Abstract
Control of mRNA stability is critical for expression of short-lived transcripts from cytokines and proto-oncogenes. Regulation involves an AU-rich element (ARE) in the 3′ untranslated region (3′UTR) and cognate trans-acting factors thought to promote either degradation or stabilization of the mRNA. In this study we present a novel approach using somatic cell genetics designed to identify regulators of interleukin-3 (IL-3) mRNA turnover. Mutant cell lines were generated from diploid HT1080 cells transfected with a reporter construct containing green fluorescent protein (GFP) linked to the IL-3 3′UTR. GFP was expressed at low levels due to rapid decay of the mRNA. Following chemical mutagenesis and selection of GFP-overexpressing cells, we could isolate three mutant clones (slowA, slowB, and slowC) with a specific, trans-acting defect in IL-3 mRNA degradation, while the stability of IL-2 and tumor necrosis factor alpha reporter transcripts was not affected. Somatic cell fusion experiments revealed that the mutants are genetically recessive and form two complementation groups. Expression of the tristetraprolin gene in both groups led to reversion of the mutant phenotype, thereby linking this gene to the IL-3 mRNA degradation pathway. The genetic approach described here should allow identification of the defective functions by gene transfer and is also applicable to the study of other mRNA turnover pathways.
Expression of a number of cytokines, including interleukin 2 (IL-2), IL-3, granulocyte-macrophage colony-stimulating-factor (GM-CSF) and tumor necrosis factor alpha (TNF-α), in response to extracellular stimuli involves transient stabilization of corresponding mRNAs, which are otherwise degraded rapidly in the cytoplasm. Transcripts of proto-oncogenes, e.g., c-myc, c-fos, and c-jun, are also very short-lived, which allows the cell to limit their expression to a narrow time window after receiving a mitogenic signal. Selective degradation of these mRNAs involves recognition of cis elements (reviewed in reference 37), is generally preceded by shortening of the poly(A) tail (1), and, in some cases, appears to occur cotranslationally (17). In c-myc and c-fos transcripts, different cis elements have been identified: rapid mRNA decay is mediated by sequences in the 5′ untranslated region (5′UTR), potent destabilizing elements in the coding region, and an equally important AU-rich element (ARE) located in the 3′UTR (19, 40, 46, 47). In cytokine transcripts, where the function of the ARE was initially revealed (5, 39), it appears to be the major destabilizing element. In the case of IL-3, point mutations in the ARE are sufficient to stabilize the full-length transcript, which implies that no additional elements mediate destabilization (41). In general, the AREs of cytokines are composed of multiple, partially overlapping AUUUA pentamers, while those of c-myc and c-fos contain only a few AUUUA motifs in a U-rich context. A correlation between these sequence features and different deadenylation kinetics has allowed the classification of AREs into types I, II, and III (7). While these cis-acting elements have been extensively studied, we are only at the beginning of understanding how the corresponding trans-acting factors target ARE-containing transcripts to the degradation machinery in a regulated fashion. A functional role in ARE-dependent turnover control has been documented at this time for a small number of genes including the HuR, AUF1, tristetraprolin (TTP), and von Hippel-Lindau (VHL) genes.
HuR, cloned as a gene with homology to HuD, is an RNA binding protein which can interact in vitro with the AREs of c-fos and IL-3 mRNA (26), as well as with synthetic, mRNA-destabilizing AREs (31). In vivo, overexpression of HuR caused stabilization of reporter transcripts containing the AREs of c-fos and GM-CSF, as well as of vascular endothelial growth factor (VEGF) mRNA (11, 22, 36).
AUF1 was first purified from a postribosomal supernatant by its ability to accelerate the decay of c-myc mRNA in an in vitro system (3, 4). Later the gene was cloned (49), but its destabilizing activity in vivo could be demonstrated only recently, as overexpression of AUF1 in K562 erythroleukemia cells antagonized the stabilizing effect of hemin on reporter transcripts bearing the AREs of c-fos and GM-CSF (24).
TTP was initially cloned from NIH3T3 cells as an immediate-early response gene (25, 45). It belongs to a small family of zinc finger proteins which have two copies of the unusual Cys-Cys-Cys-His zinc finger domain. Its function was discovered in TTP knockout mice, which develop a severe inflammatory syndrome due to an increase in TNF-α production (43). Overproduction of TNF-α by macrophages derived from TTP−/− mice appeared to be the result of increased stability of TNF-α mRNA (6). Indeed, TTP was shown to bind to the ARE of TNF-α mRNA, and binding was dependent on the integrity of the two zinc finger domains (20).
VHL has been identified as a tumor suppressor gene which is inactivated in von Hippel-Lindau tumors and in some sporadic renal carcinomas (21). VEGF production is elevated in these tumors and can be suppressed under normoxic conditions by ectopic expression of wild-type (wt) VHL. Suppression appears to occur at the posttranscriptional level (12) by promoting rapid degradation of VEGF mRNA (16). Recent experiments have identified VHL as a component of an E3 ubiquitin-protein ligase complex (23), suggesting that VHL might act upstream by inducing ubiquitination of a regulator that controls rapid mRNA turnover of hypoxia-inducible genes.
ARE-dependent control of mRNA stability involves a complex interplay between the RNA, stabilizing factors (e.g., HuR), destabilizing factors (e.g., AUF1 and TTP), upstream regulators (e.g., VHL), and probably additional proteins which remain to be identified. As an alternative to biochemical strategies based on purification of ARE-binding proteins, tumor cells with impaired mRNA turnover are potentially helpful tools for a functional approach. Tumors with trans-acting defects, due to lack of regulatory functions, could serve for genetic complementation and cDNA transfer experiments which should eventually allow the identification of the defective regulators. Indeed, Schuler and Cole have reported a mouse monocytic tumor which expressed abnormally stable GM-CSF mRNA (38). Stabilization occurred in trans, since mRNA of a reporter construct containing the 3′UTR of GM-CSF also showed a prolonged half-life. However, no further work has been reported on this model tumor. Another system with similar characteristics has been developed in our laboratory. Tumor lines derived from v-H-ras-transformed PB-3c mast cells overexpress IL-3, which serves as a growth stimulus in an autocrine fashion (32). In some of these tumors, IL-3 overproduction is the result of an IL-3 gene rearrangement that leads to enhanced transcription. In the majority of the tumors, however, IL-3 is upregulated by stabilization of the mRNA (15, 33). The corresponding mutation acts in trans, since mRNA of a heterologous IL-3 reporter construct is also abnormally stable (14). Fusion of these tumor cells with the PB-3c precursor line suppressed the tumor phenotype and restored rapid IL-3 mRNA decay, indicating that the mutation in these tumors is recessive (8). PB-3c and derivative tumor lines exhibit a rather low transfection efficiency and are therefore not suitable for identification of the mutant genes by cDNA transfer.
As an alternative, we decided to develop a system where cellular mutants with a defect in mRNA decay could be generated and isolated. While many mammalian regulators have been identified and studied in yeast mutants, this powerful approach could not be used because the IL-3 ARE is not recognized as a destabilizing element in yeast. Compared to haploid yeast cells, the use of mammalian cells is less convenient for somatic genetics. Apart from their slower generation time, the occurrence of recessive mutants requires inactivation of both alleles and hence represents a rare event. Nevertheless, the group of G. R. Stark has succeeded in isolating recessive mutants unresponsive to interferons. The mutants were generated from diploid human HT1080 fibrosarcoma cells and selected in 6-thioguanine by their failure to activate an interferon-responsive guanine phosphoribosyltransferase reporter gene (18, 28, 35). In the work presented here, we have adopted a modified strategy to study IL-3 mRNA turnover. Three mutants were isolated which display a trans-acting, recessive defect in IL-3 mRNA degradation. Somatic cell fusion experiments revealed that the mutants belong to two complementation groups. In both groups, ectopic expression of TTP restored rapid decay of IL-3 mRNA, thereby linking this gene to the IL-3 mRNA degradation pathway.
MATERIALS AND METHODS
Plasmid constructs.
To generate GFPIL3-wt, the 5′UTR and proximal promoter region of murine IL-3 were amplified by PCR using primers M1217 (5′-AGCTGCTTCTGATGCCT-3′) and M1216 (5′-GTGGATCCTGTCTCGTTCTGGTCCT-3′) and inserted as an ApaI-BamHI fragment into the multiple cloning site of pEGFP-N1 (Clontech). The genomic 1.9-kb HindIII-ApaI fragment upstream of IL-3 was placed 5′ to the proximal promoter. Green fluorescent protein (GFP) linked to the IL-3 promoter was then inserted as a HindIII-XbaI fragment into the previously described vector IL3MXh-wt (41), which was cut with HindIII and XbaI. This step placed GFP under the control of the IL-3 3′UTR and introduced the hph gene, coding for hygromycin B phosphotransferase (2), as a selection marker. For the control plasmid GFPIL3-3a, the same HindIII-XbaI fragment was inserted into IL3MXh-3a (41).
Construct HindIL3neo was made by subcloning the 2.7-kb BamHI fragment of pMAMneo (Clontech) containing a neomycin resistance gene (neoR) under the control of the simian virus 40 (SV40) promoter and polyadenylation sequences first into pGEM-3Z (Promega). From this vector (pGEMneo), neoR was excised as a KpnI-SalI fragment and inserted into pSP72 containing the 5.4-kb HindIII-EcoRI region of murine genomic IL-3 (48). MXIL3neo was generated by ligation of the 2.7-kb BamHI fragment of pMAMneo into the AatII site of MXIL3-wt, which contains the IL-3 gene driven by a 0.6-kb HindIII-SacI fragment of the Moloney murine leukemia virus (MMLV) long terminal repeat (LTR) promoter (41).
For plasmid neoMXβglobin-IL2, the 3′UTR of murine IL-2 was amplified by PCR using primers M1458 (5′-CAATAACATTGTACCTCCTGC-3′) and M1758 (5′-AGAGGAGAGCTTTATTTCTTG-3′) from cDNA of T-cell line 9-6 (10). The 388-bp fragment was blunt-end ligated into the SmaI site of pSP73 (Promega). From this vector, the insert was excised as a BamHI-BglII fragment to replace the BglII fragment of plasmid Mxh-β-globin-IL3 (29). Finally, the hph sequence between the XmnI and ClaI sites was replaced by the neoR-containing XmnI-AccI insert of pGEMneo. Similarly, the 3′UTR (763 bp) of murine TNF-α was amplified by PCR using primers M1750 (5′-AAGCGATCTTTATTTCTCTC-3′) and M1751 (5′-AGGGAATGGGTGTTCATCCA-3′) and ligated into pSP73. For neoMXβglobin-TNFα, the HindIII-BglII fragment from Mxh-β-globin-IL3 (29) was inserted into the HindIII and BamHI sites, and neoR was introduced as described above.
To generate mTTP.tag, murine TTP was cloned from cDNA of NIH3T3 cells by PCR using primers M1804 (5′-ATGAATTCGCGCCACCATGGATCTC-3′) and M1803 (5′-ATTCTAGACTCAGAGACAGAGATACG-3′). After digestion with EcoRI and XbaI, the 965-bp fragment was ligated in frame into the EcoRI and XbaI sites of pcDNA3.1/Myc-HisA (Invitrogen). The absence of mutations and presence of correct boundaries were confirmed by sequencing.
Cell culture and transfection.
HT1080 cells were grown in Iscove's modified Dulbecco medium (IMDM) supplemented with 10% fetal calf serum (FCS), 50 μM 2-mercaptoethanol, 2 mM glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Transfections were performed in 6-well plates with 1 μg of plasmid DNA and 3 μl of Lipofectamine (Gibco) following the manufacturer's protocol. Hygromycin (1 mg/ml), G418 (1 mg/ml), or puromycin (10 μg/ml) was added 48 h later for selection of stably transfected cells. Whenever needed, cells were subcloned by limiting dilution in 60-well Terazaki plates to achieve uniform expression of the transgene.
Cell fusion.
To obtain hybrids with the precursor cell line, HT1080 was first transfected with pBABEpuro (30), kindly provided by H. Hirsch. This cell line could then be directly crossed either with a trans mutant (slowA, slowB, or slowC), or with the control cis-mutant HT-GFPIL3-3a, since all these mutants express hph on the GFP reporter plasmid. For intermutant hybrids, resistance to puromycin or G418 was conferred by transfecting the fusion partners with either pBABEpuro or HindIL3neo. The following combinations were used: slowA-HindIL3neo × slowB-pBABEpuro, slowA-HindIL3neo × slowC-pBABEpuro, and slowB-HindIL3neo × slowC-pBABEpuro. In order to induce fusion, cells were trypsinized and 106 cells of each partner were mixed and centrifuged. The pellet was resuspended in 50 μl of medium, and 700 μl of 50% (wt/vol) polyethylene glycol 1500 in IMDM was added and carefully mixed. After incubation for 90 s at 37°C, 9 ml of medium was added, and cells were washed once, resuspended in 15 ml of medium, and plated in a 10 cm-dish. Twenty-four hours later, selection was started by addition of the selection markers puromycin (10 μg/ml) and hygromycin (1 mg/ml) or puromycin and G418 (1 mg/ml). Selection was completed after 2 to 3 weeks.
Mutagenesis and selection.
Prior to mutagenesis, the optimal concentration of the frameshift mutagen was determined. HT-GFPIL3-wt cells were seeded at medium density (2 × 106 cells per 10-cm dish) and exposed for 2 h to different concentrations of ICR191 (Sigma). Cell survival was estimated by measuring plating efficiencies after mitogen treatment, and 6 μg/ml was chosen, which corresponds to a survival rate of 15%. For mutagenesis, HT-GFPIL3-wt cells were seeded at a density of 5 × 106 per 15-cm dish 1 day before treatment with ICR191 (6 μg/ml) for 2 h. Cells were washed twice with medium and allowed to grow to subconfluency for 4 to 5 days before the next round of mutagenesis. Pools 9, 10, 11, and 12 underwent 8, 10, 11, and 12 rounds, respectively.
After recovery from the last round of mutagenesis, cells were either frozen or prepared for sorting by flow cytometry using a FACSorter (Becton Dickinson) and Cellquest software. During sorting, cells were kept in FCS-free medium at 4°C. A small fraction of cells (0.02 to 0.05%) displaying fluorescence emissions above an arbitrarily chosen threshold was recovered and either expanded for a second round of sorting or directly subcloned in 60-well Terazaki plates using medium supplemented with 20% FCS. Eight to 10 days later, clones were examined under the fluorescence microscope to select candidates for further analysis.
Flow cytometry.
Cells were grown in 6-well dishes, trypsinized, and resuspended in 500 μl of phosphate-buffered saline (PBS) containing 5 μg of propidium iodide/ml. A total of 104 cells were analyzed using a FACScan (Beckton Dickinson) and Cellquest software. The fluorescence of GFP was excited at 488 nm, and emission was measured using a 510-nm filter. Propidium iodide staining was detected with a 580-nm filter, which allowed exclusion of dead cells and cellular particles during data analysis.
Actinomycin D chase experiments and Northern blot analysis.
Cells were seeded in 10-cm dishes and grown to subconfluency, and fresh medium was added 12 to 16 h prior to blocking of transcription with actinomycin D (5 μg/ml). RNA was extracted 0, 1, 2, and 3 h later using the method described by N. M. Gough (13). Twenty-five micrograms of total cytoplasmic RNA was resolved in 1.1% agarose-formaldehyde gels, blotted onto Hybond N+ membranes (Amersham) for 3 h with 50 mM NaOH, hybridized at 55°C overnight in the presence of 50% formamide, and washed at 65°C as described previously (34). GFPIL3 mRNA was detected either with an SP6 RNA probe from the XbaI-EcoRI fragment of murine IL-3 cDNA directed against the 3′UTR or with an SP6 probe from the 760-bp ApaI-XbaI fragment of pEGFP-N1 which spans the coding region of GFP. Full-length IL-3 mRNA was detected using an SP6 probe from pSP65-multi-CSF containing the 368-bp HindIII-XbaI fragment located in the coding region of IL-3 cDNA. β-Globin reporter mRNA was detected with an SP6 probe generated from the 86-bp BglII-EcoRI fragment of rabbit β-globin. Endogenous human TTP (hTTP) mRNA was detected using a random-primed probe from an hTTP cDNA template. In this case, hybridization was carried out at 45°C, and washing at 55°C. hTTP cDNA was amplified by reverse transcription-PCR (RT-PCR) from HT1080 cells using primers M1933 (5′-ATGAATTCCACTCTCGGCCGACAC-3′) and M1938 (5′-ATAAGCTTCGCTGAGATCCAGCTG-3′). All filters were stripped in 0.5% sodium dodecyl sulfate (SDS) at 100°C and rehybridized with an SP6 probe from a 567-bp PstI fragment of chicken β-actin. Signal intensities were quantified using a PhosphorImager (Molecular Dynamics) and ImageQuant software.
Western blot analysis.
Cell lysates were prepared from confluent 10-cm dishes by addition of 100 μl of radioimmunoprecipitation assay (RIPA) buffer (120 mM NaCl, 50 mM Tris-HCl [pH 8.0], 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS) supplemented with 10 μg of aprotinin/ml, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μg of leupeptin/ml, and 10 μg of pepstatin/ml. Forty microliters of lysate was resolved on a 12% polyacrylamide gel, blotted onto an Immobilon-P (Millipore) membrane, equilibrated for 30 min in TBS (150 mM NaCl–50 mM Tris [pH 7.5]), blocked for 1 h with TBS containing 1% milk powder and 1% Tween 20, and incubated overnight at 4°C with the mouse anti-myc antibody 9E10. The membrane was washed twice for 10 min with 0.1% Tween 20 in TBS and twice for 10 min with blocking solution. Incubation with the secondary, alkaline phosphatase-coupled anti-mouse antibody was performed for 30 min at room temperature, and the final washing was done with 0.1% Tween 20 in TBS four times for 15 min. Proteins were visualized with CDP-tar (Roche) and subsequent autoradiography. mTTP.tag migrates at approximately 46 kDa, as calculated by addition of the molecular size of murine TTP (mTTP) (43 kDa [see reference 44]) and the 26 amino acid residues of the myc-His tag.
RESULTS
A reporter system for IL-3 mRNA turnover.
With the aim of establishing a system that allowed for isolation of mutant cell lines defective in cytokine mRNA turnover, we followed a strategy where GFP, linked to the ARE-containing 3′UTR of murine IL-3, served as a reporter gene in a pool of chemically mutagenized cells. Mutants lacking a regulator of rapid mRNA decay would overexpress GFP and hence should be selectable by flow cytometry, cloning, and subsequent analysis of reporter mRNA stability.
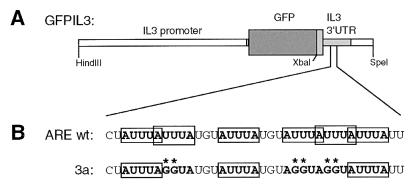
As target cells we chose the diploid human HT1080 fibrosarcoma cell line, which had previously been successfully used to isolate mutants defective in an interferon response pathway (28, 35). As depicted in Fig. 1A, the reporter construct (GFPIL3-wt) contains at its 5′ end a 1.9-kb fragment of the murine IL-3 promoter and at its 3′ end the entire 3′UTR of IL-3, including a small portion of exon 5 starting from the XbaI site. Upon stable transfection into HT1080 cells, GFP was expressed at low levels, and the corresponding mRNA showed the expected short half-life when transcription was blocked by actinomycin D (Fig. 2A). Quantitation revealed that the mRNA decayed with an apparent half-life of about 1 h due to the presence of the IL-3 ARE in the reporter transcript. When, as a control, construct GFPIL3-3a, whose destabilizing element was mutated (Fig. 1B), was used, GFP expression was elevated about eightfold and reporter mRNA was very stable (Fig. 2B), consistent with the effect of this mutation in PB-3c mast cells (41). We concluded that GFPIL3-wt-transfected HT1080 cells could serve as a reporter system to isolate trans mutants defective in mRNA decay. Such mutants could be expected to overexpress GFP, like the cis mutant HT-GFPIL3-3a used here as a control.
FIG. 1.
(A) Schematic representation of the reporter constructs GFPIL3-wt and -3a. GFP is flanked 5′ by a 1.9-kb fragment of the IL-3 promoter and 3′ by an XbaI-SpeI fragment of murine IL-3 containing part of exon 5 including the entire 3′UTR and the poly(A) site. Dark shaded box, GFP, light shaded boxes, IL-3 cDNA sequences. (B) Nucleotide sequence of the core AUUUA motif cluster within the ARE of IL-3, which contains 6 partially overlapping AUUUA pentamers. In mutation 3a, three pentamers were mutated to AGGUA, as indicated by double asterisks. This is the minimal mutation that abrogates the destabilizing effect of the ARE (41).
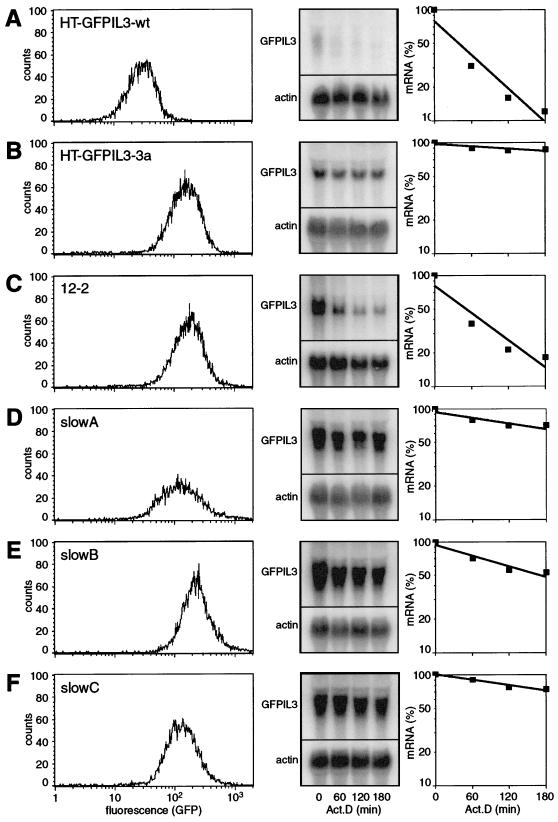
FIG. 2.
Expression of the GFP reporter construct in the parental cell line HT-GFPIL3-wt (A), the control mutant HT-GFPIL3-3a (B), and the mutants 12-2 (C), slowA (D), slowB (E), and slowC (F). (Left) Analysis of GFP expression by flow cytometry. (Center) Analysis of reporter mRNA stability by actinomycin D chase experiments and Northern blotting. RNA was isolated 0, 1, 2, and 3 h after inhibition of transcription with 5 μg of actinomycin D/ml. Twenty-five micrograms of total RNA was resolved on 1.1% agarose-formaldehyde gels and transferred onto Hybond N+ membranes, and GFPIL3 reporter mRNA was detected with a radiolabeled SP6 probe against the IL-3 3′UTR. All blots were stripped and rehybridized with a β-actin probe as a loading control. (Right) Signal intensities were quantified using a PhosphorImager and ImageQuant software. Values of GFPIL3 were normalized to actin and plotted as relative mRNA levels against time. mRNA half-lives were calculated by means of linear regression and are summarized in Table 2.
Mutagenesis and isolation of mutant cell lines.
As recessive mutations require inactivation of all alleles of a given gene, we verified that the HT1080 indicator line was diploid and not polyploid. Indeed, about half of the HT1080 cells transfected with GFPIL3-wt were tetraploid or nearly tetraploid when assayed for DNA content using propidium iodide staining and fluorescence-activated cell sorter (FACS) analysis. Cells were therefore subcloned, a suitable clone (no. 16) was selected, and its diploid chromosome number was confirmed by microscopic examination of chromosome spreads (data not shown). This clone will be referred to as HT-GFPIL3-wt.
For mutagenesis, we essentially followed the protocol published by G. R. Stark's laboratory (28), which is based on multiple rounds of treatment with the frameshift mutagen ICR191. A total of 8 × 107 HT-GFPIL3-wt cells were divided into four pools and exposed for 2 h to 6 μg of ICR191/ml. At this concentration, about 15% of the cells survived (data not shown); these were allowed to recover for 4 to 5 days and then were subjected to the next round of mutagenesis. The efficiency of mutagenesis was monitored by determining the number of cells that had become resistant to 6-thioguanine, reflecting loss of the hypoxanthin phosphoribosyltransferase (HPRT) gene. This occurred at a relatively high frequency, since HPRT is X-chromosome linked and therefore monoallelic. The frequency of colonies resistant to 6-thioguanine had continuously risen from 3 × 10−6 in unmutagenized cells to a plateau value of about 0.8 × 10−3 after 8 to 12 rounds of mutagenesis.
Selection of GFP-overexpressing cells from the four mutagenized pools was achieved by a combination of automated sorting by flow cytometry, subsequent cloning, and direct analysis by fluorescence microscopy. Table 1 gives a summary of the recovery rates at each step. From a total of 6 × 107 cells subjected to FACS sorting, the 0.03% (2 × 104) with the highest fluorescence intensities were retained. These cells were directly cloned in microtiter plates, whereby cloning efficiency was very low (∼7%). When examined by eye, roughly one-third (444) of the clones displayed increased fluorescence. About half (237) of these promising candidates could be further expanded. FACS analysis confirmed increased GFP levels (twofold or more above the basal level) for 156 clones. For further characterization, Northern blot analysis was performed and revealed increased mRNA levels corresponding to the GFP levels. However, most clones did not show a change in reporter mRNA stability. In this class of mutants, overexpression of GFP appeared to result from transcriptional activation. Clone 12-2 (Fig. 2C) is an example of a transcriptional mutant with the typical characteristics (see Table 2): increased levels of GFP (18.5-fold) and reporter mRNA (11.4-fold), and an mRNA half-life (1.3 h) similar to that in the parental cell line HT-GFPIL3-wt (1.0 h).
TABLE 1.
Selection of GFP-overexpressing clones
| Pool no. | No. of input cells (106) | No. of cells sorted (by FACS) | No. of clones:
|
||||
|---|---|---|---|---|---|---|---|
| Total | With high GFP levels (eye) | Capable of expansion | With high GFP levels (FACS) | For which mRNA was stabilized | |||
| 12 | 18.0 | 230 | 11 | 10 | 18a | 16 | 0 |
| 9 | 11.8 | 4,800 | 131 | 42 | 32 | 18 | 0 |
| 10 | 14.5 | 5,250 | 455 | 113 | 70 | 38 | 1 |
| 11 | 16.5 | 10,370 | 828 | 279 | 117 | 84 | 2 |
| Total | 60.8 | 20,650 | 1,425 | 444 | 237 | 156 | 3 |
Number increased due to subcloning of certain cell lines.
TABLE 2.
Reporter gene expression in parental and mutant cell lines
| Cell line | Mean GFP fluorescence intensity ± SE (fold)a | Mean GFPIL3 mRNA level ± SE (fold)a | Mean GFPIL3 mRNA half-life ± SE (h)a |
|---|---|---|---|
| HT-GFPIL3-wt | 1.0b | 1.0 ± 0.08 | 1.0 ± 0.04 |
| HT-GFPIL3-3a | 8.1 ± 0.3 | 2.4 ± 0.1 | >10.0c |
| 12-2 | 18.5 ± 2.8 | 11.4 ± 1.9 | 1.3 ± 0.04 |
| slowA | 7.5 ± 1.7 | 3.5 ± 0.5 | 4.3 ± 0.8 |
| slowB | 20.6 ± 1.4 | 16.3 ± 0.4 | 3.5 ± 0.1 |
| slowC | 13.2 ± 0.8 | 7.5 ± 0.4 | 8.6 ± 1.0 |
All mean and standard error calculations are based on triplicates (n = 3).
There was no error calculation, because this value was set to 1 in every experiment.
The exact value is 27.7 ± 8.2 h, based on extrapolation of 3-h time course experiments.
Nevertheless, three clones could be identified in which reporter mRNA degradation was consistently and significantly slower than in the wt precursor (Fig. 2D through F). These mutants were termed slowA, slowB, and slowC, and they displayed GFPIL3 mRNA half-lives of 4.3, 3.5, and 8.6 h, respectively (summarized in Table 2). For slowA and slowC, these values correlated well with the increased levels of mRNA expression (3.5- and 7.5-fold, respectively) and GFP fluorescence (7.5- and 13.2-fold, respectively). In slowB cells, the mRNA half-life of 3.5 h seemed to be too short for its considerably high mRNA (16.3-fold) and GFP (20.6-fold) levels, and it remained unclear whether additional transcriptional activation occurred in this mutant, or whether the more-pronounced accumulation of GFP is a consequence of its lower proliferation rate (data not shown).
The defect in slowA, slowB, and slowC acts in trans and is restricted to IL-3.
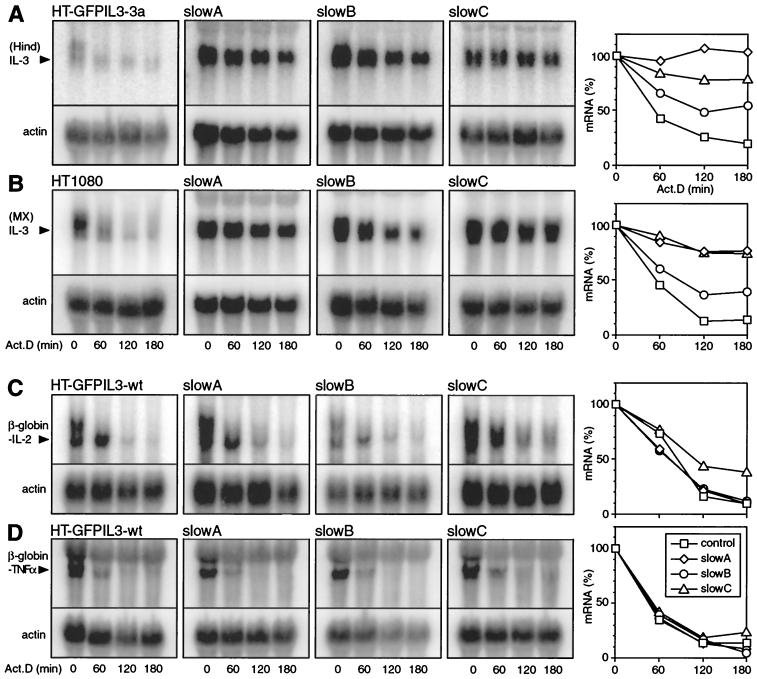
Since cytokine mRNA stabilization can occur via cis mutations located on the mRNA itself, or in trans through loss of a regulatory function, we had to distinguish between these two possibilities. For this purpose, murine genomic IL-3 was introduced as a second reporter gene into the mutants and also, as a control, into the cis mutant HT-GFPIL3-3a. As shown in Fig. 3A, IL-3 mRNA was overexpressed and clearly more stable in slowA, slowB, and slowC compared to HT-GFPIL3-3a. To visualize the mRNA turnover rates, signal intensities were quantified (right panel). Stabilization was more pronounced in slowA and slowC than in slowB, consistent with the half-lives calculated for the GFPIL3 transcripts (Table 2). The same result was obtained in a parallel set of transfections with an IL-3 gene driven by the MMLV LTR promoter (Fig. 3B), indicating that upregulation of IL-3 in the mutants occurred independently of the nature of the promoter. We concluded that all three mutants are defective in a trans-acting function essential for rapid IL-3 mRNA decay. This conclusion was confirmed by direct sequencing of the 3′UTR of reporter mRNA amplified by RT-PCR from slowA, slowB, and slowC cells, where we failed to detect any mutation (data not shown).
FIG. 3.
Decay of mRNA from stably transfected IL-3, IL-2, and TNF-α reporter constructs in control and mutant cell lines. Actinomycin D chase experiments and Northern blot analysis were performed as described for Fig. 2. For quantification (right), signal intensities were normalized to that of actin and plotted as percentages of the initial value against time. (A) Cells were transfected with HindIL3neo, a 5.3-kb genomic IL-3 construct that uses the IL-3 promoter. (B) Expression of MXIL3neo, where transcription of IL-3 is driven by an MMLV LTR promoter. For panels A and B, a HindIII-XbaI fragment of the IL-3 coding region was used to generate a radiolabeled SP6 RNA probe that specifically recognizes IL-3 mRNA but not GFPIL3 mRNA, which is also expressed in the mutants. (C) Cells were transfected with neoMXβglobin-IL2, a plasmid that contains a β-globin reporter gene linked to the entire 3′UTR of murine IL-2. (D) Expression of neoMXβglobin-TNF-α, containing the 3′UTR of murine TNF-α. In panels C and D, an SP6 probe against β-globin was used to detect reporter mRNA.
For a specificity control, we decided to examine whether mRNA turnover of two different cytokines, IL-2 and TNF-α, would also be affected in the mutants. For this purpose, a β-globin reporter gene was linked to the 3′UTRs of IL-2 and TNF-α in constructs neoMXβglobin-IL2 and neoMXβglobin-TNFα, respectively, and stably transfected into control and mutant cell lines. In slowA and slowB, the IL-2 reporter mRNA decayed as rapidly as in the parental cells (Fig. 3C). The degradation rate was slightly reduced in slowC, yet not comparable to the stabilization observed with all IL-3 transcripts. Likewise, TNF-α reporter mRNA underwent rapid decay in both the mutants and the control cell line (Fig. 3D). These results imply that the mutants do not exhibit a general lack of rapid mRNA degradation, but rather a defect restricted to IL-3.
slowA, slowB, and slowC are recessive mutants and form two complementation groups.
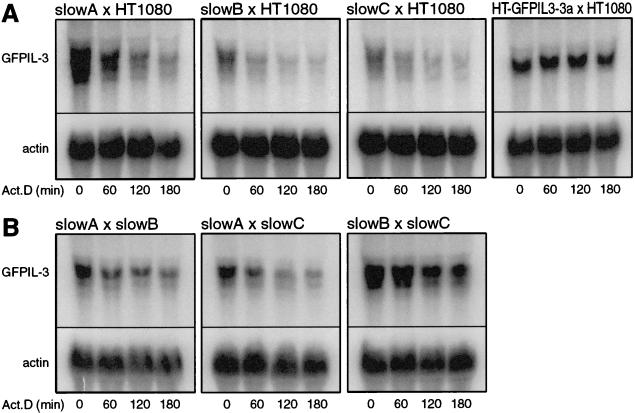
To address the question whether the decay mutants are dominant or recessive in nature, somatic cell fusion experiments were performed. The hygromycin-resistant mutants slowA, slowB, and slowC were fused with parental, pBABEpuro-expressing HT1080 cells. After selection with hygromycin and puromycin, the stability of the reporter mRNA was tested in the hybrids (Fig. 4A). Fusion led to reversion of the mutant phenotype, since rapid decay was restored in all three hybrids. To validate the approach, the dominant cis mutant HT-GFPIL3-3a was also fused with HT1080, and, as expected, the mutant phenotype was retained in this hybrid. We concluded that slowA, slowB, and slowC are recessive mutants reverting when complemented with the parental genome.
FIG. 4.
Decay of GFPIL3 mRNA in hybrid cell lines. (A) Somatic cell fusion was performed between parental HT1080 cells expressing pBABEpuro and the hygromycin-resistant trans mutants slowA, slowB, and slowC, as well as the cis mutant HT-GFPIL3-3a. After selection with puromycin and hygromycin, hybrids were analyzed by actinomycin D chase experiments and Northern blotting, as described for Fig. 2. (B) Intermutant hybrids were generated by crossing mutants slowA, slowB, and slowC with each other (as specified in Materials and Methods). GFPIL3 reporter mRNA, expressed by both fusion partners, was detected with an SP6 probe directed against the GFP coding region in order to prevent hybridization with IL-3 mRNA from the HindIL3neo-transfected fusion partner.
It was of interest now to examine whether the mutants themselves could complement for each other's defect or whether they belong to one complementation group. After transfection of each partner with a plasmid that confers resistance to either puromycin or G418 (specified in Materials and Methods), the following fusions were performed: slowA × slowB, slowA × slowC, and slowB × slowC. Again, the stability of GFPIL3 mRNA was measured in the hybrids (Fig. 4B). It appeared that rapid mRNA decay was reinstalled by fusion of slowA × slowB and slowA × slowC, although the effect was less pronounced than that observed upon fusion with parental HT-1080 cells. In hybrid slowB × slowC, however, the mutant phenotype was retained, as GFPIL3 mRNA remained stable. These results were confirmed in a repeat experiment, and we concluded that the defect in slowA is distinct from that in slowB and slowC, while slowB and slowC belong to the same complementation group. It is possible that mutants slowB and slowC are actually siblings, as they were both isolated from the same pool of mutagenized cells. But they also showed marked differences with respect to cell morphology and growth rate (data not shown), as well as a reproducible difference of about twofold in the GFPIL3 mRNA half-life (see Table 2).
TTP reverts the mutant phenotype.
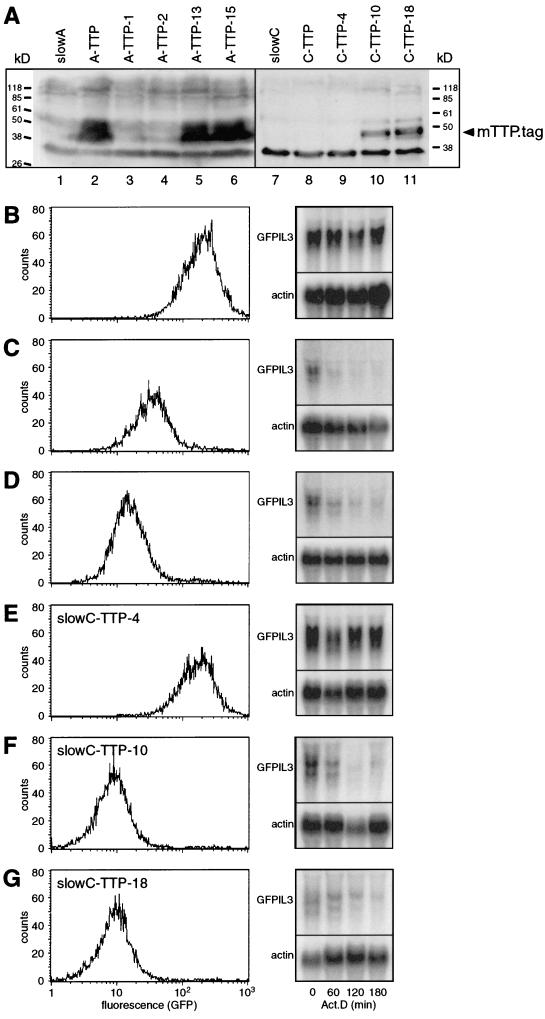
In order to characterize the defective functions in the two complementation groups, we decided to test whether expressing genes known to promote rapid degradation of ARE-containing transcripts had an effect on reporter mRNA stability in the mutants. The AUF1 (p37), TTP, and VHL genes, cloned into the pcDNA3.1/MycHis expression vector, were introduced into slowA and slowC. After selection of stable transfectants, expression was confirmed by Western blot analysis using an antibody against the myc tag. AUF1 and VHL were expressed readily in slowA and slowC, but neither GFP fluorescence nor GFPIL3 mRNA stability was altered (data not shown). Hence, AUF1 and VHL did not appear to be directly related to the defective function in slowA and slowC. As shown in Fig. 5A, TTP (mTTP.tag) was well expressed in slowA (lane 2), but hardly at all in slowC (lane 8). Therefore, both mass cultures were subcloned, and from each, one negative clone (A-TTP-1 [lane 3] and C-TTP-4 [lane 9]) and two TTP-positive clones (A-TTP-13 [lane 5], A-TTP-15 [lane 6], C-TTP-10 [lane 10], and C-TTP-18 [lane 11]) were selected. Analysis of GFP expression revealed that TTP had a pronounced effect on the mutant phenotype of both slowA and slowC. In the TTP-positive clones, GFP fluorescence was reduced to low levels and GFPIL3 mRNA decayed rapidly (Fig. 5C, D, F, and G). In contrast, the TTP-negative clones behaved identically to the untransfected mutants (Fig. 5B and E). We concluded that expression of mTTP.tag can functionally revert mutants slowA and slowC.
FIG. 5.
Transfection of tristetraprolin (mTTP.tag) into slowA and slowC, and analysis of GFP reporter gene expression in subclones. (A) Western blot analysis with untransfected slowA (lane 1), transfected slowA-TTP (lane 2), and subclones of slowA-TTP (lanes 3 to 6). Similarly, mTTP.tag expression was analyzed in untransfected slowC (lane 7), transfected slowC-TTP (lane 8), and subclones of slowC-TTP (lanes 9 to 11). A lysate of 1 × 106 to 2 × 106 cells was resolved on a 12% polyacrylamide-SDS gel and blotted onto an Immobilon-P membrane, and the anti-myc antibody 9E10 served to detect the myc- and His-tagged protein, which has an approximate molecular weight of 46 kDa. GFP reporter gene expression was analyzed in the following subclones: A-TTP-1 (B), A-TTP-13 (C), A-TTP-15 (D), C-TTP-4 (E), C-TTP-10 (F), and C-TTP-18 (G). GFP levels were assayed by flow cytometry (left panel), and reporter mRNA stability was measured by actinomycin D chase experiments (right panel). Northern blot analysis was performed as described for Fig. 2.
Endogenous TTP is not mutated in either complementation group.
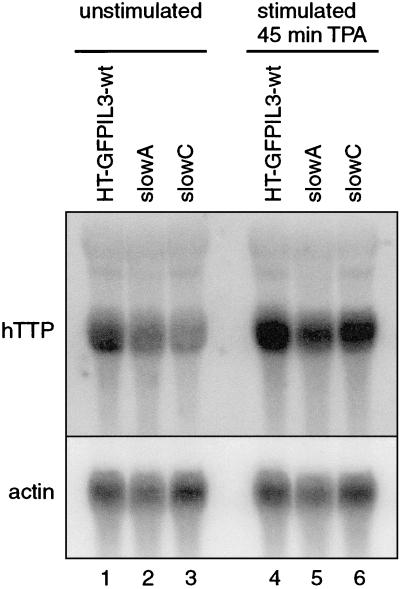
The effect of ectopically expressed TTP in slowA and slowC called for testing the possibility that the mutants had lost endogenous TTP expression. Northern blot analysis was performed using a random-primed probe against human TTP (Fig. 6). RNA was isolated from unstimulated parental HT-GFPIL3-wt, slowA, and slowC cells (Fig. 6, lanes 1 to 3). hTTP mRNA was detected in all three cell lines, albeit at a somewhat reduced level in slowC. Upon stimulation for 45 min with 10 ng of tetradecanoyl phorbol acetate (TPA)/ml, hTTP mRNA was induced to similar levels in the parental cell and the two mutants (Fig. 6, lanes 4 to 6). Thus, lack of endogenous TTP mRNA expression could not be observed in either mutant. Alternatively, a frameshift mutation might have occurred in the coding region of the TTP gene, which would lead to an aberrant protein. To check for this possibility, cDNA of endogenous TTP mRNA was amplified by RT-PCR from parental HT-GFPIL3-wt, slowA, and slowC cells. After ligation into the pGEM7Z vector and cloning, the entire 960-bp coding region was sequenced. From all three cell lines we obtained wild-type sequences identical to the human TTP sequence published under GenBank accession no. M92843 (data not shown). These results indicated that in both mutants, at least one TTP allele is intact, and they largely ruled out the possibility that mutation of TTP itself is the genetic defect in either of the complementation groups.
FIG. 6.
Northern blot analysis of endogenous TTP (hTTP) expression in parental HT-GFPIL3-wt cells and mutants slowA and slowC. Cells were grown in low concentrations of serum (0.5% FCS) for 24 h, and RNA was isolated before (lanes 1 to 3) or after stimulation with TPA (10 ng/ml) for 45 min (lanes 4 to 6). Fifty micrograms of total RNA was used for each lane, and the blot was hybridized with a random-primed probe generated from human TTP cDNA. The membrane was stripped and rehybridized with a β-actin probe as a loading control.
DISCUSSION
In this work we present a general strategy for generating mutants of a specific mRNA degradation pathway. To our knowledge, this is the first report where somatic cell genetics has been used to study mRNA turnover in a mammalian system allowing the definition of complementation groups. With the help of a reporter gene containing the ARE of the IL-3 3′UTR, changes in mRNA stability could be translated into different steady-state expression levels (Fig. 2A and B). To obtain posttranscriptional mutants, it was important to use GFP as a reporter gene instead of a selectable “all-or-nothing” resistance marker, because alteration of mRNA stability only modulates the levels of gene expression but does not completely turn it on or off. Using an efficient mutagenesis protocol adopted from that of McKendry et al. (28), four pools of 2 × 107 cells each were exposed to 8 to 12 rounds of treatment with the frameshift mutagen ICR191. The extent of random genomic mutations was estimated by the frequency of cells that had lost the monoallelic (X-linked) HPRT function, which reached a plateau value at about 0.8 in 103 cells. Based on this frequency, the probability of knocking out both alleles of any gene could be estimated to (0.8 × 10−3)2, or 0.64 × 10−6. Hence, each pool of 2 × 107 cells should contain roughly 13 mutants for an autosomal gene promoting mRNA decay. We thus could believe that isolating a mutant should be feasible, barring unexpected complications such as redundancy of the putative regulatory gene or a lethal effect of the homozygous mutation.
From the mutagenized cells, GFP-overexpressing clones were selected by a multistep procedure. Automated sorting by flow cytometry allowed us to enrich for highly fluorescent cells. These cells were immediately subcloned in order to prevent the formation of many siblings and to protect slowly dividing cells from being overgrown. An advantage of using GFP was that the clones could be directly analyzed by eye with a fluorescent microscope. Promising candidates were expanded, and GFP overexpression could finally be confirmed for 156 clones by FACS analysis. Altogether, selection was rather time-consuming, and a considerable number of clones were lost at each step (see Table 1). Increasing the cloning efficiency would certainly help to reduce the amount of work in future attempts and could perhaps be achieved by the use of conditioned medium.
The GFP-overexpressing clones were first characterized by performing actinomycin D chase experiments and Northern blot analysis. As expected, all 156 candidate clones displayed increased steady-state levels of the reporter mRNA. Surprisingly, stabilization of the mRNA was found in only three clones, whereas other events, apparently leading to transcriptional activation, were more frequent. This class of mutants, exemplified by clone 12-2 (Fig. 2C), might have undergone amplification of the reporter gene, although it is difficult to imagine how a frameshift mutagen could achieve this. Another, more likely possibility is that loss of a transcriptional repressor has occurred in these mutants. Interestingly, a repressor element, termed the nuclear inhibitory protein (NIP) region, has been identified at position −260 in the human IL-3 promoter (27). One of three complexes that bind to the NIP region in vitro is believed to represent the repressor activity (9), but the protein has not been identified so far. Although it is not known whether the NIP region, or a second repressor mapped further downstream (42), is also functional in the murine IL-3 promoter, it would be worthwhile to analyze the transcriptional mutants for promoter binding proteins by DNA footprinting or electromobility shift assays.
Our interest, however, focused on the three posttranscriptional mutants termed slowA, slowB, and slowC (Fig. 2D through F). First, the mutants were shown to have a trans-acting defect, as degradation of mRNA from two genomic IL-3 constructs was also impeded, similar to the stabilization observed with the GFP reporter transcript (Fig. 3A and B). Reporter transcripts with the ARE-containing 3′UTRs of IL-2 and TNF-α, however, were not stabilized in either of the mutants (Fig. 3C and D). This might indicate that the AREs of these two cytokines are recognized by a decay pathway different from the one which promotes IL-3 mRNA degradation. Alternatively, the (class II) AREs are targeted by one mechanism—which is defective in the mutants—yet IL-2 and TNF-α, in contrast to IL-3, harbor an additional destabilizing element in their 3′UTRs. A more extensive analysis using reporter constructs containing different AREs with and without flanking 3′UTR sequences will enable this issue to be resolved.
It was crucial to establish that the mutants displayed recessive defects, since this determines the strategy for later identification of the defective genes, as outlined below. A dominant mutation would require a different strategy, as the defective gene could be cloned directly by expressing a DNA library from the mutant in wt cells. Fusion of the parental cell line with the three mutants revealed that they are genetically recessive, since their phenotype was corrected in the hybrids (Fig. 4A). Two complementation groups could be defined by analyzing mRNA decay patterns of intermutant hybrids (Fig. 4B). slowA belongs to one group, while slowB and slowC form a second group. The two groups could not be distinguished further, either with respect to decay of β-globin–IL2 or β-globin–TNF-α mRNA or by their response to expression of TTP. In this context it would be worthwhile to generate mutants of different mRNA decay pathways, using the technique described here with GFP reporter constructs containing AREs of other cytokines or proto-oncogenes, particularly IL-2 and TNF-α, as our mutants failed to stabilize these transcripts. This should allow definition of more complementation groups and help to identify the common as well as the specific trans-acting factors that participate in these pathways.
Once recessive IL-3 mRNA turnover mutants had been obtained, it was possible to test whether expression of known regulators of mRNA degradation could rescue the mutant phenotype. With AUF1 (p37) and VHL, no effect on GFP levels or reporter mRNA stability could be observed in either complementation group. In the case of VHL, this is perhaps not surprising, since VHL is mainly involved in the regulation of hypoxia-induced genes such as the VEGF gene. Interestingly, the p37 isoform of AUF1 also had no effect on reporter mRNA decay. Recently, expression of the p42 isoform of AUF1 in K562 erythroleukemia cells has been shown to antagonize hemin-induced stabilization of β-globin transcripts containing the AREs of c-fos and GM-CSF, the latter being almost identical to the ARE of IL-3. The p37 isoform of AUF1 is also an effective destabilizer, as was demonstrated with the ARE of c-fos (24). The gene product missing in our mutants is obviously not AUF1, but perhaps a component downstream, or an essential upstream activator of AUF1. Alternatively, the destabilizing function of AUF1 could be restricted to certain cell lineages and might not be active in HT1080 fibrosarcoma cells.
TTP, on the other hand, could correct the defect in slowA and slowC, as its expression reinstalled rapid decay of GFPIL3 mRNA (Fig. 5). Originally, the mRNA turnover-promoting function of TTP was discovered in macrophages from TTP knockout mice, which overexpress TNF-α due to enhanced mRNA stability (6). Upon reintroduction of TTP into TTP−/− macrophages, reduced levels of β-globin reporter transcripts containing AREs from TNF-α, IL-3, and GM-CSF were observed. This suggested a broader role for TTP in regulating mRNA degradation for various cytokines, but destabilizing activity had been shown only with TNF-α mRNA. Our data establish that TTP is also a component of the IL-3 mRNA degradation pathway. Since it fully restored rapid decay in both complementation groups, TTP appears to be a potent mRNA destabilizer that acts downstream in the degradation pathway. The observation that TTP can directly bind to class II AREs (6, 20) supports this idea.
On the basis of this finding with TTP, experiments are in progress to investigate whether TTP can antagonize IL-3 overproduction in autocrine tumors which express abnormally stable IL-3 mRNA (32, 33). This can be tested at the levels of mRNA stability, IL-3 secretion, autocrine growth in vitro, and tumor formation in vivo. Should TTP have tumor suppressor activity, this would further emphasize its role as an important biological regulator.
More than one hypothesis could explain the capacity of ectopically expressed TTP to revert the mutant phenotypes. First, both alleles of the TTP gene itself could be mutated. This was ruled out, as wild-type cDNA sequences of the human TTP coding region were obtained from both mutants. Second, a gene dosage effect might have occurred following a TTP promoter mutation or by loss of a transcription factor. Similar levels of hTTP mRNA were found in slowA and the parental cell line (Fig. 6). Whether the modest but consistent reduction of hTTP mRNA observed in unstimulated slowC (Fig. 6 and data not shown) is of any significance for the mutant phenotype remains to be investigated. Northern blot analysis also largely excluded the possibility of incorrect splicing or 3′-end processing, since hTTP mRNA had the same size in the parental cell and the mutants. Taken together, these data indicate that the TTP gene itself is intact in both complementation groups. Therefore, we favor a model where the missing component in the IL-3 mRNA degradation pathway is an activator of decay, located upstream of TTP, which can be overruled by ectopic expression of high levels of TTP. The precise relationship between TTP and the two functions A and B/C, however, will become clear only after cloning of the corresponding genes. Taking advantage of the GFP reporter system, we plan to perform complementation by transfer of a cDNA library into the mutants and selection of revertant clones. This should hopefully allow identification of the defective genes in slowA and slowB/C, and eventually help us understand how upstream factors function in concert with known regulators of ARE-dependent mRNA degradation.
ACKNOWLEDGMENTS
We are especially grateful to Verena Jaeggin and Genaro DeLibero (Research Department, Kantonsspital Basel) for help and advice with cell sorting. We also thank Asha P. K. Nair, Adrian Wyss, and Lyndall Brennan for critical comments on the manuscript.
This work was supported by grant 31-40816.94 to C.M. from the Schweizerischer Nationalfonds zur Foerderung der Wissenschaftlichen Forschung. G.S. received a fellowship from the Schweizerische Akademie der Medizinischen Wissenschaften.
REFERENCES
- 1.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 2.Blochlinger K, Diggelmann H. Hygromycin B phosphotransferase as a selectable marker for DNA transfer experiments with higher eucaryotic cells. Mol Cell Biol. 1984;4:2929–2931. doi: 10.1128/mcb.4.12.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brewer G. An A+U-rich element RNA-binding factor regulates c-myc stability in vitro. Mol Cell Biol. 1991;11:2460–2466. doi: 10.1128/mcb.11.5.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brewer G, Ross J. Regulation of c-myc mRNA stability in vitro by a labile destabilizer with an essential nucleic acid component. Mol Cell Biol. 1989;9:1996–2006. doi: 10.1128/mcb.9.5.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carballo E, Lai W S, Blackshear P J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 7.Chen C-Y A, Shyu A-B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 8.Diamantis I D, Nair A P, Hirsch H H, Moroni C. Tumor suppression involves down-regulation of interleukin 3 expression in hybrids between autocrine mastocytoma and interleukin 3-dependent parental mast cells. Proc Natl Acad Sci USA. 1989;86:9299–9303. doi: 10.1073/pnas.86.23.9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engeland K, Andrews N C, Mathey-Prevot B. Multiple proteins interact with the nuclear inhibitory protein repressor element in the human interleukin-3 promoter. J Biol Chem. 1995;270:24572–24579. doi: 10.1074/jbc.270.41.24572. [DOI] [PubMed] [Google Scholar]
- 10.Erb P, Troxler M, Fluri M, Grogg D, Alkan S S. Functional heterogeneity of CD4-positive T-cell subsets: the correlation between effector functions and lymphokine secretion is limited. Cell Immunol. 1991;135:232–244. doi: 10.1016/0008-8749(91)90268-g. [DOI] [PubMed] [Google Scholar]
- 11.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnarra J R, Zhou S, Merrill M J, Wagner J R, Krumm A, Papavassiliou E, Oldfield E H, Klausner R D, Linehan W M. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc Natl Acad Sci USA. 1996;93:10589–10594. doi: 10.1073/pnas.93.20.10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gough N M. Rapid and quantitative preparation of cytoplasmic RNA from small numbers of cells. Anal Biochem. 1988;173:93–95. doi: 10.1016/0003-2697(88)90164-9. [DOI] [PubMed] [Google Scholar]
- 14.Hirsch H H, Nair A P, Backenstoss V, Moroni C. Interleukin-3 mRNA stabilization by a trans-acting mechanism in autocrine tumors lacking interleukin-3 gene rearrangements. J Biol Chem. 1995;270:20629–20635. doi: 10.1074/jbc.270.35.20629. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch H H, Nair A P, Moroni C. Suppressible and nonsuppressible autocrine mast cell tumors are distinguished by insertion of an endogenous retroviral element (IAP) into the interleukin 3 gene. J Exp Med. 1993;178:403–411. doi: 10.1084/jem.178.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iliopoulos O, Levy A P, Jiang C, Kaelin W G, Jr, Goldberg M A. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc Natl Acad Sci USA. 1996;93:10595–10599. doi: 10.1073/pnas.93.20.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 18.John J, McKendry R, Pellegrini S, Flavell D, Kerr I M, Stark G R. Isolation and characterization of a new mutant human cell line unresponsive to alpha and beta interferons. Mol Cell Biol. 1991;11:4189–4195. doi: 10.1128/mcb.11.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones T R, Cole M D. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3′ untranslated sequences. Mol Cell Biol. 1987;7:4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai W S, Carballo E, Strum J R, Kennington E A, Phillips R S, Blackshear P J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latif F, Tory K, Gnarra J, Yao M, Duh F M, Orcutt M L, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 22.Levy N S, Chung S, Furneaux H, Levy A P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 23.Lisztwan J, Imbert G, Wirbelauer C, Gstaiger M, Krek W. The von Hippel-Lindau tumor suppressor protein is a component of an E3 ubiquitin-protein ligase activity. Genes Dev. 1999;13:1822–1833. doi: 10.1101/gad.13.14.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loflin P, Chen C Y, Shyu A B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Q, Herschman H R. A corrected sequence for the predicted protein from the mitogen-inducible TIS11 primary response gene. Oncogene. 1991;6:1277–1278. [PubMed] [Google Scholar]
- 26.Ma W-J, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 27.Mathey-Prevot B, Andrews N C, Murphy H S, Kreissman S G, Nathan D G. Positive and negative elements regulate human interleukin 3 expression. Proc Natl Acad Sci USA. 1990;87:5046–5050. doi: 10.1073/pnas.87.13.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKendry R, John J, Flavell D, Muller M, Kerr I M, Stark G R. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc Natl Acad Sci USA. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ming X F, Kaiser M, Moroni C. c-jun N-terminal kinase is involved in AUUUA-mediated interleukin-3 mRNA turnover in mast cells. EMBO J. 1998;17:6039–6048. doi: 10.1093/emboj/17.20.6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myer V E, Fan X C, Steitz J A. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nair A P, Diamantis I D, Conscience J F, Kindler V, Hofer P, Moroni C. A v-H-ras-dependent hemopoietic tumor model involving progression from a clonal stage of transformation competence to autocrine interleukin 3 production. Mol Cell Biol. 1989;9:1183–1190. doi: 10.1128/mcb.9.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair A P, Hahn S, Banholzer R, Hirsch H H, Moroni C. Cyclosporin A inhibits growth of autocrine tumour cell lines by destabilizing interleukin-3 mRNA. Nature. 1994;369:239–242. doi: 10.1038/369239a0. [DOI] [PubMed] [Google Scholar]
- 34.Nair A P, Hirsch H H, Moroni C. Mast cells sensitive to v-H-ras transformation are hyperinducible for interleukin 3 expression and have lost tumor-suppressor activity. Oncogene. 1992;7:1963–1972. [PubMed] [Google Scholar]
- 35.Pellegrini S, John J, Shearer M, Kerr I M, Stark G R. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol Cell Biol. 1989;9:4605–4612. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng S S, Chen C Y, Xu N, Shyu A B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuler G D, Cole M D. GM-CSF and oncogene mRNA stabilities are independently regulated in trans in a mouse monocytic tumor. Cell. 1988;55:1115–1122. doi: 10.1016/0092-8674(88)90256-5. [DOI] [PubMed] [Google Scholar]
- 39.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 40.Shyu A-B, Greenberg M E, Belasco J G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 41.Stoecklin G, Hahn S, Moroni C. Functional hierarchy of AUUUA motifs in mediating rapid interleukin-3 mRNA decay. J Biol Chem. 1994;269:28591–28597. [PubMed] [Google Scholar]
- 42.Taylor D S, Laubach J P, Nathan D G, Mathey-Prevot B. Cooperation between core binding factor and adjacent promoter elements contributes to the tissue-specific expression of interleukin-3. J Biol Chem. 1996;271:14020–14027. doi: 10.1074/jbc.271.24.14020. [DOI] [PubMed] [Google Scholar]
- 43.Taylor G A, Carballo E, Lee D M, Lai W S, Thompson M J, Patel D D, Schenkman D I, Gilkeson G S, Broxmeyer H E, Haynes B F, Blackshear P J. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 44.Taylor G A, Thompson M J, Lai W S, Blackshear P J. Phosphorylation of tristetraprolin, a potential zinc finger transcription factor, by mitogen stimulation in intact cells and by mitogen-activated protein kinase in vitro. J Biol Chem. 1995;270:13341–13347. doi: 10.1074/jbc.270.22.13341. [DOI] [PubMed] [Google Scholar]
- 45.Varnum B C, Lim R W, Sukhatme V P, Herschman H R. Nucleotide sequence of a cDNA encoding TIS11, a message induced in Swiss 3T3 cells by the tumor promoter tetradecanoyl phorbol acetate. Oncogene. 1989;4:119–120. [PubMed] [Google Scholar]
- 46.Wellington C L, Greenberg M E, Belasco J G. The destabilizing elements in the coding region of c-fos mRNA are recognized as RNA. Mol Cell Biol. 1993;13:5034–5042. doi: 10.1128/mcb.13.8.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wisdom R, Lee R. The protein-coding region of c-myc mRNA contains a sequence that specifies rapid mRNA turnover and induction by protein synthesis inhibitors. Genes Dev. 1991;5:232–243. doi: 10.1101/gad.5.2.232. [DOI] [PubMed] [Google Scholar]
- 48.Wyss, A., and C. Moroni. Calcium-dependent and oncogenic IL-3 mRNA stabilization can be distinguished pharmacologically and by sequence requirements in the 3′ UTR. Growth Factors, in press. [DOI] [PubMed]
- 49.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]