Abstract
The role of human papillomavirus (HPV) in anogenital carcinogenesis is firmly established, but evidence that supports a similar role in skin remains speculative. Immunosuppressed renal transplant recipients have an increased incidence of viral warts and nonmelanoma skin cancer, and the presence of HPV DNA in these lesions, especially types associated with the condition epidermodysplasia verruciformis (EV), has led to suggestions that HPV may play a pathogenic role. However, differences in the specificities and sensitivities of techniques used to detect HPV in skin have led to wide discrepancies in the spectrum of HPV types reported. We describe a degenerate nested PCR technique with the capacity to detect a broad spectrum of cutaneous, mucosal, and EV HPV types. In a series of 51 warts from 23 renal transplant recipients, this method detected HPV DNA in all lesions, representing a significant improvement over many previously published studies. Cutaneous types were found in 84.3% of warts and EV types were found in 80.4% of warts, whereas mucosal types were detected in 27.4% of warts. In addition, the method allowed codetection of two or more distinct HPV types in 94.1% of lesions. In contrast, single HPV types were detected in all but 1 of 20 warts from 15 immunocompetent individuals. In summary, we have established a highly sensitive and comprehensive degenerate PCR methodology for detection and genotyping of HPV from the skin and have demonstrated a diverse spectrum of multiple HPV types in cutaneous warts from transplant recipients. Studies designed to assess the significance of these findings to cutaneous carcinogenesis are under way.
Human papillomaviruses (HPVs) are important human carcinogens (30). There is now overwhelming evidence from both epidemiological and functional studies that specific high-risk HPV types are one of the major etiological agents responsible for anogenital cancer. A role for HPV in nonmelanoma skin cancer (NMSC) has also been proposed (16, 17) but remains controversial with the possible exception of a role in the rare disorder epidermodysplasia verruciformis (EV). This condition is characterized by a genetic predisposition to widespread cutaneous infection with HPV types not usually pathogenic in the healthy population and the subsequent progression of these warts to squamous cell carcinoma (SCC) on sun-exposed sites (13).
There is also accumulating evidence that HPV may participate in the pathogenesis of NMSC in immunosuppressed renal transplant recipients (RTRs). Over 90% of patients develop viral warts and up to 40% of patients develop NMSC within 15 years of transplantation, a 50- to 100-fold increased risk compared with that for the general population (3). An association between viral warts and skin cancer in RTRs was first noted in Australia (29), and there is now both clinical and histological evidence which indirectly supports the progression of viral warts through increasingly dysplastic squamous lesions to invasive SCC (1, 4, 5, 11). In the early posttransplantation years, warts are usually of the common or palmoplantar type (11). With increasing time after transplantation, increasing age, and higher levels of sun exposure, flat warts on sun-exposed sites develop, and these may become confluent, particularly over the dorsum of the hands and forearms. Histologically, such warts often show cytological atypia, and it is from such areas that SCCs often arise (4).
The study of HPV in NMSC has been limited by the methods used for detection and typing of HPV from the skin. Most available methods rely on the amplification of DNA by PCR. However, the existence of at least 80 distinct HPV types and their genomic diversity dictate that only closely related HPV types will be detected if type-specific primers are used. This has led to the application of degenerate PCR (21). By this technique the combined studies from several groups suggest that diverse cutaneous, mucosal, and EV HPV types may be found in benign and malignant skin lesions, particularly from immunosuppressed individuals, and that more than one HPV type may be detected within an individual lesion (2, 8, 20, 22). Nonetheless, discrepancies still exist in the published data, which may in part reflect the different sensitivities and specificities of particular degenerate primers for the detection of HPV types (16, 17).
We have recently examined these discrepancies by evaluating the sensitivities and specificities of three degenerate primer sets which have previously been used individually to detect HPV DNA in benign and malignant skin lesions from RTRs (27). By comparing the primer sets HVP2-B5 and F15-B15 (20, 21) and MY09-MY11 (15) and the nested set CP62-CP69 and CP65-CP68 (2) in PCRs with serial dilutions of cloned HPV and with a series of mucosal and cutaneous warts, we observed that the combined panel of primers allowed detection of a broader range of HPV types than was possible with the individual primer sets. However, it was apparent that such an approach, although advantageous, would require further modification. In particular, while the sensitivity for detection of EV types was high, that for detection of mucosal types was lower and that for detection of cutaneous types was lower still. Furthermore, sequence data for HPV isolates from some lesions indicated the presence of multiple HPV types which could not be individually identified with the primer sets used.
In the present study, we describe a modified degenerate PCR technique which seeks to address the problems inherent in existing methodologies. This approach has the capacity to detect a broad spectrum of cutaneous, mucosal, and EV HPV types to a high degree of sensitivity. We have used this technique to analyze a series of viral warts from RTRs.
MATERIALS AND METHODS
HPV plasmids.
Twenty-eight plasmid clones containing HPV genomes were used as representatives of cutaneous (HPV type 1 [HPV-1], -3, -4, -10, and -41), mucocutaneous (HPV-2 and -57), mucosal (HPV-6, -11, -16, -18, -26, -31, -32, -33, -34, -66 and -72), and EV (HPV-5, -8, -14, -19, -20, -22, -23, and -36) HPV types (see Table 2).
TABLE 2.
Detection of cloned HPV DNA by each degenerate primer set
| HPV type
|
Primer paira | Plasmid testedb | Sensitivityc | |
|---|---|---|---|---|
| Clinical groupd | Phylogenetic groupe | |||
| Cutaneous | General | HVP2-B5f | 1 | 1 pg |
| 2 | 10 pg | |||
| 3 | 100 pg | |||
| 4 | NDi | |||
| 10 | 100 pg | |||
| 41 | 1 pg | |||
| 57 | 100 pg | |||
| E | CN1F-CN1Rg | 1 | 0.001 fg | |
| 41 | 0.01 fg | |||
| A4 | CN2F-CN2Rg | 2 | 0.01 fg | |
| 57 | 10 fg | |||
| A2 | CN3F-CN3Rg | 3 | 1 fg | |
| 10 | 1 fg | |||
| B2 | C4F4Rf | 4 | 10 fg | |
| Mucosal | General | MY09-MY11f | 6 | 10 fg |
| 11 | 10 fg | |||
| 16 | 1 fg | |||
| 18 | 1 fg | |||
| 26 | 1 pg | |||
| 31 | 10 fg | |||
| 32 | 100 fg | |||
| 33 | 10 fg | |||
| 34 | 10 fg | |||
| 66 | 10 fg | |||
| 72 | 100 fg | |||
| General | MY11-GP6h | |||
| A1 | 32 | 1 fg | ||
| A3 | 72 | 1 fg | ||
| A5 | 26 | 10 fg | ||
| A6 | 66 | 1 fg | ||
| A7 | 18 | 0.1 fg | ||
| A9 | 16 | 0.1 fg | ||
| 31 | 1 fg | |||
| 33 | 0.1 fg | |||
| A10 | 6 | 1 fg | ||
| 11 | 1 fg | |||
| A11 | 34 | 1 fg | ||
| EV | General | CP62-CP69g | 14 | 0.01 fg |
| 19 | 0.01 fg | |||
| 20 | 10 fg | |||
| 5 | 0.01 fg | |||
| 8 | 0.01 fg | |||
| 36 | 100 pg | |||
| 22 | 10 fg | |||
| 23 | 1 pg | |||
| Cluster a1 | EN1F-EN1Rg | 5 | 10 fg | |
| 8 | 10 fg | |||
| 36 | 10 fg | |||
| Cluster a2 | EN2F-EN2Rg | 14 | 0.1 fg | |
| 19 | 10 fg | |||
| 20 | 10 fg | |||
| Cluster b1/b2 | EN3F-EN3Rg | 22 | 10 fg | |
| 23 | 10 fg | |||
Degenerate primer pairs (see text).
Plasmids containing cloned HPV DNA of representative HPV types within the group or cluster indicated.
Tenfold serial dilutions of HPV plasmid were amplified in a background of 100 ng of human placental DNA (see text) and the numbers of copies per cell were calculated. The absolute amount of HPV plasmid DNA detected ranged from 0.001 fg (equivalent to approximately 5 × 10−6 copies/cell) to 100 pg (equivalent to approximately 500 copies/cell).
HPV types categorized according to clinical association.
HPV type categorized according to phylogenetic group (6).
First-round primer pair.
Seminested primer pair.
Second-round (nested) primer pair.
ND, not detected.
Patients.
Patients comprised 23 immunosuppressed RTRs (18 men and 4 women). Ten had a history of NMSC, 3 had a history of premalignancies only, and 10 had had no premalignant or malignant skin lesions, despite the presence of a transplanted kidney for at least 10 years.
Tissue samples.
Specimens comprised 51 cutaneous warts. All lesions were histologically confirmed to be warts and were derived from biopsy specimens collected at the time of surgery, snap frozen, and stored at −70°C. Three large warts were subdivided into between two and four sections, and each section was analyzed independently. On the basis of clinical and/or histological criteria, the warts fell into two categories. The first category comprised verrucae vulgares or common warts (n = 23), which were considered to be those warts with typical morphological features and no evidence of dysplasia histologically. The second category of warts (n = 28) were those warts that occur on sun-exposed sites with atypical clinical features and/or evidence of epithelial dysplasia histologically (4). In addition, 20 benign warts obtained from 15 immunocompetent patients with warts were also analyzed (see below).
DNA extraction.
Samples were finely minced with sterile scalpels on a petri dish, washed twice in phosphate-buffered saline, and resuspended in lysis buffer containing proteinase K to a concentration of 0.1 mg/ml. Following lysis overnight at 37°C, samples were pelleted and the supernatant was removed and placed in a fresh tube. DNA was then extracted by a standard phenol-chloroform-isoamyl alcohol technique followed by ethanol precipitation (14).
Specific precautions were taken to prevent and monitor cross-contamination during the DNA extraction process. All procedures were carried out under conditions of strict pre- and post-PCR separation (12). DNA was extracted from the warts in 30 separate procedures. DNA was simultaneously extracted from samples from immunocompetent patients and samples from non-RTR immunosuppressed patients. DNA was usually extracted from multiple samples from individual patients in at least two different batches. HPV-negative skin (as defined by the fact that the sample was negative by our methodology) was used as a negative tissue control in seven extraction procedures. Buffer extraction controls which did not contain tissue were placed in the middle or at the end of an extraction procedure. Although the identities of the samples were not masked, they were coded following extraction, and the coding was revealed only after completion of PCR and sequencing. The coding was known to one investigator only.
PCR primers.
On the basis of our preliminary data, established degenerate primer pairs were modified in order to improve the detection of cutaneous, mucosal, and EV HPV types (Table 1). These primers were located within the highly conserved L1 (major capsid protein) open reading frame (ORF) of the HPV genome. By using the multiple alignment program Clustal V and the Genetics Computer Group (GCG) package of software programs, nested primers were designed within the existing degenerate primer pairs in the case of all HPV phylogenetic groups (6) with the exception of cutaneous group B2 (see below). The resulting primers were analyzed with the Oligo 4 Primer Analysis Software program (National Biosciences Incorporated).
TABLE 1.
L1 and E6 ORF oligonucleotide primers used in study
| HPV class | HPV group (type[s]) detected | ORF | Primer | Sequence (5′-3′)a | Annealing site
|
Size (bp) | Degeneracy | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| HPV type | Bases | ||||||||
| Cutaneous | General cutaneous | L1 | HVP2 | TCNMGNGGNCANCCNYTNGG | 18 | 5934–5953 | 650 | 16,384 | 21 |
| B5 | AYNCCRTTRTTRTGNCCYTG | 6561–6580 | 512 | 20 | |||||
| Cutaneous | Group B2 (4, 48, 50, 60, 65) | C4R | GGAGATACAGAAAATCCT | 4 | 5728–5746 | 330–335 | 0 | ||
| C4R | SHATCTCCATAGATATCTTT | 6062–6082 | 6 | ||||||
| Cutaneous | Group E (1, 41, 63) | L1 | CN1F | AATARGTTWGATGATGCWGAA | 1 | 5793–5814 | 309–328 | 8 | |
| CN1R | AKRTARTCWGGATATTTGCA | 6108–6128 | 16 | ||||||
| Cutaneous | Group A2 (3, 10, 28, 29, 77) | L1 | CN3F | AACTCTAAYATWGCACATG | 3 | 6140–6159 | 273 | 4 | |
| CN3R | CAVGTRCSYTGGCAAATATC | 6407–6427 | 24 | ||||||
| Mucocutaneous | Group A4 (2, 27, 57) | L1 | CN2F | GGGGATATGGTTGAAACAGGT | 2 | 6369–6360 | 294 | 0 | |
| CN2R | CAGAGGACACCATAGAGCCA | 6661–6681 | 0 | ||||||
| EV | General EV | L1 | CP62 | GTWAATGAAAYTTGYAANTATCC | 8 | 6520–6543 | 690 | 32 | 2 |
| CP69 | GWTAGATCWACATTCCARAA | 7231–7250 | 8 | 2 | |||||
| EV | General EV | L1 | CP65 | CARGGTCAYAAYAATTGGYAT | 8 | 6832–6851 | 250 | 16 | 2 |
| CP68 | GGDACRAAACCYARYTGCCA | 7100–7120 | 48 | 27 | |||||
| EV | B1, cluster a1 (5, 8, 12, 36, 47) | L1 | EN1F | TATTTCCCWACHGTHAGTGGCTC | 8 | 6753–6776 | 254 | 18 | |
| EN1R | TCATAYTCYTCTACATGTCT | 6987–7007 | 4 | ||||||
| EV | B1, cluster a2 (14, 19, 20, 21, 25) | L1 | EN2F | CTGTCAGTGGCTCATTGGT | 14 | 6524–6543 | 314 | 0 | |
| EN2R | CATWGCATTAATTTGAGCTA | 6818–6838 | 2 | ||||||
| EV | B1, cluster b1 + b2 (9, 15, 17, 37, 22, 23, 38) | L1 | EN3F | ATGKCWAATGATGTHTATGG | 23 | 6367–6387 | 293 | 12 | |
| EN3R | TGRTTRYYCCAYAAAATRCCATT | 6637–6660 | 32 | ||||||
| Mucosal | General mucosal | L1 | MY11 | GCMCAGGGWCATAAYAAYTGG | 6 | 6722–6742 | 450 | 16 | 15 |
| MY09 | CGTCCMARRGGAWACTGATC | 7150–7170 | 16 | 15 | |||||
| Mucosal | Seminested | L1 | MY11 | GCMCAGGGWCATAAYAAYTGG | 190 | 16 | 15 | ||
| GP6 | GAAAAATAAACTGTAAATCA | 0 | 24 | ||||||
| Cutaneous | 10 (type specific) | E6b | 10E6F | ATGTCCATGGGTGCACAGGAA | 10 | 1–21 | 444 | 0 | |
| 10E6R | CTGTGGGATGCGGACCGTGCA | 424–444 | 0 | ||||||
| EV | 23 (type specific) | E6b | 23E6F | ATGCAGACTGTGCATTATTTA | 23 | 1–21 | 454 | 0 | |
| 23E6R | TCATTCTATTTCCTTACAATG | 454–474 | 0 | ||||||
| EV | 24 (type specific) | E6b | 24E6F | ATGGCTCAACCAGGTAAACCT | 24 | 1–21 | 423 | 0 | |
| 24E6R | TTATATCTGCTTACACTGCCT | 403–423 | 0 | ||||||
| Cutaneous | 27 (type specific) | E6b | 27E6F | ATGCGCACAAGGGCAGGGATG | 27 | 1–21 | 480 | 0 | |
| 27E6R | TTAATGTAATGTCCGCGAGGC | 460–480 | 0 | ||||||
Degenerate base code: N = G, A, C or T; D = G, A, or T; H = A, T, or C; K = G or T; M = A or C; R = A or G; S = C or G; V = G, A or C; W = A or T; Y = T or C.
For E6 type-specific primers, each of the forward primers had the sequence CACGGATCC at the 5′ end and each of the reverse primers had the sequence CACGAATTC at the 5′ end to facilitate subsequent cloning.
(i) Cutaneous HPV types.
The degenerate primer pair HVP2-B5 was described by Shamanin et al. (20, 21) for detection of HPV from all groups with the exception of the phylogenetic clade comprising HPV-4, -48, -50, -60, and -65, for which the primer pair F14-B15 was used. Of the primer pairs evaluated in our preliminary studies, these preferentially detected cutaneous HPV types, but only at a viral copy number of 5 to 5,000 copies/cell (27). We therefore designed the following primers nested within HVP2 and B5 to increase the sensitivity and specificity for detection of indicated cutaneous types: CN3F-CN3R, group A2 (HPV-3, -10, -28, -29, and -77); CN2F-CN2R, group A4 (HPV-2, -27, and -57); CN1F-CN1R, group E (HPV-1, -41, and -63). The single-round primer pair C4F-C4R was designed to detect the cutaneous group B2 (HPV-4, -48, -50, -60, and -65) in order to improve the sensitivity obtained with F14-B15.
(ii) Mucosal HPV types.
The primer pair MY09-MY11 was originally designed by Manos et al. (15) for the detection of HPV in genital lesions. In preliminary studies it detected mucosal HPV types at approximately 10 fg to 1 pg (0.05 to 5 copies/cell). To increase the level of sensitivity, a second established primer, GP6, was seminested within MY11 (24).
(iii) EV HPV types.
Berkhout et al. (2) have described several pairs of nested primers which preferentially detect EV HPV types. Of these, one particular set comprising CP62-CP69 as an outer primer pair and CP65-CP68 as an internal nested pair was chosen for the purposes of this study since they were highly sensitive, detecting EV HPV types at levels of 10−4 copies per cell. In order to facilitate the detection of mixed EV-associated HPV infections potentially present in skin, as predicted by our preliminary data and those of others (2, 27), additional nested primer pairs within CP62-CP69 were designed for the major EV HPV clusters, as follows: EN1F-EN1R, cluster a1 (HPV-5, -8, -12, -36, -47, and -ICPX1); EN2F-EN2R, cluster a2 (HPV-14, -19, -20, -21, and -25); and EN3F-EN3R, clusters b1 and b2 (HPV-9, -15, -22, -23, -37, -38, -RTRX1, -VS42, -VS73, -RTRX3, -RTRX6, -VS92, and -VS102).
PCR amplifications.
PCR amplifications were performed exactly as described for previously published primer pairs. For the newly designed primers, amplification reactions were performed in 50 μl of a reaction mixture containing 1 U of AmpliTaq DNA polymerase (Perkin-Elmer, Norwalk, Conn.), GeneAmp 10× PCR Buffer II and 2 mM MgCl2 (supplied by the manufacturer), a 0.2 mM concentration of each deoxynucleotide triphosphate (Advanced Biotechnologies Ltd.), and 50 pmol of each primer. These conditions were established with magnesium and annealing temperature titrations. Thirty cycles of PCR were performed (95°C for 1 min, 50°C for 1.5 min, and 72°C for 2 min), followed by extension at 72°C for 5 min. All PCRs were performed on a Perkin-Elmer 480 thermal cycler.
Initially, the sensitivity and specificity of each primer set were determined by PCR amplification of 10-fold serial dilutions of each HPV plasmid (ranging from 100 ng to 0.001 fg of plasmid DNA) in the presence of a background of 100 ng of human placental DNA (Sigma). For the subsequent analysis of clinical samples, 100 to 200 ng of cellular DNA was used as the template in each first-round PCR. Prior to amplification with the HPV-specific primers, samples were amplified with beta-globin primers PC04 and GH20 to confirm adequate preservation of DNA (18).
For each PCR amplification, negative controls for reagents and DNA (human placental DNA [Sigma]) were included and processed in the same way as the lesional samples throughout, as were the negative controls for DNA extraction. None of the negative controls was positive for HPV. HPV plasmid clones (10 fg) containing the genotype of HPV-1, -2, -3, -4, -5, or -6 served as positive controls.
Sequence analysis.
Amplified PCR products that appeared as visible bands after ethidium bromide staining were purified after separation in a 2% agarose low-melting-point gel (QIAquick Gel Extraction Kit; QIAGEN) and were directly sequenced by fluorescent dideoxynucleotide chain termination cycle sequencing on a Perkin-Elmer 2400 thermal cycler (ABI Prism Dye Terminator Cycle Sequencing Ready Reaction Kit; Perkin-Elmer) with both forward and reverse primers. The products were analyzed on a Perkin-Elmer 377 ABI Prism automated sequencer. The nucleotide sequences obtained were analyzed with the Sequence Navigator computer software (Macintosh). Sequences of 140 bases or more with fewer than 5% unidentified bases were processed. The forward and reverse complement sequences were aligned, and the homology of the consensus sequence was compared with those of known HPV types available through the GenBank database (National Center for Biotechnology Information, National Institutes of Health, Bethesda, Md.) by using the GCG Blast program. In accordance with established guidelines, a nucleotide sequence was regarded as an HPV type if it shared over 90% homology with a known type and a related type if the sequence homology was less than 90% (6).
Use of differential PCR to detect mixtures of viruses.
On the basis of our previous study, it was predicted that a proportion of lesions were likely to harbor more than one HPV type (27). In order to determine whether the panel of primer sets would detect their target when a mixture of viruses was present, HPV plasmids from three different phylogenetic groups were mixed together. The primers designed to detect these types were used sequentially to amplify the mixture. Since mixtures with EV HPV types in particular had previously emerged in clinical samples from immunosuppressed patients (27), EV HPV types 5, 14, and 23 from clusters a1, a2 and b1-b2, respectively, within group B1 (6) and the corresponding primers EN1, EN2, and EN3 were chosen for the purposes of this experiment. Each plasmid was used at a final concentration of 50 ng/ml in the plasmid mixtures.
Comparison of differential PCR versus PCR and cloning for detection of mixtures of viruses.
An alternative strategy for the detection of mixtures of HPV types within individual lesions is to clone the PCR products generated by the general degenerate primers for cutaneous, EV, and mucosal types and then to sequence a proportion of the resulting clones. We compared the ability of the multiple nested primer and direct sequencing approach adopted in this study with PCR and cloning to differentially detect mixtures of HPV types. By using as a model the mixtures of EV HPV plasmids described above, the gel-purified PCR amplification products generated by the nested PCR with CP62-CP69 and CP65-CP68 were cloned with the PCR-Script Cloning Kit (Stratagene). At least five resulting colonies were picked and amplified with primers CP65 and CP68 to identify positive clones. The PCR products were then gel purified and sequenced as described above.
Confirmation of results obtained for clinical specimens with L1 degenerate primers with E6 type-specific primers.
Type-specific primers for the E6 ORFs of HPV-10, -23, -24, and -27 (Table 1) were designed by using E6 nucleotide sequence data (Los Alamos National Laboratory HPV Sequence Database, 1997). The HPV types were randomly chosen from among those that had occurred at least once in the series of clinical specimens. By using the E6 ORF primers, PCR amplification of several representative clinical samples was used as additional confirmation of the results obtained with the L1 degenerate nested primers. Amplification reactions were performed in 50 μl of reaction mixture as described above. Forty cycles of PCR were performed (95°C for 1 min, 55°C for 1 min, and 72°C for 1 min), followed by extension at 72°C for 5 min. In plasmid titration experiments, each E6 primer set detected its relevant target plasmid to a sensitivity of at least 0.01 fg.
Comparison of results obtained by an alternative degenerate PCR-based methodology.
De Villiers et al. (9) have recently refined their PCR-based HPV DNA detection and genotyping method to include a set of degenerate EV HPV-specific nested primers first described by Berkhout et al. (2), in addition to their 16 established pairs of degenerate PCR primers (22). DNA from some of the clinical samples analyzed as described above had previously been analyzed by this methodology. Although DNA was extracted from the residual tissue for the purposes of this study, such that identical aliquots of DNA were not analyzed by the two laboratories, it was of interest to us to compare the results obtained by both methods for the same lesions as further validation of our nested primer approach.
Comparison of results with those for warts from immunocompetent individuals.
In order to exclude the possibility that our HPV detection methodology was overly sensitive or that the results were attributable to PCR artifacts, a biological control in the form of a series of warts from immunocompetent patients was also analyzed. Lesions comprised 20 benign warts from 15 patients, and the clinical spectrum was extended to include not only cutaneous warts but also examples of lesions from anogenital, oral, and conjunctival mucosae. HPV DNA detection and genotyping were performed exactly as described above for warts from RTRs.
RESULTS
Sensitivity and specificity of degenerate primer sets.
Analysis of serial dilutions of individual cloned HPV plasmids confirmed the increased specificity and sensitivity of the modified degenerate PCR approach described here (Table 2). Representative HPV types from the main cutaneous, mucosal, and EV phylogenetic groups were each detected to a sensitivity of at least 10 fg (0.05 copies per cell), although this varied (0.001 to 10 fg, equivalent to 5 × 10−6 to 0.05 copies per cell).
Detection of mixtures of EV-associated HPV plasmids by differential PCR or PCR and cloning.
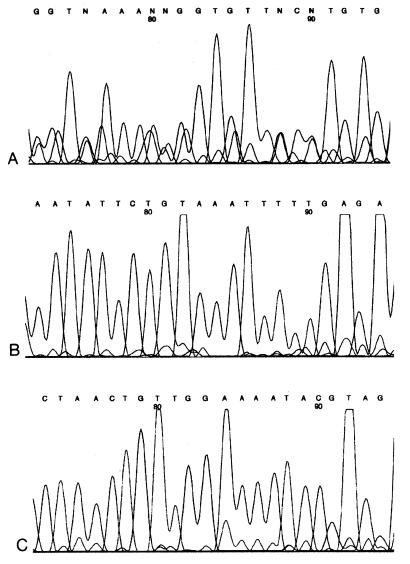
By analyzing mixtures of EV HPV types 5, 14, and 23, we observed that the respective primer sets EN1, EN2, and EN3 successfully identified the EV HPV type for which they were specifically designed. Following PCR cloning of the same plasmid mixtures, all three HPV types included were also successfully detected. It proved necessary to sequence at least four clones from each respective mixture in order to ensure detection of both HPV types present. The capacity of differential PCR to detect mixed EV HPV infections in clinical lesions is illustrated in Fig. 1. The electropherogram presented in Fig. 1A is the result of PCR amplification of lesion iv from patient 11 with the general EV HPV primers CP62-CP69 and CP65-CP68, followed by sequencing with CP68. The poor quality of the sequence is evident, in particular, the large number of ambiguous bases, and this almost certainly reflects the superimposed sequences of at least two HPV types. In contrast, Fig. 1B and C show the results for the same lesion following PCR with the nested primer pair EN2 and EN3, respectively. In both cases the sequences are clear with no ambiguous bases and represent HPV-25 and -38, respectively.
FIG. 1.
Mixed EV HPV types are detected with cluster-specific nested primers. The electropherograms in panels A to C were from lesion iv from patient 11. (A) The sequence was obtained with the general EV HPV primer CP68 and shows features suggestive of the presence of at least two distinct HPV types (see text). (B) The electropherogram was obtained with EN2R, the reverse primer for the pair designed to detect EV cluster a2. This sequence appears to be a single type and was subsequently proved to represent HPV-25. (C) The electropherogram was obtained with EN3R, the reverse primer for EV HPV clusters b1 and b2. This sequence also appears to be a single type and represents HPV-38.
Clinical specimens from RTRs.
By using the combination of primers, HPV DNA was found in all of the warts analyzed. Examples of PCR amplifications from selected clinical samples are shown in Fig. 2. Cutaneous HPV types predominated, being found in 43 of 51 (84.3%) warts; EV HPV types were found in 41 (80.4%) lesions, and mucosal types were found in 14 (27.4%) warts. Mixed infections, in which two or more HPV types were detected, occurred in 48 of 51 (94.1%) warts. The majority of mixed infections contained two or three different HPV types, but six distinct types were identified in one wart (lesion i, patient 12). Mixed infections were predominantly with cutaneous and EV HPV types.
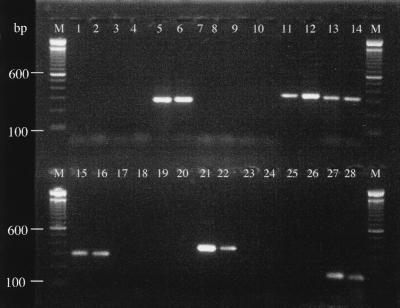
FIG. 2.
Gel image of representative PCR amplification products from clinical samples after amplification with selected primer pairs. Lanes 1 to 6, lesion iii from patient 12 with nested general EV HPV-specific primer pair (CP62-CP69 and CP65-CP68); lanes 1 and 2, PCR negative buffer controls (first and second rounds, respectively); lane 3, buffer extraction control; lane 4, PCR-negative DNA control (human placental DNA); lane 5, patient sample (HPV-RTRX5); lane 6, positive control (10 fg of HPV-5 plasmid); lanes 7 to 12, lesion iv from patient 6 with nested cutaneous primers for group E (CN1F and CN1R); lanes 6 and 7, PCR-negative buffer controls; lane 9, buffer extraction control; lane 10, PCR negative DNA control; lane 11, patient sample (HPV-41); lane 12, positive control (10 fg of HPV-1 plasmid); lanes 13 and 14, lesion i from patient 5 with nested cutaneous primers for group A4 (CN2F and CN2R); lane 13, patient sample (HPV-57); lane 14, positive control (10 fg of HPV-2 plasmid); lanes 15 and 16, lesion ii from patient 3 with nested cutaneous primers for group A2 (CN3F and CN3R); lane 15, patient sample (HPV-10); lane 16, positive control (10 fg of HPV-3 plasmid); lanes 17 to 22, lesion from immunocompetent patient 2 with cutaneous primers for group B1 (C4F and C4R); lane 17, PCR-negative buffer control; lanes 18 and 19, negative extraction controls (buffer and HPV-negative tissue); lane 20, PCR-negative DNA control; lane 21, patient sample (HPV-4); lane 22, positive control (10 fg of HPV-4 plasmid); lanes 23 to 28, lesion from immunocompetent patient 12 with mucosal seminested primers (MY11 and GP6); lanes 23 and 24, PCR-negative buffer controls; lane 25, buffer extraction control; lane 26, PCR-negative DNA control; lane 27, patient sample (HPV-11); lane 28, positive control (10 fg of HPV-16 plasmid); lanes M, molecular size markers (100-bp DNA ladder). Bands of 600 and 100 bp are indicated.
Of the cutaneous HPV types, 10 distinct types were identified (HPV-1, -2, -3, -7, -10, -27, -28, -41, -57, and -77). HPV-27 was observed most frequently, occurring in 17 (33.3%) warts. The other most prevalent cutaneous types were all members of group A2: HPV-28 and HPV-10 (15.7% each) and HPV-77 (17.6%). The HPV types usually reported as being associated with warts in immunocompetent individuals (HPV-1, -2, -3, and -4) were detected least often.
Eighteen recognized EV HPV types were identified (HPV-5, -9, -14, -20, -23, -24, -25, -36, -37, -38, -49, -75, -RTRX1, -RTRX2, -RTRX5, -RTRX9, -vs20-4, and -vs92-1) and a number of putatively novel EV-related HPV types were also found (HPV-20-related, -23-related, -24-related, -19-related, -36-related, -12-related, -RTRX1-related). No individual EV HPV type or EV HPV cluster predominated.
Some concordance in the HPV types harbored by multiple warts within the same individual was observed. Such concordance was found in warts from different anatomical sites that were removed on different occasions (up to 4 years apart) and analyzed independently (Table 3). For example, HPV-7 was found in warts from the elbow, thigh, and hand of patient 10, and patient 12 carried both HPV-28 and HPV-RTRX5 in warts on the neck, thigh, and wrist. Analysis of HPV type with respect to wart type and site showed no correlation; EV-associated and cutaneous HPV types occurred in dysplastic warts on sun-exposed skin and verrucae vulgares alike (Tables 3 and 4).
TABLE 3.
Clinical details of RTR patients and HPV types of viruses in transplant-associated wartsa
| Patient no. | Sexb | NMSCc | Site | Typed | HPV type(s) detectede
|
HPV type by DKFZ typingf | ||
|---|---|---|---|---|---|---|---|---|
| Cutaneous | EV associated | Mucosal | ||||||
| 1 | M | No | (i) Hand | SE | 27 | 23 | − | 27 |
| 2 | F | SCC | (i) Finger | W | 1, 27, 77 | RTRX2 | − | |
| (ii) Arm | SE | − | 20, RTRX9-rel | − | ||||
| (iii) Hand | SE | 2 | 20 | − | ||||
| (iv) Arm | SE | 3, 57 | − | |||||
| (v) Arm | SE | 27 | RTRX2 | − | ||||
| (vi) Arm | SE | − | 20, RTRX2 | − | ||||
| (vii) Arm | SE | 27 | PsoX1-rel | − | ||||
| 3 | M | SCC | (i) Handg | SE | 2, 10 | RTRX5 | − | |
| 10 | RTRX5,14D | 16 | ||||||
| 10, 27 | 5/8-rel | − | ||||||
| 4 | M | No | (i) Heelg | W | 27 | − | − | |
| 27 | 9 | − | ||||||
| 27 | − | − | ||||||
| 27, 77 | − | − | ||||||
| (ii) Arm | W | 7, 27 | 20 | − | ||||
| (iii) Eyelid | W | − | 38 | − | ||||
| 5 | M | SCC | (i) Wrist | SE | 3, 57 | RTRX5 | − | |
| 6 | M | SCC | (i) Finger | W | 1, 10 | 23-rel | 11 | |
| (ii) Finger | W | 1, 27, 77 | 23-rel | − | ||||
| (iii) Thumb | W | 1, 77 | RTRX1 | − | ||||
| (iv) Knee | W | 28, 41 | − | − | DL267 | |||
| 7 | F | CIS | (i) Thigh | W | − | 20 | 11 | |
| (ii) Finger | W | 77 | 23-rel | |||||
| 8 | M | No | (iii) Wrist | W | 2 | 49 | − | |
| 9 | M | SCC | (i) Neck | SE | − | 14, 38, 19-rel | − | |
| (ii) Chest | SE | − | 36 | − | ||||
| (iii) Footg | W | − | − | − | ||||
| 27 | 36 | − | ||||||
| 10 | M | No | (i) Elbow | W | 7, 10 | − | − | |
| (ii) Thigh | W | 7, 27 | − | − | ||||
| (iii) Thigh | W | 10 | 23 | 16 | ||||
| (iv) Hand | W | 7, 10 | − | − | ||||
| (v) Arm | W | 10, 27 | + | − | ||||
| 11 | M | SCC | (i) Hand | SE | 28 | 25 | 11 | |
| (ii) Arm | SE | 28 | + | 11 | ||||
| (iii) Arm | SE | 77 | + | − | ||||
| (iv) Ear | SE | − | 25, 38 | 11 | ||||
| 12 | M | SCC | (i) Neck | SE | 27, 28, 41 | 24,RTRX5,vs20-4 | − | 27, 28 |
| (ii) Neck | SE | 28 | 12-rel | 11 | 76 | |||
| (iii) Thigh | W | 2, 28, 41 | RTRX5 | 11 | ||||
| (iv) Wrist | SE | 28 | RTRX5,5/24-rel | 11 | ||||
| (v) Foot | W | 41 | − | 11 | ||||
| 13 | M | BCC | (i) Hand | SE | 27, 77 | 20 | − | |
| (ii) Arm | SE | 77 | 5 | 16 | ||||
| 14 | F | No | (i) Chest | SE | 10, 57 | − | − | 57 |
| 15 | M | No | (i) Hand | SE | 57 | − | − | 57 |
| 16 | M | SCC | (i) Chest | SE | 27 | RTRX9 | − | |
| (ii) Forehead | SE | 27, 77 | 24,RTRX9 | − | ||||
| 17 | M | SCC | (i) Neck | SE | 27, 28 | vs92-1 | 11 | 9 |
| 18 | M | SCC | (i) Arm | SE | 3 | 37 | − | |
| 19 | M | No | (i) Leg | W | 10 | 23, 24, 75 | − | |
| 20 | F | No | (i) Forehead | SE | 28, 41 | − | − | |
| 21 | M | No | (i) Neck | SE | 10, 57 | 23 | − | 10, DL347 |
| 22 | M | No | (i) Elbow | W | 2 | 24 | − | |
| (ii) Abdomen | W | − | 24 | 6 | ||||
| 23 | M | No | (i) Leg | W | 27 | − | − | |
Note that the order of samples listed is not related to the order of sample extraction.
M, male; F, female.
Past history of NMSC is described as follows: No, no history of NMSC despite receipt of a transplanted kidney at least 10 years earlier; CIS, previous squamous carcinoma in situ (premalignant); BCC, previous basal cell carcinoma; SCC, previous SCC.
Clinical classification of wart type: VV, verrucae vulgares (common cutaneous wart); SE, clinically atypical or plane wart on sun-exposed site (see text).
HPV types detected in individual lesions; rel, related; −, negative; +, positive band on PCR but PCR product uncharacterized. The following are HPV sequences available in the GenBank database for which the full genome sequence has not yet been published (locus names, accession numbers are given in parentheses): RTRx1 (HPVRTRX1,L38918), RTRx2 (HPVRTRX2, L38919), RTRx5 (HPVRTRX5, L38922), RTRx9 (HPVRTRX9, U85662), vs92-1 (HPVS921, X79949), and vs20-4 (HPVS204, X79941).
HPV genotyping was performed with these samples at the Division for Tumour Virus Characterizatio, (Deutsches Krebsforschungszentrum [DKFZ]), Heidelberg, Germany, by the degenerate PCR methodology described by de Villiers et al. (9).
Lesions which were subdivided into two or more parts for independent analysis (see text).
TABLE 4.
HPV types detected in relationship to clinical type of transplant-associated wart
| Wart typea | No. tested
|
No. (%) of warts harboring each HPV groupb
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Lesions | Cutaneous
|
EV associated
|
Mucosal | |||||||||||
| A2 | A4 | A8 | E | Totalc | a1 | a2 | b1 | b2 | Others | Unch | Totalc | ||||
| SE | 15 | 28 | 16 (57) | 15 (54) | 0 | 2 (7) | 23 (82) | 6 (21) | 9 (32) | 2 (7) | 4 (14) | 10 (36) | 2 (7) | 24 (86) | 8 (29) |
| VV | 11 | 23 | 12 (52) | 10 (43) | 3 (13) | 6 (26) | 20 (87) | 1 (4) | 2 (9) | 1 (4) | 7 (30) | 6 (26) | 1 (4) | 17 (74) | 6 (26) |
| Total | 23d | 51 | 28 (55) | 2 (49) | 3 (6) | 8 (16) | 43 (84.3) | 7 (13) | 11 (22) | 3 (6) | 11 (22) | 16 (31) | 3 (6) | 41 (80.4) | 14 (27.4) |
SE, atypical warts on sun-exposed sites; VV, verrucae vulgaris (common warts).
HPV types detected in each group: A2, HPV group A2 (HPV-3, -10, -28, and -77); A4, HPV group A4 (HPV-2, -27, -57); A8, HPV group A8 (HPV-7); E, HPV group E (HPV-1, -41); a1, EV-associated HPV cluster a1 (HPV-5, -8, -12, -36); a2, EV-associated HPV cluster a2 (HPV-14, -19, -20, -25); b1, EV-associated HPV group b1 (HPV-9, -15, -37, -vs92-1); b2, EV-associated HPV group b2 (HPV-23, -38, -RTRX1); Others, other EV HPV types (HPV-24, -49, -RTRX2, -RTRX5, -RTRX9, -VS20-4, -75, -PsoX1); Mucosal, HPV-11 detected in 9 lesions, HPV-16 in 3 lesions and HPV-6 in one lesion; Unch, uncharacterized.
Note that these figures are not additive due to the presence of mixed infections.
Note that these figures are not additive as different wart types from several individuals were analyzed.
In addition, three warts were subdivided and each portion was analyzed independently. Although HPV-27 was detected in all four sections of one plantar wart (lesion i, patient 4), in two of the sections additional viruses were found (HPV-9 and -77). Similarly, for a second wart divided into three sections (lesion i, patient 3), while HPV-10 was identified in all three sections, HPV-2, RTRX5, -14, -16, and -27 and another unidentified EV HPV type were variably found. Finally, in a third wart (lesion iii, patient 9), HPV-27 and -36 were found together in one-half of the lesion, whereas HPV DNA was not detectable in the remainder of the wart. The reason for the latter discrepancy was not clear; beta-globin amplification of DNA extracted from this section had been satisfactory, and histological reanalysis performed by reblocking of the residual frozen specimen confirmed the presence of a viral wart.
All extraction and PCR controls in this study were negative. Where there appeared to be clustering of HPV types, the possibility of cross-contamination was assessed by reference to the order of sample extraction and PCR amplification, and in this context it is important that the order of presentation of data in Table 3 is not related to the order of specimen processing. For example, although there appears to be a clustering of HPV-77 and HPV-23-related in patient 6 and 7, these five lesions were extracted in four separate extraction procedures. The lesions from patients 6 (lesion iv) and 7 (lesion ii) were the only lesions extracted in the same procedure, but the HPV types present in each lesion were entirely different. Furthermore, the three lesions containing HPV-23-related were extracted in three separate procedures and were amplified in three independent PCRs. In addition, despite coextraction of HPV DNA from lesions from immunosuppressed individuals containing multiple HPV types, all warts from immunocompetent patients harbored single HPV types (with the exception of the wart from patient 4).
Clinical specimens from immunocompetent individuals.
We were particularly interested to compare the spectrum of HPV types detected within transplant-associated warts with those in warts from immunocompetent individuals in order to exclude the possibility that the observation of multiple, diverse types within single lesions from RTRs is an artifact of highly sensitive PCR primers (Table 5). In only one lesion was more than one HPV type found; this was also the only lesion in which an EV HPV type was identified and was clinically a somewhat unusual recurrent nipple wart. For all other lesions the HPV types detected were those expected at cutaneous (HPV-2, -3, -4, -10, and -57) and mucosal (HPV-6, -11, -16, -32, and -57) sites.
TABLE 5.
Clinical details of immunocompetent individuals and HPV genotyping of warts from immunocompetent individuals
| Patient no. | Anatomical site | HPV type detected
|
|||
|---|---|---|---|---|---|
| Cutaneous | Mucocutaneous | EV associated | Mucosal | ||
| 1 | (i) Hand | 3 | |||
| (ii) Palm | 57 | ||||
| (iii) Foot | 57 | ||||
| 2 | Finger | 4 | |||
| 3 | Forearm | 10 | |||
| 4 | Chest | 15 | 6 | ||
| 5 | Hand | 2 | |||
| 6 | Finger | 57 | |||
| 7 | Anus | 57 | |||
| 8 | Anus | 6 | |||
| 9 | (i) Vagina | 6 | |||
| (ii) Urethra | 6 | ||||
| 10 | Anogenital region | 6 | |||
| 11 | (i) Vulva | 6 | |||
| (ii) Vulva | 6 | ||||
| (iii) Vulva | 6 | ||||
| 12 | Vulva | 11 | |||
| 13 | Anogenital region | 57 | |||
| 14 | Conjunctiva | 16 | |||
| 15 | Oral | 32 | |||
Confirmation of results.
DNA from three samples which were found to harbor HPV-27 with the degenerate L1 ORF primers and DNA from four samples which were HPV-27 negative were amplified with the HPV-27 E6 type specific primers, and in all cases the results confirmed the results obtained with the L1 primer. Similarly, the presence of HPV-23, -24, and -10 was confirmed in the warts tested. The HPV genotypes in eight warts were also independently tested by a second laboratory, the Deutsches Krebsforschungszentrum, Heidelberg, Germany. The technique used consisted of a degenerate PCR methodology with 16 degenerate primer pairs and a pair of nested EV HPV primers (9). In five of the warts examined, the methods were concordant for the presence of at least one HPV type (Table 2). For the remaining three lesions, the HPV types identified by the two laboratories differed. However, it is noteworthy that for one of the warts with apparently discrepant results (lesion i, patient 17), the HPV types detected by the two laboratories (HPV-VS92-1 and HPV-9, respectively) are very closely related.
DISCUSSION
Previous studies of benign warts in RTRs by degenerate PCR and other methodologies have failed to detect HPV DNA in as many as 40% of lesions, suggesting either a failure to detect known HPV types or the presence of as yet unidentified HPV types (17). By using the modified degenerate nested PCR technique described here, we have achieved an HPV DNA detection level of 100% for the 71 viral warts analyzed. These data suggest that such a technique is sufficiently sensitive and discriminatory for detection of the wide spectrum of HPV types that may be harbored by the lesions of immunosuppressed individuals. Particularly notable is the capacity of this approach to identify mixed infections, such that combinations of cutaneous, mucosal, and EV HPV types were successfully detected within individual warts. We have confirmed the validity of this technique by comparison with PCR and cloning, by using type-specific primers from a different ORF, by comparison with results obtained from a different laboratory, and finally, by comparison of the spectrum of HPV types detected with a control series of warts from immunocompetent individuals.
Comparison with previous data on warts from transplant recipients.
Over 20 case reports or series published to date have included data on the HPV types found in warts from RTRs, and the spectrum of types identified has proved to be remarkably variable. These discrepancies most likely reflect the different experimental approaches used (17). The majority of studies have used DNA hybridization techniques (Southern blotting, dot blotting, and in situ hybridization), which, in general, tend to be less sensitive and specific than PCR-based methods. Many of these series report a spectrum of HPV types in warts from RTRs similar to those found in the immunocompetent population, particularly HPV types 2 and 4 and, to a lesser extent, types 1, 3, and 10. Fewer studies have found EV or mucosal HPV types. The problem of cross-hybridization inherent in these techniques is exemplified by the discrepancies in the rates of detection of HPV-2 and -3. Previous studies may have overreported the presence of HPV-2 as a probable consequence of cross-hybridization with probes for the closely related type HPV-27, which we found to occur more frequently than HPV-2. This has also been confirmed with a recent series of warts from immunocompetent individuals, in which HPV-27 and -57 were also the most commonly detected HPV types (19). Similarly, HPV-3 may have been overreported through cross-hybridization with HPV-77 and HPV-28, which are closely related and which we found to be more prevalent than HPV-3.
When PCR-based techniques for the detection of HPV DNA have been used, discrepancies between our data and previous reports are likely to reflect the primers used. For example, some investigators have used type-specific primers which detect only a limited range of HPV types, and others have used consensus primers which were designed primarily for the detection of mucosal HPV types. Even in those studies that use degenerate primers, few have comprehensively presented the sensitivities of their primer panels, and direct comparisons between data are therefore complex. Nonetheless, PCR-based techniques have indicated that warts from RTRs may be associated with a more diverse range of HPV types than has been found in the general population. In one series (25) in which PCR with type-specific primers for HPV-5, -6/11, -16, and -18 was used, HPV DNA was detected in 11 of 18 (55%) warts; mucosal HPV types were the most prevalent types, occurring in 7 (39%) warts. In contrast, by use of primers specific for HPV-1, -2, -5, -8, -6, -11, -16, and -18 (26), 3 of 18 warts were found to contain HPV-5 and HPV-8, and the remainder of the positive lesions were found to contain cutaneous or low-risk mucosal types. Another group (20) used two pairs of degenerate primers to analyze 47 viral warts; cutaneous types HPV-10, -27, -28, and -57 were identified in 10 warts and a new HPV-29-related cutaneous type (subsequently defined as HPV-77) was identified in 4 lesions. EV HPV types were less commonly identified, although six lesions contained five putatively novel EV-related HPV types. However, HPV DNA was not detected in 40% of these warts, indicating probable deficiencies in the primers used. This group has since modified this technique by increasing the number of degenerate primer pairs used (22). Most recently, the same group (9) has combined this panel of primers with the degenerate nested EV HPV primers of Berkhout et al. (2); in this way they achieved detection of HPV DNA in all 15 warts from transplant recipients examined. However, in clear contrast to our data, cutaneous types HPV-1, -27, and -57 were the most prevalent types detected, and only 5 of 15 (33.3%) of the warts harbored two or more distinct HPV types. Differences in the sensitivity and specificity profiles of the respective primers sets are once more the most likely cause of these differences.
Finally, it is also possible that geographic or ethnicity differences could account for the variation in the spectrum of HPV types detected in published series. To our knowledge, such variations have not previously been examined for cutaneous warts.
Interlaboratory variation in HPV typing results.
It is well recognized that variations in HPV typing methodologies between different investigators may account for discrepancies in published data on warts from transplant recipients and patients with nonmelanoma skin cancers (16, 17, 27), and this is the first study to examine this possibility directly by analyzing the same lesions by two such methodologies. In the majority (five of eight lesions), there was concordance in the HPV types identified, while the results for three lesions were nonconcordant and in several lesions additional HPV types were found by our laboratory. Differences in the sensitivity and specificity profiles for the panel of primer pairs used in each laboratory are a likely explanation (9, 27), and these data highlight the desirability of developing a unified typing methodology for laboratories that study HPV in the skin. However, it is also important that for each of the eight samples, DNA was extracted independently from duplicate portions of the same lesion. Since we have observed that different portions of the same lesion processed under identical conditions may harbor different HPV types, this may also have been a factor that contributed to nonconcordance.
Detection of multiple HPV types.
The frequent existence of mixed infections in warts from transplant recipients as shown in our study is striking and is in contrast to the findings for warts from immunocompetent individuals. It is unclear whether this represents coinfection of single cells with multiple HPV types or the presence of a mixture of cells infected with single virus types. Similarly, it is not known whether only one or all of the HPV types present are responsible for the phenotype or whether lesions are polyclonal with several HPV types simultaneously being transcriptionally active. Published data relating to these issues are so far limited and inconsistent. In one study in which several genital HPV types were detected within a single anal wart from an RTR, localization by in situ hybridization suggested that each HPV type maintained regional separation within the lesion (7). Unger et al. (28) also reported the presence of multiple HPV types within anogenital warts in immunosuppressed human immunodeficiency virus-positive patients clustered in geographically distinct areas. These data may lend support to the hypothesis of a polyclonal origin for viral warts; i.e., they arise from multiple founder cells that carry distinct HPV types. In contrast, by double fluorescence hybridization, HPV-1 and -63 were both detected within nuclei from a plantar wart (10), although only the cytopathogenic effect of HPV-63 was evident in the infected cells. Our data for three warts divided for the purposes of HPV genotyping might provide further evidence of regional separation of individual HPV types within lesions. Furthermore, differences in HPV detection between our methodology and that of the Deutsches Krebsforschungszentrum might also be partly explicable on this basis since DNA was extracted from different portions of the same lesion independently. Rigorous evaluation by in situ hybridization of cutaneous warts from transplant recipients will be necessary to further our understanding of the presence of multiple HPV genotypes within single lesions in terms of the spatial localization, relative viral load, and physical state of each type; we are undertaking such a study.
HPV typing and clinical features of warts from transplant recipients.
Few previous studies have analyzed HPV in warts from transplant recipients with respect to their anatomical localization and morphological features, yet this may be particularly relevant in terms of establishing the relationship between HPV and NMSC. Malignancies are usually observed to colocalize with clinically and histologically atypical warts on sun-exposed sites rather than with common warts (verrucae vulgares) at other sites (11). In immunocompetent individuals, there seems to be a correlation between the morphology of common warts and the inducing HPV type (19). Overall, however, we found no significant differences in the prevalence of each of the major HPV groups found in common warts and the clinically or histologically atypical warts from sun-exposed sites, nor did a clear association emerge between one individual HPV type or any one group of HPVs and skin cancer risk.
Limitations of differential degenerate PCR methodology.
Our aim in this study was to develop a robust method capable of detecting HPV types across a broad spectrum of HPV groups. The resulting differential PCR methodology, despite its proven usefulness in detecting multiple HPV types within individual lesions, has a number of inherent limitations. Perhaps the most obvious is that it may not detect mixtures of HPV types within a particular group. This may be the explanation for failure to identify the HPV type(s) present in the lesions from patients 10 (lesion v) and 11 (lesions ii and iii). However, to achieve this by a PCR and direct sequencing approach, type-specific primers would have to be used (i.e., at least 80 primer pairs), and these would be unlikely to allow detection of potentially novel HPV types. In circumstances in which the presence of more than one HPV type within a particular group is suspected, PCR cloning might therefore prove to be a useful adjunct. In addition, it is also possible that this methodology may still fail to detect certain known or novel HPV types with sufficient sensitivity, a likely explanation for failure to detect HPV in a portion of a wart from patient 9 (lesion iii). Nonetheless, the rate of detection of HPV DNA in 75 of 76 (98.7%) warts or portions of warts (and 100% of individual warts) is higher than that achieved in most previously published series.
In summary, we have developed a degenerate PCR-based technique for the typing of HPV DNA in skin lesions. It has enabled us to detect HPV DNA in 100% of warts analyzed, suggesting that this method is more sensitive and comprehensive than the many previously described PCR-based approaches. The HPV types identified in warts from transplant recipients are diverse and often multiple. It remains of critical interest to determine whether a subgroup of HPV types is specifically associated with the development of malignant lesions analogous to that seen in HPV-associated anogenital cancers. The methodology described here allows sufficiently accurate and sensitive typing of HPV in the skin to address such questions.
ACKNOWLEDGMENTS
We are grateful to G. Orth for providing plasmid clones for HPV-14, -19, -20, -22, -23, and -36; E.-M. de Villiers for providing plasmid clones for HPV-1, -2, -3, -4, -5, -8, -41, -57, -66, and -72; and R. Ostrow for providing a plasmid clone for HPV-26.
C.A.H. is supported by a Medical Research Council (United Kingdom) Clinical Training Fellowship. T.S. is supported by a Joint Research Board grant, and P.J.S. is supported by a Research Advisory Committee grant from St Bartholomew's and the Royal London School of Medicine and Dentistry, Queen Mary and Westfield College.
REFERENCES
- 1.Benton C, Shahidullah H, Hunter J A A. Human papillomavirus in the immunosuppressed. Papillomavirus Rep. 1992;2:23–26. [Google Scholar]
- 2.Berkhout R J M, Tieben L M, Smits H L, Bouwes Bavinck J N, Vermeer B J, ter Schegget J. Nested PCR approach to detection and typing of epidermodysplasia verruciformis-associated human papillomavirus types in cutaneous cancers from renal transplant recipients. J Clin Microbiol. 1995;33:690–695. doi: 10.1128/jcm.33.3.690-695.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birkeland S A, Storm H H, Lamm L U, Barlow L, Blohme I, Forsberg B, Eklund B B, Fjeldborg O, Friedberg M, Frodin L. Cancer risk after renal transplantation in the Nordic countries, 1964–1986. Int J Cancer. 1995;60:183–189. doi: 10.1002/ijc.2910600209. [DOI] [PubMed] [Google Scholar]
- 4.Blessing K, McClaren K M, Benton E C, Bar B B, Bunney M H, Smith F W, Beveridge G W. Histopathology of skin lesions in renal allograft recipients: an assessment of viral features and dysplasia. Histopathology. 1989;14:129–139. doi: 10.1111/j.1365-2559.1989.tb02123.x. [DOI] [PubMed] [Google Scholar]
- 5.Boyle J, McKie R, Briggs J, Junor B J, Aitchison T C. Cancer, warts and sunshine in renal transplant recipients: a case control study. Lancet. 1984;i:702–705. doi: 10.1016/s0140-6736(84)92221-9. [DOI] [PubMed] [Google Scholar]
- 6.Chan S Y, Delius H, Halpern A L, Bernard H U. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny and taxonomy. J Virol. 1995;69:3074–3083. doi: 10.1128/jvi.69.5.3074-3083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen N D, Koltun W A, Cladel N M, Budgeon L R, Reed C A, Kreidlwee J W, Welsh P A, Patrick S D, Yang H. Coinfection of human foreskin fragments with multiple human papillomavirus types (HPV-11, -40, and -LVX82/MM7) produces regionally separate HPV infections within the same athymic mouse xenograft. J Virol. 1997;71:7337–7344. doi: 10.1128/jvi.71.10.7337-7344.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Jong-Tieben L M, Berkhout R J M, Smits H L, Bouwes Bavinck J N, Vermeer B J, van der Woude F J, ter Schegget J. High frequency of detection of epidermodysplasia verruciformis-associated human papillomavirus DNA in biopsies from malignant and premalignant skin lesions from transplant recipients. J Invest Dermatol. 1995;105:367–371. doi: 10.1111/1523-1747.ep12320803. [DOI] [PubMed] [Google Scholar]
- 9.De Villiers E-M, Lavergne D, McLaren K, Benton E C. Prevailing papillomavirus types in non-melanoma carcinomas of the skin in renal allograft recipients. Int J Cancer. 1997;73:356–361. doi: 10.1002/(sici)1097-0215(19971104)73:3<356::aid-ijc9>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 10.Egawa K, Shibasaki Y, de Villiers E-M. Double infection with human papillomavirus 1 and human papillomavirus 63 in single cells of a lesion displaying only an human papillomavirus 63-induced cytopathogenic effect. Lab Invest. 1993;69:583–588. [PubMed] [Google Scholar]
- 11.Glover M T, Proby E M, Leigh I M. Skin cancer in renal transplant patients. Cancer Bull. 1993;45:220–224. [Google Scholar]
- 12.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 13.Majewski S, Jablonska S. Epidermodysplasia verruciformis as a model of human papillomavirus-induced genetic cancer of the skin. Arch Dermatol. 1995;131:1312–1318. [PubMed] [Google Scholar]
- 14.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 15.Manos M M, Ting Y, Wright D K, Lewis A J, Broker T R, Wolinsky S M. The use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cells. 1989;7:209–214. [Google Scholar]
- 16.McGregor J M, Proby C M. The role of papillomaviruses in human nonmelanoma skin cancer. In: Leigh I M, Newton Bishop J A, Kripke M L, editors. Cancer surveys: skin cancer. Vol. 26. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 219–236. [PubMed] [Google Scholar]
- 17.Proby C, Storey A, McGregor J, Leigh I. Does human papillomavirus infection play a role in non-melanoma skin cancer? Papillomavirus Rep. 1996;7:53–60. [Google Scholar]
- 18.Resnick R M, Cornilissen M T, Wright D K, Eichenger G H, Fox H S, Manos M M. Detection and typing of human papillomavirus in archival cervical cancer specimens by DNA amplification with consensus primers. J Natl Cancer Inst. 1990;82:1477–1484. doi: 10.1093/jnci/82.18.1477. [DOI] [PubMed] [Google Scholar]
- 19.Rubben A, Kalka K, Spelten B, Grußendorf-Conen E-I. Clinical features and age distribution of patients with HPV 2/27/57-induced common warts. Arch Dermatol Res. 1997;289:337–340. doi: 10.1007/s004030050201. [DOI] [PubMed] [Google Scholar]
- 20.Shamanin V, Glover M, Rausch C, Proby C, Leigh I M, Zur Hausen H, de Villiers E-M. Specific types of human papillomavirus found in benign proliferations and carcinomas of the skin in immunosuppressed patients. Cancer Res. 1994;54:4610–4613. [PubMed] [Google Scholar]
- 21.Shamanin V, Delius H, de Villiers E-M. Development of a broad spectrum PCR assay for papillomaviruses and its application in screening lung cancer biopsies. J Gen Virol. 1994;75:1149–1156. doi: 10.1099/0022-1317-75-5-1149. [DOI] [PubMed] [Google Scholar]
- 22.Shamanin V, zur Hausen H, Lavergne D, Proby C M, Leigh I M, Neumann C, Hamm H, Goos M, Haustein U-F, Jung E G, Plewig G, Wolff H, de Villiers E-M. Human papillomavirus infections in non-melanoma skin cancers from renal transplant recipients and non-immunosuppressed patients. J Natl Cancer Inst. 1996;88:802–811. doi: 10.1093/jnci/88.12.802. [DOI] [PubMed] [Google Scholar]
- 23.Shiel A, Flavel S, Disney A, Mathew T. Cancer development in patients progressing to dialysis and renal transplantation. Transplant Proc. 1985;17:1685–1688. [PubMed] [Google Scholar]
- 24.Snijders P J F, Van den Brule A J C, Schrijnemakers H F J. The use of general primers in the polymerase chain reaction permits the detection of a broad spectrum of HPV genotypes. J Gen Virol. 1990;71:173–181. doi: 10.1099/0022-1317-71-1-173. [DOI] [PubMed] [Google Scholar]
- 25.Soler C, Chardonnet Y, Allibert P, Euvrard S, Schmitt D, Mandrand B. Detection of mucosal human papillomavirus types 6/11 in cutaneous lesions from transplant recipients. J Invest Dermatol. 1993;101:286–291. doi: 10.1111/1523-1747.ep12365211. [DOI] [PubMed] [Google Scholar]
- 26.Stark L A, Arends M J, McLaren K M, Benton E C, Shahidullah H, Hunter J A, Bird C C. Prevalence of human papillomavirus DNA in cutaneous neoplasms from renal allograft recipients supports a possible viral role in tumour promotion. Br J Cancer. 1994;69:222–229. doi: 10.1038/bjc.1994.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surentheran T, Harwood C A, Spink P J, Sinclair A, Leigh I M, Proby C M, McGregor J M, Breuer J. Detection and typing of human papillomaviruses in mucosal and cutaneous biopsies from immunosuppressed and immunocompetent patients and patients with epidermodysplasia verruciformis: a unified diagnostic approach. J Clin Pathol. 1998;51:606–610. doi: 10.1136/jcp.51.8.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Unger E R, Vernon S D, Lee D R, Miller D L, Sharma S, Clancy K A, Hart C E, Reeves W C. Human papillomavirus type in anal epithelial lesions is influenced by human immunodeficiency virus. Arch Pathol Lab Med. 1997;121:820–824. [PubMed] [Google Scholar]
- 29.Walder B K, Robertson M R, Jeremy D. Skin cancer and immunosuppression. Lancet. 1971;ii:1282–1283. doi: 10.1016/s0140-6736(71)90602-7. [DOI] [PubMed] [Google Scholar]
- 30.Zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]