Gliomas are the most common primary tumors of the central nervous system (CNS). Among low-grade gliomas, mitogen-activated protein kinase (MAPK) pathway alterations are frequent and may provide a therapeutic target. Currently, mechanism-of-action based therapeutic approaches outside the MAPK pathway are scarce. However, especially patients with subtotally resected, recurrent or highly malignant tumors may substantially benefit from the identification of additional specific oncogenic drivers that not only provide insight into disease pathogenesis, but also offer targets for personalized cancer therapies. The ROS proto-oncogene 1 (ROS1) gene encodes a receptor tyrosine kinase that is involved in chromosomal rearrangements in various cancers [6], which present an attractive therapeutic target, since specific inhibitors have been approved for several entities [4, 10]. Data on ROS1 fusions in glioma are limited to single cases or small series [3, 5, 8, 9].
Recently, an enrichment of these fusions (about 7%) was found in a small number of mostly gliomas in infants [2, 7]. Routine diagnostic assessment of ROS1 status in gliomas, however, is so far restricted to a few specialized centers or molecularly informed trials [11]. Thus, the landscape of ROS1 fusions across a broad series of glial tumors of all age groups has not been comprehensively studied so far. Consequently, the distribution among the various types of low- to high-grade glioma is unknown. Similarly, no data exist to determine whether ROS1 fusion-positive gliomas, irrespective of histology, may share further biological features, potentially supporting a ‘ROS1-subtype’ of gliomas. Here, we investigated the presence of ROS1 fusions in a large cohort of 20,723 patients encompassing different diagnostic entities within the spectrum of glioma, to elucidate the frequency of such fusions and the characteristics of the respective cases.
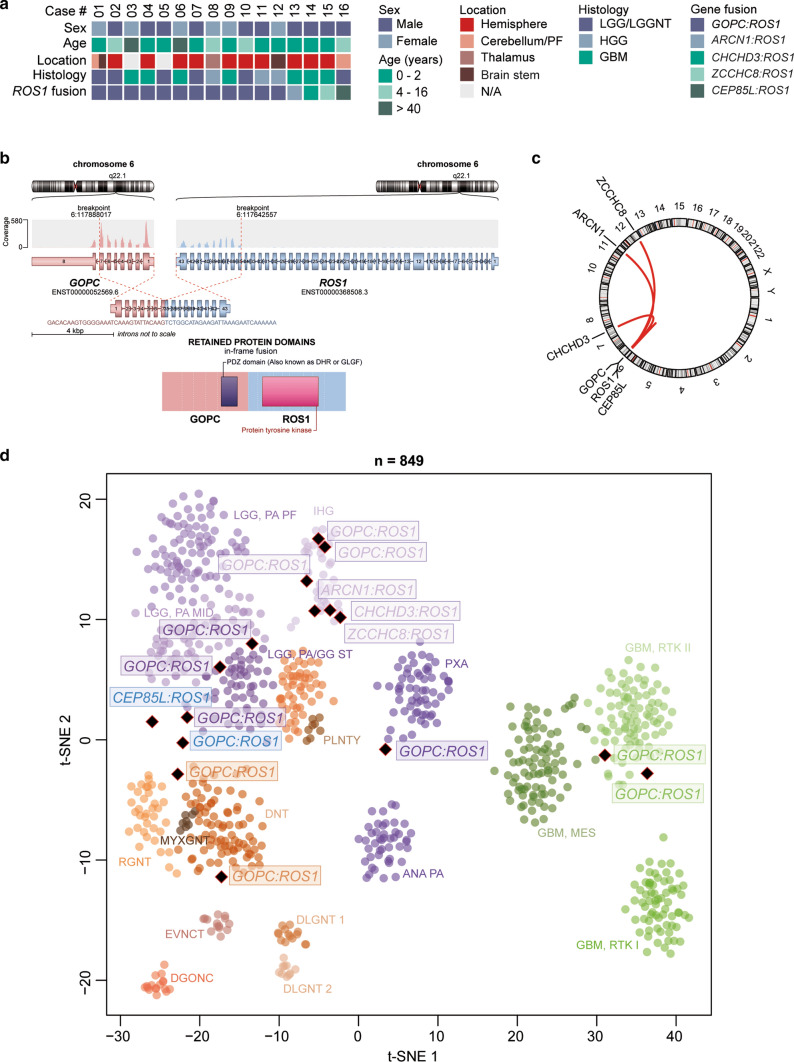
To identify gliomas with structural alterations affecting chromosome 6q (around the ROS1 locus), we systematically evaluated copy-number data of our DNA methylation dataset encompassing 20,723 gliomas, irrespective of specific entity and WHO grade (Supplementary Fig. 1 and 2, online resource). As a high proportion of ROS1 fusions (in particular the most frequent GOPC:ROS1 fusion) are accompanied by a segmental loss of chromosome 6q22 in the copy-number profile, DNA methylation data were screened for a segmental loss covering that region (Supplementary Fig. 1, online resource). Automated analysis was followed by visual inspection and led to the identification of 14 potential cases. On suspicious cases, we performed RNA and targeted exome sequencing, and confirmed the presence of ROS1 fusions in all 14 tumors (Fig. 1a and Supplementary Table 1). In the most common (n = 11) GOPC:ROS1 fusions (Fig. 1b), exons 1–7 or 1–4 of GOPC (NM_001017408) are fused in frame to exons 35–43 of ROS1 (NM_002944). Single cases of exons 36–43 of ROS1 fused downstream of ZCCHC8 exons 1–2 (NM_0017612), ARCN1 exons 1–5 (NM_001655), or CHCHD3 exons 1–2 (NM_017812) were also observed (Fig. 1c). In all fusion events, the kinase domain of ROS1 was retained (Fig. 1b and Supplementary Table 2). ROS1 transcript levels were upregulated in all ROS1-fused gliomas (Supplementary Fig. 3, online resource). Interestingly, ROS1 partners are associated with very different cellular functions, including, e.g., intracellular protein trafficking and RNA processing and degradation. In addition, two further ROS1-fused glioma samples that were already detected as such by performing RNA sequencing in a diagnostic context, after the initial screen was performed were included into subsequent analyses. One of the samples harbored a GOPC:ROS1 fusion (with exons 1–7 of GOPC fused to exons 35–43 of ROS1) and indeed showed segmental loss of chromosome 6q22, while the other case harbored a CEP85L:ROS1 fusion (with exons 1–12 of CEP85L (NM_001042475) fused to exons 35–43 of ROS1) with a segmental gain of chromosome 6q22. In addition, we analyzed RNA sequencing data from a set of > 1000 FFPE tissue samples processed in a diagnostic setting. Here, no further gliomas harboring a ROS1-fusion were detected.
Fig. 1.
Summary of clinico-pathological characteristics and key molecular findings in tumors with ROS1 gene fusion (a). Schematic illustration of the GOPC:ROS1 fusion detected in case #3 involving exons 1–7 of GOPC and exons 35–43 of ROS1 (b). Circos plot of gene fusions targeting ROS1 (lines link fusion gene partners according to chromosomal location; c). t-distributed stochastic neighbor embedding (t-SNE) analysis of DNA methylation profiles of ROS1-fused glioma alongside selected reference samples (d). Reference DNA methylation classes: posterior fossa pilocytic astrocytoma (LGG, PA PF), hemispheric pilocytic astrocytoma and ganglioglioma (LGG, PA/GG ST), midline pilocytic astrocytoma (LGG, PA MID), polymorphous low-grade neuroepithelial tumor of the young (PLNTY), diffuse leptomeningeal glioneuronal tumor subgroup 1 (DLGNT 1), diffuse leptomeningeal glioneuronal tumor subgroup 2 (DLGNT 2), infantile hemispheric glioma (IHG), extraventricular neurocytoma (EVNCT), dysembryoplastic neuroepithelial tumor (DNT), rosette-forming glioneuronal tumor (RGNT), myxoid glioneuronal tumor of the septum pellucidum and lateral ventricle (MYXGNT), diffuse glioneuronal tumor with oligodendroglioma-like features and nuclear clusters (DGONC), anaplastic astrocytoma with piloid features (ANA PA), pleomorphic xanthoastrocytoma (PXA), glioblastoma IDH wildtype subclass RTK I (GBM, RTK I), glioblastoma IDH wildtype subclass RTK II (GBM, RTK II), glioblastoma IDH wildtype subclass mesenchymal (GBM, MES). The two ROS1-fused glioma samples that were already detected as such by performing RNA sequencing in a diagnostic context are highlighted in blue. Other abbreviations: LGG/LGGNT low-grade glioma/low-grade glioneuronal tumor, HGG high-grade glioma, GBM glioblastoma, PF posterior fossa, N/A not available
A t-distributed stochastic neighbor embedding (t-SNE) analysis of DNA methylation profiles alongside a broad reference set of CNS tumors [1] revealed that the ‘ROS1 cohort’ molecularly segregated into different glioma groups (Fig. 1d). Six of the samples grouped with the DNA methylation class infantile hemispheric glioma, other tumors clustered with various reference classes of glioma from low- to high-grade (Fig. 1d). Histological re-evaluation confirmed the different histological entities and underline that ROS1 fusions are not specific to any one glioma entity. Interestingly, most of the patients harboring a fusion were children (particularly infants). Of note, however, was the finding that two classical adult IDH-wildtype glioblastomas in adult patients also harbored a GOPC:ROS1 fusion.
Our data show a high frequency of ROS1 gene fusions within the DNA methylation class infantile hemispheric glioma, which is in line with recent studies [2, 7]. This clinically distinct group of gliomas (that were initially often diagnosed as glioblastomas) carries a high prevalence of gene fusions with ROS1, ALK, NTRK1/2/3, or MET as a fusion partner. However, our finding that ROS1 fusions also occur in cases that were both histologically and epigenetically clearly pilocytic astrocytoma or IDH-wildtype glioblastoma, respectively, underscores that this event is not pathognomonic for infantile hemispheric glioma, nor limited to pediatric patients, so in that respect concerns a quite ‘promiscuous’ marker.
Although relatively rare in other gliomas, identification of ROS1 fusions is important from a treatment perspective, as there are specific inhibitors available. Screening via copy-number profiling and subsequent validation using RNA sequencing provides an efficient approach to identify patients who may benefit from this targeted therapy. However, as illustrated by one of the cases that was identified by performing RNA sequencing in a diagnostic setting, not all variants of ROS1 fusion necessarily show a deletion around the ROS1 locus. For example, copy-neutral translocations can lead to ROS1 fusions as well, and such cases would be missed by screening for segmental 6q22 loss. RNA sequencing thus remains the ‘gold standard’ for adequate detection of these rare events. However, it should be noted that tumor heterogeneity and blood–brain barrier permeability of specific ROS1-inhibitors could be one of the major problems limiting the efficacy of targeted therapies.
Our findings highlight ROS1 fusions as a rare but potentially highly relevant therapeutic target for a subset of patients with gliomas of different histological grades and biological classes. Even though these fusions have no strong diagnostic relevance, since they are not pathognomonic for a tumor type, they are in line with the increasing demand to provide predictive markers in diagnostic neuropathology. This highlights the need for expanded testing for such alterations beyond infant gliomas. It will be interesting to see whether ROS1-inhibitors will be effective in upcoming clinical trials for glioma patients.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank J. Meyer, L. Hofmann and A. Habel for skillful technical assistance and the microarray unit of the DKFZ Genomics and Proteomics Core Facility for providing Illumina DNA methylation array-related services. This study was supported by the Hertie Network of Excellence in Clinical Neuroscience. P.S. is a fellow of the Hertie Academy of Excellence in Clinical Neuroscience. D.T.W.J. is supported by the Everest Centre for Low-Grade Paediatric Brain Tumours (the Brain Tumour Charity, UK; GN-000382).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Philipp Sievers and Damian Stichel are co-first authors.
References
- 1.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555:469–474. doi: 10.1038/nature26000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clarke M, Mackay A, Ismer B, Pickles JC, Tatevossian RG, Newman S, et al. Infant high grade gliomas comprise multiple subgroups characterized by novel targetable gene fusions and favorable outcomes. Cancer Discov. 2020 doi: 10.1158/2159-8290.CD-19-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cocce MC, Mardin BR, Bens S, Stutz AM, Lubieniecki F, Vater I, et al. Identification of ZCCHC8 as fusion partner of ROS1 in a case of congenital glioblastoma multiforme with a t(6;12)(q21;q24.3) Genes Chromosomes Cancer. 2016;55:677–687. doi: 10.1002/gcc.22369. [DOI] [PubMed] [Google Scholar]
- 4.Das A, Cheng RR, Hilbert ML, Dixon-Moh YN, Decandio M, Vandergrift WA, 3rd, et al. Synergistic effects of crizotinib and temozolomide in experimental FIG-ROS1 fusion-positive glioblastoma. Cancer Growth Metastasis. 2015;8:51–60. doi: 10.4137/CGM.S32801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davare MA, Henderson JJ, Agarwal A, Wagner JP, Iyer SR, Shah N, et al. Rare but recurrent ROS1 fusions resulting from chromosome 6q22 microdeletions are targetable oncogenes in glioma. Clin Cancer Res. 2018;24:6471–6482. doi: 10.1158/1078-0432.CCR-18-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res. 2013;19:4040–4045. doi: 10.1158/1078-0432.CCR-12-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerreiro Stucklin AS, Ryall S, Fukuoka K, Zapotocky M, Lassaletta A, Li C, et al. Alterations in ALK/ROS1/NTRK/MET drive a group of infantile hemispheric gliomas. Nat Commun. 2019;10:4343. doi: 10.1038/s41467-019-12187-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakano Y, Tomiyama A, Kohno T, Yoshida A, Yamasaki K, Ozawa T, et al. Identification of a novel KLC1-ROS1 fusion in a case of pediatric low-grade localized glioma. Brain Tumor Pathol. 2019;36:14–19. doi: 10.1007/s10014-018-0330-3. [DOI] [PubMed] [Google Scholar]
- 9.Richardson TE, Tang K, Vasudevaraja V, Serrano J, William CM, Mirchia K, et al. GOPC-ROS1 fusion due to microdeletion at 6q22 is an oncogenic driver in a subset of pediatric gliomas and glioneuronal tumors. J Neuropathol Exp Neurol. 2019;78:1089–1099. doi: 10.1093/jnen/nlz093. [DOI] [PubMed] [Google Scholar]
- 10.Shaw AT, Ou SH, Bang YJ, Camidge DR, Solomon BJ, Salgia R, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wick W, Dettmer S, Berberich A, Kessler T, Karapanagiotou-Schenkel I, Wick A, et al. N2M2 (NOA-20) phase I/II trial of molecularly matched targeted therapies plus radiotherapy in patients with newly diagnosed non-MGMT hypermethylated glioblastoma. Neuro Oncol. 2019;21:95–105. doi: 10.1093/neuonc/noy161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.