Abstract
Gulf War Veterans' Illnesses (GWI) encompasses a broad range of unexplained symptomology specific to Veterans of the Persian Gulf War. Gastrointestinal (GI) distress is prominent in veterans with GWI and often presents as irritable bowel syndrome (IBS). Neurotoxins, including organophosphorus pesticides and sarin gas, are believed to have contributed to the development of GWI, at least in a subset of Veterans. However, the effects of such agents have not been extensively studied for their potential impact to GI disorders and immunological stability. Here we utilized an established murine model of GWI to investigate deleterious effects of diisopropyl fluorophosphate (DFP) exposure on the mucosal epithelium in vivo and in vitro. In vivo, acute DFP exposure negatively impacts the mucosal epithelium by reducing tight junction proteins and antimicrobial peptides as well as altering intestinal microbiome composition. Furthermore, DFP treatment reduced the expression of IL-17 in the colonic epithelium. Conversely, both IL-17 and IL-17C treatment could combat the negative effects of DFP and other cholinesterase inhibitors in murine intestinal organoid cells. Our findings demonstrate that acute exposure to DFP can result in rapid deterioration of mechanisms protecting the GI tract from disease. These results are relevant to suspected GWI exposures and could help explain the propensity for GI disorders in GWI Veterans.
Keywords: Persian Gulf Syndrome, Isoflurophate, Sarin, Intestinal mucosa, Interleukin-17a
INTRODUCTION
Around 700,000 U.S troops were deployed during The Persian Gulf War (1990–1991) (1,2). Combat-related deaths were less than 200; however, many soldiers subsequently developed an array of maladies, collectively known as Gulf War Veterans' Illnesses (GWI) (1,2,3). Symptoms include stress, fatigue, cognitive dysfunction, and gastrointestinal (GI) distress (1,2,3,4). GWI may have resulted from exposures, including toxic chemicals (e.g. sarin and pesticides), microbes, insects, and stress (2,3,4). Unique exposure conditions likely led to a particularly high incidence of symptoms among members of the US Naval Mobile Construction Battalions (5). Among self-reported and physician-diagnosed conditions, GI disorders were over 2-fold higher, which included irritable bowel syndrome (IBS) that was significantly higher compared to Veterans deployed elsewhere. GWI patients exhibiting IBS are also far more likely to have additional GWI-related symptoms including posttraumatic stress disorder (5). In a recent study incorporating all service branches, IBS was found to afflict close to 20% of Gulf War Veterans (6). Furthermore, GWI Veterans with chronic GI symptoms were found to exhibit intestinal epithelial hyperpermeability (7), which is consistent with IBS.
The development of a GI disorder such as IBS, is multi-factorial; involving genetics, psychological contributions, infection, cytokines, microbiome, and tight junction modifications among others (8,9,10,11,12,13). Moreover, military deployment and the inherent stress associated with combat aid the development of IBS in Veterans (12,14). Antimicrobial peptide (AMP) secretion by intestinal epithelial cells (IEC) is critical for regulating microbial colonization as dysregulation can lead to dysbiosis (15) and contribute to inflammatory bowel disease and IBS (16). Altered intestinal permeability has also been extensively linked to IBS, especially for post-infectious IBS (8) and for GWI Veterans (7). The formation of tight junctions between IECs in the gut barrier plays a significant role in regulating intestinal permeability (17).
IL-17, is the hallmark member of a cytokine family that also includes IL-17B-F (18,19). Multiple cell types in the mucosa have been shown to produce IL-17, including T helper 17 cells, γδ T cells, and intestinal Paneth cells (20,21). IL-17 is typically associated with pro-inflammatory outcomes during infection and autoimmune inflammation; however, in the mucosal epithelium, IL-17 can be protective by mediating tight junction localization, decreasing barrier permeability, and stimulating AMP production (22,23). Much less is known about other IL-17 family cytokine members, especially in the context of GI immunity. IL-17C was initially thought to play similar pro-inflammatory roles as IL-17 (24-26) but this cytokine can also help protect the intestinal epithelium. For example, colonic inflammation was more substantial in IL-17C−/− mice and IL-17C, along with its receptor IL-17 receptor E (IL-17RE), was restricted to IECs (27). These findings mirrored what others observed for IL-17RE−/− mice in colitis (28) and Citrobacter rodentium infection (29). Studies have also demonstrated that IL-17C is enhanced in human colon cancer and that IL-17C is involved in the development of colorectal cancer in mice through altering the microbiota (30).
In this study we analyzed the effects of acute neurotoxin exposure on protective mechanisms governed by the intestinal epithelium. This work focuses on DFP exposure, which has the benefit for modeling both organophosphorus pesticide and sarin exposure experienced by GW Veterans. We found that DFP treatment deleteriously impacts IECs cells, including modulating tight junction, AMP, and IL-17 cytokine expression.
MATERIALS AND METHODS
Mice
C57BL/6 animals were purchased from The Jackson Laboratory. IL-17−/− and IL-17RE−/− mice have been described previously (31,32). All mice used for these experiments were bred in house to ensure microbiome equivalency. Animal procedures were performed using both male and female mice (6–12 weeks old) and sex differences were not observed. All animal experiments were conducted under approval of the Institutional Animal Care and Use Committee (#B17-16) at Rosalind Franklin University of Medicine and Science and in accordance with the National Institutes of Health guide for the care and use of laboratory animals.
GWI mouse model
An established GWI mouse model was described (33) and adapted for these studies. Briefly, corticosterone (CORT; Sigma-Aldrich, St. Louis, MO, USA) was suspended in 1.2% EtOH and administered to animals for 7-day in drinking water (200 mg/L). The vehicle control for CORT was an equivalent volume 1.2% EtOH alone in drinking water. On day 8, animals were then injected i.p. with vehicle (saline) or 1 mg/kg DFP (Sigma-Aldrich) for 24 h prior to tissue harvest. For the acute DFP exposure model, mice were only subjected to the 24 h i.p. DFP injection. DFP dosing at 1 mg/kg did not result in seizures or other obvious adverse consequences to the experimental animals.
Microbiome sequencing
Genomic DNA was purified from fresh fecal samples using a QIAamp Fast DNA Stool Kit (Qiagen, Hilden, Germany). Genomic material was then PCR amplified with primers CS1_515Fa and CS2_806Ra (ACACTGACGACATGGTTCTACAGTGYCAGCMGCCGCGGTAA and TACGGTAGCAGAGACTTGGTCTGGACTACNVGGGTWTCTAAT), which target the V4 regions of microbial small subunit ribosomal RNA genes. Amplicons were generated using a 2-stage PCR amplification protocol with primers containing 5′ common sequence tags (34,35). First stage PCR amplifications were performed in 10 µl reactions using PhusionTaq (Thermo Fisher, Waltham, MA, USA). PCR conditions were 95°C for 5 min, followed by 28 cycles of 95°C for 30″, 55°C for 45″ and 72°C for 60″. A second PCR amplification was performed in 10 µl reactions using a MyTaq HS 2X mastermix. Each well received a separate primer pair with a unique 10-base barcode, obtained from the Access Array Barcode Library for Illumina (Fluidigm, South San Francisco, CA, USA). These Access Array primers contained the CS1 and CS2 linkers at the 3′ ends of the oligonucleotides. Cycling conditions were: 95°C for 5 min, followed by 8 cycles of 95°C for 30″, 60°C for 30″ and 72°C for 30″. Samples were then pooled in equal volume using an EpMotion5075 liquid handling robot (Eppendorf, Hamburg, Germany). The pooled library was purified using an AMPure XP cleanup protocol (0.6X, vol/vol; Agencourt; Beckman-Coulter, Brea, CA, USA) to remove fragments smaller than 300 bp. The pooled libraries, with a 20% phiX spike-in, were loaded onto a Miniseq flow cell, and sequenced (2×153 paired-end reads). Fluidigm sequencing primers, targeting the CS1 and CS2 linker regions, were used to initiate sequencing. De-multiplexing of reads was performed on the instrument. Library preparation, pooling, and sequencing were performed at the University of Illinois at Chicago Genome Research Core within the Research Resources Center (RRC).
In vitro organoid culture
Ileum-derived crypts were isolated from mucosal beds and differentiated into organoids by adapting established procedures (36,37). Briefly, intestinal tissue was opened longitudinally and washed with ice-cold PBS. Tissues were then incubated with Cell Recovery Solution (Corning) to release crypts. Recovered materials were filtered and centrifuged at 80 × g for 5 min at 4oC. The resulting crypt-containing pellet was suspended in Advanced DMEM/F12 media (ADF; Gibco, Waltham, MA, USA) and crypts were counted and checked for contamination. Crypts were resuspended in Geltrex (Life Technologies, Carlsbad, CA, USA) basement membrane at a concentration of 1,000 crypts/well in a 48-MW plate. Crypts were then treated with ADF media supplemented with the following: Glutamax, HEPES, Pen/Strep solution, N-acetyl cysteine, N2 supplement, and B27 supplement (Gibco). The culture media also contained the following soluble factors to induce organoid differentiation: 500 ng/ml rhRSPO-1 (R&D Systems, Minneapolis, MN, USA), 100 ng/ml rmWnt3a (Peprotech, Cranbury, NJ, USA), 100 ng/ml rmNoggin (Peprotech), and 50 ng/ml rmEGF (Gibco). After establishment, organoids were broken up and passaged into fresh media every 7—10 days as lumens became filled with sloughed cells. For chemical and cytokine treatments, organoids were treated at day 6 after passage. For quantitative PCR (qPCR) experiments, organoids were treated as indicated for 24 h prior to harvest. For Western Blotting, organoid cells were treated as indicated for 3-day prior to lysis and protein extraction.
Western blot
The 20 µg total protein from tissue (ileum or colon) or organoid cell lysates (10 µg) was loaded into 10% SDS-PAGE gels and run using a Mini-PROTEAN Tetra electrophoresis system (Bio-Rad, Hercules, CA, USA). Proteins were then transferred onto a nitrocellulose membrane using a Trans-blot SD transfer apparatus (Bio-Rad). The following Abs (anti-mouse unless indicated) were used for protein visualization according to the manufacturer's (Life Technologies) protocols: αClaudin-1, αClaudin-4, αOccludin, αβ-Actin, and αRabbit-HRP (Cell Signaling Technology, Danvers, MA, USA). Gel images and densitometry results were obtained through analysis on an AlphaImager.
Quantitative PCR
Total tissue sample and organoid cell RNA was extracted through homogenization and lysis in TRIzol reagent (Life Technologies). cDNA was then synthesized using MMLV system per manufacturer's protocol (Life Technologies). qPCR was performed using PowerUp SYBR Green Master Mix (Life Technologies) on an Applied Biosystems 7500 Real Time System or a Thermo-Fisher QuantStudio 3 instrument. All gene expression values were normalized to the expression of the housekeeping gene Gapdh. Relative gene expression values were calculated using the 2−ΔΔCT method, where vehicle control groups or the lowest value in a group was used as the expression control (value set at “1”). The primer sequences used for this study are listed in Supplementary Table 1.
Statistics
The 1-way and 2-way ANOVA statistical analyses, as indicated, were performed using GraphPad Prism software. For multiple comparisons, both Sidak's (2-way) and Tukey's (1-way) multiple comparison tests were used. The p-values <0.05 were considered statistically significant. For microbiome sequencing experiments, bioinformatical analysis was performed by the Research Informatics Core, RRC, University of Chicago Illinois.
RESULTS
Acute DFP administration reduces intestinal tight junction expression in vivo
We first investigated whether DFP impinges on intestinal integrity through analyzing the expression of the epithelial tight junction proteins claudin-1, claudin-4, and occludin. These markers were chosen based on our previous research demonstrating that all 3 are reduced in a model of colitis and that cytokines can regulate the expression of each in IECs (27). For these initial experiments we used a GWI mouse model that relies on stress priming (7-day CORT dosing) followed by 24 h injection with the organophosphate DFP (33). In this study the authors demonstrated that CORT primes mice for more severe neuroinflammation after injection of DFP. We found that DFP injection, regardless of priming mice with CORT, results in a reduction of tight junction proteins in the colon (Fig. 1A and B). Representative immunoblots from 2 individual animals per group are presented in Fig. 1A. Similar results were observed in the small intestinal tissues (not shown). Mice treated with CORT alone did not exhibit substantial changes in the protein expression of these tight junction molecules, with the exception of claudin-4. These findings suggest that stress is mostly dispensable for effects on intestinal tight junctions. Indeed, comparable results were obtained by reanalyzing pooled data that only factored vehicle (saline) or DFP injection as experimental variables (no CORT) (Fig. 1C). The reductions observed for occludin are also consistent with another study investigating the effects of permethrin on intestinal integrity (38).
Figure 1. DFP injection decreases intestinal tight junction protein expression. (A) Representative immunoblot analysis of colon tissue obtained from mice following V/S, V/D, C/S or C/D treatments in the GWI model. Representative analysis of tissues from 2 individual mice/group are presented. (B) Representative densitometry analysis of the indicated tight junction proteins from mice treated as indicated (n=4–6 animals/group). *p<0.05, **p<0.001, ***p<0.0001 for comparisons to the V/S control group (2-way ANOVA). (C) Pooled densitometry (3 independent experiments) results of mouse colons broken down into only treatment with saline (vehicle) or DFP 24 h prior to immunoblotting for the indicated tight junction proteins. **p<0.001, ***p<0.0001 for comparisons to the vehicle control group (2-way ANOVA) (D) Gene expression analysis of colon tissues from mice treated with saline or DFP for 24 h. Vehicle (saline) was used as the control (value set to “1”). Expression values were normalized to the expression of Gapdh. *p<0.05 for comparisons to the vehicle control group (2-way ANOVA). Data are representative of at least 3 independent experiments.
V/S, vehicle/saline; V/D, vehicle/DFP; C/S, CORT/saline; C/D, CORT/DFP.
Performing immunoblot analysis of total intestinal tissue lysates led to variability in tight junction observations across multiple experiments (Fig. 1A-C). Therefore, we next verified these findings using qPCR by comparing colon tissue of mice injected with saline or DFP (no CORT treatment). We found that the mRNA expression of both claudin-4 and occludin was consistently decreased following 24 h DFP treatment (Fig. 1D). The expression of claudin-1 remained variable across multiple experiments, suggesting that this molecule may not be a primary target of DFP (or CORT) action. These findings overall demonstrate that a neurotoxin, such as DFP, can have acute and likely drastic effects on the epithelial barrier.
The loss of IL-17 or IL-17C signaling exacerbates the effect of DFP on intestinal tight junctions
The expression of IL-17 and IL-17C increases following acute colonic injury and both cytokines have been shown to promote tight junction formation (23,27). Importantly, The Th17-IL-23 (IL-17 production) axis is implicated in veterans with GWI (39,40). Analysis of the colon tissues from DFP-treated mice, however, revealed that only IL-17 expression was reduced following 24 h exposure (Supplementary Fig. 1A). We also investigated the outcome of acute DFP exposure on tight junction mRNA expression using C57BL/6 (wild type [WT]), IL-17−/−, and IL-17RE−/− mice. IL-17RE is the receptor for IL-17C and previous work has demonstrated that IL-17C−/− and IL-17RE−/− mice have similar outcomes in models of GI inflammation (27,29). The loss of either signaling pathway resulted in exacerbated reductions of tight junction transcript levels in comparison to DFP-treated WT animals (Supplementary Fig. 1B). These effects were most pronounced for claudin-1 and occludin in the case of IL-17 deficiency, while IL-17RE−/− mice only exhibited a significant reduction in occludin. Although claudin-4 mRNA was reduced in both sets of deficient animals (not shown), these comparisons failed to reach statistical significance.
Our results above indicated a more substantial role for IL-17 in vivo (in comparison to IL-17C) for establishing tight junctions following the DFP insult. To further determine the source for intestinal IL-17 expression, we performed similar DFP exposures in Rag1−/− mice, which are deficient in both T and B cells. Similar to WT controls, Rag1−/− mice exhibited slightly elevated mRNA expression of IL-1β in both colon and small intestine (Supplementary Fig. 1C). However, these differences failed to reach statistical significance. Importantly, IL-17 mRNA in the intestinal tissues of Rag1−/− mice was only moderately decreased following acute DFP exposure, indicating that T lymphocytes are likely the primary source of intestinal IL-17 in this model. Overall, these results demonstrate that IL-17 and, to a lesser extent, IL-17C are important for the reestablishment of tight junctions following an intestinal insult.
DFP exposure reduces intestinal AMP expression
We investigated the expression of AMPs following acute DFP exposure given their importance for maintaining intestinal microbial stability (15). We purified mRNA from the colonic or small intestinal epithelium of mice injected 24 h with either saline or DFP (Fig. 2). We observed that DFP exposure results in a significant reduction of both RegIIIβ and RegIIIγ AMP mRNA expression in the colon (Fig.2A) and the ileum (not shown). We also thought it likely that reduced expression of IL-17 and IL-17C may potentially contribute to dysbiosis in this GWI model, considering that both cytokines regulate tight junctions, permeability, microbiome, and AMPs (28,29). Indeed, we verified that the expression of IL-17, but not IL-17C, mRNA was likewise reduced with DFP exposure (Fig. 2B). Importantly, DFP treatment alone did not induce an overtly inflammatory microenvironment as IL-1β and TNFα (Fig. 2C) was unchanged and intestinal tissues did not exhibit thickening, shortening, or other macroscopic signs of inflammation (not shown).
Figure 2. DFP down-regulates AMP and IL-17 expression in the colon. (A-C) Colonic mRNA was extracted from mice treated for 24 h with vehicle (saline) or DFP, and then gene expression was analyzed as indicated. Relative expression was calculated using the lowest value as the control (value of “1”) and normalized to the expression of the housekeeping reference gene, Gapdh (n=4 mice/group). Data represent at least 3 independent experiments.
*p<0.05, **p<0.005 for comparison to vehicle control (2-way ANOVA).
To determine if IL-17 or IL-17C signaling is critical for helping to maintain AMP expression upon DFP exposure, we performed similar experiments in WT, IL-17−/−, and IL-17RE−/− mice (Supplementary Fig. 1B). We found that the loss of IL-17 signaling was detrimental for reestablishing both RegIIIβ and RegIIIγ AMP expression. Although IL-17C expression was not affected by DFP treatment, the loss of this pathway still resulted in decreased RegIIIγ expression in comparison to DFP-treated animals. Therefore, organophosphate exposure can reduce AMP expression, which can be worsened in the case of IL-17 family cytokine dysregulation.
IECs respond directly to cholinesterase inhibitors
The dramatic effects we observed on tight junctions, AMP expression, and IL-17 family cytokines with acute (24 h) DFP treatment led us to hypothesize that DFP, via contacting intestinal tissues through the i.p. route, could directly influence the function of IECs. DFP can act as an irreversible acetylcholinesterase (AChE) inhibitor that prevents the termination of neuronal signals; however, DFP would not impact epithelial cells in this manner. Other studies have demonstrated that AChE is in fact expressed by epithelial cells and has different activities. For example, monomeric AChE was found bound to Caco-2 cell membranes (41), and inhibition of AChE negatively affected organogenesis and intestinal tissue development (42). Moreover, AChE can act a pro-apoptotic mediator outside of synapses (43). Therefore, we tested if cholinesterase inhibitors could directly impact tight junctions and AMP production in IECs in vitro by using the intestinal organoid system. We previously demonstrated that IL-17 and IL-17C could promote the expression of claudin-1, claudin-4, and occludin in colonic cell lines and ex vivo IECs (27). However, we wanted to first verify that same could be observed in ileum-derived organoid cells (Supplementary Fig. 2). Indeed, all 3 molecules can be observed at the protein level in organoid cells, and all are responsive to IL-17 and IL-17C.
Ileum-derived organoids were then treated with vehicle (saline) or 5 nM DFP for 24 h prior to qPCR analysis. We found that direct 24 h DFP treatment only slightly reduced the expression of claudin-4 and occludin in comparison to saline treatment (Fig. 3). Despite robust protein expression (Supplementary Fig. 2), claudin-1 mRNA expression was very low in all conditions and remained unchanged with all treatments (not shown). These results suggest that the tight junction effects observed for DFP in vivo are not a direct product of neurotoxin action on IECs, at least in an acute period of time. In contrast, the expression of the AMPs RegIIIβ and RegIIIγ were all significantly reduced in response to DFP exposure (Fig. 3), which indicates a direct effect on IECs.
Figure 3. Cholinesterase inhibition reduces AMP expression in IECs. Ileum-derived organoid cultures were treated 24 h with DFP (5 nM), rivastigmine (1 mg/ml), or donepezil (20 ng/ml) prior to mRNA harvest and qPCR analysis. Vehicle controls include saline (DFP), DMSO (rivastigmine), and chloroform (donepezil). Gene expression analysis of the indicated genes was performed using the 2−ΔΔCT method with each respective vehicle control serving as the control analysis group (value set at “1”). All expression values were normalized to the expression of the housekeeping gene Gapdh (n=6 individual wells/treatment analyzed in duplicate). Data are representative of 3 independent experiments.
*p<0.05, **p<0.01, and ***p<0.001 (1-way ANOVA) in comparison to respective vehicle-treated cells.
To determine if the effects on AMP expression could be attributed to the activity of AChE, we also treated organoids cells for 24 h with reversible inhibitors of AChE (donepezil, 20 ng/ml) or both AChE and butyrylcholinesterase (rivastigmine, 1 mg/ml). In these experiments we used vehicle controls of DMSO and chloroform for the rivastigmine and donepezil treatments, respectively. We found that rivastigmine substantially increased the mRNA expression of both claudin-4 and occludin. Donepezil treatment also increased the expression of claudin-4 but the effects on occludin expression did not reach statistical significance (Fig. 3). We next sought to examine whether DFP, rivastigmine, or donepezil alters AMP expression by intestinal organoid cells. All 3 of the cholinesterase inhibitors tested effectively reduced (>50%) the expression of both RegIIIβ and RegIIIγ compared to respective vehicle controls.
We investigated the expression of IL-17C mRNA in organoid cells following 24 h treatment to determine if IL-17C is induced in response to chemical toxicity/insult or cholinesterase inhibition. Notably, IECs do not express IL-17 and we could not quantify IL-17 mRNA in our organoid cultures (not shown). We found that both rivastigmine and donepezil could increase IL-17C transcript levels in these cultures (Fig. 3). DFP, on the other hand, only moderately increased IL-17C mRNA, which is consistent with what we observed in vivo. These results suggest the ability of DFP to act as a cholinesterase inhibitor is detrimental to the expression of AMPs. However, DFP may still impact additional pathways that ultimately results in a reduction of tight junction expression as we observed in vivo.
IL-17 and IL-17C can rescue AMP expression in DFP-treated IECs.
IL-17 and IL-17C are protective to mucosal homeostasis through a variety of means, including the regulation of tight junctions and AMP expression (23,28,29,44). Therefore, we investigated whether IL-17 or IL-17C treatment could negate the effects of cholinesterase inhibition on IECs, especially for AMP production. Ileum-derived organoid cells were treated as above (Fig. 3), only now we included the addition of 100 ng/ml of either rmIL-17 or IL-17C. As expected, DFP treatment only moderately reduced the expression of claudin-4 and occludin (Fig. 4A). IL-17 or IL-17C treatment, however, increased the transcript levels of both tight junctions, as previously reported by Reynolds et al. (27) and Supplementary Fig. 2. Interestingly, treatment of organoid cells with either IL-17 or IL-17C rescued DFP-dependent reductions in both RegIIIβ and RegIIIγ (Fig. 4A), similar to values observed for IL-17 or IL-17C treatment alone. The expression of IL-17C, however, remained unchanged among all of the treatment groups.
Figure 4. IL-17 and IL-17C treatment can negate cholinesterase inhibition IECs. (A) Ileum-derived organoid cultures were treated 24 h with vehicle (saline), 100 ng/ml IL-17, 100 ng/ml IL-17C, 5 nM DFP, or a combination as indicated prior to mRNA harvest and qPCR analysis. (B) Organoid cells were treated 24 h with vehicle (DMSO), 100 ng/ml IL-17, 100 ng/ml IL-17C, 5 nM rivastigmine, or a combination prior to mRNA harvest and qPCR analysis. Gene expression analysis of the indicated genes was performed using the 2−ΔΔCT method with each respective vehicle control serving as the control analysis group (value set at “1”). All expression values were normalized to the expression of the housekeeping gene Gapdh (n=4 individual wells/treatment analyzed in duplicate). Data are representative of 3 independent experiments.
*p<0.05, **p<0.001, and ***p<0.0001 (1-way ANOVA) in comparison to either DFP only (A) or rivastigmine only (B) treated cells.
We next performed a similar set of experiments on organoid cells focusing only on the effects of rivastigmine. The addition of IL-17 or IL-17C, in combination with rivastigmine, did not further increase the expression of claudin-4 or occludin. In fact, IL-17 and IL-17C treatment actually blunted the rivastigmine-induced increase of occludin mRNA (Fig. 4B). As we observed with DFP exposure, the addition of IL-17 rescued the expression of RegIIIβ and RegIIIγ from reductions observed with rivastigmine treatment. In contrast, IL-17C could not. Finally, IL-17 treatment worked in an additive fashion with rivastigmine to increase the expression of IL-17C mRNA (Fig. 4B). Collectively, these results demonstrate that exposure to an organophosphate toxin, such as DFP, directly impacts IEC functions, through both unique and similar mechanisms compared to more specific and reversible cholinesterase inhibitors.
CORT and DFP treatment alters the intestinal microbiome
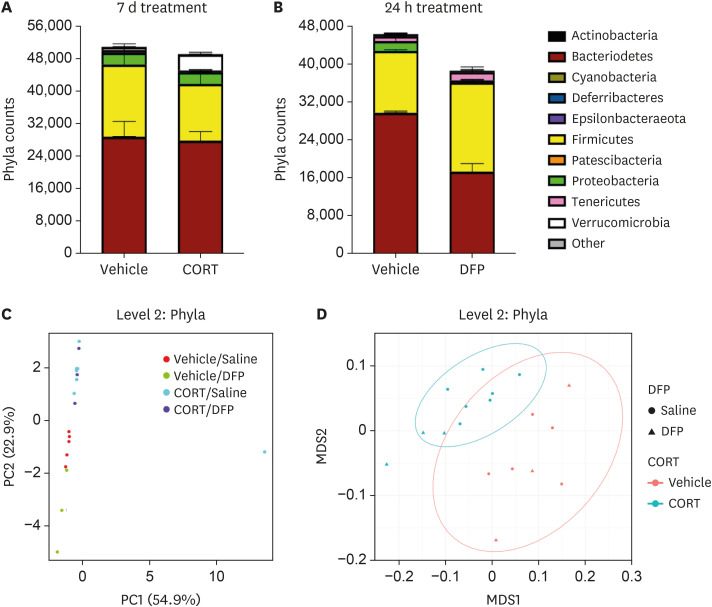
Dysregulation of both tight junction and AMP expression following acute DFP exposure (Figs. 1 and 2) are indicative of microbial dysbiosis, prompting us to examine whether exposure to such organophosphate agents could alter the microbiome. Indeed, GWI Veterans with chronic GI disease exhibit increased intestinal permebaility compared to control subjects (7). For these experiments we investigated the effects of both DFP and stress on microbiome composition, although DFP adminstration alone was sufficient to reduce intestinal tight junctions (Fig. 1). Littermate C57BL/6 WT mice were first separated, randomized, and put on CORT (200 mg/L drinking water in 1.2% ethanol) or vehicle (equivalent volume of 1.2% ethanol) for 7d prior to obtaining fecal samples for 16S amplicon sequencing. As demonstrated in Fig. 5A, at the phylum level CORT treatment resulted in reductions of Proteobacteria (p<0.01) and Tenericutes (p=2.49×10−5) while increasing the abundance of Verrucomicrobia (p=6.47×10−3). Thus, stress is indeed a potentially important variable for veterans with GWI in contributing to intestinal microbial composition.
Figure 5. Stress and DFP exposure alters the intestinal microbiota. (A) Littermate animals were treated with vehicle or CORT in drinking water for 7-day prior to fecal sample collection and 16S sequencing (n=5 per group). (B) Littermate animals were injected i.p. with vehicle (saline) or DFP for 24 h prior to fecal sample collection and 16S sequencing (n=4 per group). (C) Principal component analysis of phyla differentially expressed in the indicated groups (n=3–5 mice/group.) (D) β-diversity NMDS plots (Bray distance) of the groups presented in (C). Data represent 2 independent experiments.
We performed a similar analysis to determine if acute (24 h) DFP treatment can also shift the microbiota (Fig. 5B). Even in this short time period, we found that the intestinal microbiomes of DFP-treated animals were exhibiting altered colonization profiles (Fig. 5B). DFP-treated animals exhibited a remarkable (>2.5) increase in the ratio of Firmicutes: Bacteroidetes compared to saline-injected mice (1.08 vs 0.4387, respectively). We also observed significant increases in Firmicutes (p=0.01) and a near complete loss of Proteobacteria (p=0.029) in comparison to vehicle controls. Thus, short-term DFP toxicity is sufficient to impact intestinal microbial colonization.
We also performed principal component and diversity analysis on the microbiome of mice treated with combinations of CORT and DFP (Fig. 1). Principal component analysis of phlya (Fig. 5C) revealed different groupings of control, CORT-, and DFP-treated animals. Animals treated with DFP and CORT tended to group closer to CORT alone compared to the DFP alone animals. Indeed, all animals treated with CORT exhibited reductions in the abundance of Proteobacteria, Tenericutes, and Epsilonbacteraeota (p-values of 0.01, 9.47×10−3, and 0.03, respectively). When CORT/DFP animals were compared to Vehicle/DFP, we found significant alterations in Proteobacteria (p=3.75×10−3), Verrucomicrobia (p=2.4×10−7), and Tenericutes (p=7.01×10−4). However, only one moderate difference was observed in Proteobacteria (p=0.02) for DFP alone compared to control animals. More differences can be observed for both CORT and DFP treatments at higher levels, for example at the class level (Supplementary Fig. 3A).
We performed α- and β-diversity analysis of the data set described in Fig. 5C. α-analysis revealed no changes in diversity with the exception of CORT treatment at higher levels (level 5–6, not shown). On the other hand, β-diversity revealed that CORT treatment actually results in a higher diversity index at the phylum (p<0.001, Fig. 5D) and the class level (p<0.001, Supplementary Fig. 3B). DFP treatment decreased the overall microbial diversity, but these differences were not observed until the class (p=0.009, Supplementary Fig. 3B) and subsequently higher levels (not shown). Overall stress appears to be a more substantial driver of potential dysbiosis. However, the results obtained for DFP-treated animals cannot be discounted due to the short observational window.
DISCUSSION
In this work we demonstrated that organophosphate chemicals, such as DFP, have the ability to directly impact the GI epithelium. The exposure model used in these studies (DFP +/− CORT) is limited to acute exposure, so relevance to GWI Veterans will still need to be established in future work. A recent study demonstrated that veterans with GWI and chronic GI distress exhibit intestinal hyperpermeability (7), suggesting that mechanisms such as proper tight junction localization or microbiome composition could be key to understanding the biological mechanisms underlying GI disease in these veterans. A common problem with GWI mouse models, especially those relying on exposure agents, is translating what happened to a rodent over the course of weeks to what happened to a Veteran decades after exposure. Yet, animal models still are an important tool for understanding the potential impact of suspected GWI exposures.
DFP was chosen as a model exposure agent due to its similarities with sarin in mechanism of action. DFP is also commonly used to model both sarin and organophosphate pesticide exposures, both of which are hypothesized to contribute to GWI (45,46). DFP can act as an irreversible AChE inhibitor, which greatly contributes to the toxic properties. Indeed, DFP was found to be a driver of neuroinflammation in a GWI mouse model (33), which is logical due to a clear and apparent effect on neuromuscular junctions. However, how DFP may directly impact IECs becomes more complicated. Non-canonical roles for AChE in epithelial cell function have been discovered, which prompted us to investigate whether DFP, or other cholinesterase inhibitors for that matter, could influence protective mechanisms existing at the epithelial barrier. Using intestinal organoid cells we did find a role for DFP in forcing a down-regulation of the AMPs, RegIIIβ and RegIIIγ, which was consistent with what was observed in the GWI animal model. However, in contrast to our in vivo results, DFP did not play a substantial role in regulating tight junction molecule expression in vitro. It still remains possible that other tight junction proteins are directly affected by DFP, but we also failed to observe substantial changes when testing an additional set of molecules (not shown).
The effect of DFP on IECs in vitro and in vivo subsequently led us to investigate if AChE inhibition is the relevant mechanism. Using both donepezil and rivastigmine, we found that AMP expression was similarly reduced in intestinal organoid cells, which points to a functional role of AChE in regulating various IEC parameters. Alternatively, DFP may impinge on AMP expression through another, possibly redundant pathway. Future studies include modulating the expression of AChE in organoid cells and determining if there is an impact on AMP and tight junction expression.
We observed that rivastigmine treatment actually increases the expression of IL-17C mRNA in intestinal organoid cells. This is interesting because IL-17C also plays a role in promoting the expression of AMPs, so this finding could indicate that IL-17C is induced following this type of insult to help restore protective intestinal functions. However, treating cells with rivastigmine alone still results in a reduction of AMP expression despite this induction of IL-17C mRNA, which could suggest that that the timeframe (24 h) is not sufficient to observe secondary effects on AMP expression by the IL-17C induced by rivastigmine. Importantly, donepezil and rivastigmine are reversible AChE inhibitors, which extends the relevance of our study to other suspected exposure agents, namely the AChE inhibitor pyridostigmine bromide. The administration of pyridostigmine tablets to guard against chemical neurotoxin attacks was common in the Gulf War (4). Therefore, future studies are planned to determine if pyridostigmine bromide can act in a similar capacity as DFP, donepezil, or rivastigmine in our intestinal organoid cell exposure model.
Stress is another variable believed to be at the root of GWI, at least in a subset of veterans (4,47). Stress is also an obvious choice as a contributor to neuroinflammation in GWI. We included stress in the GWI model because we wanted to determine if there are combinatorial effects with DFP in the GI tract as previously reported for the brain and neuroinflammation (33). We did observe that stress can influence certain GI parameters, including the composition of the intestinal microbiome. These results were not surprising as other studies have demonstrated similar findings in rodents and humans (48,49). Furthermore, the effect of CORT was likely primarily in the brain, rather than directly influencing the functions of IECs or other intestinal cellular population. The probable explanation is that the stress disrupts the “gut-brain” axis, leading to changes in the GI tract. However, future studies will still need to be conducted to determine if CORT can directly influence IEC function.
In almost all experiments in this study we included assessments of IL-17 and IL-17C expression and function. Our primary reason for choosing these 2 cytokines is grounded in past studies showing the importance of both in maintaining permeability, tight junction expression, microbiome composition, and AMP production—all of which become dysregulated with DFP exposure. The Th17/IL-23 axis has been previously implicated in GWI (39,40) and the presence of both IL-17 and IL-17F are associated with increased susceptibility to ME/CFS. Other biomarker studies with GWI veterans have revealed associations with Th1 cells, inflammatory markers such as CRP, endocrine dysfunction, and neuroantigen autoantibodies (50,51,52). Thus, there clearly is a role of the immune system in GWI, albeit not an obvious one that persists across all Veterans with GWI. Moreover, other cytokines can promote as well as inhibit many of the GI parameters tested in this study. Current studies are underway to examine the contribution to the milieu of cytokines that are highly responsive to an intestinal insult.
These findings were all obtained using rodent models, the limitations of which are discussed above. Mice are an obvious choice due to the plethora of experimental tools available to conduct these types of studies. However, to better understand the relevance of our findings to human GWI, our future directions will be focused on performing these types of experiments in human intestinal organoids and examining relevant molecule expression in samples obtained from veterans with GWI.
ACKNOWLEDGEMENTS
The authors would like to thank the Center for Cancer Cell Biology, Immunology, and Infection at RFUMS and Gastroenterology at James A. Lovell FHCC for help with this study. We would also like to thank Dr. Stefan Green at the University of Illinois Chicago RRC for performing the microbiome sequencing experiments. Bioinformatics analysis (microbiome) was performed by the University of Illinois Chicago Research Informatics Core, supported in part by NCATS through Grant UL1TR002003. This study was supported by the VA grant 5 I21 BX003760 to A.G.F and J.M.R.
Abbreviations
- AChE
acetylcholinesterase
- AMP
antimicrobial peptide
- CORT
corticosterone
- DFP
diisopropyl fluorophosphate
- GI
Gastrointestinal
- GWI
Gulf War Veterans' Illnesses
- IBS
irritable bowel syndrome
- IEC
intestinal epithelial cell
- qPCR
quantitative PCR
- RegIII
regenerating islet-derived protein 3
- RRC
Research Resources Center
- WT
wild type
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Feller AG, Reynolds JM.
- Funding Acquisition: Feller AG, Reynolds JM.
- Investigation: Patterson KM, Vajdic TG.
- Methodology: Patterson KM, Martinez GJ, Reynolds JM.
- Supervision: Feller AG, Reynolds JM.
- Validation: Patterson KM, Vajdic TG.
- Visualization: Patterson KM, Vajdic TG, Reynolds JM.
- Writing - original draft: Patterson KM, Martinez GJ, Reynolds JM.
SUPPLEMENTARY MATERIALS
Primer sequences used for qPCR
IL-17 and IL-17C following to DFP exposure. (A) Littermate animals were treated with saline or 1 mg/kg DFP prior to harvesting colon tissues and analyzing gene expression by qPCR (n=4 mice per group). *p<0.05 (2-way ANOVA) for IL-17 mRNA expression in comparison to vehicle control. (B) WT, IL-17−/−, and IL-17RE−/− mice were treated with DFP as above and colon tissue was analyzed by qPCR for the indicated genes (n=4 mice/group with duplicate assessments presented). *p<0.01, **p<0.001, ***p<0.0001 for comparisons to DFP-treated WT animals. (C) C57BL/6 (WT) or Rag1−/− mice were injected with DFP for 24 h followed by similar analysis of IL-1b and IL-17 mRNA in the colon (top) or small intestinal (bottom) tissues (n=4 mice/group analyzed in duplicate). ***p<0.001 for comparison to vehicle treated animals. Data are representative of 2 independent experiments.
IL-17 and IL-17C promote tight junction protein expression in intestinal organoids. Organoids were stimulated for 24 h (250 ng/ml cytokine) and proteins were analyzed by immunoblot as indicated. Data indicate as below: US = unstimulated/vehicle, +A = IL-17A, +C = IL-17C. Numbers indicate densitometry analysis (protein/actin). Data are representative of 3 independent experiments.
Effects of stress and DFP exposure at the class level. Principal component (A) and β-diversity (B) of microbial communities from animals presented in Fig. 5C and D at the class level.
References
- 1.Office of Research and Development. Gulf War research strategic plan 2013–2017 (2015 update) [Internet] [accessed on 28 October 2021]. Available at https://www.research.va.gov/pubs/docs/gwresearch-strategicplan.pdf.
- 2.Eisen SA, Kang HK, Murphy FM, Blanchard MS, Reda DJ, Henderson WG, Toomey R, Jackson LW, Alpern R, Parks BJ, et al. Gulf War veterans' health: medical evaluation of a U.S. cohort. Ann Intern Med. 2005;142:881–890. doi: 10.7326/0003-4819-142-11-200506070-00005. [DOI] [PubMed] [Google Scholar]
- 3.Kang HK, Mahan CM, Lee KY, Magee CA, Murphy FM. Illnesses among United States veterans of the Gulf War: a population-based survey of 30,000 veterans. J Occup Environ Med. 2000;42:491–501. doi: 10.1097/00043764-200005000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Presidential advisory committee on Gulf War veterans' illnesses. Final report. Washington D.C.: U.S. Government Printing Office; 1996. [Google Scholar]
- 5.Gray GC, Reed RJ, Kaiser KS, Smith TC, Gastañaga VM. Self-reported symptoms and medical conditions among 11,868 Gulf War-era veterans: the Seabee Health Study. Am J Epidemiol. 2002;155:1033–1044. doi: 10.1093/aje/155.11.1033. [DOI] [PubMed] [Google Scholar]
- 6.Kang HK, Li B, Mahan CM, Eisen SA, Engel CC. Health of US veterans of 1991 Gulf War: a follow-up survey in 10 years. J Occup Environ Med. 2009;51:401–410. doi: 10.1097/JOM.0b013e3181a2feeb. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B, Verne ML, Fields JZ, Verne GN, Zhou Q. Intestinal hyperpermeability in Gulf War veterans with chronic gastrointestinal symptoms. J Clin Gastroenterol. 2019;53:e298–e302. doi: 10.1097/MCG.0000000000001135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camilleri M, Gorman H. Intestinal permeability and irritable bowel syndrome. Neurogastroenterol Motil. 2007;19:545–552. doi: 10.1111/j.1365-2982.2007.00925.x. [DOI] [PubMed] [Google Scholar]
- 9.Chassard C, Dapoigny M, Scott KP, Crouzet L, Del'homme C, Marquet P, Martin JC, Pickering G, Ardid D, Eschalier A, et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment Pharmacol Ther. 2012;35:828–838. doi: 10.1111/j.1365-2036.2012.05007.x. [DOI] [PubMed] [Google Scholar]
- 10.Collins SM. A role for the gut microbiota in IBS. Nat Rev Gastroenterol Hepatol. 2014;11:497–505. doi: 10.1038/nrgastro.2014.40. [DOI] [PubMed] [Google Scholar]
- 11.Öhman L, Törnblom H, Simrén M. Crosstalk at the mucosal border: importance of the gut microenvironment in IBS. Nat Rev Gastroenterol Hepatol. 2015;12:36–49. doi: 10.1038/nrgastro.2014.200. [DOI] [PubMed] [Google Scholar]
- 12.Saito YA. The role of genetics in IBS. Gastroenterol Clin North Am. 2011;40:45–67. doi: 10.1016/j.gtc.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swan C, Duroudier NP, Campbell E, Zaitoun A, Hastings M, Dukes GE, Cox J, Kelly FM, Wilde J, Lennon MG, et al. Identifying and testing candidate genetic polymorphisms in the irritable bowel syndrome (IBS): association with TNFSF15 and TNFα. Gut. 2013;62:985–994. doi: 10.1136/gutjnl-2011-301213. [DOI] [PubMed] [Google Scholar]
- 14.Savas LS, White DL, Wieman M, Daci K, Fitzgerald S, Laday Smith S, Tan G, Graham DP, Cully JA, El-Serag HB. Irritable bowel syndrome and dyspepsia among women veterans: prevalence and association with psychological distress. Aliment Pharmacol Ther. 2009;29:115–125. doi: 10.1111/j.1365-2036.2008.03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostaff MJ, Stange EF, Wehkamp J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol Med. 2013;5:1465–1483. doi: 10.1002/emmm.201201773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langhorst J, Choi KE. The role of human defensins in gastrointestinal diseases. Expert Rev Clin Immunol. 2011;7:779–787. doi: 10.1586/eci.11.62. [DOI] [PubMed] [Google Scholar]
- 17.Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr. 2011;141:769–776. doi: 10.3945/jn.110.135657. [DOI] [PubMed] [Google Scholar]
- 18.Dong C. Regulation and pro-inflammatory function of interleukin-17 family cytokines. Immunol Rev. 2008;226:80–86. doi: 10.1111/j.1600-065X.2008.00709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaffen SL. Structure and signalling in the IL-17 receptor family. Nat Rev Immunol. 2009;9:556–567. doi: 10.1038/nri2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cua DJ, Tato CM. Innate IL-17-producing cells: the sentinels of the immune system. Nat Rev Immunol. 2010;10:479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- 21.Reynolds JM, Angkasekwinai P, Dong C. IL-17 family member cytokines: regulation and function in innate immunity. Cytokine Growth Factor Rev. 2010;21:413–423. doi: 10.1016/j.cytogfr.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar P, Monin L, Castillo P, Elsegeiny W, Horne W, Eddens T, Vikram A, Good M, Schoenborn AA, Bibby K, et al. Intestinal interleukin-17 receptor signaling mediates reciprocal control of the gut microbiota and autoimmune inflammation. Immunity. 2016;44:659–671. doi: 10.1016/j.immuni.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JS, Tato CM, Joyce-Shaikh B, Gulen MF, Cayatte C, Chen Y, Blumenschein WM, Judo M, Ayanoglu G, McClanahan TK, et al. Interleukin-23-independent il-17 production regulates intestinal epithelial permeability. Immunity. 2015;43:727–738. doi: 10.1016/j.immuni.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Chen J, Huang A, Stinson J, Heldens S, Foster J, Dowd P, Gurney AL, Wood WI. Cloning and characterization of IL-17B and IL-17C, two new members of the IL-17 cytokine family. Proc Natl Acad Sci U S A. 2000;97:773–778. doi: 10.1073/pnas.97.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamaguchi Y, Fujio K, Shoda H, Okamoto A, Tsuno NH, Takahashi K, Yamamoto K. IL-17B and IL-17C are associated with TNF-alpha production and contribute to the exacerbation of inflammatory arthritis. J Immunol. 2007;179:7128–7136. doi: 10.4049/jimmunol.179.10.7128. [DOI] [PubMed] [Google Scholar]
- 26.Hurst SD, Muchamuel T, Gorman DM, Gilbert JM, Clifford T, Kwan S, Menon S, Seymour B, Jackson C, Kung TT, et al. New IL-17 family members promote Th1 or Th2 responses in the lung: in vivo function of the novel cytokine IL-25. J Immunol. 2002;169:443–453. doi: 10.4049/jimmunol.169.1.443. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds JM, Martinez GJ, Nallaparaju KC, Chang SH, Wang YH, Dong C. Cutting edge: regulation of intestinal inflammation and barrier function by IL-17C. J Immunol. 2012;189:4226–4230. doi: 10.4049/jimmunol.1103014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez-Carrozzi V, Sambandam A, Luis E, Lin Z, Jeet S, Lesch J, Hackney J, Kim J, Zhou M, Lai J, et al. IL-17C regulates the innate immune function of epithelial cells in an autocrine manner. Nat Immunol. 2011;12:1159–1166. doi: 10.1038/ni.2156. [DOI] [PubMed] [Google Scholar]
- 29.Song X, Zhu S, Shi P, Liu Y, Shi Y, Levin SD, Qian Y. IL-17RE is the functional receptor for IL-17C and mediates mucosal immunity to infection with intestinal pathogens. Nat Immunol. 2011;12:1151–1158. doi: 10.1038/ni.2155. [DOI] [PubMed] [Google Scholar]
- 30.Song X, Gao H, Lin Y, Yao Y, Zhu S, Wang J, Liu Y, Yao X, Meng G, Shen N, et al. Alterations in the microbiota drive interleukin-17C production from intestinal epithelial cells to promote tumorigenesis. Immunity. 2014;40:140–152. doi: 10.1016/j.immuni.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 31.Chang SH, Reynolds JM, Pappu BP, Chen G, Martinez GJ, Dong C. Interleukin-17C promotes Th17 cell responses and autoimmune disease via interleukin-17 receptor E. Immunity. 2011;35:611–621. doi: 10.1016/j.immuni.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XO, Chang SH, Park H, Nurieva R, Shah B, Acero L, Wang YH, Schluns KS, Broaddus RR, Zhu Z, et al. Regulation of inflammatory responses by IL-17F. J Exp Med. 2008;205:1063–1075. doi: 10.1084/jem.20071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Callaghan JP, Kelly KA, Locker AR, Miller DB, Lasley SM. Corticosterone primes the neuroinflammatory response to DFP in mice: potential animal model of Gulf War Illness. J Neurochem. 2015;133:708–721. doi: 10.1111/jnc.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naqib A, Poggi S, Wang W, Hyde M, Kunstman K, Green SJ. Making and sequencing heavily multiplexed, high-throughput 16s ribosomal RNA gene amplicon libraries using a flexible, two-stage PCR protocol. Methods Mol Biol. 2018;1783:149–169. doi: 10.1007/978-1-4939-7834-2_7. [DOI] [PubMed] [Google Scholar]
- 35.Moonsamy PV, Williams T, Bonella P, Holcomb CL, Höglund BN, Hillman G, Goodridge D, Turenchalk GS, Blake LA, Daigle DA, et al. High throughput HLA genotyping using 454 sequencing and the Fluidigm Access Array™ System for simplified amplicon library preparation. Tissue Antigens. 2013;81:141–149. doi: 10.1111/tan.12071. [DOI] [PubMed] [Google Scholar]
- 36.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 37.Xue X, Shah YM. In vitro organoid culture of primary mouse colon tumors. J Vis Exp. 2013:e50210. doi: 10.3791/50210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alhasson F, Das S, Seth R, Dattaroy D, Chandrashekaran V, Ryan CN, Chan LS, Testerman T, Burch J, Hofseth LJ, et al. Altered gut microbiome in a mouse model of Gulf War Illness causes neuroinflammation and intestinal injury via leaky gut and TLR4 activation. PLoS One. 2017;12:e0172914. doi: 10.1371/journal.pone.0172914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khaiboullina SF, DeMeirleir KL, Rawat S, Berk GS, Gaynor-Berk RS, Mijatovic T, Blatt N, Rizvanov AA, Young SG, Lombardi VC. Cytokine expression provides clues to the pathophysiology of Gulf War illness and myalgic encephalomyelitis. Cytokine. 2015;72:1–8. doi: 10.1016/j.cyto.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smylie AL, Broderick G, Fernandes H, Razdan S, Barnes Z, Collado F, Sol C, Fletcher MA, Klimas N. A comparison of sex-specific immune signatures in Gulf War illness and chronic fatigue syndrome. BMC Immunol. 2013;14:29. doi: 10.1186/1471-2172-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plageman LR, Pauletti GM, Skau KA. Characterization of acetylcholinesterase in Caco-2 cells. Exp Biol Med (Maywood) 2002;227:480–486. doi: 10.1177/153537020222700712. [DOI] [PubMed] [Google Scholar]
- 42.Pickett MA, Dush MK, Nascone-Yoder NM. Acetylcholinesterase plays a non-neuronal, non-esterase role in organogenesis. Development. 2017;144:2764–2770. doi: 10.1242/dev.149831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du A, Xie J, Guo K, Yang L, Wan Y, OuYang Q, Zhang X, Niu X, Lu L, Wu J, et al. A novel role for synaptic acetylcholinesterase as an apoptotic deoxyribonuclease. Cell Discov. 2015;1:15002. doi: 10.1038/celldisc.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolls JK, McCray PB, Jr, Chan YR. Cytokine-mediated regulation of antimicrobial proteins. Nat Rev Immunol. 2008;8:829–835. doi: 10.1038/nri2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaubey K, Alam SI, Waghmare CK, Singh L, Srivastava N, Bhattacharya BK. Differential proteome analysis of rat plasma after diisopropyl fluorophosphate (DFP) intoxication, a surrogate of nerve agent sarin. Chem Biol Interact. 2019;298:66–71. doi: 10.1016/j.cbi.2018.10.026. [DOI] [PubMed] [Google Scholar]
- 46.Coughlin SS. A neuroimmune model of Gulf War Illness. J Environ Health Sci. 2017;3:1–6. doi: 10.15436/2378-6841.17.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macht VA, Woodruff JL, Grillo CA, Wood CS, Wilson MA, Reagan LP. Pathophysiology in a model of Gulf War Illness: contributions of pyridostigmine bromide and stress. Psychoneuroendocrinology. 2018;96:195–202. doi: 10.1016/j.psyneuen.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 48.Foster JA, Rinaman L, Cryan JF. Stress & the gut-brain axis: regulation by the microbiome. Neurobiol Stress. 2017;7:124–136. doi: 10.1016/j.ynstr.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Madison A, Kiecolt-Glaser JK. Stress, depression, diet, and the gut microbiota: human-bacteria interactions at the core of psychoneuroimmunology and nutrition. Curr Opin Behav Sci. 2019;28:105–110. doi: 10.1016/j.cobeha.2019.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abou-Donia MB, Conboy LA, Kokkotou E, Jacobson E, Elmasry EM, Elkafrawy P, Neely M, Bass CR, Sullivan K. Screening for novel central nervous system biomarkers in veterans with Gulf War Illness. Neurotoxicol Teratol. 2017;61:36–46. doi: 10.1016/j.ntt.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Craddock TJ, Fritsch P, Rice MA, Jr, del Rosario RM, Miller DB, Fletcher MA, Klimas NG, Broderick G. A role for homeostatic drive in the perpetuation of complex chronic illness: Gulf War Illness and chronic fatigue syndrome. PLoS One. 2014;9:e84839. doi: 10.1371/journal.pone.0084839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson GJ, Slater BC, Leis LA, Rector TS, Bach RR. Blood biomarkers of chronic inflammation in Gulf War Illness. PLoS One. 2016;11:e0157855. doi: 10.1371/journal.pone.0157855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences used for qPCR
IL-17 and IL-17C following to DFP exposure. (A) Littermate animals were treated with saline or 1 mg/kg DFP prior to harvesting colon tissues and analyzing gene expression by qPCR (n=4 mice per group). *p<0.05 (2-way ANOVA) for IL-17 mRNA expression in comparison to vehicle control. (B) WT, IL-17−/−, and IL-17RE−/− mice were treated with DFP as above and colon tissue was analyzed by qPCR for the indicated genes (n=4 mice/group with duplicate assessments presented). *p<0.01, **p<0.001, ***p<0.0001 for comparisons to DFP-treated WT animals. (C) C57BL/6 (WT) or Rag1−/− mice were injected with DFP for 24 h followed by similar analysis of IL-1b and IL-17 mRNA in the colon (top) or small intestinal (bottom) tissues (n=4 mice/group analyzed in duplicate). ***p<0.001 for comparison to vehicle treated animals. Data are representative of 2 independent experiments.
IL-17 and IL-17C promote tight junction protein expression in intestinal organoids. Organoids were stimulated for 24 h (250 ng/ml cytokine) and proteins were analyzed by immunoblot as indicated. Data indicate as below: US = unstimulated/vehicle, +A = IL-17A, +C = IL-17C. Numbers indicate densitometry analysis (protein/actin). Data are representative of 3 independent experiments.
Effects of stress and DFP exposure at the class level. Principal component (A) and β-diversity (B) of microbial communities from animals presented in Fig. 5C and D at the class level.