 | 8.05 × 10−4 | rs16986825; rs3790844 |
0.32 |
| cg07507801 | 5 | 1291235 | 5 | intronic | 0.03 | 2.29 (1.66–3.16) | 5.14 × 10−7 | 1.30 × 10−3 | rs2736098; rs35226131; rs401681 |
0.13 |
| cg07380026 | 5 | 1296007 | 14 | upstream | 0.01 | 4.52 (2.97–6.90) | 2.39 × 10−12 | 1.67 × 10−8 | rs2736098; rs35226131; rs401681 |
4.55 × 10−3 |
| cg26603275 | 5 | 1298965 | 10 | intergenic | 0.04 | 2.24 (1.75–2.87) | 1.11 × 10−10 | 6.36 × 10−7 | rs2736098; rs35226131; rs401681 |
0.05 |
| cg11624060 | 5 | 1316038 | 25 | intergenic | 0.18 | 1.28 (1.17–1.40) | 2.49 × 10−8 | 9.23 × 10−5 | rs2736098; rs35226131; rs401681 |
0.93 |
| cg26209169 | 5 | 1316264 | 22 | intergenic | 0.24 | 1.24 (1.15–1.34) | 2.19 × 10−8 | 8.62 × 10−5 | rs2736098; rs35226131; rs401681 |
0.83 |
| cg10441424 | 5 | 1316636 | 16 | intergenic | 0.01 | 2.08 (1.52–2.86) | 5.82 × 10−6 | 1.02 × 10−2 | rs2736098; rs35226131; rs401681 |
0.65 |
| cg07493874 | 5 | 1342172 | 11 | intronic | 0.15 | 0.69 (0.61–0.77) | 8.91 × 10−11 | 5.61 × 10−7 | rs2736098; rs35226131; rs401681 |
0.93 |
| cg19915256 | 5 | 1345677 | 11 | upstream | 0.02 | 2.85 (2.00–4.04) | 5.16 × 10−9 | 2.32 × 10−5 | rs2736098; rs35226131; rs401681 |
0.52 |
| cg27028750 | 5 | 1349422 | 20 | intergenic | 0.25 | 0.79 (0.74–0.85) | 6.59 × 10−10 | 3.46 × 10−6 | rs2736098; rs35226131; rs401681 |
0.43 |
| cg03474926 | 9 | 136023407 | 24 | intronic | 0.01 | 2.72 (1.90–3.89) | 5.18 × 10−8 | 1.72 × 10−4 | rs505922 | 0.36 |
| cg01169778 | 9 | 136038690 | 14 | intronic | 0.04 | 1.98 (1.46–2.68) | 1.04 × 10−5 | 1.62 × 10−2 | rs505922 | 0.13 |
| cg14653977 | 9 | 136038692 | 20 | intronic | 0.03 | 4.27 (3.09–5.89) | 1.12 × 10−18 | 3.53 × 10−14 | rs505922 | 0.08 |
| cg13531387 | 9 | 136078657 | 13 | intergenic | 0.11 | 0.34 (0.25–0.45) | 3.16 × 10−13 | 2.49 × 10−9 | rs505922 | 0.75 |
| cg00878953 | 9 | 136129875 | 36 | downstream | 0.15 | 0.65 (0.54–0.79) | 6.83 × 10−6 | 1.16 × 10−2 | rs505922 | 0.42 |
| cg11879188 | 9 | 136149908 | 36 | intronic | 0.5 | 2.28 (1.84–2.83) | 4.84 × 10−14 | 4.36 × 10−10 | rs505922 | 0.89 |
| cg21160290 | 9 | 136149941 | 43 | intronic | 0.71 | 1.99 (1.69–2.34) | 8.87 × 10−17 | 1.12 × 10−12 | rs505922 | 0.76 |
| cg22535403 | 9 | 136150032 | 44 | intronic | 0.69 | 2.29 (1.89–2.77) | 4.63 × 10−17 | 7.29 × 10−13 | rs505922 | 0.59 |
| cg24267699 | 9 | 136151359 | 13 | upstream | 0.59 | 2.50 (2.07–3.02) | 1.33 × 10−21 | 8.38 × 10−17 | rs505922 | 0.01 |
| cg06818865 | 9 | 136151958 | 10 | intergenic | 0.3 | 1.84 (1.52–2.24) | 8.47 × 10−10 | 4.10 × 10−6 | rs505922 | 0.16 |
| cg13660174 | 9 | 136238392 | 19 | intronic | 0.07 | 1.64 (1.34–2.00) | 1.30 × 10−6 | 2.73 × 10−3 | rs505922 | 0.29 |
| cg13568213 | 9 | 136387235 | 16 | intronic | 0.03 | 7.05 (3.43–14.48) | 1.08 × 10−7 | 3.40 × 10−4 | rs505922 | 0.17 |
| cg21101465 | 13 | 28493404 | 20 | upstream | 0.04 | 0.61 (0.49–0.76) | 9.94 × 10−6 | 1..61 × 10−2 | rs9581943 | 0.06 |
| cg11853320 | 13 | 28493913 | 52 | upstream | 0.08 | 0.69 (0.61–0.79) | 3.88 × 10−8 | 1.36 × 10−4 | rs9581943 | 0.46 |
| cg26793256 | 13 | 28494004 | 55 | upstream | 0.06 | 0.72 (0.62–0.82) | 1.56 × 10−6 | 3.17 × 10−3 | rs9581943 | 0.16 |
| cg04633225 | 13 | 28494161 | 22 | upstream | 0.02 | 0.45 (0.34–0.59) | 1.09 × 10−8 | 4.58 × 10−5 | rs9581943 | 0.06 |
| cg11213248 | 13 | 28534648 | 7 | intergenic | 0.22 | 0.81 (0.75–0.88) | 1.16 × 10−6 | 2.61 × 10−3 | rs9581943 | 2.00 × 10−4 |
R2: model prediction performance (R2) derived using FHS data
OR (odds ratio) and CI (confidence interval) per one standard deviation increase in genetically predicted DNA methylation
P value: derived from association analyses of 8,282 cases and 6,728 controls; FDR-adjust p≤0.05 considered statistically significant
Candidate target genes of associated CpGs
For the 45 CpGs associated with pancreatic cancer risk, ANNOVAR annotation suggested 32 adjacent genes. Of them, we were able to build blood tissue gene expression prediction models with R2 ≥ 0.01 for nine (RPS2, STARD3, GBGT1, ABO, SURF6, ERBB2, ORMDL3, SNHG9, SOWAHC). We further assessed Spearman’s rank correlations for 17 pairs of CpG site-gene for their genetically predicted levels of DNA methylation and gene expression, respectively (Supplementary Table 2). For all genes except for STARD3, we observed significant (P < 0.001) correlations (Supplementary Table 2).
Associations of predicted expression of candidate target genes with pancreatic cancer risk
Of these eight genes showing significant correlations, six further showed a significant association with pancreatic cancer risk for their genetically predicted expression levels, namely, ABO (P = 6.72 × 10−12), RPS2 (P = 3.48 × 10−5), SURF6 (P = 8.47 × 10−3), ORMDL3 (P = 2.58 × 10−4), SNHG9 (P = 1.15 × 10−2), and SOWAHC (P = 8.30 × 10−4). Overall, a total of 12 CpGs with 6 genes showed significant associations in each pair of the relationships in the DNA methylation-gene expression-pancreatic cancer risk pathway. Encouragingly, all these associations showed consistent directions. Taken the CpG site cg24267699 located upstream of ABO as an example, its genetically predicted DNA methylation showed a positive association with pancreatic cancer risk (odds ratio (OR) = 2.50; P = 1.33 × 10−21). Meanwhile, we observed an inverse correlation between the genetically predicted DNA methylation level of cg24267699 and predicted expression of ABO (correlation coefficient = −0.62; P < 0.001), as well as an inverse association between predicted expression of ABO and pancreatic cancer risk (OR = 0.89, P = 6.72 × 10−12) (Tables 3, Supplementary Tables 2–3 and Supplementary Figure 3). Consistent three-way associations were also observed for CpGs and five other genes (RPS2, SURF6, ORMDL3, SNHG9, and SOWAHC), which have not been previously reported as pancreatic cancer susceptibility genes in GWAS or TWAS.
Table 3.
Associations showing consistent direction of effect for predicted DNA methylation-predicted gene expression-pancreatic cancer risk pathway
| CpG site | Chr | Position | Associated Gene | Classification | DNA methylation and pancreatic cancer risk | DNA methylation and gene expression | Gene expression and pancreatic cancer risk | |||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | P value | Correlation coefficient | Correlation P value | OR | P value | |||||
| cg20930114 | 2 | 110372285 | SOWAHC | exonic | 1.94 | 1.28 × 10−5 | −0.516 | <.001 | 0.64 | 8.30 × 10−4 |
| cg00878953 | 9 | 136129875 | ABO | downstream | 0.65 | 6.83 × 10−6 | 0.420 | <.001 | 0.49 | 6.72 × 10−12 |
| cg11879188 | 9 | 136149908 | intronic | 2.28 | 4.84 × 10−14 | −0.350 | <.001 | |||
| cg21160290 | 9 | 136149941 | intronic | 1.99 | 8.87 × 10−17 | −0.344 | <.001 | |||
| cg22535403 | 9 | 136150032 | intronic | 2.29 | 4.63 × 10−17 | −0.369 | <.001 | |||
| cg24267699 | 9 | 136151359 | upstream | 2.50 | 1.33 × 10−21 | −0.620 | <.001 | |||
| cg06818865a | 9 | 136151958 | intergenic | 1.84 | 8.47 × 10−10 | −0.423 | <.001 | |||
| cg06818865a | 9 | 136151958 | SURF6 | intergenic | 1.84 | 8.47 × 10−10 | −0.323 | <.001 | 0.91 | 8.47 × 10−3 |
| cg02871659 | 16 | 2014063 | RPS2 | intronic | 1.18 | 3.34 × 10−5 | −0.742 | <.001 | 0.64 | 3.48 × 10−5 |
| cg18279742 | 16 | 2015703 | upstream | 1.20 | 2.89 × 10−5 | −0.739 | <.001 | |||
| cg18279742 | 16 | 2015703 | SNHG9 | downstream | 1.20 | 2.89 × 10−5 | 0.305 | <.001 | 1.10 | 1.15 × 10−2 |
| cg22833065 | 17 | 38095691 | ORMDL3 | downstream | 0.59 | 3.14 × 10−5 | −0.831 | <.001 | 1.15 | 2.58 × 10−4 |
The same CpG site was annotated to two different genes
Directly measured levels of associated CpGs in pancreatic tumor tissue versus benign pancreatic tissue
Of the 45 CpGs, 16 were directly captured in the Reduced representation bisulfite sequencing (RRBS) of 18 pancreatic tumor tissue specimens and 18 benign pancreatic tissue specimens. Of them, significances of levels of two CpGs (cg04520704 and cg04633225) in tumor versus benign tissues could not be determined. Among the others, six demonstrated significant different levels in pancreatic tumor tissue versus benign pancreatic tissue (Table 4). Encouragingly, the effect directions for all of them are consistent with findings from analyses using genetic instruments (Table 4).
Table 4.
CpGs showing consistent direction of effect for directly measured levels in pancreas tumor versus benign tissues and genetically predicted levels in blood of pancreatic cancer cases versus controls
| CpG site | Chr | Position | Direction of association between genetically predicted levels and pancreatic cancer risk | Average levels in benign pancreatic tissue | Standard deviation of levels in benign pancreatic tissue | Average levels in pancreatic tumor tissue | Standard deviation of levels in pancreatic tumor tissue | P value comparing levels in pancreas tumor versus benign tissue |
|---|---|---|---|---|---|---|---|---|
| cg17804356 | 1 | 200009927 | + | 0.02 | 0.04 | 0.12 | 0.15 | <0.0004 |
| cg20930114 | 2 | 110372285 | + | 0.005 | 0.02 | 0.04 | 0.05 | 0.0004 |
| cg07380026 | 5 | 1296007 | + | 0.24 | 0.18 | 0.54 | 0.20 | <0.0004 |
| cg01169778 | 9 | 136038690 | + | 0.23 | 0.11 | 0.46 | 0.29 | 0.01 |
| cg22535403 | 9 | 136150032 | + | 0.35 | 0.21 | 0.48 | 0.28 | 0.05 |
| cg21101465 | 13 | 28493404 | − | 0.36 | 0.19 | 0.27 | 0.22 | 0.02 |
Discussion
The current study is by far the first large-scale study that evaluated the relationship between genetically predicted DNA methylation levels and pancreatic cancer risk. We identified 45 CpGs of which the predicted DNA methylation levels showed significant associations with pancreatic cancer risk at FDR < 0.05, including 15 CpGs located at five novel loci that have not been reported in previous GWAS. For the remaining 30 CpGs located at four known pancreatic cancer risk loci, the observed associations were substantially attenuated after adjusting for GWAS-identified risk SNPs, implying that the associations may be at least partly due to the reported risk SNPs. We found consistent direction of associations in the DNA methylation-gene expression-pancreatic cancer risk pathway for 12 CpGs with six genes. Our findings were further supported with the evidence from differentiated DNA methylation at six CpGs for their directly measured levels observed in pancreatic tumor versus benign tissue. Our study identified novel methylation biomarker candidates for pancreatic cancer, as well as provided new information in understanding etiology of pancreatic cancer, a highly lethal malignancy.
Of the 45 identified associated CpGs, we were able to assess correlations between genetically predicted DNA methylation and gene expression levels for 17 CpGs with 9 adjacent genes. Among the examined correlations, except for the one between cg19586165 and STARD3, all others were statistically significant. The possible speculation for the insignificant correlation suggested that the most proximal gene of cg19586165, STARD3, might not be the actual target gene. Additional strategies beyond the scope of simple statistical correlations are needed to verify its actual target gene. Of the eight linked genes correlated with predicted DNA methylation of the identified CpGs, six (ABO, RPS2, SURF6, ORMDL3, SNHG9 and SOWAHC) demonstrated significant associations with pancreatic cancer risk for their predicted expression. Among them, The ABO blood group gene located at 9q34.2 has already been implicated as a potential target gene of pancreatic cancer risk SNPs from previous GWAS and TWAS [17, 21]. Genotype-inferred non-O blood type was consistently suggested to be associated with an increased risk of pancreatic cancer compared to other blood types, which may be partly explained by differentiated expression of blood group antigens, or alterations in the systemic inflammatory state [41]. SURF6 has been previously suggested as a potential pancreatic cancer biomarker, as indicated by a study comparing its expression level in malignant pancreatic cells to that in normal pancreatic duct cells or human papillomavirus-immortalized pancreatic duct epithelial cells [42]. A higher expression level of SNHG9, a non-coding RNA, has been identified as a novel prognostic markers for pancreatic cancer [43]. To the best of our knowledge, our study is the first one implicating potential link between this gene and pancreatic cancer risk. Further functional studies are needed to better understand potential regulatory effects of the identified CpGs on expression of the genes, and link between expression of the genes and pancreatic cancer.
In this study we systematically assessed relationships between genetically predicted DNA methylation in blood, genetically predicted expression for putatively target genes in blood, and pancreatic cancer risk. For our analyses using genetic instruments we used data generated from white blood cells rather than pancreatic tissue for several reasons. First, it is very challenging to acquire a large sample of pancreatic tissue from healthy subjects without pancreatic cancer. Information from pancreatic tumor-adjacent normal tissue would be less desirable, due to potential influence of somatic alterations on DNA methylation. Furthermore, findings of biomarkers identified in a study design using data from white blood cell samples may confer more translational and practical utilities for future risk assessment of pancreatic cancer, compared with biomarkers in pancreas tissue as it is impractical to obtain pancreas tissue from healthy subjects. We also acknowledge that compared with pancreas specimens, a study focusing on blood samples may not be ideal for pinpointing the underlying etiology of pancreatic cancer development given possible tissue-specific DNA methylation patterns. However, it is also worth noting that, high concordance for the genetically regulated component of DNA methylation cross several tissue types has been reported for a large number of CpGs [44, 45]. In this study, we have compared the directly measured levels of a proportion of identified associated CpGs in pancreatic tumor tissue versus benign pancreatic tissue. It is worth noting that for this comparison, the overall DNA methylation levels influenced by both genetic and non-genetic factors were assessed, which is different from the analyses focusing on genetic instruments, in which case only genetically regulated components of DNA methylation levels were evaluated. Although the involved sample size is relatively small (18 vs 18), we were still able to observe significant differences for six of the CpGs among the limited associated CpGs that were captured in our measurement using RRBS. Unlike The Cancer Genome Atlas (TCGA) study, in which only methylation of pancreatic tumor and tumor-adjacent normal tissue from pancreatic cancer patients are available, in our comparison the control group focuses on histologically normal pancreas tissue from subjects without pancreatic cancer, thus representing a better design compared with other datasets such as TCGA.
Our study has several strengths. First, we used datasets with relatively large sample sizes for both methylation prediction model building (N=1,595) and main association analyses for pancreatic cancer risk (8,280 cases and 6,728 controls), which enabled us to conduct a well powered assessment of the DNA methylation-pancreatic cancer risk associations. Second, our innovative study design of using genetic instruments to predict DNA methylation decreased several biases that are commonly embedded in traditional epidemiological studies, such as residual confounding and reverse causality. In addition, by integrating multi-omics data of DNA methylation and gene expression from various resources, we were able to further verify our findings by examining the consistency of the associations in the DNA methylation-gene expression-pancreatic cancer risk pathway for the identified significant CpGs, which may further contribute to potential etiologic understanding of pancreatic cancer. The performance of our developed models were externally validated in an independent WHI dataset, which uses different genotyping platforms (Illumina vs Affymatrix used in FHS dataset), supporting the utility of our prediction models across platforms. Finally, besides evidence from analyses using genetic instruments, we found additional evidence for some of the identified CpGs using their directly measured levels in pancreatic tissue, further supporting relevance of the identified CpGs with pancreatic cancer. Although the sample size for this analysis is relatively small, it is worth noting that our study comparing tissue samples of PC cases and non-PC controls could well overcome the potential limitation of many other studies (e.g., The Cancer Genome Atlas) comparing tumor samples of cases and tumor-adjacent normal tissue samples of cases.
Several potential limitations need to be acknowledged for appropriate interpretation of our findings. First, the associated CpGs identified in this study do not necessarily imply their causal role in pancreatic cancer. Similar to TWAS, although our findings will be useful for prioritizing candidate DNA methylation biomarkers, false positive findings could exist for some of the identified associations [46]. There are several potential reasons for this, such as correlated DNA methylation across individuals, correlated predicted DNA methylation, as well as shared variants [46]. In our study, multiple identified CpGs locate at the same loci. Future functional investigation will better characterize whether the identified CpGs play a causal role in pancreatic tumorigenesis. Second, during the DNA methylation genetic prediction model building, due to a lack of data, we were not able to incorporate additional variables, including established pancreatic cancer risk factors, such as smoking, alcohol drinking, body mass index, diabetes status, etc for adjustments. Future work for developing DNA methylation genetic prediction models after adjusting for these additional variables are warranted to validate our findings. Third, although we were able to show that a proportion of the pancreatic cancer associated CpGs we identified demonstrated differential levels in pancreatic tumor versus benign tissue, further work directly comparing DNA methylation levels of these CpGs in pre-diagnosed blood of pancreatic cancer cases and controls are warranted to further validate our findings. Fourth, it is worth noting that the PanScan III data on dbGaP only contained data for cases but not for controls. In the current analysis for improving statistical power we included cases of PanScan III in the analyses. Previous work suggested that imputation of datasets genotyped by different platforms before merging could generate slightly more SNPs than imputations after combining the datasets together [47]. In the current study, we merged genotyped data across cases and controls of PanScan I, II, III along with PanC4 and then imputed the data together. Although the design of incorporating data of cases only in PanScan III could be of potentially concerning, we carefully compared the association results in different subgroups (Supplementary Table 1), and the estimates are quite robust, suggesting that this is a less concerning issue and our design should be appropriate. Lastly, in this study we evaluated ANNOVAR annotated genes as candidate target genes of associated CpG sites for correlation analysis. With the recognized chromatin interaction and long-range regulation of gene expression in the human genome, it is possible that for some CpGs the target genes may not necessarily the nearest genes. Further work is warranted to better characterize potential target genes of our identified CpGs using other approaches beyond simply statistical correlation analyses.
In summary, in a large-scale study, we identified 45 CpGs showing significant associations with pancreatic cancer risk for their genetically predicted DNA methylation, including 15 at five novel loci showing an association independent from known risk variants. We further observed consistent directions of associations in the DNA methylation-gene expression-pancreatic cancer risk pathway. We found differentiated DNA methylation at six of the identified CpGs for their measured levels in pancreatic tumor versus benign tissue. The pancreatic cancer risk associated CpGs identified in this study could be investigated in future studies with direct measurement of circulating DNA methylation levels for examining potential utility in pancreatic cancer risk assessment.
Supplementary Material
Acknowledgements
The authors would like to thank all of the individuals for their participation in the parent studies of PanScan/PanC4 consortia and all the researchers, clinicians, technicians and administrative staff for their contribution to the studies. L. Wu is supported by the University of Hawaii Cancer Center, and NCI R00 CA218892. Duo Liu is partially supported by the Harbin Medical University Cancer Hospital. Data on CpG positions in the independent case and control tissues was funded in part by Exact Sciences (Madison WI). The PanScan study was funded in whole or in part with federal funds from the National Cancer Institute (NCI), US National Institutes of Health (NIH) under contract number HHSN261200800001E. Additional support was received from NIH/NCI K07 CA140790, the American Society of Clinical Oncology Conquer Cancer Foundation, the Howard Hughes Medical Institute, the Lustgarten Foundation, the Robert T. and Judith B. Hale Fund for Pancreatic Cancer Research and Promises for Purple. A full list of acknowledgments for each participating study is provided in the Supplementary Note of the manuscript with PubMed ID: 25086665. For the PanC4 GWAS study, the patients and controls were derived from the following PANC4 studies: Johns Hopkins National Familial Pancreas Tumor Registry, Mayo Clinic Biospecimen Resource for Pancreas Research, Ontario Pancreas Cancer Study (OPCS), Yale University, MD Anderson Case Control Study, Queensland Pancreatic Cancer Study, University of California San Francisco Molecular Epidemiology of Pancreatic Cancer Study, International Agency of Cancer Research and Memorial Sloan Kettering Cancer Center. This work is supported by NCI R01CA154823 Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN2682011000111.
Role of the Funder/Sponsor: The funding organization had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Footnotes
Declaration of interests
Dr. Kisiel, Mr. Mahoney and Mr. Taylor are listed as inventors on intellectual property jointly owned by Mayo Clinic and Exact Sciences (Madison WI) and may receive royalties under Mayo Clinic policy. We declare no competing interests for other authors.
Data Availability
The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession phs000206.v5.p3 and phs000648.v1.p1 for PanScan/PanC4 data, phs000342 and phs000724 for FHS, phs000315, phs000675 and phs001335 for WHI, and phs000424.v6.p1 for GTEx.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA A Cancer J Clin 2020; 70(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R et al. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Research 2014; 74(11):2913–2921. [DOI] [PubMed] [Google Scholar]
- 3.Loosen SH, Neumann UP, Trautwein C et al. Current and future biomarkers for pancreatic adenocarcinoma. Tumour Biol. 2017; 39(6):1010428317692231. [DOI] [PubMed] [Google Scholar]
- 4.Eissa MAL, Lerner L, Abdelfatah E et al. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin Epigenetics 2019; 11(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronica A, Avagliano L, Caretti A et al. Unexpected distribution of CA19.9 and other type 1 chain Lewis antigens in normal and cancer tissues of colon and pancreas: Importance of the detection method and role of glycosyltransferase regulation. Biochim Biophys Acta Gen Subj 2017; 1861(1 Pt A):3210–3220. [DOI] [PubMed] [Google Scholar]
- 6.Schott S, Yang R, Stöcker S et al. HYAL2 methylation in peripheral blood as a potential marker for the detection of pancreatic cancer: a case control study. Oncotarget 2017; 8(40):67614–67625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mardin WA, Ntalos D, Mees ST et al. SERPINB5 Promoter Hypomethylation Differentiates Pancreatic Ductal Adenocarcinoma From Pancreatitis. Pancreas 2016; 45(5):743–747. [DOI] [PubMed] [Google Scholar]
- 8.Melson J, Li Y, Cassinotti E et al. Commonality and differences of methylation signatures in the plasma of patients with pancreatic cancer and colorectal cancer. Int. J. Cancer 2014; 134(11):2656–2662. [DOI] [PubMed] [Google Scholar]
- 9.Bell JT, Pai AA, Pickrell JK et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011; 12(1):R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannon E, Weedon M, Bray N et al. Pleiotropic Effects of Trait-Associated Genetic Variation on DNA Methylation: Utility for Refining GWAS Loci. Am. J. Hum. Genet 2017; 100(6):954–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grundberg E, Meduri E, Sandling JK et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am. J. Hum. Genet 2013; 93(5):876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McRae AF, Powell JE, Henders AK et al. Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol. 2014; 15(5):R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M, Wang Z, Obazee O et al. Three new pancreatic cancer susceptibility signals identified on chromosomes 1q32.1, 5p15.33 and 8q24.21. Oncotarget 2016; 7(41):66328–66343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolpin BM, Rizzato C, Kraft P et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat. Genet 2014; 46(9):994–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen GM, Amundadottir L, Fuchs CS et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet 2010; 42(3):224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet 2009; 41(9):986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein AP, Wolpin BM, Risch HA et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat Commun 2018; 9(1):556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Childs EJ, Mocci E, Campa D et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. Nat. Genet 2015; 47(8):911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen F, Childs EJ, Mocci E et al. Analysis of Heritability and Genetic Architecture of Pancreatic Cancer: A PanC4 Study. Cancer Epidemiol Biomarkers Prev 2019; 28(7):1238–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen F, Childs EJ, Mocci E et al. Analysis of Heritability and Genetic Architecture of Pancreatic Cancer: A PanC4 Study. Cancer Epidemiol Biomarkers Prev 2019:cebp;1055–9965.EPI-18–1235v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong J, Jermusyk A, Wu L et al. A Transcriptome-Wide Association Study (TWAS) Identifies Novel Candidate Susceptibility Genes for Pancreatic Cancer. Journal of the National Cancer Institute 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Wu L, Shu X et al. Genetic Data from Nearly 63,000 Women of European Descent Predicts DNA Methylation Biomarkers and Epithelial Ovarian Cancer Risk. Cancer Res. 2019; 79(3):505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Wu L, Shu X-O et al. Genetically predicted levels of DNA methylation biomarkers and breast cancer risk: data from 228,951 women of European descent. J. Natl. Cancer Inst 2019. doi: 10.1093/jnci/djz109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L, Yang Y, Guo X et al. An integrative multi-omics analysis to identify candidate DNA methylation biomarkers related to prostate cancer risk. Nat Commun 2020; 11(1):3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aryee MJ, Jaffe AE, Corrada-Bravo H et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014; 30(10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, Shi W, Long J et al. A transcriptome-wide association study of 229,000 women identifies new candidate susceptibility genes for breast cancer. Nat. Genet 2018; 50(7):968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu L, Wang J, Cai Q et al. Identification of Novel Susceptibility Loci and Genes for Prostate Cancer Risk: A Transcriptome-Wide Association Study in Over 140,000 European Descendants. Cancer Res 2019; 79(13):3192–3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.NBCS Collaborators, kConFab/AOCS Investigators, Wu L et al. A transcriptome-wide association study of 229,000 women identifies new candidate susceptibility genes for breast cancer. Nat Genet 2018; 50(7):968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium GTEx, Gamazon ER Wheeler HE et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 2015; 47(9):1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huan T, Joehanes R, Song C et al. Genome-wide identification of DNA methylation QTLs in whole blood highlights pathways for cardiovascular disease. Nat Commun 2019; 10(1):4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wheeler HE, Shah KP, Brenner J et al. Survey of the Heritability and Sparse Architecture of Gene Expression Traits across Human Tissues. PLoS Genet 2016; 12(11):e1006423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McRae AF, Marioni RE, Shah S et al. Identification of 55,000 Replicated DNA Methylation QTL. Sci Rep 2018; 8(1):17605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein AP, Wolpin BM, Risch HA et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat Commun 2018; 9(1):556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.the Haplotype Reference Consortium. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016; 48(10):1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods 2012; 9(2):179–181. [DOI] [PubMed] [Google Scholar]
- 36.Howie BN, Donnelly P, Marchini J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet 2009; 5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbeira AN, Dickinson SP, Bonazzola R et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun 2018; 9(1):1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Ferreira T, Morris AP et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet 2012; 44(4):369–375, S1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kisiel JB, Raimondo M, Taylor WR et al. New DNA Methylation Markers for Pancreatic Cancer: Discovery, Tissue Validation, and Pilot Testing in Pancreatic Juice. Clin. Cancer Res. 2015; 21(19):4473–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Z, Baheti S, Middha S et al. SAAP-RRBS: streamlined analysis and annotation pipeline for reduced representation bisulfite sequencing. Bioinformatics 2012; 28(16):2180–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolpin BM, Chan AT, Hartge P et al. ABO Blood Group and the Risk of Pancreatic Cancer. JNCI Journal of the National Cancer Institute 2009; 101(6):424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones S, Zhang X, Parsons DW et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008; 321(5897):1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang B, Li C, Sun Z. Long non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 are novel prognostic markers for pancreatic cancer. Am J Transl Res 2018; 10(8):2648–2658. [PMC free article] [PubMed] [Google Scholar]
- 44.Hannon E, Weedon M, Bray N et al. Pleiotropic Effects of Trait-Associated Genetic Variation on DNA Methylation: Utility for Refining GWAS Loci. Am. J. Hum. Genet 2017; 100(6):954–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stueve TR, Li W-Q, Shi J et al. Epigenome-wide analysis of DNA methylation in lung tissue shows concordance with blood studies and identifies tobacco smoke-inducible enhancers. Hum. Mol. Genet 2017; 26(15):3014–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wainberg M, Sinnott-Armstrong N, Mancuso N et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet 2019; 51(4):592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Iperen EPA, Hovingh GK, Asselbergs FW, Zwinderman AH. Extending the use of GWAS data by combining data from different genetic platforms. PLoS One 2017; 12(2):e0172082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession phs000206.v5.p3 and phs000648.v1.p1 for PanScan/PanC4 data, phs000342 and phs000724 for FHS, phs000315, phs000675 and phs001335 for WHI, and phs000424.v6.p1 for GTEx.