Abstract
Background
Circular RNAs (circRNAs) are a new kind of non-coding RNA(ncRNA). Throughout research, we see an increase in the number of studies demonstrating that circRNAs occupy a pivotal role in the growth and advancement of human tumors. Nevertheless, hsa_circ_001787′s role in the evolution of colorectal cancer (CRC) remains unclear. This current study ascertained the expression level of circRNA001787 in CRC specimens and neighboring healthy tissues, and investigated the miRNAs associate with hsa_circ_001787, as well as the relationship between hsa_circ_001787 and pathological factors.
Method
First, the expression level of hsa_circ_001787 was measured in 43 matched Tissues from CRC and normal tissues through using real-time quantitative reverse transcription PCR (qRT-PCR). Second, based on circular RNA-microRNA and microRNA-mRNA pairs, a circRNA-miRNA-mRNA network was created. The survival rate of mRNAs was investigated through the GEPIA in the network. Regarding the elucidated function analysis of hsa_circ_001787, The biological, molecular, cellular function (GO) and pathway (KEGG) enrichment was obtained.
Result
We detected that hsa_circ_001787 expression level was significantly down expressed in CRC tissue versus paired CRC histological normal tissue. The area under the curve (AUC) was 0.83. The expression level of hsa_circ_001787 was significantly associated with pathological factors such as tumor grade and the primary site of the tumor. Based on the hsa_circ_001787, a novel circRNA/miRNA/mRNA network has been built up, four miRNAs, and 24 mRNA. The pathway of mRNAs analyzed in the pathogenesis of CRC. Four genes distinguished via the GEPIA database were positively linked to the overall survival of CRC patients.
Conclusion
Our study suggested that hsa_circ_001787 was significantly down-regulated in CRC. We might be able to use this as a new biomarker in the screening of CRC. Furthermore, our finding achieves a broader understanding of the regulatory mechanisms by which hsa_circ_001787 acts as ceRNA in colorectal cancer.
Keywords: circRNA, Colorectal cancer, Biomarker, and ceRNA
1. Introduction
Colorectal cancer is the third most common type of human neoplasms and the fourth most common contributing factor in deaths worldwide (Siegel et al., 2016). Even with the advancement of different options, such as conventional tumor biomarkers, colonoscopy screening, and fecal occult blood tests, the rate of survival remains for CRC remains low (Cho, 2011, Levin, 2003). Based on the smith et al. study, 90% of the deaths would be preventable if CRC patients were diagnosed at the infancy stage (Smith, et al., 2001). The discovery of useful biological markers for the screening and treatment of CRC is essential. circRNAs are a covalent type of non-coding RNA that has a crucial role in gene regulation and function (Chalbatani, 2016, Tran, et al., 2020). In comparison with linear RNA, circRNAs are new molecules that covalently joined 3′- and 5′-ends, which causes increased stability (Chen and Yang, 2015). According to recent research, circRNAs are aberrantly expressing in a wide range of tumors such as colorectal, breast, gastric, etc. (Hansen, 2013, Saglam et al., 2019, Zhang, 2017, Hsiao, 2017, Weng, 2017, Greco, 2018, Liu, 2017). Recently, more and more research has displayed that circRNAs can serve as miRNA sponge or competitive endogenous RNA (ceRNA), binding with miRNA to regulate gene expression such as CDR1as with miR-7 in cervical cancer (Hansen, 2013, Saglam et al., 2019). The sponge function of hsa_circ_100269 for hsa_miR_630 in gastric carcinoma (Zhang, 2017). Specifically, down-regulated circRNAs has a crucial regulatory role in colorectal cancer as a miRNA sponge (Hsiao, 2017, Weng, 2017). Moreover, circRNAs can act as competitors or enhancers to regulate corresponding genes in various cancers (Greco, 2018, Liu, 2017). An enormous body of studies have shown that circRNAs can play a significant role as a diagnostic marker in CRC (Zhuo, 2017). In this present study, searching colorectal cancer-associated circRNA databases (CircBase and circ2Traits), we predicted that hsa_circ_001787, which is located at chr1:26772806–26774151, and its associated gene symbol, DHDDS, has a strong relationship with CRC (P < 0.05). We set out to find the expression of hsa_circ_001787 in tissue from CRC patients at various stages. We also considered the connections between hsa_circ_001787 expression level and clinical-pathological features to analyze the diagnostic value as a biomarker in the detection of CRC. Moreover, we created a ceRNA network which contains circRNA, miRNA (sponge), and mRNA, with analysis of the total survival incident of interact genes as well.
2. Material and method
2.1. Tissue sampling
The ethics committee of the National Cancer Institute at Tehran University of Medical Science (Tehran, Iran) confirmed the current study. The CRC specimens and marginal tissues as healthy tissue were obtained from CRC patients. The tissues were quickly snap-frozen and kept at −80 °C for more analysis. In this study samples, of patients with colon and rectum cancer were involved in the study. The patients with metastatic to liver and other organs were excluded from the study. In accordance with ethical approval number IR.TUMS.REC.1394.2197, all of the patients signed informed consent.
2.2. The expression analysis of circ001787 through qRT-PCR
Total RNA was extracted from CRC tissues using RNeasy Mini Kit (QIAGEN, USA) according to the company’s instructions. For circular RNA analysis, cDNA was synthesized via a TaKaRa kit (PrimeScript First Strand cDNA Synthesis Kit). The expression level of circRNA was determined via qRT-PCR TaKaRa SYBR GREEN master mix, with Human Beta-actin as an internal control. The divergent primers and human beta-actin were designed and synthesized by the Sinagene Company (IRAN). The primers were used for qRT-PCR as follows: Human Beta-actin primers (5′-AGAGCTACGAGCTGCCTGAC-3′, 5′-AGCACTGTGTTGGCGTACAG-3′), hsa_circ_001787 (aka hsa_circ_0000034), (5′-AGATCCAAGGGCAACAAGTG-3′ and 5′-TTGCATACACATCCCGTCAT-3′). The GraphPad Prism version 8.0 (La Jolla, CA) was used for statistical analysis. The qRT-PCR result was analyzed using the ΔCt method. The relations between circ001787 regulation and clinical pathology features was analyzed via one-way analysis of variance (ANOVA). The receiver operating characteristic (ROC) curve was created using MedCal 19.4.1. The P values < 0.05 have been selected as statistically significant.
2.3. In silico analysis
2.3.1. MRE prediction
miRNA binding sites (MREs) of hsa_circ_001787 were predicted via two databases include CircInteractome and Cancer-Specific CircRNA (CSCD). We recognized miRNAs based on the intersection of the two mentioned databases (Table 2).
Table 2.
Binding sites of miRNA with hsa_circ_001787.
| miRNA ID | Site Type | CircRNA Start | CircRNA End |
|---|---|---|---|
| hsa-miR-1204 | 8mer-1a | 97 | 104 |
| hsa-miR-450b-3p | 7mer-m8 | 212 | 218 |
| hsa-miR-769-3p | 7mer-m8 | 212 | 218 |
| hsa-miR-1270 | 7mer-m8 | 66 | 72 |
2.3.2. miRNAs expression validated by GEO data
The expression profile of miRNAs was retrieved from GEO. We searched for various terms such as : colorectal, rectal, cancer, carcinoma, neoplasm, colon, CRC, tumor, tumor, and malignant, as well as miRNA, microRNA, miR, noncoding RNA, and ncRNA. The different criteria used to screen the datasets were: 1) diagnosis of rectal and/or colon cancer and 2) minimum tumor size of 3 cm. The following information was documented for each miRNA: miRNA type, region, the number of cases, and miRNA expression. Also, the information, including the name of the initial author, year of publishing, data of resources, and a webserver.
2.3.3. Prediction of miRNA target genes
MicroRNA target genes (MiTGs) were predictedby miRWalk. In addition, only interested genes predicted by a minimum of eight algorithms were crossed for additional consideration. The research involved 12 predicted algorithms (miRWalk, Microt4, mirbridge Targetscan, RNAhybrid, RNA22, PITA, Pictar2, miRNAMap, miRDB, miRanda, and miRMap) to ensure the correctness of forecast results (Dweep and Gretz, 2015).
2.3.4. Construction of network ceRNA
The circRNA-miRNA-mRNA network was created through the convergence of miRNA target genes forecasted and DEGs. The free, open-source Cytoscape v3.8.2 has been used for the illustration of construction.
2.3.5. GO and KEGG functional enrichment analysis
DAVID is a user-friendly bioinformatics resource freely available to anyone that provides a comprehensive biological knowledge base of functional interpretation tools for lists of genes/proteins. We sequentially used this to perform GO analysis and KEGG pathway enrichment analysis about the imbricated genes with a setting P value<0.05 and a count greater than 2.
2.3.6. Survival incident rate evaluation
GEPIA is an online software tool that can implement rapid and customizable TCGA and GTEx. Additionally, overall and disease-free survival rates (OS) were calculated according to the Kaplan-Meier curve; survival analysis was evaluated by using the log-rank test. A P values below 0.05 has been settled as the threshold for significance. A ceRNA subnetwork was generated based on mRNAs that have significant associations with colorectal cancer.
3. Result
3.1. The expression of circ001787 in colorectal cancer tissues
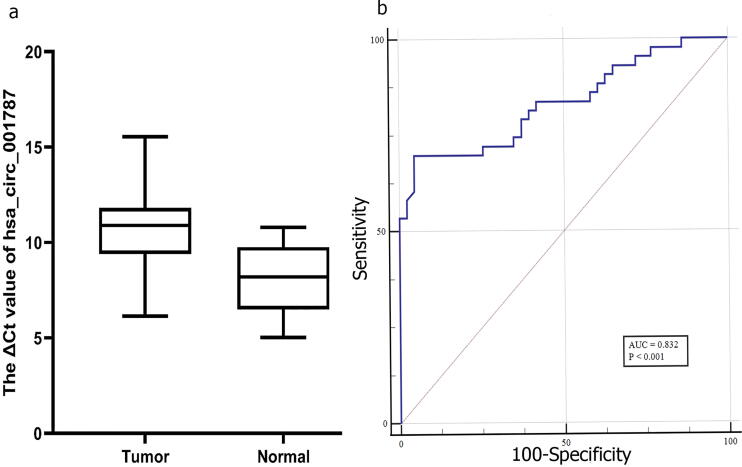
Due to the expression of circ001787 in colorectal tissues, qRT-PCR was performed in 43 paired CRC and non-tumor tissues. The result showed that circ001787 has been significantly downregulated in tissues in contrast to paired normal tissue (P values < 0.05) Fig. 1a.
Fig 1.
a) The expressions of circ001787 in colorectal cancer tissue samples and corresponding normal tissues. The box plot showed that circ001787 expressions in CRC tissue were significantly more than those in corresponding non-tumorous tissues (n = 43, P values < 0.05). (a higher ΔCt value indicates lower expression).Fig b). Diagnostic value (ROC) for the expression circ001787 in 43 paired tissue of CRC patients.
3.2. Diagnostic and prognostic value of circ001787 in colorectal cancer
Since we found there was a significantly low expression of circ001787 in CRC tissues, we further considered the relationships between circ001787 and some clinical-pathological factors. According to the data in Table 1, hsa_circ_001787 levels have been significantly associated with tumor grade (P = 0.039) and site of the primary tumor (P value = 0.040). Furthermore, hsa_circ_001787 level was negatively related to other clinic pathological features including, age and TNM. The ROC curve was created to examine the potential diagnosis value of hsa_circ_001787 Fig. 1b. We found that in CRC tissues, the area under the curve(AUC) reached 0.83, while sensitivity and specificity were 69.77% and 95.35%, respectively.
Table 1.
Correlation between hsa_circ_0001787 expression and clinicopathological parameters in CRC patients.
| Variable | Number of case (percentage) | Mean ± SD | P value |
|---|---|---|---|
| Tumor grade | |||
| I-II | 37 (86%) | 10.33 ± 1.98 | 0.0397 |
| III, VI | 6 (14%) | 12.27 ± 2.47 | |
| Location | |||
| Colon | 31 | 10.30 ± 2.07 | 0.0403 |
| Rectum | 12 | 11.36 ± 2.10 | |
| Age | |||
| ≥60 | 25 | 10.76 ± 1.94 | 0.324 |
| <60 | 18 | 9.91 ± 2.10 | |
| TNM | |||
| I-II | 21 | 10.19 ± 1.84 | 0.1346 |
| III-VI | 22 | 11.00 ± 1.42 | |
3.3. Deregulation of four miRNAs targeted by circRNA001787
Deregulation of four miRNAs including miR-1204, miR-450b-3p, miR-1205 and miR-1270 were confirmed in data sets obtained from GEO. Depending on the pooled assessment, down-regulation of miR-1204 (standardized mean difference) SMD = 0.10, 95% CI −1.47 to −1.66, p< 0.01); miR-450b-3p (SMD = 0.21, 95% CI −1.68 to 2.09 p< 0.01) (Fig. 2 a and b); miR-769 (SMD = -0.07, 95% CI −1.48 to 1.33p < 0.01) and up-regulation of miR-1270 (SMD = 1.07, 95% CI 0.60 to 1.54, p < 0.01) in CRC were confirmed (Fig. 3 c and d). The differential expression of miRNAs has shown in Supplementary Fig. 1 through dbDEMC.
Fig. 2.
Meta-analysis of gene set enrichment evaluating the expression of the four miRNAs in colorectal cancer (CRC): a) miR‑1204, b) hsa-miR‑450b-3p, c) hsa-miR-769-3p, and d) hsa-miR-1270.
Fig. 3.
circRNA–miRNA–mRNA regulatory network.
3.4. Building of a circular RNA, microRNA, and messenger RNA network
To introduce the interrelationships between circRNA, miRNA, and mRNA, a circRNA-miRNA-mRNA network was created by combining the circRNA/miRNA relationships and miRNA/miRNA interplay (Fig. 3). It represented an interpretation about the linkages between the hsa_circ_001787, 4 miRNAs and the 24 mRNAs (Fig. 4) (See Fig. 5).
Fig. 4.
Venn diagram for the intersections between DEGs and miRNA target genes.
Fig. 5.
Volcano plot of the differentially expressed genes (DEGs) in COAD (colon) and READ (Rectal) based on data from TCGA.
3.5. Gene ontology and KEGG enrichment analyses
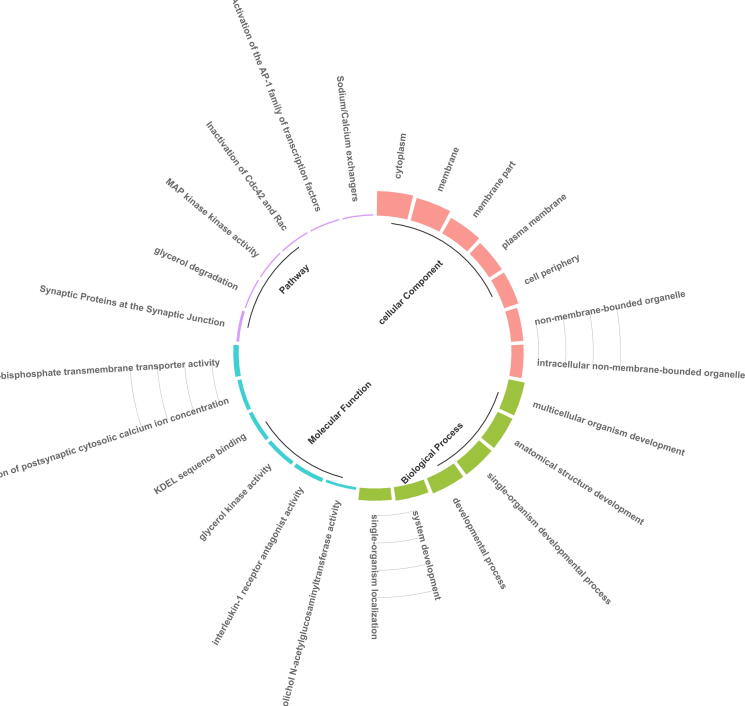
The performance illustrated that the declared mRNAs were particularly enriched in biological function, cellular components, and molecular function. In terms of MF, these overlapped genes are associated with “N-acetylglucosaminyltransferase activity” “interleukin-1 receptor antagonist activity” “glycerol kinase activity” “KDEL sequence binding” “calcium: cation antiporter activity involved in the regulation of postsynaptic cytosolic calcium ion concentration”, and “adenosine 3',5'-bisphosphate transmembrane transporter activity”. In terms of BP, these overlapped genes are associated with “multicellular organism development” “anatomical structure development” “single-organism developmental process” “developmental process” system development” “single-organism localization” “nervous system development” “protein localization” “cellular localization” and “macromolecule localization”. In terms of CC, these overlapped genes are associated with “cytoplasm” “membrane” “membrane part” “plasma membrane” “cell periphery” “intracellular non-membrane-bounded organelle” and “non-membrane-bounded organelle”. In terms of pathway, these overlapped genes are associated with “Synaptic Proteins at the Synaptic Junction” “glycerol degradation” “MAP kinase activity” “Inactivation of Cdc42 and Rac” “Activation of the AP-1 family of transcription factors” and “Sodium/Calcium exchangers”, Fig. 6.
Fig. 6.
The pie chart of hub gene enrichment analysis in overlapping genes.
3.6. Total survival analysis of mRNAs and subnetwork construction
We accomplished the total survival rate for the four mRNAs in the built-up circRNA-miRNA-mRNA network. Four mRNAs became markedly associated in the progression of CRC patients (Fig. 7 a,b,c, and d). We structured a subnetwork with four mRNAs, four miRNAs, and circRNA-001787 (Fig. 8).
Fig. 7.
The high expression level of significant genes group in colorectal cancer had a high level of positive results in screening candidates in diagnosing colorectal cancer. a) FGF13 (P = 0.032) b) KCNB1(P = 0.013), c) RNF217 (P = 0.073) and SRGAP2 (P = 0.053). (P values < 0.05 was the cut-off criterion).
Fig. 8.
The subnetwork was built using mRNAs with survival prognostic potential as circular RNA, microRNAs, and mRNAs.
4. Discussion
Circular RNAs had been initially discovered in RNA viruses (Sanger, 1976). With the rapid application development of next-generation sequencing, circRNAs have gradually become a new hotspot in cancer and RNA research (Chen, 2015). CircRNAs have a covalently closed cyclical structure without a free 5′ cap to 3′ poly(A) tail (Zaphiropoulos, 1996). CircRNAs have two significant characteristics: high stability and conspicuously conserved with a halftime of more than 48 h. circRNAs have the potential to be biomarkers in diagnosing different diseases such as neurological disorders, cardiovascular diseases, and tumors. (Rong, 2017). Recently, attention has been primarily focused on the potential function of circRNAs. Even so, their clinical diagnostic and prognostic value stays largely unexplored. Subsequent studies indicate that circRNAs act as miRNA sponges and controls splicing and transcription. For instance, hsa-circ-0000567 serves as a sponge for microRNA-421, a tumor repressor of hepatocellular carcinoma (HCC) via the circSETD3/miR-421/MAPK14 pathway (Xu, 2019). To the best of our knowledge, this is the first research to investigate the expression and diagnostic significance of hsa_circ_001787 in CRC. Importantly, we identified its potential diagnostic value for the first time. The tumor grade and primary site of the tumor are vital clinical factors impacting therapeutic outcomes in cancer patients. In the current research, We realized that low levels of hsa_circ_001787 in CRC tissues become positively associated with tumor grade and the site of the primary tumor (Table 1). There was no evidence of a clear association between the level of has circ 001,787 expressions and the number of lymph node metastases. The expression of hsa_circ_001787 is significantly less in the tumor grads. Furthermore, the low level of hsa_circ_001787 was significantly associated considerably with primer tumor site the level of hsa_circ_001787 in the colon and rectosigmoid was lower than other primary tumor sites. ROC curve analyses revealed that hsa_circ_001787 had a diagnostic accuracy of 0.832 (95% CI is from 0.735 to 0.904) with 69.77% sensitivity and 95.35% specificity. The positive and negative likelihood of AUC is 1. The ROC curve indicated that hsa_circ_001787 has a high potential for screening elevated colorectal cancer. In this analysis, we only used a small number of samples, and the findings need to be checked with a significantly larger number of samples. To examine the functionality in colorectal cancer, the circRNA-miRNA-mRNA network was constructed according to The Cancer Genome Atlas (TCGA) and Bioinformatics’ analysis.
Gene ontology enrichment evaluation established that mRNAs (genes) in this network were principally included in the “multicellular development organism” “adenosine 3',5'-bisphosphate transmembrane transporter activity” and “cytoplasm “. The pathway (KEGG) analysis suggests that these transcripts are involved in “MAP Kinas activity” “Activation AP-1 family “, and other pathways. In brief, MAPK is a family member of Ser/Thr, which triggers numerous of phosphorylation-activating kinase circles from the membrane to the core of the cells (nucleus). Three large subfamilies of MAPK are the NK/SAPK's, p38 MAPK, and MEK/ERK. MAP kinase signaling pathway has been coordinated to oncogenic activity and therefore plays the leading role in tumor genesis and promotion of CRC. Activation 1 (AP1) regulates different genes essential for proliferation, invasion, migration, and differentiation in cancer cells. The AP1 is a transcription factor that includes JUN-JUN heterodimer or JUN-FOS heterodimer. AP1 has a crucial role in CRC development and inhibition of AP1 by regulating circRNAs which can reduce the proliferation of CRC cells. These outcomes show that CircRNAs in this network plays an essential part in the incidence and progression of colorectal cancer. This achievement is worth more investigation. To further more study of which hsa_circ_001787 influences the condition's prognosis CRC, we investigated the connection between the expression of 24 mRNAs in overall survival and network construction in colorectal cancer patients authorized on the GEPIA data. The research we conducted demonstrated that four mRNAs (FGF13, KCNB1, RNF217, and SARGAP2) may impact the diagnosis and treatment of CRC. In addition, we algorithmically constructed subnetworks containing the overall survival-related mRNAs, four miRNAs (hsa-miR-1204, hsa-miR-450b-3p, hsa-miR-769-3p, and hsa-miR-1270) and hsa_circ_001787. The upregulation of FGF13 saved the inhibition impact of miR-10b on CRC cells promotion, invasion, and metastasis. Subsequently, the knockdown of FGF13 was eligible to mimic the suppression effects of miR-10b on the growth of colorectal cancer cell lines (Song and Li, 2019).
Immunotherapy plays an important role in the treatment of many different types of cancers like colorectal, liver, glioma, and other malignancies. However, finding the right target for the cancer treatment is also a challenge we face. Here, for the first time in the world, we used a biological to forecast four significant survival genes in colorectal cancer; it might be the best candidate for immunotherapy. At this moment, chimeric Antigen Receptor T cells (CARs) have started a new avenue for cancer therapy (Dana, 2020); For instance, in this study we found FGF13 has a significant association with CRC, so then it can be good candidate for CAR T cells therapy. Recently, a Creative Biolabs company constructed an FGF13 vector CAR T cells against lymphoma cancer (https://www.creative-biolabs.com/car-t/anti-fgf13-s235-22-h-cd28-cd3-car-pcdcar1-16025.htm).
Although colonoscopy is the gold standard of colorectal cancer, it serves as a labor-intensive and invasive means that cannot be applied to every patient. Therefore, there is an essential need for a cost-effective and noninvasive CRC diagnosing and screening test to improve accuracy and acceptability. The use of hsa_circ_001787 could be the next generation CRC screening biomarkers. Furthermore, the circ001787 connection with four miRNAs with indirect regulation of four significant genes in CRC has been documented for the first time in the world.
The other genes have not been discovered in the corresponding miRNAs in cancer, specifically in CRC. We have found 4 prognostic genes that have not been represented in past research regarding diagnosing colorectal cancer in different stages. Further research is needed to determine whether the deregulation of these genes affects the prognosis of CRC patients. At the moment, there have been insufficient studies on the mechanism of circular RNAs in colorectal cancer. In this study, we, for the first time, reported the possible biomarker of hsa_circ_001787 in CRC tissues and constructed a network hsa_circ_001787-miRNAs-mRNAs acting as miRNA sponges. We further demonstrated the overall survival response of prognostic genes based on the GEPIA and created a subnetwork.
5. Conclusion
All taken together, our experiments show that precise circRNAs, such as hsa_circ_001787, may be suggested as a potential noninvasive indicator for the prognosis of colorectal cancer. The sensitivity and specificity of forty-three colorectal and non-colorectal tissues were 69.77%, and 95.35%, respectively. We identified different expressions of miRNAs (meta-analysis) and mRNAs through the GO and TCGA databases. Furthermore, four mRNAs linked to the prognosis of CRC have been recognized through the GEPIA webserver. We constructed a circRNA-miRNA-mRNA subnetwork that perhaps, in the future, will be related to the prognosis of CRC. The outcomes of this research may enable a novel perspective into the pathogenesis of colorectal cancer and recommend powerful therapeutic agents that warrant additional investigation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We thank Mrs. Barbara Berwick for editing the manuscript. This research was supported by a grant from Tehran University of Medical Science.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2021.06.071.
Contributor Information
Mohammad Hossein Sanati, Email: m-sanati@nigeb.ac.ir.
Seyed Rohollah Miri, Email: Srmiri@sina.tums.ac.ir.
Habibollah Mahmoodzadeh, Email: hmahmoodzadeh@tums.ac.ir.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Supplementary Fig. 1.
The differential expression of miRNAs, Red: up-regulate, and Green: down-regulate
References
- Siegel, R.L., K.D. Miller, and A. Jemal, Cancer statistics, 2016. CA: a cancer journal for clinicians, 66(1) (2016) 7–30. [DOI] [PubMed]
- Cho W.C. Epigenetic alteration of microRNAs in feces of colorectal cancer and its clinical significance. Expert Rev. Mol. Diagnostics. 2011;11(7):691–694. doi: 10.1586/erm.11.57. [DOI] [PubMed] [Google Scholar]
- Levin, B., et al., Emerging technologies in screening for colorectal cancer: CT colonography, immunochemical fecal occult blood tests, and stool screening using molecular markers. CA: a cancer journal for clinicians, 3(1) (2005) 44–55. [DOI] [PubMed]
- Smith, R.A., et al., American Cancer Society guidelines for the early detection of cancer: update of early detection guidelines for prostate, colorectal, and endometrial cancers: Also: update 2001—testing for early lung cancer detection. CA: a cancer journal for clinicians, 51(1) (2001) 38–75. [DOI] [PubMed]
- Chalbatani G.M. BIOMARKERS & CLINICAL RESEARCH. Oncol. 2016;2:3. [Google Scholar]
- Tran, A.M., et al., A new world of biomarkers and therapeutics for female reproductive system and breast cancers: circular RNAs. Frontiers in cell and developmental biology, 2020. 8. [DOI] [PMC free article] [PubMed]
- Chen L.-L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T.B. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Saglam, A.S.Y., E. Alp, and H.I. Onen, Circular RNAs and Its Biological Functions in Health and Disease, in Gene Expression and Phenotypic Traits. 2019, IntechOpen.
- Zhang Y. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY) 2017;9(6):1585. doi: 10.18632/aging.101254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K.-Y. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77(9):2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng W. Circular RNA ciRS-7—a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin. Cancer Res. 2017;23(14):3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco S. Circular RNAs in muscle function and disease. Int. J. Mol. Sci. 2018;19(11):3454. doi: 10.3390/ijms19113454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. Circular RNAs: Isolation, characterization and their potential role in diseases. RNA Biol. 2017;14(12):1715–1721. doi: 10.1080/15476286.2017.1367886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo F. The expression profile and clinical significance of circRNA0003906 in colorectal cancer. OncoTargets Therapy. 2017;10:5187. doi: 10.2147/OTT.S147378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep, H. and N. Gretz, miRWalk2. 0: a comprehensive atlas of microRNA-target interactions. Nature methods 12(8) (2015) pp. 697–697. [DOI] [PubMed]
- Sanger H.L. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Circular RNAs in eukaryotic cells. Curr. Genomics. 2015;16(5):312–318. doi: 10.2174/1389202916666150707161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos P.G. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc. Natl. Acad. Sci. 1996;93(13):6536–6541. doi: 10.1073/pnas.93.13.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong D. Novel insights into circular RNAs in clinical application of carcinomas. OncoTargets Therapy. 2017;10:2183. doi: 10.2147/OTT.S134403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J. Exp. Clin. Cancer Res. 2019;38(1):1–15. doi: 10.1186/s13046-019-1041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Li W. MiR-10b suppresses the growth and metastasis of colorectal cancer cell by targeting FGF13. Eur. Rev. Med. Pharmacol. Sci. 2019;23(2):576–587. doi: 10.26355/eurrev_201901_16870. [DOI] [PubMed] [Google Scholar]
- Dana H. CAR-T cells: early successes in blood cancer and challenges in solid tumors. Acta Pharmaceutica Sinica B. 2020 doi: 10.1016/j.apsb.2020.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]